ABSTRACT
Rice is an important source of food for more than half of the world’s population. Bacterial panicle blight (BPB) is a disease of rice characterized by grain discoloration or sheath rot caused mainly by Burkholderia glumae. B. glumae synthesizes toxoflavin, an essential virulence factor that is required for symptoms of the disease. The products of the tox operons, ToxABCDE and ToxFGHI, are responsible for the synthesis and the proton motive force (PMF)-dependent secretion of toxoflavin, respectively. The DedA family is a highly conserved membrane protein family found in most bacterial genomes that likely function as membrane transporters. Our previous work has demonstrated that absence of certain DedA family members results in pleiotropic effects, impacting multiple pathways that are energized by PMF. We have demonstrated that a member of the DedA family from Burkholderia thailandensis, named DbcA, is required for the extreme polymyxin resistance observed in this organism. B. glumae encodes a homolog of DbcA with 73% amino acid identity to Burkholderia thailandensis DbcA. Here, we created and characterized a B. glumae ΔdbcA strain. In addition to polymyxin sensitivity, the B. glumae ΔdbcA strain is compromised for virulence in several BPB infection models and secretes only low amounts of toxoflavin (∼15% of wild-type levels). Changes in membrane potential in the B. glumae ΔdbcA strain were reproduced in the wild-type strain by the addition of subinhibitory concentrations of sodium bicarbonate, previously demonstrated to cause disruption of PMF. Sodium bicarbonate inhibited B. glumae virulence in rice, suggesting a possible non-toxic chemical intervention for bacterial panicle blight.
IMPORTANCE Bacterial panicle blight (BPB) is a disease of rice characterized by grain discoloration or sheath rot caused mainly by Burkholderia glumae. The DedA family is a highly conserved membrane protein family found in most bacterial genomes that likely function as membrane transporters. Here, we constructed a B. glumae mutant with a deletion in a DedA family member named dbcA and report a loss of virulence in models of BPB. Physiological analysis of the mutant shows that the proton motive force is disrupted, leading to reduction of secretion of the essential virulence factor toxoflavin. The mutant phenotypes are reproduced in the virulent wild-type strain without an effect on growth using sodium bicarbonate, a nontoxic buffer that has been reported to disrupt the PMF. The results presented here suggest that bicarbonate may be an effective antivirulence agent capable of controlling BPB without imposing an undue burden on the environment.
KEYWORDS: membrane protein, proton motive force, rice, bacterial panicle blight, antibiotic resistance, plant pathogens, virulence
INTRODUCTION
Rice is a main source of food for more than half of the world’s population. It is an important cash crop in the United States, and six states account for over 99% of the U.S. rice output (California, Arkansas, Louisiana, Texas, Mississippi, and Missouri). Bacterial panicle blight (BPB) has become a global disease of rice and is capable of causing losses in production approaching 75% where it strikes. A characteristic grain discoloration or sheath rot is usually an indication of BPB. BPB is caused by Burkholderia glumae or, less frequently, by the related Burkholderia gladioli (1). Both are hardy soil bacteria and, like other Burkholderia species, are highly resistant to antibiotics.
B. glumae synthesizes and secretes an essential virulence factor called toxoflavin that is required for symptoms of BPB. Toxoflavin acts as an electron carrier between NADH and oxygen and can produce hydrogen peroxide, increasing levels of reactive oxygen species and leading to toxicity to the plant (2). The products of the tox operons, ToxABCDE and ToxFGHI, are responsible for the synthesis of toxoflavin and the secretion of toxoflavin, respectively (3). Both toxABCDE and toxFGHI operons are regulated in a quorum sensing (QS)-dependent manner. The toxFGHI operon encodes the following two types of membrane transporters: a tripartite efflux pump with similarity to AcrAB of Escherichia coli belonging to the resistance-nodulation-division (RND) family of efflux pumps (ToxGHI) and an unrelated transporter belonging to the drug/metabolite transporter (DMT) superfamily (ToxF). Both the DMT and RND family transporters are proton-dependent transporters and thus are dependent upon the membrane proton motive force (PMF) for function.
Our laboratory has a longstanding interest in the conserved DedA membrane protein family. Our data show that processes requiring the PMF, including proton-dependent transporters, are compromised in certain dedA family mutants. Our work with the DedA family in Burkholderia thailandensis and Escherichia coli has shown that they are membrane transporters required for efficient operation of efflux pumps belonging to several families, including the RND family and the major facilitator superfamily (MFS). E. coli encodes two members of the DedA family (YqjA and YghB) that carry out partially redundant functions and display roughly 60% amino acid identity. Mutation of E. coli yqjA and yghB in the same strain results in antibiotic sensitivity (4, 5), temperature sensitivity, and cell division defects (6–8), as well as activation of several envelope stress responses (9). The cell division defect is caused by loss of the PMF-dependent twin arginine transport (TAT) pathway, needed for the export of periplasmic amidases (6). Antibiotic sensitivity is due to loss of activity of PMF-dependent multidrug efflux pumps belonging to the MFS and RND families (4). Expression of mdfA, a multidrug resistance protein of the MFS, or artificially restoring the PMF by growth at pH 6.0 can mitigate most of these phenotypes (4, 9). YqjA is required for growth of E. coli above pH 8.5 (10), similar to what was reported for MdfA (11) and other membrane transporters (12–14). Both YqjA and YghB possess essential membrane-embedded charged amino acids (4, 15) that are present in proton-dependent transporters belonging to the MFS and RND families of efflux pumps (16–21), suggesting key transport functions of the DedA family.
Burkholderia species, including B. glumae, are highly resistant to polymyxin (Pm) with MICs exceeding 500 μg/ml (22). A contributing factor is the expression of aminoarabinose (Ara4N)-transferase (ArnT), which transfers Ara4N to lipopolysaccharide (LPS) lipid A, neutralizing the negative charge of lipid A and weakening Pm binding (23). Pm acts in a manner mechanistically similar to the CAMPS (cationic antimicrobial peptides) of the innate immune response by disrupting negatively charged bacterial membranes, leading to cell lysis and death (24). CAMPs are found in all kingdoms of life where they play roles in immune defense, predation, and competition (25, 26). Plant CAMPS have been isolated from roots, stems, leaves, flowers, and seeds and often have antibacterial or antifungal activity (27). Most CAMPS produced in plants are cysteine-rich and include defensins, thionins, and cyclotides (28).
Mutation of a Burkholderia thailandensis dedA gene results in sensitivity to polymyxin, due to inefficient modifications to lipopolysaccharide by the cationic sugar (Ara4N) that is required for Pm resistance (5). We have renamed the gene dbcA (dedA required for Burkholderia CAMP resistance) (5, 29, 30). Burkholderia glumae, the cause of BPB, encodes a homolog of DbcA that displays 73% amino acid identity to B. thailandensis DbcA. In this work, we created and characterized a B. glumae ΔdbcA mutant. In addition to sensitivity to Pm, we demonstrate that the B. glumae ΔdbcA strain is hyperpolarized, secretes toxoflavin inefficiently, and is avirulent in several plant infection models. The membrane hyperpolarization can be chemically replicated in the wild-type (WT) strain by addition of 5 mM sodium bicarbonate, a nontoxic and inexpensive chemical. Simultaneous application of wild-type B. glumae and sodium bicarbonate causes a marked reduction of toxoflavin secretion and virulence in rice, suggesting a possible chemical intervention to prevent bacterial panicle blight.
RESULTS
B. glumae possesses a homolog of B. thailandensis DbcA.
Previously, we demonstrated that mutation of a B. thailandensis DedA family gene, renamed dbcA (dedA required for Burkholderia colistin resistance), results in sensitivity to colistin (5). DbcA is required for PMF-dependent modification of outer membrane lipid A with the cationic sugar aminoarabinose (Ara4N), thus contributing to colistin resistance (29). B. glumae possesses a DbcA homolog (GenBank accession number WP_012734729.1) of 231 amino acids with 73% amino acid identity to B. thailandensis DbcA. B. glumae DbcA also possesses 35% and 29% identity to E. coli YqjA and YghB, respectively, which are involved in antibiotic and alkaline pH tolerance (4, 10). B. glumae does possess additional genes encoding members of the DedA superfamily, possibly some with redundant functions, similar to most other bacterial species (8, 31). Alignments were performed using Needleman-Wunsch alignment (32, 33).
B. glumae DbcA is required for colistin resistance.
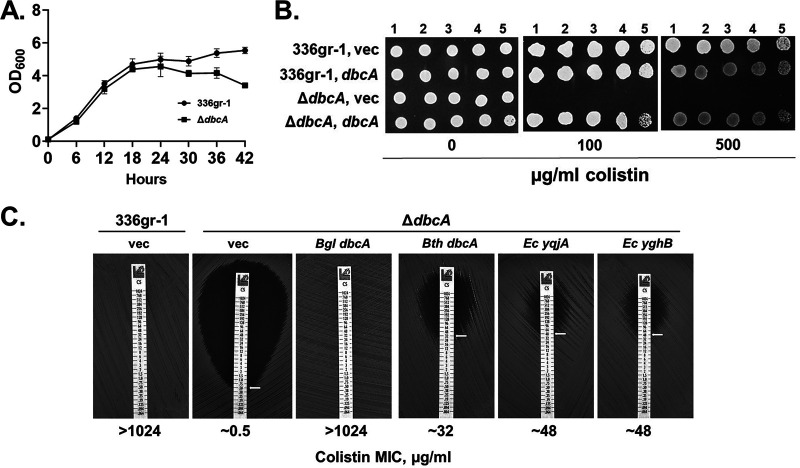
To study the function of B. glumae DbcA, we constructed a ΔdbcA in-frame deletion strain (see Fig. S1 in the supplemental material), which grows similarly to wild type, displaying a slight growth defect in late stationary phase (Fig. 1A). We also constructed a complementing plasmid with dbcA expressed behind a rhamnose-inducible promoter (pSC301). We first tested sensitivity of the B. glumae ΔdbcA strain to colistin and found that it is indeed highly sensitive to the antibiotic, with an MIC of ∼0.5 μg/ml (Fig. 1B and C), similar to that of E. coli K-12, which possesses no lipid A modifications contributing to colistin resistance (34). The MIC of the parent B. glumae strain was at least 1,000 times greater, consistent with resistance observed in other Burkholderia species (22). Complementation with a plasmid copy of dbcA completely restores colistin resistance to the ΔdbcA strain (Fig. 1B and C). In addition, we observed partial complementation of this phenotype with expression of B. thailandensis dbcA, E. coli yqjA, and E. coli yghB (Fig. 1C), indicative of an evolutionarily conserved function shared by these DedA family proteins.
FIG 1.
The B. glumae ΔdbcA strain is sensitive to colistin. (A) Growth curve of B. glumae 336gr-1 and ΔdbcA strains in LB broth buffered to pH 7.0 with 70 mM Tris. Equal amounts of cells (5 × 107 CFU/ml) were used to inoculate LB broth, and bacterial growth was analyzed at 6-h intervals. (B) Serially log10-diluted cells of B. glumae 336gr-1 and ΔdbcA strains transformed with control vector pSCrhaB2 (vec) or pSC301 (dbcA) were spotted and grown on MH2 agar medium containing 100 μg/ml trimethoprim and the indicated concentration of colistin. The 500 μg/ml colistin plate also contained 0.0004% rhamnose. (C) Colistin E-strips were used to determine the colistin MIC for B. glumae 336gr-1 transformed with pBBR1MCS-2 (vec), and the ΔdbcA strain transformed with pBBR1MCS-2, pMCS501, pRP101, pRP102, or pRP103 (see Table 1). Bacterial strains were grown in MH2 agar containing 100 μg/ml Kan. Bgl, B. glumae; Bth, B. thailandensis; Ec, E. coli.
B. glumae DbcA is required for virulence in BPB infection models.
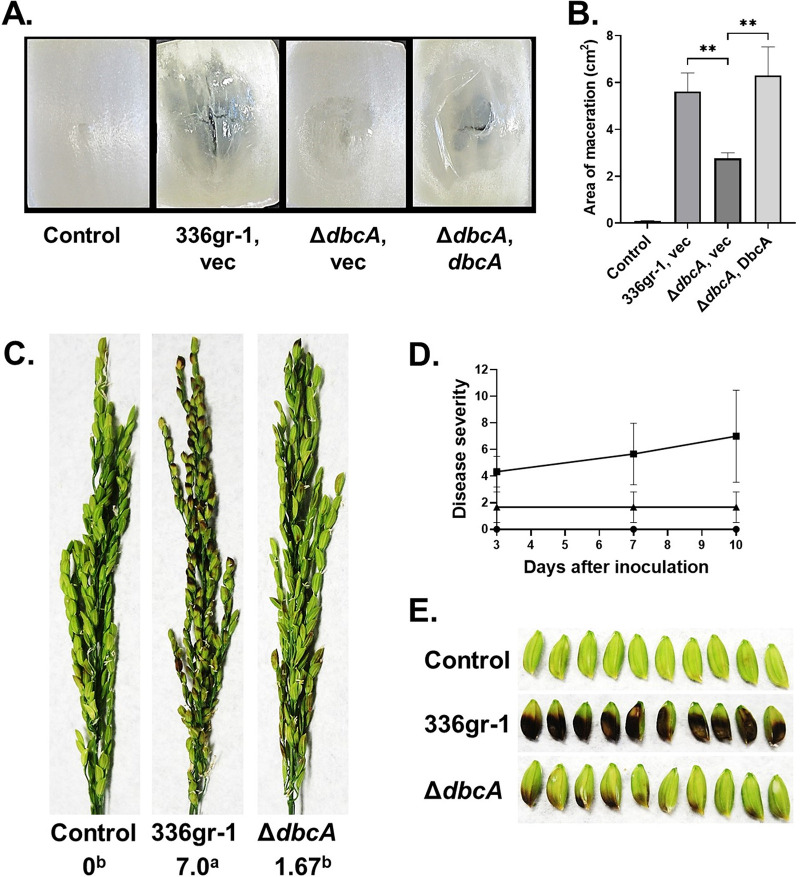
Since B. glumae is a plant pathogen and the cause of rice BPB, we were interested in measuring virulence of the wild-type and ΔdbcA strains. Virulence of B. glumae strains on rice was found to correlate to virulence on onion bulb scales, suggesting that this can be a convenient host system to measure B. glumae virulence (35). Onion scales were inoculated with growth medium alone, wild-type B. glumae, or the ΔdbcA strain (harboring control or complementing vectors) and incubated at 30°C for 72 h. The macerated area visible after inoculation with the wild-type B. glumae was found to be significantly larger than that seen with the ΔdbcA strain, and the defect was corrected with a plasmid copy of the gene (Fig. 2A and B). This suggests that DbcA may be required for virulence in rice.
FIG 2.
B. glumae ΔdbcA strain is compromised for virulence in onion slices and rice panicles. (A) The area of maceration on onion slices indicates the virulence phenotype for each bacterial strain. Onion slices were infected with equal amounts of cells from B. glumae 336gr-1 transformed with control vector (vec) and the B. glumae ΔdbcA strain transformed with control vector (vec) and pSC501 (dbcA). Onion slices were incubated at 30°C for 72 h in a humid chamber. (B) Area of maceration (cm2) produced by indicated strains. **, P < 0.01. (C) Rice seed discoloration (black necrosis) indicates the virulence phenotype for control (water), B. glumae 336gr-1, and the ΔdbcA strain. The images were taken 10 days postinoculation, and disease severity score was determined with a 0 to 9 scale. Different letters below the score indicate statistical significance. (D) The line graph shows the disease progress for rice panicles inoculated with water (circles), B. glumae 336gr-1 (squares), and the ΔdbcA strain (triangles) at 3, 7, and 10 days postinoculation. (E) Detailed view of seed discoloration produced by control, B. glumae 336gr-1, and the ΔdbcA strain at 10 days postinoculation.
We proceeded to measure virulence using mature rice plants. For this assay, rice panicles were sprayed with 5 ml of 5 × 107 CFU/ml suspensions of the wild-type B. glumae and ΔdbcA strains twice at 2-day intervals, on the panicles at the 20 to 30% flowering stage of rice plant. The panicle blight symptoms were evaluated at 3, 7, and 10 days after the first inoculation by assigning virulence scores on a 0 to 9 scale (36). We observed that the panicles sprayed with wild-type bacteria started showing symptoms within 3 days of exposure compared to the water-treated control, and these symptoms worsened for the duration of the experiment (Fig. 2C). In contrast, panicles exposed to the ΔdbcA strain displayed significantly fewer symptoms and fared not much worse than water-treated controls (Fig. 2C). At the conclusion of the experiment, virulence scores were 0, 7, and 1.7 for water, the wild-type B. glumae strain, and the ΔdbcA strain exposures, respectively (Fig. 2C and D). Rice panicles taken from plants exposed to wild-type B. glumae were distinctively discolored while ΔdbcA strain-treated panicles were only slightly browned (Fig. 2E). This experiment shows that DbcA function is required for B. glumae to cause symptoms of BPB in rice panicles.
B. glumae DbcA is required for toxoflavin production.
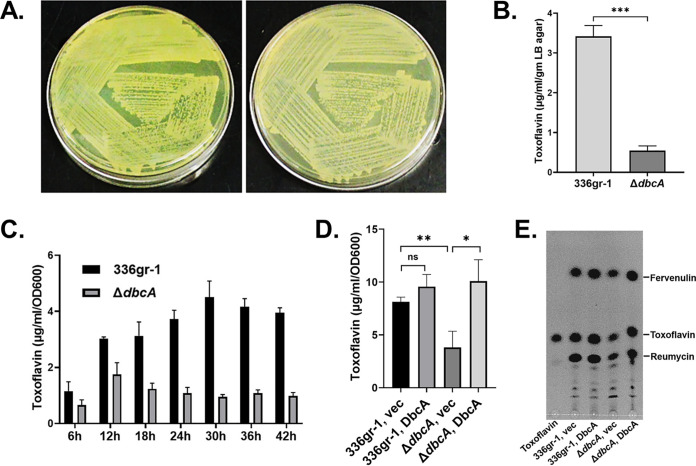
Toxoflavin is the sole yellow pigment produced by B. glumae (37) and is an essential virulence factor, the production of which is controlled by quorum sensing (1). To begin to understand the loss of virulence, we extracted and measured levels of toxoflavin produced by each strain. The B. glumae ΔdbcA strain is nearly nonpigmented (Fig. 3A), and much less toxoflavin is secreted by the ΔdbcA strain into the solid growth medium (Fig. 3B). Toxoflavin concentrations in liquid growth media were also significantly lower during growth for up to at least 42 h (Fig. 3C), and this was restored upon introduction of a complementing plasmid (Fig. 3D). Thin-layer chromatography (TLC) was used to resolve chloroform extracts of strains grown for 30 h. Levels of each secreted phytotoxin reumycin, fervenulin, and toxoflavin were lower in the uncomplemented ΔdbcA strain, and levels were restored to wild-type levels with expression from a complementing plasmid (Fig. 3E). These results collectively suggest that B. glumae DbcA is required for efficient toxoflavin synthesis and secretion and is consistent with loss of virulence of the ΔdbcA strain.
FIG 3.
Toxoflavin production is compromised in the B. glumae ΔdbcA strain. (A) Visual observation of toxoflavin (yellow pigment) produced by B. glumae 336gr-1 and the ΔdbcA strain on an LB agar plate after growth at 37°C for 72 h. Left and right panels of the figure indicate toxoflavin produced by the B. glumae 336gr-1 and ΔdbcA strains, respectively. (B) Quantification of toxoflavin production by the B. glumae 336gr-1 and ΔdbcA strains on LB agar plates. (C) Quantification of toxoflavin production in LB broth medium over time. B. glumae 336gr-1 (black bars) and the ΔdbcA strain (gray bars) were grown in LB broth medium buffered with 70 mM Tris (pH 7.0) at 37°C with shaking for 42 h. Toxoflavin levels in the culture medium were determined at 6-h intervals. (D) Complementation of toxoflavin production in the B. glumae ΔdbcA strain. Indicated strains were grown in LB broth medium buffered with 70 mM Tris (pH 7.0) without addition of antibiotic and rhamnose at 37°C with shaking for 30 h. (E) Analysis of toxoflavin produced by B. glumae 336gr-1 and the ΔdbcA strain after 30 h of growth by TLC. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
B. glumae DbcA is required for normal PMF.
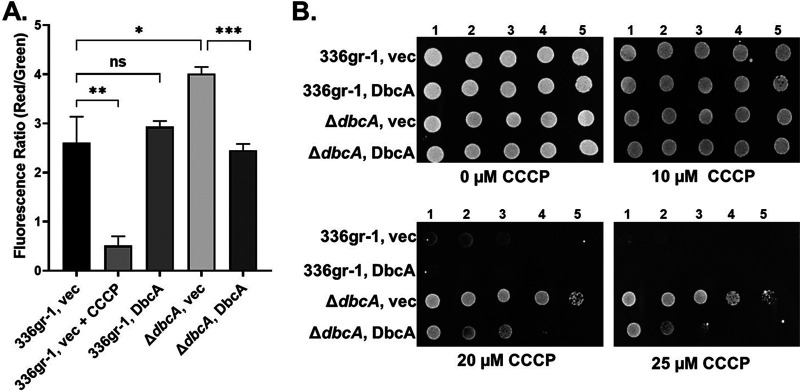
In order to understand the physiological state of B. glumae in the absence of dbcA, we analyzed the state of the PMF since the B. thailandensis ΔdbcA strain has altered PMF as do certain E. coli DedA family mutants (9, 29). According to chemiosmotic theory (38), the membrane PMF is equal to the sum of the charge difference across the membrane (ΔΨ) and the pH difference across the membrane (ΔpH). Importantly, when either ΔpH or ΔΨ is dissipated chemically or by mutation, bacteria have the ability to increase the other component of the PMF to compensate (39). To examine the PMF in more detail, we measured the ΔΨ component of the PMF using the dye JC-1. JC-1 is a membrane permeable dye that exhibits green fluorescence (530 nm) as a monomer but forms aggregates in the presence of membrane potential, shifting its emission from green to red (595 nm). Therefore, relative membrane potential can be expressed as the ratio of red to green fluorescence (40, 41). Wild-type B. glumae and the ΔdbcA strain harboring either control or complementing vector were treated with JC-1 dye. Cells treated with proton ionophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) were included as a control. B. glumae strains and the complemented ΔdbcA strain exhibited a consistent 595/530 ratio (Fig. 4A). However, the B. glumae ΔdbcA strain displayed a higher red/green ratio (Fig. 4A), suggesting partial hyperpolarization of the PMF. This result was unexpected, since we previously reported PMF depolarization of the B. thailandensis ΔdbcA strain (5). Consistent with this observation, the B. glumae ΔdbcA strain is resistant to CCCP (Fig. 4B) and tetracycline (see Fig. S2 in the supplemental material), suggesting a dissipation of the ΔpH component of the PMF of B. glumae ΔdbcA strain (42, 43). These observations suggest that any alteration of the PMF can have profound effects on bacterial physiology.
FIG 4.
Measurement of membrane potential and CCCP sensitivity. (A) Measurement of membrane potential (ΔΨ) for B. glumae strains. Fluorescence ratio in the graph represents the red (595 nm)/green (530) emission ratio of JC-1 dye. Twenty-five micromolar CCCP added to B. glumae 336gr-1 (vec) was used as a control for loss of ΔΨ. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant. (B) CCCP sensitivity of B. glumae strains. Serially log10-diluted cells of B. glumae 336gr-1 and the ΔdbcA strain transformed with control vector (vec) and pSC301 (DbcA) were spotted and grown on MH2 agar growth medium containing 100 μg/ml Tmp, no rhamnose, and either 10, 20, or 25 μM CCCP at 37°C for 48 h.
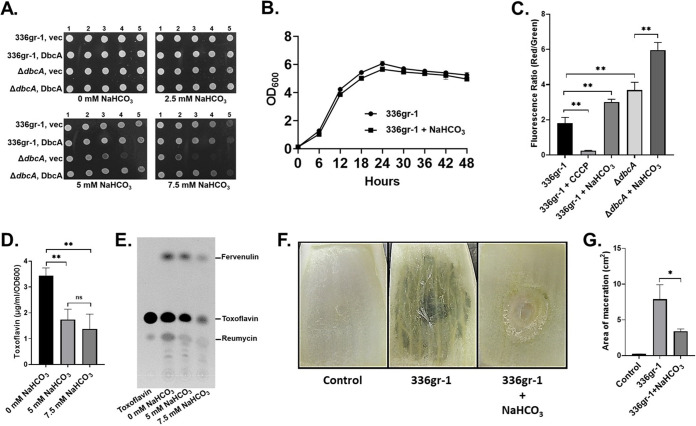
Sodium bicarbonate (NaHCO3) at physiological concentrations dissipates the ΔpH component of the PMF and can alter the sensitivity of bacteria to commonly used antibiotics (42). NaHCO3 at 5.0 mM has no effect on the growth of wild-type B. glumae with slight effects on the ΔdbcA strain (Fig. 5A and B), consistent with our previous results with B. thailandensis (30). Treatment of B. glumae and B. glumae ΔdbcA strains with 5 mM NaHCO3 results in a significant increase of ΔΨ (Fig. 5C). The effect of NaHCO3 on the ΔΨ of the wild-type strain is similar in magnitude to the effect of the ΔdbcA mutation. This was coupled with a significant decrease in toxoflavin production (Fig. 5D and E) and colistin sensitivity (see Fig. S3 in the supplemental material), showing that we can mimic the effect of the ΔdbcA mutation on the PMF using low concentrations of NaHCO3. Furthermore, the presence of 5 mM NaHCO3 significantly impacts B. glumae virulence in an onion scale model (Fig. 5F and G). Collectively, these results demonstrate that NaHCO3 can disrupt the bacterial PMF in a manner similar to the ΔdbcA mutation, and this disruption can impact B. glumae toxoflavin secretion and virulence. The additive effect of bicarbonate on growth (Fig. 5A) and membrane potential (Fig. 5C) of the ΔdbcA strain implies that this is not due to a direct effect of bicarbonate on DbcA.
FIG 5.
Bicarbonate selectively dissipates B. glumae PMF and reduces toxoflavin production and virulence without impacting growth. (A) Sodium bicarbonate (NaHCO3) sensitivity of B. glumae strains. Serially log10-diluted cells of B. glumae 336gr-1 and the ΔdbcA strain transformed with control vector (vec) and pSC301 (DbcA) were spotted and grown on MH2 agar medium containing 100 μg/ml trimethoprim and either 2.5, 5, or 7.5 mM sodium bicarbonate at 37°C for 48 h. (B) Growth curve of B. glumae 336gr-1 in LB broth buffered to pH 7.0 with 70 mM BTP with and without 5 mM NaHCO3. (C) Measurement of membrane potential (ΔΨ). Fluorescence ratio in the graph represents the red (595 nm)/green (530) emission ratio of JC-1 dye. 5 mM NaHCO3 was present when indicated. **, P < 0.01. (D) Production of toxoflavin by B. glumae 336gr-1 in growth medium containing sodium bicarbonate. LB broth medium was buffered with 70 mM BTP, pH 7.0. Equal numbers of bacterial cells (5 × 107 CFU/ml) were transferred into 250-ml conical flasks containing 25 ml buffered LB broth and either 0, 5, or 7.5 mM NaHCO3 and grown at 37°C with shaking for 30 h. (E) TLC analysis for toxoflavin produced by B. glumae 336gr-1 in growth medium containing indicated concentrations of NaHCO3. (F) Onion scales were inoculated with B. glumae preincubated with or without 5 mM NaHCO3, which were then incubated at 30°C for 6 days in a humid chamber. (G) Area of maceration (cm2) produced by indicated strains. **, P < 0.01; *, P < 0.05; NS, not significant.
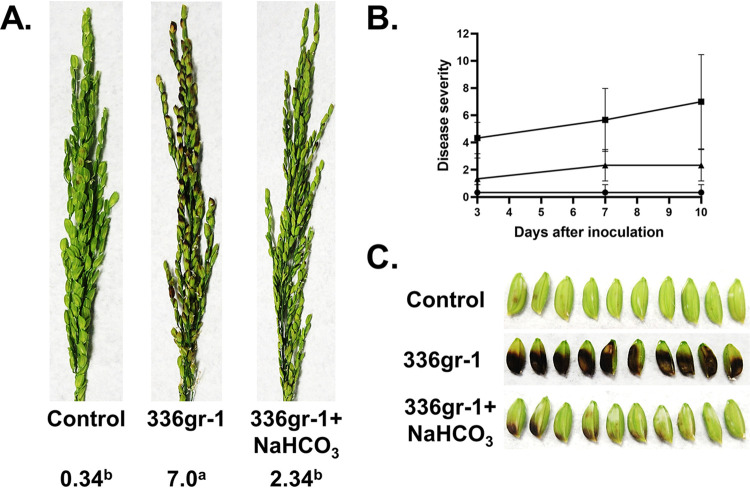
We then measured the effect of bicarbonate on B. glumae virulence using the rice panicle model. Wild-type B. glumae was grown and resuspended in the presence or absence of 5 mM NaHCO3 and applied to rice panicles as described for Fig. 2. At day 10 following exposure of panicles, B. glumae in water caused a similar pathology as described in Fig. 2. Panicles were discolored and wilted with a virulence score of 7.0 (Fig. 6A and B), while 5 mM NaHCO3 alone caused no harm to panicles. Plants that were treated with the combination of B. glumae and NaHCO3 fared much better (virulence score of 2.3) and displayed much less wilting and discoloration (Fig. 6A and B). Panicles from these plants were also less discolored than plants exposed to B. glumae delivered in water (Fig. 6C). These results suggest that the common buffer sodium bicarbonate may be capable of reversing virulence of B. glumae independently of any effect on growth and suggest a potential intervention strategy for bacterial panicle blight.
FIG 6.
Bicarbonate treatment results in loss of B. glumae virulence in rice. (A) Virulence phenotype on rice panicles for control, B. glumae 336gr-1 cells with no sodium bicarbonate, and B. glumae 336gr-1 cells with 5 mM sodium bicarbonate. For the control, 5 mM sodium bicarbonate was directly sprayed on rice panicles. The images were taken at 10 days postinoculation, and disease severity score was determined with a 0 to 9 scale. (B) The line graph shows the disease progress for rice panicles inoculated with control (circles), B. glumae 336gr-1 cells with no sodium bicarbonate (squares), and B. glumae 336gr-1 cells with 5 mM sodium bicarbonate (triangles) at 3, 7, and 10 days postinoculation. (C) Detailed view of seed discoloration observed by indicated treatments at 10 days postinoculation.
DISCUSSION
Rice is a primary source of food for more than half of the world’s population. Bacterial panicle blight (BPB), caused by Burkholderia glumae, results in extensive damage to rice (Oryza sativa) crops in the United States and worldwide. It is an important cash crop in the United States, and six states account for over 99% of the U.S. rice output (California, Arkansas, Louisiana, Texas, Mississippi, and Missouri). The U.S. rice output was about 18.6 billion pounds in 2019 with a value of $2,370,000,000 (USDA). Bacterial panicle blight (BPB) has become a global disease of rice and is capable of causing losses in production approaching 75% where it strikes. B. glumae infection is thought to originate from infected seeds. The infection can spread to upper tissues and leaves during plant growth and inoculate emerging panicles during the flowering stage (1, 44). A characteristic grain discoloration or sheath rot is usually an indication of BPB. A survey found it to be present in 60% of Louisiana fields, and yield losses may reach 40% in severely infected fields (45). Based upon annual rice production in the American South, it has been estimated that BPB caused $61 million in damage between 2003 and 2013. While the United States produces only 1.3% of the world’s rice, it is traded to 120 countries and accounted for 7.7% of the global rice trade between 2014 and 2016 (USDA). Therefore, rice shortages in the United States can have extensive impacts on global food security (46). BPB is predicted to significantly increase due to climate change (1, 47). While some rice varieties are more resistant than others to BPB, there is no completely resistant rice species, and no chemical treatment is currently recommended in the United States to combat the disease (1).
The DedA superfamily is a highly conserved membrane protein family that remains poorly characterized and for which there is little structural information. There are currently 29,230 individual sequences in the protein database across 7,915 species belonging to the “SNARE-associated PF09335” family of proteins (PFAM 34.0). DedA family proteins have been shown to be required for a number of important functions in various bacterial species, including growth and cell division (6, 7, 48), envelope integrity (9), and resistance to a number of antibiotics and biocides (4), including polymyxin and antimicrobial peptides in a number of different species (5, 49–53). Absence of certain DedA family members results in pleiotropic effects, impacting multiple pathways that are energized by PMF (8). Our lab has recently demonstrated that a B. thailandensis DedA family protein is required for resistance to colistin, an antibiotic belonging to the CAMP family (5). We named the B. thailandensis DedA protein DbcA (DedA of Burkholderia required for CAMP resistance) and note that B. glumae possesses a homolog of DbcA with ∼73% amino acid identity. Here, we show that B. glumae DbcA is required for resistance to colistin, efflux of toxoflavin, and virulence (Fig. 1 to 3).
In addition to a near absence of toxoflavin secretion, the sensitivity of the B. glumae ΔdbcA strain to polymyxin antibiotics, such as colistin, may be significant in understanding its loss of virulence. Polymyxin is thought to act in a manner similar to the CAMPs of the innate immune system of both plants and animals, by disruption of membranes possessing lipids with exofacial negatively charged headgroups (24). A number of CAMPs have been isolated from different plants tissues, including seeds, stems, leaves, flowers, and roots (27). Certain plant CAMPs do possess antibacterial and antifungal activity and are believed to play important roles in immunity (27, 28). Direct application (54) and overexpression (55) of CAMPs are being explored as strategies for disease control in rice. Whether the B. glumae ΔdbcA strain is sensitive to specific plant CAMPs is an important question that remains to be answered.
While characterizing the physiological effects of the ΔdbcA mutation on B. glumae, we observed that the ΔΨ component of the PMF was higher in the mutant than the wild type, an unexpected result considering that the opposite was observed with the B. thailandensis ΔdbcA strain (29). We took this to indicate that the ΔdbcA mutation was causing a dissipation of the ΔpH component of the PMF and was increasing ΔΨ to compensate for this loss. This further indicates that optimal maintenance of membrane potential is required and any perturbation can result in colistin sensitivity and additional phenotypes. Consistent with hyperpolarization, the ΔdbcA strain is also resistant to tetracycline (see Fig. S2 in the supplemental material), which can be caused by dissipation of ΔpH (42, 43). Recently, it was reported that a common buffer, sodium bicarbonate, could potentiate the effects of certain antibiotics specifically by dissipating the cellular ΔpH component of the PMF (42). Therefore, we tested to determine if sodium bicarbonate by itself could reproduce the effects of the ΔdbcA mutation on toxoflavin production and virulence. Indeed, this was found to be the case (Fig. 5 and 6) and at concentrations that did not cause a significant impact on bacterial growth. Importantly, these effects were not due to a trivial effect of bicarbonate on pH, as all growth media were buffered to pH 7.0 for these studies and no change in pH of the media was observed during growth (data not shown).
Sodium bicarbonate has been used for many years as a common and nontoxic household item and is an additive to foods and dental products. While it has been reported to inhibit the growth of fungi (56, 57) and bacteria including Streptococcus mutans (58), its antibacterial and antivirulence properties have not been fully exploited to date. In an agricultural setting, bicarbonate can be delivered as an ammonium or potassium salt, rather than sodium, with similar effects on PMF (42). The results presented here clearly suggest that bicarbonate may be an effective antivirulence agent capable of not just controlling BPB, but doing so in a responsible manner without imposing an undue burden on the environment typically seen with common herbicides, fungicides, and insecticides (59, 60).
In summary, we have identified a chemical treatment that mimics the effect of a mutation in a dedA family gene of Burkholderia glumae, the cause of bacterial panicle blight. Both mutation of B. glumae dbcA and exposure to sodium bicarbonate act similarly to dissipate the PMF, resulting in a loss of virulence. Since the DedA family is widespread in bacteria, similar phenotypes may be found in other bacterial species when the PMF is targeted.
MATERIALS AND METHODS
Culture conditions and chemicals.
Table 1 lists bacterial strains and plasmids used in this study. E. coli strains were grown in lysogeny broth (LB) medium (1% NaCl, 1% tryptone, 0.5% yeast extract) with appropriate antibiotics. Mobilizer strain E. coli RHO3 was grown in LB containing 200 μg/ml DAP (2,6-diaminopimelic acid; LL-, DD-, and meso-isomers; VWR). B. glumae 336gr-1 was grown in either LB or cation-adjusted Mueller-Hinton broth 2 (MH2, pH 7.3; Sigma-Aldrich). LB buffered with 70 mM Tris (pH 7.0 or 7.4) was used for toxoflavin production experiments and growth curves in liquid medium. For toxoflavin production in the presence of sodium bicarbonate, LB medium buffered with 70 mM BTP (bis-Tris propane), pH 7.0, was used. Antibiotics were purchased either from VWR or Sigma-Aldrich and used at the following concentrations: nitrofurantoin (Nit), 100 μg/ml (B. glumae); colistin (at the indicated concentrations); kanamycin (Kan), 30 μg/ml (E. coli) and 100 μg/ml (B. glumae and B. thailandensis); gentamicin (Gen), 30 μg/ml; trimethoprim (Tmp), 100 μg/ml; and tetracycline (Tet), 15 μg/ml. All bacterial strains were grown at 37°C unless otherwise indicated. Toxoflavin was purchased from Cayman Chemical. All other chemicals were purchased from VWR. Table 2 lists oligonucleotide primers used in this study (Sigma-Aldrich).
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Description | Source |
|---|---|---|
| Strain | ||
| Escherichia coli | ||
| XL1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1lac [F′ proAB lacIqZΔM15] Tn10 (Tetr) | Stratagene |
| RHO3 | SM10(λpir) Δasd::FRT ΔaphA::FRT, Kans | 64 |
| Burkholderia glumae | ||
| 336gr-1 | Wild-type Burkholderia glumae | 65 |
| ΔdbcA strain | 336gr-1 ΔdbcA::Tmpr | This study |
| ΔdbcA::FRT strain | 336gr-1 ΔdbcA::Tmps | This study |
| Plasmid | ||
| pUC18T-mini-Tn7T-Tp | Mini-Tn7 T-based vector containing a trimethoprim resistance cassette; Tmpr (GenBank accession no. DQ493875) | 69 |
| pEX18Gm | oriT+sacB+, gene replacement vector with multiple cloning site from pUC18, Gmr | 63 |
| pEX3525 | Assembled upstream and downstream fragment of dbcA gene interrupted by Tmpr fragment cloned into pEX18Gm | This study |
| pFlpTet | Rham-inducible flp, TS ori | 66 |
| pSCrhaB2 | Expression vector; oripBBR1rhaR, rhaS, PrhaBTmprmob+ | 68 |
| pSC301 | pSCrhaB2 expressing B. glumae dbcA | This study |
| pBBR1MCS-2 | Expression vector; RK2mob lacZα E. coli lac promoter, Kanr | 70 |
| pMCS501 | pBBR1MCS-2 expressing B. glumae dbcA with dbcA promoter | This study |
| pRP101 | pBBR1MCS-2 expressing B. thailandensis dbcA | 5 |
| pRP102 | pBBR1MCS-2 expressing E. coli yqjA | 5 |
| pRP103 | pBBR1MCS-2 expressing E. coli yghB | 5 |
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| K1F | ATTAGGTACCAACACGTACTGGTGGATCGAG |
| K1R | TCCCCAATTCGAGCTCATCAGCGTATCCAAGACGTTTCCTT |
| K2F | ACGTCTTGGATACGCTGATGAGCTCGAATTGGGGATCTTGAAGTA |
| K2R | TCCCACCAGCACGATCACCCACTAGTGAGCTCATGCATGAT |
| K3F | CATGAGCTCACTAGTGGGTGATCGTGCTGGTGGGAGTC |
| K3R | ATATAAGCTTCTGTTCCACAGCACCTCGTTT |
| Fw_pEX18Cont. | AGGCAAATTCTGTTTTATCAGACCGC |
| Rev_pEX18Cont. | GAGCGGATAACAATTTCACACAGGAA |
| Seq 1 | ATACTTCTTCGTCAACGGCCG |
| Seq 2 | AGGCGTGTTGAAGTACAGCTC |
| Seq 3 | GTGGAATCGGTGGAATCGGTA |
| Seq 4 | GCATAGCCTTCAGGAGTGAGT |
| Fw_Seq 5A | CGATCATGCATGAGCTCACTAGT |
| Rev_Seq 5A | TCAAGGTCGGCGACTCGATCT |
| Seq 5 | TCGCAATAGACCTGCCAGTAG |
| Seq 6 | AGGATCAGCACGATGGTCAGC |
| KO-FW | AACCGAAGGAAACGTCTTGGATACG |
| KO-REV | AGCTACTGGCAGGTCTATTGCGA |
| FW-pSCdbcA | CGCCCATATGATGGATACGCTGCTTCACTTCGTCAATC |
| REV-pSCdbcA | CGCGAAGCTTTCAGCCGCGCCCGCGCG |
| Fr-Seq-pSCdbcA | CATCATCACGTTCATCTTTCCCTG |
| Rv-Seq-pSCdbcA | GCAAATTCTGTTTTATCAGACCGC |
| pMCSdbcA-FW | ATTAGGTACCATTCGGACATGCGGGAATTATAACGACG |
| pMCSdbcA-REV | CGCGAAGCTTTCAGCCGCGCCCGCGCG |
| SeqM13-FW | TGTAAAACGACGGCCAGTGAG |
| SeqM13-REV | TCACACAGGAAACAGCTATGA |
Underlined text represents homology sequences. Bolded italic text represents restriction enzyme sites.
Deletion of B. glumae 336gr-1 dbcA gene.
Homologous recombination was used to knock out dbcA (bglu_1g06460) from the B. glumae genome (61, 62). Q5 DNA polymerase (New England BioLabs) was used for PCR amplification of DNA fragments. The 1,258-bp upstream and 1,395-bp downstream regions of the dbcA gene were PCR amplified from B. glumae genomic DNA. A trimethoprim resistance (Tmpr) cassette was PCR amplified from pUC18T-mini-Tn7-Tp plasmid (5). The 5′ end of the amplified Tmpr fragment contained a 35-bp homology with the 3′ end of amplified upstream region of dbcA. Similarly, the 3′ end of the amplified Tmpr fragment contained 35-bp homology with the 5′ end of amplified downstream region of dbcA. The Gibson Assembly kit (New England Biolabs) was used to ligate the three amplified fragments. One microliter of the ligated product (3,525 bp) was used as template for PCR amplification. The PCR product was gel purified using QIAquick gel extraction kit (Qiagen), digested with KpnI and HindIII, and ligated into a similarly digested vector, pEX18Gm (63). The plasmid construct, named pEX3525, was transformed into E. coli XL1 Blue and sequenced to confirm 100% similarity with reference sequence. Vector pEX3525 was transformed into mobilizer strain E. coli RHO3 and introduced into parental B. glumae strain by conjugation (64). Recombinant B. glumae colonies were selected on LB agar medium containing 100 μg/ml trimethoprim and 30 μg/ml gentamicin. To induce a secondary homologous recombination, the recombinant B. glumae colonies were grown in LB medium containing no antibiotic at 30°C with shaking overnight. The 10−2 and 10−3 diluted overnight cultures were spread on TY (tryptone and yeast extract) agar medium containing 30% sucrose for counter selection (65). To confirm the deletion of dbcA, sucrose-resistant colonies were screened for gentamicin sensitivity and trimethoprim resistance. Replacement of the dbcA gene with the Tmpr cassette was confirmed by PCR. The FLP recombination target (FRT) method was used to remove the Tmpr cassette from the B. glumae ΔdbcA::Tmpr strain to obtain ΔdbcA::FRT using pFlpTet (66). The genomic DNA was extracted (Easy DNA kit; Invitrogen) from parental WT, Tmpr, and Tmps. B. glumae strains were PCR amplified with KO-FW and KO-REV primers to confirm the dbcA mutant strains (see Fig. S1 in the supplemental material).
Transformation and complementation analysis.
Heat shock was used for transformation of E. coli unless otherwise indicated (67). Biparental conjugation was used for transformation of B. glumae (64). Colony PCR (OneTaq Hot Start Quick-Load 2× master mix with GC buffer; New England BioLabs) was performed to confirm the exconjugant B. glumae colonies harboring appropriate plasmid. B. glumae dbcA with a start codon changed from TTG to ATG was PCR amplified from genomic DNA of B. glumae and ligated into NdeI and HindIII restriction sites of an expression vector pSCrhaB2 under rhamnose-inducible PrhaB promoter resulting in pSC301 (68). For one experiment (Fig. 1C), the dbcA gene with its own promoter sequence was PCR amplified from genomic DNA of B. glumae 336gr-1 and ligated into KpnI and HindIII restriction sites of expression vector pBBR1MCS-2 resulting in pMCS501. Transformed E. coli and B. glumae strains were selected on 100 μg/ml Tmp (pSCrhaB2-based vectors) or 100 μg/ml Kan (pBBR1MCS-2-based vectors). All DNA sequencing was performed at the Louisiana State University (LSU) College of Science Genomic Facility.
Susceptibility of colistin and CCCP.
Colistin and CCCP susceptibility assays were performed in MH2 medium. Fresh 1:50 diluted overnight cultures in MH2 medium with appropriate antibiotics were grown to an optical density at 600 nm (OD600) of ∼0.6 at 37°C in a shaking incubator. The MH2 agar plates containing appropriate antibiotics were spotted with 5 μl of serially log10-diluted bacterial cells. The plates were incubated at 37°C, and bacterial growth was analyzed after 48 h of incubation. Colistin MIC strips (Liofilchem, Waltham, MA) were used to measure colistin MIC of B. glumae strains according to the manufacturer’s instructions.
Virulence assays.
Onion scales are a convenient host system to measure the virulence of B. glumae strains (35). B. glumae strains were grown in MH2 agar medium containing 100 μg/ml Tmp at 37°C for 48 h. Bacterial culture grown on MH2 agar plates was removed with sterile loops and resuspended in MH2. The bacterial suspension was adjusted to 5 × 109 CFU/ml. The fresh scales of yellow onion were cut into rectangular slices with a sterile razor blade. A 5-mm square wound in the center of the inner surface of each of the onion scales was formed using a sterile 200-μl micropipette tip. Ten microliters of bacterial suspension (∼5 × 107 CFU) was applied to the onion scales. For control onion scales, 10 μl sterile MH2 medium was added. The infected onion scales were incubated at 30°C for 72 h in a humid chamber. The disease severity of each B. glumae strain was determined by visual observation and measuring the area of maceration. For onion virulence assay in the presence of sodium bicarbonate, an equal amount of bacterial suspension (5 × 109 CFU/ml) was added into microcentrifuge tubes containing sterile MH2 medium supplemented with either 0 or 5 mM sodium bicarbonate. The tubes were incubated for 1.5 h at room temperature (RT) without shaking. Ten microliters of suspension was directly inoculated into onion scales. For the control, 10 μl of sterile MH2 medium containing 5 mM sodium bicarbonate was used.
For B. glumae virulence assays using rice panicles, the LSU College of Agriculture Greenhouse was used to grow rice (Oryza sativa cv. Bengal) and perform the assays as described (35). Briefly, B. glumae 336gr-1 and the ΔdbcA strain were grown in LB agar without antibiotics at 37°C for 48 h. Bacteria were removed with sterile loops and suspended in sterile water at 5 × 107 CFU/ml. Five milliliters of bacterial suspension were directly sprayed, twice at 2-day intervals, on the panicles at the 20 to 30% flowering stage of rice plant. For the control, sterile water was sprayed on the panicles. The panicle blight symptoms were evaluated at 3, 7, and 10 days after first inoculation by assigning a virulence score on a 0 to 9 scale (36). Virulence scores of 0, 1, 2, 3, 4, 5, 6, 7, 8, and 9 indicate no symptom, 1% to 10% symptomatic, 11% to 20% symptomatic, 21% to 30% symptomatic, 31% to 40% symptomatic, 41% to 50% symptomatic, 51% to 60% symptomatic, 61% to 70% symptomatic, 71% to 80% symptomatic, and more than 80% symptomatic panicle, respectively. For panicle assays in the presence of sodium bicarbonate, bacteria were grown in LB agar medium containing either 0 or 5 mM sodium bicarbonate at 37°C for 72 h. LB agar was buffered with 70 mM Tris, pH 7.0. Bacteria were removed with sterile loops and suspended in either sterile water or 5 mM sodium bicarbonate at 5 × 107 CFU/ml. Five milliliters of bacterial suspension was directly sprayed, twice at 2-day intervals, on the panicles at the 20 to 30% flowering stage of rice plant. For the control, 5 ml of sodium bicarbonate was sprayed on the panicles. The panicle blight symptoms were evaluated as described in above. All experiments were repeated three time with three replicates each.
Toxoflavin quantification and thin-layer chromatograph.
Extraction and quantification of toxoflavin from liquid and solid culture media were conducted as described (65). For toxoflavin quantification from liquid culture medium, B. glumae strains were grown shaking at 37°C in a 250-ml conical flask containing 25 ml fresh LB medium buffered with 70 mM Tris, pH 7.0. The culture supernatant was collected by centrifugation and 1 ml of culture supernatant was extracted with 1 ml of chloroform. The extracted chloroform fraction was transferred to a new microcentrifuge tube and dried in a fume hood at RT. The residue in tubes was dissolved in 1 ml 80% methanol. Absorbance was measured at 393 nm against a toxoflavin-free control and normalized to OD600. Toxoflavin concentration was calculated from a standard curve (see Fig. S4 in the supplemental material).
For toxoflavin quantification from solid medium, B. glumae strains were grown in LB agar medium without antibiotics at 37°C for 72 h. The bacterial culture on agar plates was completely removed by scraping the culture with a sterile razor blade. Five grams of agar medium was collected from the plate, cut into small pieces, and soaked in 5 ml of chloroform. The agar medium/chloroform fractions were incubated for 30 min at RT in 50-ml Falcon tubes. One milliliter of the extracted chloroform fraction was aliquoted in new microcentrifuge tubes and dried in a fume hood at RT. The residue in tubes was dissolved in 1 ml 80% methanol. The toxoflavin absorbance and concentration was measured as described above.
Thin-layer chromatography (TLC) analysis for toxoflavin produced by B. glumae strains was performed as described previously with some modifications (3). Five milliliters of culture supernatant was extracted with 5 ml of chloroform (vol/vol). The extracted chloroform fraction was transferred to new tubes and dried in a rotary evaporator (Vacufuge; Eppendorf) at 30°C. The residue in tubes was dissolved in 30 μl of 80% methanol. Ten microliters of methanol extract was spotted on a TLC plate (TLC silica gel 60; EMD Millipore Corporation). The TLC plate was developed with chloroform/methanol (95:5, vol/vol), dried, and imaged using 365-nm UV light. Purified toxoflavin was purchased from Cayman Chemical.
Membrane potential.
JC-1 dye was used to measure membrane potential (9). All B. glumae strains were treated with 6 μM JC-1 in permeabilization buffer (10 mM tris, pH 8.0, 1 mM EDTA, 10 mM glucose). Twenty-five micromolar CCCP was used as a control for loss of ΔΨ. Bacterial cells were incubated at 30°C in the dark without shaking for 1 h. A JASCO FP-6300 spectrofluorometer was used to measure the fluorescence ratio. To measure membrane potential for bicarbonate-treated bacterial cells, 5 mM sodium bicarbonate was directly added to growth media, and strains were grown until their OD600 reached ∼0.6.
Statistical analysis.
Experiments were repeated three times with three biological replicates. Representative images of virulence assays are included in the figures. The data presented in the graphs indicate the mean ± standard deviation (SD) value for three independent replicates of each treatment. GraphPad Prism 9.0 was used to produce graphs and calculate the statistical significances by unpaired Student's t test.
ACKNOWLEDGMENTS
J. H. Ham was supported by the USDA NIFA (Hatch project 1015305).
We thank Marcia Newcomer (LSU, Department of Biological Sciences) and Inderjit Barphagha (LSU, Department of Plant Pathology and Crop Physiology) for technical support.
Footnotes
Supplemental material is available online only.
Contributor Information
William T. Doerrler, Email: wdoerr@lsu.edu.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Ham JH, Melanson RA, Rush MC. 2011. Burkholderia glumae: next major pathogen of rice? Mol Plant Pathol 12:329–339. 10.1111/j.1364-3703.2010.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park J, Lee HH, Jung H, Seo YS. 2019. Transcriptome analysis to understand the effects of the toxoflavin and tropolone produced by phytopathogenic Burkholderia on Escherichia coli. J Microbiol 57:781–794. 10.1007/s12275-019-9330-1. [DOI] [PubMed] [Google Scholar]
- 3.Kim J, Kim JG, Kang Y, Jang JY, Jog GJ, Lim JY, Kim S, Suga H, Nagamatsu T, Hwang I. 2004. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol Microbiol 54:921–934. 10.1111/j.1365-2958.2004.04338.x. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S, Doerrler WT. 2014. Members of the conserved DedA family are likely membrane transporters and are required for drug resistance in Escherichia coli. Antimicrob Agents Chemother 58:923–930. 10.1128/AAC.02238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panta PR, Kumar S, Stafford CF, Billiot CE, Douglass MV, Herrera CM, Trent MS, Doerrler WT. 2019. A DedA family membrane protein is required for Burkholderia thailandensis colistin resistance. Front Microbiol 10:2532. 10.3389/fmicb.2019.02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sikdar R, Doerrler WT. 2010. Inefficient Tat-dependent export of periplasmic amidases in an Escherichia coli strain with mutations in two DedA family genes. J Bacteriol 192:807–818. 10.1128/JB.00716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompkins K, Chattopadhyay B, Xiao Y, Henk MC, Doerrler WT. 2008. Temperature sensitivity and cell division defects in an Escherichia coli strain with mutations in yghB and yqjA, encoding related and conserved inner membrane proteins. J Bacteriol 190:4489–4500. 10.1128/JB.00414-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doerrler WT, Sikdar R, Kumar S, Boughner LA. 2013. New functions for the ancient DedA membrane protein family. J Bacteriol 195:3–11. 10.1128/JB.01006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sikdar R, Simmons AR, Doerrler WT. 2013. Multiple envelope stress response pathways are activated in an Escherichia coli strain with mutations in two members of the DedA membrane protein family. J Bacteriol 195:12–24. 10.1128/JB.00762-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Doerrler WT. 2015. Escherichia coli YqjA, a member of the conserved DedA/Tvp38 membrane protein family, is a putative osmosensing transporter required for growth at alkaline pH. J Bacteriol 197:2292–2300. 10.1128/JB.00175-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewinson O, Padan E, Bibi E. 2004. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc Natl Acad Sci U S A 101:14073–14078. 10.1073/pnas.0405375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holdsworth SR, Law CJ. 2013. Multidrug resistance protein MdtM adds to the repertoire of antiporters involved in alkaline pH homeostasis in Escherichia coli. BMC Microbiol 13:113. 10.1186/1471-2180-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinner E, Kotler Y, Padan E, Schuldiner S. 1993. Physiological role of NhaB, a specific Na+/H+ antiporter in Escherichia coli. J Biol Chem 268:1729–1734. 10.1016/S0021-9258(18)53913-2. [DOI] [PubMed] [Google Scholar]
- 14.Radchenko MV, Tanaka K, Waditee R, Oshimi S, Matsuzaki Y, Fukuhara M, Kobayashi H, Takabe T, Nakamura T. 2006. Potassium/proton antiport system of Escherichia coli. J Biol Chem 281:19822–19829. 10.1074/jbc.M600333200. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Bradley CL, Mukashyaka P, Doerrler WT. 2016. Identification of essential arginine residues of Escherichia coli DedA/Tvp38 family membrane proteins YqjA and YghB. FEMS Microbiol Lett 363:fnw133. 10.1093/femsle/fnw133. [DOI] [PubMed] [Google Scholar]
- 16.Noumi T, Inoue H, Sakurai T, Tsuchiya T, Kanazawa H. 1997. Identification and characterization of functional residues in a Na+/H+ antiporter (NhaA) from Escherichia coli by random mutagenesis. J Biochem 121:661–670. 10.1093/oxfordjournals.jbchem.a021637. [DOI] [PubMed] [Google Scholar]
- 17.Adler J, Bibi E. 2004. Determinants of substrate recognition by the Escherichia coli multidrug transporter MdfA identified on both sides of the membrane. J Biol Chem 279:8957–8965. 10.1074/jbc.M313422200. [DOI] [PubMed] [Google Scholar]
- 18.Sigal N, Vardy E, Molshanski-Mor S, Eitan A, Pilpel Y, Schuldiner S, Bibi E. 2005. 3D model of the Escherichia coli multidrug transporter MdfA reveals an essential membrane-embedded positive charge. Biochemistry 44:14870–14880. 10.1021/bi051574p. [DOI] [PubMed] [Google Scholar]
- 19.Holdsworth SR, Law CJ. 2012. Functional and biochemical characterisation of the Escherichia coli major facilitator superfamily multidrug transporter MdtM. Biochimie 94:1334–1346. 10.1016/j.biochi.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Fluman N, Ryan CM, Whitelegge JP, Bibi E. 2012. Dissection of mechanistic principles of a secondary multidrug efflux protein. Mol Cell 47:777–787. 10.1016/j.molcel.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abramson J, Iwata S, Kaback HR. 2004. Lactose permease as a paradigm for membrane transport proteins (Review). Mol Membr Biol 21:227–236. 10.1080/09687680410001716862. [DOI] [PubMed] [Google Scholar]
- 22.Loutet SA, Valvano MA. 2011. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front Cell Infect Microbiol 1:6. 10.3389/fcimb.2011.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeaman MR, Yount NY. 2007. Unifying themes in host defence effector polypeptides. Nat Rev Microbiol 5:727–740. 10.1038/nrmicro1744. [DOI] [PubMed] [Google Scholar]
- 26.Lazzaro BP, Zasloff M, Rolff J. 2020. Antimicrobial peptides: application informed by evolution. Science 368:eaau5480. 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nawrot R, Barylski J, Nowicki G, Broniarczyk J, Buchwald W, Goździcka-Józefiak A. 2014. Plant antimicrobial peptides. Folia Microbiol (Praha) 59:181–196. 10.1007/s12223-013-0280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverstein KA, MoskalWA, Jr, Wu HC, Underwood BA, Graham MA, Town CD, VandenBosch KA. 2007. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J 51:262–280. 10.1111/j.1365-313X.2007.03136.x. [DOI] [PubMed] [Google Scholar]
- 29.Panta PR, Doerrler WT. 2020. A Burkholderia thailandensis DedA family membrane protein is required for proton motive force dependent lipid A modification. Front Microbiol 11:618389. 10.3389/fmicb.2020.618389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panta PR, Doerrler WT. 2021. A link between pH homeostasis and colistin resistance in bacteria. Sci Rep 11:13230. 10.1038/s41598-021-92718-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boughner LA, Doerrler WT. 2012. Multiple deletions reveal the essentiality of the DedA membrane protein family in Escherichia coli. Microbiology (Reading) 158:1162–1171. 10.1099/mic.0.056325-0. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109. 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaara M, Vaara T. 1994. Ability of cecropin B to penetrate the enterobacterial outer membrane. Antimicrob Agents Chemother 38:2498–2501. 10.1128/AAC.38.10.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karki HS, Shrestha BK, Han JW, Groth DE, Barphagha IK, Rush MC, Melanson RA, Kim BS, Ham JH. 2012. Diversities in virulence, antifungal activity, pigmentation and DNA fingerprint among strains of Burkholderia glumae. PLoS One 7:e45376. 10.1371/journal.pone.0045376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lelis T, Peng J, Barphagha I, Chen R, Ham JH. 2019. The virulence function and regulation of the metalloprotease gene prtA in the plant-pathogenic bacterium Burkholderia glumae. Mol Plant Microbe Interact 32:841–852. 10.1094/MPMI-11-18-0312-R. [DOI] [PubMed] [Google Scholar]
- 37.Lee J, Park J, Kim S, Park I, Seo YS. 2016. Differential regulation of toxoflavin production and its role in the enhanced virulence of Burkholderia gladioli. Mol Plant Pathol 17:65–76. 10.1111/mpp.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell P. 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191:144–148. 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 39.Bakker EP, Mangerich WE. 1981. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J Bacteriol 147:820–826. 10.1128/jb.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engl C, Beek AT, Bekker M, de Mattos JT, Jovanovic G, Buck M. 2011. Dissipation of proton motive force is not sufficient to induce the phage shock protein response in Escherichia coli. Curr Microbiol 62:1374–1385. 10.1007/s00284-011-9869-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jovanovic G, Lloyd LJ, Stumpf MP, Mayhew AJ, Buck M. 2006. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem 281:21147–21161. 10.1074/jbc.M602323200. [DOI] [PubMed] [Google Scholar]
- 42.Farha MA, French S, Stokes JM, Brown ED. 2018. Bicarbonate alters bacterial susceptibility to antibiotics by targeting the proton motive force. ACS Infect Dis 4:382–390. 10.1021/acsinfecdis.7b00194. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi A, Ohmori H, Kaneko-Ohdera M, Nomura T, Sawai T. 1991. Delta pH-dependent accumulation of tetracycline in Escherichia coli. Antimicrob Agents Chemother 35:53–56. 10.1128/AAC.35.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naughton LM, An SQ, Hwang I, Chou SH, He YQ, Tang JL, Ryan RP, Dow JM. 2016. Functional and genomic insights into the pathogenesis of Burkholderia species to rice. Environ Microbiol 18:780–790. 10.1111/1462-2920.13189. [DOI] [PubMed] [Google Scholar]
- 45.Nandakumar R, Shahjahan AKM, Yuan XL, Dickstein ER, Groth DE, Clark CA, Cartwright RD, Rush MC. 2009. Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the southern United States. Plant Dis 93:896–905. 10.1094/PDIS-93-9-0896. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q. 2007. Strategies for developing Green Super Rice. Proc Natl Acad Sci U S A 104:16402–16409. 10.1073/pnas.0708013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shew AM, Durand-Morat A, Nalley LL, Zhou XG, Rojas C, Thoma G. 2019. Warming increases bacterial panicle blight (Burkholderia glumae) occurrences and impacts on USA rice production. PLoS One 14:e0219199. 10.1371/journal.pone.0219199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang FT, Xu Q, Sikdar R, Xiao Y, Cox JS, Doerrler WT. 2010. BB0250 of Borrelia burgdorferi is a conserved and essential inner membrane protein required for cell division. J Bacteriol 192:6105–6115. 10.1128/JB.00571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Y, Cromie MJ, Hsu FF, Turk J, Groisman EA. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol Microbiol 53:229–241. 10.1111/j.1365-2958.2004.04107.x. [DOI] [PubMed] [Google Scholar]
- 50.Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol 187:5387–5396. 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weatherspoon-Griffin N, Zhao G, Kong W, Kong Y, Morigen Andrews-Polymenis H, McClelland M, Shi Y. 2011. The CpxR/CpxA two-component system up-regulates two Tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J Biol Chem 286:5529–5539. 10.1074/jbc.M110.200352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jana B, Cain AK, Doerrler WT, Boinett CJ, Fookes MC, Parkhill J, Guardabassi L. 2017. The secondary resistome of multidrug-resistant Klebsiella pneumoniae. Sci Rep 7:42483. 10.1038/srep42483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang L, Feng Y, Zong Z. 2019. Heterogeneous resistance to colistin in Enterobacter cloacae complex due to a new small transmembrane protein. J Antimicrob Chemother 74:2551–2558. 10.1093/jac/dkz236. [DOI] [PubMed] [Google Scholar]
- 54.Datta A, Ghosh A, Airoldi C, Sperandeo P, Mroue KH, Jimenez-Barbero J, Kundu P, Ramamoorthy A, Bhunia A. 2015. Antimicrobial peptides: insights into membrane permeabilization, lipopolysaccharide fragmentation and application in plant disease control. Sci Rep 5:11951. 10.1038/srep11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee IH, Jung YJ, Cho YG, Nou IS, Huq MA, Nogoy FM, Kang KK. 2017. SP-LL-37, human antimicrobial peptide, enhances disease resistance in transgenic rice. PLoS One 12:e0172936. 10.1371/journal.pone.0172936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montville TJ, Goldstein PK. 1987. Sodium bicarbonate reduces viability and alters aflatoxin distribution of Aspergillus parasiticus in Czapek's agar. Appl Environ Microbiol 53:2303–2307. 10.1128/aem.53.10.2303-2307.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letscher-Bru V, Obszynski CM, Samsoen M, Sabou M, Waller J, Candolfi E. 2013. Antifungal activity of sodium bicarbonate against fungal agents causing superficial infections. Mycopathologia 175:153–158. 10.1007/s11046-012-9583-2. [DOI] [PubMed] [Google Scholar]
- 58.Drake DR, Cardenzana A, Srikantha R. 1995. Enhanced bactericidal activity of Arm and Hammer Dental Care. Am J Dent 8:308–312. [PubMed] [Google Scholar]
- 59.Md Meftaul I, Venkateswarlu K, Dharmarajan R, Annamalai P, Megharaj M. 2020. Pesticides in the urban environment: a potential threat that knocks at the door. Sci Total Environ 711:134612. 10.1016/j.scitotenv.2019.134612. [DOI] [PubMed] [Google Scholar]
- 60.Bojarski B, Witeska M. 2020. Blood biomarkers of herbicide, insecticide, and fungicide toxicity to fish—a review. Environ Sci Pollut Res 27:19236–19250. 10.1007/s11356-020-08248-8. [DOI] [PubMed] [Google Scholar]
- 61.Silayeva O, Barnes AC. 2018. Gibson assembly facilitates bacterial allelic exchange mutagenesis. J Microbiol Methods 144:157–163. 10.1016/j.mimet.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 62.Schweizer HP. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol 6:1195–1204. 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 63.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 64.Lopez CM, Rholl DA, Trunck LA, Schweizer HP. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol 75:6496–6503. 10.1128/AEM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen R, Barphagha IK, Karki HS, Ham JH. 2012. Dissection of quorum-sensing genes in Burkholderia glumae reveals non-canonical regulation and the new regulatory gene tofM for toxoflavin production. PLoS One 7:e52150. 10.1371/journal.pone.0052150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia EC, Anderson MS, Hagar JA, Cotter PA. 2013. Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Mol Microbiol 89:1213–1225. 10.1111/mmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Froger A, Hall JE. 2007. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp 2007:e253. 10.3791/253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cardona ST, Valvano MA. 2005. An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid 54:219–228. 10.1016/j.plasmid.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 69.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 70.Kovach ME, Elzer PH, Steven Hill D, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download AEM.00915-21-s0001.pdf, PDF file, 0.7 MB (748.1KB, pdf)