Abstract
Klebsiella pneumoniae poses an urgent public health threat, causing nosocomial outbreaks in different continents. It has been observed to develop resistance to antimicrobials more easily than most bacteria. These days, multidrug-resistant strains are being increasingly reported from different countries. However, studies on the surveillance of multidrug-resistant Klebsiella pneumoniae are very rare in Ethiopia. This study aimed to determine the antimicrobial resistance patterns and magnitude of MDR K. pneumoniae isolates from patients attending or admitted to Tikur Anbessa Specialized Hospital (TASH). A cross-sectional study was conducted from September 2018 to February 2019 at TASH, Addis Ababa, Ethiopia. Identification of K. pneumoniae was done by examining the Gram stain, colony characteristics on MacConkey agar and 5% sheep blood agar, as well as using a series of biochemical tests. Antimicrobial susceptibility testing of the isolates for 21 antimicrobials was done by the Kirby–Bauer disc diffusion technique. Data were double entered using Epidata 3.1 and exported to SPSS version 25 software for analysis. Among the total K. pneumoniae isolates (n = 132), almost all 130 (98.5%) were MDR. Two (1.5%) isolates showed complete non-susceptibility to all antimicrobial agents tested. Moreover, a high rate of resistance was observed to cefotaxime and ceftriaxone 128 (97%), trimethoprim-sulfamethoxazole 124 (93.9%), and cefepime 111 (84.1%). High susceptibility was recorded to amikacin 123 (93.2%), imipenem 107 (81.1%), meropenem 96 (72.7%), and ertapenem 93 (70.5%). K. pneumoniae isolates showed a high rate of resistance to most of the tested antimicrobials. The magnitude of MDR K. pneumoniae was very alarming. Therefore, strengthening antimicrobial stewardship programs and antimicrobial surveillance practices is strongly recommended in TASH.
Keywords: K. pneumoniae, antimicrobial susceptibility patterns, MDR, Ethiopia
1. Introduction
Klebsiella pneumoniae is a Gram-negative, rod-shaped bacterium that belongs to the family Enterobacteriaceae. K. pneumoniae is primarily an opportunistic pathogen that attacks immune-compromised individuals who are hospitalized and suffer from severe underlying diseases. In these patients, it results in hospital-acquired infections associated with the urinary tract, blood-stream, wounds, and respiratory tract [1]. Besides this, K. pneumoniae has emerged as a cause of severe community-acquired infections, such as community-acquired pneumonia, pyogenic liver abscess, and metastatic infections such as meningitis [2]. It accounts for about one-third of all Gram-negative infections overall [3].
Antimicrobial resistance is a growing problem in modern healthcare around the world. It is estimated that antimicrobial resistance-related deaths each year will rise from currently 700,000 to 10 million and cost 100 trillion dollars to global economic output by 2050 if containment of antimicrobial resistance at a global level is not effectively implemented [4]. Multidrug-resistant (MDR) bacterial infections that pose a serious risk to patients are increasing worldwide [5]. One of the most common species of bacteria that cause problems in healthcare today is K. pneumoniae [6], which, together with other highly important MDR pathogens, comprises the ESKAPE group that stands for Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, to emphasize that these pathogens effectively “escape” the effects of antibacterial drugs [7]. Its contribution to the antimicrobial resistance crisis is impossible to quantify. It is unique amongst the Gram-negative ESKAPE pathogens because of its high diversity of acquired antimicrobial resistance genes, high plasmid load, significantly more varied DNA composition reflecting diverse horizontal gene transfer partners, and broad ecological range [3]. Nowadays, K. pneumoniae strains are recognized as an urgent threat to human health, because of the emergence of MDR strains associated with hospital outbreaks [6]. It has gained attention worldwide, especially in developed countries, due to its high drug resistance. Antimicrobial resistance rates in K. pneumoniae have steadily increased over the years, and K. pneumoniae is becoming resistant to virtually all aminoglycosides, quinolones, and β-lactams [8]. For instance, at the European level, more than one-third (36.6%) of K. pneumoniae isolates reported to the European antimicrobial resistance surveillance network for 2019 were resistant to at least one of the antimicrobial groups under regular surveillance, i.e., fluoroquinolones, third-generation cephalosporins, aminoglycosides, and carbapenems [9]. The Ethiopian annual antimicrobial surveillance report showed that 95.8% of K. pneumoniae isolates were resistance to ceftriaxone, 86.7% to ceftazidime, 95.6% to trimethoprim-sulfamethoxazole, 83.3% to cefepime, 62.7% to gentamicin, 48.1% to ciprofloxacin, 30.6% to meropenem, and 7.2% to amikacin [10]. K. pneumoniae has been observed to develop resistance to antimicrobials more easily than most bacteria [2,11]. One of the main mechanisms of resistance is through β-lactamase production that hydrolyzes β-lactam antibiotics. The most reported β-lactamase genes among K. pneumoniae worldwide were blaSHV-12, blaCTX-M-2, and blaSHV-5. Furthermore, occurrence of the blaNDM-1 gene carrying K. pneumoniae has been reported in all continents [12]. In Ethiopia, in a recent systematic review and meta-analysis, the pooled estimates of ESBL-producing K. pneumoniae was 64.3% (95% CI: 47.0–81.5) [13]. Few treatment options, such as polymyxins and tigecycline, are available for infections caused by MDR K. pneumoniae. However, there are increasing reports of K. pneumoniae isolates resistance to these drugs [3]. Thus, the best option is to control the development and spread of antimicrobial resistance. It is obvious that to better understand the extent of drug resistance in different settings and design better control strategies, research on such a type of pathogen is crucial. Continuous global dissemination of multidrug-resistant and extremely drug-resistant K. pneumoniae, with superior ability to cause multi-continent outbreaks, have been noted [3,6]. Some of K. pneumoniae strains are resistant to about 95% of antimicrobials of the pharmaceutical market in the world [12]. However, to the best of our knowledge, studies emphasizing the degree of drug resistance in K. pneumoniae in Africa, particularly in Ethiopia, are still limited. Therefore, this study was planned to determine the antimicrobial-resistance patterns and magnitude of MDR K. pneumoniae at Tikur Anbessa Specialized Hospital (TASH), Addis Ababa, Ethiopia. TASH is the largest teaching and referral hospital in Ethiopia.
2. Results
2.1. Socio-Demographic and Clinical Characteristics
A total of 132 non-repetitive K. pneumoniae isolates collected from patients who were admitted to or attended different departments of TASH were included. As shown in Table 1, among the total isolates, 83 (62.9%) were isolated from males. The majority of the isolates were recovered from blood specimens (63, 47.7%) followed by urine (34, 25.8%), wounds (21, 15.9 %), body fluids (10, 7.6 %), and sputum specimens (4, 3.0%). The average age of the study participants was 13.4 years, varying from at least 1 day to a maximum of 86 years, of which 74 (56.1%) participants were below 5 years. The majority of K. pneumoniae isolates were recovered from hospitalized patients (120, 90.9%), of which the largest number were from patients admitted in pediatric wards (53, 44.2%) and intensive care units (ICUs) (46, 38.3%). Conversely, only two isolates were from patients in orthopedics, and two were from emergency wards.
Table 1.
Distribution of K. pneumoniae isolates among demographic and clinical characteristics of study participants.
| Variables | Category | Frequency | Percentage |
| Gender | Male | 83 | 62.9 |
| Female | 49 | 37.1 | |
| Age | birth to <5 years | 74 | 56.1 |
| 5 years to <18 years | 20 | 15.2 | |
| 18 years to <45 years | 25 | 18.9 | |
| ≥45 years | 13 | 9.8 | |
| Specimen Type | Blood | 63 | 47.7 |
| Urine | 34 | 25.8 | |
| Wound | 21 | 15.9 | |
| Body fluids | 10 | 7.6 | |
| Sputum | 4 | 3.0 | |
| Patient Setting | Inpatient | 120 | 90.9 |
| Outpatient | 12 | 9.1 | |
| Ward type | Pediatric ward | 53 | 44.2 |
| ICU | 46 | 38.3 | |
| Medical ward | 9 | 7.5 | |
| Surgical ward | 8 | 6.7 | |
| Other wards | 4 | 3.3 |
Other wards: emergency (2) and orthopedics (2).
2.2. Antimicrobial Susceptibility Patterns of K. pneumoniae Isolates
In this study, the susceptibility of 132 K. pneumoniae isolates to 21 antimicrobial agents belonging to 12 antimicrobial categories was determined. As presented in Table 2, a high rate of resistance was observed to commonly used antimicrobials such as cefotaxime (128/132, 97%), ceftriaxone (128/132, 97%), trimethoprim-sulfamethoxazole (124/132, 93.9%), and cefepime (111/132, 84.1%). Besides this, a significant intermediate level of resistance was noted to tobramycin (26.5%), amoxicillin-clavulanate (24.2%), ceftazidime (21.2%), piperacillin-tazobactam (20.5%), aztreonam (22.7%), and ciprofloxacin (18.9%). On the other hand, K. pneumoniae isolates showed the highest susceptibility to amikacin (123, 93.2%), followed by carbapenem antimicrobials, which were imipenem (107, 81.1%), meropenem (96, 72.7%), and ertapenem (93, 70.5%).
Table 2.
Antimicrobial susceptibility patterns of K. pneumoniae isolates.
| Antimicrobial Categories | Antimicrobial Agents | Antimicrobial Susceptibility Pattern | |||
|
Susceptible
n (%) |
Inter
Mediate n (%) |
Resistant
n (%) |
|||
| Tetracyclines | Tetracycline | 21(15.9) | 15(11.4) | 96(72.7) | |
| Aminoglycosides | Gentamicin | 29(22.0) | 8(6) | 95(72.0) | |
| Tobramycin | 49(37.1) | 35(26.5) | 48(36.4) | ||
| Amikacin | 123(93.2) | 5(3.8) | 4(3.0) | ||
| Fluoroquinolones | Ciprofloxacin | 58(43.9) | 25(18.9) | 49(37.1) | |
| Naldixic acid | 30(22.7) | 42(31.8) | 60(45.5) | ||
| Antipseudomonal Penicillins + β-lactamase inhibitors | Piperacillin-tazobactam | 55(41.7) | 27(20.5) | 50(37.9) | |
| Penicillins + β-lactamase inhibitors | Amoxicillin-clavulanate | 18(13.6) | 32(24.2) | 82(62.1) | |
| Folate pathway inhibitors | Trimethoprim-Sulfamethoxazole | 6(4.5) | 2(1.5) | 124(94) | |
| Phenicols | Chloramphenicol | 59(44.7) | 13(9.8) | 60(45.5) | |
| Cephamycins | Cefoxitin | 62(47.0) | 12(9.1) | 58(43.9) | |
| β-lactams | 1st and 2nd generation cephalosporins | Cefuroxime | 2(1.5) | 2(1.5) | 128(97.0) |
| Cefazolin | 1(0.8) | 1(0.8) | 130(98.5) | ||
| 3rd and 4th generation cephalosporins | Cefepime | 5(3.8) | 16(12.1) | 111(84.1) | |
| Ceftriaxone | 4(3.0) | 0(0.0) | 128(97.0) | ||
| Cefotaxime | 4(3.0) | 0(0.0) | 128(97.0) | ||
| Ceftazidime | 17(12.9) | 28(21.2) | 87(65.9) | ||
| Monobactams | Aztreonam | 13(9.8) | 30(22.7) | 89(67.4) | |
| Carbapenems | Meropenem | 96(72.7) | 4(3.0) | 32(24.3) | |
| Imipenem | 107(81.1) | 9(6.8) | 16(12.1) | ||
| Ertapenem | 93(70.5) | 5(3.7) | 34(25.8) | ||
2.3. Magnitude of Multidrug Resistance among K. pneumoniae Isolates
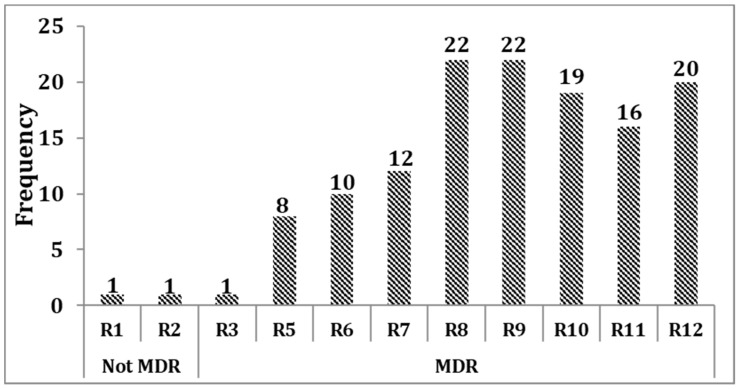
Almost all (130/132, 98.5%) of the isolates were non-susceptible to at least three antimicrobials in different categories and, hence, defined as MDR, whereas only two (1.5%) isolates were not MDR. In our case, the MDR range was wide, which encompassed resistance to three (R3) to 12 (R12) antimicrobial categories. Consequently, it was necessary to indicate the number of isolates in each level of MDR. Based on this, from the total isolates, 20/132 (15.2%) showed non-susceptibility to at least one antimicrobial agent in all categories. Of these, two isolates showed complete non-susceptibility to all antimicrobial agents. Furthermore, 16/132 (12.1%) isolates were non-susceptible to at least one antimicrobial agent in 11 antimicrobial categories. Only one isolate was non-susceptible to at least one antimicrobial agent in merely three antimicrobial categories, which was the least MDR (Figure 1). The details of multidrug-resistance patterns of K. pneumoniae isolates are presented in Figure S1.
Figure 1.
Multidrug resistance levels of K. pneumoniae isolates. MDR: multidrug-resistance; Rn: non-susceptibility to at least one antimicrobial agent in “n” antimicrobial categories, where “n” is the number of antimicrobial categories.
2.4. Distribution of MDR K. pneumoniae Isolates among Inpatient and Outpatient Wards
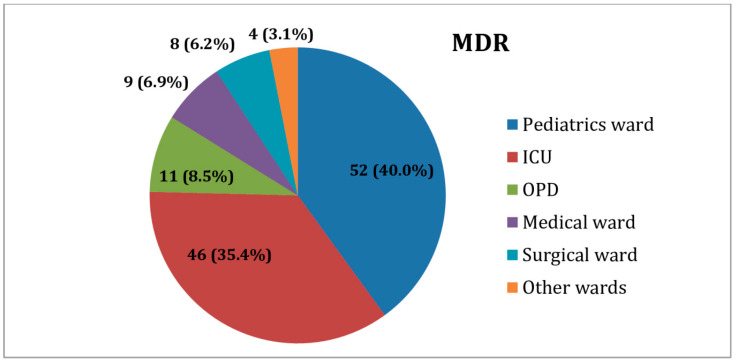
From the total MDR K. pneumoniae isolates, the large majority were isolated from patients in pediatric wards (52/130, 40%) and ICUs (46/130, 35.4%), followed by the outpatient department (11/130, 8.5%). The lowest proportion (4/130, 3.1%) of MDR isolates was obtained from those in orthopedics and emergency wards (Figure 2).
Figure 2.
Distribution of MDR K. pneumoniae isolates among inpatients and outpatients. ICUs: Intensive care units; other wards: emergency (2) and orthopedics (2) OPD: outpatient department.
3. Discussion
Klebsiella pneumoniae is one of the multidrug resistant microorganisms identified as an urgent threat to human health by the World Health Organization [5] and Centers for Disease Control and Prevention [14]. It is rapidly becoming untreatable using even last-line antimicrobials, especially in hospital settings [6], suggesting the need for continuous surveillance of antimicrobial resistance patterns. This work focused on the magnitude of antimicrobial resistance among clinical K. pneumoniae isolates.
3.1. Antimicrobial Resistance Patterns of K. pneumoniae Isolates
Third-generation cephalosporins have been the treatment of choice for Gram-negative bacteria, including K. pneumoniae, but nowadays, they are largely ineffective due to the emergence of ESBL-producing bacteria [8]. This was also true in our study, in which one of the high antimicrobial resistance was against 3rd and 4th generation cephalosporins, ceftriaxone (97%), cefotaxime (97%), cefepime (84.1%), and ceftazidime (65.9%). Likewise, high resistance rates to these antimicrobials were also observed in other recent studies in Ethiopia: 86.4% to cefotaxime and 85.4% to ceftazidime and cefepime in Addis Ababa [15] and 97.6% to ceftazidime, 94.1% to cefepime, and 88.2% to ceftriaxone in Bahir Dar [16]. However, Badamchi et al. from Iran reported a resistance rate of 60.2% to ceftriaxone, 32.6% cefotaxime, and 54.8% cefepime, which was lower than the results of our study [17].
In this study, more than half of the participants had taken 3rd and 4th generation cephalosporins prior to recruitment to the study (see Table S1). Additionally, a previous study by Gutema et al. found that three out of four patients in the hospital where we conducted our study were prescribed antimicrobials empirically, of whom almost 90% were prescribed broad-spectrum β-lactams [18]. Therefore, the high resistance to broad-spectrum β-lactams might be attributed to the indiscriminate use of these antimicrobials, which creates a selective pressure. Resistance to trimethoprim-sulfamethoxazole was 93.9%, which is comparable with different studies in Ethiopia; 86.4% in Addis Ababa [15] and 96.5% in Bahir Dar [16]; furthermore, there was a consistent report from Tunisia citing 94.06% [19]. Nevertheless, it was higher than those observed in studies conducted in Bangladesh (44%) [20] and Nepal (51.3%) [21]. High resistance to trimethoprim-sulfamethoxazole may be due to variations in prescription practices and self-medication because of affordability and availability [22]. The main mechanism of resistance to trimethoprim-sulfamethoxazole in K. pneumoniae is the acquisition of transferable dfr genes, which could mediate the overproduction of dihydrofolate reductase and the presence of drug-resistant dihydropteroate synthetase enzymes, encoded by sul genes [23].
In the current study, from all tested antimicrobials, the best susceptibility (93.2%) was observed to amikacin. A low resistance of K. pneumoniae to amikacin was also reported in previous study in Ethiopia [15]. The lower resistance to amikacin may be due to its absence of use as empirical therapy and non-existence of considerable cross-resistance with β-lactam antimicrobials [24].
K. pneumoniae isolates were more susceptible to carbapenems, following to amikacin, with an overall non-susceptibility rate of 29.6%. Imipenem was the most active carbapenem and revealed a susceptibility rate of 81.1%, followed by meropenem (72.7%) and ertapenem (70.5%). This is in accordance with a previous study conducted in Egypt, which reported susceptibility rates of 75% to meropenem and imipenem [25]. In a study in Addis Ababa, Ethiopia [15], 10.7% of K. pneumoniae isolates were resistant to meropenem, which was lower than our study. Moreover, there was also a study in which no resistance was detected to carbapenems [19], but our study showed a considerable carbapenem resistance, considering that carbapenems are the last-resort antibiotics. In the current study, 15.2% of the study participants had taken carbapenems prior to recruitment to the study (see Table S1). Empirical prescription of carbapenems, particularly meropenem, was very common in the hospital [26], which may have created resistance and furthered the emergence of carbapenemase-producing bacteria.
3.2. Multidrug Resistance among K. pneumoniae Isolates
In our study, almost all (98.5%) K. pneumoniae isolates were MDR, which is comparable with the result of a study conducted in Gondar, Ethiopia (citing 95.6%) from patients with urinary tract infections [27] and Iraq (citing 100%) [28]. On the contrary, our result was higher than studies conducted in Addis Ababa, (citing 83.5%) [15] and Bahir Dar (citing 87.6%) [16]. This high prevalence might be because the majority (90.91%) of K. pneumoniae isolates were collected from hospitalized patients, where many antimicrobials were being prescribed in the hospital, which serves as a selective pressure for drug resistance. Moreover, in this study, more antimicrobial agents were covered in the assessment based on the recommendations of Magiorakos et al., which increases the detection of MDR [29].
Generally, in our study, K. pneumoniae showed a high resistance to most of the antimicrobials tested, which is very alarming. A study on K. pneumoniae in five African and two Vietnamese major towns stated uncontrolled consumption of antimicrobial agents through self-medication, inappropriate antibiotic prescription, the substandard quality of some drugs, and a lack of effective measures to prevent nosocomial infections as key factors for facilitating the spread of antimicrobial resistance among studied countries [30]. Since these conditions could be present in Ethiopia, the high drug resistance observed in the current study could also be due to these factors.
4. Materials and Methods
4.1. Study Design
A cross-sectional study was conducted on a total of 132 study participants. K. pneumoniae were isolated from each of the study participants from September 2018 to February 2019. Participants were enrolled using a convenient sampling technique. Socio-demographic characteristics were obtained using a well-designed questionnaire.
4.2. Bacterial Isolation and Identification
Biological specimens were inoculated on appropriate culture media. Briefly, blood specimens were collected on BacT/ALERT® 3D culture bottles and incubated in automatic BacT/ALERT® 3D at 35–37 °C in 5% CO2 for 5 days or until they signaled positive for growth. The microbial growth that could be detected by the flag and audible sound of the instrument was subsequently sub-cultured on MacConkey agar (Oxoid, Basingstoke, UK), 5% sheep blood agar (Oxoid, UK), and chocolate agar (Oxoid, UK) plates. A negative result was reported by examining the flag, gram staining, and subculturing at the end of the 5th day before discarding it as negative [31]. Specimens from wound, sputum, and body fluids were cultured on MacConkey agar, 5% sheep blood agar, and chocolate agar, while inoculation of urine was done only on MacConkey agar and 5% sheep blood agar. After incubation of all inoculated culture plates at 35–37 °C for 18–24 hours, preliminary identification of K. pneumoniae was done through examining colony characteristics on MacConkey agar and blood agar. Further identification was done using a Gram stain and a series of biochemical tests, including indole, triple sugar iron agar, citrate, mannitol, malonate, lysine decarboxylase, urea, and motility medium. K. pneumoniae is Gram-negative and rod shaped, indole negative, a gas and acid producer, hydrogen sulfide negative, citrate positive, a mannitol fermenter, malonate positive, lysine decarboxylase positive, urea slow producing, and non-motile [2,32]. After identification, the isolates were sub-cultured on 5% sheep blood agar plate and incubated overnight to get fresh colonies for antimicrobial susceptibility testing (AST).
4.3. Antimicrobial Susceptibility Testing
Using a sterile wire loop, 3–5 pure colonies were picked from blood agar and emulsified in 3–4 mL normal saline to prepare a 0.5 McFarland standard using a McFarland Densitometer. From the preparation, we inoculated onto Muller–Hinton agar (Oxoid, UK) using a sterile swab for the AST [33]. The AST was performed using the Kirby–Bauer disc diffusion method, with the following antimicrobial discs: tetracycline (30 μg), gentamicin (10 μg), amikacin (30 μg), tobramycin (10 μg), ciprofloxacin (5 μg), nalidixic acid (30 μg), amoxicillin-clavulanate (20/10 μg), piperacillin-tazobactam(100/10 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), aztreonam (30 μg), chloramphenicol (30 μg), cefoxitin (30 µg), Cefazolin (30 μg), Cefuroxime (30 μg), Ceftriaxone (30 μg), Ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 µg), imipenem (10 µg), ertapenem (10 μg), and meropenem (10 µg) (Oxoid, Basingstoke, UK and BD, Franklin Lakes, NJ, USA). The results were read and interpreted after 16–18 h of incubation at 35 ± 2 °C [34]. Control strains K. pneumoniae ATCC® 700603 were used during identification, as well as Escherichia coli ATCC® 25922, Pseudomonas aeruginosa ATCC® 27853 (for carbapenems), Escherichia coli ATCC® 35218 (for β-lactam/ β-lactamase inhibitor combinations) for controlling the potency of the drugs. MDR was defined as non-susceptibility to at least one agent in three or more antimicrobial categories [29].
4.4. Data Analysis
Data were checked, cleaned, and double entered into Epidata software version 3.1 (The EpiData Association, Enghavevej, Odense, Denmark), and then it was exported to Statistical Package for Social Sciences (SPSS version 25.0, IBM Corp., Armonk, NY, USA) software for analysis. Descriptive statistics were used to present the findings.
4.5. Ethics Approval and Consent to Participate
This study was approved by the Ethics Review Committee of the Department of Microbiology, Immunology and Parasitology, School of Medicine, College of Health Sciences, Addis Ababa University (Reference number: DERC/17/18/02-N) and the AHRI/ALERT ethical review committee (Protocol number: PO12/18). A permission letter was obtained from TASH. Moreover, before commencing the study, a written informed consent/assent was obtained from each study participant. Confidentiality was maintained for all data collected.
5. Conclusions
The findings of our study revealed that K. pneumoniae isolates showed a high resistance to most of the drugs commonly used to treat infections, and the magnitude of MDR K. pneumoniae isolates was very alarming. Virtually all (98.5%) of the isolates were MDR. Higher susceptibility was observed to amikacin and carbapenems. Therefore, our data should be used as an alert for the need for prevention and control of MDR K. pneumoniae in hospital settings, most specifically within the study area.
Acknowledgments
We thank Tikur Anbessa Specialized Hospital for giving permission to conduct this study. Our deepest gratitude goes to Mequanint Mitiku and Berhanu Yitayew for their assistance. Our countless acknowledgments go to the study participants for their willingness to participate.
Supplementary Materials
The following are available online https://www.mdpi.com/article/10.3390/antibiotics10081007/s1. Figure S1: Multidrug resistance patterns of K. pneumoniae isolates; Table S1: Previous antimicrobial treatment status of study participates (n = 132)
Author Contributions
Conceptualization, T.A. (Tewachew Awoke), A.M., T.A. (Tamrat Abebe); methodology, T.A. (Tewachew Awoke); validation, T.A. (Tewachew Awoke), B.T.; formal analysis, T.A. (Tewachew Awoke), B.T.; investigation, T.A. (Tewachew Awoke), S.S., A.S.; resources, A.M.; writing—original draft preparation, T.A. (Tewachew Awoke); writing—review and editing, B.T., B.Y., A.A., A.M., T.A. (Tamrat Abebe); supervision, B.Y.; project administration, A.M.; funding acquisition, A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Addis Ababa University and Armauer Hansen Research Institute, Ethiopia.
Institutional Review Board Statement
This study was approved by the Ethics Review Committee of the Department of Microbiology, Immunology, and Parasitology, School of Medicine, College of Health Sciences, Addis Ababa University (reference number: DERC/17/18/02-N) and the AHRI/ALERT ethical review committee (protocol number: PO12/18).
Informed Consent Statement
A permission letter was obtained from TASH. Moreover, before commencing the study, written informed consent/assent was obtained from each study participant. Confidentiality was maintained for all data collected.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Podschun R., Ullmann U. Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998;11:589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Jesus M.B., Ehlers M.M., Dos Santos R.F., Kock M.M. Understanding β-lactamase producing Klebsiella pneumoniae. In: Ossiprandi M.C., editor. Antimicrobial Resistance—An Open Challenge. InTechOpen; Rijeka, Croatia: 2015. pp. 51–83. [Google Scholar]
- 3.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. HM Government and Welcome Trust; London, UK: 2018. [(accessed on 17 August 2020)]. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf. [Google Scholar]
- 5.World Health Organization . Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; Geneva, Switzerland: 2014. [(accessed on 5 May 2019)]. Available online: https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf?sequence=1. [Google Scholar]
- 6.Holt K.E., Wertheim H., Zadoks R.N., Baker S., Whitehouse C.A., Dance D., Jenney A., Connor T.R., Hsu L.Y., Severin J., et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pendleton J.N., Gorman S.P., Gilmore B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 8.Pistella E., Santini C. Risk factors and outcomes of carbapenem-resistant Klebsiella pneumoniae infections. Ital. J. Med. 2016;10:339. doi: 10.4081/itjm.2016.798. [DOI] [Google Scholar]
- 9.European Centre for Disease Prevention and Control . Antimicrobial resistance in the EU/EEA (EARS-Net)-Annual Epidemiological Report 2019. ECDC; Stockholm, Sweden: 2020. [(accessed on 12 January 2021)]. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/surveillance-antimicrobial-resistance-Europe-2019.pdf. [Google Scholar]
- 10.Ethiopian Public Health Institute 2020 Ethiopia Antimicrobial Resistance Surveillance. [(accessed on 2 March 2021)]; Annual Report (2nd Year) September 2018–October 2019. Available online: https://www.ephi.gov.et/images/files/Final-Annual-report---V.9.-Gebrie-1.pdf.
- 11.Nirwati H., Sinanjung K., Fahrunissa F., Wijaya F., Napitupulu S., Hati V.P., Hakim M.S., Meliala A., Aman A.T., Nuryastuti T. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13:1–8. doi: 10.1186/s12919-019-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Silva Y., Ferrari R., Marin V.A., Junior C.A. A Global Overview of β-lactam Resistance Genes in Klebsiella pneumoniae. Open Infect. Dis. J. 2019;11:22–34. doi: 10.2174/1874279301911010022. [DOI] [Google Scholar]
- 13.Muhie O.A. Antibiotic use and resistance pattern in Ethiopia: Systematic review and meta-analysis. Int. J. Microbiol. 2019;2019:2489063. doi: 10.1155/2019/2489063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centres for Disease Control and Prevention Antibiotic Resistance Threats in the United States. US Department of Health and Human Services, Centres for Disease Control and Prevention: Atlanta, GA, USA. [(accessed on 21 September 2020)];2019 Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- 15.Teklu D.S., Negeri A.A., Legese M.H., Bedada T.L., Woldemariam H.K., Tullu K.D. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control. 2019;8:39. doi: 10.1186/s13756-019-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moges F., Eshetie S., Abebe W., Mekonnen F., Dagnew M., Endale A., Amare A., Feleke T., Gizachew M., Tiruneh M. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS ONE. 2019;14:e0215177. doi: 10.1371/journal.pone.0215177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badamchi A., Farahani R.K., Naghadalipoor M., Reza Etemadi M., Tabatabaie A. Phenotypic and genotypic characterization of antibiotic resistance in Klebsiella pneumoniae isolated from patients admitted to a third-level hospital in Tehran, Iran. Curr. Pediatr Res. 2018;22:258–262. [Google Scholar]
- 18.Gutema G., Håkonsen H., Engidawork E., Toverud E.-L. Multiple challenges of antibiotic use in a large hospital in Ethiopia–a ward-specific study showing high rates of hospital-acquired infections and ineffective prophylaxis. BMC Health Serv. Res. 2018;18:326. doi: 10.1186/s12913-018-3107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alibi S., Ferjani A., Boukadida J. Molecular characterization of extended spectrum beta-lactamases produced by Klebsiella pneumoniae clinical strains from a Tunisian Hospital. Med. Mal. Infect. 2015;45:139–143. doi: 10.1016/j.medmal.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty S., Mohsina K., Sarker P.K., Alam M., Karim M., Sayem S. Prevalence, antibiotic susceptibility profiles and ESBL production in Klebsiella pneumoniae and Klebsiella oxytoca among hospitalized patients. Period Biol. 2016;118:53–58. doi: 10.18054/pb.2016.118.1.3160. [DOI] [Google Scholar]
- 21.Nepal K., Pant N.D., Neupane B., Belbase A., Baidhya R., Shrestha R.K., Lekhak B., Bhatta D.R., Jha B. Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Ann. Clin. Microbiol. Antimicrob. 2017;16:1–7. doi: 10.1186/s12941-017-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geresu B., Misganaw D., Beyene Y. Retrospective evaluation of cotrimoxazole use as preventive therapy in people living with HIV/AIDS in Boru Meda Hospital. BMC Pharmacol. Toxicol. 2014;15:4. doi: 10.1186/2050-6511-15-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Bi W., Dong G., Zhang Y., Wu Q., Dong T., Cao T., Zhou T. The new perspective of old antibiotic: In vitro antibacterial activity of TMP-SMZ against Klebsiella pneumoniae. J. Microbiol. Immunol. Infect. 2020;53:757–765. doi: 10.1016/j.jmii.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Magalhães M.L., Blanchard J.S. Aminoglycosides: Mechanisms of action and resistance. In: Mayers D.L., editor. Antimicrobial Drug Resistance: Mechanisms of Drug Resistance. Humana Press; New York, NY, USA: 2009. pp. 171–181. [Google Scholar]
- 25.El-Badawy M.F., Tawakol W.M., El-Far S.W., Maghrabi I.A., Al-Ghamdi S.A., Mansy M.S., Ashour M.S., Shohayeb M.M. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. Int. J. Microbiol. 2017;2017:8050432. doi: 10.1155/2017/8050432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fenta T., Engidawork E., Amogne W., Berha A.B. Evaluation of current practice of antimicrobial use and clinical outcome of patients with pneumonia at a tertiary care hospital in Ethiopia: A prospective observational study. PLoS ONE. 2020;15:e0227736. doi: 10.1371/journal.pone.0227736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eshetie S., Unakal C., Gelaw A., Ayelign B., Endris M., Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob. Resist. Infect. Control. 2015;4:1–8. doi: 10.1186/s13756-015-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aljanaby A., Alhasnawi H. Phenotypic and Molecular Characterization of Multidrug Resistant Klebsiella pneumoniae Isolated from Different Clinical Sources in Al-Najaf Province-Iraq. Pak. J. Biol. Sci. 2017;20:217–232. doi: 10.3923/pjbs.2017.217.232. [DOI] [PubMed] [Google Scholar]
- 29.Magiorakos A.P., Srinivasan A., Carey R., Carmeli Y., Falagas M., Giske C., Harbarth S., Hindle J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 30.Breurec S., Guessennd N., Timinouni M., Le T., Cao V., Ngandjio A., Randrianirinag J.M., Thibergeh A., Kinanaa A., Dufougeraya A., et al. Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: Multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol Infect. 2013;19:349–355. doi: 10.1111/j.1469-0691.2012.03805.x. [DOI] [PubMed] [Google Scholar]
- 31.Wilson M.L., Mirrett S., Reller L.B., Weinstein M.P., Reimer L.G. Recovery of clinically important microorganisms from the BacT/Alert blood culture system does not require testing for seven days. Diagn. Microbiol. Infect. Dis. 1993;16:31–34. doi: 10.1016/0732-8893(93)90127-S. [DOI] [PubMed] [Google Scholar]
- 32.Procop G.W., Church D.L., Hall G.S., Janda W.M. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. 7th ed. Wolters Kluwer; Philadelphia, PA, USA: 2017. [Google Scholar]
- 33.Cheesbrough M. Medical Laboratory Practice in Tropical Countries. Part 2. 2nd ed. Cambridge University Press; Cambridge, UK: 2006. [Google Scholar]
- 34.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. CLSI supplement M100. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.