Abstract
The purpose of this study was to analyse the prevalence and genetic characteristics of ESBL and acquired-AmpC (qAmpC)-producing Escherichia coli isolates from healthy and sick dogs in Portugal. Three hundred and sixty-one faecal samples from sick and healthy dogs were seeded on MacConkey agar supplemented with cefotaxime (2 µg/mL) for cefotaxime-resistant (CTXR) E. coli recovery. Antimicrobial susceptibility testing for 15 antibiotics was performed and the ESBL-phenotype of the E. coli isolates was screened. Detection of antimicrobial resistance and virulence genes, and molecular typing of the isolates (phylogroups, multilocus-sequence-typing, and specific-ST131) were performed by PCR (and sequencing when required). CTXR E. coli isolates were obtained in 51/361 faecal samples analysed (14.1%), originating from 36/234 sick dogs and 15/127 healthy dogs. Forty-seven ESBL-producing E. coli isolates were recovered from 32 sick (13.7%) and 15 healthy animals (11.8%). Different variants of blaCTX-M genes were detected among 45/47 ESBL-producers: blaCTX-M-15 (n = 26), blaCTX-M-1 (n = 10), blaCTX-M-32 (n = 3), blaCTX-M-55 (n = 3), blaCTX-M-14 (n = 2), and blaCTX-M-variant (n = 1); one ESBL-positive isolate co-produced CTX-M-15 and CMY-2 enzymes. Moreover, two additional CTXR ESBL-negative E. coli isolates were CMY-2-producers (qAmpC). Ten different sequence types were identified (ST/phylogenetic-group/β-lactamase): ST131/B2/CTX-M-15, ST617/A/CTX-M-55, ST3078/B1/CTX-M-32, ST542/A/CTX-M-14, ST57/D/CTX-M-1, ST12/B2/CTX-M-15, ST6448/B1/CTX-M-15 + CMY-2, ST5766/A/CTX-M-32, ST115/D/CMY-2 and a new-ST/D/CMY-2. Five variants of CTX-M enzymes (CTX-M-15 and CTX-M-1 predominant) and eight different clonal complexes were detected from canine ESBL-producing E. coli isolates. Although at a lower rate, CMY-2 β-lactamase was also found. Dogs remain frequent carriers of ESBL and/or qAmpC-producing E. coli with a potential zoonotic role.
Keywords: antimicrobial resistance, dogs, Escherichia coli, ESBL, CTX-M-15, CTX-M-1, CTX-M-32, CTX-M-55, CTX-M-14, qAmpC, CMY-2
1. Introduction
Antimicrobial resistance has become a major challenge for public health worldwide. The selective pressure, which results from the long-term use of antibiotics, allowed bacterial species to be resistant to these agents. It has been believed that this resistance is reaching alarming levels, considering that resistance rates have risen extremely, during the last two decades [1,2].
Escherichia coli, a Gram-negative bacterium belonging to the Enterobacteriaceae family, is a common member of the intestinal microbiota of humans and companion animals [3,4]. However, this opportunistic pathogen can cause intestinal and extra-intestinal diseases. It may contribute, in many cases, to antimicrobial resistance dissemination. Recently, the World Health Organization [5] published a global priority list of antibiotic-resistant bacteria, where third-generation cephalosporin- and/or carbapenem-resistant Enterobacteriaceae, including E. coli, were included in the Priority 1 group. It is important to note that first-generation cephalosporins and amoxicillin + clavulanic acid are among the most prescribed drugs for dogs [3,4,6].
During recent years, the emergence and rapid dissemination of Enterobacteriaceae carrying genes encoding the extended-spectrum-β-lactamases (ESBLs), acquired AmpC β-lactamases (qAmpC), or carbapenemases are considered of great concern [4,7]. One of the most important mechanisms is the plasmid-mediated production of extended-spectrum β-lactamases (ESBLs), which can hydrolyse broad-spectrum cephalosporins (such as cefotaxime). The horizontal gene transfer (HGT) among bacteria is driven by plasmids [8,9], which play an important role in the transference of antibiotic-resistance genes among bacteria, contributing to the spread of multidrug resistance (MDR), and limiting therapeutic options [10]. ESBLs of the CTX-M-type and the qAmpC CMY-2 are increasingly being reported in bacteria worldwide, while livestock or companion animals are potential sources, leading to the spread of β-lactam-resistant bacteria in humans [11,12].
The close proximity between dogs and their owners increases the possibility of transmitting resistant bacteria [13,14]. According to Dupouy et al. [6], dogs could transmit MDR bacteria due to their close contact with humans, the high consumption of β-lactams in small animal veterinary practice, and also the frequent occurrence of ESBL/qAmpC-producing E. coli. The occurrence of ESBL-producing E. coli has been widely reported in both healthy companion animals [12,15] and diseased ones [1,16,17,18,19]. International high-risk clones of E. coli are frequently detected worldwide, not only in human infections but also in those of companion animals [2,3,17]. Over the past 5 years, the presence of ESBL/qAmpC genes in Enterobacteriaceae strains from faeces of dogs in Europe has been reported in several studies [6,12,13,20], including Portugal [21,22]. However, knowledge about the clonality of ESBL/qAmpC-producing isolates and the potential zoonotic reservoir of human-associated STs is not well documented. Moreover, there is still a lack of data about their prevalence in sick and healthy dogs, simultaneously. In this study, we aim at characterizing the prevalence and diversity of ESBL- and qAmpC- producing E. coli faecal isolates from healthy and sick dogs in Portugal, as well as determining their genetic lineages and phylogenetic groups.
2. Materials and Methods
2.1. Animals and Sampling
A total of 361 faecal samples were recovered from 127 healthy and 234 hospitalized dogs from different cities in Portugal. All samples were collected between April andAugust 2017 (one sample/animal) using standardized procedures [23].
The hospitalized dogs came from 7 different veterinary hospitals or clinic centers; the healthy dogs came from a local kennel located in Vila Real (n = 31) and from local houses (n = 96). The seven hospitals/clinic centers were located in different centers of the Portuguese territory: Bragança (1 hospital, n = 29 dogs), Vila Real (4 hospitals, n = 62), Aveiro (1 hospital, n = 58), Leiria (1 hospital, n = 17), and Lisbon (1 hospital, n = 68) (Figure S1). It is important to note that faecal samples from unhealthy dogs were collected from the ordinary population of animals hospitalized in hospitals or veterinary clinics, not endangering their health, or causing harm or pain. In the same line, faecal samples from healthy animals were also recovered by their owners. All of them were analysed with the owner’s permission or with kennel collaboration. The faecal samples were dispatched immediately to the Microbiology Laboratory of the University of Trás-os-Montes and Alto-Douro (UTAD).
2.2. E. coli Isolation
From each faecal sample, a small portion of 2 g was diluted in Brain Heart Infusion (BHI, Condalab, Spain) and incubated in aerobic conditions for 24 h at 37 °C. After that, samples were seeded on MacConkey agar (Becton, Dickinson and Company Sparks, Le Pont de Claix, France) supplemented with cefotaxime (2 µg/mL) and incubated for 24 h at 37 °C. Colonies showing E. coli morphology were recovered (one colony per sample) and identified by a classical biochemical method named IMViC (Indol, Methyl-red, Voges–Proskauer, and Citrate).
The matrix-assisted laser desorption/ionization time-of-flight mass spectrometry method (MALDI-TOF MS, MALDI Biotyper® from Bruker Daltonik, Bremen, Germany) was applied in this study to confirm bacterial species identification. E. coli isolates were kept at −80 °C and were further characterized.
2.3. Susceptibility Testing
Antimicrobial susceptibility testing was performed using the Kirby–Bauer disk diffusion method and according to Clinical and Laboratory Standards Institute guidelines (2019) [24] for the following 15 antibiotics (μg/disk): ampicillin (10), amoxicillin + clavulanic acid (20), cefotaxime (30), cefoxitin (30), ceftazidime (30), aztreonam (30), imipenem (10), gentamicin (10), streptomycin (10), ciprofloxacin (5), trimethoprim-sulfamethoxazole (1.25 ± 23.75), amikacin (30), tobramycin (10), tetracycline (30), and chloramphenicol (30). In addition, the screening of phenotypic ESBL production was carried out by the double-disk synergy test using cefotaxime, ceftazidime, and amoxicillin/clavulanic discs in Mueller Hinton (MH) agar (Condalab, Spain) [24].
2.4. DNA Extraction and Quantification
Genomic DNA from cefotaxime-resistant (CTXR) isolates were extracted using the boiled method [25]. In order to quantify the nucleic acid concentration and the level of purity, the absorbance readings were taken at 260 and 280 nm (Spectrophotometer ND-100, Nanodrop, Thermo Fisher Scientific, Waltham, MA USA).
2.5. Antibiotic Resistance and Virulence Genes Detection
The genetic basis of resistance was investigated using PCR methods and subsequent sequencing of the obtained amplicons (specific genes). Negative and positive controls of the University of La Rioja were used in this work. Moreover, the data regarding PCR conditions for each primer (Sigma-Aldrich, Madrid, Spain) as well as the size of the obtained amplicons that were sequenced are illustrated in detail in Table S1.
The presence of blaCTX-M (Groups 1 and 9), blaCMY-2, blaDHA-1, blaTEM, blaSHV, blaVEB, blaKPC2/3, blaNDM, blaOXA-48, and blaVIM was tested by PCR/sequencing (Table S1) [26,27,28,29,30]. Furthermore, the mcr-1 gene (colistin resistance) [31], tetA/tetB (tetracycline resistance) [32], stx1,2 genes related to Shiga toxin-producing E. coli (STEC) [33], and int1 gene (integrase of class 1 integrons) and its variable region (RV int1) were also analysed by PCR/sequencing [30]. Analysis of DNA sequences was performed using the standard databases (nucleotide collection) in the BLASTN program (2021 version), available at the National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 31 January 2021).
2.6. Multilocus Sequence Typing and Phylogroup Typing of E. coli Isolates
Multilocus sequence typing (MLST), by the analysis of seven housekeeping genes (fumC, adk, purA, icd, recA, mdh, and gyrB), was carried out for thirteen representative E. coli isolates (based on the antimicrobial resistance phenotype) according to the protocol described on PubMLST (Public databases for molecular typing and microbial genome diversity) website [34]. The allele combination was determined after sequencing of the seven genes, and the sequence type (ST) and clonal complex (CC) were identified.
Phylogenetic classification of all E. coli isolates was performed according to the presence of chuA, yjaA, and TSPE4.C2 genes [35].
2.7. Statistical Analyses
All statistical analyses were performed using the JMP Statistics software (v7.0, SAS Institute). The Pearson’s Chi-square and Fisher’s exact tests were performed to understand and identify the associations between the origin of strain (healthy or sick dog) and antibiotic resistance (antibiotic and gene). In this line, we consider two categorical variables: the sick or healthy animal, and the resistance for each antibiotic/gene. A p-value < 0.05 was established as indicating statistical significance [36].
3. Results
CTXR E. coli isolates were recovered in 51/361 faecal samples tested (14.1%), originating from 36/234 sick dogs (15.4%) and 15/127 healthy dogs (11.8%). These CTXR isolates were detected among 29 male dogs (56.9%) and 22 female dogs (43.1%); most of them belonged to an undetermined breed (n = 38), followed by the Labrador/Golden Retriever breed (n = 4), while the remaining dogs belonged to different pure breeds (Table 1 and Table 2).
Table 1.
Phenotypic and molecular features of the 47 ESBL-producing E. coli isolates recovered from healthy and sick dogs in Portugal.
| Isolate Number |
Origin a | Sick/Healthy | Gender b | Age c | Breed d | Phenotype of Antibiotic Resistance e | β-Lactamases | Other Genes and Integrons f | PG g | MLST h |
|---|---|---|---|---|---|---|---|---|---|---|
| X605 | HV Lisboa | Sick | M | 15A | UD | AMP, CTX, ATM, CHL, CIP, TET | CTX-M-15 | tet(A) | B1 | ST6448 |
| X614 | HV Lisboa | Sick | F | 2A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, CHL, CIP, TET | CTX-M-15, CMY-2 | tet(A) | B1 | ST6448 |
| X607 | HV Lisboa | Sick | F | 1A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, TOB, CN, S, TET | CTX-M-15 | tet(A) | B2 | ST131 |
| X610 | HV Lisboa | Sick | F | 1,5A | UD | AMP, CTX, CAZ, ATM, CIP, S, TET | CTX-M-15 | tet(A) | B2 | ST12 |
| X603 | CV Bragança | Sick | F | 10A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, TOB, CN, S, TET | CTX-M-15 | tet(A) | B2 | ST131 |
| X602 | CV VR | Sick | F | 3A | UD | AMP, AUG, CTX, CHL, TOB, CN, S, TET | CTX-M-15 | tet(A) | B2 | ST12 |
| X558 | Kennel | Healthy | M | 2A | Labrador | AMP, AUG, CTX, CAZ, CIP, SXT, S, TET | CTX-M-15, TEM | int1 | B1 | NT |
| X562 | Kennel | Healthy | F | 4M | UD | AMP, CTX, CIP, SXT, S, TET | CTX-M-15, TEM | tet(A), int1 | B1 | NT |
| X569 | HVTM | Sick | M | 4A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, SXT, TOB, CN, S, TET | CTX-M-15, TEM | tet(A), tet(B), int1 | A | NT |
| X575 | CV Transm | Sick | M | 4A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, SXT, TOB, CN, TET | CTX-M-15, TEM | tet(A), tet(B), int1 | A | NT |
| C10151 | HV Lisboa | Sick | F | 11A | UD | AMP, AUG, CTX, TET | CTX-M-15, TEM | ND | A | NT |
| X550 | HD | Healthy | F | 8A | UD | AMP, AUG, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | B1 | NT |
| X556 | HD | Healthy | F | 14A | Yorkshire | AMP, AUG, FOX, CTX, ATM | CTX-M-15 | ND | B1 | NT |
| X563 | Kennel | Healthy | M | 5A | Rottweiler | AMP, AUG, CTX, TET | CTX-M-15 | ND | A | NT |
| X588 | HVTM | Sick | M | 5A | UD | AMP, AUG, CTX, CAZ, ATM, CHL, CN, TET | CTX-M-15 | tet(A) | D | NT |
| X598 | HVTM | Sick | M | 6A | Russell Terrier | AMP, AUG, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | ND | B1 | NT |
| X576 | HV Lisboa | Sick | M | 15A | UD | AMP, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | D | NT |
| X577 | HV Lisboa | Sick | F | 6M | UD | AMP, AUG, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | B1 | NT |
| X578 | HV Lisboa | Sick | M | 13A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, CHL, CIP, SXT, TOB, TET | CTX-M-15 | tet(A) | B1 | NT |
| X580 | HV Lisboa | Sick | F | 5A | UD | AMP, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | int1 | B1 | NT |
| X584 | HV Lisboa | Sick | M | 5A | UD | AMP, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | B1 | NT |
| X604 | HV Lisboa | Sick | M | 12A | UD | AMP, AUG, CTX, ATM | CTX-M-15 | ND | D | NT |
| X618 | HV Lisboa | Sick | F | 2A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | B1 | NT |
| X620 | HV Lisboa | Sick | M | 9A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, SXT, TOB, CN, S, TET | CTX-M-15 | tet(A), int1 | D | NT |
| X622 | HV Lisboa | Sick | M | 3A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, CIP, SXT, S, TET | CTX-M-15 | ND | A | NT |
| X599 | CV Bragança | Sick | M | 7A | Rodengo | AMP, AUG, FOX, CTX, CAZ, ATM, CHL, CIP, TET | CTX-M-15 | ND | B1 | NT |
| C10264 | CV Vouga | Sick | F | 9M | Pincher | AMP, CTX, CAZ | CTX-M-1 | ND | D | ST57 |
| X554 | HVTM | Sick | M | 1A | Labrador | AMP, CTX, CAZ, TET | CTX-M-1, TEM | ND | A | NT |
| X557 | Kennel | Healthy | F | 3A | Serra Estrela | AMP, AUG, CTX, CAZ, TOB, AK, S | CTX-M-1 | ND | B1 | NT |
| X559 | Kennel | Healthy | F | 3M | UD | AMP, AUG, CTX, CAZ | CTX-M-1 | ND | D | NT |
| X560 | Kennel | Healthy | M | 1A | Labrador | AMP, AUG, CTX, CAZ, TET | CTX-M-1 | ND | B1 | NT |
| X581 | HV Lisboa | Sick | M | 14A | UD | AMP, AUG, CTX, CAZ, TET | CTX-M-1 | tet(A) | B1 | NT |
| X611 | HV Lisboa | Sick | M | 5A | UD | AMP, CTX, CAZ, ATM, CHL, CIP, CN, TET | CTX-M-1 | tet(A) | D | NT |
| X616 | HV Lisboa | Sick | M | 3M | UD | AMP, AUG, CTX, CAZ, TET | CTX-M-1 | ND | B1 | NT |
| X617 | HV Lisboa | Sick | F | 3A | UD | AMP, CTX, CAZ, TET | CTX-M-1 | tet(A) | B1 | NT |
| C10265 | CV Bragança | Sick | M | 1A | UD | AMP, CTX, CAZ, S | CTX-M-1 | ND | A | NT |
| X555 | HD | Healthy | M | 6A | Pastor alemão | AMP, CTX, CAZ, ATM, CIP, SXT, TOB, CN, TET | CTX-M-55 | tet(B), int1 | A | ST617 |
| X568 | HD | Healthy | M | 7A | UD | AMP, AUG, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-55 | tet(A), int1 | B1 | NT |
| C10149 | HVTM | Sick | F | 1,5A | UD | AMP, CTX, CAZ, CHL, TOB, CN, S, TET | CTX-M-55, TEM | tet(A) | A | NT |
| X573 | HD | Healthy | M | 1A | UD | AMP, AUG, CTX, CAZ, ATM, CHL, SXT, S, TET | CTX-M-32 | int1 | A | ST5766 |
| X561 | Kennel | Healthy | M | 2A | Gado transm. | AMP, AUG, CTX, ATM, CHL, CIP, SXT, TOB, CN, S, TET | CTX-M-32 | tet(A), int1 | B1 | ST3078 |
| X571 | HD | Healthy | M | 1A | UD | AMP, AUG, CTX, CAZ, CHL, SXT, S, TET | CTX-M-32, TEM | tet(B), int1 | B1 | NT |
| X572 | HD | Healthy | F | 1A | UD | AMP, AUG, CTX, CAZ, S, TET | CTX-M-14 | tet(B) | A | ST542 |
| X574 | CVTransm | Sick | M | 4A | UD | AMP, CTX, CHL, SXT, TOB, CN, S, TET | CTX-M-14 | ND | A | NT |
| X565 | HD | Healthy | M | 6A | UD | AMP, CTX, ATM, CIP, SXT, TOB, CN, S, TET | CTX-M-variant | ND | A | NT |
| C10147 | HVLisboa | Sick | M | 7A | UD | AMP, CTX, CHL, SXT, CN, S, TET | TEM-1 | tet(A), int1 | B2 | NT |
| X587 | HVTM | Sick | M | 2A | Bulldog Francês | AMP, CTX, ATM, CHL, SXT, CN, S, TET | No bla genes | int1 | A | NT |
a HD- healthy dogs from their owners; HVTM- Hospital Veterinário de Trás os Montes (Vila Real); Kennel-healthy dogs from the kennel (Vila Real); CV Transm- Clínica Veterinária Transmonvete (Vila Real, Portugal); HV Lisboa- Hospital Veterinário de São Bento (Lisboa); CV Vouga- Clínica Veterinária do Vouga (Sever do Vouga, Portugal); CV Bragança- Clínica Veterinária de Macedo de Cavaleiros (Bragança, Portugal); CV VR- Clínica Veterinária dos Quinchosos (Vila Real, Portugal); b female; M-male; c A- years; M- months; d UD- undetermined dog breed; e AMP, ampicillin; AUG, amoxicillin–clavulanic acid; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; ATM, aztreonam; CHL, chloramphenicol; CIP, ciprofloxacin; TOB, tobramycin; AK, amikacin; CN, gentamicin; SXT, trimethoprim–sulfamethoxazole; S, streptomycin; TET, tetracycline; f ND: not detected; g Phylogroups; h MLST-Multilocus Sequence Typing; NT: not tested.
Table 2.
Phenotypic and molecular features of ESBL-negative E. coli isolates recovered from healthy and sick dogs in Portugal.
| Isolate Number |
Origin a | Gender b | Age c | Breed d | Antimicrobial Resistance Phenotype e | Resistance Genotype | Other Resistance Genes f | PG g | MLST h |
|---|---|---|---|---|---|---|---|---|---|
| X551 | HD | F | 24M | Golden Retriever | AMP, CTX | CMY-2 | ND | D | New ST * |
| X567 | CV Vouga | F | 8A | UD | AMP, AUG, FOX, CTX, CAZ, CIP, S, TET | CMY-2, TEM | tet(A) | D | ST115 |
| X549 | HVTM | F | 6A | Leão Rodesea | AMP, AUG, CTX | ND | ND | D | NT |
| C10266 | HV Lisboa | F | 6A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, NA, CIP, SXT, S, TET | ND | tet(B) | A | NT |
a HD- healthy dogs from their owners; HVTM- Hospital Veterinário de Trás os Montes (Vila Real); Kennel- healthy dogs from kennel (Vila Real); CV Transm- Clínica Veterinária Transmonvete (Vila Real, Portugal); HV Lisboa- Hospital Veterinário de São Bento (Lisboa); CV Vouga- Clínica Veterinária do Vouga (Sever do Vouga, Portugal); CV Bragança- Clínica Veterinária de Macedo de Cavaleiros (Bragança, Portugal); CV VR- Clínica Veterinária dos Quinchosos (Vila Real, Portugal); b F-female; M-male; c A- years; M- months; d UD- undetermined dog breed; e AMP, ampicillin; AUG, amoxicillin–clavulanic acid; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; ATM, aztreonam; NA, nalidixic acid; CIP, ciprofloxacin; SXT, trimethoprim–sulfamethoxazole; S, streptomycin; TET, tetracycline; IMP, imipenem; ETP, ertapenem. f ND: not detected; g Phylogroups; h MLST-Multilocus Sequence Typing; NT: not tested. * New ST allelic combination: fumC (26), adk (4), purA (5), icd (25), gyrB (2), recA (2), and mdh (5).
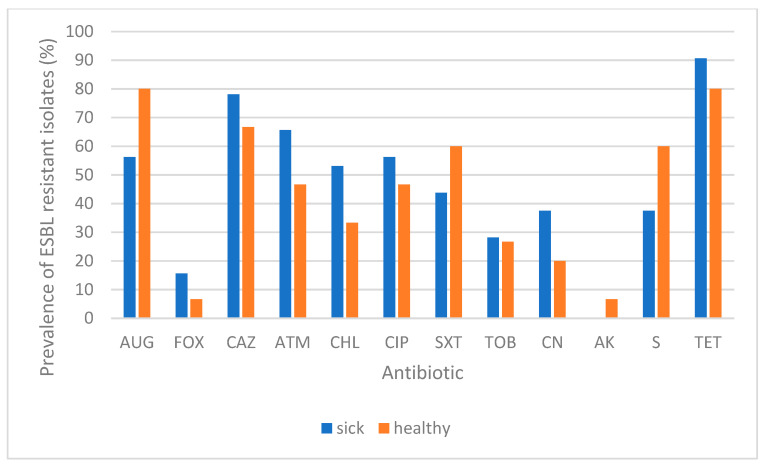
Forty-seven ESBL-producing E. coli isolates were detected among the 51 CTXR isolates, recovered from 32 sick and 15 healthy dogs (frequencies of 13.7% and 11.8%, respectively). The phenotypes of antibiotic resistance for these ESBL-producing isolates are shown in Table 1 and the rates of antibiotic resistance of these isolates depending on their origin (sick or healthy dogs) are represented in Figure 1. No statistical difference could be established between the origin of the strain (healthy or sick dog) and the resistance to different antibiotics (p > 0.05) (Figure 1).
Figure 1.
Prevalence of antibiotic-resistance among ESBL-producing E. coli isolates in sick and healthy dogs. Antibiotics tested: AUG, amoxicillin-clavulanic acid; FOX, cefoxitin; CAZ, ceftazidime; ATM, aztreonam; CHL, chloramphenicol; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; TOB, tobramycin; CN, gentamicin; AK, amikacin; S, streptomycin; TET, tetracycline. No significant association was detected between antibiotic resistance and type of animal (sick or healthy) (p > 0.05).
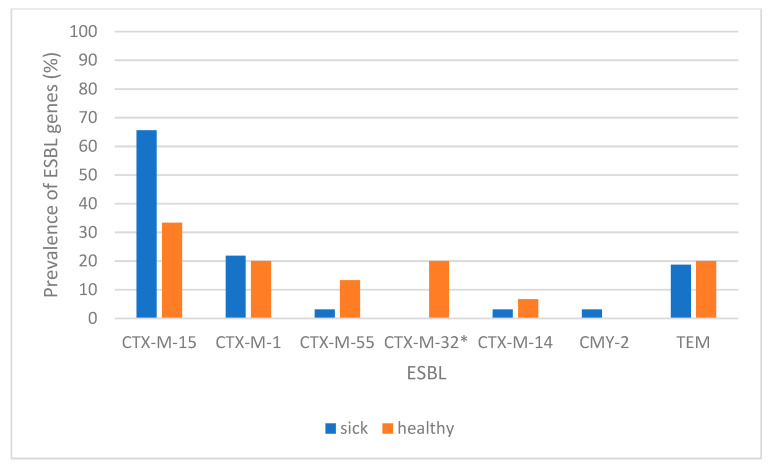
Different variants of blaCTX-M genes were detected among 45 of these 47 ESBL-producing isolates (95.4%): blaCTX-M-15 (n = 26 isolates), blaCTX-M-1 (n = 10), blaCTX-M-32 (n = 3), blaCTX-M-55 (n = 3), blaCTX-M-14 (n = 2), and blaCTX-M (n = 1, no variant determined) (Table 1). Figure 2 shows the distribution of the ESBL variants depending on the origin of the isolates; no statistical difference could be established between the origin of the strain (healthy or sick dog) and the ESBL type (p > 0.05) (Figure 2), except for CTX-M-32, in which this relation was present (it was detected just in healthy dogs).
Figure 2.
Distribution of ESBL-encoding genes from E. coli isolates in sick and healthy dogs. Gene encoding β-lactamases with p < 0.05 is indicated with (*).
The two remaining ESBL-positive isolates were revealed negative to all ESBL genes under study. Furthermore, a blaTEM gene was detected in eight blaCTX-M-producing isolates. On the other hand, six ESBL-positive isolates showed cefoxitin-resistance (FOXR), and the blaCMY-2 gene was detected in one CTX-M-15-producing isolate obtained from a sick dog; the others ESBL-positive-FOXR isolates were negative for blaCMY-2 and blaDHA genes by PCR. Among the ESBL-positive isolates, resistance to tetracycline was mediated by the tetA (24 isolates) and/or tetB genes (Table 1).
Two of the four CTXR and ESBL-negative isolates were CMY-2-producers (qAmpC type), and they were recovered from a healthy and a sick dog (one each) (Table 2). We could not detect the mechanisms of CTXR in the two remaining ESBL-negative isolates. None of the CTXR E. coli isolates carried the mcr-1 gene (related to colistin resistance).
Moreover, other β-lactamases genes such as blaVEB, blaNDM, blaOXA-48, and blaVIM were tested by PCR/sequencing but all isolates were revealed to be negative. Furthermore, the stx1,2 genes related to Shiga toxin-producing E. coli (STEC) were not detected among our isolates.
The ESBL-positive isolates were ascribed to phylogenetic groups B1 (n = 21 isolates), A (n = 14), D (n = 7), and B2 (n = 5, two of them CTX-M-15-producers, typed as ST131) (Table 1). Furthermore, the four ESBL-negative isolates belonged to phylogroups D (n = 3, including the two CMY-2 producers) and A (n = 1) (Table 2).
MLST analysis, which was performed in thirteen representative E. coli isolates (based on the antimicrobial-resistance phenotype), revealed ten different lineages (ST/phylogenetic-group/β-lactamase): ST131/B2/CTX-M-15 (n = 2, from sick dogs, one from Lisbon and another from Bragança hospitals), ST617/A/CTX-M-55 (n = 1, from a healthy dog), ST3078/B1/CTX-M-32 (n = 1, from a healthy dog from the kennel), ST57/D/CTX-M-1 (n = 1, from a sick dog from Vouga clinic), ST12/B2/CTX-M-15 (n = 2 sick dogs, one from Vila Real and another from Lisbon), ST6448/B1/CTX-M-15 (n = 2 sick dogs, one of them CMY-2 positive and both from Lisbon), ST542/A/CTX-M-14 (n = 1, from a healthy dog), ST5766/A/CTX-M-32 (n = 1, from a healthy dog), and ST115/D/CMY-2 (n = 1, from a sick dog from Vouga clinic); moreover, one CMY-2-producing E. coli isolate of phylogroup D obtained in a healthy dog, presented a new combination of alleles (fumC (26), adk (4), purA (5), icd (25), gyrB (2), recA (2) and mdh (5)), rendering a new ST (Table 1).
4. Discussion
Regarding the Portuguese situation, the prevalence of ESBL-producing E. coli isolates in healthy dogs obtained in this work is similar to previous studies performed in dogs and cats [12,22,23] in the South and the North of Portugal. Worldwide, this prevalence was lower than the ones obtained with faecal samples of healthy dogs in Germany, Brazil, or China (24–29%) [15,37,38], but it is similar to the results of previous studies performed in Tunisia and France (12.7–17%) [11,39]. These differences could be explained by differences in the epidemiology of ESBL genes among different countries, considering the year in which the studies were performed, but we cannot discard methodological effects in the different studies.
Five types of CTX-M ESBLs were detected, indicating a high diversity of CTX-M genes (mainly blaCTX-M-15 gene) among the CTXR E. coli isolates; these results are in accordance with a previous study done in Portugal on healthy dogs [12]. This blaCTX-M-15 gene was also the most frequently detected in E. coli isolated from dogs in different countries [3,15,40]. The CTX-M-1- and CTX-M-15-encoding genes were also detected among E. coli canine isolates in Italy [41] and Denmark [13], which are in agreement with our data. The same variants of CTX-M genes were observed in a recent study conducted on healthy humans in Spain [42]. Moreover, during the last few years, new variants are becoming more common, in particular CTX-M-55 [3], especially from companion animals in Asian countries [43].
In the past, the blaCTX-M-15 gene was mainly associated with strains of human origin while blaCTX-M-1 was the major CTX-M sub-type among livestock and companion animal isolates in Europe [15,41]. Actually, this close correspondence is no longer so obvious, and our results confirm these data. A further study should be implemented to determine the ESBL gene in the two uncharacterized ESBL-producing isolates.
In this study, the CMY-2 gene was the qAmpC β-lactamase type found among two CTXR-ESBL-negative isolates and one ESBL-producing isolate, and it has been previously reported among E. coli strains from healthy and sick pets worldwide [20,23,39,44]. The detection of tetA and/or tetB genes in most of our tetracycline-resistant isolates seem to be similar to the results obtained by Costa et al. [45] from dogs, in Northern Portugal.
In this work, the most common phylogenetic groups among our isolates were B1 and A, these being the phylogroups more associated with commensal E. coli both in humans and in dogs, as well as in other animals [11,13]. On the other hand, isolates belonging to phylogroup B2 and D are more likely to be recovered from extra-intestinal infections of companion animals [4]. An interesting study related to 78 dogs that visited a veterinary hospital in Northern Portugal (either for a normal checkout or in case of disease) revealed the prevalence of E. coli isolates of groups A (n = 19), D (n = 9), and B1 (n = 7) [46], similar to our observation. So, the carriage of ESBL/qAmpC producing E. coli of these phylogroups in the gastrointestinal tract suggests a potential reservoir of MDR ESBL-producing bacteria in dogs.
Regarding the MLST results, the pandemic virulent E. coli ST131-B2 clone was detected among two isolates of sick dogs tested in this study. It is important to note that this clone was widely detected in pets [47,48], including in sick dogs in Portugal [17,49].
On the other hand, we detected one E. coli strain, ST57/D/CTX-M-1, that was recently detected in Portugal (associated with CMY-2 gene) in a dog with a UTI from a Lisbon hospital [17]. Similarly, the same lineage was identified in a faecal isolate from a healthy dog in Mexico, characterized as CMY-2/ST57/D) [50].
The frequency of the ST6448 lineage, which was observed in two sick dogs in this study, is considered an infrequent clone in humans and companion animals. This lineage was also found among a vulture faecal sample from Canary Islands [51]. To our knowledge, there is only one previous report related to the detection of this clone in humans, which was recently reported in healthy children from Sweden [52].
Additionally, our data indicate the presence of E. coli ST12/B2/CTX-M-15, which should be considered an agent of high clinical relevance for humans and animals. Furthermore, the ST12 lineage (associated with CMY-2) was identified in healthy dogs from Spain [6], Brazil [2], and France [11]. Furthermore, this lineage was found among isolates from children with a febrile UTI in France [53] and in healthy humans in Spain [42]. These findings highlight the dissemination of ST12 lineage and its presence in animal and human’ isolates.
To our knowledge, the ST617 lineage (clonal complex ST10) was identified for the first time in pets from Portugal in this study. CTX-M-15-producing E. coli isolates of sequence type ST617/phylogroup A have been reported in sick dogs in France [40] and in hospitalized patients in Tunisia [54,55]. Similarly, Rocha-Gracia et al. [50] identified the same lineage among a faecal isolate from healthy dogs in Mexico (ST617/A/CTX-M-15). According to a recent study, Gauthier, et al. [56] found this lineage in four isolates from dogs in France harbouring carbapenemase genes. Furthermore, this clone was widely disseminated.
The ST542 lineage detected in one of the healthy dogs is not commonly reported; however, this clone was found in a farmworker from Germany [57] and in a pig in Australia [58]. On the other hand, an ST115/CMY-2 isolate (found in a sick dog from the Vouga clinic) was previously reported among chickens and human patients in Germany [47].
We also detected a ST5766/A/CTX-M-32 isolate in a healthy dog; this clone is unusual, and it was previously reported in broilers’ osteomyelitis in Brazil [59]. To our knowledge, this is the first report of the ST5766 clone among pets, and the first detection in Europe. In this study, we also found an E. coli isolate, ST3078/B1/CTX-M-32, recovered from a healthy dog from a kennel. To our knowledge, the only unique previous study related to the ST3078 lineage was found in wastewater in Eastern France [60]. This suggests that the environment likely plays a role in the spread of ESBL-producing E. coli isolates in the community, associated with a One Health approach (human-animals-environment). Importantly, a new combination of alleles was found in an isolate of a healthy dog, rendering a new ST.
The use of β-lactams in the clinical practice of veterinary medicine may be considered one of the reasons for the high incidence of ESBL-producers worldwide. Thus, pets can be a significant source of ESBL/qAmpC-producing E. coli isolates. Considering the prevalence of ESBLs (notably the large reservoir in dogs of E. coli isolates with genes encoding CTX-M-15 and CTX-M-1, or CMY-2 β-lactamases), there is a serious and plausible risk of future acquisition of these resistant genes by their owners.
5. Conclusions
Antimicrobial resistance can make infections difficult to treat, which represents a global public health problem, due to the negative consequences for human health. This study shows that healthy and sick dogs are frequent carriers of faecal ESBL-producing E. coli strains, harbouring different variants of blaCTX-M genes (mostly blaCTX-M-15 and blaCTX-M-1), and presenting a high genetic MLST diversity (including the ST131/B2 lineage). Although at a lower rate, the blaCMY-2 gene was also found. This fact suggests the implication of mobile genetic elements in the dissemination of this relevant mechanism of resistance. This underlies the complexity of the antimicrobial resistance of bacteria occurring in dogs and the possible interspecies transmission between humans, domestic animals, and into the environment, important knowledge given the One-Health approach.
Acknowledgments
The authors would like to thank Carla Oliveira from Transmonvete Veterinary Clinic, and also Anabela Pais from Quinchosos Veterinary center who helped us with the collection of faecal samples of pets in Vila Real (Portugal). In the same line, we also appreciate the collaboration with the local kennel located inVila Real.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10081013/s1, Figure S1: Geographic location of the different areas where the faecal samples from sick dogs were collected in Portugal. Table S1: Primers sequences and PCR conditions used for genes encoding antibiotic resistance in E. coli.
Author Contributions
I.C.: conceptualization, sampling, methodology, investigation, resources, data curation, writing and rewriting; R.C.(Rosa Capita): sampling, methodology; C.M.: sampling, methodology; S.M.-Á.: investigation, data curation; N.S.C.: investigation, data curation, writing—review and editing; P.P. (Paulo Pimenta): sampling, methodology; A.R.P.: sampling, methodology; S.R.: sampling, methodology; M.S.: investigation, data curation; Â.M.: statistical analyses; J.F.: sampling, methodology; F.R.: review and editing; R.C. (Rita Cunha): validation, funding acquisition; C.A.-C.: validation, funding acquisition; M.d.L.N.E.D.: review and editing; G.I.: conceptualization, methodology, validation, resources, data curation, writing—review and editing, visualization, supervision, project administration, funding acquisition; C.T.: conceptualization, methodology, validation, resources, data curation, writing—review and editing, visualization, supervision, project administration, funding acquisition; P.P. (Patrícia Poeta): conceptualization, methodology, validation, resources, data curation, writing—review and editing, visualization, supervision, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
I.C. gratefully acknowledges the financial support of “Fundação para a Ciência e Tecnologia” (FCT—Portugal) related to PhD grant, through the reference SFRH/BD/133266/2017 (Medicina Clínica e Ciências da Saúde), as well as MCTES (Ministério da Ciência, Tecnologia e Ensino Superior) and European Union (EU), with reference to Fundo Social Europeu (FSE). The experimental work carried out in the University of La Rioja (Spain) was financed by the project SAF2016-76571-R from the Agencia Estatal de Investigation (AEI) of Spain and FEDER of EU. N.S.C. was awarded a grant for the year 2018, from the Algerian Ministry of Higher Education and Scientific Research (The PNE Program), under the direction of Carmen Torres. This work was supported by the Ministerio de Ciencia, Innovación y Universidades (Spain; grant number RTI2018-098267-R-C33), the Junta de Castilla y León (Consejería de Educación, Spain; grant number LE018P20) and the Associate Laboratory for Green Chemistry—LAQV which is financed by national funds from FCT/MCTES (UIDB/50006/2020 and UIDP/50006/2020).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest. Preliminary data was presented as a poster entitled “Diversity of CTX-M β-lactamases in Escherichia coli isolated from dogs in Portugal”, in ECCMID 2019 Congress (Amsterdam), and as part of an awarded oral presentation named “High frequency of ESBL- E. coli producers in pets in Portugal with detection of ST131 clone carrying different variants of CTX-M genes”, in IC2AR 2019 (Portugal).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen Y., Liu Z., Zhang Y., Zhang Z., Lei L., Xia Z. Increasing Prevalence of ESBL-Producing Multidrug Resistance Escherichia coli From Diseased Pets in Beijing, China From 2012 to 2017. Front. Microbiol. 2019;10:2852. doi: 10.3389/fmicb.2019.02852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Melo L.C., Oresco C., Leigue L., Netto H.M., Melville P.A., Benites N.R., Saras E., Haenni M., Lincopan N., Madec J.-Y. Prevalence and molecular features of ESBL/pAmpC-producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet. Microbiol. 2018;221:59–66. doi: 10.1016/j.vetmic.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Bortolami A., Zendri F., Maciuca E.I., Wattret A., Ellis C., Schmidt V., Pinchbeck G., Timofte D. Diversity, Virulence, and Clinical Significance of Extended-Spectrum β-Lactamase- and pAmpC-Producing Escherichia coli From Companion Animals. Front. Microbiol. 2019;10:1260. doi: 10.3389/fmicb.2019.01260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne J.A., Chong W.L., Gordon D.M. Genetic structure, antimicrobial resistance and frequency of human associated Escherichia coli sequence types among faecal isolates from healthy dogs and cats living in Canberra, Australia. PLoS ONE. 2019;14:e0212867. doi: 10.1371/journal.pone.0212867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiot-ics. [(accessed on 15 January 2021)];2017 Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
- 6.Dupouy V., Abdelli M., Moyano G., Arpaillange N., Bibbal D., Cadiergues M.-C., Lopez-Pulin D., Sayah-Jeanne S., De Gunzburg J., Saint-Lu N., et al. Prevalence of Beta-Lactam and Quinolone/Fluoroquinolone Resistance in Enterobacteriaceae From Dogs in France and Spain—Characterization of ESBL/pAmpC Isolates, Genes, and Conjugative Plasmids. Front. Vet. Sci. 2019;6:279. doi: 10.3389/fvets.2019.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho I., Silva N., Carrola J., Silva V., Currie C., Igrejas G., Poeta P. Antibiotic Drug Resistance Chapter 11. John Wiley & Sons; Hoboken, NJ, USA: 2019. Immunity-Acquired Resistance: Evolution of Antimicrobial Resistance Among Extended-Spectrum β-Lactamases and Carbapenemases in Klebsiella pneumoniae and Escherichia coli; pp. 239–259. [DOI] [Google Scholar]
- 8.Salgado-Caxito M., Benavides J.A., Adell A.D., Paes A.C., Moreno-Switt A.I. Global prevalence and molecular characterization of extended-spectrum β-lactamase producing-Escherichia coli in dogs and cats—A scoping review and meta-analysis. One Health. 2021;12:100236. doi: 10.1016/j.onehlt.2021.100236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benavides J.A., Salgado-Caxito M., Opazo-Capurro A., González Muñoz P., Piñeiro A., Otto Medina M., Rivas L., Munita J., Millán J. ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile. Antibiotics. 2021;10:510. doi: 10.3390/antibiotics10050510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Bi Z., Ma S., Chen B., Cai C., He J., Schwarz S., Sun C., Zhou Y., Yin J., et al. Inter-host Transmission of Carbapenemase-Producing Escherichia coli among Humans and Backyard Animals. Environ. Health Perspect. 2019;127:107009. doi: 10.1289/EHP5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haenni M., Saras E., Métayer V., Médaille C., Madec J.-Y. High Prevalence of blaCTX-M-1/IncI1/ST3 and blaCMY-2/IncI1/ST2 Plasmids in Healthy Urban Dogs in France. Antimicrob. Agents Chemother. 2014;58:5358–5362. doi: 10.1128/AAC.02545-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belas A., Salazar A.S., Gama L., Couto N., Pomba C. Risk factors for faecal colonisation with Escherichia coli producing extended-spectrum and plasmid-mediated AmpC β-lactamases in dogs. Vet. Rec. 2014;175:202. doi: 10.1136/vr.101978. [DOI] [PubMed] [Google Scholar]
- 13.Damborg P., Morsing M.K., Petersen T., Bortolaia V., Guardabassi L. CTX-M-1 and CTX-M-15-producing Escherichia coli in dog faeces from public gardens. Acta Vet. Scand. 2015;57:83. doi: 10.1186/s13028-015-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gómez-Sanz E., Torres C., Ceballos S., Lozano C., Zarazaga M. Clonal Dynamics of Nasal Staphylococcus aureus and Staphylococcus pseudintermedius in Dog-Owning Household Members. Detection of MSSA ST398. PLoS ONE. 2013;8:e69337. doi: 10.1371/journal.pone.0069337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehmer T., Vogler A.J., Thomas A., Sauer S., Hergenroether M., Straubinger R.K., Birdsell D., Keim P., Sahl J.W., Williamson C.H.D., et al. Phenotypic characterization and whole genome analysis of extended-spectrum beta-lactamase-producing bacteria isolated from dogs in Germany. PLoS ONE. 2018;13:e0206252. doi: 10.1371/journal.pone.0206252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaspar U., Von Lützau A., Schlattmann A., Roesler U., Köck R., Becker K. Zoonotic multidrug-resistant microorganisms among small companion animals in Germany. PLoS ONE. 2018;13:e0208364. doi: 10.1371/journal.pone.0208364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques C., Belas A., Franco A., Aboim C., Gama L., Pomba C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2017;73:377–384. doi: 10.1093/jac/dkx401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zogg A.L., Simmen S., Zurfluh K., Stephan R., Schmitt S.N., Nüesch-Inderbinen M. High Prevalence of Extended-Spectrum β-Lactamase Producing Enterobacteriaceae Among Clinical Isolates from Cats and Dogs Admitted to a Veterinary Hospital in Switzerland. Front. Vet. Sci. 2018;5:62. doi: 10.3389/fvets.2018.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nigg A., Brilhante M., Dazio V., Clément M., Collaud A., Brawand S.G., Willi B., Endimiani A., Schuller S., Perreten V. Shedding of OXA-181 carbapenemase-producing Escherichia coli from companion animals after hospitalisation in Switzerland: An outbreak in 2018. Eurosurveillance. 2019;24:1900071. doi: 10.2807/1560-7917.ES.2019.24.39.1900071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hordijk J., Schoormans A., Kwakernaak M., Duim B., Broens E., Dierikx C., Mevius D., Wagenaar J.A. High prevalence of fecal carriage of extended spectrum β-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front. Microbiol. 2013;4:242. doi: 10.3389/fmicb.2013.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brilhante M., Menezes J., Belas A., Feudi C., Schwarz S., Pomba C., Perreten V. OXA-181-Producing Extraintestinal Pathogenic Escherichia coli Sequence Type 410 Isolated from a Dog in Portugal. Antimicrob. Agents Chemother. 2020;64:e02298-19. doi: 10.1128/AAC.02298-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa D., Poeta P., Briñas L., Sáenz Y., Rodrigues J., Torres C. Detection of CTX-M-1 and TEM-52 β-lactamases in Escherichia coli strains from healthy pets in Portugal. J. Antimicrob. Chemother. 2004;54:960–961. doi: 10.1093/jac/dkh444. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho I., Safia Chenouf N., Cunha R., Martins C., Pimenta P., Pereira A.R., Martínez-Álvarez S., Ramos S., Silva V., Igrejas G., et al. Antimicrobial Resistance Genes and Diversity of Clones among ESBL—And Acquired AmpC-Producing Escherichia coli Isolated from Fecal Samples of Healthy and Sick Cats in Portugal. Antibiotics. 2021;10:262. doi: 10.3390/antibiotics10030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI . Performed Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI Supplement; Wayne, PA, USA: 2019. Clinical and Laboratory Standards Institute. [Google Scholar]
- 25.Holmes D., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 26.Ruiz E., Sáenz Y., Zarazaga M., Rocha-Gracia R., Martínez L.M., Arlet G., Torres C. qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: Genetic environments and plasmid and chromosomal location. J. Antimicrob. Chemother. 2012;67:886–897. doi: 10.1093/jac/dkr548. [DOI] [PubMed] [Google Scholar]
- 27.Zong Z., Partridge S.R., Thomas L., Iredell J.R. Dominance of blaCTX-M within an Australian Extended-Spectrum β-Lactamase Gene Pool. Antimicrob. Agents Chemother. 2008;52:4198–4202. doi: 10.1128/AAC.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitout J.D., Thomson K.S., Hanson N.D., Ehrhardt A.F., Moland E.S., Sanders C.C. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob. Agents Chemother. 1998;42:1350–1354. doi: 10.1128/AAC.42.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porres-Osante N., Azcona-Gutiérrez J.M., Rojo-Bezares B., Undabeitia E., Torres C., Sáenz Y. Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J. Antimicrob. Chemother. 2014;69:1792–1795. doi: 10.1093/jac/dku055. [DOI] [PubMed] [Google Scholar]
- 30.Vinué L., Sáenz Y., Somalo S., Escudero E., Moreno M.A., Ruiz-Larrea F., Torres C. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 2008;62:934–937. doi: 10.1093/jac/dkn331. [DOI] [PubMed] [Google Scholar]
- 31.Hassen B., Abbassi M.S., Ruiz-Ripa L., Mama O.M., Hassen A., Torres C., Hammami S. High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int. J. Food Microbiol. 2019;318:108478. doi: 10.1016/j.ijfoodmicro.2019.108478. [DOI] [PubMed] [Google Scholar]
- 32.Ellington M.J., Kistler J.J., Livermore D.M., Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-lactamases. J. Antimicrob. Chemother. 2006;59:321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 33.Vidal R., Vidal M., Lagos R., Levine M., Prado V. Multiplex PCR for Diagnosis of Enteric Infections Associated with Diarrheagenic Escherichia coli. J. Clin. Microbiol. 2004;42:1787–1789. doi: 10.1128/JCM.42.4.1787-1789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.PubMLST Escherichia coli (Achtman) MLST Locus/Sequence Definitions Database. [(accessed on 15 January 2021)]; Available online: https://pubmlst.org/bigsdb?db=pubmlst_ecoli_achtman_seqdef.
- 35.Clermont O., Bonacorsi S., Bingen E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000;66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jara D., Bello-Toledo H., Domínguez M., Cigarroa C., Fernández P., Vergara L., Quezada-Aguiluz M., Opazo-Capurro A., Lima C., González-Rocha G. Antibiotic resistance in bacterial isolates from freshwater samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020;10:1–8. doi: 10.1038/s41598-020-60035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carvalho A., Barbosa A., Arais L., Ribeiro P., Carneiro V., Cerqueira A. Resistance patterns, ESBL genes, and genetic relatedness of Escherichia coli from dogs and owners. Braz. J. Microbiol. 2016;47:150–158. doi: 10.1016/j.bjm.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y., Zeng Z., Chen S., Ma J., He L., Liu Y., Deng Y., Lei T., Zhao J., Liu J.-H. High prevalence of blaCTX-M extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 2010;16:1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x. [DOI] [PubMed] [Google Scholar]
- 39.Ben Sallem R., Gharsa H., BEN Slama K., Rojo-Bezares B., Estepa V., Porres-Osante N., Jouini A., Klibi N., Sáenz Y., Boudabous A., et al. First Detection of CTX-M-1, CMY-2, and QnrB19 Resistance Mechanisms in Fecal Escherichia coli Isolates from Healthy Pets in Tunisia. Vector Borne Zoonotic Dis. 2013;13:98–102. doi: 10.1089/vbz.2012.1047. [DOI] [PubMed] [Google Scholar]
- 40.Dahmen S., Haenni M., Châtre P., Madec J.-Y. Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J. Antimicrob. Chemother. 2013;68:2797–2801. doi: 10.1093/jac/dkt291. [DOI] [PubMed] [Google Scholar]
- 41.Iseppi R., Di Cerbo A., Messi P., Sabia C. Antibiotic Resistance and Virulence Traits in Vancomycin-Resistant Enterococci (VRE) and Extended-Spectrum β-Lactamase/AmpC-producing (ESBL/AmpC) Enterobacteriaceae from Humans and Pets. Antibiotics. 2020;9:152. doi: 10.3390/antibiotics9040152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Navarro J., Miro E., Brown-Jaque M., Hurtado J.C., Moreno A., Muniesa M., Gonzalez-Lopez J.J., Vila J., Espinal P., Navarro F. Diversity of plasmids in Escherichia coli and Klebsiella pneumoniae: A comparison of commensal and clinical isolates. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02064-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawamura K., Sugawara T., Matsuo N., Hayashi K., Norizuki C., Tamai K., Kondo T., Arakawa Y. Spread of CTX-Type Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates of Epidemic Clone B2-O25-ST131 Among Dogs and Cats in Japan. Microb. Drug Resist. 2017;23:1059–1066. doi: 10.1089/mdr.2016.0246. [DOI] [PubMed] [Google Scholar]
- 44.Aslantaş O., Yilmaz E. Prevalence and molecular characterization of extended-spectrum β-lactamase (ESBL) and plasmidic AmpC β-lactamase (pAmpC) producing Escherichia coli in dogs. J. Vet. Med Sci. 2017;79:1024–1030. doi: 10.1292/jvms.16-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa D., Poeta P., Sáenz Y., Coelho A.C., Matos M., Vinue L., Rodrigues J., Torres C. Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Vet. Microbiol. 2008;127:97–105. doi: 10.1016/j.vetmic.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Meireles D., Leite-Martins L., Bessa L., Cunha S., Fernandes R., De Matos A., Manaia C., Da Costa P.M. Molecular characterization of quinolone resistance mechanisms and extended-spectrum β-lactamase production in Escherichia coli isolated from dogs. Comp. Immunol. Microbiol. Infect. Dis. 2015;41:43–48. doi: 10.1016/j.cimid.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Pietsch M., Irrgang A., Roschanski N., Michael G.B., Hamprecht A., Rieber H., Käsbohrer A., Schwarz S., Rösler U. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018;19:601. doi: 10.1186/s12864-018-4976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liakopoulos A., Betts J., La Ragione R., van Essen-Zandbergen A., Ceccarelli D., Petinaki E., Koutinas C.K., Mevius D.J. Occurrence and characterization of extended-spectrum cephalosporin-resistant Enterobacteriaceae in healthy household dogs in Greece. J. Med. Microbiol. 2018;67:931–935. doi: 10.1099/jmm.0.000768. [DOI] [PubMed] [Google Scholar]
- 49.Belas A., Marques C., Aboim C., Pomba C. Emergence of Escherichia coli ST131 H30/H30-Rx subclones in companion animals. J. Antimicrob. Chemother. 2018;74:266–269. doi: 10.1093/jac/dky381. [DOI] [PubMed] [Google Scholar]
- 50.Rocha-Gracia R., Cortés-Cortés G., Lozano-Zarain P., Bello F., Martínez-Laguna Y., Torres C. Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 β-lactamases. Vet. J. 2014;203:315–319. doi: 10.1016/j.tvjl.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 51.Carvalho I., Tejedor-Junco M.T., González-Martín M., Corbera J.A., Suárez-Pérez A., Silva V., Igrejas G., Torres C., Poeta P. Molecular diversity of Extended-spectrum β-lactamase-producing Escherichia coli from vultures in Canary Islands. Environ. Microbiol. Rep. 2020;12:540–547. doi: 10.1111/1758-2229.12873. [DOI] [PubMed] [Google Scholar]
- 52.Kaarme J., Riedel H., Schaal W., Yin H., Nevéus T., Melhus Å. Rapid Increase in Carriage Rates of Enterobacteriaceae Producing Extended-Spectrum β-Lactamases in Healthy Preschool Children, Sweden. Emerg. Infect. Dis. 2018;24:1874–1881. doi: 10.3201/eid2410.171842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birgy A., Madhi F., Jung C., Levy C., Cointe A., Bidet P., Hobson C.A., Bechet S., Sobral E., Vuthien H., et al. Diversity and trends in population structure of ESBL-producing Enterobacteriaceae in febrile urinary tract infections in children in France from 2014 to 2017. J. Antimicrob. Chemother. 2019;75:96–105. doi: 10.1093/jac/dkz423. [DOI] [PubMed] [Google Scholar]
- 54.Mhaya A., Trabelsi R., Begu D., Aillerie S., M’zali F., Tounsi S., Gdoura R., Arpin C. Emergence of B2-ST131-C2 and A-ST617 Escherichia coli clones producing both CTX-M-15 and CTX-M-27 and ST147 NDM-1 positive Klebsiella pneumoniae in the Tunisian community. bioRxiv. 2019:713461. doi: 10.1101/713461. [DOI] [Google Scholar]
- 55.Ben Sallem R., Ben Slama K., Estepa V., Cheikhna E.O., Mohamed A.M., Chairat S., Ruiz-Larrea F., Boudabous A., Torres C. Detection of CTX-M-15-producing Escherichia coli isolates of lineages ST410-A, ST617-A and ST354-D in faecal samples of hospitalized patients in a Mauritanian hospital. J. Chemother. 2014;27:114–116. doi: 10.1179/1973947814Y.0000000172. [DOI] [PubMed] [Google Scholar]
- 56.Gauthier L., Dortet L., Cotellon G., Creton E., Cuzon G., Ponties V., Bonnin R.A., Naas T. Diversity of Carbapenemase-Producing Escherichia coli Isolates in France in 2012–2013. Antimicrob. Agents Chemother. 2018;62:e00266-18. doi: 10.1128/AAC.00266-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dahms C., Hübner N.-O., Kossow A., Mellmann A., Dittmann K., Kramer A. Occurrence of ESBL-Producing Escherichia coli in Livestock and Farm Workers in Mecklenburg-Western Pomerania, Germany. PLoS ONE. 2015;10:e0143326. doi: 10.1371/journal.pone.0143326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reid C., Wyrsch E.R., Chowdhury P.R., Zingali T., Liu M., Darling A., Chapman T., Djordjevic S.P. Porcine commensal Escherichia coli: A reservoir for class 1 integrons associated with IS26. Microb. Genom. 2017;3:e000143. doi: 10.1099/mgen.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Braga J.F.V., Chanteloup N.K., Trotereau A., Baucheron S., Guabiraba R., Ecco R., Schouler C. Diversity of Escherichia coli strains involved in vertebral osteomyelitis and arthritis in broilers in Brazil. BMC Vet. Res. 2016;12:1–12. doi: 10.1186/s12917-016-0762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bréchet C., Plantin J., Thouverez M., Cholley P., Bertrand X., Hocquet D. The wastewater network likely plays a role in the spread of ESBL-producing Escherichia coli in the community; Proceedings of the 23rd European Society of Clinical Microbiology and Infectious Diseases (ESCMID); Berlin, Germany. 12 February 1983. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Supplementary Material.