Abstract
Gut microbes are crucial to human health, but microbial composition is often disturbed in a number of human diseases. Accumulating evidence points to nutritional modulation of the gut microbiota as a potentially beneficial therapeutic strategy. Vitamin C (ascorbic acid) may be of particular interest as it has known antioxidant and anti-inflammatory properties. In this study, we investigated whether supplementation with high-dose vitamin C may favourably affect the composition of the gut microbiota. In this pilot study, healthy human participants received 1000 mg vitamin C supplementation daily for two weeks. Gut microbiota composition was analysed before and after intervention by performing faecal 16S rRNA gene sequencing. In total, 14 healthy participants were included. Daily supplementation of high-dose vitamin C led to an increase in the relative abundances of Lachnospiraceae (p < 0.05), whereas decreases were observed for Bacteroidetes (p < 0.01), Enterococci (p < 0.01) and Gemmiger formicilis (p < 0.05). In addition, trends for bacterial shifts were observed for Blautia (increase) and Streptococcus thermophilus (decrease). High-dose vitamin C supplementation for two weeks shows microbiota-modulating effects in healthy individuals, with several beneficial shifts of bacterial populations. This may be relevant as these bacteria have anti-inflammatory properties and strongly associate with gut health.
Keywords: vitamin C, ascorbic acid, gut microbiome, antioxidant, micronutrient, pilot study
1. Introduction
The gut microbiota consists of a vast collection of bacteria colonizing the gastrointestinal (GI) tract which are pivotal to human health. A disturbed gut microbiota composition occurs in a number of human diseases (often termed “dysbiosis”), characterized by typical shifts of bacterial clusters, e.g., increases in potentially pathogenic bacteria and decreased abundances of commensal, beneficial bacteria [1,2]. Examples of such diseases include inflammatory bowel diseases (IBD), type 2 diabetes mellitus (T2DM), non-alcoholic fatty liver disease (NAFLD) and cardiovascular diseases [3,4]. Recent efforts are increasingly directed towards therapeutic and/or nutritional modulation of gut microbiota as an innovative and promising treatment strategy. Multiple substrates have already been identified as potential health-promoting modulators of the gut microbiota [5]. Interestingly, specific dietary interventions and several types of micronutrients have demonstrated pronounced effects on the gut microbiota, showing both favourable and unfavourable consequences [6,7,8,9,10].
One of the nutritional components that are believed to have potentially beneficial microbiota-modulating effects constitute vitamins. Currently, vitamins are not classified as prebiotics by the International Scientific Association for Probiotics and Prebiotics; however, their ability to alter the gut microbial composition is well-recognised [5,11].
Vitamin C (ascorbic acid) is a particularly interesting micronutrient which has known beneficial health effects. Vitamin C is an essential water-soluble vitamin that cannot be endogenously synthesized by humans, but is supplied through diet, mainly via consumption of fruits and vegetables [12,13]. This vitamin is involved in biological processes supporting the immune system, and it has been identified as a main physiological antioxidant in higher-developed organisms [14,15]. Vitamin C supplementation has been reported to exert cell-protective properties against oxidative stress and is thus of particular interest in diseases in which oxidative stress plays a pathophysiological role. However, whether vitamin C has microbiota-modulating properties still needs to be elucidated.
Here, we designed a pilot intervention study in healthy volunteers to determine whether supplementation with high-dose (1000 mg) vitamin C can favourably alter the composition of the gut microbiota. Vitamin C is proficiently absorbed in the proximal small intestine, as is generally the case with vitamins. A daily intake of 30–180 mg/day leads to 70–90% absorption of vitamin C, yet when applying doses exceeding 1000 mg/day, absorption falls to less than 50% [12,16]. This dose-dependent gastrointestinal uptake in the small intestine allows for delivery of vitamin C to the gut microbiota, which is primarily located in the ileum and colon, when orally administered in high doses of 1000 mg. The aim of this study was to investigate shifts in bacterial populations upon high-dose vitamin C supplementation, which may be a possible beneficial treatment strategy for microbiota-associated diseases.
2. Materials and Methods
2.1. Study Population
Healthy volunteers aged 18–30 years were included. This pilot study was performed at the University Medical Center Groningen (UMCG), Groningen, the Netherlands, and participants were recruited from March 2017 until May 2017. Subjects were allowed to participate if they fulfilled the following inclusion criteria: age 18–30 years, body mass index (BMI) of ≥18.5 and ≤24.9 kg/m2, and self-proclaimed health. Furthermore, it was necessary that participants showed a fine understanding of the study procedures, motivation to comply with study requirements and the willingness to provide written informed consent for participation in the study. Participants were excluded when one (or more) of the following criteria were met: Regular use of dietary supplements (e.g., vitamin C, fish oil) one month prior to screening, known allergies or sensitivities to the study product or any ingredients thereof and use of commercially available probiotic, prebiotic or other supplements that may affect the gut microbiota.
2.2. Ethical Considerations
This pilot study was approved by the Institutional Review Board (IRB) (in Dutch: ‘Medisch Ethische Toetsingscommissie’, METc) of the University Medical Center Groningen (UMCG) (IRB ref no. M21.276231 and RR201700217). All subjects provided written informed consent. The study was performed in accordance with the Declaration of Helsinki (2013).
2.3. Data Collection and Study Design
At the time of inclusion, standard demographic characteristics, including age, sex, length (cm), weight (kg), body mass index (BMI) (body weight divided by squared height, kg/m2), smoking behaviour, alcohol consumption and weekly sports activity were recorded. On days 1 and 3, enrolled participants were instructed to take a baseline faecal sample as control. Participants were instructed to store the samples in their home freezers, immediately after production. Participants were asked to ingest vitamin C 1000 mg daily, for a period of two weeks. Subjects were asked to maintain normal dietary habits during the study. Additional samples were collected on days 15 and 17 after vitamin C supplementation. Frozen faecal samples were transported to the UMCG on dry ice and stored at −80 °C. Compliance to the study procedures was recorded on a predeveloped scheme by the participant.
2.4. Vitamin C Supplement
Patients received daily vitamin C supplementation of the normal diet for a period of 2 weeks. The supplement consisted of 1000 mg of vitamin C (L-ascorbic acid, commercially available) per capsule.
2.5. Sample Processing and Intestinal Microbiota Analysis (16S rRNA Gene Sequencing)
DNA was extracted from faecal samples using the RBB + Qiagen method and quantified by a Nanodrop machine, following the exact same procedure as described previously [17]. Amplification of the genes for the 16S rRNA V3/V4 regions was performed by polymerase chain reaction (PCR), using 2% agar gel electrophoresis (TaKaRa Taq Hot start version kit, Bio Inc., Kusatsu, Japan). The 341F and 806R primers (containing a six-nucleotide barcode and an Illumina-MiSeq adapter sequence) were used (Illumina, San Diego, CA, USA) [17,18,19]. Due to failure of some PCR reactions, DNA samples were cleaned-up using AMPure XP beads (Beckman Coulter, Brea, CA, USA) before repeating the PCR. After successful amplification, concentrations of DNA were measured using Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). To ensure similar library presentations for each sample, dilutions were prepared accordingly. A library was created by pooling 5 μL of each DNA sample as input for 16S rRNA amplicon sequencing (MiSeq Benchtop Sequencer, Illumina, San Diego, CA, USA). Samples with low DNA concentrations after clean-up (quality score < 0.9) were excluded from further analysis. Taxonomic profiling was performed using PAired-eND Assembler for DNA sequences (PANDAseq) and Quantitative Insights into Microbial Ecology (QIIME) [17,20,21]. This procedure was used to assign bacterial taxonomy up to family and genus level, and ARB was used to further identify sequences to species level [22]. Finally, relevant bacterial groups and bacterial species were extracted from the data for downstream analysis, which were selected based on their relative abundance and their importance with regard to vitamin metabolism and observed alterations in human diseases.
2.6. Statistical Analysis
Descriptive data were presented as medians with interquartile ranges (IQR) for continuous variables and proportions n with corresponding percentages (%) in case of nominal variables. From the original QIIME raw data, relative abundances of bacteria and groups of bacterial families were extracted. Differences in relative abundances of bacterial groups before and after intervention were compared using Wilcoxon signed-rank tests. Hierarchical agglomerative clustering (HAC) analysis was performed by computing pairwise distances between observations using Euclidean distances and visualizing these using the clustermap function of the seaborn package in Python. Clustering was performed to identify distinct bacterial clusters and shifts of these clusters before and after vitamin C supplementation. Correlations between bacterial relative abundances were determined using Spearman’s rank correlation coefficients (ρ), presented in a correlation matrix and visualized within a heatmap. Two-tailed nominal p-values ≤ 0.05 were considered as relevant trends, whereas false discovery rates (FDR) < 0.1 after adjustment for multiple comparisons (Benjamini–Hochberg method) were considered statistically significant. Statistical analyses were performed using the Python programming language (v.3.8.5, Python Software Foundation, https://www.python.org, accessed on 18 June 2021), using the pandas (v.1.2.3), numpy (v.1.20.0) and statsmodels (v.0.12.2) modules. Data visualization was performed using the seaborn (v.0.11.1) and matplotlib (v.3.4.1) packages in Python.
3. Results
In total, 14 participants were included who completed the full study programme and whose samples passed quality control after 16S rRNA gene sequencing data processing. No adverse events occurred in this pilot study.
3.1. Baseline Characteristics of the Study Population
Baseline demographic characteristics and lifestyle habits are shown in Table 1. Median age of participants was 22 years (IQR: 21–23), and gender distributions were almost equal (males: 43% vs. females: 57%). Only two participants (14%) were smokers, and median alcohol consumption was 5 units per week. Participants were moderately physically active with a median of 3 (IQR: 2–6) hours of exercise per week.
Table 1.
Baseline demographic and lifestyle characteristics of the study population (n = 14) consisting of healthy volunteers.
| Numbers (n, %) or Median [IQR] | |
|---|---|
| Age (years) | 22 [21;23] |
| Gender | |
| Male, n (%) | 6 (43%) |
| Female, n (%) | 8 (57%) |
| Length (cm) | 177 [169;183] |
| Weight (kg) | 69 [62;78] |
| BMI (kg/m2) | 22.0 [21.7;23.3] |
| Current smoking, n (%) | 2 (14%) |
| Alcohol use (units/week) | 5 [3;6] |
| Sports activity (h/week) | 3 [2;6] |
Data are presented as median (interquartile range, IQR) or proportions n with corresponding percentages (%). Abbreviations: BMI, body mass index.
3.2. Changes in Bacterial Groups after Vitamin C Supplementation
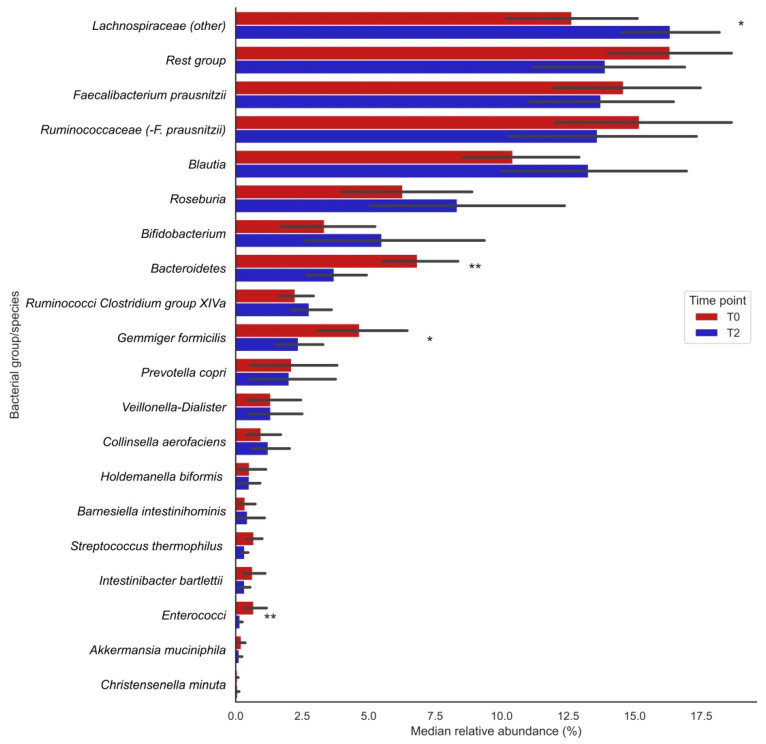
Analysis of 16S rRNA gene sequencing data showed that the relative abundances (%) of the bacterial groups Lachnospiraceae (other), Bacteroidetes, enterococci and the species Gemmiger formicilis were significantly changed after vitamin C supplementation (visualized in Figure 1; the exact numbers and corresponding P-values can be found in Table 2). The relative abundances of Lachnospiraceae significantly increased after supplementation (T0: 12.7 [9.71;16.6] vs. T2: 16.2 [13.8;17.6], p = 0.013), whereas those of Bacteroidetes (T0: 5.97 [4.88;8.87] vs. T2: 3.09 [2.14;4.77], p= 0.004), Gemmiger formicilis (T0: 3.96 [2.51;6.59] vs. T2: 1.86 [0.81;3.68], p = 0.013) and enterococci (T0: 0.40 [0.22;0.51] vs. T2: 0.13 [0.07;0.18], p = 0.009) significantly decreased after supplementation (Table 2). Furthermore, trends for bacterial shifts were observed for Blautia (nominal p = 0.035), Ruminococcaceae Clostridium group XIVa (p = 0.035) and Streptococcus thermophilus (p = 0.096). No particular trends of bacterial shifts were observed for smoking participants.
Figure 1.
Changes in median relative abundances (%) of bacterial groups before (T0) and after (T2) vitamin C supplementation. Bars indicate median relative abundances of bacterial groups with a corresponding IQR (black lines). * Nominal p-value ≤ 0.05; ** Nominal p-value ≤ 0.01.
Table 2.
Median relative abundances (%) of bacterial groups before (T0) and after (T2) two weeks of vitamin C supplementation.
| Bacterial Group | T0 | T2 | p-Value |
|---|---|---|---|
| Bacteroidetes | 5.97 [4.88;8.87] | 3.09 [2.14;4.77] | 0.004 |
| Ruminococci Clostridium group XIVa | 2.06 [1.28;2.82] | 2.66 [1.92;3.13] | 0.048 |
| Bifidobacterium | 1.86 [1.10;3.77] | 3.29 [1.65;5.61] | 0.272 |
| Blautia | 9.68 [7.67;11.8] | 11.3 [10.1;14.5] | 0.035 |
| Ruminococcaceae (-F. prausnitzii) | 16.0 [12.1;17.8] | 13.0 [7.6;15.5] | 0.035 |
| Enterococci | 0.40 [0.22;0.51] | 0.13 [0.07;0.18] | 0.009 |
| Veillonella-Dialister | 0.40 [0.04;0.96] | 0.44 [0.13;1.46] | 0.300 |
| Lachnospiraceae (other) | 12.7 [9.71;16.6] | 16.2 [13.8;17.6] | 0.013 |
| Rest group | 15.8 [14.2;20.0] | 13.2 [10.4;16.4] | 0.510 |
| Roseburia | 5.26 [2.77;8.81] | 5.54 [4.38;9.67] | 0.140 |
| Faecalibacterium prausnitzii | 14.9 [11.4;17.7] | 12.0 [10.3;17.8] | 0.551 |
| Gemmiger formicilis | 3.96 [2.51;6.59] | 1.86 [0.81;3.68] | 0.013 |
| Prevotella copri | 0.01 [0.002;3.39] | 0.003 [0.001;3.01] | 0.221 |
| Collinsella aerofaciens | 0.49 [0.26;0.98] | 0.69 [0.44;1.49] | 0.363 |
| Intestinibacter bartlettii | 0.19 [0.13;0.90] | 0.14 [0.05;0.56] | 0.158 |
| Barnesiella intestinihominis | 0.16 [0.12;0.24] | 0.11 [0.06;0.16] | 0.638 |
| Holdemanella biformis | 0.001 [0.00;0.54] | 0.004 [0.00;0.85] | 0.889 |
| Akkermansia muciniphila | 0.13 [0.02;0.28] | 0.07 [0.02;0.12] | 0.258 |
| Christensenella minuta | 0.05 [0.04;0.09] | 0.03 [0.01;0.08] | 0.683 |
| Streptococcus thermophilus | 0.51 [0.33;0.78] | 0.25 [0.16;0.49] | 0.096 |
Data are presented as median (interquartile range, IQR) of relative abundances (%) at baseline (T0) and two weeks after vitamin C supplementation (T2). Nominal p-value for Wilcoxon’s signed-rank test. Significances after adjustment for multiple comparisons (Benjamini–Hochberg method, FDR < 0.1) are indicated in bold. Abbreviations: FDR, false discovery rate; T0, baseline; T2, two weeks after vitamin C supplementation.
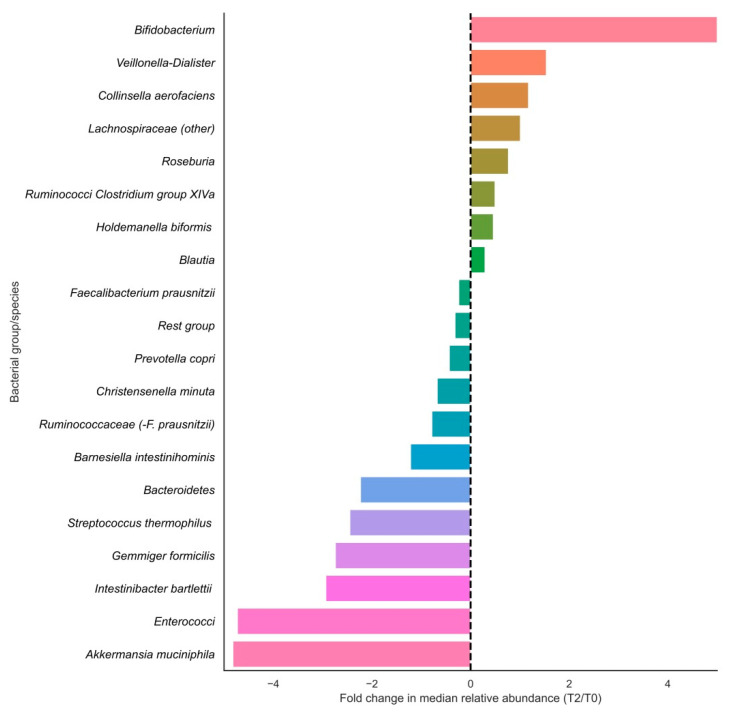
In terms of relative changes in bacterial group abundances, highest fold changes in relative abundances in the positive direction were observed for Bifidobacteriaceae, Veillonella-Dialister, Collinsella aerofaciens, Lachnospiraceae (other), Roseburia and Ruminococci from Clostridium group XIVa, and largest fold changes were observed in the negative direction for Akkermansia muciniphila, enterococci, Intestinibacter bartlettii, Gemmiger formicilis and Streptococcus thermophilus (Figure 2). Of these fold changes, only those of Lachnospiraceae (other), Gemmiger formicilis, Bacteroidetes and Enterococci were statistically significant (p < 0.05).
Figure 2.
Fold changes in relative abundances (%) of bacterial groups before and after vitamin C supplementation. Bars indicate fold changes of bacterial groups after supplementation (T2) compared to the relative abundance at baseline (T0). Relative increases in bacterial abundance are depicted on the right side of the plot, whereas decreases are shown on the left side of the plot. Bacterial groups are sorted on fold change sizes.
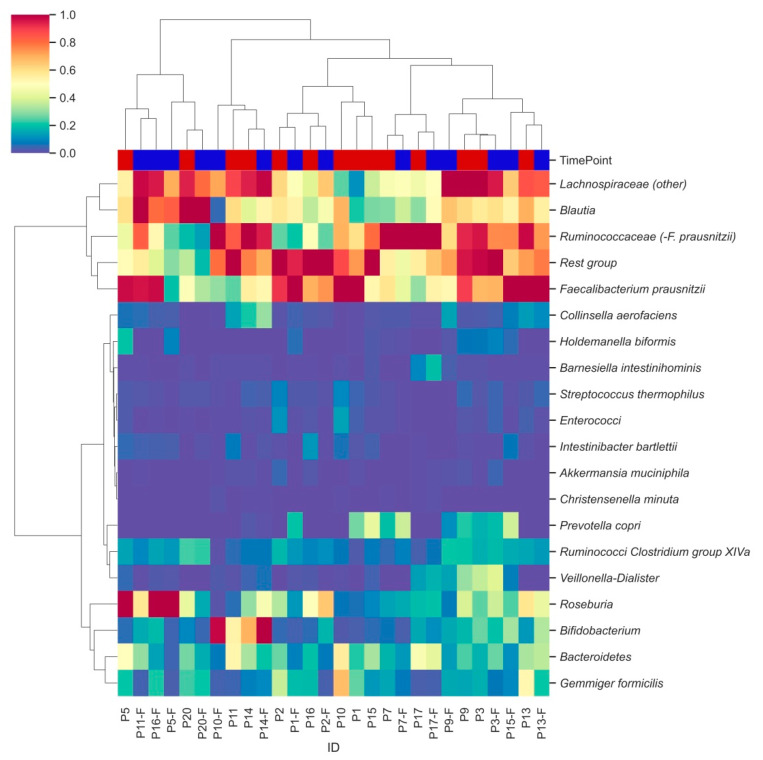
3.3. Characterization of Bacterial Clusters before and after Supplementation
Hierarchical clustering analysis identified two major clusters of bacterial groups, one of which is characterized by the combination of Lachnospiraceae, Blautia, Ruminococcaceae and Faecalibacterium prausnitzii (Figure 3, row level dendrogram). In the second cluster, subclusters were observed for Ruminococci from Clostridium group XIVa and Veillonella-Dialister, as well as for Gemmiger formicilis and Bacteroidetes. Remaining bacterial groups were less abundant and did not show distinct clustering. Moderate clustering was observed between samples from the two different time points (column labels, Figure 3).
Figure 3.
Hierarchically clustered heatmap showing distinct bacterial clusters across participants’ faecal samples. Heatmap columns are coloured by the time point (T0: red; T2: blue) from which each sample was derived. Hierarchical clustering was performed using pairwise Euclidean distances across sample observations. Colouring was based on relative abundances set on a standardized scale.
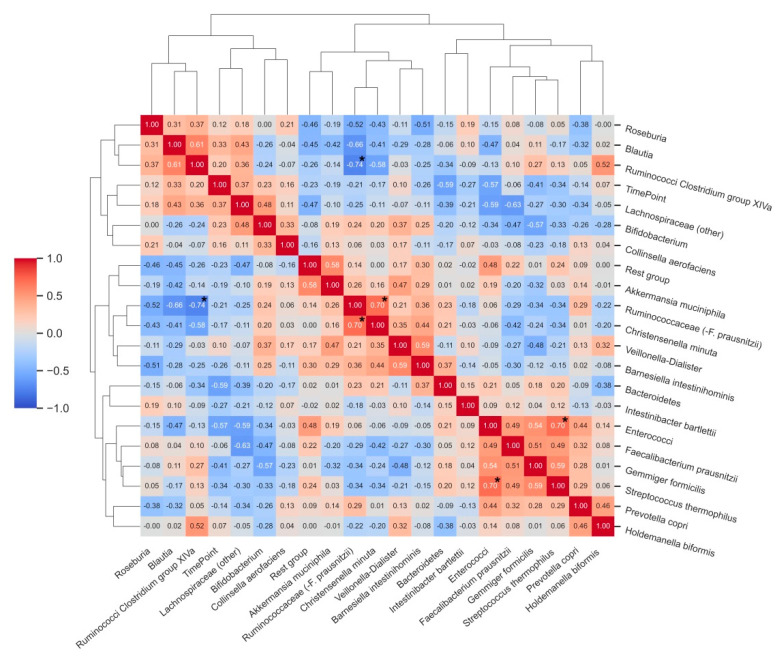
Many correlations were observed among the analysed bacterial groups, although few of them reached statistical significance after adjustment for multiple comparisons (Figure 4). Statistically significant correlations were found between the relative abundance of enterococci and Streptococcus thermophilus (ρ = 0.70, p < 0.05), Ruminococcaceae (−F. prausnitzii) and Christensenella minuta (ρ = 0.70, p < 0.05), and there was an inverse correlation between the relative abundance of Ruminococci from Clostridium group XIVa and Ruminococcaceae (−F. prausnitzii). Relative abundances of Bacteroidetes (ρ = −0.59) and enterococci (ρ= −0.57) showed strong, inverse associations with the T2 time point of the study (after vitamin C supplementation).
Figure 4.
Correlation matrix heatmap demonstrating correlations between relative abundances (%) of bacterial groups before and after vitamin C supplementation. Time point (T0 and T2) was included as an additional variable to the correlative clustering as well as to demonstrate associations between bacterial groups and changes upon intervention. Dendrograms represent hierarchical agglomerative clustering based on Euclidean distances (same as in Figure 3). * Bold asterisks indicate statistically significant correlations after adjustment for multiple comparisons.
4. Discussion
In this human pilot intervention study, high-dose vitamin C supplementation for two weeks led to substantial shifts in faecal bacterial populations. In particular, we demonstrated a significant increase in the median relative abundance of the Lachnospiraceae family and trends towards an increased relative abundance of the genus Blautia. In contrast, significant decreases in median relative abundances were observed for the phylum Bacteroidetes and genera enterococci after vitamin C supplementation. The bacterial species Gemmiger formicilis also demonstrated a significant decrease in median relative abundance. Taken together, these results suggest that two-week high-dose vitamin C supplementation may confer beneficial alterations in gut microbial populations, while leaving only a few exceptions.
The results of this study are relevant for a variety of reasons. Currently, human clinical studies investigating the effect of vitamins on the gut microbiota are sparsely available. Previous clinical studies investigating vitamin B2 and vitamin D supplementation demonstrated potential beneficial alterations in microbiota compositions [23,24]. More recently, a study in healthy volunteers investigating the effects of colon-delivered vitamins A, B2, C, D and E on the gut microbiota reported microbiota-modulating effects of vitamins B2, C and D. Vitamin C supplementation (500 mg/day for 4 weeks) demonstrated increased microbiota diversity and an increased abundance of Collinsella [25]. The advantage of oral high-dose vitamin C supplementation over controlled colon-delivery of vitamin C lies in the combination of its absorption in the upper gastrointestinal tract while concurrently establishing vitamin C delivery to the gut microbiota, as high-dose vitamin C is only partially absorbed in the small bowel [12]. This approach may be of particular value for patients with diseases that might benefit from microbiota-modulation and who are simultaneously at risk of vitamin C deficiency.
No studies have followed to investigate whether commercially available high-dose vitamin C supplementation has the capacity to alter the gut microbiota. The strength of our study primarily lies in its novelty. Apart from that, our results may be of interest to a multitude of medical disciplines. For instance, neurodegenerative diseases, such as Parkinson’s disease (PD), are characterized by disturbed gut microbial composition and functionality. A previous study in patients with PD showed that abundances of the bacterial genera Blautia and Roseburia, two bacterial groups whose presence could potentially be increased through vitamin C supplementation, were markedly decreased compared to healthy individuals [26]. Furthermore, in patients with liver cirrhosis, an increased abundance of the genus Blautia was found to be associated with favourable disease outcomes, whereas overrepresentation of the genus Enterococcus was linked to inflammation and a reduction in cognitive function [27].
At the same time, however, some limitations of this study also warrant recognition. First, this study was of explorative origin as a pilot study, in which only 14 healthy volunteers were included. Therefore, our results should be interpreted with caution as these should be confirmed and validated in larger study populations, consisting of more follow-up data. Second, although the observed changes in bacterial populations may suggest potential beneficial effects to the gut microbial community, they confer limited generalizability due to (1) our strict inclusion and exclusion criteria, (2) the fact that gut microbiota in diseased circumstances may already be in an altered state, and (3) the fact that the gut microbiota may respond differently to nutritional interventions between individuals. Another limitation of our study included the relatively short duration of follow-up, preventing us from studying long-term effects of high-dose vitamin C supplementation on the gut microbiota composition as well as the absence of additional biological health parameters (e.g., inflammatory or oxidative stress biomarkers), which may have sustained the observed potentially beneficial effects. We have not performed measurements related to the gut metabolome, which may provide valuable information regarding microbial functions [28]. However, Pham et al. [25] reported a significant increase in total faecal SCFA concentrations after vitamin C supplementation, which corresponds with the shift towards butyrate-producing bacteria as observed in our study.
The strongest effect of high-dose vitamin C supplementation was the increased relative abundance of the family Lachnospiraceae. The Lachnospiraceae bacterial family belongs to the phylum Firmicutes and is a predominant bacterial group of the gut microbiota in healthy subjects [29,30]. Lachnospiraceae form one of the main producers of short-chain fatty acids (SCFAs). These are bacterial metabolites that are known to possess several anti-inflammatory and antioxidant effects [31]. A study investigating the effect of micronutrients on the gut microbiota composition of patients with cystic fibrosis found that increased vitamin C intake positively correlated with the abundance of Firmicutes [32]. The same study found a negative correlation of vitamin C with abundance of the phylum Bacteroidetes, a finding which is also supported by the present study. The exact mechanisms of interaction between the Lachnospiraceae family and its human host remain to be determined. As is often the case with specific bacterial groups, it remains largely unclear whether an increased or decreased abundance may be a cause or consequence of the disease. For instance, a greater abundance of Lachnospiraceae is found in patients with T2DM and liver and kidney diseases [29]. Interestingly, a decreased abundance of Lachnospiraceae is found in faecal samples of Crohn’s disease (CD) and ulcerative colitis (UC) patients [29,33].
In this study, we demonstrated an approximately 4-fold decrease in the median relative abundance of A. muciniphila. This is a strictly anaerobic, mucin-degrading bacterial species that has been reported to support immunological and metabolic responses of its host [34] and is of interest due to a potential role as a probiotic [35]. However, decreased abundances of A. muciniphila are usually inversely associated with disease states, e.g., T2DM, IBD and obesity [34].
Vitamin C is particularly known for its antioxidant properties, and it belongs to the antioxidant machinery, where it is part of the pool of nonenzymatic antioxidants such as vitamin E (alpha-tocopherol), polyphenols (e.g., N-acetylcysteine, NAC) and uric acid [36]. These substances are able to modulate oxidative stress by scavenging reactive oxygen species (ROS). In a variety of disease contexts, vitamin C has been reported to decrease indicators of oxidative stress and inflammation [37,38], but further high-quality clinical trials with relevant endpoints are needed to establish the true role of vitamin C as an antioxidant supplement [39,40]. The direct effect of vitamin C on the intestinal redox balance presumably plays an important gut microbiota-modulating role. Faecal redox potential is dose-dependently related to the ratio of relative abundance of aerotolerant to strict anaerobic species [41]. Strictly anaerobic gut bacteria, such as the bacterial family Lachnospiraceae and the species Faecalibacterium prausnitzii, which belong to the Healthy Human Mature Anaerobic Gut Microbiota (HMAGM), are profoundly influenced by oxidative stress [41,42]. The presence of oxygen in the intestinal lumen can promote the growth of Enterobacteriaceae; whereas it compromises the growth of F. prausnitzii, as this bacterial species is particularly susceptible to oxygen [43,44,45].
Vitamin C supplementation could be of particular interest for patients with inflammatory bowel disease (IBD). Associations between IBD and imbalances of gut microbiota composition are becoming increasingly apparent. Homeostasis of the gut microbiota is hypothesized to play a pivotal role in pro-inflammatory and anti-inflammatory responses in the intestines [46,47]. IBD is characterized by chronic inflammation of the gastrointestinal tract. IBD is typically diagnosed in young adulthood and requires lifelong medical therapy [48,49]. The bacterial family Lachnospiraceae and the genus Blautia (increased relative abundance in our study) have been found to be reduced in abundance in patients with Crohn’s disease [50,51] and the bacterial genus Enterococcus and species G. formicilis (decreased relative abundance in our study) have been associated with recurrent and relapsing CD [52,53]. A reduction in Lachnospiraceae abundance is associated with attenuated butyrogenesis, which is linked to a possible recurrence of UC [33]. However, the literature is inconsistent about the effect of low levels of Bacteroidetes found after vitamin C supplementation. Enrichment of Bacteroidetes has been shown to be associated with low-grade systemic inflammation, yet increased Bacteroidetes abundance is also associated with decreased colitis development during cancer treatment with ipilimumab [54]. Low levels of genus Bacteroides have been associated with IBD, and a decreased abundance was more noticeable during active disease when compared to quiescent disease [55]. Contrarily, studies have associated an increased abundance of Bacteroides with inflammation of the gut mucosa and early recurrence of CD after ileocecal resection [56,57].
A trial that investigated the effects of fish oil and antioxidants (vitamins A, C, E and selenium) vs. placebo in patients with CD mainly observed decreased inflammatory markers [58]. Taking into consideration the increased risk of micronutrient deficiency (including vitamin C) [59,60], as well as the combined presence of oxidative stress and gut microbiota disturbances in the IBD population, high-dose vitamin C supplementation may be considered a highly promising nutritional intervention in this disease. Clinical trials with CD patients further investigating the microbiota-modulating role of vitamin supplements, including vitamin C, are forthcoming (www.clinicaltrials.gov, accessed on 18 June 2021; NCT04913467).
5. Conclusions
In conclusion, this human pilot study demonstrates that supplementation of high-dose vitamin C can manipulate the gut microbiota composition, leading to shifts of bacterial populations in the gut. This alteration of the gut microbiota composition and its associated anti-inflammatory effects with, for example, butyrate production may potentially be beneficial for a variety of diseases characterized by disturbed gut microbiota homeostasis, e.g., IBD, metabolic, cardiovascular and neurological diseases.
Acknowledgments
The authors would like to thank all healthy volunteers for participating in this pilot study. The authors also would like to thank Anneke Boddeke, Elisa Doldersum, Leonie Steenhof, Hendrea Tingen and Carlijn Wagenaar (medical students from the University of Groningen) for their contributions to this pilot study.
Author Contributions
Conceptualization: H.J.M.H., B.Z.A. and G.D.; methodology, A.T.O., A.R.B., H.J.M.H. and G.D.; software, H.J.M.H. and A.R.B.; validation, all authors; formal analysis, A.T.O., A.R.B. and H.J.M.H.; investigation, A.T.O., A.R.B., V.P. and H.J.M.H.; resources, B.Z.A., H.J.M.H., G.D.; data curation, A.T.O., A.R.B., V.P. and H.J.M.H.; writing—original draft preparation, A.T.O., A.R.B. and V.P.; writing—review and editing, all authors; visualization, A.R.B.; supervision, B.Z.A., H.J.M.H. and G.D.; project administration, H.J.M.H.; funding acquisition, H.J.M.H. and G.D. All authors have read and agreed to the published version of the manuscript.
Funding
The research position of ARB was supported by a JSM M.D.–Ph.D. trajectory grant (grant no. 17-57) from the Junior Scientific Masterclass (JSM) of the University of Groningen, the Netherlands. The funders had no role in the design of the study, collection, analysis, or interpretation of data, or writing of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of University Medical Center Groningen (IRB ref no. M21.276231 and RR201700217).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data is contained within the present article.
Conflicts of Interest
G.D. received an unrestricted research grant from Royal DSM and received speaker fees from Pfizer, Abbvie and Janssen Pharmaceuticals. All other authors have no conflict of interest to declare.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Power S.E., O’Toole P., Stanton C., Ross R., Fitzgerald G.F. Intestinal microbiota, diet and health. Br. J. Nutr. 2013;111:387–402. doi: 10.1017/S0007114513002560. [DOI] [PubMed] [Google Scholar]
- 2.Gensollen T., Iyer S.S., Kasper D.L., Blumberg R.S. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kho Z.Y., Lal S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch S.V., Pedersen O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 5.Gibson G.R., Hutkins R., Sanders M.E., Prescott S.L., Reimer R.A., Salminen S.J., Scott K., Stanton C., Swanson K.S., Cani P.D., et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 6.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hold G.L. Western lifestyle: A ‘master’ manipulator of the intestinal microbiota? Gut. 2014;63:5–6. doi: 10.1136/gutjnl-2013-304969. [DOI] [PubMed] [Google Scholar]
- 8.Wu G.D., Bushmanc F.D., Lewis J.D. Diet, the human gut microbiota, and IBD. Anaerobe. 2013;24:117–120. doi: 10.1016/j.anaerobe.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K., Kurilshikov A., Bonder M.J., Valles-Colomer M., Vandeputte D., et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 10.Sugihara K., Morhardt T.L., Kamada N. The Role of Dietary Nutrients in Inflammatory Bowel Disease. Front. Immunol. 2019;9:3183. doi: 10.3389/fimmu.2018.03183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinert R.E., Lee Y.-K., Sybesma W. Vitamins for the Gut Microbiome. Trends Mol. Med. 2020;26:137–140. doi: 10.1016/j.molmed.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Jacob R.A., Sotoudeh G. Vitamin C Function and Status in Chronic Disease. Nutr. Clin. Care. 2002;5:66–74. doi: 10.1046/j.1523-5408.2002.00005.x. [DOI] [PubMed] [Google Scholar]
- 13.Padayatty S., Levine M. Vitamin C: The known and the unknown and Goldilocks. Oral Dis. 2016;22:463–493. doi: 10.1111/odi.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr A.C., Maggini S. Vitamin C and Immune Function. Nutrients. 2017;9:1211. doi: 10.3390/nu9111211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padayatty S.J., Katz A., Wang Y., Eck P., Kwon O., Lee J.-H., Chen S., Corpe C., Dutta A., Dutta S.K., et al. Vitamin C as an Antioxidant: Evaluation of Its Role in Disease Prevention. J. Am. Coll. Nutr. 2003;22:18–35. doi: 10.1080/07315724.2003.10719272. [DOI] [PubMed] [Google Scholar]
- 16.Padayatty S.J., Sun H., Wang Y., Riordan H.D., Hewitt S.M., Katz A., Wesley R.A., Levine M. Vitamin C pharmacoki-netics: Implications for oral and intravenous use. Ann. Intern. Med. 2004;140:533–537. doi: 10.7326/0003-4819-140-7-200404060-00010. [DOI] [PubMed] [Google Scholar]
- 17.Heida F.H., Van Zoonen A.G.J.F., Hulscher J.B.F., Kiefte B.J.C.T., Wessels R., Kooi E.M.W., Bos A.F., Harmsen H.J.M., de Goffau M. A Necrotizing Enterocolitis-Associated Gut Microbiota Is Present in the Meconium: Results of a Prospective Study. Clin. Infect. Dis. 2016;62:863–870. doi: 10.1093/cid/ciw016. [DOI] [PubMed] [Google Scholar]
- 18.Bartram A.K., Lynch M.D., Stearns J.C., Moreno-Hagelsieb G., Neufeld J.D. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol. 2011;77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bokulich N.A., Joseph C.M.L., Allen G., Benson A.K., Mills D.A. Next-Generation Sequencing Reveals Significant Bacterial Diversity of Botrytized Wine. PLoS ONE. 2012;7:e36357. doi: 10.1371/journal.pone.0036357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masella A.P., Bartram A.K., Truszkowski J.M., Brown D.G., Neufeld J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, Buchner A., Lai T., Steppi S., Jobb G., et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Von Martels J.Z.H., Bourgonje A., Klaassen M.A.Y., A A Alkhalifah H., Sadabad M.S., Vila A.V., Gacesa R., Gabriëls R.Y., E Steinert R., Jansen B.H., et al. Riboflavin Supplementation in Patients with Crohn’s Disease [the RISE-UP study] J. Crohn’s Colitis. 2019;14:595–607. doi: 10.1093/ecco-jcc/jjz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schäffler H., Herlemann D., Klinitzke P., Berlin P., Kreikemeyer B., Jaster R., Lamprecht G. Vitamin D administration leads to a shift of the intestinal bacterial composition in Crohn’s disease patients, but not in healthy controls. J. Dig. Dis. 2018;19:225–234. doi: 10.1111/1751-2980.12591. [DOI] [PubMed] [Google Scholar]
- 25.Pham V.T., Fehlbaum S., Seifert N., Richard N., Bruins M.J., Sybesma W., Rehman A., Steinert R.E. Effects of co-lon-targeted vitamins on the composition and metabolic activity of the human gut microbiome—A pilot study. Gut Microbes. 2021;13:1875774. doi: 10.1080/19490976.2021.1875774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keshavarzian A., Green S., Engen P.A., Voigt R.M., Naqib A., Forsyth C.B., Mutlu E., Shannon K.M. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015;30:1351–1360. doi: 10.1002/mds.26307. [DOI] [PubMed] [Google Scholar]
- 27.Bajaj J.S., Hylemon P.B., Ridlon J.M., Heuman D.M., Daita K., White M.B., Monteith P., Noble N.A., Sikaroodi M., Gillevet P.M. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Liver Physiol. 2012;303:G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee-Sarwar K.A., Lasky-Su J., Kelly R.S., Litonjua A.A., Weiss S.T. Metabolome-Microbiome Crosstalk and Human Dis-ease. Metabolites. 2020;10:181. doi: 10.3390/metabo10050181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vacca M., Celano G., Calabrese F.M., Portincasa P., Gobbetti M., De Angelis M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms. 2020;8:573. doi: 10.3390/microorganisms8040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 32.Li L., Krause L., Somerset S. Associations between micronutrient intakes and gut microbiota in a group of adults with cyst-ic fibrosis. Clin. Nutr. 2017;36:1097–1104. doi: 10.1016/j.clnu.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 33.Sasaki K., Inoue J., Sasaki D., Hoshi N., Shirai T., Fukuda I., Osawa R. Construction of a model culture system of human colonic microbiota to detect decreased Lachnospiraceae abundance and butyrogenesis in the feces of ulcerative colitis patients. Biotechnol. J. 2019;14:1800555. doi: 10.1002/biot.201800555. [DOI] [PubMed] [Google Scholar]
- 34.Macchione I.G., Lopetuso L.R., Ianiro G., Napoli M., Gibiino G., Rizzatti G., Petito V., Gasbarrini A., Scaldaferri F. Akkermansia muciniphila: Key player in metabolic and gastrointestinal disorders. Eur. Rev. Med. Pharmacol Sci. 2019;23:8075–8083. doi: 10.26355/eurrev_201909_19024. [DOI] [PubMed] [Google Scholar]
- 35.Zhang T., Li Q., Cheng L., Buch H., Zhang F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019;12:1109–1125. doi: 10.1111/1751-7915.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourgonje A.R., Feelisch M., Faber K.N., Pasch A., Dijkstra G., van Goor H. Oxidative Stress and Redox-Modulating Therapeutics in Inflammatory Bowel Disease. Trends Mol. Med. 2020;26:1034–1046. doi: 10.1016/j.molmed.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Aghdassi E., Wendland B.E., Steinhart A.H., Wolman S.L., Jeejeebhoy K., Allard J.P. Antioxidant vitamin supplementa-tion in Crohn’s disease decreases oxidative stress. a randomized controlled trial. Am. J. Gastroenterol. 2003;98:348–353. doi: 10.1111/j.1572-0241.2003.07226.x. [DOI] [PubMed] [Google Scholar]
- 38.Sasazuki S., Hayashi T., Nakachi K., Sasaki S., Tsubono Y., Okubo S., Hayashi M., Tsugane S. Protective effect of vita-min C on oxidative stress: A randomized controlled trial. Int. J. Vitam. Nutr. Res. 2008;78:121–128. doi: 10.1024/0300-9831.78.3.121. [DOI] [PubMed] [Google Scholar]
- 39.Bjelakovic G., Nikolova D., Gluud L.L., Simonetti R.G., Gluud C. Mortality in randomized trials of antioxidant supple-ments for primary and secondary prevention: Systematic review and meta-analysis. JAMA. 2007;297:842–857. doi: 10.1001/jama.297.8.842. [DOI] [PubMed] [Google Scholar]
- 40.Lykkesfeldt J., Poulsen H.E. Is vitamin C supplementation beneficial? Lessons learned from randomised controlled trials. Br. J. Nutr. 2009;103:1251–1259. doi: 10.1017/S0007114509993229. [DOI] [PubMed] [Google Scholar]
- 41.Million M., Raoult D. Linking gut redox to human microbiome. Hum. Microbiome J. 2018;10:27–32. doi: 10.1016/j.humic.2018.07.002. [DOI] [Google Scholar]
- 42.Million M., Alou M.T., Khelaifia S., Bachar D., Lagier J.-C., Dione N., Brah S., Hugon P., Lombard V., Armougom F., et al. Increased Gut Redox and Depletion of Anaerobic and Methanogenic Prokaryotes in Severe Acute Malnutrition. Sci. Rep. 2016;6:26051. doi: 10.1038/srep26051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L.G., Gratadoux J.J., Blugeon S., Bridonneau C., Furet J.P., Corthier G. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan M.T., Browne W.R., van Dijl J.M., Harmsen H.J. How can Faecalibacterium prausnitzii employ riboflavin for extra-cellular electron transfer? Antioxid. Redox Signal. 2012;17:1433–1440. doi: 10.1089/ars.2012.4701. [DOI] [PubMed] [Google Scholar]
- 45.Khan M.T., Duncan S.H., Stams A., Van Dijl J.M., Flint H.J., Harmsen H.J.M. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J. 2012;6:1578–1585. doi: 10.1038/ismej.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schirmer M., Garner A., Vlamakis H., Xavier R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019;17:497–511. doi: 10.1038/s41579-019-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forbes J.D., Chen C.-Y., Knox N.C., Marrie R.-A., El-Gabalawy H., De Kievit T., Alfa M., Bernstein C.N., Van Domselaar G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases—does a common dysbiosis exist? Microbiome. 2018;6:221. doi: 10.1186/s40168-018-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres J., Mehandru S., Colombel J.F., Peyrin-Biroulet L. Crohn’s disease. Lancet. 2017;389:1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 49.Ungaro R., Mehandru S., Allen P.B., Peyrin-Biroulet L., Colombel J.F. Ulcerative colitis. Lancet. 2017;389:1756–1770. doi: 10.1016/S0140-6736(16)32126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Øyri S.F., Műzes G., Sipos F. Dysbiotic gut microbiome: A key element of Crohn’s disease. Comp. Immunol. Microbiol. Infect. Dis. 2015;43:36–49. doi: 10.1016/j.cimid.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K., Nishida A., Fujimoto T., Fujii M., Shioya M., Imaeda H., Inatomi O., Bamba S., Andoh A., Sugimoto M. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. Digestion. 2016;93:59–65. doi: 10.1159/000441768. [DOI] [PubMed] [Google Scholar]
- 52.Watson A.J., Biancheri P., Patterson A. The Mucosal Microbiome and Recurrence After Surgery for Crohn’s Disease. Gastroenterology. 2016;150:1682–1684. doi: 10.1053/j.gastro.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y., Chen H., He H., Du Y., Hu J., Li Y., Li Y., Zhou Y., Wang H., Chen Y., et al. Increased Enterococcus faecalis infection is associated with clinically active Crohn disease. Medicine. 2016;95:e5019. doi: 10.1097/MD.0000000000005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaput N., Lepage P., Coutzac C., Soularue E., Le Roux K., Monot C., Boselli L., Routier E., Cassard L., Collins M., et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y., Zhi F. Lower Level of Bacteroides in the Gut Microbiota Is Associated with Inflammatory Bowel Disease: A Me-ta-Analysis. Biomed. Res. Int. 2016;2016:5828959. doi: 10.1155/2016/5828959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neut C., Bulois P., Desreumaux P., Membré J.M., Lederman E., Gambiez L., Cortot A., Quandalle P., van Kruiningen H., Colombel J.F. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn’s disease. Am. J. Gastroenterol. 2002;97:939–946. doi: 10.1111/j.1572-0241.2002.05613.x. [DOI] [PubMed] [Google Scholar]
- 57.Sadaghian Sadabad M., Regeling A., de Goffau M.C., Blokzijl T., Weersma R.K., Penders J., Faber K.N., Harmsen H.J., Dijkstra G. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn’s disease patients. Gut. 2015;64:1546–1552. doi: 10.1136/gutjnl-2014-307289. [DOI] [PubMed] [Google Scholar]
- 58.Trebble T.M., Stroud M.A., Wootton S., Calder P., Fine D., Mullee M.A., Moniz C., Arden N.K. High-dose fish oil and antioxidants in Crohn’s disease and the response of bone turnover: A randomised controlled trial. Br. J. Nutr. 2005;94:253–261. doi: 10.1079/BJN20051466. [DOI] [PubMed] [Google Scholar]
- 59.Imes S., Dinwoodie A., Walker K., Pinchbeck B., Thomson A.B. Vitamin C status in 137 outpatients with Crohn’s disease. Effect of diet counseling. J. Clin. Gastroenterol. 1986;8:443–446. doi: 10.1097/00004836-198608000-00012. [DOI] [PubMed] [Google Scholar]
- 60.Ratajczak A., Szymczak-Tomczak A., Skrzypczak-Zielińska M., Rychter A., Zawada A., Dobrowolska A., Krela-Kaźmierczak I. Vitamin C Deficiency and the Risk of Osteoporosis in Patients with an Inflammatory Bowel Disease. Nutrients. 2020;12:2263. doi: 10.3390/nu12082263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is contained within the present article.