Abstract
Simple Summary
The genus Lycoris (Amaryllidaceae) comprises about 20 species with high ornamental and medicinal value. However, germplasm identification is still difficult due to frequent interspecific hybridization and intraspecific morphological variation within this genus. Plastid genome sequencing has been proven to be a useful tool to identify closely related species and is widely used in the field of plant evolution and phylogeny. In the present study, we provided four chloroplast genomes of Lycoris and retrieved seven published species in the genus for comparative genomics and phylogenetic analyses. All these chloroplast genomes possess the typical quadripartite structure with conserved genome arrangement and gene content, yet their lengths varied due to expansion/contraction of the IR/SC boundaries. Phylogenetic relationships within Lycoris were resolved with high resolution using complete cp genome sequences. These results could not only offer a genome-scale platform for identification and utilization of Lycoris but also provide a phylogenomic framework for future studies in this genus.
Abstract
The genus Lycoris (Amaryllidaceae) consists of about 20 species, which is endemic to East Asia. Although the Lycoris species is of great horticultural and medical importance, challenges in accurate species identification persist due to frequent natural hybridization and large-scale intraspecific variation. In this study, we sequenced chloroplast genomes of four Lycoris species and retrieved seven published chloroplast (cp) genome sequences in this genus for comparative genomic and phylogenetic analyses. The cp genomes of these four newly sequenced species were found to be 158,405–158,498 bp with the same GC content of 37.8%. The structure of the genomes exhibited the typical quadripartite structure with conserved gene order and content. A total of 113 genes (20 duplicated) were identified, including 79 protein-coding genes (PCGs), 30 tRNAs, and 4 rRNAs. Phylogenetic analysis showed that the 11 species were clustered into three main groups, and L. sprengeri locate at the base of Lycoriss. The L. radiata was suggested to be the female donor of the L. incarnata, L. shaanxiensis, and L. squamigera. The L. straminea and L. houdyshelii may be derived from L. anhuiensis, L. chinensis, or L. longituba. These results could not only offer a genome-scale platform for identification and utilization of Lycoris but also provide a phylogenomic framework for future studies in this genus.
Keywords: Lycoris, Amaryllidaceae, chloroplast genome, interspecific comparison, phylogenetic analysis
1. Introduction
The genus Lycoris Herb. is a group of perennial bulbous plants with high ornamental and medicinal values that belongs to the family Amaryllidaceae [1,2]. More than 110 Amaryllidaceae alkaloids were identified in Lycoris, which have the function of antitumor, antibacterial, cytotoxic, and cholinesterase inhibition activities [3]. Species of Lycoris are widely cultivated as ornamental plants for their large and beautiful flowers [4].
The genus contains about 20 species, mainly distributed in China (15 species recorded) and Japan, and a few in Myanmar and North Korea. It has been demonstrated that frequent interspecific hybridization and intraspecific morphological variation commonly happen in Lycoris [5], resulting in the difficulty to make a clear standard for germplasm identification at the morphological level. Moreover, new Lycoris species are being reported, such as, L. hunanensis which was published as a new species from Yuanling County in China, which showed some difference from L. straminea [6]. Lycoris × hubeiensis K. Liu was identified as a natural hybrid of putative parents L. radiata and L. aurea [7]. In the Tsinling Mountains in China, L. tsinlingensis was found and published as a new species, but it is largely similar to L. chinensis [8]. In fact, there are different opinions on whether these subtle differences could be used as a criterion for determining a new species, so a feasible evaluation standard was suggested to clarify whether it is a new species or a variant. It requires some specific sequences for germplasm identification and a clear interspecific relationship in Lycoris.
To explore the interspecific relationships and clarify the hypothesis of hybrid origin in Lycoris, molecular markers of RAPD (random amplified polymorphic DNA) [9], nuclear ITS (internal transcribed spacer) sequences [10], inter-simple sequence repeat (ISSR) [11], and SCoT (start codon targeted) [12] have been used. RAPD analysis was consistent with the classification based on chromosome karyotype, which divided 13 Lycoris species into two groups. Nuclear ITS sequences of 15 Lycoris species suggested the three infrageneric clades and the hybrid origin of L. straminea, L. caldwellii, and L. albiflora. However, the extensive sequence variation has existed in many plant genomes, the complex and unpredictable evolutionary behavior of ITS sequence reduced the utility for phylogenetic analysis [13]. Thus, more methods were developed, inter-simple sequence repeat (ISSR) analyses of 20 species and varieties indicated a high level of genetic variation among species in Lycoris, and four major groups clustered by UPGMA analysis presented a consistence with morphological and karyotype observations. SCoT markers of 14 Lycoris species were tested and clustered into four groups, in which L. squamigera, L. incarnata, and all hybrids with the characteristic of multi-colored flowers were gathered together, suggesting the possibility of the hybrid origin of these two species. Although several strategies were developed for the analysis of interspecific relationships in the genus Lycoris, each method offered limited resolution within closely related species, resulting in that they did not get unanimous conclusion. More effective molecular markers are needed to be developed for germplasm identification, conservation, utilization, and breeding of the Lycoris species.
Plastid genes are regularly utilized in biotechnology or phylogeny, but with the limitation of DNA sequencing costs, investigators always chose a dense taxon sampling, which had a small number of informative loci for molecular phylogenetic analysis in Lycoris. For example, the cpDNA trnL-F sequence of 15 Lycoris species was selected to construct a phylogeny tree, which contained three infrageneric clades and was basically consistent with the classification of morphology except for L. longituba, L. aurea, and L. straminea. Phylogenetic reconstruction was obtained using plastid markers (trnS-trnfM and trnC-ycf6), which clustered Lycoris spp. into three clades and differed from that derived using ITS sequences [14]. Considering the rapid radiations and conservative genome evolution, limited sequence variation could be detected, particularly at low taxonomic levels. More sequence information and species were often desirable to increase phylogenetic resolution. Actually, complete chloroplast genome sequences were more highly discriminating and efficient as plant DNA barcodes. The development of next-generation DNA sequencing has brought the benefits of large numbers of genome data collection and allowed the rapid obtaining of complete organellar genomes. Whole plastome sequencing has been an efficient option to increase the phylogenetic resolution for the phylogenetic analyses, especially at lower taxonomic levels [15,16]. In angiosperms, the cp genome is highly conserved in terms of structure, content, and order of genes [17]. They usually have a circular structure, where two large, inverted repeat (IR) regions were separated by a large single-copy (LSC) region and a small single-copy (SSC) region [18]. The cp genome sequences contain many noncoding and variation regions, which has provided an essential molecular source for interspecific phylogenetic and phylogeographic studies [19]. It has been successfully used in many families and genus, for example, Dracunculus (Araceae), Cardiocrinum and Amana (Liliaceae) [20,21], Artemisia (Asteraceae) [22], and Withania somnifera (Solanaceae) [23].
Since the first Lycoris complete cp genome of L. squamigera was published in 2018 [24], there are six Lycoris species that have been published, and a phylogenetic tree based on the complete cp genome sequences was constructed [25]. There is no doubt that more complete chloroplast genome sequences will provide more information and insights for phylogenetic relationship reconstruction. In this study, we sequenced complete cp genomes of four Lycoris species. Based on previous studies, we systematically analyzed the similarities and differences of global structural patterns, variations of genes, simple sequence repeats (SSRs), and inverted repeats. Then the phylogenetic relationship was constructed based on the complete chloroplast genome sequences of 11 species Lycoris. The comparative analysis has demonstrated the effectiveness and applicability of chloroplast genome sequences for Lycoris phylogeny and remarked on the potential applications for species identification, development of DNA barcoding, and future phylogenetic studies of the genus and family.
2. Materials and Methods
2.1. Plant Sample Collection, DNA Extraction and Sequencing
The bulbs of L. incarnata, L. shaanxiensis, L. straminea, and L. houdyshelii were planted in Nanjing Botanical Garden, Mem. Sun Yat-sen (E118_83, N32_06), Nanjing, China. The specimen of L. incarnata (No. SYS00024942) was stored at the herbarium of Sun Yat-Sen University, the specimen of L. straminea (No. 00110652) and L. houdyshelii (No. 00110525) were stored at the herbarium of the Institute of Botany, Chinese Academy of Sciences. The L. shaanxiensis was collected from Shanxi Province and identified in 2018, but there was no specimen record in the herbarium in China. Fresh leaves were collected, quick freezed in liquid nitrogen, then stored at −80 °C until use. Genomic DNA was extracted using the Plant Genomic DNA Kit (Huayueyang, Beijing, China). DNA integrity was examined by electrophoresis in 1% (w/v) agarose gel, and concentration was measured using a NanoDrop spectrophotometer 2000 (Thermo Scientific; Waltham, MA, USA), then accurate quantifications were completed by Qubit 2.0. High-quality DNA libraries were constructed and sequenced at Novogene Bioinformatics Technology Co., Ltd. (https://www.novogene.com/, accessed on March 2011 Tianjin, China). The strategy of Nova-PE150 was selected for high-throughput sequencing, with an insert size of 350 bp.
2.2. Complete Cp Genome Assembly, Annotation and Structure Analysis
The complete cp genomes were assembled using the organelle assembler NOVOPlasty (Version 3.3) [26] with the parameters of genome range (148,500–168,500) and k-mer (39). The complete cp genome sequence of L. radiata (GenBank accession no. MN158120) was set as a reference [27]. Assembled genome sequences were manually corrected by BLASTn comparison and circularized. GC content was calculated by Geneious software (version R11, http://www.geneious.com accessed on 3 October 2017). Correct cp genome sequences were input on web server CPGAVAS2 (http://www.herbalgenomics.org/cpgavas2, accessed on 14 October 2020.) for the cp genome annotation and visualization with the default parameters. Microsatellite sequences were identified with MISA [28], which set the unit_size/min_repeats as 1/10, 2/6, 3/5, 4/5, 5/5, and 6/5. The maximum length of a sequence between two SSRs was set as 100. MEGA [29] was performed for calculating the relative synonymous codon usage (RSCU) values. Seven previously reported Lycoris chloroplast genome sequences, i.e., L. squamigera (MH118290), L. radiata (MN158120), L. sprengeri (MN158986), L. longituba (MN096601), L. chinensis (MT700549), L. anhuiensis (MT700550, and L. aurea (NC_046752), were downloaded from the National Center of Biotechnology Information (NCBI) database. The obtained cp genome sequences in the present study were deposited in the NCBI, with the GenBank accession numbers of MW477439 (L. incarnata), MW477440 (L. shaanxiensis), MW477441 (L. straminea), and MW477442 (L. houdyshelii).
2.3. Interspecific Comparison of Chloroplast Genomes
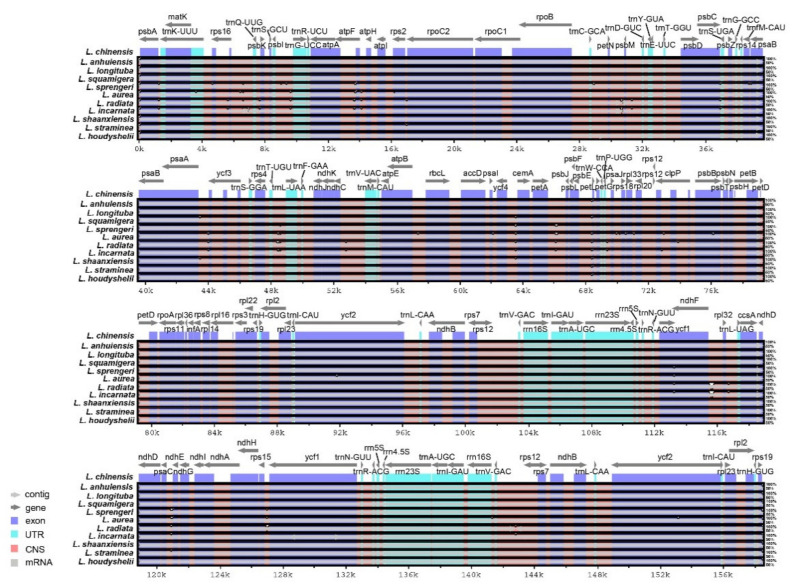
To explore the divergence regions in Lycoris, the program IRscope (https://irscope.shinyapps.io/irapp/, accessed on 20 June 2021) was used to visualize the divergence on the boundaries of the junction sites of the 11 chloroplast genome sequences in Lycoris [30]. The mVISTA program (http://genome.lbl.gov/vista/index.shtml, accessed on 23 June 2021) [31] was used to align and compare the complete cp genomes of Lycoris with the default parameters. Each annotation of the Lycoris species was selected as a reference, and Shuffle-LAGAN mode was visualized in an mVISTA plot.
2.4. Phylogenetic Analyses
A total of 11 complete cp genome sequences of Lycoris were used for phylogenetic analysis, including seven reported species and four new species in this study, Narcissus poeticus (MH706763) was selected as outgroup taxa. The reported sequences were downloaded from the NCBI database. Both the complete plastid sequences and 79 common PCGs (output by Geneious) were used for the ML tree construction. The nucleotide sequences were aligned using the MAFFT plugin [32,33] in Geneious with default settings. All gaps are treated as missing data. The complete alignment was used to reconstruct a maximum likelihood tree using PHYML [34] with 1000 bootstrap replicates. The GTR+G+I model suggested by jModelTest 2.1.4 [35] was used for each dataset.
3. Results and Discussion
3.1. General Features of the Cp Genomes of Lycoris
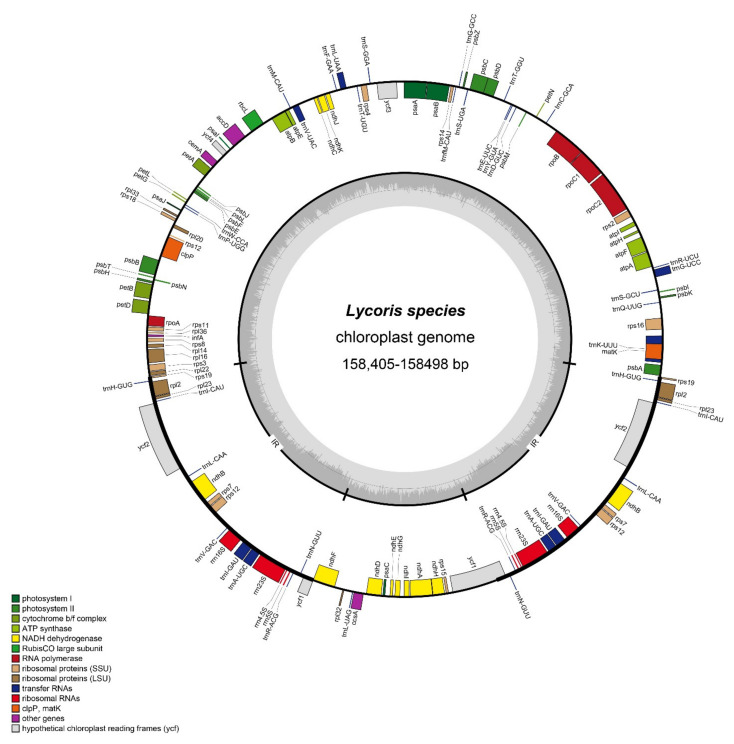
In the present study, we obtained four cp genomes of Lycoris by next-generation sequencing and de novo assembly, which were L. incarnata, L. shaanxiensis, L. straminea, and L. houdyshelii. A total of 7.2, 6.7, 5.5, and 4.8 million reads were obtained, and the average organelle coverage reached 7291×, 6761×, 5531× and 4581×, respectively (Table 1). The complete chloroplast genomes typically range from 120 to 170 kilobase pairs (kb) in length [17]. In Lycoris, the length of most species was around 158 kb [25]. In the present study, the full length of four species was 158,405, 158,498, 158,490, and 158,490 bp, respectively, with the same GC content of 37.8% (Table 1). The visualized circular map showed the typical angiosperm cp genome structure in Lycoris, which consists of one large single-copy (86,464–86,593 bp) and one small single-copy (18,352–18,499 bp) region, separated by a pair of inverted repeat (IR) (26,730–26,765) regions (Table 1 and Figure 1).
Table 1.
Summary information of four chloroplast genomes of the Lycoris species.
| Genome Features | L. incarnata | L. shaanxiensis | L. straminea | L. houdyshelii |
|---|---|---|---|---|
| Average organelle coverage | 7291× | 6761× | 5531× | 4581× |
| Genome size (bp) | 158,405 | 158,498 | 158,490 | 158,490 |
| LSC size (bp) | 86,593 | 86,469 | 86,473 | 86,464 |
| SSC size (bp) | 18,352 | 18,499 | 18,487 | 18,496 |
| IR size (bp) | 26,730 | 26,765 | 26,765 | 26,765 |
| GC content (%) | 37.8 | 37.8 | 37.8 | 37.8 |
| No. of genes | 113 | 113 | 113 | 113 |
| No. of PCGs | 79 | 79 | 79 | 79 |
| No. of tRNAs | 30 | 30 | 30 | 30 |
| No. of rRNAs | 4 | 4 | 4 | 4 |
| Duplicated genes | 20 | 20 | 20 | 20 |
Figure 1.
The plastome features of the Lycoris species. The map contains four rings, from the center going outward, the first circle means the forward and reverse repeats connected with red and green arcs, respectively. The second circle shows the tandem repeats marked with short bars. The third circle shows the microsatellites. The fourth circle shows the gene structure on the plastome. The genes were colored based on the functional categories, which were shown in the center of the map.
Compared with reported Lycoris species [25], the complete cp genomes size ranged from 158,335 (L. radiata) to 158,687 bp (L. sprengeri). Here, the genome size of these three species is between the longest and shortest; the length of LSC and SSC regions made a greater contribution to the full size. On the contrary, the IR regions were relatively conservative. In Lycoris cp genomes, a total of 113 genes were annotated, including 79 protein-coding genes (PCGs), 30 tRNAs, and 4 rRNAs (Table 1 and Figure 1). Compared with our previous reports, we found that the number of genes was highly conservative in Lycoris cp genomes. They were divided into four categories; contained genes for photosynthesis, self-replication, other genes, and function unknown (Table 2). There were 20 genes that were duplicated more than once. Four rRNAs were duplicated, which is consistent with other Lycoris species [25] and most plants, such as Allium [36] and Amomum [37].
Table 2.
Gene composition of four Lycoris chloroplast genomes.
| Category of Genes | Group of Genes | Name of Genes |
|---|---|---|
| Genes for photosynthesis | Subunits of photosystem I | psaB, psaA, psaI, psaJ, psaC, ycf4 |
| Subunits of photosystem II | psbA, psbK, psbI, psbM, psbD, psbC, psbZ, psbJ, psbL, psbF, psbE, psbB, psbT, psbN, psbH, ycf3 | |
| Subunits of NADH-dehydrogenase | ndhJ, ndhK, ndhC, ndhB a (×2), ndhF, ndhD, ndhE,ndhG, ndhI, ndhA a, ndhH | |
| Subunits of cytochrome b/f complex | petN, petA, petL, petG, petB a, petD a | |
| Subunits of ATP synthase | atpA, atpF a, atpH, atpI, atpE, atpB | |
| Subunit of rubisco | rbcL | |
| Self-replication | Large subunit of ribosome | rpl33, rpl20, rpl36, rpl14, rpl16 a, rpl22, rpl2 a (×2), rpl23 (×2), rpl32 |
| DNA dependent RNA polymerase | rpoC2, rpoC1 a, rpoB, rpoA | |
| Small subunit of ribosome | rps16 a, rps2, rps14, rps4, rps18, rps12 b (×2), rps11, rps8, rps3, rps19 (×2), rps7 (×2), rps15 | |
| Ribosomal RNAs | rrn16 (×2), rrn23 (×2), rrn4.5 (×2), rrn5 (×2) | |
| Transfer RNAs | trnK-UUU a, trnQ-UUG, trnS-GCU, trnG-GCC a, trnR-UCU,trnC-GCA, trnD-GUC, trnY-GUA, trnE-UUC, trnT-GGU,trnS-UGA, trnG-GCC, trnfM-CAU, trnS-GGA, trnT-UGU,trnL-UAA a, trnF-GAA, trnV-UAC a, trnM-CAU, trnW-CCA,trnP-UGG, trnH-GUG (×2), trnI-CAU (×2), trnL-CAA (×2),trnV-GAC (×2), trnI-GAU a (×2), trnA-UGC a (×2), trnR-ACG (×2)trnN-GUU (×2), trnL-UAG | |
| Other genes | Subunit of Acetyl-CoA-carboxylase | accD |
| c-type cytochrome synthesis gene | ccsA | |
| Envelop membrane protein | cemA | |
| Protease | clpP | |
| Translational initiation factor | infA | |
| Maturase | matK | |
| Component of TIC complex | ycf1 (x2) | |
| Unknown | Conserved open reading frames | ycf2 (x2) |
a means the genes containing a single intron; b indicates the genes containing two introns; (×2) indicates the genes duplicated in the IR regions.
Group II (G2) introns are self-splicing RNAs and mobile elements, which could provide rich characters for comparative analysis and phylogeny construction at both infrageneric and intrafamilial levels [38,39,40]. For example, the matK open reading frame (ORF) has been used as a marker for plant evolutionary studies. Tnterestingly, trnK-UUU contains a group II intron (trnKI1), which encodes the matK ORF, which attracts interest because it represents an unusual form of a group II intron [41]. In four Lycoris species, there were 18 splitting genes in L. incarnata and 17 in the other three species. There is one more ndhF located in IR and SSC regions in L. incarnata, which happened in L. radiata and L. sprengeri [25]. Most of the splitting genes contain one intron and two exons, except for ycf3 and clpP; they contained two introns and three exons (Table S1).
3.2. CpSSRs and Repeat Structures
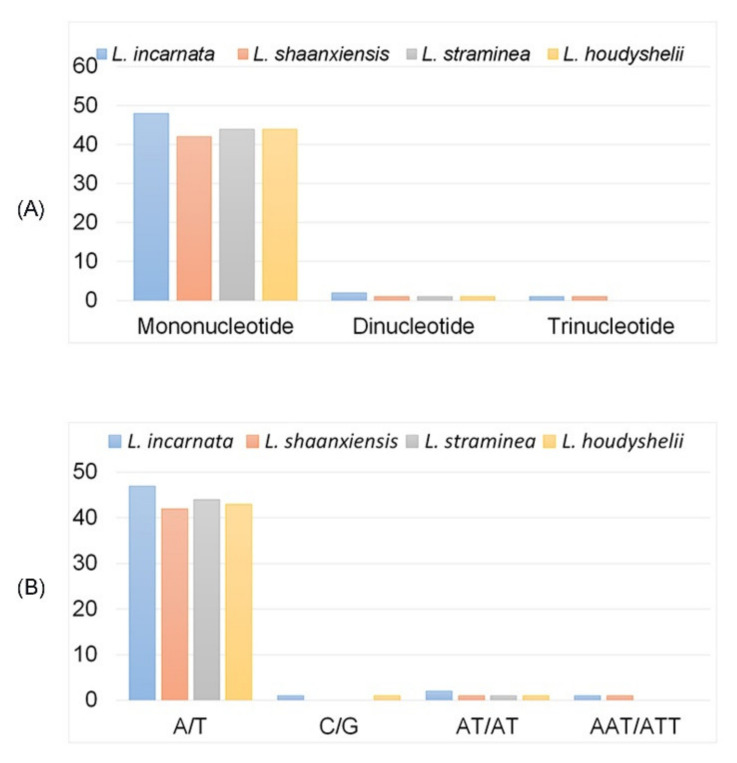
Chloroplast simple sequence repeats (cpSSRs) are microsatellites, showing typically mononucleotide tandem repeats. They commonly showed intraspecific variation in repeat numbers when they were located in the noncoding regions of the chloroplast genome [42]. Some works have proved the potential applications of variations in the noncoding regions of the chloroplast genome for phylogenetic analysis at the level of genus and species [43,44]. In the cp genomes of L. incarnata, L. shaanxiensis, L. straminea, and L. houdyshelii, there were 51, 44, 45, and 45 SSRs, respectively. The same as other reported Lycoris species [25], type of mononucleotide (A/T) was the most variable, which was 48, 42, 44, and 44 in four species (Figure 2 and Table S2). One mononucleotide (C/G) was detected in only L. incarnata and L. houdyshelii. One dinucleotide (AT/AT) was commonly detected in Lycoris species except for the L. incarnata, which contains two SSRs of AT/AT. A previous study also showed one dinucleotide (AT/AT) in seven Lycoris species [25], suggesting the conservation of dinucleotide SSRs among species. Trinucleotide (AAT/ATT) was only detected in L. incarnata and L. shaanxiensis. The trinucleotide (ATT) was only detected in L. incarnata and L. shaanxiensis, which was existed in other four species (Figure 2), L. aurea, L. radiata, L. sprengeri, and L. squamigera [25], accounting for half of the reported Lycoris species. No tetranucleotide repetition was detected in all reported Lycoris species.
Figure 2.
Statistics of simple sequence repeats of cp genomes of four Lycoris species. (A) Numbers of different repeat types; (B) Numbers of identified each SSRs motifs.
3.3. Statistics of Codon Usage
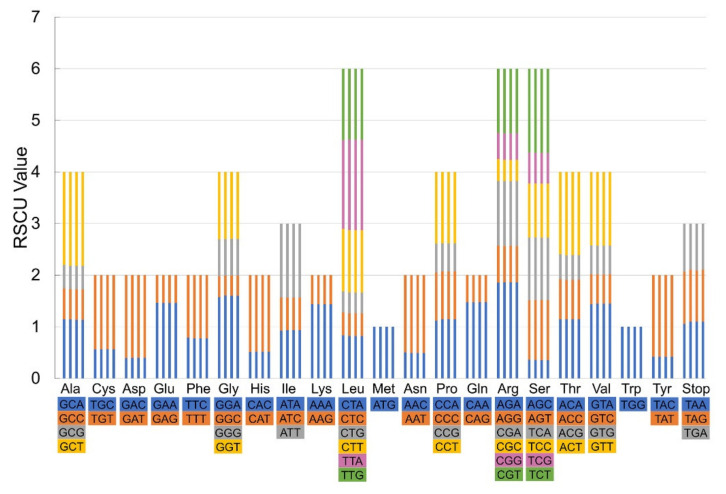
Codon usage analysis is beneficial for studies of evolution and new gene mining, which varies among different species [45]. Here, the complete cp genome sequences of four Lycoris were analyzed to investigate the amino acid frequency, the number of codon usage, the bias of codon usage, and relative synonymous codon usage (RSCU) (Table S3). Although the total number of codons was ranging from 48,207 to 49,641 in four species, showing a tiny change, the types of codons and amino acids were the same. A total of 64 codons were deduced, which were encoding 21 amino acids. Met and Trp were encoded by one codon usage, while others were encoding by multiple synonymous codons, ranging from two to six (Figure 3). The three highest frequency (AGA, GCT, and TTA) and four lowest frequency (AGC, GGC, GAC, and CTG) codons were observed in four species. It was defined as preferred codon usage when the RSCU value was >1.00 and vice versa. Except for methionine and tryptophan, there were 32 preferred and 30 non-preferred codon usages in L. incarnata, L. shaanxiensis, and L. houdyshelii, which was the same with the reported five Lycoris species, suggesting the main pattern of this codon usage in Lycoris [25]. In L. straminea, 31 preferred and 31 non-preferred codon usages were identified, which is different from any reported Lycoris species. The result will help us to understand the related patterns in Lycoris species and improve the research on codon usage in plant biology.
Figure 3.
Codon content and codon usage of 20 amino acids and stop codons in protein-coding genes of the four Lycoris cp genomes. Each histogram from left to right was L. incarnata, L. shaanxiensis, L. straminea, and L. houdyshelii, respectively.
3.4. Inverted Repeats Contraction, Expansion, and Interspecific Comparison
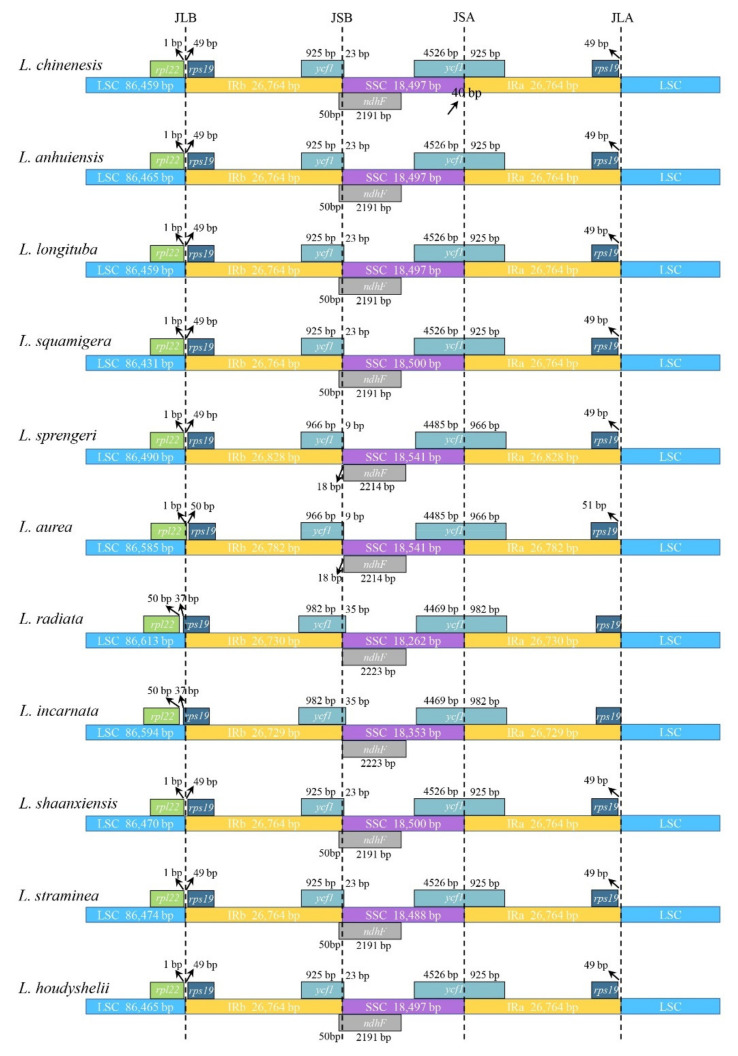
The typical circular structure of the chloroplast genome consists of regions of IR, LSC, and SSC, which makes four boundaries (IRb/LSC, IRb/SSC, IRa/SSC, and IRa/LSC). The contraction or expansion of the IR regions commonly leads to the length variation of the chloroplast genomes among different plant species [20,46,47]. In the present study, we compared the IR/SC borders and the adjacent genes among the eleven Lycoris species, including the previously reported seven species [25,48,49] and four newly sequenced species. It showed the well-conserved genomic structure, but it also exhibited divergence at the IR/SC boundary regions among eleven Lycoris chloroplast genomes (Figure 4). In the most monocot plastid genome structure, IR regions expand into rps19 [47]; there was no obvious expansion at the IRb/LSC boundary in Lycoris, except for the L. radiata and L. incarnata, their IRb regions expanded by 37 bp toward the rps19 gene. In all Lycoris species, the IRa/SSC border extended into the ycf1 genes with 925–982 bp. In addition, the ndhF gene overlapped with the IRa/SSC border by 50 bp in seven species, including L. chinensis, L. anhuiensis, L. longituba, L. squamigera, L. shaanxiensis, L. straminea, and L. houdyshelii (Figure 4).
Figure 4.
Comparison of border pattern of large single-copy regions (LSC), small single-copy regions (SSC), and an inverted repeat (IR) among 11 Lycoris chloroplast genomes.
To rapidly identify the conserved sequences in long alignments, global interspecific comparisons were performed using software mVISTA [31,50]. A total of 11 Lycoris cp genome sequences (same as IR analysis) were selected for comparative analysis. Most of the sequence variations were found in the LSC and SSC regions, which being largely consistent with our previous studies [25], in which the IR regions showed the high sequence conservation of the 11 species (Figure 5). A lot of evidence indicated that ndhF has great power in discrimination at the low taxonomic level [51]. In Lycoris, ycf1 and ndhF presented the most divergence in all species, suggesting the potential molecular markers for phylogenetic analysis and species identification. Although the length and boundary distribution characteristics of ndhF suggested that the L. radiata was a putative female parent of L. incarnata, the comparative analysis suggested the closer relationship of seven related species that have similar IR boundaries features (Figure 4 and Figure 5).
Figure 5.
Sequence identity plot using mVISTA based on the complete cp sequences of 11 Lycoris species with L. sprengeri as a reference. A 70% cut-off identity was used for the plots, and the Y-scale represents the percent identity from 50% to 100%.
3.5. Phylogenetic Analysis
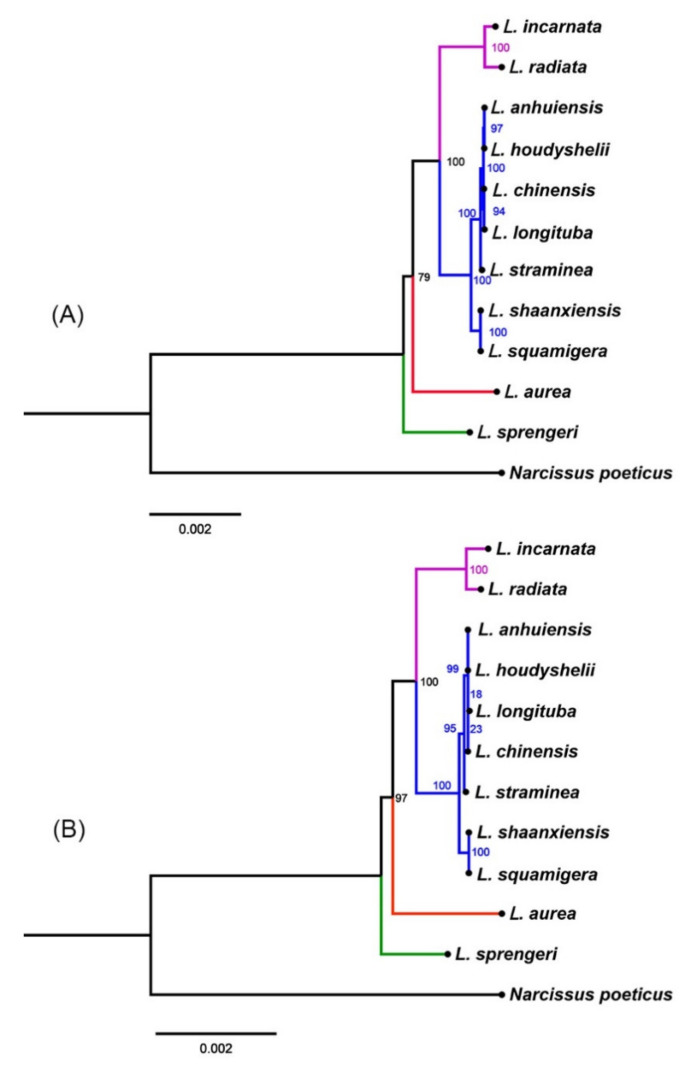
To explore the interspecific relationship and phylogeny reconstruction, a total of 11 Lycoris species with complete chloroplast genome sequences were selected for the construction of the maximum likelihood (ML) tree and Narcissus poeticus was chosen as the outgroup taxa. Both the complete plastid genome sequences and 79 common protein-coding genes were used for phylogenetic analysis, and the phylogenetic trees based on these two datasets showed the same topology (Figure 6).
Figure 6.
Phylogenetic analysis of the 11 Lycoris species by maximum likelihood (ML) analyses, Narcissus poeticus was the outgroup taxa. (A) The topology was constructed by the complete cp genome sequences. (B) The tree was constructed using 85 common protein-coding genes.
Here, 11 Lycoris species were clustered into three main groups, and L. sprengeri is basal for the other species in Lycoris; however, previous cpDNA sequences analysis showed that the L. radiata had a basal position within Lycoris [10]. It involves the discussion of the origin of hybrid species of Lycoris. Some studies have suggested that the four species of L. incarnata, L. shaanxiensis, L. straminea, and L. houdyshelii were hybrid origin species [52], plastid DNA sequences and SCoT analysis showed that natural hybrids L. incarnata and L. squamigera were located in same clade [12,14]. Here, two species, L. incarnata and L. shaanxiensis, with similar morphological characteristics, are not clustered together. L. incarnata was clustered with L. radiata, suggesting that the L. radiata may be the female donor of the L. incarnata. L. shaanxiensis showed the closest relationship to L. squamigera, which was also considered as a hybrid origin species [14], suggesting the same ancestor of these two species, and L. radiata may be their donor according to the phylogenetic analysis by complete cp genome sequences. L. straminea and L. houdyshelii showed the most similar morphological and ecological characteristics except for the flower color. L. straminea was a species with multiple ecological properties, which always exhibits a color change from light yellow to medium yellow degrees, but the flower of L. houdyshelii is white. If only based on morphological features, L. houdyshelii could be considered as a variant; however, evidence has shown that they have a totally different chromosome number and karyotype [12,53]. RAPD analysis also indicated that they were clustered into two groups [9]. Evaluation of different methods, including morphology, karyotypes, plastid sequences, and molecular marker, produced both overlap and conflict on the interspecific relationship and phylogeny because of the variant resolutions and parameters.
Actually, the complete plastid sequence has been proved as an ideal method for phylogenetic relationship reconstruction. In the genus of Lycoris, some specific plastid gene sequences [14,54] and rDNA internal transcribed spacer (ITS) sequences [55,56] and have been developed before more complete cp genome sequences were available. More complete cp genome sequences provided adequate information and foundation for the clarification of inter-specific relationships and phylogenetic analysis. In the present study, we provided four cp genome sequences of Lycoris; they not only had similar morphological features but were also considered as the natural hybrid species. The phylogenic analysis supported the same group between L. straminea and L. houdyshelii, but L. houdyshelii showed a closer relationship with L. anhuiensis, L. chinensis, and L. longituba than L. straminea, the closest relationship of these five species also suggesting that L. straminea and L. houdyshelii may be derived from one of the three species.
4. Conclusions
In this study, we provided the complete cp genome sequences of L. incarnata, L. shaanxiensis, L. straminea, and L. houdyshelii and performed the interspecific comparison and phylogenetic analysis using whole cp genome sequences of 11 Lycoris species. The results not only showed the sequence conservation of genome size, gene number, and order but also distinguish the difference between IR-SC boundary regions. The interspecific comparison analysis supported the branch of phylogenetic analysis, where the species on the same sub-branch had the same border patterns, suggesting the high resolution and reliability of phylogenetic reconstruction by the complete cp genome sequences in Lycoris. Phylogeny analysis suggested that the L. radiata may be the female donor of the L. incarnata, L. shaanxiensis, and L. squamigera. L. straminea and L. houdyshelii may be derived from L. anhuiensis, L. chinensis, or L. longituba. The results will help to make the intraspecific relationship and evolution clear and benefit the identification, protection, and utilization of Lycoris germplasm resources.
Acknowledgments
The authors thank Feng Peng, Shulan He, and Jianliang Wang, who helped to collect and plant the materials used for the experiments.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biology10080715/s1, Table S1: Splitting genes with introns and exons in the four Lycoris chloroplast genomes, Table S2: Chloroplast simple sequence repeats (cpSSRs) of four Lycoris species, Table S3: Relative synonymous codon usage (RSCU) in the four Lycoris chloroplast genomes.
Author Contributions
F.Z. and Z.W. designed the experiment; F.Z. prepared the original draft; R.L. analyzed the data, mapped the figures, and revised the manuscript; N.W. and G.C. prepared the material and performed the formal analysis; X.S., T.W. and W.Z. validated the data. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, grant number 31801900; Natural Science Foundation of Jiangsu Province, grant number BK20180310; Jiangsu Agricultural Science and Technology Innovation Fund, grant number CX(19)3033; Program of Innovation Capacity Construction of Jiangsu Province, grant number BM2018021-5; Jiangsu Key Laboratory for the Research and Utilization of Plant Resources, grant number JSPKLB202020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available at https://www.ncbi.nlm.nih.gov/ (accessed on 14 January 2021) with the access numbers of MW477439, MW477440, MW477441, and MW477442.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Meerow A., Snijman D. Amaryllidaceae. Flowering Plants·Monocotyledons. Springer; Berlin, Germany: 1998. pp. 83–110. [Google Scholar]
- 2.Jin Z. Amaryllidaceae and sceletium alkaloids. Nat. Prod. Rep. 2003;20:606–614. doi: 10.1039/b304144c. [DOI] [PubMed] [Google Scholar]
- 3.Cahlíková L., Breiterová K., Opletal L. Chemistry and biological activity of alkaloids from the genus Lycoris (Amaryllidaceae) Molecules. 2020;25:4797. doi: 10.3390/molecules25204797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tae K.H., Ko S.C. A taxonomic study of the genus Lycoris (Amaryllidaceae) based on morphological characters. Trop. Med. Int. Health. 1995;9:41–46. doi: 10.11110/kjpt.1995.25.4.237. [DOI] [Google Scholar]
- 5.Kurita S. Variation and evolution in the karyotype of Lycoris, Amaryllidaceae. Cytologia. 1986;51:803–815. doi: 10.1508/cytologia.51.803. [DOI] [Google Scholar]
- 6.Quan M.H., Ou L.J., She C.W. A new species of Lycoris (Amaryllidaceae) from Hunan, China. Novon. 2013;22:307–310. doi: 10.3417/2011013. [DOI] [Google Scholar]
- 7.Meng W., Zheng L., Shao J., Zhou S., Liu K. A new natural allotriploid, Lycoris × hubeiensis hybr. nov. (Amaryllidaceae), identified by morphological, karyological and molecular data. Nord. J. Bot. 2018;36 doi: 10.1111/njb.01780. [DOI] [Google Scholar]
- 8.Lu Y.J., Wang T., Wang Y.C., Zhang P.C. Lycoris tsinlingensis (Amaryllidaceae), a new species from Shaanxi, China. Ann. Bot. Fenn. 2020;57:193–196. doi: 10.5735/085.057.0424. [DOI] [Google Scholar]
- 9.Zhang L., Cai Y.M., Zhuge Q., Lou L.H., Zou H.Y., Huang M.R., Wang M.X. Analysis of the inter-species relationships on Lycoris (Amaryllidaceae) by use of RAPD. Yi Chuan Xue Bao. 2002;29:915–921. [PubMed] [Google Scholar]
- 10.Shi S., Qiu Y., Li E., Wu L., Fu C. Phylogenetic relationships and possible hybrid origin of Lycoris species (Amaryllidaceae) revealed by ITS sequences. Biochem. Genet. 2006;44:198–208. doi: 10.1007/s10528-006-9023-4. [DOI] [PubMed] [Google Scholar]
- 11.Shi S., Qiu Y., Wu L., Fu C. Interspecific relationships of Lycoris (Amaryllidaceae) inferred from inter-simple sequence repeat data. Sci. Hortic Amst. 2006;110:285–291. doi: 10.1016/j.scienta.2006.07.011. [DOI] [Google Scholar]
- 12.Gao Y., Zhu Y., Tong Z., Xu Z., Jiang X., Huang C. Analysis of genetic diversity and relationships among genus Lycoris based on start codon targeted (SCoT) marker. Biochem. Syst. Ecol. 2014;57:221–226. doi: 10.1016/j.bse.2014.08.002. [DOI] [Google Scholar]
- 13.Álvarez I., Wendel J.F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 2003;29:417–434. doi: 10.1016/S1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 14.Shi S., Sun Y., Wei L., Lei X., Cameron K.M., Fu C. Plastid DNA sequence data help to clarify phylogenetic relationships and reticulate evolution in Lycoris (Amaryllidaceae) Bot. J. Linn. Soc. 2014;176:115–126. doi: 10.1111/boj.12198. [DOI] [Google Scholar]
- 15.Parks M., Cronn R., Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:1–17. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delseny M., Han B., Hsing Y. High throughput DNA sequencing: The new sequencing revolution. Plant Sci. 2010;179:407–422. doi: 10.1016/j.plantsci.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 17.Shaw J., Lickey E.B., Schilling E.E., Small R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- 18.Ravi V., Khurana J., Tyagi A., Khurana P. An update on chloroplast genomes. Plant Syst. Evol. 2008;271:101–122. doi: 10.1007/s00606-007-0608-0. [DOI] [Google Scholar]
- 19.Daniell H., Lin C., Yu M., Chang W. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:1–29. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu R.S., Li P., Qiu Y.X. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: Comparative genomic and phylogenetic analyses. Front. Plant Sci. 2017;7:2054. doi: 10.3389/fpls.2016.02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P., Lu R.S., Xu W.Q., Tetsuo O.T., Cai M.Q., Qiu Y.X., Cameron K.M., Fu C.X. Comparative genomics and phylogenomics of East Asian tulips (Amana, Liliaceae) Front. Plant Sci. 2017;8:451. doi: 10.3389/fpls.2017.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahzadi I., Mehmood F., Ali Z., Ahmed I., Mirza B. Chloroplast genome sequences of Artemisia maritima and Artemisia absinthium: Comparative analyses, mutational hotspots in genus Artemisia and phylogeny in family Asteraceae. Genomics. 2020;112:1454–1463. doi: 10.1016/j.ygeno.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Mehmood F., Shahzadi I., Ahmed I., Waheed M.T., Mirza B. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 2020;112:1522–1530. doi: 10.1016/j.ygeno.2019.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Jin S.W., Park J.Y., Kang S.J., Park H.S., Shim H., Lee T.J., Kang J.H., Sung S.H., Yang T.J. The complete chloroplast genome sequence of Magic Lily (Lycoris squamigera) Mitochondrial DNA B. 2018;3:1210–1211. doi: 10.1080/23802359.2018.1527193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F., Wang T., Shu X., Wang N., Zhuang W., Wang Z. Complete chloroplast genomes and comparative analyses of L. chinensis, L. anhuiensis, and L. aurea (Amaryllidaceae) Int. J. Mol. Sci. 2020;21:5729. doi: 10.3390/ijms21165729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dierckxsens N., Mardulyn P., Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016;45:e18-e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F., Shu X., Wang T., Zhuang W., Wang Z.L. The complete chloroplast genome sequence of Lycoris radiata. Mitochondrial DNA B. 2019;4:2886–2887. doi: 10.1080/23802359.2019.1660265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beier S., Thiel T., Münch T., Scholz U., Mascher M. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amiryousefi A., Hyvonen J., Poczai P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 31.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuraku S., Zmasek C.M., Nishimura O., Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013;41:22–28. doi: 10.1093/nar/gkt389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozewicki J., Li S., Amada K.M., Standley D.M., Katoh K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019;47:W5–W10. doi: 10.1093/nar/gkz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stéphane G., Jean-François D., Vincent L., Maria A., Wim H., Olivier G. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 35.Posada D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 36.Xie D.F., Tan J.B., Yu Y., Gui L.J., Su D.M., Zhou S.D., He X.J. Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann. Bot. Lond. 2020;125:1039–1055. doi: 10.1093/aob/mcaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui Y., Chen X., Nie L., Sun W., Hu H., Lin Y., Li H., Zheng X., Song J., Yao H. Comparison and phylogenetic analysis of chloroplast genomes of three medicinal and edible Amomum species. Int. J. Mol. Sci. 2019;20:4040. doi: 10.3390/ijms20164040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jurica M.S., Stoddard B.L. Homing endonucleases: Structure, function and evolution. Cell. Mol. Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelchner S.A. Group II introns as phylogenetic tools: Structure, function, and evolutionary constraints. Am. J. Bot. 2002;89:1651–1669. doi: 10.3732/ajb.89.10.1651. [DOI] [PubMed] [Google Scholar]
- 40.Lambowitz A.M., Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 41.Hausner G., Olson R., Simon D., Johnson I., Sanders E.R., Karol K.G., McCourt R.M., Zimmerly S. Origin and evolution of the chloroplast trnK (matK) intron: A model for evolution of group II intron RNA structures. Mol. Biol. Evol. 2006;23:380–391. doi: 10.1093/molbev/msj047. [DOI] [PubMed] [Google Scholar]
- 42.Ebert D., Peakall R. Chloroplast simple sequence repeats (cpSSRs): Technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol. Ecol. Resour. 2009;9:673–690. doi: 10.1111/j.1755-0998.2008.02319.x. [DOI] [PubMed] [Google Scholar]
- 43.Kelchner S.A. The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann. Mo. Bot. Gard. 2000;87:482–498. doi: 10.2307/2666142. [DOI] [Google Scholar]
- 44.Borsch T., Quandt D. Mutational dynamics and phylogenetic utility of noncoding chloroplast DNA. Plant Syst. Evol. 2009;282:169–199. doi: 10.1007/s00606-009-0210-8. [DOI] [Google Scholar]
- 45.Feng C., Xu C.J., Wang Y., Liu W.L., Yin X.R., Li X., Chen M., Chen K.S. Codon usage patterns in Chinese bayberry (Myrica rubra) based on RNA-Seq data. BMC Genom. 2013;14:732. doi: 10.1186/1471-2164-14-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang C.C., Lin H.C., Lin I.P., Chow T.Y., Chen H.H., Chen W.H., Cheng C.H., Lin C.Y., Liu S.M., Chang C.C., et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- 47.Yang J.B., Tang M., Li H.T., Zhang Z.R., Li D.Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013;13:84. doi: 10.1186/1471-2148-13-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F., Zhuang W., Shu X., Wang T., Wang Z.L. Complete chloroplast genome of Lycoris sprengeri (Amaryllidaceae) and genetic comparison. Mitochondrial DNA B. 2019;4:3577–3578. doi: 10.1080/23802359.2019.1676673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F., Tong H., Yang H., Wang T., Zhuang W., Shu X., Wang Z.L. Characterisation of the complete chloroplast genome of Lycoris longituba (Amaryllidaceae) Mitochondrial DNA B. 2019;4:3782–3783. doi: 10.1080/23802359.2019.1681324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L., Dubchak I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 51.Amar M.H. ycf1-ndhF genes, the most promising plastid genomic barcode, sheds light on phylogeny at low taxonomic levels in Prunus persica. J. Genet. Eng. Biotechnol. 2020;18:1–10. doi: 10.1186/s43141-020-00057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng W., Zhang D., Qin H., Wang L., Zheng L., Xia Q., Liu K. Hybrid origin of Lycoris shaanxiensis revealed by karyotype survey. Cytologia. 2018;83:133–136. doi: 10.1508/cytologia.83.133. [DOI] [Google Scholar]
- 53.Yan L., Hsu P.S. A study on karyotypes of the genus Lycoris. J. Syst. Evol. 1989;27:257–264. [Google Scholar]
- 54.Hori T.A., Hayashi A., Sasanuma T., Kurita S. Genetic variations in the chloroplast genome and phylogenetic clustering of Lycoris species. Genes. Genet. Syst. 2006;81:243–253. doi: 10.1266/ggs.81.243. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y., Gao Y., Liao W., Tong Z. Analysis on rDNA-ITS sequence and research of intra-specific phylogeny of Lycoris albiflora. J. Plant. Resour. Environ. 2009;18:25–31. [Google Scholar]
- 56.Quan M., Ou L., She C., Wu X., Chen D. rDNA internal transcribed spacer sequence analysis of Lycoris Hert. Afr. J. Biotechnol. 2012;11:7361–7365. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are openly available at https://www.ncbi.nlm.nih.gov/ (accessed on 14 January 2021) with the access numbers of MW477439, MW477440, MW477441, and MW477442.