Abstract
Simple Summary
Esophageal adenocarcinoma has a poor 5-year survival rate and is among the highest mortality cancers. Changes in the esophageal microbiome have been associated with cancer pathogenesis; however, the molecular mechanism remains obscure. This review article critically analyzes the molecular mechanisms through which microbiota may mediate the development and progression of esophageal adenocarcinoma and its precursors-gastroesophageal reflux disease and Barrett’s esophagus. It summarizes changes in esophageal microbiome composition in normal and pathologic states and subsequently discusses the role of altered microbiota in disease progression. The potential role of esophageal microbiota in protecting against the development of esophageal adenocarcinoma is also discussed. By doing so, this article highlights specific directions for future research developing microbiome-mediated therapeutics for esophageal adenocarcinoma.
Abstract
Esophageal adenocarcinoma (EAC) is associated with poor overall five-year survival. The incidence of esophageal cancer is on the rise, especially in Western societies, and the pathophysiologic mechanisms by which EAC develops are of extreme interest. Several studies have proposed that the esophageal microbiome may play an important role in the pathophysiology of EAC, as well as its precursors—gastroesophageal reflux disease (GERD) and Barrett’s esophagus (BE). Gastrointestinal microbiomes altered by inflammatory states have been shown to mediate tumorigenesis directly and are now being considered as novel targets for both cancer treatment and prevention. Elucidating molecular mechanisms through which the esophageal microbiome potentiates the development of GERD, BE, and EAC will provide a foundation on which new therapeutic targets can be developed. This review summarizes current findings that elucidate the molecular mechanisms by which microbiota promote the pathogenesis of GERD, BE, and EAC, revealing potential directions for additional research on the microbiome-mediated pathophysiology of EAC.
Keywords: esophageal adenocarcinoma, microbiota, molecular pathogenesis
1. Introduction
Esophageal cancer accounts for approximately 3.2% of cancer cases worldwide [1]. It is responsible for 1 in every 20 cancer deaths and ranks sixth among the highest mortality cancers [1]. The two predominant histological subtypes of esophageal cancer are squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). Historically, ESCC accounted for a greater percentage of esophageal cancer than EAC, but recent studies anticipate increasing global EAC incidence [2]. Currently, the main histological subtype of esophageal cancer in Western societies is EAC [3]. The overall incidence of EAC increased six-fold in the United States between 1975–2001 [4]. Since 2001, EAC incidence has stabilized in the United States at an estimated 18,440 new EAC cases and 16,170 estimated deaths in 2020 alone [5]. The five-year survival rate for EAC remains poor at 21.4%, which is often attributed to the advanced stage at the diagnosis [6]. The increased incidence of EAC and low five-year survival rate have caused an increasing interest in determining the pathogenesis of EAC.
EAC develops in the distal esophagus in approximately 75% of cases [7]. Normally, the distal esophagus is lined by non-keratinized squamous epithelium. It transitions to a simple columnar epithelium at the gastroesophageal junction where protection against stomach acids is needed. Considering EAC develops from columnar glandular cells, conditions that predispose changes from this normal squamous epithelial lining of the distal esophagus to columnar metaplasia increase the risk of EAC [8]. Conditions that promote this transition include a reflux-mediated cycle from gastroesophageal reflux disease (GERD) to Barrett’s esophagus (BE), to EAC, termed the “EAC cascade” [9].
The EAC cascade begins with GERD, a pathologic condition in which gastric contents reflux into the esophagus [10,11]. Anatomic factors that mediate GERD development include crural diaphragm disruption, also known as hiatal hernias, and disruption of the lower esophageal sphincter [10]. Apart from the abovementioned anatomical factors, neurological and salivary disorders that disrupt esophageal motility can also contribute to GERD pathophysiology. Specifically, saliva contains bicarbonate that aids the neutralization of refluxed stomach acids. Rat-model studies demonstrated that removing salivary glands disrupts esophageal mucosa integrity and increases esophageal mucosa permeability to damaging hydrogen ions [12,13]. Thus, a change in saliva content may affect esophageal mucosa integrity. Dysregulation of gastrointestinal enzymes and hormones may also play an important role in the development of GERD and its related complications [10,14,15,16]. Gastric emptying disorders, increased abdominal pressure, or defective esophageal peristalsis can also contribute to GERD development [17]. Each of these mechanisms can lead to refluxate entering the esophagus where the acid, pepsin, bile salts, and pancreatic enzymes contained in the refluxate damage the normal squamous epithelial lining of the distal esophagus [18]. Acid in particular has been shown to degrade the esophageal mucosa and lead to dilated intercellular spaces [19]. When chronically exposed to this acidic refluxate, the esophagus is driven into an inflammatory cycle of epithelial cell damage and regrowth [20]. Studies have reported that weekly GERD symptoms increase the odds of EAC five-fold [21]. Furthermore, the damage-proliferation cycle of GERD can predispose esophageal tissue to BE, the second step in the reflux-mediated cascade to EAC [22,23,24].
BE is characterized by a metaplastic transition from reflux-damaged squamous epithelium to mucus-secreting columnar cells. This metaplastic change is considered a pre-cancerous epithelium that increases the risk of EAC 30 to 125-fold [20,25]. Considering this association, GERD and BE are clear risk factors for EAC; however, GERD leads to genetic alterations in BE cells and progression to EAC at a rate of only 0.12–0.6% per year [26]. EAC can also develop independently of GERD or BE. Therapeutic targets for these risk factors have yet to significantly improve the outcomes of patients with EAC. Consequently, new molecular targets for EAC therapies are of great interest.
Microbiota have been implicated as independent factors that contribute to GERD, BE, and EAC development. The term ‘microbiota’ refers to the commensal organisms, primarily bacteria, residing in the human gastrointestinal tract. Normally, microbiota play a mutualistic role within the human gastrointestinal tract. However, when microbiota and host immunity become imbalanced, or microbiota maladapt to the host environment, dysbiosis develops [27]. The relationship between dysbiosis and cancer is critical considering microbiota are capable of affecting tumor-promoting inflammation, genomic instability, mutations, proliferative signaling, and immune system evasion [27,28,29]. Dysbiosis is already implicated in colon, gastric, pancreatic, laryngeal, breast, and gallbladder cancers [30]. In the past two decades, research on the role of microbiota in EAC has surged due to the development of 16S rRNA and rDNA sequencing technologies. This review article will summarize the current findings from various studies that elucidate the molecular mechanisms by which microbiota promote the pathogenesis of GERD, BE, and EAC.
2. Microbiota Expression: Normal vs. Pathologic
2.1. Mutualistic and Dysbiotic Roles of Microbiota
Microbiota play a mutualistic role by aiding the host in digestion, vitamin synthesis, immune system development, and drug metabolism in exchange for the host’s nutrients and secure environment [31]. Microbiota can also protect the host tissue from pathogenic invasion and contribute to the structural integrity of the esophageal mucosal barrier [31]. For this mutualistic relationship to occur, the host’s immune system must tolerate the microbiota, and the commensal microbiota must adapt to changes in the host’s microenvironment. If either of these is not maintained, there is a change in the composition and function of the microbiota, a phenomenon known as dysbiosis.
Dysbiosis can arise as a result of two general mechanisms: a loss of tolerance by the host immune system or maladaptation of the microbiota to the host’s environment. A loss of tolerance by the host immune system induces dysbiosis considering the mutualistic microbiota become targets of the host innate and adaptive immune system [32]. With subsequent immune system reactions and inflammation, the composition of microbiota is altered. This leads to a loss of microbial functions that are vital to the host. Dysbiosis can also result from the inability of the microbiota to adapt to the host’s microenvironment. Variations in the host microenvironment can be induced by saliva, inflammation, infection, diet, or xenobiotics and consequently influence gastrointestinal dysbiosis [27,29,33]. Saliva can mediate the esophageal microbiome environment considering it decreases esophageal mucosa permeability to acid, restores physiologic esophagus pH through buffering, strengthens the pre-epithelial barrier of the esophageal mucosa, and contains epidermal growth factor that is essential for healing esophageal mucosal lesions [12,34,35]. Inflammation is a particularly well-established mediator of dysbiosis [33]. Inflammation mediates dysbiosis through positive selection of microbial species with characteristics advantageous for the inflamed microenvironment and negatively selects against microbiota without factors that enable them to thrive in the inflamed microenvironment. The inflammatory environment seen in the EAC cascade may initiate dysbiosis in this manner.
Once established, dysbiotic microbiota can manipulate host immune system mechanisms, such as inflammasome and TLR signaling, in order to maintain the dysbiotic composition and function of the microbiota [27]. This manipulation of the immune system, in addition to microbial mediation of metabolism, cellular proliferation, and inflammation, makes dysbiotic microbiota key factors in the initiation and development of cancer [28]. Consequently, determining the role of microbiota in the pathogenesis of the EAC cascade is of great consequence.
2.2. Microbiota Composition in Healthy Esophageal Tissue
There are an estimated 1014 bacteria in the human digestive tract [36]. In the distal esophagus alone, there are 104 bacteria per mm2 of mucosal surface [37]. The density and composition of microbiota varies from the oral cavity to the colon. Studies have demonstrated that the esophageal microbiome has distinct resident microbiota that are distinct from—but similar to—the microbiome found in the oral mucosa [37,38]. The distinct composition of the esophageal microbiome community may be explained by predominantly resident, rather than transient, microbiota. Pei et al.’s observation of a close association between bacteria and the cell surface of the mucosal epithelium lining the esophagus supports this possibility [39].
Several studies have used culture-dependent and culture-independent analysis methods to determine the composition of the healthy esophageal microbiome (Table 1). Despite differences in sample collection and analysis methods, all studies found that Streptococcus is one of the dominant genera in the normal esophagus—a genus of Gram-positive, predominantly facultative anaerobes that are catalase-negative. Two studies noted Streptococcus as the genera with the greatest relative abundance [37,40]. Peter et al. uniquely noted the dominant genus was Tissierella soehngenia—a Gram-positive anaerobe genus normally associated with fecal tracts [41,42].
Table 1.
Microbiota expressed by normal esophagus.
| Article | Publication Year | Method | Sample Size (n) | Number of Identified Genera | Dominant Phyla | Dominant Genera |
|---|---|---|---|---|---|---|
| Peter et al. [42] | 2020 | 16S rRNA gene DNA sequencing | 12 | N/A | Firmicutes (47.81%), Proteobacteria (20.67%), Bacteroidetes (16.93%), Actinobacteria (5.57%), Fusobacteria (4.76%) |
Tissierella soehngenia (16.67%) Lactobacillus (7.15%) Streptococcus (7.27%) Acinetobacter (5.80%) Prevotella (5.24%) |
| Dong et al. [43] | 2018 | 16S rRNA Illumina sequencing | 27 | 594 |
Firmicutes (37.42%) Proteobacteria (43.61%) Bacteroidetes (13.17%) Actinobacteria (2.53%) Fusobacteria (1.22%) TM7 (1.06%) |
Streptococcus
Neisseria Prevotella Actinobacillus Veillonella |
| Blackett et al. [45] | 2013 | Selective media cultures + 16S rRNA PCR |
39 | 19 | N/A |
Streptococcus
Prevotella Staphylococcus Rothia Actinomyces Bifidobacterium Staphylococcus Neisseria |
| Yang et al. [46] | 2009 | 16S rRNA Sanger Sequencing | 12 | N/A | Firmicutes |
Streptococcus (78.750%) Prevotella (4.300%) Gemella (3.400%) Veilonella (3.075%) Pasteurellaceae (2.075%) Haemophilus (1.925%) Rothia (1.025%) |
| Zilberstein et al. [40] | 2007 | Selective media cultures | 10 | N/A | N/A |
Streptococcus (40%) Staphylococcus (20%) Corynebacterium (10%) Lactobacillus (10%) Peptococcus (10%) |
| Pei et al. [37] | 2004 | Broad-range 16S rDNA PCR | 4 | 41 |
Firmicutes
Bacteroidetes Actinobacteria Fusobacteria TM7 |
Streptococcus (39%) Prevotella (17%) Veilonella (14%) |
Studies utilizing sequencing and selective media cultures demonstrate a predominantly Gram-positive microbiota inhabiting the normal esophagus. All studies found Streptococcus to be a dominant genus in normal esophageal microbiota. rDNA = ribosomal DNA; PCR = polymerase chain reaction; TM-7 = Saccharibacteria; rRNA = ribosomal ribonucleic acid; N/A = not available.
Several studies demonstrate that the esophageal microbiome did not express site-specific bacteria when comparing proximal, middle, and distal esophageal segments [43,44]. This consistent microbial pattern along the length of the normal esophagus was shown to have variable composition when comparing study subjects. Genetic and lifestyle factors may contribute to this esophageal microbiome variability between healthy study subjects [43]. Under pathologic conditions, this microbial pattern of the esophagus is altered.
2.3. Microbiota Composition in Pathological Esophageal Tissue
The microbiota composition in pathological states, including GERD, BE, and EAC, differs from healthy esophageal tissue (Table 2). Deshpande et al. reported that GERD samples showed increased Gram-negative and decreased Gram-positive bacterial composition [9]. Similar results were observed in BE samples [9]. However, there is no consistent trend observed across the studies analyzing EAC samples. Elliott et al. found that EAC did not selectively express more Gram-negative microbiota, while Lopetuso et al. found that EAC was dominated by Gram-negative anaerobes [47,48].
Table 2.
Microbiota expressed by pathological esophagus.
| Article | Publication Year | Method | Sample Size | Findings | |||
|---|---|---|---|---|---|---|---|
| Control | GERD | BE | EAC | ||||
| Peter et al. [42] | 2020 | 16S rRNA | 12 | N/A | 31 | 10 |
|
| Zhou et al. [50] | 2020 | 16S rRNA | 16 | N/A | 17 | 6 |
|
| Lopetuso et al. [48] | 2020 | 16S rRNA | 10 | N/A | 10 | 6 |
|
| Snider et al. [49] | 2019 | 16S rRNA | 16 | N/A | 25 | 4 |
|
| Deshpande et al. [9] | 2018 | 16S rRNA 18S rRNA | 59 | 29 | 5 | 1 |
|
| Elliott et al. [47] | 2017 | 16S rRNA | 16 | N/A | 17 | 15 |
|
| Gall et al. [51] | 2015 | 16S rRNA | N/A | N/A | 12 | N/A |
|
| Amir et al. [52] | 2014 | 16S rRNA | 15 | N/A | 6 | N/A |
|
| Yang et al. [46] | 2009 | 16S rRNA | 12 | N/A | 10 | N/A |
|
| Macfarlane et al. [53] | 2007 | 16S rDNA | 7 | N/A | 7 | N/A |
|
Results from nine studies that used 16s rRNA sequencing to analyze esophageal microbiota in controls, GERD, BE, and EAC. Results across studies are variable, which may be attributable to different sample collection methods and patient populations. Several studies show that GERD and BE express increased Gram-negative and decreased Gram-positive microbiota. According to two studies, EAC expresses decreased microbial diversity. A consensus regarding the microbiota composition in EAC has not been reached. Lactic acid-producing bacteria are increased in GERD, BE, and EAC. GERD = gastroesophageal reflux disease; BE = Barrett’s esophagus; EAC = esophageal adenocarcinoma; rDNA = ribosomal DNA; N/A = not available; rRNA = ribosomal ribonucleic acid.
Several studies evaluated the effect of the EAC cascade on microbial diversity. Both Elliott et al. and Snider et al. noted a decreased microbial diversity in EAC [47,49]. This observation may be due to an altered tumor environment in which only select microbes can survive. Lopetuso et al., on the other hand, noted an increased microbial diversity in BE and EAC compared to controls [48]. Furthermore, Deshpande et al. and Peter et al. noted no differences in microbial diversity between normal and pathologic samples [9,42]. Consequently, a consensus has not been reached regarding the effect of the EAC cascade on microbial diversity.
Lactic acid-producing bacteria have been shown to be in abundance in the EAC cascade microenvironment [9,50]. Increased lactic acid production as a result of this dysbiotic shift may contribute to the Warburg effect, as discussed below. Overall, however, future studies analyzing the microbiome in EAC samples in large sample populations are needed to understand the clinical impact of these findings.
2.4. Controversial Associations between Helicobacter pylori and the EAC Cascade
Helicobacter pylori (H. pylori) is not a dominant bacteria in normal or pathologic esophageal tissue. However, it plays an important but controversial role in the EAC cascade. It is a Gram-negative helical bacteria that is commonly present in the stomach. By utilizing catalase, urease, and oxidase enzymes, H. pylori is able to survive in the otherwise acidic environment of the stomach. Its association with malignancies such as gastric cancer and gastric mucosa-associated lymphoid tissue lymphoma has led to a plethora of studies investigating the relationship between H. pylori and the EAC cascade.
In regard to GERD, a recent meta-analysis of randomized controlled trials conducted from 1990–2019 demonstrated that H. pylori eradication increased the risk of new erosive GERD by two-fold [54]. This finding was not consistent across all studies, however. Contrastingly, a meta-analysis by Yaghoobi et al. demonstrated no association between H. pylori eradication and the risk of new GERD development [55]. Similar controversies have been raised regarding the relationship between H. pylori and preexisting GERD. Saad et al.’s meta-analysis demonstrated an improvement in GERD symptoms with H. pylori eradication, while Zhao et al.’s findings suggest that H. pylori does not have a significant effect on the healing or relapse rate of preexisting GERD [54,56]. It is evident from the literature review that the role of H. pylori in new GERD development as well as improvement following its eradication has been controversial and requires further research.
The effect of H. pylori on BE is also controversial. A meta-analysis of 72 studies composed of 84,717 total patients with BE found that H. pylori infection was associated with a reduced risk of dysplastic, non-dysplastic, and long-segment BE [57]. Similar results were reported in a case-control study where BE was associated with a lower prevalence rate of H. pylori infection when compared to control [58]. This association was recently analyzed in a cohort study of 81,919 patients undergoing eradication treatment for H. pylori [59]. Despite the previously established association between H. pylori and BE, the cohort study did not provide evidence of an increased risk of BE following H. pylori eradication. Consequently, future studies are essential in elucidating whether the relationship between H. pylori and BE is merely an association or a more established mediator of BE pathophysiology.
Recent studies have also evaluated H. pylori in association with EAC. A meta-analysis of 28 studies discerned that H. pylori infection is inversely associated with EAC [60]. Cytotoxin-associated gene A strains specifically were less likely to be associated with EAC when compared to strains negative for cytotoxin-associated gene A. Conversely, a literature review by Polyzos et al. remarks that meta-analysis of observational studies suggests an inverse association between H. pylori infection and EAC, whereas epidemiologic studies are collectively inconclusive [61].
Overall, the lack of consensus regarding the implications of the associations between H. pylori and the EAC warrants future studies.
3. Role of Microbiota in the Pathogenesis of the EAC Cascade
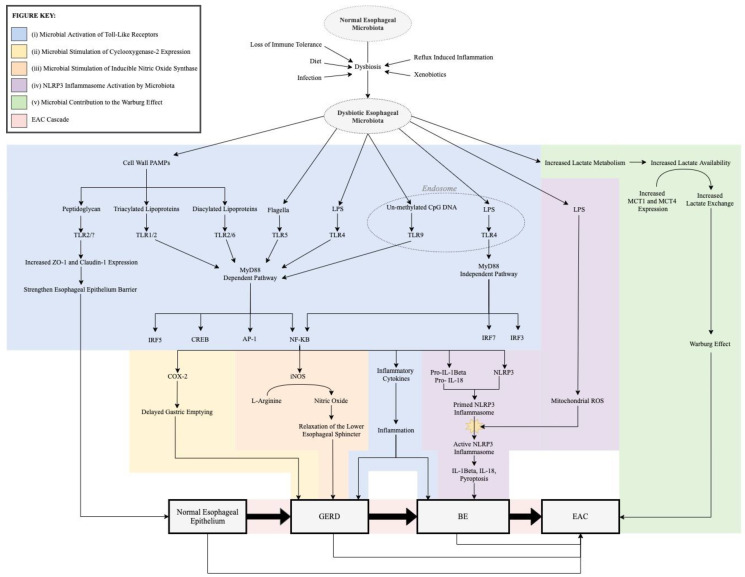
The dysbiotic shift in esophageal microbiota during the EAC cascade may promote the pathogenesis of EAC through five possible mechanisms: (i) activation of toll-like receptors, (ii) stimulation of cyclooxygenase-2 expression and subsequent delayed gastric emptying, (iii) stimulation of iNOS expression leading to relaxation of the lower esophageal sphincter, (iv) stimulation of the NLRP3 inflammasome, and (v) increased lactate availability for the Warburg effect (Figure 1).
Figure 1.
Molecular signaling in response to dysbiosis in EAC pathogenesis. The predominantly Gram-positive esophageal microbiome in the normal esophagus transforms through dysbiosis to a predominantly Gram-negative microbiome in the EAC cascade. The new microbiota can subsequently promote the pathogenesis of the EAC cascade through activation of toll-like receptors, stimulation of cyclooxygenase-2 expression, stimulation of inducible nitric oxide synthase, NLRP3 inflammasome activation, and contribution to the Warburg effect. PAMP = pathogen-associated molecular pattern; LPS = lipopolysaccharide; MCT = monocarboxylate transporter; TLR = toll-like receptor; ZO-1 = zonula occludens-1; MyD88 = myeloid differentiation primary response 88; IRF =interferon-regulator factor; CREB = cAMP-response element binding protein; AP-1 = activator protein 1; NF-kB = nuclear factor kappa B; COX-2 = cyclooxygenase-2; iNOS = inducible nitric oxide synthase; IL = interleukin; NLRP3 = nod-like receptor protein; ROS = reactive oxygen species; GERD = gastroesophageal reflux disease; BE = Barrett’s esophagus; EAC = esophageal adenocarcinoma.
3.1. Microbial Activation of Toll-Like Receptors
The observed dysbiotic microbiota may aberrantly activate TLRs and lead to NF-κB, CREB, AP-1, and IRF activity that promotes EAC cascade development.
3.1.1. Background
Toll-like receptors (TLRs) are pattern recognition receptors (PRRs) expressed by immune and epithelial cells. As a component of the innate immune system, TLRs lead to phagocytosis, inflammatory cytokine release, and complement activation [62,63,64,65]. TLRs also play an essential role in linking the innate and adaptive immune systems due to their presence on antigen-presenting cells [66]. TLRs consequently play an essential role in the gastrointestinal tract where a delicate balance between immunity against pathogens and tolerance of symbiotic bacteria must be maintained. TLRs differentiate between pathogenic and symbiotic microbiota by recognizing pathogen-associated molecular patterns (PAMPs), unique conserved structures native to both pathogenic and non-pathogenic microorganisms, or damage-associated molecular patterns (DAMPs). PAMPs expressed in gastrointestinal tract microbiota include Gram-negative bacterial lipopolysaccharide (LPS), bacterial peptidoglycan, specific bacterial RNA/DNA characteristics, as well as many others. When TLRs recognize PAMPs or DAMPs, there is a downstream activation of transcription factors that regulate cytokine gene expression (NF-κB, AP-1, CREB, IRFs, IFN-α/β); proliferation, differentiation, and apoptosis (AP-1); and cellular mechanisms involved in carcinogenesis (IFN-α/β) (Table 3).
Table 3.
| Toll-Like Receptor | TLR 1 + TLR 2 | TLR 2 + TLR 6 | TLR 3 | TLR 4 | TLR 5 | TLR 7 | TLR 9 | |
|---|---|---|---|---|---|---|---|---|
| Cellular Expression | Monocytes | + | + | + | + | + | + | |
| Macrophages | + | + | + | + | + | + | ||
| Dendritic cells | + | + | + | + | + | + | ||
| NK cells | + | |||||||
| Mast cells | + | + | ||||||
| B cells | + | + | + | + | + | |||
| T cells | + | + | ||||||
| Esophageal epithelium | + | + | + | + | + | + | + | |
| Membrane Expression | Plasma membrane | Plasma membrane | Endosome | Plasma membrane (Endosomal membrane) |
Plasma membrane | Endosomal membrane | Endosomal membrane | |
| PAMPs | Bacterial cell wall Triacyl Lipoproteins |
Bacterial cell wall Diacyl lipoproteins Lipoteichoic Acid Zymosan |
Viral dsRNA Viral ssRNA |
Lipopolysaccharide Mannan Glycoinositolphospholipids Envelope Proteins |
Flagellin | Viral ssRNA Imidazoquinolines | Unmethylated CpG DNAViral dsDNA | |
| Downstream Signaling Pathway | MyD88 Dependent | MyD88 Dependent | MyD88 Independent | MyD88 Dependent (MyD88 Independent) |
MyD88 Dependent | MyD88 Dependent | MyD88 Dependent | |
| Downstream Signaling Factors | NFKB | + | + | + | + (+) | + | + | + |
| CREB | + | + | + | + | + | |||
| AP-1 | + | + | + | + | ||||
| IRF | IRF5 | IRF5 | IRF3, IRF7 | IRF5 IRF3, IRF7 (+) |
IRF5 | IRF7 | IRF5, IRF7 | |
Parentheses in association with TLR4 denotes the endosomal membrane expression of TLR4. TLR = toll-like receptor; NK cells = natural killer cells; PAMP = pathogen-associated molecular pattern; dsRNA = double stranded ribonucleic acid; ssRNA = single stranded ribonucleic acid; dsDNA = double stranded deoxyribonucleic acid; MyD88 = Myeloid differentiation primary response 88; NF-κB = Nuclear factor kappa B; CREB = cAMP-response element binding protein; AP-1 = Activator protein 1; IRF = Interferon-regulatory factor.
Considering the role of TLRs in immunity as well as their downstream transcription factors, TLRs are capable of being anti-tumorigenic or pro-tumorigenic. Whether the TLR is anti- or pro-tumorigenic depends on the TLR, cancer subtype, and the immune cells infiltrating the tumor [68]. TLRs can be pro-tumorigenic through pro-inflammatory cytokines, anti-apoptotic signaling, proliferative signaling, and profibrogenic signals in the tumor microenvironment or tumor cells themselves [69]. This pro-tumorigenic role of TLRs has been demonstrated in colitis-associated cancer, where TLR recognition of microbiota promoted the development of invasive carcinoma [70]. On the other hand, TLR stimulation can lead to anti-tumorigenic effects through T-cell-mediated immunity and recognition of tumor-associated DAMPs [68,71]. This anti-tumorigenic effect is demonstrated in several cancers, as summarized by Dajon et al., but has yet to be demonstrated in EAC [68]. Considering both the anti-tumorigenic and pro-tumorigenic roles of TLRs, the potential activation of TLRs by dysbiotic esophageal microbiota in the EAC cascade is of significant interest.
3.1.2. TLR1, TLR2, and TLR6
The role of TLRs, including TLR2, has been studied in the EAC cascade. Mulder et al. studied TLR expression in esophageal cell lines. Their results demonstrated TLR2 expression only in EAC cell lines and not normal cell lines [72]. However, Verbeek et al. report TLR2 mRNA expression in inflammatory cells as well as epithelial cells in biopsies from patients with normal esophagus, reflux esophagitis, BE, and EAC [73]. Furthermore, TLR2 expression was increased in reflux esophagitis, BE, and EAC relative to normal esophageal epithelium [73]. Similar findings in regard to TLR2 expression in the EAC cascade have been demonstrated in additional studies [74,75]. Collectively, these studies show that TLR2 is overexpressed in inflammatory states like GERD, BE, and EAC.
Studies also demonstrate the variable location of TLR2 expression along the EAC cascade. TLR2 was expressed in basal keratinocytes of normal esophageal epithelium and in superficial epithelial cells, crypts, and lamina propria in BE [73]. EAC, on the other hand, demonstrated diffuse TLR2 expression throughout the biopsy [73]. This observation raises the question of whether progressive mucosal disruption in the EAC cascade exposes epithelial TLR2 to PAMPs of dysbiotic microbiota not tolerated by the immune system.
TLR2 may play a heightened role in dysbiotic microbial recognition along the pathogenesis of the EAC cascade due to its ability to heterodimerize with other TLRs and recognize a wider variety of ligands. Current research demonstrates that TLR2/6 and TLR1/6 heterodimers recognize several PAMPs, including components of bacterial cell walls known as diacylated and triacylated lipoproteins [74,76]. Heterodimerization, specifically in the EAC cascade, was suggested by Huhta et al.’s immunohistochemistry analysis of TLR1, TLR2, and TLR6 expression in pathologic esophageal samples [74]. Study results demonstrated a stepwise increase in TLR1 and TLR6 from normal esophageal tissue to high-grade dysplasia. The increased expression of TLR2, TLR1, and TLR6 along the EAC cascade, may consequently enable TLR2 to heterodimerize and recognize a greater diversity of dysbiotic PAMPs. Additionally, the microbiota itself may also mediate TLR2 expression. Hörmann et al. demonstrated a microbiota-dependent upregulation of TLR2 as well as TLR1 in small intestinal mucosa [77]. Even colonization with single microbes was capable of eliciting this response. This effect was reversed with a seven-day course of broad-spectrum antibiotics [77]. Consequently, TLR2 mediates microbiota, and microbiota mediate TLR2. Studies evaluating this relationship in the EAC cascade specifically are warranted.
In addition to the abovementioned heterodimerization and pro-inflammatory signaling, TLR2 can regulate esophageal epithelial barrier function and mediate proliferation of epithelial cells. Specifically, TLR2 activation with bacterial PAMP peptidoglycan and fungal zymosan in normal esophageal epithelium leads to upregulation of tight junction complexes claudin-1 and zonula occludens-1, strengthening esophageal epithelial barrier function [78]. This homeostatic regulation of esophageal epithelial barrier function by TLR2 mediates the ability of microbes and DAMPs to pass through the mucosal barrier and further activate the host immune system [78]. Consequently, TLR2 may be an essential protective mediator of the EAC cascade in which the microbial shifts and reflux-mediated damage lead to aberrant expression of PAMPs and DAMPs. The effect of TLR2 on epithelial cell proliferation was demonstrated by Hörmann et al., who showed that TLR2 agonists significantly increased proliferation of small intestinal epithelial cell lines through multiple protein kinase pathways and moderately increased apoptosis [77]. The effect of TLR2 on epithelial proliferation was further supported by decreased proliferation in TLR2 deficient mice [77]. If TRL2 elicits the same effect in the esophagus, the interplay between microbiota and TLR2 may affect the propensity for mucosal damage and tumorigenesis in the EAC cascade.
In summary, TLR2 overexpression and potential for heterodimerization, pro-inflammatory signaling, and epithelial barrier regulation in the EAC cascade have been established. Currently, there is a paucity of studies analyzing how TLR2 expression directly varies with composition of the esophageal microbiota, although TLR2 expression in the small intestine has been shown to be regulated by microbiota. Further studies examining TLR2 in relation to normal and dysbiotic esophageal microbiota are warranted. Elucidating this relationship would help establish whether the dysbiotic activation of TLR2 in the EAC cascade triggers a pro-inflammatory cascade or a protective regulation of the esophageal epithelial barrier.
3.1.3. TLR 4
TLR4 is perhaps the most well-studied TLR in relation to the EAC cascade. Kohtz et al. determined how reflux conditions, as seen in GERD, affect TLR4 expression [79]. Their analysis demonstrated that TLR4 expression increased in normal cell lines under reflux conditions simulated by acidic or bile-containing solutions [79]. Similarly, Verbeek et al. demonstrated increased TLR4 expression in EAC (3.2 folds), BE (2.7 folds), and reflux esophagitis (1.9 folds) compared to normal squamous epithelium esophageal samples [80]. The location of TLR4 expression also differed along the EAC cascade. Specifically, TLR4 expression in normal squamous epithelium samples was mainly confined to the basal layer of the squamous epithelium. In contrast, TLR4 expression in BE samples was observed in superficial epithelial cells, crypts, and lamina propria cells. EAC expression of TLR4 was diffusely distributed across superficial and infiltrating cells. Huhta et al.’s findings support Verbeek et al.’s observations regarding increased TLR4 expression in abnormal esophageal samples and elaborate that TLR4 expressed by EAC localizes in the nucleus rather than the normally observed cytoplasm [74,80]. This nuclear localization of TLR4 in EAC was found to correlate with distant metastasis and poor prognosis.
Increased expression of TLR4 in pathological states of the EAC cascade has severe implications for the role of microbiota in EAC development [74,79,80]. When TLR4 is normally confined to the basal layer in the squamous epithelium of the esophagus, it may not come into contact with PAMPs expressed by esophageal microbiota. However, gastric reflux conditions may lead to an increase in TLR4 expression and damage the superficial esophageal epithelium normally separating microbial PAMPS from basally expressed TLR4. Consequently, the reflux conditions seen in the EAC cascade may not only increase the expression of TLR4 but also expose TLR4 to microbial PAMPs.
The implications of TLR4 activation by microbial PAMPs in this manner were elucidated by Verbeek et al. [80]. Analysis was performed with ex vivo cultures of BE and normal squamous esophagus biopsies as well as non-neoplastic BE cell lines. Downstream effects of TLR4 activation by LPS were monitored by NF-κB, IL-8, and COX-2 expression analysis with western blots, ELISA, or quantitative RT PCR, respectively. Results demonstrated that LPS stimulation of TLR4 caused: (i) NF-κB activation; (ii) a dose-dependent increase in IL-8 expression and secretion in non-neoplastic BE cell lines and ex vivo cultures of squamous esophagus, BE, and duodenum biopsies; and (iii) increased COX-2 expression in ex vivo BE biopsies and BE cell lines, but not ex vivo cultures of normal squamous esophagus or duodenum. Kohtz et al. noted a similar effect of TLR4 activation by LPS in normal and EAC cell lines [79]. Specifically, LPS-activation of TLR4 led to downstream NF-κB activation and increased esophageal cell growth rates. Of note, inhibiting NF-κB attenuated the accelerated growth rates previously observed [79].
These results suggest that LPS activation of TLR4 can lead to activation of NF-κB, inflammation, and cell proliferation. Increased expression of Gram-negative bacteria, known to uniquely express LPS, are increased in the EAC cascade. Therefore, the shift in EAC cascade microbiota may enhance the probability of LPS activating TLR4 and lead to consequent activation of NF-κB, secretion of IL-8, increased COX-2 expression, and increased proliferation [80]. Each of these downstream effects may promote carcinogenesis. NF-κB, as discussed previously, is a key regulator of inflammation, proliferation, and cell survival. COX-2 can mediate cell proliferation, apoptosis inhibition, angiogenesis, tumor invasiveness, and immunosuppression, as discussed below [81]. The role of IL-8 in carcinogenesis includes regulation of angiogenesis, cancer cell growth and survival, tumor cell motion, leukocyte infiltration, and immune responses [81]. Consequently, the Gram-negative shift in BE microbiota may enable the carcinogenesis of EAC cascade through the activation of TLR4.
3.1.4. TLR 5
Helminen et al. assessed the expression of TLR5 in normal esophageal squamous epithelium, BE with and without dysplasia, and EAC biopsies by means of immunohistochemistry staining [82]. TLR5 expression was only noted in the basal third of normal squamous epithelium and non-dysplastic BE tissue. In contrast, dysplastic BE and EAC tissue demonstrated increased, diffuse TLR5 expression with no apparent polarization. Similar to Verbeek et al.’s analysis of TLR4, this suggests that esophageal epithelium damage and transformation may expose TLR5 to microbial PAMPs and lead to consequent inflammatory cascades [80].
The composition of the dysbiotic microbiota in the EAC cascade further increases the likelihood of TLR5 activation, considering Snider et al. noted an increased relative abundance of the Proteobacteria phylum in high-grade dysplasia BE and EAC [49]. Proteobacteria is a phylum predominantly composed of bacteria with flagella, a well-established PAMP for TLR5 [83]. If this dysbiotic shift triggers TLR5, it can lead to downstream inflammatory cascades via NF-κB, CREB, and AP-1.
The potential for inflammation downstream of TLR5 may make TLR5 an important marker for EAC cascade progression. Specifically, Helminen et al. found moderate to high expression of TLR5 to be a marker for low-grade dysplasia with 86% sensitivity and 83% specificity [82]. Consequently, TLR5 may serve as a diagnostic marker for esophageal columnar dysplasia and BE, a condition known for controversial diagnostic parameters. This would have positive implications for prophylactic EAC treatment and monitoring of BE. However, a study by Helminen et al. did not find TLR5 expression to correlate with EAC prognosis [82].
Although TLR5 expression, the potential for activation by a dysbiotic flagellated microbiota, and correlation with dysplasia have been elucidated, more studies are needed to identify the direct interactions between dysbiotic microbiota and TLR5. Analyzing how TLR5 expression relates to the relative abundances of dysbiotic flagellated bacteria would provide the foundation on which additional studies could analyze this relationship.
3.1.5. TLR 9
TLR9 expression in EAC was assessed by Kauppila et al. by means of immunohistochemical staining [84]. TLR9 expression was observed in all 76 EAC tumors. Furthermore, strong cytoplasmic TLR9 immunoreactivity correlated with high pathological tumor stage, positive lymph nodes, distant organ metastases, high tumor grade, tumor unresectability, and decreased 10-year survival rates. Kauppila et al.’s findings lay the foundation for future studies investigating the role of TLR9 in EAC. Specifically, while the correlation between TLR9 and tumorigenesis was determined, the downstream effects in EAC carcinogenesis were not determined. Future studies will need to analyze how TLR9 directly or indirectly mediates tumorigenesis in the EAC cascade.
Furthermore, the ligand for TLR9 in EAC was not determined. Previous studies have shown that TLR9 can recognize CpG DNA sequences present in bacteria and viruses [67,85]. The viral composition of the esophageal microbiota has yet to be fully elucidated. However, bacterial unmethylated CpG DNA sequences in the esophageal microbiota could be responsible for the observed correlations between TLR9 expression and EAC tumorigenesis. Studies analyzing the effects of dysbiotic CpG DNA expression in relation to TLR9 expression in the EAC cascade would consequently be of interest.
3.1.6. Summary of TLRs in the EAC Cascade
TLR2 expression is increased in the EAC cascade, and the location of TLR2 expression varies with the pathologic state. It is capable of heterodimerizing with TLR1 and TLR6 and regulating esophageal epithelial barrier function.
TLR4 expression is increased in a stepwise fashion along the EAC cascade, and the location of TLR4 expression varies with the pathologic state. After recognizing the Gram-negative PAMP LPS, it causes pro-inflammatory signaling through NF-κB, IL-8, and COX-2.
TLR5 expression is increased in BE and EAC, and the location of TLR5 expression changes depending on the pathologic state. TLR5 expression has been shown to be a marker for low-grade esophageal dysplasia.
TLR9 is expressed in EAC tumors, and cytoplasmic TLR9 immunoreactivity correlates with high pathological tumor stage, positive lymph nodes, distant organ metastases, high tumor grade, tumor unresectability, and decreased 10-year survival rates [84].
3.2. Microbial Stimulation of Cyclooxygenase-2 Expression
The Gram-negative, dysbiotic microbiota observed in the EAC cascade may stimulate COX-2 overexpression that promotes GERD development and neoplastic progression of BE into EAC.
Cyclooxygenase-2 (COX-2) is a rate-limiting enzyme that catalyzes the initial step of arachidonic acid metabolism into prostaglandin H2, a precursor to prostanoids such as prostaglandin, thromboxane, and prostacyclin. These products act as autocrine and paracrine lipid mediators that maintain local homeostasis by mediating vascular function, wound healing, and inflammation [86,87]. COX-2 expression is initiated in response to IL-1, IFN-γ, TNF-α, and epidermal growth factor receptor [88]. COX-2 has substantial consequences in cancers, considering it is capable of mediating cell proliferation, apoptosis inhibition, angiogenesis, invasiveness, and immunosuppression [89]. Overexpression of COX-2 has been shown to induce tumorigenesis in mammary epithelium, and COX-2/prostaglandin E2 dysregulation can promote colorectal tumorigenesis through tumor maintenance, metastatic spread, and perhaps tumor initiation [86,90]. Several studies have already demonstrated increased COX-2 protein expression in esophageal samples of patients with BE and adenocarcinoma compared to the normal esophagus; however, the inciting factor triggering increased expression of COX-2 is not well defined [88,91,92].
The dysbiotic shift towards a higher relative abundance of Gram-negative bacteria in the EAC cascade may be a source of this increased COX-2 expression. Verbeek et al. demonstrated that ex vivo cultured BE biopsies incubated with LPS, a PAMP expressed by Gram-negative bacteria, had increased expression of COX-2 downstream of TLR4 [80]. This suggests that LPS may be the mediator through which COX-2 expression is increased.
Increased COX-2 expression as a result of LPS activation may have several effects on the EAC cascade. First, dysbiotic LPS-induced COX-2 expression may mediate the development of GERD. Building on Collares’ observation, Calatayud et al. sought to determine the mechanism by which LPS induced a delay in gastric emptying, a possible GERD risk factor [93,94]. Rats were treated with LPS as well as different prostaglandin and COX-2 inhibitors. Results demonstrated that dual COX-1 and COX-2 inhibitors prevented the LPS-induced delay in gastric emptying to a similar degree as COX-2 inhibitors alone. Phospholipase-2 inhibitors did not modify the rate of gastric emptying in LPS-treated animals. These results suggest that LPS stimulates the release of prostanoids and consequently delay gastric emptying through COX-2 rather than phospholipase expression. Delayed gastric emptying distends the stomach and increases the quantity of food available for reflux into the esophagus. This also generates transient lower esophageal sphincter relaxations that may facilitate reflux into the esophagus [95]. However, other studies failed to show similar results and a consensus behind the mechanism of gastroparesis is still not well established [96,97,98,99].
The LPS-induced COX-2 expression is also implicated in the potentiation of tumorigenesis. COX-2 overexpression has been shown to be a key mediator of BE neoplastic progression into EAC by promoting angiogenesis, cell proliferation, and reduced apoptosis [100]. Morris et al. specifically noted that COX-2 expression increased from low-grade dysplasia to high-grade dysplasia in the esophagus [88]. This result implies that COX-2 expression may contribute to cancer development rather than cancer inducing COX-2 expression. Furthermore, meta-analysis shows that overexpression of COX-2 is associated with poor overall survival, depth of invasion, metastasis, and tumor, node, and metastasis (TNM) stage in esophageal cancers [101].
In summary, LPS, expressed by the dysbiotic shift to a Gram-negative microbiota in the EAC cascade, leads to an increase in COX-2 expression. Increased COX-2 expression, in turn, leads to delayed gastric emptying that may increase the risk of GERD. Furthermore, COX-2 expression may promote carcinogenesis by promoting angiogenesis, cell proliferation, and a reduction in apoptosis. With these findings as a foundation, more studies are needed to elucidate the direct relationship between COX-2 and the EAC cascade microbiota.
3.3. Microbial Stimulation of Inducible Nitric Oxide Synthase
iNOS stimulated by Gram-negative, dysbiotic microbiota observed in the EAC cascade may relax the lower esophageal sphincter and promote GERD development.
Inducible nitric oxide synthase (iNOS) is an enzyme that produces nitric oxide (NO) by oxidizing L-arginine. Although iNOS is predominantly expressed by macrophages, iNOS expression can be induced in nearly any cell type with the appropriate stimulatory factors such as bacterial LPS and cytokines. NO produced by iNOS is capable of damaging both pathogenic cells and host cells through several mechanisms. Specifically, NO has a large affinity for protein-bound iron and is consequently capable of blocking iron-dependent enzymes that play an essential role in the citric acid cycle, mitochondrial electron transport, and DNA replication [102]. High concentrations of NO can also form inflammatory radicals, directly causing single and double-stranded DNA breaks, and inhibiting DNA repair enzymes through tyrosine and cysteine nitrosation [102,103]. iNOS activation also enables radical damage through peroxynitrite, a radical formed by NO interaction with superoxide [103,104].
Each of these mechanisms can have a protective effect on the host by killing tumor cells and microorganisms. However, similar effects can be directed against host cells, causing inflammation and tissue damage. iNOS activity and consequent high concentrations of NO have been shown to be involved in several cytotoxic processes such as apoptosis, angiogenesis, and DNA damage during cancer development [104]. Consequently, iNOS can play both an anti-tumorigenic and pro-tumorigenic role in cancers. There is reason to suspect iNOS in the pathogenesis of the EAC cascade considering several studies have demonstrated that iNOS is overexpressed in BE and EAC compared to normal esophagus [91,105,106].
The role of microbiota in the observed iNOS overexpression has been evaluated by several studies. Specifically, several studies demonstrated that LPS caused a dose-dependent decrease in lower esophageal sphincter tone, an effect attenuated by selective inhibition of iNOS with aminoguanidine or L-canavanine [107,108]. Park et al. evaluated this relationship further and analyzed these changes directly related to iNOS expression [109]. The study analyzed plasma nitrite/nitrate levels and iNOS expression in opossum models by western blot and RT-PCR assays before and after LPS administration. Results showed that LPS treatment increased plasma nitrite/nitrate levels and iNOS expression in the lower esophageal sphincter. To establish the role of iNOS in this observation, a selective iNOS inhibitor was administered. iNOS inhibitor treatment significantly attenuated the increase in plasma nitrite/nitrate levels and slightly attenuated the increased iNOS expression. Consequently, LPS expressed by Gram-negative dysbiotic microbiota along the EAC cascade can induce increased iNOS expression in the lower esophageal sphincter and subsequently causing lower esophageal sphincter relaxation.
Relaxation of the lower esophageal sphincter in a chronic fashion enables the development of GERD, a known risk factor for EAC.
3.4. NLRP3 Inflammasome Activation by Microbiota
Dysbiotic microbiota in the EAC cascade may trigger NLRP3 Inflammasomes independently of TLRs.
Inflammasomes are expressed by epithelial and immune cells. Inflammasomes interact with receptors, including TLRs and nod-like receptors (NLR)s, to monitor damage or foreign microorganisms, and trigger an immune response when appropriate. The nod-like receptor protein 3 (NLRP3) inflammasome has specifically been shown to be modulated by gastrointestinal dysbiosis but also regulates gastrointestinal dysbiosis [110]. Activation of NLRP3 requires two steps. First, there is a priming signal in which microbial ligands activate TLRs or cytokines activate tumor necrosis factor receptors (TNFRs). Either the activated TLR or TNFR causes activation of NF-κB, leading to upregulation of pro-IL-1β and NLRP3 transcription [111]. A second signal, stimulated by DAMPs or PAMPs such as LPS, then promotes the assembly of the inflammasome complex, causing the activation of the NLRP3 inflammasome [111].
Activation of the NLRP3 inflammasome in this manner may regulate the composition of the esophageal microbiome. Yao et al. demonstrated that hyperactive NLRP3 inflammasomes improve the symbiotic relationship between gut microbiota and the immune system through increased regulatory T cell induction [112]. The influential role of the NLRP3 inflammasome in microbiota composition was also demonstrated by Hirota et al. [113]. Specifically, the study demonstrated impaired crypt bactericidal activity in NLRP3 knock-out mice with a subsequent dramatic change in intestinal microbiota. NLRP3 knock-out mice were also associated with decreased innate immune mechanisms combating mucosal injury by intestinal microbiota [113]. This suggests that NLRP3 may play an important protective role in maintaining symbiosis between host immune systems and gut microbiota. Li et al. alternatively demonstrated that NLRP3 activation in mice models of acute pancreatitis plays less of a regulatory role and instead disrupts gut microbiota through NLRP3 induced inflammation [114]. Collectively, these studies suggest that NLRP3 plays an influential role in gut microbiome composition and microbiota symbiosis with the host. Whether NLRP3 activation promotes symbiosis or dysbiosis in the EAC cascade will require additional studies analyzing the esophageal microbiome specifically.
On the other hand, the impact of dysbiosis on NLRP3 inflammasome activation has been evaluated in regard to the EAC cascade. Considering LPS is both an activating molecule for NLRP3 inflammasome and a PAMP with increased expression in the EAC cascade microbiota, Nadatani et al. analyzed NLRP3 in regard to LPS and the EAC cascade [115]. Normal esophageal squamous cells and BE epithelial cells were treated with LPS, and the following were measured with and without TLR4 or NLRP3 inflammasome inhibition: (i) TLR4, pro-IL1β, pro-IL18, and NLRP3 expression; (ii) caspase-1 activity; (iii) TNF-α, IL8, IL1β, and IL18 secretion; (iv) lactate dehydrogenase release (a pyroptosis marker); and (v) mitochondrial reactive oxygen species (ROS). Results demonstrated that LPS both primes and activates the NLRP3 inflammasome via two separate mechanisms. First, LPS primes for NLRP3 inflammasome activation by inducing the expression of NLRP3, pro-IL1β, and pro-IL18 downstream of TLR4. It is important to note that LPS only has this priming effect downstream of TLR4 in BE cells, not normal esophageal squamous cells. Second, independently of TLR4, LPS increases mitochondrial ROS that activate the NLRP3 inflammasome. The activated NLRP3 inflammasome subsequently enables pyroptosis and converts pro-IL1β and pro-IL18 into mature IL1β and IL18. Therefore, LPS expressed by the Gram-negative dysbiotic microbiota in the EAC cascade can both prime and activate the NLRP3 inflammasome and lead to downstream production of IL-1β and IL-18.
Activation of the NLRP3 inflammasome in this manner may have significant consequences on the development of the EAC cascade, considering NLRP3 can directly promote or suppress tumorigenesis through IL-1β and IL-18 [116,117]. Depending on the cancer, IL-1 β and IL-18 have been shown to have pro-tumorigenic roles, including angiogenesis as well as anti-tumorigenic roles through immune cell modulation [118]. Whether the pro-tumorigenic or anti-tumorigenic pathway of NLRP3 predominates in the EAC cascade has yet to be determined. Recent studies demonstrate that increased NLRP3 expression in ESCC was associated with IL-1β as well as increased tumor proliferation, migration, and invasion [119]. Similar studies will need to be conducted in regard to the EAC cascade and their relationship with microbiota.
3.5. Microbial Contribution to the Warburg Effect
Dysbiotic microbiota of the EAC cascade may increase lactate availability for EAC carcinogenesis.
Lactate is an essential metabolite and signaling molecule. It is needed for carcinogenic angiogenesis, immune system evasion, cell migration, and metastasis [120]. Lactate is particularly important for cancer metabolism in which there are increased metabolic demands. In esophageal cancers specifically, there is a marked reduction of blood glucose levels with an increased level of lactate compared to healthy controls [121]. This reflects the “Warburg effect”, in which cancers have increased glucose uptake and fermentation of glucose into lactate, enabling increased tumor survival and proliferation.
Microbiota in the EAC cascade may facilitate the development of the Warburg effect. Specifically, Deshpande et al. analyzed microbiota in 106 microbial brush samples of normal, GERD, or BE esophagus [9]. Relative to normal esophagus brush samples, GERD and BE microbiota demonstrated an overall increase in lactic acid production. Similarly, Zhou et al. observed an EAC dysbiotic microbiota composed of a high abundance of lactic acid-producing bacteria, including Staphylococcus, Lactobacillus, Bifidobacterium, and Streptococcus [50]. These findings are significant, considering increased lactate availability may potentiate the Warburg effect in the EAC cascade.
The Warburg effect is promoted by mutations that favor increased glucose uptake; increased glycolytic enzyme expression; increased lactate production, accumulation, and release; and upregulated lactate exchange between cells via monocarboxylate transporters (MCT)s. [120]. Specifically, in the EAC cascade, Huhta et al. demonstrated a linearly increasing cytoplasmic MCT1 and MCT4 expression from normal epithelium to BE to dysplasia and EAC [122]. Consequently, not only do dysbiotic microbiota in the EAC cascade increase lactate availability, but the EAC cascade expresses increased concentrations of MCTs capable of transporting lactate into the cell for utilization. Zhang et al. further supported this finding by analyzing serum metabolite profiles of patients with normal esophagus, BE, high-grade dysplasia, and EAC by LC-MS and NMR assays [123]. Results demonstrated that lactate was present at statistically increased levels in BE, high-grade dysplasia, and EAC compared to normal controls.
Whether the increased relative abundance of lactate-producing bacteria is a cause or symptom of EAC is unclear at this point in time, however. Many patients with EAC experience dysphagia which limits their diet to a primarily liquid-dairy diet. Considering diet mediates microbial composition, EAC’s altered dairy-based diet may promote this dysbiotic shift to increased relative abundance of lactate-producing microbiota.
In summary, dysbiotic microbiota in GERD, BE, and EAC are composed of increased relative amounts of lactate-producing bacteria. With increased MCT1 and MCT4 expression along the EAC cascade, the lactate produced by the dysbiotic microbiota can be transported into the cell for the Warburg effect and subsequent tumorigenesis. However, further studies are required to evaluate the role of antibiotics targeting the lactate-producing microbiota in the EAC cascade development. By doing so, the degree to which dysbiotic lactate production contributes to the development of the EAC cascade could be determined. Furthermore, tracing how the dysbiotic lactate products are used by normal, GERD, BE, and EAC cells would be of interest.
4. Therapeutic Potential of Esophageal Microbiota
As discussed above, many studies have evaluated the pathologic role of microbiota in EAC cascade pathogenesis. Recent studies are beginning to evaluate the esophageal microbiota from an alternative therapeutic perspective. Specifically, they are evaluating how supplementing the esophageal microbiome, rather than targeting it with antibiotics, may serve a therapeutic role in the EAC cascade.
Prebiotics are compounds that stimulate growth or activity of microbiota and may be a mediator through which the esophageal microbiota can play a therapeutic role in the EAC cascade. A study by Selling et al. evaluated the effect of a prebiotic for Gram-positive lactobacilli in chronic GERD [124]. Daily supplementation of maltosyl-isomaltooligosaccharides as a prebiotic for lactobacilli led to a significant improvement of symptoms in patients with chronic GERD [124]. A systematic review suggested probiotics led to a similar improvement in GERD symptoms [125]. However, it has been noted that studies with improved internal validity are warranted to confirm the efficacy of probiotics in GERD [125].
Another mechanism through which esophageal microbiota may play a therapeutic role in the EAC cascade is through the bioengineered Escherichia coli Nissle 1917 [126]. It is capable of acting as a probiotic as well as producing and delivering antitumor treatment. In colitis, Escherichia coli Nissle 1917 has been shown to positively affect microbiota balance and induce and maintain ulcerative colitis remission [127]. The potential therapeutic effect of Escherichia coli Nissle 1917 has not been analyzed in the EAC cascade at this point in time, however it certainly can be a target for future research [126].
5. Conclusions and Future Directions
In summary, the EAC cascade expresses dysbiotic microbiota relative to the normal esophagus. The composition of microbiota shifts from a predominantly Gram-positive to a predominantly Gram-negative microbiota in GERD and BE. Although similar findings have been found in EAC, there are conflicting results, and a consensus has not been reached. LPS, a PAMP expressed by Gram-negative bacteria, may promote the development of the EAC cascade through (i) activation of TLRs; (ii) stimulation of COX-2 expression and subsequent delayed gastric emptying; (iii) stimulation of iNOS expression leading to relaxation of the lower esophageal sphincter; and (iv) priming and activation of the NLRP3 inflammasome. Dysbiotic microbiota in the EAC cascade also express an increased relative abundance of bacteria that utilize lactate metabolism. This increased availability of lactate may potentiate the Warburg effect and subsequent tumorigenesis.
Amidst these findings, several key gaps in knowledge remain. Although a trend from a Gram-positive to a Gram-negative microbiota is noted in GERD and BE, studies have not reached a consensus in EAC. Furthermore, a consensus has not been reached regarding the specific bacterial species with increased expression along the EAC cascade. A portion of this variance is attributable to differences in host age, genetics, geography, environmental exposures, and diet. Consequently, additional studies investigating the composition of EAC cascade microbiota with larger sample sizes of a more diverse population are required. Another source of this variability is the diversity of microbiome analysis techniques. No consensus has been reached in regard to the most accurate technique for microbiome analysis. Furthermore, the same techniques for genomic analysis can vary with the method used to collect the sample. For example, the reproducibility of high-throughput sequencing of 16S rRNA gene amplicons is significantly dependent on the DNA extraction method [128]. Additionally, DNA extraction for high-throughput sequencing is not selective for DNA from live bacteria. DNA from dead bacteria may also be extracted from the sample. It is consequently essential to have standardized microbiome sample collection methods and analysis techniques for future studies. Similarly, standardized methods are needed to further evaluate the association and implications of H. pylori in the EAC cascade if a consensus is to be reached.
The relationship between dysbiotic microbiota, TLRs, and the EAC cascade also warrants additional studies. Although TLRs are known to interact with PAMPs expressed by dysbiotic microbiota such as LPS, the degree to which PAMPs expressed by dysbiotic microbiota directly activate TLRs and mediate the EAC cascade remains unknown. Future studies utilizing TLR receptor modifiers among dysbiotic microbiota in the EAC cascade would greatly advance our understanding of how dysbiotic microbiota interact with TLRs and mediate the development of the EAC cascade. Considering TLR4 recognizes LPS, a PAMP with increased expression along the EAC cascade, studies investigating the direct effects of TLR4 on the EAC cascade would be of particular interest. In contrast to TLR4, analysis of TLR2 in relation to the microbiota would focus on the protective effects downstream of TLR2 and whether dysbiotic microbiota promote or interfere with TLR2 regulation of epithelial tight junctions.
Microbial activation of COX-2 and iNOS may separately promote GERD development. Although LPS has been established as an activator for both, whether the observed shift towards a Gram-negative microbiota can directly activate their expression and subsequent GERD development remains unknown. Consequently, how the expression of each varies in relation to dysbiotic microbial composition warrants investigation.
Analysis of whether LPS priming and activation of the NLRP3 inflammasome protects against or promotes EAC carcinogenesis is also needed. The effects of the NLRP3 inflammasome on tumorigenesis have been shown to vary from cancer to cancer. It will be important to elucidate whether it promotes or inhibits EAC development. Furthermore, how the effect of the NLRP3 inflammasome directly mediates the esophageal microbiome in the EAC cascade and how dysbiosis manipulates NLRP3 inflammasome function have yet to be determined. Further studies investigating the role of the esophageal microbiome in EAC pathogenesis and the changed molecular mechanism leading to EAC are warranted. Investigating the cause of change in the esophageal microbiome leading to EAC will facilitate in-depth understanding and prophylactic strategies to decrease EAC incidence.
Considering both lactate-producing bacteria and MCTs are increased in GERD, BE, and EAC, how the microbial lactate products are utilized by pathologic cells in the EAC cascade is of extreme importance. If the lactate is utilized for the Warburg effect in EAC, lactate and the dysbiotic microbiota producing it may be important targets for slowing EAC tumorigenesis.
Finally, the therapeutic role of microbiota in EAC pathogenesis is a new approach through which there are many avenues for future studies. Prebiotics and probiotics show promising beginnings, but more studies with strong internal validity are needed to evaluate the potential therapeutic role of prebiotics and probiotics in the EAC cascade. Additionally, Escherichia coli Nissle 1917 is a novel microbial mechanism through which EAC cascade pathogenesis can be mediated. No studies have evaluated Escherichia coli Nissle 1917 in the EAC cascade up to this point. The analysis of Escherichia coli Nissle 1917 in the EAC cascade is subsequently a novel field that would benefit from analysis.
Abbreviations
EAC = esophageal adenocarcinoma; GERD = gastroesophageal reflux disease; BE = Barrett’s esophagus; ESCC = esophageal squamous cell carcinoma; H. pylori = Helicobacter pylori; TLR = toll like receptor; PRR = pattern recognition receptor; PAMP = pathogen-associated molecular pattern; DAMP = damage-associated molecular pattern; LPS = lipopolysaccharide; NF-κB = nuclear factor kappa beta; AP-1 = activator protein 1; CREB = cAMP-response element binding protein; IRF = interferon-regulatory factor; IFN = interferon; COX-2 = cyclooxygenase 2; TNM = tumor, node, and metastasis; iNOS = inducible nitric oxide synthase; NO = nitric oxide; NLR = nod-like receptor; NLRP3 = nod-like receptor protein 3; TNFR = tumor necrosis factor receptor; IL = interleukin; TNF-α = tumor necrosis factor alpha; ROS = reactive oxygen species; MCT = monocarboxylate transporter; ZO-1 = zonula occludens-1; MyD88 = myeloid differentiation primary response 88; TM-7 = Saccharibacteria; rRNA = ribosomal ribonucleic acid; rDNA = ribosomal DNA; PCR = polymerase chain reaction.
Author Contributions
M.R.G.: Developed the project and wrote the manuscript. V.R.: Meticulously evaluated manuscript drafts and made critical revisions. S.A.: Critically appraised the final manuscript. K.C.N.: Developed and supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M., Laversanne M., Brown L.M., Devesa S.S., Bray F. Predicting the Future Burden of Esophageal Cancer by Histological Subtype: International Trends in Incidence up to 2030. Am. J. Gastroenterol. 2017;112:1247–1255. doi: 10.1038/ajg.2017.155. [DOI] [PubMed] [Google Scholar]
- 3.Coleman H.G., Xie S.-H., Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154:390–405. doi: 10.1053/j.gastro.2017.07.046. [DOI] [PubMed] [Google Scholar]
- 4.Pohl H., Welch H.G. The Role of Overdiagnosis and Reclassification in the Marked Increase of Esophageal Adenocarcinoma Incidence. J. Natl. Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 6.SEER*Explorer: An Interactive Website for SEER Cancer Statistics. Surveillance Research Program, National Cancer Institute. [(accessed on 1 November 2020)];2020 Available online: https://seer.cancer.gov/explorer/
- 7.Zhang Y. Epidemiology of esophageal cancer. World J. Gastroenterol. 2013;19:5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giroux V., Rustgi A.K. Metaplasia: Tissue injury adaptation and a precursor to the dysplasia–cancer sequence. Nat. Rev. Cancer. 2017;17:594–604. doi: 10.1038/nrc.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshpande N.P., Riordan S.M., Castaño-Rodríguez N., Wilkins M.R., Kaakoush N.O. Signatures within the esophageal microbiome are associated with host genetics, age, and disease. Microbiome. 2018;6:1–14. doi: 10.1186/s40168-018-0611-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zachariah R.A., Goo T., Lee R.H. Mechanism and Pathophysiology of Gastroesophageal Reflux Disease. Gastrointest. Endosc. Clin. N. Am. 2020;30:209–226. doi: 10.1016/j.giec.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 11.De Giorgi F., Palmiero M., Esposito I., Mosca F., Cuomo R. Pathophysiology of gastro-oesophageal reflux disease. [(accessed on 12 June 2021)];Acta Otorhinolaryngol. Ital. 2006 26:241–246. Available online: https://pubmed.ncbi.nlm.nih.gov/17345925. [PMC free article] [PubMed] [Google Scholar]
- 12.Sarosiek J., Feng T., McCallum R.W. The Interrelationship Between Salivary Epidermal Growth Factor and the Functional Integrity of the Esophageal Mucosal Barrier in the Rat. Am. J. Med. Sci. 1991;302:359–363. doi: 10.1097/00000441-199112000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Helm J.F., Dodds W.J., Riedel D.R., Teeter B.C., Hogan W.J., Arndorfer R.C. Determinants of esophageal acid clearance in normal subjects. Gastroenterology. 1983;85:607–612. doi: 10.1016/0016-5085(83)90015-X. [DOI] [PubMed] [Google Scholar]
- 14.Takashima T., Tsukano M., Kawano H. Digestive Endocrine Cell Numbers Contribute to Contraction/Relaxation of the Lower Esophageal Sphincter. SAGE Open Nurs. 2016;2 doi: 10.1177/2377960816650446. [DOI] [Google Scholar]
- 15.Parikh A., Thevenin C. Physiology, Gastrointestinal Hormonal Control. StatPearls Publishing; Treasure Island, FL, USA: 2021. [(accessed on 13 June 2021)]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK537284/?report=classic. [PubMed] [Google Scholar]
- 16.Ceranowicz P., Warzecha Z., Dembinski A. Peptidyl hormones of endocrine cells origin in the gut-their discovery and physiological relevance. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2015;66:11–27. [PubMed] [Google Scholar]
- 17.Stefanidis D., The SAGES Guidelines Committee. Hope W.W., Kohn G., Reardon P.R., Richardson W.S., Fanelli R.D. Guidelines for surgical treatment of gastroesophageal reflux disease. Surg. Endosc. 2010;24:2647–2669. doi: 10.1007/s00464-010-1267-8. [DOI] [PubMed] [Google Scholar]
- 18.Woodland P., Sifrim D. The refluxate: The impact of its magnitude, composition and distribution. Best Pract. Res. Clin. Gastroenterol. 2010;24:861–871. doi: 10.1016/j.bpg.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Chen C.-L., Hsu P.-I. Current Advances in the Diagnosis and Treatment of Nonerosive Reflux Disease. Gastroenterol. Res. Pract. 2013;2013:1–8. doi: 10.1155/2013/653989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gokulan R.C., Garcia-Buitrago M.T., Zaika A.I. From genetics to signaling pathways: Molecular pathogenesis of esophageal adenocarcinoma. Biochim. Biophys. Acta Bioenerg. 2019;1872:37–48. doi: 10.1016/j.bbcan.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubenstein J.H., Taylor J.B. Meta-analysis: The association of oesophageal adenocarcinoma with symptoms of gastro-oesophageal reflux. Aliment. Pharmacol. Ther. 2010;32:1222–1227. doi: 10.1111/j.1365-2036.2010.04471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubenstein J.H., Scheiman J.M., Sadeghi S., Whiteman D., Inadomi J. Esophageal Adenocarcinoma Incidence in Individuals with Gastroesophageal Reflux: Synthesis and Estimates from Population Studies. Am. J. Gastroenterol. 2011;106:254–260. doi: 10.1038/ajg.2010.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haider S.H., Kwon S., Lam R., Lee A.K., Caraher E.J., Crowley G., Zhang L., Schwartz T.M., Zeig-Owens R., Liu M., et al. Predictive Biomarkers of Gastroesophageal Reflux Disease and Barrett’s Esophagus in World Trade Center Exposed Firefighters: A 15 Year Longitudinal Study. Sci. Rep. 2018;8:1–7. doi: 10.1038/s41598-018-21334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazelton W.D., Curtius K., Inadomi J., Vaughan T.L., Meza R., Rubenstein J.H., Hur C., Luebeck E.G. The Role of Gastroesophageal Reflux and Other Factors during Progression to Esophageal Adenocarcinoma. Cancer Epidemiol. Biomark. Prev. 2015;24:1012–1023. doi: 10.1158/1055-9965.EPI-15-0323-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron A.J., Ott B.J., Payne W.S. The Incidence of Adenocarcinoma in Columnar-Lined (Barrett’s) Esophagus. N. Engl. J. Med. 1985;313:857–859. doi: 10.1056/NEJM198510033131404. [DOI] [PubMed] [Google Scholar]
- 26.Rubenstein J.H., Shaheen N.J. Epidemiology, Diagnosis, and Management of Esophageal Adenocarcinoma. Gastroenterology. 2015;149:302–317. doi: 10.1053/j.gastro.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy M., Kolodziejczyk A., Thaiss C.A., Elinav E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 28.Roy S., Trinchieri S.R.G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer. 2017;17:271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- 29.Goodman B., Gardner H. The microbiome and cancer. J. Pathol. 2018;244:667–676. doi: 10.1002/path.5047. [DOI] [PubMed] [Google Scholar]
- 30.Sheflin A., Whitney A., Weir T.L. Cancer-Promoting Effects of Microbial Dysbiosis. Curr. Oncol. Rep. 2014;16:1–9. doi: 10.1007/s11912-014-0406-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jandhyala S.M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Nageshwar Reddy D. Role of the normal gut microbiota. World J. Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B., Selmi C., Tang R., Gershwin M.E., Ma X. The microbiome and autoimmunity: A paradigm from the gut–liver axis. Cell. Mol. Immunol. 2018;15:595–609. doi: 10.1038/cmi.2018.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng M.Y., Inohara N., Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10:18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarosiek J., McCallum R.W. What Role Do Salivary Inorganic Components Play in Health and Disease of the Esophageal Mucosa? Digestion. 1995;56:24–31. doi: 10.1159/000201298. [DOI] [PubMed] [Google Scholar]
- 35.Rourk R.M., Namiot Z., Sarosiek J., Yu Z., McCallum R.W. Impairment of salivary epidermal growth factor secretory response to esophageal mechanical and chemical stimulation in patients with reflux esophagitis. Am. J. Gastroenterol. 1994;89:237–244. [PubMed] [Google Scholar]
- 36.Thursby E., Juge N. Introduction to the human gut microbiota. Biochem. J. 2017;474:1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pei Z., Bini E.J., Yang L., Zhou M., Francois F., Blaser M.J. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. USA. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grusell E.N., Dahlén G., Ruth M., Ny L., Bergquist H., Bove M., Quiding-Järbrink M. Bacterial flora of the human oral cavity, and the upper and lower esophagus. Dis. Esophagus. 2012;26:84–90. doi: 10.1111/j.1442-2050.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 39.Pei Z., Yang L., Peek R.M., Levine J.S.M., Pride D.T., Blaser M.J. Bacterial biota in reflux esophagitis and Barrett’s esophagus. World J. Gastroenterol. 2005;11:7277–7283. doi: 10.3748/wjg.v11.i46.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zilberstein B., Quintanilha A.G., Santos M.A.A., Pajecki D., de Moura E., Alves P.R.A., Filho F.M., De Souza J.A.U., Gama-Rodrigues J. Digestive tract microbiota in healthy volunteers. Clin. Sci. 2007;62:47–54. doi: 10.1590/S1807-59322007000100008. [DOI] [PubMed] [Google Scholar]
- 41.Parshina S.N., Stams A.J.M. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd.; Chichester, UK: 2015. Soehngenia; pp. 1–3. [Google Scholar]
- 42.Peter S., Pendergraft A., Vanderpol W., Wilcox C.M., Baig K.R.K.K., Morrow C., Izard J., Mannon P.J. Mucosa-Associated Microbiota in Barrett’s Esophagus, Dysplasia, and Esophageal Adenocarcinoma Differ Similarly Compared with Healthy Controls. Clin. Transl. Gastroenterol. 2020;11:e00199. doi: 10.14309/ctg.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong L., Yin J., Zhao J., Ma S.-R., Wang H.-R., Wang M., Chen W., Wei W.-Q. Microbial Similarity and Preference for Specific Sites in Healthy Oral Cavity and Esophagus. Front. Microbiol. 2018;9:1603. doi: 10.3389/fmicb.2018.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin J., Dong L., Zhao J., Wang H., Li J., Yu A., Chen W., Wei W. Composition and consistence of the bacterial microbiome in upper, middle and lower esophagus before and after Lugol’s iodine staining in the esophagus cancer screening. Scand. J. Gastroenterol. 2020;55:1467–1474. doi: 10.1080/00365521.2020.1839961. [DOI] [PubMed] [Google Scholar]
- 45.Blackett K.L., Siddhi S.S., Cleary S., Steed H., Miller M.H., Macfarlane S., Macfarlane G.T., Dillon J.F. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: Association or causality? Aliment. Pharmacol. Ther. 2013;37:1084–1092. doi: 10.1111/apt.12317. [DOI] [PubMed] [Google Scholar]
- 46.Yang L., Lu X., Nossa C.W., Francois F., Peek R.M., Pei Z. Inflammation and Intestinal Metaplasia of the Distal Esophagus Are Associated with Alterations in the Microbiome. Gastroenterology. 2009;137:588–597. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elliott D.R.F., Walker A., O’Donovan M., Parkhill J., Fitzgerald R.C. A non-endoscopic device to sample the oesophageal microbiota: A case-control study. Lancet Gastroenterol. Hepatol. 2017;2:32–42. doi: 10.1016/S2468-1253(16)30086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopetuso L.R., Severgnini M., Pecere S., Ponziani F.R., Boskoski I., Larghi A., Quaranta G., Masucci L., Ianiro G., Camboni T., et al. Esophageal microbiome signature in patients with Barrett’s esophagus and esophageal adenocarcinoma. PLoS ONE. 2020;15:e0231789. doi: 10.1371/journal.pone.0231789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snider E.J., Compres G., Freedberg D.E., Khiabanian H., Nobel Y.R., Stump S., Uhlemann A.-C., Lightdale C.J., Abrams J.A. Alterations to the Esophageal Microbiome Associated with Progression from Barrett’s Esophagus to Esophageal Adenocarcinoma. Cancer Epidemiol. Biomark. Prev. 2019;28:1687–1693. doi: 10.1158/1055-9965.EPI-19-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J., Shrestha P., Qiu Z., Harman D.G., Teoh W.-C., Al-Sohaily S., Liem H., Turner I., Ho V. Distinct Microbiota Dysbiosis in Patients with Non-Erosive Reflux Disease and Esophageal Adenocarcinoma. J. Clin. Med. 2020;9:2162. doi: 10.3390/jcm9072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gall A., Fero J., McCoy C., Claywell B.C., Sanchez C.A., Blount P.L., Li X., Vaughan T.L., Matsen F.A., Reid B.J., et al. Bacterial Composition of the Human Upper Gastrointestinal Tract Microbiome Is Dynamic and Associated with Genomic Instability in a Barrett’s Esophagus Cohort. PLoS ONE. 2015;10:e0129055. doi: 10.1371/journal.pone.0129055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amir I., Konikoff F.M., Oppenheim M., Gophna U., Half E.E. Gastric microbiota is altered in oesophagitis and Barrett’s oesophagus and further modified by proton pump inhibitors. Environ. Microbiol. 2014;16:2905–2914. doi: 10.1111/1462-2920.12285. [DOI] [PubMed] [Google Scholar]
- 53.Macfarlane S., Furrie E., Macfarlane G.T., Dillon J. Microbial Colonization of the Upper Gastrointestinal Tract in Patients with Barrett’s Esophagus. Clin. Infect. Dis. 2007;45:29–38. doi: 10.1086/518578. [DOI] [PubMed] [Google Scholar]
- 54.Zhao Y., Li Y., Hu J., Wang X., Ren M., Lu G., Lu X., Zhang D., He S. The Effect of Helicobacter pylori Eradication in Patients with Gastroesophageal Reflux Disease: A Meta-Analysis of Randomized Controlled Studies. Dig. Dis. 2020;38:261–268. doi: 10.1159/000504086. [DOI] [PubMed] [Google Scholar]
- 55.Yaghoobi M., Farrokhyar F., Yuan Y., Hunt R.H. Is There an Increased Risk of GERD After Helicobacter pylori Eradication?: A Meta-Analysis. Am. J. Gastroenterol. 2010;105:1007–1013. doi: 10.1038/ajg.2009.734. [DOI] [PubMed] [Google Scholar]
- 56.Saad A.M., Choudhary A., Bechtold M.L. Effect of Helicobacter pyloritreatment on gastroesophageal reflux disease (GERD): Meta-analysis of randomized controlled trials. Scand. J. Gastroenterol. 2012;47:129–135. doi: 10.3109/00365521.2011.648955. [DOI] [PubMed] [Google Scholar]
- 57.Erőss B., Farkas N., Vincze A., Tinusz B., Szapáry L., Garami A., Balaskó M., Sarlós P., Czopf L., Alizadeh H., et al. Helicobacter pyloriinfection reduces the risk of Barrett’s esophagus: A meta-analysis and systematic review. Helicobacter. 2018;23:e12504. doi: 10.1111/hel.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonnenberg A., Turner K.O., Genta R.M. Interaction Between Ethnicity and Helicobacter pylori Infection in the Occurrence of Reflux Disease. J. Clin. Gastroenterol. 2019;54:614–619. doi: 10.1097/MCG.0000000000001274. [DOI] [PubMed] [Google Scholar]
- 59.Doorakkers E., Lagergren J., Santoni G., Engstrand L., Brusselaers N. Helicobacter pylori eradication treatment and the risk of Barrett’s esophagus and esophageal adenocarcinoma. Helicobacter. 2020;25:e12688. doi: 10.1111/hel.12688. [DOI] [PubMed] [Google Scholar]
- 60.Nie S., Chen T., Yang X., Huai P., Lu M. Association of Helicobacter pyloriinfection with esophageal adenocarcinoma and squamous cell carcinoma: A meta-analysis. Dis. Esophagus. 2014;27:645–653. doi: 10.1111/dote.12194. [DOI] [PubMed] [Google Scholar]
- 61.Polyzos S.A., Zeglinas C., Artemaki F., Doulberis M., Kazakos E., Katsinelos P., Kountouras J. Helicobacter pylori infection and esophageal adenocarcinoma: A review and a personal view. Ann. Gastroenterol. 2017;31:8–13. doi: 10.20524/aog.2017.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pasare C., Medzhitov R. Toll-Like Receptors: Linking Innate and Adaptive Immunity. Adv. Exp. Med. Biol. 2005;560:11–18. doi: 10.1007/0-387-24180-9_2. [DOI] [PubMed] [Google Scholar]
- 63.Schnare M., Barton G., Holt A.C., Takeda K., Akira S., Medzhitov R. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2001;2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- 64.El-Zayat S.R., Sibaii H., Mannaa F.A. Toll-like receptors activation, signaling, and targeting: An overview. Bull. Natl. Res. Cent. 2019;43:1–12. doi: 10.1186/s42269-019-0227-2. [DOI] [Google Scholar]
- 65.Blander J.M. Regulation of Phagosome Maturation by Signals from Toll-Like Receptors. Science. 2004;304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 66.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 67.Kawai T., Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 68.Dajon M., Iribarren K., Cremer I. Toll-like receptor stimulation in cancer: A pro- and anti-tumor double-edged sword. Immunobiology. 2017;222:89–100. doi: 10.1016/j.imbio.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 69.Pradère J.-P., Dapito D.H., Schwabe R.F. The Yin and Yang of Toll-like receptors in cancer. Oncogene. 2014;33:3485–3495. doi: 10.1038/onc.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uronis J.M., Mühlbauer M., Herfarth H.H., Rubinas T.C., Jones G., Jobin C. Modulation of the Intestinal Microbiota Alters Colitis-Associated Colorectal Cancer Susceptibility. PLoS ONE. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cen X., Liu S., Cheng K. The Role of Toll-Like Receptor in Inflammation and Tumor Immunity. Front. Pharmacol. 2018;9:878. doi: 10.3389/fphar.2018.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mulder D., Lobo D., Mak N., Justinich C.J. Expression of Toll-Like Receptors 2 and 3 on Esophageal Epithelial Cell Lines and on Eosinophils During Esophagitis. Dig. Dis. Sci. 2011;57:630–642. doi: 10.1007/s10620-011-1907-4. [DOI] [PubMed] [Google Scholar]
- 73.Verbeek R.E., Siersema P.D., Vleggaar F.P., Kate F.J.T., Posthuma G., Souza R.F., de Haan J., Van Baal J.W. Toll-like Receptor 2 Signalling and the Lysosomal Machinery in Barrett’s Esophagus. J. Gastrointest. Liver Dis. 2016;25:273–282. doi: 10.15403/jgld.2014.1121.253.rc2. [DOI] [PubMed] [Google Scholar]
- 74.Huhta H., Helminen O., Lehenkari P.P., Saarnio J., Karttunen T.J., Kauppila J.H. Toll-like receptors 1, 2, 4 and 6 in esophageal epithelium, Barrett’s esophagus, dysplasia and adenocarcinoma. Oncotarget. 2016;7:23658–23667. doi: 10.18632/oncotarget.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaidi A.H., Kelly L.A., Kreft R.E., Barlek M., Omstead A.N., Matsui D., Boyd N.H., Gazarik K.E., Heit M.I., Nistico L., et al. Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer. 2016;16:1–10. doi: 10.1186/s12885-016-2093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Oliviera Nascimento L., Massari P., Wetzler L.M. The Role of TLR2 in Infection and Immunity. Front. Immunol. 2012;3:79. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hörmann N., Brandão I., Jäckel S., Ens N., Lillich M., Walter U., Reinhardt C. Gut Microbial Colonization Orchestrates TLR2 Expression, Signaling and Epithelial Proliferation in the Small Intestinal Mucosa. PLoS ONE. 2014;9:e113080. doi: 10.1371/journal.pone.0113080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruffner M.A., Song L., Maurer K., Shi L., Carroll M.C., Wang J.X., Muir A.B., Spergel J.M., Sullivan K.E. Toll-like receptor 2 stimulation augments esophageal barrier integrity. Allergy. 2019;74:2449–2460. doi: 10.1111/all.13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kohtz P.D., Halpern A.L., Eldeiry M.A., Hazel K., Kalatardi S., Ao L., Meng X., Reece T.B., Fullerton D.A., Weyant M.J. Toll-Like Receptor-4 Is a Mediator of Proliferation in Esophageal Adenocarcinoma. Ann. Thorac. Surg. 2019;107:233–241. doi: 10.1016/j.athoracsur.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 80.Verbeek R.E., Siersema P.D., Kate F.J.T., Fluiter K., Souza R.F., Vleggaar F.P., Bus P., Van Baal J.W.P.M. Toll-like receptor 4 activation in Barrett’s esophagus results in a strong increase in COX-2 expression. J. Gastroenterol. 2014;49:1121–1134. doi: 10.1007/s00535-013-0862-6. [DOI] [PubMed] [Google Scholar]
- 81.Ang Y. The role of interleukin-8 in cancer cells and microenvironment interaction. Front. Biosci. 2005;10:853–865. doi: 10.2741/1579. [DOI] [PubMed] [Google Scholar]
- 82.Helminen O., Huhta H., Takala H., Lehenkari P.P., Saarnio J., Kauppila J.H., Karttunen T.J. Increased Toll-like receptor 5 expression indicates esophageal columnar dysplasia. Virchows Archiv. 2013;464:11–18. doi: 10.1007/s00428-013-1505-2. [DOI] [PubMed] [Google Scholar]
- 83.Hayashi F., Smith K.D., Ozinsky A., Hawn T.R., Yi E.C., Goodlett D.R., Eng J., Akira S., Underhill D., Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nat. Cell Biol. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 84.Kauppila J.H., Takala H., Selander K.S., Lehenkari P.P., Saarnio J., Karttunen T.J. Increased Toll-like receptor 9 expression indicates adverse prognosis in oesophageal adenocarcinoma. Histopathology. 2011;59:643–649. doi: 10.1111/j.1365-2559.2011.03991.x. [DOI] [PubMed] [Google Scholar]
- 85.Bauer S., Kirschning C.J., Hacker H., Redecke V., Hausmann S., Akira S., Wagner H., Lipford G.B. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA. 2001;98:9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smyth E.M., Grosser T., Wang M., Yu Y., Fitz-Gerald G.A. Prostanoids in health and disease. J. Lipid Res. 2009;50:S423–S428. doi: 10.1194/jlr.R800094-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Turman M.V., Marnett L.J. Prostaglandin Endoperoxide Synthases. Compr. Nat. Prod. II. 2010;2010:35–63. doi: 10.1016/b978-008045382-8.00028-9. [DOI] [Google Scholar]
- 88.Morris C.D., Armstrong G.R., Bigley G., Green H., Attwood S.E. Cyclooxygenase-2 Expression in The Barrettʼs Metaplasia–Dysplasia–Adenocarcinoma Sequence. Am. J. Gastroenterol. 2001;96:990–996. doi: 10.1111/j.1572-0241.2001.03599.x. [DOI] [PubMed] [Google Scholar]
- 89.Rizzo M.T. Cyclooxygenase-2 in oncogenesis. Clin. Chim. Acta. 2011;412:671–687. doi: 10.1016/j.cca.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 90.Greenhough A., Smartt H.J.M., Moore A.E., Roberts H.R., Williams A., Paraskevaand C., Kaidi A. The COX-2/PGE2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–386. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 91.Wilson K.T., Fu S., Ramanujam K.S., Meltzer S.J. Increased expression of inducible nitric oxide synthase and cyclooxygenase-2 in Barrett’s esophagus and associated adenocarcinomas. Cancer Res. 1998;58:2929–2934. [PubMed] [Google Scholar]
- 92.Kandil H.M., Tanner G., Smalley W., Halter S., Radhika A., Dubois R.N. Cyclooxygenase-2 expression in Barrett’s esophagus. Dig. Dis. Sci. 2001;46:785–789. doi: 10.1023/A:1010700400960. [DOI] [PubMed] [Google Scholar]
- 93.Collares E. Effect of bacterial lipopolysaccharide on gastric emptying of liquids in rats. Braz. J. Med. Biol. Res. 1997;30:207–211. doi: 10.1590/S0100-879X1997000200008. [DOI] [PubMed] [Google Scholar]
- 94.Calatayud S., García-Zaragozá E., Hernández C., Quintana E., Felipo V., Esplugues J.V., Barrachina M.D. Downregulation of nNOS and synthesis of PGs associated with endotoxin-induced delay in gastric emptying. Am. J. Physiol. Liver Physiol. 2002;283:G1360–G1367. doi: 10.1152/ajpgi.00168.2002. [DOI] [PubMed] [Google Scholar]
- 95.Lipham J.C., Sandhu K.K. Options to Address Delayed Gastric Emptying in Gastroesophageal Reflux Disease. Shackelford Surg. Aliment. Tract 2 Vol. Set. 2019;2019:262–268. doi: 10.1016/b978-0-323-40232-3.00022-4. [DOI] [Google Scholar]
- 96.Gourcerol G., Benanni Y., Boueyre E., Leroi A.M., Ducrotté P. Influence of gastric emptying on gastro-esophageal reflux: A combined pH-impedance study. Neurogastroenterol. Motil. 2013;25 doi: 10.1111/nmo.12181. [DOI] [PubMed] [Google Scholar]
- 97.Emerenziani S., Sifrim D. Gastroesophageal reflux and gastric emptying, revisited. Curr. Gastroenterol. Rep. 2005;7:190–195. doi: 10.1007/s11894-005-0033-x. [DOI] [PubMed] [Google Scholar]
- 98.Maddern G., Chatterton B.E., Collins P.J., Horowitz M., Shearman D.J.C., Jamieson G.G. Solid and liquid gastric emptying in patients with gastro-oesophageal reflux. BJS. 2005;72:344–347. doi: 10.1002/bjs.1800720505. [DOI] [PubMed] [Google Scholar]
- 99.Cunningham K.M., Horowitz M., Riddell P.S., Maddern G., Myers J., Holloway R.H., Wishart J.M., Jamieson G.G. Relations among autonomic nerve dysfunction, oesophageal motility, and gastric emptying in gastro-oesophageal reflux disease. Gut. 1991;32:1436–1440. doi: 10.1136/gut.32.12.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Majka J., Rembiasz K., Migaczewski M., Budzynski A., Ptak-Belowska A., Pabianczyk R., Urbanczyk K., Zub-Pokrowiecka A., Matlok M., Brzozowski T. Cyclooxygenase-2 (COX-2) is the key event in pathophysiology of Barrett’s esophagus. Lesson from experimental animal model and human subjects. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2010;61:409–418. [PubMed] [Google Scholar]
- 101.Hu Z., Yang Y., Zhao Y., Huang Y. The prognostic value of cyclooxygenase-2 expression in patients with esophageal cancer: Evidence from a meta-analysis. Onco Targets Ther. 2017;10:2893–2901. doi: 10.2147/OTT.S134599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Förstermann U., Sessa W.C. Nitric oxide synthases: Regulation and function. Eur. Heart J. 2012;33:829–837. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McAdam E., Haboubi H.N., Forrester G., Eltahir Z., Spencer-Harty S., Davies C., Griffiths A.P., Baxter J.N., Jenkins G.J.S. Inducible Nitric Oxide Synthase (iNOS) and Nitric Oxide (NO) are Important Mediators of Reflux-induced Cell Signalling in Esophageal Cells. Carcinogenesis. 2012;33:2035–2043. doi: 10.1093/carcin/bgs241. [DOI] [PubMed] [Google Scholar]
- 104.Vannini F., Kashfi K., Nath N. The dual role of iNOS in cancer. Redox Biol. 2015;6:334–343. doi: 10.1016/j.redox.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clemons N.J., Shannon N., Abeyratne L.R., Walker C.E., Saadi A., O’Donovan M.L., Lao-Sirieix P.P., Fitzgerald R.C. Nitric oxide-mediated invasion in Barrett’s high-grade dysplasia and adenocarcinoma. Carcinogenesis. 2010;31:1669–1675. doi: 10.1093/carcin/bgq130. [DOI] [PubMed] [Google Scholar]
- 106.Bae J.D., Jung K.H., Ahn W.S., Bae S.H., Jang T.J. Expression of Inducible Nitric Oxide Synthase Is Increased in Rat Barrett’s Esophagus Induced by Duodenal Contents Reflux. J. Korean Med. Sci. 2005;20:56–60. doi: 10.3346/jkms.2005.20.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park H., Calrk E., Cullen J.J., Conklin J.L. Effect of endotoxin on opossum oesophageal motor function. Neurogastroenterol. Motil. 2000;12:215–221. doi: 10.1046/j.1365-2982.2000.00202.x. [DOI] [PubMed] [Google Scholar]
- 108.Fan Y.-P., Chakder S., Gao F., Rattan S. Inducible and neuronal nitric oxide synthase involvement in lipopolysaccharide-induced sphincteric dysfunction. Am. J. Physiol. Liver Physiol. 2001;280:G32–G42. doi: 10.1152/ajpgi.2001.280.1.G32. [DOI] [PubMed] [Google Scholar]
- 109.Park H., Clark E., Cullen J.J., Koland J.G., Kim M.S., Conklin J.L. Expression of inducible nitric oxide synthase in the lower esophageal sphincter of the endotoxemic opossum. J. Gastroenterol. 2002;37:1000–1004. doi: 10.1007/s005350200169. [DOI] [PubMed] [Google Scholar]
- 110.Feng Y., Huang Y., Wang Y., Wang P., Song H., Wang F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS ONE. 2019;14:e0218384. doi: 10.1371/journal.pone.0218384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jo E.-K., Kim J.K., Shin D.-M., Sasakawa C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 2016;13:148–159. doi: 10.1038/cmi.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yao X., Zhang C., Xing Y., Xue G., Zhang Q., Pan F., Wu G., Hu Y., Guo Q., Lu A., et al. Remodelling of the gut microbiota by hyperactive NLRP3 induces regulatory T cells to maintain homeostasis. Nat. Commun. 2017;8:1–17. doi: 10.1038/s41467-017-01917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hirota S.A., Ng J., Lueng A., Khajah M., Parhar K.K.S., Li Y., Lam V., Potentier M.S., Ng K., Bawa M., et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm. Bowel Dis. 2011;17:1359–1372. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li X., He C., Li N., Ding L., Chen H., Wan J., Yang X., Xia L., He W., Xiong H., et al. The interplay between the gut microbiota and NLRP3 activation affects the severity of acute pancreatitis in mice. Gut Microbes. 2020;11:1774–1789. doi: 10.1080/19490976.2020.1770042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nadatani Y., Huo X., Zhang X., Yu C., Cheng E., Zhang Q., Dunbar K.B., Theiss A., Pham T.H., Wang D.H., et al. NOD-Like Receptor Protein 3 Inflammasome Priming and Activation in Barrett’s Epithelial Cells. Cell. Mol. Gastroenterol. Hepatol. 2016;2:439–453. doi: 10.1016/j.jcmgh.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moossavi M., Parsamanesh N., Bahrami A., Atkin S.L., Sahebkar A. Role of the NLRP3 inflammasome in cancer. Mol. Cancer. 2018;17:1–13. doi: 10.1186/s12943-018-0900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.He Q., Fu Y., Tian D., Yan W. The contrasting roles of inflammasomes in cancer. Am. J. Cancer Res. 2018;8:566–583. [PMC free article] [PubMed] [Google Scholar]
- 118.Baker K.J., Houston A., Brint E. IL-1 Family Members in Cancer; Two Sides to Every Story. Front. Immunol. 2019;10:1197. doi: 10.3389/fimmu.2019.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu S., Yin J., Miao J., Li S., Huang C., Huang N., Fan T., Li X., Wang Y., Han S., et al. Activation of NLRP3 inflammasome promotes the proliferation and migration of esophageal squamous cell carcinoma. Oncol. Rep. 2020;43:1113–1124. doi: 10.3892/or.2020.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.San-Millán I., Brooks G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis. 2016;38:119–133. doi: 10.1093/carcin/bgw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang X., Xu L., Shen J., Cao B., Cheng T., Zhao T., Liu X., Zhang H. Metabolic signatures of esophageal cancer: NMR-based metabolomics and UHPLC-based focused metabolomics of blood serum. Biochim. Biophys. Acta Mol. Basis Dis. 2013;1832:1207–1216. doi: 10.1016/j.bbadis.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 122.Huhta H., Helminen O., Palomäki S., Kauppila J.H., Saarnio J., Lehenkari P., Karttunen T. Intratumoral lactate metabolism in Barrett’s esophagus and adenocarcinoma. Oncotarget. 2017;8:22894–22902. doi: 10.18632/oncotarget.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang J., Bowers J., Liu L., Wei S., Gowda G.A.N., Hammoud Z., Raftery D. Esophageal Cancer Metabolite Biomarkers Detected by LC-MS and NMR Methods. PLoS ONE. 2012;7:e30181. doi: 10.1371/journal.pone.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Selling J., Swann P., Madsen L.R., 2nd, Oswald J. Improvement in Gastroesophageal Reflux Symptoms from a Food-grade Maltosyl-isomaltooligosaccharide Soluble Fiber Supplement: A Case Series. [(accessed on 12 June 2021)];Integr. Med. 2018 17:40–42. Available online: https://pubmed.ncbi.nlm.nih.gov/31043918/ [PMC free article] [PubMed] [Google Scholar]
- 125.Cheng J., Ouwehand A.C. Gastroesophageal Reflux Disease and Probiotics: A Systematic Review. Nutrients. 2020;12:132. doi: 10.3390/nu12010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yu X., Lin C., Yu J., Qi Q., Wang Q. Bioengineered Escherichia coli Nissle 1917 for tumour-targeting therapy. Microb. Biotechnol. 2020;13:629–636. doi: 10.1111/1751-7915.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fabrega M.J., Rodríguez-Nogales A., Mesa J.G., Algieri F., Badía J., Giménez R., Galvez J., Baldomà L. Intestinal Anti-inflammatory Effects of Outer Membrane Vesicles from Escherichia coli Nissle 1917 in DSS-Experimental Colitis in Mice. Front. Microbiol. 2017;8:1274. doi: 10.3389/fmicb.2017.01274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Teng F., Nair S.S.D., Zhu P., Li S., Huang S., Li X., Xu J., Yang F. Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-34294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.