Abstract
The alternatively spliced 290-nucleotide NS2-specific exon of the parvovirus minute virus of mice (MVM), which is flanked by a large intron upstream and a small intron downstream, constitutively appears both in the R1 mRNA as part of a large 5′-terminal exon (where it is translated in open reading frame 3 [ORF3]), and in the R2 mRNA as an internal exon (where it is translated in ORF2). We have identified a novel bipartite exon enhancer element, composed of CA-rich and purine-rich elements within the 5′ and 3′ regions of the exon, respectively, that is required to include NS2-specific exon sequences in mature spliced mRNA in vivo. These two compositionally different enhancer elements are somewhat redundant in function: either element alone can at least partially support exon inclusion. They are also interchangeable: either element can function at either position. Either a strong 3′ splice site upstream (i.e., the exon 5′ terminus) or a strong 5′ splice site downstream (i.e., the exon 3′ terminus) is sufficient to prevent skipping of the NS2-specific exon, and a functional upstream 3′ splice site is required for inclusion of the NS2-specific exon as an internal exon into the mature, doubly spliced R2 mRNA. The bipartite enhancer functionally strengthens these termini: the requirement for both the CA-rich and purine-rich elements can be overcome by improvements to the polypyrimidine tract of the upstream intron 3′ splice site, and the purine-rich element also supports exon inclusion mediated through the downstream 5′ splice sites. In summary, a suboptimal large-intron polypyrimidine tract, sequences within the downstream small intron, and a novel bipartite exonic enhancer operate together to yield the balanced levels of R1 and R2 observed in vivo. We suggest that the unusual bipartite exonic enhancer functions to mediate proper levels of inclusion of the NS2-specific exon in both singly spliced R1 and doubly spliced R2.
Recent evidence indicates that in vertebrates, exons rather than introns may be the primary units of recognition during pre-mRNA splicing. Multiple splicing factors can bind cooperatively to splice sites flanking the exon and communicate across the exon (17, 19, 23, 35, 41, 51; reviewed in references 2, 3, 12, 34, and 50). Work in a number of laboratories has suggested that a strong downstream 5′ splice site can facilitate splicing of an upstream intron by strengthening its 3′ splice site via a network of protein interactions across the intervening exon (17, 19; reviewed in references 2 and 3).
Many exons are known to also contain auxiliary splicing elements known as exonic splicing enhancers (ESEs) (9, 10, 14, 15, 18, 22, 24, 25, 36, 38, 40, 42, 46, 49, 51–54). While most ESEs have been found in exons that are associated with alternative splicing events, they can also be found in constitutive exons. ESEs are associated with introns containing weak flanking splice sites and have been shown to function by strengthening interactions at upstream 3′ splice sites (4, 24, 38, 40, 44–46, 51–53). The majority of ESEs that have been identified are purine-rich repeats of the canonical sequence GARGARGAR; however, non-purine-rich enhancers have also been identified (9, 11, 43, 44, 49, 51). Activation of purine-rich ESEs requires the binding of essential splicing factors called SR proteins (24, 33, 39; reviewed in references 12 and 26), and protein-protein interactions between SR proteins and other splicing factors are thought to contribute to the cooperative assembly of early splicing complexes (2, 12, 26, 34).
Recently, a new class of non-purine-rich enhancers with CA-rich motifs (called ACEs) was identified by in vivo selection experiments, and individual selected ACEs were found to enhance splicing both in vivo and in vitro (9). For example, the well-characterized Drosophila doublesex (dsx) splicing enhancer (dsxRE) contains a CA-rich motif (25, 44) that enhances the splicing of a vertebrate exon in vertebrate cells (9), and an ACE within cTNT exon 16 has recently been shown to promote the binding of U2AF65 to the upstream 3′ splice site of a heterologous transcript in vitro (51).
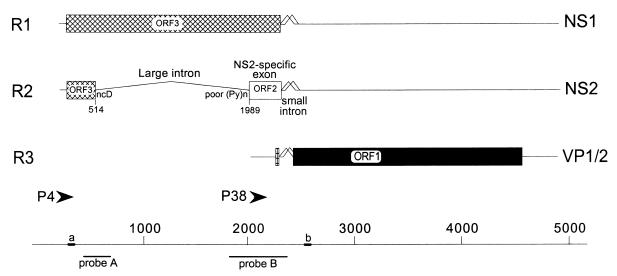
The parvovirus minute virus of mice (MVM) is organized into two overlapping transcription units, which produce three major transcript classes, R1, R2, and R3 (1, 6, 31) (Fig. 1). mRNAs R1 and R2 are generated from a promoter (P4) at map unit 4 (7, 31) and encode the viral nonstructural proteins NS1 and NS2, respectively. Both NS1 and NS2 play essential roles in viral replication and cytotoxicity (8), and so maintenance of their relative steady-state levels is critical to the MVM life cycle. P4-generated pre-mRNAs undergo alternative splicing, which determines the relative steady state ratios of mRNAs R1 and R2, thereby influencing the relative ratios of NS1 and NS2, respectively (5, 21, 28). There is approximately twice as much R2 as R1 present throughout infection of a wide variety of cell types (8, 55, 57), and all MVM mRNAs are very stable (37). The constitutive, alternative splicing of MVM pre-mRNAs must be accomplished solely by the interactions between cellular factors and viral cis-acting signals, because no viral proteins participate in this process (32).
FIG. 1.
Genetic map of MVM. The three major transcript classes and protein-encoding ORFs are shown. The two promoters (P4 and P38) are indicated by arrowheads. The large intron, small intron, and NS2-specific exon are indicated. The nonconsensus donor (ncD) and the poor polypyrimidine tract [poor (Py)n] of the large intron are also shown. The bottom diagram shows nucleotide locations, the two probes (A [nt 385 to 650] and B [nt 1854 to 2378]) used for RNase protection assays, and the two primers [a (nt 326 to 345) and b (nt 2557 to 2538)] used for RT-PCR, as described fully in Materials and Methods.
There are two types of introns in MVM P4-generated transcripts. An overlapping downstream small intron, located between nucleotides (nt) 2280 and 2399, is common to both P4-generated transcripts (R1 and R2) and P38-generated transcripts (R3). This small intron undergoes an unusual pattern of overlapping alternative splicing by using two donors (D1 and D2) and two acceptors (A1 and A2) within a region of 120 nt (28). The determinants governing excision of the small intron are complex, and the intron appears to be excised by a hybrid of intron and exon definition mechanisms (16).
An upstream large intron, located between nt 514 and 1989, is also excised from a subset of P4-generated pre-mRNAs to generate R2 mRNA. This upstream intron utilizes a nonconsensus donor and has a weak polypyrimidine tract, as defined by the presence of several interrupting purines, at its 3′ splice site. Excision of this intron from P4-generated pre-mRNA and the generation of the R2 mRNA require inclusion of the intervening NS2-specific exon as an internal exon.
The NS2-specific exon is an unusual internal exon in several ways. First, it occupies an unusual position flanked by a large intron upstream and a small intron downstream, in contrast to most vertebrate internal exons, which are flanked by large introns on both sides. Second, it is 290 nt, i.e., within the upper 5% of vertebrate exon size limits (2). Finally, the alternatively spliced NS2-specific exon is translated in two open reading frames (ORFs): when included in singly spliced R1, this exon is translated in ORF3 to encode NS1; when included in doubly spliced R2, the exon is translated in ORF2 to encode NS2.
We have previously shown that efficient excision of the upstream intron from P4-generated pre-mRNA depends upon the initial presence of downstream small intron sequences, including at least one downstream 5′ splice site and one downstream 3′ splice site, within the pre-mRNA (55, 57); prior splicing of the small intron, however, is not necessary. In the absence of both the downstream intron 5′ splice sites (which constitute the 3′ terminus of the NS2-specific exon), a new spliced product (here called the exon-skipped product [ES]) not seen during viral infection is produced, in which the NS2-specific exon is skipped and the upstream intron 5′ splice site at nt 514 is joined to a downstream intron 3′ splice site (55), predominantly A1 at nt 2377. Improvement of the polypyrimidine tract of the upstream intron 3′ splice site (which constitutes the 5′ terminus of the NS2-specific exon) not only can render its efficient excision independent of downstream intron sequences but also can suppress exon skipping caused by deletion of both the small intron donors (55). Inclusion of the NS2-specific exon also requires sequences within the exon itself; replacement of the entire exon by a heterologous exon sequence also results in uniform exon skipping (56). Finally, multiple determinants that govern excision of the small intron, including its small size and a G-rich intronic enhancer sequence, also participate in inclusion of the upstream NS2-specific exon (16).
In this study, we have performed an extensive mutational analysis of NS2-specific exon sequences, both at the termini and within the exon. We have identified a novel bipartite exonic enhancer, consisting of an unusual combination of CA-rich and purine-rich enhancer elements located at the 5′ and 3′ ends of the exon, respectively, that have at least partially redundant functions and function to mediate the proper levels of inclusion of the NS2-specific exon in both singly spliced (R1) and doubly spliced (R2) mRNA.
MATERIALS AND METHODS
Mutant construction.
Construction of pΔD1/2, p4TΔD1/2, p5A, p1989, pEx(→), and pEx(←) has been described previously (55, 56). The mutants pEx(→→) and pEx(←←) were generated by substituting two copies of NS2-specific exonic sequences from nt 2005 to 2270 in either the forward or reverse orientation, respectively, between the SmaI (nt 2005) and SmaI (nt 2270) sites of pEx(→) by standard recombinant DNA techniques. [RNA generated by the parent construct, pEx(→), which contains two SmaI sites engineered at nt 2005 and 2270, was spliced identically to that of wild type (13, 56).] All the final mutant clones were sequenced to confirm that only the desired mutations were introduced.
Mutants p1TΔD1/2, p2TΔD1/2, and pCSDΔD1/2 were constructed by subcloning fragments containing the 1T, 2T, and CSD mutations, respectively (described previously in references 55 and 57), into pΔD1/2 by standard recombinant DNA techniques and sequenced to ensure that only the desired mutations were introduced.
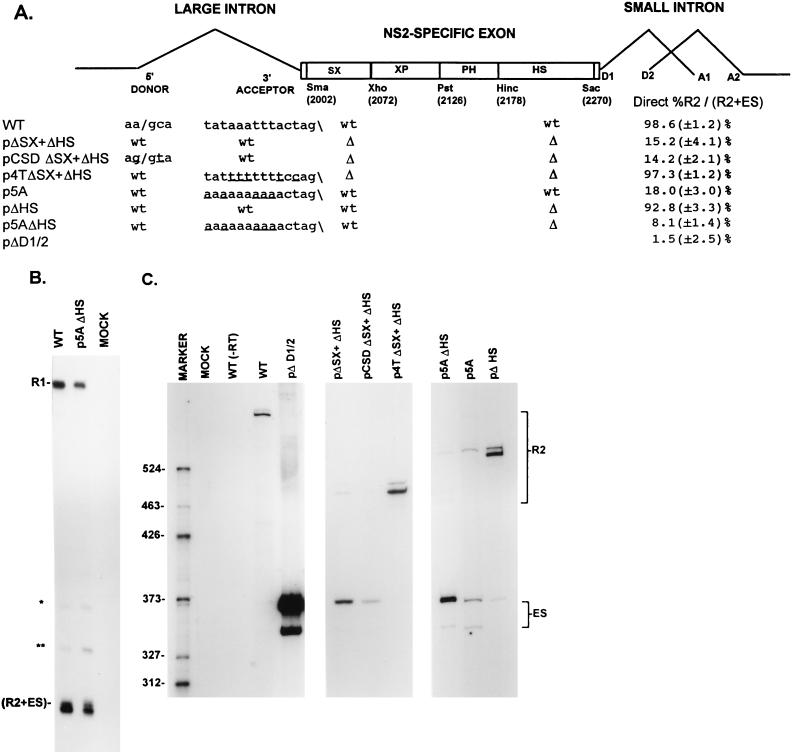
The parent construct for the pΔSX, pΔXP, pΔPH, pΔHS, pΔSma-Hinc, and pΔXho-Sac mutants was made by introducing unique SmaI (nt 2005) and SacI (nt 2270) restriction sites into the wild-type clone of MVM by M13-based site-directed mutagenesis (29), so that the highly conserved exon sequences at the 3′ splice site of the upstream large intron and those for the donor site of the downstream small intron were left intact. The deletions were performed in a pUC19 clone that contains MVM nt 1854 to 2378 with the engineered SmaI sites at nt 2005 and 2270, using the restriction sites at nt 2005 (SmaI), 2072 (XhoI), 2126 (PstI), 2178 (HincII), and 2270 (SacI (see Fig. 2A). Fragments containing the individual mutations were subcloned back into the plasmid clone of full-length MVM. RNA generated by the parent construct containing the SmaI (nt 2005) and SacI (nt 2270) sites was spliced like that of the wild type (13). pΔSX and pΔHS were combined to generate pΔSX+ΔHS, pCSD and pΔSX+ΔHS were combined to generate pCSDΔSX+ΔHS, and p5A and pΔHS were combined to generate p5AΔHS by standard recombinant DNA techniques. p4TΔSX+ΔHS was generated by initially cloning an MVM fragment containing the ΔHS deletion into the plasmid clone of MVM containing the 4T and SmaI site (nt 2005) mutations to generate p4TSmaΔHS and subsequently deleting the region between SmaI (nt 2005) and XhoI (nt 2270) in this mutant. All the final mutant clones were sequenced to confirm that only the desired mutations were introduced.
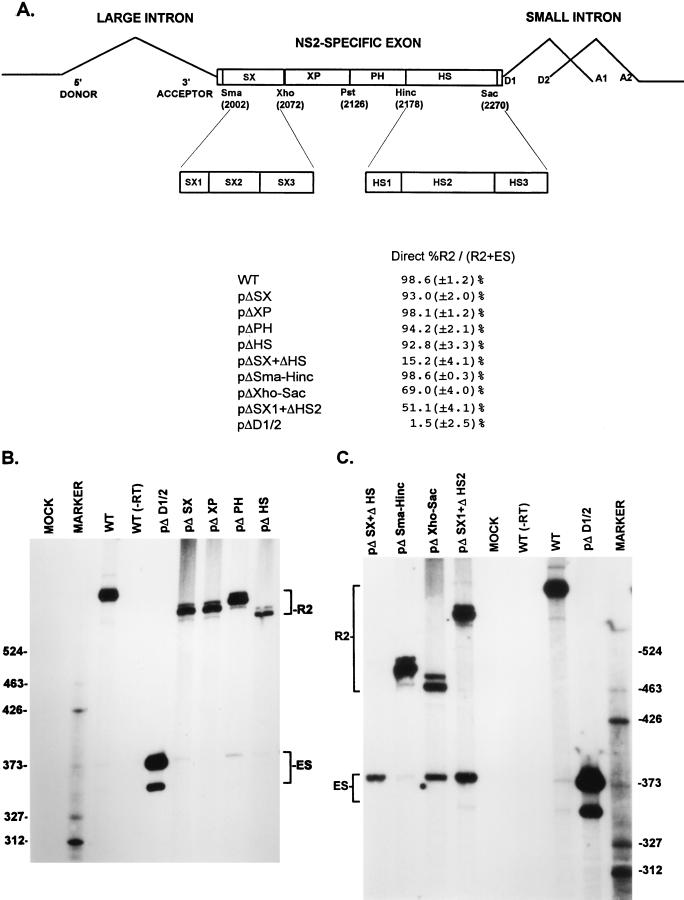
FIG. 2.
A bipartite enhancer within the NS2-specific exon is required for inclusion of this exon in mature spliced mRNA. (A) The restriction sites within the NS2-specific exon (SmaI, XhoI, PstI, HincII, and SacI) which divide the exon into four regions (SX, XP, PH, and HS) and were used to generate the different exon deletion mutants are indicated (ΔSX, for example, is a deletion between the SmaI and XhoI sites [see the text]). The SX1 to SX3 and HS1 to HS3 regions extend between the following nucleotide positions: SX1, nt 2002 to 2025; SX2, nt 2019 to 2052; SX3, nt 2050 to 2109; HS1, nt 2177 to 2196; HS2, nt 2196 to 2253; HS3, nt 2252 to 2270 (MVM nucleotide numbers as in reference 1). Quantitations of the direct percent R2/(R2+ES) ratio obtained by RT-PCR analysis for various mutants are also shown. All the values are the average of at least three separate experiments. Standard deviations are indicated in parentheses. ES values for pΔD1/2 are shown for comparison; the mutation is shown in Fig. 5A. (B) RT-PCR analysis of RNA generated by wild-type MVM (WT), mutants (described in the text), or mock transfected, as designated at the top of each lane. Samples were run on a 6% acrylamide—urea gel. pΔD1/2 was used as a control for amplification of the ES. WT(−RT) is a control reaction with wild-type RNA but excluding reverse transcriptase. An RNase protection analysis, with probe B, of RNA generated by the wild type was used as a marker (MARKER; the sizes of the marker bands are shown on the left) for the sizes of the RT-PCR-amplified bands. Wild-type RNA generated a 658-nt amplified R2 product, while RNA generated by the mutants showed R2 products of sizes consistent with the sizes of the deletions in these mutants. As explained in the legend to Fig. 5D, two kinds of amplified ES, using either A1 or A2, were observed. (C) RT-PCR analysis of RNA generated by wild-type MVM (WT), mutants (described in the text), or mock transfected, as designated at the top of each lane. Samples were run on a 6% acrylamide–urea gel. pΔD1/2 and WT(−RT) controls, and the marker (MARKER; the sizes of the marker bands are shown on the right for the sizes of the RT-PCR-amplified bands) are as in panel B. Wild-type RNA generated a 658-nt amplified R2 product, while RNA generated by the mutants showed R2 products of sizes consistent with the sizes of the deletions in these mutants. As explained in the legend to Fig. 5D, two kinds of amplified ES using either A1 or A2 were observed.
The mutants pSX1(−) and pHS2(−), as well as those with the smaller deletions within the SX and HS regions of the exon including pΔSX1 and pΔHS2, were constructed by M13-based oligonucleotide mutagenesis as previously described (29). Mutant oligonucleotides were homologous to the viral DNA except at the nucleotides which were to be altered or deleted. For the changes made in these mutants, see Fig. 2A and 3A. pSX1(−) and pHS2(−) were combined to generate pSX1(−)+HS2(−), and pΔSX1 and pΔHS2 were combined to generate pΔSX1+ΔHS2 by standard recombinant DNA techniques. All the final mutant clones were sequenced to confirm that only the desired mutations were introduced. Unlike the larger exon deletions described above, pSX1(−), pHS2(−), pSX1(−)+HS2(−), pΔSX1, pΔHS2, and pΔSX1+ΔHS2 do not contain SmaI (nt 2005) and SacI (nt 2270) sites.
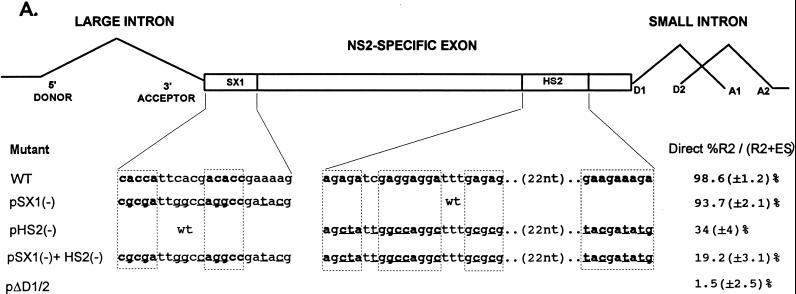
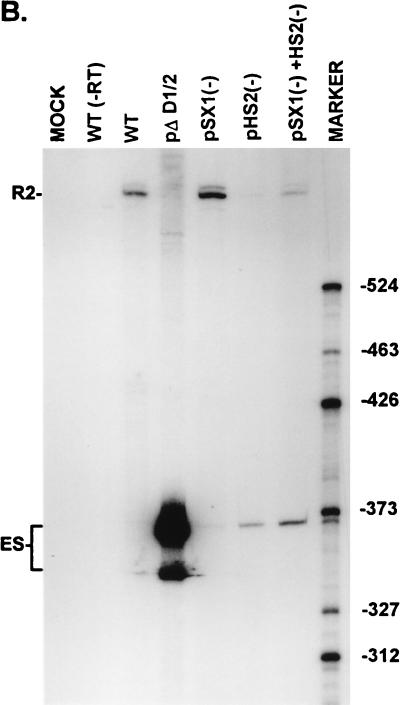
FIG. 3.
Point mutations within the bipartite enhancer result in exon skipping. (A) The sequences of the wild-type and mutant SX1 and HS2 regions are shown underneath their appropriate map positions (with deviations from the wild-type sequence underlined), together with quantitations of the direct percent R2/(R2+ES) ratio obtained by RT-PCR analysis for each mutant. The CA-rich sequences in the SX1 region and the purine-rich sequences in the HS2 region are boxed and in boldface type. All the values are the average of at least three separate experiments. Standard deviations are indicated in parentheses. ES values for pΔD1/2 are shown for comparison; the mutation is shown in Fig. 5A. wt, wild type sequence. (B) RT-PCR analysis of RNA generated by wild-type MVM (WT), mutants (described in the text), or mock transfected, as designated at the top of each lane. Samples were run on a 6% acrylamide–urea gel. pΔD1/2 and WT(−RT) controls and MARKER (the sizes of the marker bands are shown on the right) are as described in the legend to Fig. 2B. RNAs generated by either the wild type or the point mutants showed a 658-nt amplified R2 product. As explained in the legend to Fig. 5D, two kinds of amplified ES, using either A1 or A2, were observed.
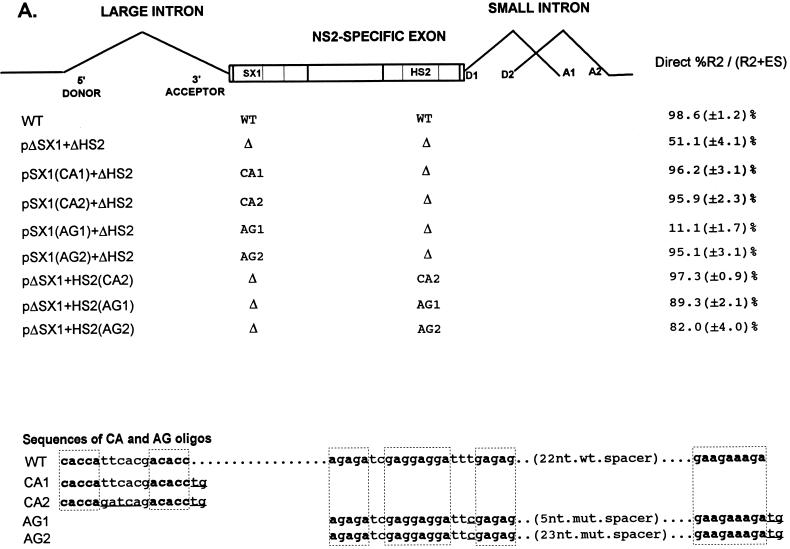
To generate the mutants pSX1(CA1)+ΔHS2, pSX1(CA2)+ΔHS2, pSX1(AG1)+ΔHS2, pΔSX1+HS2(CA1), pΔSX1+HS2(CA2), and pΔSX1+HS2(AG1), top- and bottom-strand oligonucleotides for either the CA1, CA2, or AG1 sequences were annealed and ligated into unique Msc1 sites created by either the ΔSX1 region or the ΔHS2 region of pΔSX1+ΔHS2. For the sequences of the CA1, CA2, and AG1 oligonucleotides, see Fig. 4A. To generate the mutants pSX1(AG2)+ΔHS2 and pΔSX1+HS2(AG2), an 18-nt spacer region was inserted into unique Sma sites created by either the SX(AG1) region of pSX1(AG1)+ΔHS2, or the HS2(AG1) region of pΔSX1+HS(AG1). The sequences of AG1 and AG2 are identical except for the mutant spacers which separate the purine-rich regions within these oligonucleotides (the sequence of the 5-nt mutant spacer in the AG1 oligonucleotide is 5′ CCCGG 3′, and the sequence of the 23-nt mutant spacer in the AG2 nucleotide is 5′ CCCGCTCGAATGGCTGGATTCGG 3′, compared to the 22-nt wild-type spacer sequence 5′-CGTGCTTCGGTGCGGAACCGTT 3′) (see Fig. 4A). All the final mutant clones were sequenced to confirm that only the desired mutations were introduced.
FIG. 4.
A CA-rich element and a purine-rich element together constitute the bipartite enhancer of the NS2-specific exon. (A) The sequences of the CA-rich and AG-rich oligonucleotides that were tested (deviations from the wild-type sequence are underlined; the sequences of the wild-type spacer in HS1 or the mutant spacers in AG1 and AG2 are described in Materials and Methods), and the positions at which they were replaced into pΔSX1+ΔHS2 are indicated (see the text for details). The CA-rich sequences in the SX1 region and the purine-rich sequences in the HS2 region are boxed and in boldface type. Quantitations of the direct percent R2/(R2+ES) ratio obtained by RT-PCR analysis for each mutant are also shown. All the values are the average of at least three separate experiments. Standard deviations are indicated in parentheses. WT, wild-type sequence. Δ, deletion. (B) RT-PCR analysis of RNA generated by wild-type MVM (WT), mutants (described in the text), or mock transfected, as designated at the top of each lane. Samples were run on a 6% acrylamide—urea gel. pΔD1/2 and WT(−RT) controls and MARKER (the sizes of the marker bands are shown on the left) are as described in the legend to Fig. 2B. RNA generated by WT showed a 658-nt amplified R2 product, while RNA generated by the mutants showed R2 products of sizes consistent with the sizes of the deletions and/or substitutions in these mutants. As explained in the legend to Fig. 5D, two kinds of amplified ES, using either A1 or A2, were observed.
Transfection and RNA isolation.
Murine A92L cells, the normal tissue culture host for MVM(p), were grown as previously described (31) and transfected with wild-type and mutant MVM plasmids by using either DEAE-dextran (30) or Lipofectamine-Plus reagent (as described in the Lipofectamine-Plus reagent kit [Gibco BRL]). RNA was typically isolated 48 h posttransfection, after lysis in guanidinium thiocyanate, by centrifugation through CsCl exactly as previously described (37).
RNA analysis. (i) RNase protection assays.
RNase protection assays were performed as previously described (37), with an [α-32P]UTP-labeled, SP6-generated antisense MVM RNA probe from either MVM nt 385 to 652 (probe A) or nt 1854 to 2378 (probe B) (Fig. 1). Probe A identifies all P4 products and distinguishes between R1 and the RNAs that use the nt 514 donor (R2+ES). Probe B extends from before the 3′ splice site of the large intron to within the small intron common to all MVM RNAs and distinguishes between P4 (R1 and R2) and P38 (R3) species using the large intron 3′ splice site and either of the alternative small-intron donors (designated M for the major splice donor [D1] at nt 2280 and m for the minor splice donor [D2] at nt 2317 [37]). Unspliced P4 and unspliced P38 products are designated R1un and R3un, respectively. R1, R2, and R3 RNAs using D1 at nt 2280 (Fig. 1) are designated R1M, R2M, and R3M, respectively. R1, R2, and R3 RNAs using D2 at nt 2317 (Fig. 1) are designated R1m, R2m, and R3m, respectively. Thus, probe B can distinguish between unspliced, minor, and major forms of both R1 and R2; however, it cannot detect the ES. For analysis of RNA produced after transfection with mutants containing mutations within this region covered by probe B (pΔD1/2, p1TΔD1/2, p2TΔD1/2, and p4TΔD1/2), antisense RNase protection probes homologous to the mutants being analyzed were used. RNase protection products were analyzed on a Betagen β-scanning phosphorimage analyzer, and molar ratios of MVM RNA were determined by standardization to the number of uridines in each protected fragment.
(ii) Quantitative RT-PCRs.
First-strand cDNA synthesis used 5 μg of total RNA isolated after transfection and oligonucleotide dT priming by standard techniques (20). PCR detection of R2 and ES was performed with primers a and b (Fig. 1) as described previously (56) with minor modifications (6a). The forward primer (designated a in Fig. 1) was 5′-end labeled with [γ-32P]ATP (as described in reference 27) and added to a 15-cycle PCR mixture (94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by a single extension at 72°C for 5 min). The ratio of the two products did not vary detectably over a wide range of dilutions and cycle numbers (15, 20, and 30 cycles). PCR products were run on 6% acrylamide–urea gels and analyzed on a Betagen β-scanning phosphorimage analyzer, to calculate molar ratio of R2 to ES and obtain a direct value of percent R2/(R2+ES) (see Fig. 5A and D).
FIG. 5.
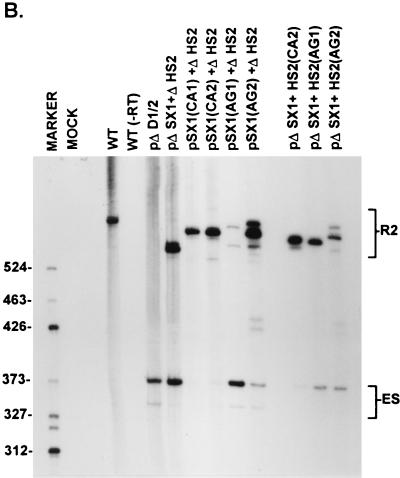
The NS2-specific exon of MVM requires only one strong exon terminus (upstream 3′ splice site or downstream 5′ splice site) to prevent exclusion from mature spliced mRNA. (A) The sequences of the large-intron 5′-splice-site and 3′-splice-site polypyrimidine tract and cleavage site in wild-type MVM and mutants are shown underneath their appropriate map positions (deviations from the wild-type sequence are underlined), with quantitations of the direct percent R2/(R2+ES) ratio obtained by quantitative RT-PCR (described fully in Materials and Methods). The presence or absence of the small-intron 5′ splice sites in these constructs is indicated by either wt (presence) or Δ (absence). All the values are the average of at least three separate experiments. Standard deviations are indicated in parentheses. wt, wild type sequence. Δ, deletion. (B) The two panels show RNase protection analyses with probe A (Fig. 1) of RNA generated by wild-type MVM (WT), mutants (as described in the text), or mock transfected, as designated at the top of each lane. The identities of the protected bands are shown on the left (in the left panel) and on the right (in the right panel). The larger species (R1) represents mRNA R1, while the major smaller species (R2+ES) represents RNA that uses the upstream large-intron 5′ splice site at nt 514 (i.e., mRNA R2 plus the ES). The identities of the bands designated ∗ and ∗∗ are unknown; however, they are probably breakdown products of the probe since they are not reproducibly seen and occasionally appear in lanes of mock-infected RNA (data not shown). (C) RNase protection analysis with probe B (Fig. 1), of RNA generated by wild-type MVM (WT), mutants (as described in the text), or mock transfected, as designated at the top of each lane. The identities of the protected bands for the wild type are shown on the left. The mutants were protected with versions of probe B homologous to the mutant region of the upstream intron 3′-splice-site polypyrimidine tract but nonhomologous in the region of D1 and D2, and therefore the RNA generated by these mutants shows fragments shorter than those generated by wild-type RNA. These mutants produce R1, R2, and R3 products that are unspliced across the small intron (designated R1un, R2un, and R3un, respectively) due to the loss of the two downstream small intron 5′ splice sites, as well as R2 and R3 products that are spliced across the downstream small intron (designated R2cryptic and R3cryptic, respectively) by using a cryptic donor within the NS2-specific exon (see Discussion). Improvements of the upstream intron 3′-splice-site polypyrimidine tract disrupted the P38 promoter, resulting in decreased production of R3cryptic as well as R3un. ∗, undigested probe B. (D) RT-PCR analysis of RNA generated by wild-type MVM (WT), mutants (as described in the text), or mock transfected, as designated at the top of each lane, with primers a and b as shown in Fig. 1 and performed as explained in Materials and Methods. Samples were run on a 6% acrylamide–urea gel. WT(−RT) is a control reaction with wild-type RNA but excluding reverse transcriptase. An RNase protection analysis with probe B of RNA generated by the wild type was used as a marker (MARKER; the sizes of the marker bands are shown on the right) for the sizes of the RT-PCR-amplified bands. WT RNA generated a 658-nt amplified R2 product. RNA generated by the mutants showed two kinds of amplified ES, both of which were considered in the quantitations: a larger (368-nt) product and a smaller (346-nt) product which represent exon skipping to 3′ splice sites A1 and A2, respectively, of the downstream small intron. RNAs generated by pΔD1/2, p1TΔD1/2, p2TΔD1/2, and p4TΔD1/2 also show two kinds of amplified R2 products, which were considered in the quantitations; the largest of these is unspliced across the small intron, and the smaller two products (designated ∗) probably utilized a donor within the NS2-specific exon (see Discussion). RNAs generated by p5A and p1989 also showed the smaller R2 product that probably utilized a donor within the exon. The ES has previously been sequenced across the splice junction to confirm its identity (56).
To demonstrate that our reverse transcription PCR (RT-PCR) assay was quantitative and accurately reflected the true ratio of R2 to ES in total RNA, direct percent R2/(R2+ES) values obtained from RT-PCR for several mutants were compared to indirect percent R2/(R2+ES) values obtained by using quantitative RNase protection analyses with probes A and B. For an example of these analyses, see Fig. 5. First, RNase protection probe A (Fig. 1) was used to determine the ratio of molecules using the upstream intron donor at nt 514 (R2+ES) relative to R1 [(R2+ES)/R1; see Fig. 5B and Table 1]. RNase protection probe B (Fig. 1) was then used to establish a quantitative ratio of R2 molecules relative to R1 molecules (R2/R1; see Fig. 5C and Table 1). These analyses then enabled the indirect determination of the ratio between R2 and (R2+ES) [indirect percent R2/(R2+ES); Table 1]. Direct percent R2/(R2+ES) values obtained by quantitative RT-PCR (see Fig. 5D and summary in Fig. 5A) varied by no more than 7% from the indirect percent R2/(R2+ES) values obtained from comparison of quantitative RNase protection assays (Table 1) when tested multiple times with a panel of 15 different mutants with different percent R2/(R2+ES) values (13). For example, compare RT-PCR and RNase protection values for wild-type pΔD1/2, p1TΔD1/2, p2TΔD1/2, and p4TΔD1/2 constructs (see Fig. 5A and Table 1). RNase protection assays also showed that mutant RNAs accumulated to wild-type levels. Furthermore, the direct percent R2/(R2+ES) value obtained from RT-PCR varied by no more than 4% over a series dilution of template cDNA and over a range of 15, 20 or 30 cycles (13). Such analyses allowed us to obtain an indirect percent R2/(R2+ES) value (Table 1) which varied by no more than 7% from the direct percent R2/(R2+ES) value obtained by the RT-PCR assay (Fig. 5A).
TABLE 1.
Indirect measure of the percent R2 relative to (R2+ES) molecules in mRNA generated by either wild-type MVM or the mutants indicated, as determined by RNase protection assays with probes A and B
| Mutant | (R2+ES)/R1a | R2/R1b | Indirect %R2/(R2+ES)c |
|---|---|---|---|
| WT | 1.91 ± 0.05 | 1.89 ± 0.08 | 98.3 ± 1.1 |
| pΔD1/2 | 5.26 ± 0.07 | 0.091 ± 0.003 | 1.6 ± 2.1 |
| p1TΔD1/2 | 7.10 ± 0.26 | 0.43 ± 0.04 | 6.0 ± 1.4 |
| p2TΔD1/2 | 5.40 ± 0.16 | 1.78 ± 0.07 | 33.0 ± 3.0 |
| p4TΔD1/2 | 7.90 ± 0.22 | 7.10 ± 0.10 | 89.6 ± 4.3 |
| p5Ad | 0.09 ± 0.01 | 0.010 ± 0.007 | 11.0 ± 2.4 |
| p1989d | 0.05 ± 0.01 | 0.011 ± 0.010 | 22.0 ± 7.2 |
Ratio of RNAs using the upstream intron 5′ splice site at nt 514 (R2+ES) relative to R1, as determined by RNase protection assays with probe A (Fig. 1). The values shown are the averages of at least three separate experiments, an example of which is shown in Fig. 5B. Standard deviations are indicated.
Ratio of R2 relative to R1, as determined by RNase protection assays with probe B (Fig. 1). The values shown are the averages of at least three separate experiments, an example of which is shown in Fig. 5C. Standard deviations are indicated.
The indirect percent R2/(R2+ES) value was determined from the (R2+ES)/R1 ratio (column 2) and the R2/R1 ratio (column 3), as described in Materials and Methods. The values shown are the averages of at least three separate experiments. Standard deviations are indicated.
Although the indirect percent R2/(R2+ES) value was determined from the (R2+ES)/R1 ratio and the R2/R1 ratio with probes A and B, respectively, for these mutants, only an example of RNase protection analysis with probe A is shown in Fig. 5B.
For some of the mutants, the relative amounts of R1, R2, and ES were also calculated as a percentage of the total P4-generated product (see Table 2). These calculations, for all mutants except p5AΔHS, were done by using the (R2+ES)/R1 ratio (obtained by RNase protection analysis with probe A), the R2/R1 ratio (obtained by RNase protection analysis with probe B), and the indirect %R2/(R2+ES) value (obtained as explained above). For p5AΔHS, these calculations were done by using the (R2+ES)/R1 ratio (obtained by RNase protection analysis with probe A) and the direct percent R2/(R2+ES) value (obtained from quantitative RT-PCR).
TABLE 2.
Relative amounts of R1, R2, and ES as a percentage of total P4-generated product, calculated from RNase protection analysis (with probes A and B) of RNA generated by either wild-type MVM or the mutants indicated, as described in Materials and Methods
| Mutant | % of P4 producta
|
||
|---|---|---|---|
| R1 | R2 | ES | |
| WT | 34.1 ± 2.1 | 65.2 ± 1.9 | 1.2 ± 0.8 |
| p5A | 92.6 ± 1.5 | 1.5 ± 0.8 | 6.2 ± 1.1 |
| pΔHS | 32.2 ± 2.5 | 63.1 ± 1.3 | 5.4 ± 1.5 |
| p5AΔHSb | 25.6 ± 1.7 | 9.2 ± 2.5 | 66.1 ± 3.2 |
| p1989 | 99.7 ± 0.2 | 0.10 ± 0.05 | 0.4 ± 0.1 |
Values represent the averages of at least three separate experiments. Standard deviations are indicated.
For this mutant, the values of R1, R2, and ES, as a percentage of total P4 product, were calculated from RNase protection analysis with probe A (Fig. 1) and quantitative RT-PCR analysis, as described in Materials and Methods.
RESULTS
A bipartite enhancer within the NS2-specific exon is required for inclusion of this exon in mature spliced mRNA.
To assess the relative accumulated levels of the P4-generated R2 and ES, we first established a quantitative RT-PCR assay, whose results were validated indirectly by quantitative RNase protection assays as described in Materials and Methods. We have previously shown that sequences within the NS2-specific exon are required for its inclusion in mature spliced mRNA (56). To further define the sequences required for efficient exon inclusion, RNA generated from mutants containing single deletions of regions within the exon was examined. Efficient inclusion of the NS2-specific exon was maintained following deletion of any of four regions alone (pΔSX, pΔXP, pΔPH, and pΔHS [Fig. 2A], as detected by quantitative RT-PCR in Fig. 2B [summarized in Fig. 2A]). RNase protection analysis confirmed that P4 RNA generated by all these mutants exhibited a wild-type ratio of R1 to R2 (13). Deletion of the SX and HS regions together (pΔSX+ΔHS, Fig. 2A), however, resulted in substantial skipping of the NS2-specific exon (Fig. 2A and C). RNase protection assays showed that pΔSX+ΔHS generated little R1 mRNA (13), demonstrating that the majority of the P4 product generated by the pΔSX+HS mutant skipped the NS2-specific exon. Deletions of approximately the same size as (pΔSma-Hinc) or somewhat larger than (pΔXho-Sac) pΔSX+ΔHS (Fig. 2A), which contain the HS and SX elements alone, respectively, still allowed substantial exon inclusion (Fig. 2A and C). These data suggested a model in which the SX and HS regions function as two essential elements that have significantly redundant functions in governing the inclusion of the NS2-specific exon into mature mRNA. The difference in exon inclusion between pΔSma-Hinc (which retains the HS region) and pΔXho-Sac (which retains the SX region), however, suggested that at least in the context of these smaller mutant exons, the HS region plays a more critical role in exon inclusion, an observation which is further supported by point mutagenesis of the HS region described below.
The SX and HS elements were each further subdivided into three regions, designated SX1 to SX3 and HS1 to HS3, respectively (Fig. 2A). All single mutants and all possible combinations of single mutants were tested, and all permitted near-wild-type levels of exon inclusion (13), except a mutant with both the SX1 and HS2 regions deleted (pΔSX1+ΔHS2; Fig. 2A and C). These results suggested that the 22-nt SX1 and the 53-nt HS2 regions contained the elements important for exon inclusion found in the larger SX and HS regions, respectively. The SX1 region contains two CA-rich elements separated by 6 nt and followed by a purine-rich sequence, while the HS2 region contains two purine-rich elements separated by a 22-nt spacer (see Fig. 3A). The NS2-specific exon was retained more efficiently in RNA generated by pΔSX1+ΔHS2 than it was for pΔSX+ΔHS (Figs. 2A and C); however, the level of exon inclusion in RNA generated by pΔSX1+ΔHS2 was similar to that seen for a mutant in which both the SX1 and the complete HS regions were deleted together (13). This suggested either that the larger SX region contained additional exon-defining elements or that in construction of the pΔSX1+ΔHS2 mutant an exon enhancer-like element was inadvertently created. Upon inspection, it was noted that the deletion junction generated in ΔSX1 formed a new 6-nt CA-rich motif (CCACTC) that could potentially function as such an enhancer element.
As expected, disruption of the SX1 element alone by point mutation [pSX1(−); Fig. 3A] sustained efficient inclusion of the NS2-specific exon (Fig. 3). Surprisingly, in contrast to results seen with deletion of either the complete HS region (pΔHS; Fig. 2A and B) or the HS2 region alone (13), disruption of the HS2 element alone by point mutation [pHS2(−); Fig. 3A] resulted in inefficient inclusion of the NS2-specific exon (to 34% as seen in Fig. 3). Thus, it appeared that the role of the purine-rich HS2 element was more significant for inclusion of the wild-type length exon; however, the possibility that a negatively acting element was created in pHS2(−) cannot be ruled out. It is unlikely that the deletion junction in pΔHS2 created a positively acting element, however, because the exon was similarly included in both pΔHS (Fig. 2) and pΔHS1+pΔHS2 (data not shown), which have different deletion junctions. Disruption of both the SX1 and HS2 elements together by point mutations [pSX1(−)+HS2(−); Fig. 3A] resulted in NS2-specific exon skipping (Fig. 3) to levels similar to that seen with deletion of the larger SX and HS regions together (pΔSX+ΔHS; Fig. 2A and B).
A CA-rich element and a purine-rich element together constitute the bipartite enhancer of the NS2-specific exon.
To determine which sequences within the SX1 and HS2 regions were the exon-defining motifs, we added both wild-type and mutant variants of the CA-rich element (CA1 and CA2, Fig. 4A) or the purine-rich element (AG1 and AG2; Fig. 4A) back to a mutant in which the SX1 and HS2 regions had been deleted (pΔSX1+ΔHS2; Fig. 4A). Replacement of either the SX1 or HS2 regions of pΔSX1+ΔHS2 with an oligonucleotide containing the two native CA-rich motifs separated by a 6-nt mutant spacer (CA2) [pSX1(CA2)+ΔHS2, pΔSX1+HS2(CA2); Fig. 4A] restored exon inclusion to the parent construct (as shown by quantitative RT-PCR [Fig. 4]) to the same level as achieved by replacement with the authentic wild-type CA-rich element (CA1) [pSX1(CA1)+ΔHS2 (Fig. 4); pΔSX1+HS2(CA1) (13)]. These results demonstrate that the CA-rich motifs within the SX1 region (and not the linker region) confer enhancer activity and that these sequences can function at either enhancer position, independent of the local sequence environment.
Replacement of the 3′ HS2 region of pΔSX1+ΔHS2 with oligonucleotides containing the native purine-rich sequences separated by a mutant spacer of the native length (AG2) or a shortened 5-nt mutant spacer (AG1) [pΔSX1+HS2(AG2) and pΔSX1+HS2(AG1) respectively; Fig. 4A] also resulted in efficient exon inclusion (Fig. 4), demonstrating that the purine-rich motifs within the HS2 element can confer enhancer activity. The correctly spaced purine rich element (AG2) was also capable of restoring efficient exon inclusion when it replaced the 5′ SX1 region in pΔSX1+ΔHS2 [pSX1(AG2)+ΔHS; Fig. 4]. Replacement of the SX1 region with the purine-rich sequence with a short spacer (AG1) [pSX1(AG1)+ΔHS; Fig. 4A], however, resulted in an even greater loss of exon inclusion (Fig. 4), suggesting that perhaps the native configuration was required to function at a location previously occupied by a CA-rich element.
These results demonstrated that the compositionally different CA and purine-rich elements were at least partially redundant and were able to function in a heterologous sequence context. These elements could functionally substitute for each other in inclusion of the NS2-specific exon at either their original locations or each other’s locations in a manner independent of the local sequence environment.
The requirement of either the downstream 5′ splice sites or the bipartite exonic enhancer for exon inclusion can be overcome by improvement of the upstream 3′-splice-site polypyrimidine tract.
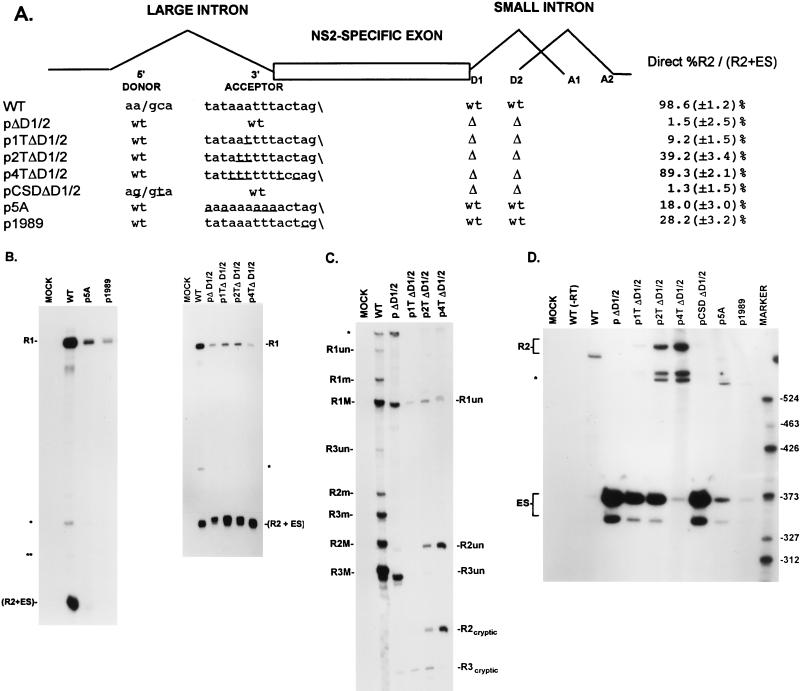
The native NS2-specific exon is flanked upstream by a poor 3′ splice site (the exon 5′ terminus) and downstream by an overlapping small intron with fairly consensus, although closely spaced, 5′ splice sites (the exon 3′ terminus). Deletion of the two downstream 5′ splice sites, which leaves the NS2-specific exon without a strong terminus at either end, resulted in almost uniform skipping of this exon. This skipping could be suppressed by making even subtle improvements of the upstream 3′-splice-site polypyrimidine tract (pΔD1/2, p2TpptΔD1/2, p4TpptΔD1/2 [Fig. 5A]; shown by quantitative RT-PCR analysis and comparative RNase protection analysis in Fig. 5 and Table 1, as explained above) but not by changing the upstream 5′ splice site to consensus (pCSDΔD1/2 [Fig. 5A]; shown by RT-PCR analysis in Fig. 5A and D]. When the downstream 5′ splice sites were disabled, a cryptic donor within the NS2-specific exon was used (R2cryptic; Fig. 5C). Improvement of the upstream 3′-splice-site polypyrimidine tract also substantially restored exon inclusion in RNA generated by a mutant lacking the bipartite exon enhancer (p4TΔSX+ΔHS [Fig. 6A]; shown by RT-PCR analysis in Fig. 6A and C), and increased utilization of the cryptic donor (R2cryptic; Fig. 5C and D), while improvement of the upstream 5′ splice site in the same mutant (pCSDΔSX+ΔHS; Fig. 6A) again had no effect (shown by RT-PCR analysis in Fig. 6A and C). RNase protection analyses with probe A (which detects all P4 products) revealed that the majority of the P4 product in p4TΔSX+ΔHS were R2 mRNAs (13). Thus, the requirement for the bipartite exon enhancer, as well as for the downstream 5′ splice site, could be overcome by improving the upstream 3′-splice-site polypyrimidine tract, perhaps by strengthening interactions at this weak polypyrimidine tract.
FIG. 6.
Improvement of the upstream intron 3′-splice-site polypyrimidine tract can overcome exon skipping caused by deletion of the bipartite exon enhancer. (A) The restriction sites within the NS2-specific exon (SmaI, XhoI, PstI, HincII, and SacI) which were used to generate the exon deletion mutants and the sequences of the large-intron 5′-splice-site and 3′-splice-site polypyrimidine tract and cleavage site in the wild type and mutants are shown underneath their appropriate map positions (deviations from the wild-type sequence are underlined), together with quantitations of the direct percent R2/(R2+ES) ratio obtained by RT-PCR analysis for each mutant. All the values are the averages of at least three separate experiments. Standard deviations are indicated in parentheses. ES values for pΔD1/2 are shown for comparison; the mutation is shown in Fig. 5A. wt, wild-type sequence. Δ, deletion. (B) RNase protection analyses with probe A (Fig. 1) of RNA generated by wild-type MVM (WT), mutants (as described in the text), or mock transfected, as designated at the top of each lane. The identities of the protected bands are shown on the left and explained in the legend to Fig. 5B. (C) RT-PCR analysis of RNA generated by wild-type MVM (WT), mutants (as described in the text), or mock transfected, as designated at the top of each lane. Samples were run on a 6% acrylamide–urea gel. pΔD1/2 and WT(−RT) controls and MARKER (the sizes of the marker bands are shown on the left) are as described in the legend to Fig. 2B. RNA generated by the wild type showed a 658-nt amplified R2 product, while RNA generated by the mutants showed R2 products of sizes consistent with the sizes of the deletions in these mutants. As explained in the legend to Fig. 5D, two kinds of amplified ES, using either A1 or A2, were observed.
Surprisingly, however, when the already poor upstream 3′ splice site was completely disabled by mutation of either its cleavage site (p1989; Fig. 5A) or polypyrimidine tract (p5Appt; Fig. 5A), leaving the downstream 5′ splice sites intact, almost all the steady-state P4-generated mRNAs were R1 molecules, which included the NS2-specific exon but from which the large intron was not excised. Very little of either R2 or ES was generated (quantitative RT-PCR [Fig. 5A and B and 6A and C] and RNase protection analyses [Fig. 5B and Table 1] allowed the determination of the relative amounts of R1, R2 and ES product as a percentage of the total P4 product [Table 2], as explained in Materials and Methods). These results suggested that while either a strong upstream 3′ splice site or downstream 5′ splice site was sufficient to prevent skipping of the NS2-specific exon, a functional upstream intron 3′ splice site was required for inclusion as an internal exon into mature, doubly spliced R2 mRNA.
Interestingly, however, when the purine-rich element was deleted from a mutant in which the upstream 3′-splice-site polypyrimidine tract had been disabled (p5AΔHS; Fig. 6A), the NS2-specific exon was substantially skipped, as was the case for RNA generated by the exon enhancer double mutant pΔSX+ΔHS (quantitative RT-PCR [Fig. 6A and C] and RNase protection analysis [Fig. 6B] allowed the determination of the relative amounts of R1, R2 and ES as a percentage of total P4 product [Table 2], as explained in Materials and Methods).
That P4 RNAs generated by p5A included NS2-specific exon sequences as R1 (Table 2) while P4 RNAs generated by p5AΔHS predominantly skipped the NS2-specific exon (Table 2) suggested further that the purine-rich element could also functionally strengthen the downstream 5′ splice site and thereby prevent exon skipping when the upstream 3′ splice site was disabled (p5A). Disabling of both the HS element and the upstream 3′ splice site (p5AΔHS) resulted in loss of inclusion, presumably due to a functional inactivation of both exon termini.
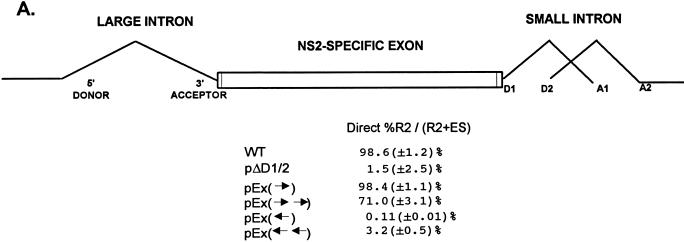
Surprisingly, when the NS2-specific exon was doubled in length by inserting two copies of the wild-type exon between engineered SmaI sites at nt 2005 and 2270, [pEx(→→); Fig. 7A], the larger exon was still included to 70% of wild-type levels (Fig. 7). Thus, inclusion of the NS2-specific exon was less sensitive to a dramatic increase in its length than might be expected for an internal exon.
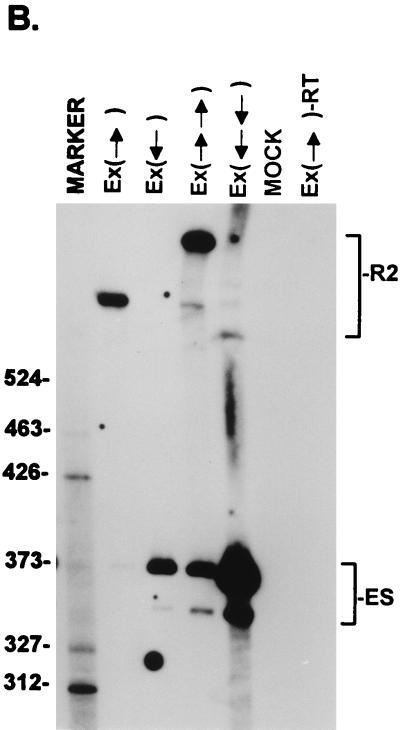
FIG. 7.
Inclusion of the NS2-specific exon tolerates doubling its length. (A) Quantitations of the direct percent R2/(R2+ES) ratio obtained by RT-PCR analysis for each mutant are shown. Construction of the mutants is explained in Materials and Methods. All the values are the average of at least three separate experiments. Standard deviations are indicated in parentheses. (B) RT-PCR analysis of RNA generated by wild-type MVM (WT), mutants (as described in the text), or mock transfected, as designated at the top of each lane. Samples were run on a 6% acrylamide–urea gel. MARKER is as described in the legend to Fig. 2B (the sizes of the marker bands are shown on the left). (Ex→)-RT is a control reaction with p(Ex→)-generated RNA but excluding reverse transcriptase. RNA generated by the wild type showed a 658-nt amplified R2 product, while RNA generated by the mutants showed R2 products of sizes consistent with the sizes of the substitutions in these mutants. As explained in the legend to Fig. 5D, two kinds of amplified ES, using either A1 or A2, were observed. The parent mutant bearing only the Sma sites at nt 2005 and 2270 [pEx(→)] is indistinguishable from the wild type with respect to splicing of the upstream intron and inclusion of the NS2-specific exon (56). Inclusion of the exon that is doubled in length still required wild-type exon sequences; when either one copy or two copies of the exon in the reverse orientation were inserted between the Sma sites [pEx(←) and pEx(←←), respectively (56)], the exon was almost uniformly skipped.
DISCUSSION
Identification and characterization of a partially redundant bipartite enhancer within the NS2-specific exon.
The NS2-specific exon bipartite enhancer was first identified by deletion and point mutagenesis (Fig. 2 and 3). This analysis indicated that the enhancer was composed of CA-rich and purine-rich elements within the 5′ and 3′ regions of the exon, respectively. Addition of both wild-type and mutant motifs to an exon mutant lacking the 5′ and 3′ enhancer regions resulted in reinclusion of the mutant exon (Fig. 4). These experiments demonstrated that the critical sequences within the upstream enhancer were two 5-nt CA-rich motifs separated by a 6-nt spacer and that the critical sequences within the downstream enhancer were two purine-rich motifs separated by a 22-nt spacer (Fig. 4A). These experiments also demonstrated that the CA-rich and purine-rich elements were able to function in a heterologous sequence context, i.e., at each other’s positions within the exon, to efficiently include the NS2-specific exon. This confirmed that these two elements were bona fide ESEs which could function independently of the local sequence environment and also implied that these elements did not form critical secondary structures with the other exon sequences. Experiments to determine whether the CA-rich and purine-rich enhancer elements might stimulate the splicing of other heterologous 3′ splice sites and to determine which cellular factors they interact with are in progress. The sequences of the NS2-specific exon enhancer elements are very highly conserved among the rodent parvoviruses, and these viruses have a wide host range (8). This suggests that these enhancer sequences may interact with prevalent cellular factors. It has been previously shown that tissue-specific alternative splicing of exon 4 of the calcitonin gene in thyroid C cells is also governed by both purine-rich and non-purine-rich exon enhancer elements (49).
Analysis of deletion mutants in which one of the two enhancer elements was left intact, together with the add-back experiments described above, suggested that the CA-rich and purine-rich elements were significantly redundant in the context of an exon of less than wild-type length. In the context of the full-length exon, however, while point mutagenesis of the CA-rich element alone permitted wild-type exon inclusion by the purine-rich element, point mutagenesis of the purine-rich element alone resulted in significant exon skipping (Fig. 3). These results suggested that the two elements are not fully redundant in function and that the purine-rich element, which may play a more dominant role under native conditions, may not be as strictly required in a smaller exon that permits more efficient exon bridging. An alternative interpretation would be that the HS2(−) mutation created a negatively acting element, which would then imply that the HS2 region is also significantly redundant in the wild-type-length exon, as suggested by deletion analysis.
In the context of wild-type virus, the CA-rich region may have a modifying effect on the more dominant purine-rich region; however, the importance of the CA-rich element was underscored by the observations that the CA-rich motif alone was shown to support significant exon inclusion in a mutant with the HS region deleted (pΔHS; Fig. 2) and that replacement of the 3′ HS2 region in pΔSX1+ΔHS2 with a CA-rich motif recovered exon inclusion to near-wild-type levels [pΔSX1+HS2(CA2); Fig. 4]. That the CA-rich and purine-rich elements are partially redundant and show significant position independence suggests that they may bind a common subset of cellular proteins. It has been previously determined that a CA-rich enhancer substrate as well as the cTNT purine-rich enhancer substrate can titrate factors required for CA-rich enhancer activity in vitro, suggesting that at least one of the factors that binds to the purine-rich enhancer is required for CA-rich enhancer activity (9).
The spacer sequence within the purine-rich element contains two potential 5′ splice donor sites. When both the downstream small intron donors (3′ terminus of the exon) were disabled, one of these sites was, in fact, used as a cryptic donor to splice out the small intron (R2cryptic generated [Fig. 5C]). Perhaps one consequence of this arrangement of the purine-rich motifs is to prevent the U1 small nuclear ribonucleoprotein (U1snRNP) and other splicing factors from assembling at these cryptic donors.
The NS2-specific exon is unusual in that it is flanked by a large intron upstream that has both a nonconsensus 5′ splice site and a poor 3′ splice site and by a small overlapping intron downstream. This exon requires both a bipartite exonic enhancer as well as a downstream 5′ splice site for efficient exon inclusion, suggesting that the NS2-specific exonic enhancer requires exon bridging involving the downstream 5′ splice site in order to function efficiently in vivo. The existence of multiple exon bridging interactions between the two exon-defining elements at both ends of the exon, as well as the downstream small intron, may also be reflected in the observation that the NS2-specific exon is less sensitive to a doubling of its length than might be expected for an internal exon.
Improvement of the upstream intron 3′ splice site polypyrimidine tract substantially restored exon inclusion to a mutant in which both enhancer elements had been deleted (Fig. 6) and to a mutant in which both downstream intron 5′ splice sites had been deleted (Fig. 5). The simplest explanation for this observation is that both the bipartite enhancer and the downstream 5′ splice sites strengthen interactions at the upstream 3′-splice-site polypyrimidine tract. This explanation was supported by the observation that when the 3′ purine-rich element was absent, either disabling the upstream 3′ splice site polypyrimidine tract or deleting the CA-rich element had similar effects (i.e., exon skipping), suggesting that one function of the CA-rich element was to strengthen interactions at the upstream 3′-splice-site polypyrimidine tract.
Disabling the upstream 3′ splice site of the NS2-specific exon does not result in exon skipping.
It has been reported that mutations of the AG dinucleotide at 3′ splice sites disrupt downstream exon inclusion, leading either to the utilization of a downstream AG or to skipping of the exon following the mutation, as predicted by the exon definition model (47, 48 reviewed in references 2 and 3). Surprisingly, when the already poor 3′ splice site upstream of the NS2-specific exon was completely disabled, by mutation of either its cleavage site (p1989) or its polypyrimidine tract (p5A), almost all the steady-state P4-generated mRNAs were R1 molecules which included the NS2-specific exon but from which the large intron was not excised. Very little of either R2 or ES was generated (see Results and Table 2). Perhaps this is related to the presence of the exon-defining elements at each exon terminus and strong downstream 5′ splice sites which together function to include the NS2-specific exon in such a way that it can be included at required levels in both R1 and R2.
The observation that the NS2-specific exon was included as R1 in P4 RNAs generated by p5A but was skipped in RNAs generated by p5AΔHS (see Results and Table 2) is consistent with a model in which the HS element also functioned downstream to improve the downstream 5′ splice sites. Deletion of the HS element in p5A (p5AΔHS) resulted in a loss of exon inclusion, presumably because both exon termini were disabled. The NS2-specific exon sequences in R1 mRNA are part of a very long (2,300-nt) 5′-terminal exon, which may necessitate the presence of an exon-defining element (HS) at the 3′ end of this exon. Since the CA-rich enhancer probably functions to strengthen the upstream 3′ splice site whereas the purine-rich enhancer probably functions to strengthen both the upstream 3′ splice site and the downstream 5′ splice sites, we suggest that these two enhancer elements may function to control, in a constitutive manner, the relative efficiency with which the NS2-specific exon sequences are included in both R1 and R2 and thereby govern the appropriate relative steady-state levels of R1 and R2 that accumulate during viral infection.
ACKNOWLEDGMENTS
This work was supported by PHS grant RO1 AI21302 from NIAID and a grant from the Council for Tobacco Research. A.G. was partially supported by the University of Missouri Molecular Biology Program during a portion of this work.
We thank members of our laboratory for helpful discussions, especially Don Haut for a critical review of the manuscript. We thank Lisa Burger for expert technical assistance. We are grateful to Tom Cooper and Andrew McCullough for help in developing our quantitative RT-PCR assay.
REFERENCES
- 1.Astell C R, Gardiner E M, Tattersall P. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J Virol. 1986;57:656–669. doi: 10.1128/jvi.57.2.656-669.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2444. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 3.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 4.Buvoli M, Mayer S A, Patton J G. Functional crosstalk between exon enhancers, polypyrimidine tracts and branchpoint sequences. EMBO J. 1997;16:7174–7183. doi: 10.1093/emboj/16.23.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemens K E, Cerutis D R, Burger L R, Yang G Q, Pintel D. Cloning of minute virus of mice cDNAs and preliminary analysis of individual viral proteins expressed in murine cells. J Virol. 1990;64:3967–3973. doi: 10.1128/jvi.64.8.3967-3973.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens K E, Pintel D J. Minute virus of mice (MVM) mRNAs predominantly polyadenylate at a single site. Virology. 1987;160:511–514. doi: 10.1016/0042-6822(87)90028-6. [DOI] [PubMed] [Google Scholar]
- 6a.Cooper, T. Personal communication.
- 7.Cotmore S F, Tattersall P. Organization of nonstructural genes of the autonomous parvovirus minute virus of mice. J Virol. 1986;58:724–732. doi: 10.1128/jvi.58.3.724-732.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 9.Coulter L R, Landree M A, Cooper T A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dirksen W P, Hampson R K, Sun Q, Rottman F M. A purine-rich exon sequence enhances alternative splicing of bovine growth hormone pre-mRNA. J Biol Chem. 1994;269:6431–6436. [PubMed] [Google Scholar]
- 11.Dominski Z, Kole R. Identification of exon sequences involved in splice site selection. J Biol Chem. 1994;269:23590–23596. [PubMed] [Google Scholar]
- 12.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 13.Gersappe, A., and D. J. Pintel. 1998. Unpublished data
- 14.Gontarek R R, Derse D. Interactions among SR proteins, an exonic splicing enhancer, and a lentivirus Rev protein regulate alternative splicing. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham I R, Hamshere M, Eperon I C. Alternative splicing of a human α-tropomyosin muscle-specific exon: identification of determining sequences. Mol Cell Biol. 1992;12:3872–3882. doi: 10.1128/mcb.12.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haut D D, Pintel D J. Intron definition is required for excision of the minute virus of mice small intron and definition of the upstream exon. J Virol. 1998;72:1834–1843. doi: 10.1128/jvi.72.3.1834-1843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman B E, Grabowski P J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 18.Humphrey M D, Bryan J, Cooper T A, Berget S M. A 32-nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol Cell Biol. 1995;15:3979–3988. doi: 10.1128/mcb.15.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang D Y, Cohen J B. Base pairing at the 5′ splice site with U1 small nuclear RNA promotes splicing of the upstream intron but may be dispensable for splicing of the downstream intron. Mol Cell Biol. 1996;16:3012–3022. doi: 10.1128/mcb.16.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Innis M A, GelFand D H, Sninski J J, White T J. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. [Google Scholar]
- 21.Jongeneel C V, Sahli R, McMaster G K, Hirt B. A precise map of splice junctions in the mRNAs of minute virus of mice, an autonomous parvovirus. J Virol. 1986;59:564–573. doi: 10.1128/jvi.59.3.564-573.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz R A, Skalka A M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990;10:696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo H C, Nasim F H, Grabowski P J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991;251:1045–1051. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- 24.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 25.Lynch K W, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 26.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 27.McCullough A J, Berget S M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan W R, Ward D C. Three splicing patterns are used to excise the small intron common to all minute virus of mice RNAs. J Virol. 1986;60:1170–1174. doi: 10.1128/jvi.60.3.1170-1174.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naeger L K, Cater J, Pintel D J. The small nonstructural protein (NS2) of MVM is required for efficient DNA replication and infectious virus production in a cell-type specific manner. J Virol. 1990;64:6166–6175. doi: 10.1128/jvi.64.12.6166-6175.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naeger L K, Schoborg R V, Zhao Q, Tullis G E, Pintel D J. Nonsense mutation inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992;6:1107–1111. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- 31.Pintel D J, Dadachanji D, Astell C R, Ward D C. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 1983;11:1019–1038. doi: 10.1093/nar/11.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pintel D J, Gersappe A, Haut D, Pearson J. Determinants that govern alternative splicing of parvovirus pre-mRNAs. Semin Virol. 1995;6:283–290. [Google Scholar]
- 33.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 35.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan K J, Cooper T A. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoborg R V, Pintel D J. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- 38.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 41.Talerico M, Berget S M. Effect of 5′ splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990;10:6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian H, Kole R. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian M, Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992;256:237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- 45.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 46.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 47.Tromp G, Prockop D J. Single base mutation in the proalpha2(I) collagen gene that causes efficient splicing of RNA from exon 27 to exon 29 and synthesis of a shortened but in-frame proalpha2(I) chain. Proc Natl Acad Sci USA. 1988;85:5254–5258. doi: 10.1073/pnas.85.14.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulfendahl J P, Kreivi J, Askujarvi G. Role of the branch/3′-splice site region in adenovirus-2 EIA pre-mRNA alternative splicing: evidence for 5′ and 3′-splice site cooperation. Nucleic Acids Res. 1989;17:925–938. doi: 10.1093/nar/17.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Oers C C M, Adema G J, Zandberg H, Moen T C, Baas P D. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol Cell Biol. 1994;14:951–960. doi: 10.1128/mcb.14.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Manley J L. Regulation of pre-mRNA splicing in metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, Hoffmann H M, Grabowski P J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 52.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 53.Xu R, Teng J, Cooper T A. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeakley J M, Hedjran F, Morfin J-P, Merillat N, Rosenfeld M G, Emeson R B. Control of calcitonin/calcitonin gene-related peptide pre-mRNA processing by constitutive intron and exon elements. Mol Cell Biol. 1993;13:5999–6011. doi: 10.1128/mcb.13.10.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Q, Gersappe A, Pintel D J. Efficient excision of the upstream large intron from P4-generated pre-mRNA of the parvovirus minute virus of mice requires at least one donor and the 3′ splice site of the small downstream intron. J Virol. 1995;69:6170–6179. doi: 10.1128/jvi.69.10.6170-6179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Q, Mathur S, Burger L R, Pintel D J. Sequences within the parvovirus minute virus of mice NS2-specific exon are required for inclusion of this exon into spliced steady-state RNA. J Virol. 1995;69:5864–5868. doi: 10.1128/jvi.69.9.5864-5868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Q, Schoborg R V, Pintel D J. Alternative splicing of pre-mRNAs encoding the nonstructural proteins of minute virus of mice is facilitated by sequences within the downstream intron. J Virol. 1994;68:2849–2859. doi: 10.1128/jvi.68.5.2849-2859.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]