Abstract
Background and Aims
Endogenous pararetroviruses (EPRVs) are widespread components of plant genomes that originated from episomal DNA viruses of the Caulimoviridae family. Due to fragmentation and rearrangements, most EPRVs have lost their ability to replicate through reverse transcription and to initiate viral infection. Similar to the closely related retrotransposons, extant EPRVs were retained and often amplified in plant genomes for several million years. Here, we characterize the complete genomic EPRV fraction of the crop sugar beet (Beta vulgaris, Amaranthaceae) to understand how they shaped the beet genome and to suggest explanations for their absent virulence.
Methods
Using next- and third-generation sequencing data and genome assembly, we reconstructed full-length in silico representatives for the three host-specific EPRVs (beetEPRVs) in the B. vulgaris genome. Focusing on the endogenous caulimovirid beetEPRV3, we investigated its chromosomal localization, abundance and distribution by fluorescent in situ and Southern hybridization.
Key Results
Full-length beetEPRVs range between 7.5 and 10.7 kb in size, are heterogeneous in structure and sequence, and occupy about 0.3 % of the beet genome. Although all three beetEPRVs were assigned to the florendoviruses, they showed variably arranged protein-coding domains, different fragmentation, and preferences for diverse sequence contexts. We observed small RNAs that specifically target the individual beetEPRVs, indicating stringent epigenetic suppression. BeetEPRV3 sequences occur along all sugar beet chromosomes, preferentially in the vicinity of each other and are associated with heterochromatic, centromeric and intercalary satellite DNAs. BeetEPRV3 members also exist in genomes of related wild species, indicating an initial beetEPRV3 integration 13.4–7.2 million years ago.
Conclusions
Our study in beet illustrates the variability of EPRV structure and sequence in a single host genome. Evidence of sequence fragmentation and epigenetic silencing implies possible plant strategies to cope with long-term persistence of EPRVs, including amplification, fixation in the heterochromatin, and containment of EPRV virulence.
Keywords: Beta vulgaris, sugar beet, endogenous pararetrovirus, Caulimoviridae, Florendovirus, retrotransposon, in situ hybridization
INTRODUCTION
Endogenous pararetroviruses (EPRVs) are viral double-stranded nucleic acids that permanently reside in the genome of their host. In plants, the ancestral EPRV progenitors are exogenous viruses of the Caulimoviridae family (caulimovirids) that integrated into the host nuclear genome through illegitimate recombination several million years ago (Jakowitsch et al., 1999; Diop et al., 2018). Caulimoviruses are reverse-transcribing viruses (Ortervirales; reviewed by Krupovic et al., 2018; Teycheney et al., 2020), although, in contrast to the closely related retroviruses (e.g. HIV; International Committee on Taxonomy of Viruses, 2019), integration into the host genome is not obligatory for their replication. Over time, nearly all EPRVs underwent fragmentation and rearrangements within the host genome, thus losing their activity. However, in some cases these ancient integrated viral sequences can be activated through reverse transcription and recombination, thereby forming virulent episomes often associated with diseases (reviewed by Staginnus and Richert-Pöggeler, 2006; Chabannes and Iskra-Caruana, 2013; Kuriyama et al., 2020).
As EPRVs exist in a broad range of vascular plants (Diop et al., 2018; Gong and Han, 2018) and cover a wider spectrum of genera than the exogenous caulimovirids, an extinction of several homologous episomal counterparts is repeatedly assumed (reviewed by Chen and Kishima, 2016). In general, the caulimoviral genera are characterized by differences in their nucleotide sequence and the organization of the viral genome, e.g. the number of open reading frames (ORFs) and the arrangement of essential protein domains within them. For instance, petuviruses typically have one single ORF (Richert-Pöggeler et al., 2003), whereas caulimoviruses and solendoviruses differ in the allocation of the protein domains in their four ORFs (Geering et al., 2010). Using gene order and ORF arrangements, further genera were defined: among these, the florendoviruses (Flora endogenous viruses; FEVs) encode the characteristic movement protein (MP), the coat protein with a zinc finger motif (ZF), the aspartic protease (AP), the reverse transcriptase (RT) and the ribonuclease H1 (RH) on the first of two overlapping ORFs (Geering et al., 2014). The FEVs are among the most abundant EPRVs in the plant kingdom (Geering et al., 2014; Bombarely et al., 2016; Diop et al., 2018), occurring in economically important plants such as Elaeis guineensis (oil palm), Gossypium raimondii (cotton), Citrus × sinensis (orange), Glycine max (soybean), Petunia sp. and Beta vulgaris (sugar beet). Although EPRVs have been detected in many plant genomes, it has not yet been possible to resolve their complex organization in the deep heterochromatin.
In sugar beet, EPRVs contribute ~0.4–0.5 % of the genome (Dohm et al., 2014; Diop et al., 2018). As we do not know of any outbreaks of associated diseases, beet’s endogenous viral sequences have likely been assimilated by the host. Therefore, beet may represent a suitable organism to study how the host genome buries, disassembles and inactivates potentially destructive sequences.
Sugar beet is one of the most important crops of the moderate climate zones, contributing ~14 % of the world’s sugar production (FAOSTAT, 2017). Cultivated beet species and related wild beets belong to the sister genera Beta and Patellifolia within the Amaranthaceae. According to Ulbrich (1934) and Frese et al. (2000), the genus Beta can be further subdivided into the three sections Beta, Corollinae and Nanae. A comparison with wild beet genomes may offer an insight into the acquisition of EPRVs in the Beta genus.
Reference genome sequences and long-read information are already available for two B. vulgaris genotypes (Dohm et al., 2014; Funk et al., 2018; McGrath et al., 2020). Similar to the euchromatic genic regions of beet, its heterochromatin is well studied (Schmidt and Heslop-Harrison, 1998) and consists in large part of satellite DNA (satDNA), such as the centromeric satDNA family pBV (Schmidt and Metzlaff, 1991; Zakrzewski et al., 2013) and the intercalary satDNA family pEV (Schmidt et al., 1991).
Here, a combination of bioinformatics, advanced genomics and molecular cytogenetics is used to investigate how the genome of beet may repress and disassemble EPRVs. For this, we characterize the EPRV landscape in the sugar beet genome and resolve the highly repetitive environment. Finally, we test whether beetEPRVs are targeted for silencing by small RNAs (smRNAs), presumably involved in protecting the genome from subsequent beetEPRV infection. Thus, we aim to illustrate the variety EPRVs can attain in a single host and within the same genus, and we provide a possible explanation for the ability of EPRVs to escape elimination after ancient infection events – by integration into preserved heterochromatic genomic environments and by contribution to the host’s defence against EPRV-derived pathogens.
MATERIALS AND METHODS
Bioinformatic identification of B. vulgaris-specific EPRVs
To enable a targeted EPRV detection, we collected 13 publicly available EPRVs, including the nine sequences from gydb.org (Llorens et al., 2009, 2011), FriEPRV (Becher et al., 2014) and the FEVs AtrichBV, GmaxV and LjapAV (Geering et al., 2014). Subsequently, representative sequences of beetEPRV1, beetEPRV2 and beetEPRV3 were added (Supplementary Data S1–S5). The EPRV reference set therefore contained 16 sequences in total, representing the caulimovirid genera Petuvirus, Badnavirus, Caulimovirus, Cavemovirus, Solendovirus, Soymovirus, Tungrovirus and Florendovirus (Supplementary Data Table S1). After identification of their MP and RT domains, we aligned the respective nucleic acid sequences using MAFFT (Katoh and Standley, 2013) followed by manual refinement to build nucleotide hidden Markov models (nHMMs).
Using these nHMMs with the nhmmer tool (Wheeler and Eddy, 2013), we identified the EPRV RT and MP sequences from the sugar beet EL10.1 assembly (Funk et al., 2018; McGrath et al., 2020) as well as the corresponding single-molecule real-time (SMRT) reads (GenBank accession number SRX3402137). The results were parsed to analyse the hits, choose cut-off parameters and extract the corresponding sequences. After parameter analysis (Supplementary Data Fig. S1), we selected all detected 262 (assembly) and 350 (SMRT reads) MP hits for further analysis. In contrast, to avoid cross-detection of similar Ty3-gypsy RTs, 125 assembly-derived and 320 SMRT read-derived RT sequences with an nHMM coverage of at least 200 bp and an nHMM coverage/bitscore quotient between 1.5 and 2.5 were considered. From the assembly RT hits, another six candidates were excluded: five showed a high degree of fragmentation and one represented a Ty3-gypsy sequence. Therefore, 119 assembly RT hits remained for our analysis. Visualizations of the nhmmer search results were created using Python v. 2.7 with the seaborn package (Waskom et al., 2018).
To identify potentially intact beetEPRV members, we screened the flanking region (±8 kb) of the beetEPRV RTs for adjacent MP domains, subsequently excluding RT fragments.
Sequence analyses and comparisons
Multiple sequence alignments were calculated with MUSCLE and MAFFT (Edgar, 2004; Katoh and Standley, 2013), followed by manual refinement. To generate representative reference sequences for each beetEPRV sequence cluster (Supplementary Data S1), we built consensuses from alignments of 11–42 sequences, respectively (Supplementary Data S2–5, Table S2). Secondary structures for the beetEPRV consensus elements were predicted with JPred 4.0 (Drozdetskiy et al., 2015). The presumed weights of the encoded proteins were determined by the Protein Molecular Weight Calculator (sciencegateway.org/tools/proteinmw.htm).
To assign the beetEPRVs to a caulimovirid genus, we initially used the neighbour-joining algorithm (Saitou and Nei, 1987) embedded in Geneious 6.1.8 (https://www.geneious.com; Kearse et al., 2012) and for confirmation we used the maximum likelihood method, the UPGMA method, and the minimum evolution method integrated in MEGA X (Kumar et al., 2018). Here, beetEPRV amino acid sequences were compared with the EPRV references already used for the nhmmer analysis, which were later complemented with the 31 remaining FEVs described by Geering et al. (2014). As outgroup the two sugar beet long terminal repeat (LTR) retrotransposons, Beetle7 and Elbe2 of the Ty3-gypsy family, were selected (Wollrab et al., 2012; Weber et al., 2013).
To assess the genomic environment of beetEPRV copies individually, we visually inspected self dotplots of all members identified from the sugar beet assembly and the SMRT reads. Dotplots for a total of 514 sequences containing either MP or RT nhmmer hits or both of them with up to 8000 bp of flanking regions were generated automatically using the tool FlexiDot (Seibt et al., 2018) with a wordsize of 9. With this method, we were able to identify and analyse characteristic repetitive regions that appear up- and downstream of the full-length sequences, hereinafter called terminal repeats (TRs).
We manually refined annotations of the beetEPRV members detected in the sugar beet assembly and the SMRT reads. Boxplots illustrating the sequence lengths of the beetEPRV members (derived from their chromosomal position; Supplementary Data Table S2) were generated using ggplot2 (Wickham, 2009) implemented in R (R Core Team, 2018). The whiskers comprise all underlying data points.
Search for beetEPRV transcripts and smRNA mapping
A publicly available cDNA library of B. vulgaris (GenBank accession number SRX674050) was searched for beetEPRV transcripts using blastn. Small RNA reads (Zakrzewski et al., 2011) were mapped to the consensus sequences of the three beetEPRV sequence clusters using the built-in mapping tool in Geneious 6.1.8 (https://www.geneious.com; Kearse et al., 2012) with medium-low sensitivity and up to five iterations. Reads harbouring insertions or deletions were discarded using a custom Python script. Read position, length, orientation and counts were scored for graphical illustration by Python 2.7 using NumPy (Oliphant, 2006) and Matplotlib (Tosi, 2009).
Plant material and genomic DNA extraction
Seeds of the B. vulgaris ssp. vulgaris genotype KWS 2320 were obtained from KWS Saat, Einbeck, Germany. Five other Beta and Patellifolia accessions as well as two further genera of the Amaranthaceae were analysed: B. vulgaris ssp. vulgaris convar. cicla (chard ‘Vulkan’), B. maritima (BETA 1233), B. patula (BETA 548), B. lomatogona (BETA 674), B. nana (BETA 541), Patellifolia patellaris (BETA 534), Chenopodium quinoa (CHEN 125) and Spinacia oleracea (‘Matador’). The seeds were obtained from the Leibniz Institute of Plant Genetics and Crop Plant Research Gatersleben, Germany. The plants were grown under long-day conditions in a greenhouse. Genomic DNA was isolated from young leaves using the cetyltrimethylammonium bromide (CTAB) standard protocol (Saghai-Maroof et al., 1984).
PCR amplification, cloning and sequencing of beetEPRV3 sequences
Standard PCR reactions of genomic B. vulgaris DNA were performed using primer pairs designed for the RT and MP sequences of beetEPRV3 (Supplementary Data Table S3). The PCR conditions were 94 °C for 3 min followed by 35 cycles of 94 °C for 1 min, primer-specific annealing temperature for 30 s, 72 °C for 45 s, and a final incubation at 72 °C for 5 min. PCR fragments were purified, cloned and commercially sequenced. Sequenced inserts with an identity of at least 99.5 % to the reference beetEPRV3 element were used as probes for the following hybridization experiments.
Southern hybridization
Genomic DNA of sugar beet and related species was restricted with different enzymes, separated on 1.2 % agarose gels and transferred onto membranes using alkaline transfer. We used random priming to radioactively label the beetEPRV3 probes (GenBank accession numbers LR812097 and LR812098), followed by hybridization according to Sambrook et al. (1989). Filters were hybridized at 60 °C and washed at the same temperature in 2 × saline sodium citrate (SSC)/0.1 % sodium dodecyl sulphate (SDS) and 1 × SSC/0.1 % SDS for 10 min each. Signals were detected by autoradiography.
Preparation of chromosome spreads
The meristem of young leaves was used for the preparation of mitotic chromosomes. For this, the leaves were treated for 3 h in 2 mm 8-hydroxyquinoline to accumulate metaphases, followed by fixation in 100 % methanol:glacial acetic acid (3:1). Fixed plant material was digested at 37 °C in the PINE enzyme mixture, consisting of 2 % (w/v) cellulase from Aspergillus niger (Sigma C-1184), 4 % (w/v) cellulase Onozuka R10 (Sigma 16419), 2 % (w/v) cytohelicase from Helix pomatia (Serva C-8274), 0.5 % (w/v) pectolyase from Aspergillus japonicus (Sigma P3026) and 20 % (v/v) pectinase from A. niger (Sigma P4716) in citrate buffer (4 mm citric acid and 6 mm sodium citrate). After maceration, the mix was incubated for another 30 min and centrifuged at 2200 × g for 5 min. The nuclei pellet was washed and resuspended in citrate buffer. To spread the chromosomes, 20 μL of the solution was dropped onto an ethanol-cleaned slide from a height of ~50 cm, as published by Heslop-Harrison et al. (1991) and modified for beet by Schmidt et al. (1994). Finally, the chromosomes were rinsed in methanol:glacial acetic acid fixative.
Fluorescent in situ hybridization
The beetEPRV3 probes (RT, GenBank accession number LR812097; MP, GenBank accession number LR812098) were labelled by PCR in the presence of digoxigenin-11-dUTP detected by antidigoxigenin-fluorescein isothiocyanate (FITC; both from Roche Diagnostics) and biotin-16-dUTP (Roche Diagnostics) detected by streptavidin-Cy3 (Sigma–Aldrich), respectively. The probe pZR18S, containing a part of the sugar beet 18S-5.8S-26S rRNA gene (HE578879; Dechyeva and Schmidt, 2009), and the probe pEV I, marking an intercalary sat DNA family (Schmidt et al., 1991; Kubis et al., 1998), were labelled with DY415-dUTP (Dyomics). The probe pXV1 for the 5S rRNA gene (Schmidt et al., 1994) and the probe pBV I for the centromeric satDNA family (Schmidt and Metzlaff, 1991; Kubis et al., 1998) were labelled with DY647-dUTP (Dyomics). Chromosomes were counterstained with DAPI (4′,6′-diamidino-2-phenylindole; Böhringer, Mannheim) and mounted in antifade solution (CitiFluor).
The hybridization and rehybridization procedure was performed as described previously (Schmidt et al., 1994) with a stringency of 82 %. Slides were examined with a fluorescent microscope (Zeiss Axioplan 2 imaging) equipped with appropriate filters. Images were acquired directly with the Applied Spectral Imaging v. 3.3 software coupled to a high-resolution CCD camera (ASI BV300-20A). After separate capture for each fluorochrome, the individual images were combined computationally and processed using Adobe Photoshop CS5 software (Adobe Systems, San Jose, CA, USA). We used only contrast optimization, Gaussian and channel overlay functions affecting all pixels of the image equally. Chromosomes were identified and numbers assigned following Paesold et al. (2012). For this, we considered the position of the rRNA genes (pairs 1 and 4) and the distribution and density of the sat DNAs pBV I and pEV I.
RESULTS
beetEPRVs can be grouped into three clusters according to their RT sequences
In order to identify endogenous caulimovirid sequences in the genome of sugar beet, we queried the high-quality B. vulgaris assembly EL10.1 (Funk et al., 2018) for caulimovirid MPs and RTs (Hansen and Heslop-Harrison, 2004) using individual nHMMs. As the RT is the key enzyme of all retroviral lineages (Xiong and Eickbush, 1990), we closely inspected all 119 RT matches. On nucleotide level, they showed identities of at least 50 % to the EPRV RT reference sequences (Llorens et al., 2009, 2011; Becher et al., 2014; Geering et al., 2014). We also included truncated RTs with at least two of the seven conserved RT domains as defined by Xiong and Eickbush (1988) and Hansen and Heslop-Harrison (2004) in our dataset. To allow a sequence comparison, the EPRV RT hits were aligned to each other; as outgroup, two Ty3-gypsy LTR retrotransposons from B. vulgaris were considered, Beetle7 and Elbe2 (Weber et al., 2013; Wollrab et al., 2012). A neighbour-joining tree (Fig. 1A) confirms a separation of all detected beet EPRV hits from the known Ty3-gypsy retrotransposons (validated by a maximum likelihood clustering; Supplementary Data Fig. S2). The beetEPRV RTs form three distinct clusters marked by nucleotide sequence identities of <79 % between each other. In contrast, they can attain up to 100 % identity within one cluster, often reflected by short branch lengths (Fig. 1A, insets). We named the three clusters beetEPRV1, beetEPRV2 and beetEPRV3 according to the abundance of the endogenous caulimovirid sequences: the majority (50.4 %; n = 60) of the 119 beetEPRV RTs belong to beetEPRV1, followed by beetEPRV2 with 30.3 % (n = 36) of the sequences. Cluster beetEPRV3 has the fewest members, with 19.3 % (n = 23) of the assigned sequences.
Fig. 1.
The diversity in sequence and structure of beetEPRVs leads to their classification into three clusters. (A) Dendrogram showing the relationships among the 119 EPRV RT hits found in the B. vulgaris genome. Two Ty3-gypsy RT sequences were used as outgroup (black), the chromovirus Beetle7 (GenBank accession number JX455085) and the errantivirus Elbe2 (GenBank accession HE598759). The dendrogram is drawn to scale, with branch length units corresponding to the evolutionary distance (p-distance). Circles with zoomed dendrogram segments are shown for the densely packed branches. (B) Element structure of full-length beetEPRV consensus sequences from each cluster. Grey boxes mark ORFs; changes in the vertical position indicate frameshifts. White boxes within the ORFs represent conserved protein domains. The PBSs complementary to the initiator tRNA of methionine are displayed. In addition, TRs are marked at the beginning and end of the sequences as hatched boxes scaled according to their length. The conservation of the sequence between the PBS and the first ORF of both beetEPRV2 components is indicated as dotted boxes. As beetEPRV1 and beetEPRV3 sequences are usually terminated by TA microsatellites, the TA-rich sites are shown as (TA)n. Black bars above the MP and RT of the beetEPRV3 scheme indicate probes used for FISH.
To identify full-length beetEPRV members, we searched the RT hits for flanking upstream MP domains and further EPRV protein domains. In total, we detected 22 full-length sequences (Supplementary Data Table S2, asterisks), 14 beetEPRV1 and 8 beetEPRV3 members with the EPRV-specific arrangement of the protein domains (MP-ZF-AP-RT-RH). Strikingly, no canonical beetEPRV2 sequences were found, either in the genome assembly or in the SMRT read data: nearby beetEPRV2 MP domains (±8 kb) were always separated from beetEPRV2 RT hits by additional primer binding sites (PBSs; Supplementary Data Fig. S3B).
The three beetEPRVs are characterized by distinct properties regarding their element structure, with beetEPRV1 and beetEPRV3 differing strongly from the structural organization of beetEPRV2 (Fig. 1B). Both beetEPRV1 and beetEPRV3 harbour all characteristic protein domains in an uninterrupted structure. BeetEPRV1 encodes a single, continuous ORF harbouring all protein domains, whereas beetEPRV3 has two overlapping ORFs. The large beetEPRV1 ORF is terminated by a poly(A) region, while none of the identified beetEPRV3 ORFs have a poly(A) stretch at the 3′ end (Fig. 1B). BeetEPRV1 and beetEPRV3 each contain a specific conserved region (~500 bp) downstream of their ORF(s) that is often repeated in a fragmented manner upstream of the PBS (Fig. 1B, Supplementary Data Fig. S3; hatched boxes). Due to its repeated nature and terminal position, we refer to it as a TR. All 20 beetEPRV1 members with an intact 5′ region (Supplementary Data Table S2, column ‘PBS’) also contain at least five TR nucleotides upstream of the PBS, thus creating a conserved 5′-TATCC-3′ motif.
In contrast, beetEPRV2 members are organized differently: they exhibit a bipartite structure with the MP and ZF domain on one entity (component A, beetEPRV2-A) and the AP–RT–RH complex on the other (component B, beetEPRV2-B). As a consequence, most of the beetEPRV2 sequences including the AP–RT–RH polyprotein are located apart from the dedicated MP–ZF domain, although co-occurrence of both entities was also found (Supplementary Data Fig. S3B). Both beetEPRV2 components also contain a non-functional ORF (ORF3 and ORF2, respectively; Fig. 1B). Strikingly, both beetEPRV2 components are characterized by independent PBS motifs. Their underlying sequence 5′-TGGTATC(A/C)GAGC-3′ is homologous to the initiator tRNA of methionine (tRNAMet) and the PBS of other EPRV genera (Hohn et al., 1985; Verver et al., 1987; Richert-Pöggeler and Shepherd, 1997). Similar to the poly(A) region of beetEPRV1, the 3′ TR of the beetEPRV2 components starts with an A-rich region that includes up to 31 adenines within a 34-bp window, potentially acting as a polyadenylation signal.
All detected B. vulgaris EPRVs belong to the FEVs
To exactly position the beetEPRVs within the caulimovirids, we compared their consensus coding sequence of key domains (in particular RT, MP and RT–RH) with selected representatives from eight caulimovirid genera, as well as the chromovirus Beetle7 and the errantivirus Elbe2 retrotransposons as outgroups (Fig. 2). We detected all seven conserved RT domains described by Xiong and Eickbush (1988) within each beetEPRV consensus sequence (Fig. 2A). The beetEPRVs also showed the seven β-strands within the MP core (Fig. 2B; Mushegian and Elena, 2015), which together form a secondary structure similar in all viral MPs (validated by the internet tool JPred 4.0; Drozdetskiy et al., 2015). We found a conserved HX25D motif that extends to the second and third β-strands of all analysed EPRV MPs (Fig. 2B, asterisks). Remarkably, an insertion of three amino acids between the sixth and seventh β-strands of the MP (Fig. 2B) is unique for beetEPRV2 and sets it apart from beetEPRV1 and beetEPRV3. Generally, all beetEPRVs show the highest pairwise amino acid identity to the FEVs (Supplementary Data Fig. S4). In both alignments, we detected discriminatory amino acids present in all analysed FEVs, including the beetEPRVs, which distinguished them from the other EPRV genera (Fig. 2, triangles; Supplementary Data Table S4).
Fig. 2.
Comparative amino acid alignments of conserved EPRV protein domains compared with reference sequences. (A) EPRV and Ty3-gypsy RT and (B) EPRV MP sequence alignments accentuate high similarities between the beetEPRVs and the FEVs. The shading reflects the similarity of the amino acids according to their physiochemical characteristics, and FEV-characteristic amino acids are distinguished by triangles. (A) For the RT alignment, the Ty3-gypsy retrotransposons Beetle7 and Elbe2 were chosen as outgroup. Clamps above the sequences indicate the seven conserved RT domains. (B) Clamps above the sequences indicate the seven regions forming β-strands in the MP secondary structure. Asterisks mark the conserved HX25D motif spanning the second and third β-strands. Abbreviations of the reference elements: cauliflower mosaic virus (CaMV) and figwort mosaic virus (FMV) from the genus Caulimovirus, soybean chlorotic mottle virus (SbCMV; Soymovirus), cassava vein mosaic virus (CSVMV; Cavemovirus) and tobacco vein clearing virus (TVCV; Solendovirus), banana streak VA virus (BSVAV) and Commelina yellow mottle virus (ComYMV) from the genus Badnavirus, rice tungro bacilliform virus (RTBV; Tungrovirus), petunia vein clearing virus (PVCV) and Fritillaria imperialis EPRV (FriEPRV) from the genus Petuvirus, and the FEVs Amborella trichopoda B virus (AtrichBV), Glycine max virus (GmaxV) and Lotus japonicus A virus (LjapAV)
Neighbour-joining dendrograms based on the RT and MP alignments are similar. They demonstrate an affiliation of the beetEPRVs to the FEVs, with a maximum bootstrap support of 100 % (Fig. 3). This assignment is validated by calculations based on the maximum likelihood method, the UPGMA method and the minimum evolution method (Supplementary Data Figs S5 and S6). If we follow the viral classification system as laid out in Teycheney et al. (2020), the beetEPRVs may represent three novel species within the Florendovirus genus as they exhibit RT–RH nucleotide identities of <80 % to each other and to known FEVs (Supplementary Data Fig. S7). However, despite their common host, beetEPRV1 is not grouped on the same branch with the other two beetEPRVs in all calculated dendrograms, indicating a high structural diversity.
Fig. 3.
Dendrograms grouping the beetEPRVs with the FEVs based on the protein sequence of RT (A) and MP (B). Dendrograms were constructed with the neighbour-joining method. Bootstrap values (1000 replicates) are shown above the nodes. The dendrograms are drawn to scale, with branch length units corresponding to the evolutionary distance (p-distance). This analysis includes the RT and MP domains of the representative beetEPRV sequences (Fig. 1B) and the homologous region of the caulimoviruses and Ty3-gypsy retrotransposons as references (for abbreviations see legend of Fig. 2). The neighbour-joining clustering of beetEPRVs with FEVs was validated using maximum likelihood, UPGMA and minimum evolution methods (Supplementary Data Fig. S5).
Apart from the sequence similarities of the RT and MP, a number of further beetEPRV hallmarks support their assignment to the FEVs: with 7.6 and 7.5 kb, respectively (Supplementary Data Table S2; Fig. 4A), the mean length of beetEPRV1 and beetEPRV3 members corresponds to the length of FEVs (7.2–8.5 kb; Geering et al., 2014). Due to an additional ORF3, composite beetEPRV2 elements were much longer (10.7 kb; component A 5.5 kb, component B 5.2 kb). The additional ORF2 in beetEPRV2-B and beetEPRV3 is also characteristic of FEVs and is assumed to encode an FEV-specific protein. Its estimated molecular weight of 50–54 kDa is well in line with the ORF2 of other FEVs (45–58 kDa; Geering et al., 2014). Although there is no clear ORF2 in the beetEPRV1 reference element due to the accumulation of frameshifts, there are several AUG start codons within a short interval in the 3′ region of ORF1 that enable reconstruction of a putative ORF of an appropriate molecular weight (51–55 kDa, depending on the start codon position).
Fig. 4.
Structural characteristics of the three beetEPRVs, including mean lengths (A) and genomic contexts (B–D). (A) Mean length of beetEPRV1, the two components beetEPRV2-A and beetEPRV2-B, and beetEPRV3. The underlying sequences refer to elements marked by an asterisk in Supplementary Data Table S2. (B–D) Analysis of the genomic context of beetEPRV1 (B), beetEPRV2 (C) and beetEPRV3 (D) using two sequence data sets of the sugar beet genotype EL10 (assembly, dark colour; SMRT reads, light colour). As beetEPRV1 (B) and beetEPRV3 (D) showed greater variation in properties compared with beetEPRV2-A (C left) and beetEPRV2-B (C right), the corresponding bar charts highlight different aspects. (B, D) The first bar represents the total beetEPRV number (100 %). The second bar shows an association with various microsatellites. The third bar demonstrates the amount of clustered/nested (filled) and tandemly arranged (hatched) beetEPRV sequences. The fourth bar shows an association with known beet tandem repeats, namely the centromeric pBV (filled) and the intercalary pEV (hatched) satellite repeat. (C) The first bar represents the total beetEPRV2-A number (100 %). The second bar demonstrates the amount of clustered/nested (filled) and tandemly arranged (hatched) beetEPRV2-A sequences. The third bar represents the total beetEPRV2-B number (100 %). The fourth bar demonstrates the amount of clustered/nested (filled) and tandemly arranged (hatched) beetEPRV2-B sequences. The sample size is given above the box plots (A) and next to the bar charts (B–D). The percentages (B–D) refer to the total number of analysed beetEPRV sequences for each sequence cluster.
Taken these results together, based on the sequence similarities in the key protein domains, the conserved element length and the presence of an additional ORF, we conclude with confidence that all detected EPRVs in beet belong to the FEV genus.
beetEPRVs are embedded in a repeat-rich environment
To assess the genomic context, we manually extracted 514 individual candidate beetEPRV sequences (full-length as well as partial) from the sugar beet genotype EL10 reference genome assembly and the corresponding SMRT read data (Funk et al., 2018). Using our nHMMs, we extracted:
-
•
161 sequences from the EL10 assembly with 60 beetEPRV1, 42 beetEPRV2-A, 36 beetEPRV2-B and 23 beetEPRV3 sequences; and
-
•
353 sequences from the raw EL10 PacBio long (SMRT) reads with 31 beetEPRV1, 7 beetEPRV2-A, 9 beetEPRV2-B and 306 beetEPRV3 sequences.
Differences in the relative abundance of the three beetEPRVs, such as the high abundance of beetEPRV3 in the long reads as opposed to its rareness in the assembled genome, may reflect biases in these two datasets.
We compared these endogenous caulimovirid elements with the respective consensus sequences (Fig. 1B; Supplementary Data S1), examined self dotplots to investigate each element’s structure and organization, annotated the beginning and the end of each integrated FEV (Fig. 4A) and investigated the flanking regions (~8 kb for each site). The majority (73 % from the assembly; 71 % from the SMRT reads) of the beetEPRV1 sequences as well as several beetEPRV3 sequences (52 and 27 %, respectively) are directly flanked by AT-rich low-complexity regions at one or both ends. These are often arranged as (TA)n microsatellites (Fig. 4B, 4D) that frequently harbour short stretches of CA or TG dinucleotides. In some cases, beetEPRV3 elements on the SMRT reads are flanked by longer motifs, such as TATC (n = 1), TATACA (n = 5) and TTTCGGGG (n = 1). In contrast, we did not detect any beetEPRV2 members associated with low-complexity motifs.
BeetEPRV1, beetEPRV2 and beetEPRV3 members without low-complexity (TA)n flanking regions were also detected in a highly repetitive neighbourhood characterized by fragmental duplications, rearrangements and juxtapositions of further truncated beetEPRV copies. In particular, beetEPRV elements of the same cluster often localized close to each other as fragments or full-length copies. Noteworthy, the intact, adjacent beetEPRV sequences were connected by TRs (Supplementary Data Fig. S2). Taking into account the assembly and the SMRT reads, the frequency of such arrangements in tandem-like arrays varied: it was 3 % for beetEPRV1, 19 % for beetEPRV2-A, 11–22 % for beetEPRV2-B and 13–28 % for beetEPRV3 (Fig. 4B–D). Regarding the bipartite nature of beetEPRV2, a tandem-like arrangement of the beetEPRV2 components A and B in the same orientation was found frequently (assembly, 33 %; SMRT, 63 %), in which the A–B arrangement was half as common as the B–A arrangement (Supplementary Data Fig. S2).
Some beetEPRV1 sequences (2–3 %) localize adjacent to units of the intercalary sat DNA family pEV I described by Schmidt et al. (1991), while more (16 %; Fig. 4B) border the centromeric pBV sat DNA arrays of Schmidt and Metzlaff (1991). BeetEPRV3 was also found to be associated with pEV I and pBV (0.7 and 13–14 %, respectively; Fig. 4D). For two instances along the assembly and five instances on the SMRT reads, beetEPRV3 was flanked by pBV arrays on both sides. In addition, combinations of a pBV array on one end and a (TA)n microsatellite on the other were also detected. All six described pBV subfamilies (Zakrzewski et al., 2013) were observed in the associations with beetEPRV1 and beetEPRV3.
In summary, the sugar beet EPRVs are embedded in highly repetitive genomic contexts. They form complex arrays, often containing multiple, rearranged elements of the same beetEPRV cluster. Co-occurrences with heterochromatic sat DNAs as well as low-complexity microsatellites were observed frequently.
beetEPRVs show a high, locally focused coverage with small RNAs
It is assumed that most repeats are transcribed at a basal level regulated by the host through epigenetic silencing (reviewed by Lippman and Martienssen, 2004). We detected transcripts for all three beetEPRVs in a cDNA library (GenBank accession number SRX674050) that could potentially lead to virus activation. To investigate how the beet host genome may prevent such virus activation, we analysed the potential silencing by smRNAs. Publicly available smRNA reads from B. vulgaris were mapped against the consensus sequences of the three beetEPRVs (Fig. 1B; Supplementary Data S1). Out of 20 091 021 smRNA reads in total, 1051 reads matched to beetEPRV1, 381 reads to the concatenated consensus sequence of both beetEPRV2 components, and 13 235 reads to beetEPRV3 (Fig. 5). Among the smRNAs matching the three beetEPRVs, only beetEPRV3-derived smRNAs covered the wide spectrum from 18 to 30 nt (Fig. 5A). However, for all three beetEPRVs, smRNAs with a length of 20–26 nt contributed >99 % of all mapping smRNAs. These smRNAs were divided into smRNAs that induce posttranscriptional gene silencing (PTGS, 20–23 nt; Rosa et al., 2018) and transcriptional gene silencing (TGS, 24–26 nt; Ghoshal and Sanfaçon, 2015). In beetEPRV1 and beetEPRV3, about two-thirds of the smRNAs potentially mediate PTGS (65.9 and 64.7 % respectively), whereas TGS-associated smRNAs contribute the smaller fraction (33.7 and 34.8 % respectively). Strikingly, the opposite applies for beetEPRV2: only 13.4 % of the smRNAs potentially induce PTGS, while 85.8 % may lead to TGS.
Fig. 5.
Evaluation of smRNAs mapping to the consensus sequences of the three beetEPRVs. Small RNAs inducing PTGS (20–23 nt) are marked by black bars and those inducing TGS (24–26 nt) by white bars. (A) Length distribution of smRNAs matching the respective beetEPRV consensus sequences (colour-coded bars). Note the logarithmic scale. (B–D) Read depths of smRNAs mapped against the beetEPRV consensus sequences. Positive peaks represent sense RNAs and negative peaks indicate anti-sense RNAs. Only smRNAs mediating PTGS (black; 20–23 nt) and TGS (white; 24–26 nt) are considered. The respective beetEPRV element structure is shown below the diagram, as introduced in Fig. 1B.
We observed peaks with high (>100 reads, beetEPRV2) and very high smRNA read abundances (>200 reads, beetEPRV1; >6000 reads, beetEPRV3) at particular positions along the three consensus sequences (Fig. 5B–D). These peaks are preferentially located in regions or ORFs that do not carry any known protein domains: This refers to ORF2 in beetEPRV3 and the 3′ region of the beetEPRV1-ORF, as well as to beetEPRV2’s internal region between ORF1A and ORF1B. Moderately sized peaks were also found in the terminal repeats.
beetEPRV3 is highly methylated in the B. vulgaris genome and occurs in closely related beet species
As beetEPRV3 is characterized by the highest average pairwise identity of its members and many intact copies with continuous ORFs, and as we found an extraordinarily high copy number in the SMRT read data set, we used beetEPRV3 as reference for experimental studies on EPRVs in beet. In order to gain information about cytosine methylation of beetEPRV3 sequences, we restricted genomic B. vulgaris DNA with the methylation-sensitive enzymes MspI and HpaII and hybridized the RT and MP probe to the membranes (Fig. 6A, B, lanes 6 and 7; Supplementary Data Table S3). Whereas HpaII only cuts unmethylated CCGG sequences, MspI is able to tolerate methylation of the internal cytosine (Waalwijk and Flavell, 1978). In the beetEPRV3 reference sequence, the CCGG restriction site is present three times; point mutations may also lead to further or fewer CCGG sites. As beetEPRV3 was cut by neither HpaII nor MspI, we conclude that methylation of the outer or both cytosines occurred within the CCGG motifs.
Fig. 6.
Autoradiograms of Southern hybridization to estimate the abundance of beetEPRV3 in sugar beet and related species. As probes we used RT (A, C) and MP (B, D). In (A) and (B), DNA of B. vulgaris was restricted by different enzymes: AluI (lane 1), FspBI (lane 2), BseGI (lane 3), BspCNI (lane 4), HindIII (lane 5), MspI (lane 6) and HpaII (lane 7). In (C) and (D), AluI-restricted DNA of different Amaranthaceae species was used: from the section Beta the B. vulgaris cultivars KWS 2320 (lane 1) and Swiss chard (lane 2), B. maritima (lane 3) and B. patula (lane 4); from the section Corollinae (C) B. lomatogona (lane 5); from the section Nanae (N) B. nana (lane 6); and from the sister genus Patellifolia patellaris (P, lane 7); quinoa (Chenopodium quinoa; 8) and spinach (Spinacia oleracea; lane 9) were selected as outgroup (O). Hybridization was carried out with a stringency of 79 %.
The genomic B. vulgaris DNA (KWS 2320) was also restricted by five additional restriction enzymes to estimate the sequence conservation and abundance of beetEPRV3 in sugar beet (Fig. 6A, B; lanes 1–5). The Southern hybridization required a long exposure time (11 d), a sign of low beetEPRV3 abundance in B. vulgaris. The clear bands point to a strong conservation of the restriction sites within the beetEPRV3 sequence, confirming the high similarity of beetEPRV3 members to each other.
In order to investigate the beetEPRV3 abundance in related genomes, we comparatively hybridized both probes (RT and MP) to AluI-restricted DNA of the B. vulgaris cultivars KWS 2320 and Swiss chard, B. maritima, B. patula, B. lomatogona, and B. nana, as well as P. patellaris, a member of the Beta sister genus Patellifolia (Fig. 6C, D). As outgroup we chose C. quinoa and S. oleracea, both also belonging to the Amaranthaceae. The two probes hybridized to all four genomes of the section Beta, producing the expected AluI patterns (Fig. 6C, D, lanes 1–4). This included the cultivated beet and chard species as well as the wild beet B. maritima. In the further sections of the genus Beta represented by B. lomatogona and B. nana, RT hybridization signals also became visible, but in a divergent pattern with lower intensity. Signals for the MP probe, which were less conserved than the RT in EPRVs (Fig. 2), were not observed, even after an extended exposure time. This may indicate either beetEPRV3 presence in much lower abundance and/or higher divergence in these species or, more likely, cross-hybridization of the RT probe to a related EPRV sequence. In the sister genus Patellifolia as well as in the outgroups no signals were detected, either for the RT or for the MP probe. This indicates no significant sequence homologies between beetEPRV3 from B. vulgaris and possible EPRVs from P. patellaris, quinoa and spinach.
beetEPRV3 occurs on all B. vulgaris chromosomes
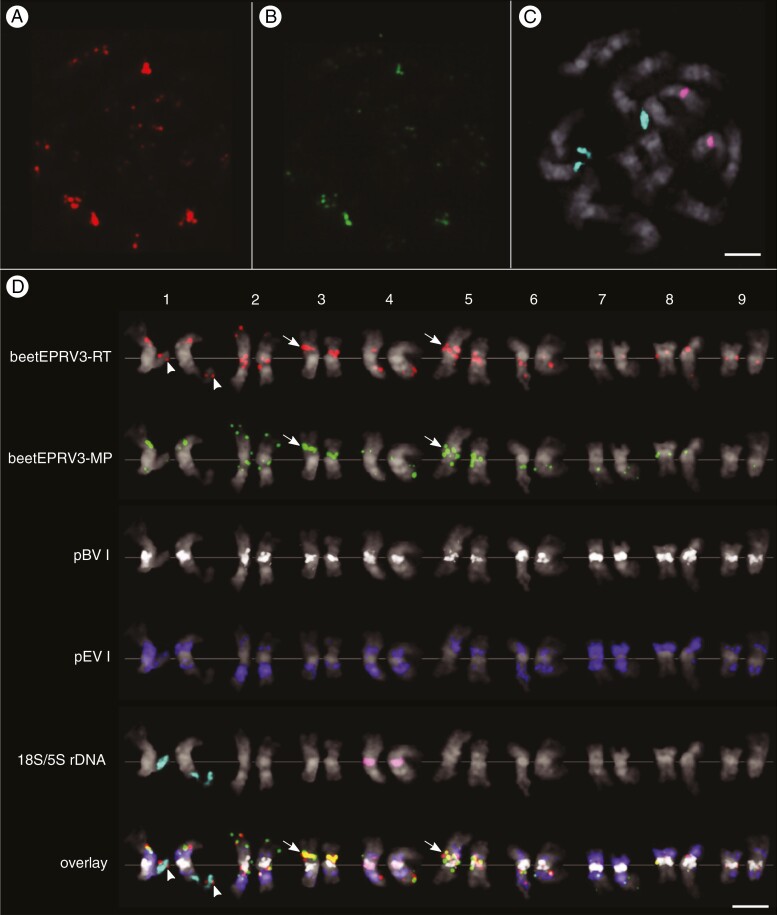
To determine the chromosomal localization of beetEPRV3, mitotic chromosomes of B. vulgaris (KWS 2320) were prepared and hybridized with the biotin-labelled beetEPRV3 RT probes (red) and digoxigenin-labelled beetEPRV3 MP probes (green; Figs 1B and 7A, B; Supplementary Data Fig. S8). The RT and MP signals for beetEPRV3 often co-localize, shown by the merging of red and green signals to yellow ones in the overlay (Fig. 7D). Nevertheless, the observation of distinct signals may point to the presence of truncated beetEPRV3 sequences. This observation is corroborated by the bioinformatic analyses, in which the reshuffling of beetEPRV sequences was also detected.
Fig. 7.
Localization of beetEPRV3 along mitotic metaphase chromosomes of B. vulgaris. DAPI-stained mitotic chromosomes of B. vulgaris are shown in grey. Cloned sequences of the RT (red) and the MP (green) domain of beetEPRV3 were used as probes. (A–D) Multicolour FISH of beetEPRV3-RT (red), beetEPRV3-MP (green), centromeric pBV I satellite (white), intercalary pEV I satellite (blue), 18S rDNA genes (turquoise) and 5S rDNA genes (magenta). Information on probe labelling and detection can be found in the Materials and methods section. (D) Sorted chromosomes from Fig. 6A–C additionally showing pBV and pEV signals. Paired chromosomes represent the homologous chromosomes. The assignment of chromosome numbers is based on the rDNA genes (pairs 1 and 4) and on the distribution and density of the satellites pBV I and pEV I according to Paesold et al. (2012). The strongest beetEPRV3 clusters on chromosomes 3 and 5 are highlighted (arrows), as well as the co-localization of beetEPRV3 fragments, including the RT, with the 18S-5.8S-26S rDNA (arrowheads). Scale bars = 2 µm.
The diploid chromosome set of B. vulgaris consists of 18 chromosomes and all chromosomes can be identified using rDNA and repetitive sequences (Schmidt et al., 1994; Paesold et al., 2012). Therefore, the beetEPRV probes were hybridized together with additional probes for the 18S-5.8S-26S and 5S rDNA (turquoise and magenta in Fig. 7C, 7D; Supplementary Data Fig. S8) to identify the homologous chromosomes 1 and 4, respectively. To designate the remaining chromosomes, rehybridization with probes marking the centromeric sat DNA pBV I and the intercalary sat DNA pEV I (white and blue in Fig. 7D and Supplementary Data Fig. S8) was carried out. The intensity and co-occurrence of these sat DNA arrays allowed assignment to the respective chromosome pairs according to Paesold et al. (2012).
Hybridization of beetEPRV3 RT and beetEPRV3 MP probes show the localization of beetEPRV3 on all 18 B. vulgaris chromosomes (Fig. 7D; Supplementary Data Fig. S8). Signal strengths differ strongly between chromosomes, ranging from very strong to faint fluorescent, indicating the presence of large and small accumulations of beetEPRV3 sequences. The signals were often detected in heterochromatic regions, which was revealed by the co-localization with the heterochromatic sat DNAs (pBV I, pEV I) and densely stained DAPI signals (Supplementary Data Fig. S8). This is in accordance with the association with these sequences found during the analysis of the flanking regions. We detected pericentromeric, intercalary and distal positions with similar localization patterns along the two homologous chromosomes. Thus, the strongest signals reside in the intercalary region of chromosome 3 and the pericentromeric region of chromosome 5 (Fig. 7D, arrows). Fluorescent in situ hybridization (FISH) images of chromosomes 1 and 2 show the most distal signals, where we detected co-localization of beetEPRV3 RT with the 18S-5.8S-25S rDNA (Fig. 7D, arrowheads).
DISCUSSION
EPRVs are a widespread component of plant genomes that often accumulate over time and become a noteworthy part of the repetitive fraction of the genome (Hohn et al., 2008; Diop et al., 2018; Gong and Han, 2018). Our analyses in sugar beet show that EPRVs constitute at least 2 Mbp, which are equal to 0.3 % of the B. vulgaris genome. This value is within the EPRV proportion range of 0–2 % in other host plants (Geering et al., 2014; Duroy et al., 2016). Although we find some essentially intact representative EPRV sequences, the majority does not comprise all EPRV-specific protein domains due to fragmentation and/or truncation. For this reason, it is likely that some beetEPRV fragments with strongly diverged or missing RT or MP protein domains (the major criteria used to search for EPRVs and applied here) may have escaped our computational detection and that our beetEPRV quantification is probably an underrepresentation. Nevertheless, the resulting, relatively large amount of integrated EPRV sequences was unexpected given that no exogenous viral sequences have been reported for beets so far.
Based on their sequence and structural characteristics, we subdivided the sugar beet EPRVs into three clusters: beetEPRV1, beetEPRV2 and beetEPRV3 (Figs 1 and 3; Supplementary Data Figs S2–S7). Considering their overall length, their structure with a putative additional ORF and their amino acid sequence homologies, all beetEPRVs represent typical members of the genus Florendovirus (FEV) as described by Geering et al. (2014).
Specific element structures indicate different evolutionary beetEPRV origins
Certain features in beetEPRV structure and sequence provide evidence for potential infections and subsequent amplifications at multiple time points during beet evolution.
For our reference beetEPRV3, we detected high nucleotide identities between the individual beetEPRV3 members and multiple genomic copies with intact ORFs. Comparative Southern hybridizations with MP and RT probes led to conserved patterns within the section Beta, indicative of a single or only few integration events that were subsequently amplified within the beet genome.
The species in the sister sections Corollinae and Nanae do not seem to harbour beetEPRV3-related sequences as they do not produce MP signals and only weak, dissimilar signals for the RT, likely the result of cross-hybridization rather than true homology. Together, these findings may point to an initial beetEPRV3 integration into the beet genome after the split of the sections Corollinae/Nanae from Beta ~13.4–7.2 million years ago (mya; Hohmann et al., 2006) and before speciation within the section Beta. The initial beetEPRV3 integration could be much younger and/or could have happened more than once as the individual exposure to the same FEV ancestor may have resulted in independent infection events among the respective wild beets. Yet, the most parsimonious scenario would support an estimated ancestral infection in the common ancestor 13.4–7.2 mya. Given the estimated age of FEVs (34–20 mya; Geering et al., 2014) and EPRVs in general (320 mya; Diop et al., 2018), this seems to be an evolutionarily young infection history. Nevertheless, in comparison with the time of invasion of EPRVs into other host species [e.g. eBSV into the genome of Musa sp. 640 000 years ago (Gayral et al., 2010; Duroy et al., 2016) and eRTBVL-D into the genome of Oryza sp. 2.4–15 mya (Chen et al., 2018)] or compared with the estimated integration time points of other retroelements into the sugar beet genome [chromovirus Beetle2, 130 000 years ago (Weber and Schmidt, 2009) and Cassandra TRIMs, 0.1–8 mya (Maiwald et al., 2020)], the assumed beetEPRV3 endogenization event ranges at a similar timeline.
Apart from the distribution across the beet genera, we suggest that the overall structural organization of the beetEPRVs may also point to their evolutionary origin. Across the angiosperms, FEVs usually harbour two overlapping ORFs, but in some instances structural variations have been reported as well, such as a single continuous ORF, three ORFs or bipartitely organized structures (Geering et al., 2014). Interestingly, despite residing in a single host, the three beetEPRVs correspond to three of the four structural variants (Fig. 1B).
With a single continuous ORF, members of beetEPRV1 have the most compact, least conserved element structure. As that may be a sign of an early stage in FEV evolution, we believe that beetEPRV1 may comprise the oldest beetEPRV members. Although the beetEPRV1 ORF structure is rare, the FEV AtrichBV has a similar organization (Geering et al., 2014). This FEV is hosted by the evolutionarily old, basal angiosperm Amborella trichopoda and harbours only low sequence identities to beetEPRV1. Nevertheless, despite its low complexity, we do not think that beetEPRV1 served as precursor for beetEPRV2 and beetEPRV3. Instead, as the beetEPRV1 ORF differs considerably from those of the other beetEPRVs and all dendrograms place beetEPRV1 separately (Figs 1 and 3; Supplementary Data Figs S2, S5 and S6), we argue for an independent beetEPRV1 infection event.
In contrast to beetEPRV1 and beetEPRV3, elements of beetEPRV2 are characterized by a bipartite structure with two components, beetEPRV2-A and beetEPRV2-B. While the beetEPRV2 components fully share their 5′ end sequences, the protein domains are only present once, on either component A or component B. The overall beetEPRV2 structure resembles other bipartitely organized FEVs (VvinBV, VvinDV, OsatBV, SbicV) from rice, sorghum and grapevine (Geering et al., 2014). Of these, only VvinDV shows a complete separation of the protein domains into two complementary components as observed for beetEPRV2, whereas the components of VvinBV, OsatBV and SbicV rather show a redundancy in their protein sets. Comparing the coding regions of all three beetEPRVs, we found that beetEPRV2 ORFs exhibit a high overall similarity to the respective regions of beetEPRV1 and beetEPRV3 (Figs 1B and 8A, dotted boxes). Thus, for beetEPRV2’s origin several scenarios may be possible.
Fig. 8.
Evolutionary hypothesis to explain the bipartite structure of beetEPRV2 based on sequence similarities to the other beetEPRVs. (A) Similarity of the coding regions of beetEPRV2 to beetEPRV1 and beetEPRV3. The nucleic acid sequence identity between the ORF sections is given in the respective grey-shaded regions. BeetEPRV2’s internal region between ORF1A and ORF1B does not have an equivalent in beetEPRV1 and beetEPRV3. (B) We propose two evolutionary scenarios that may have led to the bipartite structure of beetEPRV2. In scenario A, we assume the nested integration of two beetEPRV2 precursor sequences. The internal beetEPRV2 element may have piggybacked an additional ORF (ORF3). To yield the bipartite element observed today, internal domains were subsequently deleted. Scenario B may have involved the recombination of two independently reshuffled and partly deleted beetEPRV2 entities. gDNA, genomic DNA. Dotted boxes represent homologous sequence regions.
First, beetEPRV2 may have originated from an ancestral virus with a bipartite genome. Genome segmentation into two or more components is quite common among RNA viruses [e.g. Secoviridae (positive-sense ssRNA; Thompson et al., 2017); Fimoviridae (negative-sense ssRNA; Elbeaino et al., 2018); Chrysoviridae (dsRNA; Kotta-Loizou et al., 2020)], but is rare among DNA viruses [Begomovirus/Geminiviridae (ssDNA; Zerbini et al., 2017)]. The examples named here are so-called multipartite viruses as they encapsulate their respective genomic components in separate virions. These virions thereby contain either a single genomic component (Begomovirus, Secoviridae) or several genomic components (Chrysoviridae, Fimoviridae). ‘Segmented’ viruses, on the other hand, package all of their genomic components into a single particle. Genome segmentation is assumed to facilitate a rapid evolution of the virus (reviewed by Sicard et al., 2016; Newburn and White, 2019).
However, bipartite FEVs have only been reported from four rather distantly related FEV species (Geering et al., 2014), which may point to an independent beetEPRV2 emergence. Moreover, beetEPRV2 and beetEPRV3 share high sequence conservation (Fig. 8A; Supplementary Data Figs S4–7), supporting an origin from a precursor containing both A and B components in a single element. Therefore, we consider the emergence of the bipartite beetEPRV2 from an undivided beetEPRV3-like precursor as likely and present two possible scenarios how this emergence may have happened (Fig. 8B).
On the one hand, nested integration of one beetEPRV2 precursor into another may have resulted in the separation of ORF1 (Fig. 8B, scenario A). Retroelements like retrotransposons and EPRVs tend to integrate in a nested manner (SanMiguel et al., 1996; Jakowitsch et al., 1999), and in sugar beet this was observed for several LTR retrotransposons as well (Weber and Schmidt, 2009; Wollrab et al., 2012). The transduction of flanking DNA during this process presumably led to the acquisition of ORF3 between ORF1A and ORF1B, followed by the deletion of most of the integrated sequence, thus creating the same 5′-end in both components.
On the other hand, internal deletions may have happened first, forming two differently truncated beetEPRV2 entities (Fig. 8B, scenario B). One of these entities gained an additional ORF, possibly by xenologous recombination (McClure, 2000) with a foreign caulimovirid sequence, as ORF3 bears no resemblance to any other sequence in beet. Many viruses and repeats have modularly acquired domains and ORFs (Smyshlyaev et al., 2013; Koonin et al., 2015), and some instances were also reported in beet (Heitkam et al., 2014). Such an acquisition of ORFs likely contributed to the evolution of EPRVs, potentially leading to the speciation into virus families (McClure, 2000). In beet, we detected the resulting beetEPRV2 components A and B as both independent and combined insertions. We assume that recombination with a foreign caulimovirid sequence, as well as between the two beetEPRV2 components, may have been enabled by similarities between their terminal repeats. One way or another, the shared sequences between the two beetEPRV2 components may have facilitated their mobilization as separate entities.
Targeted integration or toleration: beetEPRVs accumulate in repetitive environments
Most of the detected beetEPRV members were embedded in repeat-rich environments, as evidenced by sequence analysis (Fig. 4) and FISH (Fig. 7). On the nucleotide level, the flanking regions (usually several hundred bases) of beetEPRV1 and beetEPRV3 contained a high amount of TA dinucleotides present as simple repeats. EPRVs with adjacent (TA)n repeats were detected in various plant species [Oryza sp. (Kunii et al., 2004; Liu et al., 2012); various species (Geering et al., 2014)], indicating a potential caulimovirid integration preference and retention. Besides the weaker electrostatic attraction between A and T nucleotides, with only two hydrogen bonds, TA dinucleotide-rich sites can form secondary structures that disturb DNA replication and thereby lead to instability within the chromosomes (Dillon et al., 2013). Hence, the frequency of double strand breaks increases, as does the likelihood of a caulimovirid insertion during DNA repair, as already suggested by Liu et al. (2012) and Geering et al. (2014). Consequently, beetEPRV integration may critically depend on the TA frequency in a potential genomic target.
On a macro-scale, 3–16 % of the beetEPRV sequences were flanked by sat DNA arrays (Fig. 4), i.e. the major centromeric pBV and the intercalary pEV families (Schmidt and Metzlaff, 1991; Schmidt et al., 1991; Zakrzewski et al., 2013). This was corroborated by our beetEPRV3 FISH (Fig. 7). The high TA content of the centromeric satDNA in beet (59–69 %; Schmidt and Metzlaff, 1991) has presumably provided a suitable target for beetEPRV insertions. Although centromeres are generally reduced in meiotic recombination (Bennetzen, 2000), centromeric repetitive DNA still evolves by unequal recombinatorial exchange (Ma and Jackson, 2006; Talbert and Henikoff, 2010). Thus, mitotic and meiotic DNA breaks may have been filled by beetEPRVs as observed for other retroelements (reviewed by Schubert and Vu, 2016).
A potential accumulation in DNA breaks may also explain the beetEPRV tendency to form arrays. Generally, arrays may result from the simultaneous involvement of several EPRV copies (e.g. EPRV concatemers) in a single break repair event, from a nested integration or from the recombination of episomal viral genomes with integrated forms (Hohn et al., 2008). EPRV accumulations were observed in various plant genomes [e.g. tobacco (Lockhart et al., 2000), petunia (Richert-Pöggeler et al., 2003), rice (Liu et al., 2012), Citrinae sp. (Yu et al., 2019)]. In B. vulgaris, EPRV accumulations are scattered throughout the entire genome and usually only contain beetEPRV members of the same cluster. This may be explained by rolling circle amplification or integration through recombination (e.g. Jakowitsch et al., 1999; Kirik et al., 2000; Gayral et al., 2008), which depends on the sequence similarity that is present within the respective beetEPRV cluster.
The specific distribution of beetEPRV3 on particular chromosome pairs and its association with specific host genome sequences may result from selective integration, retention or removal, potentially also dictated by the 3-D structure of the genome (Bousios et al., 2020). Regarding a selective integration, EPRVs do not encode an integrase and are, to our knowledge, unable to recognize specific target sequences. Nevertheless, EPRVs may be mobilized together with adjacent transposable elements, thus hitch-hiking to a location preferred by the transposable element (Staginnus and Richert-Pöggeler, 2006). Regarding removal and retention, we have observed EPRV depletion in the euchromatin: as euchromatic EPRVs are more likely to reduce plant fitness by interfering with gene expression, there would be negative selective pressure in favour of EPRV removal. EPRVs in the heterochromatin, on the other hand, would likely be suppressed by their inactive chromatin environment, limiting detrimental effects and allowing EPRV retention (Hohn et al., 2008). The survival of repeats in the heterochromatin, which form so-called safe havens, has been described for a number of repeats, mostly retrotransposons (Boeke and Devine, 1998; Gao et al., 2008). Thus, the accumulation of beetEPRVs in repeat-rich environments is presumably the result of active selective targeting or passive retention in the heterochromatin or a combination of both.
beetEPRV endurance: low-level transcription despite silencing through RNA interference
Similar to LTR retrotransposons, the spreading of EPRVs requires reverse transcription of the RNA intermediate derived from the endogenous sequence. The arising episomal caulimovirids are potentially virulent and can cause diseases such as banana streaks induced by the badnavirus BSV (Harper et al., 1999), or vein clearing in tobacco induced by the solendovirus TVCV (Lockhart et al., 2000) and in petunia induced by the petuvirus PVCV (Richert-Pöggeler et al., 2003). For both LTR retrotransposons and EPRVs, the transcriptional activity depends on the required sequence motifs in the genomic copy to facilitate transcription by the host RNA polymerase, but may be counteracted by the host through epigenetic regulatory mechanisms (Ghoshal and Sanfaçon, 2015).
The potentially active endogenous PVCV sequence is characterized by flanking quasi-long tandem repeats (QTRs) that are assumed to have an LTR-like function facilitating its transcription. The QTRs comprise promoter and polyadenylation sequences and usually separate two adjacent PVCV sequences from another (Richert-Pöggeler et al., 2003). Thus, transcription may start at the promoter of the upstream copy and terminate at the polyadenylation site of the consecutive one. PVCV also harbours a polypurine tract (5′-TTGATAAAAGAAAGGGGT-3′; Richert-Pöggeler and Shepherd, 1997) that is supposed to function as primer binding site for plus-strand synthesis during reverse transcription. The TRs that we detected at the ends of all three beetEPRVs do not have an upstream polypurine tract and in the case of beetEPRV3 also lack the poly(A) region that might act as a polyadenylation signal. Thus, beetEPRVs do not contain canonical QTRs as described for PVCV and hence beetEPRV transcription and reverse transcription might be impaired. Nevertheless, we detected beetEPRV sequences in the published cDNA library of beet (GenBank accession number SRX674050), implying that basal EPRV transcription takes place.
As we detected smRNAs matching the beetEPRV consensus sequences (Fig. 5) and infrequent cutting with methylation-sensitive restriction enzymes (Fig. 6), we assume that beetEPRV transcription is under epigenetic control. The beetEPRV-matching smRNAs target both coding and non-coding domains. However, the highest peaks were found in beetEPRV3 ORF2 and the region homologous to ORF2 of beetEPRV1, highlighting that the presumed FEV-specific protein encoded by ORF2 (Geering et al., 2014) may be important for beetEPRV replication.
The three beetEPRVs are specifically targeted by smRNAs of variable lengths between 18 and 30 nt, indicating that beetEPRVs could be silenced by both TGS and PTGS. TGS is based on RNA-dependent DNA methylation, which is mostly mediated by 24-nt smRNAs, while the hallmarks of PTGS are predominantly 21- to 22-nt smRNAs that lead to mRNA degradation and translation suppression of the targeted sequence (Ghoshal and Sanfaçon, 2015; Rosa et al., 2018). We found that the beetEPRVs differ in the amount of matching smRNAs, as well as in their classification as either TGS- or PTGS-mediating smRNAs.
For beetEPRV2, we found only few smRNAs, predominantly those that are involved in TGS. The TGS pathway is also predominant for transposable element silencing in sugar beet (Zakrzewski et al., 2013; Dohm et al., 2014), characterized by high DNA methylation levels and contributing to the formation of large heterochromatic regions (Weber and Schmidt, 2009; Weber et al., 2010; Zakrzewski et al., 2011, 2017). For EPRVs in particular, this silencing route was also observed in other host plants, such as petunia (Richert-Pöggeler et al., 2003), rice (Kunii et al., 2004), tomato (Staginnus et al., 2007) and rapeseed (Omae et al., 2020). The Fritillaria imperialis-specific EPRV (FriEPRV) was also found to be targeted by mostly TGS inducing 24-nt smRNAs (Becher et al., 2014) and, similar to beetEPRV2, no complete EPRV sequence could be identified for FriEPRV. However, as recombination of EPRV fragments has been observed to lead to the generation of complete and active viral genomes (Chabannes et al., 2013), simple sequence rearrangements that do not lose vital parts of the virus genome, as in the case of beetEPRV2, might not be sufficient to avoid viral activation.
In stark contrast, for beetEPRV1 and beetEPRV3, we detected mostly PTGS-specific lengths, supporting our assumption of a basal beetEPRV transcription, possibly by read-through mechanisms. Therefore, silencing has to take place at the posttranscriptional level to be effective. Strikingly, for beetEPRV3, the number of smRNAs is 13 and 36 times as high as for beetEPRV1 and beetEPRV2, respectively. This goes along with the observation that beetEPRV3 members are highly conserved and not fragmented, whereas beetEPRV1 and beetEPRV2 are usually marked by truncation and reshuffling. Consequently, we assume that (1) smRNA silencing is used to suppress a possible activation of beetEPRV3, and (2) beetEPRV3 may be able to become active and to replicate, and could be potentially infectious, if not hindered by the host.
As we detected beetEPRV1 and beetEPRV3 adjacent to the centromeric sat DNA family pBV, and as in situ hybridization also revealed beetEPRV3 signals close to the centromeres of four chromosomes, the epigenetic centromere maintenance processes may also play a role in beetEPRV regulation. To initiate and maintain the centromere by incorporation of the centromere-specific histone H3 (CENH3) into the nucleosomes, transcription of centromeric DNA is required (Jiang et al., 2003); during transcription, chromatin is disrupted and nucleosomes are destabilized and removed, providing an ideal opportunity for histone replacement. Any centromere-embedded transcription unit that comprises a promoter, including transposable elements, can initiate transcription. This may explain why we found transcriptionally active beetEPRV1 and beetEPRV3 members, and we speculate that these are the copies associated with the centromeric repeats. Further, integration near transcriptionally active regions, such as the ribosomal genes, may also result in unavoidable transcription of the adjacent EPRV sequences and subsequent silencing through PTGS. It is not uncommon that other repetitive elements are indeed adjacent to or even embedded in the 18S-5.8S-26S rDNA (Balint-Kurti et al., 2000; Jo et al., 2009; Weber et al., 2013). In the case of beetEPRV3, we noticed an association with the 18S-5.8S-26S rRNA genes, localized distally on chromosome 1 (Schmidt et al., 1994; Dechyeva and Schmidt, 2006), that would allow such transcription.
As our smRNA quantification relies on the mapping to beetEPRV consensus sequences, beetEPRV members with a deviating sequence may potentially differ in smRNA coverage. Nevertheless, we can confidently state that there is a high abundance of beetEPRV-derived smRNAs. These potentially contribute to the resistance of sugar beet to infection with exogenous viruses of related sequences (Huang and Li, 2018), i.e. beetEPRV-derived smRNAs may play a role in the suppression of infectious, potentially pathogenic EPRVs. Therefore, beetEPRVs may represent a beneficial component of the host’s genome.
Conclusions
The three beetEPRVs in sugar beet are characterized by differences in their structure, their genomic context, and the way in which they are silenced. Nevertheless, their common affiliation to the genus Florendovirus demonstrates their close relationship. Based on the low-complexity organization of beetEPRV1 members and the high sequence similarity between beetEPRV2 and beetEPRV3, we postulate that the three beetEPRVs originated from at least two independent integration events 13.4–7.2 mya, with subsequent diversification of beetEPRV2 from beetEPRV3. During this process, the beetEPRV2 precursor probably underwent structure-changing mechanisms leading to the bipartite nature that we observe today.
BeetEPRVs likely favour integration into genomic regions of low complexity, such as (AT)n microsatellites. The accumulation of beetEPRV sequences next to each other points to active selective targeting and/or passive retention in primary heterochromatic regions, while selective beetEPRV removal from the euchromatin of the host genome may also play a role. The observed embedding of beetEPRV sequences in the deep AT-rich heterochromatin of the host may ensure that the EPRV sequences remain inaccessible for several DNA-binding factors, thus reinforcing their silencing by the epigenetic control machinery. While beetEPRV2 is mostly targeted by smRNAs inducing DNA methylation, beetEPRV1 and beetEPRV3 show a high coverage with smRNAs inducing mRNA degradation and translation suppression, presumably as a result of basal transcription of the latter two beetEPRVs.
Taken these findings together, the sugar beet host employs three strategies to shut down the beetEPRV copies, thus preventing re-infection: heterochromatic burial, epigenetic silencing and structural disassembly. As a result, EPRVs in beet provide an example of complete assimilation and inactivation of a plant virus in the host genome.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: specificity of EPRV nHMMs for the MP and RT domains and considerations for parameter choice. Figure S2: maximum likelihood dendrogram showing the relationships among 119 EPRV RT hits found in the B. vulgaris genome by an nhmmer analysis. Figure S3: schematic representation of beetEPRV sequence variants and their occurrence in the EL10 assembly and on the SMRT reads. Figure S4: pairwise amino acid identity of the RT and MP between several members of the caulimovirids and two Ty3-gypsy retrotransposons. Figure S5: dendrograms grouping the beetEPRVs with the FEVs based on the protein sequence of RT and MP. Figure S6: maximum likelihood dendrograms showing the relationship of the beetEPRVs to the different FEVs based on the protein sequence of RT and MP. Figure S7: pairwise nucleotide identity of the RT–RH domain between the closest FEV relatives of the beetERVs and the petuviruses PVCV and FriEPRV as outgroup. Figure S8: localization of beetEPRV3 along mitotic prometaphase and metaphase chromosomes of B. vulgaris. Table S1: EPRV reference sequences used for beetEPRV identification and their sources. Table S2: chromosomal position of beetEPRV sequences along the EL10 sugar beet assembly. Table S3: primer sequences for the amplification of beetEPRV3-specific probes. Table S4: FEV-specific amino acids in the RT and MP revealed by the alignment of 16 caulimovirid sequences. Additional sequence data for beetEPRV reference sequences are accessible at http://doi.org/10.5281/zenodo.3888270 and comprise the following. Data S1: beetEPRV reference sequences in fasta format. Data S2: multiple sequence alignment of 27 beetEPRV1 sequences in fasta format. Data S3: multiple sequence alignment of 42 beetEPRV2-A sequences in fasta format. Data S4: multiple sequence alignment of 23 beetEPRV2-B sequences in fasta format. Data S5: multiple sequence alignment of 11 beetEPRV3 sequences in fasta format.
ACKNOWLEDGEMENTS
We want to dedicate this manuscript to Prof. Dr. rer. nat. Thomas Schmidt, our group leader, PhD supervisor, mentor and colleague. The identification of beetEPRVs has been on the mind of Thomas, his group and his collaborators for a long time, dating back 20 years. A number of people have helped us to get to this point and we thank them for their initial work. We thank Pat Heslop-Harrison, University of Leicester, UK, for advice and stimulating discussions, and Katja Richert-Pöggeler, JKI Braunschweig, Germany, for advice regarding caulimovirid taxonomy. We wish to thank the scientists involved in beet genome sequencing for providing early and prepublication access to their data, thereby speeding up our research process. We acknowledge the gene bank at the IPK Gatersleben for providing plant seeds.
LITERATURE CITED
- Balint-Kurti PJ, Clendennen SK, Dolezelová M, et al. 2000. Identification and chromosomal localization of the monkey retrotransposon in Musa sp. Molecular & General Genetics 263: 908–915. [DOI] [PubMed] [Google Scholar]
- Becher H, Ma L, Kelly LJ, Kovarik A, Leitch IJ, Leitch AR. 2014. Endogenous pararetrovirus sequences associated with 24 nt small RNAs at the centromeres of Fritillaria imperialis L. (Liliaceae), a species with a giant genome. Plant Journal 80: 823–833. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. 2000. Transposable element contributions to plant gene and genome evolution. Plant Molecular Biology 42: 251–269. [PubMed] [Google Scholar]
- Boeke JD, Devine SE. 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93: 1087–1089. [DOI] [PubMed] [Google Scholar]
- Bombarely A, Moser M, Amrad A, et al. 2016. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nature Plants 2: 1–9. [DOI] [PubMed] [Google Scholar]
- Bousios A, Nuetzmann HW, Buck D, Michieletto D. 2020. Integrating transposable elements in the 3D genome. Mobile DNA 11; doi: 10.1186/s13100-020-0202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabannes M, Iskra-Caruana ML. 2013. Endogenous pararetroviruses – a reservoir of virus infection in plants. Current Opinion in Virology 3: 615–620. [DOI] [PubMed] [Google Scholar]
- Chabannes M, Baurens FC, Duroy PO, et al. 2013. Three infectious viral species lying in wait in the banana genome. Journal of Virology 87: 8624–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Kishima Y. 2016. Endogenous pararetroviruses in rice genomes as a fossil record useful for the emerging field of palaeovirology. Molecular Plant Pathology 17: 1317–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Saito N, Encabo JR, Yamada K, Choi IR, Kishima Y. 2018. Ancient endogenous pararetroviruses in Oryza genomes provide insights into the heterogeneity of viral gene macroevolution. Genome Biology and Evolution 10: 2686–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechyeva D, Schmidt T. 2006. Molecular organization of terminal repetitive DNA in Beta species. Chromosome Research 14: 881–897. [DOI] [PubMed] [Google Scholar]
- Dechyeva D, Schmidt T. 2009. Molecular cytogenetic mapping of chromosomal fragments and immunostaining of kinetochore proteins in Beta. International Journal of Plant Genomics 2009: 721091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon LW, Pierce LC, Ng MC, Wang YH. 2013. Role of DNA secondary structures in fragile site breakage along human chromosome 10. Human Molecular Genetics 22: 1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diop SI, Geering ADW, Alfama-Depauw F, Loaec M, Teycheney PY, Maumus F. 2018. Tracheophyte genomes keep track of the deep evolution of the Caulimoviridae. Scientific Reports 8: 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgräwe D, et al. 2014. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505: 546–549. [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Research 43: W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroy PO, Perrier X, Laboureau N, Jacquemoud-Collet JP, Iskra-Caruana ML. 2016. How endogenous plant pararetroviruses shed light on Musa evolution. Annals of Botany 117: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbeaino T, Digiaro M, Mielke-Ehret N, Muehlbach HP, Martelli GP, ICTV Report Consortium . 2018. ICTV Virus Taxonomy Profile: Fimoviridae. Journal of General Virology 99: 1478–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT, Food and Agriculture Organization of the United Nations . 2017. Food and agriculture data: crops. http://www.fao.org/faostat/en/#data/QC(13 November 2019, date last accessed).
- Frese L, Desprez B, Ziegler D. 2000. Potential of genetic resources and breeding strategies for base-broadening in Beta. In: Cooper HD, Spillane C, Hodgkin T, eds. Broadening the genetic base of crop production. Rome: FAO and IBPRGI jointly with CABI Publishing, 295–309. [Google Scholar]
- Funk A, Galewski P, McGrath JM. 2018. Nucleotide-binding resistance gene signatures in sugar beet, insights from a new reference genome. Plant Journal 95: 659–671. [DOI] [PubMed] [Google Scholar]
- Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. 2008. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Research 18: 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayral P, Noa-Carrazana JC, Lescot M, et al. 2008. A single Banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus. Journal of Virology 82: 6697–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayral P, Blondin L, Guidolin O, et al. 2010. Evolution of endogenous sequences of banana streak virus: what can we learn from banana (Musa sp.) evolution? Journal of Virology 84: 7346–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geering AD, Scharaschkin T, Teycheney PY. 2010. The classification and nomenclature of endogenous viruses of the family Caulimoviridae. Archives of Virology 155: 123–131. [DOI] [PubMed] [Google Scholar]
- Geering AD, Maumus F, Copetti D, et al. 2014. Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution. Nature Communications 5: 5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal B, Sanfaçon H. 2015. Symptom recovery in virus-infected plants: revisiting the role of RNA silencing mechanisms. Virology 479–480: 167–179. [DOI] [PubMed] [Google Scholar]
- Gong Z, Han G-Z. 2018. Euphyllophyte paleoviruses illuminate hidden diversity and macroevolutionary mode of Caulimoviridae. Journal of Virology 92: e02043-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C, Heslop-Harrison JS. 2004. Sequences and phylogenies of plant pararetroviruses, viruses, and transposable elements. Advances in Botanical Research 41: 165–193. [Google Scholar]
- Harper G, Osuji JO, Heslop-Harrison JS, Hull R. 1999. Integration of banana streak badnavirus into the Musa genome: molecular and cytogenetic evidence. Virology 255: 207–213. [DOI] [PubMed] [Google Scholar]
- Heitkam T, Holtgräwe D, Dohm JC, et al. 2014. Profiling of extensively diversified plant LINEs reveals distinct plant-specific subclades. Plant Journal 79: 385–397. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T, Anamthawat-Jónsson K, Leitch AR, Shi M, Leitch IJ. 1991. In situ hybridization with automated chromosome denaturation. Technique3: 109–116. [Google Scholar]
- Hohmann S, Kadereit JW, Kadereit G. 2006. Understanding Mediterranean-Californian disjunctions: molecular evidence from Chenopodiaceae-Betoideae. Taxon55: 67–78. [Google Scholar]
- Hohn T, Hohn B, Pfeiffer P. 1985. Reverse transcription in CaMV. Trends in Biochemical Sciences 5: 205–209. [Google Scholar]
- Hohn T, Richert-Pöggeler KR, Staginnus C, et al. 2008. Evolution of integrated plant viruses. In: Roossinck MJ, ed. Plant virus evolution. Berlin: Springer, 53–81. [Google Scholar]
- Huang Y, Li Y. 2018. Secondary siRNAs rescue virus-infected plants. Nature Plants 4: 136–137. [DOI] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses . 2019. Taxonomy. https://talk.ictvonline.org/taxonomy/. (2 November 2020, date last accessed).
- Jakowitsch J, Mette MF, van Der Winden J, Matzke MA, Matzke AJ. 1999. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proceedings of the National Academy of Sciences of the USA 96: 13241–13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Birchler JA, Parrott WA, Dawe RK. 2003. A molecular view of plant centromeres. Trends in Plant Science 8: 570–575. [DOI] [PubMed] [Google Scholar]
- Jo SH, Koo DH, Kim JF, et al. 2009. Evolution of ribosomal DNA-derived satellite repeat in tomato genome. BMC Plant Biology 9: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Salomon S, Puchta H. 2000. Species-specific double-strand break repair and genome evolution in plants. EMBO Journal 19: 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479-480: 2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotta-Loizou I, Castón JR, Coutts RHA, et al. 2020. ICTV Virus Taxonomy Profile: Chrysoviridae. Journal of General Virology 101: 143–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Blomberg J, Coffin JM, et al. 2018. Ortervirales: new virus order unifying five families of reverse-transcribing viruses. Journal of Virology 92: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Schmidt T, Heslop-Harrison JS. 1998. Repetitive DNA elements as a major component of plant genomes. Annals of Botany 82: 45–55. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunii M, Kanda M, Nagano H, Uyeda I, Kishima Y, Sano Y. 2004. Reconstruction of putative DNA virus from endogenous rice tungro bacilliform virus-like sequences in the rice genome: implications for integration and evolution. BMC Genomics 5: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama K, Tabara M, Moriyama H, et al. 2020. Disturbance of floral colour pattern by activation of an endogenous pararetrovirus, petunia vein clearing virus, in aged petunia plants. Plant Journal 103: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Martienssen R. 2004. The role of RNA interference in heterochromatic silencing. Nature 431: 364–370. [DOI] [PubMed] [Google Scholar]
- Liu R, Koyanagi KO, Chen S, Kishima Y. 2012. Evolutionary force of AT-rich repeats to trap genomic and episomal DNAs into the rice genome: lessons from endogenous pararetrovirus. Plant Journal 72: 817–828. [DOI] [PubMed] [Google Scholar]
- Llorens C, Muñoz-Pomer A, Bernad L, Botella H, Moya A. 2009. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biology Direct 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens C, Futami R, Covelli L, et al. 2011. The Gypsy Database (GyDB) of mobile genetic elements: Release 2.0. Nucleic Acids Research 39: 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart BE, Menke J, Dahal G, Olszewski NE. 2000. Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. Journal of General Virology 81: 1579–1585. [DOI] [PubMed] [Google Scholar]
- Ma J, Jackson SA. 2006. Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Research 16: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiwald S, Weber B, Seibt KM, Schmidt T, Heitkam T. 2020. The Cassandra retrotransposon landscape in sugar beet (Beta vulgaris) and related Amaranthaceae: recombination and re-shuffling lead to a high structural variability. Annals of Botany 127: 91–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure MA. 2000. The complexities of genome analysis, the Retroid agent perspective. Bioinformatics 16: 79–95. [DOI] [PubMed] [Google Scholar]
- McGrath JMM, Funk A, Galewski P, et al. 2020. A contiguous de novo genome assembly of sugar beet EL10 (Beta vulgaris L.). bioRxiv doi: 10.1101/2020.09.15.298315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AR, Elena SF. 2015. Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology 476: 304–315. [DOI] [PubMed] [Google Scholar]
- Newburn LR, White KA. 2019. Trans-acting RNA-RNA interactions in segmented RNA viruses. Viruses 11: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant TE. 2006. Guide to NumPy. Provo, UT: Brigham Young University. [Google Scholar]
- Omae N, Suzuki M, Ugaki M. 2020. The genome of the Cauliflower mosaic virus, a plant pararetrovirus, is highly methylated in the nucleus. FEBS Letters 594: 1974–1988. [DOI] [PubMed] [Google Scholar]
- Paesold S, Borchardt D, Schmidt T, Dechyeva D. 2012. A sugar beet (Beta vulgaris L.) reference FISH karyotype for chromosome and chromosome-arm identification, integration of genetic linkage groups and analysis of major repeat family distribution. Plant Journal 72: 600–611. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2018. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Richert-Pöggeler KR, Shepherd RJ. 1997. Petunia vein-clearing virus: a plant pararetrovirus with the core sequences for an integrase function. Virology 236: 137–146. [DOI] [PubMed] [Google Scholar]
- Richert-Pöggeler KR, Noreen F, Schwarzacher T, Harper G, Hohn T. 2003. Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia. EMBO Journal 22: 4836–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa C, Kuo YW, Wuriyanghan H, Falk BW. 2018. RNA interference mechanisms and applications in plant pathology. Annual Review of Phytopathology 56: 581–610. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. 1984. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences of the USA 81: 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. 1998. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends in Plant Science 3: 195–199. [Google Scholar]
- Schmidt T, Metzlaff M. 1991. Cloning and characterization of a Beta vulgaris satellite DNA family. Gene 101: 247–250. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Jung C, Metzlaff M. 1991. Distribution and evolution of two satellite DNAs in the genus Beta. Theoretical and Applied Genetics 82: 793–799. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Schwarzacher T, Heslop-Harrison JS. 1994. Physical mapping of rRNA genes by fluorescent in-situ hybridization and structural analysis of 5S rRNA genes and intergenic spacer sequences in sugar beet (Beta vulgaris). Theoretical and Applied Genetics 88: 629–636. [DOI] [PubMed] [Google Scholar]
- Schubert I, Vu GTH. 2016. Genome stability and evolution: attempting a holistic view. Trends in Plant Science 21: 749–757. [DOI] [PubMed] [Google Scholar]
- Seibt KM, Schmidt T, Heitkam T. 2018. FlexiDot: highly customizable, ambiguity-aware dotplots for visual sequence analyses. Bioinformatics 34: 3575–3577. [DOI] [PubMed] [Google Scholar]
- Sicard A, Michalakis Y, Gutiérrez S, Blanc S. 2016. The strange lifestyle of multipartite viruses. PLoS Pathogens 12: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyshlyaev G, Voigt F, Blinov A, Barabas O, Novikova O. 2013. Acquisition of an Archaea-like ribonuclease H domain by plant L1 retrotransposons supports modular evolution. Proceedings of the National Academy of Sciences of the USA 110: 20140–20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staginnus C, Richert-Pöggeler KR. 2006. Endogenous pararetroviruses: two-faced travelers in the plant genome. Trends in Plant Science 11: 485–491. [DOI] [PubMed] [Google Scholar]
- Staginnus C, Gregor W, Mette MF, et al. 2007. Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species. BMC Plant Biology 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2010. Centromeres convert but don’t cross. PLoS Biology 8: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teycheney PY, Geering ADW, Dasgupta I, et al. 2020. ICTV Virus Taxonomy Profile: Caulimoviridae. Journal of General Virology 101: 1025–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Dasgupta I, Fuchs M, et al. 2017. ICTV Virus Taxonomy Profile: Secoviridae. Journal of General Virology 98: 529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi S. 2009. Matplotlib for Python developers. Birmingham, UK: Packt Publishing. [Google Scholar]
- Ulbrich E. 1934. Chenopodiaceae. In: Engler A, Prantl K, eds. Natürliche Pflanzenfamilien, 2nd edn. Leipzig: Wilhelm Engelmann, 379–584. [Google Scholar]
- Verver J, Schijns P, Hibi T, Goldbach R. 1987. Characterization of the genome of soybean chlorotic mottle virus. Journal of General Virology 68: 159–167. [Google Scholar]
- Waalwijk C, Flavell RA. 1978. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Research 5: 3231–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskom M, Botvinnik O, O’Kane Det al. 2018. mwaskom/seaborn: v0.9.0 (July 2018). Zenodo; doi: 10.5281/zenodo.1313201. [DOI]
- Weber B, Schmidt T. 2009. Nested Ty3-gypsy retrotransposons of a single Beta procumbens centromere contain a putative chromodomain. Chromosome Research 17: 379–396. [DOI] [PubMed] [Google Scholar]
- Weber B, Wenke T, Frömmel U, Schmidt T, Heitkam T. 2010. The Ty1-copia families SALIRE and Cotzilla populating the Beta vulgaris genome show remarkable differences in abundance, chromosomal distribution, and age. Chromosome Research 18: 247–263. [DOI] [PubMed] [Google Scholar]
- Weber B, Heitkam T, Holtgräwe D, et al. 2013. Highly diverse chromoviruses of Beta vulgaris are classified by chromodomains and chromosomal integration. Mobile DNA 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TJ, Eddy SR. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics 29: 2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. 2009. ggplot2: elegant graphics for data analysis, 1st edn. New York: Springer. [Google Scholar]
- Wollrab C, Heitkam T, Holtgräwe D, et al. 2012. Evolutionary reshuffling in the Errantivirus lineage Elbe within the Beta vulgaris genome. Plant Journal 72: 636–651. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. 1988. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Molecular Biology and Evolution 5: 675–690. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO Journal 9: 3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Wang X, Lu Z, Xu Y, Deng X, Xu Q. 2019. Endogenous pararetrovirus sequences are widely present in Citrinae genomes. Virus Research 262: 48–53. [DOI] [PubMed] [Google Scholar]
- Zakrzewski F, Weisshaar B, Fuchs J, et al. 2011. Epigenetic profiling of heterochromatic satellite DNA. Chromosoma 120: 409–422. [DOI] [PubMed] [Google Scholar]
- Zakrzewski F, Weber B, Schmidt T. 2013. A molecular cytogenetic analysis of the structure, evolution, and epigenetic modifications of major DNA sequences in centromeres of Beta species. In: Jiang J, Birchler JA, eds. Plant centromere biology. Oxford: John Wiley & Sons, 39–55. [Google Scholar]
- Zakrzewski F, Schmidt M, Van Lijsebettens M, Schmidt T. 2017. DNA methylation of retrotransposons, DNA transposons and genes in sugar beet (Beta vulgaris L.). Plant Journal 90: 1156–1175. [DOI] [PubMed] [Google Scholar]
- Zerbini FM, Briddon RW, Idris A, et al. 2017. ICTV Virus Taxonomy Profile: Geminiviridae. Journal of General Virology 98: 131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.