Abstract
Free-living amoeba (FLA) is widely distributed in the natural environment. Since these amoebae are widely found in various waters, they pose an important public health problem. The aim of this study was to detect the presence of Acanthamoeba, B. mandrillaris, and N. fowleri in various water resources by qPCR in Izmir, Turkey. A total of (n = 27) 18.24% Acanthamoeba and (n = 4) 2.7% N. fowleri positives were detected in six different water sources using qPCR with ITS regions (ITS1) specific primers. The resulting concentrations varied in various water samples for Acanthamoeba in the range of 3.2x105-1.4x102 plasmid copies/l and for N. fowleri in the range of 8x103-11x102 plasmid copies/l. The highest concentration of Acanthamoeba and N. fowleri was found in seawater and damp samples respectively. All 27 Acanthamoeba isolates were identified in genotype level based on the 18S rRNA gene as T4 (51.85%), T5 (22.22%), T2 (14.81%) and T15 (11.11%). The four positive N. fowleri isolate was confirmed by sequencing the ITS1, ITS2 and 5.8S rRNA regions using specific primers. Four N. fowleri isolates were genotyped (three isolate as type 2 and one isolate as type 5) and detected for the first time from water sources in Turkey. Acanthamoeba and N. fowleri genotypes found in many natural environments are straightly related to human populations to have pathogenic potentials that may pose a risk to human health. Public health professionals should raise awareness on this issue, and public awareness education should be provided by the assistance of civil authorities. To the best of our knowledge, this is the first study on the quantitative detection and distribution of Acanthamoeba and N. fowleri genotypes in various water sources in Turkey.
Introduction
Free-living amoeba (FLA) are unicellular protozoa that commonly find in soil and water throughout the world. Free-living amoeba could be found in tap water, well water, seawater, streams, river, swimming pools, dams, lakes, and air-conditioning systems [1]. Among the numerous FLA species present in nature, the most common species are Acanthamoeba, Balamuthia mandrillaris (B. mandrillaris), and Naegleria species, which play a role in human and animal infections [2]. Acanthamoeba spp. and B. mandrillaris may cause granulomatous amoebic encephalitis (GAE), cutaneous lesions, lung infections, and also Acanthamoeba keratitis (AK) in immunocompetent persons. Acanthamoeba genus is divided into 22 different genotypes based on the 18S rRNA gene, and genotype T4 is one of the most common in the environment and the most common genotype causing human infection [3, 4]. N. fowleri causes primary amoebic meningoencephalitis (PAM) in immunocompetent children and young adults [1, 5].
This pathogenic FLA enters the body via nasal mucosa and/or the skin lesions and then disseminate along the olfactory neuroepithelial route or by following the hematogenous spread route they gain entry into the brain to occur infection [6]. Quantitative screening of these amoebae in various water sources is crucial since they pose risk to human health. To date, in vitro culture methods were used to quantitatively assessment of aquatic FLA, but there are some limitations such as time-consuming procedure, precision, and accuracy [7]. Quantitative real-time PCR (qPCR) assay is a method with high specificity and sensitivity useful for detecting the presence of the amoebae in water resources [8, 9].
The aim of this study was to identify rapidly and accurately the presence of Acanthamoeba spp., B. mandrillaris, and N. fowleri in various water sources by qPCR assay. Moreover, the sensitivity, specificity, and efficiency of the qPCR were also evaluated. Finally, the water quality parameters were measured and Acanthamoeba culture-positive samples were subjected to osmo/thermo-tolerance test to measure their pathogenicity. In the light of the results obtained, identified isolates were evaluated as potential risks for humans.
Material and methods
The geographical location of the study area
The study is conducted in the province of Izmir, which is the third-largest city in western Turkey. Izmir is located between the northern latitudes 37° 45’ and 39° 15’ and 26° 15’ and the east longitudes 28° 20’ and have a surface area of 12.012 km2. Izmir city is in Mediterranean climate zone and summers are also hot and dry and followed by mild and wet winters. According to National Meteorological Service average temperature in summertime was higher than 30°C and highest temperature might be higher than 40°C. While the approximate population for 2020 was 4.394.694, this number increases even more during the summer months, since the region is one of the important touristic areas in Turkey. There are many irrigation dams, lakes, and ponds in this region.
Water sample collection and processing
A total of 148 water samples were collected from places within the boundaries of Izmir that could pose a risk to people and where human contact was high. Tap water (TW), pool water (PW), well water (WW), lake water (LW), dam water (DW), stream water (StW), seawater (SeW), and thermal spring water (TsW) were collected from various water sources and the geographic coordinates were shown (S1 Table). Water samples were collected in approximately one liter (lt) of the sterile glass bottles and stored at 4°C for subsequent analyses within 24 hr.
For culture and DNA isolation one liter of water sample was concentrated by filtration using a nitrocellulose membrane with a pore size of 0.22 μm (Sartorius Stedim Biotech, Göttingen, Germany). The filter membrane was divided into two equal parts with the help of a sterile scalpel and forceps, then half of the filter was transferred into the center of the NNA plate [10].
Analysis of water quality parameters
Water quality parameters, including total dissolved solids content (TDS), electrical conductivity (EC), and temperature were measured in situ using the portable thermometer (TDS&EC meter hold). The temperature, TDS, and EC of the parameters indicate sensitivity in degrees between 0.1 and 80.0°C (Celsius), 0 and 5000 ppm (parts per million), 0 and 9990 μs/cm, respectively. The chlorine level of the water samples was evaluated in situ using the chlorine test kit (Sutest Liquid Test Kit). The pH of the collected water samples was determined using a pH-meter (HI 2211–02, Hanna Instruments Inc., Woonsocket, MA, USA) in Ege University Parasitology Department Laboratory.
Culture of free-living amoeba
After filtration of each water sample, the half of the cut filter was placed in the center of 2% non-nutrient agar (NNA) plates previously seeded with heat killed 100 μl Escherichia coli (ATCC 25922) bacterial suspension and the edges of the plates were sealed around with parafilm (Heathrow Scientific, Vernon Hills, IL, USA). The plates were incubated in the inverted position at 30–32°C and exanimated daily with the inverted microscope for 10 days. Plates without proliferation were considered negative after a check of at least two weeks. FLA-positive plates were then sub-cultured by cutting off small pieces circled with a pen under the microscope and transferring them to new fresh NNA plates to purifying them from other organisms, especially fungi and yeasts. The grown of Acanthamoeba trophozoites and cysts were characterized from other free-living amoebas. Besides, the cyst shape has been easily identified by the double wall of the cyst and typical star shape [11, 12].
Tolerance assays for Acanthamoeba positive samples
Pathogenicity tests were repeated three times for each positive sample. The pathogen strains of Acanthamoeba castellanii and Acanthamoeba spp. (EU266547 –T4) from Cumhuriyet and Dokuz Eylül Universities were used as reference strains.
Osmo-tolerance assay
To investigate the effect of osmolarity of each isolate on the trophozoites of Acanthamoeba (approximately 103 trophozoites/plate), trophozoites were coated with mannitol-free E. coli and transferred to the center of NNA plates (as a control). Positive isolates (approximately 103 trophozoites/plate) were transferred to the center of the plates by coating the NNA plate with E. coli suspension prepared at 0.5 M and 1 M mannitol concentration. The plates were then incubated at 30°C for 10 days and the growth of amoebae at 24, 48, and 72 hours was evaluated. Trophozoites or cysts were counted at a microscopic magnification at x100 of five microscopic areas of approximately 20 mm from the center of each plate. The presence of proliferation was evaluated as (+) positive, and the absence of growth (-) as negative [13, 14].
Thermo-tolerance assay
For the thermo-tolerance assay, the trophozoites of Acanthamoeba spp. (approximately 103 trophozoites/plate) were transferred to the center of E. coli coated NNA plates. These plates were incubated at 30°C (as a control), 37°C, and 42°C for 10 days; It was evaluated after 24, 48, and 72 hours of the incubation. During this period, proliferation was evaluated under the microscope as mentioned in the osmo-tolerance test [13, 14].
DNA extraction from culture and filter membranes
For the DNA isolation of the amoeba from the NNA plates, which were identified as Acanthamoeba spp. by microscopy, 2 ml of 1xPBS (Thermo Fisher Scientific, Phosphate-Buffered Saline (PBS), pH: 7.4) buffer solution was dropped onto the plate. The amoebas from agar plates were collected into the tube using the sterile swab after waiting for approximately 5 minutes. The tubes were centrifuged at 2500 rpm for 10 min and washed with PBS buffer solution. Acanthamoeba genomic DNA was extracted with the QIAamp DNA mini kit (Qiagen GmbH, Germany) according to the manufacturer’s recommendations.
Half of the 0.22 μm diameter membrane of the filtered water sample was cut into several pieces with sterile scissors and transferred to the bead tube. Total genomic DNA was extracted using the Norgen Biotek Water RNA/DNA Purification Kit (Water RNA / DNA Norgen Biotek Corp., Canada) following the manufacturer’s protocol. Briefly, after placing the filter into the tube with beads, 500 μl Lysis buffer E was added. The tubes were vortexed for 30 sec using FastPrep®-24 instrument (MP Biomedical). The tubes were then incubated in the thermal block at 65°C for 10 min. and then centrifuged at 20 000 g for 1 min. After centrifugation, 600 μl ethanol was added to the mixture and transferred to filter tubes. The filter tubes were then washed twice with 400 μl of wash solution A and genomic DNA sample was obtained by adding 100 μl of elution buffer H. DNA concentration and purity were measured using the NanoDrop® 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). DNA samples were kept at -20°C until the PCR experiments.
Positive control plasmid for PCR
The positive control of Acanthamoeba spp. was obtained from a reference strain, which was isolated from a human case with Acanthamoeba keratitis (GenBank No: EU266547 –T4). DNA samples of N. fowleri and B. mandrillaris strains were obtained from the Center for Disease Control and Prevention (CDC).
The 18S rRNA gene for Acanthamoeba spp., and B. mandrillaris and the 5.8S rRNA and ITS (ITS1 and ITS2) regions for N. fowleri were selected as targets to determine the presence of the plasmid copy quantification in water samples [15–17]. The primers and conditions were used for amplification are the same as those used for the LightCycler 480 PCR test described below. A plasmid containing the PCR amplified product was commercially synthesized by Letgen Biotechnology Laboratory (Letgen Biotechnology, İzmir, Turkey) using the pGEM-T Vector cloning kit (Promega Corporation, Madison, WI) following the manufacturer’s instructions. The number of copies in the plasmid solution was calculated using a NanoDrop ND1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Serial dilutions (plasmid controls ranging from 1x109 to 1x100 copies plasmid/μl) on the order of 10-fold of 18S rRNA gene and ITS region fragment were used to generate the standard curve of concentrations expressed in log units (log10) versus the values obtained in amplification cycles. Quantification analysis for each plasmid control was performed with the Light Cycler 480 II® Thermal Cycler (Roche Diagnostic) on 96-well white LightCycler 480® multiwell plates (Roche Diagnostics Ltd, Switzerland).
Quantitative real-time PCR (qPCR) assay
The qPCR was performed using DNA obtained from cultures and direct water filters. The quantification of Acanthamoeba spp., B. mandrillaris, and N. fowleri DNA was performed by using a LightCycler 480 II (Roche Diagnostics, Mannheim, Germany) Real-Time PCR Systems. The targets sequence of the 180-bp (Acanthamoeba spp), 171-bp (B. mandrillaris), and 123-bp (N. fowleri) fragments of the 18S rRNA and ITS gene regions were amplified. The primers and the respective probes used in this study were described in S2 Table [15, 17]. Reaction was performed in a final volume of a 20 μl in 96-well plates, containing 5 μl DNA template or controls, 5 x TaqMan Master Mix (Roche), 0.25 μM each of primer and 0.2 μM of each probe and then centrifuged at 1500 g for 2 min at 4°C. The PCR conditions were conducted using the following calculated control protocol: 10 min pre-incubation step at 95°C, followed by 45 cycles of 10 sec at 95°C, 1 min at 60°C and 1 sec at 72°C and followed by a final cooling step at 40°C for 30 sec.
All qPCR reactions were run using positive control (plasmid DNA), negative control DNA (using double distilled water) and were tested in triplicate in each reaction. The Ct value (cycle threshold) was defined as the number of cycles required for the fluorescence signal to cross the threshold. The Ct value is inversely correlated with the quantity of DNA. A standard curve was constructed using a series of dilutions with a known quantity of plasmid DNA standards. The slope (S) of the standard curve was adopted as an indication of the efficiency of the real-time PCR amplification. The efficiency (E) of the qPCR amplification was calculated according to the equation of E = 10 (-1 /Slope)-1.
Assessment of possible the qPCR inhibition in water samples
Duplicate PCR reactions were performed to eliminate possible PCR inhibition that might arise from water samples. One of the reactions contained only purified water sample DNA, while the other reaction contained ten plasmid copies with the spiked into the purified water DNA sample. To generate a cycle threshold value, three reactions containing ten plasmid control DNA copies in distilled water were performed. The Ct values observed for the spiked water sample with a positive control plasmid differed from the mean >40 Ct value were considered to be indicative of PCR inhibition. The inhibited samples were diluted 10-fold and PCR was analyzed again as described above.
DNA sequencing and phylogenetic analysis
To genotype qPCR positive samples, conventional PCR was performed for Acanthamoeba spp. and Naegleria spp., with primers specific for 18S rRNA gene (JDP1-JDP2) and ITS region (FW2-RV2), respectively (S2 Table) [18, 19]. PCR was performed in a total volume of 50 μl including 25 μl of 2x PCR Master Mix, 10 pmol of each primer and 5 μl template DNA. reaction was carried out in a Techne TC-3000 Thermal Cycle (Techne, Staffordshire, UK) by following program: an initial denaturation at 95°C for 2 min followed by 35 cycles 95°C for 35 s, annealing step at 60°C and 58°C for 40 s for Acanthamoeba spp. and Naegleria spp. respectively and final extension 3 min at 72°C. All PCR products were separated by electrophoresis on 2% agarose gel, stained with Safeview Classic (Applied Biological Materials Inc., Richmond, Canada) and were photographed using an Alpha Imager HP (Alpha Innotech), on a UV transilluminator. All the PCR products were purified with the QIAquick PCR purification kit (QIAGEN, Germany) according to the manufacturer’s instructions. Sequencing was performed by a commercial company using an ABI automated sequencing system (Microsynth, Balgach, Switzerland). The resulting sequences were alimented using ClustalW based on sequence analysis of DF3 and ITS region as previously described [20, 21] by comparing to the available Acanthamoeba and N. fowleri DNA sequences in GenBank database (Department of Molecular Genetics, The Ohio State University, OH, USA). Phylogenetic relationship among the sequences was made using the neighbor-joining with molecular distances under the Kimura two-parameter distance model with the Molecular Evolutionary Genetics Analysis (MEGA X) software program [22]. The accuracy of the phylogenetic tree was assessed by 1000 bootstrap replicate data sets. The root of the tree was established using an outgroup (Saccamoeba lacustris GenBank No: JN112797.1). Sequence data obtained for Acanthamoeba and N. fowleri isolates were deposited in the GenBank database under the accession numbers MW689474–MW689500 and MW677627-MW677629, MW676178, respectively.
Statistical analysis
The data obtained were analyzed using Mac OSX SPSS 25.0 version (SPSS Inc, Chicago, IL, USA) software. Numerical variables were summarized using the mean and standard error of the mean. Kolmogorov-Smirnov test was used to determine the importance of normality. The Mann-Whitney U test was used to compare the relationship between water quality parameters and the presence/absence of Acanthamoeba spp., and N. fowleri in environmental water samples. A value of p≤0.05 was considered statistically significant.
Results
Isolation of Acanthamoeba spp. in culture
A total of 148 water samples were collected from 19 different districts of Izmir including tap water (n = 44), well water (n = 31), pool water (n = 26), lake water (n = 18), dam water (n = 10), stream (n = 9), seawater (n = 8) and thermal spring water (n = 2). From the total of samples, 71 (47.97%) were found to be positive for FLA. Eighteen out of 148 water samples (12.16%) included in this study were found positive for Acanthamoeba spp. according to Page’s morphological analysis criteria (Fig 1). Acanthamoeba were detected in various water sources including 12.9% (4/31) WW, 44.44% (4/9) StW, 6.81% (3/44) TW, 11.53% (3/26) PW, 11.11% (2/18) LW, and 25% (2/8) SeW (Table 1).
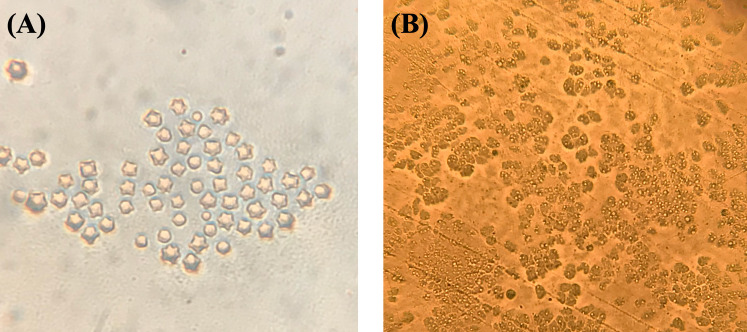
Fig 1. Acanthamoeba cysts and trophozoites in non-nutrient agar plate found in various water sources collected from in İzmir, Turkey.
A) Acanthamoeba cysts, B) Acanthamoeba trophozoites.
Table 1. Result of the qPCR assay and culture for Acanthamoeba spp. in various waters sources.
| Water sources | Sample No. | Culture | qPCR | Mean Ct value | Acanthamoeba spp. (plasmid copies/l) | ||
|---|---|---|---|---|---|---|---|
| N | (%) | N | (%) | ||||
| StW | 9 | 4 | 44.44 | 4 | 44.44 | 32.9 | 8.8x104-7.3x102 |
| LW | 18 | 2 | 11.11 | 5 | 27.77 | 37.2 | 1.2x103-1.4x102 |
| SeW | 8 | 2 | 25 | 2 | 25 | 31.2 | 3.2x105-5.3x103 |
| PW | 26 | 3 | 11.53 | 6 | 23.07 | 36.8 | 5.3x103-1.6x102 |
| WW | 31 | 4 | 12.9 | 6 | 19.35 | 34.4 | 1.6x104-1.4x103 |
| TW | 44 | 3 | 6.81 | 4 | 9.09 | 36.1 | 1.9x104-1.8x102 |
| Total | 136 | 18 | 13.23 | 27 | 19.85 | ||
TW: Tap water, PW: Pool water, WW: Well water, LW: Lake water, StW: Stream water, SeW: Seawater
Pathogenic potential of Acanthamoeba spp. in positive water samples
The results of the tolerance assay of 18 Acanthamoeba isolates grown in culture were shown in Table 2. All of the 18 Acanthamoeba isolates investigated were grown at 37°C and 0.5 M mannitol. Ten (55.5%) of the isolates were grown both at 42°C, and 1 M mannitol. Only eight (44.4%) of the isolates were not grown at 42°C and 1 M mannitol. Therefore, ten isolates (55.5%) were considered potentially pathogenic, and the rest (44.4%) were classified as low pathogenic potential.
Table 2. Thermo/osmo-tolerance assay of Acanthamoeba positive strains isolated from water samples in different districts of İzmir province.
| Tolerance Assay | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain No. | Sampling area | Locality | NNA | qPCR | PCR (JDP) | Species | Genotype | Osmo-tolerance (M, mannitol) | Thermo-tolerance (°C) |
| 0.5 / 1 | 37 / 42 | ||||||||
| IWS3 | Tap water | Torbalı | + | + | + | Acanthamoeba sp. | T4 | +/+ | +/+ |
| IWS24 | Tap water | Torbalı | + | + | + | Acanthamoeba sp. | T4 | +/+ | +/+ |
| IWS30 | Tap water | Çeşme | + | + | + | Acanthamoeba sp. | T5 | +/+ | +/+ |
| IWS47 | Pool water | Bornova | + | + | + | Acanthamoeba sp. | T2 | +/- | +/- |
| IWS56 | Pool water | Bornova | + | + | + | Acanthamoeba sp. | T5 | +/- | +/- |
| IWS60 | Pool water | Bornova | + | + | + | Acanthamoeba sp. | T4 | +/+ | +/+ |
| IWS78 | Well water | Bayraklı | + | + | + | Acanthamoeba sp. | T4 | +/+ | +/+ |
| IWS80 | Well water | Dikili | + | + | + | Acanthamoeba sp. | T15 | +/- | +/- |
| IWS93 | Well water | Bayraklı | + | + | + | Acanthamoeba sp. | T4 | +/+ | +/+ |
| IWS95 | Well water | Dikili | + | + | + | Acanthamoeba sp. | T2 | +/+ | +/+ |
| IWS103 | Lake water | Menderes | + | + | + | Acanthamoeba sp. | T5 | +/+ | +/+ |
| IWS114 | Lake water | Ödemiş | + | + | + | Acanthamoeba sp. | T4 | +/- | +/- |
| IWS130 | Stream water | Menderes | + | + | + | Acanthamoeba sp. | T2 | +/- | +/- |
| IWS132 | Stream water | Menderes | + | + | + | Acanthamoeba sp. | T5 | +/+ | +/+ |
| IWS134 | Stream water | Menderes | + | + | + | Acanthamoeba sp. | T4 | +/+ | +/+ |
| IWS136 | Stream water | Menderes | + | + | + | Acanthamoeba sp. | T4 | +/- | +/- |
| IWS142 | Seawater | Çeşme | + | + | + | Acanthamoeba sp. | T15 | +/- | +/- |
| IWS146 | Seawater | Çeşme | + | + | + | Acanthamoeba sp. | T4 | +/- | +/- |
(+): Positive samples, (-): Negative samples
The growth ability was evaluated by exposure at different temperatures (37°-42°C) and osmolarity ranges (0.5-1M mannitol) to test the pathogenicity potential of 18 Acanthamoeba positive samples in NNA plate culture. The isolates that can grow at high temperature (at 42°C) and high osmolarity (1 M mannitol) were considered as potential pathogenic strains. However, the other isolates that were able to grow at 37°C temperature and 0.5 M osmolarity were considered as low pathogens.
PCR amplification efficiency (E) and standard curve
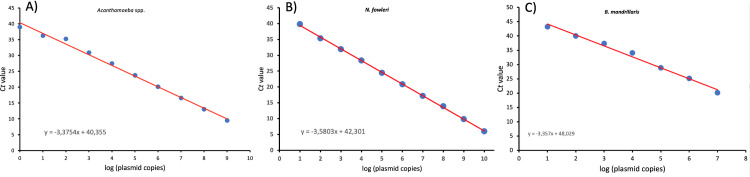
A standard curve was developed by serial dilutions of known amounts of plasmid DNA ranging 109 to 100 plasmids copy from Acanthamoeba, N. fowleri, and B. mandrillaris. The standard curve of the known plasmid concentrations and Ct values of Acanthamoeba, N. fowleri, and B. mandrillaris was shown in Fig 2. A slope (S) of -3.37, -3.58, and -3.35 equals a PCR amplification efficiency (E) of 97.8%, 90.2% and 98.5% for Acanthamoeba, N. fowleri and B. mandrillaris, respectively. Moreover, the determination coefficient (R2) of 0,99, 0,99, and 0,98 were observed for Acanthamoeba, N. fowleri, and B. mandrillaris, respectively. The lowest detection limit for Acanthamoeba, and N. fowleri of the qPCR assay was determined as one plasmid copy per reaction. However, the lowest detection limit for B. mandrillaris was set at 10 plasmid control DNA copies.
Fig 2.
Standard curves were generated by linear regression of the cycle threshold (Ct) versus (A) Acanthamoeba, (B) N. fowleri, and (C) B. mandrillaris plasmid control DNAs.
Quantification of Acanthamoeba and N. fowleri from various water
A total of 27/148 (18.24%) Acanthamoeba spp. and 4/148 (2.7%) N. fowleri positives were detected in six different water sources by the qPCR assay. However, all water samples were found negative for B. mandrillaris. The qPCR assay was applied to confirm positive samples in terms of FLA in culture, and 18 (12.16%) Acanthamoeba and four (2.7%) N. fowleri were found to be positive. The overall presence of Acanthamoeba spp. in various water sources were 4/9 (44.44%) in StW, 5/18 (27.77%) in LW, 2/8 (25%) in SeW, 6/26 (23.07%) in PW, 6/31 (19.35%) in WW, and 4/44 (9.09%) in TW (Table 1).
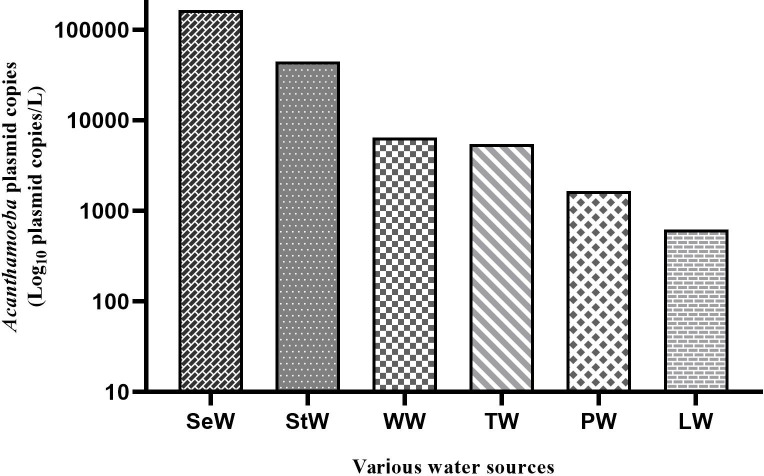
Acanthamoeba spp. were found more frequent in streams samples than tap waters. However, it could not detect in dam water and thermal spring water. The Ct value of PCR amplification of Acanthamoeba in StW, LW, SeW, PW, WW, and TW in the ranges from 28.2 to 39.4, and also plasmid copies concentrations of Acanthamoeba 18S rRNA gene were detected in the range of 3.2x105-1.4x102 plasmid copies/l. In our study, the highest concentration of Acanthamoeba was found quantitatively in seawater samples, while the lowest concentration was found in lake water (Fig 3).
Fig 3. DNA quantity of Acanthamoeba plasmid copies determined by qPCR assay in various water sources from İzmir, Turkey.
N. fowleri was detected in various water sources, 2/26 (7.69%) in PW, 1/18 (5.5%) in LW, and 1/10 (10%) in DW samples. For various water sources in PW, LW, and DW, means of Ct values was ranged from 35.13 to 38.13, and plasmid copies of N. fowleri were between 8x103 and 11x102 plasmid copies/l. The average highest concentration of N. fowleri was shown in dam water (Table 3). Two water samples which sample code PW12 and LW4 were detected positive both Acanthamoeba and N. fowleri.
Table 3. Result of the qPCR assay for Naegleria fowleri in various waters.
| Strain No. | Sample code | Mean Ct value | N. fowleri (plasmid copies/l) |
|---|---|---|---|
| IWS52 | PW8 | 38.13 | 11 x102 |
| IWS56 | PW12 | 37.71 | 15 x102 |
| IWS105 | LW4 | 36.68 | 3 x103 |
| IWS120 | DW1 | 35.13 | 8 x103 |
IWS: Izmir water sample, PW: Pool water, LW: Lake water, DW: Dam water
Genotyping of Acanthamoeba spp. and N. fowleri-positives in water samples
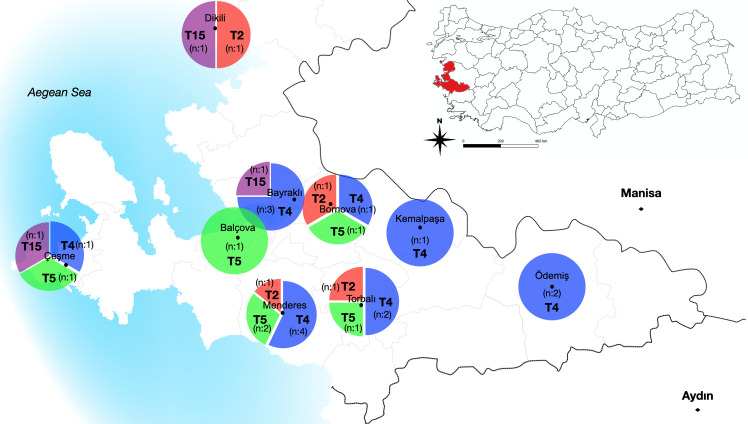
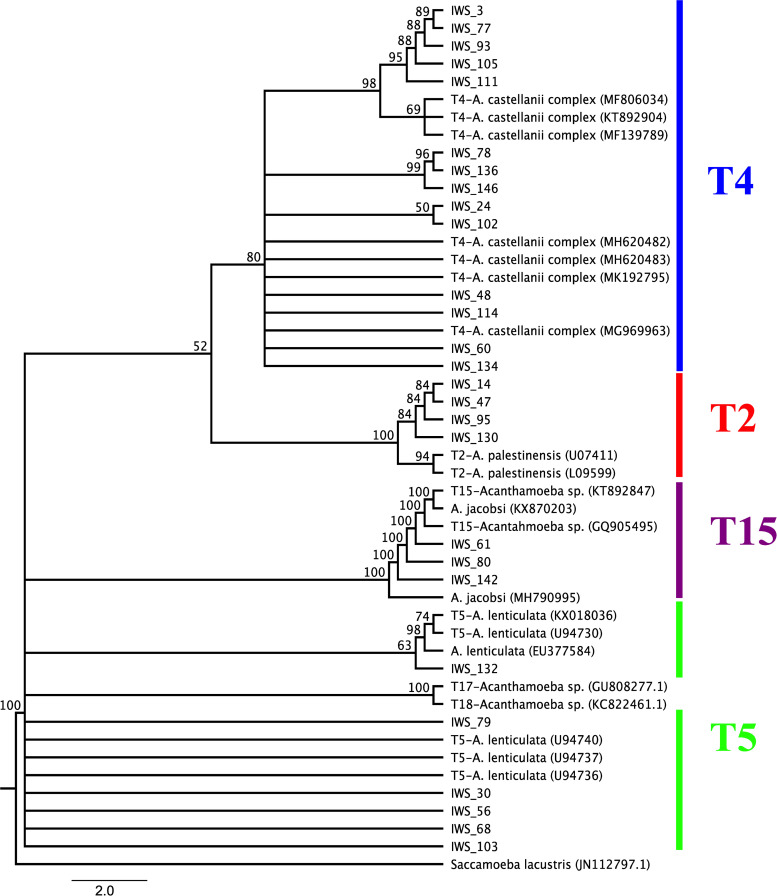
All 27 Acanthamoeba isolates detected positive by qPCR were subjected for PCR amplification using JDP primer sets, which is specific for the DF3 region of the 18S rRNA gene sequences. The sequence data obtained from Acanthamoeba isolates were aligned using Clustal W software and were used to construct the phylogenetic tree to illustrate the relationships between the isolates obtained and reference sequences of Acanthamoeba genotypes retrieved from GenBank (A. palestinensis genotype T2 accession nos: U07411 and L09599; A. castellanii genotype T4 accession nos: MF806034, KT892904, MF139789,MH620482, MH620483, MK192795 and MG969963; A. lenticulate genotype T5 accession nos: KX018036, U94730, EU377584, U94740, U94737 and U94736; A. jacobsi genotype T15 accession nos:KX870203, KT892847, GQ905495 and MH790995; Acanthamoeba genotype T17 accession no:GU808277; Acanthamoeba genotype T18 accession no:KC822461). All samples showed nucleotide identity between 98% and 100% with reference strains deposited in GenBank (BLASTn) (www.ncbi.nlm.nih.gov/BLAST). In this study, four different genotypes (T2, T4, T5, and T15) were detected in water samples. Fourteen (51.85%) of 27 Acanthamoeba isolates were belonged to T4 genotype. The remaining isolates were belonging to T5 genotype 6/27 (22.22%), T2 genotype 4/27 (14.81%) and T15 genotype 3/27 (11.11%). The distribution of Acanthamoeba genotypes according to different districts of İzmir province and various water resources was showed in Fig 4 and Table 4. According to the phylogenetic tree, Acanthamoeba isolates T4 obtained from various water sources were grouped within the clade including the other sequences of Acanthamoeba castellanii complex available from Genbank. Six isolates (IWS_132, IWS_79, IWS_30, IWS_56, IWS_68, IWS_103) were found to be Acanthamoeba genotype T5 revealing 98% sequence identity to various T5 reference strain. Phylogenetic tree showed that three isolates (IWS_61, IWS_80 and IWS_142) were strictly related with Acanthamoeba T15 genotype chosen as references with 100% of identity, four isolates (IWS_14, IWS_47, IWS_95 and IWS_130) T2 genotype with 98% of identity with the T2 sequence references (Fig 5).
Fig 4. The distribution of Acanthamoeba genotypes according to different districts of İzmir province and various water resources.
Table 4. Distribution of Acanthamoeba genotype in various water sources.
| Genotype | Total | LW | WW | TW | StW | PW | SeW | |
|---|---|---|---|---|---|---|---|---|
| % | n | |||||||
| T4 | 51.85 | 14 | 4 | 3 | 2 | 2 | 2 | 1 |
| T5 | 22.22 | 6 | 1 | 1 | 1 | 1 | 1 | |
| T2 | 14.81 | 4 | 1 | 1 | 1 | 1 | ||
| T15 | 11.11 | 3 | 1 | 1 | 1 | |||
| Total | 100 | 27 | 5 | 6 | 4 | 4 | 5 | 2 |
TW: Tap water, PW: Pool water, WW: Well water, LW: Lake water, StW: Stream water, SeW: Seawater
Fig 5. Phylogenetic tree inferred using neighbor-joining models of the 18S rRNA gene DF3 region sequence data for the Acanthamoeba genotypes by MEGA X.
Bootstrap values are based on 1000 replicates and the root of the tree evaluated by an outgroup Saccamoeba lacustris.
Four positive samples detected N. fowleri by qPCR were shown to be 99–100% nucleotide identity with references isolates in GenBank (N. fowleri accession nos: AJ132028, X96565, AJ132019 and X96564). Genotypic differences of N. fowleri can be distinguished based on a one bp transition in 5.8S rRNA and the length of the internally transcribed spacer (1) [23]. Among the N. fowleri positive water samples (DW1, LW1and PW12), three of them do have T at position 31 in the 5.8S rRNA, and the ITS1 length was 42 bp, so these were identified as belonging to type 2. However, in one N. fowleri (PW8) sample, the sequence C has not changed to T at position 31 in 5.8S rRNA and the ITS1 length was found to be 84 bp with a two bp deletion in the sequence of repeat. According to obtained data this sample was identified as type 5 (Table 5). The phylogenetic tree was shown using neighbor-joining models of 5.8S rRNA, ITS1, and ITS2 sequence data for Naegleria spp. (Fig 6). According to the phylogenetic tree, N. fowleri isolates obtained from various water sources were grouped within the clade including the other sequences of N. fowleri genotypes available from GenBank. Three isolates (LW1, DW1 and PW12) were found to be N. fowleri type 2 revealing 100% sequence identity to various N. fowleri type 2 reference strains (N. fowleri type 2 accession nos: AJ132019 and X96564). However, phylogenetic tree showed that one isolates (PW8) were strictly related with N. fowleri type 5 chosen as references (N. fowleri type 5 accession nos: AJ132028 and X96565) with 99% of identity. In the present study, N. fowleri type 2 and type 5 were identified in water samples for the first in Turkey.
Table 5. Base length and position of ITS1, ITS2 and 5.8S rRNA sequences of references strains and all the positive isolates in this study.
| Isolate | Genotype | ITS1 | 5.8S | Position 31 in 5.8S | ITS2 | Total length | Accession number | Reference |
|---|---|---|---|---|---|---|---|---|
| 7853 | T1 | 42 | 175 | C | 106 | 323 | AY376149 | [1] |
| AR12 | T2 | 42 | 175 | T | 106 | 323 | X96564 | [2] |
| LEE | T3 | 86 | 175 | T | 106 | 367 | X96562 | [2] |
| Ch2-1-f2 | T4 | 86 | 175 | T | 106 | 367 | AJ132030 | [3] |
| Na 420c | T5 | 84 | 175 | C | 106 | 365 | AJ132028 | [3] |
| J2B2 | T6 | 114 | 175 | C | 106 | 395 | FR875287 | [4] |
| M4E | T7 | 142 | 175 | T | 106 | 423 | X96563 | [2] |
| C0504 | T8 | 130 | 175 | T | 106 | 411 | FR875288 | [4] |
| DW1 | T2 | 42 | 175 | T | 106 | 323 | MW677629 | This study |
| LW1 | T2 | 42 | 175 | T | 106 | 323 | MW677627 | This study |
| PW12 | T2 | 42 | 175 | T | 106 | 323 | MW676178 | This study |
| PW8 | T5 | 84 | 175 | C | 106 | 365 | MW677628 | This study |
[1] Zhou et al., 2003.
[2] De Jonckheere, 1998.
[3] Pélandakis et al., 2000.
[4] De Jonckheere, unpublished.
Fig 6. Phylogenetic tree of Naegleria isolates for the ITS sequences.

Phylogenetic tree inferred using neighbor-joining cluster analysis of the sequence obtained and the sequences from ITS1, 5.8S rRNA and ITS2 sequences of various Naegleria strains, with special reference to N. fowleri produced in MEGA X. Bootstrap values are based on 1000 replicates.
Associations between Acanthamoeba and water quality parameter variables
A non-parametric test was conducted to determine the relationship between five different water quality parameters and the presence/absence of Acanthamoeba in various water sources. The result of the non-parametric statistical test was shown in Table 6. A significant relationship with Mann-Whitney U test was found between the presence/absence of Acanthamoeba spp. and pH values from well water and also EC-TDS values from pool water, lake water, and sea waters (p<0.05).
Table 6. Result of non-parametric test showing the relationship between five different water quality parameters and the presence/absence of Acanthamoeba spp. in various water sources.
| Water quality parameters | Mann-Whitney U test | |||||
|---|---|---|---|---|---|---|
| TW | PW | WW | LW | StW | SeW | |
| pH value | P = 0,390 | P = 0,223 | P = 0,004 * | P = 0,692 | P = 0,135 | P = 0,495 |
| Temperature (°C) | P = 0,744 | P = 0,271 | P = 0,515 | P = 0,235 | P = 0,618 | P = 0,495 |
| Free Cl (ppm (mg/l)) | P = 0,163 | P = 1,000 | P = 0,118 | P = 1,000 | P = 0,091 | P = 1,000 |
| a EC (0–9990 μs/cm) | P = 0,624 | P = 0,002 * | P = 0,146 | P = 0,002 * | P = 0,135 | P = 0,008 * |
| b TDS (0–9990 ppm) | P = 0,540 | P = 0,002 * | P = 0,051 | P = 0,002 * | P = 0,618 | P = 0,040 * |
a Electrical conductivity,
b Total dissolved solids
* Statistically significant p < 0.05
TW: Tap water, PW: Pool water, WW: Well water, LW: Lake water, StW: Stream water, SeW: Seawater
Discussion
Among the FLA, Acanthamoeba spp., N. fowleri, and B. mandrillaris are eukaryotic protists widely found in many places of the world. These FLAs can potentially cause opportunistic/non-opportunistic infections in humans and animals [1, 24]. Recently, they have received increasing attention in the medical and scientific world due to the serious fatal infections in humans. The impact of Acanthamoeba and N. fowleri on human health is associated with the genotypes of these pathogens and their reproduction in water and soil resources, a natural reservoir location. Therefore, preventive, and investigational monitoring programs for measuring the density of Acanthamoeba and N. fowleri are important in aquatic environments with human exposure, which can be achieved with real-time qPCR [3]. This study was occurred the quantify presence of Acanthamoeba and N. fowleri from various water sources in province İzmir, Turkey. Acanthamoeba spp. were found positive ranging from 4.4% to 50% in various water sources such as tap water, ponds, rivers, streams, and water wells in Turkey [25–29]. Acanthamoeba was detected positively 17.3%, 28.8%, 15.9% and 42.9% in Jamaica, Iran, Thailand, and Uganda in tap water sources, respectively [30–33]. The results of this study which indicate that the Acanthamoeba spp. occurrence in various water sources (18.24%) are similar to those results obtained in the America, Brazil, and Japan Jamaica, Iran, Thailand, Turkey. In previous reports, Naegleria spp. was found in various water sources worldwide at 0.6%–60.9% all over the world, and 0.7–10% in Turkey. [26, 34, 35]. We conclude that Naegleria spp. and Acanthamoeba spp. are free-living amoebas that have suitable growth in various water sources worldwide, but detection rates at different regions may be influenced by water types and geographical conditions.
The qPCR assay is a method that can effectively detect and quantify amoeba even it was failed to culture due to low sample. However, standard curves characterizing the relationship between plasmid copy number and qPCR data can facilitate the quantification of microorganisms in environmental water samples [8, 17]. In this study, the lowest detection limit (one plasmid copy) was detected for Acanthamoeba and N. fowleri for each reaction by qPCR method. In previous reports, similar results was identified the lowest detection limit of one plasmid copy DNA [9]. In this study, the quantitative amount of Acanthamoeba spp. DNA obtained from direct water samples was ranged from 3.2x105 to 1.4x102 plasmid copies/l. In previous studies performed in Germany and Taiwan, Acanthamoeba spp. was reported to range from 2.0–3.0×103 and 2.0×102–9.0×104 and 3.4–5.0×103−2.2–1.4×103 amoebae/l, respectively [8, 36]. We thought that might be the reason why Acanthamoeba is detected in high concentrations in seawater samples is due to the mixing of sewage wastes or industrial wastewater to the beaches. Moreover, The World Health Organization (WHO 2006) reported the presence of Acanthamoeba in seawater to be associated with sewage and waste effluent outlets. Lorenzo-Morales et al. reported the high rate of isolation (49.6% and 64.0%) for Acanthamoeba in seawater due to the sewage-waste and industrial effluent [30, 37]. In our study, the quantitative amount of N. fowleri in dam, lake, and pool water samples was determined as between 11x102 and 8.0x103 plasmid copies/l. Recent study performed in Belgium was reported that Naegleria spp. in cooling water samples was 6.3x102–4.1x103 cells/l [7]. In Australia, Naegleria spp. have been reported in the range of 4–3.4×102 cells/l in the drinking water sample [38]. Naegleria spp. concentration has been reported in the range of 1.1–24.2 cells/l in hot spring and drinking water samples in Taiwan [39].
Up to date, 22 genotypes (T1-22) of Acanthamoeba have been identified as a result of the sequence analysis of the DF3 region of the 18S rRNA gene, but this classification containing both pathogens and non-pathogens genotypes [3, 40]. Among these genotypes, it has been reported that the T4 genotype was found the most common in environmental and clinical samples and is the pathogen of different diseases (AK, GAE, skin lesions) [5, 41]. Furthermore, the other genotypes including T2, T3, T5, T6, T10, T11, T12, T15, and T18 was reported the association with human infections [1, 42, 43]. In our study, genotype T4 (51.85%) was determined as the most common genotype in water samples. Apart from that, T5 (22.22%), T2 (14.81%), and T15 (11.11%) genotypes were detected in water samples. In earlier studies, genotypes T1, T2, T3, T4, and T7 of Acanthamoeba spp. were detected in freshwater resources in Egypt [44]. In another study, the genotype T4 (51%), as well as T14 (18%), T5 (11%), T3, T15, T16 and T10 (4% per each), T11 (3%) and T7, T9 (1% per each) was reported the most common genotype in water samples in Tunisia [45]. The most common genotypes, T4 (93.7%) and T2 (6.25%) were determined in the geothermal river in the southwest of Iran by Niyyati et al. [46]. The most common T4 and T2, T3, T5, T11, T15 genotypes of Acanthamoeba were detected in environmental water samples in Turkey [34, 47–50]. In the previous study T4 and T5 genotypes were reported in keratitis wild birds in İzmir [51]. Moreover, Acanthamoeba T4 genotypes were detected in corneal scraping samples taken from patients with suspected Acanthamoeba keratitis in Turkey [52, 53]. Acanthamoeba genotypes T4 and T5 commonly found in environmental samples pose a greater risk to humans and animals.
N. fowleri, which is the pathogen type of the Naegleria genus for humans, causes fatal PAM. N. fowleri is also a protist pathogen widely found in the environment including rivers, lakes, dams, hot springs, geothermal springs, untreated and treated domestic water sources, and swimming pools [54, 55]. In this study, N. fowleri was found positive for the first time in environmental water resources collected in İzmir province, Turkey. N. fowleri was found positively in various water sources including in 7.69% (2/26) pool water, 10% (1/10) dam water and 5.5% (1/18) lake water. The presence of Naegleria species has been reported to be positively detected in environmental water samples at a rate of 13.2–60.9% in Europe, 3–46% in the USA and 0.6–56.9% in Asia [7, 56–58]. Up to date, there are 47 species in the genus of Naegleria and also eight genotype of N. fowleri [58, 59] were identified. N. fowleri genotypes are characterized that difference 1, 2, 3, and 4 types are the only C to T transition at position 31 in the 5.8S rRNA sequence, because of the equal of the ITS lengths. However, N. fowleri type 5 has not identical the ITS1 lengths with the T at position 31 in the 5.8S rRNA sequence [23, 60, 61]. In this study N. fowleri type 2 and type 5 were isolated for the first time from water sources in Turkey. N. fowleri type 2 and 3 have been reported in many patients and water samples worldwide. Type 2, 3, 4, 5, 6, 7, and 8 in Europe, types 1, 2, and 3 in the USA, types 2 and 3 in Asia and only one type of type 5 in the Western Pacific (Oceania and Japan) has been found in human specimens and water samples [18, 23, 62]. Since there are a limited number of studies typing N. fowleri, the information about the pathogenicity of the types is insufficient. However, there is not yet conclusive evidence of any difference in virulence for any of the detected N. fowleri types. It is likely to be detected in humans, as types 2, 3, and 5 are the most common in waters.
Conclusions
In conclusion, the present study reports both the presence and the concentration of Acanthamoeba and N. fowleri in various water sources and demonstrates their rapidly determination by qPCR. Although the presence of Acanthamoeba was found higher in stream samples, the quantitative value of Acanthamoeba was detected higher in seawater samples. Therefore, it should be kept in mind that it may pose a risk for people during sea activities in summer in Izmir, which is a holiday region. Acanthamoeba T4 and T5 genotypes, which are commonly detected as causal agents of AK and GAE infection, were found at a high rate in various water sources in our study. In this study, N. fowleri type 2 and type 5 were isolated the first time from water sources in Turkey. Since the genotypes of Acanthamoeba spp. and N. fowleri types can be detected in many environments, they have pathogenic potentials that may pose a risk to human health. For this reason, a mandatory inspection is necessary, especially in potable waters, as it may pose a risk to people in swimming and recreational waters. In this study, the presence of pathogenic potential of the identified strains has been revealed. However, further studies needs to be done using environmental samples across Turkey. Also, clinicians and public health professionals should increase awareness about these issues by the help of civilian authorities.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
We are grateful to Ibne Karim M. Ali who provided N. fowleri and B. mandrillaris positive control DNA from Free-Living and Intestinal Amebas (FLIA) Lab, CDC, Atlanta, Georgia, USA. We would like to thank the colleague Dr. Mehmet Karakus for editing an earlier draft of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research was supported by a grant from the Budget of the Academic Staff Training Program by The Council of Higher Education and the Scientific Research Projects Branch Directorate of Ege University, Turkey (Project No: 18-TIP-025). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Khan NA. Acanthamoeba: biology and increasing importance in human health. FEMS microbiology reviews. 2006;30(4):564–95. doi: 10.1111/j.1574-6976.2006.00023.x . [DOI] [PubMed] [Google Scholar]
- 2.Schuster FL, Visvesvara GS. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. International journal for parasitology. 2004;34(9):1001–27. doi: 10.1016/j.ijpara.2004.06.004 . [DOI] [PubMed] [Google Scholar]
- 3.Angelici MC, Walochnik J, Calderaro A, Saxinger L, Dacks JB. Free-living amoebae and other neglected protistan pathogens: Health emergency signals? European journal of protistology. 2020;77:125760. Epub 2020/12/20. doi: 10.1016/j.ejop.2020.125760. [DOI] [PubMed] [Google Scholar]
- 4.Fuerst PA, Booton GC, Crary M. Phylogenetic analysis and the evolution of the 18S rRNA gene typing system of Acanthamoeba. The Journal of eukaryotic microbiology. 2015;62(1):69–84. doi: 10.1111/jeu.12186 . [DOI] [PubMed] [Google Scholar]
- 5.Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS immunology and medical microbiology. 2007;50(1):1–26. doi: 10.1111/j.1574-695X.2007.00232.x . [DOI] [PubMed] [Google Scholar]
- 6.Marciano-Cabral F, Cabral G. Acanthamoeba spp. as agents of disease in humans. Clinical microbiology reviews. 2003;16(2):273–307. doi: 10.1128/CMR.16.2.273-307.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behets J, Declerck P, Delaedt Y, Verelst L, Ollevier F. A duplex real-time PCR assay for the quantitative detection of Naegleria fowleri in water samples. Water research. 2007;41(1):118–26. Epub 2006/11/14. doi: 10.1016/j.watres.2006.10.003 . [DOI] [PubMed] [Google Scholar]
- 8.Chang CW, Wu YC, Ming KW. Evaluation of real-time PCR methods for quantification of Acanthamoeba in anthropogenic water and biofilms. Journal of applied microbiology. 2010;109(3):799–807. doi: 10.1111/j.1365-2672.2010.04708.x . [DOI] [PubMed] [Google Scholar]
- 9.Kao PM, Tung MC, Hsu BM, Tsai HL, She CY, Shen SM, et al. Real-time PCR method for the detection and quantification of Acanthamoeba species in various types of water samples. Parasitology research. 2013;112(3):1131–6. Epub 2013/01/12. doi: 10.1007/s00436-012-3242-x . [DOI] [PubMed] [Google Scholar]
- 10.Tawfeek GM, Bishara SA-H, Sarhan RM, Taher E, Khayyal A. Genotypic, physiological, and biochemical characterization of potentially pathogenic Acanthamoeba isolated from the environment in Cairo, Egypt. Parasitology research. 2016:1–11. doi: 10.1007/s00436-016-4927-3 [DOI] [PubMed] [Google Scholar]
- 11.Pussard M, Pons R. Morphology of cystic wall and taxonomy of genus Acanthamoeba (Protozoa, Amoebida). Protistologica. 1977;13(4):557–98. [Google Scholar]
- 12.Page FC. A new key to freshwater and soil gymnamoebae: with instructions for culture. 1988. [Google Scholar]
- 13.Todd CD, Reyes-Batlle M, Martin-Navarro CM, Dorta-Gorrin A, Lopez-Arencibia A, Martinez-Carretero E, et al. Isolation and Genotyping of Acanthamoeba Strains from Soil Sources from Jamaica, West Indies. Journal of Eukaryotic Microbiology. 2015;62(3):416–21. doi: 10.1111/jeu.12197 PubMed PMID: WOS:000353551300015. [DOI] [PubMed] [Google Scholar]
- 14.Caumo K, Frasson AP, Pens CJ, Panatieri LF, Frazzon AP, Rott MB. Potentially pathogenic Acanthamoeba in swimming pools: a survey in the southern Brazilian city of Porto Alegre. Annals of tropical medicine and parasitology. 2009;103(6):477–85. doi: 10.1179/136485909X451825 . [DOI] [PubMed] [Google Scholar]
- 15.Mull BJ, Narayanan J, Hill VR. Improved Method for the Detection and Quantification of Naegleria fowleri in Water and Sediment Using Immunomagnetic Separation and Real-Time PCR. Journal of parasitology research. 2013;2013:608367. doi: 10.1155/2013/608367; PubMed Central PMCID: PMC3818898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Streby A, Mull BJ, Levy K, Hill VR. Comparison of real-time PCR methods for the detection of Naegleria fowleri in surface water and sediment. Parasitology research. 2015;114(5):1739–46. Epub 2015/04/10. doi: 10.1007/s00436-015-4359-5 ; PubMed Central PMCID: PMC4631115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qvarnstrom Y, Visvesvara GS, Sriram R, da Silva AJ. Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J Clin Microbiol. 2006;44(10):3589–95. doi: 10.1128/JCM.00875-06 ; PubMed Central PMCID: PMC1594764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelandakis M, Serre S, Pernin P. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. The Journal of eukaryotic microbiology. 2000;47(2):116–21. Epub 2000/04/06. doi: 10.1111/j.1550-7408.2000.tb00020.x . [DOI] [PubMed] [Google Scholar]
- 19.Schroeder JM, Booton GC, Hay J, Niszl IA, Seal DV, Markus MB, et al. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J Clin Microbiol. 2001;39(5):1903–11. doi: 10.1128/JCM.39.5.1903-1911.2001 ; PubMed Central PMCID: PMC88046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Booton GC, Kelly DJ, Chu YW, Seal DV, Houang E, Lam DSC, et al. 18S Ribosomal DNA Typing and Tracking of Acanthamoeba Species Isolates from Corneal Scrape Specimens, Contact Lenses, Lens Cases, and Home Water Supplies of Acanthamoeba Keratitis Patients in Hong Kong. Journal of Clinical Microbiology. 2002;40(5):1621–5. doi: 10.1128/JCM.40.5.1621-1625.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panda A, Khalil S, Mirdha BR, Singh Y, Kaushik S. Prevalence of Naegleria fowleri in Environmental Samples from Northern Part of India. PloS one. 2015;10(10):e0137736. Epub 2015/10/21. doi: 10.1371/journal.pone.0137736; PubMed Central PMCID: PMC4618853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution. 2018;35(6):1547–9. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Jonckheere JF. Origin and evolution of the worldwide distributed pathogenic amoeboflagellate Naegleria fowleri. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2011;11(7):1520–8. Epub 2011/08/17. doi: 10.1016/j.meegid.2011.07.023 . [DOI] [PubMed] [Google Scholar]
- 24.Cope JR, Ali IK, Visvesvara GS. 107—Pathogenic and Opportunistic Free-Living Ameba Infections. In: Ryan ET, Hill DR, Solomon T, Aronson NE, Endy TP, editors. Hunter’s Tropical Medicine and Emerging Infectious Diseases (Tenth Edition). London: Content Repository Only!; 2020. p. 814–20. [Google Scholar]
- 25.Coskun KA, Ozcelik S, Tutar L, Elaldi N, Tutar Y. Isolation and identification of free-living amoebae from tap water in Sivas, Turkey. BioMed research international. 2013;2013:675145. doi: 10.1155/2013/675145; PubMed Central PMCID: PMC3736494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozcelik S, Coskun KA, Yunlu O, Alim A, Malatyali E. The prevalence, isolation and morphotyping of potentially pathogenic free-living amoebae from tap water and environmental water sources in Sivas. Turkiye parazitolojii dergisi / Turkiye Parazitoloji Dernegi = Acta parasitologica Turcica / Turkish Society for Parasitology. 2012;36(4):198–203. doi: 10.5152/tpd.2012.48 . [DOI] [PubMed] [Google Scholar]
- 27.Kuk S, Yazar S, Dogan S, Çetinkaya Ü, Sakalar Ç. Molecular characterization of Acanthamoeba isolated from Kayseri well water. Turkish Journal of Medical Sciences. 2013;43(1):12–7. [Google Scholar]
- 28.Yazar S, Gurbuz E, Sonmez MF, Cetinkaya U, Kuk S. Investigation of Potentially Pathogenic Free-Living Amoebae and Their In Vivo Pathogenicity in Water Supplies of Turkey. Mikrobiyoloji Bulteni. 2016;50(3):449–59. PubMed PMID: WOS:000381942800010. doi: 10.5578/mb.27898 [DOI] [PubMed] [Google Scholar]
- 29.Ustunturk-Onan M, Walochnik J. Identification of free-living amoebae isolated from tap water in Istanbul, Turkey. Experimental parasitology. 2018;195:34–7. Epub 2018/10/20. doi: 10.1016/j.exppara.2018.10.002 . [DOI] [PubMed] [Google Scholar]
- 30.Todd CD, Reyes-Batlle M, Pinero JE, Martinez-Carretero E, Valladares B, Streete D, et al. Isolation and molecular characterization of Acanthamoeba genotypes in recreational and domestic water sources from Jamaica, West Indies. Journal of water and health. 2015;13(3):909–19. doi: 10.2166/wh.2015.232 PubMed PMID: WOS:000360793100028. [DOI] [PubMed] [Google Scholar]
- 31.Thammaratana T, Laummaunwai P, Boonmars T, Neglected z, vector-borne disease research g. Isolation and identification of Acanthamoeba species from natural water sources in the northeastern part of Thailand. Parasitology research. 2016. doi: 10.1007/s00436-016-4911-y. [DOI] [PubMed] [Google Scholar]
- 32.Sente C, Erume J, Naigaga I, Magambo PK, Ochwo S, Mulindwa J, et al. Occurrence and genetic characterisation of Acanthamoeba spp. from environmental and domestic water sources in Queen Elizabeth Protected Area, Uganda. Parasites & vectors. 2016;9:127. doi: 10.1186/s13071-016-1411-y; PubMed Central PMCID: PMC4776447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohammadi Manesh R, Niyyati M, Yousefi HA, Eskandarian AA. Isolation of Acanthamoeba spp. from different water sources in Isfahan, central Iran, 2014. Journal of Parasitic Diseases. 2016. doi: 10.1007/s12639-015-0716-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Değerli S, Değerli N, Çamur D, Doğan Ö, İlter H. Genotyping by Sequencing of Acanthamoeba and Naegleria Isolates from the Thermal Pool Distributed Throughout Turkey. Acta Parasitologica. 2019. doi: 10.2478/s11686-019-00148-3 [DOI] [PubMed] [Google Scholar]
- 35.Kao PM, Hsu BM, Hsu TK, Chiu YC, Chang CL, Ji WT, et al. Application of TaqMan qPCR for the detection and monitoring of Naegleria species in reservoirs used as a source for drinking water. Parasitology research. 2014;113(10):3765–71. Epub 2014/08/01. doi: 10.1007/s00436-014-4042-2 . [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann R, Michel R. Distribution of free-living amoebae (FLA) during preparation and supply of drinking water. Int J Hyg Environ Health. 2001;203(3):215–9. Epub 2001/03/31. doi: 10.1078/S1438-4639(04)70031-0 . [DOI] [PubMed] [Google Scholar]
- 37.Lorenzo-Morales J, Ortega-Rivas A, Foronda P, Martinez E, Valladares B. Isolation and identification of pathogenic Acanthamoeba strains in Tenerife, Canary Islands, Spain from water sources. Parasitology research. 2005;95(4):273–7. doi: 10.1007/s00436-005-1301-2 . [DOI] [PubMed] [Google Scholar]
- 38.Puzon GJ, Lancaster JA, Wylie JT, Plumb IJ. Rapid detection of Naegleria fowleri in water distribution pipeline biofilms and drinking water samples. Environmental science & technology. 2009;43(17):6691–6. Epub 2009/09/22. doi: 10.1021/es900432m . [DOI] [PubMed] [Google Scholar]
- 39.Kao PM, Tung MC, Hsu BM, Hsueh CJ, Chiu YC, Chen NH, et al. Occurrence and distribution of Naegleria species from thermal spring environments in Taiwan. Letters in applied microbiology. 2013;56(1):1–7. doi: 10.1111/lam.12006 PubMed PMID: WOS:000312650200001. [DOI] [PubMed] [Google Scholar]
- 40.Corsaro D, Venditti D. Unusual 18S rDNA of Acanthamoeba containing intron turned out to be a T5/T4 chimera. Parasitology research. 2018. doi: 10.1007/s00436-018-6136-8 [DOI] [PubMed] [Google Scholar]
- 41.Lorenzo-Morales J, Khan NA, Walochnik J. An update on Acanthamoeba keratitis: diagnosis, pathogenesis and treatment. Parasite. 2015;22:10. doi: 10.1051/parasite/2015010; PubMed Central PMCID: PMC4330640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Booton GC, Visvesvara GS, Byers TJ, Kelly DJ, Fuerst PA. Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J Clin Microbiol. 2005;43(4):1689–93. Epub 2005/04/09. doi: 10.1128/JCM.43.4.1689-1693.2005 ; PubMed Central PMCID: PMC1081337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qvarnstrom Y, Nerad TA, Visvesvara GS. Characterization of a new pathogenic Acanthamoeba species, A. byersi n. sp., isolated from a human with fatal amoebic encephalitis. Journal of Eukaryotic Microbiology. 2013;60(6):626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorenzo-Morales J, Ortega-Rivas A, Martinez E, Khoubbane M, Artigas P, Periago MV, et al. Acanthamoeba isolates belonging to T1, T2, T3, T4 and T7 genotypes from environmental freshwater samples in the Nile Delta region, Egypt. Acta tropica. 2006;100(1–2):63–9. doi: 10.1016/j.actatropica.2006.09.008 . [DOI] [PubMed] [Google Scholar]
- 45.Dendana F, Trabelsi H, Neiji S, Sellami H, Kammoun S, Makni F, et al. Isolation and molecular identification of Acanthamoeba spp from oasis water in Tunisia. Experimental parasitology. 2018;187:37–41. Epub 2018/03/05. doi: 10.1016/j.exppara.2018.02.009 . [DOI] [PubMed] [Google Scholar]
- 46.Niyyati M, Saberi R, Latifi A, Lasjerdi Z. Distribution of Acanthamoeba Genotypes Isolated from Recreational and Therapeutic Geothermal Water Sources in Southwestern Iran. Environmental health insights. 2016;10:69–74. doi: 10.4137/EHI.S38349 ; PubMed Central PMCID: PMC4838054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evyapan G, Koltas IS, Eroglu F. Genotyping of Acanthamoeba T15: the environmental strain in Turkey. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2015;109(3):221–4. doi: 10.1093/trstmh/tru179 . [DOI] [PubMed] [Google Scholar]
- 48.Kilic A, Tanyuksel M, Sissons J, Jayasekera S, Khan NA. Isolation of Acanthamoeba isolates belonging to T2, T3, T4 and T7 genotypes from environmental samples in Ankara, Turkey. Acta Parasitologica. 2004;49(3):246–52. [Google Scholar]
- 49.Koltas IS, Eroglu F, Erdem E, Yagmur M, Tanir F. The role of domestic tap water on Acanthamoeba keratitis in non-contact lens wearers and validation of laboratory methods. Parasitology research. 2015;114(9):3283–9. doi: 10.1007/s00436-015-4549-1 . [DOI] [PubMed] [Google Scholar]
- 50.Koyun I, Koloren Z, Karaman U, Tsiami A, Karanis P. Acanthamoeba spp. in river water samples of Black Sea, Turkey. Journal of water and health. 2020. doi: 10.2166/wh.2020.170 [DOI] [PubMed] [Google Scholar]
- 51.Karakavuk M, Aykur M, Sahar E, Karakus M, Aldemir D, Donduren O, et al. First time identification of Acanthamoeba genotypes in the cornea samples of wild birds; Is Acanthamoeba keratitis making the predatory birds a target? Experimental parasitology. 2017;183:137–42. doi: 10.1016/j.exppara.2017.08.007 PubMed PMID: WOS:000418107300022. [DOI] [PubMed] [Google Scholar]
- 52.Ertabaklar H, Turk M, Dayanir V, Ertug S, Walochnik J. Acanthamoeba keratitis due to Acanthamoeba genotype T4 in a non-contact-lens wearer in Turkey. Parasitology research. 2007;100(2):241–6. doi: 10.1007/s00436-006-0274-0 . [DOI] [PubMed] [Google Scholar]
- 53.Ozkoc S, Tuncay S, Delibas SB, Akisu C, Ozbek Z, Durak I, et al. Identification of Acanthamoeba genotype T4 and Paravahlkampfia sp. from two clinical samples. Journal of medical microbiology. 2008;57(Pt 3):392–6. doi: 10.1099/jmm.0.47650-0 . [DOI] [PubMed] [Google Scholar]
- 54.Marciano-Cabral F. Biology of Naegleria spp. Microbiological reviews. 1988;52(1):114–33. doi: 10.1128/mr.52.1.114-133.1988 ; PubMed Central PMCID: PMC372708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siddiqui R, Ali IKM, Cope JR, Khan NA. Biology and pathogenesis of Naegleria fowleri. Acta tropica. 2016;164:375–94. Epub 2016/10/30. doi: 10.1016/j.actatropica.2016.09.009 . [DOI] [PubMed] [Google Scholar]
- 56.Ettinger MR, Webb SR, Harris SA, McIninch SP, G CG, Brown BL. Distribution of free-living amoebae in James River, Virginia, USA. Parasitology research. 2003;89(1):6–15. doi: 10.1007/s00436-002-0707-3 . [DOI] [PubMed] [Google Scholar]
- 57.Garcia A, Goñi P, Cieloszyk J, Fernandez MT, Calvo-Beguería L, Rubio E, et al. Identification of Free-Living Amoebae and Amoeba-Associated Bacteria from Reservoirs and Water Treatment Plants by Molecular Techniques. Environmental science & technology. 2013;47(7):3132–40. doi: 10.1021/es400160k [DOI] [PubMed] [Google Scholar]
- 58.Maciver SK, Pinero JE, Lorenzo-Morales J. Is Naegleria fowleri an Emerging Parasite? Trends in parasitology. 2020;36(1):19–28. Epub 2019/11/21. doi: 10.1016/j.pt.2019.10.008 . [DOI] [PubMed] [Google Scholar]
- 59.Jahangeer M, Mahmood Z, Munir N, Waraich UE, Tahir IM, Akram M, et al. Naegleria fowleri: Sources of infection, pathophysiology, diagnosis, and management; a review. Clin Exp Pharmacol Physiol. 2020;47(2):199–212. Epub 2019/10/16. doi: 10.1111/1440-1681.13192 . [DOI] [PubMed] [Google Scholar]
- 60.De Jonckheere JF. Isolation and molecular identification of free-living amoebae of the genus Naegleria from Arctic and sub-Antarctic regions. European journal of protistology. 2006;42(2):115–23. doi: 10.1016/j.ejop.2006.02.001 . [DOI] [PubMed] [Google Scholar]
- 61.De Jonckheere JF. Sequence Variation in the Ribosomal Internal Transcribed Spacers, Including the 5.8S rDNA, of Naegleria spp. Protist. 1998;149(3):221–8. Epub 1998/09/01. doi: 10.1016/S1434-4610(98)70030-6 . [DOI] [PubMed] [Google Scholar]
- 62.Zhou L, Sriram R, Visvesvara GS, Xiao L. Genetic variations in the internal transcribed spacer and mitochondrial small subunit rRNA gene of Naegleria spp. The Journal of eukaryotic microbiology. 2003;50Suppl:522–6. Epub 2004/01/23. doi: 10.1111/j.1550-7408.2003.tb00617.x . [DOI] [PubMed] [Google Scholar]