Abstract
Many Caenorhabditis elegans genes exist in operons in which polycistronic precursors are processed by cleavage at the 3′ ends of upstream genes and trans splicing 100 to 400 nucleotides away, at the 5′ ends of downstream genes, to generate monocistronic messages. Of the two spliced leaders, SL1 is trans spliced to the 5′ ends of upstream genes, whereas SL2 is reserved for downstream genes in operons. However, there are isolated examples of what appears to be a different sort of operon, in which trans splicing is exclusively to SL1 and there is no intercistronic region; the polyadenylation signal is only a few base pairs upstream of the trans-splice site. We have analyzed the processing of an operon of this type by inserting the central part of mes-6/cks-1 into an SL2-type operon. In this novel context, cks-1 is trans spliced only to SL1, and mes-6 3′-end formation occurs normally, demonstrating that this unique mode of processing is indeed intrinsic to this kind of operon, which we herein designate “SL1-type.” An exceptionally long polypyrimidine tract found in the 3′ untranslated regions of the three known SL1-type operons is shown to be required for the accumulation of both upstream and downstream mRNAs. Mutations of the trans-splice and poly(A) signals indicate that the two processes are independent and in competition, presumably due to their close proximity, raising the possibility that production of upstream and downstream mRNAs is mutually exclusive.
Caenorhabditis elegans is unusual among eukaryotes in that a significant number of its genes are arranged in operons (23, 29). Typically, an operon may contain from two to more than six genes, and the intercistronic space is 100 to 400 nucleotides (nt). Polycistronic precursor RNAs are processed by a combination of trans splicing at the 5′ ends of genes and cleavage and polyadenylation at the 3′ ends to generate monocistronic mRNAs (23). There are two 22-nt spliced leaders, SL1 and SL2, which are trans spliced to the 5′ ends of mRNAs. SL1 is the more abundant spliced leader and is trans spliced primarily to the 5′ ends of monocistronic RNAs and to upstream genes in operons, although it is also sometimes found on the 5′ ends of downstream genes in operons as well. In contrast, SL2 is trans spliced exclusively to downstream genes in operons (29). The trans-splice signal is identical to the signal for a 3′ splice site (UUUUCAG/R) and differs only in that it lacks a complementary upstream 5′ splice signal (8). The signals for SL1 and SL2 trans splicing differ only in their contexts; SL2 trans-splice signals lie downstream of another gene, and SL1 trans-splice signals lie downstream of AU-rich intron-like sequences called outrons (7).
While little is known about 3′-end formation in C. elegans, it seems likely to occur by a mechanism similar to that in other animals. The vast majority of C. elegans genes have a match to the AAUAAA polyadenylation signal at their 3′ ends (2), and homologs of many of the mammalian factors, including subunits of the cleavage and polyadenylation specificity factor (CPSF) and the cleavage stimulation factor (CstF), have been identified by the genome sequencing project (28). In mammals, CPSF has been shown to recognize the AAUAAA signal, while CstF binds to a GU-rich sequence downstream of the cleavage site. CPSF is required for both cleavage and polyadenylation in vitro, while CstF is necessary only for the cleavage step. These two factors interact and recruit the cleavage factors and poly(A) polymerase to the cleavage site (see reference 6 for a review).
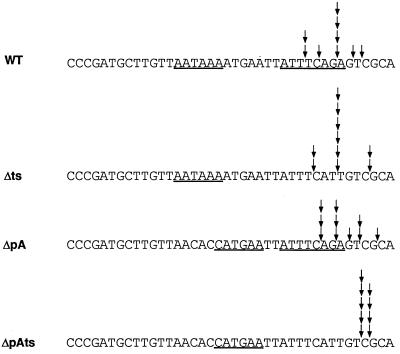
In 1994 Hengartner and Horvitz (10) reported cloning the cyt-1/ced-9 operon. Intriguingly, although ced-9 is the downstream gene in this operon, it is trans spliced exclusively to SL1. Also, the trans-splice site of ced-9 lies at the same position as the 3′ end of cyt-1, and there is an AAUAAA signal within a few base pairs of the trans-splice site (Fig. 1). This gene organization is conserved in C. briggsae (10). A second operon with this type of gene arrangement was recently cloned by Korf et al. (12). The cks-1 gene is trans spliced exclusively to SL1 even though it lies immediately downstream of mes-6. In both examples, polycistronic precursor RNA as well as both monocistronic mRNAs are clearly detected on Northern blots. One possible explanation is that the downstream gene in each case is expressed from an unidentified promoter within the coding sequence of the upstream gene instead of from a shared upstream promoter. Thus, these would not be examples of a novel class of operons, but of overlapping transcription units. When the 3′ end of such an upstream gene was formed, the trans-splice site of the downstream gene would be destroyed, resulting in rapid decay of the uncapped downstream RNA. If this hypothesis is correct, then the downstream mRNAs are expressed exclusively from the unidentified promoter and not by processing of the polycistronic precursor.
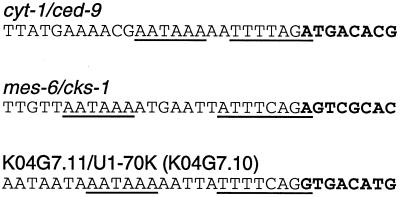
FIG. 1.
Sequences of the polyadenylation/trans-splice site regions of three SL1-type operons. Polyadenylation signals and trans-splice signals are underlined. The sequences are aligned at the sites of trans splicing. Upstream mRNA sequences are in plain type, while downstream mRNA sequences are in boldface. cyt-1/ced-9 sequences are from Hengartner and Horvitz (10), and mes-6/cks-1 sequences are from Korf et al. (12). The U1-70K sequence is from cosmid K04G7, which was sequenced by the C. elegans Genome Center (accession no. U21320).
If both upstream and downstream RNAs are expressed only from the same promoter, then the mechanism of processing of the precursor is not clear. In order for the downstream gene to be expressed, trans splicing must occur on the polycistronic precursor RNA. This would leave a free 3′ end on the upstream RNA, which could either be polyadenylated or degraded. CPSF is able to promote efficient polyadenylation of a free 3′ end in vitro (19). An alternative model is that each pre-mRNA could produce the mRNA from either the upstream or the downstream gene, but not both. In this model a pre-mRNA could either be trans spliced to form the downstream mRNA (with the upstream, presumably branched, portion discarded) or be cleaved and polyadenylated by CPSF-CstF, which would destroy the trans-splice site. Thus, 3′-end formation and trans splicing could be in direct competition, or trans splicing might be able to create a free 3′ end that could then be polyadenylated. These two mechanisms are not mutually exclusive.
We present here the identification of a third example of this SL1-type operon. We have also created an artificial SL1-type operon, which exhibits the same properties in vivo as the endogenous examples. There is no space between the genes, and the downstream gene is trans spliced to SL1 instead of SL2. We have analyzed processing of this artificial operon by mutating the signals predicted to be involved.
MATERIALS AND METHODS
Worm culture and RNA preparation.
Worms were grown and maintained as described elsewhere (4, 24). Transgenic worms bearing extrachromosomal arrays were made by the method of Mello et al. (18) and Spieth et al. (23) by using the pRF5 rol-6 plasmid as a marker. The rol-6 null strain MT2597 was the parent strain in all cases. Mixed-stage populations of worms were heat shocked at 30°C for 1 h in water. Total RNA was prepared as described elsewhere (9).
Plasmids.
A 739-bp PCR product amplified from genomic DNA by using Pfu DNA polymerase (Stratagene) and oligonucleotides mes-6-Sal and cks-1PR was inserted into the StuI site of the vector pΔHSgpdvit (the plasmid pHS-1496 [23] with the SalI site in the vector deleted [16]) to make pMGV-WT. This plasmid was used as a template for recombinant PCRs (11) to generate mutant plasmids pMGV-ΔpA, pMGV-Δts, pMGV-ΔpA2, pMGV-ΔpAts, pMGV-ΔUtr, and pMGV-ΔUtrDS. Pfu DNA polymerase was used for the first round of PCR; Taq DNA polymerase (Gibco-BRL) was used for the recombinant PCR. The following oligonucleotides were used (mismatches from the wild-type sequence are lowercased): mes-6-Sal, 5′-CCTAATGACAATCGATAGCAACTTAC-3′; cks-1PR, 5′-GCACGTGACGCTCGGGAAGACTTC-3′; mes-6ΔpA(+), 5′-GATGCTTGTTAAcAccATGAATTATTTCAG-3′; mes-6ΔpA(−), 5′-AAATAATTCATggTgTTAACAAGCATCGGG-3′; cks-1Δts(+), 5′-TAATAAATGAATTATTTCAttGtCGCACCTTCTCACG-3′; cks-1Δts(−), 5′-GAAGGTGCGaCttAGAAATAATTCATTTTATTAACAAGC-3′; mes-6ΔpA2(+), 5′-GATGCTTGTTAAcAccATGccTTaTTTCAG-3′; mes-6ΔpA2(−), 5′-AAATAAggCATggTgTTAACAAGCATCGGG-3′; ΔpAts(+), 5′-CTTGTTAAcAccATGAATTATTTCAttGTCGCACCTTCTC-3′; ΔpAts(−), 5′-GAAGGTGCGACaaTGAAATAATTCATggTgTTAACAAGCATC-3′; mes-6ΔUtr(+), 5′-CTCACTCTTGTCTCATTCCCACCCGATGCTTGTTAATAAAATG-3′; mes-6ΔUtr(−), 5′-CATTT TATTAACAAGCATCGGGTGGGAATGAGACAAGAGTGAG-3′; mes-6ΔUtrDS(+), 5′-CTTTTTTTTCAATATTTTTTACCCGATGCTTGTTAATAAAATG-3′; and mes-6ΔUtrDS(−), 5′-CATTTTATTAACAAGCATCGGGTAAAAAATATTGAAAAAAAAGAG-3′. The mutant constructs were all sequenced before transformation. Due to the infidelity of the Taq polymerase, several unwanted single nucleotide mismatches were observed, but unless they were in regions predicted to be important for processing (e.g., splice site mutations) or stability (e.g., creation of a premature stop codon), these were ignored.
RNase protection and primer extension analysis.
RNase protection was carried out as described elsewhere (13) except that 20 μg of total RNA was treated with RNase-free DNase I prior to hybridization to the probe. Primer extension was also carried out as described elsewhere (13) with the primer cks1-PE (5′-GGCGTGAGAAGGTGCGACTC-3′) in the presence of dideoxycytidine (ddC).
RT-PCR.
Reverse transcription-PCR (RT-PCR) to determine the trans splicing specificity of U1-70K was carried out as described previously (29); the downstream primer was to nt 17906 to 17927 of cosmid K04G7, which is complementary to a sequence near the 5′ end of the predicted U1-70K mRNA. The resulting RT-PCR product with SL1 or SL2 trans spliced at the predicted trans-splice site, 18233, would be 349 bp. RT-PCR products were electrophoresed, blotted, and probed with an oligonucleotide from 18100 to 18121 of K04G7.
For rapid amplification of 3′ cDNA ends (3′ RACE), total RNA was denatured at 65°C for 3 min prior to addition of reverse transcriptase (RT) buffer, nucleoside triphosphates, 3′20 primer [5′-GCGGCCGCAGATCTCGAG(T)20(G/A/C)-3′] and avian myeloblastosis virus RT (Promega). The 20-μl reaction mixture was incubated at 42°C for 1 h, then diluted 10-fold with water. Five microliters was used in a PCR with 3′20 as the downstream primer and mes/gpd-5′ (5′-GCCACCAAGGCCTAATGACAGTCG-3′) as the upstream primer. mes/gpd-5′ crosses the border between gpd-2 and mes-6 in the vector and hence is unable to prime off the products of either endogenous gene. The products from 3′ RACE were cloned into the pGEM-T vector (Promega) and sequenced by using primer SP6.
RESULTS
A third SL1-type operon.
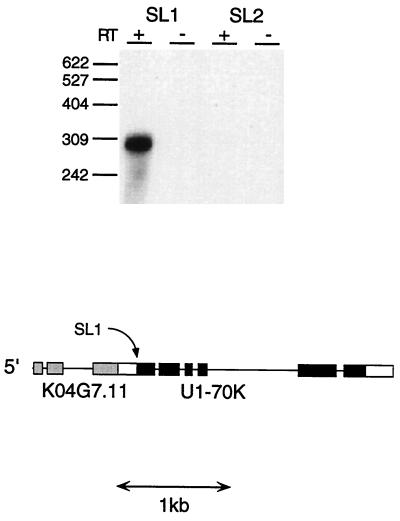
In scanning the output of the C. elegans genome project for proteins involved in splicing, we discovered a presumptive operon containing the gene for the U1-70K snRNP protein. This gene is the third gene in a three-gene cluster; the two genes upstream are not clearly related to any other genes in the database. We noticed that the trans-splice site of the U1-70K gene (K04G7.10) is just downstream of an AAUAAA site and that there is no other 3′-end formation signal in the predicted 3′ untranslated region (3′ UTR) of the upstream gene (K04G7.11). Since this is the hallmark of the other two operons of this type (Fig. 1), we performed two experiments to determine the nature of the trans splicing of U1-70K and the site of 3′-end formation of K04G7.11. When we performed RT-PCR with an oligonucleotide equivalent either to the SL1 or to the SL2 spliced leader and an oligonucleotide in the U1-70K gene, we found that all trans splicing is to SL1 (Fig. 2). When we performed 3′ RACE RT-PCR with an oligonucleotide in the K04G7.11 gene (data not shown), we found that all three clones sequenced represented instances of polyadenylation either 1 or 2 nt upstream of the trans-splice site and at an appropriate distance downstream of the AAUAAA signal (2) (Fig. 1).
FIG. 2.
U1-70K is a downstream gene in an operon and is trans spliced to SL1. RT-PCR was performed with U1-70K-Pro (5′-GCAACTCCAGTCATCGGAGCCC-3′) as the downstream oligonucleotide and either SL1 or SL2 as the upstream oligonucleotide, as described in Materials and Methods. + and −, presence and absence, respectively, of RT. PCR products were blotted and probed with the U1-70K-PR oligonucleotide (5′-GCTGGATTCCACGTGGCGATTCC-3′). The SL2 oligonucleotide was also used in an RT-PCR with a known SL2-accepting gene at the same time, and the band of the expected size was found (data not shown). The structure of the two downstream genes in the U1-70K operon, as predicted by the genome sequence center and as determined by us, is depicted. The gene upstream of the U1-70K gene (K04G7.11) is of unknown function. The location of its 3′ end was determined by RT-PCR (data not shown). Solid boxes, coding regions; open boxes, noncoding regions. The site of SL1 trans splicing of U1-70K (and of 3′-end formation of K04G7.11) is indicated by an arrow.
Examination of the sequences immediately upstream of the polyadenylation signal in these novel operons revealed that all have extensive polypyrimidine tracts in the 3′ UTRs of the upstream genes (data not shown). The cyt-1 3′ UTR has a poly(Y) tract of 131 nt (80% pyrimidines) ending 42 nt upstream of the ced-9 trans-splice site; the mes-6 3′ UTR has a poly(Y) tract of 84 nt (86% pyrimidines) ending 31 nt upstream of the cks-1 trans-splice site; and the K04G7.11 3′ UTR has a poly(Y) tract of 107 nt (82% pyrimidines) ending 42 nt upstream of the U1-70K trans-splice site. Such extensive pyrimidine tracts are not generally found within 3′ UTRs of other C. elegans genes (1). Thus, the three examples of this type of operon share the following unique characteristics: (i) the mRNAs from the downstream genes are trans spliced exclusively to SL1, rather than to SL2; (ii) the mRNAs of the upstream genes are polyadenylated at, or a few nucleotides upstream of, the trans-splice site of the downstream gene, rather than the usual 100 to 400 nt upstream; (iii) they each contain a CPSF binding site 8 to 14 nt upstream of the site of trans splicing; and (iv) the upstream genes have extensive polypyrimidine tracts in their 3′ UTRs.
An artificial SL1 operon.
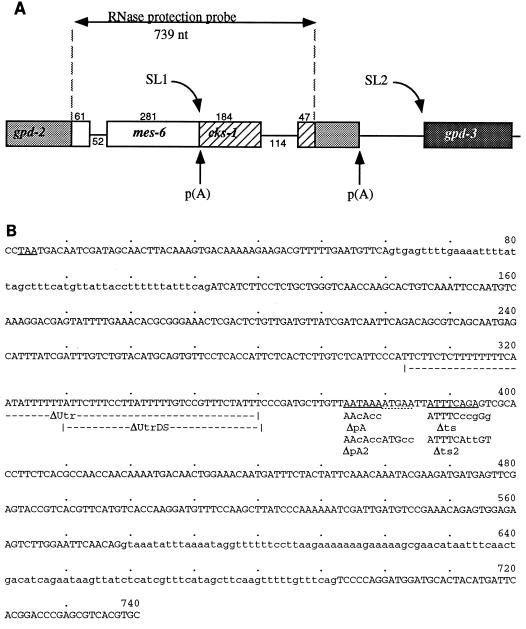
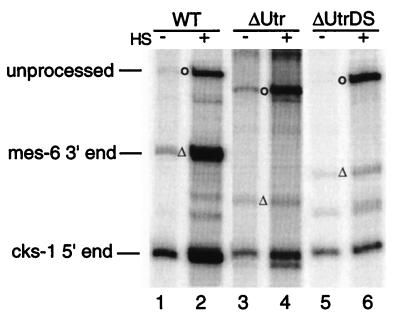
An artificial SL1-type operon was constructed and tested in vivo. This construct, depicted in Fig. 3A, is based on the gpd-2/gpd-3 operon described previously (13, 23). A 739-bp region of the mes-6/cks-1 operon, containing the 3′ 394 bp of mes-6 and the 5′ 345 bp of cks-1, was inserted into the 3′ UTR of gpd-2 immediately after the stop codon (Fig. 3B). Thus, the 3′ UTR of gpd-2 is replaced with that of mes-6, and the cks-1 coding region terminates within the 3′ UTR of gpd-2. The gpd-2–gpd-3 intercistronic space and gpd-3 itself are unaffected. The entire three-gene operon is expressed under the control of the hsp-16-41 heat-shock promoter. The construct, pMGV-WT, was used to generate stable transgenic lines, and expression was induced by heat shock as described in Materials and Methods. Total RNA from these strains, before and after heat shock, was analyzed for expression of the operon as described previously (13). An RNAse protection probe covering the region shown in Fig. 3A protected the expected fragments (Fig. 4, lanes 1 and 2). The probe detects both endogenous and transgenic products, which are distinguishable by heat inducibility. Both mes-6 and cks-1 products were heat shock inducible, indicating that they are both derived from the polycistronic precursor RNA. Unprocessed precursor RNA, neither trans spliced nor cleaved, was also readily detectable.
FIG. 3.
(A) Map of the gpd-2–mes-6–cks-1–gpd-3 fusion operon. Exons of gpd-2 (light shading), gpd-3 (dark shading), mes-6 (open), and cks-1 (stripes) are shown as boxes, and introns and the gpd-2–gpd-3 intercistronic region are shown as lines. Positions of trans-splice and poly(A) sites are indicated by arrows. Numbers above exons and below introns give their sizes in base pairs. The region covered by the RNAse protection probe is also shown. (B) Sequence of the mes-6–cks-1 region inserted into the 3′ end of gpd-2, with the exon sequence uppercased and the intron sequence lowercased. The extents of deletions and the positions of mutations, with mismatches from the wild-type sequence in lowercase, are shown below the wild-type sequence. The gpd-2 stop codon, the mes-6 poly(A) signal, and the cks-1 trans-splice signals are underlined. The dotted underline indicates the nonconsensus poly(A) signal.
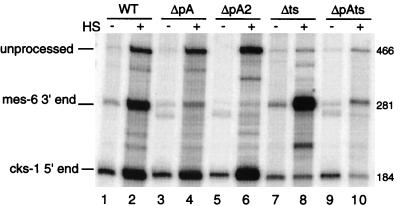
FIG. 4.
RNase protection analysis of wild-type (WT), poly(A) mutant, and trans-splice site mutant genes. The positions of unprocessed, mes-6 3′-end, and cks-1 5′-end protected fragments are indicated. − and +, RNA from untreated and heat-shocked (HS) populations, respectively. The mutants tested are described in Fig. 3A and in the text.
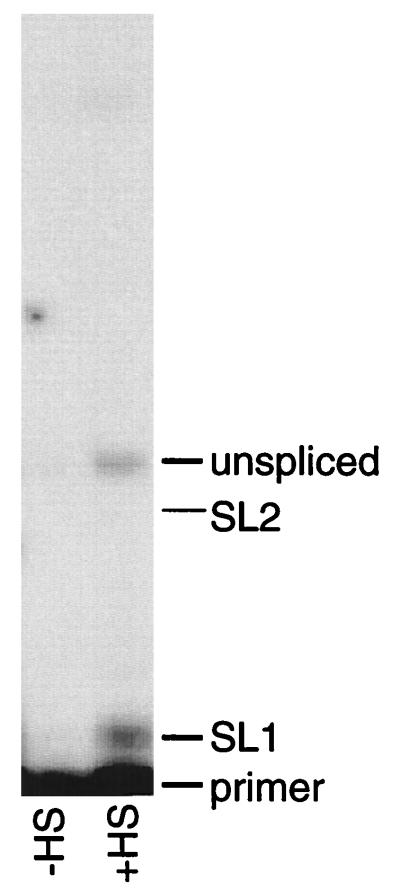
The trans splicing specificity of cks-1 was tested by a ddC primer extension assay (Fig. 5). A primer was annealed to the extreme 5′ end of cks-1 and extended in the presence of ddC. The size of the extension product differs depending on whether the RNA is un-trans-spliced (+11), trans spliced to SL1 (+2), or trans spliced to SL2 (+9). In this case, the only products detected were un-trans- spliced and trans spliced to SL1, as occurs in the endogenous mes-6/cks-1 operon (12). Two other strains bearing the same plasmid gave the same results (data not shown). In addition, we confirmed that the band at +11 really represents un-trans-spliced product, rather than a +9 SL2 band, by performing ddG primer extension (data not shown). In this case, we saw only the bands expected for RNA trans spliced to SL1 (+9) and un-trans-spliced RNA (+2), and no band at the SL2 position (+3). Thus, even in this novel context, this trans-splice site was spliced to SL1. Since its precursor was transcribed from the heat shock promoter 1.5 kb upstream, this result demonstrates that this type of operon is a class distinct from the usual C. elegans operon. It is clear that the downstream product can be made from the polycistronic precursor, rather than from an internal promoter, and that this results in SL1-specific trans splicing, even though the trans-splice site is now at a location normally occupied by an SL2-accepting trans-splice site.
FIG. 5.
Primer extension analysis of RNA from worms bearing the pMGV-WT construct. Primer extension was performed in the presence of ddC as described in Materials and Methods. Positions of un-trans-spliced, SL1-trans-spliced, and SL2-trans-spliced cks-1 primer extension products are indicated. RNA was isolated from untreated worms (left lane) or heat-shocked (HS) worms (right lane).
3′ RACE was used to amplify and clone cDNAs from the 3′ end of mes-6. The site of polyadenylation was found to lie at or upstream of the trans-splice site in seven of nine cDNAs (Fig. 6). The remaining two cDNAs ended downstream of the consensus trans-splice site and hence must have resulted in destruction of the site. However, a weak match to the trans-splice consensus TCAGAG/T is also present 2 bp downstream. The experiments show that the artificial operon is mimicking 3′-end formation as observed in vivo for operons of this type: the upstream gene is polyadenylated at or just upstream of the trans-splice site in most cases. Since these locations are appropriate distances downstream of the AAUAAA, they could result either from CPSF-CstF-dependent cleavage or from polyadenylation of polycistronic precursors cleaved by trans splicing.
FIG. 6.
The position of the mes-6 3′ end can vary depending on the presence of a trans-splice site downstream. RT-PCR was performed on RNA from transgenic strains carrying the wild-type (WT) gene and the indicated mutant constructs by using an oligo(dT) oligonucleotide and an internal oligonucleotide as described in Materials and Methods. Individual cDNAs were cloned and sequenced. Sequences of the wild-type and mutant construct 3′-end regions are shown, with positions of the mes-6 3′ ends (i.e., sites at which nontemplated A’s begin in the individual cDNA sequences) marked by arrows. Where it cannot be determined whether an A is templated or not, the arrows are placed to the left of the A. Active poly(A) signals and trans-splice signals are underlined.
3′-end formation occurs in the absence of trans splicing.
The close apposition of the sites of trans splicing and 3′-end formation suggests that the two processes could occur in concert on the same pre-mRNA, resulting in production of both mature mRNAs from a single processing event. In order to determine whether 3′-end formation was dependent on trans splicing, the trans-splice site was mutated to create pMGV-Δts. The effects of the trans-splice site mutation on expression of both the upstream and downstream genes were examined. Transgenic strains were analyzed by RNAse protection (Fig. 4, lanes 7 and 8). To our surprise, 3′-end formation was not dependent on trans splicing. In strains bearing the trans-splice site mutant construct, no heat-inducible band at the position of the normal trans-spliced product was found. Although we do not know what the novel band in lane 8 at about 210 bp is, it is almost certainly not an alternative trans-spliced product, since no sequence even distantly related to a trans-splice site is present in that region of the mes-6 3′ UTR (Fig. 3B). Even though no cks-1 product above background level was detected, the mes-6 3′ end appeared to form normally. In fact, 3′-end formation appears to be more efficient, since polycistronic precursor RNA is significantly reduced in these strains. We also determined the range of sites used for 3′-end formation in the absence of a trans-splice site by sequencing RT-PCR products. The distribution of sites was indistinguishable from that seen with the wild-type construct (Fig. 6). We conclude that 3′-end formation can occur in the absence of trans splicing in this operon and that in an otherwise wild-type construct, the trans-splice site does not influence the site of 3′-end formation. Furthermore, it appears that RNA that would otherwise have been trans spliced or not processed at all is efficiently cleaved by the 3′-end formation machinery when the trans-splice site is eliminated.
trans splicing without 3′-end formation.
In order to determine whether trans splicing is influenced by the presence of a closely juxtaposed 3′-end formation signal, the mes-6 polyadenylation signal, AAUAAA, was mutated (to AACACC) to create pMGV-ΔpA (Fig. 3B). Strangely, the poly(A) signal mutation reduced, but did not abolish, 3′-end formation of the upstream gene (Fig. 4, lanes 3 and 4). RNase protection analysis showed that trans splicing occurred at normal levels in the ΔpA mutant strains, and primer extension showed that cks-1 was still trans spliced exclusively to SL1 (data not shown). In the ΔpA mutant we identified a possible cryptic polyadenylation signal, CAUGAA, immediately downstream of the AAUAAA (Fig. 3B), which could have been used for residual 3′-end formation. To test this idea, we constructed a double poly(A) site mutation (ΔpA2) (Fig. 3B), which did effectively eliminate 3′-end formation (Fig. 4, lanes 5 and 6). trans splicing still occurred at approximately the same level as that in the wild type, and polycistronic precursor RNA still accumulated. This shows that trans splicing is not significantly influenced by a nearby 3′-end formation signal.
Influence of the trans-splice site on the site of 3′-end formation.
To test whether mutation of the primary AAUAAA resulted in a new site of 3′-end formation directed by the hypothetical remaining signal 5 bp downstream, we sequenced 10 RT-PCR clones from RNA isolated from transgenic strains carrying the ΔpA mutant construct. If 3′-end formation was occurring by the normal CPSF-dependent mechanism in the strain with only a weak CPSF binding site 5 bp downstream of the normal site, we would perhaps expect these 3′ ends to be located about 5 bp downstream of the location found in the wild-type strain. However, when we measured the locations of mes-6 3′ ends in the ΔpA mutant, we found a distribution similar to that shown by the wild-type construct (Fig. 6). One possible explanation for this distribution would be that these 3′ ends were actually generated by trans splicing rather than by the conventional CPSF-CstF-dependent mechanism. In this case we would expect the site of cleavage to have remained the same. Thus, this result suggests that the site of cleavage may have been determined, or at least influenced, by the trans-splice site.
To test the idea that trans splicing might be generating the free 3′ ends for polyadenylation, we combined the ΔpA and Δts mutations. Simultaneous mutation of the AAUAAA and the trans-splice signal (ΔpAts) resulted in the accumulation of precursor RNA and polyadenylated mes-6 mRNA, but no cks-1 mRNA (Fig. 4, lanes 9 and 10). Sequencing of the 3′ ends of mes-6 RT-PCR clones from this mutant revealed a change in the distribution of poly(A) sites. All nine cDNAs now lay 3 or 4 nt downstream of the original site of polyadenylation (Fig. 6). This is consistent with 3′-end formation occurring in the regular fashion, with cleavage directed by the nonconsensus polyadenylation site when the trans-splice site is mutated. The difference in poly(A) site distribution between the ΔpA and the ΔpAts mutant suggests that in ΔpA, the trans-splice site is influencing the site of cleavage. Since trans splicing does result in cleavage, our data are consistent with a model in which the free 3′ end created by trans splicing is polyadenylated, at least for 6 of the 10 cDNAs we sequenced. Although the close proximity of the trans-splice site and polyadenylation signal in this system makes it difficult to mutate the trans-splice site without risking a direct effect on sequences required for polyadenylation, the Δts mutation did not change the site of 3′-end formation in the absence of the ΔpA mutation. Therefore, it is very unlikely that there is a direct effect of the Δts mutation on the choice of cleavage sites.
Competition between trans splicing and 3′-end formation.
A comparison of the wild type with the Δts mutant (Fig. 4) demonstrates clearly a strong competition between 3′-end formation and trans splicing. The level of 3′-end formation goes up dramatically, and the level of unprocessed precursor is significantly reduced, when the trans-splice site is mutated. This suggests that trans splicing reduces the level of substrate available for 3′-end formation. The fact that unprocessed precursor accumulates when all sites are intact but does not when the trans-splice site is mutated suggests that the presence of a trans-splice site somehow interferes with 3′-end formation even when it does not lead to the formation of a trans-spliced product.
A comparison of the levels of trans-spliced product in the wild type and the ΔpA2 mutant does not show an increase in trans splicing or a decrease in unprocessed precursor when the poly(A) sites are eliminated, suggesting that 3′-end formation is not reducing the level of substrate available for trans splicing. We interpret these results to indicate that the trans-splice site is normally occupied first, but not all of the precursor is trans spliced. The formation of 3′ ends then occurs on a fraction of the substrate that was not trans spliced.
Functions of the 3′ UTR polypyrimidine tract.
As all three examples of SL1-type operons have extensive polypyrimidine tracts in the 3′ UTRs of the upstream gene, we deleted a portion of this region in the artificial operon (ΔUtr [Fig. 3B]) and measured the effect on processing (Fig. 7). A 58-bp deletion in the mes-6 3′ UTR results in accumulation of polycistronic precursor RNA but no mes-6 3′-end product (Fig. 7). Thus, it appears that there are sequences within the long poly(Y) tract that are required for the accumulation of 3′-end product. These sequences could be needed either for 3′-end formation or for the stability of the product. trans splicing further downstream was also significantly reduced by the ΔUtr mutation. (The band just below the trans-spliced product in Fig. 7, lane 4, is of unknown origin, but it is not a trans-spliced product, based on primer extension results which are not shown.) In order to further define the sequences required, another deletion, half the size of the original, was made (ΔUtrDS [Fig. 3B]). This mutant apparently has a phenotype similar to that of the mutant with the larger deletion—most of the RNA appears to be unprocessed precursor (Fig. 7, lanes 5 and 6), although ΔUtrDS does show a small accumulation of mes-6 3′ end.
FIG. 7.
RNase protection analysis of the wild type and of polypyrimidine tract mutants. RNAse protection with RNA isolated from the indicated mutant strains was performed as described for Fig. 4. ○, positions expected for unprocessed precursor for each construct; ▵, positions expected for mes-6 3′ ends for each construct. The size of the cks-1 mRNA (184 bp) is not affected by the mutations.
DISCUSSION
A second class of operon in C. elegans.
In the major class of C. elegans operons, there is an intercistronic region of 100 to 400 bp between the site of 3′-end formation of the upstream gene and the trans-splice site of the downstream gene. In this type of operon, SL2 or a mixture of SL2 and SL1 is trans spliced to the downstream mRNA. Furthermore the pre-mRNAs are processed in such a way as to produce mature mRNAs from a single polycistronic precursor. Here we demonstrate the existence of a novel type of C. elegans operon in which there is no space between the genes and the downstream gene is trans spliced exclusively to SL1. This type of operon is also different from the major class in that our results suggest the possibility that a single precursor may produce either the upstream or the downstream product, but not both. This is because the trans-splice site and the 3′-end formation site are so close that when 3′-end formation occurs, the precursor for trans splicing is destroyed. One possibility had been that the downstream mRNA was made from an internal promoter within the upstream gene. However, our results demonstrate that the downstream mRNA, at least, can be made from a polycistronic precursor when it is expressed from a heat shock promoter more than 1.5 kb upstream. When it is produced from such an upstream promoter, at a distance which in the majority class of C. elegans operon results in SL2-specific trans splicing (13), we found that it was nevertheless trans-spliced exclusively to SL1 (Fig. 5). This result argues strongly that the SL1-trans-spliced products are indeed generated from polycistronic precursor RNAs.
Relationship between 3′-end formation and trans splicing.
Our data are most consistent with a model in which most pre-mRNAs from this type of operon produce either an upstream mRNA or a downstream mRNA, and the rest of the pre-mRNA is destroyed. In order for the downstream RNA to be made, trans splicing must occur before cleavage and polyadenylation of the upstream gene, because cleavage downstream of the AAUAAA would destroy the trans-splice site of the downstream gene. Our results suggest that trans splicing does indeed interfere with formation of the upstream mRNA, presumably because it reduces the level of precursor available for CPSF-CstF-dependent 3′-end formation. Polyadenylation levels increase significantly and unprocessed precursor decreases significantly when the trans-splice site is mutated, indicating an inhibitory effect of the trans-splicing event on 3′-end formation (Fig. 4). Interestingly, the converse does not appear to be true. When the poly(A) signals are mutated, the level of trans splicing does not increase, and precursor that has been neither trans spliced nor cleaved and polyadenylated remains (Fig. 4). This suggests a model in which the precursor is first subjected to trans splicing, but some of it somehow escapes this process. Then a portion of the remaining precursor is cleaved and polyadenylated in a CPSF-dependent process.
Why is SL1 used for trans splicing at these internal trans- splice sites?
In most operons, trans splicing occurs 100 to 400 bp downstream of a conventional 3′-end formation site. The structure of the actual precursor for this trans-splicing event, which utilizes the SL2 snRNP as a donor, is not known. It may already be cleaved, or trans splicing and cleavage-polyadenylation may occur cooperatively and simultaneously. In the new class of operons studied here, SL1 snRNP is used as a splice donor. This snRNP has previously been shown to be used at outrons, intron-like sequences with a cap at the 5′ end and a trans-splice site at the 3′ end (2). We argue above that in the SL1-type operons, trans splicing occurs on the polycistronic RNA, so the entire upstream gene may be recognized as an outron, resulting in SL1 trans splicing.
Does the free 3′ end created by trans splicing sometimes get polyadenylated?
While there is undeniably competition between the 3′-end formation machinery and the trans-splicing machinery for processing at the sites where the two genes meet, our data are also consistent with a role for trans splicing in generating a free 3′ end, which can then be polyadenylated as long as it has a CPSF binding site. There is evidence in mammalian systems that both CPSF and CstF specify the site of 3′-end formation. A change in the relative positions of the CPSF and CstF binding sites generally results in a concomitant shift in the site of cleavage (5). This is apparently due to a requirement for a minimal distance between the CPSF binding site and the cleavage site. In the ΔpA mutant, the CPSF binding site has effectively been moved downstream by 5 nt, but we see essentially no alteration in the site of cleavage in the presence of an active trans-splice signal (Fig. 6). However, upon mutation of the trans-splice signal, there is an accompanying shift in the site of polyadenylation. This is consistent with CPSF allowing polyadenylation of the free 3′ end generated by trans splicing. We propose that in these operons polyadenylation may occur both by the usual CstF-dependent mechanism in competition with trans splicing and also by a second, trans-splicing-dependent and perhaps CstF-independent mechanism.
The importance of a long upstream polypyrimidine tract.
We have identified an extensive polypyrimidine tract in the 3′ UTR of each of the three operons of this type, and we have shown that this region of mes-6 is required for processing. When either the entire sequence or just its downstream portion is deleted, levels of both cks-1 and mes-6 mRNAs are dramatically reduced (Fig. 7). The effect on mes-6 mRNA is not due to a secondary effect of inhibition of trans splicing, since inhibition of trans splicing by elimination of the trans-splice site did not reduce the level of 3′-end formation (Fig. 4). While the lack of downstream product must be attributed to a direct effect on trans splicing, the dramatic reduction of mes-6 mRNA levels could be due either to decreased stability of the message or to failure to form 3′ ends.
Polypyrimidine tracts have previously been implicated in both cis and trans splicing. cis splicing in mammalian systems requires a polypyrimidine tract downstream of the branch site, which interacts with U2AF (21). In C. elegans no such element has been identified, but the 3′ splice site has an unusually long and well-conserved consensus, UUUUCAG/R, which has been hypothesized to substitute for the polypyrimidine tract (30). In trypanosomes, trans splicing and polyadenylation of polycistronic transcripts are directly coupled (14). trans splicing requires a polypyrimidine tract in the intercistronic region (17), and 3′-end formation upstream requires trans splicing (15). Deletion analysis of part of the 84-nt polypyrimidine tract of the mes-6/cks-1 operon indicates that, as in trypanosomes, a polypyrimidine tract in this type of C. elegans operon plays a crucial role in trans splicing and possibly in 3′-end formation as well. It should be emphasized here that in no other case has such a far-upstream polypyrimidine tract been shown to play a role in trans splicing, nor does an upstream polypyrimidine tract generally perform a role in 3′-end formation in animals. However, pyrimidine-rich elements have been identified as upstream efficiency elements required for polyadenylation of viral mRNAs and a few cellular RNAs (3, 20, 22, 25), and the polypyrimidine tract in the mes-6 3′ UTR could be performing an analogous function in its 3′-end formation. The mes-6 polyadenylation signal is not optimal by mammalian standards, as it lacks any U-rich or GU-rich downstream sequence element at an appropriate position to act as a CstF binding site. Also, trans splicing would result in cleavage and loss of any downstream sequences. One hypothesis to explain the lack of a downstream sequence element is that the upstream pyrimidine tract is acting as an upstream efficiency element to promote mes-6 3′-end formation. In the case of the mammalian C2 complement poly(A) signal, there is an upstream polypyrimidine tract which has been shown to interact both with polypyrimidine tract binding protein and with CstF, which binds cooperatively with CPSF (19a). A similar role could be performed by the extensive poly(Y) tracts in the 3′ UTRs of the SL1-type operons.
We cannot rule out the possibility that the pyrimidine-rich region in the 3′ UTR is required for mes-6 mRNA stability. A pyrimidine-rich element has been identified as a constitutive stabilizer of the human β-globin mRNA (26, 27). Deletion of the stabilizer element in this mRNA results in its becoming highly unstable. Further experiments are required to elucidate the role of the pyrimidine tract in trans splicing of cks-1 and to determine whether it is required for the stability of the mes-6 message or is acting as an upstream polyadenylation element.
Conclusions.
We have analyzed a novel class of C. elegans operon, which we call SL1-type. These operons are different from the majority, SL2-type operons in several interesting ways: (i) the downstream genes are trans spliced exclusively by SL1; (ii) there is no intercistronic region, and the 3′-end formation signal, AAUAAA, is only a few base pairs upstream of the trans-splice site, so the two processes compete; (iii) there is an unusually long polypyrimidine tract in the 3′ UTR of the upstream gene, which is required for accumulation of both upstream and downstream mRNAs; (iv) these operons appear to be designed to produce only upstream mRNA or only downstream mRNA, but usually not both, from a single precursor. It is not yet clear, given the small number of SL1-type operons that we know of, whether the genes in these operons have a relationship with one another that makes this a functionally significant gene arrangement.
ACKNOWLEDGMENTS
We are grateful to Susan Strome and Ian Korf for communication of their results prior to publication and to our colleagues in the lab and Susan Strome for their helpful comments on the manuscript.
This work was supported by research grant GM42432 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Blumenthal, T. Unpublished data.
- 2.Blumenthal T, Steward K. RNA processing and gene structure. In: Riddle D, Blumenthal T, Meyer B, Priess J, editors. C. elegans II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 117–145. [PubMed] [Google Scholar]
- 3.Brackenridge S, Ashe H L, Giacca M, Proudfoot N J. Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res. 1997;25:2326–2335. doi: 10.1093/nar/25.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F, MacDonald C C, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 7.Conrad R, Lea K, Blumenthal T. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA. 1995;1:164–170. [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad R, Liou R F, Blumenthal T. Functional analysis of a C. elegans trans-splice acceptor. Nucleic Acids Res. 1993;21:913–919. doi: 10.1093/nar/21.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad R C, Thomas J, Spieth J, Blumenthal T. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol. 1991;11:1921–1926. doi: 10.1128/mcb.11.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengartner M O, Horvitz H R. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi R, Krummel B, Saiki R R. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korf, I., Y. Fan, and S. Strome. The polycomb group in Caenorhabditis elegans and maternal control of germline development. Development 13:2469–2478. [DOI] [PubMed]
- 13.Kuersten R S, Lea K, MacMorris M, Spieth J, Blumenthal T. Relationship between 3′ end formation and trans-splicing in polycistronic Caenorhabditis elegans pre-mRNA processing. RNA. 1997;3:269–278. [PMC free article] [PubMed] [Google Scholar]
- 14.LeBowitz J H, Smith H Q, Rusche L, Beverly S M. Coupling of poly(A) site selection trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Estrano C, Tschudi C, Ullu E. Exonic sequences in the 5′ untranslated region of α-tubulin mRNA modulate trans splicing in Trypanosoma brucei. Mol Cell Biol. 1998;18:4620–4628. doi: 10.1128/mcb.18.8.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacMorris, M. Unpublished data.
- 17.Matthews K R, Tschudi C, Ullu E. A common pyrimidine rich motif governs trans-splicing and polyadenylation of polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 18.Mello C, Kramer J, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore C L, Sharp P A. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985;41:845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- 19a.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. The upstream sequence of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreira A, Wollerton M, Monks J, Proudfoot N J. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruskin B, Zamore P D, Green M R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1987;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 22.Schek N, Cooke C, Alwine J C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 24.Sulston J, Hodgkin J. Methods. In: Wood W B, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- 25.Valsamakis A, Schek N, Alwine J C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss I, Liebhaber S A. Erythroid cell-specific determinants of α-globin mRNA stability. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss I, Liebhaber S A. Erythroid cell-specific mRNA stability elements in the α2-globin 3′ nontranslated region. Mol Cell Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams, C. J., and T. Blumenthal. Unpublished data.
- 29.Zorio D A R, Cheng N N, Blumenthal T, Spieth J. Operons represent a common form of chromosomal organization in C. elegans. Nature (London) 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]
- 30.Zorio D A R, Lea K, Blumenthal T. Cloning of Caenorhabditis U2AF65: an alternatively spliced RNA containing a novel exon. Mol Cell Biol. 1997;17:946–953. doi: 10.1128/mcb.17.2.946. [DOI] [PMC free article] [PubMed] [Google Scholar]