Abstract
Programmed −1 ribosomal frameshifting is utilized by a number of RNA viruses as a means of ensuring the correct ratio of viral structural to enzymatic proteins available for viral particle assembly. Altering frameshifting efficiencies upsets this ratio, interfering with virus propagation. We have previously demonstrated that compounds that alter the kinetics of the peptidyl-transfer reaction affect programmed −1 ribosomal frameshift efficiencies and interfere with viral propagation in yeast. Here, the use of a genetic approach lends further support to the hypothesis that alterations affecting the ribosome’s peptidyltransferase activity lead to changes in frameshifting efficiency and virus loss. Mutations in the RPL3 gene, which encodes a ribosomal protein located at the peptidyltransferase center, promote approximately three- to fourfold increases in programmed −1 ribosomal frameshift efficiencies and loss of the M1 killer virus of yeast. The mak8-1 allele of RPL3 contains two adjacent missense mutations which are predicted to structurally alter the Mak8-1p. Furthermore, a second allele that encodes the N-terminal 100 amino acids of L3 (called L3Δ) exerts a trans-dominant effect on programmed −1 ribosomal frameshifting and killer virus maintenance. Taken together, these results support the hypothesis that alterations in the peptidyltransferase center affect programmed −1 ribosomal frameshifting.
Programmed −1 ribosomal frameshifting is a mode of regulating gene expression used predominantly by RNA viruses and by a subset of bacterial genes to induce elongating ribosomes to shift the reading frame in response to specific mRNA signals (reviewed in references 16, 24, 27, and 30). Many viruses of clinical, veterinary, and agricultural importance utilize programmed frameshifting for the production of their structural and enzymatic gene products (reviewed in references 5, 6, 24, 27, 30, and 51). Thus, ribosomal frameshifting is a unique target with which to identify and develop antiviral agents (20, 41). Programmed −1 ribosomal frameshifting causes the ribosome to slip one base in the 5′ direction and requires two cis-acting mRNA signals. The first sequence element is called the “slippery site,” which, in eukaryotic viruses, consists of a heptamer sequence spanning three amino acid codons, X XXY YYZ (the gag reading frame is indicated by spaces), where XXX can be any three identical nucleotides, YYY can be AAA or UUU, and Z is A, U, or C (8, 17, 21, 31). The second frameshift-promoting signal is usually a sequence that forms a defined RNA secondary structure, such as an RNA pseudoknot (7, 17, 36). This element is located approximately 4 to 8 nucleotides 3′ of the slippery site and is thought to increase the probability that the ribosome will slip from the original reading frame in the −1 direction, in part by inducing ribosomes to pause at the slippery site (48, 53).
Based on the repetitive nature of the heptamer slippery sequence required for efficient programmed −1 ribosomal frameshifting, a simultaneous slippage model has been proposed to explain how ribosomes can be induced to change reading frames (31). A translating ribosome in which the A- and P-sites are occupied by tRNAs is forced to pause over the slippery site as a consequence of the RNA pseudoknot. The increased pause time over this sequence is thought to give an opportunity for the ribosome and bound tRNAs to slip 1 base in the 5′ direction. Because of the nature of the slippery site, this still leaves their non-wobble bases correctly paired with the mRNA in the new reading frame. Following the slip in the −1 direction, the ribosome continues translation in the new reading frame, producing the Gag-Pol polyprotein.
Since the simultaneous slippage model of programmed −1 ribosomal frameshifting requires that both the ribosomal A- and P-sites be occupied by tRNAs, it is implicit that this mechanism must occur after insertion of cognate aminoacyl-tRNA into the A-site, but prior to the translocation step of the translation elongation cycle. Furthermore, since programmed ribosomal frameshifting is driven by ribosomal pause events, mutations or agents that would serve to alter the amount of time that ribosomes are paused with both A- and P-sites occupied by tRNAs should specifically have an impact on the efficiency of programmed −1 ribosomal frameshifting. Since the peptidyl-transfer step in translation occurs while both the ribosomal A- and P-sites are occupied by tRNAs, we predicted that agents and mutations which alter the rate of this reaction would promote changes in programmed −1 ribosomal frameshift efficiencies and consequently would have antiviral properties.
In the yeast Saccharomyces cerevisiae, the L-A double-stranded RNA (dsRNA) virus utilizes a −1 ribosomal frameshift event for the production of a Gag-Pol fusion protein and has been an excellent model system with which to investigate this process (reviewed in references 12 and 20). M1, a satellite dsRNA virus of L-A that encodes a secreted killer toxin, is encapsidated and replicated by using the Gag and Gag-Pol gene products synthesized by the L-A virus (reviewed in reference 58). Maintaining the appropriate ratio of Gag to Gag-Pol is critical for maintenance of the M1 virus (21). Alteration of the frameshift process by as little as two- to threefold promotes rapid loss of M1 (21, 22). Genetic and biochemical analyses have identified a number of factors involved in determining the efficiency of programmed −1 ribosomal frameshifting (11, 12, 18, 19, 21–23, 34, 44). Based on this analysis, we have hypothesized that there is a surveillance complex which functions to monitor the A- and P-sites on the ribosomes to ensure that translation elongation occurs with high fidelity (15, 44). We suspect that this surveillance complex monitors the ribosome’s peptidyltransferase activity to ensure maximal translational fidelity. Mutations that alter or inactivate the activity of this complex reduce translational fidelity at the A- and P-sites and allow increased levels of ribosomal frameshifting to occur. A set of mof (maintenance of frame) alleles were shown to increase programmed −1 ribosomal frameshifting efficiencies and promote loss of the killer virus (22). Furthermore, compounds that bind to the peptidyltransferase center on the ribosome and reduce translation fidelity can also modulate ribosomal frameshifting (19). Anisomycin and sparsomycin were shown to alter programmed −1 ribosomal frameshifting efficiencies both in cells and in in vitro translation extracts and to promote loss of the yeast L-A and its satellite dsRNA virus, M1 (19). Taken together, these results indicate that modulation of the ribosomal peptidyltransferase center can alter the efficiency of programmed −1 ribosomal frameshifting and lead to inefficient virus propagation.
In the current study, we have genetically investigated the role of a ribosomal protein that is located at the ribosomal peptidyl-transfer center in modulating programmed frameshifting efficiencies. Previous results have shown that the yeast RPL3 gene encoding the ribosomal protein L3 participates in the formation of the peptidyltransferase center (reviewed in references 38 and 39). Mutations in the RPL3 gene (called TCM1) were initially identified by conferring resistance to the peptidyltransferase inhibitors trichodermin and anisomycin (32, 45). Independently, the MAK8 gene (MAK, maintenance of killer) was identified by the inability of mutant alleles to maintain the M1 satellite virus (59). Subsequent analysis demonstrated that MAK8 is allelic to RPL3 (60). Thus, a mutation in a ribosomal protein located in the peptidyltransferase center that cannot maintain the killer virus has been identified. We hypothesized that the underlying cause of killer virus loss observed in these cells may be a consequence of increased programmed −1 ribosomal frameshifting efficiency (i.e., that the mak8 alleles may demonstrate a Mof− phenotype). The results presented here demonstrate that strains harboring the mak8-1 allele have increased programmed frameshifting efficiencies and strongly suggest that the loss of the killer virus is a due to alteration in translation fidelity. Furthermore, a trans-dominant RPL3 allele has been identified that both increases programmed −1 frameshifting and interferes with the ability of yeast cells to maintain the M1 dsRNA virus. Taken together, these results support the notion that modulation of the peptidyltransferase center results in alteration of programmed −1 ribosomal frameshifting efficiencies, promoting loss of the killer virus.
MATERIALS AND METHODS
Strains, media, enzymes, oligonucleotides, and drugs.
Escherichia coli DH5α and MV1190 were used to amplify plasmid DNA. The yeast strains used in this study are listed in Table 1. Transformations of yeasts and E. coli were performed as described previously (13). YPAD, YPG, SD, synthetic complete medium (H−), and 4.7MB plates for testing the killer phenotype were used as previously reported (22). Restriction enzymes were obtained from Promega, MBI Fermentas, Bethesda Research Laboratories, and Boehringer Mannheim. T4 DNA ligase and T4 DNA polymerase were obtained from Boehringer Mannheim, and precision Taq polymerase was obtained from Stratagene. Radioactive nucleotides were obtained from NEN. Oligonucleotides used in these studies were purchased from IDT, and DNA sequence analysis was performed by the UMDNJ-RWJ DNA synthesis center. Anisomycin was purchased from Sigma, and sparsomycin was a generous gift from S. Pestka.
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea | Source |
|---|---|---|
| 1906 | MATa leu2 mak8-1 K− MKT+ | R. Wickner |
| 5X47 | MATa/MAT α his1/+ trp1/+ ura3/+ K−R− | R. Wickner |
| 2373 | MATa ura3 ski4-1 mkt1 K++ | R. Wickner |
| 2898 | MATa ura3 ade3 his(5,6) ski6-2 K++ | R. Wickner |
| 2413 | MATa ura3 cyh2 ski7-2 K++ | R. Wickner |
| JD100 | MATa ura3-52 his3 trp1-δ1 K1+ | This study |
| JD973 | MATα ura3-SK1 LEU2::hisG TRP1::hisG lys2-SK1 ho::LYS2 ade3-210S | This study |
| Cross JD980 | JD100 × JD973 with RPL3::hisG on 1 chromosome | This study |
| JD980-10C | MATα lys2 his3 ura3 LEU2::hisG trp1-δ1 RPL3::hisG + pRPL3 or pmak8-1 | This study |
| JD13 | MATa his3 leu2 PEP4::HIS3 NUC1::LEU2 ura3 K1+ | This study |
| JD111 | MATα ura3-52 lys2-801 trp1-δ1 leu2−− his3−− K1+ | This study |
| JD890 | MATα ura3-52 trp1 leu2Δ1 his3Δ300 pbrΔ1-6 can1r pep4::HIS3 SKI1::LEU2 [L-AHN M1] K++ | This study |
| JD2 | MATα ura3 trp1 ade8 ski2-2 K++ | This study |
−−, molecularly undefined allele that at the genetic level does not revert, i.e., is a very strong mutant; ++, phenotypically strong killer strain.
Plasmid constructs and programmed ribosomal frameshift assays.
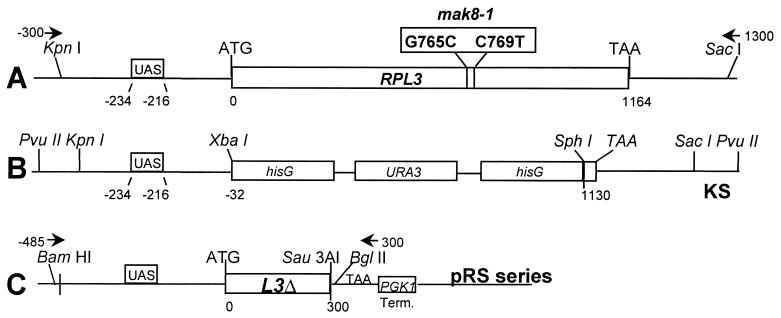
BlueScript KS plasmid was obtained from Stratagene. The pRS series of plasmids (10, 47) and pAS134 (1) have been previously described. Full-length RPL3 and mak8-1 were amplified from genomic DNA by PCR using the oligonucleotide primers −300KpnI (5′C CCCCGGTACCTCACGCACACTGGAATGAAT 3′) and +1300SacI (5′ CCCCGAGCGCAACCTCCATTTTGGACTTGG 3′) and were cloned into the pRS300 series (pRS314, pRS315, and pRS316) digested with KpnI and SacI to make the pRPL3 and the pmak8-1 series of plasmids (Fig. 1A). To construct an RPL3 gene disruption plasmid, the KpnI-SacI RPL3 clone was subcloned into BlueScript KS (KS-RPL3), digested with SphI, the overhanging ends were filled with deoxynucleoside triphosphates using T4 DNA polymerase, and the clone was then digested with XbaI. Subsequently, pAS134 was digested with XbaI and PvuII to liberate the hisG-URA3 cassette, which was subcloned into the XbaI/blunt-ended KS-RPL3 to create pJD168 (Fig. 1B).
FIG. 1.
(A) The full-length RPL3 gene is located on chromosome XV in yeast cells and consists of an upstream activation signal (UAS) at bp −234 through bp −216 relative to the ATG start codon. The RPL3 coding sequence is 1,164 bp long, and the gene encodes the 388-amino acid L3 protein. Restriction endonuclease sites are indicated. Full-length RPL3 and mutant mak8-1 alleles were cloned into the pRS series of plasmids by PCR with primers −300KpnI and 1300SacI as described in Materials and Methods. The sequence of mak8-1 was determined from three independently isolated clones as described in Materials and Methods. The mak8-1-specific G765C (encoding a Trp-to-Cys change at amino acid residue 255) and C769T (encoding a Pro-to-Ser change at amino acid residue 257, which is a potentially significant change) mutations are indicated. (B) Map of the RPL3-hisG-URA3 disruption plasmid. Based on BlueScript KS, a hisG::URA3 marker was integrated into the RPL3 locus between the XbaI site at −32 and an SphI site at 1130. (C) Map of the L3Δ fragment. L3Δ contains the 5′ noncoding sequence from position −485 plus the first 300 bp of RPL3 (to the Sau3AI site). L3Δ was cloned into the pRS series of vectors by using primers −485BamHI and 300BglII. A PGK1 terminator sequence is located downstream of the L3Δ coding region.
The original clone (YPF2-9) harboring the L3Δ fragment was obtained as a high-copy suppressor of a upf2 deletion strain as described previously (13). The active L3Δ fragment was amplified and subcloned into the pRS series of vectors by using Taq DNA polymerase and the −485BamHI (5′ ATAGGATCCTTAACCGGCCGGACAGTAATA 3′) and +300BglII (5′ ATAGGATCCTTGTCATCGTCGTCCTTGTAGTCTCTCAAACCTCTTGGGGT 3′) oligonucleotide primers to create pJD138 (low copy number, pRS315 based) and pJD139 (high copy number, pRS426 based) (Fig. 1C). The PGK1 transcriptional terminator was subcloned 3′ of the L3Δ fragments as previously described (12).
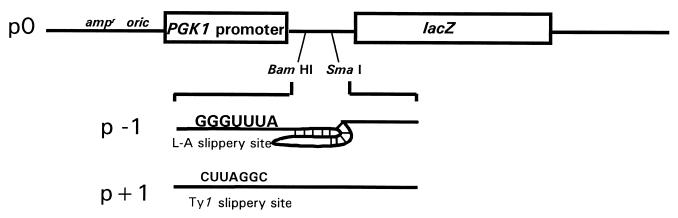
The plasmids used to monitor programmed ribosomal frameshifting were previously described (11, 12, 18, 19, 54). Briefly, in all of these plasmids, transcription is driven from the yeast PGK1 promoter into an AUG translational start site (Fig. 2). The E. coli lacZ gene serves as the reporter, and transcription termination utilizes the yeast PGK1 transcriptional terminator. In the p0 plasmids, lacZ is in the 0-frame with respect to the translational start site, and measurement of β-galactosidase activity generated from cells transformed with these plasmids serves to represent the 0-frame controls. In the p-1 series, lacZ is in the −1 frame with respect to the translational start site and is 3′ of the L-A-programmed −1 ribosomal frameshift signal, such that β-galactosidase can only be produced as a consequence of a programmed −1 ribosomal frameshift. Similarly, in the p+1 series, lacZ is in the +1 frame with respect to the translational start site and is 3′ of the Ty1-programmed +1 ribosomal frameshift signal, such that β-galactosidase can only be produced as a consequence of a programmed +1 ribosomal frameshift. The efficiency of programmed ribosomal frameshifting is calculated by determining the ratio of β-galactosidase activity produced by cells harboring either p−1 or p+1 divided by the β-galactosidase activity produced by cells harboring p0 and multiplying the result by 100%. Measurement of programmed −1 ribosomal frameshifting in the presence of anisomycin and sparsomycin was performed as described previously (19).
FIG. 2.
Vectors used to measure programmed ribosomal frameshifting efficiencies. The 0-frame control reporter plasmid p0 and the −1 ribosomal frameshift test plasmid p−1 are described in references 12 and 17, and the +1 ribosomal frameshift test plasmid p+1 is described in references 3 and 58. In these constructs, transcription is driven from the constitutive phosphoglycerol kinase 1 (PGK1) promoter. The efficiencies of programmed ribosomal frameshifting are determined by dividing the β-galactosidase activities produced from the frameshift reporters (p−1 or p+1) by those produced from the p0 control and multiplying the resulting ratios by 100%.
Construction of isogenic mak8-1 and RPL3 strains.
Yeast strains JD100 and JD973 were mated, and the diploids were transformed with PvuII-linearized pJD168 and selected on H-Ura medium (22). Disruption of the RPL3 locus on one chromosome was confirmed by Southern analysis as described below. Diploids were selected for loss of the chromosomal URA3 insert by growth on 5-flouroorotic acid (5-FOA). Ura− cells were transformed with pRPL3-Ura3, sporulated, and dissected onto YPAD medium. The resulting tetrads are from cross JD980. rpl3Δ status was confirmed by the inability of spore clones to grow in the presence of 5-FOA. To construct isogenic mak8-1 strains, cells were transformed with pmak8-1-TRP1 and were subsequently grown in the presence of 5-FOA to select for loss of the wild-type pRPL3-Ura3 plasmid.
Killer assay.
The killer virus assay was carried out as previously described (21). Briefly, yeast colonies were replica plated to 4.7MB plates (22) with a newly seeded lawn of strain 5X47 (0.5 ml of a suspension at 1 Unit of optical density at 550 nm per ml per plate). After 2 to 3 days at 20°C, killer activity was observed as a clear zone around the killer colonies. Loss-of-killer assays were performed with multiple wild-type and mutant strains.
Nucleic acid analyses.
dsRNAs of L-A and M1 viruses were prepared as described previously (25), separated by electrophoresis through 1.2% agarose gels, denatured in the gels in two changes of 30 min each of 50% formamide–9.25% formaldehyde–1 × Tris-acetate-EDTA at room temperature, and transferred to nitrocellulose in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). L-A and M1 negative strand RNA probes were labeled with [α-32P]UTP and hybridized to blots and washed as described in reference 22. RNase protection assays to determine the relative abundances of the lacZ −1 frameshift reporter mRNAs and U3 small nuclear RNA in the isogenic wild-type, mak8-1, and L3Δ strains were carried out as described previously (44).
RESULTS
The mak8-1 allele of RPL3 promotes increased programmed −1 ribosomal frameshifting efficiencies.
Previous studies have demonstrated that peptidyltransferase inhibitors specifically affect programmed −1 ribosomal frameshifting efficiencies (19). Based on these and other observations (11, 14, 15, 43, 44, 55–57), we hypothesized that the surveillance complex monitors the ribosomal peptidyltransferase center and that mutations that affect either the surveillance complex or the peptidyltransferase center will affect programmed −1 ribosomal frameshifting efficiencies (19, 44). Thus, we predicted that yeast strains harboring chromosomal mutations affecting the peptidyltransferase center may also have defects in programmed −1 ribosomal frameshifting and killer virus maintenance. The mak8-1 allele of ribosomal protein L3 initially presents a logical candidate with which to test this hypothesis, since strains harboring this mutation promoted loss of the killer virus. Programmed ribosomal frameshifting efficiencies were measured in vivo by using a series of lacZ reporter plasmids as described previously (12, 17, 19, 54) (see Fig. 2 for constructs). The p0 series of plasmids serve as the 0-frame controls, since lacZ is in the 0-frame with respect to the translational start site (Fig. 2). In the p−1 plasmid series, an L-A-derived programmed −1 ribosomal frameshift signal is cloned into the polylinker, and the lacZ gene is in the −1 frame with respect to the translational start site (Fig. 2). Therefore, in these constructs, the lacZ gene can only be translated if the ribosome shifts the frame in the −1 direction. Similarly, the p+1 plasmid series contains at Ty1 a programmed +1 ribosomal frameshift signal cloned into the polylinker, and the lacZ gene is in the +1 frame with respect to the translational start site (Fig. 2). The Ty1 +1 reporter plasmid is used as a control to determine the specificity of the effect of the mutation on translation. The efficiencies of −1 and +1 ribosomal frameshifting are calculated by determining the ratio of β-galactosidase activities measured in cells harboring p−1 or p+1 to those harboring p0 and multiplying the result by 100%.
After cells (strain 1906 [Table 1]) harboring the mak8-1 allele were transformed with p0, p−1, or p+1, the efficiencies of programmed ribosomal frameshifting were determined. The results demonstrated that the programmed −1 frameshifting efficiency in the mak8-1 strain was 5.2%, approximately threefold greater than the 1.7 to 2.0% normally observed in wild-type strains (Table 2). To confirm that the change in programmed −1 ribosomal frameshifting efficiency was solely due to the mak8-1 allele, isogenic wild-type and mak8-1 strains were constructed, and programmed −1 frameshifting in these cells was determined as described above (cross JD980 [Table 1]). In isogenic backgrounds, the mak8-1 allele of RPL3 promotes an approximately 2.5-fold increase in programmed −1 ribosomal frameshift efficiency (≈4.9% in mak8-1 compared to ≈1.9% in the isogenic wild-type strain [Table 2]). The mak8-1 allele was also unable to maintain the M1 killer virus (Table 2). However, mak8-1 had no effect on programmed +1 ribosomal frameshifting (Table 2). Taken together, these results demonstrate that the mak8-1 allele causes an alteration in programmed −1 ribosomal frameshift efficiencies. Thus, the mak8-1 allele is also a mof mutant, in that these strains demonstrate increased programmed −1 ribosomal frameshifting efficiencies and loss of the killer virus (11, 12).
TABLE 2.
Assays of programmed −1 ribosomal frameshifting and the killer phenotype in yeast cells harboring the wild-type RPL3 gene or the mak8-1 allele
| Strain | % Ribosomal frameshifta
|
Killer phenotypeb | |
|---|---|---|---|
| −1 | +1 | ||
| 1906 (mak8-1) | 5.18 ± 0.12 | 4.12 ± 0.16 | − |
| 980-10C pRPL3 | 1.93 ± 0.18 | 5.34 ± 0.18 | + |
| 980-10C + pmak8-1 | 4.85 ± 0.12 | 5.47 ± 0.11 | − |
The percent −1 ribosomal frameshifting was calculated by multiplying the ratio of p−1/p0 β-galactosidase activities by 100%. Absolute error is shown.
Killer phenotype was determined as described in Materials and Methods.
Characterization of the mak8-1 lesion.
The mak8-1 allele was amplified by PCR from genomic DNA harvested from strain 1906, and the DNA sequence was obtained from three independently isolated clones (see Materials and Methods). The results demonstrated that the mak8-1 allele harbors two separate mutations spaced four nucleotides apart (Fig. 1A). The G765C mutation encodes a Trp-to-Cys change at amino acid residue 255. The C769T mutation changes a proline at residue 257 to serine, a potentially significant structural change.
Strains harboring the mak8-1 allele are resistant to the effects of peptidyltransferase inhibitors on programmed −1 ribosomal frameshifting.
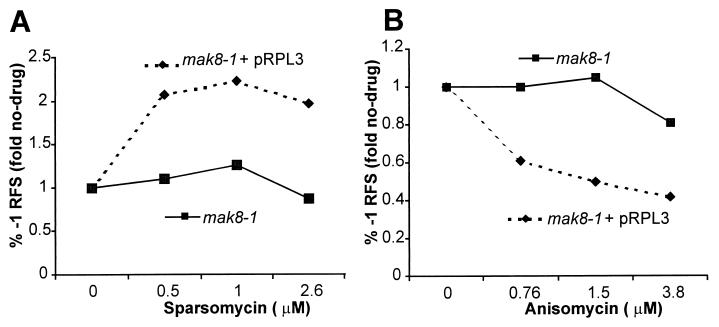
We previously demonstrated that peptidyltransferase inhibitors specifically alter programmed −1 ribosomal frameshifting efficiencies (19). It has been previously demonstrated that cells harboring mutant alleles of rpl3 are resistant to the cytotoxic effects of peptidyltransferase inhibitors (28, 32, 45, 60). These include strains harboring the mak8 and the tcm1 classes of RPL3 alleles. Thus, we asked whether members of this class of agents affect programmed −1 ribosomal frameshifting in strains harboring mak8-1. To examine this, mak8-1 and wild-type cells harboring either p0 or p−1 frameshift indicator plasmids were grown in the presence of various concentrations of either anisomycin or sparsomycin for 4 h, and programmed ribosomal frameshifting efficiencies were determined as described above. The results demonstrated that both anisomycin and sparsomycin altered ribosomal frameshifting in wild-type cells (Fig. 3). In contrast, neither anisomycin nor sparsomycin had any further effect on programmed −1 ribosomal frameshifting in mak8-1 strains (Fig. 3). These results provide strong evidence that a defect affecting the peptidyltransferase center is responsible for the observed increase in programmed −1 ribosomal frameshifting in mak8-1 cells.
FIG. 3.
Programmed −1 ribosomal frameshifting (−1 RFS) in a mak8-1 strain is not further affected by peptidyltransferase inhibitors. Isogenic wild-type and mak8-1 cells harboring either p0 or p−1 frameshift indicator plasmids were grown in the presence of the indicated concentrations of sparsomycin (A) or anisomycin (B) for 4 h, after which programmed −1 ribosomal frameshifting efficiencies were determined as described in Materials and Methods. In the absence of drugs, wild-type cells promote approximately 2% efficiency of programmed −1 ribosomal frameshifting, whereas this value is approximately 5% in cells harboring the mak8-1 allele. The fold changes in programmed −1 ribosomal frameshifting efficiencies are plotted on the y axis.
Episomal expression of the N-terminal 100 amino acids of ribosomal protein L3 increases programmed frameshifting and loss of the killer virus.
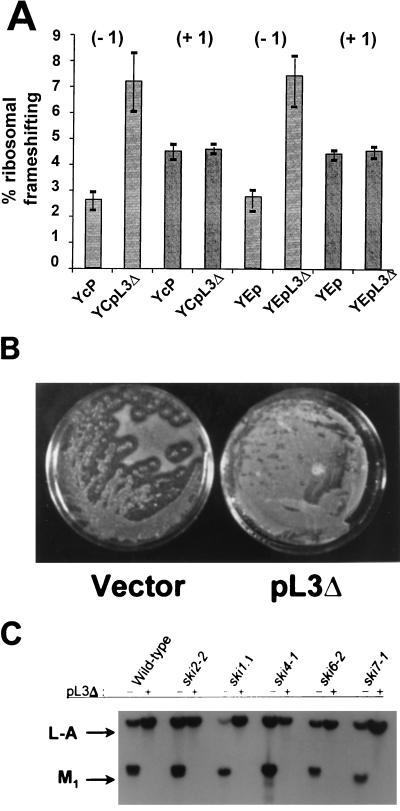
We have recently identified a second RPL3 allele that affects both programmed −1 ribosomal frameshifting and virus maintenance. This allele was isolated by its ability to abrogate the capability of cells harboring the upf2-1 allele to allow the expression of nonsense-containing mRNAs (13). Previous results have demonstrated that mutations in the UPF1, UPF2, and UPF3 genes can affect several aspects of translational fidelity (12, 15, 44, 55, 56). In particular, a specific mutation in the UPF1 gene (mof4-1) and a deletion of the UPF3 gene both increase programmed −1 ribosomal frameshifting efficiencies and lead to loss of the killer virus (12, 44). Characterization of the antisuppressor of upf2-1, YPF2-9, demonstrated that it encodes the N-terminal 100 amino acids of the RPL3 (see Materials and Methods). This allele was renamed L3Δ (Fig. 1C). Based on this connection between the upf mutants and L3, we examined the effect of episomal expression of L3Δ on programmed ribosomal frameshifting. Episomal expression of L3Δ protein in wild-type cells from either low- or high-copy plasmids promoted an approximately threefold increase in the efficiency of programmed −1 ribosomal frameshifting, but had no effect on programmed +1 ribosomal frameshifting (Fig. 4A). Expression of L3Δ in wild-type cells also promoted high rates of killer phenotype loss (Fig. 4B). RNA hybridization analysis revealed that loss of the killer phenotype was a consequence of failure to maintain the M1 satellite dsRNA virus (Fig. 4C).
FIG. 4.
The L3Δ allele of RPL3 confers a dominant-negative Mof− phenotype. (A) Wild-type cells were transformed with L3Δ cloned into either low-copy-number (YCp) or high-copy-number (YEp) vectors, and programmed −1 and +1 ribosomal frameshifting efficiencies were monitored with the following results: programmed −1 ribosomal frameshifting, YCp = 2.63% ± 0.34%, YCpL3Δ = 7.22% ± 1.13%, YEp = 2.74% ± 0.64%, and YEpL3Δ = 7.42% ± 1.13%; programmed +1 ribosomal frameshifting, YCp = 4.52% ± 0.29%, YCpL3Δ = 4.60% ± 0.14%, YEp = 4.42% ± 0.18%, and YEpL3Δ = 4.53% ± 0.22%. (B) Killer phenotypes of wild-type cells (JD890) transformed with YCpL3Δ or vector. (C) Total nucleic acids were extracted from cells transformed with vector alone (−) or from cells transformed with YCpL3Δ (+), and RNA was transferred to nitrocellulose and hybridized with L-A- and M1-specific negative-strand probes. L-A- and M1-hybridizing bands are indicated.
Although the +1 frameshift and killer loss data strongly suggest that programmed −1 ribosomal frameshifting is elevated in cells harboring the mak8-1 and L3Δ alleles, the formal possibility exists that specific stabilization of the LacZ −1 mRNA relative to the 0-frame control mRNA may account for an apparent increase in programmed −1 ribosomal frameshift efficiencies and that M1 loss is a consequence of some other defect. The level of the LacZ −1 frameshift reporter mRNA was not affected either by cells harboring the mak8-1 allele or by expression of L3Δ (data not shown). These data are consistent with previous results demonstrating that expression of L3Δ (YPF2-9) did not affect the levels of a nonsense-containing Cyh2 precursor mRNA in a upf2-1 strain (13). Thus, L3Δ also behaves like a mof mutant in that it inhibits virus propagation as a consequence of increased programmed −1 ribosomal frameshifting.
L3Δ is dominant to the ski mutants.
The L-A and M1 mRNAs present poor translational substrates because they do not possess either the 5′ m7G5′ppp5′Xp cap or 3′ poly(A) tails (9, 52). The lack of these structures does not prevent their expression in wild-type cells, but does make their expression sensitive to mutations which affect translation (e.g., the mak mutants) (4, 35, 40). Conversely, L-A and M1 copy numbers are increased in cells harboring mutations in chromosomal genes which are involved in recognition of 5′ caps or 3′ poly(A) tails and in the degradation of mRNAs lacking these structures (2, 4, 35). However, since the ratio of Gag to Gag-Pol is critical for viral particle morphogenesis, we predicted that mutations which change this ratio by altering frameshift efficiencies should be dominant to the effects of the ski mutants, because although the L-A and M1 mRNAs are either stabilized or translated more efficiently by the ski mutations, the ratios of Gag to Gag-Pol should still be altered as a consequence of the mof mutations.
To test this hypothesis, a plasmid harboring the L3Δ allele was introduced into cells harboring mutations in a series of SKI genes. These included the SKI1/XRN1 gene, which encodes the major 5′→3′ exoribonuclease that degrades uncapped RNAs (29, 33, 37, 49, 50), the SKI2 and SKI6 genes, which play a role in ribosome biogenesis, and which are both required for efficient translation of poly(A) mRNAs, and the 3′→5′ exonuclease activity of the exosome (2, 4, 35), SKI4, and SKI7 (42). The results demonstrated that episomal expression of L3Δ promoted rapid loss of the killer phenotype, which was due to loss of the M1 virus (Fig. 4C).
DISCUSSION
Mutations affecting ribosomal protein L3 promote loss of the M1 killer virus by altering the efficiency of programmed −1 ribosomal frameshifting.
The mechanism governing programmed −1 ribosomal frameshifting suggests that drugs and mutations which affect the peptidyl-transfer reaction may alter programmed −1 ribosomal frameshift efficiencies and have antiviral effects (19). We previously used peptidyltransferase inhibitors to demonstrate the validity of this model (19). The results presented here have shown that two alleles encoding mutant forms of ribosomal protein L3, which was previously implicated in formation of the peptidyltransferase center, also alter programmed −1 ribosomal frameshift efficiencies and have antiviral effects. Taken together, these results support the hypothesis that the peptidyltransferase center may present a novel target for antiretroviral therapeutic agents.
It has long been known that cells harboring mak8 alleles cannot propagate the M1 satellite virus (59). Additional alleles of RPL3, named tcm1, were also characterized based on their resistance to the peptidyltransferase inhibitor trichodermin (26, 28, 32, 45, 46). These alleles also have the Mak− phenotype (60). However, the precise mechanism responsible for killer virus loss in this class of mutants was not determined. Previous results suggested that mak8-1 did not affect programmed −1 ribosomal frameshifting efficiencies (22). However, the interpretation of those results was incorrect in that only changes in overall β-galactosidase activities generated from a frameshift reporter construct by using sister spore clones were examined. The present study rectifies those defects by directly measuring programmed ribosomal frameshifting efficiencies in isogenic strains. The results presented here demonstrate that alterations in programmed −1 ribosomal frameshifting efficiencies are responsible for the inability of cells harboring this mutation to maintain the M1 dsRNA virus. Given the previous demonstration that peptidyltransferase inhibitors promote virus loss by altering programmed −1 ribosomal frameshift efficiencies, as well as the role of the L3 protein in peptidyltransferase center formation, our results indicating that mutations in RPL3 affect programmed −1 ribosomal frameshifting are consistent with the view that alteration of peptidyl-transfer activity affects this process. In addition, the finding that episomal expression of the L3Δ allele confers a dominant negative Mof− phenotype provides a novel tool that can be used to probe the contribution of L3 to translational fidelity. The trans-dominance of the L3 peptide illuminates the importance of programmed −1 ribosomal frameshifting in the viral life cycle. The fact that L3Δ is dominant to the ski mutants demonstrates that even when viral RNAs and proteins are in excess, these mutants cannot overcome the imbalance in the ratio of Gag to Gag-Pol proteins as a consequence of altered programmed −1 ribosomal frameshifting efficiency. Thus, the trans-dominance of the L3Δ allele with respect to the ski mutants supports the hypothesis that the efficiency of programmed ribosomal frameshifting plays a critical role in the viral particle morphogenetic process by ensuring the correct ratio of viral structural to enzymatic proteins.
We envision two models to explain the role of the L3 protein in programmed −1 ribosomal frameshifting. In one, we suggest that the incorporation of defective L3 protein (either Mak8-1p or L3Δp) into ribosomes would result in suboptimal L3 function, yielding the observed translational fidelity defect. Alternatively, it is possible that expression of these alleles results in a subpopulation of L3-deficient ribosomes. Since it is thought that the large rRNA is responsible for peptidyltransferase activity (38, 61), these L3-deficient ribosomes would retain a small amount of peptidyltransferase activity. In both scenarios, defects in peptidyltransferase activity are predicted to slow the rate of translation elongation while both the ribosomal A- and P-sites are occupied. In the context of frameshifting, this would result in a longer ribosomal pause at the programmed −1 ribosomal frameshift signal, increasing the likelihood of a successful frameshift. If this model is true, then the observed increases in programmed −1 ribosomal frameshifting efficiencies promoted by these alleles should represent the sum of programmed frameshifting promoted by normal plus defective ribosomes.
In sum, we have demonstrated that two genetically defined alleles encoding L3, a ribosomal protein which has been demonstrated to participate in formation of the peptidyltransferase center, both affect the efficiency of programmed −1 ribosomal frameshifting and promote loss of the killer virus. These studies are consistent with our pharmacologically based observations that peptidyltransferase inhibitors specifically affect programmed −1 ribosomal frameshifting efficiencies and demonstrate the utility of using programmed ribosomal frameshifting as an assay to probe the mechanisms which regulate the process of protein translation.
ACKNOWLEDGMENTS
We thank Reed Wickner for strain 1906 and Michael Leibowitz for helpful discussions.
This work was supported by grants to J.D.D. by the Foundation of UMDNJ (#16-98), the New Jersey Commission on Cancer Research (97-60-CCR), and the National Science Foundation (MCB-9807890) and to S.W.P. by the National Institutes of Health (GM48631). S.W.P. was also supported by an American Heart Established Investigator Award. A.B.H. was supported in part by a training grant from the National Institutes of Health (T32 AI07403-07), and L.P. was supported in part by the Henry Rutgers Scholars Program (RPO 6183).
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson J S J, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasundaram D, Dinman J D, Wickner R B, Tabor C W, Tabor H. Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:172–176. doi: 10.1073/pnas.91.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benard L, Carroll K, Valle R C P, Wickner R B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop J M. Retroviruses. Annu Rev Biochem. 1978;47:35–88. doi: 10.1146/annurev.bi.47.070178.000343. [DOI] [PubMed] [Google Scholar]
- 6.Brierley I. Ribosomal frameshifting on viral RNAs. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 7.Brierley I A, Dingard P, Inglis S C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brierley I A, Jenner A J, Inglis S C. Mutational analysis of the “slippery sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruenn J, Keitz B. The 5′ ends of yeast killer factor RNAs are pppGp. Nucleic Acids Res. 1976;3:2427–2436. doi: 10.1093/nar/3.10.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Yeast. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 11.Cui Y, Dinman J D, Kinzy T G, Peltz S W. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol Cell Biol. 1998;18:1506–1516. doi: 10.1128/mcb.18.3.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui Y, Dinman J D, Peltz S W. mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 14.Cui, Y., T. G. Kinzy, J. D. Dinman, and S. W. Peltz. Unpublished data.
- 15.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1667. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinman J D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinman J D, Icho T, Wickner R B. A −1 ribosomal frameshift in a double-stranded RNA virus forms a Gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinman J D, Kinzy T G. Translational misreading: mutations in translation elongation factor 1α differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- 19.Dinman J D, Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Peptidyl transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: development of model systems. Proc Natl Acad Sci USA. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinman J D, Ruiz-Echevarria M J, Peltz S W. Translating old drugs into new treatments: identifying compounds that modulate programmed −1 ribosomal frameshifting and function as potential antiviral agents. Trends Biotechnol. 1998;16:190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinman J D, Wickner R B. Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered −1 ribosomal frameshifting efficiencies. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinman J D, Wickner R B. 5S rRNA is involved in fidelity of translational reading frame. Genetics. 1995;141:95–105. doi: 10.1093/genetics/141.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fried H M, Fink G R. Electron microscopic heteroduplex analysis of “killer” double-stranded RNA species from yeast. Proc Natl Acad Sci USA. 1978;75:4224–4228. doi: 10.1073/pnas.75.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fried H M, Warner J R. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc Natl Acad Sci USA. 1981;78:238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 28.Grant P G, Schindler D, Davies J E. Mapping of trichodermin resistance in Saccharomyces cerevisiae: a genetic locus for a component of the 60S ribosomal subunit. Genetics. 1976;83:667–673. doi: 10.1093/genetics/83.4.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1996;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- 31.Jacks T, Madhani H D, Masiraz F R, Varmus H E. Signals for ribosomal frameshifting in the Rous Sarcoma Virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jimenez A, Sanchez L, Vazquez D. Simultaneous ribosomal resistance to trichodermin and anisomycin in Saccharomyces cerevisiae mutants. Biochim Biophys Acta. 1975;383:427–434. doi: 10.1016/0005-2787(75)90312-3. [DOI] [PubMed] [Google Scholar]
- 33.Johnson A W, Kolodner R D. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S I, Umen J G, Varmus H E. A genetic screen identifies cellular factors involved in retroviral −1 frameshifting. Proc Natl Acad Sci USA. 1995;92:6587–6591. doi: 10.1073/pnas.92.14.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Decoying the Cap− mRNA degradation system by a double-stranded RNA virus and poly(A)− mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morikawa S, Bishop D H L. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 38.Noller H F. Peptidyl transferase: protein, ribonucleoprotein, or RNA? J Bacteriol. 1993;175:5297–5300. doi: 10.1128/jb.175.17.5297-5300.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noller H F. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 40.Ohtake Y, Wickner R B. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peltz S W, Trotta C, He F, Brown A, Donahue J, Welch E, Jacobson A. Identification of the cis-acting sequences and trans-acting factors involved in nonsense-mediated mRNA decay. In: Tuite M, McCarthy J, Brown A, Sherman F, editors. Protein synthesis and targeting in yeast. Berlin, Germany: Springer-Verlag; 1993. pp. 1–10. [Google Scholar]
- 42.Ridley S P, Sommer S S, Wickner R B. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz-Echevarria M J, Yasenchak J M, Han X, Dinman J D, Peltz S W. The Upf3p is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral maintenance. Proc Natl Acad Sci USA. 1998;95:8721–8726. doi: 10.1073/pnas.95.15.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindler D, Grant P, Davies J. Trichodermin resistance—mutation affecting eukaryotic ribosomes. Nature. 1974;248:535–536. doi: 10.1038/248535a0. [DOI] [PubMed] [Google Scholar]
- 46.Schultz L D, Friesen J D. Nucleotide sequence of the tcm1 gene (ribosomal protein L3) of Saccharomyces cerevisiae. J Bacteriol. 1983;155:8–14. doi: 10.1128/jb.155.1.8-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somogyi P, Jenner A J, Brierley I, Inglis S C. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to 3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 50.Stevens A, Maupin M K. A 5′—3′ exoribonuclease of Saccharomyces cerevisiae: size and novel substrate specificity. Arch Biochem Biophys. 1987;252:339–347. doi: 10.1016/0003-9861(87)90040-3. [DOI] [PubMed] [Google Scholar]
- 51.TenDam E, Pleij K, Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiele D J, Hannig E M, Leibowitz M J. Genome structure and expression of a defective interfering mutant of the killer virus of yeast. Virology. 1984;137:20–31. doi: 10.1016/0042-6822(84)90004-7. [DOI] [PubMed] [Google Scholar]
- 53.Tu C, Tzeng T-H, Bruenn J A. Ribosomal movement impeded at a pseudoknot required for ribosomal frameshifting. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tumer N E, Parikh B A, Li P, Dinman J D. Pokeweed antiviral protein specifically inhibits Ty1-directed +1 ribosomal frameshifting and Ty1 retrotransposition in Saccharomyces cerevisiae. J Virol. 1998;72:1036–1042. doi: 10.1128/jvi.72.2.1036-1042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weng Y, Czaplinski K, Peltz S W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng Y, Czaplinski K, Peltz S W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biochem. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weng Y, Czaplinski K, Peltz S W. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- 58.Wickner R B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickner R B, Leibowitz M J. Chromosomal and non-chromosomal mutations affecting the “killer character” of Saccharomyces cerevisiae. Genetics. 1974;76:423–432. doi: 10.1093/genetics/76.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickner R B, Porter-Ridley S, Fried H M, Ball S G. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1982;79:4706–4708. doi: 10.1073/pnas.79.15.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang B, Cech T R. Peptide bond formation by in vitro selected ribozymes. Nature. 1997;390:96–100. doi: 10.1038/36375. [DOI] [PubMed] [Google Scholar]