Abstract
Simple Summary
Talaromyces species are distributed all around the world and occur in various environments, e.g., soil, air, living or rotten plants, and indoors. Some of them produce enzymes and pigments of industrial importance, while some cause Talaromycosis. Talaromyces marneffei, a well-known and important human pathogen, is endemic to Southeast Asia and causes high mortality, especially in HIV/AIDS patients and those with other immunodeficiencies. China covers 3 of the 35 global biodiversity hotspots. During the explorations of fungal diversity in soil samples collected at different sites of southwestern China, two new Talaromyces species, T. chongqingensis X.C. Wang and W.Y. Zhuang and T. wushanicus X.C. Wang and W.Y. Zhuang, were discovered based on phylogenetic analyses and morphological comparisons. They are described and illustrated in detail. Six phylogenetic trees of the sections Talaromyces and Trachyspermi were constructed based on three-gene datasets and revealed the phylogenetic positions of the new species. This work provided a better understanding of biodiversity and phylogeny of the genus. The results make the concepts of the two sections of Talaromyces well-established. The discovery will be beneficial for future evaluation of the potential usages and functions of the new species.
Abstract
Southwestern China belongs among the global biodiversity hotspots and the Daba Mountains are recognized as one of the priority conservation areas. During the exploration of fungal biodiversity from soil samples collected from Mount Daba, two species of Talaromyces were discovered as new to science based on phylogenetic analyses and morphological comparisons. Talaromyces chongqingensis sp. nov. is a sister taxon of T. minioluteus and T. minnesotensis in the section Trachyspermi; and T. wushanicus sp. nov., affiliated to the section Talaromyces, is closely related to T. cnidii and T. siamensis. The new species differ from their sisters in DNA sequences, growth rates, and morphological characteristics. Descriptions and illustrations of them are provided in detail.
Keywords: Ascomycota, biodiversity hotspot, DNA barcodes, Eurotiales, phylogeny, taxonomy, Trichocomaceae
1. Introduction
Talaromyces C.R. Benj. is a cosmopolitan genus occurring in various environments, e.g., soil, air, living or rotten plants, and indoors. Its beneficial and harmful effects on humans have been well documented. Enzymes and pigments produced by some species of the genus are of industrial importance, such as β-glucosidase produced by T. amestolkiae N. Yilmaz et al. [1] and T. cellulolyticus T. Fujii et al. [2], and red pigments by T. atroroseus N. Yilmaz et al. [3,4]. Talaromycosis caused by several species were also reported [5,6]. Among them, T. marneffei (Segretain et al.) Samson et al., endemic to Southeast Asia, is a well-known and important human pathogen causing high mortality in the absence of proper diagnosis and prompt treatment, especially in HIV/AIDS patients and those with other immunodeficiencies [7].
A total of 170 Talaromyces species were accepted and classified into seven sections according to a recent monographic study [8]. Moreover, T. albisclerotius B.D. Sun et al., T. aspriconidius B.D. Sun et al., T. aureolinus L. Wang, T. bannicus L. Wang, T. brevis B.D. Sun et al., T. guizhouensis B.D. Sun et al., T. penicillioides L. Wang, T. pulveris Crous, T. rufus B.D. Sun et al., T. sparsus L. Wang, T. tenuis B.D. Sun et al., and T. yunnanensis Doilom and C.F. Liao were later described [9,10,11,12]. In the section (sect.) Trachyspermi Yaguchi and Udagawa, 30 species are commonly accepted; and in the sect. Talaromyces, the largest part of the genus, 75 species have been recognized.
During the explorations of fungal diversity in soil samples collected at different sites of Chongqing and Sichuan in southwestern China, two Talaromyces species belonging to the sections Talaromyces and Trachyspermi were further discovered as new to science based on phylogenetic analyses and morphological comparisons. They are described and illustrated in detail.
2. Materials and Methods
2.1. Fungal Materials
Cultures were isolated from soil samples collected from Chongqing and areas nearby in Sichuan Province in October 2020. Dried cultures were deposited in the Herbarium Mycologicum Academiae Sinicae (HMAS), and the living ex-type strains were preserved in the China General Microbiological Culture Collection Center (CGMCC).
2.2. Morphological Observations
Morphological characterization was conducted following the standardized methods [13]. Four standard growth media were used: Czapek yeast autolysate agar (CYA, yeast extract Oxoid, Hampshire, UK), malt extract agar (MEA, Amresco, Solon, OH, USA), yeast extract agar (YES) and potato dextrose agar (PDA). The methods for inoculation, incubation, microscopic examinations, and digital recordings were following our previous studies [14,15,16].
2.3. DNA Extraction, PCR Amplification, and Sequencing
DNA was extracted from the cultures grown on PDA for 7 days using the Plant Genomic DNA Kit (DP305, TIANGEN Biotech, Beijing, China). Polymerase chain reaction (PCR) amplifications of the internal transcribed spacer (ITS), beta-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) gene regions were conducted with routine methods [14,15,16]. The products were purified and subject to sequencing on an ABI 3730 DNA Sequencer (Applied Biosystems, Bedford, MA, USA). Although the ITS region, the recommended standard DNA barcode for fungi, is not sufficient to discriminate the species of this genus, the sequences provided here will be helpful for other researchers in case of need.
2.4. Phylogenetic Analyses
The forward and reverse sequences newly generated in this study were assembled using Seqman v. 7.1.0 (DNASTAR Inc., Madison, WI, USA). The assembled sequences were deposited in GenBank. Previously described species from the corresponding sections, which were used for phylogenetic analyses, are listed in Table 1 and Table 2. Newly generated sequences of this study are shown in Table 3. For each section, three datasets of BenA, CaM, and RPB2 were compiled. Sequences of each dataset (35 species for sect. Trachyspermi and 79 species for sect. Talaromyces) were aligned using MAFFT v. 7.221 [17], and then manually edited in BioEdit v. 7.1.10 [18] and MEGA v. 6.0.6 [19]. Maximum likelihood (ML) analyses were performed using RAxML-HPC2 [20] on XSEDE 8.2.12 on CIPRES Science Gateway v. 3.3 [21] with the default GTRCAT model. Bayesian Inference (BI) analyses were performed with MrBayes v. 3.2.5 [22]. Appropriate nucleotide substitution models and parameters were determined by Modeltest v. 3.7 [23]. The consensus trees were viewed in FigTree v. 1.3.1 (Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 September 2015)). The type species of section Talaromyces served as outgroup taxon of the Trachyspermi tree and vice versa.
Table 1.
Previously described Talaromyces species used in phylogenetic analyses of the sect. Trachyspermi.
| Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. aerius A.J. Chen et al. 2016 | CGMCC 3.18197 T | China: Beijing | indoor air | KU866647 | KU866835 | KU866731 | KU866991 |
| T. affinitatimellis Rodr.-Andr. et al. 2019 | CBS 143840 T | Spain | honey | LT906543 | LT906552 | LT906549 | LT906546 |
| T. albisclerotius B.D. Sun et al. 2020 | CBS 141839 T | China: Guizhou | soil | MN864276 | MN863345 | MN863322 | MN863334 |
| T. albobiverticillius (H.M. Hsieh et al.) Samson et al. 2011 | CBS 133440 T | China: Taiwan | decaying leaves | HQ605705 | KF114778 | KJ885258 | KM023310 |
| T. amyrossmaniae Rajeshkumar et al. 2019 | NFCCI 1919 T | India | decaying fruits of Terminalia bellerica | MH909062 | MH909064 | MH909068 | MH909066 |
| T. assiutensis Samson and Abdel-Fattah 1978 | CBS 147.78 T | Egypt | soil | JN899323 | KJ865720 | KJ885260 | KM023305 |
| T. atroroseus N. Yilmaz et al. 2013 | CBS 133442 T | South Africa | house dust | KF114747 | KF114789 | KJ775418 | KM023288 |
| T. austrocalifornicus Yaguchi and Udagawa 1993 | CBS 644.95 T | USA | soil | JN899357 | KJ865732 | KJ885261 | MN969147 |
| T. basipetosporus Stchigel et al. 2019 | CBS 143836 T | Argentina | honey | LT906542 | LT906563 | n.a. | LT906545 |
| T. brasiliensis R.N. Barbosa et al. 2018 | CBS 142493 T | Brazil | honey | MF278323 | LT855560 | LT855563 | MN969198 |
| T. catalonicus Guevara-Suarez et al. 2020 | CBS 143039 T | Spain | herbivore dung | LT899793 | LT898318 | LT899775 | LT899811 |
| T. clemensii Visagie and N. Yilmaz 2019 | PPRI 26753 T | South Africa | wood in mine | MK951940 | MK951833 | MK951906 | MN418451 |
| T. convolutus Udagawa 1993 | CBS 100537 T | Nepal | soil | JN899330 | KF114773 | MN969316 | JN121414 |
| T. diversus (Raper and Fennell) Samson et al. 2011 | CBS 320.48 T | USA | mouldy leather | KJ865740 | KJ865723 | KJ885268 | KM023285 |
| T. erythromellis (A.D. Hocking) Samson et al. 2011 | CBS 644.80 T | Australia | soil | JN899383 | HQ156945 | KJ885270 | KM023290 |
| T. guatemalensis A. Nováková et al. 2019 | CCF 6215 T | Guatemala | soil | MN322789 | MN329687 | MN329688 | MN329689 |
| T. halophytorum Y.H. You and S.B. Hong 2020 | KACC 48127 T | South Korea | roots of Limonium tetragonum | MH725786 | MH729367 | MK111426 | MK111427 |
| T. heiheensis X.C. Wang and W.Y. Zhuang 2017 | CGMCC 3.18012 T | China: Heilongjiang | rotten wood | KX447526 | KX447525 | KX447532 | KX447529 |
| T. minioluteus (Dierckx) Samson et al. 2011 | CBS 642.68 T | unknown | unknown | JN899346 | MN969409 | KJ885273 | JF417443 |
| T. minnesotensis Guevara-Suarez et al. 2017 | CBS 142381 T | USA | human ear | LT558966 | LT559083 | LT795604 | LT795605 |
| T. pernambucoensis R. Cruz et al. 2019 | URM 6894 T | Brazil | soil | LR535947 | LR535945 | LR535946 | LR535948 |
| T. resinae (Z.T. Qi and H.Z. Kong) Houbraken and X.C. Wang 2020 | CGMCC 3.4387 T | China: Guizhou | resin of Eucalyptus tereticornis | MT079858 | MN969442 | MT066184 | MN969221 |
| T. rubrifaciens W.W. Gao 2016 | CGMCC 3.17658 T | China: Beijing | hospital air | KR855658 | KR855648 | KR855653 | KR855663 |
| T. solicola Visagie and K. Jacobs 2012 | DAOM 241015 T | South Africa | soil | FJ160264 | GU385731 | KJ885279 | KM023295 |
| T. speluncarum Rodr.-Andr. et al. 2020 | CBS 143844 T | Spain | sparkling wine | LT985890 | LT985901 | LT985906 | LT985911 |
| T. subericola Rodr.-Andr. et al. 2020 | CBS 144322 T | Spain | sparkling wine | LT985888 | LT985899 | LT985904 | LT985909 |
| T. systylus S.M. Romero et al. 2015 | BAFCcult 3419 T | Argentina | soil | KP026917 | KR233838 | KR233837 | n.a. |
| T. trachyspermus (Shear) Stolk and Samson 1973 | CBS 373.48 T | USA | unknown | JN899354 | KF114803 | KJ885281 | JF417432 |
| T. ucrainicus (Panas.) Udagawa 1966 | CBS 162.67 T | Ukraine | potato starch | JN899394 | KF114771 | KJ885282 | KM023289 |
| T. udagawae Stolk and Samson 1972 | CBS 579.72 T | Japan | soil | JN899350 | KF114796 | KX961260 | MN969148 |
| T. flavus (Klöcker) Stolk and Samson 1972 | CBS 310.38 T | New Zealand | unknown | JN899360 | JX494302 | KF741949 | JF417426 |
Table 2.
Previously described Talaromyces species used in phylogenetic analyses of the sect. Talaromyces.
| Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. aculeatus (Raper and Fennell) Samson et al. 2011 | CBS 289.48 T | USA | textile | KF741995 | KF741929 | KF741975 | MH793099 |
| T. adpressus A.J. Chen et al. 2016 | CGMCC 3.18211 T | China: Beijing | indoor air | KU866657 | KU866844 | KU866741 | KU867001 |
| T. alveolaris Guevara-Suarez et al. 2017 | CBS 142379 T | USA | human bronchoalveolar lavage | LT558969 | LT559086 | LT795596 | LT795597 |
| T. amazonensis N. Yilmaz et al. 2016 | CBS 140373 T | Colombia | leaf litter | KX011509 | KX011490 | KX011502 | MN969186 |
| T. amestolkiae N. Yilmaz et al. 2012 | CBS 132696 T | South Africa | house dust | JX315660 | JX315623 | KF741937 | JX315698 |
| T. angelicae S.H. Yu et al. 2013 | KACC 46611 T | South Korea | dried root of Angelica gigas | KF183638 | KF183640 | KJ885259 | KX961275 |
| T. annesophieae Houbraken 2017 | CBS 142939 T | Netherlands | soil | MF574592 | MF590098 | MF590104 | MN969199 |
| T. apiculatus Samson et al. 2011 | CBS 312.59 T | Japan | soil | JN899375 | KF741916 | KF741950 | KM023287 |
| T. argentinensis Jurjević and S.W. Peterson 2019 | NRRL 28750 T | Ghana | soil | MH793045 | MH792917 | MH792981 | MH793108 |
| T. aurantiacus (J.H. Mill. et al.) Samson et al. 2011 | CBS 314.59 T | USA | soil | JN899380 | KF741917 | KF741951 | KX961285 |
| T. aureolinus L. Wang 2021 | CGMCC 3.15865 T | China: Yunnan | soil | MK837953 | MK837937 | MK837945 | MK837961 |
| T. australis Visagie et al. 2015 | CBS 137102 T | Australia | soil under pasture | KF741991 | KF741922 | KF741971 | KX961284 |
| T. bannicus L. Wang 2021 | CGMCC 3.15862 T | China: Yunnan | soil | MK837955 | MK837939 | MK837947 | MK837963 |
| T. beijingensis A.J. Chen et al. 2016 | CGMCC 3.18200 T | China: Beijing | indoor air | KU866649 | KU866837 | KU866733 | KU866993 |
| T. calidicanius (J.L. Chen) Samson et al. 2011 | CBS 112002 T | China: Taiwan | soil | JN899319 | HQ156944 | KF741934 | KM023311 |
| T. californicus Jurjević and S.W. Peterson 2019 | NRRL 58168 T | USA | air | MH793056 | MH792928 | MH792992 | MH793119 |
| T. cnidii S.H. Yu et al. 2013 | KACC 46617 T | South Korea | dried roots of Cnidium sp. | KF183639 | KF183641 | KJ885266 | KM023299 |
| T. coprophilus Guevara-Suarez et al. 2020 | CBS 142756 T | Spain | herbivore dung | LT899794 | LT898319 | LT899776 | LT899812 |
| T. cucurbitiradicus L. Su and Y.C. Niu 2018 | ACCC 39155 T | China: Beijing | endophyte from root of pumpkin (Cucurbita moschata) | KY053254 | KY053228 | KY053246 | n.a. |
| T. derxii Takada and Udagawa 1988 | CBS 412.89 T | Japan | cultivated soil | JN899327 | JX494306 | KF741959 | KM023282 |
| T. dimorphus X.Z. Jiang and L. Wang 2018 | CGMCC 3.15692 T | China: Hainan | forest soil | KY007095 | KY007111 | KY007103 | KY112593 |
| T. domesticus Jurjević and S.W. Peterson 2019 | NRRL 58121 T | USA | floor swab | MH793055 | MH792927 | MH792991 | MH793118 |
| T. duclauxii (Delacr.) Samson et al. 2011 | CBS 322.48 T | France | canvas | JN899342 | JX091384 | KF741955 | JN121491 |
| T. euchlorocarpius Yaguchi et al. 1999 | CBM PF1203 T | Japan | soil | AB176617 | KJ865733 | KJ885271 | KM023303 |
| T. flavovirens (Durieu and Mont.) Visagie et al. 2012 | CBS 102801 T | Spain | unknown | JN899392 | JX091376 | KF741933 | KX961283 |
| T. flavus (Klöcker) Stolk and Samson 1972 | CBS 310.38 T | New Zealand | unknown | JN899360 | JX494302 | KF741949 | JF417426 |
| T. francoae N. Yilmaz et al. 2016 | CBS 113134 T | Colombia | leaf litter | KX011510 | KX011489 | KX011501 | MN969188 |
| T. funiculosus (Thom) Samson et al. 2011 | CBS 272.86 T | India | Lagenaria vulgaris | JN899377 | MN969408 | KF741945 | KM023293 |
| T. fuscoviridis Visagie et al. 2015 | CBS 193.69 T | Netherlands | soil | KF741979 | KF741912 | KF741942 | MN969156 |
| T. fusiformis A.J. Chen et al. 2016 | CGMCC 3.18210 T | China: Beijing | indoor air | KU866656 | KU866843 | KU866740 | KU867000 |
| T. galapagensis Samson and Mahoney 1977 | CBS 751.74 T | Ecuador | soil under Maytenus obovata | JN899358 | JX091388 | KF741966 | KX961280 |
| T. indigoticus Takada and Udagawa 1993 | CBS 100534 T | Japan | soil | JN899331 | JX494308 | KF741931 | KX961278 |
| T. intermedius (Apinis) Stolk and Samson 1972 | CBS 152.65 T | UK | swamp soil | JN899332 | JX091387 | KJ885290 | KX961282 |
| T. kabodanensis Houbraken et al. 2016 | CBS 139564 T | Iran | hypersaline soil | KP851981 | KP851986 | KP851995 | MN969190 |
| T. kendrickii Visagie et al. 2015 | CBS 136666 T | Canada | forest soil | KF741987 | KF741921 | KF741967 | MN969158 |
| T. lentulus X.Z. Jiang and L. Wang 2018 | CGMCC 3.15689 T | China: Shandong | soil | KY007088 | KY007104 | KY007096 | KY112586 |
| T. liani (Kamyschko) N. Yilmaz et al. 2014 | CBS 225.66 T | China | soil | JN899395 | JX091380 | KJ885257 | KX961277 |
| T. louisianensis Jurjević and S.W. Peterson 2019 | NRRL 35823 T | USA | air | MH793052 | MH792924 | MH792988 | MH793115 |
| T. macrosporus (Stolk and Samson) Frisvad et al. 1990 | CBS 317.63 T | South Africa | apple juice | JN899333 | JX091382 | KF741952 | KM023292 |
| T. mae X.Z. Jiang and L. Wang 2018 | CGMCC 3.15690 T | China: Shanghai | forest soil | KY007090 | KY007106 | KY007098 | KY112588 |
| T. malicola Jurjević and S.W. Peterson 2019 | NRRL 3724 T | Italy | rhizosphere of an apple tree | MH909513 | MH909406 | MH909459 | MH909567 |
| T. mangshanicus X.C. Wang and W.Y. Zhuang 2016 | CGMCC 3.18013 T | China: Hunan | soil | KX447531 | KX447530 | KX447528 | KX447527 |
| T. marneffei (Segretain et al.) Samson et al. 2011 | CBS 388.87 T | Vietnam | bamboo rat (Rhizomys sinensis) | JN899344 | JX091389 | KF741958 | KM023283 |
| T. muroii Yaguchi et al. 1994 | CBS 756.96 T | China: Taiwan | soil | MN431394 | KJ865727 | KJ885274 | KX961276 |
| T. mycothecae R.N. Barbosa et al. 2018 | CBS 142494 T | Brazil | nest of stingless bee (Melipona scutellaris) | MF278326 | LT855561 | LT855564 | LT855567 |
| T. neofusisporus L. Wang 2016 | CGMCC 3.15415 T | China: Tibet | leaf sample | KP765385 | KP765381 | KP765383 | MN969165 |
| T. oumae-annae Visagie et al. 2014 | CBS 138208 T | South Africa | house dust | KJ775720 | KJ775213 | KJ775425 | KX961281 |
| T. panamensis (Samson et al.) Samson et al. 2011 | CBS 128.89 T | Panama | soil | JN899362 | HQ156948 | KF741936 | KM023284 |
| T. penicillioides L. Wang 2021 | CGMCC 3.15822 T | China: Guizhou | soil | MK837956 | MK837940 | MK837948 | MK837964 |
| T. pinophilus (Hedgc.) Samson et al. 2011 | CBS 631.66 T | France | PVC | JN899382 | JX091381 | KF741964 | KM023291 |
| T. pratensis Jurjević and S.W. Peterson 2019 | NRRL 62170 T | USA | effluent of water treatment plant | MH793075 | MH792948 | MH793012 | MH793139 |
| T. primulinus (Pitt) Samson et al. 2011 | CBS 321.48 T | USA | unknown | JN899317 | JX494305 | KF741954 | KM023294 |
| T. pseudofuniculosus Guevara-Suarez et al. 2020 | CBS 143041 T | Spain | herbivore dung | LT899796 | LT898323 | LT899778 | LT899814 |
| T. purgamentorum N. Yilmaz et al. 2016 | CBS 113145 T | Colombia | leaf litter | KX011504 | KX011487 | KX011500 | MN969189 |
| T. purpureogenus (Stoll) Samson et al. 2011 | CBS 286.36 T | unknown | unknown | JN899372 | JX315639 | KF741947 | JX315709 |
| T. qii L. Wang 2016 | CGMCC 3.15414 T | China: Tibet | leaf sample | KP765384 | KP765380 | KP765382 | MN969164 |
| T. rapidus Guevara-Suarez et al. 2017 | CBS 142382 T | USA | human bronchoalveolar lavage | LT558970 | LT559087 | LT795600 | LT795601 |
| T. ruber (Stoll) N. Yilmaz et al. 2012 | CBS 132704 T | UK | aircraft fuel tank | JX315662 | JX315629 | KF741938 | JX315700 |
| T. rubicundus (J.H. Mill. et al.) Samson et al. 2011 | CBS 342.59 T | USA | soil | JN899384 | JX494309 | KF741956 | KM023296 |
| T. sayulitensis Visagie et al. 2014 | CBS 138204 T | Mexico | house dust | KJ775713 | KJ775206 | KJ775422 | MN969146 |
| T. siamensis (Manoch and C. Ramírez) Samson et al. 2011 | CBS 475.88 T | Thailand | forest soil | JN899385 | JX091379 | KF741960 | KM023279 |
| T. soli Jurjević and S.W. Peterson 2019 | NRRL 62165 T | USA | soil | MH793074 | MH792947 | MH793011 | MH793138 |
| T. sparsus L. Wang 2021 | CGMCC 3.16003 T | China: Beijing | soil | MT077182 | MT083924 | MT083925 | MT083926 |
| T. stellenboschiensis Visagie and K. Jacobs 2015 | CBS 135665 T | South Africa | soil | JX091471 | JX091605 | JX140683 | MN969157 |
| T. stipitatus (Thom) C.R. Benj. 1955 | CBS 375.48 T | USA | rotting wood | JN899348 | KM111288 | KF741957 | KM023280 |
| T. stollii N. Yilmaz et al. 2012 | CBS 408.93 T | Netherlands | AIDS patient | JX315674 | JX315633 | JX315646 | JX315712 |
| T. striatoconidium (R.F. Castañeda and W. Gams) Houbraken et al. 2020 | CBS 550.89 T | Cuba | leaf litter of Pachyanthus poirettii | MN431418 | MN969441 | MN969360 | MT156347 |
| T. thailandensis Manoch et al. 2013 | CBS 133147 T | Thailand | forest soil | JX898041 | JX494294 | KF741940 | KM023307 |
| T. tumuli Jurjević and S.W. Peterson 2019 | NRRL 62151 T | USA | soil from prairie | MH793071 | MH792944 | MH793008 | MH793135 |
| T. veerkampii Visagie et al. 2015 | CBS 500.78 T | Columbia | soil | KF741984 | KF741918 | KF741961 | KX961279 |
| T. verruculosus (Peyronel) Samson et al. 2011 | NRRL 1050 T | USA | soil | KF741994 | KF741928 | KF741944 | KM023306 |
| T. versatilis Bridge and Buddie 2013 | IMI 134755 T | UK | unknown | MN431395 | MN969412 | MN969319 | MN969161 |
| T. viridis (Stolk and G.F. Orr) Arx 1987 | CBS 114.72 T | Australia | soil | AF285782 | JX494310 | KF741935 | JN121430 |
| T. viridulus Samson et al. 2011 | CBS 252.87 T | Australia | soil | JN899314 | JX091385 | KF741943 | JF417422 |
| T. xishaensis X.C. Wang et al. 2016 | CGMCC 3.17995 T | China: Hainan | soil | KU644580 | KU644581 | KU644582 | MZ361364 |
| T. trachyspermus (Shear) Stolk and Samson 1973 | CBS 373.48 T | USA | unknown | JN899354 | KF114803 | KJ885281 | JF417432 |
Table 3.
New species and newly generated sequences reported in this study.
| Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|
| T. chongqingensis X.C. Wang and W.Y. Zhuang sp. nov. | CS26-67 T | China: Chongqing | soil | MZ358001 | MZ361343 | MZ361350 | MZ361357 |
| CS26-63 | China: Chongqing | soil | MZ358002 | MZ361344 | MZ361351 | MZ361358 | |
| CS26-73 | China: Chongqing | soil | MZ358003 | MZ361345 | MZ361352 | MZ361359 | |
| CS26-75 | China: Chongqing | soil | MZ358004 | MZ361346 | MZ361353 | MZ361360 | |
| T. wushanicus X.C. Wang and W.Y. Zhuang sp. nov. | CS17-05 T | China: Chongqing | soil | MZ356356 | MZ361347 | MZ361354 | MZ361361 |
| CS17-04 | China: Chongqing | soil | MZ356357 | MZ361348 | MZ361355 | MZ361362 | |
| CS17-06 | China: Chongqing | soil | MZ356358 | MZ361349 | MZ361356 | MZ361363 |
3. Results
3.1. Phylogenetic Analysis
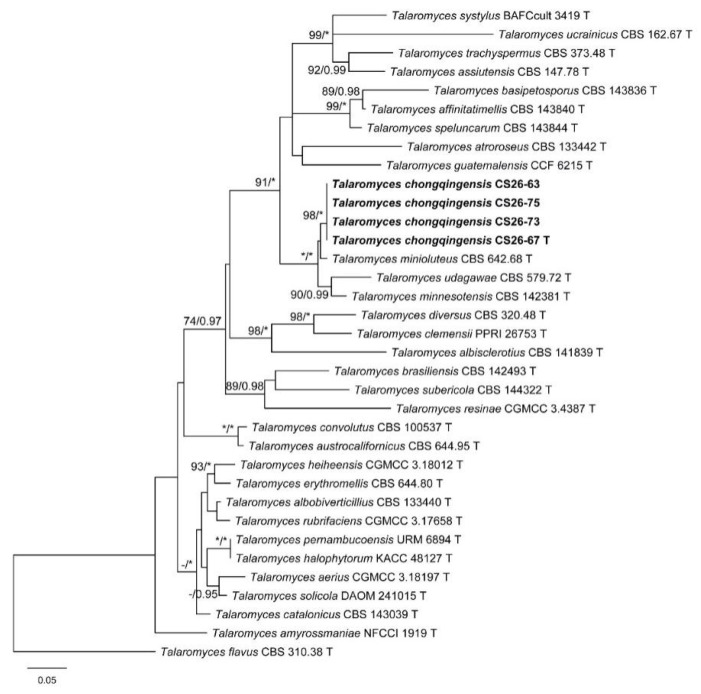
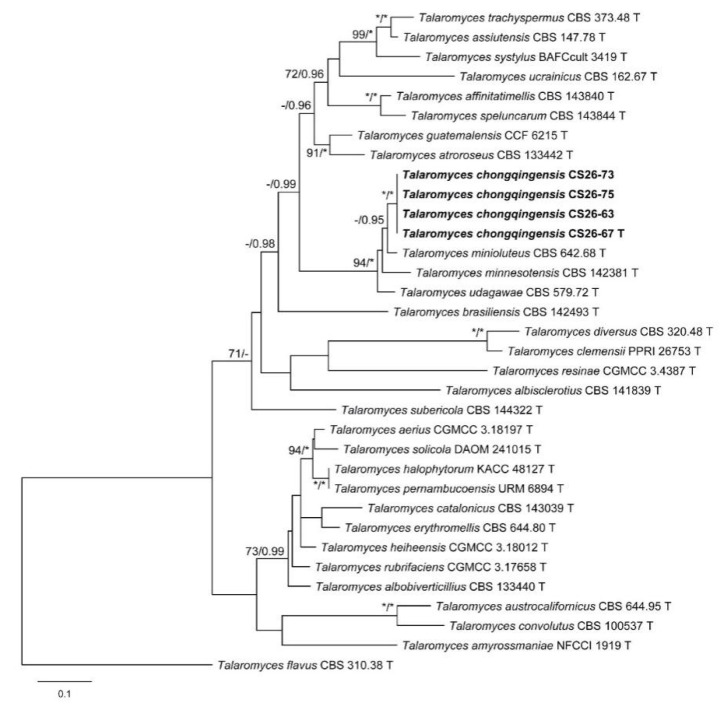
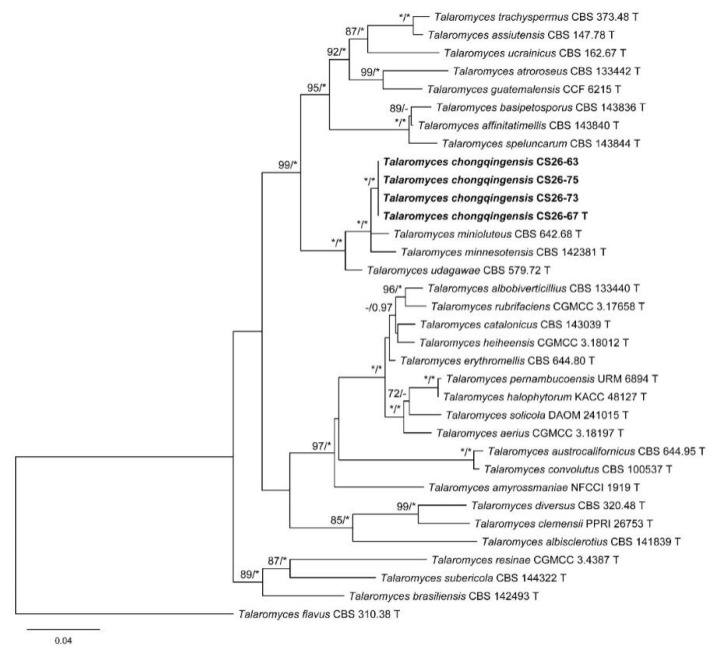
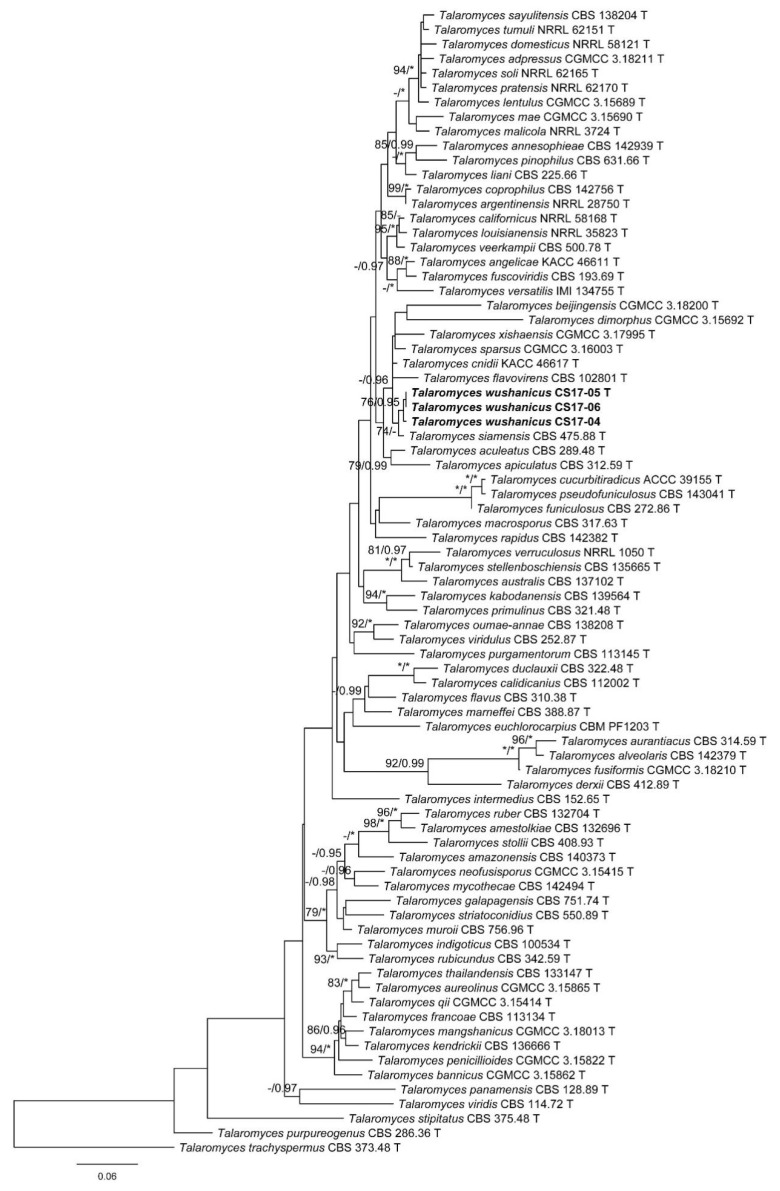
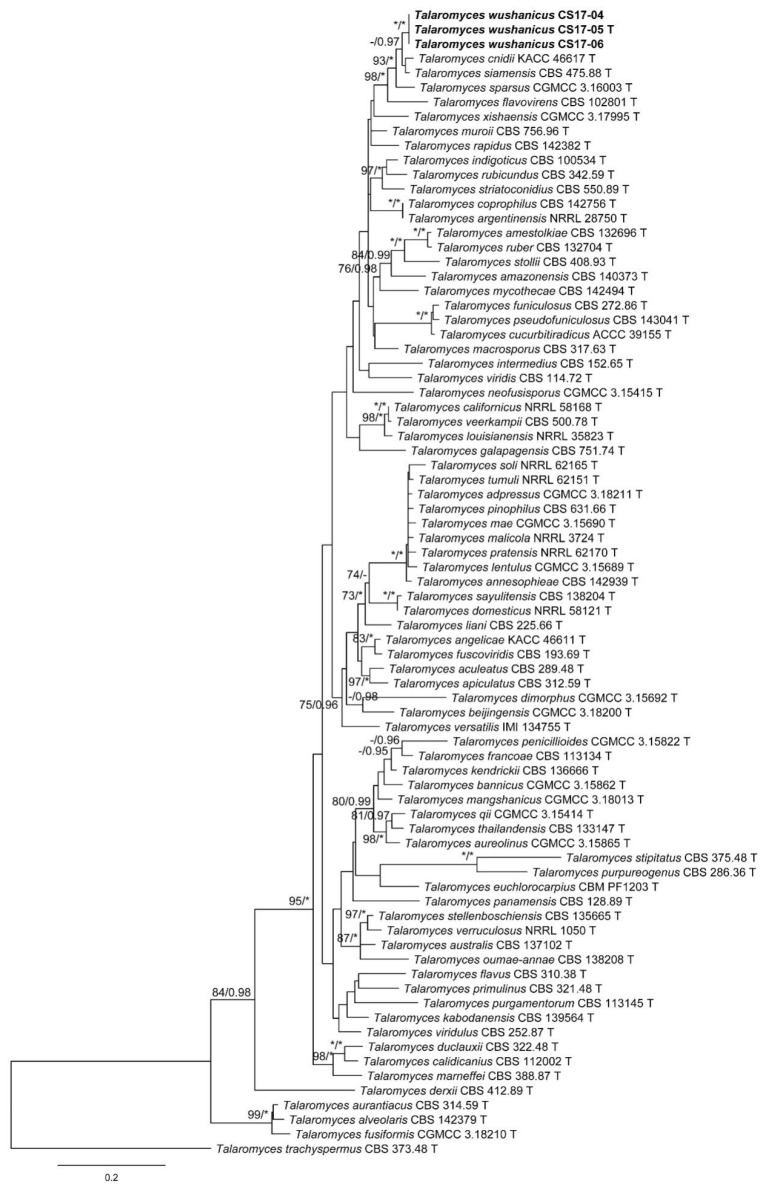
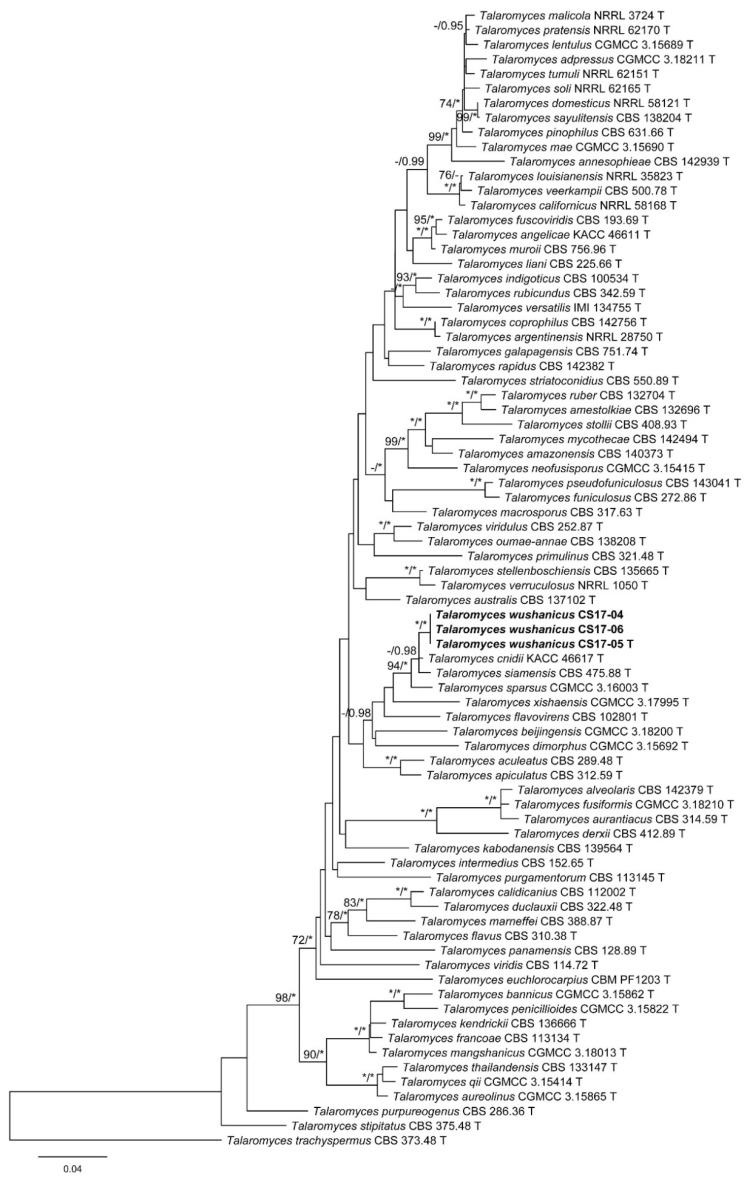
The characteristics of datasets used in the phylogenetic analyses are presented in Table 4. Phylogenetic analyses of the section Trachyspermi revealed that T. chongqingensis always grouped with T. minioluteus, T. minnesotensis, and T. udagawae, having strong statistic supports. In the BenA and CaM analyses (Figure 1 and Figure 2), T. minioluteus was the closest sister of the new species; while T. minioluteus and T. minnesotensis were both closely related to T. chongqingensis in the RPB2 tree (Figure 3). In the phylogenetic analysis of section Talaromyces based on the BenA dataset, T. wushanicus clustered with T. siamensis (Figure 4); while T. cnidii and T. siamensis were closely related to the new species in the CaM and RPB2 analyses (Figure 5 and Figure 6).
Table 4.
Detailed characteristics of the datasets.
| Section | Loci | No. of Seq. | Length of Alignment | Model for BI |
|---|---|---|---|---|
| Trachyspermi | BenA | 35 | 533 | TVM+I+G |
| CaM | 34 | 656 | SYM+I+G | |
| RPB2 | 34 | 920 | GTR+I+G | |
| Talaromyces | BenA | 79 | 490 | TrN+I+G |
| CaM | 79 | 565 | SYM+I+G | |
| RPB2 | 78 | 978 | TVM+I+G |
Full names of the used models: GTR+I+G (General Time Reversible with Invariant sites and Gamma distribution); SYM+I+G (Symmetrical model with Invariant sites and Gamma distribution); TrN+I+G (Tamura–Nei model with Invariant sites and Gamma distribution); TVM+I+G (Transversion model with Invariant sites and Gamma distribution).
Figure 1.
Maximum likelihood phylogeny of Talaromyces sect. Trachyspermi inferred from the BenA dataset. Bootstrap values ≥ 70% (left) or posterior probability values ≥ 0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability.
Figure 2.
Maximum likelihood phylogeny of Talaromyces sect. Trachyspermi inferred from the CaM dataset. Bootstrap values ≥ 70% (left) or posterior probability values ≥ 0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability.
Figure 3.
Maximum likelihood phylogeny of Talaromyces sect. Trachyspermi inferred from the RPB2 dataset. Bootstrap values ≥ 70% (left) or posterior probability values ≥ 0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability.
Figure 4.
Maximum likelihood phylogeny of Talaromyces sect. Talaromyces inferred from the BenA dataset. Bootstrap values ≥ 70% (left) or posterior probability values ≥ 0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability.
Figure 5.
Maximum likelihood phylogeny of Talaromyces sect. Talaromyces inferred from the CaM dataset. Bootstrap values ≥ 70% (left) or posterior probability values ≥ 0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability.
Figure 6.
Maximum likelihood phylogeny of Talaromyces sect. Talaromyces inferred from the RPB2 dataset. Bootstrap values ≥ 70% (left) or posterior probability values ≥ 0.95 (right) are indicated at nodes. Asterisk denotes 100% bootstrap or 1.00 posterior probability.
3.2. Taxonomy
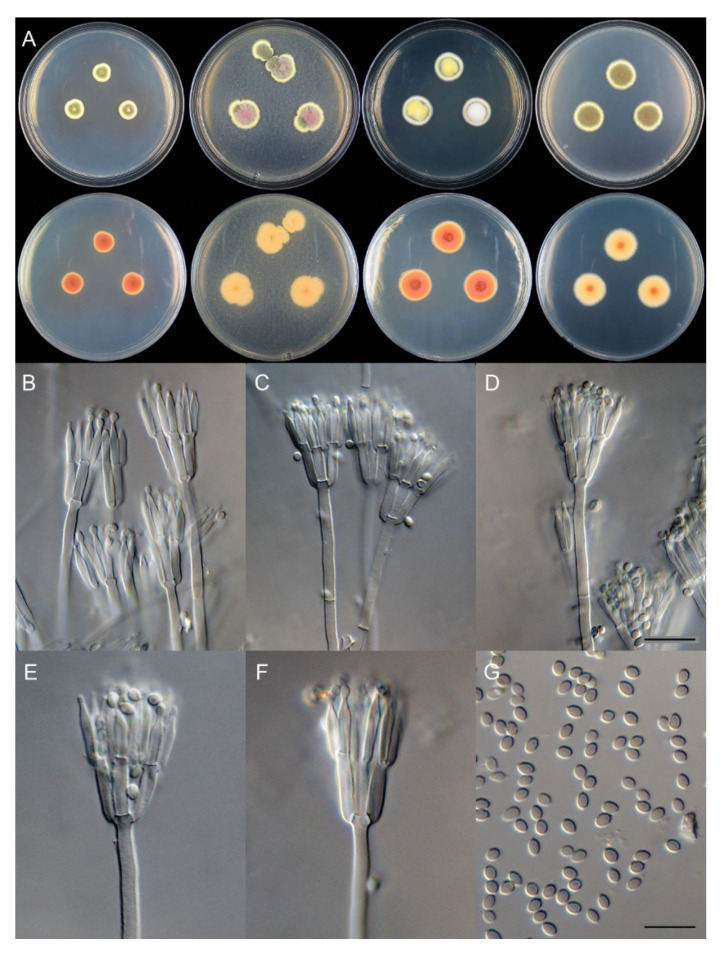
Talaromyces chongqingensis X.C. Wang and W.Y. Zhuang, sp. nov., Figure 7.
Figure 7.
Talaromyces chongqingensis (CS26-67). (A) Colonies: top row left to right, obverse CYA, MEA, YES, and PDA; bottom row left to right, reverse CYA, MEA, YES, and PDA; (B–F) Conidiophores; (G) Conidia. Bars: (D) 15 µm, applies also to (B,C); (G) 10 µm, applies also to (E,F).
Fungal Names: FN570851
Etymology: The specific epithet refers to the type locality.
in Talaromyces sect. Trachyspermi
Typification: China, Chongqing City, Chengkou County, Daba Mountain National Nature Reserve, Gaoguan Town, at the riverside of River Ren, 31°49′40′′ N 109°0′24′′ E, in soil under a palm tree, 30 October 2020, Xin-Cun Wang, Huan-Di Zheng and Chang Liu, culture, Zhi-Kang Zhang, CS26-67 (holotype HMAS 247849, ex-type strain CGMCC 3.20482).
DNA barcodes: ITS MZ358001, BenA MZ361343, CaM MZ361350, RPB2 MZ361357.
Colony diam: after 7 days at 25 °C (unless stated otherwise): on CYA, 12–13 mm; on CYA at 37 °C, no growth; on CYA at 5 °C, no growth; on MEA, 17–18 mm; on YES 18–19 mm; on PDA, 18–19 mm.
Colony characteristics:
On CYA at 25 °C, after 7 days: colonies nearly circular, protuberant in centers; margins moderately wide, entire; mycelia white and yellow; texture velutinous; sporulation dense; conidia en masse yellowish green to dull green; soluble pigments light brown; exudates small, clear; reverse orange, buff at the margins but dark orange at centers.
On MEA at 25 °C, after 7 days: Colonies irregular, protuberant in centers, pink hyphae growing at centers; margins moderately wide, irregular; mycelia white and yellow; texture floccose; sporulation dense; conidia en masse greyish green; soluble pigments absent; exudates absent; reverse buff.
On YES at 25 °C, after 7 days: Colonies nearly circular, strongly protuberant in centers; margins moderately wide, entire; mycelia white and yellow; texture velutinous; sporulation moderately dense; conidia en masse pale green; soluble pigments light brown; exudates absent; reverse orange, yellow brown at the margins but dark orange at centers.
On PDA at 25 °C, after 7 days: Colonies nearly circular, plain, slightly protuberant in centers; margins moderately wide, irregular; mycelia white and yellow; texture velutinous; sporulation very dense; conidia en masse yellowish green; soluble pigments absent; exudates absent; reverse yellow brown, buff at the margins but orange at centers.
Micromorphology: Conidiophores biverticillate; stipes smooth-walled, 90–250 × 2.5–3.0 μm; metulae 4–5, 10–13 × 2.5–3.5 μm; phialides acerose, tapering into very thin neck, 3–5 per metula, 10–13.5 × 2.0–2.5 μm; conidia ellipsoidal to broad fusiform, smooth-walled, 2.5–3.5 × 2.0–2.5 μm.
Additional strains examined: China, Chongqing City, Chengkou County, Daba Mountain National Nature Reserve, Gaoguan Town, at the riverside of River Ren, 31°49′40″ N 109°0′24″ E, in soil under a palm tree, 30 October 2020, Xin-Cun Wang, Huan-Di Zheng and Chang Liu, culture, Zhi-Kang Zhang, CS26-63; ibid., CS26-73; ibid., CS26-75.
Notes: This species is phylogenetically close to T. minioluteus and T. minnesotensis, but differs from them in growth rate on CYA and MEA at 25 °C (Table 5) and pink hyphae present at the centers of colonies on MEA. The sequence data of the four cultures of this fungus are completely identical.
Table 5.
Cultural and morphological comparisons of new species and their closely related species.
| Species | CYA 25 °C (mm) | CYA 37 °C (mm) | MEA (mm) | YES (mm) | Conidia Shape | Conidia Wall | Conidia Size (μm) | Reference |
|---|---|---|---|---|---|---|---|---|
| T. chongqingensis | 12–13 | no growth | 17–18 | 18–19 | ellipsoidal to broad fusiform | smooth | 2.5–3.5 × 2–2.5 | This study |
| T. minioluteus | 17–18 | no growth | 21–22 | 18 | ellipsoidal | smooth | 2.5–4 × 1.5–2.5 | [24] |
| T. minnesotensis | 24–26 | no growth | 13–15 | 21–24 | ellipsoidal | smooth | 2.5–3.5 × 2–3 | [5] |
| T. udagawae | 6–8 | no growth | 10–11 | 8–9 | subglobose to ellipsoidal | smooth | 3–4 × 2–3 | [24] |
| T. cnidii | 30–35 | 17–20 | 38–43 | 40–45 | ellipsodial | smooth to finely rough | 3–4 × 2–2.5 | [25] |
| T. siamensis | 20–22 | 15 | 32–33 | 27–28 | ellipsoidal to fusiform | smooth to finely rough | 3–4 × 2–3 | [24] |
| T. wushanicus | 21–24 | 17–19 | 40–44 | 24–28 | ellipsoidal to broad fusiform | smooth to finely rough | 3–4 × 2.5–3 | This study |
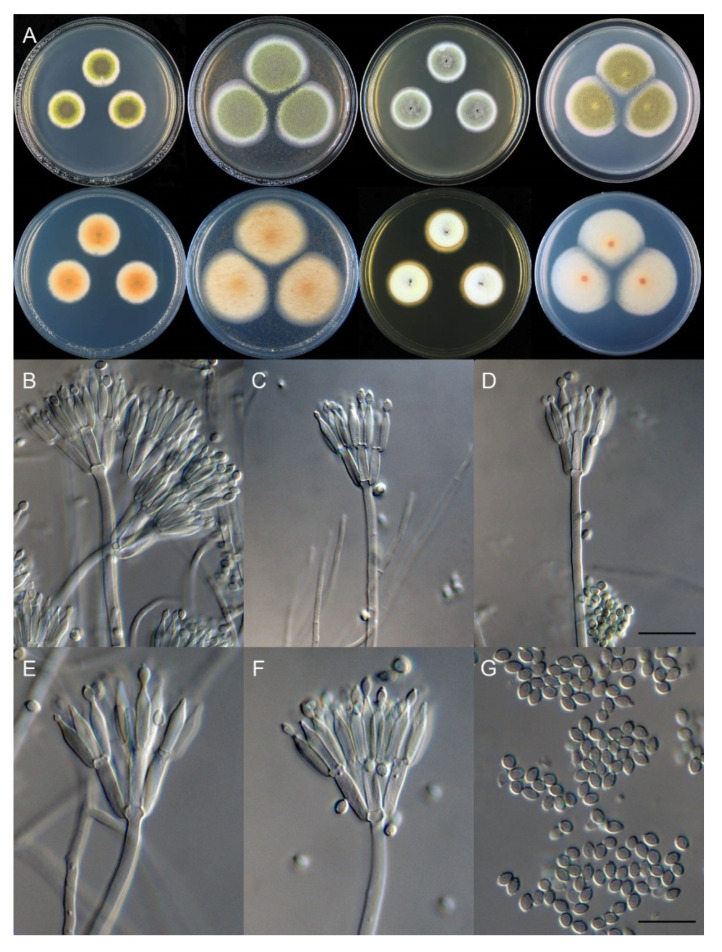
Talaromyces wushanicus X.C. Wang and W.Y. Zhuang, sp. nov., Figure 8.
Figure 8.
Talaromyces wushanicus (CS17-05). (A) Colonies: top row left to right, obverse CYA, MEA, YES, and PDA; bottom row left to right, reverse CYA, MEA, YES, and PDA; (B–F) Conidiophores; (G) Conidia. Bars: (D) 15 µm, applies also to (B,C); (G) 10 µm, applies also to (E,F).
Fungal Names: FN570852
Etymology: The specific epithet refers to the type locality.
in Talaromyces sect. Talaromyces
Typification: China, Chongqing City, Wushan County, Dachang Town, Yanghe Village, 31°17′33′′ N 109°50′44′′ E, in soil, 29 October 2020, Xin-Cun Wang, Huan-Di Zheng and Chang Liu, culture, Zhi-Kang Zhang, CS17-05 (holotype HMAS 247848, ex-type strain CGMCC 3.20481).
DNA barcodes: ITS MZ356356, BenA MZ361347, CaM MZ361354, RPB2 MZ361361.
Colony diam: after 7 days at 25 °C (unless stated otherwise): on CYA, 21–24 mm; on CYA at 37 °C, 17–19 mm; on CYA at 5 °C, no growth; on MEA, 40–44 mm; on YES, 24–28 mm; on PDA, 37–38 mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, protuberant in centers; margins narrow to moderately wide, nearly entire; mycelia white; texture velutinous; sporulation moderately dense; conidia en masse yellowish green; soluble pigments absent; exudates almost absent, sometimes very tiny, red, clear; reverse buff, orange to light brown at centers, but white and pink at periphery.
On CYA at 37 °C, after 7 days: Colonies irregular or nearly circular, protuberant in centers; margins moderately wide, nearly entire; mycelia white; texture velutinous; sporulation moderately dense; conidia en masse dull green to greyish green; soluble pigments absent; exudates absent; reverse buff.
On MEA at 25 °C, after 7 days: Colonies nearly circular, plain; margins wide, entire; mycelia yellow; texture velutinous; sporulation dense; conidia en masse yellowish green; soluble pigments absent; exudates almost absent, sometimes very tiny, hyaline, clear; reverse buff, but yellow to orange in centers.
On YES at 25 °C, after 7 days: Colonies nearly circular, deep, wrinkled, highly protuberant in centers; margins narrow to moderately wide, entire; mycelia white; texture velutinous; sporulation dense; conidia en masse yellowish green to dark green; soluble pigments absent; exudates absent, rarely red and clear; reverse white, yellow brown to light brown, rimose, or deeply concave in centers.
On PDA at 25 °C, after 7 days: Colonies nearly circular, plain, slightly protuberant in centers; margins moderately wide, entire; mycelia white; texture velutinous; sporulation dense; conidia en masse yellowish green; soluble pigments absent; exudates hyaline, clear, present at centers; reverse greyish white to greenish white, reddish brown at centers.
Micromorphology: Conidiophores biverticillate, rarely terverticillate; stipes smooth-walled, 85–225 × 2.0–3.0 μm; metulae 5, 9.5–11.5 × 2.5–3.0 μm; phialides acerose, tapering into very thin neck, 3–4 per metula, 10–11 × 2.0–2.5 μm; conidia ellipsoidal to broad fusiform, smooth to finely rough, 3–4 × 2.5–3 μm.
Additional strains examined: China, Chongqing City, Wushan County, Dachang Town, Yanghe Village, 31°17′33′′ N 109°50′44′′ E, in soil, 29 October 2020, Xin-Cun Wang, Huan-Di Zheng and Chang Liu, culture, Zhi-Kang Zhang, CS17-04; ibid., CS17-06.
Notes: This species is closely related to T. cnidii and T. siamensis in the phylogenetic trees (Figure 4, Figure 5 and Figure 6), but it differs from T. cnidii in obviously slower growth rate on CYA and YES at 25 °C and from T. siamensis by an obviously faster growth on MEA at 25 °C (Table 5). Sequence comparisons indicate that the isolate CS17-04 has a one-base difference in ITS and a two-base difference in BenA from the other two strains. No morphological diversification was found among the strains.
4. Discussion
Of the 35 global biodiversity hotspots, 3 are located in southwestern China, consisting of Chongqing, Guizhou, Sichuan, Tibet, and Yunnan provinces [26]. Eight hotspot regions in the southwest of China were identified as priority conservation areas, including the Daba Mountains [27] where materials used for this study were gathered. Soil samples for floristic studies of fungi were collected from Chengkou, Wushan, and Wuxi counties in Chongqing and Wanyuan City in Sichuan. Although Talaromyces is a widespread genus and distributed in more than 27 provinces, cities, or regions of China [14], it has never been reported from the above areas.
In recent years, the number of new species of Talaromyces increased dramatically. There were 12 species recorded in Talaromyces sect. Trachyspermi and 36 ones in Talaromyces sect. Talaromyces in 2014 [24]. From 2018 to 2021, 13 additional species were discovered in the former section, and 20 new members were described in the latter. We are witnessing a trend: new fungal species are described at an accelerated rate.
Talaromyces species occur in diversified environments. When the information about the extype strains of more than 100 species in these two sections is gathered (Table 1 and Table 2), it is found that soil is commonly the substrate. Fifty or so species were isolated from different kinds of soil, e.g., forest, cultivated, and swamp soil. Plant debris appears to be the second frequent source, which nearly 20 species inhabited. Four species were from humans and one, the well-known T. marneffei, from bamboo rat. Surprisingly, T. pinophilus was discovered on PVC, the third widely used plastic in the world, which is hard to biodegrade.
Among the 30 species accepted in Talaromyces sect. Trachyspermi, 6 were originally reported from China (Table 1). Moreover, 18 of the 75 species known in Talaromyces sect. Talaromyces were described based on the Chinese samples or specimens (Table 2). These data surely demonstrate that China has a high fungal diversity. With more investigations conducted, we expect to discover more new species of this group of fungi.
5. Conclusions
The present work provides a better understanding of biodiversity and phylogeny of the genus. The results make the concepts of the two sections of Talaromyces well-established and more sophisticated. The discovery will be beneficial for future evaluation of the potential usages and functions of the new species.
Acknowledgments
The authors would like to thank Huan-Di Zheng, Zhao-Qing Zeng, and Chang Liu of Institute of Microbiology, Chinese Academy of Sciences for collecting jointly the samples for this study.
Author Contributions
Conceptualization, W.-Y.Z. and P.Z.; methodology, Z.-K.Z. and X.-C.W.; software, X.-C.W.; validation, X.-C.W.; formal analysis, X.-C.W. and Z.-K.Z.; investigation, X.-C.W.; resources, W.-Y.Z., P.Z. and X.-H.C.; data curation, X.-C.W.; writing—original draft preparation, X.-C.W.; writing—review and editing, W.-Y.Z. and X.-C.W.; visualization, X.-C.W.; supervision, X.-C.W., P.Z. and W.-Y.Z.; project administration, W.-Y.Z.; funding acquisition, P.Z., W.-Y.Z. and X.-C.W. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the National Natural Science Foundation of China (31750001) and Key Research Program of Frontier Science, Chinese Academy of Sciences (QYZDY-SSW-SMC029).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mendez-Liter J.A., Nieto-Dominguez M., Fernandez de Toro B., Gonzalez Santana A., Prieto A., Asensio J.L., Canada F.J., de Eugenio L.I., Martinez M.J. A glucotolerant β-glucosidase from the fungus Talaromyces amestolkiae and its conversion into a glycosynthase for glycosylation of phenolic compounds. Microb. Cell Fact. 2020;19:127. doi: 10.1186/s12934-020-01386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inoue H., Decker S.R., Taylor L.E., Yano S., Sawayama S. Identification and characterization of core cellulolytic enzymes from Talaromyces cellulolyticus (formerly Acremonium cellulolyticus) critical for hydrolysis of lignocellulosic biomass. Biotechnol. Biofuels. 2014;7:151. doi: 10.1186/s13068-014-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morales-Oyervides L., Ruiz-Sanchez J.P., Oliveira J.C., Sousa-Gallagher M.J., Mendez-Zavala A., Giuffrida D., Dufosse L., Montanez J. Biotechnological approaches for the production of natural colorants by Talaromyces/Penicillium: A review. Biotechnol. Adv. 2020;43:107601. doi: 10.1016/j.biotechadv.2020.107601. [DOI] [PubMed] [Google Scholar]
- 4.Frisvad J.C., Yilmaz N., Thrane U., Rasmussen K.B., Houbraken J., Samson R.A. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE. 2013;8:e84102. doi: 10.1371/journal.pone.0084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guevara-Suarez M., Sutton D.A., Gene J., Garcia D., Wiederhold N., Guarro J., Cano-Lira J.F. Four new species of Talaromyces from clinical sources. Mycoses. 2017;60:651–662. doi: 10.1111/myc.12640. [DOI] [PubMed] [Google Scholar]
- 6.Li L., Chen K., Dhungana N., Jang Y., Chaturvedi V., Desmond E. Characterization of clinical isolates of Talaromyces marneffei and related species, California, USA. Emerg. Infect. Dis. 2019;25:1765–1768. doi: 10.3201/eid2509.190380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao C., Xi L., Chaturvedi V. Talaromycosis (Penicilliosis) due to Talaromyces (Penicillium) marneffei: Insights into the clinical trends of a major fungal disease 60 years after the discovery of the pathogen. Mycopathologia. 2019;184:709–720. doi: 10.1007/s11046-019-00410-2. [DOI] [PubMed] [Google Scholar]
- 8.Houbraken J., Kocsube S., Visagie C.M., Yilmaz N., Wang X.C., Meijer M., Kraak B., Hubka V., Bensch K., Samson R.A., et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doilom M., Guo J.W., Phookamsak R., Mortimer P.E., Karunarathna S.C., Dong W., Liao C.F., Yan K., Pem D., Suwannarach N., et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: Four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front. Microbiol. 2020;11:585215. doi: 10.3389/fmicb.2020.585215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun B.D., Chen A.J., Houbraken J., Frisvad J.C., Wu W.P., Wei H.L., Zhou Y.G., Jiang X.Z., Samson R.A. New section and species in Talaromyces. MycoKeys. 2020;68:75–113. doi: 10.3897/mycokeys.68.52092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crous P.W., Cowan D.A., Maggs-Kolling G., Yilmaz N., Larsson E., Angelini C., Brandrud T.E., Dearnaley J.D.W., Dima B., Dovana F., et al. Fungal Planet description sheets: 1112–1181. Persoonia. 2020;45:251–409. doi: 10.3767/persoonia.2020.45.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei S., Xu X., Wang L. Four new species of Talaromyces section Talaromyces discovered in China. Mycologia. 2021;113:492–508. doi: 10.1080/00275514.2020.1853457. [DOI] [PubMed] [Google Scholar]
- 13.Visagie C.M., Houbraken J., Frisvad J.C., Hong S.B., Klaassen C.H., Perrone G., Seifert K.A., Varga J., Yaguchi T., Samson R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X.C., Chen K., Xia Y.W., Wang L., Li T.H., Zhuang W.Y. A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China. Phytotaxa. 2016;267:187–200. doi: 10.11646/phytotaxa.267.3.2. [DOI] [Google Scholar]
- 15.Wang X.C., Chen K., Qin W.T., Zhuang W.Y. Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycol. Prog. 2017;16:73–81. doi: 10.1007/s11557-016-1251-3. [DOI] [Google Scholar]
- 16.Wang X.C., Chen K., Zeng Z.Q., Zhuang W.Y. Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci. Rep. 2017;7:8233. doi: 10.1038/s41598-017-08697-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- 19.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 21.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees; Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 22.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posada D., Crandall K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 24.Yilmaz N., Visagie C.M., Houbraken J., Frisvad J.C., Samson R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sang H., An T.J., Kim C.S., Shin G.S., Sung G.H., Yu S.H. Two novel Talaromyces species isolated from medicinal crops in Korea. J. Microbiol. 2013;51:704–708. doi: 10.1007/s12275-013-3361-9. [DOI] [PubMed] [Google Scholar]
- 26.Marchese C. Biodiversity hotspots: A shortcut for a more complicated concept. Glob. Ecol. Conserv. 2015;3:297–309. doi: 10.1016/j.gecco.2014.12.008. [DOI] [Google Scholar]
- 27.Zhang Y.B., Wang G.Y., Zhuang H.F., Wang L.H., Innes J.L., Ma K.P. Integrating hotspots for endemic, threatened and rare species supports the identification of priority areas for vascular plants in SW China. Forest Ecol. Manag. 2021;484 doi: 10.1016/j.foreco.2021.118952. [DOI] [Google Scholar]