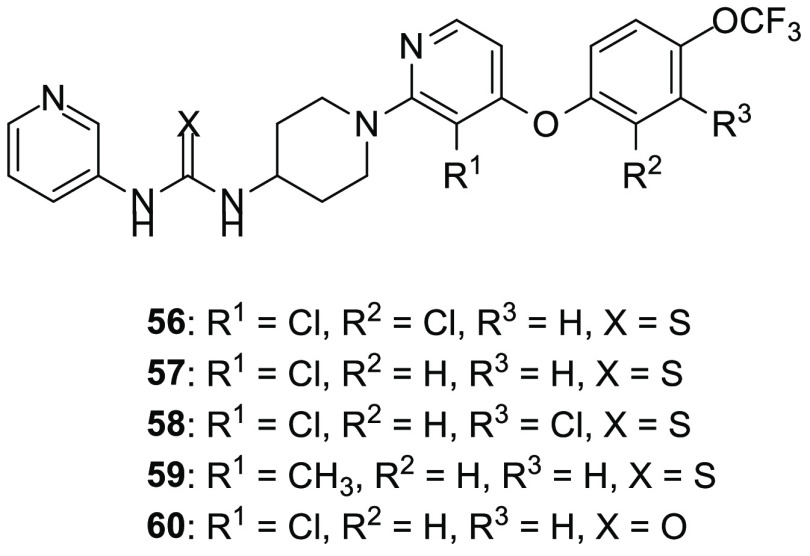
Abstract
Much of the experimental evidence in the literature has linked altered lipid metabolism to severe diseases such as cancer, obesity, cardiovascular pathologies, diabetes, and neurodegenerative diseases. Therefore, targeting key effectors of the dysregulated lipid metabolism may represent an effective strategy to counteract these pathological conditions. In this context, α/β-hydrolase domain (ABHD) enzymes represent an important and diversified family of proteins, which are involved in the complex environment of lipid signaling, metabolism, and regulation. Moreover, some members of the ABHD family play an important role in the endocannabinoid system, being designated to terminate the signaling of the key endocannabinoid regulator 2-arachidonoylglycerol. This Perspective summarizes the research progress in the development of ABHD inhibitors and modulators: design strategies, structure–activity relationships, action mechanisms, and biological studies of the main ABHD ligands will be highlighted.
1. Introduction
Endocannabinoids 2-arachidonoylglycerol (2-AG) and N-arachidonoylethanolamine (anandamide, AEA) are endogenous lipid molecules activating the two G protein-coupled cannabinoid receptors 1 and 2 (CB1R and CB2R). 2-AG and AEA are produced following stimulation from phospholipid precursors present in the cell membranes and immediately metabolized after their activation of specific signaling pathways by specific lipases.1 Therefore, the endocannaboinoid system (ECS) includes also enzymes controlling endocannabinoid levels and the most important is fatty acid amide hydrolase (FAAH), mainly responsible for the hydrolysis of AEA and monoacylglycerol lipase (MAGL), which is designated for 2-AG inactivation.2 In this context, it is noteworthy to introduce a family of endocannabinoid-degrading enzymes which is progressively attracting more interest by the scientific community: the α/β-hydrolase domain (ABHD) enzymes. Muccioli et al. provided the first evidence that not only does MAGL hydrolyze 2-AG, since they found that MAGL was not expressed in the mouse microglial cell line, BV-2, but also a 2-AG hydrolyzing activity was present.3 In the same year, ABHD6 and ABHD12 were identified by activity-based protein profiling (ABPP).2 In particular, 85% of brain 2-AG hydrolase activity can be ascribed to MAGL, and the remaining 15% is mostly performed by ABHD6 and ABHD12 (4% and 9%, respectively).
Besides ABHD6 and ABHD12 which are related to ECS, many other ABHD enzymes have been identified and they showed specific physiological functions as regulators of lipid metabolism and signal transduction. Their association to human diseases of altered lipid metabolism will be explained in detail in the following specific sections.
Importantly, all ABHD enzymes belong to the α/β-hydrolase fold superfamily,4 which includes many different hydrolytic enzymes and shares a common three-dimensional feature since members of this family contain eight β-strands with the second antiparallel strand. The β sheets are surrounded on both sides by α helices and loops connecting the eight sheets. Each member of this family derives its hydrolytic activity from a highly conserved catalytic triad, characterized by the sequence: (a) nucleophile residue (serine, cysteine, or aspartate) located in the nucleophilic elbow in the loop following strand β5; (b) acid residue (glutamate or aspartate) after strand β7; (c) histidine residue located after the last β strand. The active site can be covered by a dynamic lid. Most of the ABHD enzymes are also endowed with acyltransferase activity due to the conserved His-XXXX-Asp region (X is any amino acid).5,6
It is noteworthy to underline that the α/β-hydrolase fold superfamily is a very large multifaceted protein family which includes more than 50 enzymes possessing different names. Nevertheless, the present Perspective is focused on those members of this superfamily which are usually named ABHD enzymes, with the aim of highlighting the therapeutic potential of this group of proteins.
Despite the fact that ABHD enzymes are attractive targets for novel therapies targeting cancer and metabolic diseases,7,8 the research field concerning the development of inhibitors/modulators of these ABHDs is still quite unexplored. A greater interest has been devoted to ABHD6 and ABHD12 inhibitors, due to their involvement in the ECS. In fact, CB1R and CB2R are involved in many physiological and pathological processes; therefore, beneficial effects derive from their modulation. Nevertheless, it is well-known that their direct activation is associated with many drawbacks such as receptor desensitization and abuse potential. For this reason, more recent therapeutic approaches are directed toward their indirect stimulation by the inhibition of endocannabinoid degradation.9 While a growing number of selective and potent inhibitors of FAAH and MAGL have been published or patented in the last decades, the discovered ABHD6 and ABHD12 inhibitors are still in their beginning, since the amount of inhibitors is limited and few of them have been the object of extensive studies.
Many developed ABHD inhibitors reported in the literature and reviewed here were characterized by activity-based protein profiling (ABPP), because ABPP is a proteomic technology used to determine not only the activity in cells and tissues but also the selectivity of ABHD inhibitors in an unbiased proteome-wide fashion. A variety of applications of ABPP have been developed in the last decades, since ABPP combines different scientific disciplines. In order to speed up the drug discovery process, ABPP is able to test inhibitors against many enzymes in parallel, and thus, potency and selectivity can be determined in a saving-time approach.10 ABPP relies on the design of small-molecule probes that covalently label the active site of families of enzymes in complex proteomes. In particular, these probes possess (a) a “warhead” that is a chemical portion targeting conserved structural features present in active sites of an enzyme family, such as electrophilic groups binding conserved active-site nucleophile serine of serine hydrolase enzymes, and (b) a reporter tag, to facilitate target characterization, i.e., fluorophores, biotin, and alkynes or azides (which can be modified by Huisgen 1,3-dipolar cycloaddition).11 Experimental read-out techniques such as gel-based methods or LC-MS approaches are usually adopted for analyzing probe-treated proteomes.
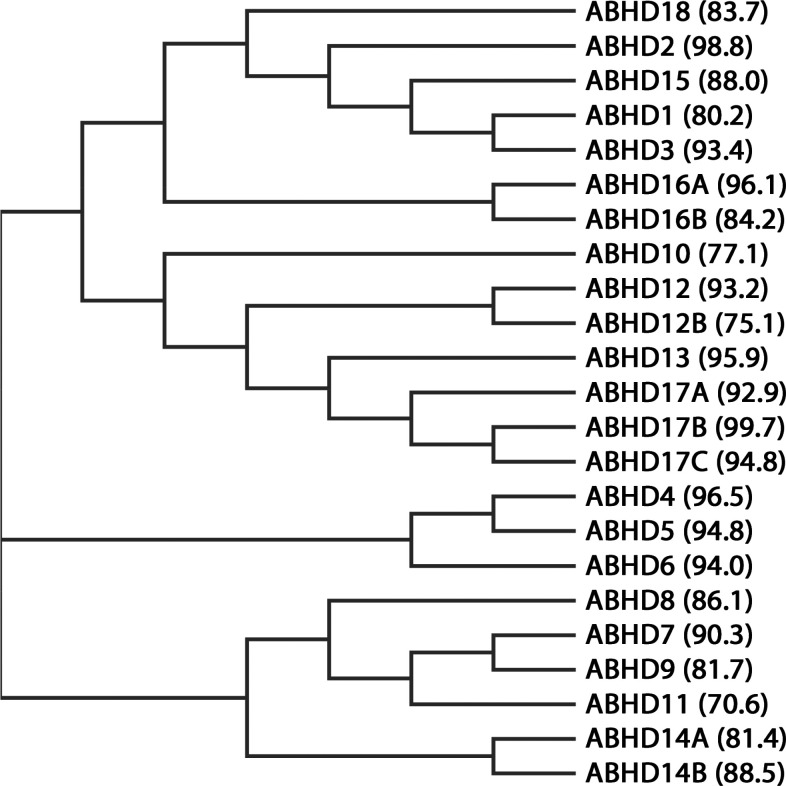
All ABHD proteins are reviewed in Table 1 and Figure 1, in which their main features are summarized. In this Perspective, inhibitors and modulators of ABHDs will be reviewed, classifying the compounds on the basis of the specific inhibited ABHD enzyme and on the different chemical families, with a special focus on the specific roles of each ABHD enzyme. Additionally, specific attention will be dedicated to the patented ligands of ABHDs, in particular to those that have not been reviewed elsewhere as dual MAGL/ABHDs inhibitors.12,13
Table 1. Overview of ABHD Proteins: Main Expression Pattern in Humans and Substrates of Each Protein Are Reported.
| ABHD protein | main expression pattern | main substrates |
|---|---|---|
| ABHD1 | testis | –a |
| ABHD2 | ubiquitous expression, liver, stomach | triacylglycerols, esters |
| ABHD3 | appendix, colon, gall bladder, lymph nodes, stomach, thyroid, small intestine, duodenum | medium-chain phospholipids, phosphatidylcholines containing C14 acyl chain, oxidatively truncated phospholipids |
| ABHD4 | testis, gall bladder | –a |
| ABHD5 | bone marrow, fat, skin | arachidonoyl-CoA, oleoyl-CoA, 1-oleoyl-lysophosphatidic, triacylglycerols |
| ABHD6 | small intestine, duodenum, spleen, brain, brown adipose tissue, kidney, liver, skin, ovary | diacylglycerols, 1(3)-monoacylglycerols with saturated medium or long acyl chains, 2-arachidonoylglycerol, lysophosphatidylinositols, bis(monoacylglycero)phosphate |
| ABHD7 | brain | –a |
| ABHD8 | brain, testis | –a |
| ABHD9 | skin, esophagus | epoxyeicosatrienoic acids, 9,10-epoxyoctadecamonoenoic acids, leukotoxin, linoleate-derived epoxy-alcohols |
| ABHD10 | kidney, thyroid | S-palmitoyl substrates |
| ABHD11 | skeletal muscle, colon, prostate, small intestine, thyroid | triacylglycerols, 2-oxoglutarate |
| ABHD12 | ubiquitous expression, brain | 2-arachidonoylglycerol, 1(3)-isomer of arachidonoylglycerol, unsaturated C20:4 monoacylglycerols, lysophosphatidylserine lipids |
| ABHD12B | skin | –a |
| ABHD13 | ubiquitous expression | –a |
| ABHD14A | adrenal glands, brain, kidney, thyroid | –a |
| ABHD14B | ubiquitous expression | p-nitrophenyl butyrate |
| ABHD15 | fat, liver | –a |
| ABHD16A | ubiquitous expression, skeletal muscle, brain and platelets | medium-chain saturated monoacylglycerols, 1-linoleylglycerol, 15-deoxy-Δ12,14-prostaglandin J2-2-glycerol ester |
| ABHD16B | testis | –a |
| ABHD17A | bone marrow, fat, lung, skin, spleen | S-palmitoyl-l-cysteine residue |
| ABHD17B | brain | S-palmitoyl-l-cysteine residue |
| ABHD17C | colon, esophagus, stomach, small intestine, brain, duodenum, lung, prostate, urinary bladder | S-palmitoyl-l-cysteine residue |
| ABHD18 | ubiquitous expression | –a |
Not determined.
Figure 1.
Phylogenetic relationship of the human ABHD proteins. For each protein, the percentage of residue identity between human and mouse species is highlighted between brackets.
2. ABHD2
2.1. Biochemical Features and Biological Roles
The serine hydrolase ABHD2 is a 425-residue protein (48 kDa) possessing a typical Ser207-His376-Asp345 catalytic triad, and it is ubiquitously expressed, mainly in liver and stomach. ABHD2 is considered a triacylglycerol lipase and an ester hydrolase.14 It is overexpressed in human androgen-sensitive prostate cancer tissues since lipid metabolism plays a key role in the development and progression of this type of tumor. Moreover, high ABHD2 expression is correlated with resistance to docetaxel-based chemotherapy.15 Deletion of the ABHD2 gene was correlated to anoikis resistance in high-grade serous ovarian cancer (HGSOC), thus promoting a malignant phenotype and poor prognosis.16 Furthermore, ABHD2 was shown to be involved in many diseases such as Hepatitis B virus propagation,17 since its downregulation using antisense oligonucleotides blocked Hepatitis B virus replication and expression without affecting host cell physiology. ABHD2 plays a key role in monocyte/macrophage recruitment, therefore influencing the development of chronic diseases such as atherosclerosis and emphysema. In particular, ABHD2 deficiency induced emphysema, due to increased macrophage infiltration, increased inflammatory cytokines and enhanced apoptosis because ABHD2 is important to maintain lung structural integrity.18 With regard to its involvement in the pathogenesis of atherosclerosis, ABHD2 genetic deficiency enhances the migration of vascular smooth muscle cells, which is one of the causes of this vascular disease.19 In addition, ABHD2 expression was significantly increased in parallel with the differentiation from monocyte into macrophage, and macrophages of atherosclerotic lesions abundantly expressed ABHD2.20 High expression of ABHD2 in spermatozoa revealed the ability of this protein to bind progesterone, triggering 2-AG degradation, thus revealing that progesterone-mediated activation of ABHD2 finally stimulates sperm activation.21 The same mechanism was induced by pregnenolone sulfate: similarly to progesterone, it activated calcium channel of sperm by ABHD2 binding.22 Finally, ABHD2 proved to be involved in the regulation of calcium release from the endoplasmic reticulum (ER).23
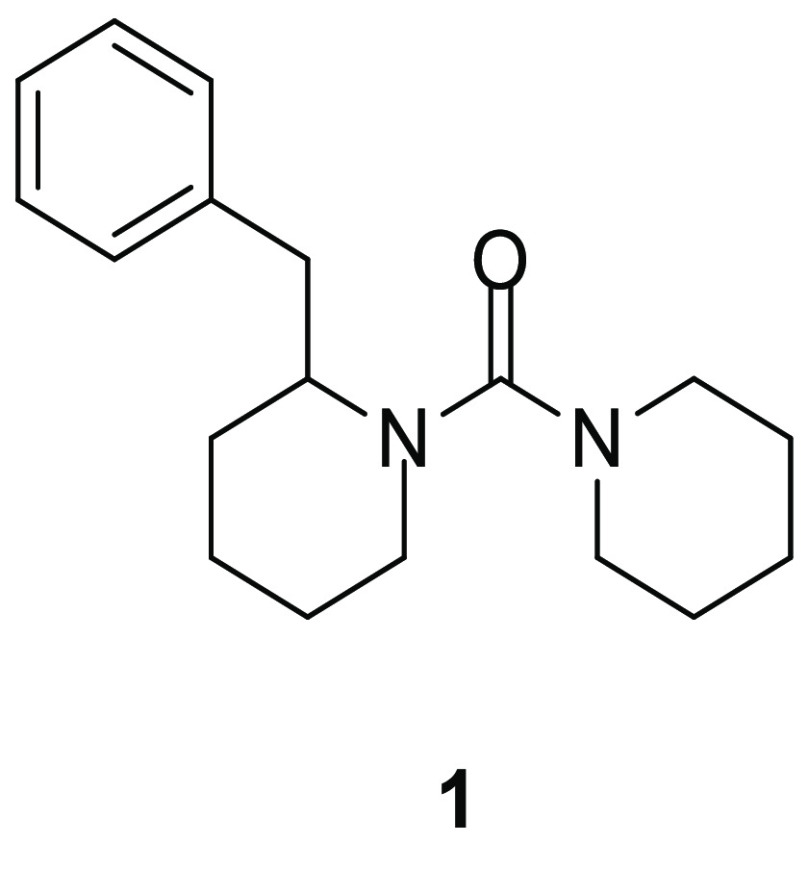
2.2. Inhibitors
Very recently, Baggelaar and his research group conducted an ABPP screening based on ABHD proteins and a library composed of 207 lipase inhibitors to identify selective ABHD2 inhibitors.24 Urea derivative 1 (Figure 2) exerted a notable activity on ABHD2 (pIC50 = 5.50) with no other off-targets in mouse testis proteome. This selectivity assay was performed in this specific proteome, since ABHD2 has an important role in sperm fertility. In order to analyze this aspect, inhibitor 1 was evaluated for its capacity to reduce progesterone-induced acrosome reaction (AR) in vitro, which is an important calcium-dependent process for the fertilization of mammalian eggs by spermatozoa, and it is stimulated by many molecules including progesterone. Compound 1 reduced progesterone-induced AR in vitro in a concentration-dependent manner, by blocking calcium increase induced by progesterone, thus confirming that ABHD2 finely tunes intracellular calcium levels in mouse sperm. These results suggest that urea derivative 1 could represent an interesting starting compound to develop new ABHD2 inhibitors as perspective novel contraceptives.
Figure 2.
ABHD2 inhibitor.
3. ABHD3
3.1. Biochemical Features and Biological Roles
ABHD3, previously known as lung α/β-hydrolase 3 (LABH3),25 is a poorly characterized 409-residue (46 kDa) serine hydrolase highly expressed in appendix, colon, gall bladder, lymph nodes, stomach, thyroid, small intestine, duodenum, whose biochemical or physiological functions are still scarcely known. ABHD3 showed a multifaceted role in the catabolism of medium-chain phospholipids, that is distinct from those of other known phospholipases, as demonstrated in metabolomic studies.26 In fact, ABHD3 showed a good specificity toward phosphatidylcholines (PCs) containing C14 acyl chain and oxidatively truncated phospholipids over other phospholipids. ABHD3 was shown to be upregulated in a series of pathological conditions: in human ovarian cancer cell lines exposed to standard chemotherapeutic drugs (cisplatin, paclitaxel or topotecan),27 in breast cancer tumors, as a pro-apoptotic gene,28 in a human osteosarcoma cell line overexpressing the tumor suppressor gene HIC1 (Hypermethylated in Cancer 1)29 and in mice hippocampus after low-intensity exercise alone and/or in combination with the natural antioxidant carotenoid astaxanthin, revealing an antioxidant function of ABHD3.30 Conversely, ABHD3 is downregulated in peripheral blood mononuclear cells of patients affected by Crohn’s disease31 and in a rat model of glaucoma characterized by early optic nerve head, which is the principal site of initial axonal injury.32
3.2. Inhibitors
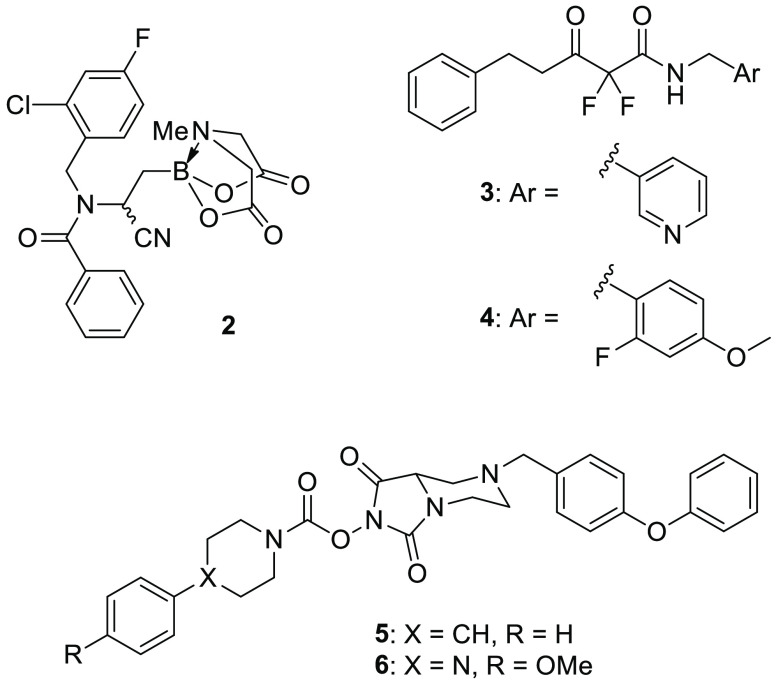
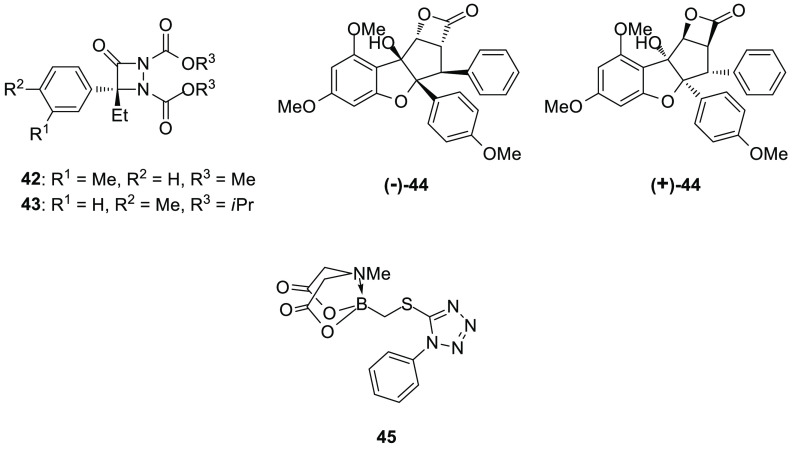
Tan and collaborators performed a competitive ABPP screening on a library of synthesized α- and β-aminocyano N-methyliminodiacetic acid-containing (MIDA) boronates in mouse brain proteome.33 Several compounds belonging to this class exhibited ABHD3 inhibition, but further studies on HEK293T (human embryonic kidney cells) lysates overexpressing ABHD3 showed that the most active and selective ABHD3 inhibitor was β-aminocyano(MIDA)boronate 2 (Figure 3), with an IC50 value of 0.14 μM in vitro. With regard to 2 selectivity, SDS-PAGE analysis of tissue proteomes was able to identify only a limited number of serine hydrolases. Consequently, the authors further investigated the selectivity of 2 by MS-based ABPP using stable isotope labeling with amino acids in cell culture (SILAC). This technique allowed to confirm the selectivity of boronate 2 on ABHD3 (>95% of blockade at 0.5 μM) without detecting any activity over 60 additional serine hydrolases in human colon cancer cell line SW620. A structure–activity relationship analysis revealed the importance of the phenylamide portion, the cyano group, and the fluorine atom of 2 for inhibition potency. Importantly, the boron atom is fundamental for ABHD3 covalent inhibition, and the MIDA boronate portion seemed to increase cell permeability or stability in cells when compared to the free boronic acid analogue, proving to be resistant to hydrolytic cleavage under neutral conditions during the ABPP experiments. Metabolomic studies of 2 confirmed the previous findings that ABHD3 inhibition leads to an increase of medium-chain PCs in human cells.
Figure 3.
ABHD3 inhibitors.
It is also noteworthy to cite 1,3-dicarbonyl derivatives 3 and 4 (Figure 3) identified in the previously mentioned screening aimed at finding new ABHD2 inhibitors:24 these two compounds proved to selectively inhibit ABHD3 over the other tested ABHD enzymes.
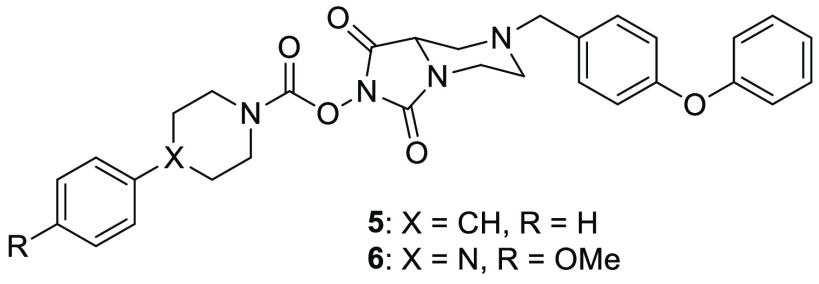
The research group of Cravatt synthesized a small library of N-hydroxyhydantoin carbamates and screened them through competitive ABPP on serine hydrolases.34 Compounds 5 (ABC47, Figure 3) and 6 (ABC34, Figure 3) demonstrated a good activity on ABHD3 (IC50 = 0.1 and 7.6 μM, respectively), but a more potent inhibition was observed on ABHD4 (IC50 = 0.03 and 0.1 μM, respectively) in mouse brain membrane proteome. However, ABPP-SILAC experiments in human PC3 cells highlighted that 5 and 6 inhibited not only ABHD3 and ABHD4, but they also had four additional off-targets: ABHD6, hormone-sensitive lipase (HSL), phospholipase A2 Group VII (PLA2G7), and carboxylesterase 2 (CES2). This study suggests that the N-hydroxyhydantoin carbamate scaffold could be finely optimized to achieve the inhibition activity toward the desired serine hydrolase.
4. ABHD4
4.1. Biochemical Features and Biological Roles
Human ABHD4 is composed of 342 residues (39 kDa) and is prevalently expressed in testis and gall bladder. ABHD4 is a lysophospholipase/phospholipase B first identified in 2006 as the enzyme responsible for the deacylation of N-acyl phosphatidylethanolamines and lyso-N-acyl phosphatidylethanolamines to generate glycerophospho-N-acyl ethanolamines, which are intermediates for the biosynthesis of N-acyl ethanolamines, an important group of signaling lipids including anandamide.35 Later, biochemical and in vivo studies revealed that brain N-acyl lysophosphatidylserines are also substrates of ABHD4.36 ABHD4 has a beneficial role in a fibrosarcoma model, limiting cell proliferation.37 ABHD4 is a regulator of anoikis, which is a programmed cell death of anchorage-dependent cells when they detach from the extracellular matrix, and resistance to anoikis usually leads to cancer metastases. Genetic deletion of ABHD4 induced anoikis resistance in prostate cells as well as nasopharyngeal and ovarian cancer cells; however, the exact mechanism was not yet elucidated.38 Very recently, László et al. found that ABHD4 is a necessary mediator for the elimination of pathologically detached cells in embryonic brain, confirming that downregulation of ABHD4 may induce resistance to anoikis.39
4.2. Inhibitors
Very few ABHD4 inhibitors are reported in literature: the most potent are the previously mentioned compounds 5 and 6 (Figure 4) identified by Cognetta et al. These N-hydroxyhydantoin carbamates displayed IC50 values in the submicromolar range (IC50 = 0.03 and 0.1 μM for 5 and 6, respectively) in mouse brain membrane proteome analyzed by gel-based ABPP, although they are not highly selective for ABHD4, because of their additional inhibition activity on ABHD3 (subsection 3.2). Interestingly, some analogues of compound 6 were further developed as probes for gel-based detection of ABHD4 in ABPP experiments; however, their discussion is out the scope of this perspective. Cognetta et al. identify other ABHD4 ligands, unfortunately none of them were selective for ABHD4 nor reached a greater inhibition potency than compounds 5 and 6.34
Figure 4.
ABHD4 inhibitors.
5. ABHD5
5.1. Biochemical Features and Biological Roles
ABHD5 or Comparative Gene Identification 58 (CGI-58) is a well characterized member of this class of ABHDs. It is a 349-residue protein (39 kDa) mainly expressed in bone marrow, fat, and skin. The mutation of ABHD5 gene causes the human Chanarin-Dorfman Syndrome or Neutral Lipid Storage Disease with Ichthyosis (NLSDI), which is a rare autosomal recessive disorder characterized by the presence of intracellular accumulation of triacylglycerol (TG) droplets in many tissues. Multiple organs and tissues are affected by this syndrome, since patients suffering of NLSDI manifest ichthyosis and sometimes liver steatosis with hepatomegaly, muscle weakness (or myopathy), ataxia, neurosensory hearing loss, subcapsular cataracts, nystagmus, strabismus, and mental retardation.40,41 ABHD5 mutation is also related to a rare heritable form of nonalcoholic fatty liver disease (NAFLD), a severe health disease associated with significant morbidity and mortality.42,43 In ABHD5, the nucleophilic serine is substituted by asparagine; therefore, ABHD5 itself is not able to hydrolyze triacylglycerols, but it coactivates adipose triglyceride lipase (ATGL), an important TG hydrolase which catalyzes the formation of glycerol and free fatty acids.44 Mutations in both ATGL and ABHD5 cause the “neutral lipid storage disease” characterized by massive accumulation of TG in various tissues. Knockout of ABHD5 in mice resulted in an excessive lipid storage due to defective activation of ATGL-mediated TG hydrolysis. In fact, newborn mice showed a condition similar to human NLSDI, with severe hepatic steatosis and a defective skin permeability barrier. These studies have highlighted that ABHD5 exhibits a crucial role in cellular TG catabolism by its regulation on ATGL activity.45 Differently, the use of antisense oligonucleotides to inhibit ABHD5 expression in adult mice induced severe hepatic steatosis, but at the same time prevented high-fat diet-induced obesity and insulin resistance.46 Conversely, when mice were genetically deprived of ATGL, they showed a massive accumulation of lipids in several tissues and the inability to mobilize these fat stores, along with an increase in insulin sensitivity, glucose use, and tolerance.47 A further study confirmed that ABHD5 knockdown by antisense oligonucleotides paradoxically improved hepatic insulin signaling, reducing diet-induced stress kinase activation, thus highlighting an important role of ABHD5 in mediating inflammatory responses.48 ABHD5 overexpression in mice did not prevent the development of diet-induced obesity; therefore, the ATGL activation induced by ABHD5 is not a determining factor for lipolysis.49 Therefore, despite the involvement of both ABHD5 and ATGL in TG hydrolysis, experimental evidence suggests distinct roles of these two proteins.50 ABHD5 displayed acyl-CoA-dependent acyltransferase activity to lysophosphatidic acid, showing a preference for unsaturated species of acyl-CoA, such as arachidonoyl-CoA, oleoyl-CoA, and 1-oleoyl-lysophosphatidic acid.6 ABHD5 was found to be located in the lipid droplets in adipocytes, thanks to the interaction with perilipin-1 (PLIN1 or perilipin-A), which is expressed almost exclusively in adipocytes,51 and it is designated to the breakdown of TG in lipid droplets via its phosphorylation. A mutation of ABHD5, as in Chanarin-Dorfman syndrome, determines a weakening of the ABHD5 binding to PLIN1, suggesting that the loss of this interaction could induce this syndrome.52 Lipolytic stimulation by catecholamines triggers the phosphorylation of PLIN1,53 disrupts the complex ABHD5/PLIN1, thus inducing release and translocation of ABHD5 from the lipid droplets surface into the cytosol, enabling it to activate ATGL-mediated lipolysis.54 The structure of C-terminal moiety of PLIN1 is of crucial importance, because mutations affecting this region proved to make PLIN1 unable to sequester ABHD5, thus triggering ATGL activation and resulting in increased basal lipolysis.55 Another isoform of this protein, perilipin-5 (PLIN5 or Mldp), is highly expressed in tissues characterized by high rates of fatty acid oxidation, such as heart, skeletal muscle, and liver, and PLIN5 was able to bind both ABHD5 and ATGL, but not both the protein at the same time.56 Both PLIN5 and ABHD5 were observed on the surface of cardiomyocyte lipid droplets, and their interaction was promoted by lipid loading.57 Cardiac PLIN5 overexpression regulated ATGL-mediated TG catabolism under regulation of protein kinase A, but PLIN5 does not constantly impair cardiac lipolysis.58 Patatin Like Phospholipase Domain Containing 3 (PNPLA3, also known as adiponutrin) interacts with ABHD5 competing with ATGL, so preventing its activation and their binding was much stronger than the interaction of ABHD5 with ATGL. Importantly, PNPLA3 suppressed ABHD5-dependent lipolysis in brown adipocytes.59,60 ABHD5 is involved in cancer development: its reduced expression was detected in metastatic castration-resistant prostate cancer and colorectal tumors, in which ABHD5 deficiency induced epithelial to mesenchymal transition and promoted Warburg effect; thus ABHD5 acts as a tumor suppressor.61,62 Differently, ABHD5 expression was increased in tumor-associated macrophages in colorectal cancer, and ABHD5 facilitated cancer growth by suppression of spermidine synthase-dependent spermidine production, since spermidine exerts an inhibitory effect on the growth of colorectal cells.63 Later, the same authors proved that ABHD5 expressed in macrophages displayed an antimetastatic effect mediated by matrix metalloproteinases, and this opposite finding was justified by the observation that tumor-associated macrophages exhibited heterogeneous expression of ABHD5 and that subgroup of macrophages with low ABHD5 expression was found to be correlated with the invasive behavior of the tumor.64 However, the role of ABHD5 in tumors is quite controversial: other studies reported that ABHD5 was overexpressed in prostate cancer cells and ABHD5 genetic deletion decreased growth of prostate cancer cells by inducing apoptosis.65 Recent studies demonstrated that overexpression of ABHD5 induces cell cycle arrest at the G1 phase and blocks cell proliferation in prostate cancer cells by inhibition of protein synthesis mediated by mTOR complex 1 (mTORC1); therefore, activation of ABHD5 by ligands may represent a promising therapeutic option against cancer.66 ABHD5 was found to be overexpressed and exerted a protumorigenic role in endometrial cancer by involving the AKT signaling pathway.67 Travers et al. provided the first evidence of serine protease activity of ABHD5. Histone deacetylases (HDACs) act as repressors of cardiomyocyte hypertrophy through association with the pro-hypertrophic transcription factor myocyte enhancer factor-2 (MEF2). Catecholamine-induced stimulation of β-adrenergic receptors leads to activation of protein kinase A, which triggers the cleavage of HDAC4, with the subsequent production of an amino-terminal polypeptide of HDAC4, and ABHD5 was identified as the one responsible for HDAC4 proteolysis. This series of events ultimately ends with the inhibition of MEF2 transcriptional activity, with resulting protective effects in cultured cardiomyocytes and diabetic hearts, in turn identifying a cardioprotective role for ABHD5. In vivo studies confirmed that ABHD5 lacking mice displayed cardiomyopathy typically associated with neutral lipid storage disease.68,69
5.2. Modulators
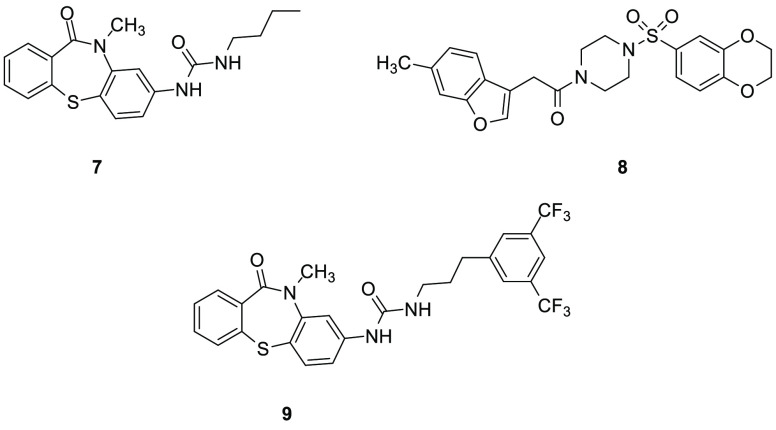
In 2015, Sanders et al. developed the only existing synthetic ABHD5 ligands, which may be useful to target lipid disorders such as obesity, diabetes, and cardiovascular diseases, because of their ability to promote fat catabolism.70 The authors considered previous studies assessing that PLIN1 suppresses lipolysis by binding ABHD5, thus preventing ABHD5-mediated activation of ATGL. On the other side, phosphorylation of PLIN1 by protein kinase A led to ABHD5 release, which activates ATGL, thus promoting lipolysis in adipocytes.53 High-throughput screening identified two compounds able to disrupt the interaction between ABHD5 and PLIN1 or PLIN5 in the absence of protein kinase A activation: the thiaza-tricyclic urea 7 (SR-4995, Figure 5) and the sulfonyl piperazine 8 (SR-4559, Figure 5). These two derivatives prevented the binding of ABHD5 to PLIN1, with IC50 values of 200 and 510 nM, respectively. The newly developed ligands 7 and 8 directly bound to ABHD5 and were shown to be potent and specific allosteric modulators of this enzyme. In brown adipocytes, 7 quickly disrupted the complex between ABHD5 and PLIN5. Inhibitors 7 and 8 were also tested in adipocytes and muscles to evaluate their effects on lipolysis, and they rapidly stimulated lipolysis, displaying EC50 values of 4–7 μM. ABHD5 knockdown experiments highlighted that ABHD5 deletion abolished the efficacy of synthetic ligands 7 and 8 of stimulating lipolysis. Moreover, these two compounds promoted dissociation of ABHD5 from PLIN1 or PLIN5, without affecting the ABHD5 capacity to activate ATGL. These two compounds, together with 9 (SR-3420, Figure 5), another thiaza-tricyclic urea derivative differing from 7 only in the presence of the 1,3-(trifluoromethyl)benzene substituent at the end of the alkyl-urea chain, were subjected to further biological experiments.71 Compound 9 was more effective in inducing lipolysis than 7 or 8 in white and brown adipocytes. Activation of ABHD5 by 9 potently inhibited mTORC1, thus blocking mTORC1 signaling and inhibiting the anabolism of cancer cells.66 Inhibitor 9 regulated the interaction between ABHD5 and PNPLA3 by increasing this interaction.59
Figure 5.
ABHD5 allosteric modulators.
Compounds 7 and 8 were also the object of a patent dating from 2016, claiming small molecules as modulators of cellular lipolysis.72 The authors declared that these modulators, by facilitating fat catabolism, may be used as therapeutic agents to cure diabetes, obesity, cardiovascular diseases but also cancer. Moreover, these derivatives could increase the content of skin barrier lipids upon topical application. Structure–activity relationship (SAR) studies focusing on the thiaza-tricyclic urea scaffold highlighted that the urea at C4 of the tricyclic ring was fundamental for the activity, while the shift at C3 caused a loss of activity; moreover, the replacement of the urea group with esters, amides and N-dialkyl ureas at C4 was detrimental for ABHD5 activity. The activity against this enzyme increased by introducing long alkyl chains on the urea group (i.e., n-butyl chain in 7), in particular those ending with aryl groups, while the presence of an oxygen atom in this side chain decreased activity. In addition, small substituents were preferred on the nitrogen atom of the amidic group (i.e., methyl group of 7). SAR studies on the sulfonyl-piperazine scaffold demonstrated that the length of the linker between the sulfonyl-piperazine moiety in 8 and the benzofuran ring could be slightly increased but this modification decreased the inhibition potency. For what concerns the benzofuran ring, it tolerated alkyl substituents such as methyl group as in 8 and a benzothiazole ring without electron-donating substituents was also allowed. Moreover, if the benzofuran was connected at C2 to the rest of the molecule, the potency was reduced.
6. ABHD6
6.1. Biochemical Features and Biological Roles
Serine hydrolase ABHD6 is a 337-amino acid protein (38 kDa) with its catalytic triad composed of Ser148-Asp278-His306. It is an integral membrane protein possessing a N-terminal transmembrane helix73 and is ubiquitously expressed, in particular in brain (cerebellum, frontal cortex, hippocampus, and striatum),74 small intestine (duodenum), brown adipose tissue,75 spleen, skin liver, kidney, and ovary.76 Moreover, female hormones such as estradiol and progesterone can promote the overexpression of ABHD6 in immune cells.77 ABHD6 is an important enzyme not only in the central nervous system but also in peripheral tissues, and it is involved in many physiological and pathological states.78−80 ABHD6 is significantly expressed in several cancer cell lines, such as bone, prostate, and leukocyte tumor cell lines.76 ABHD6 expression is increased in Ewing family tumors (EFT), thus representing a possible diagnostic and/or therapeutic target for this disease, although ABHD6 knockdown in EFT cell lines did not result in a decreased proliferative activity or increased apoptosis rate.81 Human pancreatic ductal adenocarcinoma (PDAC) cell lines displayed an high expression of ABHD6, and this enzyme was considered the driving force for the metastatic potential of PDAC cells.82 ABHD6 is an important oncogene in non-small-cell lung carcinoma (NSCLC) cells,83 since ABHD6 silencing reduced migration and invasion in vitro as well as metastatic potential and tumor growth in vivo. Differently, ABHD6 was identified as an antioncogene in hepatocellular carcinoma (HCC).84 A recent study revealed a diacylglycerol lipase (DAGL) activity for ABHD6 in Neuro-2a cells.85 A study identified ABHD6 as the main monoacylglycerol lipase present in pancreatic islet β-cells, in which glucose-stimulated insulin secretion is amplified by ABHD6 inhibition. This effect was ascribed to reduced hydrolysis of 1-monoacylglycerols, which activated the protein Munc13-1 (a key exocytotic effector), thus triggering insulin secretion.86,87 Deprivation of ABHD6 in mice fed with a high-fat diet induced a reduction of weight gain and liver steatosis, an improved glucose tolerance and insulin sensitivity, an enhanced locomotor activity, and browning of white adipose tissues. In particular, the mechanism of adipose browning behind ABHD6 suppression seems to involve an increase in 1-monoacylglycerols (MAGs), which causes peroxisome proliferator-activated receptors α and γ (PPARα and PPARγ) activation.88 A study was focused on the role of ABHD6 in the central control of energy homeostasis. ABHD6 knockdown in neurons of the ventromedial hypothalamus in mice led to impaired adaptive responses to high-fat feeding, dieting, and cold exposure, thus underlining the importance of ABHD6 in maintaining a good flexibility in energetic metabolism.89 Some studies highlighted the correlation between ABHD6 expression and the pathogenesis of Epstein–Barr virus (EBV)-related diseases90 and the autoimmune disease systemic lupus erythematosus.91 As anticipated, the main substrate of ABHD6 is 2-AG:2,92,93 ABHD6 controls 2-AG at the site of 2-AG production (postsynaptic), differently MAGL exerts the control at the site of CB1R (presynaptic). The intracellular orientation of ABHD6 is strategic to regulate 2-AG production at the site of its formation. ABHD6 preferentially cleaves MAGs possessing saturated acyl chains, with medium or long chains, with a preference for 1(3)-isomers compared to 2-isomers.94 Considering that ABHD6 increases the formation of arachidonic acid by hydrolyzing 2-AG, it is easy to explain its involvement in inflammatory processes. ABHD6 inhibition reduces lipopolysaccharide (LPS)-induced macrophage activation by increasing 2-AG levels in vitro, since 2-AG oxygenation by cyclooxygenase-2 (COX-2) led to the formation of anti-inflammatory prostaglandin D2-glycerol ester (PGD2-G). ABHD6 was also able to reduce LPS-induced inflammation in mice without provoking the typical central effects of MAGL inhibition95,96 (cannabinoid behavioral and functional antagonism of the endocannabinoid system due to chronic MAGL inhibition).97 The role of ABHD6 in peripheral tissues was established by Thomas et al. using antisense oligonucleotides to knock down the enzyme in vivo. ABHD6 proved to be implicated in lipid metabolism, since ABHD6 inhibition resulted in the accumulation of lysophosphatidylglycerol (LPG) and phosphatidylglycerol (PG). It exerted a protecting activity from high-fat-diet-induced obesity, hepatic steatosis, hyperglycemia, hyperinsulinemia and it improved both glucose and insulin tolerance in mice. Therefore, ABHD6 contributes to the development of the metabolic syndrome.75 ABHD6 is implicated in lysophosphatidylinositols (LPI) metabolism in J774 macrophages as ABHD6 inhibition led to an increase of the levels of all LPI. The effect of ABHD6 inhibition was investigated in LPS-activated J774 cells to study the role of this enzyme in the response of an inflammatory setting. The authors of this study observed an increase in 20:4 LPI levels, therefore ABHD6 could be involved in the hydrolysis of 20:4 LPI; however, extensive studies are still needed to clarify the complex metabolic pathways of LPI.98 Bis(monoacylglycero)phosphate (BMP), a phospholipid present in the intraluminal vesicles of late endosomes and lysosomes exerting a fundamental role in degradation and sorting of lipids, was identified as a substrate of ABHD6, thus revealing a role for ABHD6 in the late endosomal/lysosomal lipid sorting.99 A more recent study pointed out that ABHD6 affected circulating BMP levels both in mice and humans; consequently deletion of ABHD6 led to increased BMP concentrations without provoking lysosomal storage disorders (LSDs).100 These studies suggest that ABHD6 is a key regulator of different classes of lipids. High expression of ABHD6 was detected in an animal model of multiple sclerosis (cuprizone model of nonimmune dependent demyelination) and pharmacological blockade of this hydrolase partially attenuated demyelination and astrogliosis.101 The role of ABHD6 was investigated in another animal model of multiple sclerosis, the experimental autoimmune encephalomyelitis (EAE): the use of an ABHD6 inhibitor remarkably ameliorated the clinical signs of EAE, exerting an anti-inflammatory and neuroprotective action.102 However, more recently, the therapeutic efficacy of the pharmacological blockade of ABHD6 in improving the clinical signs of EAE was discredited, considering that the ABHD6 inhibition resulted only in a modest slowdown of EAE progression.103 Wei and co-workers performed studies regarding the involvement of ABHD6 and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors (AMPARs), which are tetrameric receptors formed by GluA1–4 subunits. ABHD6 inhibited the glutamate-induced currents of GluA1-, GluA2-, and GluA3-containing AMPARs, by binding to GluA1–3 C-terminal regions.104,105 Pharmacological ABHD6 inhibition in a mouse model of traumatic brain injury had multiple positive effects: it improved motor coordination and working memory performance due to a reduction of brain lesions, neuroinflammation, neurodegeneration and blood-brain dysfunctions.106 An antiepileptic role was reported for ABHD6: ABHD6 pharmacological inhibition reduced pentylenetetrazole-induced seizures and also blocked spontaneous seizures in R6/2 mice, a genetic model of Huntington’s disease characterized by dysregulated endocannabinoid signaling. This study suggests that the observed anticonvulsive effect was independent of cannabinoid receptors, but it involved GABAA receptors; however, further experiments are needed to confirm the above-mentioned mechanism of action.107
6.2. Inhibitors
6.2.1. Carbamate Derivatives
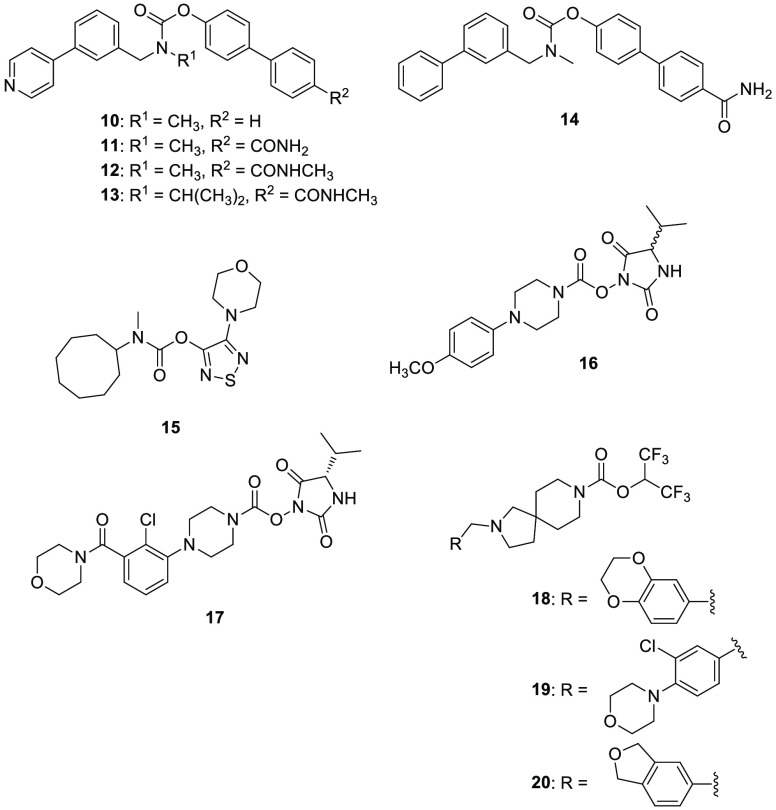
Cravatt and collaborators performed a competitive ABPP in COS-7 cells transfected with the human ABHD6 and a library of known carbamate serine hydrolase inhibitors, with the aim of demonstrating that ABPP can be applied for the identification of potent and selective inhibitors for serine hydrolases.108 On the basis of this strategy, carbamate 10 (Figure 6) was the most potent and selective inhibitor of this library on ABHD6 (IC50 = 350 nM). Compound 10 was further optimized to improve ABHD6 inhibition potency and among the 20 newly synthesized derivatives compound 11 (WWL70, Figure 6), which differs from compound 10 only for the presence of a p-carboxamide group in the para position on the terminal phenyl ring, showed the highest ABHD6 inhibitory activity, with an IC50 value of 70 nM, still maintaining an excellent selectivity. Compound 11 was widely investigated in further pharmacological studies. It inhibited of about 50% the [3H]-2-AG hydrolysis in homogenates prepared from neurons in primary culture, whereas the inhibition of [3H]-2-AG hydrolysis was reduced to about 20% in homogenates prepared from adult mouse brain, without exerting significant effects in homogenates prepared from microglia in primary culture. These findings are consistent with the fact that ABHD6 activity is greater in neurons in primary culture than in adult mouse brain and ABHD6 expression is very low in microglia in primary culture.93
Figure 6.
Carbamate-based ABHD6 inhibitors.
Further pharmacological evaluations highlighted the potential therapeutic role of 11 in animal models of traumatic brain injury and experimental autoimmune encephalomyelitis, as previously described.102,106 Kiritoshi et al. tested 11 in an arthritis pain model: 11, by increasing 2-AG levels and hence activating CB1R, rescued the metabotropic glutamate receptor 5 (mGluR5) activity with a consequent restore of the medial prefrontal cortex output and cognitive function, in addition it reduced pain in the animal model.109 Tanaka et al. reported that the anti-inflammatory and neuroprotective properties of 11 were not attributable to ABHD6 inhibition but to its interference with the metabolic pathway from arachidonic acid to prostaglandin E2 (PGE2).110,111 In particular, derivative 11 blocked PGE2 production and the expression of COX-2 and microsomial prostaglandin E synthase-1/2 (mPGES-1/2), the metabolic enzymes necessary for PGE2 production from arachidonic acid, in LPS-activated microglia cells and in an animal model of neuropathic pain (chronic constriction injury of the mouse sciatic nerve), thus proving its possible use for the treatment of inflammatory diseases and neuropathic pain.
Madiraju et al. deposited three patents showing that ABHD6 activity is tightly correlated to insulin secretion and to conversion of white into brown adipose tissue.112−114 In particular, the three patents described ABHD6 inhibitors which promoted insulin secretion by increasing the accumulation of MAGs and that may be useful for the treatment of type-2 diabetes, insulin resistance and metabolic syndrome. In a cell-based model for insulin secretion, regulation, and pancreatic islet β-cell function studies, the carbamate derivative 11 and the related analogues 12 and 13 (Figure 6) showed 95%, 98%, and 95% of ABHD6 inhibition, respectively, when tested at 10 μM. Moreover, they displayed an increased percentage of insulin secretion compared to control. Finally, compound 11 exerted a benefic effect on mice blood glucose level by increasing plasma insulin concentrations, thus confirming its potential application for treating type-2 diabetes and any other conditions associated with a low level of insulin secretion/production.113
A novel carbamate-based compound, 14 (WWL123, Figure 6), was discovered by Cravatt’s research group in 2010 by an ABPP screening.115 Compound 14 is a selective ABHD6 inhibitor (IC50 = 0.43 μM), which also maintained its selective inhibitory activity on ABHD6 in vivo (mice treated with 5–20 mg/kg, i.p., 4 h). Carbamate 14, thanks to its high permeability to the blood-brain barrier, exerted an antiepileptic activity in vivo, as previously described.107
The 1,2,5-thiadiazole carbamate scaffold, present in potent inhibitors of lysosomal acid lipase, was properly optimized by Patel et al. to develop selective ABHD6 inhibitors, considering that many carbamate-based compounds were found to efficiently inhibit enzymes of the ECS.116 The most potent ABHD6 inhibitor of this class was 15 (JZP-430, Figure 6), possessing a carbamate moiety linked to a saturated eight-membered ring in position 3 of the thiadiazole ring and a morpholine ring in position 4 of the central heterocycle, in order to balance the increased lipophilicity determined by the big ring size on the other position. Compound 15 showed an IC50 value of 44 nM in lysates of HEK293 cells transiently expressing human ABHD6 and it was also able to inhibit ABHD6 in competitive ABPP of the mouse brain membrane proteome. Derivative 15 was endowed with a good selectivity for ABHD6 over FAAH (only 18% inhibition when tested at 10 μM concentration), and it maintained only a negligible residual activity on lysosomal acid lipase (<20% when tested at 10 μM concentration), without exerting any appreciable activity on cannabinoid receptors, ABHD12 and MAGL. As expected, 15 inhibits ABHD6 by an irreversible mechanism of action. The class of 1,2,5-thiadiazole carbamates was subjected to comparative molecular field analysis (CoMFA) and molecular dynamic (MD) studies on a homology model of ABHD6.117 This study highlighted that the most important bond was the hydrogen bond established between the carbonyl group of 15 and the Phe80 backbone, one of the two residues forming the oxyanion hole, thus demonstrating the proper fitting of the compound in this region of the enzyme. During MD simulations, the ABHD6-15 complex was quite stable; however, the distance between the carbonyl group of the ligand and the Phe80 backbone increased during the simulation, thus weakening the hydrogen bond. On the other hand, the formation of an additional hydrogen bond between Ser148 and the carbonyl group of the inhibitor promoted the covalent bond necessary for the irreversible inhibition of the enzyme.
In the previously mentioned screening of Cognetta et al., N-hydroxyhydantoin carbamate 16 (MJN193, Figure 6), characterized by an isopropyl group and a N-substituted piperazine on the hydantoin moiety, showed a considerable activity and selectivity for ABHD6.34
In 2017, Abide Therapeutics, Inc. patented a series of dual lipoprotein-associated phospholipase A2 (Lp-PLA2) and ABHD6 inhibitors for the treatment of several pathological conditions such as multiple sclerosis, ischemia, traumatic brain injury, Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, cancer, and diabetes.118 These newly developed Lp-PLA2/ABHD6 inhibitors show the common chemical structure of 2,5-dioxoimidazolidin-1-yl phenylpiperazine-1-carboxylates, resembling N-hydroxyhydantoin 16. They were tested in vitro (ABPP assays) to evaluate their inhibition potency on both enzymes, and they did not show selectivity for ABHD6. Representative compound 17 (Figure 6) showed an IC50 value lower than 100 nM for ABHD6 and between 100 nM and 1 μM for Lp-PLA2.
In the same year, a series of spirocyclic-fused carbamates as modulators of MAGL/ABHD6 was reported by the same company for the treatment of pain.119 These dual MAGL/ABHD6 inhibitors were characterized by a hexafluoropropan-2-yl piperidine-1-carboxylate moiety. The most promising derivatives for what concerns ABHD6 inhibition potency were compounds 18, 19, and 20 (Figure 6). All the three spirocyclic-fused carbamates proved to be slightly selective for ABHD6 versus MAGL and FAAH. Indeed, they showed IC50 values lower than 100 nM on ABHD6, between 100 and 1000 nM on MAGL and greater than 1000 nM on FAAH. Compounds 18, 19, and 20 displayed a MAGL and ABHD6 inhibition activity greater than or equal to 75% at 1 μM, on the contrary FAAH inhibition activity was lower than 25% when tested in competitive ABPP assays in mouse brain membrane fraction. No in vivo data were available for these three inhibitors.
6.2.2. Triazole Urea Derivatives
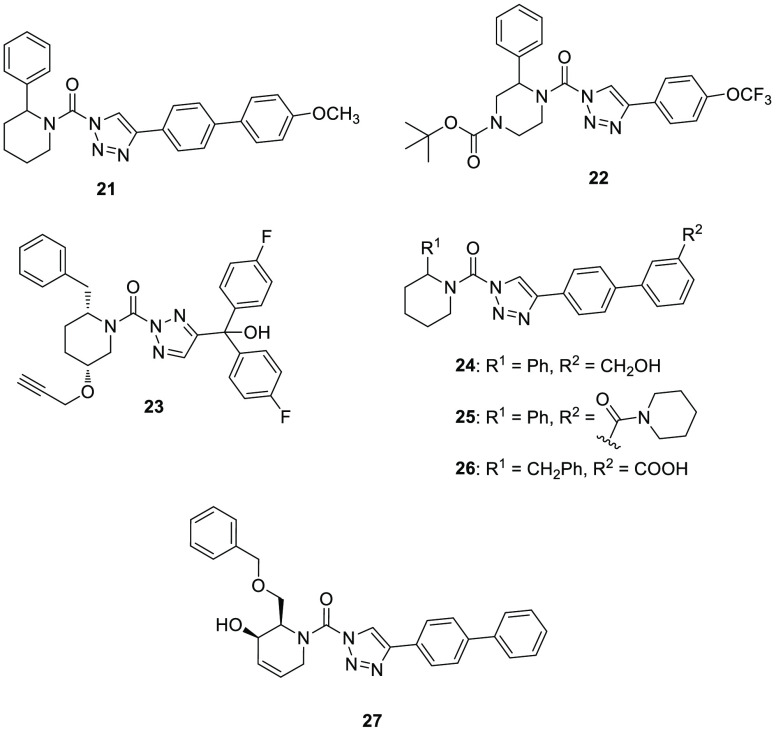
The 1,2,3-triazole urea scaffold is a typical feature of serine hydrolase inhibitors,120 in particular in 2012, the research group of Prof. Cravatt focused on this scaffold to develop new DAGL inhibitors. In this screening campaign, the piperidyl-1,2,3-triaziole urea 21 (KT195, Figure 7) was identified as a selective ABHD6 inhibitor (IC50 = 10 nM) in competitive ABPP with a marginal cross-reactivity against DAGLβ,121,122 and it was predicted to irreversibly bind to the enzyme, by carbamoylating the enzyme’s serine nucleophile. In Neuro-2a cells, 21 confirmed its inhibition activity by fully blocking ABHD6 with an IC50 value of 1 nM and a negligible inhibition of DAGLβ. Similarly, in peritoneal macrophages from inhibitor-treated mice, 21 inhibited ABHD6 and lowered interleukin-1β secretion from LPS-treated macrophages; however, two carboxylesterases (CES3 and CES2G) and lysosomal phospholipase A2 group XV (PLA2G15) were identified as off targets of this compound. Compound 21 was further studied to evaluate its potential role to block necrotic cell death.123 This ABHD6 inhibitor was able to attenuate necrotic cell death of cultured fibroblasts by preventing mitochondrial calcium uptake and permeability transition pore formation. In addition to the above-mentioned off-targets, 21 also blocked ER calcium release and cell death by targeting the nucleophilic serine in ABHD2.23
Figure 7.
Triazole urea-based ABHD6 inhibitors.
On the basis of ABHD6 inhibitor 21 and with the aim of obtaining new selective and central nervous system (CNS)-active inhibitors of DAGLα and β, Ogasawara and collaborators synthesized a new triazole urea 22 (DO53, Figure 7), characterized by a 2-phenyl-piperazine moiety instead of the 2-phenyl-piperidine group of 21.124 After intraperitoneal administration to C57BL/6 mice, it showed a good selectivity on ABHD6 together with inhibition of PLA2G7, and a low potency on the original DAGL targets. The selectivity profile of 22 was more extensively elucidated by ABPP experiments coupled to quantitative high-resolution mass spectrometry: it confirmed a negligible activity against DAGLs, but it showed notable cross-reactivity with many other targets, such as ABHD2, ABHD3, carboxylesterase CES1C, and the platelet activating factor acetylhydrolase 2 (PAFAH2). In the same research program, the potent DAGLα and β inhibitor 23 (DH376, Figure 7) showed an undesired ABHD6 inhibition activity both in vitro and in vivo and also a cross-reactivity with carboxylesterase CES1C and HSL.124,125 Later, 23 was used to identify the enzymes responsible for 2-AG production during retinoic acid (RA)-induced neurite outgrowth of murine neuroblastoma Neuro-2a cells.85 The terminal alkyne group present in the chemical structure of 23 was used in a “click chemistry” approach to introduce reporter tags, which allowed one to visualize by a chemical proteomic strategy the targets of 23 in Neuro-2a cells. ABHD6 and DAGLβ were identified as the only targets and ABHD6 was found to hydrolyze diacylglycerols, thus contributing to the production of 2-AG during RA-induced differentiation of Neuro-2a cells, since 23 blocked 2-AG production and reduced neuronal differentiation.
Compound 21 was structurally optimized by Hsu et al. to improve its potency, selectivity and in vivo activity toward ABHD6. In this new series of irreversible piperidyl-1,2,3-triazole urea inhibitors, compounds 24 (KT182, Figure 7), 25 (KT185, Figure 7), and 26 (KT203, Figure 7) showed a remarkable inhibitory activity against ABHD6, with IC50 values of 1.7, 1.3, and 0.82 nM, respectively, corresponding to 0.24, 0.21, and 0.31 nM, when their potencies were measured in situ in Neuro-2a cells. None of them exerted any significant off-target activity.126 In these compounds, polar substituents were added in meta position of the biphenyl moiety (R2 group, Figure 5), such as hydroxymethyl (24), piperidine-amide (25), or carboxylic acid (26). The quantitative mass-spectrometry-based proteomic method ABBP-SILAC was applied to verify their activities: both 24 and 26 inhibited >90% of ABHD6 activity, while 24 blocked >80% of ABHD6 activity in Neuro-2a cells. In addition, the three developed inhibitors did not show any considerable cross-reactivity toward a panel of serine hydrolases present in Neuro-2a cell line, confirming their selectivity for ABHD6 in living cells. Compounds 24 and 26 were also tested in vivo when intraperitoneally administered in mice: both compounds were effective in blocking ABHD6 in the liver at the higher tested dose (1 mg/kg), and only 24 reached the same effect in the brain, probably due to the carboxylic acid of 26 which hinders its brain penetration. A mild systemic inhibitory effect on a plasma esterase carboxylesterase-1 (CES1) was detected only for 24. Encouraging results were observed with compound 25, that proved to be an orally bioavailable and selective ABHD6 inhibitor in vivo, even if complete ABHD6 inhibition was only observed at higher dose (40 mg/kg). As anticipated in subsection 6.1, Manterola and co-workers used 24 in the cuprizone model of nonimmune dependent demyelination,101 because of its ability to cross the blood-brain barrier and its selectivity in vivo after intraperitoneal administration. After the promising results of this first evaluation, the use of ABHD6 inhibitors was reassessed in multiple sclerosis by testing compounds 24 and 26, showing different CNS permeability.103 The administration of systemically active inhibitor 24 modestly attenuated the neurological disability of the EAE; on the contrary, the peripherally active inhibitor 26 was not effective in ameliorating the clinical signs of EAE. Both compounds 24 and 26 did not attenuate inflammatory responses associated with tissue damage in the chronic phase of EAE, and the chronic treatment with 24 caused the desensitization of brain CB1R. All together, these results suggest that ABHD6 blockade has only a moderate therapeutic effect in this model of demyelination.
A series of dual ABHD6 and DAGLα inhibitors were recently published by Deng et al.127 Their strategy aimed at finding dual inhibitors as potential therapeutic agents to treat metabolic and neurodegenerative diseases. This series of dual inhibitors bear the chiral hydroxylated 2-benzylpiperdine scaffold with a triazole urea moiety. Surprisingly, some of them including compound 27 (Figure 7), showed a good combination of inhibition activity of ABHD6 (pIC50 = 6.6 in membranes from HEK293T cells expressing recombinant human ABHD6; 83% inhibition in ABPP experiments) and selectivity for ABHD6 versus DAGLα (4-fold) and other serine hydrolases such as FAAH and MAGL.
In conclusion, 1,2,3-triazole urea represents a suitable scaffold to design irreversible inhibitors of serine hydrolases, thanks to the electrophilic carbonyl group which promotes the binding to the nucleophile active site serine. To date, all DAGL inhibitors reported in literature also inhibit ABHD6,128 and this aspect may be exploited as the starting point to develop new selective ABHD6 inhibitors, by conveniently modifying this chemotype. Moreover, some activity-based probes (i.e., compounds binding to the enzyme covalently, useful to detect the amount of the enzyme present in a biological system) based on the triazole-urea scaffold were developed to target ABHD6, thus highlighting the high versatility of this chemical core.129
6.2.3. Other ABHD6 Inhibitors
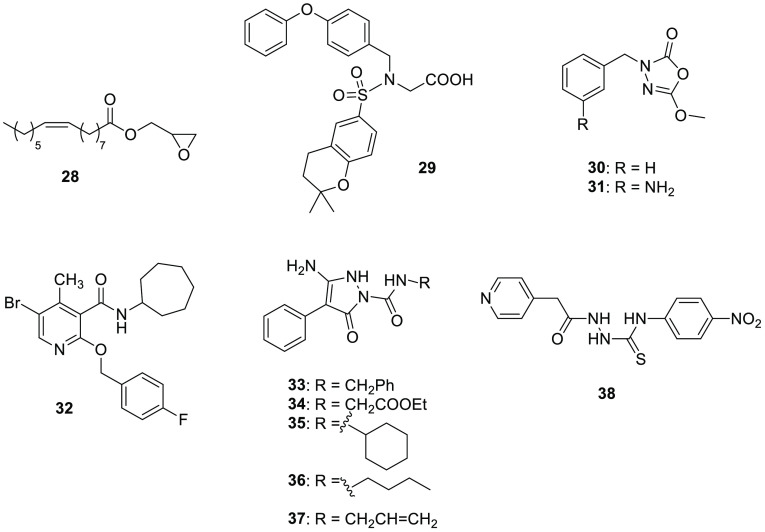
In 2011, Marrs and co-workers designed a series of esters by replacing the glycerol polar head of 2-AG with various oxygenated heterocycles.130 The ester derivative 28 (UCM710, Figure 8), characterized by an oxirane moiety, proved to be a potent dual inhibitor of ABHD6/FAAH (IC50 values of 2.4 and 4.0 μM, respectively) when tested in neuron homogenates, without inhibiting MAGL nor binding to cannabinoid receptors. Additionally, it was able to efficiently inhibit 2-AG and EAE hydrolysis also in intact neurons, although without reaching the maximum activity (60% and 30% inhibition of AEA and 2-AG hydrolysis, respectively). The unique pharmacological profile of 28 may be determined by its chemical structure, which mimics the natural substrates of the target enzymes, thus likely the oxirane group cannot fit into the active site of MAGL which is covered by the cap domain, differently from ABHD6 and FAAH that lack the cap domain necessary for the substrate recognition and interaction.
Figure 8.
Various ABHD6 inhibitors.
In 2014, Janssen and collaborators developed a series of glycine sulfonamides as novel DAGLα inhibitors. A member of this chemical class, compound 29 (LEI106, Figure 8), acts as a submicromolar dual ABHD6/DAGLα inhibitor.131 In the colorimetric biochemical assay performed in HEK293 membranes overexpressing human DAGLα, the sulfonamide 29 showed an IC50 value of 18 nM and inhibited the hydrolysis of DAGLα natural substrate, [14C]-sn-1-oleoyl-2-arachidonoyl-glycerol, with a Ki value of 0.7 μM. After observing an off target in brain membrane homogenate-based assays, a specific biochemical human ABHD6 activity assay revealed that 29 inhibited ABHD6 with a Ki value of 0.8 μM.
The 1,3,4-oxadiazol-2-one scaffold is widely adopted for the discovery of serine hydrolase inhibitors. Patel et al. optimized the 1,3,4-oxadiazol-2-one 30 (Figure 8),132 in order to develop new potent and selective ABHD6 inhibitors. Compound 30 was previously synthesized by their research group in a discovery campaign of FAAH inhibitors, but 30 selectively inhibited human ABHD6 (about 40% inhibition at 1 μM), without affecting FAAH or MAGL. An extensive structure–activity analysis led to the identification of the meta-amino analogue of the lead compound 30, compound 31 (JZP-169, Figure 8), which exerted a notable AHBD6 inhibition with an IC50 value of 216 nM.133 The free amino group in the meta position seemed to be essential for the activity on ABHD6, since its protection or shift led to detrimental decreases of inhibition activity. Compound 31 was selective for ABHD6 when tested at 10 μM concentration, with any notable activity on other members of the ECS (FAAH, MAGL, ABHD12, and cannabinoid receptors). This novel and selective ABHD6 inhibitor interacts with the enzyme through an irreversible mechanism, as suggested by dilution assays and further confirmed by molecular docking studies. Docking of 31 underlined that the compound was located in the oxyanion hole, thus the carbonyl group of the inhibitor was suggested in proximity of the nucleophilic Ser148. Additionally, the importance of the free amino group on the benzyl moiety was explained by considering its involvement in hydrogen bonds with the side chains of Glu190 and Glu253.
1,2-Dihydro-2-oxo-pyridine-3-carboxamides were developed as potential CB2R ligands; however, this scaffold furnished a very promising ABHD6 inhibitor.134 4-Methyl-5-bromo-2-substituted pyridine 32 (Figure 8) bound not only to both cannabinoid receptors as expected (Ki values of 113 and 606 nM for CB1R and CB2R, respectively) but exhibited a remarkable inhibition activity of ABHD6 enzyme with an IC50 value of 530 nM, exerting also inhibitory activity against anandamide cell uptake (IC50 = 620 nM), without affecting FAAH.
In 2021, a study about the role of 2-AG protection of the retina against the excitatory amino acid AMPA involved two ABHD6 inhibitors: AM12100 which was selective for ABHD6 (IC50 = 8 nM) and AM11920 which was a dual MAGL and ABHD6 inhibitor (IC50 values of 12.1 and 6.0 nM, respectively).135 The structures of both inhibitors are not disclosed yet. Interestingly, both compounds exerted a neuroprotective effect in the animal retinal model of AMPA excitotoxicity, but the selective ABHD6 inhibitor was less effective, thus leading to the conclusion that the dual inhibition exerted by AM11920 induced a more evident 2-AG increase and therefore it showed a better pharmacological profile.
It is noteworthy to add in this section a series of dual inhibitors of human ABHD6 and ABHD12 (ABHD12 will be analyzed in detail in section 10) discovered in 2014 by Kaczor et al. The authors screened an in-house library of heterocyclic compounds,136 leading to six weak inhibitors, pyrazole-based derivatives 33–37 (Figure 8) and thiosemicarbazide compound 38 (Figure 8). The remaining enzymatic activity on each enzyme was measured as a percentage compared to control and ranged from 65.3 to 84.2% for ABHD6 and 78.4 to 85.4% for ABHD12. Despite their low inhibition activity on both ABHDs, these heterocycles could represent a starting point for further structural modifications to tune their activity selectively on ABHD6 or ABHD12.
7. ABHD9
7.1. Biochemical Features and Biological Roles
ABHD9, also named epoxide hydrolase 3 (EPHX3), is a 360-amino acid protein (41 kDa) characterized by the presence of a nucleophilic aspartate in place of a serine. ABHD9 is prevalently expressed in skin and esophagus. ABHD9 was renamed EPHX3 after studies in which it displayed epoxide hydrolase activity against epoxyeicosatrienoic acids and 9,10-epoxyoctadecamonoenoic acids in vitro.137 Nevertheless, in a more recent in vivo study, genetic silencing of ABHD9 had no significant effects on the metabolism of fatty acid epoxides and did not alter LPS-induced lung inflammation or functional recovery after ischemia/reperfusion injury, that are two models regulated by epoxyeicosatrienoic acids.138 ABHD9-mediated hydrolysis of leukotoxin led to the production of a metabolite which was identified as a strong mediator of acute respiratory distress syndrome (ARDS).137 ABHD9 seems to be involved in cancer, since ABHD9 expression has been reported to be downregulated in tumors, such as prostate cancer,139,140 melanoma,141 B cell tumor,142 gastric cancer,143 salivary gland adenoid cystic carcinoma,144 oral squamous cell carcinoma,145 head and neck squamous cell carcinoma,146 and colorectal carcinoma.147 ABHD9 was considered a potential ichthyosis-related gene.148 The role of ABHD9 in the regulation of skin barrier function was confirmed by other studies; in fact, ABHD9 was found to be involved in the production of epidermis-related linoleate triols, considering that it is highly expressed in the external cells of human epidermis.149 Moreover, ABHD9 hydrolyzes linoleate-derived epoxy-alcohols esterified in skin ceramides in vivo.150
7.2. Inhibitors
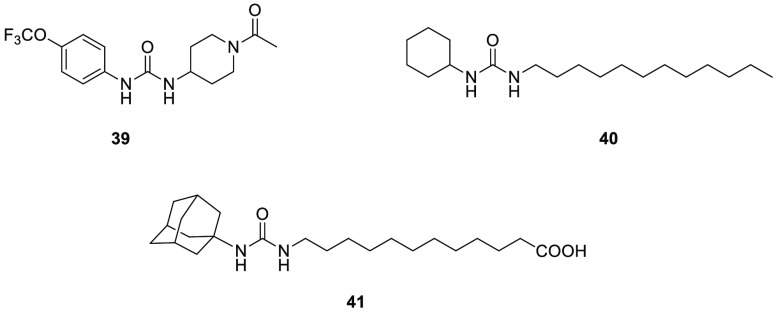
In 2012 Decker and colleagues tested a class of N,N′-disubstituted urea derivatives, which previously were considered inhibitors of mammalian soluble epoxide hydrolase, on ABHD9.137 Among these N,N’-disubstituted urea derivatives, 1-(1-acetylpiperidin-4-yl)-3-(4-(trifluoromethoxy)phenyl)urea 39 (TPAU, Figure 9), 1-cyclohexyl-3-dodecylurea 40 (CDU, Figure 9), and 12-(3-adamantan-1-yl-ureido)-dodecanoic acid 41 (AUDA, Figure 9) were the most active inhibitors on ABHD9, with IC50 values of 75, 80, and 100 nM, respectively. These findings could be a starting point for the development of new ABHD9 inhibitors able to better elucidate their possible use as new therapeutic agents.
Figure 9.
ABHD9 inhibitors.
8. ABHD10
8.1. Biochemical Features and Biological Roles
ABHD10 is a 306-residue protein (34 kDa), ubiquitously expressed yet prevalent in kidney and thyroid. Proteomic studies located ABHD10 in the mitochondria.151 Some studies in the literature described the involvement of ABHD10 in drug metabolism.152 ABHD10 plays a key role in the metabolism of the immunosuppressant mycophenolate mofetil (MMF), because it led to the deglucuronidation in human liver of acyl glucuronide metabolite (AcMPAG), potentially responsible for some MMF-induced adverse effects such as leucopenia or gastrointestinal toxicity; therefore, ABHD10 exerted a detoxifying effect.153 A similar detoxifying activity was observed in the case of probenecid acyl glucuronide (PRAG), which is the main metabolite of the uricosuric agent probenecid, that can provoke severe allergic or anaphylactic reactions, as ABHD10 catalyzed PRAG deglucuronidation in human liver.154S-Depalmitoylase activity was observed for ABHD10; in particular ABHD10 acts on peroxiredoxin-5 (PRDX5), a key antioxidant protein and therefore ABHD10 can be included in the acyl protein thioesterases (APT) family of regulatory proteins.155
8.2. Inhibitors
Cravatt and his research group, with the aim to identify new serine hydrolase inhibitors, discovered a series of aza-β-lactams (ABLs), which efficiently inhibited the mammalian serine hydrolase protein-phosphatase methylesterase-1 (PME-1).156 Further structural optimization led to the identification of 42 (R enantiomer, ABL117, Figure 10), which inhibited both PME-1 and ABHD10 with IC50 values of 250 and 210 nM, respectively. Thereafter, in order to improve ABHD10 inhibition, the authors performed a SAR evaluation of the ABL scaffold. Using bulky substituents as O-alkyl groups on the carbamates or shifting the methyl group to the para position of the benzene ring increased potency for ABHD10, as demonstrated by compound 43 (R enantiomer, ABL303, also named ML257,157,158Figure 10), which showed an augmented inhibition potency on ABHD10 (IC50 = 30 nM) and a marked selectivity over ABHD6, prolyl endopeptidase (PREP), PME-1, and other serine hydrolases. Aza-β-lactam 43 maintained a notable activity in living Neuro-2a cells with an IC50 value of 21 nM, without exhibiting off-targets when tested at 1 μM concentration and no appreciable inhibition of PME-1 at 10 μM. The quantitative mass spectrometry-based proteomic method ABPP-SILAC was employed to further test 43 in Neuro-2a cells: it selectively and near-completely inhibited ABHD10 (>95%), and thus, 43 was the first discovered potent ABHD10 inhibitor. Compound 43 is an irreversible inhibitor, acting via aza-β-lactam ring opening and subsequent serine acylation.157
Figure 10.
ABHD10 inhibitors.
The same research group combined the flavagline rocaglate core, typical of natural compounds isolated from the genus Aglaia, characterized by a cyclopenta[b]-benzofuran structure, with a β-lactone scaffold, to give a class of rocaglate-derived β-lactones as potential serine hydrolases inhibitors.159 The most interesting derivative of this series is compound 44 (both enantiomers (+)-44 and (−)-44 are reported in Figure 10). Unfortunately, ABPP in proteomes deriving from human cancer cell lines (PC3 and LNCaP) and mouse tissues (brain, liver, and testes) and ABPP-SILAC analysis on PC3 cells pointed out that β-lactone 44 inhibited different serine hydrolases including not only ABHD10 (IC50 value of about 100 nM) but also cathepsin A (CTSA), retinoid-inducible serine carboxypeptidase 1 (SCPEP1), and acyl-CoA thioesterase 1/2 (ACOT1/2). In particular, the pure (−)-44 enantiomer was shown to be responsible for most of the ABHD10 and ACOT1/2 inhibition activity in competitive APBB assay on PC3 cells. The authors hypothesized that compound (−)-44 irreversibly inhibits the target hydrolase, by acylation of the active site nucleophilic serine, as has been reported for other β-lactones.
In 2012, Adachi et al. employed MIDA-boronates to identify new inhibitors of ABHD10 and serine carboxypeptidase (CPVL).160 Alkyl(MIDA)boronate 45 (Figure 10) was tested in ABPP experiments in PC3 cell proteome: its conversion into the corresponding boronic acid was evident in buffer after 2 h of incubation. Therefore, the inhibition required the decomposition of the (MIDA)boronate portion to the free boronic acid. Compound 45 induced a complete inhibition of ABHD10 with few off-targets and showed a near-complete inactivation of ABHD10 at 10 μM and of ACOT1/2 at 100 μM. CPVL inhibition was confirmed in ABPP-SILAC assays, in which 45 inhibited by more than 95% both ABHD10 and CPVL, at 25 μM.
9. ABHD11
9.1. Biochemical Features and Biological Roles
ABHD11 or Williams-Beuren syndrome chromosomal region 21 protein (WBSCR21) or PP1226 is a 315-amino acid protein (35 kDa). ABHD11 is a mitochondrial protein, mainly found in skeletal muscle,161 but it is an ubiquitous protein with higher expression in colon, prostate, small intestine, and thyroid. Its alternative name WBSCR21 originates from the fact that ABHD10 is among the deleted genes in Williams-Beuren syndrome, a severe neurodevelopmental disorder characterized by several diseases and abnormalities, concerning both physical and cognitive aspects.162 ABHD11 expression was reduced in white adipose tissue in mice fed with a high-fat diet as well as in HSL knockout mice. On the contrary, treatment with the antidiabetic drug rosiglitazone increased its expression; however, other analyzed lipases and esterases were unaffected. Therefore, the importance of these changes needs further elucidation.163 ABHD11 is involved in cancer aggressiveness, since increased ABHD11 is a predictive biomarker of metastases in lung adenocarcinoma.164 In breast cancer, ABHD11 was downregulated in paclitaxel-resistant MCF7/PacR cells (68% compared to MCF7 cells),165 but it was also related to breast cancer malignancy.166 Arya et al. expressed human ABHD11 in budding yeast, Saccharomyces cerevisiae, to further elucidate the role of this protein in lipid metabolism: ABHD11 overexpression decreased triacylglycerol content in yeast, thus playing a key role in lipid hydrolysis.167 ABHD11 involvement in the regulation of the metabolic state was confirmed by knockout ABHD11 mice, which did not gain weight when fed a high-fat diet, maintaining a lean phenotype, normal biochemical plasma parameters, and reduced fat intestinal absorption.168 ABHD11 regulates 2-oxoglutarate (2-OG) metabolism: genetic deletion of ABHD11 led to the accumulation of 2-OG, resulting in inhibition of 2-OG dependent dioxygenases which are involved in the hypoxia inducible factor (HIF) response, DNA methylation, and histone modifications. Moreover, ABHD11 proved to be fundamental for functional lipoylation of the 2-oxoglutarate dehydrogenase complex (OGDHc), the enzyme of the tricarboxylic acid cycle that decarboxylates 2-OG to succinyl-CoA.169 Recently, a role for ABHD11 in embryonic stem cell (ESC) maintenance was highlighted, determining that ABHD11 is important for self-renewal and metabolic homeostasis of ESC.170 The ABHD11 locus also encodes for long noncoding RNA, named ABHD11-antisense (ABHD11-AS1), whose increased expression was observed in gastric,171 colorectal,172 pancreatic,173 endometrial,174 nonsmall-cell lung,175 papillary thyroid,176 and ovarian cancer.177
9.2. Inhibitors
9.2.1. Carbamate Derivatives
As mentioned before, the carbamate scaffold is very common among serine hydrolase inhibitors. During the screening study performed by Cravatt’s group to identify new inhibitors of serine hydrolases, during the discovery of compound 14, compound 46 (WWL151, Figure 11) was identified as a mild inhibitor of ABHD11 (IC50 = 5.3 μM), however highly selective, likely due to the unicity of its seven-membered azepane ring, compared to other carbamate derivatives with broad spectrum activity on the panel of serine hydrolases. The substitution with a 2-ethylpiperidine ring proved to be successful, giving rise to a more potent inhibitor 47 (WWL222, Figure 11) which selectively blocked ABHD11 (IC50 = 170 nM) without any activity against other serine hydrolases.115 Carbamate 47 was also very efficacious and selective in vivo when administered intraperitoneally in mice at 10 mg/kg.
Figure 11.
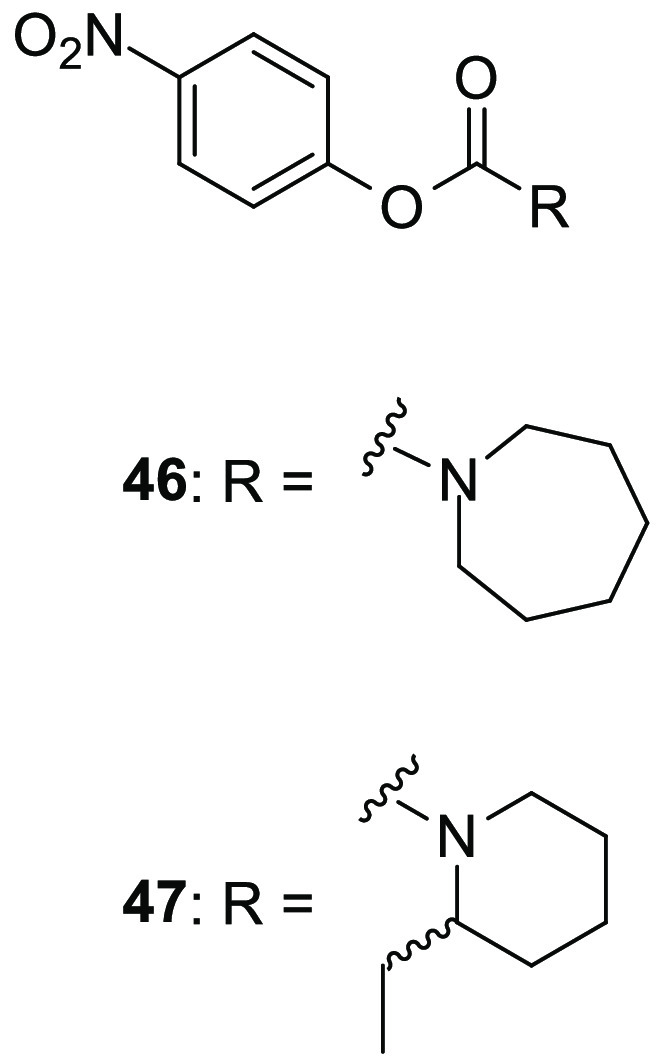
Carbamate-based ABHD11 inhibitors.
9.2.2. Urea Derivatives
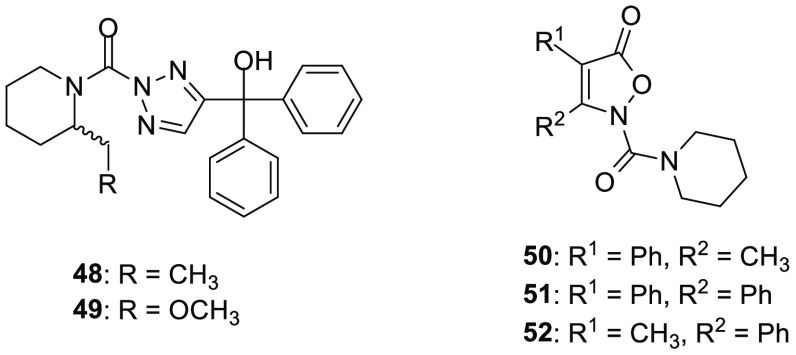
In 2010, Cravatt and colleagues carried out a fluorescence polarization-based competitive ABPP high throughput screening study to discover new inhibitors of lysophospholipase 1 and 2 (LYPLA1 and LYPLA2).178 During this study, performed on a library of triazole urea-based compounds, the authors serendipitously identified the racemic compound 48 (ML226, Figure 12) as a remarkably potent (IC50 = 15 nM) and selective (≥100-fold over more than 20 serine hydrolases) ABHD11 inhibitor, with a residual activity on N-acylaminoacyl-peptide hydrolase (APEH, 50% inhibition at 1.5 μM). The mode of action of 48 was assessed by LC-MS/MS studies, which revealed a covalent modification of the catalytic Ser141 of ABHD11, in which the triazole ring acts as the leaving group. A close analogue of 48, derivative 49(178) (AA44-2, Figure 12), bearing a bulkier methoxymethyl group instead of the ethyl group in 2-position of the piperidine ring, showed an improved ABHD11 inhibition with an IC50 value of 1 nM, still maintaining a high selectivity versus other serine hydrolases and no activity on APEH.120 These properties were confirmed by ABPP-SILAC analysis in living mouse T-cells: treatment with 49 resulted in a blockade greater than 95% of ABHD11 activity at the concentration of 3 nM with no cross-reactivity over other 40 serine hydrolases observed in T-cells.
Figure 12.
Urea-based ABHD11 inhibitors.
In 2016, Navia-Paldanius and collaborators screened more than 200 in-house synthesized compounds designed to target serine hydrolases by using competitive ABPP tests.179 This screening led to the identification of three isoxazol-5(2H)-one-containing urea derivatives exerting a nanomolar potency against human ABHD11: 50 (JZP-228), 51 (JZP-245), and 52 (JZP-249) reported in Figure 12 showed IC50 values of 2.4, 3.4, and 2.3 nM, respectively. The three compounds were assessed in a competitive ABPP assay among the serine hydrolases of mouse whole brain membrane in order to evaluate their selectivity. All of them completely blocked ABHD11 activity when tested at 100 nM concentration; however, 50 inhibited an additional protein band, migrating at ∼60 kDa, attributable to FAAH. Moreover, 50 was previously found to be a HSL inhibitor with a reported IC50 value of 14 nM.180 The three inhibitors were tested in competitive ABPP with lysates of prostate cancer LNCaP and VCaP cells (both expressing FAAH) and PC3 cells (not expressing FAAH). Predictably, compounds 51 and 52 selectively blocked ABHD11, differently from 50 which confirmed its activity on FAAH in LNCaP and VCaP cells. Inhibitor 51 was further investigated in competitive ABPP: at 0.1 μM, 51 inhibited ABHD11 in all tested proteomes (mouse whole brain membranes, prostate cancer cell lysates, and mitochondrial fraction of brown fat and testicle), but at higher concentrations (1–10 μM) it also inhibited FAAH. Additionally, at 10 μM, 51 showed as off-targets ABHD6 and the serine hydrolase KIAA1363 in mouse whole brain membrane proteome. The cytotoxic effect of urea 51 was evaluated in prostate cancer cells: it reduced proliferation of the nonaggressive cell line LNCaP, but it was poorly effective on the aggressive cell line PC3. Nevertheless, in LNCaP and VCaP cells, 51 acted as a dual inhibitor targeting both ABHD11 and FAAH with a similar potency. Navia-Paldanius et al. built an ABHD11 homology model in order to better understand the interactions between these urea-based compounds in the enzyme active site. The docking studies suggested that the inhibitors properly fitted the active site of ABHD11, where they established π–π interactions. It was postulated a possible irreversible inhibition mechanism, through active site serine acylation, in which the isoxazol-5(2H)-one ring behaves as the leaving group.
10. ABHD12
10.1. Biochemical Features and Biological Roles
ABHD12 is also known as ABHD12A, c20orf22, or 2-arachidonoylglerol hydrolase, and it is a 398-residue protein (45 kDa). From a structural point of view, ABHD12 is a single-pass integral membrane protein, possessing a N-terminal transmembrane helix, which points its active site toward the extracellular space, and its catalytic triad is Ser246-Asp333-His372, as discovered by site-directed mutagenesis studies.73,92 The ubiquitously expressed ABHD12 has the highest expression in the brain (especially in microglia), and it is localized to the ER membrane in the mammalian brain,181 where it is responsible for about 9% of 2-AG hydrolysis, together with MAGL and ABHD6.2 ABHD12 is also present in macrophages and osteoclasts. Studies of substrate specificity revealed that ABHD12 prefers the 1(3)-isomer of arachidonoylglycerol over 2-AG and unsaturated C20:4 MAGs over C18:2 MAGs.73 It was found that ABHD12 required glycosylation for optimal activity and it showed a strong preference for very-long-chain lipid substrates, such as lysophosphatidylserine (lysoPS) lipids.181 Furthermore, in the brain, ABHD12 hydrolyzes oxidizedphosphatidylserine, which is considered an apoptotic signal, under severe inflammatory stress.182 Mutations of ABHD12 were found to be related to the etiology of some pathologies, such as the neurodegenerative disorder called polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract “PHARC”, likely due to impaired 2-AG metabolism.183,184 Other studies suggest that PHARC may be induced by a dysregulated lysoPS lipase activity which is typical of ABHD12, since ABHD12 deficient mice displayed increased proinflammatory lysoPS lipid levels and neurobehavioral abnormalities similar to those of the PHARC phenotype.185 Together with ABHD16A, ABHD12 dynamically regulates lysoPS metabolism: ABHD16A contributes to the production of both cellular and secreted lysoPS starting from phosphatidylserine (PS), and ABHD12 preferentially controls degradation of secreted lysoPS to glycerophosphoserine, thus exerting complementary roles.186,187 A recent study in ABHD12 knockout mice reveals an upregulation of lipids deriving from arachidonic acid in the brain, thus suggesting that neuroinflammation may contribute to the development of PHARC-like symptoms.188 Dysfunctional ABHD12 has been linked to a variant of PHARC named Usher syndrome 3 (USH3), an autosomal recessive genetically heterogeneous disorder, characterized by congenital sensorineural hearing impairment and retinitis pigmentosa.189,190 Some tumor types showed an increased ABHD12 expression, such as in colorectal cancer191 and in breast cancer MCF7 and MDA-MB-231 cell lines ABHD12 knockdown reduced cell growth, proliferation, and invasiveness.192
10.2. Inhibitors
10.2.1. Natural Compounds
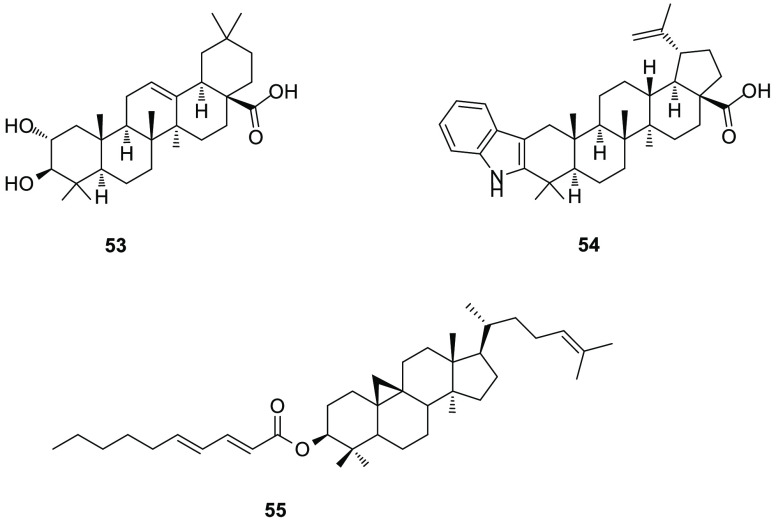
Encouraged by the fact that some natural triterpenes exerted a certain inhibition activity on hydrolases (i.e., MAGL and ABHD6), such as pristimerin 69(193) (Figure 17), Parkkari et al. performed a screening of triterpene and triterpenoid derivatives by purchasing 15 commercially available compounds. The inhibition data were determined in lysates of HEK293 cells transiently overexpressing human ABHD12 and revealed that the oleanane derivative maslinic acid 53 (Figure 13) was the most potent ABHD12 inhibitor of this series, showing an IC50 value of 1.3 μM.194 A preliminary SAR study revealed that the presence of a carboxylate in position 17 in combination with small hydrophobic groups such as the methyl groups at position 4 determined a good inhibition activity. The screening of triterpene derivatives continued with a series of synthetic betulinic acid derivatives: among them, triterpene 54 (Figure 13), bearing an indole heterocycle fused with the central core in the place of the two hydroxyl groups of maslinic acid 53, showed good inhibition of ABHD12 (IC50 = 0.9 μM). The authors enriched the SAR relative to this class of derivatives, since it was evident that the presence of hydrogen bond donors or acceptors at position 3 was required for an optimal inhibition activity. Later, the inhibition mechanism for the best two compounds was investigated: they proved to inhibit ABHD12 in a reversible manner, as tested by a dilution assay of the enzyme–inhibitor complex. Moreover, compounds 53 and 54 were tested in ABPP of HEK293 cell lysates and mouse brain membrane preparations and proved to be selective for ABHD12 over ABHD6, MAGL, FAAH, CB1R, and CB2R.
Figure 17.
Unselective ABHDs inhibitors.
Figure 13.
Natural ABHD12 inhibitors.
A cycloartane-type triterpene derivative 55 (Figure 13) isolated from Euphorbia pterococca proved to be a moderate ABHD12 inhibitor (IC50 = 11.6 μM); however, it was surprisingly selective, since it did not affect ABHD6, MAGL, and FAAH enzymes.195
10.2.2. Synthetic Compounds
In 2019, the research group of Cravatt developed a thiourea derivative, 56 (DO264, Figure 14), which proved to efficiently and selectively inhibit ABHD12 in vitro and in vivo.196,197 An initial HTS procedure based on an innovative fluorescence assay, which measures the ABHD12-mediated hydrolysis of lysophosphatidic acid, was used to screen the Maybridge HitFinder library. Afterward, two further screenings, LC-MS-based lysoPS hydrolysis and ABPP assays, were pursued in order to identify new ABHD12 inhibitors and remove false positive compounds. After the identification of a hit compound based on a thiourea central core, a detailed SAR exploration of this scaffold led to the discovery of N-3-pyridyl-N′-4-piperidinylthiourea 56, that competitively and selectively inhibited ABHD12, with and IC50 value of 11 nM in ABPP assays, in a competitive fashion, and in spite of its thiourea-based structure, 56 reversibly inhibited ABHD12. Compound 56 blocked lysoPS hydrolysis of recombinant mouse and human ABHD12 in transfected HEK293T cell lysates (IC50 values of about 30 and 90 nM against mouse and human ABHD12, respectively) and the lysoPS lipase activity of membrane lysates from mouse brain (IC50 = 2.8 nM) and human monocytic THP-1 cells (IC50 = 8.6 nM), confirming its activity. Considering that ABHD12 is highly expressed in macrophages, 56 was tested in THP-1 cells and primary human macrophages, in which it induced a significant increase in lysoPS and polyunsaturated 20:4 PS lipids. Inhibitor 56 provoked a high cytokine production in THP-1 cells; however, at concentrations of at least 5 μM, 56 exerted an undesired cytotoxic effect on this cell line. The authors excluded any cytotoxic effect generated by ABHD12 inhibition, since the block of the enzyme occurred at a lower concentration of the inhibitor, and this fact was confirmed testing 56 on different cell lines, when exposed to 1 μM 56. An excellent ABHD12 inhibition was confirmed in C57BL/6 mice treated with 56 by intraperitoneal or oral administration, observing only a low inhibition of ABHD2 and phospholipase A2 group VI (PLA2G6). 56-treated mice exhibited increased levels of brain lysoPS and 20:4 PS lipids, similarly to the changes observed in ABHD12 knockout mice, although they did not show any auditory defects, which are typical symptoms of PHARC disease. Moreover, 56-treated and ABHD12 knockout mice exhibited a heightened immunological responses in a lymphocytic choriomeningitis virus (LCMV) clone 13 infection animal model, thus highlighting that ABHD12 may have an immunosuppressive function. A recently discovered effect of compound 56 is its ability to enhance ferroptotic death, a particular form of cell death defined by peroxidation of membrane phospholipids, in a similar fashion to what observed in ABHD12 knockout mice.198
Figure 14.
Synthetic ABHD12 inhibitors.
Inhibitor 56 and structurally similar thiourea derivatives are also reported in a patent of the Scripps Research Institute, in which these ABHD12 inhibitors were claimed as useful for the treatment of cancer, neuropsychiatric disorders, and neurodegenerative, autoimmune, neuroinflammatory, and infectious diseases.199 Besides 56, the most promising compounds are compounds 57, 58, and 59 (Figure 14), which displayed IC50 values lower than or equal to 100 nM for ABHD12 inhibition in competitive ABPP assays and in a substrate-based assay by using HEK293T cells overexpressing ABHD12.
In 2020, Lundbeck La Jolla Research Center Inc. published a patent including pyridinyl urea derivatives as ABHD12 inhibitors, which may be useful for the treatment of cancer and infectious, neurodegenerative, autoimmune, and neuroinflammatory diseases.200 The most potent inhibitor of this series was compound 60 (Figure 14), possessing the same structure of compound 57 (Figure 14) with the exception of the urea instead of the thiourea moiety. This pyridinyl urea inhibited mouse brain ABHD12 with an IC50 value lower than or equal to 100 nM, and it showed an ABHD12 inhibition activity greater than or equal to 75% when tested at 1 μM in mouse brain membrane proteomes.
11. ABHD16A
11.1. Biochemical Features and Biological Roles
ABHD16A is also named Human Lymphocyte Antigen B-associated transcript 5 (BAT5), and it is composed of 558 residues (63 kDa). It is a poorly known serine hydrolase, whose highest expression was observed in skeletal muscle and brain.201 ABHD16A is localized in the plasma membranes in human platelets and mouse megakaryocytes.202 It is palmitoylated; however, further investigation about this modification was not performed.203 The substrate preference for ABHD16A was investigated by Savinainen et al., and it was found that ABHD16A acts as a hydrolase for medium-chain saturated MAGs, long-chain unsaturated MAGs (in particular 1-linoleylglycerol, 1-LG) as well as the 15-deoxy-Δ12,14-prostaglandin J2-2-glycerol ester (15d-PGJ2-G).204 Polymorphisms of ABHD16A are correlated to Kawasaki syndrome, a disease characterized by vascular inflammation, which may cause coronary artery aneurysm formations and cardiac complications.205 In pigs, the polymorphism of ABHD16A was related with back fat thickness, thus suggesting its potential role as a marker associated with obesity.206 As already anticipated in subsection 9.1, ABHD16A is implicated in immunoregulation together with ABHD12, since both regulate lysoPS metabolism in vivo. In particular, ABHD16A regulates the lysoPS-induced release of proinflammatory cytokines from macrophages.186 The involvement of ABHD16 in immunoregulation originates from studies regarding its gene location, considering that ABHD16A belongs to a cluster of genes within the human major histocompatibility complex class III.207,208 Moreover, the expression of ABHD16A could influence the immunogenicity of bone marrow cells in mice.209
11.2. Inhibitors
In 2014, the first ABHD16A inhibitor was reported, the β-lactone palmostatin B 61 (Figure 15), which inhibited the hydrolysis of the MAG substrate 1-LG inhibitor in HEK293 lysates transfected with human ABHD16A, with an IC50 value of 100 nM.204 Considering thatpalmostatin B 61 was first developed as a LYPLA1 inhibitor (IC50 = 670 nM),210 further assays to determine its selectivity disclosed that compound 61 dose-dependently inactivated not only LYPLA1/2 and ABHD16A, but also ABHD12 (IC50 = 2 nM), ABHD6 (IC50 = 50 nM) and MAGL (IC50 = 90 nM). Considering the low selectivity of 61 and the potent activity of the HSL inhibitor 62 (C7600, Figure 15) on human ABHD16A (IC50 = 8.3 nM), Savinainen et al. decided to modify the 1,3,4-oxadiazol-2(3H)-one scaffold of 62 with the purpose to develop new selective ABHD16A inhibitors.204 Nevertheless, none of the modified derivatives of 62 showed an improved activity on the desired target. Two representative 1,3,4-oxadiazol-2(3H)-one derivatives are 63 and 64 (IC50 values of 63 and 32 nM for ABHD16A inhibition, respectively) reported in Figure 15. They differ from the lead compound because of the presence of a m-nitrophenyl (63) or a p-fluorophenyl ring (64) in the place of 3-phenoxyphenyl moiety of 62. Competitive ABPP assays on mouse brain membrane proteome suggested that, at 1 μM concentration, 1,3,4-oxadiazol-2(3H)-ones 63 selectively inhibited ABHD16A with no cross-reactivity over other serine hydrolases such as FAAH, KIAA1363, MAGL, and LYPLA1/2 as they were off-targets of 62, yet analogue 64 showed only activity against KIAA1363.
Figure 15.
ABHD16A inhibitors.
Considering the electrophilic nature of the β-lactone group, which is suitable to bind to the active site nucleophilic serine, a screening of α-alkylidene-β-lactone-based library of compounds211 led to the identification of derivative 65 (KC01, Figure 15) as an inhibitor of human and mouse ABHD16A, with IC50 values in the range 0.2–0.5 μM in competitive ABPP assays.186 A comparable inhibition activity was detected when a PS substrate was used in the membrane proteome of ABHD16A-transfected HEK293T cells, obtaining IC50 values of 90 and 520 nM for human and mouse ABHD16A, respectively. Further assays were performed in the membrane fraction of COLO205 (colon cancer), K562 (leukemia), and MCF7 (breast cancer) cancer cells, confirming the in situ inhibition of ABHD16A by 65. Quantitative ABPP-MS experiments revealed that compound 65 also inhibited ABHD2 (94% at a concentration of 1 μM) as well as many other targets such as ABHD3, Patatin Like Phospholipase Domain Containing 4 (PNPLA4), PAFAH2, ABHD6, ABHD13, ABHD11, and LYPLA1 although to a minor extent (14–80%), thus revealing a low selectivity for ABHD16A. Furthermore, treatment of COLO205, K562, and MCF7 cells with 65 showed significant reductions in the levels of cellular lysoPSs compared to untreated cells, whereas different lipids were unaffected. Importantly, pretreatment of macrophages with 65 reduced the lysoPS-induced cytokine release, thus affecting immune response. Additionally, inhibitor 65 lowered the elevated lysoPS secretion observed in ABHD12-null cells derived from a PHARC subject. Compound 65 was also patented in 2016 by the Scripps Research Institute and the University of Connecticut.212 Pharmacological investigation of 65 suggested that ABHD16A plays a key role in the production of lysoPSs in both mammalian cells and in vivo and it should be a suitable target for the development of therapeutic agents to treat PHARC and other neuroinflammatory disorders.
The most recently discovered ABHD16A inhibitor is a diterpenoid of the abietane family, compound 66 (Figure 15), which was selected as the most promising inhibitor from a screening of an in-house library of 50 similar derivatives.213 Compound 66 led to a 23% remaining activity of ABHD16A when tested in lysates of HEK293 cells transfected with ABHD16A (IC50 value of 3.4 μM). It was selective over ABHD12 and demonstrated a reversible inhibition, as proved by dilution assays. Interestingly, the authors hypothesized allosteric binding of the compound because 66 reached an incomplete ABHD16A inhibition in all used assay conditions.
12. ABHD17A, ABHD17B, and ABHD17C
12.1. Biochemical Features and Biological Roles
Very little information is known about ABHD17 enzymes, which are broadly expressed in several cell types. They are localized in the plasma membrane and are identified as proteins able to depalmitoylate N-Ras.214 Palmitoylation is a biological process in which proteins are modified by the addition of palmitate, thus directing them to the cellular compartments where they play their specific functions. N-Ras is a protein that can promote the development of cancer, so the protein depalmitoylases ABHDs may influence tumor growth.
12.2. Inhibitors
Few inhibitors are reported in the literature for ABHD17 proteins, and among them the first discovered compounds are the unselective palmostatin B 61 (Figure 15),210 the analogue palmostatin M 67 (Figure 16),215 and hexadecylfluorophosphonate HDFP 68 (Figure 16).216 Palmostatin M 67 has the same β-lactone-based scaffold of palmostatin B, and it is metabolically unstable; moreover, it is unselective, since it also inhibits APT.215 Hexadecylfluorophosphonate HDFP 68 lacks selectivity, since it inhibits many different targets, above all fatty acid synthase (FASN), FAAH, neutral cholesterol ester hydrolase 1 (AADACL1), MAGL, and LYPLA1/2.216 In 2021, Remsberg et al. performed a gel-based ABPP screening in a native mouse brain proteome of a serine hydrolase-directed compound library developed at Lundbeck La Jolla Research Center, Inc., followed by a chemical optimization process aimed at improving selectivity and potency. This strategy led to the identification of a more selective pan-ABHD17 inhibitor ABD957 69 (Figure 16).217 Compound 69 blocked covalently (likely due to its urea-containing moiety) more than 90% all the ABHD17 enzymes in MS-based ABPP experiments in OCI-AML3 (leukemia) cells, with a reported IC50 value against ABHD17B of 0.21 μM in lysates of HEK293T cells. Nevertheless, compound 69 cannot be defined as highly selective, due to a residual inhibition activity on CES1/2, ABHD6, and ABHD13. The importance of 69 derives from its contribution to the palmitoylation/depalmitoylation cycle, because it partially impaired N-Ras depalmitoylation in human acute myeloid leukemia (AML) cells, mainly affecting plasma membrane-associated dynamically palmitolylated proteins. Its therapeutic significance is linked to the impairment of NRAS mutant cancer cell growth, since the antiproliferative effect was abrogated in cells lacking ABHD17A and ABHD17B, thus confirming the ABHD17 inhibition by 69.
Figure 16.
ABHD17 inhibitors.
13. Unselective ABHD Inhibitors
Some well-known nonselective ABHD inhibitors that deserve to be mentioned are included in this section. The most important are methylarachidonoyl fluorophosphonate (MAFP 70, Figure 17), the approved antiobesity drug tetrahydrolipstatin (THL, Orlistat, 71 in Figure 17), the natural compound triterpene pristimerin 72 (Figure 17), and compound RHC-80267 73 (Figure 17), and, with the exception of 71, these compounds are generally used as pharmacological probes for biological studies rather than hit compounds to be developed as potential new drugs, because of their broad-spectrum activity. MAFP 70, a FAAH inhibitor (IC50 = 2.5 nM),218 inhibited not only ABHD6 (IC50 = 0.017 μM) but also ABHD12 (IC50 = 0.087 μM)73 as well as other serine hydrolases (i.e., KIAA1363 and MAGL), and it was a cannabinoid receptor modulator. This compound was also reported in a patent dated 2017 concerning the progesterone activation of ABHD2.219 Treatment with compound 70 completely removed progesterone-dependent activation of calcium channel CatSper, without exerting any effect on basal CatSper activity. Moreover, ABHD2 inhibition by 70 blocked progesterone-activated calcium influx into sperm flagella. In this patent, it was demonstrated that the ABHD2 enzyme played a major role in mouse AR finely tuning intracellular calcium influx in mouse sperm. Compounds 71 and 73 were originally developed as DAGL-α/β inhibitors; however, they proved to be nonselective for these enzymes, targeting also other serine hydrolases.220 In fact, 71 inhibited ABHD6 (IC50 = 0.048 μM), ABHD12 (IC50 = 0.19 μM),73 and ABHD16A (IC50 = 0.03 μM).220 Derivative 70 inhibited ABHD6 (IC50 = 0.66 μM) and ABHD16A (IC50 = 23 μM).220 The reversible MAGL inhibitor (IC50 = 93 nM)193 pristimerin 72 blocked ABHD6 (IC50 = 1.3 μM73 or 98 nM,193 according to different experimental data reported in literature).
14. Conclusions and Future Perspectives
The family of serine hydrolases is one of the most diversified and numerous existing classes of enzymes. More than 200 enzymes belong to this class and all share a typical feature that is the presence of an active site serine, which is fundamental to catalyze the hydrolysis of substrates. Among serine hydrolases, ABHD proteins play many important roles in a wide range of pathophysiological processes and, thanks to their multifaceted roles, they have progressively acquired more importance in the scientific community. In this Perspective, the main members of the ABHD family are described for what concerns their biochemical role as well as their involvement in human diseases. Many ABHDs are involved in lipid metabolism, thus affecting obesity and fat-related diseases; moreover, some ABHD mutations are often correlated to rare genetic diseases. ABHD6 and ABHD12 are strictly connected to ECS since they are implicated in 2-AG metabolism. In addition, immunoregulation, cancer, and neurodegeneration are affected by downregulation or overexpression of ABHD enzymes.
Since 2007, many compounds targeting ABHD enzymes have been developed with the aim of blocking ABHD enzymes. Considering all the enzymes of the ABHD family, the research efforts led to a quite limited number of inhibitors, restricted to ABHD2, ABHD3, ABHD4, ABHD5, ABHD6, ABHD9, ABHD10, ABHD11, ABHD12, ABHD16A, and ABHD17A-C likely due to a scarce characterization of the remaining ABHD enzymes. An exception is represented by modulators of ABHD5 7–9 which are efficient modulators of cellular lipolysis; however, they do not block ABHD catalytic activity but rather they are allosteric ABHD5 ligands. Future research lines should be aimed at characterizing the other less known members of the ABHD family, that are ABHD1, ABHD7, ABHD8, ABHD12B, ABHD13, ABHD14A, ABHD14B, ABHD15, ABHD16B, and ABHD18, to completely understand the complex biological roles of ABHD proteins. The identification of selective ligands for these proteins may help to fill a knowledge gap on endogenous lipid biosynthesis and metabolism as well as physiological and pathophysiological functions of these proteins in humans.
Other ABHD enzymes play key biological roles, although no selective ligands have yet been developed, and therefore, they are not extensively reviewed in this Perspective. For instance, ABHD1, whose biochemical function is not fully discovered, is ubiquitously expressed in murine and human tissues, reaching the highest expression in testis.221 ABHD1 overexpression was observed in a renal cell line: it contributed to the reduction of reactive oxygen species formation by NADPH oxidase, thus contributing to the protection against oxidative stress. For this reason, upregulation of ABHD1 in D5 dopamine receptor deficient mice, which develop hypertension and increased systemic oxidative stress, may contribute to the protective mechanism against oxidative stress.222
The herein reported ABHD-targeting compounds belong to several chemical classes (Figure 18). It is evident that most of them are urea- or carbamate-based molecules, since they exploit the intrinsic reactivity of active-site nucleophilic serine by promoting its acylation. Nevertheless, also lactones and lactams were expected to irreversibly bind to ABHD enzymes. Differently, compounds derived from natural sources, MIDA-boronates, and heterocycle-containing derivatives in general act as reversible inhibitors.
Figure 18.
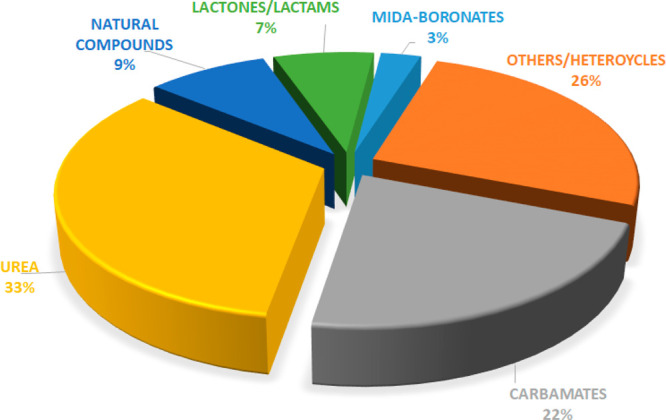
Percentages of the different chemical classes of ABHD targeting compounds, based on the representative ABHD inhibitors or modulators herein reported.
The structural similarity among ABHD proteins increases the risk of developing compounds inhibiting more ABHD enzymes, thus losing selectivity, and this may represent an obstacle to the development of an ideal clinical candidate. Nevertheless, considering the overlapping roles of some ABHD enzymes, the low selectivity of most ABHD inhibitors could be considered an opportunity to simultaneously interfere with a pathophysiological process by blocking different strategic targets.
In this Perspective, it is evident how many ligands were identified and profiled for their selectivity thanks to ABBP experiments. ABPP technology shows an undeniable utility in profiling ABHD inhibitor selectivity, because it allows one to determine the binding ability of a compound to different hydrolase enzymes at the same time in complex biological systems. The convenience and versatility of ABPP techniques were also exploited to identify potential off-targets, as it occurred for the FAAH inhibitor BIA 10-2474, which unfortunately provoked deleterious problems (death or neurological symptoms) in some volunteers during a phase 1 clinical trial. It was found that BIA 10-2474 and its main metabolite BIA 10-2639 inhibited many other enzymes, such as ABHD6 (inhibition greater than 90% at the tested concentrations) and ABHD11, in MS-based ABPP studies. The cross-reactivity with some serine hydrolases and the consequent marked alteration of the lipid metabolism may represent one of the causes of the compound’s neurotoxicity.223
The use of broad-spectrum probes in ABPP technology has accelerated the identification of new clinical candidates, finding new serine hydrolase inhibitors and assessing their selectivity both in vitro and in living systems, as in the case of ABHD inhibitors.224 The great improvement in the discovery of serine hydrolase inhibitors as new potential drugs made by application of ABPP assays is confirmed by two compounds which are currently studied in human clinical trials: MAGL inhibitor ABX-1431225 and FAAH inhibitor PF-04457845.226 In the future, further efforts should be directed toward the development of novel approaches enabling the identification of selective enzyme modulators, in order to speed up the drug discovery process. For example, the production of functional, pure recombinant ABHD enzymes and the development of reliable biochemical assays227 would be an important goal for a fast screening of large libraries of compounds aimed at finding new ABHD modulators.
Acknowledgments
We are grateful to MIUR (PRIN 2017, Project 2017SA5837) and the Italian Ministry of Health – Ricerca Finalizzata 2016 - NET-2016-02363765 for funding.
Glossary
Abbreviations
- AADACL1
neutral cholesterol ester hydrolase 1
- ABHD
α/β-hydrolase domain
- ABHD11-AS1
ABHD11-antisense
- ABPP
activity-based protein profiling
- AcMPAG
acyl glucuronide metabolite
- ACOT1/2
acyl-CoA thioesterase 1/2
- anandamide
AEA)N-arachidonoylethanolamine
- 2-AG
2-arachidonoylglycerol
- AMPARs
(RS-α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors
- APEH
N-acylaminoacyl-peptide hydrolase
- APT
acyl protein thioesterases
- AR
acrosome reaction
- ARDS
acute respiratory distress syndrome
- ATGL
adipose triglyceride lipase
- BMP
bis(monoacylglycero)phosphate
- CB1R and CB2R
cannabinoid receptors 1 and 2
- CES1
carboxylesterase 1
- CES2
carboxylesterase 2
- CoMFA
comparative molecular field analysis
- COX-2
cyclooxygenase-2
- CPVL
predicted serine carboxypeptidase
- CTSA
cathepsin A
- DAGL
diacylglycerol lipase
- 15d-PGJ2-G
15-deoxy-Δ12,14-prostaglandin J2–2-glycerol ester
- EAE
experimental autoimmune encephalomyelitis
- EBV
Epstein–Barr virus
- ECS
endocannabinoid system
- EFT
Ewing family tumors
- ER
endoplasmic reticulum
- ESC
embryonic stem cell
- FAAH
fatty acid amide hydrolase
- FASN
fatty acid synthase
- HCC
hepatocellular carcinoma
- HDAC
histone deacetylase
- HGSOC
high-grade serous ovarian cancer
- HIC1
hypermethylated in cancer 1
- HIF
hypoxia inducible factor
- HSL
hormone-sensitive lipase
- LCMV
lymphocytic choriomeningitis virus
- 1-LG
1-linoleylglycerol
- LPG
lysophosphatidylglycerol
- LPI
lysophosphatidylinositol
- Lp-PLA2
lipoprotein-associated phospholipase A2
- LPS
lipopolysaccharide
- LSD
lysosomal storage disorder
- LYPLA1 and LYPLA2
lysophospholipase 1 and 2
- lysoPS
lysophosphatidylserine
- MAG
1-monoacylglycerol
- MAGL
monoacylglycerol lipase
- MD
molecular dynamic
- MEF2
myocyte enhancer factor-2
- mGluR5
metabotropic glutamate receptor 5
- MIDA
N-methyliminodiacetic acid
- MMF
mycophenolate mofetil
- mPGES-1/2
microsomial prostaglandin E synthase-1/2
- mTORC1
mTOR complex 1
- NAFLD
nonalcoholic fatty liver disease
- NLSDI
neutral lipid storage disease with ichthyosis
- NSCLC
nonsmall-cell lung carcinoma
- 2-OG
2-oxoglutarate
- OGDHc
2-oxoglutarate dehydrogenase complex
- PAFAH2
platelet activating factor acetylhydrolase 2
- PC
phosphatidylcholine
- PDAC
pancreatic ductal adenocarcinoma
- PG
phosphatidylglycerol
- PGD2-G
prostaglandin D2-glycerol ester
- PGE2
prostaglandin E2
- PHARC
polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract
- PLA2G6
phospholipase A2 group VI
- PLA2G7
phospholipase A2 group VII
- PLA2G15
lysosomal phospholipase A2 group XV
- PLIN1
perilipin-1
- PLIN5
perilipin-5
- PME-1
protein-phosphatase methylesterase-1
- PNPLA3
patatin like phospholipase domain containing 3
- PNPLA4
patatin like phospholipase domain containing 4
- PPARα and PPARγ
peroxisome proliferator-activated receptors α and γ
- PRAG
probenecid acyl glucuronide
- PRDX5
peroxiredoxin-5
- PREP
prolyl endopeptidase
- PS
phosphatidylserine
- RA
retinoic acid
- SAR
structure–activity relationship
- SCPEP1
retinoid-inducible serine carboxypeptidase 1
- SILAC
stable isotope labeling with amino acids in cell culture
- TG
triacylglycerol
- USH3
Usher syndrome 3
Biographies
Giulia Bononi graduated cum laude in Medicinal Chemistry and Pharmaceutical Technology in 2017 at the University of Pisa (Italy). She is a third year PhD student in Science of Drug and Bioactive Substances at the Department of Pharmacy of the University of Pisa under the supervision of Professor Filippo Minutolo and Professor Carlotta Granchi. In 2020, she conducted a research period in Amsterdam (Netherlands) at the Cancer Center of Vrije Universiteit in the research group of Professor Elisa Giovannetti, working on the pharmacological evaluation of MAGL inhibitors on pancreatic cancer cells. Her research project is focused on the synthesis of therapeutic and diagnostic agents targeting tumor lipid and sugar metabolism.
Tiziano Tuccinardi Tiziano is Associate Professor of Medicinal Chemistry at the Department of Pharmacy of the University of Pisa. Since July 2016, he is also Adjunct Associate Professor at the Department of Biology, Temple University’s College of Science and Technology, Philadelphia, PA, USA. He has published more than 180 papers and 6 patents. His research interests include drug and lead discovery, with a focus on computer-assisted approaches and medicinal chemistry, including synthesis of small molecules, virtual and biomolecular screening.
Flavio Rizzolio is an associate professor of molecular biology in the Department of Molecular Sciences and Nanosystems at Ca’ Foscari University of Venice. The scientific career began in 2002 with the thesis focused on the molecular, genetic, and epigenetic analyses of two X chromosome genes associated with infertility. During the PhD program at University of Siena (Italy) and in collaboration with Temple University (USA), the scientific interest was focused on the study of different proteins involved in cell cycle control of cancer cells. In 2012, at the Centro di Riferimento Oncologico of Aviano (Italy) and later at Ca’ Foscari University, the focus of the research was on novel therapeutic approaches to cancer with the development of new nanosystems based on biocompatible materials (e.g., liposomes).
Carlotta Granchi received her PhD in Medicinal Chemistry in 2011 at the University of Pisa (IT). In 2009, she spent a period in the group of Prof. P. J. Hergenrother in the Department of Chemistry at the University of Illinois at Urbana–Champaign (USA). After the PhD, she was a postdoctoral research fellow under the supervision of Prof. F. Minutolo, and in 2016 she joined the Department of Pharmacy of the University of Pisa as Assistant Professor, becoming Associate Professor in 2019. Her research interests are focused on small molecules able to interfere with the altered metabolism of invasive tumors.
The authors declare no competing financial interest.
References
- Muccioli G. G. Endocannabinoid Biosynthesis and Inactivation, from Simple to Complex. Drug Discovery Today 2010, 15 (11–12), 474–483. 10.1016/j.drudis.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Blankman J. L.; Simon G. M.; Cravatt B. F. A Comprehensive Profile of Brain Enzymes That Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem. Biol. 2007, 14 (12), 1347–1356. 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muccioli G. G.; Xu C.; Odah E.; Cudaback E.; Cisneros J. A.; Lambert D. M.; Lopez Rodriguez M. L.; Bajjalieh S.; Stella N. Identification of a Novel Endocannabinoid-Hydrolyzing Enzyme Expressed by Microglial Cells. J. Neurosci. 2007, 27 (11), 2883–2889. 10.1523/JNEUROSCI.4830-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M.; Dijkstra B. W. α/β Hydrolase Fold Enzymes: The Family Keeps Growing. Curr. Opin. Struct. Biol. 1999, 9 (6), 732–737. 10.1016/S0959-440X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Ghosh A. K.; Ramakrishnan G.; Chandramohan C.; Rajasekharan R. CGI-58, the Causative Gene for Chanarin-Dorfman Syndrome, Mediates Acylation of Lysophosphatidic Acid. J. Biol. Chem. 2008, 283 (36), 24525–24533. 10.1074/jbc.M801783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Moran G.; Caviglia J. M.; McMahon D.; Rothenberg A.; Subramanian V.; Xu Z.; Lara-Gonzalez S.; Storch J.; Carman G. M.; Brasaemle D. L. CGI-58/ABHD5 Is a Coenzyme A-Dependent Lysophosphatidic Acid Acyltransferase. J. Lipid Res. 2010, 51 (4), 709–719. 10.1194/jlr.M001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C. C.; Thomas G.; Brown J. M. Mammalian Alpha Beta Hydrolase Domain (ABHD) Proteins: Lipid Metabolizing Enzymes at the Interface of Cell Signaling and Energy Metabolism. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2013, 1831 (4), 792–802. 10.1016/j.bbalip.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind L.; Kursula P. Structural Properties and Role of the Endocannabinoid Lipases ABHD6 and ABHD12 in Lipid Signalling and Disease. Amino Acids 2019, 51 (2), 151–174. 10.1007/s00726-018-2682-8. [DOI] [PubMed] [Google Scholar]
- Long J. Z.; Nomura D. K.; Vann R. E.; Walentiny D. M.; Booker L.; Jin X.; Burston J. J.; Sim-Selley L. J.; Lichtman A. H.; Wiley J. L.; Cravatt B. F. Dual Blockade of FAAH and MAGL Identifies Behavioral Processes Regulated by Endocannabinoid Crosstalk in Vivo. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (48), 20270–20275. 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.; Lei Q.; Wu Y.; He Y.; Li W. Activity-Based Protein Profiling: Recent Advances in Medicinal Chemistry. Eur. J. Med. Chem. 2020, 191, 112151. 10.1016/j.ejmech.2020.112151. [DOI] [PubMed] [Google Scholar]
- Cravatt B. F.; Wright A. T.; Kozarich J. W. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem. 2008, 77 (1), 383–414. 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Granchi C.; Caligiuri I.; Minutolo F.; Rizzolio F.; Tuccinardi T. A Patent Review of Monoacylglycerol Lipase (MAGL) Inhibitors (2013–2017). Expert Opin. Ther. Pat. 2017, 27 (12), 1341–1351. 10.1080/13543776.2018.1389899. [DOI] [PubMed] [Google Scholar]
- Bononi G.; Poli G.; Rizzolio F.; Tuccinardi T.; Macchia M.; Minutolo F.; Granchi C. An Updated Patent Review of Monoacylglycerol Lipase (MAGL) Inhibitors (2018-Present). Expert Opin. Ther. Pat. 2021, 31, 153–168. 10.1080/13543776.2021.1841166. [DOI] [PubMed] [Google Scholar]
- Naresh Kumar M.; Thunuguntla V. B. S. C.; Veeramachaneni G. K.; Chandra Sekhar B.; Guntupalli S.; Bondili J. S. Molecular Characterization of Human ABHD2 as TAG Lipase and Ester Hydrolase. Biosci. Rep. 2016, 36 (4), e00358 10.1042/BSR20160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obinata D.; Takada S.; Takayama K.; Urano T.; Ito A.; Ashikari D.; Fujiwara K.; Yamada Y.; Murata T.; Kumagai J.; Fujimura T.; Ikeda K.; Horie-Inoue K.; Homma Y.; Takahashi S.; Inoue S. Abhydrolase Domain Containing 2, an Androgen Target Gene, Promotes Prostate Cancer Cell Proliferation and Migration. Eur. J. Cancer 2016, 57, 39–49. 10.1016/j.ejca.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Yamanoi K.; Matsumura N.; Murphy S. K.; Baba T.; Abiko K.; Hamanishi J.; Yamaguchi K.; Koshiyama M.; Konishi I.; Mandai M. Suppression of ABHD2, Identified through a Functional Genomics Screen, Causes Anoikis Resistance, Chemoresistance and Poor Prognosis in Ovarian Cancer. Oncotarget 2016, 7 (30), 47620–47636. 10.18632/oncotarget.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.; Yang J.; Wang S. Antisense Oligonucleotides Targeting Abhydrolase Domain Containing 2 Block Human Hepatitis B Virus Propagation. Oligonucleotides 2011, 21 (2), 77–84. 10.1089/oli.2011.0280. [DOI] [PubMed] [Google Scholar]
- Jin S.; Zhao G.; Li Z.; Nishimoto Y.; Isohama Y.; Shen J.; Ito T.; Takeya M.; Araki K.; He P.; Yamamura K. Age-Related Pulmonary Emphysema in Mice Lacking α/β Hydrolase Domain Containing 2 Gene. Biochem. Biophys. Res. Commun. 2009, 380 (2), 419–424. 10.1016/j.bbrc.2009.01.098. [DOI] [PubMed] [Google Scholar]
- Miyata K.; Oike Y.; Hoshii T.; Maekawa H.; Ogawa H.; Suda T.; Araki K.; Yamamura K. Increase of Smooth Muscle Cell Migration and of Intimal Hyperplasia in Mice Lacking the α/β Hydrolase Domain Containing 2 Gene. Biochem. Biophys. Res. Commun. 2005, 329 (1), 296–304. 10.1016/j.bbrc.2005.01.127. [DOI] [PubMed] [Google Scholar]
- Miyata K.; Nakayama M.; Mizuta S.; Hokimoto S.; Sugamura K.; Oshima S.; Oike Y.; Sugiyama S.; Ogawa H.; Yamamura K. Elevated Mature Macrophage Expression of Human ABHD2 Gene in Vulnerable Plaque. Biochem. Biophys. Res. Commun. 2008, 365 (2), 207–213. 10.1016/j.bbrc.2007.10.127. [DOI] [PubMed] [Google Scholar]
- Miller M. R.; Mannowetz N.; Iavarone A. T.; Safavi R.; Gracheva E. O.; Smith J. F.; Hill R. Z.; Bautista D. M.; Kirichok Y.; Lishko P. V. Unconventional Endocannabinoid Signaling Governs Sperm Activation via the Sex Hormone Progesterone. Science 2016, 352 (6285), 555–559. 10.1126/science.aad6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannowetz N.; Miller M. R.; Lishko P. V. Regulation of the Sperm Calcium Channel CatSper by Endogenous Steroids and Plant Triterpenoids. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (22), 5743–5748. 10.1073/pnas.1700367114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B.; Lee H.; Powell R.; Reisdorph N.; Ewing H.; Gelb M. H.; Hsu K.-L.; Cravatt B. F.; Leslie C. C. Regulation of Calcium Release from the Endoplasmic Reticulum by the Serine Hydrolase ABHD2. Biochem. Biophys. Res. Commun. 2017, 490 (4), 1226–1231. 10.1016/j.bbrc.2017.06.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggelaar M. P.; Den Dulk H.; Florea B. I.; Fazio D.; Bernabò N.; Raspa M.; Janssen A. P. A.; Scavizzi F.; Barboni B.; Overkleeft H. S.; Maccarrone M.; Van Der Stelt M. ABHD2 Inhibitor Identified by Activity-Based Protein Profiling Reduces Acrosome Reaction. ACS Chem. Biol. 2019, 14, 2943. 10.1021/acschembio.9b00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar A. J.; Polak J. M. Cloning and Tissue Distribution of Three Murine α/β Hydrolase Fold Protein CDNAs. Biochem. Biophys. Res. Commun. 2002, 292 (3), 617–625. 10.1006/bbrc.2002.6692. [DOI] [PubMed] [Google Scholar]
- Long J. Z.; Cisar J. S.; Milliken D.; Niessen S.; Wang C.; Trauger S. A.; Siuzdak G.; Cravatt B. F. Metabolomics Annotates ABHD3 as a Physiologic Regulator of Medium-Chain Phospholipids. Nat. Chem. Biol. 2011, 7 (11), 763–765. 10.1038/nchembio.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Espérance S.; Bachvarova M.; Tetu B.; Mes-Masson A.-M.; Bachvarov D. Global Gene Expression Analysis of Early Response to Chemotherapy Treatment in Ovarian Cancer Spheroids. BMC Genomics 2008, 9 (1), 99. 10.1186/1471-2164-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B.; Huntley D.; AbuAli G.; Langley S. R.; Sindelar G.; Petretto E.; Butcher S.; Grimm S. Determining Signalling Nodes for Apoptosis by a Genetic High-Throughput Screen. PLoS One 2011, 6 (9), e25023 10.1371/journal.pone.0025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rechem C.; Rood B. R.; Touka M.; Pinte S.; Jenal M.; Guérardel C.; Ramsey K.; Monté D.; Bégue A.; Tschan M. P.; Stephan D. A.; Leprince D. Scavenger Chemokine (CXC Motif) Receptor 7 (CXCR7) Is a Direct Target Gene of HIC1 (Hypermethylated in Cancer 1). J. Biol. Chem. 2009, 284 (31), 20927–20935. 10.1074/jbc.M109.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook J. S.; Rakwal R.; Shibato J.; Takahashi K.; Koizumi H.; Shima T.; Ikemoto M. J.; Oharomari L. K.; McEwen B. S.; Soya H. Leptin in Hippocampus Mediates Benefits of Mild Exercise by an Antioxidant on Neurogenesis and Memory. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (22), 10988–10993. 10.1073/pnas.1815197116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Becker N. Q.; Moss A. C. In Silico Analysis of T-Bet Activity in Peripheral Blood Mononuclear Cells in Patients with Inflammatory Bowel Disease (IBD). In Silico Biol. 2009, 9 (5,6), 355–363. 10.3233/ISB-2009-0410. [DOI] [PubMed] [Google Scholar]
- Johnson E. C.; Doser T. A.; Cepurna W. O.; Dyck J. A.; Jia L.; Guo Y.; Lambert W. S.; Morrison J. C. Cell Proliferation and Interleukin-6–Type Cytokine Signaling Are Implicated by Gene Expression Responses in Early Optic Nerve Head Injury in Rat Glaucoma. Invest. Ophthalmol. Visual Sci. 2011, 52 (1), 504–518. 10.1167/iovs.10-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.; Cognetta A. B. III; Diaz D. B.; Lum K. M.; Adachi S.; Kundu S.; Cravatt B. F.; Yudin A. K. Multicomponent Mapping of Boron Chemotypes Furnishes Selective Enzyme Inhibitors. Nat. Commun. 2017, 8 (1), 1–8. 10.1038/s41467-017-01319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognetta A. B.; Niphakis M. J.; Lee H.-C.; Martini M. L.; Hulce J. J.; Cravatt B. F. Selective N-Hydroxyhydantoin Carbamate Inhibitors of Mammalian Serine Hydrolases. Chem. Biol. 2015, 22 (7), 928–937. 10.1016/j.chembiol.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G. M.; Cravatt B. F. Endocannabinoid Biosynthesis Proceeding through Glycerophospho-N-Acyl Ethanolamine and a Role for α/β-Hydrolase 4 in This Pathway. J. Biol. Chem. 2006, 281 (36), 26465–26472. 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- Lee H. C.; Simon G. M.; Cravatt B. F. ABHD4 Regulates Multiple Classes of N -Acyl Phospholipids in the Mammalian Central Nervous System. Biochemistry 2015, 54 (15), 2539–2549. 10.1021/acs.biochem.5b00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady C. A.; Jiang D.; Mello S. S.; Johnson T. M.; Jarvis L. A.; Kozak M. M.; Broz D. K.; Basak S.; Park E. J.; McLaughlin M. E.; Karnezis A. N.; Attardi L. D. Distinct P53 Transcriptional Programs Dictate Acute DNA-Damage Responses and Tumor Suppression. Cell 2011, 145 (4), 571–583. 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C. D.; Hurren R.; Kasimer D.; MacLean N.; Eberhard Y.; Ketela T.; Moffat J.; Schimmer A. D. A Genome Wide ShRNA Screen Identifies α/β Hydrolase Domain Containing 4 (ABHD4) as a Novel Regulator of Anoikis Resistance. Apoptosis 2012, 17 (7), 666–678. 10.1007/s10495-012-0723-4. [DOI] [PubMed] [Google Scholar]
- László Z. I.; Lele Z.; Zöldi M.; Miczán V.; Mógor F.; Simon G. M.; Mackie K.; Kacskovics I.; Cravatt B. F.; Katona I. ABHD4-Dependent Developmental Anoikis Safeguards the Embryonic Brain. Nat. Commun. 2020, 11 (1), 4363. 10.1038/s41467-020-18175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre C.; Jobard F.; Caux F.; Bouadjar B.; Karaduman A.; Heilig R.; Lakhdar H.; Wollenberg A.; Verret J.-L.; Weissenbach J.; Özgüc M.; Lathrop M.; Prud’homme J.-F.; Fischer J. Mutations in CGI-58, the Gene Encoding a New Protein of the Esterase/Lipase/Thioesterase Subfamily, in Chanarin-Dorfman Syndrome. Am. J. Hum. Genet. 2001, 69 (5), 1002–1012. 10.1086/324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missaglia S.; Coleman R.; Mordente A.; Tavian D. Neutral Lipid Storage Diseases as Cellular Model to Study Lipid Droplet Function. Cells 2019, 8 (2), 187. 10.3390/cells8020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian L.; Vahidnezhad H.; Saeidian A. H.; Pajouhanfar S.; Sotoudeh S.; Mansouri P.; Amirkashani D.; Zeinali S.; Levine M. A.; Peris K.; Colombo R.; Uitto J. Inherited Non-Alcoholic Fatty Liver Disease and Dyslipidemia Due to Monoallelic ABHD5Mutations. J. Hepatol. 2019, 71 (2), 366–370. 10.1016/j.jhep.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adant I.; Declercq M.; Bird M.; Bauters M.; Boeckx N.; Devriendt K.; Cassiman D.; Witters P. Two Cases of Non-Alcoholic Fatty Liver Disease Caused by Biallelic ABHD5Mutations. J. Hepatol. 2020, 72 (5), 1030–1032. 10.1016/j.jhep.2019.12.013. [DOI] [PubMed] [Google Scholar]
- Lass A.; Zimmermann R.; Haemmerle G.; Riederer M.; Schoiswohl G.; Schweiger M.; Kienesberger P.; Strauss J. G.; Gorkiewicz G.; Zechner R. Adipose Triglyceride Lipase-Mediated Lipolysis of Cellular Fat Stores Is Activated by CGI-58 and Defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006, 3 (5), 309–319. 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Radner F. P. W.; Streith I. E.; Schoiswohl G.; Schweiger M.; Kumari M.; Eichmann T. O.; Rechberger G.; Koefeler H. C.; Eder S.; Schauer S.; Theussl H. C.; Preiss-Landl K.; Lass A.; Zimmermann R.; Hoefler G.; Zechner R.; Haemmerle G. Growth Retardation, Impaired Triacylglycerol Catabolism, Hepatic Steatosis, and Lethal Skin Barrier Defect in Mice Lacking Comparative Gene Identification-58 (CGI-58). J. Biol. Chem. 2010, 285 (10), 7300–7311. 10.1074/jbc.M109.081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. M.; Betters J. L.; Lord C.; Ma Y.; Han X.; Yang K.; Alger H. M.; Melchior J.; Sawyer J.; Shah R.; Wilson M. D.; Liu X.; Graham M. J.; Lee R.; Crooke R.; Shulman G. I.; Xue B.; Shi H.; YuYu L. CGI-58 Knockdown in Mice Causes Hepatic Steatosis but Prevents Diet-Induced Obesity and Glucose Intolerance. J. Lipid Res. 2010, 51 (11), 3306–3315. 10.1194/jlr.M010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G. Defective Lipolysis and Altered Energy Metabolism in Mice Lacking Adipose Triglyceride Lipase. Science 2006, 312 (5774), 734–737. 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- Lord C. C.; Betters J. L.; Ivanova P. T.; Milne S. B.; Myers D. S.; Madenspacher J.; Thomas G.; Chung S.; Liu M.; Davis M. A.; Lee R. G.; Crooke R. M.; Graham M. J.; Parks J. S.; Brasaemle D. L.; Fessler M. B.; Brown H. A.; Brown J. CGI-58/ABHD5-Derived Signaling Lipids Regulate Systemic Inflammation and Insulin Action. Diabetes 2012, 61 (2), 355–363. 10.2337/db11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviglia J. M.; Betters J. L.; Dapito D.-H.; Lord C. C.; Sullivan S.; Chua S.; Yin T.; Sekowski A.; Mu H.; Shapiro L.; Brown J. M.; Brasaemle D. L. Adipose-Selective Overexpression of ABHD5/CGI-58 Does Not Increase Lipolysis or Protect against Diet-Induced Obesity. J. Lipid Res. 2011, 52 (11), 2032–2042. 10.1194/jlr.M019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C. C.; Brown J. M. Distinct Roles for α-β Hydrolase Domain 5 (ABHD5/CGI-58) and Adipose Triglyceride Lipase (ATGL/PNPLA2) in Lipid Metabolism and Signaling. Adipocyte 2012, 1 (3), 123–131. 10.4161/adip.20035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V.; Rothenberg A.; Gomez C.; Cohen A. W.; Garcia A.; Bhattacharyya S.; Shapiro L.; Dolios G.; Wang R.; Lisanti M. P.; Brasaemle D. L. Perilipin A Mediates the Reversible Binding of CGI-58 to Lipid Droplets in 3T3-L1 Adipocytes. J. Biol. Chem. 2004, 279 (40), 42062–42071. 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T.; Omatsu N.; Matsushita S.; Osumi T. CGI-58 Interacts with Perilipin and Is Localized to Lipid Droplets. J. Biol. Chem. 2004, 279 (29), 30490–30497. 10.1074/jbc.M403920200. [DOI] [PubMed] [Google Scholar]
- Granneman J. G.; Moore H.-P. H.; Krishnamoorthy R.; Rathod M. Perilipin Controls Lipolysis by Regulating the Interactions of AB-Hydrolase Containing 5 (Abhd5) and Adipose Triglyceride Lipase (Atgl). J. Biol. Chem. 2009, 284 (50), 34538–34544. 10.1074/jbc.M109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T.; Omatsu N.; Morimoto E.; Nakashima H.; Ueno K.; Tanaka T.; Satouchi K.; Hirose F.; Osumi T. CGI-58 Facilitates Lipolysis on Lipid Droplets but Is Not Involved in the Vesiculation of Lipid Droplets Caused by Hormonal Stimulation. J. Lipid Res. 2007, 48 (5), 1078–1089. 10.1194/jlr.M600493-JLR200. [DOI] [PubMed] [Google Scholar]
- Gandotra S.; Lim K.; Girousse A.; Saudek V.; O’Rahilly S.; Savage D. B. Human Frame Shift Mutations Affecting the Carboxyl Terminus of Perilipin Increase Lipolysis by Failing to Sequester the Adipose Triglyceride Lipase (ATGL) Coactivator AB-Hydrolase-Containing 5 (ABHD5)*. J. Biol. Chem. 2011, 286 (40), 34998–35006. 10.1074/jbc.M111.278853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman J. G.; Moore H.-P. H.; Mottillo E. P.; Zhu Z.; Zhou L. Interactions of Perilipin-5 (Plin5) with Adipose Triglyceride Lipase. J. Biol. Chem. 2011, 286 (7), 5126–5135. 10.1074/jbc.M110.180711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman J. G.; Moore H.-P. H.; Mottillo E. P.; Zhu Z. Functional Interactions between Mldp (LSDP5) and Abhd5 in the Control of Intracellular Lipid Accumulation. J. Biol. Chem. 2009, 284 (5), 3049–3057. 10.1074/jbc.M808251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak N. M.; Jaeger D.; Kolleritsch S.; Zimmermann R.; Zechner R.; Lass A.; Haemmerle G. The Interplay of Protein Kinase A and Perilipin 5 Regulates Cardiac Lipolysis. J. Biol. Chem. 2015, 290 (3), 1295–1306. 10.1074/jbc.M114.604744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A.; Mottillo E. P.; Mladenovic-Lucas L.; Zhou L.; Granneman J. G. Dynamic Interactions of ABHD5 with PNPLA3 Regulate Triacylglycerol Metabolism in Brown Adipocytes. Nat. Metab. 2019, 1 (5), 560–569. 10.1038/s42255-019-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S.; Savage D. B. Lipase Tug of War: PNPLA3 Sequesters ABHD5 from ATGL. Nat. Metab. 2019, 1 (5), 505–506. 10.1038/s42255-019-0067-2. [DOI] [PubMed] [Google Scholar]
- Chen G.; Zhou G.; Aras S.; He Z.; Lucas S.; Podgorski I.; Skar W.; Granneman J. G.; Wang J. Loss of ABHD5 Promotes the Aggressiveness of Prostate Cancer Cells. Sci. Rep. 2017, 7 (1), 13021. 10.1038/s41598-017-13398-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J.; Miao H.; Ma Y.; Guo F.; Deng J.; Wei X.; Zhou J.; Xie G.; Shi H.; Xue B.; Liang H.; Yu L. Loss of Abhd5 Promotes Colorectal Tumor Development and Progression by Inducing Aerobic Glycolysis and Epithelial-Mesenchymal Transition. Cell Rep. 2014, 9 (5), 1798–1811. 10.1016/j.celrep.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H.; Ou J.; Peng Y.; Zhang X.; Chen Y.; Hao L.; Xie G.; Wang Z.; Pang X.; Ruan Z.; Li J.; Yu L.; Xue B.; Shi H.; Shi C.; Liang H. Macrophage ABHD5 Promotes Colorectal Cancer Growth by Suppressing Spermidine Production by SRM. Nat. Commun. 2016, 7 (1), 11716. 10.1038/ncomms11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang S.; Ji X.; Zhang L.; Chen J.; Li C.; Shi R.; Xiang W.; Kang X.; Zhang D.; Yang F.; Dai R.; Chen P.; Chen S.; Chen Y.; Li Y.; Miao H. Macrophage ABHD5 Suppresses NF-ΚB-Dependent Matrix Metalloproteinase Expression and Cancer Metastasis. Cancer Res. 2019, 79 (21), 5513–5526. 10.1158/0008-5472.CAN-19-1059. [DOI] [PubMed] [Google Scholar]
- Mitra R.; Le T. T.; Gorjala P.; Goodman O. B. Positive Regulation of Prostate Cancer Cell Growth by Lipid Droplet Forming and Processing Enzymes DGAT1 and ABHD5. BMC Cancer 2017, 17 (1), 631. 10.1186/s12885-017-3589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Zhou G.; Lotvola A.; Granneman J. G.; Wang J. ABHD5 Suppresses Cancer Cell Anabolism through Lipolysis-Dependent Activation of the AMPK/MTORC1 Pathway. J. Biol. Chem. 2021, 296, 100104. 10.1074/jbc.RA120.014682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q.; Wang F.; Zhou K.; Huang K.; Zhu Q.; Luo X.; Yu J.; Shi Z. Oncogenic Role of ABHD5 in Endometrial Cancer. Cancer Manage. Res. 2019, 11, 2139–2150. 10.2147/CMAR.S188648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers J. G.; McKinsey T. A. ABHD5 Cleaves HDAC4 to Benefit the Heart. Nat. Metab. 2019, 1 (11), 1034–1035. 10.1038/s42255-019-0141-9. [DOI] [PubMed] [Google Scholar]
- Jebessa Z. H.; Shanmukha K. D.; Dewenter M.; Lehmann L. H.; Xu C.; Schreiter F.; Siede D.; Gong X. M.; Worst B. C.; Federico G.; Sauer S. W.; Fischer T.; Wechselberger L.; Müller O. J.; Sossalla S.; Dieterich C.; Most P.; Gröne H. J.; Moro C.; Oberer M.; Haemmerle G.; Katus H. A.; Tyedmers J.; Backs J. The Lipid-Droplet-Associated Protein ABHD5 Protects the Heart through Proteolysis of HDAC4. Nat. Metab. 2019, 1 (11), 1157–1167. 10.1038/s42255-019-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. A.; Madoux F.; Mladenovic L.; Zhang H.; Ye X.; Angrish M.; Mottillo E. P.; Caruso J. A.; Halvorsen G.; Roush W. R.; Chase P.; Hodder P.; Granneman J. G. Endogenous and Synthetic ABHD5 Ligands Regulate ABHD5-Perilipin Interactions and Lipolysis in Fat and Muscle. Cell Metab. 2015, 22 (5), 851–860. 10.1016/j.cmet.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondini E. A.; Mladenovic-Lucas L.; Roush W. R.; Halvorsen G. T.; Green A. E.; Granneman J. G. Novel Pharmacological Probes Reveal ABHD5 as a Locus of Lipolysis Control in White and Brown Adipocytes. J. Pharmacol. Exp. Ther. 2017, 363 (3), 367–376. 10.1124/jpet.117.243253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Scripps Research Institute; Wayne State University . Small molecule modulators of cellular lipolysis. WO 2017053510, 2017.
- Navia-Paldanius D.; Savinainen J. R.; Laitinen J. T. Biochemical and Pharmacological Characterization of Human α/β-Hydrolase Domain Containing 6 (ABHD6) and 12 (ABHD12). J. Lipid Res. 2012, 53 (11), 2413–2424. 10.1194/jlr.M030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggelaar M. P.; van Esbroeck A. C. M.; van Rooden E. J.; Florea B. I.; Overkleeft H. S.; Marsicano G.; Chaouloff F.; van der Stelt M. Chemical Proteomics Maps Brain Region Specific Activity of Endocannabinoid Hydrolases. ACS Chem. Biol. 2017, 12 (3), 852–861. 10.1021/acschembio.6b01052. [DOI] [PubMed] [Google Scholar]
- Thomas G.; Betters J. L.; Lord C. C.; Brown A. L.; Marshall S.; Ferguson D.; Sawyer J.; Davis M. A.; Melchior J. T.; Blume L. C.; Howlett A.; Ivanova P. T.; Milne S. B.; Myers D. S.; Mrak I.; Leber V.; Heier C.; Taschler U.; Blankman J. L.; Cravatt B. F.; Lee R. G.; Crooke R. M.; Graham M. J; Zimmermann R.; Brown H.; Brown J. M. The Serine Hydrolase ABHD6 Is a Critical Regulator of the Metabolic Syndrome. Cell Rep. 2013, 5 (2), 508–520. 10.1016/j.celrep.2013.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Fei X.; Xu J.; Ji C. An Unannotated α/β Hydrolase Superfamily Member, ABHD6 Differentially Expressed among Cancer Cell Lines. Mol. Biol. Rep. 2009, 36 (4), 691–696. 10.1007/s11033-008-9230-7. [DOI] [PubMed] [Google Scholar]
- Drehmer M. N.; Muniz Y. C. N.; Marrero A. R.; Löfgren S. E. Gene Expression of ABHD6, a Key Factor in the Endocannabinoid System, Can Be Modulated by Female Hormones in Human Immune Cells. Biochem. Genet. 2019, 57 (1), 35–45. 10.1007/s10528-018-9871-8. [DOI] [PubMed] [Google Scholar]
- Poursharifi P.; Madiraju S. R. M.; Prentki M. Monoacylglycerol Signalling and ABHD6 in Health and Disease. Diabetes, Obes. Metab. 2017, 19, 76–89. 10.1111/dom.13008. [DOI] [PubMed] [Google Scholar]
- Cao J. K.; Kaplan J.; Stella N. ABHD6: Its Place in Endocannabinoid Signaling and Beyond. Trends Pharmacol. Sci. 2019, 40 (4), 267–277. 10.1016/j.tips.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.; Li W. Therapeutic Potential of Targeting α/β-Hydrolase Domain-Containing 6 (ABHD6). Eur. J. Med. Chem. 2020, 198, 112353. 10.1016/j.ejmech.2020.112353. [DOI] [PubMed] [Google Scholar]
- Max D.; Hesse M.; Volkmer I.; Staege M. S. High Expression of the Evolutionarily Conserved α/β Hydrolase Domain Containing 6 (ABHD6) in Ewing Tumors. Cancer Sci. 2009, 100 (12), 2383–2389. 10.1111/j.1349-7006.2009.01347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüner B. M.; Schulze C. J.; Yang D.; Ogasawara D.; Dix M. M.; Rogers Z. N.; Chuang C.-H.; McFarland C. D.; Chiou S.-H.; Brown J. M.; Cravatt B. F.; Bogyo M.; Winslow M. An in Vivo Multiplexed Small-Molecule Screening Platform. Nat. Methods 2016, 13 (10), 883–889. 10.1038/nmeth.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.; Xie H.; Heier C.; Huang J.; Zheng Q.; Eichmann T. O.; Schoiswohl G.; Ni J.; Zechner R.; Ni S.; Hao H. Enhanced Monoacylglycerol Lipolysis by ABHD6 Promotes NSCLC Pathogenesis. EBioMedicine 2020, 53, 102696. 10.1016/j.ebiom.2020.102696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-B.; Zhu L.-Y.; Wang Y.-G.; Li F.; Zhang X.-Y.; Dai W.-J. Systemic Transcriptome Analysis of Hepatocellular Carcinoma. Tumor Biol. 2016, 37 (10), 13323–13331. 10.1007/s13277-016-5286-5. [DOI] [PubMed] [Google Scholar]
- van Esbroeck A. C. M.; Kantae V.; Di X.; van der Wel T.; den Dulk H.; Stevens A. F.; Singh S.; Bakker A. T.; Florea B. I.; Stella N.; Overkleeft H. S.; Hankemeier T.; van der Stelt M. Identification of α,β-Hydrolase Domain Containing Protein 6 as a Diacylglycerol Lipase in Neuro-2a Cells. Front. Mol. Neurosci. 2019, 12, 286. 10.3389/fnmol.2019.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.; Mugabo Y.; Iglesias J.; Xie L.; Delghingaro-Augusto V.; Lussier R.; Peyot M.-L.; Joly E.; Taïb B.; Davis M. A.; Brown J. M.; Abousalham A.; Gaisano H.; Madiraju S. R. M.; Prentki M. α/β-Hydrolase Domain-6-Accessible Monoacylglycerol Controls Glucose-Stimulated Insulin Secretion. Cell Metab. 2014, 19 (6), 993–1007. 10.1016/j.cmet.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Zhao S.; Poursharifi P.; Mugabo Y.; Levens E. J.; Vivot K.; Attane C.; Iglesias J.; Peyot M.; Joly E.; Madiraju S. R. M.; Prentki M. α/β-Hydrolase Domain-6 and Saturated Long Chain Monoacylglycerol Regulate Insulin Secretion Promoted by Both Fuel and Non-Fuel Stimuli. Mol. Metab. 2015, 4 (12), 940–950. 10.1016/j.molmet.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.; Mugabo Y.; Ballentine G.; Attane C.; Iglesias J.; Poursharifi P.; Zhang D.; Nguyen T. A.; Erb H.; Prentki R.; Peyot M.-L.; Joly E..; Tobin S.; Fulton S.; Brown J. M.; Madiraju S. R. M; Prentki M. α/β-Hydrolase Domain 6 Deletion Induces Adipose Browning and Prevents Obesity and Type 2 Diabetes. Cell Rep. 2016, 14 (12), 2872–2888. 10.1016/j.celrep.2016.02.076. [DOI] [PubMed] [Google Scholar]
- Fisette A.; Tobin S.; Décarie-Spain L.; Bouyakdan K.; Peyot M.-L.; Madiraju S. R. M.; Prentki M.; Fulton S.; Alquier T. α/β-Hydrolase Domain 6 in the Ventromedial Hypothalamus Controls Energy Metabolism Flexibility. Cell Rep. 2016, 17 (5), 1217–1226. 10.1016/j.celrep.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Maier S.; Staffler G.; Hartmann A.; Höck J.; Henning K.; Grabusic K.; Mailhammer R.; Hoffmann R.; Wilmanns M.; Lang R.; Mages J.; Kempkes J. Cellular Target Genes of Epstein-Barr Virus Nuclear Antigen 2. J. Virol. 2006, 80 (19), 9761–9771. 10.1128/JVI.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparina N. Y.; Delgado-Vega A. M.; Martinez-Bueno M.; Magro-Checa C.; Fernández C.; Castro R. O.; Pons-Estel B. A.; D’Alfonso S.; Sebastiani G. D.; Witte T.; Lauwerys B. R.; Endreffy E.; Kovács L.; Escudero A.; López-Pedrera C.; Vasconcelos C.; da Silva B. M.; Frostegård J.; Truedsson L.; Martin J.; Raya E.; Ortego-Centeno N.; de los Angeles Aguirre M.; de Ramón Garrido E.; Palma M.-J. C.; Alarcon-Riquelme M. E.; Kozyrev S. V. PXK Locus in Systemic Lupus Erythematosus: Fine Mapping and Functional Analysis Reveals Novel Susceptibility Gene ABHD6. Ann. Rheum. Dis. 2015, 74 (3), e14 10.1136/annrheumdis-2013-204909. [DOI] [PubMed] [Google Scholar]
- Savinainen J. R.; Saario S. M.; Laitinen J. T. The Serine Hydrolases MAGL, ABHD6 and ABHD12 as Guardians of 2-Arachidonoylglycerol Signalling through Cannabinoid Receptors. Acta Physiol. 2012, 204 (2), 267–276. 10.1111/j.1748-1716.2011.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs W. R.; Blankman J. L.; Horne E. A.; Thomazeau A.; Lin Y. H.; Coy J.; Bodor A. L.; Muccioli G. G.; Hu S. S.-J.; Woodruff G.; Fung S.; Lafourcade M.; Alexander J. P.; Long J. Z.; Li W.; Xu C.; Möller T.; Mackie K.; Manzoni O. J.; Cravatt B. F.; Stella N. The Serine Hydrolase ABHD6 Controls the Accumulation and Efficacy of 2-AG at Cannabinoid Receptors. Nat. Neurosci. 2010, 13 (8), 951–957. 10.1038/nn.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navia-Paldanius D.; Aaltonen N.; Lehtonen M.; Savinainen J. R.; Taschler U.; Radner F. P. W.; Zimmermann R.; Laitinen J. T. Increased Tonic Cannabinoid CB1R Activity and Brain Region-Specific Desensitization of CB1R Gi/o Signaling Axis in Mice with Global Genetic Knockout of Monoacylglycerol Lipase. Eur. J. Pharm. Sci. 2015, 77, 180–188. 10.1016/j.ejps.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Long J. Z.; Li W.; Booker L.; Burston J. J.; Kinsey S. G.; Schlosburg J. E.; Pavón F. J.; Serrano A. M.; Selley D. E.; Parsons L. H.; Lichtman A.; Cravatt B. F. Selective Blockade of 2-Arachidonoylglycerol Hydrolysis Produces Cannabinoid Behavioral Effects. Nat. Chem. Biol. 2009, 5 (1), 37–44. 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosburg J. E.; Blankman J. L.; Long J. Z.; Nomura D. K.; Pan B.; Kinsey S. G.; Nguyen P. T.; Ramesh D.; Booker L.; Burston J. J.; Thomas E. A.; Selley D. E.; Sim-Selley L. J.; Liu Q. S.; Lichtman A. H.; Cravatt B. F. Chronic Monoacylglycerol Lipase Blockade Causes Functional Antagonism of the Endocannabinoid System. Nat. Neurosci. 2010, 13 (9), 1113–1119. 10.1038/nn.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhouayek M.; Masquelier J.; Cani P. D.; Lambert D. M.; Muccioli G. G. Implication of the Anti-Inflammatory Bioactive Lipid Prostaglandin D2-Glycerol Ester in the Control of Macrophage Activation and Inflammation by ABHD6. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (43), 17558–17563. 10.1073/pnas.1314017110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquelier J.; Alhouayek M.; Terrasi R.; Bottemanne P.; Paquot A.; Muccioli G. G. Lysophosphatidylinositols in Inflammation and Macrophage Activation: Altered Levels and Anti-Inflammatory Effects. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2018, 1863 (12), 1458–1468. 10.1016/j.bbalip.2018.09.003. [DOI] [PubMed] [Google Scholar]
- Pribasnig M. A.; Mrak I.; Grabner G. F.; Taschler U.; Knittelfelder O.; Scherz B.; Eichmann T. O.; Heier C.; Grumet L.; Kowaliuk J.; Romauch M.; Holler S.; Anderl F.; Wolinski H.; Lass A.; Breinbauer R.; MarscheMarsche G.; Brown J. M.; Zimmermann R. α/β Hydrolase Domain-Containing 6 (ABHD6) Degrades the Late Endosomal/Lysosomal Lipid Bis(Monoacylglycero)Phosphate. J. Biol. Chem. 2015, 290 (50), 29869–29881. 10.1074/jbc.M115.669168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner G. F.; Fawzy N.; Pribasnig M. A.; Trieb M.; Taschler U.; Holzer M.; Schweiger M.; Wolinski H.; Kolb D.; Horvath A.; Breinbauer R.; Rülicke T.; Rabl R..; LassLass A.; Stadlbauer V.; Hutter-Paier B.; Stauber R. E.; Fickert P.; Zechner R.; Marsche G..; Eichmann T. O.; Zimmermann R. Metabolic Disease and ABHD6 Alter the Circulating Bis(Monoacylglycerol)Phosphate Profile in Mice and Humans. J. Lipid Res. 2019, 60 (5), 1020–1031. 10.1194/jlr.M093351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola A.; Bernal-Chico A.; Cipriani R.; Canedo-Antelo M.; Moreno-García Á.; Martín-Fontecha M.; Pérez-Cerdá F.; Sánchez-Gómez M. V.; Ortega-Gutiérrez S.; Brown J. M.; Hsu K.-L; CravattCravatt B.; Matute C.; Mato S. Deregulation of the Endocannabinoid System and Therapeutic Potential of ABHD6 Blockade in the Cuprizone Model of Demyelination. Biochem. Pharmacol. 2018, 157, 189–201. 10.1016/j.bcp.2018.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J.; Ribeiro R.; Tanaka M.; Zhang Y. Activation of CB2 Receptor Is Required for the Therapeutic Effect of ABHD6 Inhibition in Experimental Autoimmune Encephalomyelitis. Neuropharmacology 2015, 99, 196–209. 10.1016/j.neuropharm.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Manterola A.; Bernal-Chico A.; Cipriani R.; Ruiz A.; Pérez-Samartín A.; Moreno-Rodríguez M.; Hsu K. L.; Cravatt B. F.; Brown J. M.; Rodríguez-Puertas R.; Matute C.; Mato S. Re-Examining the Potential of Targeting ABHD6 in Multiple Sclerosis: Efficacy of Systemic and Peripherally Restricted Inhibitors in Experimental Autoimmune Encephalomyelitis. Neuropharmacology 2018, 141, 181–191. 10.1016/j.neuropharm.2018.08.038. [DOI] [PubMed] [Google Scholar]
- Wei M.; Zhang J.; Jia M.; Yang C.; Pan Y.; Li S.; Luo Y.; Zheng J.; Ji J.; Chen J.; Hu X.; Xiong J.; Shi Y.; Zhang C. α/β-Hydrolase Domain-Containing 6 (ABHD6) Negatively Regulates the Surface Delivery and Synaptic Function of AMPA Receptors. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (19), E2695–E2704. 10.1073/pnas.1524589113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M.; Jia M.; Zhang J.; Yu L.; Zhao Y.; Chen Y.; Ma Y.; Zhang W.; Shi Y. S.; Zhang C. The Inhibitory Effect of α/β-Hydrolase Domain-Containing 6 (ABHD6) on the Surface Targeting of GluA2- and GluA3-Containing AMPA Receptors. Front. Mol. Neurosci. 2017, 10, 55. 10.3389/fnmol.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchantchou F.; Zhang Y. Selective Inhibition of Alpha/Beta-Hydrolase Domain 6 Attenuates Neurodegeneration, Alleviates Blood Brain Barrier Breakdown, and Improves Functional Recovery in a Mouse Model of Traumatic Brain Injury. J. Neurotrauma 2013, 30 (7), 565–579. 10.1089/neu.2012.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenov A. V.; Horne E. A.; Cheah C. S.; Swinney K.; Hsu K.-L.; Cao J. K.; Marrs W. R.; Blankman J. L.; Tu S.; Cherry A. E.; Fung S.; Wen A.; Li W.; Saporito M. S.; Selley D. E.; Cravatt B. F.; Oakley J. C.; Stella N. ABHD6 Blockade Exerts Antiepileptic Activity in PTZ-Induced Seizures and in Spontaneous Seizures in R6/2 Mice. Neuron 2014, 83 (2), 361–371. 10.1016/j.neuron.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Blankman J. L.; Cravatt B. F. A Functional Proteomic Strategy to Discover Inhibitors for Uncharacterized Hydrolases. J. Am. Chem. Soc. 2007, 129 (31), 9594–9595. 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- Kiritoshi T.; Ji G.; Neugebauer V. Rescue of Impaired MGluR5-Driven Endocannabinoid Signaling Restores Prefrontal Cortical Output to Inhibit Pain in Arthritic Rats. J. Neurosci. 2016, 36 (3), 837–850. 10.1523/JNEUROSCI.4047-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M.; Moran S.; Wen J.; Affram K.; Chen T.; Symes A. J.; Zhang Y. WWL70 Attenuates PGE2 Production Derived from 2-Arachidonoylglycerol in Microglia by ABHD6-Independent Mechanism. J. Neuroinflammation 2017, 14 (1), 7. 10.1186/s12974-016-0783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J.; Jones M.; Tanaka M.; Selvaraj P.; Symes A. J.; Cox B.; Zhang Y. WWL70 Protects against Chronic Constriction Injury-Induced Neuropathic Pain in Mice by Cannabinoid Receptor-Independent Mechanisms. J. Neuroinflammation 2018, 15 (1), 9. 10.1186/s12974-017-1045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centre Hospitalier de l’Universite de Montreal . Screening assays based on MAG and/or ABHD6 for selecting insulin promoting agents. WO 2011140659, 2011.
- Centre Hospitalier de l’Universite de Montreal . Insulin secretion promoting agents. WO 2013016807, 2013.
- Val-Chum Limited Partnership . ABHD6 antagonists for promoting browning of white adipose tissue and brown adipose tissue functionality. WO 2015127559, 2015.
- Bachovchin D. A.; Ji T.; Li W.; Simon G. M.; Blankman J. L.; Adibekian A.; Hoover H.; Niessen S.; Cravatt B. F. Superfamily-Wide Portrait of Serine Hydrolase Inhibition Achieved by Library-versus-Library Screening. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (49), 20941–20946. 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J. Z.; Nevalainen T. J.; Savinainen J. R.; Adams Y.; Laitinen T.; Runyon R. S.; Vaara M.; Ahenkorah S.; Kaczor A. A.; Navia-Paldanius D.; Gynther M.; Aaltonen N.; Joharapurkar A. A.; Jain M. R.; Haka A. S.; Maxfield F. R.; Laitinen J. T.; Parkkari T. Optimization of 1,2,5-Thiadiazole Carbamates as Potent and Selective ABHD6 Inhibitors. ChemMedChem 2015, 10 (2), 253–265. 10.1002/cmdc.201402453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczor A. A.; Targowska-Duda K. M.; Patel J. Z.; Laitinen T.; Parkkari T.; Adams Y.; Nevalainen T. J.; Poso A. Comparative Molecular Field Analysis and Molecular Dynamics Studies of α/β Hydrolase Domain Containing 6 (ABHD6) Inhibitors. J. Mol. Model. 2015, 21 (10), 250. 10.1007/s00894-015-2789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abide Therapeutics Inc. Lp-PLA2 inhibitors. WO 2017059135, 2017.
- Abide Therapeutics Inc. Spyrocycle compounds and methods of making and using same. WO 2017197192, 2017.
- Adibekian A.; Martin B. R.; Wang C.; Hsu K. L.; Bachovchin D. A.; Niessen S.; Hoover H.; Cravatt B. F. Click-Generated Triazole Ureas as Ultrapotent in Vivo-Active Serine Hydrolase Inhibitors. Nat. Chem. Biol. 2011, 7 (7), 469–478. 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K. L.; Tsuboi K.; Adibekian A.; Pugh H.; Masuda K.; Cravatt B. F. DAGLβ Inhibition Perturbs a Lipid Network Involved in Macrophage Inflammatory Responses. Nat. Chem. Biol. 2012, 8 (12), 999–1007. 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K.-L.; Tsuboi K.; Whitby L. R.; Speers A. E.; Pugh H.; Inloes J.; Cravatt B. F. Development and Optimization of Piperidyl-1,2,3-Triazole Ureas as Selective Chemical Probes of Endocannabinoid Biosynthesis. J. Med. Chem. 2013, 56 (21), 8257–8269. 10.1021/jm400898x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B.; Lee H.; Ghosh M.; Cravatt B. F.; Hsu K. L.; Bonventre J. V.; Ewing H.; Gelb M. H.; Leslie C. C. Serine Hydrolase Inhibitors Block Necrotic Cell Death by Preventing Calcium Overload of the Mitochondria and Permeability Transition Pore Formation. J. Biol. Chem. 2014, 289 (3), 1491–1504. 10.1074/jbc.M113.497651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara D.; Deng H.; Viader A.; Baggelaar M. P.; Breman A.; Den Dulk H.; Van Den Nieuwendijk A. M. C. H.; Soethoudt M.; Van Der Wel T.; Zhou J.; Overkleeft H. S.; Sanchez-Alavez M.; Mo S.; Nguyen W.; Conti B.; Liu X.; Chen Y.; Liu Q. S.; Cravatt B. F.; Van Der Stelt M. Rapid and Profound Rewiring of Brain Lipid Signaling Networks by Acute Diacylglycerol Lipase Inhibition. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (1), 26–33. 10.1073/pnas.1522364112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooden E. J.; Florea B. I.; Deng H.; Baggelaar M. P.; van Esbroeck A. C. M.; Zhou J.; Overkleeft H. S.; van der Stelt M. Mapping in Vivo Target Interaction Profiles of Covalent Inhibitors Using Chemical Proteomics with Label-Free Quantification. Nat. Protoc. 2018, 13 (4), 752–767. 10.1038/nprot.2017.159. [DOI] [PubMed] [Google Scholar]
- Hsu K. L.; Tsuboi K.; Chang J. W.; Whitby L. R.; Speers A. E.; Pugh H.; Cravatt B. F. Discovery and Optimization of Piperidyl-1,2,3-Triazole Ureas as Potent, Selective, and in Vivo-Active Inhibitors of α/β-Hydrolase Domain Containing 6 (ABHD6). J. Med. Chem. 2013, 56 (21), 8270–8279. 10.1021/jm400899c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.; van der Wel T.; van den Berg R. J. B. H. N.; van den Nieuwendijk A. M. C. H.; Janssen F. J.; Baggelaar M. P.; Overkleeft H. S.; van der Stelt M. Chiral Disubstituted Piperidinyl Ureas: A Class of Dual Diacylglycerol Lipase-α and ABHD6 Inhibitors. MedChemComm 2017, 8 (5), 982–988. 10.1039/C7MD00029D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.; Kooijman S.; van den Nieuwendijk A. M. C. H.; Ogasawara D.; van der Wel T.; van Dalen F.; Baggelaar M. P.; Janssen F. J.; van den Berg R. J. B. H. N.; den Dulk H.; Cravatt B. F.; Overkleeft H. S.; Rensen P. C. N.; van der Stelt M. Triazole Ureas Act as Diacylglycerol Lipase Inhibitors and Prevent Fasting-Induced Refeeding. J. Med. Chem. 2017, 60 (1), 428–440. 10.1021/acs.jmedchem.6b01482. [DOI] [PubMed] [Google Scholar]
- van Rooden E. J.; Kohsiek M.; Kreekel R.; van Esbroeck A. C. M.; van den Nieuwendijk A. M. C. H.; Janssen A. P. A.; van den Berg R. J. B. H. N.; Overkleeft H. S.; van der Stelt M. Design and Synthesis of Quenched Activity-Based Probes for Diacylglycerol Lipase and α,β-Hydrolase Domain Containing Protein 6. Chem. - Asian J. 2018, 13 (22), 3491–3500. 10.1002/asia.201800452. [DOI] [PubMed] [Google Scholar]
- Marrs W. R.; Horne E. A.; Ortega-Gutierrez S.; Cisneros J. A.; Xu C.; Lin Y. H.; Muccioli G. G.; Lopez-Rodriguez M. L.; Stella N. Dual Inhibition of α/β-Hydrolase Domain 6 and Fatty Acid Amide Hydrolase Increases Endocannabinoid Levels in Neurons. J. Biol. Chem. 2011, 286 (33), 28723–28728. 10.1074/jbc.M110.202853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen F. J.; Deng H.; Baggelaar M. P.; Allarà M.; Van Der Wel T.; Den Dulk H.; Ligresti A.; Van Esbroeck A. C. M.; McGuire R.; Di Marzo V.; Overkleeft H. S.; Van Der Stelt M. Discovery of Glycine Sulfonamides as Dual Inhibitors of Sn-1-Diacylglycerol Lipase α and α/β-Hydrolase Domain 6. J. Med. Chem. 2014, 57 (15), 6610–6622. 10.1021/jm500681z. [DOI] [PubMed] [Google Scholar]
- Patel J. Z.; Parkkari T.; Laitinen T.; Kaczor A. A.; Saario S. M.; Savinainen J. R.; Navia-Paldanius D.; Cipriano M.; Leppänen J.; Koshevoy I. O.; Poso A.; Fowler C. J.; Laitinen J. T.; Nevalainen T. Chiral 1,3,4-Oxadiazol-2-Ones as Highly Selective FAAH Inhibitors. J. Med. Chem. 2013, 56 (21), 8484–8496. 10.1021/jm400923s. [DOI] [PubMed] [Google Scholar]
- Patel J. Z.; Van Bruchem J.; Laitinen T.; Kaczor A. A.; Navia-Paldanius D.; Parkkari T.; Savinainen J. R.; Laitinen J. T.; Nevalainen T. J. Revisiting 1,3,4-Oxadiazol-2-Ones: Utilization in the Development of ABHD6 Inhibitors. Bioorg. Med. Chem. 2015, 23 (19), 6335–6345. 10.1016/j.bmc.2015.08.030. [DOI] [PubMed] [Google Scholar]
- Chicca A.; Arena C.; Bertini S.; Gado F.; Ciaglia E.; Abate M.; Digiacomo M.; Lapillo M.; Poli G.; Bifulco M.; Macchia M.; Tuccinardi T.; Gertsch J.; Manera C. Polypharmacological Profile of 1,2-Dihydro-2-Oxo-Pyridine-3-Carboxamides in the Endocannabinoid System. Eur. J. Med. Chem. 2018, 154, 155–171. 10.1016/j.ejmech.2018.05.019. [DOI] [PubMed] [Google Scholar]
- Kokona D.; Spyridakos D.; Tzatzarakis M.; Papadogkonaki S.; Filidou E.; Arvanitidis K. I.; Kolios G.; Lamani M.; Makriyannis A.; Malamas M. S.; Thermos K. The Endocannabinoid 2-Arachidonoylglycerol and Dual ABHD6/MAGL Enzyme Inhibitors Display Neuroprotective and Anti-Inflammatory Actions in the in Vivo Retinal Model of AMPA Excitotoxicity. Neuropharmacology 2021, 185, 108450. 10.1016/j.neuropharm.2021.108450. [DOI] [PubMed] [Google Scholar]
- Kaczor A.; Poso A.; Pitucha M. In Vitro Screening of Some Heterocyclic Compounds Against Human ABHD6 and ABHD12 Hydrolases. Lett. Drug Des. Discovery 2014, 11 (7), 944–952. 10.2174/1570180811666140313233940. [DOI] [Google Scholar]
- Decker M.; Adamska M.; Cronin A.; Di Giallonardo F.; Burgener J.; Marowsky A.; Falck J. R.; Morisseau C.; Hammock B. D.; Gruzdev A.; Zeldin D. C.; Arand M. EH3 (ABHD9): The First Member of a New Epoxide Hydrolase Family with High Activity for Fatty Acid Epoxides. J. Lipid Res. 2012, 53 (10), 2038–2045. 10.1194/jlr.M024448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes S. L.; Gruzdev A.; Edin M. L.; Graves J. P.; Bradbury J. A.; Flake G. P.; Lih F. B.; DeGraff L. M.; Zeldin D. C. Generation and Characterization of Epoxide Hydrolase 3 (EPHX3)-Deficient Mice. PLoS One 2017, 12 (4), e0175348 10.1371/journal.pone.0175348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell S.; Jung K.; Kristiansen G.; Eltze E.; Semjonow A.; Ittmann M.; Hartmann A.; Stamey T.; Haefliger C.; Weiss G. Discovery and Validation of 3 Novel DNA Methylation Markers of Prostate Cancer Prognosis. J. Urol. 2007, 177 (5), 1753–1758. 10.1016/j.juro.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Stott-Miller M.; Zhao S.; Wright J. L.; Kolb S.; Bibikova M.; Klotzle B.; Ostrander E. A.; Fan J.-B.; Feng Z.; Stanford J. L. Validation Study of Genes with Hypermethylated Promoter Regions Associated with Prostate Cancer Recurrence. Cancer Epidemiol., Biomarkers Prev. 2014, 23 (7), 1331–1339. 10.1158/1055-9965.EPI-13-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta J.; Nobeyama Y.; Umebayashi Y.; Otsuka F.; Kikuchi K.; Ushijima T. Silencing of Peroxiredoxin 2 and Aberrant Methylation of 33 CpG Islands in Putative Promoter Regions in Human Malignant Melanomas. Cancer Res. 2006, 66 (12), 6080–6086. 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- Micci F.; Panagopoulos I.; Tjønnfjord G. E.; Kolstad A.; Delabie J.; Beiske K.; Heim S. Molecular Cytogenetic Characterization of t(14;19)(Q32;P13), a New Recurrent Translocation in B Cell Malignancies. Virchows Arch. 2007, 450 (5), 559–565. 10.1007/s00428-007-0407-6. [DOI] [PubMed] [Google Scholar]
- Yamashita S.; Tsujino Y.; Moriguchi K.; Tatematsu M.; Ushijima T. Chemical Genomic Screening for Methylation-Silenced Genes in Gastric Cancer Cell Lines Using 5-Aza-2’-Deoxycytidine Treatment and Oligonucleotide Microarray. Cancer Sci. 2006, 97 (1), 64–71. 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A.; Bell D.; Weber R. S.; El-Naggar A. K. CpG Island Methylation Profiling in Human Salivary Gland Adenoid Cystic Carcinoma. Cancer 2011, 117 (13), 2898–2909. 10.1002/cncr.25818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissi D. B.; Fabbri V. P.; Gabusi A.; Lenzi J.; Morandi L.; Melotti S.; Asioli S.; Tarsitano A.; Balbi T.; Marchetti C.; Montebugnoli L. Pre-Operative Evaluation of Dna Methylation Profile in Oral Squamous Cell Carcinoma Can Predict Tumor Aggressive Potential. Int. J. Mol. Sci. 2020, 21 (18), 6691. 10.3390/ijms21186691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.; Song Y.; Cheng L.; Xu H.; Liu J. Analysis of Methylation-driven Genes for Predicting the Prognosis of Patients with Head and Neck Squamous Cell Carcinoma. J. Cell. Biochem. 2019, 120 (12), 19482–19495. 10.1002/jcb.29252. [DOI] [PubMed] [Google Scholar]
- Øster B.; Thorsen K.; Lamy P.; Wojdacz T. K.; Hansen L. L.; Birkenkamp-Demtröder K.; Sørensen K. D.; Laurberg S.; Ørntoft T. F.; Andersen C. L. Identification and Validation of Highly Frequent CpG Island Hypermethylation in Colorectal Adenomas and Carcinomas. Int. J. Cancer 2011, 129 (12), 2855–2866. 10.1002/ijc.25951. [DOI] [PubMed] [Google Scholar]
- Ala U.; Piro R. M.; Grassi E.; Damasco C.; Silengo L.; Oti M.; Provero P.; Di Cunto F. Prediction of Human Disease Genes by Human-Mouse Conserved Coexpression Analysis. PLoS Comput. Biol. 2008, 4 (3), e1000043 10.1371/journal.pcbi.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanashi H.; Boeglin W. E.; Morisseau C.; Davis R. W.; Sulikowski G. A.; Hammock B. D.; Brash A. R. Catalytic Activities of Mammalian Epoxide Hydrolases with Cis and Trans Fatty Acid Epoxides Relevant to Skin Barrier Function. J. Lipid Res. 2018, 59 (4), 684–695. 10.1194/jlr.M082701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edin M. L.; Yamanashi H.; Boeglin W. E.; Graves J. P.; DeGraff L. M.; Lih F. B.; Zeldin D. C.; Brash A. R. Epoxide Hydrolase 3 (Ephx3) Gene Disruption Reduces Ceramide Linoleate Epoxide Hydrolysis and Impairs Skin Barrier Function. J. Biol. Chem. 2021, 296, 100198. 10.1074/jbc.RA120.016570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Vande Velde C.; Israelson A.; Xie J.; Bailey A. O.; Dong M.-Q.; Chun S.-J.; Roy T.; Winer L.; Yates J. R.; Capaldi R. A.; Cleveland D. W.; Miller T. M. ALS-Linked Mutant Superoxide Dismutase 1 (SOD1) Alters Mitochondrial Protein Composition and Decreases Protein Import. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (49), 21146–21151. 10.1073/pnas.1014862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T.; Yokoi T. The Emerging Role of Human Esterases. Drug Metab. Pharmacokinet. 2012, 27 (5), 466–477. 10.2133/dmpk.DMPK-12-RV-042. [DOI] [PubMed] [Google Scholar]
- Iwamura A.; Fukami T.; Higuchi R.; Nakajima M.; Yokoi T. Human α/β Hydrolase Domain Containing 10 (ABHD10) Is Responsible Enzyme for Deglucuronidation of Mycophenolic Acid Acyl-Glucuronide in Liver. J. Biol. Chem. 2012, 287 (12), 9240–9249. 10.1074/jbc.M111.271288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y.; Fukami T.; Yokoi T.; Nakajima M. An Orphan Esterase ABHD10 Modulates Probenecid Acyl Glucuronidation in Human Liver. Drug Metab. Dispos. 2014, 42 (12), 2109–2116. 10.1124/dmd.114.059485. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Qiu T.; Kathayat R. S.; Azizi S.-A.; Thorne A. K.; Ahn D.; Fukata Y.; Fukata M.; Rice P. A.; Dickinson B. C. ABHD10 Is an S-Depalmitoylase Affecting Redox Homeostasis through Peroxiredoxin-5. Nat. Chem. Biol. 2019, 15 (12), 1232–1240. 10.1038/s41589-019-0399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhl A. M.; Mohr J. T.; Bachovchin D. A.; Niessen S.; Hsu K. L.; Berlin J. M.; Dochnahl M.; López-Alberca M. P.; Fu G. C.; Cravatt B. F. Competitive Activity-Based Protein Profiling Identifies Aza-β-Lactams as a Versatile Chemotype for Serine Hydrolase Inhibition. J. Am. Chem. Soc. 2012, 134 (11), 5068–5071. 10.1021/ja300799t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhl A. M.; Mohr J. T.; Speers A. E.; Bachovchin D. A.; Berlin J. M.; Spicer T.; Fernandez-Vega V.; Brown S. J.; Ferguson J.; Fu G. C.; Cravatt B. F.; Hodder P.; Rosen H.. Probe Development Efforts to Identify Novel Inhibitors of ABHD10. Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information (US): Bethesda, MD. 2011. [PubMed]
- The Scripps Research Institute and The Massachusets Institute of Technology . Aza-β-lactam compounds and methods of using. WO 2013152272, 2013.
- Lajkiewicz N. J.; Cognetta A. B.; Niphakis M. J.; Cravatt B. F.; Porco J. A. Remodeling Natural Products: Chemistry and Serine Hydrolase Activity of a Rocaglate-Derived β-Lactone. J. Am. Chem. Soc. 2014, 136 (6), 2659–2664. 10.1021/ja412431g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi S.; Cognetta A. B.; Niphakis M. J.; He Z.; Zajdlik A.; St. Denis J. D.; Scully C. C. G.; Cravatt B. F.; Yudin A. K. Facile Synthesis of Borofragments and Their Evaluation in Activity-Based Protein Profiling. Chem. Commun. 2015, 51 (17), 3608–3611. 10.1039/C4CC09107H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort N.; Yi Z.; Bowen B.; Glancy B.; De Filippis E. A.; Mapes R.; Hwang H.; Flynn C. R.; Willis W. T.; Civitarese A.; Højlund K.; Mandarino L. J. Proteome Profile of Functional Mitochondria from Human Skeletal Muscle Using One-Dimensional Gel Electrophoresis and HPLC-ESI-MS/MS. J. Proteomics 2009, 72 (6), 1046–1060. 10.1016/j.jprot.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merla G.; Ucla C.; Guipponi M.; Reymond A. Identification of Additional Transcripts in the Williams-Beuren Syndrome Critical Region. Hum. Genet. 2002, 110 (5), 429–438. 10.1007/s00439-002-0710-x. [DOI] [PubMed] [Google Scholar]
- Shen W.-J.; Patel S.; Yu Z.; Jue D.; Kraemer F. B. Effects of Rosiglitazone and High Fat Diet on Lipase/Esterase Expression in Adipose Tissue. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2007, 1771 (2), 177–184. 10.1016/j.bbalip.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedl T.; Arni S.; Roschitzki B.; Grossmann J.; Collaud S.; Soltermann A.; Hillinger S.; Aebersold R.; Weder W. Activity-Based Proteomics: Identification of ABHD11 and ESD Activities as Potential Biomarkers for Human Lung Adenocarcinoma. J. Proteomics 2011, 74 (10), 1884–1894. 10.1016/j.jprot.2011.04.030. [DOI] [PubMed] [Google Scholar]
- Daniel P.; Halada P.; Jelínek M.; Balušíková K.; Kovář J. Differentially Expressed Mitochondrial Proteins in Human Mcf7 Breast Cancer Cells Resistant to Paclitaxel. Int. J. Mol. Sci. 2019, 20 (12), 2986. 10.3390/ijms20122986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen H.; Lepikhova T.; Sahu B.; Pehkonen H.; Pihlajamaa P.; Louhimo R.; Gao P.; Wei G.; Hautaniemi S.; Jänne O. A.; Monni O. Identification of Several Potential Chromatin Binding Sites of HOXB7 and Its Downstream Target Genes in Breast Cancer. Int. J. Cancer 2015, 137 (10), 2374–2383. 10.1002/ijc.29616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya M.; Srinivasan M.; Rajasekharan R. Human Alpha Beta Hydrolase Domain Containing Protein 11 and Its Yeast Homolog Are Lipid Hydrolases. Biochem. Biophys. Res. Commun. 2017, 487 (4), 875–880. 10.1016/j.bbrc.2017.04.145. [DOI] [PubMed] [Google Scholar]
- Escoubet J.; Kenigsberg M.; Derock M.; Yaligara V.; Bock M.-D.; Roche S.; Massey F.; de Foucauld H.; Bettembourg C.; Olivier A.; Berthemy A.; Capdevielle J..; Legoux R.; Perret E.; Buzy A.; Chardenot P.; Destelle V.; Leroy A.; Cahours C.; Teixeira S.; Juvet P.; Gauthier P.; Leguet M.; Rocheteau-Beaujouan L.; Chatoux M.-A.; Deshayes W.; Clement M.; Kabiri M.; Orsini C.; Mikol V.; Didier M.; Guillemot J.-C. ABHD11, a New Diacylglycerol Lipase Involved in Weight Gain Regulation. PLoS One 2020, 15 (6), e0234780 10.1371/journal.pone.0234780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P. S. J.; Ortmann B. M.; Martinelli A. W.; Houghton J. W.; Costa A. S. H.; Burr S. P.; Antrobus R.; Frezza C.; Nathan J. A. ABHD11 Maintains 2-Oxoglutarate Metabolism by Preserving Functional Lipoylation of the 2-Oxoglutarate Dehydrogenase Complex. Nat. Commun. 2020, 11 (1), 4046. 10.1038/s41467-020-17862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.; Ruan Y.; Zhang J.; Wang X.; Wu W.; He P.; Wang J.; Xiong J.; Cheng Y.; Liu L.; Yang Y.; Tian Y.; Jian R. ABHD11 Is Critical for Embryonic Stem Cell Expansion, Differentiation and Lipid Metabolic Homeostasis. Front. Cell Dev. Biol. 2020, 8, 570. 10.3389/fcell.2020.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Shao Y.; Zhu M.; Li Q.; Yang F.; Lu X.; Xu C.; Xiao B.; Sun Y.; Guo J. Using Gastric Juice LncRNA-ABHD11-AS1 as a Novel Type of Biomarker in the Screening of Gastric Cancer. Tumor Biol. 2016, 37 (1), 1183–1188. 10.1007/s13277-015-3903-3. [DOI] [PubMed] [Google Scholar]
- Lei X.; Li L.; Duan X. Long Non-Coding RNA ABHD11-AS1 Promotes Colorectal Cancer Development through Regulation of MiR-133a/SOX4 Axis. Biosci. Rep. 2018, 38 (6), BSR20181386. 10.1042/BSR20181386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X.; Lv S. X.; Qiao Y.; Li Q. P.; Ye B.; Wang C. C.; Miao L. Long Noncoding RNA ABHD11-AS1 Predicts the Prognosis of Pancreatic Cancer Patients and Serves as a Promoter by Activating the PI3K-AKT Pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22 (24), 8630–8639. 10.26355/eurrev_201812_16627. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Wang L.-L.; Chen S.; Zong Z.-H.; Guan X.; Zhao Y. LncRNA ABHD11-AS1 Promotes the Development of Endometrial Carcinoma by Targeting Cyclin D1. J. Cell. Mol. Med. 2018, 22 (8), 3955–3964. 10.1111/jcmm.13675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L.; Li J.; Lin Y.; Liu D.; Yang Q.; Jian J.; Peng J. m6A Transferase METTL3-induced LncRNA ABHD11-AS1 Promotes the Warburg Effect of Non-small-cell Lung Cancer. J. Cell. Physiol. 2021, 236 (4), 2649–2658. 10.1002/jcp.30023. [DOI] [PubMed] [Google Scholar]
- Zhuang X.; Tong H.; Ding Y.; Wu L.; Cai J.; Si Y.; Zhang H.; Shen M. Long Noncoding RNA ABHD11-AS1 Functions as a Competing Endogenous RNA to Regulate Papillary Thyroid Cancer Progression by MiR-199a-5p/SLC1A5 Axis. Cell Death Dis. 2019, 10 (8), 620. 10.1038/s41419-019-1850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X.; Jiang X.; Yong J.; Xie H.; Yuan J.; Zeng D.; Dou Y.; Xiao S. LncRNA ABHD11-AS1, Regulated by the EGFR Pathway, Contributes to the Ovarian Cancer Tumorigenesis by Epigenetically Suppressing TIMP2. Cancer Med. 2019, 8 (16), 7074–7085. 10.1002/cam4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Adibekian A.; Hsu K.-L.; Speers A. E.; Brown S. J.; Spicer T.; Fernandez-Vega V.; Ferguson J.; Cravatt B. F.; Hodder P.; Rosen H.. Optimization and Characterization of a Triazole Urea Inhibitor for Alpha/Beta Hydrolase Domain-Containing Protein 11 (ABHD11): Anti-Probe for LYPLA1/LYPLA2 Dual Inhibitor ML211. Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information (US): Bethesda, MD, 2011. [PubMed]
- Navia-Paldanius D.; Patel J. Z.; López Navarro M.; Jakupović H.; Goffart S.; Pasonen-Seppänen S.; Nevalainen T. J.; Jääskeläinen T.; Laitinen T.; Laitinen J. T.; Savinainen J. R. Chemoproteomic, Biochemical and Pharmacological Approaches in the Discovery of Inhibitors Targeting Human α/β-Hydrolase Domain Containing 11 (ABHD11). Eur. J. Pharm. Sci. 2016, 93, 253–263. 10.1016/j.ejps.2016.08.031. [DOI] [PubMed] [Google Scholar]
- Taha M. O.; Dahabiyeh L. A.; Bustanji Y.; Zalloum H.; Saleh S. Combining Ligand-Based Pharmacophore Modeling, Quantitative Structure-Activity Relationship Analysis and in Silico Screening for the Discovery of New Potent Hormone Sensitive Lipase Inhibitors. J. Med. Chem. 2008, 51 (20), 6478–6494. 10.1021/jm800718k. [DOI] [PubMed] [Google Scholar]
- Joshi A.; Shaikh M.; Singh S.; Rajendran A.; Mhetre A.; Kamat S. S. Biochemical Characterization of the PHARC-Associated Serine Hydrolase ABHD12 Reveals Its Preference for Very-Long-Chain Lipids. J. Biol. Chem. 2018, 293 (44), 16953–16963. 10.1074/jbc.RA118.005640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar D. S.; Ravikumar G.; Mehendale N.; Singh S.; Joshi A.; Sharma A. K.; Mhetre A.; Rajendran A.; Chakrapani H.; Kamat S. S. A Chemical–Genetic Screen Identifies ABHD12 as an Oxidized-Phosphatidylserine Lipase. Nat. Chem. Biol. 2019, 15 (2), 169–178. 10.1038/s41589-018-0195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskerstrand T.; H’mida-Ben Brahim D.; Johansson S.; M’zahem A.; Haukanes B. I.; Drouot N.; Zimmermann J.; Cole A. J.; Vedeler C.; Bredrup C.; Assoum M.; Tazir M.; Klockgether T.; Hamri A.; Steen V. M.; Boman H.; Bindoff L. A.; Koenig M.; Knappskog P. M. Mutations in ABHD12 Cause the Neurodegenerative Disease PHARC: An Inborn Error of Endocannabinoid Metabolism. Am. J. Hum. Genet. 2010, 87 (3), 410–417. 10.1016/j.ajhg.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingaud-Sequeira A.; Raldúa D.; Lavie J.; Mathieu G.; Bordier M.; Knoll-Gellida A.; Rambeau P.; Coupry I.; André M.; Malm E.; Möller C.; Andreasson S.; Andreasson N. D.; Tranebjærg L.; Koenig M.; Lacombe D.; Goizet C.; Babin P. J. Functional Validation of ABHD12 Mutations in the Neurodegenerative Disease PHARC. Neurobiol. Dis. 2017, 98, 36–51. 10.1016/j.nbd.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Blankman J. L.; Long J. Z.; Trauger S. A.; Siuzdak G.; Cravatt B. F. ABHD12 Controls Brain Lysophosphatidylserine Pathways That Are Deregulated in a Murine Model of the Neurodegenerative Disease PHARC. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (4), 1500–1505. 10.1073/pnas.1217121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat S. S.; Camara K.; Parsons W. H.; Chen D.-H.; Dix M. M.; Bird T. D.; Howell A. R.; Cravatt B. F. Immunomodulatory Lysophosphatidylserines Are Regulated by ABHD16A and ABHD12 Interplay. Nat. Chem. Biol. 2015, 11 (2), 164–171. 10.1038/nchembio.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.; Joshi A.; Kamat S. S. Mapping the Neuroanatomy of ABHD16A, ABHD12, and Lysophosphatidylserines Provides New Insights into the Pathophysiology of the Human Neurological Disorder PHARC. Biochemistry 2020, 59 (24), 2299–2311. 10.1021/acs.biochem.0c00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leishman E.; Mackie K.; Bradshaw H. B. Elevated Levels of Arachidonic Acid-Derived Lipids Including Prostaglandins and Endocannabinoids Are Present Throughout ABHD12 Knockout Brains: Novel Insights Into the Neurodegenerative Phenotype. Front. Mol. Neurosci. 2019, 12, 142. 10.3389/fnmol.2019.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberger T.; Slim R.; Mansour A.; Nauck M.; Nürnberg G.; Nürnberg P.; Decker C.; Dafinger C.; Ebermann I.; Bergmann C.; Bolz H. Targeted Next-Generation Sequencing Identifies a Homozygous Nonsense Mutation in ABHD12, the Gene Underlying PHARC, in a Family Clinically Diagnosed with Usher Syndrome Type 3. Orphanet J. Rare Dis. 2012, 7 (1), 59. 10.1186/1750-1172-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Feng Y.; Liu Y.; He C.; Liu J.; Chen H.; Deng Y.; Li M.; Li W.; Song J.; Niu Z.; Sang S.; Wen J.; Men M.; Chen X.; Li J.; Liu X.; Ling J. A Novel ABHD12 Nonsense Variant in Usher Syndrome Type 3 Family with Genotype-Phenotype Spectrum Review. Gene 2019, 704, 113–120. 10.1016/j.gene.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Yoshida T.; Kobayashi T.; Itoda M.; Muto T.; Miyaguchi K.; Mogushi K.; Shoji S.; Shimokawa K.; Iida S.; Uetake H.; Ishikawa T.; Sugihara K.; Mizushima H.; Tanaka H. Clinical Omics Analysis of Colorectal Cancer Incorporating Copy Number Aberrations and Gene Expression Data. Cancer Inf. 2010, 9, 147–161. 10.4137/CIN.S3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S.; Kim S. W.; Lim J.-Y.; Park S.-J. ABHD12 Knockdown Suppresses Breast Cancer Cell Proliferation, Migration and Invasion. Anticancer Res. 2020, 40 (5), 2601–2611. 10.21873/anticanres.14231. [DOI] [PubMed] [Google Scholar]
- King A. R.; Dotsey E. Y.; Lodola A.; Jung K. M.; Ghomian A.; Qiu Y.; Fu J.; Mor M.; Piomelli D. Discovery of Potent and Reversible Monoacylglycerol Lipase Inhibitors. Chem. Biol. 2009, 16 (10), 1045–1052. 10.1016/j.chembiol.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkkari T.; Haavikko R.; Laitinen T.; Navia-Paldanius D.; Rytilahti R.; Vaara M.; Lehtonen M.; Alakurtti S.; Yli-Kauhaluoma J.; Nevalainen T.; Savinainen J. R.; Laitinen J. T. Discovery of Triterpenoids as Reversible Inhibitors of α/β-Hydrolase Domain Containing 12 (ABHD12). PLoS One 2014, 9 (5), e98286 10.1371/journal.pone.0098286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdelaziz I.; Gómez-Ruiz S.; Benkhaled M.; Carralero S.; Schenker P.; Salm A.; Gertsch J.; Haba H. New Cycloartane-Type Ester Triterpenes from Euphorbia Pterococca and Biological Evaluation. Fitoterapia 2018, 127, 271–278. 10.1016/j.fitote.2018.02.027. [DOI] [PubMed] [Google Scholar]
- Ogasawara D.; Ichu T.-A.; Vartabedian V. F.; Benthuysen J.; Jing H.; Reed A.; Ulanovskaya O. A.; Hulce J. J.; Roberts A.; Brown S.; Rosen H.; Teijaro J. R.; Cravatt B. F. Selective Blockade of the Lyso-PS Lipase ABHD12 Stimulates Immune Responses in Vivo. Nat. Chem. Biol. 2018, 14 (12), 1099–1108. 10.1038/s41589-018-0155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara D.; Ichu T.-A.; Jing H.; Hulce J. J.; Reed A.; Ulanovskaya O. A.; Cravatt B. F. Discovery and Optimization of Selective and in Vivo Active Inhibitors of the Lysophosphatidylserine Lipase α/β-Hydrolase Domain-Containing 12 (ABHD12). J. Med. Chem. 2019, 62 (3), 1643–1656. 10.1021/acs.jmedchem.8b01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathman S. G.; Boshart J.; Jing H.; Cravatt B. F. Blockade of the Lysophosphatidylserine Lipase ABHD12 Potentiates Ferroptosis in Cancer Cells. ACS Chem. Biol. 2020, 15 (4), 871–877. 10.1021/acschembio.0c00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Scripps Research Institute . ABHD12 inhibitors and methods of making and using same. WO 2019222267, 2019.
- Lundbeck La Jolla Research Center Inc . ABHD12 inhibitors and methods of making and using same. WO 2020232153, 2020.
- Xu J.; Gu W.; Ji K.; Xu Z.; Zhu H.; Zheng W. Sequence Analysis and Structure Prediction of ABHD16A and the Roles of the ABHD Family Members in Human Disease. Open Biol. 2018, 8 (5), 180017. 10.1098/rsob.180017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senis Y. A.; Tomlinson M. G.; García Á.; Dumon S.; Heath V. L.; Herbert J.; Cobbold S. P.; Spalton J. C.; Ayman S.; Antrobus R.; Zitzmann N.; Bicknell R.; Frampton J.; Authi K. S.; Martin A.; Wakelam M. J. O.; Watson S. P. A Comprehensive Proteomics and Genomics Analysis Reveals Novel Transmembrane Proteins in Human Platelets and Mouse Megakaryocytes Including G6b-B, a Novel Immunoreceptor Tyrosine-Based Inhibitory Motif Protein. Mol. Cell. Proteomics 2007, 6 (3), 548–564. 10.1074/mcp.D600007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B. R.; Cravatt B. F. Large-Scale Profiling of Protein Palmitoylation in Mammalian Cells. Nat. Methods 2009, 6 (2), 135–138. 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savinainen J. R.; Patel J. Z.; Parkkari T.; Navia-Paldanius D.; Marjamaa J. J. T.; Laitinen T.; Nevalainen T.; Laitinen J. T. Biochemical and Pharmacological Characterization of the Human Lymphocyte Antigen B-Associated Transcript 5 (BAT5/ABHD16A). PLoS One 2014, 9 (10), e109869 10.1371/journal.pone.0109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y.-Y.; Lin Y.-J.; Chang C.-C.; Chen D.-Y.; Hsu C.-M.; Lo M.-M.; Hsu K.-H.; Tsai F.-J. Human Lymphocyte Antigen B-Associated Transcript 2, 3, and 5 Polymorphisms and Haplotypes Are Associated with Susceptibility of Kawasaki Disease and Coronary Artery Aneurysm. J. Clin. Lab. Anal. 2010, 24 (4), 262–268. 10.1002/jcla.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L.; Galimberti G.; Calò D. G.; Fronza R.; Martelli P. L.; Scotti E.; Colombo M.; Schiavo G.; Casadio R.; Buttazzoni L.; Russo V. Identification and Association Analysis of Several Hundred Single Nucleotide Polymorphisms within Candidate Genes for Back Fat Thickness in Italian Large White Pigs Using a Selective Genotyping Approach1. J. Anim. Sci. 2012, 90 (8), 2450–2464. 10.2527/jas.2011-4797. [DOI] [PubMed] [Google Scholar]
- Sargent C. A.; Dunham I.; Campbell R. D. Identification of Multiple HTF-Island Associated Genes in the Human Major Histocompatibility Complex Class III Region. EMBO J. 1989, 8 (8), 2305–2312. 10.1002/j.1460-2075.1989.tb08357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies T.; Blanck G.; Bresnahan M.; Sands J.; Strominger J. A New Cluster of Genes within the Human Major Histocompatibility Complex. Science 1989, 243 (4888), 214–217. 10.1126/science.2911734. [DOI] [PubMed] [Google Scholar]
- Mathew P. A.; Kumar V.; Bennett M.; Flaherty L. Identification of a Recombinational Breakpoint at the BAT5 Locus in Three Intra-H-2 Recombinant Inbred Mouse Strains. Exp. Clin. Immunogenet. 1995, 12 (4), 261–267. [PubMed] [Google Scholar]
- Dekker F. J.; Rocks O.; Vartak N.; Menninger S.; Hedberg C.; Balamurugan R.; Wetzel S.; Renner S.; Gerauer M.; Schölermann B.; Rusch M.; Kramer J. W.; Rauh D.; Coates G. W.; Brunsveld L.; Bastiaens P. I. H.; Waldmann H. Small-Molecule Inhibition of APT1 Affects Ras Localization and Signaling. Nat. Chem. Biol. 2010, 6 (6), 449–456. 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- Camara K.; Kamat S. S.; Lasota C. C.; Cravatt B. F.; Howell A. R. Combining Cross-Metathesis and Activity-Based Protein Profiling: New β-Lactone Motifs for Targeting Serine Hydrolases. Bioorg. Med. Chem. Lett. 2015, 25 (2), 317–321. 10.1016/j.bmcl.2014.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Scripps Research Institute and The University of Connecticut . Lactone compounds and methods of making and using same. WO 2016069542, 2016.
- Ahonen T. J.; Savinainen J. R.; Yli-Kauhaluoma J.; Kalso E.; Laitinen J. T.; Moreira V. M. Discovery of 12-Thiazole Abietanes as Selective Inhibitors of the Human Metabolic Serine Hydrolase HABHD16A. ACS Med. Chem. Lett. 2018, 9 (12), 1269–1273. 10.1021/acsmedchemlett.8b00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. T. S.; Conibear E. ABHD17 Proteins Are Novel Protein Depalmitoylases That Regulate N-Ras Palmitate Turnover and Subcellular Localization. eLife 2015, 4, e11306 10.7554/eLife.11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg C.; Dekker F. J.; Rusch M.; Renner S.; Wetzel S.; Vartak N.; Gerding-Reimers C.; Bon R. S.; Bastiaens P. I. H.; Waldmann H. Development of Highly Potent Inhibitors of the Ras-Targeting Human Acyl Protein Thioesterases Based on Substrate Similarity Design. Angew. Chem., Int. Ed. 2011, 50 (42), 9832–9837. 10.1002/anie.201102965. [DOI] [PubMed] [Google Scholar]
- Martin B. R.; Wang C.; Adibekian A.; Tully S. E.; Cravatt B. F. Global Profiling of Dynamic Protein Palmitoylation. Nat. Methods 2012, 9 (1), 84–89. 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remsberg J. R.; Suciu R. M.; Zambetti N. A.; Hanigan T. W.; Firestone A. J.; Inguva A.; Long A.; Ngo N.; Lum K. M.; Henry C. L.; Richardson S. K.; Predovic M.; Huang B.; Dix M. M.; Howell A. R.; Niphakis M. J.; Shannon K.; Cravatt B. F. ABHD17 Regulation of Plasma Membrane Palmitoylation and N-Ras-Dependent Cancer Growth. Nat. Chem. Biol. 2021, 10.1038/s41589-021-00785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch D. G.; Omeir R.; Arreaza G.; Salehani D.; Prestwich G. D.; Huang Z.; Howlett A. Methyl Arachidonyl Fluorophosphonate: A Potent Irreversible Inhibitor of Anandamide Amidase. Biochem. Pharmacol. 1997, 53 (3), 255–260. 10.1016/S0006-2952(96)00830-1. [DOI] [PubMed] [Google Scholar]
- The Regents of the University of California . Compositions and methods for modulating ABHD2 activity. WO 2017023861, 2017.
- Hoover H. S.; Blankman J. L.; Niessen S.; Cravatt B. F. Selectivity of Inhibitors of Endocannabinoid Biosynthesis Evaluated by Activity-Based Protein Profiling. Bioorg. Med. Chem. Lett. 2008, 18 (22), 5838–5841. 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar A. J. The Gene Structure and Expression of Human ABHD1: Overlapping Polyadenylation Signal Sequence with Sec12. BMC Genomics 2003, 4 (1), 18. 10.1186/1471-2164-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoelting M.; Geyer M.; Reuter S.; Reichelt R.; Bek M. J.; Pavenstädt H. α/β Hydrolase 1 Is Upregulated in D5 Dopamine Receptor Knockout Mice and Reduces O2- Production of NADPH Oxidase. Biochem. Biophys. Res. Commun. 2009, 379 (1), 81–85. 10.1016/j.bbrc.2008.12.008. [DOI] [PubMed] [Google Scholar]
- van Esbroeck A. C. M.; Janssen A. P. A.; Cognetta A. B.; Ogasawara D.; Shpak G.; van der Kroeg M.; Kantae V.; Baggelaar M. P.; de Vrij F. M. S.; Deng H.; Allarà M.; Fezza F.; Lin Z.; van der Wel T.; Soethoudt M.; Mock E. D.; den Dulk H.; Baak I. L.; Florea B. I.; Hendriks G.; De Petrocellis L.; Overkleeft H. S.; Hankemeier T.; De Zeeuw C. I.; Di Marzo V.; Maccarrone M.; Cravatt B. F.; Kushner S. A.; van der Stelt M. Activity-Based Protein Profiling Reveals off-Target Proteins of the FAAH Inhibitor BIA 10–2474. Science 2017, 356 (6342), 1084–1087. 10.1126/science.aaf7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher F.; Bennett J. M.; Bogyo M.; Lovell S. Strategies for Tuning the Selectivity of Chemical Probes That Target Serine Hydrolases. Cell Chem. Biol. 2020, 27 (8), 937–952. 10.1016/j.chembiol.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar J. S.; Weber O. D.; Clapper J. R.; Blankman J. L.; Henry C. L.; Simon G. M.; Alexander J. P.; Jones T. K.; Ezekowitz R. A. B.; O’Neill G. P.; Grice C. A. Identification of ABX-1431, a Selective Inhibitor of Monoacylglycerol Lipase and Clinical Candidate for Treatment of Neurological Disorders. J. Med. Chem. 2018, 61 (20), 9062–9084. 10.1021/acs.jmedchem.8b00951. [DOI] [PubMed] [Google Scholar]
- Ahn K.; Smith S. E.; Liimatta M. B.; Beidler D.; Sadagopan N.; Dudley D. T.; Young T.; Wren P.; Zhang Y.; Swaney S.; Van Becelaere K.; Blankman J. L.; Nomura D. K.; Bhattachar S. N.; Stiff C.; Nomanbhoy T. K.; Weerapana E.; Johnson D. S.; Cravatt B. F. Mechanistic and Pharmacological Characterization of PF-04457845: A Highly Potent and Selective Fatty Acid Amide Hydrolase Inhibitor That Reduces Inflammatory and Noninflammatory Pain. J. Pharmacol. Exp. Ther. 2011, 338 (1), 114–124. 10.1124/jpet.111.180257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields C. M.; Zvonok N.; Zvonok A.; Makriyannis A. Biochemical and Proteomic Characterization of Recombinant Human α/β Hydrolase Domain 6. Sci. Rep. 2019, 9 (1), 890. 10.1038/s41598-018-36633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]