Abstract
Background
SARS-CoV-2 antigen rapid diagnostic tests (Ag-RDTs) are increasingly being integrated in testing strategies around the world. Studies of the Ag-RDTs have shown variable performance. In this systematic review and meta-analysis, we assessed the clinical accuracy (sensitivity and specificity) of commercially available Ag-RDTs.
Methods and findings
We registered the review on PROSPERO (registration number: CRD42020225140). We systematically searched multiple databases (PubMed, Web of Science Core Collection, medRvix, bioRvix, and FIND) for publications evaluating the accuracy of Ag-RDTs for SARS-CoV-2 up until 30 April 2021. Descriptive analyses of all studies were performed, and when more than 4 studies were available, a random-effects meta-analysis was used to estimate pooled sensitivity and specificity in comparison to reverse transcription polymerase chain reaction (RT-PCR) testing. We assessed heterogeneity by subgroup analyses, and rated study quality and risk of bias using the QUADAS-2 assessment tool. From a total of 14,254 articles, we included 133 analytical and clinical studies resulting in 214 clinical accuracy datasets with 112,323 samples. Across all meta-analyzed samples, the pooled Ag-RDT sensitivity and specificity were 71.2% (95% CI 68.2% to 74.0%) and 98.9% (95% CI 98.6% to 99.1%), respectively. Sensitivity increased to 76.3% (95% CI 73.1% to 79.2%) if analysis was restricted to studies that followed the Ag-RDT manufacturers’ instructions. LumiraDx showed the highest sensitivity, with 88.2% (95% CI 59.0% to 97.5%). Of instrument-free Ag-RDTs, Standard Q nasal performed best, with 80.2% sensitivity (95% CI 70.3% to 87.4%). Across all Ag-RDTs, sensitivity was markedly better on samples with lower RT-PCR cycle threshold (Ct) values, i.e., <20 (96.5%, 95% CI 92.6% to 98.4%) and <25 (95.8%, 95% CI 92.3% to 97.8%), in comparison to those with Ct ≥ 25 (50.7%, 95% CI 35.6% to 65.8%) and ≥30 (20.9%, 95% CI 12.5% to 32.8%). Testing in the first week from symptom onset resulted in substantially higher sensitivity (83.8%, 95% CI 76.3% to 89.2%) compared to testing after 1 week (61.5%, 95% CI 52.2% to 70.0%). The best Ag-RDT sensitivity was found with anterior nasal sampling (75.5%, 95% CI 70.4% to 79.9%), in comparison to other sample types (e.g., nasopharyngeal, 71.6%, 95% CI 68.1% to 74.9%), although CIs were overlapping. Concerns of bias were raised across all datasets, and financial support from the manufacturer was reported in 24.1% of datasets. Our analysis was limited by the included studies’ heterogeneity in design and reporting.
Conclusions
In this study we found that Ag-RDTs detect the vast majority of SARS-CoV-2-infected persons within the first week of symptom onset and those with high viral load. Thus, they can have high utility for diagnostic purposes in the early phase of disease, making them a valuable tool to fight the spread of SARS-CoV-2. Standardization in conduct and reporting of clinical accuracy studies would improve comparability and use of data.
Lukas Brümmer and co-workers report on the accuracy of rapid antigen tests for SARS-CoV-2.
Author summary
Why was this study done?
Antigen rapid diagnostic tests (Ag-RDTs) are considered an important diagnostic tool to fight the spread of SARS-CoV-2.
An increasing number of Ag-RDTs are offered on the market, and a constantly growing body of literature evaluating their performance is available.
To inform decision makers about the best test to choose, an up-to-date summary of their performance is needed.
What did the researchers do and find?
On a weekly basis, we search multiple databases for evaluations of Ag-RDTs detecting SARS-CoV-2 and post the results on https://www.diagnosticsglobalhealth.org.
Based on the search results up until 30 April 2021, we conducted a systematic review and meta-analysis, including a total of 133 clinical and analytical accuracy studies.
Across all meta-analyzed studies, when Ag-RDTs were performed according to manufacturers’ recommendations, they showed a sensitivity of 76.3% (95% CI 73.1% to 79.2%), with LumiraDx (sensitivity 88.2% [95% CI 59.0% to 97.5%]) and, of the instrument-free Ag-RDTs, Standard Q nasal (74.9% sensitivity [95% CI 69.3% to 79.7%]) performing best.
Across all Ag-RDTs, sensitivity increased to 95.8% (95% CI 92.3% to 97.8%) when we restricted the analysis to samples with high viral loads (i.e., a Ct value < 25) and to 83.8% (95% CI 76.3% to 89.2%) when tests were performed on patients within the first week after symptom onset.
What do these findings mean?
Ag-RDTs detect the vast majority of cases within the first week of symptom onset and those with high viral load. Thus, they can have high utility for diagnostic purposes in the early phase of disease.
Out of all assessed tests, LumiraDx showed the highest accuracy. Standard Q was the best-performing test when only considering those that do not require an instrument.
A standardization of reporting methods for clinical accuracy studies would enhance future test comparisons.
Introduction
As the COVID-19 pandemic continues around the globe, antigen rapid diagnostic tests (Ag-RDTs) for SARS-CoV-2 are seen as an important diagnostic tool to fight the virus’s spread [1,2]. The number of Ag-RDTs on the market is increasing constantly [3]. Initial data from independent evaluations suggest that the performance of SARS-CoV-2 Ag-RDTs may be lower than what is reported by the manufacturers. In addition, Ag-RDT accuracy seems to vary substantially between tests [4–6].
With the increased availability of Ag-RDTs, an increasing number of independent validations have been published. Such evaluations differ widely in their quality, methods, and results, making it difficult to assess the true performance of the respective tests [7]. To inform decision makers on the best choice of individual tests, an aggregated, widely available, and frequently updated assessment of the quality, performance, and independence of the data is urgently needed. While other systematic reviews have been published, they include data only up until November 2020 [8–11], exclude preprints [12], or were industry sponsored [13]. In addition, only 1 assessed the quality of studies in detail, with data up until November 2020 [7,11].
With our systematic review and meta-analysis, we aim to close this gap in the literature and link to a website (https://www.diagnosticsglobalhealth.org) that is regularly updated.
Methods
We developed a study protocol following standard guidelines for systematic reviews [14,15], which is available in S1 Text. We also completed the PRISMA checklist (S1 PRISMA Checklist). Furthermore, we registered the review on PROSPERO (registration number: CRD42020225140).
Search strategy
We performed a search of the databases PubMed, Web of Science, medRxiv, and bioRxiv using search terms that were developed with an experienced medical librarian (M. Grilli) using combinations of subject headings (when applicable) and text-words for the concepts of the search question. The main search terms were “Severe Acute Respiratory Syndrome Corona-virus 2,” “COVID-19,” “Betacoronavirus,” “Coronavirus,” and “Point of Care Testing.” The full list of search terms is available in S2 Text. We also searched the Foundation for Innovative New Diagnostics (FIND) website (https://www.finddx.org/sarscov2-eval-antigen/) for relevant studies manually. We performed the search up until 30 April 2021. No language restrictions were applied.
Inclusion criteria
We included studies evaluating the accuracy of commercially available Ag-RDTs to establish a diagnosis of SARS-CoV-2 infection, against reverse transcription polymerase chain reaction (RT-PCR) or cell culture as reference standard. We included all study populations irrespective of age, presence of symptoms, or study location. We considered cohort studies, nested cohort studies, case–control or cross-sectional studies, and randomized studies. We included both peer-reviewed publications and preprints.
We excluded studies in which patients were tested for the purpose of monitoring or ending quarantine. Also, publications with a population size smaller than 10 were excluded. Although the size threshold of 10 is arbitrary, such small studies are more likely to give unreliable estimates of sensitivity and specificity.
Index tests
Ag-RDTs for SARS-CoV-2 aim to detect infection by recognizing viral proteins. Most Ag-RDTs use specific labeled antibodies attached to a nitrocellulose matrix strip, to capture the virus antigen. Successful binding of the antibodies to the antigen either is detected visually (through the appearance of a line on the matrix strip [lateral flow assay]) or requires a specific reader for fluorescence detection. Microfluidic enzyme-linked immunosorbent assays have also been developed. Ag-RDTs typically provide results within 10 to 30 minutes [6].
Reference standard
Viral culture detects viable virus that is relevant for transmission but is available in research settings only. Since RT-PCR tests are more widely available and SARS-CoV-2 RNA (as reflected by RT-PCR cycle threshold [Ct] value) highly correlates with SARS-CoV-2 antigen quantities, we considered it an acceptable reference standard for the purposes of this systematic review [16]. It is of note that there is currently no international standard for the classification of viral load available.
Study selection and data extraction
Two reviewers (LEB and CE, LEB and SS, or LEB and MB) reviewed the titles and abstracts of all publications identified by the search algorithm independently, followed by a full-text review for those eligible, to select the articles for inclusion in the systematic review. Any disputes were solved by discussion or by a third reviewer (CMD).
A full list of the parameters extracted is included in S1 Table, and the data extraction file is available at https://zenodo.org/record/4924035#.YOlzWS223RZ. Studies that assessed multiple Ag-RDTs or presented results based on differing parameters (e.g., various sample types) were considered as individual datasets.
At first, 4 authors (SK, CE, SS, and MB) extracted 5 randomly selected papers in parallel to align data extraction methods. Afterwards, data extraction and the assessment of methodological quality and independence from test manufacturers (see below) was performed by 1 author per paper (SK, CE, SS, or MB) and controlled by a second (LEB, SK, SS, or MB). Any differences were resolved by discussion or by consulting a third author (CMD).
Study types
We differentiated between clinical accuracy studies (performed on clinical samples) and analytical accuracy studies (performed on spiked samples with a known quantity of virus). Analytical accuracy studies can differ widely in methodology, impeding an aggregation of their results. Thus, while we extracted the data for both kinds of studies, we only considered data from clinical accuracy studies as eligible for the meta-analysis. Separately, we summarized the results of analytical studies and compared them with the results of the meta-analysis for individual tests.
Assessment of methodological quality
The quality of the clinical accuracy studies was assessed by applying the QUADAS-2 tool [17]. The tool evaluates 4 domains: patient selection, index test, reference standard, and flow and timing. For each domain, the risk of bias is analyzed using different signaling questions. Beyond the risk of bias, the tool also evaluates the applicability of each included study to the research question for every domain. The QUADAS-2 tool was adjusted to the needs of this review and can be found in S3 Text.
Assessment of independence from manufacturers
We examined whether a study received financial support from a test manufacturer (including the free provision of Ag-RDTs), whether any study author was affiliated with a test manufacturer, and whether a respective conflict of interest was declared. Studies were judged not to be independent from the test manufacturer if at least 1 of these aspects was present; otherwise, they were considered to be independent.
Statistical analysis and data synthesis
We extracted raw data from the studies and recalculated performance estimates where possible based on the extracted data. The raw data can be found in S2 Table. We prepared forest plots for the sensitivity and specificity of each test and visually evaluated the heterogeneity between studies. If 4 or more datasets were available with at least 20 positive RT-PCR samples per dataset for a predefined analysis, a meta-analysis was performed. We report point estimates of sensitivity and specificity for SARS-CoV-2 detection compared to the reference standard along with 95% confidence intervals (CIs) using a bivariate model (implemented with the “reitsma” command from the R package “mada,” version 0.5.10). When there were fewer than 4 studies for an index test, only a descriptive analysis was performed, and accuracy ranges are reported. In subgroup analyses where papers presented data only on sensitivity, a univariate random-effects inverse variance meta-analysis was performed (using the “metagen” command from the R package “meta,” version 4.11–0). We predefined subgroups for meta-analysis based on the following characteristics: Ct value range, sampling and testing procedure in accordance with manufacturer’s instructions as detailed in the instructions for use (IFU) (henceforth called IFU-conforming) versus not IFU-conforming, age (<18 versus ≥18 years), sample type, presence or absence of symptoms, symptom duration (<7 days versus ≥7 days), viral load, and type of RT-PCR used.
In an effort to use as much of the heterogeneous data as possible, the cutoffs for the Ct value groups were relaxed by 2–3 points within each range. The <20 group included values reported up to ≤20, the <25 group included values reported as ≤24 or <25 or 20–25, and the <30 group included values from ≤29 to ≤33 and 25–30. The ≥25 group included values reported as ≥25 or 25–30, and the ≥30 group included values from ≥30 to ≥35. For the same reason, when categorizing by age, the age group <18 years (children) included samples from persons whose age was reported as <16 or <18 years, whereas the age group ≥18 years (adults) included samples from persons whose age was reported as ≥16 years or ≥18.
For categorization by sample type, we assessed (1) nasopharyngeal (NP) alone or combined with other (e.g., oropharyngeal [OP]), (2) OP alone, (3) anterior nasal (AN) or mid-turbinate (MT), (4) a combination of bronchoalveolar lavage and throat wash (BAL/TW), or (5) saliva. Analyses were preformed using R 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria).
We aimed to do meta-regression to examine the impact of covariates including symptom duration and Ct value range. We also performed the Deeks test for funnel-plot asymmetry as recommended to investigate publication bias for diagnostic test accuracy meta-analyses [18] (using the “midas” command in Stata, version 15); a p-value < 0.10 for the slope coefficient indicates significant asymmetry.
Sensitivity analysis
Two types of sensitivity analyses were planned: estimation of sensitivity and specificity excluding case–control studies, and estimation of sensitivity and specificity excluding non-peer-reviewed studies. We compared the results of each sensitivity analysis against the overall results to assess the potential bias introduced by considering case–control studies and non-peer-reviewed studies.
Results
Summary of studies
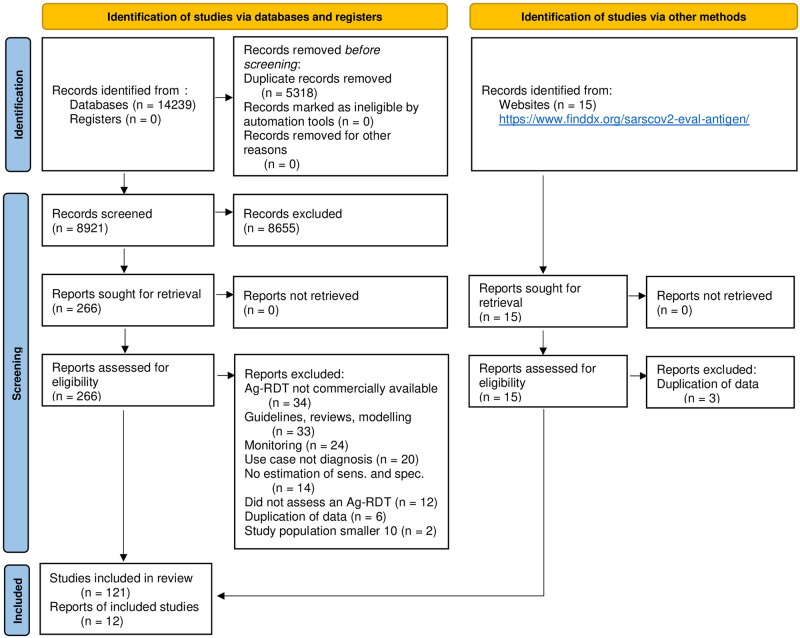
The systematic search resulted in 14,254 articles. After removing duplicates, 8,921 articles were screened, and 266 papers were considered eligible for full-text review. Of these, 148 were excluded because they did not present primary data [13,19–131] or the Ag-RDT was not commercially available [16,132–164], leaving 133 studies to be included in the systematic review (Fig 1) [4,165–296].
Fig 1. PRISMA flow diagram.
Based on Page et al. [297]. Ag-RDT, antigen rapid diagnostic test; IFU, instructions for use; sens., sensitivity; spec., specificity.
At the end of the data extraction process, 37 studies were still in preprint form [4,171,173,174,177,180,190,192,201,204,205,207,211,214–216,218,220,222,223,225,227,231,233,234,238,240,244,247,253,257,265,267,284,287,290,293]. All studies were written in English, except for 2 in Spanish [175,280]. Out of the 133 studies, 9 reported analytical accuracy [173,191,198,208,227,256,274,275,282], and the remaining 124 reported clinical accuracy.
The clinical accuracy studies were divided into 214 datasets, while the 9 analytical accuracy studies accounted for 63 datasets. A total of 61 different Ag-RDTs were evaluated (48 lateral flow with visual readout and 12 requiring an automated reader), with 56 being assessed in a clinical accuracy study. Thirty-nine studies reported data for more than 1 test, and 19 of these studies conducted a head-to-head assessment, i.e., testing at least 2 Ag-RDTs on the same sample or participant. The reference method was RT-PCR in all except 1 study, which used viral culture [281].
The most common reasons for testing were the occurrence of symptoms (55/19.9% of datasets), screening independent of symptoms (19/6.9%), and close contact with a SARS-CoV-2 confirmed case (10/3.6%). In 79 (28.6%) of the datasets, persons were tested due to more than 1 of these reasons, and for 163 datasets (59.1%), the reason for testing was unclear.
In total, 113,242 Ag-RDTs were performed, 112,323 (99.2%) in clinical accuracy studies and 919 (0.8%) in analytical accuracy studies. In the clinical accuracy studies, the mean number of samples per study was 525 (range 16 to 6,954). Only 4,752 (4.2%) tests were performed on pediatric (age group <18 years) samples, and 21,351 (18.9%) on samples from adults (age group ≥18 years). For the remaining 87,139 (76.9%) samples, the age of the persons tested was not specified. Symptomatic patients comprised 36,981 (32.7%) samples; 32,799 (29.0%) samples originated from asymptomatic patients, and for 42,462 (38.4%) samples, the patient’s symptom status could not be identified. The most common sample type evaluated was NP and mixed NP/OP (67,036 samples, 59.2%), followed by AN/MT (27,045 samples, 23.9%). There was substantially less testing done for the other sample types, with 6,254 (5.5%) tests done from OP samples, 1,351 (1.2%) from saliva, and 219 (0.2%) from BAL/TW, and for 11,337 (10.0%) tests, we could not identify the type of sample.
Of the datasets assessing clinical accuracy, 89 (41.6%) involved testing according to the manufacturers’ recommendations (i.e., IFU-conforming), while 100 (46.7%) were not IFU-conforming, and for 25 (11.7%) it was unclear. The most common deviations from the IFU were (1) use of samples that were prediluted in transport media not recommended by the manufacturer (80 datasets; 7 unclear), (2) use of banked samples (60 datasets; 14 unclear), and (3) use of a sample type that was not recommended for the Ag-RDT (17 datasets; 8 unclear).
A summary of the tests evaluated in clinical accuracy studies, including study identification, sample size, sample type, sample condition, and IFU conformity, can be found in Table 1. The Panbio test by Abbott (Germany; henceforth called Panbio) was reported the most frequently, with 39 (18.2%) datasets and 28,089 (25.0%) tests, while the Standard Q test by SD Biosensor (South Korea; distributed in Europe by Roche, Germany; henceforth called Standard Q) was assessed in 37 (17.3%) datasets, with 16,820 (15.0%) tests performed. Detailed results for each clinical accuracy study are available in S1 Fig.
Table 1. Clinical accuracy data for Ag-RDTs against SARS-CoV-2.
| Reference, first author, dataset ID | Study location | Sample type | Sample condition | IFU-conforming | Sample size | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|---|---|---|---|---|
| AAZ, COVID-VIRO (LFA) | |||||||
| [287] Schwob, a35.3 | Switzerland | NP | Fresh | Yes | 324 | 84.1% (76.9%, 89.7%) | 100% (98.0%*, 100%*) |
| Abbott, BinaxNOW (LFA) | |||||||
| [224] Pollock, f17.1 | US | AN | Fresh | Yes | 2,308 | 77.4% (72.2%, 82.1%) | 99.4% (99.0%, 99.7%) |
| [283] Pilarowski, a29.1 | US | AN/MT | Fresh | Yes | 878 | 57.7% (36.9%*, 76.6%*) | 100%* (99.6%*, 100%*) |
| [197] James, f23.1 | US | AN | Fresh | Yes | 2,339 | 56.6% (48.3%*, 64.6%*) | 99.9% (99.6%*, 100%) |
| [217] Okoye, f51.1 | US | MT | Fresh | Yes | 2,645 | 53.3% (37.9%*, 68.3%*) | 100% (99.9%, 100%) |
| Abbott, Panbio (LFA) | |||||||
| [175] Domínguez Fernández, f49.1 | Spain | Unclear | Fresh | Unclear | 30 | 95.0% (75.1%*, 99.9%*) | 100% (69.2%*, 100%*) |
| [250] Alemany, a02.1 | Spain | NP | Banked | No | 919 | 93.4% (91.5%, 95.0%) | 100% (95.8%, 100%) |
| [184] FIND, f42.2 | Germany | NP | Fresh | Yes | 281 | 90.9% (78.3%*, 97.5%*) | 99.2% (97.0%, 99.9%*) |
| [276] Merino-Amador, a25.1 | Spain | NP | Fresh | Yes | 958 | 90.5% (87.0%*, 93.4%*) | 98.8% (97.6%*, 99.5%*) |
| [267] Krüger, a52.1 | Germany | NP | Fresh | Yes | 1,034 | 87.5% (79.6%*, 93.2%*) | 99.9% (99.4%, 100%) |
| [287] Schwob, a35.2 | Switzerland | NP | Fresh | Yes | 271 | 86.1% (78.6%, 91.7%) | 100% (97.6%*, 100%*) |
| [235] Stokes, f65.1 | Canada | NP | Fresh | Yes | 1,641 | 86.2%* (81.5%*, 90.1%*) | 99.9% (99.5%, 100%) |
| [252] Berger, a05.1 | Switzerland | NP | Fresh | Yes | 535 | 85.5% (78.0%, 91.2%) | 100% (99.1%, 100%) |
| [177] Faíco-Filho, f63.1 | Brazil | NP | Fresh | Yes | 127 | 84.3% (73.6%*, 91.9%*) | 98.2%* (90.6%*, 100%*) |
| [196] Jääskeläinen, f50.3 | Finland | NP | Banked | No | 190 | 82.9%* (76.0%*, 88.5%*) | 100% (90.7%*, 100%*) |
| [247] Abdulrahman, a01.1 | Bahrain | AN/MT | Fresh | No | 4,183 | 82.1% (79.2%, 84.8%) | 99.1% (98.8%, 99.4%) |
| [263] Gremmels, a12.2 | Netherlands | NP | Fresh | Yes | 208 | 81.0% (69.1%*, 89.8%*) | 100% (97.5%, 100%) |
| [214] Ngo Nsoga, f28.1 | Switzerland | OP | Fresh | No | 402 | 81.0% (74.2%, 86.6%) | 99.1% (96.9%, 99.9%) |
| [245] Yin, f82.2 | Belgium | NP | Fresh | Yes | 101 | 80.8% (68.1%, 89.2%) | Not provided |
| [249] Albert, a03.1 | Spain | NP | Fresh | Yes | 412 | 79.6% (66.5%*, 89.4%*) | 100% (99.0%, 100%) |
| [250] Alemany, a02.2 | Spain | AN/MT | Banked | No | 487 | 79.5% (71.0%, 86.4%) | 98.6%* (96.9%, 99.6%) |
| [258] Fenollar, a11.1 | France | NP | Fresh | Yes | 341 | 75.5% (69.0%*, 81.2%*) | 94.9% (89.8%*, 97.9%*) |
| [270] Linares, a20.1 | Spain | NP | Fresh | Unclear | 255 | 73.3% (60.3%*, 83.9%*) | 100% (98.1%*, 100%*) |
| [263] Gremmels, a12.1 | Netherlands | NP | Fresh | Yes | 1,367 | 72.7%* (64.5%, 79.9%) | 100% (99.7%, 100%) |
| [192] Halfon, f18.1 | France | NP | Unclear | No | 200 | 72.0% (62.1%*, 80.5%*) | 99.0% (94.6*, 100%) |
| [253] Bulilete, a07.1 | Spain | NP | Fresh | Yes | 1,362* | 71.4% (63.2%*, 78.7%) | 99.8% (99.4%, 99.9%) |
| [165] Akingba, f30.1 | South Africa | NP | Fresh | Unclear | 657* | 69.7%* (61.5%*, 77.0%*) | 99.4%* (98.3%*, 99.9%*) |
| [174] Del Vecchio, f66.1 | Italy | Unclear | Fresh | Unclear | 1,441 | 68.9% (55.7%, 80.1%) | 99.9% (99.6%, 100%) |
| [178] Favresse, f31.2 | Belgium | NP | Fresh | No | 188 | 67.7% (57.4%, 76.9%) | 100% (96.1%, 100%) |
| [257] Drevinek, a10.1 | Czech Republic | NP | Fresh | Yes | 591 | 66.4% (59.8%*, 72.5%*) | 100% (99.0%, 100%) |
| [205] L’Huillier, f72.1 | Switzerland | NP | Fresh | Yes | 822 | 65.5%* (56.3%*, 74.0%) | 99.9%* (99.2%*, 100%) |
| [221] Pérez-García, f52.2 | Spain | NP | Banked | No | 320 | 60.0% (52.2%, 67.4%) | 100% (97.6%, 100%) |
| [248] Agulló, a56.1 | Spain | NP | Fresh | Yes | 652* | 57.6%* (48.7%*, 66.1%*) | 99.8% (98.9%*, 100%) |
| [267] Krüger, a52.2 | Germany | OP | Fresh | No | 74 | 50.0% (1.3%, 98.7%) | 100% (94.9%, 100%) |
| [286] Schildgen, a33.2 | Germany | BAL/TW | Unclear | No | 73 | 50.0% (34.2%*, 65.8%*) | 77.4% (58.9%*, 90.4%*) |
| [292] Torres, a37.1 | Spain | NP | Fresh | Yes | 634 | 48.1% (36.7%*, 59.6%*) | 100% (99.3%, 100%) |
| [244] Wagenhäuser, f89.2 | Germany | OP | Fresh | No | 1,029 | 46.7% (24.8%, 69.9%) | 99.6% (99.0%, 99.9%) |
| [243] Villaverde, f55.1 | Spain | NP | Fresh | Yes | 1,620 | 45.4% (34.1%, 57.2%) | 99.8% (99.4%, 99.9%) |
| [248] Agulló, a56.2 | Spain | AN/MT | Fresh | No | 659 | 44.7% (36.1%, 53.6%) | 100% (99.3%*, 100%) |
| [279] Olearo, a54.2 | Germany | OP | Unclear | No | 184 | 44.0%* (33.2%*, 55.3%*) | 100% (96.4%*, 100%) |
| [170] Caruana, f34.2 | Switzerland | NP | Fresh | No | 532 | 41.2% (32.1%*, 50.8%*) | 99.5% (98.3%*, 99.9%*) |
| [167] Baro, f33.1 | Spain | NP | Banked | No | 286 | 38.6% (29.1%, 48.8%) | 99.5% (97.0%, 100%) |
| [248] Agulló, a56.3 | Spain | Saliva | Fresh | No | 610 | 23.1% (16.0%*, 31.7%*) | 100% (99.2%*, 100%) |
| [213] Muhi, f90.1 | Australia | NP | Fresh | Yes | 2,413 | Not provided | 100% (99.7%, 100%) |
| Abbott, Panbio (nasal sampling) (LFA) | |||||||
| [184] FIND, f42.1 | Germany | AN/MT | Fresh | Yes | 281 | 86.4% (72.6%*, 94.8%*) | 99.2% (97.0%, 99.9%*) |
| Access Bio, CareStart COVID-19 Antigen Test (LFA) | |||||||
| [225] Pollock, f59.1 | US | AN | Fresh | Yes | 1,498 | 57.7% (51.1%, 64.1%) | 98.3% (97.5%, 99.0%) |
| Assure Tech, Ecotest COVID-19 Antigen Rapid Test (LFA) | |||||||
| [194] Homza, f87.1 | Czech Republic | NP | Fresh | Yes | 318 | 75.7% (66.5%, 83.5%) | 96.7% (93.3%, 98.7%) |
| Becton, Dickinson and Company, BD Veritor (requires reader) | |||||||
| [281] Pekosz, a28.1 | US | NP | Fresh | No | 251 | 96.4% (81.7%*, 99.9%*) | 98.7% (96.1%, 99.7%) |
| [293] Van der Moeren, a39.1 | Netherlands | MT/OP | Banked | No | 351* | 94.1% (71.1%, 100%) | 100% (98.9%, 100%) |
| [190] Gomez Marti, f46.2 | US | AN | Fresh | Unclear | Unknown | 93.8% (79.2%*, 99.2%*) | Not provided |
| [245] Yin, f82.1 | Belgium | NP | Fresh | Yes | 177 | 87.7% (80.0%, 92.7%) | Not provided |
| [296] Young, a43.1 | US | NP | Banked | No | 251 | 76.3%* (59.8%*, 88.6%*) | 99.5%* (97.4%*, 99.9%*) |
| [202] Kilic, f71.1 | US | AN | Fresh | Yes | 1,384 | 66.4% (57.0%, 74.9%) | 98.8% (98.1%, 99.3%) |
| [231] Schuit, f64.1 | Netherlands | NP | Fresh | No | 2,678 | 63.9% (57.4%, 70.1%) | 99.6% (99.3%, 99.8%) |
| [170] Caruana, f34.4 | Switzerland | NP | Fresh | No | 532 | 41.2% (32.1%*, 50.8%*) | 99.8%* (98.7%*, 100%*) |
| Becton, Dickinson and Company, Hometest (LFA) | |||||||
| [234] Stohr, f45.1 | Netherlands | AN | Fresh | Unclear | 1,604 | 48.9% (41.3%*, 56.5%*) | 99.9% (99.5%, 100%) |
| Beijing Savant Biotechnology, SARS-CoV-2 detection kit (LFA) | |||||||
| [295] Weitzel, a41.3 | Chile | NP/OP | Banked | No | 109 | 16.7% (9.2%*, 26.8%*) | 100% (88.8%*, 100%) |
| Biotime, COVID-19 Antigen Test Cassette (LFA) | |||||||
| [232] Seitz, f68.1 | Austria | Saliva | Fresh | Yes | 40 | 44.4% (21.5%*, 69.2%*) | 100% (84.6%*, 100%*) |
| Bionote, NowCheck (LFA) | |||||||
| [185] FIND, f91.1 | Brazil | AN | Fresh | Yes | 218 | 89.9% (81.0%*, 95.5%*) | 98.6% (94.9%, 99.8%*) |
| [185] FIND, f91.2 | Brazil | NP | Fresh | Yes | 218 | 89.9% (81.0%*, 95.5%*) | 98.6% (94.9%, 99.8%*) |
| [259] FIND, a61.1 | Brazil | NP | Fresh | Yes | 400 | 89.2% (81.5%*, 94.5%*) | 97.3% (94.8%, 98.8%*) |
| [228] Rottenstreich, f53.1 | Israel | NP | Unclear | Unclear | 1,326 | 55.6% (21.2%, 86.3%) | 100% (99.7%, 100%) |
| Biotical Health, SARS-CoV-2 Ag Card (LFA) | |||||||
| [178] Favresse, f31.1 | Belgium | NP | Fresh | No | 188 | 66.7% (56.3%, 76.0%) | 98.9% (94.1%, 99.9%) |
| Boditech Medical, iChroma COVID-19 Ag Test (requires reader) | |||||||
| [181] FIND, f39.1 | Switzerland | NP | Fresh | Yes | 232 | 73.2% (57.1%*, 85.8%*) | 100% (98.0%, 100%) |
| CerTest Biotec, SARS-CoV-2 one step test card (LFA) | |||||||
| [221] Pérez-García, f52.1 | Spain | NP | Banked | No | 320 | 53.5% (45.7%, 61.2%) | 100% (97.6%, 100%) |
| Coris BioConcept, COVID-19 Ag Respi-Strip (LFA) | |||||||
| [245] Yin, f82.3 | Belgium | NP | Fresh | Yes | 135 | 80.0% (69.2%, 87.7%) | Not provided |
| [277] Mertens, a48.1 | Belgium | NP | Banked | No | 328 | 57.6% (48.7%*, 66.1%*) | 99.5% (97.2%*, 100%*) |
| [269] Lambert-Niclot, a18.1 | France | NP | Fresh | No | 138 | 50.0% (39.5%, 60.5%) | 100% (92.0%*, 100%) |
| [4] Krüger, a17.3 | Germany/England | NP/OP | Unclear | No | 417 | 50.0% (21.5%*, 78.5%) | 95.8% (93.4%, 97.4%) |
| [172] Ciotti, f24.1 | Italy | NP | Fresh | Unclear | 50 | 30.8% (17.0%, 47.6%) | 100% (71.5%, 100%) |
| [288] Scohy, a34.1 | Belgium | NP | Fresh | No | 148 | 30.2% (21.7%, 39.9%) | 100% (91.6%*, 100%*) |
| [294] Veyrenche, a40.1 | France | NP | Fresh | No | 65 | 28.9%* (16.4%*, 44.3%*) | 100% (83.2%, 100%) |
| Denka, Quick Navi (LFA) | |||||||
| [237] Takeuchi, f12.1 | Japan | NP | Fresh | Unclear | 1,186 | 86.7% (78.6%, 92.5%) | 100% (99.7%, 100%) |
| [238] Takeuchi, f60.1 | Japan | AN | Fresh | Unclear | 862 | 72.5% (58.3%, 84.1%) | 100% (99.5%*, 100%) |
| DiaSorin, LIAISON SARS-CoV-2 Ag (LFA) | |||||||
| [206] Lefever, f70.1 | Belgium | NP | Banked | No | 414 | 67.6%* (60.8%*, 74.0%*) | 100% (98.3%*, 100%) |
| Dräger, Antigen Test SARS-CoV-2 (LFA) | |||||||
| [218] Osmanodja, f79.1 | Germany | NP/OP | Fresh | Yes | 379 | 88.6% (78.7%, 94.9%) | 99.7% (98.2%, 100%) |
| E25Bio, Rapid Diagnostic Test (LFA) | |||||||
| [223] Pickering, f73.2 | UK | AN/OP | Banked | No | 200 | 75.0% (65.3%*, 83.1%*) | 86% (77.6%*, 92.1%*) |
| ECO Diagnóstica, COVID-19 Ag (LFA) | |||||||
| [180] Filgueiras, f14.1 | Brazil | NP | Fresh | Unclear | 150 | 69.1% (55.2%*, 80.9%*) | 98.8% (93.5%*, 100%) |
| Fujirebio, ESPLINE SARS-CoV-2 (LFA) | |||||||
| [290] Takeda, a50.1 | Japan | NP | Unclear | No | 162 | 80.6%* (68.6%*, 89.6%*) | 100%* (96.4%*, 100%*) |
| [186] FIND, f92.1 | Germany | NP/OP | Fresh | No | 723 | 78.6% (69.8%*, 85.8%*) | 100% (99.4%, 100%) |
| [230] Sberna, f83.1 | Italy | Saliva | Unclear | Unclear | 136 | 8.1% (2.7%, 17.8%) | 100% (95.1%, 100%) |
| Fujirebio, Lumipulse G SARS-CoV-2 Ag (requires reader) | |||||||
| [189] Gili, f57.2 | Italy | NP | Banked | No | 226 | 100% (96.0%*, 100%*) | 92.1% (90.7%*, 93.4%*) |
| [189] Gili, f57.1 | Italy | NP | Fresh | No | 1,738 | 90.5% (82.8%*, 95.6%*) | 91.6% (85.5%*, 95.7%*) |
| [193] Hirotsu, f47.1 | Japan | NP | Banked | No | 1,033 | 92.5% (79.6%*, 98.4%*) | 100%* (99.6%*, 100%*) |
| [168] Basso, f10.1 | Italy | NP | Fresh | Yes | 234 | 81.6% (71.9%*, 89.1%*) | 93.9%* (88.7%*, 97.2%*) |
| [166] Asai, f74.1 | Japan | Saliva | Unclear | Yes | 305 | 77.8% (65.5%*, 87.3%*) | 98.3% (95.8%*, 99.5%*) |
| [168] Basso, f10.2 | Italy | Saliva | Fresh | Yes | 223 | 41.3% (30.4%, 52.8%) | 98.6% (95.0%, 99.8%) |
| Guangzhou Wondfo Biotech, 2019-nCoV Antigen Test (LFA) | |||||||
| [183] FIND, f41.1 | Switzerland | NP | Fresh | Yes | 328 | 85.7% (73.8%*, 93.6%*) | 100% (98.7%*, 100%*) |
| Humasis, COVID-19 Ag Test (LFA) | |||||||
| [169] Bruzzone, f86.2 | Italy | Unclear | Banked | No | 21 | 85.7% (63.7%*, 97%*) | Not provided |
| Healgen, Rapid COVID-19 Ag Test (LFA) | |||||||
| [178] Favresse, f31.3 | Belgium | NP | Fresh | No | 188 | 77.1% (67.4%, 85.1%) | 96.7% (90.8%, 99.3%) |
| Innova Medical Group, INNOVA SARS-CoV-2 Antigen Rapid Qualitative Test (LFA) | |||||||
| [223] Pickering, f73.1 | UK | AN/OP | Banked | No | 200 | 89.0% (81.2%*, 94.4%*) | 99.0% (94.6%, 100%) |
| [195] Houston, f25.1 | UK | NP | Fresh | Yes | 242 | 86.4% (81.9%*, 90.2%*) | 95.1% (92.7%*, 96.9%*) |
| [223] Pickering, f73.10 | UK | AN/OP | Banked | No | 23 | 82.6% (61.2%*, 95.0%*) | Not provided |
| [223] Pickering, f73.11 | UK | AN/OP | Banked | No | 23 | 82.6% (61.2%*, 95.0%*) | Not provided |
| [222] Peto, f21.1 | UK | Unclear | Unclear | Unclear | 6,954 | Not provided | 99.7% (99.5%*, 99.8%*) |
| [222] Peto, f21.4 | UK | Unclear | Unclear | Unclear | 198 | 78.8% (72.4%, 84.3%) | Not provided |
| [223] Pickering, f73.12 | UK | AN/OP | Banked | No | 23 | 78.3% (56.3%*, 92.5%*) | Not provided |
| [223] Pickering, f73.8 | UK | AN/OP | Banked | No | 110 | 78.2% (69.3%*, 85.5%*) | Not provided |
| [222] Peto, f21.3 | UK | Unclear | Unclear | Unclear | 223 | 70.0% (63.5%, 75.9%) | Not provided |
| [246] Young, f56.1 | UK | NP | Fresh | Unclear | 803 | 62.1%*(55.3%*, 68.7%*) | 100% (99.4%, 100%) |
| [222] Peto, f21.2 | UK | Unclear | Unclear | Unclear | 372 | 57.5% (52.3%, 62.6%) | Not provided |
| [179] Ferguson, f85.1 | UK | AN | Fresh | Yes | 720 | 3.2% (0.6%, 15.6%) | 100% (99.5%, 100%) |
| JOYSBIO Biotechnology, COVID-19 Antigen Rapid Test Kit (LFA) | |||||||
| [182] FIND, f40.1 | Switzerland | NP | Fresh | Yes | 265 | 70.5% (54.8%*, 83.2%*) | 99.1% (96.8%*, 99.9%*) |
| [194] Homza, f87.2 | Czech Republic | NP | Fresh | Yes | 225 | 57.8% (46.9%, 68.1%) | 98.5% (94.8%, 99.8%) |
| Lab Care Diagnostics, PathoCatch/ACCUCARE SARS-CoV-2 Antigen Test (LFA) | |||||||
| [239] Thakur, f88.1 | India | NP | Fresh | Yes | 677 | 34.5% (24.5%, 45.6%) | 99.8% (99.1%, 100%) |
| Lepu Medical Technology, SARS-CoV-2 Antigen Rapid Test Kit (LFA) | |||||||
| [167] Baro, f33.4 | Spain | NP | Banked | No | 286 | 45.5% (35.6%, 55.8%) | 89.2% (83.8%, 93.3%) |
| Liming Bio, SARS-CoV-2 Ag-RDT (LFA) | |||||||
| [295] Weitzel, a41.2 | Chile | NP/OP | Banked | No | 19 | 0% (0%, 29.9%) | 90.0% (59.6%, 98.2%) |
| LumiraDx, COVID-19 SARS-CoV-2 Antigen Test (requires reader) | |||||||
| [176] Drain, f43.1 | UK/US | AN | Fresh | Yes | 257 | 97.6% (91.6%*, 99.7%*) | 96.6% (92.6%*, 98.7%*) |
| [176] Drain, f43.2 | UK/US | NP | Fresh | Yes | 255 | 97.5% (86.8%*, 99.9%*) | 97.7% (94.7%, 99.2%*) |
| [204] Krüger, f58.1 | Germany | MT | Fresh | Yes | 761 | 82.2% (75.0%*, 88.0%*) | 99.3% (98.3%, 99.7%) |
| [169] Bruzzone, f86.6 | Italy | Unclear | Banked | No | 23 | 69.6% (47.1%*, 86.8%*) | Not provided |
| [203] Kohmer, f32.4 | Germany | NP | Fresh | No | 100 | 50.0% (38.1%, 61.9%) | 100% (86.8%, 100%) |
| [211] Micocci, f77.1 | UK | NP | Fresh | Unclear | 241 | 75.0%* (34.9%*, 96.8%*) | 96.1%* (92.7%*, 98.2%*) |
| MEDsan, SARS-CoV-2 Antigen Rapid Test (LFA) | |||||||
| [279] Olearo, a54.3 | Germany | OP | Unclear | No | 184 | 45.2%* (34.3%*, 56.5%) | 97.0% (91.5%, 99.4%*) |
| [244] Wagenhäuser, f89.3 | Germany | OP | Fresh | Yes | 3,221 | 36.5% (24.7%*, 49.6%*) | 99.6% (99.3%, 99.8%) |
| Mologic, COVID-19 Rapid Antigen Test (LFA) | |||||||
| [187] FIND, f93.1 | Germany | AN/MT | Fresh | Yes | 665 | 90.7% (85.7%*, 94.4%*) | 100% (99.2%, 100%) |
| nal von minden, NADAL (LFA) | |||||||
| [188] FIND, f94.1 | Switzerland | NP | Fresh | Yes | 462 | 88.4% (78.4%*, 94.9%*) | 99.2% (97.8%, 99.7%) |
| [236] Strömer, f11.1 | Germany | NP | Banked | No | 124 | 63.7%* (54.6%*, 72.2%*) | Not provided |
| [244] Wagenhäuser, f89.1 | Germany | OP | Fresh | Yes | 806 | 56.5% (34.5%*, 76.8%*) | 100% (99.5%, 100%) |
| [203] Kohmer, f32.3 | Germany | NP | Fresh | No | 100 | 24.3% (15.1%, 35.7%) | 100% (86.8%, 100%) |
| NanoEntek, FREND COVID-19 Ag (requires reader) | |||||||
| [169] Bruzzone, f86.7 | Italy | Unclear | Banked | No | 60 | 93.3% (83.8%*, 98.2%*) | Not provided |
| NDFOS, ND COVID-19 Ag Test (LFA) | |||||||
| [194] Homza, f87.3 | Czech Republic | NP | Fresh | Yes | 191 | 70.1% (58.6%, 80.0%) | 56.1% (46.4%, 65.4%) |
| Ortho Clinical Diagnostics, VITROS SARS-CoV-2 Antigen Test (requires reader) | |||||||
| [178] Favresse, f31.5 | Belgium | NP | Fresh | No | 188 | 83.3% (74.4%, 90.2%) | 100% (96.1%, 100%) |
| Precision Biosensor, Exdia COVID-19 Ag (requires reader) | |||||||
| [170] Caruana, f34.3 | Switzerland | NP | Fresh | No | 532 | 48.3% (38.8%*, 57.8%*) | 99.5% (98.3%*, 99.9%*) |
| PRIMA Lab, COVID-19 Antigen Rapid Test (LFA) | |||||||
| [169] Bruzzone, f86.3 | Italy | Unclear | Banked | No | 50 | 66.0% (51.2%*, 78.8%*) | Not provided |
| Quidel, Sofia SARS Antigen FIA (requires reader) | |||||||
| [284] Porte, a32.1 | Chile | NP/OP | Banked | No | 64 | 93.8% (79.2%*, 99.2%*) | 96.9% (83.8%*, 99.9%*) |
| [196] Jääskeläinen, f50.1 | Finland | NP | Banked | No | 188 | 80.4% (73.1%*, 86.5%*) | 100% (91.2%*, 100%*) |
| [251] Beck, a04.1 | US | NP | Fresh | Yes | 346 | 77.0% (64.5%*, 86.8%*) | 99.6% (98.1%*, 100%*) |
| [265] Herrera, a46.1 | US | Unclear | Unclear | Unclear | 1,172 | 76.8% (72.6%, 80.5%) | 99.2% (98.2%, 99.7%) |
| [190] Gomez Marti, f46.1 | US | MT | Fresh | Unclear | 427 | 72.0% (56.3%*, 84.7%*) | 99.7%* (98.6%*, 100%*) |
| RapiGEN, Biocredit Covid-19 Ag (LFA) | |||||||
| [289] Shrestha, a36.1 | Nepal | NP | Fresh | Yes | 113 | 85.0% (71.7%*, 93.8%*) | 100% (94.6%*, 100%*) |
| [260] FIND, a62.1 | Brazil | NP | Fresh | Yes | 476 | 74.4% (65.5%*, 82.0%*) | 98.9%* (97.2%, 99.7%*) |
| [295] Weitzel, a41.1 | Chile | NP/OP | Banked | No | 109 | 62.0% (50.4%*, 72.7%*) | 100% (88.4%*, 100%) |
| [233] Shidlovskaya, f61.1 | Russia | NP | Fresh | Yes | 106 | 56.4% (44.7%, 67.6%) | 100% (87.7%, 100%) |
| [260] FIND, a62.2 | Germany | NP | Fresh | Yes | 1,239 | 52.0% (31.3%*, 72.2%*) | 100% (99.7%, 100%) |
| [169] Bruzzone, f86.4 | Italy | Unclear | Banked | No | 23 | 39.1% (19.7%*, 61.5%*) | Not provided |
| [286] Schildgen, a33.1 | Germany | BAL/TW | Unclear | No | 73 | 33.3% (19.6%*, 49.6%*) | 87.1% (70.2%*, 96.4%*) |
| [200] Kenyeres, f84.1 | Hungary | NP | Fresh | No | 37 | 8.1% (1.7%*, 21.9%*) | Not provided |
| R-Biopharm, RIDA QUICK SARS-CoV-2 Antigen (LFA) | |||||||
| [291] Toptan, a55.1 | Germany | NP/OP | Banked | No | 67 | 77.6% (64.7%*, 87.5%*) | 100% (66.4%*, 100%*) |
| [291] Toptan, a55.2 | Germany | Unclear | Banked | No | 70 | 50.0% (31.9%*, 68.1%*) | 100% (90.8%*, 100%*) |
| [203] Kohmer, f32.1 | Germany | NP | Fresh | No | 100 | 39.2% (28.0%, 51.2%) | 96.2% (80.4%, 99.9%) |
| Roche, Elecsys SARS-CoV-2 Antigen Test (requires reader) | |||||||
| [216] Nörz, f78.1 | Germany | NP/OP | Banked | No | 3,139 | 60.2% (55.2%, 65.1%) | 99.9% (99.6%, 100%) |
| Roche, SARS-CoV-2 Rapid Antigen Test (LFA) | |||||||
| [240] Thell, f81.1 | Austria | Unclear | Fresh | Unclear | 591 | 80.3% (74.3%, 85.4%) | 99.1% (97.4%, 99.8%) |
| Salofa Oy, Sienna COVID-19 Antigen Rapid Test Cassette (LFA) | |||||||
| [209] Mboumba Bouassa, f67.1 | France | NP | Banked | No | 100 | 90.0% (82.4%*, 95.1%*) | 100% (92.9%*, 100%) |
| SD Biosensor, Standard F (requires reader) | |||||||
| [284] Porte, a32.2 | Chile | NP/OP | Banked | No | 64 | 90.6% (75.0%*, 98.0%*) | 96.9% (83.8%*, 99.9%*) |
| [169] Bruzzone, f86.5 | Italy | Unclear | Banked | No | 60 | 86.7% (75.4%*, 94.1%*) | Not provided |
| [261] FIND, a63.1 | Brazil | NP | Fresh | Yes | 453 | 77.5% (69.0%*, 84.6%*) | 97.9% (95.7%, 99.2%*) |
| [261] FIND, a63.2 | Germany | NP | Fresh | Yes | 676 | 69.2% (52.4%*, 83.0%*) | 96.9% (95.2%, 98.0%) |
| [257] Drevinek, a10.2 | Czech Republic | NP | Fresh | Yes | 591 | 62.3% (55.6%*, 68.7%*) | 99.5% (98.0%, 99.9%) |
| [219] Osterman, f20.1 | Germany | NP/OP | Unclear | No | 360 | 60.9% (53.5%*, 67.8%*) | 97.8% (95.7%, 99.0%*) |
| [273] Liotti, a22.1 | Italy | NP | Banked | No | 359 | 47.1% (37.1%, 57.1%) | 98.4% (96.0%, 99.6%) |
| SD Biosensor/Roche, Standard Q (LFA) | |||||||
| [255] Chaimayo, a57.1 | Thailand | NP/OP | Banked | No | 454 | 98.3% (91.1%, 100%) | 98.7% (97.1%, 99.6%) |
| [169] Bruzzone, f86.1 | Italy | Unclear | Banked | No | 16 | 93.8% (71.7%, 98.9%) | Not provided |
| [201] Kernéis, f69.1 | France | NP | Fresh | Unclear | 1,109 | 94.2%* (87.0%*, 98.1%*) | 99.0% (98.2%*, 99.5%*) |
| [287] Schwob, a35.1 | Switzerland | NP | Fresh | Yes | 333 | 92.9% (86.4%, 96.9%) | 100% (98.3%*, 100%*) |
| [215] Nikolai, f35.3 | Germany | NP | Fresh | Yes | 96 | 91.2% (76.3%*, 98.1%*) | 100% (94.2%, 100%) |
| [252] Berger, a05.2 | Switzerland | NP | Fresh | Yes | 529 | 89.0% (83.7%, 93.1%) | 99.7% (98.4%, 100%) |
| [262] FIND, a64.1 | Brazil | NP | Fresh | Yes | 400 | 88.7% (81.1%*, 94.0%*) | 97.6% (95.2%, 99.0%*) |
| [286] Schildgen, a33.3 | Germany | BAL/TW | Unclear | No | 73 | 88.1% (74.4%*, 96.0%*) | 19.4% (7.5%*, 37.5%*) |
| [207] Lindner, f15.1 | Germany | NP | Fresh | Yes | 139 | 85.0% (70.2%*, 94.3%*) | 99.1% (94.9%*, 100%*) |
| [266] Iglὁi, a15.1 | Netherlands | NP | Fresh | Yes | 970 | 84.9% (79.0%*, 89.8%*) | 99.5% (98.7%, 99.9%*) |
| [264] Gupta, a13.1 | India | NP | Fresh | Yes | 330 | 81.8% (71.4%*, 89.7%*) | 99.6% (97.8%, 99.9%) |
| [196] Jääskeläinen, f50.2 | Finland | NP | Banked | No | 198 | 81.0% (74.0%*, 86.8%*) | 100% (91.2%*, 100%*) |
| [242] Turcato, f09.1 | Italy | NP | Fresh | Unclear | 3,410 | 80.3% (74.4%*, 85.3%*) | 99.1% (98.7%*, 99.4%*) |
| [272] Lindner, a21.2 | Germany | NP | Fresh | Yes | 289 | 79.5% (63.5%*, 90.7%*) | 99.6% (97.8%, 100%) |
| [245] Yin, f82.4 | Belgium | NP | Fresh | Yes | 65 | 78.3% (58.1%, 90.3%) | Not provided |
| [4] Krüger, a17.1 | Germany/England | NP/OP | Unclear | No | 1,263 | 76.6% (62.0%*, 87.7%*) | 99.3% (98.6%, 99.7%*) |
| [212] Möckel, f19.1 | Germany | NP/OP | Fresh | Yes | 271 | 75.3% (65.0%*, 83.8%*) | 100% (98.0%*, 100%) |
| [272] Lindner, a21.1 | Germany | AN/MT | Fresh | No | 289 | 74.4% (57.9%*87.0%*) | 99.2% (97.1%, 99.9%*) |
| [271] Lindner, a53.1 | Germany | NP | Fresh | Yes | 180 | 73.2%* (57.1%*, 85.8%*) | 99.3% (96.0%, 100%) |
| [229] Salvagno, f54.1 | Italy | NP | Unclear | No | 321 | 72.5% (64.6%, 79.5%) | 99.4% (96.8%, 100%) |
| [254] Cerutti, a08.1 | Italy | NP | Unclear | No | 185 | 72.1% (62.5%*, 80.5%*) | 100% (95.6%*, 100%*) |
| [212] Möckel, f19.2 | Germany | NP/OP | Fresh | Yes | 2,020 | 72.0% (50.6%*, 87.9%*) | 99.4% (96.9%*, 100%*) |
| [268] Krüttgen, a16.1 | Germany | NP | Banked | No | 150 | 70.7% (59.0%*, 80.6%*) | 96.0% (88.8%*, 99.2%*) |
| [278] Nalumansi, a27.1 | Uganda | NP | Fresh | Yes | 262 | 70.0% (59.4%*, 79.2%*) | 92.4%* (87.4%*, 95.9%*) |
| [220] Pena, f36.1 | Chile | NP | Fresh | Yes | 842 | 69.9% (58.0%*, 80.1%*) | 99.6% (98.9%, 99.9%) |
| [178] Favresse, f31.4 | Belgium | NP | Fresh | No | 188 | 69.8% (59.6%, 78.8%) | 100% (96.1%, 100%) |
| [219] Osterman, f20.2 | Germany | NP/OP | Unclear | No | 386 | 64.5% (58.3%*, 70.3%*) | 97.7% (95.6%, 98.9%*) |
| [194] Homza, f87.4 | Czech Republic | NP | Fresh | Yes | 139 | 61.9% (45.6%, 76.4%) | 99.0% (94.4%, 100%) |
| [231] Schuit, f64.2 | Netherlands | NP | Fresh | Yes | 1,596 | 62.9% (54.0%, 71.1%) | 99.5% (98.9%, 99.8%) |
| [226] Ristić, f44.1 | Serbia | NP | Fresh | Unclear | 120 | 58.1% (42.1%, 73.0%) | 100% (95.3%*, 100%*) |
| [199] Kannian, f26.1 | India | Saliva | Unclear | No | 37 | 55.6%* (35.3%*, 74.5%*) | 100% (69.2%*, 100%*) |
| [279] Olearo, a54.1 | Germany | OP | Unclear | No | 184 | 48.8%* (37.7%*, 60.0%*) | 100% (96.4%*, 100%) |
| [167] Baro, f33.3 | Spain | NP | Banked | No | 286 | 43.6% (33.7%, 53.8%) | 96.2% (92.4%, 98.5%) |
| [203] Kohmer, f32.2 | Germany | NP | Fresh | No | 100 | 43.2% (31.8%*, 55.3%) | 100% (86.8%, 100%) |
| [170] Caruana, f34.1 | Switzerland | NP | Fresh | No | 532 | 41.2% (32.1%*, 50.8%*) | 99.8%* (98.7%*, 100%*) |
| [254] Cerutti, a08.2 | Italy | NP | Fresh | No | 145 | 40.0% (5.3%*, 85.3%*) | 100% (97.4%*, 100%*) |
| [171] Caruana, f75.1 | Switzerland | NP | Fresh | Unclear | 116 | 28.6% (3.7%*, 71.0%*) | 98.2% (93.5%*, 99.8%*) |
| SD Biosensor/Roche, Standard Q (nasal sampling) (LFA) | |||||||
| [215] Nikolai, f35.4 | Germany | MT | Fresh | Yes | 96 | 91.2% (76.3%*, 98.1%*) | 98.4% (91.3%*, 100%*) |
| [215] Nikolai, f35.2 | Germany | MT | Fresh | Yes | 132 | 86.1% (70.5%*, 95.3%*) | 100% (96.2%*, 100%*) |
| [215] Nikolai, f35.1 | Germany | AN | Fresh | Yes | 132 | 86.1% (70.5%*, 95.3%*) | 100% (96.2%*, 100%*) |
| [207] Lindner, f15.2 | Germany | MT | Fresh | Yes | 180 | 82.5% (67.2%*92.7%*) | 100% (96.5%, 100%) |
| [234] Stohr, f45.2 | Netherlands | AN | Fresh | Unclear | 1,611 | 61.5% (54.2%*, 68.4%*) | 99.7% (99.3%, 99.9%) |
| [271] Lindner, a53.2 | Germany | AN | Fresh | Yes | 179 | 80.5% (65.1%*, 91.2%*) | 98.6% (94.9%, 99.8%*) |
| Shenzhen Lvshiyuan Biotechnology, Green Spring SARS-CoV-2-Antigen-Schnelltest-Set (LFA) | |||||||
| [223] Pickering, f73.4 | UK | AN/OP | Banked | No | 200 | 77.0% (67.5%*, 84.8%*) | 98.0% (93.0%, 99.8%*) |
| Shenzhen Bioeasy Biotechnology, 2019-nCov Antigen Rapid Test Kit (requires reader) | |||||||
| [285] Porte, a31.1 | Chile | NP/OP | Banked | No | 127 | 93.9% (86.3%*, 98.0%*) | 100% (92.1%*, 100%*) |
| [295] Weitzel, a41.4 | Chile | NP/OP | Banked | No | 111 | 85.0% (75.3%*, 92.0%*) | 100% (88.8%*, 100%) |
| [280] Parada-Ricart, a58.1 | Spain | NP | Fresh | Yes | 172 | 73.1%* (52.2%*, 88.4%*) | 85.6%* (78.9%*, 90.9%*) |
| [4] Krüger, a17.2 | Germany | NP/OP | Fresh | No | 727* | 66.7% (41.7%, 84.8%) | 93.1% (91.0%, 94.8%) |
| Siemens Healthineers, CLINITEST Rapid COVID-19 Antigen Test (LFA) | |||||||
| [241] Torres, f29.1 | Spain | NP | Fresh | Yes | 178 | 80.2% (70.6%*, 87.8%*) | 100% (95.8%, 100%) |
| [241] Torres, f29.2 | Spain | NP | Fresh | Yes | 92 | 60.0% (38.7%*, 78.9%*) | 100% (94.6%, 100%) |
| [279] Olearo, a54.4 | Germany | OP | Unclear | No | 170 | 54.8%* (43.5%*, 65.7%*) | 100% (95.8%*, 100%) |
| [167] Baro, f33.2 | Spain | NP | Banked | No | 286 | 51.5% (41.3%, 61.6%) | 98.4% (95.3%, 99.7%*) |
| Sugentech, SGTi-flex COVID-19 Ag (LFA) | |||||||
| [233] Shidlovskaya, f61.2 | Russia | NP | Fresh | Yes | 106 | 52.6% (40.9%, 64.0%) | 96.4% (81.7%, 99.9%) |
| SureScreen Diagnostics, COVID-19 Rapid Antigen Visual Read (LFA) | |||||||
| [223] Pickering, f73.14 | UK | AN/OP | Banked | No | 23 | 74.0%* (51.6%*, 89.8%*) | Not provided |
| [223] Pickering, f73.3 | UK | AN/OP | Banked | No | 200 | 65.0% (54.8%*, 74.3%*) | 100% (96.4%*, 100%*) |
| [223] Pickering, f73.15 | UK | AN/OP | Banked | No | 23 | 65.2% (42.7%*, 83.6%*) | Not provided |
| [223] Pickering, f73.13 | UK | AN/OP | Banked | No | 23 | 61.0%* (38.5%*, 80.3%*) | Not provided |
| [167] Baro, f33.5 | Spain | NP | Banked | No | 286 | 28.8% (20.2%, 38.6%) | 97.8% (94.5%, 99.4%) |
| SureScreen Diagnostics, COVID-19 Rapid Antigen Fluorescent (requires reader) | |||||||
| [223] Pickering, f73.6 | UK | AN/OP | Banked | No | 200 | 69.0% (59.0%*, 77.9%*) | 98.0% (93%, 99.8%*) |
| [223] Pickering, f73.7 | UK | AN/OP | Banked | No | 141 | 60.3% (51.7%*, 68.4%*) | Not provided |
| VivaCheck, VivaDiag SARS-CoV-2 Ag Rapid Test (LFA) | |||||||
| [194] Homza, f87.5 | Czech Republic | NP | Fresh | Yes | 268 | 41.8% (31.5%, 52.6%) | 96.0% (92.0%, 98.4%) |
| Zhuhai Encode Medical Engineering, SARS-CoV-2 Antigen Rapid Test (LFA) | |||||||
| [223] Pickering, f73.5 | UK | AN/OP | Banked | No | 200 | 74.0% (64.3%*, 82.3%*) | 100% (96.4%*, 100%) |
| [223] Pickering, f73.9 | UK | AN/OP | Banked | No | 90 | 74.4% (64.2%*, 83.1%*) | Not provided |
Datasets with an underlined reference and first author had not undergone peer-review yet at the time of data extraction (1 May 2021). In datasets with an underlined sample size, the samples were used in head-to-head studies, i.e., performing different Ag-RDTs on the same patient.
*Values differ from those provided in the respective paper due to missing or contradictory data. A list including the original data can be found in S2 Table.
AN, anterior nasal; BAL/TW, bronchoalveolar lavage and throat wash; CI, confidence interval; IFU, instructions for use; FIND, Foundation for Innovative New Diagnostics; LFA, lateral flow assay; MT, mid-turbinate; NP, nasopharyngeal; OP, oropharyngeal.
Methodological quality of studies
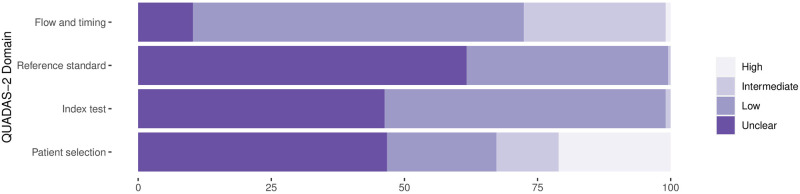
The findings on study quality using the QUADAS-2 tool are presented in Figs 2 and 3. In 190 (88.8%) datasets a relevant patient population was assessed. However, for only 44 (20.6%) of the datasets was patient selection considered representative of the setting and population chosen (i.e., they avoided inappropriate exclusions and a case–control design, and enrollment occurred consecutively or randomly).
Fig 2. Methodological quality of the clinical accuracy studies: Risk of bias.
Proportion of studies with low, intermediate, high, or unclear risk of bias (percent).
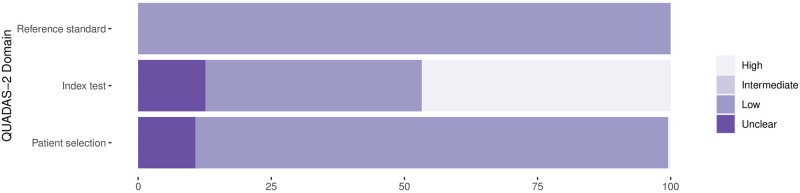
Fig 3. Methodological quality of the clinical accuracy studies: Applicability.
Proportion of studies with low, intermediate, high, or unclear concerns regarding applicability (percent).
The conduct and interpretation of the index tests was considered to have low risk for introduction of bias in 113 (52.8%) datasets (through, e.g., appropriate blinding of persons interpreting the visual readout). However, for 99 (46.3%) datasets, sufficient information to clearly judge the risk of bias was not provided. In only 89 (41.6%) datasets were the Ag-RDTs performed according to IFU, while 100 (46.7%) were not IFU-conforming, potentially impacting the diagnostic accuracy (for 25 [11.7%] datasets the IFU status was unclear).
In 81 (37.9%) datasets, the reference standard was performed before the Ag-RDT, or the operator conducting the reference standard was blinded to the Ag-RDT results, resulting in a low risk of bias. In almost all other datasets (132/61.7%), this risk could not be assessed due to missing data. The applicability of the reference test was judged to be of low concern for all datasets, as cell culture and RT-PCR are expected to adequately define the target condition.
In 209 (97.7%) datasets, the sample for the index test and reference test were obtained at the same time, while this was unclear in 5 (2.3%) datasets. All samples included in a dataset were subjected to the same type of RT-PCR in 145 (67.8%) datasets, while different types of RT-PCR were used within the same dataset in 50 (23.4%) datasets. For 19 (8.9%) datasets, it was unclear. Furthermore, for 11 (5.1%) datasets, there was a concern that not all selected patients were included in the analysis.
Finally, 32 (24.1%) of the studies received financial support from the Ag-RDT manufacturer, and in another 9 (6.8%) studies, employment of the authors by the manufacturer of the Ag-RDT studied was indicated. Overall, a competing interest was found in 33 (24.8%) of the studies.
Detection of SARS-CoV-2 infection
Out of 214 clinical datasets (from 124 studies), 20 were excluded from the meta-analysis because they included fewer than 20 RT-PCR positive samples. A further 21 datasets were missing either sensitivity or specificity and were only considered for univariate analyses. Across the remaining 173 datasets, including any test and type of sample, the pooled sensitivity and specificity were 71.2% (95% CI 68.2% to 74.0%) and 98.9% (95% CI 98.6% to 99.1%), respectively. If testing was performed in conformity with IFU, sensitivity increased to 76.3% (95% CI 73.1% to 79.2%), while non-IFU-conforming testing had a sensitivity of 65.9% (95% CI 60.6% to 70.8%). Pooled specificity was similar in both groups (99.1% [95% CI 98.8–99.4%] and 98.3% [95% CI 97.7% to 98.8%], respectively).
Analysis of specific tests
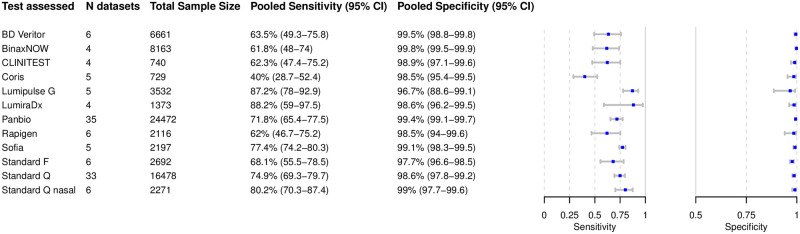
Based on 119 datasets with 71,424 tests performed, we were able to perform bivariate meta-analysis of the sensitivity and specificity for 12 different Ag-RDTs (Fig 4). Across these, the pooled estimates of sensitivity and specificity on all samples were 72.1% (95% CI 68.8% to 75.3%) and 99.0% (95% CI 98.7% to 99.2%), respectively, which were very similar to the overall pooled estimates across all meta-analyzed datasets (71.2% and 98.9%, respectively, above).
Fig 4. Bivariate analysis of 12 antigen rapid diagnostic tests.
Pooled sensitivity and specificity were calculated based on reported sample sizes, true positives, true negatives, false positives, and false negatives.
The highest pooled sensitivity was found for the SARS-CoV-2 Antigen Test by LumiraDx (UK; henceforth called LumiraDx) and the Lumipulse G SARS-CoV-2 Ag by Fujirebio (Japan; henceforth called Lumipulse G), with 88.2% (95% CI 59.0% to 97.5%) and 87.2% (95% CI 78.0% to 92.9%), respectively. The Sofia SARS Antigen FIA by Quidel (California, US; henceforth called Sofia) had a pooled sensitivity of 77.4% (95% CI 74.2% to 80.3%). Of the non-instrument tests, the Standard Q and the Standard Q nasal test by SD Biosensor (South Korea; distributed in Europe by Roche, Germany; henceforth called Standard Q nasal) performed best, with a pooled sensitivity of 74.9% (95% CI 69.3% to 79.7%) and 80.2% (95% CI 70.3% to 87.4%), respectively. The pooled sensitivity for Panbio was 71.8% (95% CI 65.4% to 77.5%). Of all Ag-RDTs, the COVID-19 Ag Respi-Strip by Coris BioConcept (Belgium; henceforth called Coris) had the lowest pooled sensitivity, 40.0% (95% CI 28.7% to 52.4%).
The pooled specificity was above 98% for all of the tests, except for the Standard F by SD Biosensor (South Korea) and Lumipulse G, with specificities of 97.7% (95% CI 96.6% to 98.5%) and 96.7% (95% CI 88.6% to 99.1%), respectively. Hierarchical summary receiver operating characteristic values for Standard Q and LumiraDx are available in S2 Fig.
Three Ag-RDTs did not have sufficient data to allow for a bivariate meta-analysis, so a univariate analysis was conducted (Fig 5). For the INNOVA SARS-CoV-2 Antigen Rapid Qualitative Test by Innova Medical Group (California, US), this resulted in a pooled sensitivity and specificity of 76.1% (95% CI 68.1% to 84.1%) and 99.4% (95% CI 98.7% to 100%), respectively. For the NADAL by nal von minden (Germany) and the COVID-19 Rapid Antigen Visual Read by SureScreen Diagnostics (UK), sufficient data were available to analyze only sensitivity, resulting in pooled sensitivity estimates of 58.4% (95% CI 29.2% to 87.6%) and 58.0% (95% CI 38.3% to 77.6%), respectively.
Fig 5. Univariate analysis of 3 antigen rapid diagnostic tests.
Pooled sensitivity and specificity were calculated based on reported sensitivity, specificity, and confidence intervals. SureScreen V, SureScreen Diagnostics COVID-19 Rapid Antigen Visual Read.
The remaining 35 Ag-RDTs did not present sufficient data for univariate or bivariate meta-analysis. However, 9/35 had results presented in more than 1 dataset, and these are summarized in Table 2. Herein, the widest ranges of sensitivity were found for the ESPLINE SARS-CoV-2 by Fujirebio (Japan), with sensitivity reported between 8.1% and 80.7%, and the RIDA QUICK SARS-CoV-2 Antigen by R-Biopharm (Germany), with sensitivity between 39.2% and 77.6%, both with 3 datasets each. In contrast, 2 other tests with 2 datasets each showed the least variability in sensitivity: The Zhuhai Encode Medical Engineering SARS-CoV-2 Antigen Rapid Test (China) reported sensitivity between 74.0% and 74.4%, and the COVID-19 Rapid Antigen Fluorescent by SureScreen Diagnostics (UK) reported sensitivity between 60.3% and 69.0%. However, for both tests, both datasets originated from the same studies. Overall, the lowest sensitivity range was reported for the SARS-CoV-2 Antigen Rapid Test by MEDsan (Germany): 36.5% to 45.2% across 2 datasets. The specificity ranges were above 96% for most of the tests. A notable outlier was the 2019-nCov Antigen Rapid Test Kit by Shenzhen Bioeasy Biotechnology (China; henceforth called Bioeasy), reporting the worst, with a specificity as low as 85.6% in 1 study. Forest plots for the datasets for each Ag-RDT are provided in S3 Fig. The remaining 26 Ag-RDTs that were evaluated in 1 dataset only are included in Table 1 S3 Fig.
Table 2. Summary clinical accuracy data for major Ag-RDTs not included in the meta-analysis.
| Manufacturer, Ag-RDT | Number of datasets | Sensitivity range | Specificity range | Comments |
|---|---|---|---|---|
| Bionote, NowCheck (LFA) | 3 | 55.6% to 89.9% | 97.3% to 100% |
|
| Denka, Quick Navi (LFA) | 2 | 72.5% to 86.7% | 100%* |
|
| Fujirebio, ESPLINE SARS-CoV-2 (LFA) | 3 | 8.1% to 80.7% | 100%* |
|
| JOYSBIO Biotechnology, COVID-19 Antigen Rapid Test Kit (LFA) | 2 | 57.8% to 70.5% | 98.5% to 99.1% |
|
| MEDsan, SARS-CoV-2 Antigen Rapid Test (LFA) | 2 | 36.5% to 45.2% | 97% to 99.6% |
|
| R-Biopharm, RIDA QUICK SARS-CoV-2 Antigen (LFA) | 3 | 39.2% to 77.6% | 96.2% to 100% |
|
| Shenzhen Bioeasy Biotechnology, 2019-nCov Antigen Rapid Test Kit (requires reader) | 4 | 66.7% to 93.9% | 85.6% to 100% |
|
| SureScreen Diagnostics, COVID-19 Rapid Antigen Fluorescent (requires reader) | 2 | 60.3% to 69.0% | 98%* |
|
| Zhuhai Encode Medical Engineering, SARS-CoV-2 Antigen Rapid Test (LFA) | 2 | 74.0% to 74.4% | 100%* |
|
*Only 1 dataset for specificity was provided.
Ag-RDT, antigen rapid diagnostic test; AN, anterior nasal; Ct, cycle threshold; IFU, instructions for use; LFA, lateral flow assay; MT, mid-turbinate; NP, nasopharyngeal; OP, oropharyngeal.
In total, 16 studies, accounting for 53 datasets, conducted head-to-head clinical accuracy evaluations of different tests using the same samples from the same participants. These datasets have underlined sample sizes in Table 1; 15 such studies included more than 100 samples, and 1 study included too few samples to draw clear conclusions [286]. Four studies performed their head-to-head evaluation as per manufacturers’ instructions and on symptomatic patients. Across 3 of them, Standard Q (sensitivity 73.2% to 91.2%) and Standard Q nasal (sensitivity 82.5% to 91.2%) showed a similar range of sensitivity [207,215,271]. The fourth reported a sensitivity of 56.4% (95% CI 44.7% to 67.6%) for the Biocredit Covid-19 Ag by RapiGEN (South Korea; henceforth called Rapigen) and 52.6% (95% CI 40.9% to 64.0%) for the SGTi-flex COVID-19 Ag by Sugentech (South Korea) [233].
All other head-to-head comparisons were not IFU-conforming. In one of these, the Rapid COVID-19 Ag Test by Healgen (sensitivity 77.1%) performed better than Standard Q and Panbio (sensitivity 69.8% and 67.7%, respectively) [178]. In contrast to the overall findings of the meta-analysis above, 2 other head-to-head studies found that both Standard Q (sensitivity 43.6% and 49.4%) and Panbio (sensitivity 38.6% and 44.6%) had lower performance than the CLINITEST Rapid COVID-19 Antigen Test by Siemens Healthineers (Germany; henceforth called Clinitest), with reported sensitivity of 51.5% and 54.9% [167,279]. However, another study found both Standard Q and Panbio (sensitivity 81.0% and 82.9%, respectively) to have a higher accuracy than Sofia (sensitivity 80.4%) [196].
Subgroup analyses
The results are presented in Figs 6–10. Detailed results for the subgroup analyses are available in S4–S9 Figs.
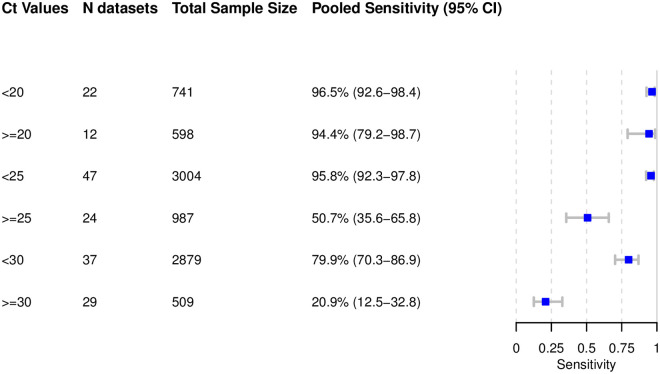
Fig 6. Pooled sensitivity by cycle threshold (Ct) values.
Low Ct values are the reverse transcription PCR semi-quantitative correlate for a high virus concentration.
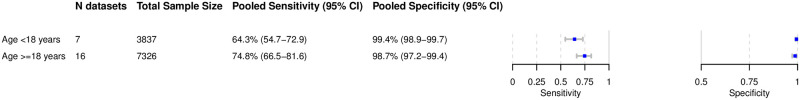
Fig 10. Pooled sensitivity and specificity by age.
Subgroup analysis by Ct values
High sensitivity was achieved for Ct value < 20, at 96.5% (95% CI 92.6% to 98.4%). The pooled sensitivity for Ct value < 25 was markedly better, at 95.8% (95% CI 92.3% to 97.8%), compared to the group with Ct value ≥ 25, at 50.7% (95% CI 35.6% to 65.8%). A similar pattern was observed when the Ct values were analyzed using the cutoffs <30 and ≥30, resulting in a sensitivity of 79.9% (95% CI 70.3% to 86.9%) and 20.9% (95% CI 12.5% to 32.8%), respectively (Fig 6).
In addition, it was possible to meta-analyze test-specific pooled sensitivity for Panbio: 97.7% sensitivity (95% CI 95.3% to 98.9%) for Ct value < 20, 95.8% (95% CI 92.3% to 97.8%) for Ct value < 25, and 83.4% (95% CI 69.1% to 91.9%) for Ct value < 30. Sensitivity was 61.2% (95% CI 38.8% to 79.7%) for Ct value ≥ 25 and 30.5% (95% CI 16.0% to 50.4%) for Ct value ≥ 30. For the other Ag-RDTs only limited data were available, which are presented in S5 Fig.
Subgroup analysis by IFU conformity
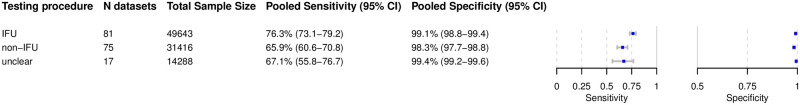
The summary results are presented in Fig 7. When assessing only studies with IFU-conforming testing, pooled sensitivity from 81 datasets with 49,643 samples was 76.3% (95% CI 73.1% to 79.2%). When non-IFU-conforming sampling (75 datasets, 31,416 samples) was performed, sensitivity decreased to 65.9% (95% CI 60.6% to 70.8%).
Fig 7. Pooled sensitivity and specificity by instructions for use (IFU) conformity.
For 5 tests it was possible to calculate pooled sensitivity estimates including only datasets with IFU-conforming testing: Panbio (sensitivity 76.5% [95% CI 69.5% to 82.3%]; 17 datasets, 12,856 samples), Standard Q (sensitivity 79.3% [95% CI 73.5% to 84.1%]; 15 datasets, 6,584 samples), BinaxNOW (sensitivity 61.8% [95% CI 48.0% to 74.0%]; 4 datasets, 8,163 samples), Rapigen (sensitivity 67.1% [95% CI 50.4% to 80.4%]; 4 datasets, 1,934 samples), and Standard Q nasal (sensitivity 83.8% [95% CI 77.8% to 88.4%]; 5 datasets, 683 samples). Specificity was above 98.6% for all tests.
In contrast, when the Panbio (14 datasets, 9,233 samples) and Standard Q (14 datasets, 4,714 samples) tests were not performed according to IFU, pooled sensitivity decreased to 64.3% (95% CI 50.9% to 75.8%) and 67.4% (95% CI 57.2% to 76.2%), respectively.
Subgroup analysis by sample type
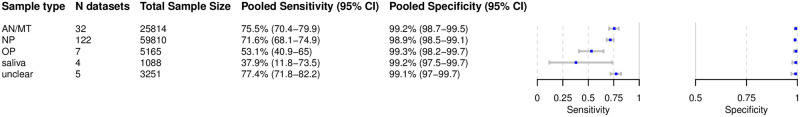
Most datasets evaluated NP or combined NP/OP swabs (122 datasets and 59,810 samples) as the sample type for the Ag-RDT. NP or combined NP/OP swabs achieved a pooled sensitivity of 71.6% (95% CI 68.1% to 74.9%). Datasets that used AN/MT swabs for Ag-RDTs (32 datasets and 25,814 samples) showed a summary estimate for sensitivity of 75.5% (95% CI 70.4% to 79.9%). This was confirmed by 2 studies that reported direct head-to-head comparison of NP and MT samples from the same participants using the same Ag-RDT (Standard Q), where the 2 sample types showed equivalent performance [271,272]. Analysis of performance with an OP swab (7 datasets, 5,165 samples) showed a pooled sensitivity of only 53.1% (95% CI 40.9% to 65.0%). Saliva swabs (4 datasets, 1,088 samples) showed the lowest pooled sensitivity, at only 37.9% (95% CI 11.8% to 73.5%) (Fig 8).
Fig 8. Pooled sensitivity and specificity by sample type.
AN, anterior nasal; MT, mid-turbinate; NP, nasopharyngeal; OP, oropharyngeal.
We were not able to perform a subgroup meta-analysis for BAL/TW due to insufficient data: There was only 1 study with 73 samples evaluating Rapigen, Panbio, and Standard Q [286]. However, BAL/TW would in any case be considered an off-label use.
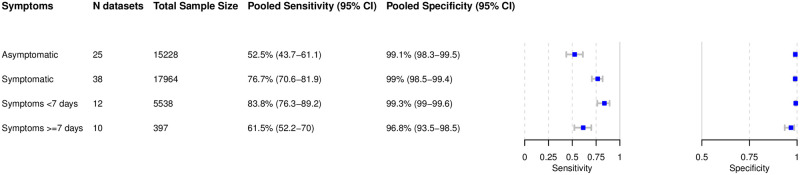
Subgroup analysis in symptomatic and asymptomatic patients
Within the datasets possible to meta-analyze, 17,964 (54.1%) samples were from symptomatic, and 15,228 (45.9%) from asymptomatic, patients. The pooled sensitivity for symptomatic patients was markedly different from that of asymptomatic patients: 76.7% (95% CI 70.6% to 81.9%) versus 52.5% (95% CI 43.7% to 61.1%). Specificity was 99% for both groups (Fig 9). Median Ct values differed in symptomatic and asymptomatic patients. For those studies where it was possible to extract a median Ct value, it ranged from 20.5 to 27.0 in symptomatic patients [170,207,226,258,271,272] and from 27.2 to 30.5 in asymptomatic patients [170,201,258].
Fig 9. Pooled sensitivity and specificity by presence of symptoms and symptom duration.
Subgroup analysis comparing symptom duration
Data were analyzed for 5,538 patients with symptoms less than 7 days, but very limited data were available for patients with symptoms ≥7 days (397 patients). The pooled sensitivity for patients with onset of symptoms <7 days was 83.8% (95% CI 76.3% to 89.2%), which is markedly higher than the 61.5% (95% CI 52.2% to 70.0%) sensitivity for individuals tested ≥7 days from onset of symptoms (Fig 9).
Subgroup analysis by age
For adult patients (age ≥ 18 years), it was possible to pool estimates across 3,837 samples, whereas the pediatric group (age < 18 years) included 7,326 samples. Sensitivity and specificity were 64.3% (95% CI 54.7% to 72.9%) and 99.4% (95% CI 98.9% to 99.7%), respectively, in mostly symptomatic patients aged <18 years. In patients aged ≥18 years, sensitivity increased to 74.8% (95% CI 66.5% to 81.6%), while the specificity was similar (98.7%, 95% CI 97.2% to 99.4%) (Fig 10).
Subgroup analysis by type of RT-PCR and viral load
We were not able to perform a meta-analysis for the subgroups by type of RT-PCR or viral load (viral copies/mL) due to insufficient data.
In 152 (71.0%) of the datasets only 1 type of RT-PCR was used, whereas 37 (17.3%) of the datasets tested samples in the same dataset using different RT-PCR methods. For 25 (11.7%) of the datasets, the type of RT-PCR was not reported. The Cobas SARS-CoV-2 Test from Roche (Germany) was used most frequently, in 63 (29.4%) of the datasets, followed by the Allplex 2019-nCoV Assay from Seegene in 41 (19.2%) and the SARS-CoV-2 assay from TaqPath in 20 (9.3%) of the datasets.
Median sensitivity was 72.4% (range 46.9% to 100%) in samples with viral load > 5 log10 copies/mL, 97.8% (range 71.4% to 100%) for >6 log10 copies/mL, and 100% (range 93.8% to 100%) for >7 log10 copies/mL, showing that the sensitivity increases with increasing viral load.
Meta regression
We were not able to perform a meta-regression due to the considerable heterogeneity in reporting subgroups, which resulted in too few studies with sufficient data for comparison.
Publication bias
The result of the Deeks test (p = 0.001) shows significant asymmetry in the funnel plot for all datasets with complete results. This indicates there may be publication bias from studies with small sample sizes. The funnel plot is presented in S10 Fig.
Comparison with analytical studies
The 9 analytical studies were divided into 63 datasets, evaluating 23 different Ag-RDTs. Only 7 studies reported a sample size, for which 833 (90.6%) samples originated from NP swabs, while for 86 (9.4%) the sample type was unclear. One of the 2 studies not reporting sample size used saliva samples [198], while the other used the sample type specified in the respective Ag-RDT’s IFU [173].
Overall, the reported analytical sensitivity (limit of detection [LOD]) in the studies resembled the results of the meta-analysis presented above. Rapigen (LOD, in log10 copies per swab: 10.2) and Coris (LOD 7.46) were found to perform worse than Panbio (LOD 6.6 to 6.1) and Standard Q (LOD 6.8 to 6.0), whereas Clinitest (LOD 6.0) and BinaxNOW by Abbott (LOD 4.6 to 4.9) performed better [191,256,282]. Similar results were found in another study, where Standard Q showed the lowest LOD (detecting virus up to what is an equivalent Ct value of 26.3 to 28.7), compared to that of Rapigen and Coris (detecting virus up to what is an equivalent Ct value of only 18.4 for both) [208,274,275]. However, another study found Panbio, Standard Q, Coris, and BinaxNOW to have a similar LOD values of 5.0 × 103 plaque forming units (PFU)/mL, but the ESPLINE SARS-CoV-2 by Fujirebio (Japan), the COVID-19 Rapid Antigen Test by Mologic (UK), and the Sure Status COVID-19 Antigen Card Test by Premier Medical Corporation (India) performed markedly better (LOD 2.5 × 102 to 5.0 × 102 PFU/mL) [173]. An overview of all LOD values reported in the studies can be found in S3 Table.
Sensitivity analysis
When the datasets from case–control studies (25/173) were excluded, the estimated sensitivity did not differ greatly, with a value of 70.9% (95% CI 67.7% to 73.9%), compared to 71.2% (95% CI 68.2% to 74.0%) in the overall analysis, with no change in pooled specificity. When the datasets from preprints (64/173) were excluded, sensitivity decreased slightly, to 67.2% (95% CI 62.9% to 71.3%), compared to the overall analysis.
Discussion
In this comprehensive systematic review and meta-analysis, we have summarized the data of 133 studies evaluating the accuracy of 61 different Ag-RDTs. Across all meta-analyzed samples, our results show a pooled sensitivity and specificity of 71.2% (95% CI 68.2% to 74.0%) and 98.9% (95% CI 98.6% to 99.1%), respectively. Over half of the studies did not perform the Ag-RDT in accordance with the test manufacturers’ recommendation, or the performance was unknown, which negatively impacted the sensitivity. When we considered only IFU-conforming studies, the sensitivity increased to 76.3% (95% CI 73.1% to 79.2%). While we found the sensitivity to vary across specific tests, the specificity was consistently high.
The 2 Ag-RDTs that have been approved through the WHO emergency use listing procedure, Abbott Panbio and SD Biosensor Standard Q (distributed by Roche in Europe), have not only drawn the largest research interest, but also perform at or above average when their pooled accuracy is compared to that of all Ag-RDTs (sensitivity of 71.8% for Panbio and 74.9% for Standard Q). Standard Q nasal demonstrated an even higher pooled sensitivity (80.2% compared to the NP test), although this is likely due to variability in the populations tested, as head-to-head performance showed a comparable sensitivity. Three other Ag-RDTs showed an even higher accuracy, with sensitivities ranging from 77.4% to 88.2% (namely Sofia, Lumipulse G, and LumiraDx), but were only assessed on relatively small samples sizes (ranging from 1,373 to 3,532), and all required an instrument/reader.
Not surprisingly, lower Ct values, the RT-PCR semi-quantitative correlate for high virus concentration, resulted in significantly higher Ag-RDT sensitivity than higher Ct values (pooled sensitivity 96.5% and 95.8% for Ct value < 20 and <25, respectively, versus 50.7% and 20.9% for Ct value ≥ 25 and ≥30, respectively). This confirms prior data that suggested that antigen concentrations and Ct values were highly correlated in NP samples [16]. Ag-RDTs also showed higher sensitivity in patients within 7 days after symptom onset compared to patients later in the course of the disease (pooled sensitivity 83.8% versus 61.5%), which is to be expected given that samples from patients within the first week after symptom onset have been shown to contain the highest virus concentrations [298]. In line with this, studies reporting an unexpectedly low overall sensitivity either shared a small population size with an on average high Ct value [230,273,288] or performed the Ag-RDT not as per IFU, e.g., using saliva or prediluted samples [167,170,203,248,279]. In contrast, studies with an unusually high Ag-RDT sensitivity were based on study populations with a low median Ct value, between 18 and 22 [189,255,284].
Our analysis also found that the accuracy of Ag-RDTs is substantially higher in symptomatic patients than in asymptomatic patients (pooled sensitivity 76.7% versus 52.5%). This is not surprising as studies that enrolled symptomatic patients showed a lower range of median Ct values (i.e., higher viral load) than studies enrolling asymptomatic patients. Given that other studies found symptomatic and asymptomatic patients to have comparable viral loads [299,300], the differences found in our analysis are likely explained by the varied time in the course of the disease at which testing is performed in asymptomatic patients presenting for one-time screening testing. Because symptoms start in the early phase of the disease, when viral load is still high, studies testing only symptomatic patients have a higher chance of including patients with high viral loads. In contrast, study populations drawn from only asymptomatic patients have a higher chance of including patients at any point of disease (i.e., including late in disease, when PCR is still positive, but viable virus is rapidly decreasing) [301].
With regards to the sampling and testing procedure, we found Ag-RDTs to perform similarly across upper respiratory swab samples (e.g., NP and AN/MT), particularly when considering the most reliable comparisons from head-to-head studies.
Similar to previous assessment [7], the methodological quality of the included studies revealed a very heterogenous picture. In the future, aligning the design of clinical accuracy studies with common agreed-upon minimal specifications (e.g., by WHO or the European Centre for Disease Control and Prevention) and reporting the results in a standardized way [302] would improve data quality and comparability.
The main strengths of our study lie in its comprehensive approach and continuous updates. By linking this review to our website, https://www.diagnosticsglobalhealth.org, we strive to equip decision makers with the latest research findings on Ag-RDTs for SARS-CoV-2 and, to the best of our knowledge, are the first in doing so. At least once per week the website is updated by continuing the literature search and process described above. We plan to update the meta-analysis on a monthly basis and publish it on the website. Furthermore, our study used rigorous methods as both the study selection and data extraction were performed by one author and independently validated by a second, we conducted blinded pilot extractions before of the actual data extraction, and we prepared a detailed interpretation guide for the QUADAS-2 tool.
The study may be limited by the inclusion of both preprints and peer-reviewed literature, which could affect the quality of the data extracted. However, we aimed to balance this potential effect by applying a thorough assessment of all clinical studies included, utilizing the QUADAS-2 tool, and performing a sensitivity analysis excluding preprint manuscripts. In addition, the studies included in our analysis varied widely in the reported range of viral loads, limiting the comparability of their results. To control for this, we analyzed the Ag-RDTs’ performance at different levels of viral load. Finally, even though we are aware that further data exist from other sources, for example from governmental research institutes [303], such data could not be included because sufficiently detailed descriptions of the methods and results are not publicly available.
Conclusion
In summary, it can be concluded that there are Ag-RDTs available that have high sensitivity for the detection of a SARS-CoV-2 infection—particularly when performed in the first week of illness, when viral load is high—and excellent specificity. However, our analysis also highlights the variability in results between tests (which is not reflected in the manufacturer-reported data), indicating the need for independent validations. Furthermore, the analysis highlights the importance of performing tests in accordance with the manufacturers’ recommended procedures, and in alignment with standard diagnostic evaluation and reporting guidelines. The accuracy achievable by the best-performing Ag-RDTs, combined with the rapid turnaround time compared to RT-PCR, suggests that these tests could have a significant impact on the pandemic if applied in thoughtful testing and screening strategies.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
Abbreviations
- Ag-RDT
antigen rapid diagnostic test
- AN
anterior nasal
- BAL/TW
bronchoalveolar lavage and throat wash
- CI
confidence interval
- Ct
cycle threshold
- IFU
instructions for use
- LOD
limit of detection
- MT
mid-turbinate
- NP
nasopharyngeal
- OP
oropharyngeal
- PFU
plaque forming units
- RT-PCR
reverse transcription polymerase chain reaction
Data Availability
All raw data is publicly available under https://zenodo.org/record/4924035#.YMSDOS2230o.
Funding Statement
The study was supported by the Ministry of Science, Research and Arts of the State of Baden-Wuerttemberg, Germany (no grant number; https://mwk.baden-wuerttemberg.de/de/startseite/) and internal funds from the Heidelberg University Hospital (no grant number; https://www.heidelberg-university-hospital.com/de/) to CMD. Further, this project was funded by United Kingdom (UK) aid from the British people (grant number: 300341-102; Foreign, Commonwealth & Development Office (FCMO), former UK Department of International Development (DFID); www.gov.uk/fcdo), and supported by a grant from the World Health Organization (WHO; no grant number; https://www.who.int) and a grant from Unitaid (grant number: 2019-32-FIND MDR; https://unitaid.org) to Foundation of New Diagnostics (FIND; JAS, SC, SO, AM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays: interim guidance, 11 September 2020. WHO/2019-nCoV/Antigen_Detection/2020.1. Geneva: World Health Organization; 2020.
- 2.Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity—a strategy for containment. N Engl J Med. 2020;383(22):e120. doi: 10.1056/NEJMp2025631 [DOI] [PubMed] [Google Scholar]
- 3.Federal Institute for Drugs and Medical Devices. Antigen-Tests auf SARS-CoV-2. Bonn: Federal Institute for Drugs and Medical Devices; 2021 [cited 2021 Jun 7]. https://www.bfarm.de/DE/Medizinprodukte/Antigentests/_node.html.
- 4.Krüger LJ, Gaeddert M, Köppel L, Brümmer LE, Gottschalk C, Miranda IB, et al. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv. 2020Oct4. doi: 10.1101/2020.10.01.20203836 [DOI] [Google Scholar]
- 5.Denkinger CM, Grenier J, Minion J, Pai M. Promise versus reality: optimism bias in package inserts for tuberculosis diagnostics. J Clin Microbiol. 2012;50(7):2455–61. doi: 10.1128/JCM.00842-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Advice on the use of point-of-care immunodiagnostic tests for COVID-19: scientific brief, 8 April 2020. WHO/2019-nCoV/Sci_Brief/POC_immunodiagnostics/2020.1. Geneva: World Health Organization; 2020.
- 7.Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;8(8):CD013705. doi: 10.1002/14651858.CD013705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olalekan A, Iwalokun B, Akinloye OM, Popoola O, Samuel TA, Akinloye O. COVID-19 rapid diagnostic test could contain transmission in low- and middle-income countries. Afr J Lab Med. 2020;9(1):1255. doi: 10.4102/ajlm.v9i1.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro R, Luz PM, Wakimoto MD, Veloso VG, Grinsztejn B, Perazzo H. COVID-19: a meta-analysis of diagnostic test accuracy of commercial assays registered in Brazil. Braz J Infect Dis. 2020;24(2):180–7. doi: 10.1016/j.bjid.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.La Marca A, Capuzzo M, Paglia T, Roli L, Trenti T, Nelson SM. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod Biomed Online. 2020;41(3):483–99. doi: 10.1016/j.rbmo.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinnes J, Deeks JJ, Berhane S, Taylor M, Adriano A, Davenport C, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2021;3:CD013705. doi: 10.1002/14651858.CD013705.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Walle I, Leitmeyer K, Broberg E. Meta-analysis of the clinical performance of commercial SARS-CoV-2 nucleic acid, antigen and antibody tests up to 22 August 2020. medRxiv. 2020Sep18. doi: 10.1101/2020.09.16.20195917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayer J, Kasapic D, Zemmrich C. Real-world clinical performance of commercial SARS-CoV-2 rapid antigen tests in suspected COVID-19: a systematic meta-analysis of available data as per November 20, 2020. medRxiv. 2020Dec4. doi: 10.1101/2020.12.22.20248614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leeflang MM, Deeks JJ, Gatsonis C, Bossuyt PM, Cochrane Diagnostic Test Accuracy Working Goup. Systematic reviews of diagnostic test accuracy. Ann Intern Med. 2008;149(12):889–97. doi: 10.7326/0003-4819-149-12-200812160-00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9, W64. doi: 10.7326/0003-4819-151-4-200908180-00135 [DOI] [PubMed] [Google Scholar]
- 16.Pollock N, Savage T, Wardell H, Lee R, Mathew A, Stengelin M, et al. Correlation of SARS-CoV-2 nucleocapsid antigen and RNA concentrations in nasopharyngeal samples from children and adults using an ultrasensitive and quantitative antigen assay. medRxiv. 2020Nov13. doi: 10.1101/2020.11.10.20227371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–36. doi: 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 18.van Enst WA, Ochodo E, Scholten RJ, Hooft L, Leeflang MM. Investigation of publication bias in meta-analyses of diagnostic test accuracy: a meta-epidemiological study. BMC Med Res Methodol. 2014;14:70. doi: 10.1186/1471-2288-14-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courtellemont L, Guinard J, Guillaume C, Giaché S, Rzepecki V, Seve A, et al. Real-life performance of a novel antigen detection test on nasopharyngeal specimens for SARS-CoV-2 infection diagnosis: a prospective study. medRxiv. 2020Nov3. doi: 10.1101/2020.10.28.20220657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diao B, Wen K, Chen J, Liu Y, Yuan Z, Han C, et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv. 2020Mar13. doi: 10.1016/j.cmi.2020.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foundation for Innovative New Diagnostics. FIND evaluation of Abbott Panbio COVID-19 Ag Rapid Test Device. External Report Version 21, 10 December 2020. Geneva: Foundation for Innovative New Diagnostics; 2020.
- 22.Foundation for Innovative New Diagnostics. FIND evaluation of Coris BioConcept COVID-19 Ag Respi-Strip. External Report Version 12, 10 December 2020. Geneva: Foundation for Innovative New Diagnostics; 2020.
- 23.Foundation for Innovative New Diagnostics. FIND evaluation of Shenzhen Bioeasy Biotechnology Co. Ltd. 2019-nCoV Ag Rapid Test Kit (Fluorescence). External Report Version 10, 11 February 2021. Geneva: Foundation for Innovative New Diagnostics; 2021.
- 24.Ikeda M, Imai K, Tabata S, Miyoshi K, Mizuno T, Murahara N, et al. Clinical evaluation of self-collected saliva by RT-qPCR, direct RT-qPCR, RT-LAMP, and a rapid antigen test to diagnose COVID-19. medRxiv. 2020Jun8. doi: 10.1101/2020.06.06.20124123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein JAF, Krüger L, Tobian F, Gaeddert M, Lainati F, Schnitzler P, et al. Head-to-head performance comparison of self-collected nasal versus professional-collected nasopharyngeal swab for a WHO-listed SARS-CoV-2 antigen-detecting rapid diagnostic test. medRxiv. 2021Mar24. doi: 10.1007/s00430-021-00710-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masiá M, Fernández-González M, Sánchez M, Carvajal M, García JA, Gonzalo N, et al. Nasopharyngeal Panbio COVID-19 antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. medRxiv. 2020Nov17. doi: 10.1101/2020.11.16.20230003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Araos R, et al. Head-to-head comparison of four antigen-based rapid detection tests for the diagnosis of SARS-CoV-2 in respiratory samples. bioRxiv. 2020May30. doi: 10.1101/2020.05.27.119255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peeling RW, Olliaro P. Rolling out COVID-19 antigen rapid diagnostic tests: the time is now. Lancet Infect Dis. 2021;21(8):1052–3. doi: 10.1016/S1473-3099(21)00152-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Z, Chang H, Wang H, Lim B, Hsu CC, Yu Y, et al. Development of a rapid test kit for SARS-CoV-2: an example of product design. Biodes Manuf. 2020May11. doi: 10.1007/s42242-020-00075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ndwandwe D, Mathebula L, Kamadjeu R, Wiysonge CS. Cochrane corner: rapid point-of-care antigen and molecular-based tests for the diagnosis of COVID-19 infection. Pan Afr Med J. 2020;37(Suppl 1):10. doi: 10.11604/pamj.supp.2020.37.10.25982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavelka M, Van-Zandvoort K, Abbott S, Sherratt K, Majdan M, Jarčuška P, et al. The effectiveness of population-wide, rapid antigen test based screening in reducing SARS-CoV-2 infection prevalence in Slovakia. medRxiv. 2020Dec4. doi: 10.1101/2020.12.02.20240648 [DOI] [Google Scholar]
- 32.Ebrahimi M, Harmooshi NN, Rahim F. Diagnostic utility of antigen detection rapid diagnostic tests for Covid- 19: a systematic review and meta-analysis. medRxiv. 2021Apr5. doi: 10.1101/2021.04.02.21254714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Everitt ML, Tillery A, David MG, Singh N, Borison A, White IM. A critical review of point-of-care diagnostic technologies to combat viral pandemics. Anal Chim Acta. 2021;1146:184–99. doi: 10.1016/j.aca.2020.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hledík M, Polechová J, Beiglböck M, Herdina AN, Strassl R, Posch M. Analysis of the specificity of the SD Biosensor Standard Q Ag-Test based on Slovak mass testing data. medRxiv. 2020Dec11. doi: 10.1101/2020.12.08.20246090 [DOI] [Google Scholar]
- 35.Huergo MAC, Thanh NTK. Current advances in the detection of COVID-19 and evaluation of the humoral response. Analyst. 2021;146:382–402. doi: 10.1039/d0an01686a [DOI] [PubMed] [Google Scholar]
- 36.Laghrib F, Saqrane S, El Bouabi Y, Farahi A, Bakasse M, Lahrich S, et al. Current progress on COVID-19 related to biosensing technologies: new opportunity for detection and monitoring of viruses. Microchem J. 2021;160:105606. doi: 10.1016/j.microc.2020.105606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddali H, Miles CE, Kohn J, O’Carroll DM. Optical biosensors for virus detection: prospects for SARS-CoV-2/COVID-19. Chembiochem. 2021;22(7):1176–89. doi: 10.1002/cbic.202000744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchán-López Á, García BA. Diagnostic performance of antigen testing for SARS-CoV-2. J Pediatr. 2021;233:283. doi: 10.1016/j.jpeds.2021.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDermott JH, Stoddard D, Ellingford JM, Gokhale D, Reynard C, Black G, et al. Utilizing point-of-care diagnostics to minimize nosocomial infection in the 2019 novel coronavirus (SARS-CoV-2) pandemic. QJM. 2020;113(12):851–3. doi: 10.1093/qjmed/hcaa185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreira V, Mascarenhas P, Machado V, Botelho J, Mendes JJ, Taveira N, et al. Diagnosis of SARS-Cov-2 infection using specimens other than naso- and oropharyngeal swabs: a systematic review and meta-analysis. Diagnostics. 2021;11:363. doi: 10.3390/diagnostics11020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nimmo C, Agbetile J, Bhowmik A, Capocci S, Rajakulasingam RK. Implementing rapid diagnostics for COVID-19. Lancet Respir Med. 2021;9(1):e7. doi: 10.1016/S2213-2600(20)30526-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raimann FJ, Piekarski F, Adam EH, Zacharowski K, Neef V. Safety considerations for the use of Point-Of-Care diagnostics during SARS-CoV-2 pandemic. J Clin Lab Anal. 2021;35(1):e23631. doi: 10.1002/jcla.23631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razmy AM, Junaideen SM. Issues of random sampling with rapid antigen tests for COVID-19 diagnosis: a special reference to Kalmunai RDHS Division. medRxiv. 2021Jan20. doi: 10.1101/2021.01.11.21249636 [DOI] [Google Scholar]
- 44.Salcedo N, Harmon A, Herrera BB. Pooling of samples for SARS-CoV-2 detection using rapid antigen tests. medRxiv. 2021Feb12. doi: 10.1101/2021.02.09.21250610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanderlin JS, Golding JD, Wilcox T, Mason DH, McKelvey KS, Pearson DE, et al. Occupancy modeling and resampling overcomes low test sensitivity to produce accurate SARS-CoV-2 prevalence estimates. BMC Public Health. 2021;21(1):577. doi: 10.1186/s12889-021-10609-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheier T, Schibli A, Eich G, Rüegg C, Kube F, Schmid A, et al. Universal admission screening for SARS-CoV-2 infections among hospitalized patients, Switzerland, 2020. Emerg Infect Dis. 2021;27(2):404–10. doi: 10.3201/eid2702.202318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stovitz SD. In suspected SARS-CoV-2, rapid antigen detection tests had 67% to 73% sensitivity and 98% to 100% specificity. Ann Intern Med. 2021;174:JC56. doi: 10.7326/ACPJ202105180-056 [DOI] [PubMed] [Google Scholar]
- 48.van Beek J, Igloi Z, Boelsums T, Fanoy E, Gotz H, Molenkamp R, et al. From more testing to smart testing: data-guided SARS-CoV-2 testing choices. medRxiv. 2020Oct14. doi: 10.1101/2020.10.13.20211524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss G, Bellmann-Weiler R. Rapid antigen testing and non-infectious shedding of SARS-Cov2. Infection. 2021;49(4):789–90. doi: 10.1007/s15010-020-01570-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ehrenberg A, Moehle E, Brook C, Doudna Cate A, Witkowsky L, Sachdeva R, et al. Launching a saliva-based SARS-CoV-2 surveillance testing program on a university campus. medRxiv. 2021Jan30. doi: 10.1371/journal.pone.0251296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boďová K, Kollár R. Characteristic spatial scales of SARS-CoV-2 pandemics: lessons from mass rapid antigen testing in Slovakia. medRxiv. 2020Dec24. doi: 10.1101/2020.12.23.20248808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Canadian Public Health Laboratory Network. Interim guidance on the use of the Abbott Panbio™ COVID-19 Antigen Rapid Test. Can Commun Dis Rep. 2021;47(1):17–22. doi: 10.14745/ccdr.v47i01a04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crozier A, Rajan S, Buchan I, McKee M. Put to the test: use of rapid testing technologies for covid-19. BMJ. 2021;372:n208. doi: 10.1136/bmj.n208 [DOI] [PubMed] [Google Scholar]
- 54.D’Agostino McGowan L, Lee E, Grantz K, Kucirka L, Gurley E, Lessler J. Testing out of quarantine. medRxiv. 2021Feb1. doi: 10.1101/2021.01.29.21250764 [DOI] [Google Scholar]
- 55.Rapid and frequent testing. Nat Biomed Eng. 2020;4(12):1121–2. doi: 10.1038/s41551-020-00670-0 [DOI] [PubMed] [Google Scholar]
- 56.Fitzpatrick MC, Pandey A, Wells CR, Sah P, Galvani AP. Buyer beware: inflated claims of sensitivity for rapid COVID-19 tests. Lancet. 2021;397(10268):24–5. doi: 10.1016/S0140-6736(20)32635-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frnda J, Durica M. On pilot massive COVID-19 testing by antigen tests in Europe. Case study: Slovakia. Infect Dis Rep. 2021;13(1):45–57. doi: 10.3390/idr13010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghaffari A, Meurant R, Ardakani A. COVID-19 point-of-care diagnostics that satisfy global target product profiles. Diagnostics. 2021;11(1):115. doi: 10.3390/diagnostics11010115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruhan A, Wang H, Wang W, Tan W. Summary of the detection kits for SARS-CoV-2 approved by the National Medical Products Administration of China and their application for diagnosis of COVID-19. Virol Sin. 2020;35:699–712. doi: 10.1007/s12250-020-00331-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdelrazik AM, Elshafie SM, Abdelaziz HM. Potential use of antigen-based rapid test for SARS-CoV-2 in respiratory specimens in low-resource settings in Egypt for symptomatic patients and high-risk contacts. Lab Med. 2020;52:e46–9. doi: 10.1093/labmed/lmaa104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aoki K, Nagasawa T, Ishii Y, Yagi S, Okuma S, Kashiwagi K, et al. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J Infect Chemother. 2020;27(4):613–6. doi: 10.1016/j.jiac.2020.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonde J, Ejegod D, Pedersen H, Smith B, Cortes D, Leding C, et al. Clinical validation of point-of-care SARS-COV-2 BD Veritor antigen test by a single throat swab for rapid COVID-19 status on hospital patients predominantly without overt COVID symptoms. medRxiv. 2021Apr17. doi: 10.1101/2021.04.12.21255299 [DOI] [Google Scholar]
- 63.Boum Y, Fai KN, Nicolay B, Mboringong AB, Bebell LM, Ndifon M, et al. Performance and operational feasibility of antigen and antibody rapid diagnostic tests for COVID-19 in symptomatic and asymptomatic patients in Cameroon: a clinical, prospective, diagnostic accuracy study. Lancet Infect Dis. 2021;21(8):1089–96. doi: 10.1016/S1473-3099(21)00132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Courtellemont L, Guinard J, Guillaume C, Giaché S, Rzepecki V, Seve A, et al. High performance of a novel antigen detection test on nasopharyngeal specimens for diagnosing SARS-CoV-2 infection. J Med Virol. 2021;93(5):3152–7. doi: 10.1002/jmv.26896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eshghifar N, Busheri A, Shrestha R, Beqaj S. Evaluation of analytical performance of seven rapid antigen detection kits for detection of SARS-CoV-2 virus. Int J Gen Med. 2021;14:435–40. doi: 10.2147/IJGM.S297762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hingrat QL, Visseaux B, Laouenan C, Tubiana S, Bouadma L, Yazdanpanah Y, et al. Detection of SARS-CoV-2 N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect. 2020;27(5):789.e1–5. doi: 10.1016/j.cmi.2020.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kobayashi R, Murai R, Asanuma K, Fujiya Y, Takahashi S. Evaluating a novel, highly sensitive, and quantitative reagent for detecting SARS-CoV-2 antigen. J Infect Chemother. 2021;27(6):800–7. doi: 10.1016/j.jiac.2021.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kritikos A, Caruana G, Brouillet R, Miroz J-P, Samia A-M, Geraldine S, et al. Sensitivity of rapid antigen testing and RT-PCR performed on nasopharyngeal swabs versus saliva samples in COVID-19 hospitalized patients: results of a prospective comparative trial (RESTART). medRxiv. 2021Apr15. doi: 10.1101/2021.04.09.21255105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lanser L, Bellmann-Weiler R, Öttl KW, Huber L, Griesmacher A, Theurl I, et al. Evaluating the clinical utility and sensitivity of SARS-CoV-2 antigen testing in relation to RT-PCR Ct values. Infection. 2021;49(3):555–7. doi: 10.1007/s15010-020-01542-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lv Y, Ma Y, Si Y, Zhu X, Zhang L, Feng H, et al. Rapid SARS-CoV-2 antigen detection potentiates early diagnosis of COVID-19 disease. Biosci Trends. 2021;15(2):93–9. doi: 10.5582/bst.2021.01090 [DOI] [PubMed] [Google Scholar]
- 71.Miyakawa K, Funabashi R, Yamaoka Y, Jeremiah SS, Katada J, Wada A, et al. SARS-CoV-2 antigen rapid diagnostic test enhanced with silver amplification technology. medRxiv. 2021Jan31. doi: 10.1101/2021.01.27.21250659 [DOI] [Google Scholar]
- 72.Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol. 2020;58(9):e01438–20. doi: 10.1128/JCM.01438-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oh SM, Jeong H, Chang E, Choe PG, Kang CK, Park WB, et al. Clinical application of the standard Q COVID-19 Ag Test for the detection of SARS-CoV-2 infection. J Korean Med Sci. 2021;36(14):e101. doi: 10.3346/jkms.2021.36.e101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peña-Rodrígez M, Viera-Segura O, García-Chagollán M, Zepeda-Nuño JS, Muñoz-Valle JF, Mora-Mora J, et al. Performance evaluation of a lateral flow assays for nasopharyngeal antigen detection for SARS-CoV-2 diagnosis. J Clin Lab Anal. 2021;35:e23745. doi: 10.1002/jcla.23745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Regev-Yochay G, Kriger O, Beni S, Rubin C, Mina M, Mechnik B, et al. Real world performance of SARS-CoV-2 antigen rapid diagnostic tests in various clinical settings. medRxiv. 2021Mar5. doi: 10.1101/2021.03.02.21252400 [DOI] [PubMed] [Google Scholar]
- 76.Ren A, Sohaei D, Zacharioudakis I, Sigal G, Stengelin M, Matthew A, et al. Ultrasensitive assay for saliva-based SARS-CoV-2 antigen detection. medRxiv. 2021Feb19. doi: 10.1101/2021.02.17.21251863 [DOI] [PubMed] [Google Scholar]
- 77.Rodrigues J, Gouveia C, Santos MA, Costa O, Côrte-Real R, Brito MJ. Comparison of nasopharyngeal samples for SARS-CoV-2 detection in a paediatric cohort. J Paediatr Child Health. 2021;57(7):1078–81. doi: 10.1111/jpc.15405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saeed U, Uppal SR, Piracha ZZ, Rasheed A, Aftab Z, Zaheer H, et al. Evaluation of SARS-CoV-2 antigen-based rapid diagnostic kits in Pakistan: formulation of COVID-19 national testing strategy. Virol J. 2021;18(1):34. doi: 10.1186/s12985-021-01505-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith R, Gibson L, Martinez P, Ke R, Mirza A, Conte M, et al. Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. medRxiv. 2021Mar22. doi: 10.1101/2021.03.19.21253964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stokes W, Berenger B, Portnoy D, Scott B, Szelewicki J, Singh T, et al. Real-world clinical performance of the Abbott Panbio with nasopharyngeal, throat and saliva swabs among symptomatic individuals with COVID-19. medRxiv. 2021Jan21. doi: 10.1101/2021.01.02.21249138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thommes L, Burkert FR, Öttl KW, Goldin D, Loacker L, Lanser L, et al. Comparative evaluation of four SARS-CoV-2 antigen tests in hospitalized patients. Int J Infect Dis. 2021;105:144–6. doi: 10.1016/j.ijid.2021.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yokota I, Sakurazawa T, Sugita J, Iwasaki S, Yasuda K, Yamashita N, et al. Performance of qualitative and quantitative antigen tests for SARS-CoV-2 in early symptomatic patients using saliva. medRxiv. 2020Nov10. doi: 10.1101/2020.11.06.20227363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zacharias M, Stangl V, Thüringer A, Loibner M, Wurm P, Wolfgruber S, et al. Rapid antigen test for postmortem evaluation of SARS-CoV-2 carriage. Emerg Infect Dis. 2021;27(6):1734–7. doi: 10.3201/eid2706.210226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Apostolou T, Kyritsi M, Vontas A, Loizou K, Hadjilouka A, Speletas M, et al. Development and performance characteristics evaluation of a new Bioelectric Recognition Assay (BERA) method for rapid Sars-CoV-2 detection in clinical samples. J Virol Methods. 2021;293:114166. doi: 10.1016/j.jviromet.2021.114166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arnaout R, Lee RA, Lee GR, Callahan C, Cheng A, Yen CF, et al. The limit of detection matters: the case for benchmarking severe acute respiratory syndrome coronavirus 2 testing. Clin Infect Dis. 2021Feb3. doi: 10.1093/cid/ciaa1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheah PK, Ongkili DF, Zaharuddin FS, Hashim MI, Ho CV, Lee HG, et al. Discrepancy in screening performances of different rapid test kits for SARS-CoV-2; a letter to editor. Arch Acad Emerg Med. 2021;9(1):e9. doi: 10.22037/aaem.v9i1.1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doron S, Ingalls R, Beauchamp A, Boehm J, Boucher H, Chow L, et al. Weekly SARS-CoV-2 screening of asymptomatic students and staff to guide and evaluate strategies for safer in-person learning. medRxiv. 2021Mar22. doi: 10.1101/2021.03.20.21253976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grossi E, Agnoli B, Baldini M, Illari S, Bonini R, Scagnelli G. Universal Sars-Cov-2 screening in pregnant women: experience from the Italian epidemic outbreak. Acta Biomed. 2021;92(S2):e2021001. doi: 10.23750/abm.v92iS2.11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hicks SM, Pohl K, Neeman T, McNamara HA, Parsons KM, He JS, et al. A dual-antigen enzyme-linked immunosorbent assay allows the assessment of severe acute respiratory syndrome coronavirus 2 antibody seroprevalence in a low-transmission setting. J Infect Dis. 2021;223(1):10–4. doi: 10.1093/infdis/jiaa623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kiyasu Y, Akashi Y, Sugiyama A, Takeuchi Y, Notake S, Naito A, et al. A prospective evaluation of the analytical performance of GENECUBE HQ SARS-CoV-2 and GENECUBE FLU A/B. medRxiv. 2021Feb25. doi: 10.1007/s40291-021-00535-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lagi F, Trevisan S, Piccica M, Graziani L, Basile G, Mencarini J, et al. Use of the FebriDx point-of-care test for the exclusion of SARS-CoV-2 diagnosis in a population with acute respiratory infection during the second (COVID-19) wave in Italy. Int J Infect Dis. 2021;108:231–6. doi: 10.1016/j.ijid.2021.04.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Latiano A, Tavano F, Panza A, Palmieri O, Niro GA, Andriulli N, et al. False positive results of IgM/IgG antibodies against antigen of the SARS-CoV-2 in sera stored before the 2020 Endemia in Italy. Int J Infect Dis. 2020;104:159–63. doi: 10.1016/j.ijid.2020.12.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li J, Hu X, Wang X, Yang J, Zhang L, Deng Q, et al. A novel one-pot rapid diagnostic technology for COVID-19. Anal Chim Acta. 2021;1154:338310. doi: 10.1016/j.aca.2021.338310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marsic T, Ali Z, Tehseen M, Mahas A, Hamdan S, Mahfouz M. Vigilant: an engineered VirD2-Cas9 complex for lateral flow assay-based detection of SARS-CoV2. Nano Lett. 2021;21(8):3596–603. doi: 10.1021/acs.nanolett.1c00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schleicher T, Spenlinhauer T, Amadei M, Dasch N, Gordon J, Macleod G, et al. Development of a multiplexed synthetic control for rapid detection of SARS-CoV-2 and other respiratory pathogens using a nucleic acid syndromic testing panel. J Mol Diagnostics. 2020;22(11):S37. [Google Scholar]
- 96.Bello-Chavolla OY, Antonio-Villa NE, Fernández-Chirino L, Guerra E, Fermín-Martínez C, Márquez-Salinas A, et al. Diagnostic performance and clinical implications of rapid SARS-CoV-2 antigen testing in Mexico using real-world nationwide COVID-19 registry data. medRxiv. 2021Jan4. doi: 10.1101/2021.01.02.21249141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dalal A, Sonika U, Kumar M, George R, Kumar A, Srivastava S, et al. COVID-19 rapid antigen test: role in screening prior to gastrointestinal endoscopy. Clin Endosc. 2021Mar4. doi: 10.5946/ce.2020.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Downs LO, Eyre DW, O’Donnell D, Jeffery K. Home-based SARS-CoV-2 lateral flow antigen testing in hospital workers. J Infect. 2021;82(2):282–327. doi: 10.1016/j.jinf.2021.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haage VC, Moreira-Soto A, Sacks J, Corman V, Drosten C, Drexler JF. Limited specificity of SARS-CoV-2 antigen-detecting rapid diagnostic tests at low temperatures. medRxiv. 2021Feb3. doi: 10.1101/2021.02.01.21250904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoehl S, Schenk B, Rudych O, Göttig S, Foppa I, Kohmer N, et al. At-home self-testing of teachers with a SARS-CoV-2 rapid antigen test to reduce potential transmissions in schools. medRxiv. 2020Dec7. doi: 10.1101/2020.12.04.20243410 [DOI] [Google Scholar]
- 101.Marco A, Solé C, Abdo IJ, Turu E. Low sensitivity of rapid antigenic tests as a screening method in an outbreak of SARS-CoV-2 infection in prison. Enferm Infecc Microbiol Clin (Engl Ed). 2021Feb11. doi: 10.1016/j.eimc.2021.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matsuda EM, de Campos IB, de Oliveira IP, Colpas DR, Carmo A, Brígido LFM. Field evaluation of COVID-19 antigen tests versus RNA based detection: potential lower sensitivity compensated by immediate results, technical simplicity and low cost. J Med Virol. 2021;93(7):4405–10. doi: 10.1002/jmv.26985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moreno G, Braun K, Pray I, Segaloff H, Lim A, Poulson K, et al. SARS-CoV-2 transmission in intercollegiate athletics not fully mitigated with daily antigen testing. medRxiv. 2021Mar6. doi: 10.1101/2021.03.03.21252838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamayoshi S, Sakai-Tagawa Y, Koga M, Akasaka O, Nakachi I, Koh H, et al. Comparison of rapid antigen tests for COVID-19. Viruses. 2020;12(12):1420. doi: 10.3390/v12121420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yokota I, Shane P, Teshima T. Logistic advantage of two-step screening strategy for SARS-CoV-2 at airport quarantine. medRxiv. 2021Jan26. doi: 10.1016/j.tmaid.2021.102127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Häuser F, Sprinzl MF, Dreis KJ, Renzaho A, Youhanen S, Kremer WM, et al. Evaluation of a laboratory-based high-throughput SARS-CoV-2 antigen assay for non-COVID-19 patient screening at hospital admission. Med Microbiol Immunol. 2021;201(2–3):165–71. doi: 10.1007/s00430-021-00706-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Colavita F, Vairo F, Meschi S, Valli MB, Lalle E, Castilletti C, et al. COVID-19 Rapid antigen test as screening strategy at points of entry: experience in Lazio region, central Italy, August–October 2020. Biomolecules. 2021;11(3):425. doi: 10.3390/biom11030425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kotsiou OS, Pantazopoulos I, Papagiannis D, Fradelos EC, Kanellopoulos N, Siachpazidou D, et al. Repeated antigen-based rapid diagnostic testing for estimating the coronavirus disease 2019 prevalence from the perspective of the workers’ vulnerability before and during the lockdown. Int J Environ Res Public Health. 2021;18(4):1638. doi: 10.3390/ijerph18041638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blairon L, Wilmet A, Beukinga I, Tre-Hardy M. Implementation of rapid SARS-CoV-2 antigenic testing in a laboratory without access to molecular methods: experiences of a general hospital. J Clin Virol. 2020;129:104472. doi: 10.1016/j.jcv.2020.104472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kashiwagi K, Ishii Y, Aoki K, Yagi S, Maeda T, Miyazaki T, et al. Immunochromatographic test for the detection of SARS-CoV-2 in saliva. medRxiv. 2020May25. doi: 10.1016/j.jiac.2020.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Itoh K, Kawamitsu T, Osaka Y, Sato K, Suzuki Y, Kiriba C, et al. False positive results in severe acute respiratory coronavirus 2 (SARS-CoV-2) rapid antigen tests for inpatients. J Infect Chemother. 2021;27(7):1089–91. doi: 10.1016/j.jiac.2021.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prince-Guerra JL, Almendares O, Nolen LD, Gunn JKL, Dale AP, Buono SA, et al. Evaluation of Abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection at two community-based testing sites—Pima County, Arizona, November 3–17, 2020. MMWR Morb Mortal Wkly Rep. 2021;70(3):100–5. doi: 10.15585/mmwr.mm7003e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aoki K, Nagasawa T, Ishii Y, Yagi S, Kashiwagi K, Miyazaki T, et al. Evaluation of clinical utility of novel coronavirus antigen detection reagent, Espline SARS-CoV-2. J Infect Chemother. 2021;27(2):319–22. doi: 10.1016/j.jiac.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caputo V, Bax C, Colantoni L, Peconi C, Termine A, Fabrizio C, et al. Comparative analysis of antigen and molecular tests for the detection of Sars-CoV-2 and related variants: a study on 4266 samples. Int J Infect Dis. 2021;108:187–9. doi: 10.1016/j.ijid.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ford L, Lee C, Pray IW, Cole D, Bigouette JP, Abedi GR, et al. Epidemiologic characteristics associated with SARS-CoV-2 antigen-based test results, rRT-PCR cycle threshold values, subgenomic RNA, and viral culture results from university testing. Clin Infect Dis. 2021Apr13. doi: 10.1093/cid/ciab303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ishii T, Sasaki M, Yamada K, Kato D, Osuka H, Aoki K, et al. Immunochromatography and chemiluminescent enzyme immunoassay for COVID-19 diagnosis. J Infect Chemother. 2021;27(6):915–8. doi: 10.1016/j.jiac.2021.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kiyasu Y, Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, et al. Prospective analytical performance evaluation of the QuickNavi™-COVID19 Ag for asymptomatic individuals. medRxiv. 2021Apr7. doi: 10.1016/j.jiac.2021.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Korenkov M, Poopalasingam N, Madler M, Vanshylla K, Eggeling R, Wirtz M, et al. Reliable assessment of in vitro SARS-CoV-2 infectivity by a rapid antigen detection test. medRxiv. 2021Apr6. doi: 10.1101/2021.03.30.21254624 [DOI] [Google Scholar]
- 120.Kronberg Jakobsen K, Schmidt Jensen J, Todsen T, Lippert F, Martel CJ, Klokker M, et al. Detection of SARS-CoV-2 infection by rapid antigen test in comparison with RT-PCR in a public setting. medRxiv. 2021Jan25. doi: 10.1101/2021.01.22.21250042 [DOI] [Google Scholar]
- 121.Kweon OJ, Lim YK, Kim HR, Choi Y, Kim MC, Choi SH, et al. Evaluation of rapid SARS-CoV-2 antigen tests, AFIAS COVID-19 Ag and ichroma COVID-19 Ag, with serial nasopharyngeal specimens from COVID-19 patients. PLoS ONE. 2021;16(4):e0249972. doi: 10.1371/journal.pone.0249972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Landaas ET, Storm ML, Tollånes MC, Barlinn R, Kran AB, Bragstad K, et al. Diagnostic performance of a SARS-CoV-2 rapid antigen test in a large, Norwegian cohort. J Clin Virol. 2021;137:104789. doi: 10.1016/j.jcv.2021.104789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McAulay K, Kaleta EJ, Grys TE. Rapid detection of SARS-CoV-2 antigen from serum in a hospitalized population. medRxiv. 2020Dec22. doi: 10.1101/2020.12.21.20248140 [DOI] [Google Scholar]
- 124.McKay SL, Tobolowsky FA, Moritz ED, Hatfield KM, Bhatnagar A, LaVoie SP, et al. Performance evaluation of serial SARS-CoV-2 rapid antigen testing during a nursing home outbreak. Ann Intern Med. 2021;174(7):945–51. doi: 10.7326/M21-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pray IW, Ford L, Cole D, Lee C, Bigouette JP, Abedi GR, et al. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses—Wisconsin, September–October 2020. MMWR Morb Mortal Wkly Rep. 2021;69(5152):1642–7. doi: 10.15585/mmwr.mm695152a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rastawicki W, Gierczyński R, Juszczyk G, Mitura K, Henry BM. Evaluation of PCL rapid point of care antigen test for detection of SARS-CoV-2 in nasopharyngeal swabs. J Med Virol. 2021;93(4):1920–2. doi: 10.1002/jmv.26765 [DOI] [PubMed] [Google Scholar]
- 127.Shah M, Salvatore P, Ford L, Kamitani E, Whaley M, Mitchell K, et al. Performance of repeat BinaxNOW SARS-CoV-2 antigen testing in a community setting, Wisconsin, November–December 2020. Clin Infect Dis. 2021;73(Suppl 1):S54–7. doi: 10.1093/cid/ciab309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Uwamino Y, Nagata M, Aoki W, Nakagawa T, Inose R, Yokota H, et al. Accuracy of rapid antigen detection test for nasopharyngeal swab specimens and saliva samples in comparison with RT-PCR and viral culture for SARS-CoV-2 detection. J Infect Chemother. 2021;27(7):1058–62. doi: 10.1016/j.jiac.2021.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Winkel BMF, Schram E, Gremmels H, Debast S, Schuurman R, Wensing AMJ, et al. Screening for SARS-CoV-2 infection in asymptomatic individuals using the Panbio™ COVID-19 Antigen Rapid Test (Abbott) compared to RT-qPCR. medRxiv. 2020Dec4. doi: 10.1101/2020.12.03.20243311 [DOI] [Google Scholar]
- 130.Yamamoto K, Suzuki M, Yamada G, Sudo T, Nomoto H, Kinoshita N, et al. Utility of the antigen test for coronavirus disease 2019: factors influencing the prediction of the possibility of disease transmission. Int J Infect Dis. 2021;104:65–72. doi: 10.1016/j.ijid.2020.12.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ahava M, Kurkela S, Kuivanen S, Lappalainen M, Jarva H, Jaaskelainen A. Detection of SARS-CoV-2 nucleocapsid antigen from serum can aid in timing of COVID-19 infection. medRxiv. 2021Jan13. doi: 10.1101/2021.01.08.20248771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Azzi L, Baj A, Alberio T, Lualdi M, Veronesi G, Carcano G, et al. Rapid salivary test suitable for a mass screening program to detect SARS-CoV-2: a diagnostic accuracy study. J Infect. 2020;81(3):E75–8. doi: 10.1016/j.jinf.2020.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Barauna VG, Singh MN, Barbosa LL, Marcarini WD, Vassallo PF, Mill JG, et al. Ultrarapid on-site detection of SARS-CoV-2 infection using simple ATR-FTIR spectroscopy and an analysis algorithm: high sensitivity and specificity. Anal Chem. 2021;93(5):2950–8. doi: 10.1021/acs.analchem.0c04608 [DOI] [PubMed] [Google Scholar]
- 134.Barlev-Gross M, Weiss S, Ben-Shmuel A, Sittner A, Eden K, Mazuz N, et al. Spike vs nucleocapsid SARS-CoV-2 antigen detection: application in nasopharyngeal swab specimens. Anal Bioanal Chem. 2021;413:3501–10. doi: 10.1007/s00216-021-03298-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cardozo KHM, Lebkuchen A, Okai GG, Schuch RA, Viana LG, Olive AN, et al. Establishing a mass spectrometry-based system for rapid detection of SARS-CoV-2 in large clinical sample cohorts. Nat Commun. 2020;11(1):6201. doi: 10.1038/s41467-020-19925-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cazares LH, Chaerkady R, Samuel Weng SH, Boo CC, Cimbro R, Hsu HE, et al. Development of a parallel reaction monitoring mass spectrometry assay for the detection of SARS-CoV-2 spike glycoprotein and nucleoprotein. Anal Chem. 2020;92(20):13813–21. doi: 10.1021/acs.analchem.0c02288 [DOI] [PubMed] [Google Scholar]
- 137.Chen H, Li Z, Feng S, Wang A, Richard-Greenblatt M, Hutson E, et al. Femtomolar SARS-CoV-2 antigen detection using the microbubbling digital assay with smartphone readout enables antigen burden quantitation and dynamics tracking. medRxiv. 2021Mar26. doi: 10.1101/2021.03.17.21253847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Colavita F, Vairo F, Meschi S, Valli B, Lalle E, Castilletti C, et al. COVID-19 antigen rapid test as screening strategy at the points-of-entry: experience in Lazio region, central Italy, August–October 2020. medRxiv. 2020Nov30. doi: 10.1101/2020.11.26.20232728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Conzelmann C, Gilg A, Groß R, Schütz D, Preising N, Ständker L, et al. An enzyme-based immunodetection assay to quantify SARS-CoV-2 infection. Antiviral Res. 2020;181:104882. doi: 10.1016/j.antiviral.2020.104882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Di Domenico M, De Rosa A, Boccellino M. Detection of SARS-COV-2 proteins using an ELISA test. Diagnostics (Basel). 2021;11(4):698. doi: 10.3390/diagnostics11040698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Diao B, Wen K, Zhang J, Chen J, Han C, Chen Y, et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect. 2020;27(2):289.e1–4. doi: 10.1016/j.cmi.2020.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ducrest PJ. Development and evaluation of a new Swiss made SARS-CoV-2 antigen-detecting rapid test. medRxiv. 2021Mar26. doi: 10.1101/2021.03.25.21252280 [DOI] [Google Scholar]
- 143.Grant BD, Anderson CE, Williford JR, Alonzo LF, Glukhova VA, Boyle DS, et al. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal Chem. 2020;92(16):11305–9. doi: 10.1021/acs.analchem.0c01975 [DOI] [PubMed] [Google Scholar]
- 144.Huang L, Ding L, Zhou J, Chen S, Chen F, Zhao C, et al. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens Bioelectron. 2021;171:112685. doi: 10.1016/j.bios.2020.112685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kyosei Y, Namba M, Yamura S, Takeuchi R, Aoki N, Nakaishi K, et al. Proposal of de novo antigen test for COVID-19: ultrasensitive detection of spike proteins of SARS-CoV-2. Diagnostics (Basel). 2020;10(8):594. doi: 10.3390/diagnostics10080594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee JH, Choi M, Jung Y, Lee SK, Lee CS, Kim J, et al. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2). Biosens Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee L, Liu F, Chen Y, Roma G. Quantitative and ultrasensitive in-situ immunoassay technology for SARS-CoV-2 detection in saliva. Research Square. 2021Jan18. doi: 10.21203/rs.3.rs-138025/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li Y, Peng Z, Holl NJ, Hassan MR, Pappas JM, Wei C, et al. MXene-graphene field-effect transistor sensing of influenza virus and SARS-CoV-2. ACS Omega. 2021;6(10):6643–53. doi: 10.1021/acsomega.0c05421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu D, Ju C, Han C, Shi R, Chen X, Duan D, et al. Ultra-sensitive nanozyme-based chemiluminescence paper test for rapid diagnosis of SARS-CoV-2 infection. bioRxiv. 2020Jun5. doi: 10.1101/2020.06.05.131748 [DOI] [Google Scholar]
- 150.Mahari S, Roberts A, Shahdeo D, Gandhi S. eCovSens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCovid-19. bioRxiv. 2020May11. doi: 10.1101/2020.04.24.059204 [DOI] [Google Scholar]
- 151.Nash B, Badea A, Reddy A, Bosch M, Salcedo N, Gomez AR, et al. The impact of high frequency rapid viral antigen screening on COVID-19 spread and outcomes: a validation and modeling study. medRxiv. 2020Nov4. doi: 10.1101/2020.09.01.20184713 [DOI] [Google Scholar]
- 152.Renuse S, Vanderboom P, Maus A, Kemp J, Gurtner K, Madugundu A, et al. Development of mass spectrometry-based targeted assay for direct detection of novel SARS-CoV-2 coronavirus from clinical specimens. medRxiv. 2020Aug6. doi: 10.1101/2020.08.05.20168948 [DOI] [Google Scholar]
- 153.Ricks S, Kendall E, Dowdy D, Sacks J, Schumacher S, Arinaminpathy N. Quantifying the potential value of antigen-detection rapid diagnostic tests for COVID-19: a modelling analysis. BMC Med. 2020;19:75. doi: 10.1101/2020.11.20.20235317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–42. doi: 10.1021/acsnano.0c02823 [DOI] [PubMed] [Google Scholar]
- 155.Shao W, Shurin MR, Wheeler SE, He X, Star A. Rapid detection of SARS-CoV-2 antigens using high-purity semiconducting single-walled carbon nanotube-based field-effect transistors. ACS Appl Mater Interfaces. 2021;13(8):10321–7. doi: 10.1021/acsami.0c22589 [DOI] [PubMed] [Google Scholar]
- 156.Singh N, Ray P, Carlin A, Magallanes C, Morgan S, Laurent L, et al. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. medRxiv. 2020Oct2. doi: 10.1101/2020.09.24.20200394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Singh P, Chakraborty R, Marwal R, Radhakrishan VS, Bhaskar AK, Vashisht H, et al. A rapid and sensitive method to detect SARS-CoV-2 virus using targeted-mass spectrometry. J Proteins Proteom, 2020Aug31. doi: 10.1007/s42485-020-00044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Torrente-Rodríguez RM, Lukas H, Tu J, Min J, Yang Y, Xu C, et al. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3(6):1981–98. doi: 10.1016/j.matt.2020.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vadlamani BS, Uppal T, Verma S, Misra M. Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors. 2020;20(20):5871. doi: 10.3390/s20205871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang H, Hogan CA, Verghese M, Solis D, Sibai M, Huang C, et al. Ultra-sensitive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen detection for the diagnosis of coronavirus disease 2019 (COVID-19) in upper respiratory samples. Clin Infect Dis. 2021Apr8. doi: 10.1093/cid/ciab063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yakoh A, Pimpitak U, Rengpipat S, Hirankarn N, Chailapakul O, Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zakashansky J, Imamura A, Salgado D, Romero Mercieca H, Aguas RFL, Lao A, et al. Detection of the SARS-CoV-2 spike protein in saliva with Shrinky-Dink electrodes. medRxiv. 2020Nov18. doi: 10.1101/2020.11.14.20231811 [DOI] [PubMed] [Google Scholar]
- 163.Zhang CY, Zhou L, Du K, Zhang Y, Wang J, Chen LJ, et al. Foundation and clinical evaluation of a new method for detecting SARS-CoV-2 antigen by fluorescent microsphere immunochromatography. Front Cell Infect Microbiol. 2020;10:553837. doi: 10.3389/fcimb.2020.553837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zou M, Su F, Zhang R, Jiang X, Xiao H, Yan X, et al. rapid point-of-care testing for SARS-CoV-2 virus nucleic acid detection by an isothermal and nonenzymatic signal amplification system coupled with a lateral flow immunoassay strip. Sens Actuators B Chem. 2021;342:129899. doi: 10.1016/j.snb.2021.129899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Akingba OL, Sprong K, Hardie DR. Field performance evaluation of the PanBio rapid SARS-CoV-2 antigen assay in an epidemic driven by 501Y.v2 (lineage B.1.351) in the Eastern Cape, South Africa. J Clin Virol Plus. 2021;1(1–2):100013. doi: 10.1016/j.jcvp.2021.100013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Asai N, Sakanashi D, Ohashi W, Nakamura A, Kawamoto Y, Miyazaki N, et al. Efficacy and validity of automated quantitative chemiluminescent enzyme immunoassay for SARS-CoV-2 antigen test from saliva specimen in the diagnosis of COVID-19. J Infect Chemother. 2021;27(7):1039–42. doi: 10.1016/j.jiac.2021.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Baro B, Rodo P, Ouchi D, Bordoy A, Saya Amaro E, Salsench S, et al. Performance characteristics of five antigen-detecting rapid diagnostic test (Ag-RDT) for SARS-CoV-2 asymptomatic infection: a head-to-head benchmark comparison. J Infect. 2021;82(6):269–75. doi: 10.1016/j.jinf.2021.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Basso D, Aita A, Padoan A, Cosma C, Navaglia F, Moz S, et al. Salivary SARS-CoV-2 antigen rapid detection: a prospective cohort study. Clin Chim Acta. 2020;517:54–9. doi: 10.1016/j.cca.2021.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bruzzone B, De Pace V, Caligiuri P, Ricucci V, Guarona G, Pennati BM, et al. Comparative diagnostic performance of different rapid antigen detection tests for COVID-19 in the real-world hospital setting. Int J Infect Dis. 2021;107:215–8. doi: 10.1016/j.ijid.2021.04.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Caruana G, Croxatto A, Kampouri E, Kritikos A, Opota O, Foerster M, et al. ImplemeNting SARS-CoV-2 Rapid antigen testing in the Emergency wArd of a Swiss univErsity hospital: the INCREASE study. Microorganisms. 2021;9(4):798. doi: 10.3390/microorganisms9040798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Caruana G, Lebrun L-L, Aebischer O, Opota O, Urbano L, DeRham M, et al. The dark side of SARS-CoV-2 rapid antigen testing: screening asymptomatic patients. medRxiv. 2021Apr27. doi: 10.1016/j.nmni.2021.100899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ciotti M, Maurici M, Pieri M, Andreoni M, Bernardini S. Performance of a rapid antigen test in the diagnosis of SARS-CoV-2 infection. J Med Virol. 2021;93:2988–91. doi: 10.1002/jmv.26830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cubas Atienzar A, Kontogianni K, Edwards T, Wooding D, Buist K, Thompson C, et al. Limit of detection in different matrices of nineteen commercially available rapid antigen tests for the detection of SARS-CoV-2. medRxiv. 2021Mar22. doi: 10.1101/2021.03.19.21253950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Del Vecchio C, Brancaccio G, Brazzale AR, Lavezzo E, Onelia F, Franchin E, et al. Emergence of N antigen SARS-CoV-2 genetic variants escaping detection of antigenic tests. medRxiv. 2021Mar26. doi: 10.1101/2021.03.25.21253802 [DOI] [Google Scholar]
- 175.Domínguez Fernández M, Peña Rodríguez MF, Lamelo Alfonsín F, Bou Arévalo G. Experience with Panbio™ rapid antigens test device for the detection of SARS-CoV-2 in nursing homes. Enferm Infecc Microbiol Clin (Engl Ed). 2021Jan19. doi: 10.1016/j.eimc.2020.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Drain PK, Ampajwala M, Chappel C, Gvozden AB, Hoppers M, Wang M, et al. A rapid, high-sensitivity SARS-CoV-2 nucleocapsid immunoassay to aid diagnosis of acute COVID-19 at the point of care: a clinical performance study. Infect Dis Ther. 2021;10(2):753–61. doi: 10.1007/s40121-021-00413-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Faíco-Filho KS, Finamor Júnior FE, Moreira LVL, Lins PRG, Justo AFO, Bellei N. Evaluation of the Panbio™ COVID-19 Ag Rapid Test at an emergency room in a hospital in São Paulo, Brazil. medRxiv. 2021Mar24. doi: 10.1101/2021.03.15.21253313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Favresse J, Gillot C, Oliveira M, Cadrobbi J, Elsen M, Eucher C, et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. J Clin Med. 2021;10(2):265. doi: 10.3390/jcm10020265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ferguson J, Dunn S, Best A, Mirza J, Percival B, Mayhew M, et al. Validation testing to determine the sensitivity of lateral flow testing for asymptomatic SARS-CoV-2 detection in low prevalence settings: testing frequency and public health messaging is key. PLoS Biol. 2021;19(4):e3001216. doi: 10.1371/journal.pbio.3001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Filgueiras P, Corsini C, Almeida NBF, Assis J, Pedrosa ML, de Oliveira A, et al. COVID-19 rapid antigen test at hospital admission associated to the knowledge of individual risk factors allow overcoming the difficulty of managing suspected patients in hospitals COVID-19 rapid antigen test facilitates the management of suspected patients on hospital admission. medRxiv. 2021Jan8. doi: 10.1101/2021.01.06.21249282 [DOI] [Google Scholar]
- 181.Foundation for Innovative New Diagnostics. FIND evaluation of Boditech Medical, Inc. iChroma COVID-19 Ag Test. External Report Version 10, 23 February 2021. Geneva: Foundation for Innovative New Diagnostics; 2021.
- 182.Foundation for Innovative New Diagnostics. FIND evaluation of Joysbio (Tianjin) Biotechnology Co., Ltd. SARS-CoV-2 Antigen Rapid Test Kit (Colloidal Gold). External Report Version 10, 11 February 2021. Geneva: Foundation for Innovative New Diagnostics; 2021.
- 183.Foundation for Innovative New Diagnostics. FIND evaluation of Guangzhou Wondfo Biotech Co., Ltd Wondfo 2019-nCoV Antigen Test (Lateral Flow Method). Public Report Version 10, 25 February 2021. Geneva: Foundation for Innovative New Diagnostics; 2021.
- 184.Foundation for Innovative New Diagnostics. FIND evaluation of Abbott Panbio COVID-19 Ag Rapid Test Device (NASAL). External Report Version 10. Geneva: Foundation for Innovative New Diagnostics; 11 February 2021, 2021.
- 185.Foundation for Innovative New Diagnostics. FIND evaluation of Bionote, Inc. NowCheck COVID-19 Ag Test, nasal swab. External Report Version 10, 30 March 2021. Geneva: Foundation for Innovative New Diagnostics; 2021.
- 186.Foundation for Innovative New Diagnostics. FIND evaluation of Fujirebio Inc. Espline SARS-CoV-2. External Report Version 10, 29 March 2021. Geneva: Foundation for Innovative New Diagnostics; 2021.
- 187.Foundation for Innovative New Diagnostics. FIND evaluation of Mologic Ltd, COVID 19 RAPID ANTIGEN TEST. External Report Version 10, 23 April 2021. Geneva: Foundation for Innovative New Diagnostics; 2021.
- 188.Foundation for Innovative New Diagnostics. FIND evaluation of NADAL COVID-19 Ag Rapid Test. External Report Version 10, 26 April 2021. Geneva: Foundation for Innovative New Diagnostics; 2021.
- 189.Gili A, Paggi R, Russo C, Cenci E, Pietrella D, Graziani A, et al. Evaluation of automated test Lumipulse G SARS-CoV-2 antigen assay for detection of SARS-CoV-2 nucleocapsid protein (NP) in nasopharyngeal swabs for community and population screening. Int J Infect Dis. 2021;105:P391–6. doi: 10.1016/j.ijid.2021.02.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gomez Marti JLG, Gribschaw J, McCullough M, Mallon A, Acero J, Kinzler A, et al. Differences in detected viral loads guide use of SARS-CoV-2 antigen-detection assays towards symptomatic college students and children. medRxiv. 2021Feb1. doi: 10.1101/2021.01.28.21250365 [DOI] [Google Scholar]
- 191.Haage V, Ferreira de Oliveira-Filho E, Moreira-Soto A, Kühne A, Fischer C, Sacks JA, et al. Impaired performance of SARS-CoV-2 antigen-detecting rapid diagnostic tests at elevated and low temperatures. J Clin Virol. 2021;138:104796. doi: 10.1016/j.jcv.2021.104796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Halfon P, Penaranda G, Khiri H, Garcia V, Drouet H, Philibert P, et al. An optimized stepwise algorithm combining rapid antigen and RT-qPCR for screening of COVID-19 patients. medRxiv. 2021Jan15. doi: 10.1101/2021.01.13.21249254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hirotsu Y, Maejima M, Shibusawa M, Amemiya K, Nagakubo Y, Hosaka K, et al. Prospective study of 1,308 nasopharyngeal swabs from 1,033 patients using the LUMIPULSE SARS-CoV-2 antigen test: comparison with RT-qPCR. Int J Infect Dis. 2021;105:7–14. doi: 10.1016/j.ijid.2021.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Homza M, Zelena H, Janosek J, Tomaskova H, Jezo E, Kloudova A, et al. Five antigen tests for SARS-CoV-2: virus viability matters. Viruses. 2021;13(4):684. doi: 10.3390/v13040684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Houston H, Gupta-Wright A, Toke-Bjolgerud E, Biggin-Lamming J, John L. Diagnostic accuracy and utility of SARS-CoV-2 antigen lateral flow assays in medical admissions with possible COVID-19. J Hosp Infect. 2021;110:203–5. doi: 10.1016/j.jhin.2021.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jääskeläinen AE, Ahava MJ, Jokela P, Szirovicza L, Pohjala S, Vapalahti O, et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. J Clin Virol. 2021;137:104785. doi: 10.1016/j.jcv.2021.104785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.James AE, Gulley T, Kothari A, Holder K, Garner K, Patil N. Performance of the BinaxNOW COVID-19 Antigen Card test relative to the SARS-CoV-2 real-time reverse transcriptase polymerase chain reaction assay among symptomatic and asymptomatic healthcare employees. Infect Control Hosp Epidemiol. 2021Jan25. doi: 10.1017/ice.2021.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jungnick S, Hobmaier B, Mautner L, Hoyos M, Haase M, Baiker A, et al. Detection of the new SARS-CoV-2 variants of concern B.1.1.7 and B.1.351 in five SARS-CoV-2 rapid antigen tests (RATs), Germany, March 2021. Euro Surveill. 2021;26(16):2100413. doi: 10.2807/1560-7917.Es.2021.26.16.2100413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kannian P, Lavanya C, Ravichandran K, Gita JB, Mahanathi P, Ashwini V, et al. SARS-CoV2 antigen in whole mouth fluid may be a reliable rapid detection tool. Oral Dis. 2021Feb3. doi: 10.1111/odi.13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kenyeres B, Ánosi N, Bányai K, Mátyus M, Orosz L, Kiss A, et al. Comparison of four PCR and two point of care assays used in the laboratory detection of SARS-CoV-2. J Virol Methods. 2021;293:114165. doi: 10.1016/j.jviromet.2021.114165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kernéis S, Elie C, Fourgeaud J, Choupeaux L, Delarue SM, Alby M-L, et al. Accuracy of antigen and nucleic acid amplification testing on saliva and naopharyngeal samples for detection of SARS-CoV-2 in ambulatory care. medRxiv. 2021Apr11. doi: 10.1101/2021.04.08.21255144 [DOI] [Google Scholar]
- 202.Kilic A, Hiestand B, Palavecino E. Evaluation of performance of the BD Veritor SARS-CoV-2 chromatographic immunoassay test in patients with symptoms of COVID-19. J Clin Microbiol. 2021;59(5):e00260–21. doi: 10.1128/JCM.00260-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kohmer N, Toptan T, Pallas C, Karaca O, Pfeiffer A, Westhaus S, et al. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. J Clin Med. 2021;10(2):328. doi: 10.3390/jcm10020328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Krüger L, Klein JAF, Tobian F, Gaeddert M, Lainati F, Klemm S, et al. Evaluation of accuracy, exclusivity, limit-of-detection and ease-of-use of LumiraDx™-Antigen-detecting point-of-care device for SARS-CoV-2. medRxiv. 2021Mar5. doi: 10.1101/2021.03.02.21252430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.L’Huillier A, Lacour M, Sadiku D, Gadiri M, De Siebenthal L, Schibler M, et al. Diagnostic accuracy of SARS-CoV-2 rapid antigen detection testing in symptomatic and asymptomatic children in the clinical setting. medRxiv. 2021Apr20. doi: 10.1128/JCM.00991-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lefever S, Indevuyst C, Cuypers L, Dewaele K, Yin N, Cotton F, et al. Comparison of the quantitative DiaSorin Liaison antigen test to RT-PCR for the diagnosis of COVID-19 in symptomatic and asymptomatic outpatients. J Clin Microbiol. 2021;59(7):e0037421. doi: 10.1128/JCM.00374-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lindner A, Nikolai O, Rohardt C, Kausch F, Wintel M, Gertler M, et al. SARS-CoV-2 patient self-testing with an antigen-detecting rapid test: a head-to-head comparison with professional testing. medRxiv. 2021Jan8. doi: 10.1101/2021.01.06.20249009 [DOI] [Google Scholar]
- 208.Mak GCK, Lau SSY, Wong KKY, Chow NLS, Lau CS, Lam ETK, et al. Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS-CoV-2. J Clin Virol. 2021;134:104712. doi: 10.1016/j.jcv.2020.104712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mboumba Bouassa RS, Veyer D, Péré H, Bélec L. Analytical performances of the point-of-care SIENNA™ COVID-19 Antigen Rapid Test for the detection of SARS-CoV-2 nucleocapsid protein in nasopharyngeal swabs: a prospective evaluation during the COVID-19 second wave in France. Int J Infect Dis. 2021;106:8–12. doi: 10.1016/j.ijid.2021.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Menchinelli G, Bordi L, Marzialiotti F, Palucci I, Capobianchi M, Sberna G, et al. Lumipulse G SARS-CoV-2 Ag Assay evaluation for SARS-CoV-2 antigen detection using 594 nasopharyngeal swab samples from different testing groups. Clin Chem Lab Med. 2021;59(8):1468–76. doi: 10.1515/cclm-2021-0182 [DOI] [PubMed] [Google Scholar]
- 211.Micocci M, Buckle P, Hayward G, Allen J, Davies K, Kierkegaard P, et al. Point of care testing using rapid automated antigen testing for SARS-COV-2 in care homes—an exploratory safety, usability and diagnostic agreement evaluation. medRxiv. 2021Apr26. doi: 10.1101/2021.04.22.21255948 [DOI] [Google Scholar]
- 212.Möckel M, Corman VM, Stegemann MS, Hofmann J, Stein A, Jones TC, et al. SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers. 2021;26(3):213–20. doi: 10.1080/1354750X.2021.1876769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Muhi S, Tayler N, Hoang T, Ballard SA, Graham M, Rojek A, et al. Multi-site assessment of rapid, point-of-care antigen testing for the diagnosis of SARS-CoV-2 infection in a low-prevalence setting: a validation and implementation study. Lancet Reg Health West Pac. 2021;9:100115. doi: 10.1016/j.lanwpc.2021.100115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ngo Nsoga M-T, Kronig I, Perez Rodriguez FJ, Sattonnet-Roche P, Da Silva D, Helbling J, et al. Diagnostic accuracy of Panbio rapid antigen tests on oropharyngeal swabs for detection of SARS-CoV-2. medRxiv. 2021Feb1. doi: 10.1371/journal.pone.0253321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Nikolai O, Rohardt C, Tobian F, Junge A, Corman V, Jones T, et al. Anterior nasal versus nasal mid-turbinate sampling for a SARS-CoV-2 antigen-detecting rapid test: does localisation or professional collection matter? medRxiv. 2021Feb16. doi: 10.1101/2021.02.09.21251274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nörz D, Olearo F, Perisic S, Bauer M, Riester E, Schneider T, et al. Multicenter evaluation of a fully automated high-throughput SARS-CoV-2 antigen immunoassay. medRxiv. 2021Apr15. doi: 10.1101/2021.04.09.21255047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Okoye NC, Barker AP, Curtis K, Orlandi RR, Snavely EA, Wright C, et al. Performance characteristics of BinaxNOW COVID-19 Antigen Card for screening asymptomatic individuals in a university setting. J Clin Microbiol. 2021;59(4):e03282–20. doi: 10.1128/JCM.03282-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Osmanodja B, Budde K, Zickler D, Naik M, Hofmann J, Gertler M, et al. Diagnostic accuracy of a novel SARS-CoV-2 antigen-detecting rapid diagnostic test from standardized self-collected anterior nasal swabs. medRxiv. 2021Apr23. doi: 10.3390/jcm10102099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Osterman A, Baldauf HM, Eletreby M, Wettengel JM, Afridi SQ, Fuchs T, et al. Evaluation of two rapid antigen tests to detect SARS-CoV-2 in a hospital setting. Med Microbiol Immunol. 2021;210(1):65–72. doi: 10.1007/s00430-020-00698-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pena M, Ampuero M, Garces C, Gaggero A, Garcia P, Velasquez MS, et al. Performance of SARS-CoV-2 rapid antigen test compared with real-time RT-PCR in asymptomatic individuals. medRxiv. 2021Feb13. doi: 10.1016/j.ijid.2021.04.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Pérez-García F, Romanyk J, Gómez-Herruz P, Arroyo T, Pérez-Tanoira R, Linares M, et al. Diagnostic performance of CerTest and Panbio antigen rapid diagnostic tests to diagnose SARS-CoV-2 infection. J Clin Virol. 2021;137:104781. doi: 10.1016/j.jcv.2021.104781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Peto T, UK COVID-19 Lateral Flow Oversight Team. COVID-19: rapid antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation for mass-testing. medRxiv. 2021Jan26. doi: 10.1016/j.eclinm.2021.100924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pickering S, Batra R, Snell L, Merrick B, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests demonstrates their utility for high sensitivity detection of infectious virus in clinical specimens. medRxiv. 2021Mar2. doi: 10.1101/2021.02.27.21252427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pollock N, Jacobs J, Tran K, Cranston A, Smith S, O’Kane C, et al. Performance and implementation evaluation of the Abbott BinaxNOW Rapid Antigen Test in a high-throughput drive-through community testing site in Massachusetts. J Clin Microbiol. 2021;59(5):e00083–21. doi: 10.1128/JCM.00083-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pollock N, Tran K, Jacobs J, Cranston A, Smith S, O’Kane C, et al. Performance and operational evaluation of the Access Bio CareStart Rapid Antigen Test in a high-throughput drive-through community testing site in Massachusetts. medRxiv. 2021Mar9. doi: 10.1101/2021.03.07.21253101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ristić M, Nikolić N, Čabarkapa V, Turkulov V, Petrović V. Validation of the STANDARD Q COVID-19 antigen test in Vojvodina, Serbia. PLoS ONE. 2021;16(2):e0247606. doi: 10.1371/journal.pone.0247606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rodgers M, Batra R, Snell L, Daghfal D, Roth R, Huang S, et al. Detection of SARS-CoV-2 variants by Abbott molecular, antigen, and serological tests. medRxiv. 2021Apr26. doi: 10.1101/2021.04.24.21256045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Rottenstreich A, Zarbiv G, Kabiri D, Porat S, Sompolinsky Y, Reubinoff B, et al. Rapid antigen detection testing for universal screening for severe acute respiratory syndrome coronavirus 2 in women admitted for delivery. Am J Obstet Gynecol. 2021;224(5):539–40. doi: 10.1016/j.ajog.2021.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Salvagno GL, Gianfilippi G, Bragantini D, Henry BM, Lippi G. Clinical assessment of the Roche SARS-CoV-2 rapid antigen test. Diagnosis (Berl). 2021Jan18. doi: 10.1515/dx-2020-0154 [DOI] [PubMed] [Google Scholar]
- 230.Sberna G, Lalle E, Capobianchi MR, Bordi L, Amendola A. Letter of concern re: “Immunochromatographic test for the detection of SARS-CoV-2 in saliva. J Infect Chemother. 2021 Feb;27(2):384–386. 10.1016/j.jiac.2020.11.016”. J Infect Chemother. 2021;27(7):1129–30. doi: 10.1016/j.jiac.2021.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Schuit E, Veldhuijzen IK, Venekamp RP, van den Bijllaardt W, Pas SD, Lodder EB, et al. Diagnostic accuracy of rapid antigen tests in pre-/asymptomatic close contacts of individuals with a confirmed SARS-CoV-2 infection. medRxiv. 2021Mar23. doi: 10.1101/2021.03.18.21253874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Seitz T, Schindler S, Winkelmeyer P, Zach B, Wenisch C, Zoufaly A, et al. Evaluation of rapid antigen tests based on saliva for the detection of SARS-CoV-2. J Med Virol. 2021;93(7):4161–2. doi: 10.1002/jmv.26983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Shidlovskaya E, Kuznetsova N, Divisenko E, Nikiforova M, Siniavin A, Ogarkova D, et al. The value of rapid antigen tests to identify carriers of viable SARS-CoV-2. medRxiv. 2021Mar12. doi: 10.1101/2021.03.10.21252667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Stohr JJM, Zwart VF, Goderski G, Meijer A, Nagel-Imming RS, Kluytmans-van den Bergh MFQ, et al. Self-testing for the detection of SARS-CoV-2 infection with rapid antigen tests. medRxiv. 2021Feb23. doi: 10.1101/2021.02.21.21252153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Stokes W, Berenger BM, Portnoy D, Scott B, Szelewicki J, Singh T, et al. Clinical performance of the Abbott Panbio with nasopharyngeal, throat, and saliva swabs among symptomatic individuals with COVID-19. Eur J Clin Microbiol Infect Dis. 2021;40(8):1721–6. doi: 10.1007/s10096-021-04202-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Strömer A, Rose R, Schäfer M, Schön F, Vollersen A, Lorentz T, et al. Performance of a point-of-care test for the rapid detection of SARS-CoV-2 antigen. Microorganisms. 2020;9(1):58. doi: 10.3390/microorganisms9010058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, Ueda A, et al. The evaluation of a newly developed antigen test (QuickNavi™-COVID19 Ag) for SARS-CoV-2: a prospective observational study in Japan. J Infect Chemother. 2021;27(6):890–4. doi: 10.1016/j.jiac.2021.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, Ueda A, et al. Diagnostic performance and characteristics of anterior nasal collection for the SARS-CoV-2 antigen test: a prospective study. Sci Rep. 2021;11:10519. doi: 10.1038/s41598-021-90026-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Thakur P, Saxena S, Manchanda V, Rana N, Goel R, Arora R. Utility of antigen-based rapid diagnostic test for detection of SARS-CoV-2 virus in routine hospital settings. Lab Med. 2021Apr30. doi: 10.1093/labmed/lmab033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Thell R, Kallab V, Weinhappel W, Mueckstein W, Heschl L, Heschl M, et al. Evaluation of a novel, rapid antigen detection test for the diagnosis of SARS-CoV-2. medRxiv. 2021Apr22. doi: 10.1101/2021.04.22.21255637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Torres I, Poujois S, Albert E, Álvarez G, Colomina J, Navarro D. Point-of-care evaluation of a rapid antigen test (CLINITEST® Rapid COVID-19 Antigen Test) for diagnosis of SARS-CoV-2 infection in symptomatic and asymptomatic individuals. J Infect. 2021;82(5):e11–2. doi: 10.1016/j.jinf.2021.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Turcato G, Zaboli A, Pfeifer N, Ciccariello L, Sibilio S, Tezza G, et al. Clinical application of a rapid antigen test for the detection of SARS-CoV-2 infection in symptomatic and asymptomatic patients evaluated in the emergency department: a preliminary report. J Infect. 2020;82:E14–6. doi: 10.1016/j.jinf.2020.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Villaverde S, Domínguez-Rodríguez S, Sabrido G, Pérez-Jorge C, Plata M, Romero MP, et al. Diagnostic accuracy of the Panbio SARS-CoV-2 Antigen Rapid Test compared with RT-PCR testing of nasopharyngeal samples in the pediatric population. J Pediatr. 2021;232:P287–9.E4. doi: 10.1016/j.jpeds.2021.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wagenhäuser I, Knies K, Rauschenberger V, Eisenmann M, McDonogh M, Petri N, et al. Clinical performance evaluation of SARS-CoV-2 rapid antigen testing in point of care usage in comparison to RT-qPCR. medRxiv. 2021Mar29. doi: 10.1016/j.ebiom.2021.103455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yin N, Debuysschere C, Decroly M, Bouazza FZ, Collot V, Martin C, et al. SARS-CoV-2 diagnostic tests: algorithm and field evaluation from the near patient testing to the automated diagnostic platform. Front Med. 2021;8:380. doi: 10.3389/fmed.2021.650581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Young BC, Eyre DW, Jeffery K. Use of lateral flow devices allows rapid triage of patients with SARS-CoV-2 on admission to hospital. J Infect. 2021;82:276–316. doi: 10.1016/j.jinf.2021.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Abdulrahman A, Mustafa F, AlAwadhi AI, Alansari Q, AlAlawi B, AlQahtani M. Comparison of SARS-COV-2 nasal antigen test to nasopharyngeal RT-PCR in mildly symptomatic patients. medRxiv. 2020Dec8. doi: 10.1101/2020.11.10.20228973 [DOI] [Google Scholar]
- 248.Agulló V, Fernández-González M, Ortiz de la Tabla V, Gonzalo-Jiménez N, García JA, Masiá M, et al. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs: a population-based point-of-care study. J Infect. 2021;82(5):186–230. doi: 10.1016/j.jinf.2020.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernandez-Fuentes MA, et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect. 2020;27(3):472.e7–10. doi: 10.1016/j.cmi.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Alemany A, Baro B, Ouchi D, Ubals M, Corbacho-Monné M, Vergara-Alert J, et al. Analytical and clinical performance of the Panbio COVID-19 antigen-detecting rapid diagnostic test. J Infect. 2020;82(5):186–230. doi: 10.1016/j.jinf.2020.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Beck ET, Paar W, Fojut L, Serwe J, Jahnke RR. Comparison of Quidel Sofia SARS FIA Test to Hologic Aptima SARS-CoV-2 TMA Test for diagnosis of COVID-19 in symptomatic outpatients. J Clin Microbiol. 2020;59(2):e02727–20. doi: 10.1128/jcm.02727-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Berger A, Ngo Nsoga M-T, Perez Rodriguez FJ, Abi Aad Y, Sattonnet P, Gayet-Ageron A, et al. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS ONE. 2020;16(3):e0248921. doi: 10.1371/journal.pone.0248921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Bulilete O, Lorente P, Leiva A, Carandell E, Oliver A, Rojo E, et al. Evaluation of the Panbio™ rapid antigen test for SARS-CoV-2 in primary health care centers and test sites. medRxiv. 2020Nov16. doi: 10.1101/2020.11.13.20231316 [DOI] [Google Scholar]
- 254.Cerutti F, Burdino E, Milia MG, Allice T, Gregori G, Bruzzone B, et al. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J Clin Virol. 2020;132:104654. doi: 10.1016/j.jcv.2020.104654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Chaimayo C, Kaewnaphan B, Tanlieng N, Athipanyasilp N, Sirijatuphat R, Chayakulkeeree M, et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol J. 2020;17(1):177. doi: 10.1186/s12985-020-01452-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Corman V, Haage V, Bleicker T, Schmidt ML, Muehlemann B, Zuchowski M, et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests. Lancet Microbe. 2021;2(7):e311–9. doi: 10.1016/S2666-5247(21)00056-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Drevinek P, Hurych J, Kepka Z, Briksi A, Kulich M, Zajac M, et al. The sensitivity of SARS-CoV-2 antigen tests in the view of large-scale testing. medRxiv. 2020Nov24. doi: 10.1101/2020.11.23.20237198 [DOI] [PubMed] [Google Scholar]
- 258.Fenollar F, Bouam A, Ballouche M, Fuster L, Prudent E, Colson P, et al. Evaluation of the Panbio Covid-19 rapid antigen detection test device for the screening of patients with Covid-19. J Clin Microbiol. 2020;59:e02589–20. doi: 10.1128/JCM.02589-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Foundation for Innovative New Diagnostics. FIND evaluation of Bionote, Inc. NowCheck COVID-19 Ag Test. External Report Version 15, 20 April 2021. Geneva: Foundation for Innovative New Diagnostics; 2020.
- 260.Foundation for Innovative New Diagnostics. FIND evaluation of RapiGEN Inc. BIOCREDIT COVID-19 Ag. External Report Version 21, 10 December 2020. Geneva: Foundation for Innovative New Diagnostics; 2020.
- 261.Foundation for Innovative New Diagnostics. FIND evaluation of SD Biosensor, Inc. STANDARD™ F COVID-19 Ag FIA. External Report Version 21, 10 December 2020. Geneva: Foundation for Innovative New Diagnostics; 2020.
- 262.Foundation for Innovative New Diagnostics. FIND evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test. External Report Version 21, 10 December 2020. Geneva: Foundation for Innovative New Diagnostics; 2020.
- 263.Gremmels H, Winkela BMF, Schuurmana R, Rosinghb A, Rigterc NAM, Rodriguezd O, et al. Real-life validation of the Panbio COVID-19 Antigen Rapid Test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine. 2020;31:100677. doi: 10.1016/j.eclinm.2020.100677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Gupta A, Khurana S, Das R, Srigyan D, Singh A, Mittal A, et al. Rapid chromatographic immunoassay-based evaluation of COVID-19: a cross-sectional, diagnostic test accuracy study & its implications for COVID-19 management in India. Indian J Med Res. 2020;153(1):126. doi: 10.4103/ijmr.IJMR_3305_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Herrera V, Hsu V, Adewale A, Hendrix T, Johnson L, Kuhlman J, et al. Testing of healthcare workers exposed to COVID19 with rapid antigen detection. medRxiv. 2020Aug18. doi: 10.1101/2020.08.12.20172726 [DOI] [Google Scholar]
- 266.Iglὁi Z, Velzing J, van Beek J, van de Vijver D, Aron G, Ensing R, et al. Clinical evaluation of the Roche/SD Biosensor rapid antigen test with symptomatic, non-hospitalized patients in a municipal health service drive-through testing site. Emerg Infect Dis. 2020;27(5):1323–9. doi: 10.3201/eid2705.204688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Krüger LJ, Gaeddert M, Tobian F, Lainati F, Gottschalk C, Klein JAF, et al. Evaluation of the accuracy and ease-of-use of Abbott PanBio—a WHO emergency use listed, rapid, antigen-detecting point-of-care diagnostic test for SARS-CoV-2. medRxiv. 2020Dec d. doi: 10.1101/2020.11.27.20239699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Krüttgen A, Cornelissen CG, Dreher M, Hornef MW, Imöhl M, Kleinesa M. Comparison of the SARS-CoV-2 rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J Virol Methods. 2020;288:114024. doi: 10.1016/j.jviromet.2020.114024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso AM, et al. Evaluation of a rapid diagnostic assay for detection of SARS-CoV-2 antigen in nasopharyngeal swabs. J Clin Microbiol. 2020;58(8):e00977–20. doi: 10.1128/JCM.00977-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-García F, Gómez-Herruz P, et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133:104659. doi: 10.1016/j.jcv.2020.104659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lindner A, Nikolai O, Rohardt C, Burock S, Hülso C, Bölke A, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with professional-collected nasal versus nasopharyngeal swab. Eur Respir J. 2020;57(5):2004430. doi: 10.1183/13993003.04430-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lindner AK, Nikolai O, Kausch F, Wintel M, Hommes F, Gertler M, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. Eur Respir J. 2021;57(4):2003961. doi: 10.1183/13993003.03961-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Liotti FM, Menchinelli G, Lalle E, Palucci I, Marchetti S, Colavita F, et al. Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clin Microbiol Infect. 2020;27:487–8. doi: 10.1016/j.cmi.2020.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mak GC, Lau SS, Wong KK, Chow NL, Lau CS, Lam ET, et al. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J Clin Virol. 2020;133:104684. doi: 10.1016/j.jcv.2020.104684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Mak GCK, Cheng PKC, Lau SSY, Wong KKY, Lau CS, Lam ETK, et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Merino-Amador P, Guinea J, Muñoz-Gallego I, González-Donapetry P, Galán J-C, Antona N, et al. Multicenter evaluation of the Panbio™ COVID-19 Rapid Antigen-Detection Test for the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect. 2020;27(5):758–61. doi: 10.1016/j.cmi.2021.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Mertens P, De Vos N, Martiny D, Jassoy C, Mirazimi A, Cuypers L, et al. Development and potential usefulness of the COVID-19 Ag Respi-Strip diagnostic assay in a pandemic context. Front Med. 2020;7:225. doi: 10.3389/fmed.2020.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Nalumansi A, Lutalo T, Kayiwa J, Watera C, Balinandi S, Kiconco J, et al. Field evaluation of the performance of a SARS-CoV-2 antigen rapid diagnostic test in Uganda using nasopharyngeal samples. Int J Infect Dis. 2020;104:282–6. doi: 10.1016/j.ijid.2020.10.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Olearo F, Nörz D, Heinrich F, Sutter JP, Rödel K, Schultze A, et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. J Clin Virol. 2020;137:104782. doi: 10.1016/j.jcv.2021.104782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Parada-Ricart E, Gomez-Bertomeu F, Picó-Plana E, Olona-Cabases M. Usefulness of the antigen for diagnosing SARS-CoV-2 infection in patients with and without symptoms. Enferm Infecc Microbiol Clin (Engl Ed). 2020Oct1. doi: 10.1016/j.eimc.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Pekosz A, Cooper C, Parvu V, Li M, Andrews J, Manabe YCC, et al. Antigen-based testing but not real-time PCR correlates with SARS-CoV-2 virus culture. Clin Infect Dis. 2020Jan20. doi: 10.1093/cid/ciaa1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Perchetti GA, Huang ML, Mills MG, Jerome KR, Greninger AL. Analytical sensitivity of the Abbott BinaxNOW COVID-19 Ag CARD. J Clin Microbiol. 2021;59(3):e02880–20. doi: 10.1128/JCM.02880-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Pilarowski G, Lebel P, Sunshine S, Liu J, Crawford E, Marquez C, et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. J Infect Dis. 2020;223(7):1139–44. doi: 10.1101/2020.11.02.20223891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Porte L, Legarraga P, Iruretagoyena M, Vollrath V, Pizarro G, Munita J, et al. Rapid SARS-CoV-2 antigen detection by immunofluorescence—a new tool to detect infectivity. medRxiv. 2020Oct6. doi: 10.1101/2020.10.04.20206466 [DOI] [Google Scholar]
- 285.Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020;99:328–33. doi: 10.1016/j.ijid.2020.05.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Schildgen V, Demuth S, Lüsebrink J, Schildgen O. Limits and opportunities of SARS-CoV-2 antigen rapid tests—an experience based perspective. Pathogens. 2020;10(1):38. doi: 10.3390/pathogens10010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Schwob J-M, Miauton A, Petrovic D, Perdrix J, Senn N, Jaton K, et al. Antigen rapid tests, nasopharyngeal PCR and saliva PCR to detect SARS-CoV-2: a prospective comparative clinical trial. medRxiv. 2020Nov24. doi: 10.1101/2020.11.23.20237057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Scohy A, Anantharajah A, Bodeus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Shrestha B, Neupane A, Pant S, Shrestha A, Bastola A, Rajbhandari B, et al. Sensitivity and specificity of lateral flow antigen test kits for COVID-19 in asymptomatic population of quarantine centre of province 3. Kathmandu Univ Med J (KUMJ). 2020;18(2):36–9. [PubMed] [Google Scholar]
- 290.Takeda Y, Mori M, Omi K. SARS-CoV-2 qRT-PCR Ct value distribution in Japan and possible utility of rapid antigen testing kit. medRxiv. 2020Jun19. doi: 10.1101/2020.06.16.20131243 [DOI] [Google Scholar]
- 291.Toptan T, Eckermann L, Pfeiffer A, Hoehl S, Ciesek S, Drosten C, et al. Evaluation of a SARS-CoV-2 rapid antigen test: potential to help reduce community spread? J Clin Virol. 2020;135:104713. doi: 10.1016/j.jcv.2020.104713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Torres I, Poujois S, Albert E, Colomina J, Navarro D. Real-life evaluation of a rapid antigen test (Panbio COVID-19 Ag Rapid Test Device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin Microbiol Infect. 2020;27(4):636.E1–4. doi: 10.1016/j.cmi.2020.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Van der Moeren N, Zwart V, Lodder E, Van den Bijllaardt W, Van Esch H, Stohr J, et al. Performance evaluation of a SARS-CoV-2 rapid antigentest: test performance in the community in the Netherlands. medRxiv. 2020Oct21. doi: 10.1101/2020.10.19.20215202 [DOI] [Google Scholar]
- 294.Veyrenche N, Bollore K, Pisoni A, Bedin AS, Mondain AM, Ducos J, et al. Diagnosis value of SARS-CoV-2 antigen/antibody combined testing using rapid diagnostic tests at hospital admission. J Med Virol. 2020;39(5):3069–76. doi: 10.1002/jmv.26855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Araos R, et al. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Med Infect Dis. 2020;39:101942. doi: 10.1016/j.tmaid.2020.101942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Young S, Taylor SN, Cammarata CL, Varnado KG, Roger-Dalbert C, Montano A, et al. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS Antigen point-of-care test. J Clin Microbiol. 2020;59(1):e02338–20. doi: 10.1128/JCM.02338-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kissler SM, Fauver JR, Mack C, Olesen SW, Tai C, Shiue KY, et al. SARS-CoV-2 viral dynamics in acute infections. medRxiv. 2020Dec2. doi: 10.1101/2020.10.21.20217042 [DOI] [Google Scholar]
- 299.Lee S, Kim T, Lee E, Lee C, Kim H, Rhee H, et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern Med. 2020;180(11):1447–52. doi: 10.1001/jamainternmed.2020.3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kociolek LK, Muller WJ, Yee R, Dien Bard J, Brown CA, Revell PA, et al. Comparison of upper respiratory viral load distributions in asymptomatic and symptomatic children diagnosed with SARS-CoV-2 infection in pediatric hospital testing programs. J Clin Microbiol. 2020;59(1):e02593–20. doi: 10.1128/JCM.02593-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Turner F, Vandenberg A, Slepnev VI, Car S, Starritt RE, Seger MV, et al. Post-disease divergence in SARS-CoV-2 RNA detection between nasopharyngeal, anterior nares and saliva/oral fluid specimens—significant implications for policy & public health. medRxiv. 2021Jan26. doi: 10.1101/2021.01.26.21250523 [DOI] [Google Scholar]
- 302.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Paul Ehrlich Institute. Comparative evaluation of the sensitivities of SARSCoV-2 antigen rapid tests Langen: Federal Institute for Vaccines and Biomedicines; 2020.