Abstract
The mechanisms by which transforming growth factor β (TGF-β) and related ligands regulate transcription remain poorly understood. The winged-helix (WH) transcription factor fork head activin signal transducer 1 (FAST-1) was identified as a mediator of activin signaling in Xenopus embryos (X. Chen, M. J. Rubock, and M. Whitman, Nature 383:691–696, 1996). We have cloned a novel WH gene from the mouse which shares many properties with FAST-1. We find that this gene, which we call FAST-2, is able to mediate transcriptional activation by TGF-β. FAST-2 also interacts directly with Smad2, a cytoplasmic protein which is translocated to the nucleus in response to TGF-β, and forms a multimeric complex with Smad2 and Smad4 on the activin response element, a high-affinity binding site for FAST-1. Analysis of the sequences of FAST-1 and FAST-2 reveals substantial protein sequence divergence compared to known vertebrate orthologs in the WH family. This suggests that FAST-2 represents a new WH gene related to FAST-1, which functions to mediate TGF-β signals in mammals. We have also examined the structure of the FAST-2 gene and find that it overlaps with a kinesin motor protein gene. The genes are transcribed in opposite orientations, and their transcripts overlap in the 3′ untranslated region.
Winged-helix (WH) proteins are a large family of putative transcription factors characterized by the unique three-dimensional structure of their DNA binding domain (6). Members of WH family are expressed in a wide range of tissues during different developmental stages (12; for a review, see reference 16). Targeted disruptions of a number of WH genes have revealed the essential functions of WH proteins in development and demonstrated their critical roles in the regulation of cell fate determination, cell proliferation, and cell differentiation (1, 2, 7, 10, 13, 15, 23, 24, 25).
Fork head activin signal transducer 1 (FAST-1) is a recently discovered member of the WH family identified by its ability to mediate transcriptional induction by activin, a member of the transforming growth factor β (TGF-β) family of polypeptide ligands in Xenopus embryos (4). TGF-β ligands also play important roles during development. Transcriptional induction by TGF-β and activin has been shown to involve cytoplasmic Smad proteins, which are phosphorylated and translocated to the nucleus in response to the binding of ligand to the receptor (for a recent review, see reference 19). FAST-1 was shown to interact directly with Smad2 to form a transcriptionally active complex on the promoter of the Xenopus mix.2 gene, at a site called the activin response element (ARE) (5, 18). These findings established a new function of WH proteins, i.e., as transcriptional partners for Smad proteins in the TGF-β signaling pathway.
The discovery of FAST-1 raised the possibility that other WH genes may function as mediators of TGF-β family signaling. However, no other WH genes have been found to date to serve in this role. Comparison of the amino acid sequence of FAST-1 with the 60 to 70 members of the WH family reveals that FAST-1 is distantly related to all other known WH genes. The WH domain is only approximately 40% identical to that of HNF-3β and several other family members. No homology to any WH protein is observed outside of WH domain 4. Postulating that FAST-1 may represent the first member of a new subfamily of WH proteins which function as effectors of the TGF-β signal transduction pathway, we searched for additional FAST-1-like proteins in mammals. In this paper, we describe the cloning of a novel mouse cDNA that is highly homologous to FAST-1 in the WH domain and also shares sequence similarity in other domains. Functional studies show that the protein product of this new gene shares many of the activities of FAST-1. However, sequence comparison with FAST-1 suggests that this protein, which we call FAST-2, may be a novel related member of the WH family rather than the mouse homolog of Xenopus FAST-1.
MATERIALS AND METHODS
Screening of cDNA and genomic libraries.
Fast-2 cDNAs were isolated from a mouse embryonic carcinoma lambda cDNA library (Stratagene) by using an EcoRI-DraI fragment (∼350 bp) from a mouse expressed sequence tag (EST) clone (AA144428) as a probe (see Fig. 2C). Several positive clones with overlapping inserts were sequenced in both strands by using DNA Sequenase2 (United States Biochemical). To isolate the FAST-2 gene, a mouse genomic DNA library (Stratagene) was screened with a probe that spans ∼500 nucleotides (nt) of cDNA downstream from the WH domain. One of the positive genomic clones was partially sequenced.
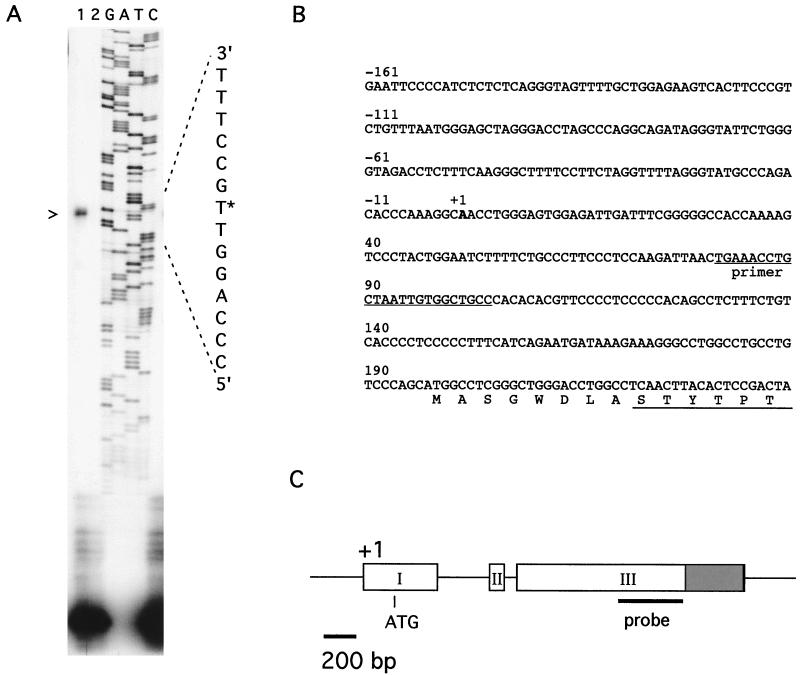
FIG. 2.
Structure of the FAST-2 gene. (A) Determination of the transcription initiation site of the FAST-2 gene by primer extension. Primers were annealed to total RNA isolated from P19 cells (lane 1) or to tRNA (lane 2). Sequences of the FAST-2 gene extended from the same primer are shown in the adjacent lanes. The arrowhead indicates the major extension product, and the asterisk marks the transcription initiation site in the nucleotide sequence. (B) Nucleotide and deduced amino acid sequences of the FAST-2 gene in the region surrounding the transcription initiation site. +1 indicates the predicted first nucleotide of the FAST-2 transcript. The P240 primer sequence is indicated, and the 5′ end of cDNA clone 1.2 is underlined. (C) Schematic diagram of the FAST-2 gene based on a comparison of genomic and cDNA sequences. Exons are labeled I to III. The shaded area in exon III represents the region that overlaps the 3′ UTR of the mouse KIFC2 cDNA (D49545 and MMU92949). The position of the EcoRI-DraI fragment from the mouse EST clone (AA144428) in exon III which was used as a probe is indicated.
Primer extension.
The transcription start site was determined by primer extension with 24-nt oligonucleotides complementary to genomic sequences in the predicted 5′ untranslated region (UTR). The oligonucleotide primers were labeled with [γ-32P]ATP at the 5′ end with T4 polynucleotide kinase (New England Biolabs). The labeled primers were annealed to 30 μg of total RNA from P19 cells or 30 μg of tRNA. The hybridization and extension conditions were as previously described (17). The extension products were analyzed on a DNA sequencing gel.
Northern blotting.
Samples (10 μg) of total RNAs from P19 and OBL21a cells were fractionated on a 1% agarose gel containing 6.7% formaldehyde. The EcoRI-DraI fragment of the mouse EST clone (see Fig. 2C) was radiolabeled by the random-priming method and used as a probe. Prehybridization and hybridization were conducted in a 5× SSC (1× SSC is 0.15 M NaCl plus 0.015% sodium citrate) solution, and membranes were washed twice with 1× SSC at 55°C for 30 min.
Constructs.
Myc-tagged FAST-2 constructs were prepared by inserting different fragments of FAST-2 cDNA clones into the CS2 vector downstream of Myc epitopes. The Myc-tagged FAST-1 construct was kindly provided by M. Whitman. Flag and hemagglutinin (HA) epitope-tagged Smad constructs and the A3-luc reporter were generously provided by F. Liu and J. Massague.
Cell culture and transfection.
COS1 cells were maintained in high-glucose Dulbecco’s modified Eagle (DME) medium, and Mv1Lu cells were maintained in DME medium supplemented with 10% fetal calf serum (FCS), nonessential amino acids, 100 U of penicillin per ml, 100 μg of streptomycin sulfate per ml, and 2 mM l-glutamine. P19 cells were maintained in a mixture of DME and F12 media (1:2) supplemented with 10% FCS and antibiotics and l-glutamine at the same concentrations. COS1 cells were transfected with DEAE-dextran 22, and Mv1Lu cells were transfected with Lipofectamine (Gibco BRL) unless otherwise specified.
Immunoprecipitation and immunoblotting.
COS1 cells were cotransfected with Myc-tagged FAST-2 and one of the Flag-tagged Smad proteins. TβR-I (T204) was cotransfected in the TGF-β-treated group for TGF-β stimulation. Forty to 48 h after transfection, cells were treated with low-serum medium (high-glucose DME medium plus 0.2% FCS) in the presence or absence of 0.5 nM TGF-β for 1 h and then lysed in 1 ml of TNE buffer (10 mM Tris [pH 8.0], 0.15 M NaCl, 1 mM EDTA, 1% Nonidet P-40) plus protease inhibitors. Cell lysates were precleared with protein A- and G-coupled agarose beads and incubated with M2 flag monoclonal antibody (Eastman Kodak) for 1 to 3 h. Immunoprecipitates and aliquots of cell lysates before immunoprecipitation were separated by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and transferred to an Immobilon P membrane. The membrane was then probed with Flag (Eastman Kodak), Myc (9E10 monoclonal antibody; Santa Cruz), or HA (12CA5 antibody; Boehringer Mannheim) mouse monoclonal antibodies. Primary antibodies were detected with a horseradish peroxidase-conjugated goat anti-mouse antibody and a chemiluminescent substrate (Amersham).
Gel mobility shift and supershift assays.
COS1 cells were transfected with Myc-tagged FAST-2 by using the Lipofectamine method or with FAST-1 constructs by using the DEAE-dextran method. In all experiments, Flag-tagged Smad2 and HA-tagged Smad4 were cotransfected and TβR-I (T204) was only cotransfected into TGF-β-treated cells. After treatment with 0.5 nM TGF-β for 18 h, nuclear extracts were prepared as described in reference 18. Two-microliter volumes of nuclear extracts (∼10 μg) were incubated with 1 ng of radiolabeled ARE probe 4 for 15 min on ice and then incubated for 10 min with antibodies in the antibody supershift assay. DNA-protein complexes were then separated on 4% polyacrylamide gels (37.5:1 acrylamide-bisacrylamide ratio) containing 1% glycerol.
Luciferase assay.
Mv1Lu cells were cotransfected with A3-luc, Rous sarcoma virus β-galactosidase β-(gal), and Myc-tagged FAST-2 or FAST-1 constructs and treated with 0.1 or 0.5 nM TGF-β for 24 to 36 h. In some experiments, Smad2 or Smad3 was also cotransfected. Luciferase activity was measured with a luciferase assay kit (Promega), and β-gal activity was determined by using a Galacto-light Plus system (Tropix). Luciferase activity values were normalized to β-gal activity.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been assigned GenBank accession no. AF079514.
RESULTS
Cloning and characterization of FAST-2 cDNA clones.
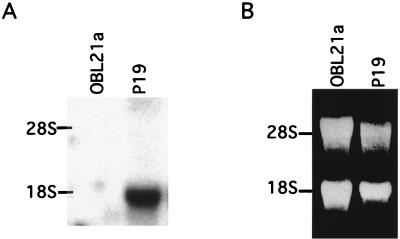
In order to find novel FAST-1-like WH genes, we searched the EST databases for sequences with homology to the Xenopus FAST-1 gene. One human EST clone was identified, from the NT2 embryonal carcinoma cell line. Additional searches for sequences related to the human EST identified a mouse EST from the P19 embryonal carcinoma cell line. Both EST clones contained sequences which were homologous to the C-terminal FAST-1 sequences, but neither encoded a WH domain. We used a fragment of the mouse EST clone to screen a mouse cDNA library from P19 cells and isolated numerous overlapping clones encoding a single cDNA. The longest cDNA clone isolated, clone 1.2, was 1.75 kb long and contained an open reading frame starting from the 5′ end of the sequence and encoding a polypeptide of 392 amino acids (aa). This cDNA, which we called FAST-2, encoded a domain with 68% identity to the WH domain of Xenopus FAST-1 (4). Northern analysis with a FAST-2 probe reveals a single major transcript in P19 cells, migrating at an apparent size of 1.9 kb. FAST-2 transcripts were not found in cells of OBL21a, another neural progenitor line (Fig. 1). The size of the FAST-2 transcript, together with the continuous open reading frame from the 5′ end, raised the possibility that clone 1.2 was not a full-length cDNA clone.
FIG. 1.
A single major transcript of FAST-2 is expressed in P19 cells. Total RNAs from OBL21a and P19 cells were fractionated on an agarose gel and transferred to a membrane. The blot was hybridized to a probe derived from the mouse EST clone (see Fig. 2C). (A) Northern blot. (B) Ethidium bromide-stained RNA gel prior to transfer.
Cloning and characterization of the FAST-2 gene.
Despite isolating over 40 overlapping cDNA clones from the P19 library and a mouse embryo library, we were unable to identify any cDNA clones longer than 1.2. Attempts to isolate additional 5′ sequence by rapid amplification of cDNA ends (8) were also unsuccessful. Therefore, we isolated mouse genomic clones containing the FAST-2 gene by using a fragment from the cDNA as a probe. One of the genomic clones contained an ∼15-kb insert and was further analyzed by restriction enzyme mapping, Southern blotting, and partial sequencing. The FAST-2 gene was found to contain three exons and two small introns (Fig. 2C). Primer extension analysis was used to define the transcription start site. Several oligonucleotides from the predicted 5′ UTR were synthesized and annealed to mRNA isolated from P19 cells. A single transcription start site was identified (Fig. 2A). Comparison of the genomic DNA sequence with that of the cDNA clones shows that the major transcription start site is located 220 bp upstream of the 5′ end of cDNA clone 1.2 (Fig. 2B). The predicted translation start site is 197 nt from the cap site, yielding a protein of 401 aa. This suggests that the longest cDNA clone which we had identified (clone 1.2) is missing only 8 aa at the N terminus. The structure of the FAST-2 gene is schematically depicted in Fig. 2C.
FAST-2 has extensive sequence similarity to FAST-1.
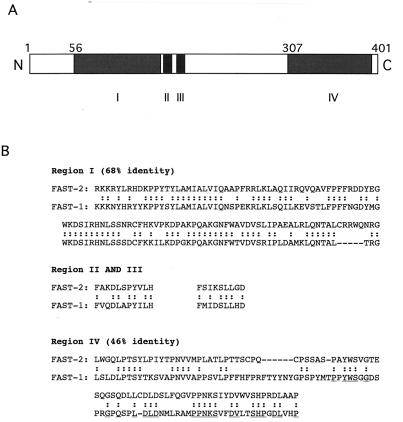
Comparison of the predicted sequence of FAST-2 with that of FAST-1 identifies several regions of homology (Fig. 3). The WH domain (region I) is the region with the highest sequence homology (68% identity over 100 aa). Besides the DNA binding domain, there are several additional regions with significant homology to FAST-1, which are designated regions II to IV (Fig. 3A). Regions II and III are two short sequences near the WH domain. Their function is unknown. Region IV is located at the C terminus of FAST-2 and has 46% identity to a C-terminal domain of FAST-1. This domain has been shown to be required for the association of FAST-1 with Smad2 and Smad4 (5) and is called the Smad interaction domain (SID). The C-terminal half of the SID has been designated the Smad2 interaction region because it is both necessary and sufficient for the coimmunoprecipitation of Smad2 with FAST-1. Deletion of the N-terminal half of the SID eliminates the ability of FAST-1 to coimmunoprecipitate with Smad4. This region has been designated a putative Smad4 interaction domain. However, this region alone is not sufficient for the association of FAST-1 with Smad4 (5). The sequence similarity between FAST-1 and FAST-2 within region IV raised the possibility that FAST-2 might also be able to interact with Smad proteins and to mediate transcriptional activation by TGF-β or related ligands.
FIG. 3.
Mouse FAST-2 has sequence similarity to Xenopus FAST-1 both in the WH domain and in the C-terminal region. (A) Schematic of the FAST-2 protein indicating the regions (I to IV) with homology to FAST-1. (B) Amino acid sequence alignments of FAST-2 and FAST-1 in these regions. The residues within region IV that are likely to be required for Smad2 interaction are underlined.
The FAST-2 transcript overlaps that of another gene.
GenBank searches with the FAST-2 sequence revealed that 366 nt of the 3′ UTR of FAST-2 is 99% identical to the antisense strand of the 3′ UTR of KIFC2, a neural tissue-specific kinesin motor protein (D49545 [21], MMU92949 [9]). This does not appear to be due to a cDNA library artifact, as all of the numerous independent cDNA clones we isolated from two different cDNA libraries contained this sequence at the 3′ end. We also isolated multiple cDNA clones encoding KIFC2 from two libraries and confirmed that the antisense FAST-2 sequence is also present in this cDNA. We next sequenced the FAST-2 genomic DNA extending past the 3′ end of the cDNA sequence and found the KIFC2 sequence in the antisense strand. Thus, the FAST-2 and KIFC2 genes appear to be transcribed in opposite orientations with an overlap in the 3′ UTR of each gene. The functional significance of this genomic arrangement is not known. While overlapping of transcription units is commonplace in prokaryotes and in yeast, there are relatively few examples of its occurrence in the vertebrate genome (3, 20). In prokaryotes, antisense transcripts often play a role in gene regulation. Their potential function in vertebrates is less well understood.
FAST-2 interacts with Smad2 and Smad3.
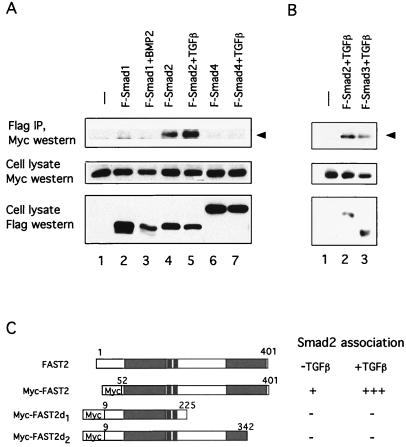
To test whether FAST-2 can associate with components of the TGF-β signal transduction pathway, we examined the ability of FAST-2 to form a complex with Smad proteins in mammalian cells by coimmunoprecpitation. Myc-tagged FAST-2 was coexpressed with various Flag-tagged Smad proteins in COS1 cells in the presence or absence of TGF-β stimulation. Lysates from cells that expressed epitope-tagged FAST-2 and Smads were immunoprecipitated with an anti-Flag antibody and blotted with an anti-Myc antibody. We found that some Smad2 coprecipitated with FAST-2 in the absence of TGF-β stimulation (Fig. 4A). This interaction between FAST-2 and Smad2 was enhanced by ligand treatment (Fig. 4A). The specificity of the interaction between FAST-2 and Smad2 was demonstrated by the absence of significant interaction between FAST-2 and Smad1 or Smad4 with or without ligand stimulation (Fig. 4A). We also observed an interaction between FAST-2 and Smad3 (Fig. 4B). Western blots of the cell extracts showed that comparable amounts of Myc-tagged FAST-2 and Flag-tagged Smad1, Smad2, Smad3, or Smad4 were expressed in these studies.
FIG. 4.
FAST-2 specifically associates with the Smad2 and Smad3 proteins. (A) COS1 cells were cotransfected with Myc-tagged FAST-2 and Flag-tagged Smad1, Smad2, or Smad4 as indicated. A TGF-β receptor, TβR-I (T204D), was cotransfected to facilitate TGF-β stimulation. Cell lysates were prepared after 1 h with or without TGF-β treatment, immunoprecipitated with an anti-Flag antibody, and analyzed by Western blotting with an anti-Myc antibody. The expression levels of the tagged constructs were determined by Western analysis with corresponding antibodies. The arrowhead identifies the Myc-tagged FAST-2 which was coimmunoprecipitated with the Flag antibody. IP, immunoprecipitate. (B) COS1 cells were cotransfected with Myc–FAST-2, TβR-I (T204D), and Flag-Smad2 or Flag-Smad3 with Lipofectamine and then treated with TGF-β for 1 h. Cell lysates were prepared and analyzed as described for panel A. (C) Diagram of two FAST-2 mutants, one with a C-terminal half deletion (Myc–FAST-2d1) and the other with a partial SID deletion (Myc–FAST-2d2), shown in comparison with Myc–FAST-2 and wild-type FAST-2. The Myc-tagged FAST-2 mutants were cotransfected with Flag-Smad2 and analyzed by coimmunoprecipitation assay. Neither mutant showns association with Smad2.
Because the C terminus of FAST-2 (region IV) is similar to the FAST-1 SID, we next constructed two C-terminal deletion mutant forms of Myc-tagged FAST-2, one with a deletion of the C-terminal half and the other with a partial deletion of region IV (Fig. 4C). Both mutations abrogate the FAST-2 interaction with Smad2, in the absence or presence of TGF-β (Fig. 4C), indicating that amino acids C terminal to position 342 are essential for Smad2 interaction. Several clusters of amino acids in this region are identical between FAST-2 and FAST-1, suggesting that these residues are required for this interaction (underlined in Fig. 3B).
FAST-2 binds to the ARE and forms a complex similar to the activin responsive factor.
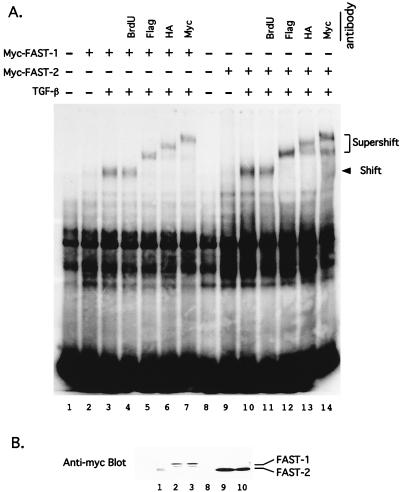
It has been shown that binding of activin or TGF-β to its cognate receptor leads to the formation of a multimeric complex containing FAST-1, Smad2, and Smad4 which associates with the ARE. We tested the ability of FAST-2 to form a similar DNA-protein complex by using the gel mobility shift assay. COS1 cells were cotransfected with TβR-I (T204), Flag-tagged Smad2, HA-tagged Smad4, and either Myc–FAST-1 or Myc–FAST-2(9-401) (Fig. 5A). Treatment of the cells with TGF-β leads to the formation of DNA binding complexes that are similar in mobility with both FAST-1 and FAST-2. The molecular masses of the two Myc-tagged proteins are similar, as shown on a Western blot (Fig. 5B). Neither of the two C-terminal deletion mutants (Fig. 4B) was able to form this high-molecular-weight DNA-protein complex (data not shown). To examine whether Smad2 or Smad4 was present in this DNA binding complex, we tested the effect of adding antibodies to the Flag, HA, or Myc epitope to the nuclear extract. Figure 5A shows that the addition of either an anti-Flag, an anti-HA, or an anti-Myc antibody leads to further retardation of the DNA binding complex. Addition of other antibodies, such as an anti-bromodeoxyuridine antibody, does not have this effect. These results show that FAST-2, like FAST-1, is able to form a complex in response to TGF-β treatment which binds to the ARE. This complex includes at least three proteins, FAST-2, Smad2, and Smad4.
FIG. 5.
FAST-2 forms a TGF-β-inducible DNA-binding complex with Smad2 and Smad4. (A) Gel mobility shift assay. Myc-tagged FAST-1 and FAST-2(9-401) constructs were cotransfected into COS1 cells with Flag-tagged Smad2 or HA-tagged Smad4. TβR-I (T204D) was transfected into TGF-β-treated cells. Nuclear extracts were incubated with the 50-bp ARE probe in the presence or absence of individual antibodies as indicated. Aliquots of nuclear extracts used in gel shift lane 3 and lane 10 were used in lanes 4 to 7 and lanes 11 to 14, respectively. BrdU, bromodeoxyuridine. (B) Western blot assay. Nuclear extracts used in the gel mobility shift assay (lanes 1 to 3 and 8 to 10) were separated by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and blotted with an anti-Myc antibody.
FAST-2 activates ARE-luc expression.
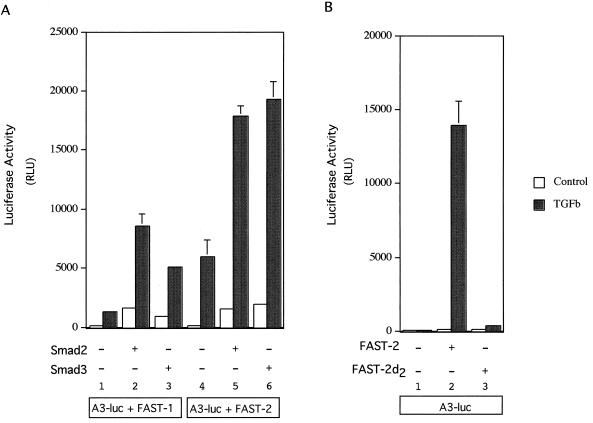
To determine the transcriptional activity of FAST-2, we examined its ability to stimulate the expression of a reporter gene, A3-luc, which contains three copies of ARE from the mix.2 promoter. Cells cotransfected with FAST-2(9-401) and A3-luc responded to the addition of 100 pM TGF-β with an increase in luciferase reporter expression (Fig. 6A). No response to TGF-β was observed in the absence of cotransfected FAST-2 (Fig. 6B). FAST-1-cotransfected cells also show TGF-β induction, as previously demonstrated by others (18). FAST-2 also is capable of mediating transcription activation in response to TGF-β on another reporter, MIX-CAT (11), which contains 5.5 kb of the mix.2 promoter, with a single ARE site (data not shown). Cotransfection of either Smad2 or Smad3 enhances TGF-β stimulated transcription mediated through FAST-1 and FAST-2 (Fig. 6A). Because the C-terminal region (aa 342 to 401) of FAST-2 is essential for its association with Smad2, we tested whether this region is also required for the TGF-β-induced transcriptional activation of a reporter gene. When cells were cotransfected with A3-luc and a C-terminal deletion mutant form of FAST-2 [Myc–FAST-2(9-342)], no significant TGF-β-stimulated luciferase activity was found (Fig. 6B). These results demonstrate that FAST-2 is able to mediate transcriptional activation through the ARE upon treatment of cells with TGF-β. This activation requires the same region of the protein that mediates association with Smad2.
FIG. 6.
FAST-2 activates the A3-luc reporter gene in response to TGF-β (A) Mv1Lu cells were cotransfected with A3-luc and FAST-1 or FAST-2(9-401) together with Smad2 or Smad3 as indicated. Luciferase activity was determined in cell extracts following 24 h with or without TGF-β (0.1 nM) treatment. Luciferase activity is presented as relative light units (RLU). Open bars; control; filled bars, TGF-β-treated cells. Shown are the means of duplicate samples from a typical experiment. (B) Mv1Lu cells were cotransfected with A3-luc and FAST-2(9-401) or FAST-2d(9-342). Luciferase activity was determined after 36 h with or without TGF-β (0.5 nM) treatment. Luciferase activity is presented in the same fashion as in panel A.
DISCUSSION
TGF-β and related ligands regulate many important cellular processes, including cell differentiation and cell proliferation, by their ability to regulate gene expression. Over the past few years, enormous progress has been made in the elucidation of the signal transduction pathway which links the binding of the ligand at the cell surface to the control of gene transcription. TGF-β ligands interact with cell membrane receptors, leading to activation of their serine-threonine kinase. Cytoplasmic Smad proteins are phosphorylated by the receptor kinase, leading to their translocation to the nucleus and the activation of transcription. The mechanisms by which Smad proteins regulate transcription are being actively investigated. An important advance was the discovery of FAST-1, a WH transcription factor which binds specifically to the ARE of the mix.2 gene. FAST-1 was found to interact directly with Smad2 in an activin-dependent fashion and to form a DNA binding complex with Smad2 and Smad4 on the ARE. These findings suggested that FAST-1 serves as a DNA binding partner which targets the Smad coactivator to specific promoter sequences.
We have cloned a new WH gene from the mouse, FAST-2, through its sequence homology to the Xenopus FAST-1 gene. Functional studies show that the protein encoded by this gene shares many of the properties of FAST-1. FAST-2 associates specifically with Smad2, forms a multimeric DNA binding complex in response to TGF-β stimulation, and mediates transcriptional activation by TGF-β through the ARE. Taken together, these results suggest that FAST-2 serves as a mediator of TGF-β and/or activin signals in mammalian cells. Despite these similarities, sequence comparison of FAST-2 with FAST-1 suggests that there may be additional genes in mammals which are more closely related to FAST-1. WH genes comprise a large family of putative transcriptional regulators. The 100-aa DNA binding WH is very highly conserved during evolution. For example, Xenopus WH proteins XFH1 and XFD-1 are 91% identical in the WH domain to their mouse homolog, HNF-3β. The WH domain of chicken gene c-qin is 99% identical to that of its human homolog, BF-1. By contrast, FAST-1 and FAST-2 are only 68% identical in the WH domain. This degree of sequence divergence raises the possibility that FAST-2 is not the mouse homolog of Xenopus FAST-1 but represents a new member of a subfamily of WH proteins which function as effectors of TGF-β family signals. The identification of FAST-2 will facilitate the discovery of additional WH genes with similar functions and lead to a better understanding of the mechanisms by which TGF-β signals regulate gene expression. Our initial studies comparing the properties of FAST-2 and FAST-1 provide evidence for the requirement of specific residues within the SID for the interaction with Smad2. Further comparisons of FAST-1 and FAST-2 should provide additional insight into how these proteins function to modulate transcriptional activity.
While this report was under review, studies describing the cloning of mouse FAST-2, as well as a related human gene, called hFAST-1, were published (14, 26). The mouse FAST-2 protein reported by Labbe et al. is identical in sequence to that reported here, except for a 1-aa difference at residue 394. FAST-2 expression is detected early in mouse development, with mRNA levels declining from E6.5 to undetectable levels at E11.5. FAST-2 is shown to bind to a site in the goosecoid promoter, called the TGF-β/activin response element (TARE). Labbe et al. demonstrated that FAST-2 forms a transcriptionally active complex containing FAST-2/Smad2/Smad4 on the TARE. Furthermore, these investigators report that Smad3 negatively regulates TARE-dependent transcription on the gsc-lux reporter, while Smad2 enhances transcription together with FAST-2.
We have investigated the activity of FAST-2 on the ARE from the mix.2 gene. We find that FAST-2 also forms a transcriptionally active complex containing FAST-2/Smad2/Smad4 on the ARE. In addition, we also show that FAST-2 interacts with Smad3, as well as Smad2, but not with Smad1, and that transcriptional activation by FAST-2 requires its C-terminal domain. TARE-stimulated transcription from the A3-luc reporter is enhanced by the cotransfection of either Smad2 or Smad3. Thus, in contrast to the goosecoid reporter, Smad3 functions as a positive regulator of transcription on the A3-luc reporter. These observations suggest that the FAST-2/Smad3/Smad4 complex may have distinct activities on different promoters.
ACKNOWLEDGMENTS
We thank Dana Benhaim, Chetna Thayyulathil, Yasmin Khakoo, and Robert Johnson for technical assistance. We are grateful to Joan Massague for Smad constructs and the A3-luc reporter and to Malcolm Whitman for Myc–FAST-1. We thank Suzanne Li, Fang Liu, and J. Massague for helpful discussions and critical comments on the manuscript.
B. Liu and C.-L. Dou contributed equally to this work.
This work was supported by National Institutes of Health grants HD29584 (E.L.), F32NS10035 (B.L.), and F32NS10313 (C.-L.D.) and a Cancer Center Support Grant to MSKCC.
REFERENCES
- 1.Ang S, Rossant J. HNF-3beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 2.Brissette J, Li J, Kamimura J, Lee D, Dotto G. The product of the mouse nude locus, Whn, regulates the balance between epithelial cell growth and differentiation. Genes Dev. 1996;10:2212–2221. doi: 10.1101/gad.10.17.2212. [DOI] [PubMed] [Google Scholar]
- 3.Bristow J, Tee M K, Gitelman S E, Mellon S H, Miller W L. Tenascin-X: a novel extracellular matrix protein encoded by the human XB gene overlapping P450c21B. J Cell Biol. 1993;122:265–278. doi: 10.1083/jcb.122.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Rubock M, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 6.Clark K, Halay E, Lai E, Burley S. Cocrystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 7.Dou C, Ye X, Stewart C, Lai E, Li S. TWH regulates the development of subsets of spinal cord neurons. Neuron. 1997;18:539–551. doi: 10.1016/s0896-6273(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 8.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanlon D W, Yang Z, Goldstein L S B. Characterization of KIFC2, a neuronal kinesin superfamily member in mouse. Neuron. 1997;18:439–451. doi: 10.1016/s0896-6273(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 10.Hatini V, Huh S, Herzlinger D, Soares V, Lai E. Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of winged-helix transcription factor, BF-2. Genes Dev. 1996;10:1467–1478. doi: 10.1101/gad.10.12.1467. [DOI] [PubMed] [Google Scholar]
- 11.Huang H-C, Murtaugh L C, Vize P D, Whitman M. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 1995;14:5965–5973. doi: 10.1002/j.1460-2075.1995.tb00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufman E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 13.Kume T, Deng K-Y, Winfrey V, Gould D B, Walter M A, Hogan B L M. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998;93:985–996. doi: 10.1016/s0092-8674(00)81204-0. [DOI] [PubMed] [Google Scholar]
- 14.Labbe E, Silvestri C, Hoodless P, Wrana J, Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 15.Labosky P, Winnier G, Jetton T, Hargett L, Ryan A, Rosenfeld M, Parlow A, Hogan B. The winged helix gene, Mf3, is required for normal development of the diencephalon and midbrain, postnatal growth and the milk-ejection reflex. Development. 1997;124:1263–1274. doi: 10.1242/dev.124.7.1263. [DOI] [PubMed] [Google Scholar]
- 16.Lai E, Clark K, Burley S, Darnell J. Hepatocyte nuclear factor-3/fork head or “winged helix” proteins: a family of transcription factors of diverse biologic function. Proc Natl Acad Sci USA. 1993;90:10421–10423. doi: 10.1073/pnas.90.22.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Tao W, Lai E. Characterization of the structure and function of the gene for transcription factor BF-1, an essential regulator of forbrain development. Mol Brain Res. 1996;37:96–104. doi: 10.1016/0169-328x(95)00276-x. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Pouponot C, Massague J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massague J, Hata A, Liu F. TGFb singalling through the Smad pathway. Trends Cell Biol. 1997;7:187–192. doi: 10.1016/S0962-8924(97)01036-2. [DOI] [PubMed] [Google Scholar]
- 20.Miyajima N, Horiuchi R, Shibuya Y, Fukushige S-I, Matsubara K-I, Toyoshima K, Yamamoto T. Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell. 1989;57:31–39. doi: 10.1016/0092-8674(89)90169-4. [DOI] [PubMed] [Google Scholar]
- 21.Saito N, Okada Y, Noda Y, Kinoshita Y, Kondo S, Hirokawa N N. KIFC2is a novel neuron-specific C-terminal type kinesin superfamily motor for dendritic transport of multivesicular body-like organelles. Neuron. 1997;18:425–438. doi: 10.1016/s0896-6273(00)81243-x. [DOI] [PubMed] [Google Scholar]
- 22.Sussman D, Milman G. Short-term, high-efficiency expression of transfected DNA. Mol Cell Biol. 1984;4:1641–1643. doi: 10.1128/mcb.4.8.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinstein D, Altaba I A, Chen W, Hoodless P, Prezioso V, Jessell T, Darnell J. The winged-helix transcription factor HNF-3gamma is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 24.Winnier G, Hargett L, Hogan B. The winged helix transcription factor MFH1 is required for proliferation and patterning of paraxial mesoderm in the mouse embryo. Genes Dev. 1997;11:926–940. doi: 10.1101/gad.11.7.926. [DOI] [PubMed] [Google Scholar]
- 25.Xuan S, Baptista C, Balas G, Tao W, Soares V, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 26.Zhou S, Zawel L, Lengauer C, Kinzler K, Vogelstein B. Characterization of human FAST-1, a TGFβ and activin signal transducer. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]