Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has resulted in the global coronavirus disease 2019 (COVID-19) pandemic. Despite several single-cell RNA sequencing (RNA-seq) studies, conclusions cannot be reached owing to the small number of available samples and the differences in technology and tissue types used in the studies. To better understand the cellular landscape and disease severity in COVID-19, we performed a meta-analysis of publicly available single-cell RNA-seq data from peripheral blood and lung samples of COVID-19 patients with varying degrees of severity. Patients with severe disease showed increased numbers of M1 macrophages in lung tissue, while the number of M2 macrophages was depleted. Cellular profiling of the peripheral blood showed a marked increase of CD14+, CD16+ monocytes and a concomitant depletion of overall B cells and CD4+, CD8+ T cells in severe patients when compared with moderate patients. Our analysis indicates the presence of faulty innate-to-adaptive switching, marked by a prolonged innate immune response and a dysregulated adaptive immune response in severe COVID-19 patients. Furthermore, we identified cell types with a transcriptome signature that can be used as a prognostic biomarker for disease state prediction and the effective therapeutic management of COVID-19 patients.
Keywords: COVID-19 patients, Single-cell RNA-Seq, Meta-analysis, Immune cell landscape, Phenotypic switching
Graphical abstract
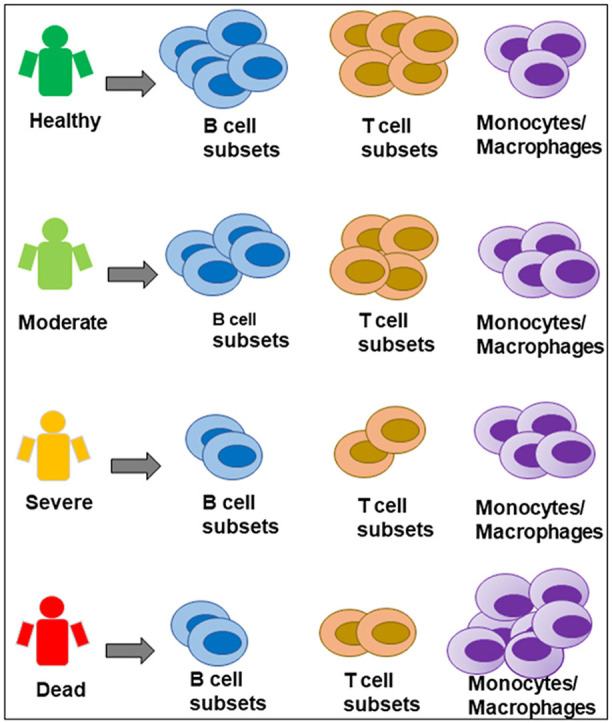
Differential cellular landscape based on disease severity observed in COVID-19 patients. Healthy controls had high numbers of B-cells and T-cells. Moderate COVID-19 patients also had high numbers of some T-cell and B-cell subsets, with a slight increase in monocytes. Severe and deceased patients had significantly fewer T-cells and B-cells and had an abundant presence of inflammatory monocytes and macrophages in their blood and lung, respectively. Thus, innate-to-adaptive switching may be key in determining COVID-19 outcomes, and the lymphoid-to-myeloid cell ratio can be an early biomarker for predicting outcomes and designing effective therapeutic regimens.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected millions of people and caused more than 2 million fatalities worldwide, resulting in the coronavirus disease 2019 (COVID-19) pandemic. Most COVID-19 patients have mild to moderate symptoms, but approximately 20% develop more severe systemic inflammation accompanied by acute respiratory distress syndrome (ARDS), often leading to death [1,2]. Multiple studies have implicated the immune response in severe cases, which plays a crucial role in determining the outcome of COVID-19 patients [[3], [4], [5], [6], [7], [8], [9], [10], [11]]. However, these studies were heavily reliant on either the local or peripheral response and included limited numbers of patient samples. Thus, an understanding of the local and peripheral immune landscape in relation to disease severity and progression is required and would help improve the therapeutic management of COVID-19 symptoms.
Recent single-cell RNA sequencing (RNA-seq) studies of severe COVID-19 patients have provided evidence of a reduced lymphocyte count and higher numbers of inflammatory myeloid cells [[3], [4], [5]]. Other studies have found that severe COVID-19 patients have abundant pro-inflammatory monocytes or monocyte-derived macrophages in bronchoalveolar lavage fluid (BALF) [[6], [7], [8]]. Older COVID-19 patients exhibit more severe symptoms compared with children and young adults. However, the molecular mechanisms that underlie the imbalanced host response leading to disease severity remain unclear.
Innate immune cells act as the first line of defense against invading viruses and clear the infection by producing type I interferon (IFN) [12]. Severe and terminally ill patients exhibit higher levels of cytokines and chemokines, such as IL6, IL36G, CXCL2, CXCL10, CCL2, CCL3, and CCL5. This finding points toward an overactive innate immune cell-mediated cytokine storm, leading to the development of ARDS. However, the type I IFN-mediated innate immune response is reportedly impaired and delayed [13], suggesting a dynamic shift in the overall immune response.
The adaptive immune response controls the excessive innate immune response; thus, lower lymphocyte counts in severe COVID-19 patients partly explain the observed hyperactivated innate immune response [14]. Furthermore, recent studies reported that a lower lymphocyte count and a reduced ratio of lymphocytes in the blood contribute to the increase in myeloid cells in the blood and BALF [15,16]. Indeed, older patients aged 60 years or more exhibit low numbers of CD4+ and CD8+ T cells [17], whereas children with SARS-CoV-2 infection have normal lymphocyte counts [18]. Therefore, we posit that this weakened adaptive response, along with a hyperactivated innate immune inflammatory response, may increase the fatality rate.
To understand the immune landscape in the lungs and blood of COVID-19 patients, we collated single-cell RNA-seq datasets from healthy controls and moderate, severe, and fatal cases. Our meta-analysis revealed phenotypic switching of immune cell types and proportionality in the lung and blood of severe cases. Principal component analysis (PCA) of important cell types could discriminate disease severity. Furthermore, cells that were strongly associated with severe disease or mortality had high expression levels of inflammatory genes.
2. Materials and methods
2.1. Ethics statement
All required ethical guidelines were followed, and ethics committee approvals were obtained by the original study groups.
2.2. Datasets
Single-cell RNA seq raw datasets were collected from the gene expression omnibus (GEO) of the National Center for Biotechnology Information database. GSE163668 [19] and GSE166992 [20] were used for the analysis of blood samples, while GSE168215 [21], GSE169471 [22], and GSE145926 [6] were used for lung samples. GSE157344 [23] was used for both blood and lung samples. Briefly, patients who were not admitted to an ICU for longer than 3 days and did not require mechanical ventilation/intubation were designated as moderate patients. Patients admitted to an ICU for a longer period and/or required mechanical ventilation/intubation were designated as severe patients.
2.3. Single-cell data analysis
Seurat (version 4.0.2) in the R program environment (4.0.1) was used for the data quality control and analysis, as comprehensively outlined by the package developer [24] and others, owing to its fast processing time and ability to integrate multiple datasets across platforms [25]. Briefly, Seurat objects were created from individual expression matrices. Unique molecular identifier (UMI) counts were scaled by library size and a natural log transformation. Gene counts for each cell were divided by the total UMI count of that cell, scaled by a factor of 10,000, and then transformed via the natural log plus 1 function, “NormalizeData”. The normalized data were further scaled with the “ScaleData” function so that the mean expression across cells was 0 and the variance was 1. PCA was used with the “RunPCA” function in Seurat to reduce the dimensionality of the data by clustering similar cells from different datasets. Next, we identified anchors across datasets using the “FindIntegrationAnchors” function in Seurat by embedding cells in a k-nearest neighborhood-based approach to identify mutual neighbors from different datasets and scored them based on their mutual nearest neighbors. Noise and batch effect variances among the datasets were taken into consideration by using reference cells from each dataset. These anchors were then used to integrate data across datasets using the “IntegrateData” function. The uniform manifold approximation and projection (UMAP) method [26] was used to visualize high-dimensional cellular data in an easy and comprehensive manner. Differentially expressed genes (DEGs) were identified using the default “FindMarkers” function in Seurat based on the non-parametric Wilcoxon rank–sum test.
2.4. Cell type annotation
The automated cell annotation program scCATCH (version 2.1) [27] was used in the R environment. scCATCH uses paired comparisons and evidence-based scoring to identify potential marker genes and annotate clustered cells.
2.5. Gene ontology analysis
Metascape [28] (web version) was used to assess the functional enrichment of DEGs. Metascape queries publicly available databases, such as Gene Ontology, Kyoto Encyclopedia of Genes and Genomes [29], and assigns DEGs to their respective enriched pathways by calculating the pairwise similarity between any two terms. The hypergeometric test and Benjamini–Hochberg p-value correction algorithm were used to identify significantly enriched ontology terms.
2.6. Principal component analysis
We performed PCA on selected gene expression changes (log2 fold change) of target cell types to distinguish the COVID-19 disease states (i.e., control, moderate, severe, deceased). The PCA involved six steps. In step 1, we standardized the expression changes in the target feature gene list by calculating the mean and standard deviation. In step 2, the covariance matrix for the feature genes was calculated. In step 3, the eigenvalues and eigenvectors for the covariance matrix were calculated. In step 4, eigenvalues and their corresponding eigenvectors were sorted. In step 5, k = 2 eigenvalues were selected to form a matrix of eigenvectors. In step 6, the original matrix was transformed (i.e., feature matrix * top k eigenvectors = transformed data). In this analysis, we selected the first two PCs because the variances explained by them were significant for evaluating the disease state. Loading plots of the first two PC coefficients for the various disease states showed that the gene expression changes (log2 fold change) widely varied among the moderate, severe, and deceased COVID-19 patients and healthy controls.
3. Results
3.1. Indications of lymphopenia and an abundance of monocytes in blood
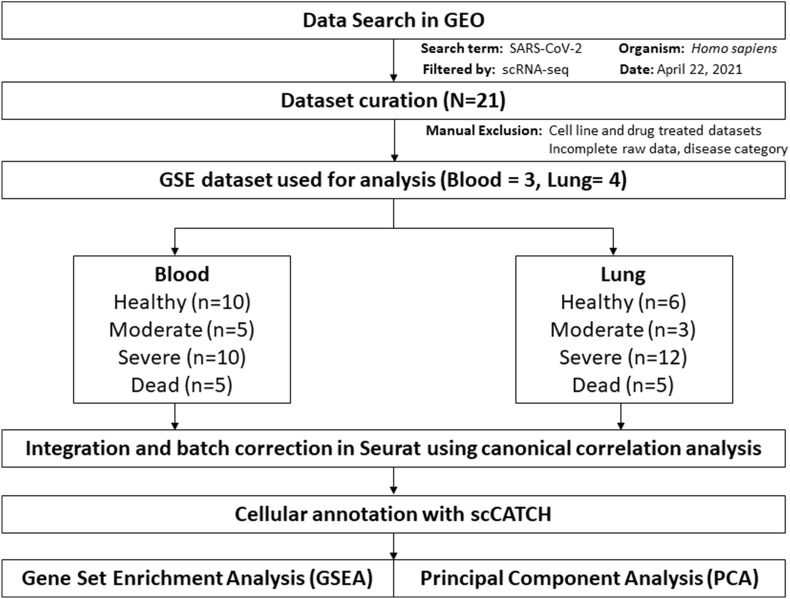
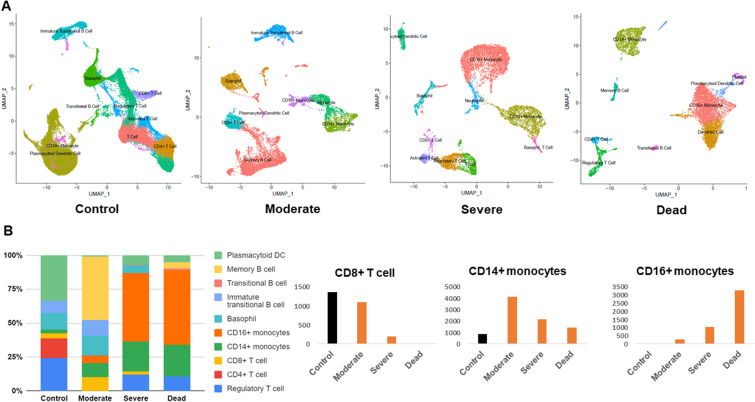
Fig. 1 presents an overview of the dataset curation and analysis. We assigned the datasets into healthy control, moderate, severe, and deceased groups. The datasets, comprising several individuals, were combined and integrated for each group (Table 1 and Table 2 ). Visualization using UMAP showed distinct differences between the COVID-19 patient groups and the healthy controls. Moderate COVID-19 cases were comparable to the controls in terms of the presence of immature transitional B cells and CD8+ T cells, whereas these cell types were depleted in severe and fatal cases (Fig. 2 A and Fig. 2B). Moderate cases had a slight increase of CD14+ monocytes and memory B cells (Fig. 2A and B). We did not observe any non-classical CD16+ monocytes in healthy controls or moderate cases; however, CD16+ monocytes were increased in severe cases and constituted approximately 50% of the total cell ratio in fatal cases. These data indicate an imbalance between lymphocytes and monocytes in the blood of severe COVID-19 cases. Our analysis also revealed significant alterations in B-cell subsets. In moderate cases, we detected transitional B cells and memory B cells, which constituted approximately 50% of the total cell population. In contrast, these B-cell subsets accounted for only 1%–4% of the total cell population in severe and fatal cases (Fig. 2B). Interestingly, we observed platelets only in fatal cases (Fig. 2A), suggesting a role of platelets in blood clot formation, which may be a decisive factor in the outcome of SARS-CoV-2 infection.
Fig. 1.
Schematic flow diagram of the dataset curation and meta-analysis process. N denotes the number of GEO datasets, n denotes the number of patients.
Table 1.
Clinical information of patients and healthy controls who donated blood samples.
| GSE | Type | Status | GSM ID | Sex | Age | ICU | Symptoms onset to sampling days | Publication status |
|---|---|---|---|---|---|---|---|---|
| GSE163668 | Blood | Healthy | GSM4995449 | Male | 49 | N/A | N/A | Yes (ref:19) |
| Healthy | GSM4995451 | Male | 52 | N/A | N/A | |||
| Healthy | GSM4995453 | Male | 30 | N/A | N/A | |||
| Healthy | GSM4995454 | Male | 34 | N/A | N/A | |||
| Healthy | GSM4995455 | Female | 28 | N/A | N/A | |||
| Healthy | GSM4995456 | Male | 41 | N/A | N/A | |||
| Healthy | GSM4995459 | Female | 34 | N/A | N/A | |||
| GSE166992 | Blood | Healthy | GSM5090446 | – | – | N/A | N/A | Yes (ref: 20) |
| Healthy | GSM5090448 | – | – | N/A | N/A | |||
| Healthy | GSM5090454 | – | – | N/A | N/A | |||
| GSE163668 | Blood | Moderate | GSM4995431 | Male | 41.1 | General ward | Not available | Yes (ref: 19) |
| Moderate | GSM4995432 | Male | 58.6 | ICU | 4 | |||
| Moderate | GSM4995434 | Male | 73.2 | General ward | Not available | |||
| Moderate | GSM4995435 | Female | 45.5 | General ward | 6 | |||
| Moderate | GSM4995436 | Female | 85 | General ward | 2 | |||
| GSE157344 | Blood | Severe | GSM4762161 | Female | 68 | ICU | Not available | Yes (ref: 23) |
| Severe | GSM4762168 | Male | 67 | ICU | Not available | |||
| Severe | GSM4762163 | Male | 77 | ICU | Not available | |||
| Severe | GSM4762164 | Male | 65 | ICU | Not available | |||
| Severe | GSM4762165 | Female | 51 | ICU | Not available | |||
| GSE163668 | Blood | Severe | GSM4995437 | Male | 44.2 | ICU | 20 | Yes (ref: 19) |
| Severe | GSM4995438 | Female | 61.4 | ICU | 14 | |||
| Severe | GSM4995442 | Female | 48.3 | ICU | 6 | |||
| Severe | GSM4995446 | Male | 62.4 | ICU | Not available | |||
| Severe | GSM4995447 | Male | 44.1 | ICU | 6 | |||
| GSE157344 | Blood | Dead | GSM4762162 | Male | 58 | ICU | Not available | Yes (ref: 23) |
| Dead | GSM4762167 | Male | 80 | ICU | Not available | |||
| Dead | GSM4762169 | Male | 69 | ICU | Not available | |||
| Dead | GSM4762171 | Male | 71 | ICU | Not available | |||
| Dead | GSM4762172 | Male | 68 | ICU | Not available |
Table 2.
Clinical information of patients and healthy controls who donated lung tissue samples.
| GSE | Type | Status | GSM ID | Sex | Age | ICU | Symptoms onset to sampling days | Publication status |
|---|---|---|---|---|---|---|---|---|
| GSE169471 | Lung | Healthy | GSM5206782 | Male | 76 | N/A | N/A | Yes (ref: 22) |
| Healthy | GSM5206783 | Male | 56 | N/A | N/A | |||
| Healthy | GSM5206784 | Male | 56 | N/A | N/A | |||
| Healthy | GSM5206785 | Male | 55 | N/A | N/A | |||
| Healthy | GSM5206786 | Female | 57 | N/A | N/A | |||
| Healthy | GSM5206787 | Male | 18 | N/A | N/A | |||
| GSE145926 | Lung | Moderate | GSM4339769 | Male | 36 | General ward | 11 | Yes (ref: 6) |
| Moderate | GSM4339770 | Female | 37 | General ward | 9 | |||
| Moderate | GSM4339772 | Male | 35 | General ward | 13 | |||
| Severe | GSM4339771 | Male | 66 | ICU | 11 | |||
| Severe | GSM4339773 | Male | 62 | ICU | 18 | |||
| Severe | GSM4339774 | Male | 63 | ICU | 14 | |||
| GSE168215 | Lung | Severe | GSM5134112 | Not available | – | Intubated | Not available | Preprint (ref: 21) |
| Severe | GSM5134116 | Not available | – | Intubated | Not available | |||
| Severe | GSM5134117 | Not available | – | Intubated | Not available | |||
| Severe | GSM5134119 | Not available | – | Intubated | Not available | |||
| GSE157344 | Lung | Severe | GSM4762139 | Female | 68 | ICU | Not available | Yes (ref: 23) |
| Severe | GSM4762146 | Male | 67 | ICU | Not available | |||
| Severe | GSM4762141 | Male | 77 | ICU | Not available | |||
| Severe | GSM4762142 | Male | 65 | ICU | Not available | |||
| Severe | GSM4762143 | Female | 51 | ICU | Not available | |||
| Dead | GSM4762140 | Male | 58 | ICU | Not available | |||
| Dead | GSM4762145 | Male | 80 | ICU | Not available | |||
| Dead | GSM4762147 | Male | 69 | ICU | Not available | |||
| Dead | GSM4762149 | Male | 71 | ICU | Not available | |||
| Dead | GSM4762150 | Male | 68 | ICU | Not available |
Fig. 2.
Cross-sectional analysis of single-cell RNA-seq blood datasets. (A) Uniform manifold approximation and projection (UMAP) of blood cells obtained from control, moderate, severe, and deceased subjects. Cells were clustered based on similar gene expression levels and colored by cell type. (B) Stacked bar chart showing the cell type proportion based on disease severity. Significantly altered cell types (CD4+ T cells, CD14+ monocytes, CD16+ monocytes) are presented in separate graphs along with their respective numbers in the different COVID-19 severities.
3.2. Macrophage class switching was evident in severe COVID-19 patients
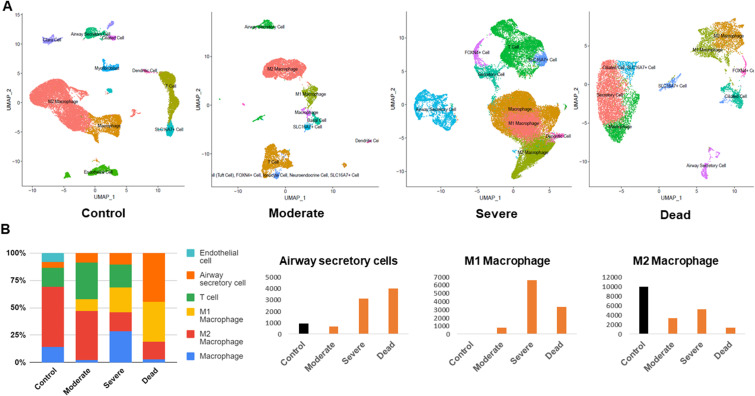
The lungs are the most severely affected organ in SARS-CoV-2 infection, and therefore we focused our analysis on the variability of the cellular landscape in lung. M2 macrophages resolve inflammation, and are known as anti-inflammatory macrophages, and were abundant in the lungs of healthy controls (Fig. 3 A and Fig. 3B). The total number of M2 macrophages was decreased in moderate, severe, and fatal cases, and the number of M1 (pro-inflammatory) macrophages increased with the progression of disease severity (Fig. 3A and B). The number of airway secretory cells was increased in severe cases and accounted for approximately 45% of the total cell ratio in fatal cases (Fig. 3B).
Fig. 3.
Cross-sectional analysis of single-cell RNA-seq lung datasets. (A) Uniform manifold approximation and projection (UMAP) of lung cells obtained from control, moderate, severe, and deceased subjects. Cells were clustered based on similar gene expression levels and colored by cell type. (B) Stacked bar chart showing the cell type proportion based on disease severity. Significantly altered cell types (airway secretory cells, M1 macrophages, M2 macrophages) are presented in separate graphs along with their respective numbers in the different COVID-19 severities.
3.3. Gene set enrichment analysis showed a lack of adaptive response in the blood of fatal cases
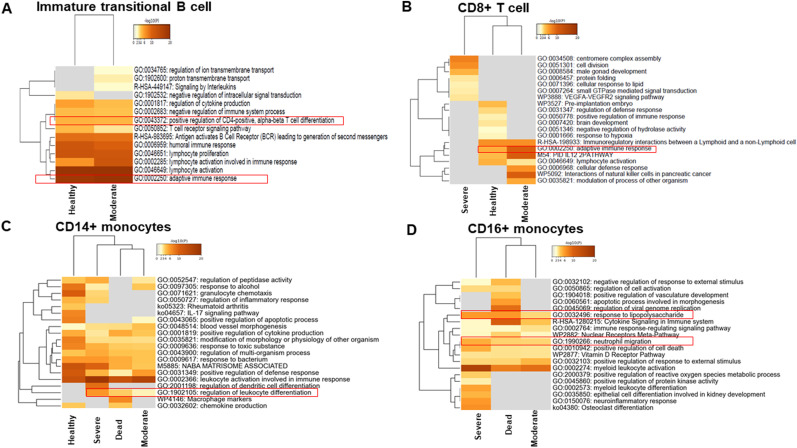
The cellular landscape was altered in SARS-CoV-2 infection, and the dynamic shift was associated with the degree of COVID-19 severity. We next analyzed whether the functional properties of the investigated cells were also altered. Gene set enrichment analysis (GSEA) was performed with the top 30 genes of the cells that demonstrated a large dynamic shift, either positively or negatively. GSEA showed that moderate cases had an adaptive response in immature transitional B cells, whereas these cells were absent in severe and fatal cases (Fig. 4 A). In moderate cases, the CD8+ T cells showed a similar pattern as in healthy controls, with a nearly identical adaptive response (Fig. 4B). However, the CD8+ T cells in severe and fatal cases did not express genes involved in the adaptive immune response. These data demonstrate the requirement for a lymphocyte-mediated adaptive response for the successful resolution of SARS-CoV-2 infection.
Fig. 4.
Hierarchical clustering of immune cells in blood based on gene set enrichment analysis. (A) Hierarchical clustering of the regulated pathways of significantly modulated genes in CD4+ T cells from different disease states. (B) Hierarchical clustering of the regulated pathways of significantly modulated genes in CD8+ T cells from different disease states. (C) Hierarchical clustering of the regulated pathways of significantly modulated genes in CD14+ monocytes from different disease states. (D) Hierarchical clustering of the regulated pathways of significantly modulated genes in CD16+ monocytes from different disease states. The dendrograms are colored according to the p values; gray cells indicate a lack of significant enrichment.
Interestingly, CD16+ monocytes were not found in healthy controls. However, CD14+ monocytes did not exhibit much difference among the disease states, apart from a milder response in terms of positive regulation of the defense response in severe and fatal cases (Fig. 4C). The GSEA of CD16+ monocytes closely clustered severe and fatal cases and indicated a strong response of the lipopolysaccharide-like phenotype and neutrophil migration (Fig. 4D). This could lead to prolonged inflammation and the development of a sepsis-like condition.
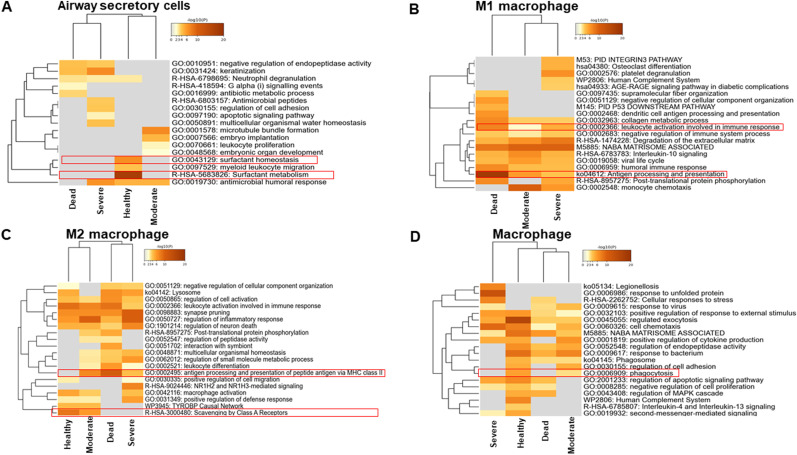
3.4. Fatal cases showed dysfunctional surfactant metabolism in the lung
Intriguingly, airway secretory cells accounted for almost half of the total cell population in the lung of fatal cases (Fig. 3B). The GSEA of airway secretory cells identified dysfunctional surfactant metabolism in all COVID-19 patient groups, whereas airway secretory cells from healthy controls were enriched with genes involved in surfactant metabolism (Fig. 5 A). We were unable to detect M1 macrophages in healthy controls; by contrast, M1 macrophages were increased in severe cases. The GSEA showed that M1 macrophages activate antigen processing and presentation (Fig. 5B). Interestingly, M2 macrophages demonstrated a similar phenotype of antigen processing and presentation via MHC class II in moderate, severe, and fatal cases (Fig. 5C). Macrophages that were positive for FCGR3A and FCGR3B were unable to initiate phagocytosis in severe and fatal cases (Fig. 5D). This not only indicates that the cellular landscape was altered but that cellular function was also impaired by SARS-CoV-2 infection.
Fig. 5.
Hierarchical clustering of lung cells based on gene set enrichment analysis. (A) Hierarchical clustering of the regulated pathways of significantly modulated genes in airway secretory cells from different disease states. (B) Hierarchical clustering of the regulated pathways of significantly modulated genes in M1 macrophages from different disease states. (C) Hierarchical clustering of the regulated pathways of significantly modulated genes in M2 macrophages from different disease states. (D) Hierarchical clustering of the regulated pathways of significantly modulated genes in macrophages from different disease states. The dendrograms are colored according to the p values; gray cells indicate a lack of significant enrichment.
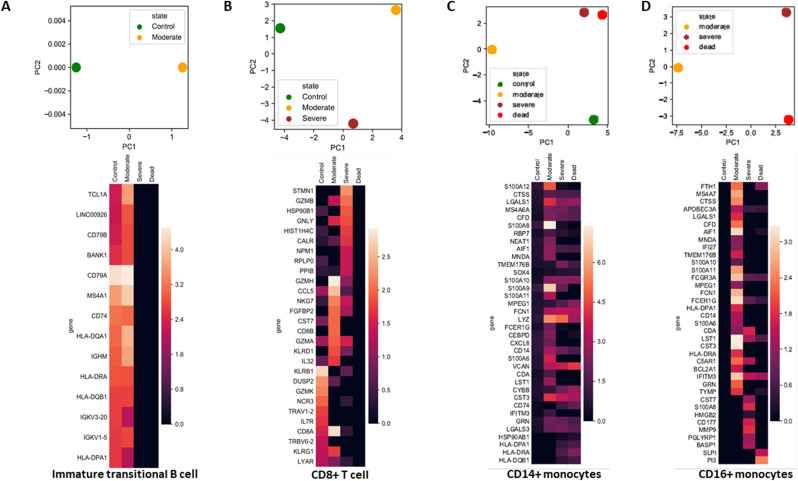
3.5. Principal component analysis could discriminate COVID-19 disease states
Functional alterations of the cellular landscape were evident and may decide the outcome of patients infected with SARS-CoV-2. We next investigated whether differential cellular landscapes and gene expression patterns could distinguish between disease states. We performed PCA on significantly altered cell types individually. The PCA of immature transitional B cells shifted directions in accordance with gene expression (Fig. 6 A). Immature transitional B cells exhibited almost identical gene expression in healthy controls and the moderate patient group (Fig. 6A). CD8+ T cells were not present in fatal cases, but the gene expression patterns could discriminate between the healthy controls, moderate cases, and severe cases (Fig. 6B). CD14+ monocytes were clearly discriminatory among the groups. Severe and fatal cases were in close proximity to each other and distinct from healthy controls and moderate cases (Fig. 6C). Auto-regulatory genes such as LAGLS1, LGALS3, and S100A4 were upregulated in the CD14+ monocytes of COVID-19 patients but not in healthy controls. Notably, the classical monocyte markers S100A8 and S100A9 were upregulated in moderate cases, but their expression levels were reduced in severe and fatal cases (Fig. 6C). Non-classical CD16+ monocytes were not present in healthy controls, but their gene expression pattern could distinguish moderate cases from severe and fatal cases. Compared with severe cases, CD16+ monocytes from moderate cases had higher expression levels of complement factors such C1QA/B/C; they also showed higher interferon-induced transmembrane protein 3 expression compared with these cells from severe and fatal cases. Severe and fatal cases distinctively expressed inflammatory chemokine CXCL8 and other inflammatory marker genes such as PTGS2, S100A12, S100A8, and MMP9 (Fig. 6D).
Fig. 6.
Principal component analysis (PCA) and heatmap of blood immune cells. (A) PCA and heatmap of significantly modulated genes along with their expression in CD4+ T cells from different disease states. (B) PCA and heatmap of significantly modulated genes along with their expression in CD8+ T cells from different disease states. (C) PCA and heatmap of significantly modulated genes along with their expression in CD14+ monocytes from different disease states. (D) PCA and heatmap of significantly modulated genes along with their expression in CD16+ monocytes from different disease states. The heatmaps are colored according to the gene expression values.
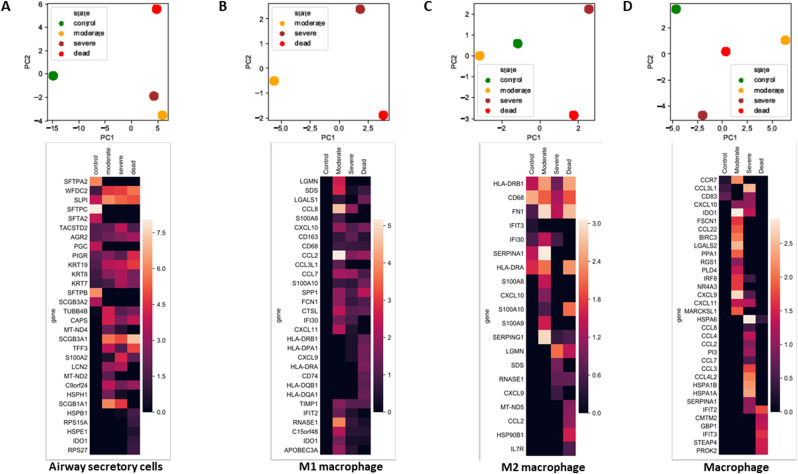
The airway secretory cells of healthy controls expressed numerous S100 fused-type proteins (SFTPs). The PCA of airway secretory cells clearly demonstrated that COVID-19 cases did not express these SFTP genes (Fig. 7 A). M1 macrophages from moderate cases expressed IF130, but this expression diminished with disease severity. Notably, M1 macrophages from all disease states expressed several chemokines, such as CCL2, CCL7, and CXCL10 (Fig. 7B). Additionally, genes responsible for inflammation, such as APOE, CD68, GRN, human leukocyte antigen (HLA)-DQA1, HLA-DQB1, HLA-DRB1, SERPINA1, and CCL18, were induced in M1 and M2 macrophages (Fig. 7B and C), indicating that macrophages play a predominantly inflammatory role. The PCA of macrophages revealed distinct clustering among groups. Macrophages from severe and fatal cases expressed HSPA1A, HSPB1, and HSPA6, indicating that an unfolded protein response was triggered. Furthermore, the severe and fatal cases showed higher expression levels of CCL3L1 and CCL4L2, indicating a chemokine-mediated action or neutrophil chemotaxis activation (Fig. 7D).
Fig. 7.
Principal component analysis (PCA) and heatmap of lung cells. (A) PCA and heatmap of significantly modulated genes along with their expression in airway secretory cells from different disease states. (B) PCA and heatmap of significantly modulated genes along with their expression in M1 macrophages from different disease states. (C) PCA and heatmap of significantly modulated genes along with their expression in M2 macrophages from different disease states. (D) PCA and heatmap of significantly modulated genes along with their expression in macrophages from different disease states. The heatmaps are colored according to the gene expression values.
4. Discussion
Mapping the immunological cell landscape of SARS-CoV-2 infection is of great importance to help combat the COVID-19 pandemic. An enormous effort by scientists across the globe has enabled us to understand the pathological nature of this virus. However, many investigations have been directed toward specific target tissues, and a lack of synchronized effort in collecting different tissues across the disease severity spectrum makes it difficult to form a complete picture. Here, we provide a comprehensive understanding of the varying disease states based on the cellular landscape of blood and lung in moderate, severe, and fatal COVID-19 cases.
Cross-sectional data analysis of moderate, severe, and fatal COVID-19 cases enabled us to identify links between the cellular landscape and disease outcome. For instance, we observed the abundant presence of lymphocytes in the blood of moderate cases, while B-cell subtypes and CD8+ T cells were largely missing in severe and fatal cases. This was also evident in previous studies [[30], [31], [32]] and in cases of SARS-CoV-1 infection [33]. T cells control hyperactive innate immune responses [34]. The loss of B-cell subtypes and CD4+ and CD8+ T cells may heighten the innate immune response for a prolonged period. This is in accordance with Zhou et al. [35], who identified a heightened innate immune response in the BALF of COVID-19 patients.
We observed that increased numbers of non-classical CD14+ and CD16+ monocytes were present in severe and fatal cases. Monocytes from older adults exhibited a higher proportion of non-classical monocytes but expressed a basal level of cytokines [36,37]. Another study reported that COVID-19 patients with ARDS have a higher percentage of intermediate CD14+ and CD16+ monocytes, which constitute almost 50% of the total cell population [38]. In our study, CD14+ and CD16+ monocytes comprised approximately 13% of the total cell ratio; however, this was increased to 31% and 57% in severe and fatal cases, respectively. Interestingly, the CD14+ and CD16+ monocytes expressed the macrophage marker FCN1, suggesting monocyte differentiation into macrophages upon SARS-CoV-2 infection. Moreover, we noted that the expression of HLA-DRA/DPA1 was absent in CD16+ monocytes isolated from severe and fatal cases. Such downregulation may hamper antigen presentation to CD4+ T cells and is often associated with a sepsis-like condition [39]. We also observed that immature transitional B cells in moderate patients activated CD4+ T-cell differentiation (Fig. 4A), and convalescent COVID-19 patients exhibited a virus-specific CD4+ T-cell response [40]. This is an indication that moderate cases may have undergone proper switching from the innate to adaptive response, whereas this switching did not take place in severe cases.
To date, most severe and fatal cases occur in older adults, and the clinical symptoms in children are mostly mild [41], raising concerns of immunosenescence. Indeed, naïve CD4+ T cells and CD8+ T cells decrease as we age [42], whereas naïve CD4+ T cells obtained from gestational age 18–22 weeks differentiate into the FoxP3+, CD25+ Treg phenotype [43]. This may explain the high fatality rate in older people and supports the idea, in line with our findings, that the depletion of T cells is linked to COVID-19 severity.
We also investigated lung epithelial cells and their immune connection using cross-sectional datasets obtained from lung or BALF. A dramatic increase of airway secretory cells and M1-like macrophages was evident. In severe and fatal cases, airway secretory cells were devoid of SFTPs. This indicates a surfactant dysfunction-like disorder that may induce breathing difficulties, leading to ARDS, which often requires mechanical ventilation. Indeed, surfactant therapy is effective in reversing the impaired oxygenation in COVID-19 patients with ARDS [44]. Macrophages accounted for 50% of the total cell population in severe and fatal cases in our analysis, which is in line with previous observations in which post-mortem lungs from Middle East respiratory syndrome (MERS) and SARS-CoV-1 patients exhibited a predominant presence of macrophages [45]. Macrophages exert their effect as innate immune cells by triggering the adaptive immune response, initiating the phagocytosis process [46,47]. However, in our data analysis, macrophages from severe and fatal cases exhibited a complete decline in phagocytosis. This suggests not only quantitative changes but also qualitative changes in the immune landscape of COVID-19 patients. Notably, CD163+ macrophages have been reported as intermediate macrophages and are increased in severe COVID-19 patients [48,49]. This is in agreement with our analysis because M1 macrophages that exhibited high CD163 expression in severe COVID-19 cases accounted for 20% of the total cell population. We found that macrophages obtained from severe and fatal COVID-19 cases expressed various neutrophil and monocyte attractant chemokines, such as CCL2, CCL3, CCL4, CCL7, CCL8, CXCL8, CXCL9, CXCL10, and CXCL11. Patients requiring mechanical ventilation exhibited an extensive occurrence of these chemokines [[50], [51], [52]]. These chemokines may conceivably attract more inflammatory cells to local sites in the lung and prolong local inflammation, thereby initiating pulmonary dysfunction [53]. Thus, these chemokines can be used as prognostic biomarkers of severity, and the early detection of these markers may help in designing effective therapeutic interventions.
To the best of our knowledge, this is the first cross-sectional study using single-cell RNA-seq datasets from COVID-19 patients with various degrees of severity. We identified that the immunological cellular landscape in blood was altered in severe and fatal cases. Notably, T-cell and B-cell subsets were reduced, and the number of monocytes was increased (see graphical abstract). This resulted in an aberrant peripheral adaptive immune response. We also found that macrophages in lung acquired an inflammatory phenotype, with a high abundance of CD163+ M1-like macrophages. Our data revealed a qualitative alteration because these immune cells adopted a pro-inflammatory phenotype and secreted an extensive set of chemokines that could continue to recruit additional inflammatory immune cells to local sites. Despite these distinct findings, our study had some limitations. Owing to the limited number of severe and fatal cases in children and young adults, we were unable to draw direct comparisons between T-cell depletion and aging. Because the number of T cells may be the determining factor for successful viral clearance, future studies should focus on the relationship between T-cell numbers and disease severity among children, young adults, and those over 65 years of age. This will shed light on the differential outcomes between children and older adults with SARS-CoV-2 infection.
Data and code availability
The datasets and code used in this study are publicly available and comprehensively described in the methods section. Any other necessary information will be provided upon reasonable request.
Author contributions
M.Z.H. and T.K. conceptualized the study. M.Z.H., S.I., and K.M. analyzed the data and interpreted the results. M.Z.H. and T.K. wrote the manuscript. K.M. and T.K. supervised the data collection and overall project.
Declaration of competing interest
The authors declare that no conflicts of interest exist.
Acknowledgments
This work was supported by KAKENHI Grants-in-Aid for Research Activity 20H03468 (T. Kawai). We thank Katherine Thieltges, from Edanz Group (https://jp.edanz.com/ac) for editing a draft of this manuscript.
References
- 1.Chen G., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. J. Am. Med. Assoc. 2020 Apr 7;323(13):1239–1242. doi: 10.1001/jama.2020.2648. PMID: 32091533. [DOI] [PubMed] [Google Scholar]
- 3.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y.…Wang H. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell discovery. 2020;6(1):1–18. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J.Y., Wang X.M., Xing X., Xu Z., Zhang C., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020 Sep;21(9):1107–1118. doi: 10.1038/s41590-020-0762-x. Epub 2020 Aug 12. PMID: 32788748. [DOI] [PubMed] [Google Scholar]
- 5.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B.…Ziebuhr J. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182(6):1419–1440. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao M., Liu Y., Yuan J., Wen Y., Xu G., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020 Jun;26(6):842–844. doi: 10.1038/s41591-020-0901-9. Epub 2020 May 12. PMID: 32398875. [DOI] [PubMed] [Google Scholar]
- 7.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H.…Shin E.C. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Science immunology. 2020;5(49) doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F.…Eils R. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 9.Guo C., Li B., Ma H., Wang X., Cai P., et al. Single-cell analysis of two severe COVID-19 patients reveals a monocyte-associated and tocilizumab-responding cytokine storm. Nat. Commun. 2020 Aug 6;11(1):3924. doi: 10.1038/s41467-020-17834-w. PMID: 32764665; PMCID: PMC7413381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y., Su B., Guo X., Sun W., Deng Y., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020 Jul 9;182(1):73–84. doi: 10.1016/j.cell.2020.05.025. e16, Epub 2020 May 18. PMID: 32425270; PMCID: PMC7231725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J.Y., Wang X.M., Xing X., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21(9):1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7(2):131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 13.Hadjadj J., Yatim N., Barnabei L., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z., et al. Overly exuberant innate immune response SARS-CoV-2 infect. Cell Host Microbe. 2020 doi: 10.2139/ssrn.3551623. [DOI] [Google Scholar]
- 15.Ciccullo A., Borghetti A., Zileri Dal Verme L., et al. Neutrophil-to-lymphocyte ratio and clinical outcome in COVID-19: a report from the Italian front line. Int. J. Antimicrob. Agents. 2020;56(2):106017. doi: 10.1016/j.ijantimicag.2020.106017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao M., Liu Y., Yuan J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 17.Diao B., Wang C., Tan Y., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. Published 2020 May 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, Z., Zhou, Q., Wang, C., Shi, Q., Lu, S, et al. Clinical characteristics of children with COVID-19: a rapid review and meta-analysis. Ann. Transl. Med., 8(10), 620. 10.21037/atm-20-3302. [DOI] [PMC free article] [PubMed]
- 19.Combes A.J., Courau T., Kuhn N.F., Hu K.H., Ray A., et al. Global absence and targeting of protective immune states in severe COVID-19. Nature. 2021 Mar;591(7848):124–130. doi: 10.1038/s41586-021-03234-7. Epub 2021 Jan 25. PMID: 33494096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson E.A., Cascino K., Ordonez A.A., et al. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021;34(11):108863. doi: 10.1016/j.celrep.2021.108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida Masahiro, Worlock Kaylee B., Huang Ni, Rik G., Lindeboom H., Meyer Kerstin B., et al. medRxiv; 2021. The Local and Systemic Response to SARS-CoV-2 Infection in Children and Adults. 03.09.21253012. [DOI] [Google Scholar]
- 22.Saygin D., Tabib T., Bittar H.E.T., Valenzi E., Sembrat J., et al. Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension. Pulm. Circ. 2020 Feb 28;(1):10. doi: 10.1177/2045894020908782. PMID: 32166015; PMCID: PMC7052475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bost P., De Sanctis F., Canè S., Ugel S., Donadello K., et al. Deciphering the state of immune silence in fatal COVID-19 patients. Nat. Commun. 2021 Mar 5;12(1):1428. doi: 10.1038/s41467-021-21702-6. PMID: 33674591; PMCID: PMC7935849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuart T., Butler A., Hoffman P., et al. Comprehensive integration of single-cell data. Cell. 2019 Jun 13;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran H.T.N., Ang K.S., Chevrier M., et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21:12. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becht E., McInnes L., Healy J., et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018 Dec 3 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 27.Shao Xin, Liao Jie, Lu Xiaoyan, Xue Rui, Ni Ai, Fan Xiaohui. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience. 2020;23(Issue 3):100882. doi: 10.1016/j.isci.2020.100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R., Wang Y., Li J., Han H., Xia Z., et al. Decreased T cell populations contribute to the increased severity of COVID-19. Clinica chimica acta. international journal of clinical chemistry. 2020;508:110–114. doi: 10.1016/j.cca.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W., Berube J., McNamara M., Saksena S., Hartman M., et al. Lymphocyte subset counts in COVID-19 patients: a meta-analysis. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2020;97(8):772–776. doi: 10.1002/cyto.a.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Long W., Tu M., Chen S., Huang Y., et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong R.S., Wu A., To K.F., Lee N., Lam C.W., et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;21(7403):1358–1362. doi: 10.1136/bmj.326.7403.1358. 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim K.D., Zhao J., Auh S., Yang X., Du P., Tang H., Fu Y.X. Adaptive immune cells temper initial innate responses. Nat. Med. 2007;13(10):1248–1252. doi: 10.1038/nm1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z., Ren L., Zhang L., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–890. doi: 10.1016/j.chom.2020.04.017. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hearps A.C., Martin G.E., Angelovich T.A., Cheng W.J., Maisa A., Landay A.L., et al. Aging is associated with chronic innate immune activation and dysregulation of monocyte phenotype and function. Aging Cell. 2012;11:867–875. doi: 10.1111/j.1474-9726.2012.00851.x. [DOI] [PubMed] [Google Scholar]
- 37.Nyugen J., Agrawal S., Gollapudi S., Gupta S. Impaired functions of peripheral bloodmonocyte subpopulations in aged humans. J. Clin. Immunol. 2010;30:806–813. doi: 10.1007/s10875-010-9448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., et al. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl Sci Rev. 2020 doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim O.Y., Monsel A., Bertrand M., Coriat P., Cavaillon J.M., Adib-Conquy M. Differential down-regulation of HLA-DR on monocyte subpopulations during systemic inflammation. Crit. Care. 2010;14:R61. doi: 10.1186/cc8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020 doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feldstein L.R., Rose E.B., Horwitz S.M., Collins J.P., Newhams M.M., et al. Multisystem inflammatory syndrome in U.S. Children and adolescents. N. Engl. J. Med. 2020 Jul 23;383(4):334–346. doi: 10.1056/NEJMoa2021680. Epub 2020 Jun 29. PMID: 32598831; PMCID: PMC7346765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saule P., Trauet J., Dutriez V., Lekeux V., Dessaint J.P., Labalette M. Accumulation of memory T cells from childhood to old age: central and effector memory cells in CD4(+) versus effector memory and terminally differentiated memory cells in CD8(+) compartment. Mech. Ageing Dev. 2006 Mar;127(3):274–281. doi: 10.1016/j.mad.2005.11.001. Epub 2005 Dec 13. PMID: 16352331. [DOI] [PubMed] [Google Scholar]
- 43.Mold J.E., Venkatasubrahmanyam S., Burt T.D., Michaëlsson J., Rivera J.M., et al. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science. 2010 Dec 17;330(6011):1695–1699. doi: 10.1126/science.1196509. Erratum in: Science. 2011 Feb 4;331(6017):534. PMID: 21164017; PMCID: PMC3276679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heching Moshe, Lev Shaul, Shitenberg Dorit, Dicker Dror, Kramer Mordechai R. Surfactant for the treatment of ARDS in a patient with coronavirus disease 2019. Chest. 2021 doi: 10.1016/j.chest.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fels A., Cohn Z. The alveolar macrophage. J. Appl. Physiol. 1986;60(2):353–369. doi: 10.1152/jappl.1986.60.2.353. [DOI] [PubMed] [Google Scholar]
- 47.Brieland J.K., Kunkel R.G., Fantone J.C. Pulmonary alveolar macrophage function during acute inflammatory lung injury. Am. Rev. Respir. Dis. 1987;135(6):1300–1306. doi: 10.1164/arrd.1987.135.6.1300. [DOI] [PubMed] [Google Scholar]
- 48.Wang J., Jiang M., Chen X., Montaner L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020 doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 53.Nicholls J.M., Poon L.L., Lee K.C., et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773–1778. doi: 10.1016/s0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and code used in this study are publicly available and comprehensively described in the methods section. Any other necessary information will be provided upon reasonable request.