Abstract
The pheromone response pathway of the yeast Saccharomyces cerevisiae is initiated in MATa cells by binding of α-factor to the α-factor receptor. MATa cells in which the a-factor receptor is inappropriately expressed exhibit reduced pheromone signaling, a phenomenon termed receptor inhibition. In cells undergoing receptor inhibition, activation of the signaling pathway occurs normally at early time points but decreases after prolonged exposure to pheromone. Mutations that suppress the effects of receptor inhibition were obtained in the STE4 gene, which encodes the β-subunit of the G protein that transmits the pheromone response signal. These mutations mapped to the N terminus and second WD repeat of Ste4p in regions that are not part of its Gα binding surface. A STE4 allele containing several of these mutations, called STE4SD13, reversed the signaling defect seen at late times in cells undergoing receptor inhibition but had no effect on the basal activity of the pathway. Moreover, the signaling properties of STE4SD13 were indistinguishable from those of STE4 in wild-type MATa and MATα cells. These results demonstrate that the effect of the STE4SD13 allele is specific to the receptor inhibition function of STE4. STE4SD13 suppressed the signaling defect conferred by receptor inhibition in a MATa strain containing a deletion of GPA1, the G protein α-subunit gene; however, STE4SD13 had no effect in a MATα strain containing a GPA1 deletion. Suppression of receptor inhibition by STE4SD13 in a MATa strain containing a GPA1 deletion was unaffected by deletion of STE2, the α-factor receptor gene. The results presented here are consistent with a model in which an a-specific gene product other than Ste2p detects the presence of the a-factor receptor and blocks signaling by inhibiting the function of Ste4p.
Cells respond to their external environment by recognizing an extracellular signal, transmitting the signal across the cell membrane, and eliciting a response through activation of the appropriate signal transduction pathway. The binding of a secreted peptide pheromone to its cell surface receptor initiates mating in the budding yeast Saccharomyces cerevisiae. Mating occurs between cells of opposite mating types; haploid yeast cells may be either a or α mating type and produce the secreted peptide pheromone a-factor or α-factor, respectively (reviewed in references 20 and 33). These pheromone ligands bind to the appropriate receptor located on the surface of cells of the opposite mating type; the a-factor receptor (encoded by STE3) is present on the surface of MATα cells, and the α-factor receptor (encoded by STE2) is present on the surface of MATa cells. The pheromone receptors are members of the G protein-coupled receptor family and are coupled to a heterotrimeric G protein composed of α-, β-, and γ-subunits (encoded by GPA1, STE4, and STE18, respectively). In addition to the G protein, the yeast pheromone response pathway utilizes another common eukaryotic signaling module, a mitogen-activated protein (MAP) kinase cascade (13). Transmission of the signal from the G protein βγ complex to the downstream kinase cascade probably occurs through activation of the PAK kinase homologue Ste20p (21). Specificity of the kinases that are sequentially activated during pheromone signaling is thought to be maintained by the scaffold protein Ste5p (38). The ultimate responses to pheromone signaling include arrest in the G1 phase of the cell cycle, which is mediated by the cyclin-dependent kinase inhibitor Far1p (3, 27, 28). Other responses to pheromone signaling include morphological changes leading to projection formation and transcriptional induction of genes involved in mating.
The differential expression of mating type-specific genes is controlled by regulatory proteins encoded by the mating-type (MAT) locus (reviewed in reference 32). Haploid MATa cells normally express Ste2p, the α-factor receptor, and undergo cell cycle arrest and transcriptional induction in response to α-factor stimulation. However, MATa cells containing the STE3DAF mutation inappropriately express the a-factor receptor and exhibit resistance to pheromone-induced cell cycle arrest, a phenomenon termed receptor inhibition (14). The STE3DAF mutation, originally named DAF2 (for dominant α-factor resistance), was isolated in a screen for mutations that resulted in resistance to α-factor-induced cell cycle arrest in MATa cells (7). The STE3DAF allele contains a rearrangement in the 5′ flanking region of the STE3 gene which permits expression of wild-type STE3 in all cell types (14). The abundance of STE3 RNA in cells containing STE3DAF is comparable to the normal level of STE3 RNA in MATα cells, so the STE3DAF phenotype is not the result of overexpression of the receptor. In addition to conferring resistance to cell cycle arrest, the STE3DAF allele also causes an increase in the basal expression of a pheromone-inducible gene, FUS1. The increase in FUS1 basal expression is eliminated in STE3DAF cells that contain deletions of the genes that encode a-factor. This finding demonstrates that the increase in FUS1 basal expression is caused by the presence of both a-factor and the a-factor receptor in the same cell. Thus, expression of a pheromone receptor and its ligand in the same cell causes autocrine stimulation of the pheromone signaling pathway. However, inhibition of cell cycle arrest by the STE3DAF allele is not affected by deletion of the a-factor genes, indicating that autocrine stimulation does not play a role in this phenotype. STE3DAF-mediated receptor inhibition can suppress the constitutive cell cycle arrest caused by deletion of GPA1, the G protein α-subunit gene, and thus does not require the α-subunit for function (7, 14). Expression of STE2, which encodes the α-factor receptor, is not required for receptor inhibition; deletion of the STE2 gene does not affect the ability of STE3DAF to suppress constitutive cell cycle arrest in cells containing null alleles of GPA1 (14). In addition to inhibition of cell cycle arrest, cells containing the STE3DAF mutation exhibit a block in pheromone-mediated signaling at late time points after pheromone treatment (6). In STE3DAF cells, initial activation of the pheromone response pathway is similar to that observed in wild-type cells, as measured by Fus3p MAP kinase activity and FUS1 RNA levels. However, at later time points after pheromone induction, STE3DAF cells display a decrease in signaling when compared to wild-type levels (6). Furthermore, epistasis experiments suggest that STE3DAF acts at the level of either STE5 or STE4 (6).
In this work, we have isolated mutations in the G protein β-subunit gene, called STE4SD mutations, that suppress receptor inhibition resulting from expression of STE3DAF in MATa cells. The STE4SD mutations encode Ste4p proteins that reverse the effects of STE3DAF on both pheromone-mediated cell cycle arrest and transcriptional activation. The effects of the STE4SD mutations are specific to cells undergoing receptor inhibition, suggesting that these mutations may define a region of Ste4p that is the target of the signaling block that occurs as a result of receptor inhibition.
MATERIALS AND METHODS
Plasmid construction.
A centromeric URA3 plasmid containing STE4 was constructed by cloning the 5-kb SphI-BamHI fragment from plasmid M81p12 (5) into YCplac33 (11) to create YCpSTE4. A pUC19 plasmid containing STE4 was constructed by cloning the 5-kb SphI-BamHI fragment from plasmid M81p12 into pUC19 to create pUC-STE4.1. A centromeric LEU2 plasmid containing STE4 was constructed by cloning the 5-kb SphI-BamHI fragment from plasmid M81p12 into YCplac111 (11) to create YCpLSTE4. Centromeric LEU2 plasmids containing other STE4SD alleles (see Table 2) were constructed by cloning the 5-kb SphI-BamHI fragment from pUC-STE4.1 plasmids that had been subjected to site-directed mutagenesis (Transformer Site-Directed Mutagenesis kit; Clontech) into YCplac111. The FUS1-lacZ reporter plasmid was constructed by cloning the 6-kb PstI fragment from pSB234 (kindly provided by E. Elion) into YCplac111 to create YCpF1-LZ.
TABLE 2.
Level of G1 arrest conferred by mutations in STE3DAF cells and degree of supersensitivity conferred by mutations in wild-type cells
| STE4 allele | Plasmid | Mutation(s) present | % Unbudded cellsa | Halo size (mm)b |
|---|---|---|---|---|
| STE4 | YCpSTE4 | 60 (56, 63) | 39.0 | |
| STE4SD1 | YCpSD1 | R162G, C182R, I195V | 92 (91, 93) | 44.0 |
| STE4SD2 | YCpSD2 | Q17L, Q21R, M283V | 70 (68, 72) | 42.5 |
| STE4SD3 | YCpSD3 | L132I, N155D, D270G | 66 (66, 66) | 44.0 |
| STE4SD13 | YCpSD13 | Q17L, Q21R, R162G | 71 (67, 75) | 40.5 |
| STE4 | YCpLSTE4 | 51 (47, 55) | 35.5 | |
| STE4SD4 | YCpLSD4 | R162G | 43 (43, 44) | 36.0 |
| STE4SD5 | YCpLSD5 | C182R | 50 (52, 48) | 43.0 |
| STE4SD7 | YCpLSD7 | Q17L | 51 (50, 52) | 37.0 |
| STE4SD8 | YCpLSD8 | Q21R | 49 (46, 52) | 35.5 |
| STE4SD9 | YCpLSD9 | R162G, I195V | 62 (65, 59) | 35.0 |
| STE4SD10 | YCpLSD10 | Q17L, R162G | 65 (61, 69) | 36.0 |
| STE4SD11 | YCpLSD11 | Q21R, R162G | 61 (63, 59) | 36.5 |
| STE4SD12 | YCpLSD12 | Q17L, Q21R | 59 (62, 56) | 37.0 |
| STE4SD13 | YCpLSD13 | Q17L, Q21R, R162G | 71 (69, 72) | 36.5 |
| STE4SD14 | YCpLSD14 | Q17L, Q21R, R162G, I195V | 62 (60, 64) | 37.5 |
MATa STE3DAF strains (AC17-2B) containing the indicated plasmids were treated with 0.1 μM α-factor for 3 h, and the percentage of unbudded cells was determined. The experiment was performed with two independent transformants, and the average of the two determinations is presented, followed by the values for each experiment in parentheses.
Halo assays were performed on a single transformant each of a MATa strain (AC17-7B) containing the indicated plasmids using 5 μl of 1 mM α-factor. Boldfacing indicates supersensitive responses.
Strains and media.
The strains used in this study are listed in Table 1. The gpa1::TRP1 null allele was made by transformation of a strain that contains a gpa1::URA3 allele (10) with a 3.8-kb SmaI fragment from marker swap plasmid pUT11 (8). The FAR1 gene was disrupted by transformation with a 3.8-kb XhoI-SacI fragment from pfar1-U1 (6) to create far1::URA3. The ste2::LEU2 allele was made by transformation with a BamHI fragment from pAB506. All strain constructions involving transformations were confirmed by Southern blotting.
TABLE 1.
Strains used in the study
| Strain | Genotype | Source |
|---|---|---|
| W3031A | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | R. Rothstein |
| Strains are isogenic to W3031A | ||
| AC17-7B | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 sst1::hisG ste4::HIS3 | This study |
| AC17-2B | MATa mfa1-Δ3::HIS3 mfa2-Δ2::HIS3 STE3DAF2.5 sst1::hisG ste4::HIS3 | This study |
| AC18-9C | MATα mfα1::LEU2 mfα2::LEU2 ste4::HIS3 | |
| K39-23B | MATa ste4::HIS3 STE3DAF2.5 gpa1::TRP1 | This study |
| K39-23B.f | MATa ste4::HIS3 STE3DAF2.5 gpa1::TRP1 far1::URA3 | This study |
| K39-23D.f | MATα ste4::HIS3 STE3DAF2.5 gpa1::TRP1 far1::URA3 | This study |
| K39-23B.s2 | MATa ste4::HIS3 STE3DAF2.5 gpa1::TRP1 ste2::LEU2 | This study |
Strains were grown on yeast extract-peptone-dextrose (2% glucose) or yeast extract–peptone–3% galactose, and strains under selection were grown on synthetic dropout medium, as described previously (30).
PCR mutagenesis and screen for STE4SD alleles.
STE4SD alleles were isolated by cotransformation of yeast with a pool of PCR-generated mutagenized linear fragments containing STE4 and with a gapped STE4 plasmid, allowing recombination to occur in vivo as described previously (2, 24). Error-prone PCR was performed with YCpSTE4 as a template in 30 cycles of PCR with oligonucleotide primers oMSTE4.1 (5′-AAGAGTACACTAGATCCATTC-3′) and oMSTE4.3 (5′-AAAGGAAGCAAATGACAATGC-3′). Error-prone incorporation of nucleotides was achieved by performing PCRs under a nucleotide imbalance (80 μM dATP, 400 μM dCTP, 400 μM dGTP, and 400 μM dTTP or 400 μM dATP, 400 μM dCTP, 80 μM dGTP, and 400 μM dTTP) and by using modified Taq polymerase buffer containing 100 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, and 0.25 mM MnCl2. Approximately 10% of the mutated fragments produced nonfunctional Ste4p proteins as determined by their ability to complement a Δste4 mutation.
Strain AC17-2B(YCpF1-LZ) was cotransformed with the pooled products of the error-prone PCRs and with the 9-kb XhoI-AflII fragment of YCpSTE4, which lacks the STE4 coding region. Transformants were replica plated to selective plates (pH 7.0) that had been spread with 15 μl of 1 mM α-factor (Sigma) and 80 μl of a 40-mg/ml concentration of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Jersey Lab Supply). The strain used in this screen contains a deletion of the SST1 gene, which encodes an extracellular protease that degrades α-factor, to ensure that the α-factor on the plates remained intact. A total of approximately 24,000 replicated colonies were scored for the absence of growth and for the development of blue color. Plasmids that retained the ability to confer the phenotype after retransformation were sequenced by the dideoxy chain termination method (Sequenase kit; Amersham). Mutagenized plasmids YCpSD1, YCpSD2, and YCpSD3 were isolated by this procedure.
Yeast methods.
Yeast transformations were performed by the lithium acetate method (16), modified as described previously (14). Yeast RNA was extracted from cells as described previously (9).
Halo assays were performed by plating a lawn of cells to be tested and placing a filter paper disk containing 5 μl of 1 mM α-factor onto the plate. The plates were then incubated at 30°C for 1 to 2 days.
The percentage of unbudded cells was determined by growing cells to log phase, treating them with α-factor (0.1 μM) for 3 h, and then fixing them for 1 h with 3.7% formaldehyde. The cell suspension was then sonicated, and the number of budded cells in a total of approximately 200 cells was counted.
Northern (RNA) blots.
Cells were treated with either 0.1 μM α-factor (Sigma) or 40 ng of a-factor (generously provided by Fred Naider) per ml for various periods of time, and RNA was isolated. RNA was transferred to a nitrocellulose membrane after formaldehyde-agarose gel electrophoresis as described previously (22). The membranes were UV cross-linked by using a Stratalinker UV box. Prehybridization and hybridization were done at 65°C in a buffer containing 0.9 M NaCl, 0.09 M sodium citrate, 0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin, 33 mM sodium pyrophosphate, and 50 mM sodium phosphate monobasic. The probes used were gel-purified DNA restriction fragments 32P-labeled by random primer labeling with a Prime-It kit (Stratagene). The fragments used were as follows: for FUS1, a 1.4-kb EcoRI-HindIII fragment from plasmid pSL589 (26); for phosphoglycerate kinase gene PGK1, a 0.5-kb BamHI-XbaI fragment from pPGK1.
RESULTS
The pheromone response signal transduction pathway is inhibited in MATa cells that contain the STE3DAF allele, which causes inappropriate expression of STE3, the a-factor receptor gene (6). Receptor inhibition is specific for the late phase of the response and only occurs at times after 1 h of exposure to pheromone. Under conditions of receptor inhibition, the signaling pathway is blocked at a step upstream of MAP kinase cascade activation and at or downstream of activation of the Ste4p Gβ subunit. These results suggest that Ste4p is a likely target for the inhibitory effect of STE3DAF. A screen was therefore performed to identify altered versions of Ste4p that can signal normally but are insensitive to receptor inhibition.
Mutations in STE4 suppress receptor inhibition.
A screen was performed to obtain mutations in STE4 that have the potential to be specific for its receptor inhibition function. This screen was designed to isolate alleles of STE4 that suppress the signaling defect of MATa cells containing the STE3DAF allele. MATa STE3DAF cells exposed to α-factor do not arrest and do not sustain pheromone-inducible transcription. Therefore, mutations in STE4 that caused cell cycle arrest and sustained transcription to occur in a MATa STE3DAF strain were sought. This phenotype requires that the altered versions of Ste4p retain the ability to transmit the pheromone response signal. Thus, this screen has the potential to generate mutations in STE4 that are specific to receptor inhibition.
The strain used to screen for STE4 mutations contains the MATa allele and the STE3DAF mutation. Previous studies have shown that MATa STE3DAF cells, which express both a-factor and the a-factor receptor, display a low level of constitutive signaling due to autocrine stimulation (14). Therefore, the strain used in this screen (AC17-2B) was constructed to contain deletions of the a-factor genes, MFA1 and MFA2. Deletion of these genes eliminates any contribution to signaling due to autocrine stimulation of the a-factor receptor (14). A STE3DAF strain containing MFA1 and MFA2 deletions thus displays the normal basal level of signaling, which facilitates the ability to score for increased signaling.
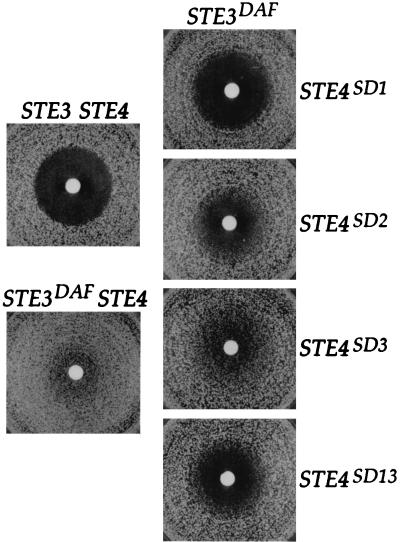
A fragment containing the STE4 gene was subjected to error-prone PCR, and the PCR products were cotransformed with a gapped STE4 plasmid into MATa STE3DAF cells to allow recombination in vivo (2, 24). Transformed cells were transferred to plates containing α-factor and X-Gal to assay their ability to undergo cell cycle arrest and to activate a pheromone-inducible lacZ reporter construct. Three STE4 alleles that caused cells to arrest and activate the reporter construct in response to α-factor were isolated, and each of them suppressed the STE3DAF phenotype to a different degree. The STE4SD alleles (designated STE4SD for suppressor of STE3DAF) were tested for their ability to confer cell cycle arrest by a halo assay, which measures the density of cell growth in an area surrounding a filter disk containing α-factor. The STE4SD1 allele caused STE3DAF cells to arrest at a level comparable to that of wild-type cells; the STE4SD2 and STE4SD3 alleles caused STE3DAF cells to undergo partial arrest (Fig. 1). To quantify the level of G1 arrest conferred by these mutations, STE3DAF strains carrying each of the STE4SD alleles were treated with pheromone and the percentage of unbudded cells was determined. After treatment with α-factor for 3 h, STE3DAF cells carrying the STE4SD1 allele were 92% unbudded, whereas STE3DAF cells carrying wild-type STE4 were 60% unbudded (Table 2). The STE4SD2 allele conferred a modest increase in the percentage of unbudded cells, in agreement with the halo assays; the STE4SD3 allele had little or no effect on cell cycle arrest in this assay.
FIG. 1.
Mutations in STE4 suppress receptor inhibition. Halo assays were performed with 5 μl of 1 mM α-factor. Top left, a MATa STE3 ste4::HIS3 strain (AC17-7B) containing a wild-type STE4 plasmid (YCpLSTE4); bottom left, a MATa STE3DAF ste4::HIS3 strain (AC17-2B) containing a wild-type STE4 plasmid (YCpLSTE4); right, top to bottom, a MATa STE3DAF ste4::HIS3 strain (AC17-2B) containing plasmids with STE4SD1, STE4SD2, STESD3, and STE4SD13 alleles (YCpSD1, YCpSD2, YCpSD3, and YCpSD13), respectively.
Isolation of these suppressors demonstrates that the block to cell cycle arrest seen in STE3DAF cells can be reversed by mutations in STE4. However, these mutations could alter residues of Ste4p that are unrelated to receptor inhibition. For example, if they confer an increase in the ability of Ste4p to transmit the pheromone response signal, then the increased signal might overcome the inhibition caused by expression of STE3. Further tests of the phenotype conferred by the STE4SD alleles were therefore necessary to determine if they specifically affect the receptor inhibition function of STE4.
Mutations conferring supersensitivity enhance the STE4SD phenotype.
To determine whether the STE4SD alleles cause increased signaling in wild-type cells, plasmids containing each of the three alleles were transformed into MATa Δste4 cells that did not contain the STE3DAF mutation. Cells carrying each of the STE4SD alleles displayed increased sensitivity to α-factor, as shown by production of halos larger than those produced by cells containing a wild-type STE4 plasmid (Table 2). Therefore, some component of the suppression of STE3DAF by the STE4SD mutations is probably due to an increase in the signaling capacity of the encoded Ste4p proteins.
The STE4SD alleles were sequenced to identify the nucleotide changes that were responsible for the STE4SD phenotype. Each allele harbored three nucleotide changes that resulted in three alterations in the coding sequence (Table 2). Because the STE4SD3 allele displayed the smallest degree of STE3DAF suppression and conferred a supersensitive phenotype, it was not studied further. Mutations present in the STE4SD1 and STE4SD2 alleles were tested individually to assess their contribution to the STE4SD phenotype. STE3DAF cells containing STE4SD alleles with single mutations did not display significant pheromone-induced cell cycle arrest, as demonstrated by the finding that only 43 to 51% of the cells were unbudded after α-factor treatment (Table 2). This percentage is similar to the fraction of G1 cells in cycling populations. The C182R mutation (STE4SD5) originally encoded by STE4SD1 was found to confer supersensitivity to α-factor in wild type cells but did not suppress the STE3DAF phenotype. Therefore, it is likely that this mutation contributed to the phenotype of the STE4SD1 allele, although it is not specific to the receptor inhibition function of STE4.
To obtain a STE4SD allele that conferred a high degree of STE3DAF suppression but did not cause supersensitivity to α-factor in wild-type cells, STE4 genes containing different combinations of STE4SD mutations were tested in both STE3DAF and wild-type cells. STE4SD alleles encoding the R162G mutation and either the I195V, Q17L, or Q21R mutation conferred a partial cell cycle arrest response to STE3DAF cells, resulting in 61 to 65% unbudded cells after α-factor treatment (Table 2). The greatest degree of arrest was seen in cells containing the STE4SD13 allele, which were 71% unbudded after α-factor treatment. The product of this allele contains the Q17L and Q21R mutations encoded by STE4SD2 and the R162G mutation encoded by STE4SD1. The STE4SD13 allele did not confer a complete cell cycle arrest response on STE3DAF cells, because wild-type cells become 90 to 97% unbudded under these conditions (17). However, this allele clearly suppressed the phenotype of STE3DAF cells to a significant degree and did not cause supersensitivity in wild-type cells, suggesting that it does not encode a version of Ste4p that has an increased ability to transmit the pheromone response signal. The STE4SD13 allele was therefore chosen for further studies because of the high probability that it encodes a version of Ste4p that has a specific defect in receptor inhibition.
STE4SD-encoded mutations map to the N terminus and second WD repeat.
The crystal structure of mammalian G protein β-subunits has revealed that their seven WD domains fold into a symmetric structure in the form of a seven-bladed β-propeller (31, 35). The yeast Ste4p β-subunit also has seven WD domains that are expected to form a similar structure. Mutations in STE4 that suppress the STE3DAF phenotype were positioned on a diagram of a β-subunit that is based on the crystal structure (Fig. 2). The Q17L and Q21R mutations map to the extreme N terminus of Ste4p, which is composed of a 30-amino-acid extension that is present only in the yeast protein and thus is not represented in the crystal structure. The R162G mutation maps to the turn between the third and fourth strands of the β-sheet of the second blade, at the end of WD repeat 2. This residue is not in either of the two regions of the β-subunit that are in direct contact with the α-subunit (19, 35). The region of the β-subunit that makes the most extensive contacts with the α-subunit is the base of the β-propeller domain (Fig. 2, surface facing away from viewer). Mutations that alter amino acids on this surface, such as changes at residue 136, disrupt the interaction between the α- and β-subunits and result in constitutive signaling (36). The other region of contact between the two subunits is along the sides of the first and seventh blades of the β-propeller. A mutation that changes the amino acid at position 124, which is in the first blade, also disrupts the interaction between the α- and β-subunits and results in constitutive signaling (36). The finding that the changes encoded by the STE4SD13 allele are not in the regions of the β-subunit that contact the α-subunit suggests that the altered residues could constitute part of a binding site for another protein that interacts with the β-subunit.
FIG. 2.
Location of mutations on Gβ structure. The diagram is a schematic drawing representing the complete Ste4p sequence of 423 amino acids as a seven-bladed β-propeller structure. The product of the STE4SD13 allele contains the Q17L and Q21R mutations encoded by STE4SD2 and the R162G mutation encoded by STE4SD1, all of which are represented by filled diamonds in the diagram. Mutations that disrupt the interaction between the α- and β-subunits and result in constitutive signaling (36) are represented as grey circles. The amino acid sequence shows amino acids 1 to 162 of Ste4p.
STE4SD13 promotes sustained transcriptional activation in STE3DAF cells.
Expression of STE3 in MATa cells inhibits signaling during the late phase of the pheromone response (6). It was therefore of interest to determine whether the STE4SD13 allele specifically affects the late phase of the response or whether it affects signaling at other times. To assess the duration of signaling in cells containing STE4SD13, a time course of RNA accumulation from the pheromone-inducible FUS1 gene was performed. The strains used in this experiment contain deletions of the MFA1 and MFA2 genes, which encode a-factor, to eliminate any contribution to signaling due to autocrine stimulation of the a-factor receptor in MATa cells.
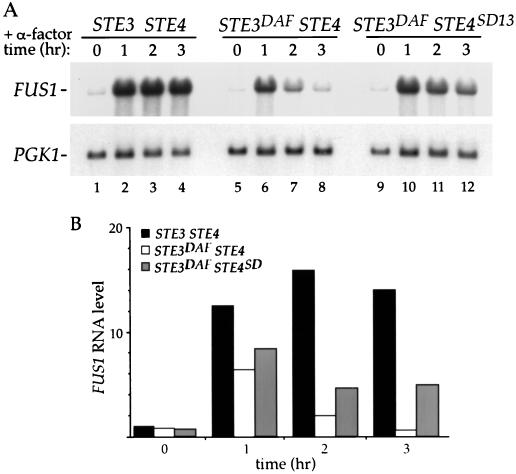
Wild-type cells treated with α-factor exhibited an increase in FUS1 RNA levels that remained high for 3 h (Fig. 3A, lanes 1 to 4). As observed previously (6), FUS1 RNA levels in STE3DAF cells increased the first hour and then decreased gradually to basal level at 3 h (Fig. 3A, lanes 5 to 8). In STE3DAF cells containing STE4SD13, the FUS1 RNA level was higher at late time points than it was in STE3DAF cells containing wild-type STE4 (Fig. 3A, lanes 8 and 12). Quantification of the results from duplicated experiments showed that there was an approximately sevenfold increase in FUS1 RNA at the 3-h time point in a STE3DAF strain containing STE4SD13 compared to the same strain containing wild-type STE4 (Fig. 3B, 3 h). Thus, although the FUS1 RNA level in STE3DAF STE4SD13 cells was not equal to the induced level seen in wild-type cells, it was significantly higher than that seen in STE3DAF STE4 cells. The STE4SD13 allele did not affect the basal level of FUS1 RNA in STE3DAF cells (Fig. 3B, 0 h), indicating that it does not cause constitutive activation of the pheromone response pathway. In addition, the STE4SD13 allele did not have a significant effect at 1 h of pheromone treatment in STE3DAF cells (Fig. 3B, 1 h), indicating that it does not cause a supersensitive response to pheromone. These results demonstrate that the STE4SD13 allele specifically suppresses the late inhibition of signaling caused by STE3DAF and does not alter other characteristics of the response.
FIG. 3.
Effect of STE4SD13 on pheromone-induced transcription. (A) The following strains were treated with α-factor (0.1 μM) for the indicated periods of time: a MATa STE3 ste4::HIS3 strain (AC17-7B) containing a wild-type STE4 plasmid (YCpLSTE4) (lanes 1 to 4) and a MATa STE3DAF ste4::HIS3 strain (AC17-2B) containing either a wild-type STE4 plasmid (YCpLSTE4) (lanes 5 to 8) or a plasmid containing the STE4SD13 allele (YCpLSD13) (lanes 9 to 12). RNA was isolated, transferred to nitrocellulose, and hybridized with a FUS1 probe. The blot was rehybridized with PGK1 to determine the amount of RNA per lane. (B) The data were quantified by PhosphorImager analysis, and the level of FUS1 RNA was normalized to the control PGK1 RNA level. Values from the STE3 STE4 strain are represented by black bars; values from the STE3DAF STE4 strain are represented by open bars; values from the STE3DAF STE4SD13 strain are represented by grey bars. The graph shows the average values from duplicate experiments.
STE4SD13 has no effect in wild-type cells.
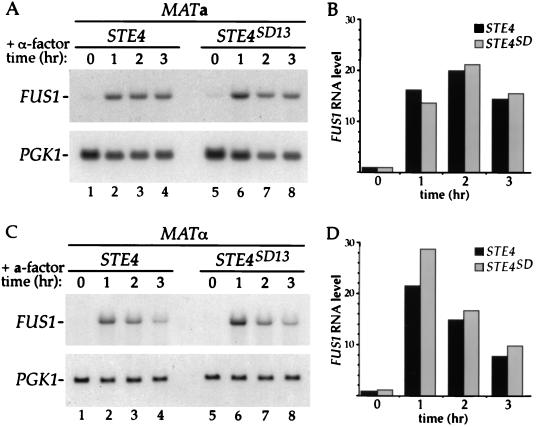
If the STE4SD mutations only affect the receptor inhibition function of STE4, then they should have no effect in MATa cells that do not express the a-factor receptor. This idea was tested by expressing STE4SD13 as the only copy of STE4 in a wild-type strain and assaying its ability to respond to pheromone. Accumulation of FUS1 RNA in cells treated with α-factor was very similar at all time points in MATa cells expressing either STE4SD13 (Fig. 4A, lanes 5 to 8) or STE4 (Fig. 4A, lanes 1 to 4). Quantification of these results showed that the levels of FUS1 RNA differed by less than 20% in MATa cells expressing STE4SD13 and STE4, even after 3 h of pheromone treatment (Fig. 4B). These experiments demonstrate that the STE4SD13 allele is capable of transmitting the pheromone signal in a manner indistinguishable from that of wild-type STE4 in MATa cells that do not express STE3.
FIG. 4.
Ability of STE4SD13 to signal in wild-type MATa and MATα cells. (A) A MATa STE3 ste4::HIS3 strain (AC17-7B) containing either a wild-type STE4 plasmid (YCpLSTE4) (lanes 1 to 4) or a plasmid containing the STE4SD13 allele (YCpLSD13) (lanes 5 to 8) was treated with α-factor (0.1 μM) for the indicated periods of time, and RNA was isolated. RNA blots were prepared and hybridized as described in the legend to Fig. 3. (B) The data from the experiment shown in panel A were quantified by PhosphorImager analysis, and the level of FUS1 RNA was normalized to the control PGK1 RNA level. Values from the STE4 strain are represented by black bars; values from the STE4SD13 strain are represented by grey bars. (C) A MATα STE3 ste4::HIS3 strain (AC18-9C) containing either a wild-type STE4 plasmid (YCpLSTE4) (lanes 1 to 4) or a plasmid containing the STE4SD13 allele (YCpLSD13) (lanes 5 to 8) was treated with a-factor (40 ng/ml) for the indicated periods of time, and RNA was isolated. RNA blots were prepared and hybridized as described in the legend to Fig. 3. (D) The data from the experiment shown in panel C were quantified by PhosphorImager analysis, and the level of FUS1 RNA was normalized to the control PGK1 RNA level. Values from the STE4 strain are represented by black bars; values from the STE4SD13 strain are represented by grey bars.
The STE4SD13 allele affects signaling in MATa cells that inappropriately express STE3 but does not affect signaling in wild-type MATa cells. It was therefore of interest to determine the effect of STE4SD13 in MATα cells, which normally express STE3. MATα cells expressing either STE4SD13 or STE4 were treated with a-factor for different lengths of time, and the level of FUS1 RNA was determined. Basal FUS1 RNA levels were very similar in MATα cells expressing either STE4 or STE4SD13 (Fig. 4C, lanes 1 and 5). In MATα STE4 cells treated with a-factor, FUS1 RNA was induced to a high level at 1 h of pheromone treatment and then gradually decreased for the next 2 h (Fig. 4C, lanes 1 to 4). A similar result was seen in MATα STE4SD13 cells (Fig. 4C, lanes 5 to 8). Quantification of these results showed that the levels of FUS1 RNA differed by less than 35% in MATα cells expressing STE4SD13 and STE4 (Fig. 4D). The decrease in signaling at late time points in wild-type cells may be due to degradation of a-factor by an extracellular protease (25). However, the observation that FUS1 RNA levels are essentially the same in MATα cells expressing either STE4 or STE4SD13 demonstrates that STE4SD13 does not affect pheromone signaling in the presence of the a-factor receptor when it is expressed in the appropriate cell type.
The STE4SD13 phenotype is independent of GPA1 and is specific to MATa cells.
Deletion of GPA1, which encodes the Gα subunit that functions in the pheromone response pathway, causes constitutive signaling due to free βγ-subunits. Therefore, cells containing a Δgpa1 mutation undergo cell cycle arrest and induction of FUS1 expression. The constitutive signaling seen in a MATa Δgpa1 strain is blocked by the STE3DAF allele, demonstrating that receptor inhibition is independent of GPA1 (7, 14). This result supports the idea that inappropriate STE3 expression inhibits a step that is downstream of α-subunit activation. If the STE4SD13 allele produces a β-subunit that is only partially inhibited by expression of STE3, then the STE4SD13 allele should increase signaling in a MATa Δgpa1 strain that expresses STE3. To test this idea, the level of FUS1 RNA was determined in the absence of pheromone in a MATa Δgpa1 STE3DAF strain expressing either STE4SD13 or wild-type STE4. A MATa Δgpa1 STE3DAF strain does not undergo cell cycle arrest due to inhibition of signaling by the STE3DAF allele; however, to prevent the possibility of cell cycle arrest under conditions where STE3DAF is suppressed by STE4SD13, this strain was constructed to contain a Δfar1 mutation. The FAR1 gene encodes a cyclin-dependent kinase inhibitor that is required for cell cycle arrest in response to pheromone (3, 27, 28).
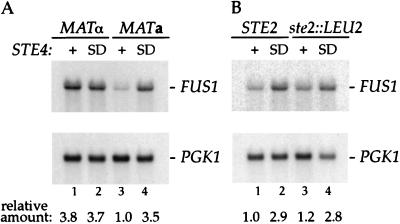
STE4SD13 caused an increase in the level of FUS1 RNA of about 3.5-fold compared to wild-type STE4 in a MATa Δgpa1 STE3DAF strain (Fig. 5A, lanes 3 and 4). This experiment rules out the possibility that the STE4SD13 allele causes an increase in signaling due to decreased binding of its encoded β-subunit to the Gpa1p α-subunit. This cannot be the case because STE4SD13 has an effect in the absence of Gpa1p. Expression of STE4SD13 was not able to confer cell cycle arrest on a MATa Δgpa1 STE3DAF FAR1 strain (17). This finding is consistent with results presented above indicating that suppression of STE3DAF by STE4SD13 is not complete.
FIG. 5.
Effect of STE4SD13 in cells lacking the Gα and α-factor receptor genes. (A) RNA was isolated from the following strains: a MATα STE3DAF ste4::HIS3 gpa1::TRP1 far1::URA3 strain (K39-23D.f) containing either a wild-type STE4 plasmid (YCpLSTE4) (lane 1) or a plasmid containing the STE4SD13 allele (YCpLSD13) (lane 2), and a MATa STE3DAF ste4::HIS3 gpa1::TRP1 far1::URA3 strain (K39-23B.f) containing either a wild-type STE4 plasmid (YCpLSTE4) (lane 3) or a plasmid containing the STE4SD13 allele (YCpLSD13) (lane 4). (B) RNA was isolated from the following strains: a MATa STE3DAF ste4::HIS3 gpa1::TRP1 strain (K39-23B) containing either a wild-type STE4 plasmid (YCpSTE4) (lane 1) or a plasmid containing the STE4SD13 allele (YCpSD13) (lane 2), and a MATa STEDAF ste4::HIS3 gpa1::TRP1 ste2::LEU2 strain (K39-23B.s2) containing either a wild-type STE4 plasmid (YCpSTE4) (lane 3) or a plasmid containing the STE4SD13 allele (YCpSD13) (lane 4). RNA blots were prepared and hybridized as described in the legend to Fig. 3. The data were quantified by PhosphorImager analysis, and the level of FUS1 RNA was normalized to the control PGK1 RNA level. The relative level of FUS1 RNA is shown below each lane.
The increase in signaling conferred by STE4SD13 in a Δgpa1 strain provides an opportunity to test whether the a-factor receptor can have an inhibitory effect in MATα cells, where it is normally expressed. Because deletion of GPA1 activates the pathway in a pheromone-independent manner, the pheromone receptors and their ligands are not required for generating the signal under these circumstances. Therefore, the negative role of STE3 can be assayed in the absence of its positive role in signal generation. If expression of STE3 causes equivalent inhibitory effects in MATa and MATα cells, then the STE4SD13 allele would be expected to increase signaling in Δgpa1 strains of both cell types. For this experiment, a MATα strain that contains the STE3DAF allele was constructed to ensure that expression of STE3 in this strain was comparable to its expression in a MATa STE3DAF strain. The strain was also constructed to contain a Δfar1 mutation to prevent the possibility of constitutive cell cycle arrest. The MATα Δgpa1 STE3DAF strain expressing wild-type STE4 contained about 3.8-fold more FUS1 RNA than the MATa Δgpa1 STE3DAF strain expressing wild-type STE4 (Fig. 5A, lanes 1 and 3). This result suggests that STE3DAF inhibits signaling to a greater degree in MATa cells than it does in MATα cells. Moreover, expression of either STE4SD13 or wild-type STE4 resulted in essentially identical levels of FUS1 RNA in a MATα Δgpa1 STE3DAF strain (Fig. 5A, lanes 1 and 2). Therefore, expression of STE3 in a MATα strain probably does not have an inhibitory effect on signaling, because all of the STE3DAF phenotypes in MATa cells are affected by STE4SD13. One interpretation of these results is that there is a cell type-specific factor in MATa cells that is required for the inhibitory function of STE3. The absence of this factor in MATα cells would explain why the normal expression of STE3 in these cells does not cause receptor inhibition.
One difference between MATa and MATα strains is that MATa strains express STE2, which encodes the α-factor receptor. It was therefore of interest to determine whether STE2 is the cell type-specific gene that allows STE3 to function as an inhibitor in MATa cells. To test this idea, FUS1 expression was assayed in MATa Δgpa1 STE3DAF cells that contained either a STE2 or a Δste2 allele. Quantification of FUS1 RNA isolated from these strains showed that expression of STE4SD13 caused an approximately threefold increase in FUS1 RNA abundance in both the STE2 and Δste2 strains (Fig. 5B, lanes 1 to 4). These results indicate that expression of the α-factor receptor has no effect on the inhibitory function of STE3. This finding is in agreement with a previous result showing that STE3DAF suppression of the cell cycle arrest phenotype of a Δgpa1 strain is unaffected by STE2 expression (14). Therefore, a different cell type-specific gene is probably responsible for the receptor inhibition function of STE3.
DISCUSSION
The STE4SD mutations were isolated based on their ability to restore pheromone-induced cell cycle arrest in MATa cells expressing STE3. The following evidence confirms the idea that the STE4SD13 allele is specific for the receptor inhibition function of STE4. First, expression of STE3DAF in MATa cells inhibits signaling only at late times during the response, and STE4SD13 causes an increase in FUS1 RNA levels only at late times after pheromone treatment in MATa STE3DAF cells. Second, STE4SD13 does not cause an increase in either the basal or induced level of FUS1 RNA in wild-type MATa or MATα cells. Third, the phenotype conferred by STE4SD13 is independent of GPA1, the Gα subunit gene. And, fourth, the effect of STE4SD13 is cell type specific. These findings rule out the possibility that the Ste4p protein encoded by STE4SD13 increases signaling by a mechanism that is unrelated to receptor inhibition, such as that it has an increased affinity for a downstream activator of the signaling pathway. Isolation of an allele of STE4 that is specific for suppression of receptor inhibition confirms that this phenomenon is an active cellular process that plays a physiological role in some aspect of the yeast life cycle.
G protein βγ-subunits have been shown to interact with a wide variety of other proteins (4). Structural and functional studies of these interactions demonstrate that the binding surfaces for α-subunits and downstream effectors are partially overlapping, suggesting that these interactions are mutually exclusive. The Ste4p β-subunit has been shown to interact with the Gpa1 α-subunit (36) and with the potential downstream effectors Ste20p (21) and Ste5p (15, 37). The binding surfaces on Ste4p that mediate these interactions are probably not altered by the mutations present in STE4SD13 because the protein encoded by this allele behaves identically to wild-type Ste4p when it is expressed in wild-type cells. It is therefore likely that the mutations present in STE4SD13 identify a binding surface for a novel β-subunit binding partner. Two of the mutations in STE4SD13 change residues in the N-terminal extension of Ste4p that is not present in the mammalian β-subunits that have been crystalized; the other mutation changes an amino acid that faces outward at the turn between the third and fourth strands of the second blade of the propeller. Because the structure of the Ste4p N terminus is unknown, the two regions of Ste4p identified by the STE4SD13 mutations could be quite close together, thus forming a unique binding surface for a protein that has not yet been identified.
STE4 alleles that confer prolonged signaling resulting in a defect in adaptation to pheromone have been isolated by Li et al. (23). These alleles contain mutations in the first, second, and seventh WD repeats of Ste4p. The mutations present in the STE4 Adp− alleles have been proposed to identify a target site for a negative regulator other than the Gpa1p α-subunit. The STE4 Adp− alleles, like the STE4SD alleles described here, are thought to confer an increase in signaling at late times during the pheromone response. However, it appears that the STE4 Adp− and STE4SD alleles do not affect the same process, for the following reasons. First, whereas the effects of STE4 Adp− alleles are observed in wild-type MATa cells, the effects of the STE4SD13 allele are only observed in MATa cells containing a STE3DAF mutation. Second, whereas all of the STE4 Adp− alleles confer an increase in the basal and induced levels of signaling, resulting in supersensitivity to pheromone, the STE4SD13 allele confers normal basal and induced levels of signaling in wild-type cells. And finally, the effects of STE4 Adp− alleles are dependent on a process initiated by Gpa1p, but the effects of the STE4SD13 allele are independent of Gpa1p function.
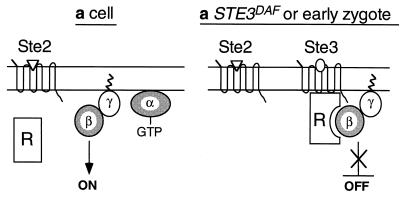
Some evidence suggests that β-subunits have the potential to bind directly to their associated receptors in the absence of an α-subunit. For example, fluorescence energy transfer experiments have demonstrated potential interactions between βγ-subunits and the β-adrenergic receptor (12) and between βγ-subunits and rhodopsin (29). In addition, one study has demonstrated direct photoaffinity labeling of a β-subunit by a peptide derived from the third cytoplasmic loop of the α-adrenergic receptor (34). Direct binding of the Ste4p β-subunit to the a-factor receptor may play a role in receptor inhibition; however, a model in which inhibition of signaling is caused by binding of Ste4p to Ste3p does not account for the observation that Ste3p only inhibits signaling in MATa cells. One explanation for the cell type specificity of receptor inhibition is that a component required for this process is expressed only in MATa cells, as shown in the model presented in Fig. 6 and described below.
FIG. 6.
Model for receptor inhibition. See text for details.
The results presented here are consistent with a model in which the activity of the βγ-subunit complex is inhibited by a MATa cell type-specific regulatory protein (factor R) that binds to or is activated by Ste3p, the a-factor receptor (Fig. 6). The observation that STE3DAF only affects signaling at late times after pheromone treatment could be due to the fact that factor R is induced by pheromone. Thus, factor R would not be present before pheromone treatment but would gradually increase in abundance during the time course of the response. In a wild-type MATa cell, the presence of factor R would have no effect because Ste3p is not expressed (Fig. 6, a cell). In a MATa STE3DAF cell, detection of Ste3p by factor R would produce a change in factor R that would cause it (or another factor activated by it) to block signaling by the βγ-subunits (Fig. 6, a STE3DAF). In the normal life cycle, factor R and Ste3p would be present in the same cell immediately after the fusion of a and α haploid cells that are in the process of mating (Fig. 6, early zygote) because factor R is an a-specific gene product and Ste3p is an α-specific gene product. Detection of Ste3p by factor R in early zygotes would result in inhibition of pheromone signaling by the same mechanism as that seen in MATa STE3DAF cells. Factor R could function by directly binding to the βγ complex and preventing the activation of downstream effectors or it could affect βγ complex activity by an indirect mechanism, such as altering its association with the plasma membrane. In this model, the effect of the STE4SD mutations would be to reduce the affinity of Ste4p for factor R, causing less Ste4p to interact with factor R. Thus, more Ste4p would be available to activate the pheromone response pathway, resulting in a higher signal. Preliminary studies on a newly identified a-specific gene suggest that its product is a good candidate for factor R (18).
The putative regulatory factor is expected to be specific to MATa cells because inhibition of signaling by STE3 occurs in a MATa Δgpa1 strain but does not occur in a MATα Δgpa1 strain. However, factor R cannot be the α-factor receptor, which is specific to MATa cells, because a null allele of STE2 does not affect inhibition of signaling by STE3. Previous studies by Bender and Sprague support the concept of a novel inhibitor of mating that is active in MATa cells that express STE3 (1). These studies showed that expression of a given combination of receptor and pheromone had different effects depending on the MAT allele of the cell in which they were expressed. The experiment was performed by expressing STE3 and MFα1 from mating type-independent promoters in either matα1 cells or MATa ste2 ste6 cells. Both of these strains are designed to express only α-factor and the a-factor receptor (in the MATa strain, the ste2 mutation eliminates expression of the α-factor receptor and the ste6 mutation prevents secretion of a-factor). However, the matα1 strain mated with 10-fold greater efficiency than the MATa strain did. The only difference between the two strains is that a-specific genes are not expressed in matα1 cells due to the presence of the Matα2p inhibitor. Therefore, this result supports the existence of an a-specific component that inhibits mating when STE3 is expressed. The a-specific component cannot be Ste2p and therefore is likely to be our proposed factor R.
MATa cells do not express STE3 at any time during their normal life cycle, so the function of this putative regulatory factor is probably not relevant to vegetative haploid growth. However, a potential physiological function of this regulatory factor is to inhibit signaling in mating cells that have recently undergone cell fusion. Fusion of cells would allow factor R to come into contact with Ste3p, which would block the signaling function of Ste4p. This process may function to promote recovery from mating and allow cell cycle progression to resume. Our attempts to demonstrate an effect of the STE4SD13 allele on recovery from mating have been hampered by an inability to obtain cultures that undergo synchronous mating. It is difficult to observe short-term effects in unsynchronized mating mixtures because recovery from mating is aided by the long-term process of transcriptional inhibition of haploid-specific genes by the Mata1p/Matα2p complex. Further experiments in which cell fusion and recovery can be precisely controlled will allow definitive testing of the model of receptor inhibition.
ACKNOWLEDGMENTS
We thank F. Cross, E. Elion, and I. Karpichev for providing plasmids used in this work and F. Naider for providing synthetic a-factor.
This project was supported by a Research Project Grant from the American Cancer Society (VM-182).
REFERENCES
- 1.Bender A, Sprague G F., Jr Pheromones and pheromone receptors are the primary determinants of mating specificity in the yeast Saccharomyces cerevisiae. Genetics. 1989;121:463–476. doi: 10.1093/genetics/121.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caplan A J, Cyr D M, Douglas M G. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 3.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 4.Clapham D E, Neer E J. G protein βγ subunits. Annu Rev Pharmacol Toxicol. 1997;37:167–203. doi: 10.1146/annurev.pharmtox.37.1.167. [DOI] [PubMed] [Google Scholar]
- 5.Clark K L, Dignard D, Thomas D Y, Whiteway M. Interactions among the subunits of the G protein involved in Saccharomyces cerevisiae mating. Mol Cell Biol. 1993;13:1–8. doi: 10.1128/mcb.13.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couve A, Hirsch J P. Loss of sustained Fus3p kinase activity and the G1 arrest response in cells expressing an inappropriate pheromone receptor. Mol Cell Biol. 1996;16:4478–4485. doi: 10.1128/mcb.16.8.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross F R. The DAF2-2 mutation, a dominant inhibitor of the STE4 step in the α-factor signalling pathway of Saccharomyces cerevisiae MATa cells. Genetics. 1990;126:301–308. doi: 10.1093/genetics/126.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross F R. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 9.Cross F R, Tinkelenberg A H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 10.Dietzel C, Kurjan J. The yeast SCG1 gene: a Gα-like protein implicated in the a- and α-factor response pathway. Cell. 1987;50:1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- 11.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 12.Heithier H, Fröhlich M, Dees C, Baumann M, Häring M, Gierschik P, Schiltz E, Vaz W L C, Hekman M, Helmreich E J M. Subunit interactions of GTP-binding proteins. Eur J Biochem. 1992;204:1169–1181. doi: 10.1111/j.1432-1033.1992.tb16744.x. [DOI] [PubMed] [Google Scholar]
- 13.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 14.Hirsch J P, Cross F R. The pheromone receptors inhibit the pheromone response pathway in Saccharomyces cerevisiae by a process that is independent of their associated Gα protein. Genetics. 1993;135:943–953. doi: 10.1093/genetics/135.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- 16.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J., A. Couve, and J. P. Hirsch. Unpublished data.
- 18.Kim, J., H. Zhong, A. Vershon, and J. P. Hirsch. Unpublished data.
- 19.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 20.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 21.Leeuw T, Wu C, Schrag J D, Whiteway M, Thomas D Y, Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- 22.Lehrach H, Diamond D, Wozney J M, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 23.Li E, Meldrum E, Stratton H F, Stone D E. Substitutions in the pheromone-responsive Gβ protein of Saccharomyces cerevisiae confer a defect in recovery from pheromone treatment. Genetics. 1998;148:947–961. doi: 10.1093/genetics/148.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H, Kunes S, Schatz P J, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 25.Marcus S, Xue C-B, Naider F, Becker J M. Degradation of a-factor by a Saccharomyces cerevisiae α-mating-type-specific endopeptidase: evidence for a role in recovery of cells from G1 arrest. Mol Cell Biol. 1991;11:1030–1039. doi: 10.1128/mcb.11.2.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCaffrey G, Clay F J, Kelsay K, Sprague G F., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 28.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 29.Phillips W J, Cerione R A. Rhodopsin/transducin interactions. I. Characterization of the binding of the transducin-βγ subunit complex to rhodopsin using fluorescence spectroscopy. J Biol Chem. 1992;267:17032–17039. [PubMed] [Google Scholar]
- 30.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Plainview, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Crystal structure of a GA protein βγ dimer at 2.1 Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 32.Sprague G F., Jr Combinatorial associations of regulatory proteins and the control of cell type in yeast. Adv Genet. 1990;27:33–63. doi: 10.1016/s0065-2660(08)60023-1. [DOI] [PubMed] [Google Scholar]
- 33.Sprague G F, Jr, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 34.Taylor J M, Jacob-Mosier G G, Lawton R G, VanDort M, Neubig R R. Receptor and membrane interaction sites on Gβ. J Biol Chem. 1996;271:3336–3339. doi: 10.1074/jbc.271.7.3336. [DOI] [PubMed] [Google Scholar]
- 35.Wall M A, Coleman D E, Lee E, Iñiguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. The structure of the G protein Giα1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 36.Whiteway M, Clark K L, Leberer E, Dignard D, Thomas D Y. Genetic identification of residues involved in association of α and β G-protein subunits. Mol Cell Biol. 1994;14:3223–3229. doi: 10.1128/mcb.14.5.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whiteway M S, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas D Y, Leberer E. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- 38.Yashar B, Irie K, Printen J A, Stevenson B J, Sprague G F, Jr, Matsumoto K, Errede B. Yeast MEK-dependent signal transduction: response thresholds and parameters affecting fidelity. Mol Cell Biol. 1995;15:6545–6553. doi: 10.1128/mcb.15.12.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]