Abstract
During sexual development, mycelial cells from most filamentous fungi differentiate into typical fruiting bodies. Here, we describe the isolation and characterization of the Sordaria macrospora developmental mutant per5, which exhibits a sterile phenotype with defects in fruiting body maturation. Cytological investigations revealed that the mutant strain forms only ascus precursors without any mature spores. Using an indexed cosmid library, we were able to complement the mutant to fertility by DNA-mediated transformation. A single cosmid clone, carrying a 3.5-kb region able to complement the mutant phenotype, has been identified. Sequencing of the 3.5-kb region revealed an open reading frame of 2.1 kb interrupted by a 66-bp intron. The predicted polypeptide (674 amino acids) shows significant homology to eukaryotic ATP citrate lyases (ACLs), with 62 to 65% amino acid identity, and the gene was named acl1. The molecular mass of the S. macrospora ACL1 polypeptide is 73 kDa, as was verified by Western blot analysis with a hemagglutinin (HA) epitope-tagged ACL1 polypeptide. Immunological in situ detection of the HA-tagged polypeptide demonstrated that ACL is located within the cytosol. Sequencing of the mutant acl1 gene revealed a 1-nucleotide transition within the coding region, resulting in an amino acid substitution within the predicted polypeptide. Further evidence that ACL1 is essential for fruiting body maturation comes from experiments in which truncated and mutated versions of the acl1 gene were used for transformation. None of these copies was able to reconstitute the fertile phenotype in transformed per5 recipient strains. ACLs are usually involved in the formation of cytosolic acetyl coenzyme A (acetyl-CoA), which is used for the biosynthesis of fatty acids and sterols. Protein extracts from the mutant strain showed a drastic reduction in enzymatic activity compared to values obtained from the wild-type strain. Investigation of the time course of ACL expression suggests that ACL is specifically induced at the beginning of the sexual cycle and produces acetyl-CoA, which most probably is a prerequisite for fruiting body formation during later stages of sexual development. We discuss the contribution of ACL activity to the life cycle of S. macrospora.
The fruiting body maturation of filamentous ascomycetes is an attractive model system to study multicellular development in eukaryotes. It involves the formation of the outer structures of the fruiting body but also development of mature ascospores within the fruiting body itself (for reviews, see references 31 and 47). Ascus development starts with the formation of female gametangia called ascogonia. The ascogenous cells are enveloped by sterile hyphae to form fruiting body precursors. Subsequent tissue differentiation gives rise to an outer pigmented peridial tissue, and following caryogamy, inner ascus initials embedded in sterile paraphyses are formed. Mature fruiting bodies from most ascomycetes harbor 200 to 400 asci, which after meiosis and postmeiotic divisions contain eight ascospores each. In many cases, ascospores are discharged through an apical pore (ostiole) at the neck of the fruiting body. Thus, fruiting body development requires the differentiation of the mycelia into several specialized tissues, and regulation of these morphological and physiological changes will require a number of different genes. However, so far only a limited set of data about the genetic control of fruiting body development is available.
Recently the mating type genes of several species have been characterized at the molecular level (for a review, see reference 7). They regulate different stages of sexual development and encode putative transcription factors that control the expression of developmental genes. Besides these, other genes involved in morphogenesis have been cloned, most of them from the closely related pyrenomycetous fungi Neurospora crassa and Podospora anserina. For example, the asd-1 gene from N. crassa encodes a putative rhamnogalacturonase which is essential for ascospore wall formation (32). Another example concerns the P. anserina car1 gene, which encodes a peroxisomal membrane protein that is essential for peroxisomal assembly (3). car1 mutants show an impaired caryogamy leading to a sterile phenotype. From these data the link between intact peroxisomes and fruiting body maturation becomes evident.
It has been demonstrated for a number of ascomycetes that several genes control not only sexual development but also asexual sporulation and vegetative growth. In N. crassa, macroconidia can serve as asexual spores or as male gametes. Among other factors, their formation is dependent on the nutritional state of the fungus and is controlled by a glucose transporter protein, the product of the rco-3 gene (26). In P. anserina, development of female gametangia and senescence are affected by the grisea gene (35). A failure in the expression of grisea leads to a prolonged life span and to defects in gametangium formation. Similarly, genes such as het-c in P. anserina and the mating type gene mt A-1 in N. crassa are responsible for both vegetative incompatibility and sexual reproduction (53, 54).
To isolate additional developmental genes from filamentous ascomycetes, we have used UV mutagenesis to generate Sordaria macrospora mutants with defects in fruiting body formation. This homothallic pyrenomycetous fungus is closely related to P. anserina and N. crassa, but in contrast to these heterothallic species, single strains of S. macrospora produce fruiting bodies (perithecia) without the presence of a mating partner. S. macrospora has already served as a model organism for the investigation of meiotic pairing and recombination (69), and several mutants with defects in perithecium development have long been reported (15). The development of molecular tools makes S. macrospora a suitable organism for studying fruiting body maturation. Transformation to hygromycin B resistance is feasible (67), and an indexed cosmid library, allowing gene isolation, has been established (42). S. macrospora mating type genes are among the genes which have already been cloned and characterized, providing some insight into fruiting body development of homothallic ascomycetes (43).
In this paper we report on the molecular investigation of the S. macrospora sterile mutant per5. We succeeded in restoring fertility by genomic complementation by using an indexed S. macrospora cosmid library. The complementing factor was found to be ATP citrate lyase (ACL), and to our knowledge, this is the first molecular analysis of a fungal ACL gene. ACL gene expression in S. macrospora is developmentally regulated, and we discuss the correlation between ACL expression and fruiting body development.
MATERIALS AND METHODS
Strains and growth conditions.
S. macrospora S 1957 and 3346 from our laboratory collection have a wild-type phenotype. The mutant per5 (strain S 10938) was isolated from wild-type strain 3346 after UV mutagenesis (27a). Strains were propagated on BMM fructification medium (14), and spore germination was achieved on BMM with 0.5% sodium acetate. For transformation and DNA isolation, S. macrospora was cultivated in CM medium [1% glucose, 0.2% tryptone, 0.2% yeast extract, 0.15% KH2PO4, 0.05% KCl, 0.05% MgSO4, 0.37% NH4Cl, and 10 mg each of ZnSO4, Fe(II)Cl2, and MnCl2 per liter].
Transformation of S. macrospora.
Formation of protoplasts was done by previously described procedures (42) with the following modifications. Inoculated Fernbach flasks were incubated for 2 days at 27°C. Protoplasts were kept in protoplast buffer (13 mM Na2HPO4, 45 mM KH2PO4, 600 mM KCl, pH 6.0) throughout the whole procedure. Transformation of S. macrospora was performed as described by Walz and Kück (67) with the following modifications. Four hours after transformation, plates were overlaid with hygromycin B-top agar to a final concentration of 110 U of hygromycin B/ml. Transformants appeared within 2 to 3 days after transformation. In order to transfer the transformants to fructification medium, plates were covered with filter paper and incubated for 12 h. The filter papers were then transferred to BMM plates with hygromycin B (110 U/ml) and incubated for another 12 h. Transformants were transferred from the hygromycin plates to BMM plates without hygromycin B by repeating the filter paper inoculation.
Preparation of RNA and genomic DNA and hybridization analysis.
Preparation of DNA was done as described by Pöggeler et al. (42). Total RNA was isolated from S. macrospora, using the method of Hoge et al. (19). Southern and Northern blotting were performed as described by Sambrook et al. (51). DNA gels were soaked in 0.1 M HCl prior to denaturation.
Construction of plasmids.
Cloning of S. macrospora DNA fragments, using vectors pBluescriptII/KS(+) (Stratagene), pANsCos1 (34), and pBCHygro (59), was done by standard techniques (51). Cosmids and plasmids used in this investigation are listed in Table 1. Construction of plasmid p85.1 was carried out as follows. A fragment of 422 nucleotides (nt) was amplified from wild-type DNA by PCR with oligonucleotide 1037 (5′ TTCGACAAGGGCCTAAGCC 3′) and the mutated oligonucleotide 1038 (5′ TTCATCCCAAGGATGACGG 3′) as primers. The fragment was cloned into plasmid p59.3 which had previously been digested with EcoRV and HindIII and was subsequently treated with Klenow polymerase to fill in the HindIII overlap. The correct orientation and sequence of the cloned fragment were checked by sequence analysis. Plasmid p94.1 was constructed by first hybridizing oligonucleotides 1049 (5′ ATACCCCTACGACGTCCCCGATTACGCCTTGCA 3′) and 1050 (5′ AGGCGTAATCGGGGACGTCGTAGGGGTATTGCA 3′), which encode the hemagglutinin (HA) epitope (58), and this double-stranded molecule was then ligated into the PstI site of the acl1 open reading frame (ORF) found in plasmid p58.1 (Table 1). The resulting plasmid was digested with ApaI, and the 3.5-kb DNA fragment containing the complete acl1 ORF was cloned into the vector pBCHygro. The construction of a cosmid library was described previously (42).
TABLE 1.
Cosmids and plasmids used for transformation or hybridization experiments
| Plasmid | Vector | Insert | Reference |
|---|---|---|---|
| B3 | pANsCos1 | S. macrospora genomic DNA | 42 |
| p27.7 | pANsCos1 | Deletion derivative of cosmid clone B3 containing the complete acl1 ORF | This work |
| p41.1 | pBCHygro | 3-kb SalI restriction fragment of cosmid clone B3 containing the N-terminal part of the acl1 ORF | This work |
| p49.4 | pANsCos1 | Deletion derivative of cosmid clone B3 containing none of the acl1 ORF | This work |
| p52.9 | pBCHygro | 1.8-kb EcoRV restriction fragment of cosmid clone B3 containing C-terminal parts of the acl1 ORF | This work |
| p58.1 | pBluescriptII/KS(+) | 4.8-kb EcoRV-HindIII fragment of cosmid clone B3 containing the complete acl1 ORF; p58.1 was used for construction of plasmids p59.3, p61.2, and p94.1 | This work |
| p59.3 | pBCHygro | 3.5-kb ApaI restriction fragment of cosmid clone B3 containing the complete acl1 ORF cloned into the ApaI and SmaI sites of pBCHygro | This work |
| p61.2 | pBCHygro | 3.5-kb ApaI restriction fragment of cosmid clone B3 containing the complete acl1 ORF cloned into the ApaI site of pBCHygro | This work |
| p85.1 | p59.3 | Nucleotide exchange from T to A at the EcoRV restriction site within the acl1 ORF | This work |
| p94.1 | pBCHygro | 3.5-kb ApaI restriction fragment of cosmid clone B3 containing the complete acl1 ORF with the HA epitope introduced into the PstI site of the ORF | This work |
DNA sequencing and sequence comparison.
The dideoxynucleotide chain termination procedure (52) was carried out with the T7 polymerase sequencing kit (Pharmacia, Freiburg, Germany). Sequencing products were separated by electrophoresis on 4% polyacrylamide gels as described by Lang and Burger (25). Wild-type cDNA and parts of the per5 mutant acl1 allele were sequenced by MWG-Biotech Customer Service (Ebersberg, Germany). Comparisons of nucleotide and amino acid sequences were performed with FASTA (36) and with programs from the HUSAR/Genius server, Heidelberg, Germany.
Preparation of crude extracts from S. macrospora.
Fernbach flasks containing 150 ml of BMM medium were inoculated with five or six 0.5-cm3 agar plugs taken from an S. macrospora BMM plate culture and incubated for 1 to 6 days at 27°C. The mycelium was filtered, washed with distilled water, and homogenized with 1 to 2 ml of extraction buffer (0.02 M Tris-HCl, 20% [wt/vol] glycerol, 2 mM MgCl2, 1 mM EDTA, 5 mM β-mercaptoethanol, and 1 mM dithiothreitol [pH 8.0 for the glucose-6-phosphate dehydrogenase test and pH 8.4 for the ACL test]). After centrifugation (15,000 × g, 10 min, 4°C) the protein content of the supernatant was determined as described by Bradford (5).
Enzyme activities. (i) Malate dehydrogenase-coupled ACL test.
The ACL (EC 4.1.3.8) activity was measured as described by Srere (62) with the following modifications. Four hundred microliters of 0.5 M Tris-HCl (pH 8.4), 100 μl of 2 mM acetyl coenzyme A (acetyl-CoA), 20 μl of 10 mM NADH, 50 μl of 0.2 M MgCl2, 0.7 μl of β-mercaptoethanol, 100 μl of 0.2 M sodium citrate, 10 μl of 1 M NaN3, 1 U of malate dehydrogenase (Boehringer, Mannheim, Germany), and S. macrospora crude extract containing 0.2 mg of protein were mixed in a reaction tube. Distilled water was added to a volume of 900 μl. The decrease of absorption at 340 nm was measured at 25°C for 5 min at 30-s intervals. The reaction was started by the addition of 100 μl of 0.2 M ATP, and measurements were taken for another 5 min at 30-s intervals. The net decrease of absorption was calculated from the difference between the values obtained before and after the addition of ATP. The average NADH oxidation before the addition of ATP was about 1 to 2 nmol per min per mg of protein. For each crude extract, at least three independent measurements were carried out. The average of the absorption decrease is directly proportional to the ACL activity.
(ii) CAT-coupled ACL test.
The chloramphenicol acetyltransferase (CAT)-coupled ACL test was done as described by Pentyala and Benjamin (37) with the following modifications. A 255-μl volume of reaction buffer (59 mM Tris-HCl [pH 8.4], 12 mM dithiothreitol, 24 mM MgCl2, 0.39 mM CoA, 3.5 mM sodium citrate, 21 μM [1,5-14C]citric acid [0.6 μCi per assay], 0.24 mM NADH, 1.4 U of malate dehydrogenase per ml, 70 U of CAT per ml, 1.4 mM chloramphenicol) was mixed with 15 μl of crude protein extract (2 μg/μl). The reaction was started by the addition of 30 μl of 25 mM ATP or of 30 μl of distilled water in control samples. Incubation was done for 5 min at 25°C, and then the reaction was stopped by heating to 65°C for 3 min. Twenty microliters of 0.1 M Tris-HCl (pH 8.7) and 900 μl of ice-cold ethyl acetate were added and mixed. After centrifugation (3 min, 12,000 × g), the upper phase (ethyl acetate) was removed and the lower phase again was extracted with 900 μl of ethyl acetate. The resulting upper phase was combined with the upper phase from the first extraction step, added to 10 ml of scintillation fluid (LSC Cocktail Hydroluma; Baker), and assayed for radioactivity. For each crude extract, at least three independent measurements were carried out. ACL activity was calculated from the differences between values obtained with and without addition of ATP. The average value without addition of ATP was 0.1 nmol per min per mg of protein.
(iii) Glucose-6-phosphate dehydrogenase test.
The glucose-6-phosphate dehydrogenase test was done as described by Scott (55) and Shepherd (57) with the following modifications. An 840-μl volume of reaction buffer (0.1 M Tris-HCl, 10 mM MgCl2, pH 8.0), 50 μl of 50 mM NADP, and 50 μl of 50 mM glucose-6-phosphate were added to a reaction tube. The reaction was started by the addition of S. macrospora crude extract containing 0.2 mg of protein. The increase of absorption at 340 nm was measured at 25°C for 5 min at 30-s intervals. For each crude extract, at least three independent measurements were done. The initial slopes of absorption increase are a measurement of the glucose-6-phosphate dehydrogenase activity.
SDS-PAGE and Western blot analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Laemmli (24). The immunological detection of the recombinant ACL1 polypeptide was done with a polyclonal anti-HA antibody (Santa Cruz Biotechnology), a peroxidase-coupled antirabbit antibody, and a chemiluminescent substrate (Boehringer) according to the manufacturers’ protocols.
Oligonucleotides.
Oligonucleotides were synthesized by the β-cyano-ethyl-phosphoamidite method (61) with an Applied Biosystems (Weiterstadt, Germany) 318A DNA synthesizer. High-pressure liquid chromatography purification was described previously (23).
PCR and RT-PCR amplification.
PCR and reverse transcription-PCR (RT-PCR) were performed as described by Kempken and Kück (20) with some modifications. A DNA template (10 to 100 ng) was amplified by using 40 ng of each primer and 1.25 U of Goldstar polymerase (Eurogentec, Cologne, Germany) in a total volume of 50 μl. The amplification reaction consisted of 40 cycles of 1 min at 92°C, 1 min at 50 to 55°C (depending on the primers used), and 1 to 1.5 min at 72°C (depending on the length of the amplification product). For RT-PCR, 5 μg of RNA was treated with 20 U of DNase (Boehringer) in an appropriate buffer for 1 h at 37°C. After phenol treatment and precipitation of the RNA, the pellet was redissolved in 20 μl of distilled water. The primer (20 ng of oligonucleotide 917 [5′ CATGATTGTAACCGCTCCG 3′] or 946 [5′ ATGGCAACACCCTCATAAACACC 3′]) was added to 10 μl of the RNA solution, and the sample was denatured for 10 min at 85°C. Reverse transcription was done with 80 U of avian myeloblastosis virus (AMV) reverse transcriptase (Boehringer) in the presence of 20 U of RNasin (Boehringer) at 45°C for 1 h in a final volume of 20 μl. After reverse transcription, 30 μl of distilled water was added, and 5 μl was used for PCR as described above. In order to detect any DNA contamination, reverse transcription was also done without AMV reverse transcriptase. Aliquots of these samples gave no PCR product, showing the complete degradation of DNA by the previous DNase treatment.
Primer extension.
Primer extension was carried out as described by Krug and Berger (22) and Kennell and Pring (21) with some modifications. Twenty nanograms of oligonucleotide 1035 (5′ GTTATGTGAATTGGTGACTCTCCC 3′) was 5′ labeled with [γ-32P]dATP and precipitated with 50 μg of S. macrospora RNA. The pellet was resuspended in distilled water and denatured at 85°C for 5 min, and reverse transcription was undertaken at 45°C for 1 h with 25 U of AMV reverse transcriptase (Boehringer) in the presence of 25 U of RNasin (Boehringer). After phenol treatment and precipitation, the pellet was dissolved in 5 μl of distilled water, and 3.6 μl of stop solution from the T7 polymerase sequencing kit (Pharmacia) was added. One to four microliters of each sample was separated on a polyacrylamide gel (see “DNA sequencing and sequence comparison” above); a sequencing reaction mixture containing just the oligonucleotide primer was used for reference.
Fluorescence microscopy.
For observations of nuclei, asci were fixed in carnoy fixative (49) and stained with DAPI (4′,6′-diamidino-2-phenylindole) (0.5 μg/ml). Immunological detection of ACL1::HA within S. macrospora hyphae was performed as described by Oakley et al. (33) with the following modifications. Strains were grown on cover slides for 48 h. For cell wall digestion, specimens were incubated for 60 min at 27°C in a solution containing 10 mg of Novozyme 234 (Novo Industrie AIS, Bagsvaerd, Denmark) per ml, 50% egg white, 25 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.7), 12.5 mM EGTA, and 2.5 mM MgSO4. Incubation in antibody solutions was performed in 50 mM Tris (pH 7.5)–150 mM NaCl. Polyclonal anti-HA antibody was used as the primary antibody; as a secondary antibody fluorescein isothiocyanate-labeled antirabbit antibody (Santa Cruz Biotechnology) was used. Mounting of specimens was performed in 50% glycerol–25 mM PIPES (pH 6.7)–12.5 mM EGTA–2.5 mM MgSO4. Observations were performed with a Zeiss Axiophot microscope with the appropriate Zeiss filter combinations for DAPI or fluorescein isothiocyanate. Photographs were taken with T-Max 400 (Kodak) or Provia 1600 (Fuji).
RESULTS
The developmental mutant per5 shows a defect in fruiting body maturation.
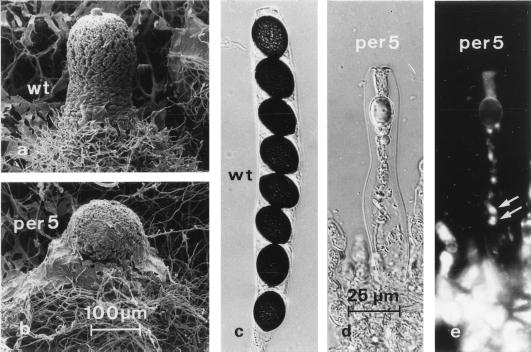
The sterile mutant per5, isolated from the wild-type strain after UV mutagenesis, displays normal vegetative growth. The growth rates of the mutant and wild-type strains are identical, and there seems to be no general impairment in essential vegetative functions (data not shown). When inoculated on fructification medium, the mutant strain shows a fivefold reduction in the number of perithecia compared to the wild-type strain. The fruiting body neck is shorter in mutant per5 than in the wild type (Fig. 1a and b). Most importantly, in comparison with those of the wild-type strain, the fruiting bodies of the mutant strain harbor only immature asci containing no ascospores (Fig. 1c to e). However, there seems to be no impairment in karyogamy or meiotic and postmeiotic divisions. As shown in Fig. 1e, we observed up to eight nuclei in immature asci after DAPI staining. In order to investigate whether a single gene is responsible for the mutant phenotype, per5 was crossed against the wild-type strain. A total of 119 tetrads were analyzed and showed a Mendelian segregation (4:4) of the mutant phenotype. These data indicate the involvement of a single gene locus in the mutant phenotype and lead to a calculated distance between the per5 locus and the centromere of 26 centimorgans.
FIG. 1.
Phenotypes of the S. macrospora wild-type strain (wt) and mutant per5. Strains were grown for 7 days at 27°C. (a and b) Scanning electron micrographs of perithecia from the wild-type strain (a) and mutant per5 (b). The magnifications in panels a and b are the same. (c and d) Differential interference contrast light micrographs of a wild-type ascus (c) and a mutant ascus (d). (e) Fluorescence micrograph of the mutant ascus in panel d stained with DAPI. Eight nuclei can be distinguished within the ascus. Two pairs of nuclei, not yet completely separated after postmeiotic mitosis, are marked by arrows. The magnifications in panels c, d, and e are the same.
Complementation of the sterile mutant per5 by using an indexed cosmid library.
Mutant per5 was complemented to fertility by transformation with an indexed cosmid library representing the S. macrospora genome. The cosmid library consists of 96 cosmid pools, each containing 48 individual cosmid clones (42). Seventy cosmid pools were used in transformation experiments, and a total of 5,100 transformants were screened for restoration of fertility. Fertile transformants, identified by their ability to eject mature ascospores, occurred after transformation with five of the cosmid pools. In order to prove genomic complementation, transformations were repeated with these five putative complementing pools. In addition, spores of the fertile transformants were genetically analyzed for linkage between fertility and hygromycin B resistance. These investigations showed that one of the five cosmid pools contained a complementing cosmid clone. Tetrad analysis demonstrated that fertile transformants obtained by transformation with the four other pools were due to suppressor mutations (data not shown).
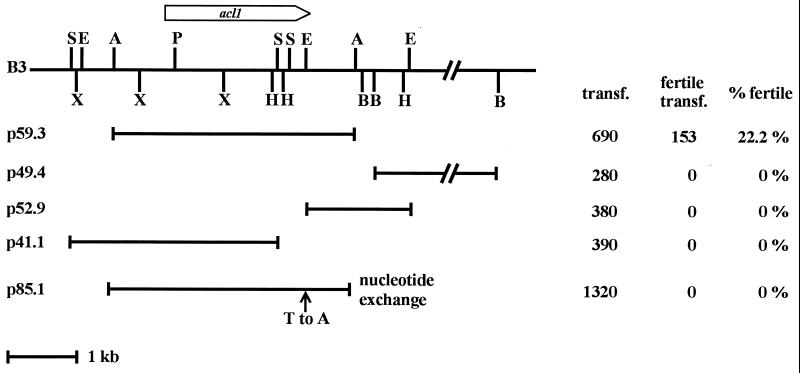
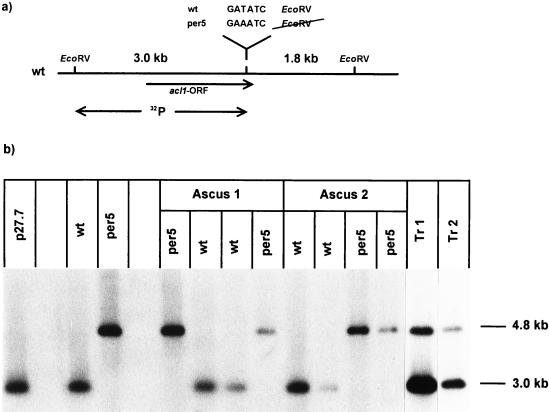
From the complementing cosmid pool, a single complementing cosmid clone was isolated and designated B3. Southern hybridization analysis of cosmid B3 and wild-type S. macrospora DNA confirmed that the cosmid clone B3 contains a native 41-kb fragment of genomic DNA showing no DNA rearrangements (data not shown). In order to identify the complementing region of clone B3, restriction fragments were cloned into vector pBCHygro (59) and used in transformation experiments. In addition, DNA fragments from cosmid B3 were eluted from agarose gels and cotransformed with plasmid pBCHygro as described by Timberlake et al. (64). As a result, we identified a complementing 3.5-kb ApaI DNA fragment which was cloned into the recombinant plasmid p59.3 as shown in Fig. 2.
FIG. 2.
Partial map of cosmid clone B3 together with derivatives used in transformation experiments. Cosmid clone B3 complements mutant per5 and contains the gene for ACL (acl1). The ORF of the acl1 gene and the direction of transcription are indicated by an arrow. Plasmids p59.3, p49.4, p52.9, p41.1, and p85.1 are derivatives of cosmid clone B3 (Table 1). The site of mutation within plasmid p85.1 is shown by an arrow. The values on the right give the total number of transformants (transf.) obtained with the corresponding plasmid and the number and corresponding percentage of fertile transformants. Abbreviations for restriction enzymes: A, ApaI; B, BamHI; E, EcoRV; H, HindIII; P, PstI; S, SalI; X, XhoI.
Sequence analysis of the complementing DNA fragment.
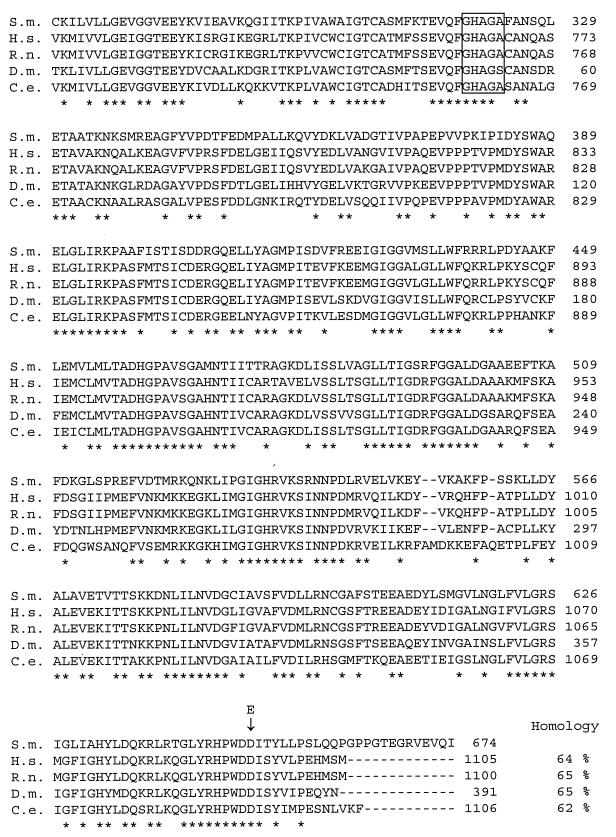
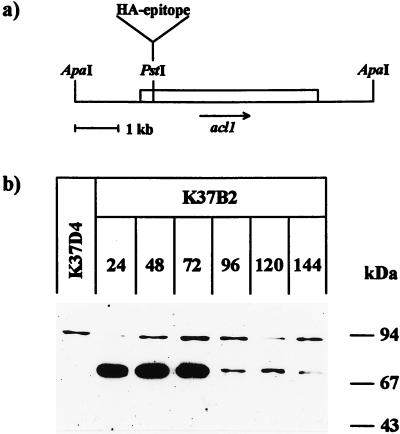
Sequencing of a total of 4.8 kb containing the 3.5-kb complementing fragment and adjacent regions revealed an ORF of 2.1 kb with a single intron of 66 nt (Fig. 3). The ORF encodes a predicted 674-amino acid-protein having significant homology with higher eukaryotic ACLs. As shown in Fig. 4, amino acid sequence homologies to animal ACLs vary between 62 and 65% over a length of about 600 amino acids. Translation is most probably initiated at the first ATG (position 1357 [Fig. 3]) of the ORF, and flanking sequences show the highest level of similarity with translation initiation sites from other S. macrospora genes (41). The putative polypeptide from S. macrospora has a calculated molecular mass of 73 kDa and thus has only about two-thirds of the molecular mass of animal ACL polypeptides (11, 12). The S. macrospora ACL1 polypeptide has homology with the C-terminal part of the corresponding animal polypeptides, which carries the enzyme’s proposed catalytic center, including the histidine residue that is autophosphorylated during the catalyzed reaction (Fig. 4). Southern hybridization analysis indicates that the acl1 gene is a single-copy gene (data not shown). The length of the ORF (2.1 kb) is consistent with data from Northern hybridizations showing a 2.7-kb transcript when the acl1 ORF is used as a probe (data not shown). Thus, the ACL1 polypeptide of S. macrospora seems to be shorter than the corresponding animal polypeptide, since there are no indications of trans-splicing events or any other acl1-containing regions elsewhere in the genome. In order to confirm that the ACL1 polypeptide has the expected size, the HA epitope of the human influenza virus was introduced into the single PstI restriction site of the acl1 gene (Fig. 5a). The resulting plasmid, p94.1, was able to complement mutant per5 in transformation experiments; thus, the HA epitope did not significantly influence the physiological activity of the protein. For immunological detection of the ACL1 polypeptide, crude protein extracts from transformants K37B2 and K37D4, carrying plasmid p94.1 and the nontagged plasmid p61.2, respectively, were separated by SDS-PAGE. Both transformants contain multiple copies of either plasmid p94.1 or p61.2. By using polyclonal anti-HA antibodies for Western blot analysis, a single specific 74-kDa band in protein extracts of K37B2 was detected (Fig. 5b). The calculated molecular mass of 74 kDa corresponds to the expected size of 75 kDa for the recombinant polypeptide carrying the HA epitope.
FIG. 3.
Nucleotide and derived amino acid sequences for the S. macrospora acl1 gene and its flanking regions. Intron sequences are indicated in lowercase, and characteristic intron sequences are underlined. The transcription initiation site is marked by an arrow; a putative CAAT box is marked by a line above the sequence. The catalytic center is indicated by a box around the sequence (11). The single nucleotide exchange present in mutant per5 at position 3372 (T to A) and the resulting amino acid exchange (aspartic to glutamic acid) are shown in boldface above and below the corresponding wild-type sequences, respectively. The nucleotide and deduced polypeptide sequence are numbered on the left, starting with nucleotide 626 according to the numbering of the complete sequence (4,847 bp) deposited in the EMBL sequence database under accession no. AJ224922.
FIG. 4.
Comparison of the C-terminal sequences of ACL polypeptides from different eukaryotes. S.m., S. macrospora (this work); H.s., human (accession no. U18197), R.n., rat (accession no. J05210); D.m., Drosophila melanogaster (accession no. U87317); C.e., Caenorhabditis elegans (accession no. U58727). Alignments were made by using the MULTalign program provided by the HUSAR/Genius computer software package. Dashes represent gaps introduced into the sequence to obtain the best consensus. Asterisks under the sequence represent amino acid residues conserved in all sequences. The catalytic center is indicated by a box around the sequences (11). The amino acid exchange (D to E) in mutant per5 is indicated by an arrow above the corresponding aspartic acid residue of the S. macrospora wild-type sequence. Homologies between the S. macrospora ACL1 sequence and those from other eukaryotes are given.
FIG. 5.
Immunological detection of the ACL1 polypeptide in crude protein extracts. (a) Introduction of the HA epitope into the acl1 gene. The insert of plasmid p94.1, which carries the complete ORF of the acl1 gene, is shown. The HA epitope was introduced into the PstI site within the ORF. (b) Detection of ACL1::HA in crude protein extracts of the transformed strain K37B2 during different stages of development. Forty micrograms of total protein per lane was separated by SDS–7% PAGE and blotted onto a nitrocellulose membrane. The HA-tagged protein was visualized by using polyclonal anti-HA antibodies, antirabbit secondary antibodies, and a chemiluminescence detection system. The time of growth in hours is given above each lane. As a control, a protein extract of strain K37D4 (48 h of growth) was used. This strain was transformed with plasmid p61.2, containing the acl1 gene without the HA epitope. Sizes of marker polypeptides are given on the right.
By using RT-PCR technology, cDNA fragments spanning a region of 2.5 kb (nt 1073 to 3499 [Fig. 3]) were amplified, with unfractionated RNA as a template. Sequencing of the cDNA fragments confirmed the presence of a single intron of 66 nt that interrupts the acl1 ORF (Fig. 3). The intron has consensus sequences typically found in introns from S. macrospora and other filamentous fungi (41). As indicated in Fig. 3, the transcription initiation site was mapped by primer extension and found to be 325 nt upstream of the putative translation start codon. Within the promoter region, a putative CAAT box (65) can be found at position −196 relative to the transcription initiation site (Fig. 3).
Analysis of the mutant acl1 gene.
In order to prove that the ACL is the complementing factor, two different strategies were used. The acl1 allele of mutant per5 was compared with the wild-type allele, while transformation experiments were carried out with clones containing either a truncated version of the acl1 gene or a mutated acl1 ORF (Fig. 2).
The Southern analysis shown in Fig. 6 revealed a change in the restriction fragment pattern within the complementing region between the wild-type and mutant per5. The mutant DNA lacks a wild-type EcoRV restriction site. Southern analysis with the wild-type 3-kb EcoRV fragment as a probe reveals a 3-kb band in the wild type and a 4.8-kb band in mutant per5. As expected, both bands were detected in complemented transformants. Analysis of the progeny derived from crosses of the wild type with mutant per5 proved that the strain-specific restriction pattern is transmitted in a Mendelian fashion (Fig. 6). The acl1 ORF and 5′ and 3′ flanking regions were amplified by PCR from mutant per5 DNA (nt 706 to 3499 [Fig. 3]). Sequencing of the amplified PCR fragments revealed only a single nucleotide exchange (T to A) leading to the change in the restriction pattern as mentioned above (Fig. 6). As a consequence, a codon for aspartic acid is changed into one for glutamic acid (Fig. 3). In order to verify that the single nucleotide exchange is responsible for the sterile phenotype of mutant per5, plasmid p59.3, carrying the wild-type acl1 gene, was subjected to an in vitro mutagenesis in which the T residue at nucleotide position 3372 was replaced by an A residue. The resulting plasmid, p85.1, was used in transformation experiments, and a total of 1,320 transformants showed no genomic complementation (Fig. 2), proving that the single nucleotide exchange within the ORF of the acl1 gene is responsible for the sterile phenotype. In addition, transformation with DNA fragments carrying either truncated or no copies of the acl1 ORF did not complement the mutant (p41.1, p49.4, and p52.9 in Fig. 2). In summary, the analysis of the mutant acl1 allele and data from the transformation experiments provide strong evidence that ACL complements the developmental defect in mutant per5.
FIG. 6.
Restriction analysis of the acl1 gene. (a) Map of the wild-type (wt) acl1 gene, giving the site of the nucleotide exchange present in mutant per5. The EcoRV fragment used as a hybridization probe is indicated by a double arrow. (b) Autoradiograph from a Southern hybridization experiment. Nucleic acids were isolated from the wild type, mutant per5, two complemented transformants (Tr 1 and Tr 2), and spore isolates. The latter were obtained from crosses between the wild type and mutant per5 (ascus 1 and ascus 2). The phenotypes of the spore isolates (wt or per5) are indicated above the lanes. EcoRV digests of genomic DNAs were separated in a 0.8% agarose gel. Hybridization was done with the radiolabeled 3-kb EcoRV fragment carrying parts of the acl1 gene ORF (panel a). Plasmid p27.7, carrying a fragment of the complementing cosmid clone, was used as a control. The sizes of the hybridization signals are given.
Physiological analysis of ACL expression in wild-type and mutant strains.
ACL has been shown to be involved in lipid metabolism in animals and some fungi (4, 12, 27, 38). It catalyzes the formation of acetyl-CoA and oxaloacetate from CoA and citrate, with concomitant hydrolysis of ATP. The enzyme activity can be assayed in crude extracts of various tissues (4, 38, 62).
We examined ACL activity in mycelial extracts from the S. macrospora wild-type strain and mutant per5, as well as in those from complemented transformants of the mutant strain, by using the malate dehydrogenase-coupled ACL test as described by Srere (62) (Table 2). The ACL activity of wild-type mycelia varies over a period of 6 days, being highest at 48 h after inoculation. Complemented transformants Tr1 and Tr2 have even higher activity than the wild type, due to multicopy integration of the complementing DNA fragment as evidenced by Southern analysis (data not shown). As shown in Table 2, no activity can be detected in crude extracts from mutant per5 by using the malate dehydrogenase-coupled test. In order to determine if there is residual activity within crude extracts from the mutant strain, the CAT-coupled test described by Pentyala and Benjamin (37) was used. When applied to protein extracts from animals, this assay is at least 10 times more sensitive than the malate dehydrogenase-coupled test (37). Using this method, we detected an activity of 10.00 ± 1.18 U/mg of protein in protein extracts from wild-type mycelia grown for 48 h, while only 0.39 ± 0.08 U/mg protein was obtained for mutant per5. Thus, mutant per5 exhibits about 4% of the wild-type activity. In order to demonstrate that the mutant does not show a general repression of enzyme activity but has a specific reduction of ACL activity, the enzyme glucose-6-phosphate dehydrogenase was assayed as a cytosolic marker. As shown in Table 2, wild-type and mutant strains display similar activities. The data confirm that a reduction of ACL activity in per5 leads to defects in fruiting body maturation.
TABLE 2.
Enzyme activity assays for ACL and glucose-6-phosphate dehydrogenase (G6PDH) of different strains
| Strain | Growth time (h) | Activitya (U/mg of protein)
|
|
|---|---|---|---|
| ACL | G6PDH | ||
| Wild type | 24 | 5.3 ± 2.0 | 196 ± 19 |
| 48 | 14.3 ± 3.0 | 273 ± 20 | |
| 72 | 7.4 ± 3.6 | 171 ± 10 | |
| 96 | 4.2 ± 3.7 | 236 ± 25 | |
| 120 | 0.7 ± 1.2 | 190 ± 5 | |
| 144 | 1.0 ± 0.9 | 235 ± 9 | |
| per5 | 24 | −1.3 ± 2.0 | 145 ± 14 |
| 48 | −0.8 ± 1.6 | 225 ± 5 | |
| 72 | −1.6 ± 0.6 | 193 ± 0 | |
| 96 | −0.5 ± 1.4 | 197 ± 18 | |
| 120 | −2.1 ± 1.5 | 187 ± 10 | |
| 144 | −1.6 ± 0.9 | 166 ± 10 | |
| Tr 1b | 48 | 58.5 ± 3.6 | 161 ± 46 |
| Tr 2b | 48 | 87.9 ± 8.2 | 201 ± 8 |
One unit represents the conversion of 1 nmol of each substrate per min. Results are means and standard deviations from at least three independent measurements. ACL activity was determined by the malate dehydrogenase-coupled test. Negative values for ACL activity indicate that the decrease of absorption due to NADH oxidation was higher before than after the addition of ATP.
Complemented transformant.
In order to determine the putative correlation between ACL activity and perithecium development, we investigated the time course of ACL activity by using the malate dehydrogenase-coupled assay (Table 2). Activity was monitored from 24 to 144 h after inoculation, covering the time from the beginning of vegetative growth to the ejection of mature spores from wild-type perithecia. As can be seen in Table 2, maximum activity in the wild-type strain occurred 48 h after the beginning of mycelial growth and was much lower before and after this time point. In parallel, mutant per5 was examined and showed no detectable ACL activity at any time during the investigation. The observed variations in wild-type ACL activity are confirmed by Western blot hybridizations with crude protein extracts of transformant K37B2, which carries the HA-tagged copy of the acl1 gene (Fig. 5). The amounts of protein detected by probing with an anti-HA antibody were highest during the first 72 h of development and were then reduced (Fig. 5). A similar time course was observed when the wild-type acl1 transcript was detected by Northern analysis (data not shown). Therefore, we suggest that acl1 expression is regulated mainly at the transcriptional level.
Under our experimental conditions, maturation of perithecia begins 72 to 96 h after Fernbach flasks are inoculated with mycelial plugs (see Materials and Methods), whereas maximum ACL expression appears after 48 h. Thus, the question arises as to whether a correlation between ACL activity and fruiting body development exists. In S. macrospora, the formation of perithecia starts at a critical mycelial density (29). This stage is reached when the surface of the plate or Fernbach flask is completely covered with mycelium. In the time course shown in Table 2, this is the case at 48 h after inoculation. Therefore, we suggest that maximum ACL expression is correlated with the transition from vegetative to sexual development. In order to confirm this assumption, Fernbach flasks were inoculated with just a single mycelial plug. As a consequence, the surface of the flask was covered at 72 h instead of 48 h after inoculation. ACL activity was monitored after 48, 72, and 96 h (data not shown). In this case, it was highest at 72 h after inoculation and thus seems to be correlated with the physiological changes at the beginning of sexual development.
Immunological detection of the ACL1 polypeptide in S. macrospora hyphae.
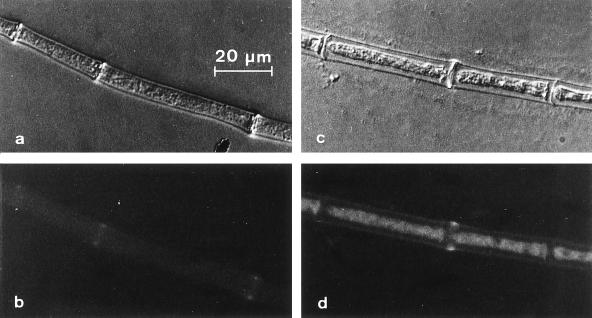
In order to determine the localization of the ACL1 polypeptide in S. macrospora, hyphae from strains K37B2 and K37D4 were processed for secondary immunofluorescence. Strain K37B2 carries an HA-tagged copy of the acl1 gene (Fig. 5), while strain K37D4, carrying the corresponding nontagged plasmid, was used as a control. As shown in Fig. 7c and d, staining of the cytoplasm was observed in strain K37B2, thus demonstrating the localization of the ACL1 polypeptide within the cytosol. As expected, in the control strain K37D4 only autofluorescence of the septa was observed (Fig. 7a and b). By this method, we cannot exclude the possibility that ACL also resides within other subcellular compartments. In general, however, our data are in accordance with subcellular fractionation experiments that detected ACL within the cytosol in some yeasts and Aspergillus niger (4, 38).
FIG. 7.
Immunological detection of the ACL1 polypeptide in S. macrospora hyphae. Hyphae from strain K37D4 (a and b) (transformed with plasmid p61.2, without the HA epitope) and strain K37B2 (c and d) (transformed with plasmid p94.1, carrying the HA epitope) were grown on cover slides and processed for immunofluorescence. (a and c) Differential interference contrast light micrographs; (b and d) fluorescence of ACL1::HA in the same hyphae. Magnifications are all the same.
DISCUSSION
The ACL gene is highly conserved.
In this paper we describe the isolation and characterization of per5, a sterile mutant of S. macrospora which was complemented to fertility by the gene encoding ACL. ACL produces acetyl-CoA, which in eukaryotes is used mainly in fatty acid and sterol biosynthesis. ACL is localized in the cytosol in animals and fungi (4, 38), whereas in plants it resides in the chloroplasts, which are the sites of fatty acid biosynthesis within photoautotrophic organisms (46). In S. macrospora, ACL also resides within the cytoplasm, as was demonstrated by immunofluorescence analysis (Fig. 7).
So far, the acl genes of several vertebrates have been sequenced, among them the genes from humans and rats (11, 12, 30). To our knowledge, we report the first molecular analysis of an acl gene from a lower eukaryote. The S. macrospora ACL1 polypeptide corresponds to the C-terminal part of the animal ACL1 polypeptides. These polypeptides are 62 to 65% identical over a length of 600 amino acids, including the proposed catalytic center (Fig. 4). Epitope tagging demonstrated that the 73-kDa S. macrospora ACL1 polypeptide is smaller than its 120-kDa animal counterparts (11). In animals and in the yeast Rhodotorula gracilis, the ACL protein is a homotetramer of four identical subunits (56, 60), whereas in the filamentous fungi Aspergillus nidulans and Penicillium spiculisporum, it seems to consist of two 55- and 70-kDa subunits forming a hexamer of about 380 kDa (1, 27). Thus, the 73-kDa ACL1 polypeptide encoded by the S. macrospora acl1 gene could be part of a multimeric protein, additional subunits of which might be encoded by other genes.
ACL is functional in S. macrospora, and its activity is detectable in crude protein extracts (Table 2). As can be concluded from the site of mutation in the mutant acl1 allele and by transformation with in vitro-mutagenized plasmid DNA, the highly conserved aspartic acid in the C-terminal part of the polypeptide is important for ACL activity (Fig. 4). Besides the histidine residue in the catalytic center, which is autophosphorylated during the catalyzed reaction, the human and rat acl genes contain three additional phosphorylation sites (39, 44). Phosphorylation of these sites is dependent on development or physiological state (2, 45), and enzymatic activity is influenced by phosphorylation (37). There are no sequences homologous to these three sites in the Sordaria polypeptide, indicating that regulation of ACL expression is not conserved among these organisms.
Actually, ACL seems to perform quite different functions in animals and fungi. In S. macrospora, it is important for sexual development, whereas its full activity is not required for vegetative growth, since mutant per5 displays wild-type vegetative growth. In animals, the highest levels of ACL expression are found in the liver. However, ACL inhibitors have no toxic effect (63), suggesting that ACL may be a putative target for hypolipidemic intervention in humans (17). In Saccharomyces cerevisiae and some other yeasts, no ACL has been detected (4), indicating that corresponding enzymatic activities are performed by other enzymes such as acetyl-CoA synthetases.
So far, no equivalent genes have been cloned from prokaryotes. However, ACL activity has been detected in some archaeal and bacterial species (66). In eukaryotes ACL is involved in lipid and sterol biosynthesis, whereas in prokaryotes it appears to be part of the reverse tricarboxylic acid cycle (66). This pathway is an alternative route for carbon fixation used in some archaea and bacteria (for a review, see reference 48). A comparison of prokaryotic and eukaryotic acl gene sequences should prove interesting, particularly with respect to any conserved amino acids essential for catalytic functions. In general, ACL seems to be an evolutionarily ancient enzyme which has achieved quite different physiological functions within diverse organisms.
ACL is essential for fruiting body development.
Analysis of the S. macrospora mutant per5 has demonstrated that although a drastic reduction of ACL activity does not impair vegetative growth, ACL is an essential requirement for fruiting body maturation. ACL produces acetyl-CoA and oxaloacetate, and so far no further function has been attributed to the protein. The acetyl-CoA produced by ACL is used mainly in fatty acid and sterol biogenesis. Fatty acids and sterols play important roles in many cellular processes, such as the generation of biomembranes, hormones, and secondary messengers. Besides these general functions, many developmental processes in different organisms are dependent on fatty acid metabolism. In plants, the formation of pollen grains and seeds is closely correlated with lipid production. Several genes of the fatty acid biosynthesis pathway of Brassica napus are tightly regulated in a spatiotemporal manner, e.g., those for acyl carrier proteins and stearoyl-acyl carrier protein desaturases (8, 40). In animals, enzymes for lipid biosynthesis and fatty acid beta-oxidation are both regulated during morphogenesis, as can be seen in developing rats (9, 13). In fungi, fatty acid biosynthesis has been well studied at the cellular level, but only in a few cases have more specialized functions been attributed to lipid metabolism (for a review, see reference 6). For example, in N. crassa fatty acids were shown to be involved in the circadian rhythm, and they are also needed for mitosis in Schizosaccharomyces pombe (28, 50). Lipids as well as sterol derivatives serve as growth factors or pheromones in some fungal species (for a review, see reference 10).
Investigation of mutant per5 indicates that fatty acids and sterols are essential for fruiting body development in S. macrospora. As the mutant displays a normal vegetative growth rate, it can be concluded that sufficient acetyl-CoA and lipids are produced for this process. This can be achieved either by the residual ACL activity present in mutant per5 or by enzymes other than ACL, such as acetyl-CoA synthetase. Although this work demonstrates a specific role for ACL, producing acetyl-CoA for fruiting body development, it is worth noting that the wild-type strain displayed ACL activity at every time point during development (Table 2), not just during perithecium formation. Nevertheless, the crucial role of ACL seems to be in fruiting body maturation, and it can be assumed that the sterility of mutant per5 is due to the fact that a certain amount of acetyl-CoA and its derivatives is a prerequisite for perithecium maturation. This is consistent with the finding that a partial restoration of the wild-type phenotype can be achieved by supplementation of growth media with fatty acids, such as oleate (unpublished results). Obviously, other enzymes producing acetyl-CoA cannot compensate for the reduced ACL activity during sexual development, indicating that ACL is a specific and probably the only relevant enzyme producing acetyl-CoA for fruiting body development. Our findings support the view that some housekeeping functions might be circumvented to a certain degree but are essential under special physiological conditions such as sexual reproduction.
The expression of acl1 is developmentally regulated, being highest during the transition from vegetative to sexual development. One of the reasons for this expression pattern might involve the demand for energy. Different biosynthetic routes for generating cytosolic acetyl-CoA influence the metabolic costs for biosynthesis of macromolecules (18). Thus, the importance of ACL for S. macrospora fruiting body development might be due to the fact that acetyl-CoA production has to meet certain energetic demands which cannot be fulfilled by other metabolic pathways. Another reason for the observed expression pattern might be that metabolites for the formation of fungal fruiting bodies are at least partially supplied by the vegetative mycelium. Therefore, the mycelium has to gain a certain competence before fruiting body formation is induced (for a review, see reference 68). As was recently shown for N. crassa, asci within perithecia contain far more oleate than perithecial wall tissues (16). It may be speculated that the lipid composition is the same in the closely related species N. crassa and S. macrospora. As oleate is a metabolic derivative of acetyl-CoA, this may explain why mutant per5 is able to form perithecial walls but no mature asci (Fig. 1).
In S. macrospora, ACL activity is highest at 48 h after inoculation, when mycelial density reaches a critical value and sexual development is induced (Table 2). We propose the existence of a yet-unidentified signal that regulates acl1 gene expression, which delivers acetyl-CoA that is required during perithecium formation. In general, our findings support the view that not only is the basic metabolism of cells regulated according to the developmental requirements, but different proteins are involved in producing the same metabolic intermediates at different developmental stages.
ACKNOWLEDGMENTS
We thank S. Schlewinski for performing the S. macrospora crosses, H. J. Rathke for the artwork, and T. Stützel for help with the scanning electron microscopy.
This work was supported by a grant from the Graduiertenförderung des Landes Nordrhein-Westfalen (NRW) (Germany) and by the Deutsche Forschungsgemeinschaft, Bonn-Bad Godesberg.
REFERENCES
- 1.Adams I P, Dack S, Dickinson F M, Midgley M, Ratledge C. ATP: citrate lyase from Aspergillus nidulans. Biochem Soc Trans. 1997;25:670. doi: 10.1042/bst025s670. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin W B, Pentyala S N, Woodgett J R, Hod Y, Marshak D. ATP citrate-lyase and glycogen synthase kinase-3 beta in 3T3-L1 cells during differentiation into adipocytes. Biochem J. 1994;300:477–482. doi: 10.1042/bj3000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berteaux-Lecellier V, Picard M, Thompson-Coffe C, Zickler D, Panvier-Adoutte A, Simonet J M. A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell. 1995;81:1043–1051. doi: 10.1016/s0092-8674(05)80009-1. [DOI] [PubMed] [Google Scholar]
- 4.Boulton C A, Ratledge C. Correlation of lipid accumulation in yeasts with possession of ATP:citrate lyase. J Gen Microbiol. 1981;127:169–176. [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Chopra A, Khuller G K. Lipid metabolism in fungi. Crit Rev Microbiol. 1984;11:209–271. doi: 10.3109/10408418409105904. [DOI] [PubMed] [Google Scholar]
- 7.Coppin E, Debuchy R, Arnaise S, Picard M. Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev. 1997;61:411–428. doi: 10.1128/mmbr.61.4.411-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Silva J, Robinson S J, Safford R. The isolation and functional characterisation of a B. napus acyl carrier protein 5′ flanking region involved in the regulation of seed storage lipid synthesis. Plant Mol Biol. 1992;18:1163–1172. doi: 10.1007/BF00047719. [DOI] [PubMed] [Google Scholar]
- 9.Djouadi F, Riveau B, Merlet-Benichou C, Bastin J. Tissue-specific regulation of medium-chain acyl-CoA dehydrogenase gene by thyroid hormones in the developing rat. Biochem J. 1997;324:289–294. doi: 10.1042/bj3240289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyer P S, Ingram D S, Johnstone K. The control of sexual morphogenesis in the ascomycotina. Biol Rev. 1992;67:421–458. [Google Scholar]
- 11.Elshourbagy N A, Near J C, Kmetz P J, Sathe G M, Southan C, Strickler J E, Gross M, Young J F, Wells T N C, Groot P H E. Rat ATP citrate-lyase. J Biol Chem. 1990;265:1430–1435. [PubMed] [Google Scholar]
- 12.Elshourbagy N A, Near J C, Kmetz P J, Wells T N C, Groot P H E, Saxty B A, Hughes S A, Franklin M, Gloger I S. Cloning and expression of a human ATP-citrate lyase cDNA. Eur J Biochem. 1992;204:491–499. doi: 10.1111/j.1432-1033.1992.tb16659.x. [DOI] [PubMed] [Google Scholar]
- 13.Eritani N, Fukuda H, Matsumura Y. Lipogenic enzyme gene expression in rat liver during development after birth. J Biochem. 1993;113:519–525. doi: 10.1093/oxfordjournals.jbchem.a124076. [DOI] [PubMed] [Google Scholar]
- 14.Esser K. Cryptogams—cyanobacteria, algae, fungi, lichens. London, United Kingdom: Cambridge University Press; 1982. [Google Scholar]
- 15.Esser K, Straub J. Genetische Untersuchungen an Sordaria macrospora Auersw., Kompensation und Induktion bei genbedingten Entwicklungsdefekten. Z Vererbungsl. 1958;89:729–746. [PubMed] [Google Scholar]
- 16.Goodrich-Tanrikulu M, Howe K, Stafford A, Nelson M A. Changes in fatty acid composition of Neurospora crassa accompany sexual development and ascospore germination. Microbiology. 1998;144:1713–1720. doi: 10.1099/00221287-144-7-1713. [DOI] [PubMed] [Google Scholar]
- 17.Gribble A D, Dolle R E, Shaw A, McNair D, Novelli R, Novelli C E, Slingsby B P, Shah V P, Tew D, Saxty B A, Allen M, Goot P H, Pearce N, Yates J. ATP-citrate lyase as a target for hypolipidemic intervention. Design and synthesis of 2-substituted butanedioic acids as novel, potent inhibitors of the enzyme. J Med Chem. 1996;39:3569–3584. doi: 10.1021/jm960167w. [DOI] [PubMed] [Google Scholar]
- 18.Henriksen C M, Christensen L H, Nielsen J, Villadsen J. Growth energetics and metabolic fluxes in continuous cultures of Penicillium chrysogenum. J Biotechnol. 1996;45:149–164. doi: 10.1016/0168-1656(95)00164-6. [DOI] [PubMed] [Google Scholar]
- 19.Hoge J H C, Springer J, Zantige B, Wessels J G H. Absence of differences in polysomal RNA from vegetative monokaryotic and dikaryotic cells of the fungus Schizophyllum commune. Exp Mycol. 1982;6:225–232. [Google Scholar]
- 20.Kempken F, Kück U. restless, an active Ac-like transposon from the fungus Tolypocladium inflatum: structure, expression, and alternative RNA splicing. Mol Cell Biol. 1996;16:6563–6572. doi: 10.1128/mcb.16.11.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennell J C, Pring D R. Initiation and processing of atp6, T-urf13 and ORF221 transcripts from mitochondria of T cytoplasm maize. Mol Gen Genet. 1989;216:16–24. [Google Scholar]
- 22.Krug M S, Berger S L. First strand cDNA synthesis primed with oligo (dT) Methods Enzymol. 1987;152:316–323. doi: 10.1016/0076-6879(87)52036-5. [DOI] [PubMed] [Google Scholar]
- 23.Kück U, Choquet Y, Schneider M, Dron M, Bennoun P. Structural and transcription analysis of two homologous genes for the P700 chlorophyll a-apoproteins in Chlamydomonas reinhardtii: evidence for in vivo trans-splicing. EMBO J. 1987;6:2185–2195. doi: 10.1002/j.1460-2075.1987.tb02489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lang B F, Burger G. A rapid, high resolution DNA sequencing gel system. Anal Biochem. 1990;188:176–180. doi: 10.1016/0003-2697(90)90548-n. [DOI] [PubMed] [Google Scholar]
- 26.Madi L, McBride S A, Bailey L A, Ebbole D J. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics. 1997;146:499–508. doi: 10.1093/genetics/146.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Måhlén A. Purification and some properties of ATP citrate lyase from Penicillium spiculisporum. Eur J Biochem. 1973;36:342–346. doi: 10.1111/j.1432-1033.1973.tb02918.x. [DOI] [PubMed] [Google Scholar]
- 27a.Masloff, S. Unpublished data.
- 28.Mattern D L. Unsaturated fatty acid isomers: effects on the circadian rhythm of a fatty-acid-deficient Neurospora crassa mutant. Arch Biochem Biophys. 1985;237:402–407. doi: 10.1016/0003-9861(85)90292-9. [DOI] [PubMed] [Google Scholar]
- 29.Molowitz R, Bahn M, Hock B. The control of fruiting body formation in the ascomycete Sordaria macrospora Auersw. by arginine and biotin: a two-factor analysis. Planta. 1976;128:143–148. doi: 10.1007/BF00390315. [DOI] [PubMed] [Google Scholar]
- 30.Moon Y A, Kim K S, Park S W, Kim Y S. Cloning and identification of exon-intron organization of the rat ATP-citrate lyase gene. Biochim Biophys Acta. 1996;1307:280–284. doi: 10.1016/0167-4781(96)00067-x. [DOI] [PubMed] [Google Scholar]
- 31.Nelson M A. Mating systems in ascomycetes: a romp in the sac. Trends Genet. 1996;12:69–74. doi: 10.1016/0168-9525(96)81403-x. [DOI] [PubMed] [Google Scholar]
- 32.Nelson M A, Merino S T, Metzenberg R L. A putative rhamnogalacturonase required for sexual development of Neurospora crassa. Genetics. 1997;146:531–540. doi: 10.1093/genetics/146.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oakley B R, Oakley C E, Yoon Y, Jung M K. γ-Tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- 34.Osiewacz H D. A versatile shuttle cosmid vector for the efficient construction of genomic libraries and for the cloning of fungal genes. Curr Genet. 1994;26:87–90. doi: 10.1007/BF00326309. [DOI] [PubMed] [Google Scholar]
- 35.Osiewacz H D, Nuber U. GRISEA, a putative copper-activated transcription factor from Podospora anserina involved in differentiation and senescence. Mol Gen Genet. 1996;252:115–124. doi: 10.1007/BF02173211. [DOI] [PubMed] [Google Scholar]
- 36.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 37.Pentyala S N, Benjamin W B. Effect of oxaloacetate and phosphorylation on ATP-citrate lyase activity. Biochemistry. 1995;34:10961–10969. doi: 10.1021/bi00035a001. [DOI] [PubMed] [Google Scholar]
- 38.Pfitzner A, Kubicek C P, Röhr M. Presence and regulation of ATP:citrate lyase from the citric acid producing fungus Aspergillus niger. Arch Microbiol. 1987;147:88–91. doi: 10.1007/BF00492910. [DOI] [PubMed] [Google Scholar]
- 39.Pierce M W, Palmer J L, Keutmann H T, Avruch J. ATP-citrate lyase. Structure of a tryptic peptide containing the phosphorylation site directed by glycagon and the cAMP-dependent protein kinase. J Biol Chem. 1981;256:8867–8870. [PubMed] [Google Scholar]
- 40.Piffanelli P, Ross J H E, Murphy D J. Intra- and extracellular lipid composition and associated gene expression patterns during pollen development in Brassica napus. Plant J. 1997;11:549–562. doi: 10.1046/j.1365-313x.1997.11030549.x. [DOI] [PubMed] [Google Scholar]
- 41.Pöggeler S. Sequence characteristics within nuclear genes from Sordaria macrospora. Fungal Genet Newsl. 1997;44:41–44. [Google Scholar]
- 42.Pöggeler S, Nowrousian M, Jacobsen S, Kück U. An efficient procedure to isolate fungal genes from an indexed cosmid library. J Microbiol Methods. 1997;29:49–61. [Google Scholar]
- 43.Pöggeler S, Risch S, Kück U, Osiewacz H D. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics. 1997;147:567–580. doi: 10.1093/genetics/147.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramakrishna S, D’Angelo G, Benjamin W B. Sequence of sites on ATP-citrate lyase and phosphatase inhibitor 2 phosphorylated by multifunctional protein kinase (a glycogen synthase kinase 3 like kinase) Biochemistry. 1990;29:7617–7624. doi: 10.1021/bi00485a011. [DOI] [PubMed] [Google Scholar]
- 45.Ramakrishna S, Murthy K S, Benjamin W B. Effect of insulin on ATP-citrate lyase phosphorylation: regulations of peptide A and peptide B phosphorylations. Biochemistry. 1989;28:856–860. doi: 10.1021/bi00428a067. [DOI] [PubMed] [Google Scholar]
- 46.Ratledge C, Bowater M D V, Taylor P N. Correlation of ATP/citrate lyase activity with lipid accumulation in developing seeds of Brassica napus L. Lipids. 1997;32:7–12. doi: 10.1007/s11745-997-0002-7. [DOI] [PubMed] [Google Scholar]
- 47.Read N D, Beckett A. Ascus and ascospore morphogenesis. Mycol Res. 1996;100:1281–1314. [Google Scholar]
- 48.Romano A H, Conway T. Evolution of carbohydrate metabolic pathways. Res Microbiol. 1996;147:448–455. doi: 10.1016/0923-2508(96)83998-2. [DOI] [PubMed] [Google Scholar]
- 49.Romeis B. Mikroskopische Technik. Munich, Germany: R Oldenbourg; 1968. [Google Scholar]
- 50.Saitoh S, Takahashi K, Nabeshima K, Yamashita Y, Nakaseko Y, Hirata Y, Yanagida M. Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J Cell Biol. 1996;134:949–961. doi: 10.1083/jcb.134.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5436–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saupe S, Descamps C, Turcq B, Bégueret J. Inactivation of the Podospora anserina vegetative incompatibility locus het-c, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proc Natl Acad Sci USA. 1994;91:5927–5931. doi: 10.1073/pnas.91.13.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saupe S, Stenberg L, Shiu K T, Griffiths A J F, Glass N L. The molecular nature of mutations in the mt A-1 gene of the Neurospora crassa A idiomorph and their relation to mating-type function. Mol Gen Genet. 1996;250:115–122. doi: 10.1007/BF02191831. [DOI] [PubMed] [Google Scholar]
- 55.Scott W A. Glucose-6-phosphate dehydrogenase from Neurospora crassa. Methods Enzymol. 1975;41:177–182. doi: 10.1016/s0076-6879(75)41043-6. [DOI] [PubMed] [Google Scholar]
- 56.Shashi K, Bachhawat A K, Joseph R. ATP:citrate lyase of Rhodotorula gracilis: purification and properties. Biochim Biophys Acta. 1990;1033:23–30. doi: 10.1016/0304-4165(90)90189-4. [DOI] [PubMed] [Google Scholar]
- 57.Shepherd M G. Glucose-6-phosphate dehydrogenase from Penicillium duponti. Methods Enzymol. 1975;41:201–205. doi: 10.1016/s0076-6879(75)41047-3. [DOI] [PubMed] [Google Scholar]
- 58.Shiio Y, Itoh M, Inoue J I. Epitope tagging. Methods Enzymol. 1995;254:497–502. doi: 10.1016/0076-6879(95)54035-0. [DOI] [PubMed] [Google Scholar]
- 59.Silar P. Two new easy to use vectors for transformations. Fungal Genet Newsl. 1995;42:73. [Google Scholar]
- 60.Singh M, Richards E G, Mukherjee A, Srere P A. Structure of ATP citrate lyase from rat liver. Physicochemical studies and proteolytic modification. J Biol Chem. 1976;251:5242–5250. [PubMed] [Google Scholar]
- 61.Sinha N D, Biernat J, McManus J, Köster H. Polymer support oligonucleotide synthesis. XVIII. Use of β-cyanoethyl-N,N-diacylamino-/N-morpholino-phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res. 1984;12:4539–4557. doi: 10.1093/nar/12.11.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srere P A. Citrate-cleavage enzyme. Methods Enzymol. 1962;5:641–644. [Google Scholar]
- 63.Sullivan A C, Triscari J, Hamilton J G, Miller O N, Wheatley V R. Effect of (−)-hydroxy-citrate upon the accumulation of lipid in the rat. I. Lipogenesis. Lipids. 1973;9:121–128. doi: 10.1007/BF02532136. [DOI] [PubMed] [Google Scholar]
- 64.Timberlake W E, Boylan M T, Cooley M B, Mirabito P M, O’Hara E B, Willett C. Rapid identification of mutation-complementing restriction fragments from Aspergillus nidulans cosmids. Exp Mycol. 1985;9:351–355. [Google Scholar]
- 65.Unkles S E. Gene organization in industrial filamentous fungi. In: Kinghorn J R, Turner G, editors. Applied molecular genetics of filamentous fungi. Glasgow, United Kingdom: Blackie Academic & Professional; 1992. pp. 28–53. [Google Scholar]
- 66.Wahlund T M, Tabita F R. The reductive tricarboxylic acid cycle of carbon dioxide assimilation: initial studies and purification of ATP-citrate lyase from the green sulfur bacterium Chlorobium tepidum. J Bacteriol. 1997;179:4859–4867. doi: 10.1128/jb.179.15.4859-4867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walz M, Kück U. Transformation of Sordaria macrospora to hygromycin B resistance: characterization of transformants by electrophoretic karyotyping and tetrad analysis. Curr Genet. 1995;29:88–95. doi: 10.1007/BF00313198. [DOI] [PubMed] [Google Scholar]
- 68.Wessels J G H. Fruiting in the higher fungi. Adv Microb Physiol. 1993;34:147–202. doi: 10.1016/s0065-2911(08)60029-6. [DOI] [PubMed] [Google Scholar]
- 69.Zickler D. Development of the synaptonemal complex and the “recombination nodules” during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora Auersw. Chromosoma. 1977;61:289–316. doi: 10.1007/BF00288615. [DOI] [PubMed] [Google Scholar]