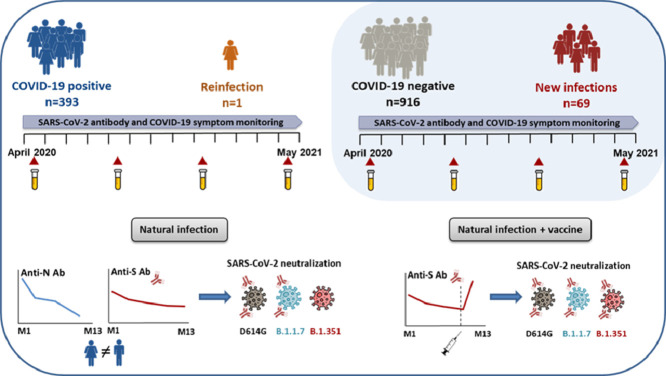
Abstract
Background
Assessment of the kinetics of SARS-CoV-2 antibodies is essential in predicting risk of reinfection and durability of vaccine protection.
Methods
This is a prospective, monocentric, longitudinal, cohort clinical study. Healthcare workers (HCW) from Strasbourg University Hospital were enrolled between April 6th and May 7th, 2020 and followed up to 422 days. Serial serum samples were tested for antibodies against the Receptor Binding Domain (RBD) of the spike protein and nucleocapsid protein (N) to characterize the kinetics of SARS-CoV-2 antibodies and the incidence of reinfection. Live-neutralization assays were performed for a subset of samples before and after vaccination to analyze sensitivity to SARS-CoV-2 variants.
Findings
A total of 4290 samples from 393 convalescent COVID-19 and 916 COVID-19 negative individuals were analyzed. In convalescent individuals, SARS-CoV-2 antibodies followed a triphasic kinetic model with half-lives at month (M) 11–13 of 283 days (95% CI 231–349) for anti-N and 725 days (95% CI 623–921) for anti-RBD IgG, which stabilized at a median of 1.54 log BAU/mL (95% CI 1.42–1.67). The incidence of SARS-CoV-2 infections was 12.22 and 0.40 per 100 person-years in COVID-19-negative and COVID-19-positive HCW, respectively, indicating a relative reduction in the incidence of SARS-CoV-2 reinfection of 96.7%. Live-virus neutralization assay revealed that after one year, variants D614G and B.1.1.7, but less so B.1.351, were sensitive to anti-RBD antibodies at 1.4 log BAU/mL, while IgG ≥ 2.0 log BAU/mL strongly neutralized all three variants. These latter anti-RBD IgG titers were reached by all vaccinated HCW regardless of pre-vaccination IgG levels and type of vaccine.
Interpretation
Our study demonstrates a long-term persistence of anti-RBD antibodies that may reduce risk of reinfection. By significantly increasing cross-neutralizing antibody titers, a single-dose vaccination strengthens protection against variants.
Fun1ding
None.
Keywords: SARS-CoV-2, COVID-19, Immunity, Neutralizing antibodies, Reinfection
Graphical abstract
Research in context.
Evidence before this study
Data on persistence and long-term efficacy of the immune response are crucial in understanding the overall evolution of the COVID-19 pandemic and post-pandemic dynamics, especially in the era of emerging SARS-CoV-2 variants. We searched PubMed using the terms “SARS-CoV-2”, “antibody”, “kinetics”, and “one year” for relevant articles published up until June 2, 2021. We found two articles that have analyzed anti-SARS-CoV-2 antibodies one year after COVID-19, including one longitudinal study focusing on anti-spike (S) antibodies only and one on a small cohort of 52 individuals. By using terms “SARS-CoV-2” and “reinfection”, only one article reported risk of reinfection in Italy up to one year after infection. However, the observation ended before new SARS-CoV-2 variants began to spread.
Added value of this study
Our prospective longitudinal study assesses over more than one year: (i) the anti-SARS-CoV-2 antibody persistence (both against anti-RBD and anti-N) after primary infection, (ii) the neutralizing capacity of these antibodies against live virus variants of concern (iii) the influence of host factors on antibody kinetics, (iv) the impact of vaccination on humoral responses against SARS-CoV-2 variants after COVID-19, and (v) long-term risk of reinfection during SARS-CoV-2 variants spread. We found that anti-N antibodies dramatically decreased whereas anti-RBD IgG persist for up to 13 months at a level that neutralizes infectious variants D614G, B.1.1.7 but less B. 1.351 and that they decline faster in men than in women over time. We also showed that vaccination of convalescent COVID-19 increases anti-RBD IgG to a level that strongly neutralizes all three variants regardless of pre-vaccine IgG levels and vaccine type. Moreover, this study offered a unique opportunity to evaluate risk of reinfection following previous COVID-19 with a longitudinal follow-up of convalescent and seronegative individuals during the same period, which encompasses the three COVID-19 waves experienced in France. Analysis of clinical and virological data revealed that the risk of reinfection was reduced by 96.7% over one year. Finally, this study revealed a strong correlation between the levels of anti-RBD IgG measured by a commercial quantitative test and the titers measured by live-virus neutralization.
Implications of all the available evidence
In conclusion, our study provides crucial information on the persistence of circulating antibodies against SARS-CoV-2 more than one year after COVID-19, and on the long-term risk of reinfection. By increasing the levels of cross-neutralizing antibodies, SARS-CoV-2 vaccination may strengthen protection, especially against variants harboring antibody escape mutations like B1.351. The strong correlation of antibody levels and their neutralizing capacity against variants may be of great help for the interpretation of serological results and for the future determination of a protective anti-RBD IgG level.
Alt-text: Unlabelled box
1. Introduction
Since the beginning of the pandemic, the hypothesis of waning humoral immunity in coronavirus disease 2019 (COVID-19) convalescent patients has raised many concerns about the reliability of population-based seroprevalence studies, and more critically about long-term antibody protection against reinfection and, by extension, the durability of vaccine protection. COVID-19 leads to the development of protective neutralizing antibodies in the vast majority of cases [1–4]. Several reports suggested a rapid decline of SARS-CoV-2 antibodies as early as 3 months after infection [3,5], while others reported persistence of antibody responses for up to eight months [4,6,7]. A recent rigorous study investigating T and B cell responses in convalescent COVID-19 patients reported that substantial immune memory is generated after COVID-19, and that 95% of subjects retained immune memory around 6 months after infection [8]. Furthermore, the presence of SARS-CoV-2 anti-spike (S) and anti-nucleocapsid (N) IgG antibodies were associated with a reduced risk of SARS-CoV-2 reinfection up to 7 months after initial infection [[9], [10]–11]. The recent emergence of SARS-CoV-2 variants with high transmissibility such as variant B.1.1.7, or decreased susceptibility to antibodies such as variant B.1.351, has raised the question of whether antibodies still protect against reinfection[12]. Data on persistence and long-term efficacy of the immune response are therefore of vital importance in understanding the overall evolution of the pandemic and post-pandemic dynamics, especially in the era of emerging variants [[13], [14], [15]–16].
Here, using validated serological assays [17–19] on a large cohort of healthcare workers (HCW) who have recovered from COVID-19, we described the dynamics of SARS-CoV-2 humoral response up to one year after infection, and analyzed the incidence of reinfection within this period. Secondly, we used the S-Fuse live-virus neutralization assay [20], to assess the sensitivity of infectious SARS-CoV-2 variants to HCW sera before and after vaccination, several months after primary infection.
2. Methods
2.1. Study design and participants
This is a prospective, monocentric, longitudinal, cohort clinical study (ClinicalTrials.gov Identifier: NCT04441684). A total of 1,496 HCW recruited among all Strasbourg University Hospital staff were initially screened by SARS-CoV-2 serology between 6th April and 7th May 2020 in our institution. All participants with a COVID-19 history, proven either by serology at screening or by a previous RT-PCR, were recruited as COVID-19 positive HCW. They were followed at month (M)3-6, M7-9 and/or M11-13 after infection. In parallel, participants displaying negative serology without a history of positive RT-PCR for SARS-CoV-2 were recruited as COVID-19 negative HCW to evaluate the incidence of infection, and were followed by the same visit schedule as the COVID-19 positive cohort. HCW without follow-up were excluded from the study. Participants completed a questionnaire at each visit in reference to sociodemographic characteristics, COVID-19 exposure, symptoms, virological findings and eventually vaccination. The investigators were not blinded to allocation during experiments and outcome assessment. However, different teams performed the clinical sampling, biological measurement and data analysis. Only the final assembly of the data revealed a global view of the results.
2.2. RT-PCR assay
RT-PCR for SARS-CoV-2 RNA detection was previously performed on nasopharyngeal swab samples at the time of diagnosis. All except six RT-PCR positive samples were analyzed in our laboratory with SARS-CoV-2 specific primers and probes targeting two regions on the viral RNA-dependent RNA polymerase (RdRp) gene (Institut Pasteur, Paris, France; WHO technical guidance). Ct values obtained in each sample were considered for statistical analyses.
2.3. Serological assays
Screening assays: All sera were initially screened for SARS-CoV-2 antibodies using the Biosynex® (COVID-19 BSS IgG/IgM) Lateral Flow Assay (LFA), and the EDITM Novel coronavirus COVID-19 IgG ELISA. These assays were the only ones available in sufficient quantities in our laboratory when we started the study in April 6th, 2020. The LFA detects separately IgM and IgG directed against the Receptor Binding Domain (RBD) of the SARS-CoV-2 S protein, with estimated overall sensitivity and specificity of 96 and 99% at 22 days since symptoms onset (DSO), respectively [18]. The ELISA detects anti-N IgG at 22 DSO [18], which, in our hands, displayed a sensitivity of 81% and a specificity of 96%. The results rely on a ratio of specimen absorbance reported to the cut off (S/CO) value defined by the manufacturer.
2.4. Confirmation assay
All sera that screened positive or were associated with a history of positive SARS-CoV-2 RT-PCR were analyzed retrospectively with the Abbott Architect SARS-CoV-2 IgG II Quant assay (Abbott, Sligo, Ireland), if possible given the remaining volume of serum. This allowed to confirm positive serological status and to measure the anti-RBD IgG titer. Results in Arbitrary Units (AU)/mL specific of this assay were converted into Binding Antibody Units (BAU)/ml adapted to the WHO standard for SARS-CoV-2 immunoglobulin by multiplying them by the factor 0.142. This assay is an automated chemiluminescence microparticle immunoassay (CMIA) that quantifies anti-RBD IgG, with 7.1 BAU/mL (50 AU/mL) as a positive cut-off and 5,680 BAU/mL (40,000 AU/mL) as an upper threshold of quantification (11,360 BAU/mL at 1:2 dilution). According to the manufacturer, this CMIA displays clinical sensitivity and specificity of 98.81 and 99.55% at 15 DSO, respectively. According to the manufacturer, antibody titers measured by this assay correlate, with a high probability (>95%), to neutralizing antibody titers assessed by plaque reduction assay on SARS-CoV-2 reference strain. This correlation was confirmed by previous studies [17,21].
2.5. S-Fuse live-virus neutralization assay
A neutralization assay was performed on sera collected at M11-13 from a panel of 28 COVID-19 positive HCW, including 13 who had received a single dose of COVID-19 vaccine. Live-virus neutralization was analyzed using the S-Fuse reporter cells, as previously reported [20]. Briefly, S-Fuse reporter cells correspond to U2OS cells (ATCC Cat# HTB-96, RRID: CVCL_0042) engineered to express ACE2 and either GFP1–10 or GFP11. When mixed, these cells produce GFP upon syncytia formation which occurs during productive infection with SARS-CoV-2. Neutralization of infectious D614G, B.1.1.7, and B.1.351 variants grown in Vero E6 cell lines (ATCC Cat# CRL-1586, RRID: CVCL_0574) was assessed for each serum using limiting dilutions. Infection was quantified by measuring the number of GFP+ syncytia 18 h after infection. The percentage of neutralization was calculated using the number of syncytia as the value with the following formula: . Neutralizing activity of each serum was expressed as the half-maximal inhibitory concentration (IC50).
2.6. Statistics
Chi-squared test, Kruskal-Wallis rank sum test and Fisher's exact test were conducted to identify any significant changes in categorical variables over time and between groups. Non-parametric Wilcoxon paired tests and Mann-Whitney tests were conducted to compare quantitative data over time or between groups, respectively. All tests were two-sided with an α level of 0.05. Missing data were excluded for analysis. Anti-RBD (log-transformed) and anti-N IgG titers over time were modelled, with the best fitting model assessed by an F Test, selecting the simplest model between simple linear regression, one-, two-, or three-phase decays of log data in the two settings. The half-life (t1/2) of each decay phase was also calculated.
To assess characteristics of patients with a faster/slower decay in anti-RBD and anti-N IgG titers, non-parametric tests were used for univariable analyses (Wilcoxon and Fisher's exact tests). Variables included in the univariable analysis were as follows: age, sex, BMI, blood group, Rhesus, SARS-CoV-2 RT-qPCR Ct, COVID-19 symptoms, DSO, hospitalization, anti-RBD or anti-N IgG titers at M3-6. Variables achieving a p value <0.17 in the univariable analysis were entered into a multivariable linear regression model (with a backward stepwise method based on the likelihood ratio test) [22]. Multivariable analyses were performed with R software version 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria). Factor effects in multivariable linear regression are given with a 95% CI. A quality control of the models was performed: the linearity assumption and the quality of the variance of the dependent variable over the full range of values of the independent variable were assessed with scatter plots and the assumption that the dependent variable is normally distributed was assessed with a normal probability plot (data not shown). All other data were analyzed and represented using Graphpad Prism version 9.0.0.
We used the Kaplan–Meier method to estimate the cumulative probability of SARS-CoV-2 infection or reinfection per group (COVID-19 negative or COVID-19 positive, respectively) and used the log-rank test to perform between-group comparisons. Time of exposure begins from the first negative serology for the COVID-19 negative HCW and from two months after initial SARS-CoV-2 infection (date of first symptoms, positive RT-PCR, or first positive serology) for the COVID-19 positive group, as described previously [11]. SARS-CoV-2 infection/reinfection was documented by RT-PCR or serological testing among COVID-19 negative and positive HCW, respectively. A sensitivity analysis was performed considering RT-PCR positive HCW only to verify that the reinfection rate was different than infection rate. Vaccinated individuals were censored at the time of the first vaccine dose. We also calculated the incidence of SARS-CoV-2 infection/reinfection per month, according to at-risk days during the follow-up of COVID-19 positive and COVID-19 negative individuals.
2.7. Ethics
This analysis was conducted on data from an on-going prospective, monocentric, longitudinal, cohort study enrolling healthcare workers from Strasbourg University Hospital (ClinicalTrials.gov Identifier: NCT04441684). All participants received written information about the study and were asked to give written informed consent for participation. The protocol was approved by the institutional review board of CPP Sud Méditerranée III (Ref: 2020.04.15 bis_ 20.04.10.66856).
2.8. Role of the funding source
The funders had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
3.1. Cohort characteristics
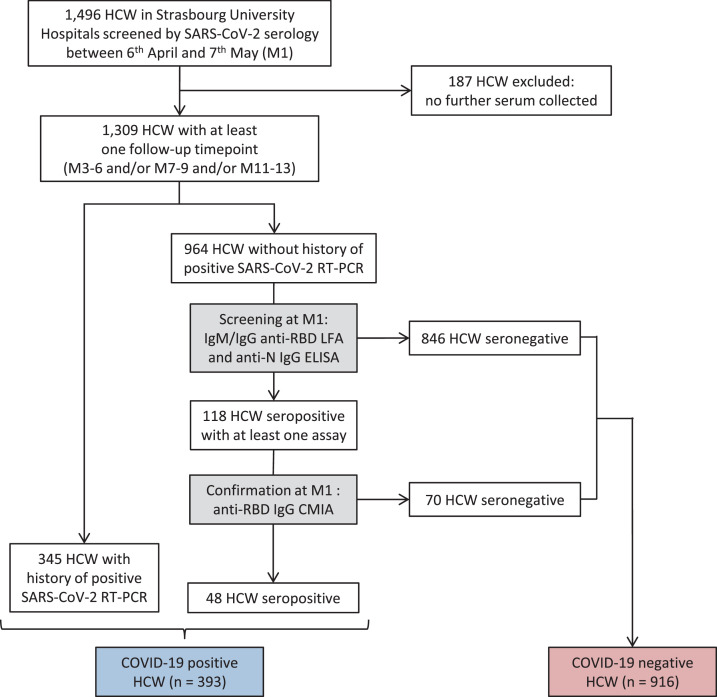
This study involved 4290 samples from 1309 HCW, including 393 convalescent COVID-19 (here called COVID-19 positive) HCW and 916 COVID-19 negative HCW (Fig. 1). The COVID-19 positive HCW included 345 with a history of positive SARS-CoV-2 RT-PCR and 48 with positive serology only. Both COVID-19 positive and COVID-19 negative cohorts included various professional groups (nurses, doctors, caregivers and administrative staff), both with a median age of 39 (Interquartile Range (IQR) 30–51 and 30–50, respectively), and a predominance of females (76.8 and 78.5%). In COVID-19 positive HCW, a history of contact with a COVID-19 case was reported in 66% of participants. COVID-19 consistent symptoms were reported by 383 participants (97.5%), including 367 (93.4%) and 16 (4.1%) with mild or moderate disease, respectively (Table 1). Ten participants were asymptomatic. No severe cases were reported. The first serum sample collected from the 383 symptomatic COVID-19 positive participants during the screening step corresponds to a median of 31 DSO (IQR: 24–38) and was considered as M1. Since the pandemic started about a month before our study, the first serum sample from the ten asymptomatic individuals was also arbitrarily considered as M1. A total of 383 were sampled at M3-6 (median: 107 DSO; IQR: 92–131), 346 at M7-9 (median: 215 DSO; IQR: 195–243) and lastly 233 at M11-13 (median: 373 DSO; IQR: 347–396), of which 93 were vaccinated against SARS-CoV-2 before M11-13 sampling. Only one asymptomatic reinfection was reported after nine months in this cohort. Conversely, among the 916 COVID-19 negative HCW, 69 (7.5%) reported a SARS-CoV-2 infection including 49 with symptoms (8 before M3-6, 29 before M7-9 and 32 before M11-13). This was confirmed by a positive RT-PCR test and by seroconversion in 62% and 100% of these cases, respectively.
Fig. 1.
Flow chart of survey recruitment and serum sampling among the healthcare workers (HCW) at Strasbourg University Hospital. First line serological screening was performed using two commercial assays: the Biosynex® (COVID-19 BSS IgG/IgM) Lateral Flow Assay (LFA) detecting anti-Receptor Binding Domain (RBD) antibodies and the EDITM Novel coronavirus COVID-19 IgG ELISA detecting the anti-nucleocapsid protein (N) IgG. A third assay, the Abbott SARS-CoV-2 IgG II Quant assay, measuring the anti-RBD IgG, was used to confirm seropositive samples. Serological testing on the first serum sample of each participant between 6 April and 7 May 2020 (M1) and on further sera collected at M3-6, M7-9, and M11-13 led to the establishment of two separate cohorts of COVID-19 positive or negative HCW, both with serological follow-up. Anti-RBD: anti-Receptor Binding Domain; Anti-N: anti-nucleocapsid protein; CMIA: chemiluminescent microparticle immunoassay; ELISA: enzyme-linked immunosorbent assay; HCW: healthcare workers; LFA: lateral flow assay; RT-PCR: real-time reverse transcriptase PCR.
Table 1.
Characteristics of the 393 COVID-19 positive healthcare workers.
| Total | Females | Males | p value | |
|---|---|---|---|---|
| Number of COVID-19 positive HCW | 393 | 302 | 91 | NA |
| Age (years), median (IQR) | 39.0 (29.6-50.5) | 40.0 (30.1-51.1) | 34.3 (28.6-44.6) | 0.0965 |
| BMI (kg/m²), median (IQR) | 23.6 (21.3-26.9) | 23.6 (21.0-27.8) | 24.0 (21.9-25.6) | 0.8157 |
| Blood group | ||||
| A, n (%) | 162 (41.2) | 125 (41.4) | 37 (40.7) | 0.2112 |
| B, n (%) | 33 (8.4) | 23 (7.6) | 10 (11.0) | |
| AB, n (%) | 16 (4.1) | 14 (4.6) | 2 (2.2) | |
| O, n (%) | 129 (32.8) | 108 (35.8) | 21 (23.1) | |
| Rhesus negative, n (%) | 60 (15.3) | 50 (16.6) | 10 (11.0) | 0.2448 |
| Unknown, n (%) | 53 (13.5) | 32 (10.6) | 21 (23.1) | 0.0045 |
| COVID-19 history | ||||
| Contact with COVID-19 case, n (%) | 259 (65.9) | 200 (66.2) | 59 (64.8) | 0.8021 |
| Previous positive SARS-CoV-2 RT-PCR, n (%) | 345 (87.8) | 263 (87.1) | 82 (90.1) | 0.5837 |
| COVID-19 symptoms, n (%) | 383 (97.4) | 294 (97.4) | 89 (97.8) | 1.0 |
| Known date of symptoms onset, n (%) | 378 (96.2) | 289 (95.7) | 89 (97.8) | 0.5359 |
| Hospitalization, n (%) | 16 (4.1) | 10 (3.3) | 6 (6.6) | 0.2211 |
| Serum collection | ||||
| HCW sampled at M1, n (%) | 393 (100) | 302 (100) | 91 (100) | 0.8042 |
| HCW sampled at M3-6, n (%) | 383 (97.5) | 294 (97.4) | 89 (97.8) | |
| HCW sampled at M7-9, n (%) | 346 (88.0) | 275 (91.1) | 71 (78.0) | |
| HCW sampled at M11-13, n (%) | 233 (59.3) | 181 (59.9) | 52 (57.1) | |
| Time from symptom onset to serum collection | ||||
| M1 (days), median (IQR; range) | 31 (24-38; 6-58) | 32 (24-38; 6-53) | 29 (24-37; 13-58) | 0.2087 |
| M3-6 (days), median (IQR; range) | 107 (92-131; 78-172) | 107.5 (92.3-131; 78-172) | 105 (90-130; 78-164) | 0.6583 |
| M7-9 (days), median (IQR; range) | 215 (195-243; 161-284) | 217 (196-246; 161-284) | 210 (194-237; 169-281) | 0.3388 |
| M11-13 (days), median (IQR; range) | 373 (347-396; 321-422) | 369 (346-396; 321-421) | 384 (348-396; 332-422) | 0.2053 |
| Vaccination | ||||
| Single-dose vaccination between M7-9 and M11-13, n (% of M11-13) | 59 (25.3) | 43 (23.8) | 16 (30.8) | 0.3655 |
| Double dose vaccination between M7-9 and M11-13, n (% of M11-13) | 34 (14.6) | 27 (14.9) | 7 (13.5) | 1.0 |
BMI: body mass index; HCW: Healthcare workers; IQR: Interquartile range; NA: not applicable; NS: not significant; RT-PCR: Real-time reverse transcriptase PCR. P values were calculated with Mann-Whitney, Chi-square and Fisher exact tests using the Graphpad Prism version 9.0.0 software.
3.2. Natural history of humoral response up to one year after COVID-19
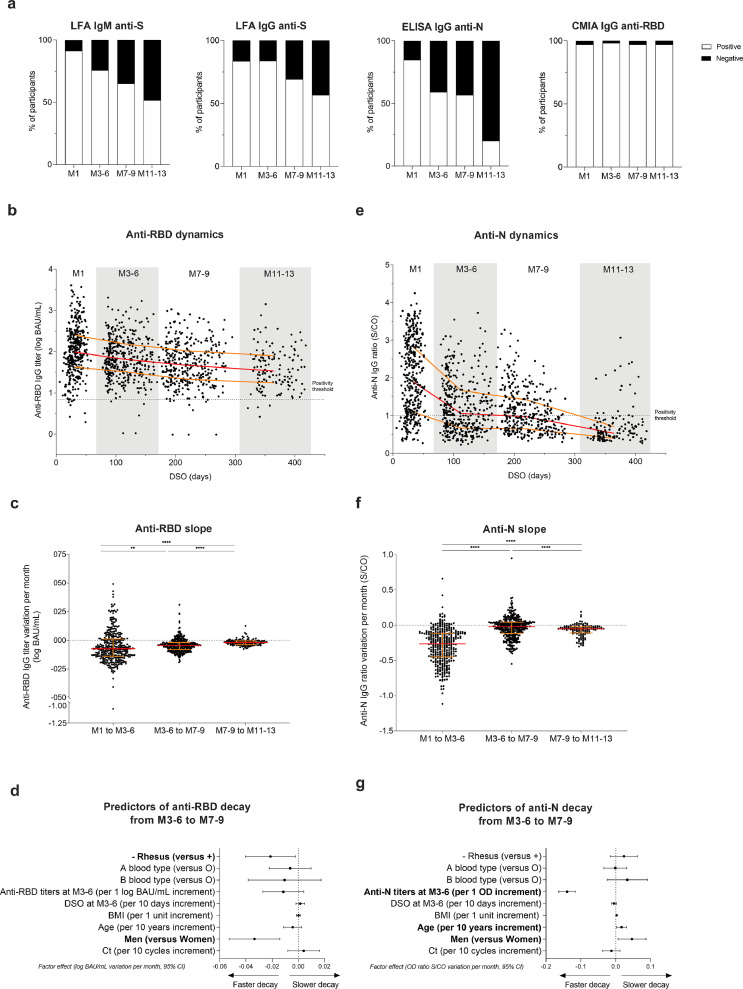
Using LFA, 91.3 and 83.7% of COVID-19 positive HCW were tested positive at M1 for anti-RBD IgM and IgG, respectively, versus 51.8% IgM and 56.8% IgG at M11-13, showing a significant decrease in the rates of LFA-detected antibodies one year after COVID-19 (both p < 0.0001). Using CMIA, 97.1% of individuals were tested positive both at M1 and M11-13 (p = 0.76). Positivity rates of anti-N IgG response as measured by ELISA significantly decreased from M1 (85.0%) to M11-13 (20.1%) (p < 0.0001).
Anti-RBD titers assessed by CMIA significantly decayed by 0.07, 0.04 and 0.02 log BAU/mL per month from M1 to M3-6, M3-6 to M7-9 and M7-9 to M11-13, respectively (all p < 0.01) (Fig. 2b, c). The estimated half-life (t1/2) of each phase was of 202 (95% CI 170–234), 306 (95% CI 292–368) and 725 (95% CI 623–921) days, respectively. At M11-13, the median titer of anti-RBD IgG was 1.54 log BAU/mL (95% CI 1.42–1.67), with 60.0% of participants showing IgG > 1.4 log BAU/mL.
Fig. 2.
Dynamics and determinants of SARS-CoV-2 humoral responses after COVID-19. (a) SARS-CoV-2 seropositive rate over time among participants (M1 (n = 393), M3-6 (n = 383), M7-9 (n = 346), M11-13 (n = 139)) according to the serological assays. (b) Dynamics of anti-RBD IgG titers expressed in log BAU/mL over time among HCW with known first date of symptoms (M1 (n = 369), M3-6 (n = 369), M7-9 (n = 332), M11-13 (n = 137). (c) Variation of anti-RBD IgG titers per month expressed in log BAU/mL between each time point (M1 to M3-6 (n = 374), M3-6 to M7-9 (n = 337), M7-9 to M11-13 (n = 128)). (d) Associations between anti-RBD IgG titers decay between M3-6 to M7-9 in log BAU/mL per month and demographic, biological, and virological data, and time of sampling at M3-6 expressed in days post symptoms onset (DSO) (n = 337). (e) Dynamics of anti-N IgG expressed in ratio optical density (OD) signal/Cut-Off (S/CO) over time among HCW with known first date of symptoms (M1 (n = 378), M3-6 (n = 369), M7-9 (n = 332), M11-13 (n = 137)). (f) Anti-N IgG ratios variation per month expressed in ratio OD Sample/CO between each timepoint (M1 to M3-6 (n = 383), M3-6 to M7-9 (n = 337), M7-9 to M11-13 (n = 128)). (g) Associations between anti-N IgG ratio decay between M3-6 to M7-9 in S/CO per month and demographic, biological, and virological data, and time of serum sampling at M3-6. (b, c, e, and f) Red lines represent median values and yellow lines the interquartile range for each population. *p value < 0.05; **p value < 0.01; ***p value < 0.001, ****p value <0.0001; calculated with non-parametric Wilcoxon paired tests using the Graphpad Prism version 9.0.0 software. (d, g) Data are depicted as factor effects in multivariable linear regression, with a 95% confidence interval given (95% CI). Multivariable analyses were performed with R software version 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Next, we investigated the effect of age, sex, body mass index (BMI), blood group, rhesus status, DSO, COVID-19 symptoms, hospitalization, and initial Ct values obtained by real-time reverse transcriptase PCR (RT-PCR) in nasal swabs, on CMIA anti-RBD IgG titers at M7-9 and on decay speed between M3-6 and M7-9 by univariable (Supplementary Table 1) and multivariable analyses. No significant difference in the absolute values of anti-RBD IgG titers was found in univariable analysis according to sex. However, antibody titers declined faster in men in univariable analysis between M3-6 and M7-9. By multivariable analysis (Fig. 2d), anti-RBD IgG titers also decayed faster in men than in women with an acceleration in this decrease of a median of -0.033 log BAU/mL per month (95% confidence interval (95%CI): -0.053 to -0.014; p = 0.0008). Another factor significantly associated with faster decay was the rhesus-negative (Rh-) status, affecting decay by a factor of -0.021 log BAU/mL per month (95% CI: - 0.002 to - 0.040; p = 0.0008). Notably, no significant effect of age, BMI, blood group, DSO or initial Ct values on the anti-RBD titer slope was observed (Fig. 2d).
As shown in Fig. 2e, triphasic kinetic dynamics of anti-N IgG ratio over time was observed, with an initial steep decay between M1 and M3-6 (median: -0.26 S/CO per month), followed by a slower decay up to M7-9 (-0.02) before a second drop up to M11-13 (-0.05; all p < 0.0001) (Fig. 2f). The t1/2 of each phase was therefore 58 (95% CI 51–66), 682 (95% CI 417–2500) and 283 (95% CI 231–349) days, respectively. In univariable analysis, higher antibody ratios were found in men at M7-9 compared to women (Supplementary Table 1). Moreover, a slower decay from M3-6 to M7-9 was revealed by multivariable analysis in men (0.046 S/CO per month; 95% CI 0.007–0.087; p = 0.02) and in older participants (0.017 per 10-year age; 95% CI 0.002–0.032; p = 0.03) (Fig. 2g). Thus, male participants displayed a faster decay of anti-RBD antibodies and, conversely, a slower decay of anti-N antibodies.
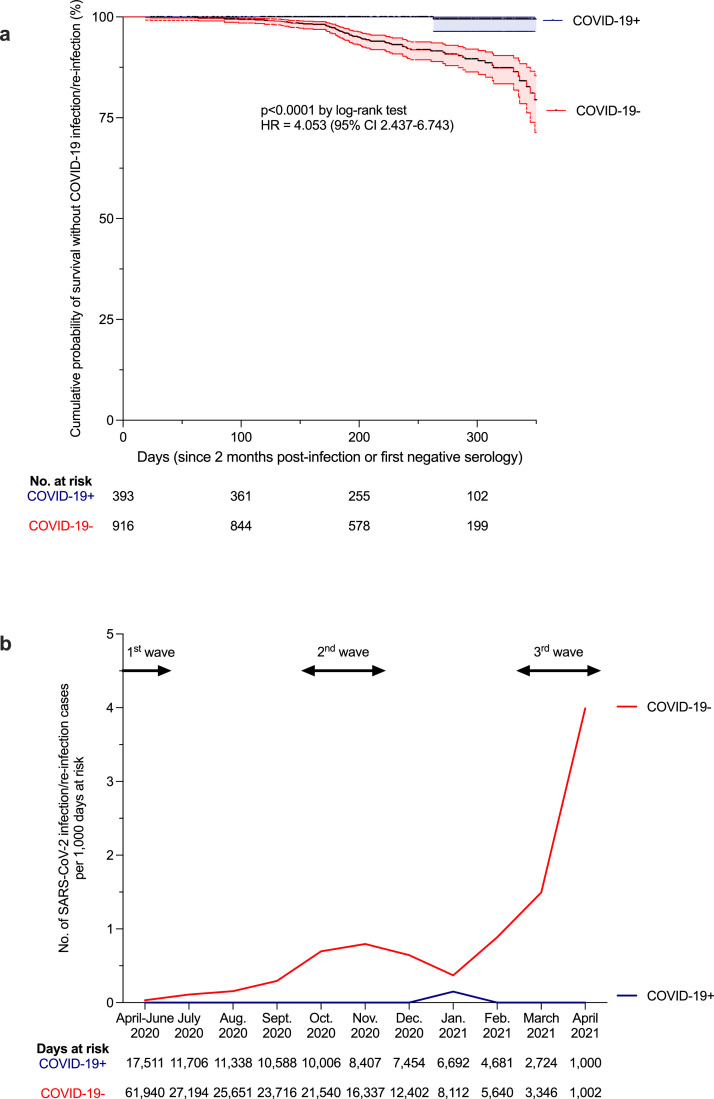
We then assessed the relative incidence of SARS-CoV-2 infection in COVID-19 positive and COVID-19 negative HCW during follow-up. Overall, 70 SARS-CoV-2 infections developed after enrollment: one RT-PCR proven reinfection in the COVID-19 positive group (incidence of 0.40 per 100 person-years) and 69 seroconversions in the COVID-19 negative group (incidence of 12.22 per 100 person-years), indicating a relative reduction in the incidence of SARS-CoV-2 reinfection in the previously infected group of 96.7% (hazard ratio (HR): 4.053 (95% CI: 2.437-6.743); p < 0.0001, Fig. 3a, b). To verify that the reinfection rate was different than infection rate, we also performed a sensitivity analysis focusing on RT-PCR tests only. Thus 43 infected participants in the COVID-19 negative group and one reinfected in the COVID-19 positive were taken into account, still providing a HR of 3.966 (95%CI: 2.099-7.494; p < 0.0001). The only case of reinfection occurred in a 23 year old female medical student. She first developed a symptomatic, mild COVID-19 in March 2020 with a high viral load, identified by nasopharyngeal swab (Ct=17), leading to an anti-RBD and anti-N IgG seroconversion (1.8 log BAU/mL and 1.0 OD S/CO after 96 DSO, respectively). The second episode in January 2021 was asymptomatic and revealed by a low viral load (Ct=34), detected six days after non-professional COVID-19 exposure. The reinfection case showed a positive anti-RBD IgM and a concomitant rebound of anti-RBD and anti-N IgG (+1.0 log BAU/mL and +0.7 S/Co, respectively) without vaccination, 22 days after a second positive RT-PCR. Since no systematic RT-PCR test was performed during follow-up, we examined whether other convalescent HCWs had an increase of anti-RBD and anti-N titers of at least +0.5 log BAU/mL and +0.5 S/Co, respectively. Only one HCW showed such increase of anti-N titer (+0.7 S/Co) but stable anti-RBD titer between M3-6 and M7-9. This HCW subsequently showed a decrease of anti-N titer (-1.1 S/Co) and an increase of anti-RBD titer (+0.6 log BAU/mL) between M7-9 and M11-13. This case was not considered here as a reinfection. Altogether, our findings indicate that although anti-SARS-CoV-2 antibody titers decline, the risk of reinfection within a year post-infection remains low.
Fig. 3.
The risk of reinfection after a first COVID-19 episode. (a) Kaplan-Meier estimates of the probability of SARS-CoV-2 infection protection. The cumulative probabilities of remaining free of SARS-CoV-2 infection among COVID-19 negative (COVID-19-) participants (red curve) and reinfection among former COVID-19 positive (COVID-19+) participants (blue curve) are shown at one year of follow-up (with 95% confidence interval (95%CI), dotted lines). The number of exposed participants is defined under the x axis. Comparison of survival curves was performed using log-rank test. A hazard ratio (HR) with 95% CI of reinfection in the COVID-19 positive group versus infection in the COVID-19 negative group was also calculated. (b) Calculated incidence of SARS-CoV-2 infection/reinfection per month, according to at-risk days during the follow-up of COVID-19+ (blue curve) and COVID-19- (red curve) individuals. Data are represented according to calendar months to allow comparison with national epidemic dynamics (epidemic waves depicted at the top of the graph with arrows). The number of at-risk days is shown under the x axis. (a, b) Exposure starts at the first negative serology for the COVID-19- group and at two months after initial SARS-CoV-2 infection for the COVID-19+ group. SARS-CoV-2 infection/reinfection was assessed either by RT-PCR or/and serology. To verify that the reinfection rate was different than infection rate, we performed a sensitivity analysis focusing on RT-PCR tests only. Thus 43 infected participants in the COVID-19 negative group and one reinfected in the COVID-19 positive were taken into account, still providing a HR of 3.966 (95%CI: 2.099-7.494; p < 0.0001 by log-rank test). Censoring criteria were: end of follow-up and anti-SARS-CoV-2 vaccination (at the time of the first vaccine dose). Comparison and p value were computed using the Graphpad Prism version 9.0.0 software. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Impact of SARS-CoV-2 vaccination on humoral response in COVID-19 positive HCW
To investigate how SARS-CoV-2 antibodies evolve after COVID-19 vaccination in COVID-19 positive HCW, serological results of the 93 COVID-19 positive HCW who received at least one dose of vaccine between M7-9 and M11-13 visits were compared to those of the 139 unvaccinated participants with a M11-13 follow-up. Among vaccinated-participants, 59 received a single-dose from 1 to 99 days before M11-13 sampling, including 27 HCW vaccinated with ChAdOx1 nCoV-19 vaccine (AstraZeneca), 4 with mRNA-1273 vaccine (Moderna) and 28 with BNT162b2 vaccine (Pfizer-BioNTech). The 34 other participants received two doses of BNT162b2 vaccine (n = 33) or mRNA-1273 vaccine (n = 1) and their M11-13 sera were collected from 3 to 94 days after the second dose of vaccine.
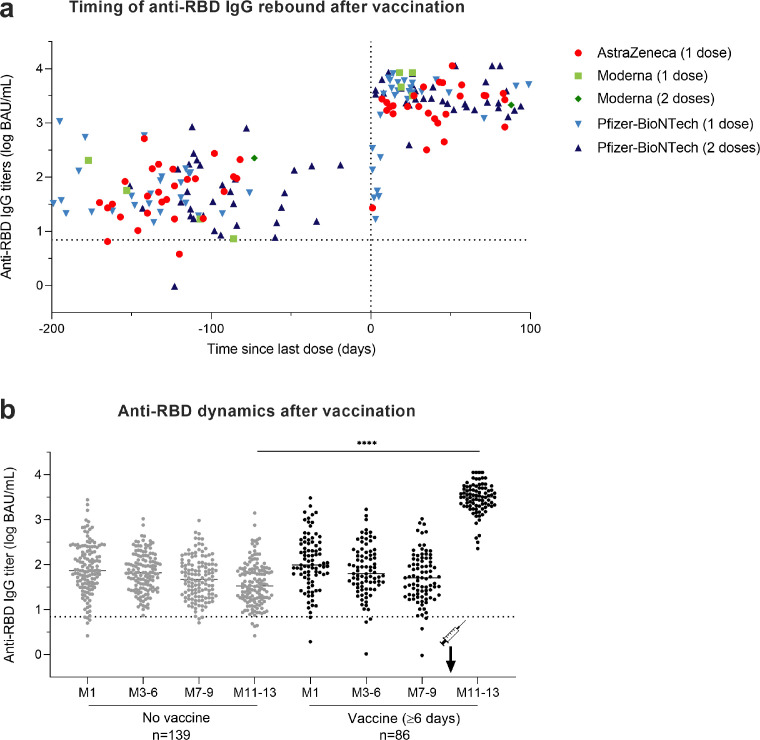
Five out of the seven participants sampled earlier than 6 days after a single dose vaccination still displayed anti-RBD antibody titers < 2.0 log BAU/mL at M11-13 (Fig. 4a). Conversely, a rebound of anti-RBD IgG titers was observed in all the 86 samples collected at least 6 days after vaccination with a median increase of 1.80 log BAU/mL between M7-9 and M11-13 (IQR, 1.38 to 2.17; p < 0.0001). Indeed, post-vaccination titers reached at least double the values measured at M1 post-infection for 98% of vaccinated HCW. Antibody titers > 3.0 log BAU/mL were found at M11-13 in 80 of these 86 vaccinated HCW (93.0%) (Fig. 4a, b).
Fig. 4.
Impact of SARS-CoV-2 vaccination on humoral response in COVID-19 positive HCW. (a) Timing of the rebound in anti-RBD IgG titers following vaccination among the 93 COVID-19 positive HCW who received at least one dose of vaccine against SARS-CoV-2 between M7-9 and M11-13 visits. The dotted vertical black line corresponds to the day of first injection (D0). Anti-RBD IgG titers among HCW vaccinated with one dose of AstraZeneca vaccine are depicted in red circles, with one or two doses of Moderna vaccine in light green squares and dark green diamonds, respectively, and with one or two doses of Pfizer vaccine in light blue upwards and dark blue downwards triangles, respectively. (b) Comparison of anti-RBD IgG titer dynamics over time between 139 unvaccinated HCW in grey dots and 86 HCW vaccinated for at least six days in black dots. *p value < 0.05; **p value < 0.01; ***p value < 0.001, ****p value <0.0001; calculated with non-parametric Wilcoxon paired tests and Mann-Whiney tests using the Graphpad Prism version 9.0.0 software. The dotted horizontal black line corresponds to the anti-RBD IgG titer positivity threshold. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Sensitivity of infectious SARS-CoV-2 variants to anti-S antibodies at M11-13
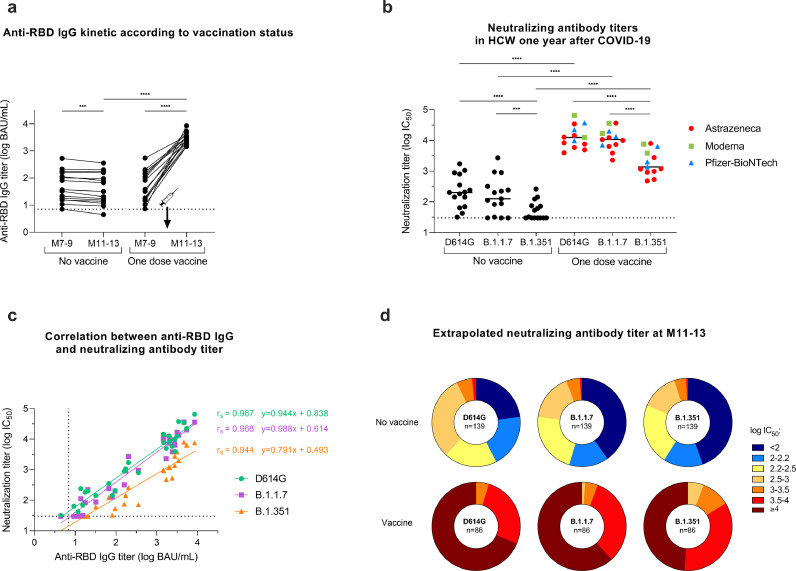
To assess whether SARS-CoV-2 variants are sensitive to anti-S antibodies that persist at M11-13 with or without prior vaccination, sera collected at M11-13 from 28 COVID-19 positive HCW (13 vaccinated and 15 unvaccinated) were analyzed with the S-Fuse live-virus neutralization assay [20] (Fig. 5a). The 13 vaccinated HCW had received a single dose, including eight with ChAdOx1 nCoV-19 vaccine (AstraZeneca), three with BNT162b2 vaccine (Pfizer-BioNTech) and two with mRNA-1273 vaccine (Moderna). Sera collected from unvaccinated participants showed median neutralizing antibody titers of 2.31 log IC50 (IQR: 2.03–2.76), 2.10 log IC50 (IQR: 1.76–2.45) and 1.51 log IC50 (IQR: 1.48–1.87) against D614G, B.1.1.7 and B.1.351 live-strains, respectively. Sera from vaccinated participants showed a median neutralizing antibody titer of 4.01 log IC50 (IQR: 3.88–4.35), 4.03 log IC50 (IQR: 3.85–4.23) and 3.14 log IC50 (IQR: 2.99–3.58) against the same viral strains, respectively (Fig. 5b). Strong correlation was observed at M11-13 between neutralizing antibody titers assessed by S-Fuse neutralization assay and anti-RBD IgG titers measured by CMIA with Spearman correlation coefficients of 0.967, 0.968 and 0.944 for variants D614G, B.1.1.7, and B.1.351, respectively (p values<0.0001) (Fig. 5c). Anti-RBD titers around 1.4 log BAU/mL neutralized D614G, B.1.1.7 but not B.1.351 variants at more than 2.0 log IC50. Anti-RBD IgG titers >2.0 log BAU/mL neutralized D614G, B.1.1.7 at > 2.5 log IC50, and B.1.351 at ≥ 2.0 log IC50. These latter anti-RBD IgG titers were reached by all HCW vaccinated at least six days before regardless of pre-vaccination anti-RBD IgG titers, type of vaccine or number of vaccine doses. Based on the strong correlation between CMIA and neutralization assays, neutralizing titers were extrapolated to the remaining 124 unvaccinated HCW and the 73 single-dose or double-dose vaccinated HCW (those with serum collected at least 6 days post-vaccination) to predict the strength of neutralization at M11-13 in all participants (Fig. 5d). Altogether, our findings suggest that former COVID-19 positive individuals benefit from a single-dose vaccine and are able to strongly neutralize current SARS-CoV-2 variants.
Fig. 5.
Neutralization efficiency after single-dose vaccination among the COVID-19 positive HCW. Neutralizing antibody titers against live-strains of D614G, B.1.1.7, and B.1.351 variants of SARS-CoV-2 were measured in sera collected at M11-13 for 13 single-dose vaccinated HCW and 15 unvaccinated HCW. (a) Anti-RBD IgG (log BAU/mL) kinetics between M7-9 and M11-13 according to vaccination status. (b) Neutralizing antibody titers (log IC50) against the D614G, B.1.1.7, and B.1.351 variants measured at M11-13. The dotted black line corresponds to the positivity threshold of neutralization assay. Neutralizing antibody titers measured in HCW vaccinated with one dose of AstraZeneca vaccine (red circles), Moderna vaccine (green squares) or Pfizer-BioNtech vaccine (light blue triangles). (c) Spearman correlation between anti-RBD IgG titers (log BAU/mL) and neutralizing antibody titer (log IC50) against D614G (green circles), B.1.1.7 (purple squares), and B.1.351 (orange triangles) variants measured at M11-13 in vaccinated (n = 13) and unvaccinated (n = 15) HCW. The calculated correlation coefficients (r) and linear regression equations are depicted. (d) Pie charts depicting the frequency of log IC50 neutralization titer categories (extrapolated from CMIA anti-S titers) for all participants at M11-13 according to the viral strain and the vaccination status. The number of participants is included at the center of the pies (139 unvaccinated HCW and 86 HCW vaccinated for at least six days). *p value < 0.05; **p value < 0.01; ***p value < 0.001, ****p value <0.0001; calculated with non-parametric Wilcoxon paired tests or Spearman correlation. P values and correlation coefficients computed using the Graphpad Prism version 9.0.0 software. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The duration and effectiveness of adaptive immunity directed against SARS-CoV-2 after primary infection are key questions in understanding the COVID-19 pandemic. The present study, involving a large cohort of HCW followed prospectively over one year, provides crucial information on the persistence of circulating SARS-CoV-2 antibodies after mild COVID-19. Serological monitoring of convalescent COVID-19 individuals up to 422 DSO shows that anti-SARS-CoV-2 antibodies directed against the S protein are well maintained over time up to more than one year, consistently with other recent studies [[23], [24], [25], [26]–27]. These antibody titers follow a tri-phasic decay, potentially reflective of B cell turnover after infection [2]. From M7, anti-RBD antibodies stabilize at a level that neutralizes infectious variants D614G and B.1.1.7, but less B.1.351, suggesting that most COVID-19 positive patients may be protected from reinfection by the former variants. Our hospital faced three waves of COVID-19, from March to June 2020, September 2020 – January 2021 and from March 2021 to present, with the current wave due to the B1.1.7 variant. During the period April 2020 – April 2021, 69 new infections were reported in COVID-19 negative participants, while only one case of reinfection, which was asymptomatic, was reported in the COVID-19 positive participants. Although antibodies represent only a part of the immune response, this strongly suggests that COVID-19 positive patients develop a robust humoral immune response that reduces the risk of SARS-CoV-2 reinfection within at least one year. Interestingly, anti-RBD levels increase as early as 6 days after vaccination. This suggests that a robust memory B cell response is established in COVID-19 convalescents, including those with low antibody titers. This is in line with the study of Dan et al. who performed an extensive characterization of memory B cells, revealing that the slight antibody decline occurring in convalescent individuals does not reflect a real waning of humoral immunity, but rather a contraction of the immune response, whilst antibody affinity maturation occurs, and anti-S memory B cells persist [8]. Recently, Wang et al. reported that memory B cell clones expressing broad and potent anti-S antibodies are selectively retained in the repertoire at least one year after infection, and expand after vaccination [28]. These observations are very hopeful for the durability of humoral responses developed after COVID-19, and suggest that protection against SARS-CoV-2 infection may last for years [28,29].
Unlike anti-RBD antibody titers that stabilize over time, we observed a steep decay of anti-N IgG titers after seven to nine months post-infection, with only 20% of COVID-19 positive HCW remaining seropositive after one year. These differences could be explained by an increased avidity or affinity that compensates antibody loss, or by changes in recognized epitopes over time [30].
We evaluated several host factors as potential predictors of antibody titers and of their kinetics up to 7–9 months after primary infection. While no differences in SARS-CoV-2 IgG titers were observed, their kinetics were influenced by sex and rhesus factor. Sex differences in the SARS-CoV-2 immune response were previously described early after infection. Takahashi and colleagues reported that female patients had more robust T cell activation than male patients in the early phase of SARS-CoV-2 infection [31]. The sex differences in immune responses may be multifactorial, notably based on sex steroid concentrations, on transcriptional factors, and on incomplete inactivation of immunoregulatory genes on the second X chromosome in females [32,33].
Our study presents some limitations. Neutralization experiments were performed on a small subset of the cohort due to insufficient volume of remaining sera. However, the strong correlation between CMIA IgG levels and neutralizing titers observed in this study, which is also reported by the manufacturer as well as by other studies [4,17,21], allows an extrapolation of the results to the entire cohort. Assessment of reinfection was based on participant reports during visits, as no RT-PCR surveillance was planned in the study. Therefore, it cannot be excluded that the COVID-19 positive participants had unnoticed asymptomatic reinfection during follow-up. However, no COVID-19 positive HCW, except the case of reinfection, had an increase of both anti-RBD and anti-N levels during follow-up. Another limitation is the unbalanced sex distribution, with a female predominance, which reflects the sex distribution of the healthcare workers in our hospital. Nevertheless, the sex difference in immune response was observed by using univariable and multivariable analysis. Furthermore, we were not able to investigate the kinetics of memory B cells because of the lack of peripheral blood mononuclear cells. Finally, our results were obtained in participants with a median age of 39 years (IQR 30–51), hence we cannot exclude that older individuals may experience a different evolution of humoral response over time.
However, taken together our data demonstrate a long-term persistence of anti-RBD IgG titers that may reduce risk of reinfection in convalescent COVID-19 patients by variants D614G and B.1.1.7. By increasing the levels of cross-neutralizing antibodies, SARS-CoV-2 vaccination may strengthen protection, especially against variants harboring antibody escape mutations like B1.351. Future work will help to determine whether vaccine-induced antibodies evolve in the same manner, and whether their kinetics differ between the sexes.
Contributors
SFK, CSM, MG, YH and NM conceived and designed the study. CSM, JDS, NC, DR, MP AF, LKP and MG recruited participants. SB, NR, AS, LG, BP, and JML carried out LFA experiments. TB, DP and OS carried out S-Fuse neutralization experiments. FG, AV, MJW, MS and EL analyzed serological data. FG and PG performed statistical analyses. SFK, FG and PG wrote the manuscript with substantial input from all co-authors.
Data sharing
All of the data generated or analyzed during this study are included in this published article. All data supporting the findings in this study are also available from the corresponding author upon request.
Declaration of Competing Interest
The authors have declared no potential conflicts of interest.
Acknowledgments
This work was supported by Strasbourg University Hospital (SeroCoV-HUS; PRI 7782), the Agence Nationale de la Recherche (ANR-18-CE17-0028), Laboratoire d'Excellence TRANSPLANTEX (ANR-11-LABX-0070_TRANSPLANTEX), Strasbourg University and Institut National de la Santé et de la Recherche Médicale (UMR_S 1109). We are grateful to all of the study participants who donated blood, the team of Cellule d'Appel, Pôle des Ressources Humaines who scheduled the planning of participants, the DRCI team including Eric Demonsant, Hélène Soavelo, Caroline Bouvrais and Evelyne Acacie who prepared the IRB protocol, the ECRF and the administrative part of the protocol, the CIC Inserm 1434 team, including Alexandre Bolle who coordinated recruitment of participants and Noelle Huber, Christine Toulouse, Sophie Wohlgemuth who collected samples, the team of Service de Medecine du Travail who managed the cohort, Carole Weber, Elodie Kleiber and Corinne Renault for data management, and the technical staff of Pôle de Biologie and Laboratoire de Virologie including Anne Moncolin, Veronique Sohn, Axelle Grub, Nathalie Durand and Nadège Frey for management and distribution of samples. Manuscript edition and proofreading was provided by Dr. Kate Dunning (HUS).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103561.
Appendix. Supplementary materials
References
- 1.Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532. doi: 10.1016/j.immuni.2020.07.020. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol. 2020;5(52) doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild COVID-19. N Engl J Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anand S.P., Prevost J., Nayrac M., Beaudoin-Bussieres G., Benlarbi M., Gasser R. Longitudinal analysis of humoral immunity against SARS-CoV-2 Spike in convalescent individuals up to eight months post-symptom onset. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall V.J., Foulkes S., Charlett A., Atti A., Monk E.J.M., Simmons R. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicenter, prospective cohort study (SIREN) The Lancet. 2021;397:1459–1469. doi: 10.1016/S0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen C.H., Michlmayr D., Gubbels S.M., Mølbak K., Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397:1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lumley S.F., O'Donnell D., Stoesser N.E., Matthews P.C., Howarth A., Hatch S.B. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreano E., Rappuoli R. SARS-CoV-2 escaped natural immunity, raising questions about vaccines and therapies. Nat Med. 2021;27:759–761. doi: 10.1038/s41591-021-01347-0. [DOI] [PubMed] [Google Scholar]
- 13.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021:372. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J., Peng P., Wang K., Fang L., Luo F.Y, Jin A.S. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell Mol Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Crispim M.A.E., Fraiji N.A. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 17.Prendecki M., Clarke C., Brown J., Cox A., Gleeson S., Guckian M. Effect of previous SARS-CoV-2 infection on humoral and T-cell responses to single-dose BNT162b2 vaccine. Lancet. 2021;397:1178–1181. doi: 10.1016/S0140-6736(21)00502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velay A., Gallais F., Benotmane I., Wendling M.J., Danion F., Collange O. Evaluation of the performance of SARS-CoV-2 serological tools and their positioning in COVID-19 diagnostic strategies. Diagn Microbiol Infect Dis. 2020;98 doi: 10.1016/j.diagmicrobio.2020.115181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fafi-Kremer S., Bruel T., Madec Y., Grant R., Tondeur L., Grzelak L. Serologic responses to SARS-CoV-2 infection among hospital staff with mild disease in eastern France. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 21.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mickey R.M., Greenland S. The impact of confounder selection criteria on effect estimation. Am J Epidemiol. 1989;129:125–137. doi: 10.1093/oxfordjournals.aje.a115101. [DOI] [PubMed] [Google Scholar]
- 23.Capetti A.F., Borgonovo F., Mileto D., Gagliardi G., Mariani C., Lupo A. One-year durability of anti-spike IgG to SARS-CoV-2: preliminary data from the anticrown prospective observational study one year durability of COVID-19 anti-spike IgG. J Infect. 2021;83:237–279. doi: 10.1016/j.jinf.2021.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choe P.G., Kang C.K., Kim K.H., Yi J., Kim E.U., Park S.W. Persistence of neutralizing antibody response up to one year after asymptomatic or symptomatic SARS-CoV-2 infection. J Infect Dis. 2021:339. doi: 10.1093/infdis/jiab339. Published online June 24jiab. [DOI] [PubMed] [Google Scholar]
- 25.Masiá M., Fernández-González M., Telenti G., Agulló V., García J.A., Padilla S. Durable antibody response one year after hospitalization for COVID-19: a longitudinal cohort study. J Autoimmun. 2021;123 doi: 10.1016/j.jaut.2021.102703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C., Yu D., Wu X., Liang H., Zhou Z., Xie Y. Twelve-month specific IgG response to SARS-CoV-2 receptor-binding domain among COVID-19 convalescent plasma donors in Wuhan. Nat Commun. 2021;12:4144. doi: 10.1038/s41467-021-24230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobaño C., Ramírez-Morros A., Alonso S., Vidal-Alaball J., Ruiz-Olalla G., Vidal M. Persistence and baseline determinants of seropositivity and reinfection rates in health care workers up to 12.5 months after COVID-19. BMC Med. 2021;19:155. doi: 10.1186/s12916-021-02032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muecksch F., Wise H., Batchelor B., Squires M., Semple E., Richardson C. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis. 2021;223:389–398. doi: 10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischinger S., Boudreau C.M., Butler A.L., Streeck H., Alter G. Sex differences in vaccine-induced humoral immunity. Semin Immunopathol. 2019;41:239–249. doi: 10.1007/s00281-018-0726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scully E.P., Haverfield J., Ursin R.L., Tannenbaum C., Klein S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat Rev Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.