Abstract
Vaccines represent preventative interventions amenable to immunogenetic prediction of how human variability will influence their safety and efficacy. The genetic polymorphism among individuals within any population can render possible that the immunity elicited by a vaccine is variable in length and strength. The same immune challenge (virus and/or vaccine) could provoke partial, complete or even failed protection for some individuals treated under the same conditions. We review genetic variants and mechanistic relationships among chemokines, chemokine receptors, interleukins, interferons, interferon receptors, toll‐like receptors, histocompatibility antigens, various immunoglobulins and major histocompatibility complex antigens. These are the targets for variation among macrophages, dendritic cells, natural killer cells, T‐ and B‐lymphocytes, and complement. The technology platforms (mRNA, viral vectors, proteins) utilized to produce vaccines against SARS‐CoV‐2 infections may each trigger genetically distinct immune reactogenic profiles. With DNA biobanking and immunoprofiling of recipients, global COVID‐19 vaccinations could launch a new era of personalized healthcare.
Keywords: COVID‐19, HLA, human polymorphism, immunogenetics, reactogenicity, vaccines
SARS‐CoV‐2 vaccines are amenable to immunogenetic response prediction. The same challenge (virus, vaccine) could provoke complete, partial, or failed protection and variable reactogenicity among individuals. We review genetic variation in immunity and HLA. Production platforms (mRNA, vectors, and proteins) may elicit genetically distinct reactogenic profiles. Vaccine recipient bioprofiling advances personalized health.
INTRODUCTION
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2), the virus causing COVID‐19 (Coronavirus Disease 2019), is the new entity in an ecosystem of several airborne respiratory viruses such as influenza virus, Rhinovirus and Respiratory Syncytial Virus among others. Being hosts of this diverse virome is the reality of our daily life.[ 1, 2 ] At an unprecedented speed, studies to develop COVID‐19 vaccines were conducted, propelled by the current coronavirus crisis worldwide. Three vaccines were given an emergency use of approval (EUA) by the U.S. Food and Drug Administration (FDA) due to their favorable balance of reactogenicity and immunogenicity profile as well as no serious safety concerns observed to date. The vaccine candidates approved by the FDA are the Pfizer/BioNTech SE (BNT162b2) Moderna (mRNA‐1273) and the Johnson and Johnson (Ad26.COV2.S) with ∼95%,∼94.5% and 65% of effectiveness in protection, respectively.[ 3, 4, 5 ]
As of this date, COVID‐19 vaccines from Novavax (NVX‐CoV2373) and AstraZeneca (ChAdOx1), among others, are under final Phase III trials, or have already been approved in other countries.[ 6, 7 ] According to the World Health Organization (WHO), there are up to fifty‐one vaccines in clinical trials, and many more in a pre‐clinical stage of assessment.[ 8 ] Although the mRNA‐based coronavirus vaccines, approved by the FDA, appear to be safe and help trigger an immune response in most of the individuals who have been enrolled so far in the trials, there are still a few who might face some adverse reactions or even a failure in protection.[ 9 ] However, this might also raise the question of whether everyone will benefit from this or any other future COVID‐19 vaccine under development
The key behind any vaccine effectiveness relies on its capacity to induce both neutralizing antibodies and T‐cells response that can fight any pathogen, in this case the SARS‐CoV‐2 virus. Unlike the passive immunization, the success of a vaccine‐induced immunization is influenced by several individual parameters that regulate our immune response.[ 10, 11, 12 ] Studies have demonstrated that 5–10% and 2–10% of healthy individuals that went through standard immunization of Hepatitis B and Measles failed to produce an immune response[ 1, 13 ] exemplifying the singularity of our very own immune system response to vaccination. A multifactorial hypothesis needs to be postulated, and one of the elements that need to be included as part of the equation are the host genetic variations since they have been reported to be one of the main factors for the variable vaccine responsiveness.[ 12 ]
The genetic variability among individuals within any given population can make possible that the immunity elicited to a determined vaccine is variable, meaning that the same viral insult or challenge (either in the form of a vaccine or the virus itself) will result in many different responses. Thus, the response heterogeneity could provoke that a vaccine can either elicit partial, complete, or even fail to protect individuals treated under the same conditions. We know that approximately 5–10% of vaccines fail to induce long‐term antibody protective levels,[ 14 ] a phenomenon that has been associated with the role of genetic factors in vaccine response. In this article, we will review and explore advances in our understanding of SARS‐CoV‐2 and our body's response to infection, with emphasis on genomics features, as well as how these findings can impact the development of effective vaccines against COVID‐19.
UNDERSTANDING THE IMMUNE SYSTEM
To understand the host‐pathogen relationship it is necessary to appreciate how our immune system functions and the evasion strategies those pathogens have developed. The immune system possesses two arms that coexist and complement each other: the innate immune system and the adaptive immune system. The first line of defense against microbial invading pathogens is the innate immune system. The innate immune system is activated when the pattern‐recognition receptors (PRRs) existing within immune cells detect conserved structures on pathogens termed pathogen‐associated molecular patterns (PAMPs).[ 15 ] The prototype of PRRs are the Toll‐like receptors (TLRs) (see Box 1), which are expressed on different immune cells including macrophages (MO), dendritic cells (DCs) and natural killer cells (NK). The innate immunity is nonspecific, rapid, short, and lacks memory. By contrast, the activation and priming of an adaptive system relies on specific recognition of antigenic epitopes, a process that develops more slowly but retains memory and capacity to develop anamnestic recall for effector cells. Nevertheless, SARS‐Cov‐2 is effectively evading the innate immune response associated with the type 1 and 2 IFN therefore delaying the priming and activation of the adaptive immune response risking a severe COVID‐19 illness.[ 16, 17, 18 ] This deviation may be linked to some immunosuppressive phenotypes that inhibit a proper antigen presentation.[ 19 ]
Box 1: Toll Like Receptors (TLR)
Human TLRs comprise 10 members (TLR1‐TLR10), which localize to the cell surface or to intracellular compartment such as endosome. Each TLR recognize distinct or overlapping PAMPs such as lipids, nucleic acid or lipoprotein. Upon TLRs recognize PAMPs, TLRs recruit adaptor proteins such as MyD88 and TRIF, which initiate a complex signal transduction pathway that culminate in the activation of the nuclear factor‐κB (NF‐κB), IRF or MAP kinases to regulate the expression of cytokines, chemokines and Type‐I interferon (IFN‐I) that protect the host from microbial infection
The protagonists of the adaptive immunity are lymphocytes B and T (B‐cells and T‐cells) although ultimately the cells that carry out the clearance and destruction of microbial agents are the cells of innate immunity. The messengers among all cells of innate and adaptive immunity belong to a large family of proteins called cytokines. The complement system (see Box 2) is another component of the immune system, which enhances the ability of phagocytic cells and antibodies secreted by B‐cells to uptake and destroy pathogens. Thus, the complement system participates in both arms of the immune system. Table 1 lists some of these potential candidates of clinical relevance. Individual variation in the genes that are involved in the HLA recognition between T‐cells and antigen presenting cells or in the complement cascade may alter the host response to pathogens. Understanding the role and the importance of the adaptive immune response in the clearance of the SARS‐CoV‐2 virus and its immune memory generated is crucial for the success of all COVID‐19 vaccines.
TABLE 1.
List of potential relevant markers for immune response
| Marker | Natural defense |
|---|---|
| BY55 | Natural killer cell receptor, immunoglobulin superfamily member |
| CCR2 | Chemokine C‐C motif receptor 2 |
| CCR5 | Chemokine C‐C motif receptor 5 |
| CCR6 | Chemokine C‐C motif receptor 6 |
| CD7 | CD7 antigen p41 |
| CD8A, CD8B1 | CD8 antigen, alpha polypeptide and beta polypeptide 1 |
| GNLY | Granulysin |
| HLA‐A, ‐C, ‐E, ‐G | Major histocompatibility complex (class I, A, C, E, and G) |
| IFNB1 | Interferon, beta 1, fibroblast |
| IFNG | Interferon gamma |
| INFAR1 | Interferon (alpha, beta, and omega) receptor 1 |
| IFNGR2 | Interferon gamma receptor 2 |
| IL‐12A | Interleukin 12A, natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35 |
| IL‐12B | Interleukin 12B natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40 |
| ITGB1 | Integrin, beta 1 fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12 |
| KIR2DL4 | Killer cell immunoglobulin‐like receptor, two domains, long cytoplasmic tail, 4 |
| KLRC3 | Killer cell lectin‐like receptor subfamily C, member 3 |
| LGALS3BP | Lectin, galactoside‐binding, soluble, 3 binding protein |
| LILRB4 | Leukocyte immunoglobulin‐like receptor, subfamily B with TM and ITIM domains, member 4 |
| MICB | MHC class I polypeptide‐related sequence B |
| PRFl | perforin 1 |
| TCRA, TCRB | T‐cell antigen receptor, alpha and beta subunits GZMA, GZMB—granzyme A and B |
| TNFRSF6 | Tumor necrosis factor receptor superfamily, member 6 (Fas) |
Box 2: Complement System
The complement system is formed by a large number of serum proteins that remain in circulation in inactive form (zymogen form). The complement system can be activated through different pathways, which although differ in the molecules that promote the initiation, converge to generate the same set of effector molecules. The classical way of complement activation is effective at late phase of infection in the presence of antibodies and thereby is part of host's defense during adaptive immunity.
MACROPHAGES AND DCs : THE LINK BETWEEN INNATE AND ADAPTIVE IMMUNITY
Dendritic cells and MO are phagocytic cells also known as polymorphonuclear leukocytes. DCs are the prototype of sentinels’ cells. They are the first cells responsible to sense and capture microbes and process microbial antigens to effectively present these antigens to naive T‐cells within lymphoid tissues. During their migration to lymphoid tissues DCs undergo extensive stimulus‐dependent irreversible differentiation, a process termed “maturation,” which influence the type of differentiation (Th1 or Th2) that undergoes the CD4+ T‐cells. [ 3, 20, 21 ] Thus, DCs allow a link between the two arms of immune response (innate and adaptive) and play key roles in the T‐cell mediated immunity. This seems to be critical for some COVID‐19 vaccines, as a strong cellular response mediated by CD4+ T‐ and CD8+T‐cells) elicited by Pfizer/ BioNTech SE (BNT162b2) has been reported in clinical trials.[ 22 ]
ACTIVATION AND MATURATION OF T‐ AND B‐CELLS: A CRITICAL STEP IN ANTIBODY PRODUCTION
T and B‐cells are lymphocytes involved in adaptive immunity. T‐cells are responsible for cell‐mediated immunity and B‐cells are responsible for antibody‐mediated immunity. The successful activation and differentiation of naive T‐cells occur only if three signals are present: 1) interaction with the antigenic peptides presented by the antigen‐presenting cells by the human leukocyte antigen (HLA) molecule, 2) the presence of stimulatory signals involved in binding of CD80 and CD86 molecules on the surface of antigen‐presenting cells (APC) to the CD28 receptor on the T‐cells, and 3) the secretion of cytokines that regulate the immune response.[ 23 ] Depending on the nature of antigen, and the activation status of APCs, the T‐cell can differentiate to CD4+ T or CD8+ T subsets. If the T cell expresses CD4, it is differentiated into T‐helper cell (Th), which produces cytokines and stimulates the production of antibodies from B cells . When the MO or DCs secrete IL‐12, IL‐18, type‐1 IFN‐α and IFN‐β, the T‐cell is differentiated towards Th1, which secrete IFNg, IL‐2, IL‐10, and TNF.[ 24 ] These cytokines enhance the microbicidal action of macrophages leading phagocytosis and destruction of invading pathogens. The differentiation of CD4+ T‐cells to Th2 is generated by anti‐inflammatory cytokines such as IL‐4, IL‐25, IL‐33 secreted by mast cells and eosinophils, which are important for induction and development of humoral (antibody) immune responses.
Naive T‐cells that express CD8 develop their effector functions converting into cytotoxic T lymphocyte that can attack and destroy cells infected with viruses. Cytotoxic T cells also produce IFN‐γ and TNFα, which play roles t in the defense against viral infections.[ 24 ] Depending on the nature of antigen encountered memory T‐cells (Tm) can be either CD4‐ or CD8 T‐cell type. Memory T‐cells cells are antigen‐specific T‐cells that remain long‐term after an infection has been eliminated. Upon re‐exposure to the specific invading pathogen that has occurred, Tm‐cells are quickly converted into large numbers of effector T‐ cells; thus, providing a rapid immune response to reinfections. Current COVID‐19 vaccines seem to elicit both, antibody and cellular antiviral immune responses characterized by strong production of CD4+ Th1 and CD8+T cell‐mediated responses. [ 22 ]
B‐cells participate in the humoral adaptive immune system and are responsible for mediating the production of antigen‐specific antibodies against invading pathogens . B‐cells originate in the bone marrow and after the encounter with the pathogen migrates to the spleen and other lymphoid organs where they mature and differentiate into immunocompetent antibody producer cells. Direct binding of the microbial antigen to receptors on its surface causes cell division and proliferation. Some stimulated B‐cells become plasma cells, which produce antibodies and others become in long‐life memory B‐cells, which can be stimulated later and differentiate into plasma cells.
HUMAN LEUKOCYTE ANTIGEN (HLA) POLYMORPHISMS AND INDIVIDUAL VACCINE EFFICACY
The immune system is diverse, with person‐ to‐ person variability, and the mosaic of human leukocyte antigens (HLA) is the best example of its human polymorphism. Humans have different allelic versions of the HLA genes, and certain variants at these loci encode for cell receptors that can bind less reliably to some viral peptides and blunt the immune system's normal defenses against the virus in vulnerable patients. Based on prior predictions, the receptor binding domain (RBD) subunit appears to have no MHC class II peptides displayed in ∼15% of the worldwide population, ranging from 0.8% in self‐reported Whites to 37% for Asians.[ 25 ] Notably, such a predicted uncovered population for RBD with no peptide‐MHC hits might be reduced to 0% (MHC class I) and 0.31% (MHC class II) by considering some computed sets of augmentation peptides encompassing all filtered peptides from SARS‐CoV‐2, according to computer‐aided predictions.[ 26 ]
Following a genomic combinatorial approach, MIT's OptiVax and EvalVax programs evaluate a host of possible combinations of common alleles (e.g., HLA haplotypes) in each ancestry group to find the most likely combo to design a vaccine with better coverage in every single population. The two algorithms work in tandem in a feedback loop, but EvalVax takes relevant population data from different individuals across the three main ancestry groups to feed the beam search that OptiVax conducts over peptide‐receptor pairs (i.e., by mapping the immune response to the unique biochemistry of each population by genetic ancestral status), and therefore ensure population coverage.[ 26 ]
SPECIFIC HLA GENETIC VARIATIONS AND DISEASE IMPLICATIONS
Significant differences among HLA alleles can define the susceptibility for a disease or the effectiveness of a vaccine. Studies have been conducted to investigate the HLA genetic variation and the immune response towards the SARS‐CoV‐2 (Figure 1). These studies have demonstrated that HLA‐B*15:03 have a high capacity for presenting peptides suggesting that this allele may be widely protective and could enable a cross‐protective T‐cell based immunity, whereas the HLA‐B* 46:01 was found to bind to fewer peptides of the SARS‐CoV‐2, suggesting that persons who hold this allele may produce a weak immune response therefore developing severe symptoms.[ 27 ] The HLA‐B*46:01 was previously predicted as a susceptibility marker of SARS‐CoV and associated with its severity in Asian populations.[ 28 ] These findings provide a means of identifying individuals at risk of developing life‐threatening COVID‐19 and ensuring their enrolment in vaccine trials.
FIGURE 1.
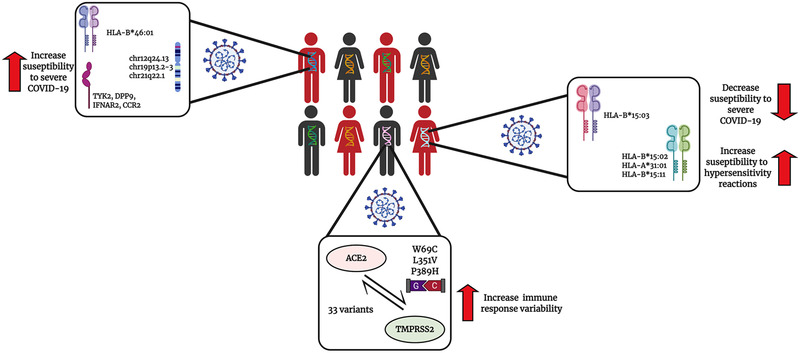
The influence of the host genetics on the SARS‐CoV‐2 infection and COVID‐19 severity susceptibility. In a population, many individuals may carry different single nucleotide polymorphisms (SNPs) in different genes. Individuals that carry specific polymorphisms near or in genes that codify for TYK2, DPP9, IFNAR2, CCR2 or carry the HLA‐B*46:01 are believe to be more susceptible to develop severe COVID‐19 symptoms. In contrast individuals that carry the HLA‐B*15:03 are believe have a more protective phenotype and unlikely will develop severe COVID‐19 symptoms. Individuals may have different variants in their ACE2 and TMPRSS2 receptors, the two main receptors that the SARS‐CoV‐2 virus use to infect the host. These variants may increase the immune response variability between individuals therefore it may affect the efficacy of both treatments and vaccines
Different genetic factors or risk loci, mostly related to key host antiviral defense mechanisms and mediators of inflammation, have been reported since the beginning of the pandemic.[ 29, 30 ] Among them, a gene cluster on chromosome 3 inherited from Neanderthals has been identified as a potential predictor a COVID‐19 severity.[ 31 ] Likewise, some novel GWAS significant hits on chr12q24.13 (rs10735079, p = 1.6 × 10–8) in a gene cluster encoding antiviral restriction enzyme activators (OAS1‐3); on chr19p13.2‐3 (rs2109069, p = 2.3 × 10–12 and rs2109069, p = 3.9 × 10–12) near the gene encoding tyrosine kinase 2 (TYK2) and within the gene encoding dipeptidyl peptidase 9 (DPP9), respectively, as well as on chr21q22.1 (rs2236757, p = 5 × 10–8) in the interferon receptor gene (IFNAR2) and the monocyte/macrophage chemotactic receptor (CCR2), have also been postulated as potential predictors of critical illness caused by COVID‐19.[ 32 ]
Notably, a significant number of patients with severe COVID‐19 carried rare genetic variants in 13 genes known to be critical in the body's defense against influenza virus, and more than 3.5% were completely missing a functioning gene to produce any detectable type I interferons (IFNs) in response to SARS‐CoV‐2.[ 33 ] A recent report by Bastard and co‐workers from the COVID Human Genetic Effort found that neutralizing autoantibodies against type I IFNs might underlie some critical COVID‐19 cases (∼10%) by impairing the binding to their receptors and the activation of the downstream responsive pathway (IFN‐stimulated genes).[ 34, 35 ] Indeed, B cell autoimmune phenocopy of inborn errors of type I IFN immunity seems to account for life‐threatening COVID‐19 events (e.g., pneumonia) in up to 12.5% patients, mostly men. Consequently, adaptive autoimmunity might be able to impair innate and intrinsic antiviral immunity in these patients.
SARS‐CoV‐2 PROTEIN MUTATIONS AND VACCINE EFFICACY
Mutations in the ACE2 and the TMPRSS2, the primary and the second host proteins involved in the SARS‐CoV‐2 infection have been identified.[ 36 ] Benetti et al. identified 33 ACE2 variations in approximately 7000 Italian persons, with one of the variations (N720D) being adjacent to the TMPRSS2 cleavage site and three other mutations: W69C, L351V and P389H were estimated to cause conformational changes altering the interactions with the receptor binding domains (RBD) of the S glycoprotein.[ 36 ] Recent reports of mutations in the spike S‐protein of the SARS‐CoV‐2 virus, and the corresponding receptor binding domains (RBD). New SARS‐CoV‐2 genetic variants have been identified. The first mutation described was identified in the early months of the pandemic and was a mutation located in the 614 amino acid position of the Spike protein.[ 10 ] Three other variants have been identified, one in South Africa designated B.1.351 or 501Y.V2,[ 10, 11, 31 ] one in the United Kingdom, designated as B.1.1.7 or 501Y.V1.[ 10, 11 ] and one in Brazil designated as P‐1.[ 11 ] The B.1.1.7 variant has caused concern since it demonstrates to be more transmissible.[ 10, 11, 18 ] Although these significant mutations in the spike protein have not proven to critically affect the efficacy of the vaccine, it is possible that further mutations can enhance the capacity of the virus to evade the immune system therefore reducing the effectiveness of the vaccines.
The rare vaccine‐elicited disease enhancement could be an example of this kind of immunity errors when already vaccinated subjects encounter circulating SARS‐CoV‐2 viruses. Such an event invariably involves antibody‐mediated immune “aberrant” mechanisms from direct antibody‐dependent enhancement (ADE) to immune complex formation by antibodies, albeit accompanied by various coordinated cellular responses such as Th2 T‐cell skewing.[ 37 ] It is like to the clinical course of COVID‐19 patients, in whom severe COVID‐19 disease is associated with the development of abnormal anti‐SARS‐CoV‐2 serum antibodies, with titers correlating directly with the severity of disease.[ 38, 39 ] Like the risk of some idiosyncratic systemic adverse events, a genetic trigger might certainly be involved in these episodes of vaccine‐elicited disease worsening. However, ADE is usually linked to viruses attacking macrophages such as dengue and Zika viruses.
POPULATION DIVERSITY AND RELATIONSHIPS TO VACCINE EFFICACY
BNT162b2 and mRNA‐1273 COVID‐19 vaccines are based on messenger RNA (mRNA) technology. mRNA vaccines consist of a single‐stranded RNA encoding key virus proteins. In the case of SARS‐CoV‐2 vaccine mRNA contains the transcript for proteins that help viruses to infect cells. Once injected, cells receive mRNA and use it as a template to make viral proteins. These proteins trigger T‐ and B‐cells, which activate, and B‐cells produce antibodies. If a person gets exposed to SARS‐CoV‐2 the T‐cells as well as the antibodies will recognize the proteins on the virus, which helps the immune system to detect and destroy the virus before it causes illness. A safe and effective mRNA vaccine for COVID‐19 is the first of its kind with an authorization granted since such a technology has yet to be used for an approved vaccine.
Using machine learning (ML)‐assisted in silico prediction modeling, researchers from the Massachusetts Institute of Technology suggested that COVID‐19 vaccines developed by Moderna, Pfizer, AstraZeneca and others, may not protect individuals of non‐European genetic backgrounds (e.g., African or Asian descent) as well as they are expected to do for white people.[ 25, 40 ] According to these authors, individuals of mostly African or Asian ancestry seemed to have on average a slightly increased risk of vaccine ineffectiveness. It is likely a consequence of the lack of a sufficiently diverse set of viral particles within the vaccine preparation to stimulate the immune response at the same level across all individuals from different populations. Indeed, depending on their genetic makeup, current vaccines could leave gaps in population coverage. However, recently released data and publications on the BNT162b2 and AZD1222 vaccine clinical trials suggest a certain degree of diversity. For BNT162b2, 26% participants self‐identified as Hispanics were enrolled whereas participants from Brazil, South Africa and UK populations were enrolled in the AZD1222 trial.[ 41, 42 ]
Concerning the development of novel vaccines for the prevention of COVID‐19, the NIH‐Wide Strategic Plan for COVID‐19 Research stated on page 19, Objective 4.1, that it is paramount to make efforts for ensuring the participation of a broad range of populations in clinical testing, including high‐risk groups as a major priority. Accordingly, efficacy studies are designed to also include more genetically diverse underserved populations along with older individuals, people with comorbidities, and other high‐risk groups, as earlier described in the NIAID Strategic Plan for COVID‐19 Research.[ 43 ]
The genetic variability among individuals within any given population can make possible that the immunity elicited to a determined vaccine is variable, meaning that the same viral insult or challenge (either in the form of a vaccine or the virus itself) will result in many different responses. Thus, the response heterogeneity could provoke that a vaccine can either elicit partial, complete, or even fail to protect individuals treated under the same conditions. We know that approximately, 5 to 10% of vaccines fail to induce long‐term antibody protective levels,[ 26 ] a phenomenon that has been associated with the role of genetic factors in vaccine response. For example, twins’ studies have revealed variations of 89%, 39% and 46% in the IgG antibody titers elicited by individuals vaccinated against measles, mumps, and rubella vaccines.59 Moreover, high heritability (40–70%) has been also observed in oral polio, tetanus, diphtheria, and hepatitis B vaccines.[ 44 ] Since these variations have been observed in vaccines that have been used worldwide for dozens of years, we can expect to see them in the vaccines against SARS‐CoV‐2 in development. Based on immunogenicity findings from conducted clinical trials, higher doses of the COVID‐19 vaccine might be necessary to elicit optimal protection in those with lower antigen‐binding IgG and virus‐neutralizing responses due to a weakened immune system.[ 45 ]
We still do not know how long immunity lasts, or whether these vaccines can only prevent the illness or also prevent the infection. To these questions are also added the fact that this is the first time that mRNA vaccines could be authorized and there are no previous experiences on how to produce, preserve and distribute it on a huge scale a vaccine of this type without affecting their stability.
REACTOGENIC TRIGGERS FROM VACCINE DELIVERY VECTORS
The variety of technology platforms (mRNA, viral vectors, protein) utilized currently to produce vaccines against SARS‐CoV‐2 infections may each also trigger genetically distinct immune reactogenic profiles against chemical or genetic components of the product.
Lipid nanoparticles (LNPs) are the vectors used for RNA delivery of BNT162b2 and mRNA‐1273 vaccines.[ 46, 47 ] Its most commonly used formulation components are ionizable lipids, phospholipids, cholesterol and lipid‐anchored polyethylene glycol (PEG), each conferring specific properties.[ 47 ] LNPs with novel ionizable lipids and lipid‐like materials maintain a neutral or mildly cationic surface charge at physiological pH. This property impacts specificity and fluidity at once, reducing lipid–protein non‐specific interactions while facilitating cytosolic RNA oligonucleotide discharge.[ 48 ] Phospholipids in turn are structural to the LNPs, supporting the formation and disruption of the lipid bilayer to facilitate endosomal fusion. Cholesterol serves as a stabilizer and supports transfection of cells. Finally, surface PEG serves as a barrier, stabilizing the LNP sterically and reducing nonspecific protein binding. PEG in the LNP coating is suspected to have led to reactogenic sequelae and anaphylaxis in some individuals.[ 35, 49 ].
Replication‐incompetent adenoviral vectors have been under investigation as a platform to carry a variety of transgenes, and express them as a basis for vaccine development.[ 50 ] A replication‐incompetent adenoviral vector based on human adenovirus type 26 (Ad26) is the basis of the ChAdOx1, Ad26.COV2.S, and Gam‐COVID‐Vac vaccines. Little is known about the mechanisms of immunity to the vector. However, neutralizing antibodies and cellular responses are induced after Ad26 vector administration to humans and non‐human species. Vector specific neutralizing antibodies can specifically inhibit vector entry. A strategy to avoid reactogenicity is to construct vaccines with initial and booster immunizations in different adenovirus vectors (e.g., rAd26 followed by rAd5) to minimize cross reactivity. This approach has been successfully implemented for the Gam‐COVID‐Vac vaccine.[ 13 ] Individuals will have heterogeneous levels of reactogenicity to the vector depending on prior adenovirus exposure and recipient immunogenetics.[ 39 ] In contrast to the RNA and adenovirus technologies, conventional protein and adjuvant formulations under development have evidenced minimal reactogenicity. There may be advantages to strategies not involving genetic programming of recipient cells, relying only on immune response to exogenous antigens.[ 51 ]
There is precedent for variability in immune responses to exogenous chemicals manifested as cutaneous adverse drug reactions (CADR). This immunogenetic link is best exemplified by the association between necrolyzing hypersensitivity to anticonvulsant drugs and HLA antigens.[ 52 ] Carbamazepine‐associated hypersensitivity can occur in up to 10% of patients with dermal reactions, and may also involve the eye. Stevens–Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), maculopapular exanthema (MPE), and drug reaction with eosinophilia and systemic symptoms (DRESS) are serious adverse reactions, even potentially life threatening. The mechanism may be a reaction of CD8 or CD14 T cells, which produces tissue injury.[ 53 ]
CADR are associated with specific HLA alleles. HLA‐B*15:02 is strongly associated with carbamazepine‐induced SJS/TEN in Southeast Asian populations where the allele is most prevalent. The FDA has established a black box warning on the carbamazepine label advising on testing for HLA‐B*15:02 for Asian patients prior to initiating therapy, and advises against using the drug in carriers for HLA‐B*15:02 unless benefit outweighs risk. HLA‐based dosing guidelines for carbamazepine have been published by various consortia.[ 54 ] HLA‐B*15:02 is also associated with phenytoin adverse reactions and increased risks of SJS and TEN.[ 42, 52, 53 ] HLA‐A*31:01 may also be a risk factor for SJS/TEN but more strongly associated with DRESS and MPE. HLA‐A*31:01 is found in most populations, worldwide. HLA‐B*15:11 is another allele that has been linked with SJS/TEN. As a counterpart, HLA‐B*0702 has been associated as a protective marker to SJS.[ 42 ]
LONGER‐TERM EFFECTS
The COVID‐19 vaccines have been launched with a median observation time of 3 months and it is expected that observation of seroconversion will be longer lasting through the end of 2021. These seroconversion studies will unveil whether periodic “boosters shots” are required and shed some light on some clinical endpoints regarding disease protection and reduction of infectiousness. These seroprevalence studies will constitute fertile ground for population genetics research as well during this period with global vaccination efforts to diverse populations.
Immunogenetics, ancestry, and other ethnicity‐specific factors need to be taken seriously into account in the acceptability of foreign clinical data by regulatory agencies, given the substantial amount of critical information collected from volunteers who participated in these clinical trials of COVID‐19 vaccines globally and its international development perspective.[ 55 ] The inter‐ethnic differences in treatment responses are well known and have been reviewed previously.[ 56 ]
The response to COVID‐19 mRNA vaccine in immunologically dysregulated individuals will require close monitoring and study. With vaccines to influenza and Hepatis B viruses, cross‐reactions have been observed between vaccine and self, leading to auto‐immune neurological sequelae (e.g., Guillain‐Barré syndrome, demyelination). Potential cross‐reactive interactions and molecular mimicry between the spike protein of SARS‐CoV‐2 and autoimmune target proteins have been demonstrated.[ 57 ]
CONCLUSIONS AND OUTLOOK
This article has attempted to encompass the range of possible genetic polymorphism that could underlie immune response to vaccines. Public health policy on vaccinations may commonly incorporate individual characteristics of age and disease comorbidity, but rarely includes genetic polymorphisms, an addressable problem. This human genomic diversity could pinpoint individuals best served by nuanced or stratified recommendations, a paradigm of personalized health. The current COVID‐19 pandemic represents an opportunity for personalized health.
The acute nature of allergic reactions to vaccination is reminiscent of the time course of adverse drug reaction mediated by the immune system. Chemical features of modified RNA or lipid coating may be triggering these hypersensitivity reactions. Here, HLA antigens could be examined first, as these have traceable ethnogeography frequencies. There are also reactogenic features of the vaccination that would be amenable to genetic analysis. The most common side effects (fatigue, chills, myalgia, arthralgia, fever) are stronger after the second dose, and were felt by one‐third to two‐thirds of recipients in clinical trials. The variable reactions constitute an early sign the vaccines are prompting a variable immune response.
At a point when antibody titers have declined yet disease resistance prevails it would be appropriate to assess the function of memory B‐cells and memory T‐cells that might retain information about the coronavirus for years or even decades. This will be a far more difficult task spanning the corresponding longer time interval of observation. Certainly, the realization that booster vaccinations may be necessary if antibody and immune protection wane would only elevate the relevance of the findings during that first‐year post‐vaccination. It is critical that vaccination efforts encompass parallel biobanking of recipient genomic DNA and serum immunoprofiling. The wonder of the novel COVID‐19 vaccines could also elicit a new era of research and application of immunogenetics and personalized health.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
DISCLAIMER
The authors are solely responsible for the drafting and editing of the paper, and its final contents. The contents of this manuscript do not represent the views of the National Institutes of Health (NIH) or the United States Government. No funded writing assistance was utilized in the production of this manuscript.
ACKNOWLEDGMENTS
The authors would like to thank The Center for Collaborative Research in Health Disparities (CCRHD‐RCMI) and The Research Initiative for Scientific Enhancement (RISE). Their funding support enables the opportunity to conduct these academic and research activities. Figures in this manuscript were created using Bio Render.
Valdés‐Fernández, B. N., Duconge, J., Espino, A. M., & Ruaño, G. (2021). Personalized health and the coronavirus vaccines—Do individual genetics matter? BioEssays, 43, e2100087. 10.1002/bies.202100087
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1.Ruaño, G., & Ha, T. (2021). Living with respiratory viruses: The next saga in human/viral coexistence? BioEssays, 43(4), 2020–2021. 10.1002/bies.202000321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kollathodi, N., Moorkoth, A., George, K., Narayanan, M., Balakrishnan, S., & LelithaBai, S. (2017). Hepatitis B vaccination—immune response and persistence of protection in susceptible population. Journal of The Academy of Clinical Microbiologists, 19(1), 42–46. 10.4103/jacm.jacm_63_16 [DOI] [Google Scholar]
- 3.Walsh, E. E., Frenck, R. W., Falsey, A. R., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., Neuzil, K., Mulligan, M. J., Bailey, R., Swanson, K. A., Li, P., Koury, K., Kalina, W., Cooper, D., Fontes‐Garfias, C., Shi, P.‐Y., Türeci, Ö., Tompkins, K. R., … Gruber, W. C. (2020). Safety and immunogenicity of two RNA‐based Covid‐19 vaccine candidates. New England Journal of Medicine, 383(25), 2439–2450. 10.1056/nejmoa2027906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novavax . (2021). Annoucement of UK and South Africa Trial Results.
- 5.Pfizer . (n.d.). Vaccines and Related Biological Products Advisory Committee Meeting, FDA Briefing. https://www.fda.gov/media/144245/download
- 6.Arunachalam, P. S., Wimmers, F., Mok, C. K. P., Perera, R. A. P. M., Scott, M., Hagan, T., Sigal, N., Feng, Y., Bristow, L., Tsang, O. T. Y., Wagh, D., Coller, J., Pellegrini, K. L., Kazmin, D., Alaaeddine, G., Leung, W. S., Chan, J. M. C., Chik, T. S. H., Choi, C. Y. C., … Pulendran, B. (2020). Systems biological assessment of immunity to mild versus severe COVID‐19 infection in humans. Science, 369(6508), 1210–1220. 10.1126/SCIENCE.ABC6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berlin, D. A., Gulick, R. M., & Martinez, F. J. (2020). Severe Covid‐19. The New England Journal of Medicine, 383(25), 2451–2460. 10.1056/NEJMcp2009575 [DOI] [PubMed] [Google Scholar]
- 8.Gandhi, R. T., Lynch, J. B., & Del Rio, C. (2020). Mild or Moderate Covid‐19. The New England Journal of Medicine, 383(18), 1757–1766. 10.1056/NEJMcp2009249 [DOI] [PubMed] [Google Scholar]
- 9.Triggle, N. (2020). Covid‐19 vaccine: Allergy warning over new jab. BBC News. https://www.bbc.com/news/health-55244122
- 10.Grubaugh, N. D., Hanage, W. P., & Rasmussen, A. L. (2020). Making sense of mutation: What D614G means for the COVID‐19 pandemic remains unclear. Cell, 182(4), 794–795. 10.1016/j.cell.2020.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grubaugh, N. D., Hodcroft, E. B., Fauver, J. R., Phelan, A. L., & Cevik, M. (2021). Public health actions to control new SARS‐CoV‐2 variants. Cell, 184(5), 1127–1132. 10.1016/j.cell.2021.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah, R. R. (2015). Inter‐ethnic differences in drug response: Implications for drug development and complying with drug regulation. Clinical Research and Regulatory Affairs, 32(3), 88–98. 10.3109/10601333.2015.1064131 [DOI] [Google Scholar]
- 13.Voysey, M., Clemens, S. A. C., Madhi, S. A., Weckx, L. Y., Folegatti, P. M., Aley, P. K., Angus, B., Baillie, V. L., Barnabas, S. L., Bhorat, Q. E., Bibi, S., Briner, C., Cicconi, P., Collins, A. M., Colin‐Jones, R., Cutland, C. L., Darton, T. C., Dheda, K., Duncan, C. J. A., … Pollard, A. J. (2021). Safety and efficacy of the ChAdOx1 nCoV‐19 vaccine (AZD1222) against SARS‐CoV‐2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet, 397(10269), 99–111. 10.1016/S0140-6736(20)32661-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, Q., Bastard, P., Liu, Z., Le Pen, J., Moncada‐Velez, M., Chen, J., Ogishi, M., Sabli, I. K. D., Hodeib, S., Korol, C., Rosain, J., Bilguvar, K., Ye, J., Bolze, A., Bigio, B., Yang, R., Arias, A. A., Zhou, Q., Zhang, Y., … Casanova, J.‐L. (2020). Inborn errors of type I IFN immunity in patients with life‐threatening COVID‐19. Science, 370(6515), eabd4570. 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai, T., & Akira, S. (2010). The role of pattern‐recognition receptors in innate immunity: Update on Toll‐like receptors. Nature Immunology, 11(5), 373–384. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 16.Netea, M. G., Domínguez‐Andrés, J., Barreiro, L. B., Chavakis, T., Divangahi, M., Fuchs, E., Joosten, L. A. B., van der Meer, J. W. M., Mhlanga, M. M., Mulder, W. J. M., Riksen, N. P., Schlitzer, A., Schultze, J. L., Benn, C. S., Sun, J. C., Xavier, R. J., & Latz, E. (2020). Defining trained immunity and its role in health and disease. Nature Reviews. Immunology, 20(6), 375–388. 10.1038/s41577-020-0285-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sette, A., & Crotty, S. (2021). Adaptive immunity to SARS‐CoV‐2 and COVID‐19. Cell, 184(4), 861–880. 10.1016/j.cell.2021.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou, D., Dejnirattisai, W., Supasa, P., Liu, C., Mentzer, A. J., Ginn, H. M., Zhao, Y., Duyvesteyn, H. M. E., Tuekprakhon, A., Nutalai, R., Wang, B., Paesen, G. C., Lopez‐Camacho, C., Slon‐Campos, J., Hallis, B., Coombes, N., Bewley, K., Charlton, S., Walter, T. S., … Screaton, G. R. (2021). Evidence of escape of SARS‐CoV‐2 variant B.1.351 from natural and vaccine‐induced sera. Cell, 184(9), 2348–2361.e6.e6. 10.1016/j.cell.2021.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray, P. J., & Wynn, T. A. (2011). Protective and pathogenic functions of macrophage subsets. Nature Reviews Immunology, 11(11), 723–737. 10.1038/nri3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer, C. D., & Genco, C. A. (2017). Microbiota, immune subversion, and chronic inflammation. Frontiers in Immunology, 8, 255. 10.3389/fimmu.2017.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul, W. E., & Zhu, J. (2010). How are T(H)2‐type immune responses initiated and amplified? Nature Reviews. Immunology, 10(4), 225–235. 10.1038/nri2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabanillas, B., Akdis, C. A., & Novak, N. (2021). Allergic reactions to the first COVID‐19 vaccine: A potential role of polyethylene glycol? Allergy, 76(6), 1617–1618. 10.1111/all.14711 [DOI] [PubMed] [Google Scholar]
- 23.MacLeod, M. K. L., Kappler, J. W., & Marrack, P. (2010). Memory CD4 T cells: Generation, reactivation and re‐assignment. Immunology, 130(1), 10–15. 10.1111/j.1365-2567.2010.03260.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox, M. A., Kahan, S. M., & Zajac, A. J. (2013). Anti‐viral CD8 T cells and the cytokines that they love. Virology, 435(1), 157–169. 10.1016/j.virol.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, G., Carter, B., & Gifford, D. K. (2021). Predicted cellular immunity population coverage gaps for SARS‐CoV‐2 subunit vaccines and their augmentation by compact peptide sets. Cell Systems, 12(1), 102–107.e4.e4. 10.1016/j.cels.2020.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Posteraro, B., Pastorino, R., Di Giannantonio, P., Ianuale, C., Amore, R., Ricciardi, W., & Boccia, S. (2014). The link between genetic variation and variability in vaccine responses: Systematic review and meta‐analyses. Vaccine, 32(15), 1661–1669. 10.1016/j.vaccine.2014.01.057 [DOI] [PubMed] [Google Scholar]
- 27.Nguyen, A., David, J. K., Maden, S. K., Wood, M. A., Weeder, B. R., Nellore, A., & Thompson, R. F. (2020). Human leukocyte antigen susceptibility map for SARS‐CoV‐2. Journal of Virology, 94(13), 1–12. 10.1128/JVI.00510-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovsyannikova, I. G., Haralambieva, I. H., Crooke, S. N., Poland, G. A., & Kennedy, R. B. (2020). The role of host genetics in the immune response to SARS‐CoV‐2 and COVID‐19 susceptibility and severity. Immunological Reviews, 296(1), 205–219. 10.1111/imr.12897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobson, G. P., Biros, E., Letson, H. L., & Morris, J. L. (2021). Living in a hostile world: Inflammation, new drug development, and coronavirus. Frontiers in Immunology, 11, 3424. https://www.frontiersin.org/article/10.3389/fimmu.2020.610131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccolo, V., Curina, A., Genua, M., Ghisletti, S., Simonatto, M., Sabò, A., Amati, B., Ostuni, R., & Natoli, G. (2017). Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross‐talk. Nature Immunology, 18(5), 530–540. 10.1038/ni.3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Netea, M. G., Giamarellos‐Bourboulis, E. J., Domínguez‐Andrés, J., Curtis, N., van Crevel, R., van de Veerdonk, F. L., & Bonten, M. (2020). Trained immunity: A tool for reducing susceptibility to and the severity of SARS‐CoV‐2 infection. Cell, 181(5), 969–977. 10.1016/j.cell.2020.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piccoli, L., Park, Y.‐J., Tortorici, M. A., Czudnochowski, N., Walls, A. C., Beltramello, M., Silacci‐Fregni, C., Pinto, D., Rosen, L. E., Bowen, J. E., Acton, O. J., Jaconi, S., Guarino, B., Minola, A., Zatta, F., Sprugasci, N., Bassi, J., Peter, A., De Marco, A., … Veesler, D. (2020). Mapping neutralizing and immunodominant sites on the SARS‐CoV‐2 spike receptor‐binding domain by structure‐guided high‐resolution serology. Cell, 183(4), 1024‐1042.e21. 10.1016/j.cell.2020.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peck, R. W., Weiner, D., Cook, J., & Powell R., J. (2020). A real‐world evidence framework for optimizing dosing in all patients with COVID‐19. Clinical Pharmacology and Therapeutics, 108(5), 921–923. 10.1002/cpt.1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bastard, P., Rosen, L. B., Zhang, Q., Michailidis, E., Hoffmann, H.‐H., Zhang, Y., Dorgham, K., Philippot, Q., Rosain, J., Béziat, V., Manry, J., Shaw, E., Haljasmägi, L., Peterson, P., Lorenzo, L., Bizien, L., Trouillet‐Assant, S., Dobbs, K., de Jesus, A. A., … Casanova, J.‐L. (2020). Autoantibodies against type I IFNs in patients with life‐threatening COVID‐19. Science, 370(6515), eabd4585. 10.1126/science.abd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodge, M. J., MacNeil, K. M., Tessier, T. M., Weinberg, J. B., & Mymryk, J. S. (2021). Emerging antiviral therapeutics for human adenovirus infection: Recent developments and novel strategies. Antiviral Research, 188, 105034. 10.1016/j.antiviral.2021.105034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benetti, E., Tita, R., Spiga, O., Ciolfi, A., Birolo, G., Bruselles, A., Doddato, G., Giliberti, A., Marconi, C., Musacchia, F., Pippucci, T., Torella, A., Trezza, A., Valentino, F., Baldassarri, M., Brusco, A., Asselta, R., Bruttini, M., Furini, S., … Pinto, A. M. (2020). ACE2 gene variants may underlie interindividual variability and susceptibility to COVID‐19 in the Italian population. European Journal of Human Genetics : EJHG, 28(11), 1602–1614. 10.1038/s41431-020-0691-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan, P. L., Jacobson, R. M., Poland, G. A., Jacobsen, S. J., & Pankratz, V. S. (2001). Twin studies of immunogenicity—Determining the genetic contribution to vaccine failure. Vaccine, 19(17–19), 2434–2439. 10.1016/S0264-410X(00)00468-0 [DOI] [PubMed] [Google Scholar]
- 38.NIH . (n.d.). Scientists discover genetic and immunologic underpinning of some cases of severe COVID‐19. https://www.nih.gov/news-events/news-releases/scientists-discover-genetic-immunologic-underpinnings-some-cases-severe-covid-19
- 39.Poland, G. A., Kennedy, R. B., McKinney, B. A., Ovsyannikova, I. G., Lambert, N. D., Jacobson, R. M., & Oberg, A. L. (2013). Vaccinomics, adversomics, and the immune response network theory: Individualized vaccinology in the 21st century. Seminars in Immunology, 25(2), 89–103. 10.1016/j.smim.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, G., Carter, B., Bricken, T., Jain, S., Viard, M., Carrington, M., & Gifford, D. K. (2020). Computationally optimized SARS‐CoV‐2 MHC Class I and II vaccine formulations predicted to target human haplotype distributions. Cell Systems, 11(2), 131–144.e6.e6. 10.1016/j.cels.2020.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banerji, A., Wickner, P. G., Saff, R., Stone, C. A. J., Robinson, L. B., Long, A. A., Wolfson, A. R., Williams, P., Khan, D. A., Phillips, E., & Blumenthal, K. G. (2021). mRNA vaccines to prevent COVID‐19 disease and reported allergic reactions: Current evidence and suggested approach. The Journal of Allergy and Clinical Immunology. In Practice, 9(4), 1423–1437. 10.1016/j.jaip.2020.12.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butt, G. F., Hassan, A., Wallace, G. R., Kinoshita, S., Ahmad, S., Ueta, M., & Rauz, S. (2021). Human leukocyte antigen B*0702 is protective against ocular Stevens–Johnson syndrome/toxic epidermal necrolysis in the UK population. Scientific Reports, 11(1), 2928. 10.1038/s41598-021-82400-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.NIH . (2020). NIH Wide Strategic Plan for COVID‐19 Research. https://www.nih.gov/sites/default/files/research-training/initiatives/covid-19-strategic-plan/coronavirus-strategic-plan-20200713.pdf
- 44.Newport, M. J., Goetghebuer, T., Weiss, H. A., Whittle, H., Siegrist, C.‐A., & Marchant, A. (2004). Genetic regulation of immune responses to vaccines in early life. Genes and Immunity, 5(2), 122–129. 10.1038/sj.gene.6364051 [DOI] [PubMed] [Google Scholar]
- 45.Voysey, M., Costa Clemens, S. A., Madhi, S. A., Weckx, L. Y., Folegatti, P. M., Aley, P. K., Angus, B., Baillie, V. L., Barnabas, S. L., Bhorat, Q. E., Bibi, S., Briner, C., Cicconi, P., Clutterbuck, E. A., Collins, A. M., Cutland, C. L., Darton, T. C., Dheda, K., Dold, C., … Zuidewind, P. (2021). Single‐dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV‐19 (AZD1222) vaccine: A pooled analysis of four randomised trials. The Lancet, 397(10277), 881–891. 10.1016/S0140-6736(21)00432-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reichmuth, A. M., Oberli, M. A., Jaklenec, A., Langer, R., & Blankschtein, D. (2016). mRNA vaccine delivery using lipid nanoparticles . Therapeutic Delivery, 7(5), 319–334. 10.4155/tde-2016-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samaridou, E., Heyes, J., & Lutwyche, P. (2020). Lipid nanoparticles for nucleic acid delivery: Current perspectives. Advanced Drug Delivery Reviews, 154–155, 37–63. 10.1016/j.addr.2020.06.002 [DOI] [PubMed] [Google Scholar]
- 48.Schultze, J. L., & Aschenbrenner, A. C. (2021). COVID‐19 and the human innate immune system. Cell, 184(7), 1671–1692. 10.1016/j.cell.2021.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machhi, J., Shahjin, F., Das, S., Patel, M., Abdelmoaty, M. M., Cohen, J. D., Singh, P. A., Baldi, A., Bajwa, N., Kumar, R., Vora, L. K., Patel, T. A., Oleynikov, M. D., Soni, D., Yeapuri, P., Mukadam, I., Chakraborty, R., Saksena, C. G., Herskovitz, J., … Kevadiya, B. D. (2021). Nanocarrier vaccines for SARS‐CoV‐2. Advanced Drug Delivery Reviews, 171, 215–239. 10.1016/j.addr.2021.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Custers, J., Kim, D., Leyssen, M., Gurwith, M., Tomaka, F., Robertson, J., Heijnen, E., Condit, R., Shukarev, G., Heerwegh, D., van Heesbeen, R., Schuitemaker, H., Douoguih, M., Evans, E., Smith, E. R., & Chen, R. T. (2021). Vaccines based on replication incompetent Ad26 viral vectors: Standardized template with key considerations for a risk/benefit assessment. Vaccine, 39(22), 3081–3101. 10.1016/j.vaccine.2020.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollet, J., Chen, W., & Strych, U. (2021). Recombinant protein vaccines, a proven approach against coronavirus pandemics. Advanced Drug Delivery Reviews, 170, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan, W.‐L., Shiao, M.‐S., Hui, R. C.‐Y., Su, S.‐C., Wang, C.‐W., Chang, Y.‐C., & Chung, W.‐H. (2017). HLA association with drug‐induced adverse reactions. Journal of Immunology Research, 2017, 1. 10.1155/2017/3186328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips, E. J., Sukasem, C., Whirl‐Carrillo, M., Müller, D. J., Dunnenberger, H. M., Chantratita, W., Goldspiel, B., Chen, Y.‐T., Carleton, B. C., George, A. L. J., Mushiroda, T., Klein, T., Gammal, R. S., & Pirmohamed, M. (2018). Clinical Pharmacogenetics Implementation Consortium Guideline for HLA genotype and use of carbamazepine and oxcarbazepine. 2017 update. Clinical Pharmacology and Therapeutics, 103(4), 574–581. 10.1002/cpt.1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karnes, J. H., Rettie, A. E., Somogyi, A. A., Huddart, R., Fohner, A. E., Formea, C. M., Ta Michael Lee, M., Llerena, A., Whirl‐Carrillo, M., Klein, T. E., Phillips, E. J., Mintzer, S., Gaedigk, A., Caudle, K. E., & Callaghan, J. T. (2021). Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA‐B genotypes and phenytoin dosing: 2020 update. Clinical Pharmacology and Therapeutics, 109(2), 302–309. 10.1002/cpt.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.World Health Organization (2021). COVID‐19 vaccine tracker and landscape.
- 56.Ethnic factors in the acceptability of foreing clinical data (ICH E5 R1). https://www.fda.gov/regulatory-information/search-fda-guidance-documents/e5-ethnic-factors-acceptability-foreign-clinical-data
- 57.Vojdani, A., & Kharrazian, D. (2020). Potential antigenic cross‐reactivity between SARS‐CoV‐2 and human tissue with a possible link to an increase in autoimmune diseases. Clinical Immunology, 217, 108480. 10.1016/j.clim.2020.108480 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


