Abstract
Background
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection-induced inflammatory responses are largely responsible for the death of novel coronavirus disease 2019 (COVID-19) patients. However, the mechanism by which SARS-CoV-2 triggers inflammatory responses remains unclear. Here, we aimed to explore the regulatory role of SARS-CoV-2 spike protein in infected cells and attempted to elucidate the molecular mechanism of SARS-CoV-2-induced inflammation.
Methods
SARS-CoV-2 spike pseudovirions (SCV-2-S) were generated using the spike-expressing virus packaging system. Western blot, mCherry-GFP-LC3 labeling, immunofluorescence, and RNA-seq were performed to examine the regulatory mechanism of SCV-2-S in autophagic response. The effects of SCV-2-S on apoptosis were evaluated by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), Western blot, and flow cytometry analysis. Enzyme-linked immunosorbent assay (ELISA) was carried out to examine the mechanism of SCV-2-S in inflammatory responses.
Results
Angiotensin-converting enzyme 2 (ACE2)-mediated SCV-2-S infection induced autophagy and apoptosis in human bronchial epithelial and microvascular endothelial cells. Mechanistically, SCV-2-S inhibited the PI3K/AKT/mTOR pathway by upregulating intracellular reactive oxygen species (ROS) levels, thus promoting the autophagic response. Ultimately, SCV-2-S-induced autophagy triggered inflammatory responses and apoptosis in infected cells. These findings not only improve our understanding of the mechanism underlying SARS-CoV-2 infection-induced pathogenic inflammation but also have important implications for developing anti-inflammatory therapies, such as ROS and autophagy inhibitors, for COVID-19 patients.
Keywords: SARS-CoV-2, Inflammation, Autophagy, Apoptosis, Reactive oxygen species
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; COVID-19, coronavirus disease 2019; ACE2, angiotensin-converting enzyme 2; ROS, reactive oxygen species; MERS-CoV, middle east respiratory syndrome coronavirus; IL, interleukin; TNF-α, tumor necrosis factor-alpha; Co-IP, Co-immunoprecipitation; RT-PCR, reverse transcription polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; DEGs, differentially expressed genes; KEGG, kyoto encyclopedia of genes and genomes; 3-MA, 3-methyladenine
1. Introduction
Since its outbreak in January 2020, the novel coronavirus disease 2019 (COVID-19), caused by the highly contagious severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has spread worldwide and become a serious public health problem [1,2]. SARS-CoV-2 mainly infects the respiratory tract and lungs and leads to a new type of coronavirus pneumonia [3,4]. As of August 2021, over 198 million confirmed cases and 4 million deaths had been reported worldwide. The rapid spread and high mortality rate have generated huge concerns to find effective methods to reduce SARS-CoV-2 transmission and develop specific treatment options for its infection. Currently, although many vaccines are being used to prevent the spread of SARS-CoV-2, the therapeutic methods for treating SARS-CoV-2 infections are mainly conventional antiviral drugs. Thus, no specific therapeutic agent is available for the resulting complications, such as dysregulated immune and inflammatory responses. Therefore, exploring the pathogenic mechanism of SARS-CoV-2 infection is important for developing effective drugs for treating COVID-19.
Autophagy, triggered by endoplasmic reticulum stress and the unfolded protein response, is a cellular degradation process in which cytoplasmic materials such as proteins, lipids, and organelles are degraded by lysosomes [5,6]. Autophagy has been recognized as a cell defense and survival mechanism since cellular components are engulfed by double membrane autophagosomes and transported to lysosomes for degradation to generate nutrients for deprived cells [7,8]. Alternatively, autophagy can induce irreversible autophagic cell death and autophagy-dependent apoptosis [9]. Excessive autophagy may lead to cell death, and it has been regularly observed in dying cells, suggesting that autophagy may be a mode of cell death or simply a failed attempt to rescue stressed cells from death [8,10]. Therefore, moderate levels of autophagy can protect against cell death but excessive autophagy may result in autophagic cell death [[11], [12], [13]]. However, considering the complexity of autophagy in different physiological and pathological conditions, further quantitative studies are needed to determine the selected boundary threshold of autophagy and its role in cell survival and death.
Viral infections frequently cause severe inflammatory responses that may play protective or destructive roles in innate immune responses against viruses and tissue damage [[14], [15], [16]]. Similarly, SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) infections are all associated with dysregulated immune, inflammatory responses, and multi-organ failure [17,18]. Clinically, SARS-CoV-2 primarily infects the respiratory tract and causes severe lung injury and inflammatory responses, ultimately leading to severe systemic inflammatory responses [19,20]. Substantial evidence has revealed that dysregulated host immune responses and inflammation-induced cytokine storms, including the production of interleukin (IL)-6, tumor necrosis factor-alpha (TNF-α), granulocyte colony-stimulating factor, IL-1β, and IL-7, are associated with worsening clinical outcomes in COVID-19 patients [21,22]. However, the molecular mechanisms underlying the uncontrolled release of inflammatory cytokines in SARS-CoV-2 infection have not been fully elucidated. Notably, autophagy performs an important role in the regulation of inflammatory signaling [23]. Some studies have reported that autophagy plays a negative regulatory role in inflammatory responses, whereas a few others have suggested that autophagy can promote the release of inflammatory factors [[24], [25], [26], [27], [28]]. Thus, autophagy may play a balancing role in supporting inflammatory responses while simultaneously preventing excessive inflammatory responses [29]. As most RNA viral infections, including respiratory syncytial virus, hepatitis C virus, and human immunodeficiency virus type-1 (HIV), can induce autophagy [[30], [31], [32]], it is therefore valuable to further explore whether SARS-CoV-2 infection can also cause autophagy, and the regulatory role of autophagy in SARS-CoV-2 infection-induced inflammation.
In this study, we demonstrated that SARS-CoV-2 spike induced autophagy through up-regulating the intracellular reactive oxygen species (ROS) and subsequently inhibiting PI3K/AKT/mTOR axis, leading to the inflammatory response and apoptosis in infected cells. These findings reveal that the autophagy induced by SARS-CoV-2 spike destroys cell homeostasis and results in autophagic death. Since the proliferative process is highly similar across different RNA virus, this study might advance our understanding of the pathogenic mechanisms of SARS-CoV-2 infections as well as provide insight into multiple viral infections and therapeutic strategies.
2. Materials and methods
2.1. Cell culture and transfection
HEK293T, Vero E6, and 16HBE cells were obtained from ATCC, and were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Gibco, USA). HMEC-1 cell line was obtained from ATCC, and it was cultured in endothelial cell medium supplemented with 10% FBS and 1% endothelial cell growth factor (ScienCell, USA). The identities of all cell lines were confirmed using short tandem repeat (STR) profiling analysis. LipoFilter (Hanbio, China) was used for transfection according to manufacturer's protocol. HEK293T-hACE2 cells were generated by transfecting HEK293T cells with hACE2-expressing lentiviruses obtained from HEK293T cells co-transfected with pCDH-CMV-hACE2-EF1-Puro, psPAX2, and pMD2.G.
2.2. Western blotting and co-immunoprecipitation (Co-IP)
Total cell proteins were extracted and boiled in RIPA buffer (Abcam, USA), and 10 μg protein samples were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins were electrophoretically blotted onto polyvinylidene fluoride membranes (Millipore, Germany) and probed with anti-ACE2, anti-LC3, anti-p62, anti-Bcl-2, anti-Bax, anti-phospho-mTOR, anti-mTOR, anti-phospho-STAT3, anti-STAT3, anti-phospho-ERK1/2, anti-ERK1/2, anti-phospho-AKT, anti-AKT, anti-Flag, and anti-GAPDH antibodies (Abcam). After incubating with an HRP-labeled goat anti-rabbit IgG or an HRP-labeled goat anti-mouse IgG, the protein expression levels were visualized using the ECL chemiluminescence reagent (Millipore). Detailed information on the antibodies is summarized in Table S1. All Western blot bands were evaluated using Gel-Pro Analyzer 4.0 and quantified by normalization to GAPDH. For Co-IP, whole-cell lysates of Vero E6, 16HBE, and HMEC-1 cells transfected with pCDH-CMV-SARS-CoV-2-S-Flag were successively incubated with ACE2 monoclonal antibodies for 6 h, followed by capturing with protein A/G agarose beads for 16 h. Bound proteins were then washed with lysis buffer, resuspended in protein sample buffer, and analysed using Western blotting.
2.3. Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells using an E.Z.N.A. HP Total RNA Kit (Omega, USA) according to the manufacturer's instructions. RNA was reverse transcribed into cDNA using the M-MLV Reverse Transcriptase (Omega). PCR was performed with KOD-Plus-Neo enzyme (TOYOBO, Japan) following manufacturer's instructions. Briefly, a total of 10 ng cDNA was used as template for each PCR amplification. All PCR reactions were carried out at an annealing temperature of 58 °C and cycle number of 35. The ACE2 primers were 5′-ATGTCAAGCTCTTCCTGGCTCC-3′ and 5′-AAAGGAGGTCTGAACATCATCAGTG-3′. The GAPDH primers were 5’-CGTCTTCACCACCATGGAGA-3′ and 5′-CGGCCATCACGCCACAGTTT-3′. The primer sequences of IL-6, IL-8, and TNF-α used in the study have been published previously [33].
2.4. SARS-CoV-2 spike pseudovirions production and infection
SARS-CoV-2 spike pseudovirions were generated and used to infect cells as described previously [34]. Briefly, HEK293T cells were cultured in 10-cm plates and incubated with DMEM supplemented with 10% FBS. The following day, cells were co-transfected with package plasmids pLP1 and pLP2, envelop plasmid pCAG-SARS-CoV-2-S-Flag or pCAG-MCS-Flag, and transfer plasmid pCDH-CMV-Luciferase-CopGFP or pCDH-CMV-Luciferase using LipoFilter according to the manufacturer's instructions. The supernatants were collected at 48 h post transfection and centrifuged at 1000g for 10 min to obtain pellet cell debris, filtered through a 0.45 μm filter, aliquoted, and stored at −80 °C.
The viral titers were determined using plaque assays as described previously [35]. To transduce target cells with SARS-CoV-2 spike pseudovirions, HEK293T, HEK293T-hACE2, Vero E6, 16HBE, or HMEC-1 cells were seeded into 12-well plates. After adding 10 μg/mL polybrene to the viral dilution, cells were infected with the SARS-CoV-2 spike pseudovirions (multiplicity of infection, MOI = 5) for 24 h. The infection efficiency of SARS-CoV-2 spike pseudovirions was viewed using a Nikon Eclipse Ti2-E fluorescence microscope (Nikon, Japan) at an excitation wavelength of 488 nm or it was measured using a luciferase substrate that determines firefly luciferase activity using a microplate reader (TECAN, Germany).
2.5. Total RNA isolation and expression analysis
HEK293T-hACE2 and Vero E6 cells were infected with or without SARS-CoV-2 spike pseudovirions (MOI = 5) for 48 h. Total RNA was extracted using TRIzol (Invitrogen, USA) according to the manufacturer's instructions. RNA-seq libraries were constructed, and the transcriptome was sequenced on an Illumina HiSeq 2500 platform by Beijing Biomarker Technology Corporation as our previously report [36]. All the raw data have been deposited under the Gene Expression Omnibus (GEO) accession number GSE169370.
2.6. Trypan blue staining
HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells were infected with or without SARS-CoV-2 spike pseudovirions (MOI = 5) for 48 h. Cells (1 × 105) were collected and treated with 0.4% trypan blue probe for 2 min. The number of trypan blue-positive cells was calculated using a hemocytometer (Cellometer, USA).
2.7. Enzyme-linked immunosorbent assay (ELISA)
Culture media were collected from HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells infected with or without SARS-CoV-2 spike pseudovirions (MOI = 5) and cells treated with SARS-CoV-2 spike pseudovirions plus 3-MA (10 mM). The IL-6, IL-8, and TNF-α concentrations were evaluated using ELISA kits (Novus, USA) according to the manufacturer's instructions. Absorbance was measured using a microplate reader (TECAN), and the concentration was calculated according to the standard concentration curve. Detailed information on the inhibitors and activator is summarized in Table S2.
2.8. Intracellular ROS detection
Intracellular ROS levels were detected as previously described [36]. Briefly, adherent HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells treated with SARS-CoV-2 spike pseudovirions for 24 h (MOI = 5) were incubated with a DCFH-DA probe at 37 °C for 30 min and washed with serum-free medium in the dark. Intracellular ROS levels were visualized under a Nikon EclipseTi2-E fluorescence microscope (Nikon) or detected using a fluorescent reader (TECAN) at an excitation wavelength of 488 nm.
2.9. Flow cytometry analysis
For flow cytometry analysis of cell surface ACE2, 100 μL of HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells (1 × 106) was digested to a single-cell suspension, and incubated with primary rabbit anti-human ACE2 antibodies followed by FITC-conjugated IgG secondary antibodies. Rabbit IgG was used as the isotype control. Cells were then washed thrice with centrifugation at 400g and analysed using a FC500 flow cytometer (Beckman, Germany) or CytoFLEX flow cytometer (Beckman). For apoptosis analysis, HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells (1 × 105) treated with or without SARS-CoV-2 spike pseudovirions or recombinant spike proteins (10 μg/mL) in parallel with the addition of an equal volume of DMSO or 3-MA (10 mM) for 24 h were labeled with FITC-conjugated Annexin V and 7-AAD (Biolegend, USA) in the dark for 15 min and analysed using an FC500 flow cytometer (Beckman) or CytoFLEX flow cytometer (Beckman).
2.10. Immunofluorescence assay
Immunofluorescence assay was performed as described previously [36]. Briefly, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells were grown on μ-Dish (Ibidi, Germany) and infected with or without SARS-CoV-2 spike pseudovirions in parallel with addition of 5 mM ROS inhibitor (Sigma, USA) or 10 μM MHY 1485 (MCE, China) treatment. Cells were then fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, blocked in 5% BSA-PBS solution, and incubated with primary anti-human LC3 antibodies. FITC-conjugated IgG secondary antibodies were used for LC-3 labeling. Cell nuclei were stained with Hoechst 33342, and imaged with a 63× objective using an LSM 880 confocal microscope (Zeiss, Germany).
2.11. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells were seeded onto μ-Dish (Ibidi) and infected with or without SARS-CoV-2 spike pseudovirions in parallel with the addition of an equal volume of DMSO or 3-MA (10 mM) for 24 h. Cells were then fixed with 4% paraformaldehyde, permeabilized in 0.2% Triton X-100, and subjected to a TUNEL assay using an in situ TUNEL apoptosis detection kit (Abbkine, USA), according to the manufacturer's instructions. Finally, cell nuclei were stained with DAPI and visualized with a 40× objective using an LSM 880 confocal microscope (Zeiss). The apoptotic cells were calculated by counting the total number of TUNEL-stained nuclei (FITC-positive).
2.12. Immunohistochemistry
Five-week-old wild-type BALB/c mice were maintained in the Laboratory Animal Center of Sun Yat-sen University. All animal studies were approved by the Institutional Animal Care and Use Committees of the Sun Yat-sen University (approval number: SYSU-IACUC-2020-B0778). Mice were sacrificed, and the trachea, lung, and liver were harvested for immunohistochemical analysis according to standard procedures [37]. The images from tissue sections were obtained and analysed using a Nikon Eclipse Ti2-E microscope (Nikon).
2.13. Statistical analysis
All results are presented as the mean ± SD of three entirely independent experiments derived from separate transfection and treatment rounds. The differences among groups were analysed using Student's t-test when only two groups were compared. For comparison between more than two groups, all data were first analysed for adherence to a normal distribution and then subjected to one-way ANOVA followed by Tukey's post hoc test. *P < 0.05, **P < 0.01, and ***P < 0.001 were considered statistically significant. All data were analysed using SPSS (version 22.0) statistical software.
3. Results
3.1. ACE2 is expressed in human bronchial epithelial and microvascular endothelial cells and mediates SARS-CoV-2 infection
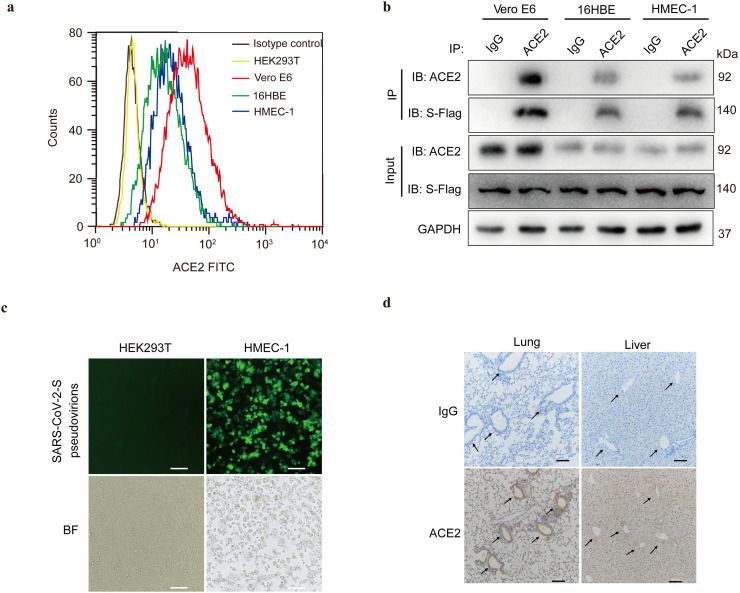
ACE2, first identified in 2000, functions as a cellular receptor for the spike protein of coronaviruses to facilitate viral entry into target cells [38,39]. To investigate the underlying pathogenic mechanisms of SARS-CoV-2, which is considered to be the third most pathogenic coronavirus [3], we first examined the expression of ACE2 in HEK293T, Vero E6, human bronchial epithelial cells 16HBE, human microvascular endothelial cells (HMEC-1), nasopharyngeal carcinoma cells (6-10B, 5-8F, and S26), and megakaryocytic leukemia cells (Dami). The protein expression of ACE2 was observed in Vero E6, 16HBE, and HMEC-1 cells but not in HEK293T, 6-10B, 5-8F, S26, and Dami cells (Fig. S1a), which corresponded to the ACE2 transcripts in Vero E6, 16HBE, and HMEC-1 cells (Fig. S1b). Importantly, flow cytometry revealed ACE2 expression on the cell surfaces of Vero E6, 16HBE, and HMEC-1 cells (Fig. 1 a). To further verify the binding interaction between SARS-CoV-2 spike protein and ACE2 in ACE2-expressing cells, we transfected the cells with Flag-labeled spike protein. Co-IP analysis revealed that the spike protein of SARS-CoV-2 co-immunoprecipitated with endogenous ACE2 in Vero E6, 16HBE, and HMEC-1 cells (Fig. 1b). Meanwhile, ACE2 expression mediated SARS-CoV-2 spike pseudovirions infection in Vero E6, 16HBE, and HMEC-1 cells (Fig. 1c and Fig. S1c). Notably, immunohistochemistry further proved that ACE2 was expressed in the tracheal and bronchial epithelial cells and vascular endothelial cells of the lungs and liver of mice (Fig. 1d and Fig. S1d).
Fig. 1.
ACE2 mediates SARS-CoV-2 spike pseudovirions entry into human bronchial epithelial and microvascular endothelial cells. (a) Cell surface expression of ACE2 on HEK293T, Vero E6, 16HBE, and HMEC-1, determined using flow staining. Rabbit IgG was used as isotype control. (b) Vero E6, 16HBE, and HMEC-1 cells were transfected with Flag-tagged SARS-CoV-2 spike protein for 48 h. The interaction between SARS-CoV-2 spike protein and ACE2 was detected by IP with an ACE2 antibody, and this was followed by detection with the Flag and ACE2 Western blotting antibodies. (c) HEK293T and HMEC-1 cells were infected with GFP-labeled SARS-CoV-2 spike pseudovirions. Fluorescence microscope imaging was performed to evaluate the infection efficiency after 24 h of infection (Scale bar, 40 μm). BF, bright field. (d) Immunohistochemical staining showing the expression and distribution of ACE2 in different tissues of mice (Scale bar, 100 μm). Arrows indicate blood vessels. Rabbit IgG was used as isotype control. All results were determined from three independent experiments.
3.2. SARS-CoV-2 spike activates autophagy and pro-apoptotic responses in ACE2-expressing cells
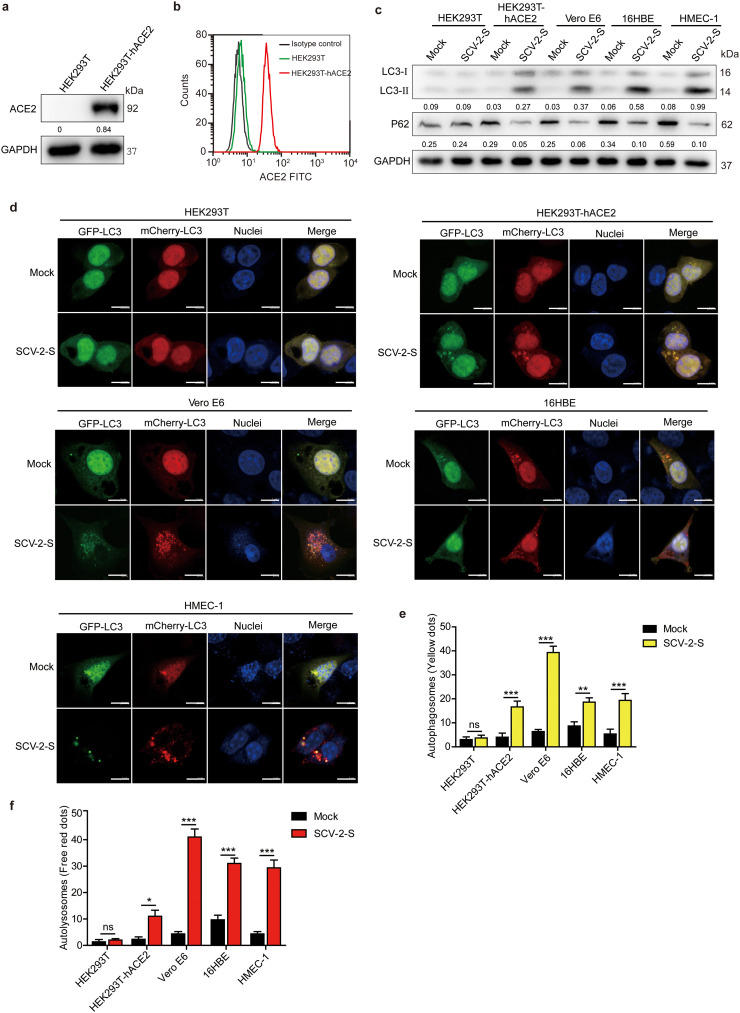
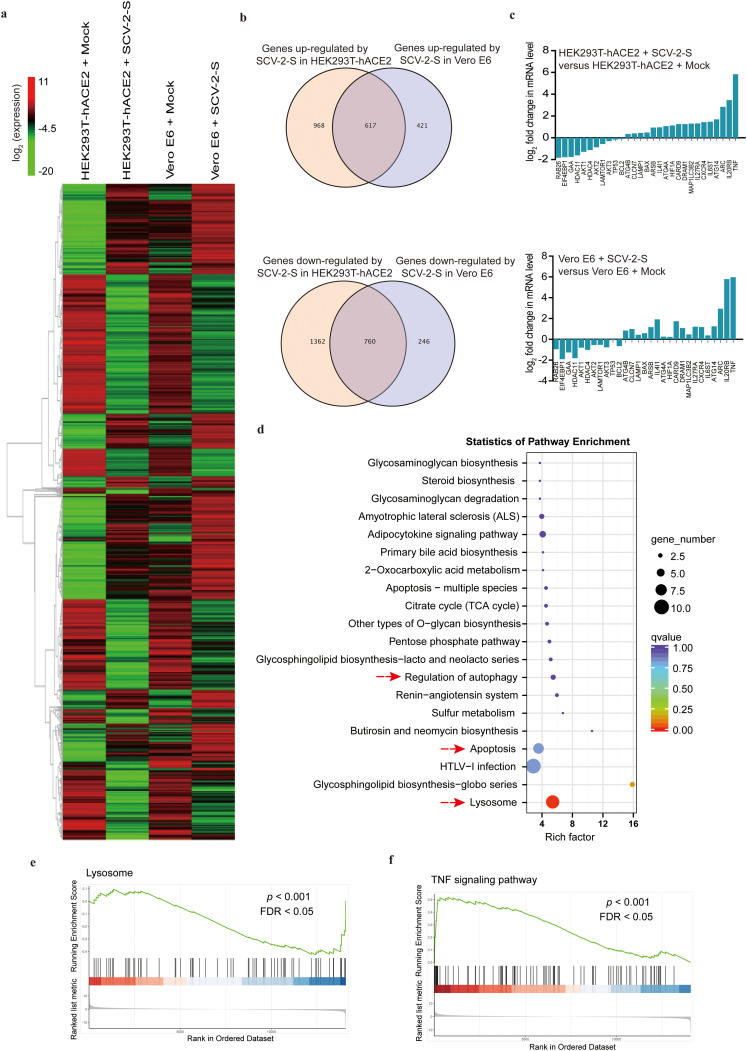
Several viral infections have been reported to activate autophagy, which is beneficial to viral replication [40,41]. To determine whether autophagy is triggered upon SARS-CoV-2 infection, HEK293T cells were transfected with human ACE2-expressing lentivirus to construct stable HEK293T-hACE2 cells (Fig. 2 a and Fig. 2b). We observed that the autophagosome marker LC3-II, proteolytically cleaved and lipidated from LC3, was increased in HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells that were infected with SARS-CoV-2 spike pseudovirions or treated with recombinant spike protein for 24 h (Fig. 2c and Fig. S2a). Consistently, SARS-CoV-2 spike pseudovirions infection and recombinant spike protein treatment reduced p62 expression in ACE2-expressing HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells, but not in control HEK293T cells (Fig. 2c and Fig. S2a). Subsequently, the cells were transfected with a double-tagged pmCherry-GFP-LC3 plasmid to visualize the progression of autophagy after SARS-CoV-2 spike pseudovirions infection. We found that following SARS-CoV-2 spike pseudovirions infection, both GFP-LC3 and mCherry-LC3 were significantly upregulated in HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells, but not in HEK293T control cells (Fig. 2d-f), indicating that SARS-CoV-2 spike induces the occurrence of autophagic responses in infected cells. Additionally, high throughput RNA-Seq analyses of differentially expressed genes (DEGs) revealed a significant overlap in the expression of altered autophagy-regulatory genes in HEK293T-hACE2 and Vero E6 cells upon SARS-CoV-2 spike pseudovirions infection (Fig. 3 a and b). The expression levels of the majority of autophagy-promoting genes, including ATG4B and MAP1LC3B2, were significantly increased in HEK293T-hACE2 and Vero E6 cells infected with SARS-CoV-2 spike pseudovirions (Fig. 3a and c). Conversely, the subset of autophagy-suppressing genes was significantly decreased in SARS-CoV-2 spike pseudovirions-treated HEK293T-hACE2 and Vero E6 cells (Fig. 3a and c). Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and gene set enrichment analysis (GSEA) of DEGs revealed that the regulation of autophagy and lysosome-related pathways were significantly enriched in SARS-CoV-2 spike pseudovirions-infected cells (Fig. 3d and e).
Fig. 2.
ACE2-mediated SARS-CoV-2 spike pseudovirions infection induces autophagic responses. (a) Western blot analysis of ACE2 expression in HEK293T-hACE2 stable cells. GAPDH was used as the loading control. ACE2/GAPDH densitometric ratios were recorded. (b) Flow cytometric analysis of cell surface expression of ACE2 on HEK293T-hACE2 stable cells. Rabbit IgG was used as an isotype control. (c) Western blotting for autophagy markers LC3 and p62 after treatment with SARS-CoV-2 spike pseudovirions for 24 h. LC3-II/GAPDH and p62/GAPDH densitometric ratios were recorded. (d) HEK293T, HEK293T-hACE2, Vero E6, 16HBE and HEMC-1 were transfected with mCherry-GFP-LC3 followed by SARS-CoV-2 spike pseudovirions treatment for 24 h. GFP-LC3 and mCherry-LC3 puncta were visualized using confocal microscopy. Hoechst 33342 was used to stain the nuclei of cells (blue) (Scale bar, 10 μm). (e, f) Statistical bar charts of autophagosome and autolysosome, as observed in SARS-CoV-2-S pseudovirions-infected HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells. SCV-2-S, SARS-CoV-2 spike pseudovirions. All results were determined from three independent experiments. Data represent mean ± SD (*P < 0.05; **P < 0.01; ***P < 0.001).
Fig. 3.
SARS-CoV-2 spike modulates the expression of a subset of autophagy and apoptotic genes. (a) Heatmap displaying the hierarchical clustering of genes in HEK293T-hACE2 and Vero E6 cells in response to SARS-CoV-2 spike pseudovirions infection. Gene expression values are indicated by the color intensities of red (upregulated) or green (downregulated). (b) Alteration of the expression of potential genes following SARS-CoV-2 spike pseudovirions infection; numbers indicate the quantity of genes in each DEG subset. (c) Column diagram represents the expression of genes correlated with autophagy, apoptosis, and inflammation following SARS-CoV-2 spike pseudovirions infection in HEK293T-hACE2 and Vero E6 cells. (d) KEGG pathway enrichment analysis of the DEGs revealed the top 20 enriched pathways in SARS-CoV-2 spike pseudovirions-treated HEK293T-hACE2 and Vero E6 cells. (e, f) Select enrichment plots highlighting the enriched lysosome-related gene (e) and TNF signaling pathways (f) identified by GSEA. SCV-2-S, SARS-CoV-2 spike pseudovirions.
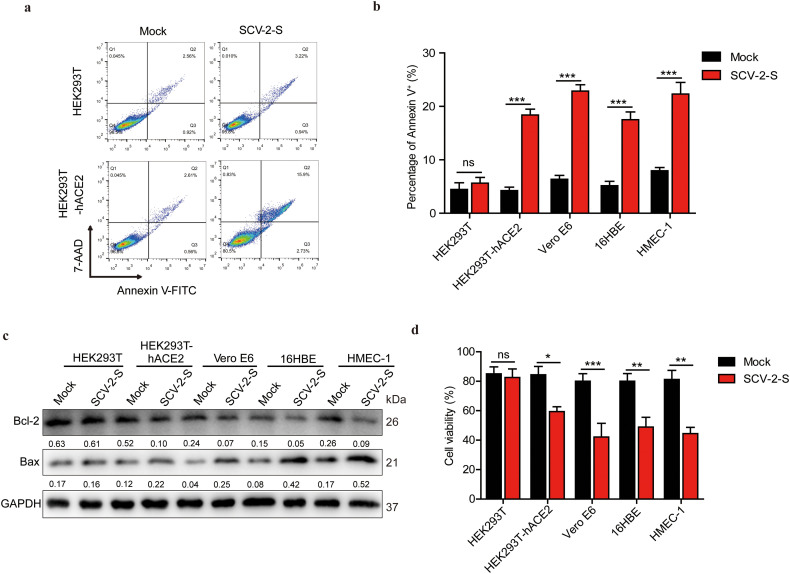
Meanwhile, flow cytometry analysis demonstrated that SARS-CoV-2 spike pseudovirions and spike protein did not alter apoptosis in HEK293T control cells, which was in striking contrast to the result that late apoptotic (FITC+/7-AAD+) populations were significantly enhanced in SARS-CoV-2 spike pseudovirions-infected and recombinant spike protein-treated HEK293T-hACE2 cells on the stable expression of hACE2 (Fig. 4 a and Fig. S2b). Annexin V-FITC staining also confirmed that SARS-CoV-2 spike pseudovirions increased the apoptosis of Vero E6, 16HBE, and HMEC-1 cells (Fig. 4b). Consistently, RNA-Seq analysis indicated that SARS-CoV-2 spike pseudovirions upregulated the majority of genes correlated with pro-apoptotic responses, including Bax and ARC, and downregulated the pro-survival gene Bcl-2 in HEK293T-hACE2 and Vero E6 cells (Fig. 3a and c), which was further confirmed by Western blot analysis in HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells (Fig. 4c). KEGG pathway enrichment analysis of DEGs also revealed that the apoptosis-related pathway was significantly enriched in SARS-CoV-2 spike pseudovirions-infected cells (Fig. 3d). Moreover, trypan blue staining indicated that SARS-CoV-2 spike pseudovirions infection resulted in a significantly decreased cell viability in ACE2-expressing cells (Fig. 4d). Thus, these findings suggest that SARS-CoV-2 spike may induce autophagy and therefore play a pro-death regulatory role in ACE2-expressing cells.
Fig. 4.
ACE2-mediated SARS-CoV-2 spike pseudovirions infection induces pro-apoptotic responses. (a-c) Cell apoptosis of HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells after 24 h of treatment with SARS-CoV-2 spike pseudovirions, measured using flow cytometry (a) Annexin V-FITC/7-AAD double binding, (b) Annexin V-FITC single staining, and (c) Western blot analysis of Bcl-2 and Bax expression. Bcl-2/GAPDH and Bax/GAPDH densitometric ratios were recorded. (d) Cell viabilities of HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells after 24 h of treatment with SARS-CoV-2 spike pseudovirions were determined with trypan blue staining. SCV-2-S, SARS-CoV-2 spike pseudovirions. All results were collected in three independent experiments. Data represent mean ± SD (*P < 0.05; **P < 0.01; ***P < 0.001).
3.3. SARS-CoV-2 spike induces ROS upregulation to inhibit the PI3K/AKT/mTOR pathway to promote autophagy
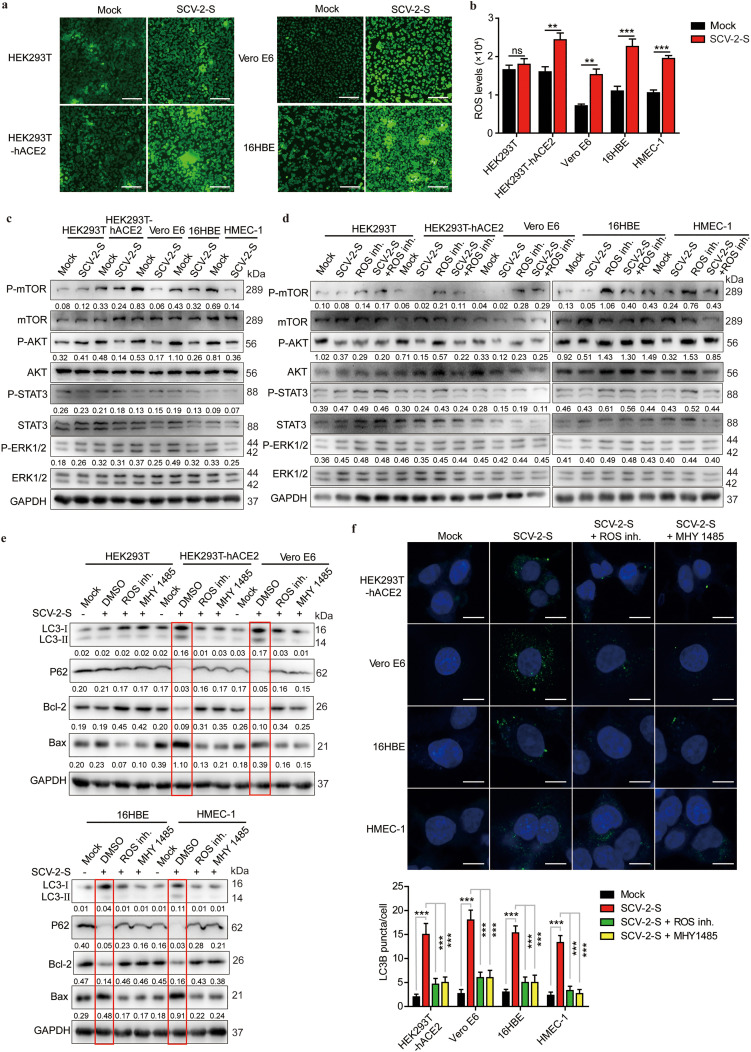
Many studies have shown that ROS levels are aberrantly upregulated in virus-infected cells, which may be responsible for cell apoptosis [42,43]. To further investigate the mechanism by which SARS-CoV-2 spike promotes autophagy and pro-apoptotic responses in ACE2-expressing cells, we evaluated the intracellular ROS levels using DCFH-DA probe, and we observed that SARS-CoV-2 spike pseudovirions treatment did not significantly alter the intracellular ROS levels in HEK293T control cells. However, the intracellular ROS levels of HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells were significantly increased after SARS-CoV-2 spike pseudovirions treatment (Fig. 5 a and b). Notably, consistent with previous results showing that ROS upregulation induced autophagy through PI3K/AKT/mTOR inhibition [44,45], ROS inhibition induced AKT and mTOR activation in all cells (Fig. 5c and d). However, on treatment with SARS-CoV-2 spike pseudovirions, phosphorylation/ activation of mTOR and AKT, rather than STAT3 and ERK1/2 signaling, was markedly inhibited in HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells, but this inhibition was reversed by an ROS inhibitor in ACE2-expressing cells (Fig. 5c and d), indicating that SARS-CoV-2 spike-induced ROS upregulation inhibited PI3K/AKT/mTOR activation. Since both ROS generation and mTOR suppression are positively associated with enhanced autophagy [46,47], we treated ACE2-expressing cells with SARS-CoV-2 spike pseudovirions along with an ROS inhibitor or MHY1485, an mTOR selective agonist. Thus, we further validated that SARS-CoV-2 spike pseudovirions increased LC3-II levels and decreased p62 expression in ACE2-expressing cells (Fig. 5e). Nevertheless, it was remarkable that SARS-CoV-2 spike pseudovirions infection-induced LC3-II increase and P62 decrease were abolished by an ROS inhibitor and MHY1485 (Fig. 5e). Meanwhile, SARS-CoV-2 spike pseudovirions infection-induced enhancement of apoptosis (Bcl-2 downregulation and Bax upregulation) was also reversed by an ROS inhibitor and MHY1485 (Fig. 5f), suggesting that SARS-CoV-2 spike pseudovirions may promote autophagy by upregulating intracellular ROS levels and inhibiting the PI3K/AKT/mTOR pathway. Furthermore, immunofluorescence confirmed that the ability of SARS-CoV-2 spike pseudovirions infection to induce an increase in LC3 punctas was considerably reduced in the presence of an ROS inhibitor and MHY1485 (Fig. 5f). Collectively, these data indicate that SARS-CoV-2 spike inhibits the PI3K/AKT/mTOR pathway via inducing ROS upregulation to promote autophagy.
Fig. 5.
SARS-CoV-2 spike promotes autophagy through the ROS-suppressed PI3K/AKT/mTOR pathway. (a, b) Intracellular ROS levels in HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells treated with or without SARS-CoV-2 spike pseudovirions using (a) fluorescence microscopy (Scale bar, 200 μm) and (b) fluorescence microplate reader. (c) Western blot analysis of phospho-mTOR, mTOR, phospho-AKT, AKT, phospho-STAT3, STAT3, phospho-ERK1/2, and ERK1/2 in HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells treated with or without SARS-CoV-2 spike pseudovirions. (d) Western blot analysis of phospho-mTOR, mTOR, phospho-AKT, AKT, phospho-STAT3, STAT3, phospho-ERK1/2, and ERK1/2 in SARS-CoV-2-S pseudovirions-treated, ROS-inhibited, and SARS-CoV-2 spike pseudovirions and ROS inhibitor (5 mM)-co-treated HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells. GAPDH was used as the loading control. Phospho-mTOR/GAPDH, phospho-AKT/GAPDH, phospho-STAT3/GAPDH, and phospho-ERK1/2/GAPDH densitometric ratios were recorded. (e) Western blot analysis of LC3, p62, Bcl-2, and Bax in SARS-CoV-2 spike pseudovirions-treated HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells pretreated with DMSO, ROS inhibitor (5 mM), or MHY 1485 (10 μM). GAPDH was used as the loading control. LC3-II/GAPDH, p62/GAPDH, Bcl-2/GAPDH, and Bax/GAPDH densitometric ratios were recorded. (f) LC3 expression using confocal microscopy in SARS-CoV-2 spike pseudovirions-treated HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells pretreated with DMSO, ROS inhibitor (5 mM), or MHY 1485 (10 μM). Protein expression was determined using LC3-specific antibodies followed by FITC-labeled secondary antibodies (green). Hoechst 33342 was used to stain the nuclei of cells (blue). (Scale bar, 5 μm). SCV-2-S, SARS-CoV-2 spike pseudovirions. All results were collected in three independent experiments. Data represent the mean ± SD (**P < 0.01; ***P < 0.001).
3.4. SARS-CoV-2 spike-induced autophagy promotes inflammation and apoptosis in infected cells
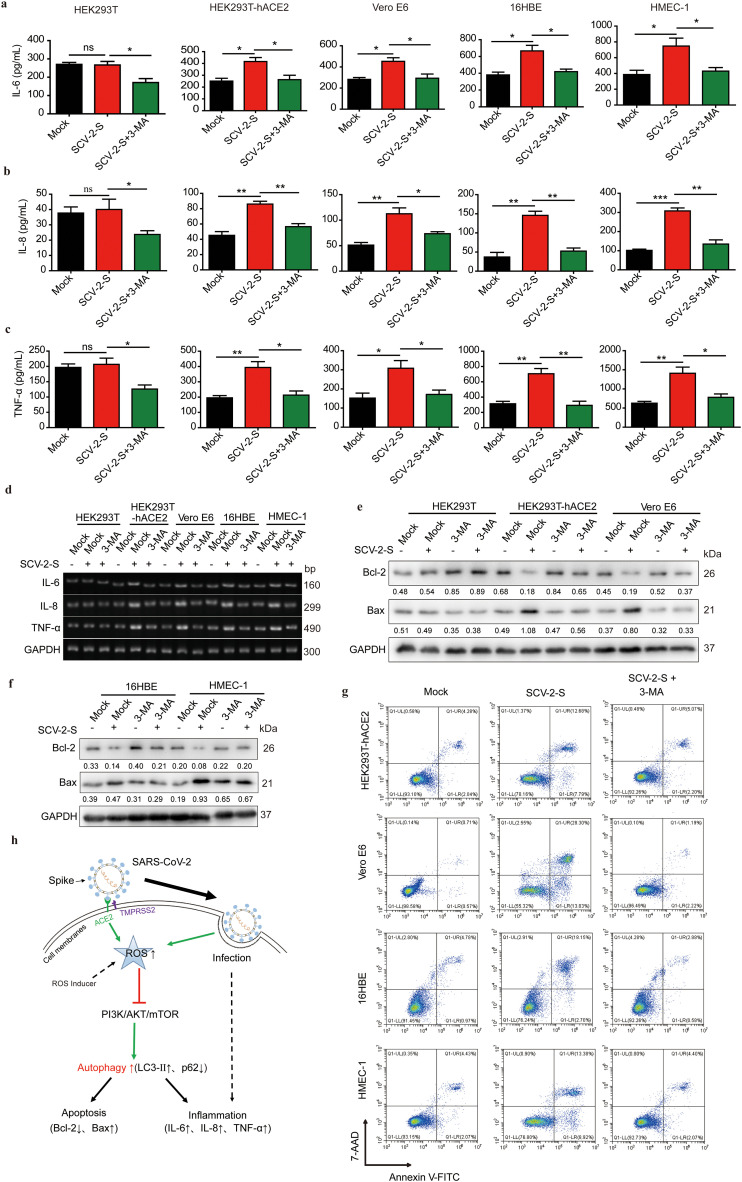
Respiratory infection caused by SARS-CoV-2 usually result in viral pneumonia and acute respiratory distress syndrome [48,49]. Meanwhile, SARS-CoV-2 infection can also trigger inflammatory responses, which in turn promote the production of pro-inflammatory cytokines, including TNF-α, IL-6, and IL-1β [49,50]. Since ACE2 was expressed in 16HBE and HMEC-1 cells and SARS-CoV-2 spike-mediated ROS upregulation induced autophagy by inhibiting the PI3K/AKT/mTOR pathway in ACE2-expressing cells, we sought to investigate whether SARS-CoV-2 spike-induced autophagy was involved in inflammatory responses in bronchial epithelial and microvascular endothelial cells. ELISA data indicated that SARS-CoV-2 spike pseudovirions treatment significantly promoted the production of pro-inflammatory cytokines, IL-6, IL-8, and TNF-α, in HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells. However, treatment with 3-methyladenine (3-MA), an autophagy inhibitor, led to a significant inhibition of SARS-CoV-2 spike pseudovirions-induced production of pro-inflammatory cytokines IL-6, IL-8, and TNF-α in theses ACE2-expressing cells (Fig. 6 a-c). These results were further confirmed by performing GSEA and RT-PCR analyses of IL-6, IL-8, and TNF-α transcripts (Fig. 3f and Fig. 6d). Similarly, RNA-Seq analyses also showed that SARS-CoV-2 spike pseudovirions infection enhanced the expression of pro-inflammatory cytokines, including TNF and IL6ST, in infected cells (Fig. 3a and c). Notably, inflammation is strongly linked to apoptosis, and it can induce apoptosis to promote cell death [51]. Autophagy can inhibit apoptosis to maintain cell survival. Alternatively, autophagy can also work with apoptosis to induce cell death [52,53]. To investigate the regulatory effects of SARS-CoV-2 spike-induced autophagy on apoptosis, we treated SARS-CoV-2 spike pseudovirions-treated cells with 3-MA, and we found that SARS-CoV-2 spike pseudovirions upregulated and downregulated Bax and Bcl-2 expression, respectively. However, this modulation was reversed by 3-MA treatment (Fig. 6e and f). Annexin V-FITC/7-AAD double staining and TUNEL staining also verified that SARS-CoV-2 spike pseudovirions-induced apoptosis (the percentage of TUNEL-positive cells) was reversed by 3-MA (Fig. 6g and Fig. S3). Altogether, these findings indicate that SARS-CoV-2 spike-induced autophagy via the ROS-suppressed PI3K/AKT/mTOR axis promotes inflammation and apoptosis in infected cells, which highlights the pathogenic regulatory role of autophagy in SARS-CoV-2 infection (Fig. 6h).
Fig. 6.
SARS-CoV-2 spike induces inflammation and apoptosis through enhanced autophagy. (a-c) The protein expression levels of IL-6, IL-8, and TNF-α in SARS-CoV-2 spike pseudovirions-infected HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells pretreated with DMSO or 3-MA (10 mM) were quantified using ELISA. (d) Semiquantitative RT-PCR analysis of IL-6, IL-8, and TNF-α expression in SARS-CoV-2 spike pseudovirions-infected HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells pretreated with DMSO or 3-MA (10 mM). (e, f) Western blot analysis of Bcl-2 and Bax in SARS-CoV-2 spike pseudovirions-treated, 3-MA-treated, and SARS-CoV-2 spike pseudovirions and 3-MA (10 mM)-treated HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells. GAPDH was used as the loading control. Bcl-2/GAPDH and Bax/GAPDH densitometric ratios were recorded. (g) Apoptotic cells were determined using Annexin V-FITC/7-AAD double staining in SARS-CoV-2 spike pseudovirions-treated, 3-MA (10 mM)-treated, and SARS-CoV-2 spike pseudovirions and 3-MA (10 mM)-treated HEK293T, HEK293T-hACE2, Vero E6, 16HBE, and HMEC-1 cells. (h) Schematic illustrating the mechanism of SARS-CoV-2 spike to promote inflammation and apoptosis in infected cells. SCV-2-S, SARS-CoV-2 spike pseudovirions. All results were collected in three independent experiments. Data represent the mean ± SD (*P < 0.05; **P < 0.01; ***P < 0.001).
4. Discussion
SARS-CoV-2 infects humans through the respiratory tract, ultimately resulting in dysfunctions of a number of systems, including the respiratory, immune, nervous, and cardiovascular systems [[54], [55], [56], [57]]. SARS-CoV-2-mediated inflammatory responses are largely responsible for multiple organ dysfunction and death of COVID-19 patients [35]. Although a recent study showed that SARS-CoV-2 spike protein stimulates IL-6 and soluble IL-6R secretion by activating the angiotensin II type 1 receptor axis in spike-transfected human hepatoma Huh7.5 and lung adenocarcinoma A549 cells [58], further exploration in normal human epithelial cells perhaps more helpful for uncovering the molecular mechanisms underlying SARS-CoV-2-triggered inflammatory responses. Here, we found that SARS-CoV-2 spike suppressed PI3K/AKT/mTOR signaling by upregulating intracellular ROS levels to enhance autophagy, thus promoting apoptotic and inflammatory responses in ACE2-expressing human bronchial epithelial and microvascular endothelial cells. This suggests that SARS-CoV-2 spike-induced autophagy does not function as a pro-survival mechanism but rather accelerates pro-apoptotic inflammatory responses and therefore disrupts cellular homeostasis. Notably, both SARS-CoV-2 spike pseudovirions and recombinant SARS-CoV-2 spike protein treatment induced apoptotic and autophagic responses in ACE2-expressing cells, suggesting that the interaction between spike protein ectodomain and ACE2 receptor initiated apoptotic and autophagic responses before viral entry. Further studies may be warranted to identify the alterations in the ACE2 molecular structure and signaling mechanisms of apoptosis and autophagy when spike protein binds to the ACE2 receptor.
Several RNA viruses can induce autophagic responses in infected cells, which are closely related to viral replication and pathogenesis [32,41,59]. Viruses have learned to manipulate the autophagic pathway, exploiting autophagosomes to facilitate viral replication [[60], [61], [62]]. Meanwhile, some studies have demonstrated that some viral proteins block the fusion of autophagosomes with lysosomes, thus inhibiting autophagic processes [63,64]. In viral infections, a balanced mechanism of self-interest may exist, that is, pathogenic proteins, such as spike proteins, trigger autophagy, whereas other accessory proteins carried by the virus may prevent the final step of autophagy and lysosome fusion, thus utilizing autophagy to accumulate viral components. Previous studies suggested that the accessory protein ORF3a encoded by SARS-CoV-2 inhibits the formation of autophagolysosomes by blocking the assembly of the SNARE complex mediated by the HOPS complex [65]. In the present study, we found that SARS-CoV-2 spike proteins induced autophagy through the ROS-suppressed PI3K/AKT/mTOR pathway in infected cells (Fig. 2 and Fig. 5), implying that spike protein may induce autophagy initiation and progression, whereas accessory protein ORF3a block the fusion of autophagosomes with lysosomes in SARS-CoV-2-infected cells. However, considering that the current research only focuses on the regulation of autophagic processes by specific proteins of SARS-CoV-2, further studies are required to determine whether multiple proteins of SARS-CoV-2, such as ORF3a and spike proteins, have a balanced mechanism to promote the accumulation of autophagosomes, which is conducive to viral replication.
As microvascular endothelial cells play a crucial role in maintaining body homeostasis, viral infection via ACE-2 receptor usually leads to inflammation and causes vascular dysfunction through microvascular thrombosis, which in turn results in organ failure [66,67]. In this study, we reported that ACE2 was expressed in human microvascular endothelial cells and that ACE2-mediated SARS-CoV-2 spike pseudovirions infection enhanced the inflammatory responses and apoptosis of microvascular endothelial cells (Fig. 1 and Fig. 6), indicating that SARS-CoV-2 infection may lead to an extensive apoptosis of microvascular endothelial cells and vascular leakage, eventually causing organ dysfunction. Most importantly, although ACE2 structure is different between humans and mice and cannot mediate SARS-CoV-2 infection in mice, the tissue specificity of ACE2 in humans and mice may be similar [68,69]. Using immunohistochemistry, we demonstrated that ACE2 was specifically enriched in epithelial cells of the trachea and pulmonary bronchus and vascular endothelial cells of the lungs and liver (Fig. 1d and Fig. S1d). This implies that SARS-CoV-2 infection not only causes an inflammation of the respiratory system but also affects the functions of many organs by destroying microvascular endothelial cells.
ROS are formed by an incomplete one-electron reduction of oxygen and mainly encompass a range of small, short-lived, and highly reactive oxygen-containing molecules, including oxygen anions, free radicals, and peroxides [70,71]. A majority of viral infections cause an increase in ROS levels in infected cells, and an increasing number of evidence suggests the role of ROS in the pathogenesis of viral infections as a factor for endothelial damage and inflammation [[72], [73], [74]]. Consistent with previous studies, we found that SARS-CoV-2 spike upregulated intracellular ROS levels and promoted the inflammatory responses in infected cells by inducing autophagy (Fig. 5 and Fig. 6), indicating that SARS-CoV-2 spike can induce cell damage by upregulating ROS. Notably, as several studies have suggested that autophagy reduces oxidative damage by decreasing ROS accumulation [75,76], future studies should determine whether the activation of autophagy can reverse ROS accumulation in SARS-CoV-2 infection. Additionally, recent studies have reported that ROS production leads to the activation of signaling cascades, including JAK/STATs, PI3K/AKT, and MAPK/ERK [77,78]. Alternatively, elevated ROS levels can also inhibit the PI3K/AKT/mTOR pathway, thus enhancing autophagy levels [79,80]. However, due to different metabolic processes in different cells, there may be a delicate balance between the promotion and inhibition of ROS for the downstream signaling pathways of autophagic responses in different cells. More precisely, ROS levels below a certain limit may activate autophagy inhibition-related pathways, while an excess ROS may induce autophagy by activating or inhibiting autophagy-related signaling in cells with different metabolic levels. Further quantitative determination of the selected boundary threshold of ROS for pathway activation and autophagy may be helpful in increasing our understanding of the pathological role of ROS upregulation induced by SARS-CoV-2 spike in infected cells.
5. Conclusion
This study is the first to reveal that SARS-CoV-2 spike induces autophagy through the intracellular ROS-suppressed PI3K/AKT/mTOR axis, which then leads to inflammatory responses and apoptosis in infected cells. Although it is clear that SARS-CoV-2 infection can induce obvious inflammatory reactions in COVID-19 patients, the presently-used anti-inflammatory drugs are not effective as the mechanisms underlying the inflammation remain unclear. Thus, our results may be helpful to develop ROS and autophagy inhibitors as adjuvant therapeutic drugs for COVID-19 patients. Due to the similarities in the infection mechanisms of viruses, these findings not only unveil the pathological mechanism of SARS-CoV-2 infection but also have broad implications in viral biology that can advance the treatment of multiple viral infections.
CRediT authorship contribution statement
FL planned and carried out experiments, analysed data, and prepared the manuscript. JL and PHW carried out some experiments, analysed data, and prepared the manuscript. NY, JH, JO, TX, XZ, TL, XH, QW, ML, LY, YL, YC, and HC performed the research and analysed data. QZ supervised the research, planned experiments, analysed data, and prepared the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 31771273), grants from Guangdong Province (No. 2017A030313875), Guangdong Provincial-level Special Funds for Promoting High-quality Economic Development (No. 2020032), and Research Funds for the Shenzhen of China (No. 20180129171138130 and JCYJ20180307163444601).
Data sharing statment
All data generated or analysed during this study are included in this article. Additional raw data may be available from the corresponding author for reasonable reasons. The RNA-seq raw data have been deposited under the GEO accession number GSE169370.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbadis.2021.166260.
Appendix A. Supplementary data
Supplementary material
References
- 1.Li Q., Guan X., Wu P., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu L., She Z.G., Cheng X., et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020;31(6) doi: 10.1016/j.cmet.2020.04.021. (1068-1077 e1063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noy-Porat T., Makdasi E., Alcalay R., et al. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020;11(1):4303. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng H., Huang L., Liao Z., Liu M., Li Q., Xu R. Itraconazole inhibits the hedgehog signaling pathway thereby inducing autophagy-mediated apoptosis of colon cancer cells. Cell Death Dis. 2020;11(7):539. doi: 10.1038/s41419-020-02742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathai B.J., Meijer A.H., Simonsen A. Studying autophagy in zebrafish. Cells. 2017;6(3) doi: 10.3390/cells6030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortes C.J., Qin K., Cook J., Solanki A., Mastrianni J.A. Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann-Straussler-Scheinker disease. J. Neurosci. 2012;32(36):12396–12405. doi: 10.1523/JNEUROSCI.6189-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sridharan S., Jain K., Basu A. Regulation of autophagy by kinases. Cancers (Basel) 2011;3(2):2630–2654. doi: 10.3390/cancers3022630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy J.M.M., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B., Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115(10):2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang W., Bai J., Tian S., et al. Autophagy protects gastric mucosal epithelial cells from ethanol-induced oxidative damage via mTOR signaling pathway. Exp. Biol. Med. 2017;242(10):1025–1033. doi: 10.1177/1535370216686221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J., Feng Y., Wang Y., et al. alpha-hederin induces autophagic cell death in colorectal cancer cells through reactive oxygen species dependent AMPK/mTOR signaling pathway activation. Int. J. Oncol. 2019;54(5):1601–1612. doi: 10.3892/ijo.2019.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores-Bellver M., Bonet-Ponce L., Barcia J.M., et al. Autophagy and mitochondrial alterations in human retinal pigment epithelial cells induced by ethanol: implications of 4-hydroxy-nonenal. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.High M., Cho H.Y., Marzec J., et al. Determinants of host susceptibility to murine respiratory syncytial virus (RSV) disease identify a role for the innate immunity scavenger receptor MARCO gene in human infants. EBioMedicine. 2016;11:73–84. doi: 10.1016/j.ebiom.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung P.S., Huang T.F., Hsieh S.L. Extracellular vesicles from CLEC2-activated platelets enhance dengue virus-induced lethality via CLEC5A/TLR2. Nat. Commun. 2019;10(1):2402. doi: 10.1038/s41467-019-10360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta P., McAuley D.F., Brown M., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan W.Y.T., Young B.E., Lye D.C., Chew D.E.K., Dalan R. Statin use is associated with lower disease severity in COVID-19 infection. Sci. Rep. 2020;10(1):17458. doi: 10.1038/s41598-020-74492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P., Luo R., Zhang M., et al. A cross-talk between epithelium and endothelium mediates human alveolar-capillary injury during SARS-CoV-2 infection. Cell Death Dis. 2020;11(12):1042. doi: 10.1038/s41419-020-03252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fragoso-Saavedra S., Iruegas-Nunez D.A., Quintero-Villegas A., et al. A parallel-group, multicenter randomized, double-blinded, placebo-controlled, phase 2/3, clinical trial to test the efficacy of pyridostigmine bromide at low doses to reduce mortality or invasive mechanical ventilation in adults with severe SARS-CoV-2 infection: the Pyridostigmine In Severe COvid-19 (PISCO) trial protocol. BMC Infect. Dis. 2020;20(1):765. doi: 10.1186/s12879-020-05485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020;53(1):19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manjili R.H., Zarei M., Habibi M., Manjili M.H. COVID-19 as an acute inflammatory disease. J. Immunol. 2020;205(1):12–19. doi: 10.4049/jimmunol.2000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitoh T., Akira S. Regulation of innate immune responses by autophagy-related proteins. J. Cell Biol. 2010;189(6):925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai Y., Lin P., Feng Z., et al. TNFAIP3-DEPTOR complex regulates inflammasome secretion through autophagy in ankylosing spondylitis monocytes. Autophagy. 2018;14(9):1629–1643. doi: 10.1080/15548627.2018.1458804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J., Bae J., Choi C.Y., et al. Autophagy induced by AXL receptor tyrosine kinase alleviates acute liver injury via inhibition of NLRP3 inflammasome activation in mice. Autophagy. 2016;12(12):2326–2343. doi: 10.1080/15548627.2016.1235124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maycotte P., Jones K.L., Goodall M.L., Thorburn J., Thorburn A. Autophagy supports breast cancer stem cell maintenance by regulating IL6 secretion. Mol. Cancer Res. 2015;13(4):651–658. doi: 10.1158/1541-7786.MCR-14-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30(23):4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M., Kenny S.J., Ge L., Xu K., Schekman R. Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. Elife. 2015:4. doi: 10.7554/eLife.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deretic V., Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14(2):243–251. doi: 10.1080/15548627.2017.1402992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M., Li J., Zeng R., et al. Respiratory syncytial virus replication is promoted by autophagy-mediated inhibition of apoptosis. J. Virol. 2018;92(8) doi: 10.1128/JVI.02193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H., Kang R., Wang J., Luo G., Yang W., Zhao Z. Hepatitis C virus inhibits AKT-tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy. 2013;9(2):175–195. doi: 10.4161/auto.22791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang G., Luk B.T., Wei X., et al. Selective cell death of latently HIV-infected CD4(+) T cells mediated by autosis inducing nanopeptides. Cell Death Dis. 2019;10(6):419. doi: 10.1038/s41419-019-1661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park K., Lee J.H., Cho H.C., Cho S.Y., Cho J.W. Down-regulation of IL-6, IL-8, TNF-alpha and IL-1beta by glucosamine in HaCaT cells, but not in the presence of TNF-alpha. Oncol. Lett. 2010;1(2):289–292. doi: 10.3892/ol_00000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou X., Liu Y., Lei X., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Y., Zhuang M.W., Han L., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target Ther. 2020;5(1):299. doi: 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F., Zhao X., Sun R., et al. EGFR-rich extracellular vesicles derived from highly metastatic nasopharyngeal carcinoma cells accelerate tumour metastasis through PI3K/AKT pathway-suppressed ROS. J. Extracell. Vesicles. 2020;10(1) doi: 10.1002/jev2.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang H., Ma Z., Peng H., He L., Hu Z., Wang Y. CXCL16 deficiency attenuates renal injury and fibrosis in salt-sensitive hypertension. Sci. Rep. 2016;6:28715. doi: 10.1038/srep28715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W., Moore M.J., Vasilieva N., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. (271-280 e278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou L., Wei L., Zhu S., et al. Avian metapneumovirus subgroup C induces autophagy through the ATF6 UPR pathway. Autophagy. 2017;13(10):1709–1721. doi: 10.1080/15548627.2017.1356950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng C., Zhou Z., Jiang K., et al. Newcastle disease virus triggers autophagy in U251 glioma cells to enhance virus replication. Arch. Virol. 2012;157(6):1011–1018. doi: 10.1007/s00705-012-1270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marullo R., Werner E., Zhang H., Chen G.Z., Shin D.M., Doetsch P.W. HPV16 E6 and E7 proteins induce a chronic oxidative stress response via NOX2 that causes genomic instability and increased susceptibility to DNA damage in head and neck cancer cells. Carcinogenesis. 2015;36(11):1397–1406. doi: 10.1093/carcin/bgv126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai Z., Zhao X., Li C., Sheng C., Li H. EV71 virus reduces Nrf2 activation to promote production of reactive oxygen species in infected cells. Gut Pathog. 2020;12:22. doi: 10.1186/s13099-020-00361-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Lin Y.M., Chen C.I., Hsiang Y.P., et al. Chrysin attenuates cell viability of human colorectal cancer cells through autophagy induction unlike 5-Fluorouracil/Oxaliplatin. Int. J. Mol. Sci. 2018;19(6) doi: 10.3390/ijms19061763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y.Q., Kou J.Y., Han X.B., et al. ROS-dependent activation of autophagy through the PI3K/Akt/mTOR pathway is induced by Hydroxysafflor yellow A-Sonodynamic therapy in THP-1 macrophages. Oxidative Med. Cell. Longev. 2017:2017. doi: 10.1155/2017/8519169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vucicevic L., Misirkic M., Janjetovic K., et al. Compound C induces protective autophagy in cancer cells through AMPK inhibition-independent blockade of Akt/mTOR pathway. Autophagy. 2011;7(1):40–50. doi: 10.4161/auto.7.1.13883. [DOI] [PubMed] [Google Scholar]
- 47.Underwood B.R., Imarisio S., Fleming A., et al. Antioxidants can inhibit basal autophagy and enhance neurodegeneration in models of polyglutamine disease. Hum. Mol. Genet. 2010;19(17):3413–3429. doi: 10.1093/hmg/ddq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishiga M., Wang D.W., Han Y., Lewis D.B., Wu J.C. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020;17(9):543–558. doi: 10.1038/s41569-020-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song J., Li N., Xia Y., et al. Arctigenin treatment protects against brain damage through an anti-inflammatory and anti-apoptotic mechanism after needle insertion. Front. Pharmacol. 2016;7:182. doi: 10.3389/fphar.2016.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenberg-Lerner A., Bialik S., Simon H.U., Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16(7):966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 53.Peng J., Zhu S., Hu L., et al. Wild-type rabies virus induces autophagy in human and mouse neuroblastoma cell lines. Autophagy. 2016;12(10):1704–1720. doi: 10.1080/15548627.2016.1196315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Islam A., Khan M.A. Lung transcriptome of a COVID-19 patient and systems biology predictions suggest impaired surfactant production which may be druggable by surfactant therapy. Sci. Rep. 2020;10(1):19395. doi: 10.1038/s41598-020-76404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Z., Ren L., Zhang L., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6) doi: 10.1016/j.chom.2020.04.017. (883-890 e882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao L., Jin H., Wang M., et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patra T., Meyer K., Geerling L., et al. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020;16(12) doi: 10.1371/journal.ppat.1009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dreux M., Chisari F.V. Autophagy proteins promote hepatitis C virus replication. Autophagy. 2009;5(8):1224–1225. doi: 10.4161/auto.5.8.10219. [DOI] [PubMed] [Google Scholar]
- 60.Dong X., Levine B. Autophagy and viruses: adversaries or allies? J. Innate Immun. 2013;5(5):480–493. doi: 10.1159/000346388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu X., Zhang C., Wang Y., et al. Autophagy induced by avian reovirus enhances viral replication in chickens at the early stage of infection. BMC Vet. Res. 2019;15(1):173. doi: 10.1186/s12917-019-1926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gannage M., Dormann D., Albrecht R., et al. Matrix protein 2 of influenza a virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6(4):367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Z., Jiang X., Liu D., et al. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5(3):321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]
- 65.Miao G., Zhao H., Li Y., et al. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell. 2021;56(4) doi: 10.1016/j.devcel.2020.12.010. (427-442 e425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pons S., Fodil S., Azoulay E., Zafrani L. The vascular endothelium: the cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Crit. Care. 2020;24(1):353. doi: 10.1186/s13054-020-03062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huertas A., Montani D., Savale L., et al. Endothelial cell dysfunction: a major player in SARS-CoV-2 infection (COVID-19)? Eur. Respir. J. 2020;56(1) doi: 10.1183/13993003.01634-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bao L., Deng W., Huang B., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 69.Sun J., Zhuang Z., Zheng J., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182(3) doi: 10.1016/j.cell.2020.06.010. (734-743 e735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Signorello M.G., Ravera S., Leoncini G. Lectin-induced oxidative stress in human platelets. Redox Biol. 2020;32:101456. doi: 10.1016/j.redox.2020.101456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castro S.M., Guerrero-Plata A., Suarez-Real G., et al. Antioxidant treatment ameliorates respiratory syncytial virus-induced disease and lung inflammation. Am. J. Respir. Crit. Care Med. 2006;174(12):1361–1369. doi: 10.1164/rccm.200603-319OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Radi R. Oxygen radicals, nitric oxide, and peroxynitrite: redox pathways in molecular medicine. Proc. Natl. Acad. Sci. U. S. A. 2018;115(23):5839–5848. doi: 10.1073/pnas.1804932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spengler G., Kincses A., Mosolygo T., et al. Antiviral, antimicrobial and antibiofilm activity of selenoesters and selenoanhydrides. Molecules. 2019;24(23) doi: 10.3390/molecules24234264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li L., Tan J., Miao Y., Lei P., Zhang Q. ROS and autophagy: interactions and molecular regulatory mechanisms. Cell. Mol. Neurobiol. 2015;35(5):615–621. doi: 10.1007/s10571-015-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guan B., Lin Z., Liu D., et al. Effect of waterlogging-induced autophagy on programmed cell death in arabidopsis roots. Front. Plant Sci. 2019;10:468. doi: 10.3389/fpls.2019.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong W.F., Wang X.H., Pan B., Li F., Kuang L., Su Z.X. Eupatilin induces human renal cancer cell apoptosis via ROS-mediated MAPK and PI3K/AKT signaling pathways. Oncol. Lett. 2016;12(4):2894–2899. doi: 10.3892/ol.2016.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Panga V., Kallor A.A., Nair A., Harshan S., Raghunathan S. Mitochondrial dysfunction in rheumatoid arthritis: a comprehensive analysis by integrating gene expression, protein-protein interactions and gene ontology data. PLoS One. 2019;14(11) doi: 10.1371/journal.pone.0224632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hwang J., Lee S., Lee J.T., et al. Gangliosides induce autophagic cell death in astrocytes. Br. J. Pharmacol. 2010;159(3):586–603. doi: 10.1111/j.1476-5381.2009.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang H., Cheng X., Zhang L., Xu S., Zhang Q., Lu R. A surface-layer protein from lactobacillus acidophilus NCFM induces autophagic death in HCT116 cells requiring ROS-mediated modulation of mTOR and JNK signaling pathways. Food Funct. 2019;10(7):4102–4112. doi: 10.1039/c9fo00109c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material