Abstract
The long noncoding RNA called MIR22 host gene (MIR22HG) was previously identified as a tumor suppressor in several cancers. However, the biological function of MIR22HG in breast cancer remains unknown. In this study, we aimed to determine the function and molecular mechanism of MIR22HG in breast cancer progression using transcriptomics and biotechnological techniques. Our results showed that MIR22HG expression was lower in the cancerous tissues than in the paired adjacent normal breast tissues. Additionally, MIR22HG was found to be mainly located in the cytoplasm and acted as a miR-629-5p sponge. Notably, MIR22HG stabilized the expression of large tumor suppressor 2 (LATS2), which promoted the LATS2-dependent phosphorylation of YAP1 and suppressed the expression of its downstream target oncogenes, thereby inhibiting the proliferation and migration of breast cancer cells. Therefore, our findings reveal the MIR22HG-dependent inhibition of breast cancer cell proliferation and migration via the miR-629-5p/LATS2 pathway, providing new insights and identifying novel therapeutic targets for breast cancer treatment.
Subject terms: Breast cancer, Long non-coding RNAs
Introduction
As the most commonly diagnosed cancer worldwide, female breast cancer had an estimated incidence rate of 11.7% in 2020 and was known to greatly influence women’s health [1]. Previous studies have successfully identified several biomarkers and therapeutic targets for breast cancer treatment. In addition, increasing evidences show that long noncoding RNAs (lncRNAs) play important roles in breast cancer progression [2].
LncRNAs are defined as noncoding RNA molecules with more than 200 nucleotides [3]. Several lncRNAs, such as MALAT1 [4], RAB11B-AS1 [5], TROJAN [6], and HOST2 [7], have been found to facilitate the proliferation and metastasis of breast cancer cells. MALAT1 binds to the transcription factor TEAD and inactivates the transcriptional activity of the YAP/TEAD complex, subsequently inhibiting the metastatic ability of breast cancer cells [4]. The HIF2-induced RAB11B-AS1 recruits RNA polymerase II to upregulate VEGFA and ANGPTL4 expression, consequently promoting tumor angiogenesis and distant metastasis of breast cancer [5]. In triple-negative breast cancer, TROJAN interacts with metastasis-repressing factor ZMYND8, subsequently increasing ZMYND8 degradation via the ubiquitin-proteasome pathway by repelling ZNF592, and thus enhancing cancer progression [6]. In our previous research, HOST2 was revealed to act as a competitive endogenous RNA via decoying of let-7b, thereby promoting STAT3-mediated cell proliferation and migration in triple-negative breast cancer [7].
Notably, the lncRNA called MIR22 host gene (MIR22HG) has also been observed in several tumors. The tumor suppression function of MIR22HG has been described in hepatocellular carcinoma [8], colorectal cancer [9], and glioblastoma [10]. However, the function of MIR22HG in breast cancer remains unknown. Hence, this study aimed to determine the function and molecular mechanism of MIR22HG in breast cancer progression using transcriptomics and biotechnological techniques.
Results
MIR22HG expression is downregulated in breast cancer tissues
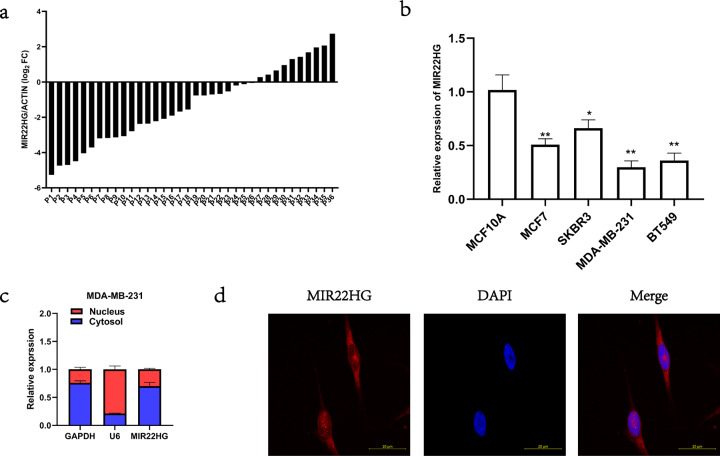
To investigate MIR22HG expression in breast cancer tissues, we collected 36 pairs of cancerous tissues and adjacent normal breast tissues for real-time PCR analysis. Our results showed that in most paired tissues (26/36), MIR22HG expression was lower in cancerous tissues than in normal tissues (Fig. 1a). Similarly, the MIR22HG expression was lower in the studied breast cancer cell lines, namely MCF7, MDA-MB-231, BT549, and SKBR3, than in the normal breast epithelial cell line MCF10A (Fig. 1b). In addition, to determine the subcellular localization of MIR22HG in breast cancer cells, we separated the nuclear and cytoplasmic RNA of MDA-MB-231 cells. The real-time PCR results indicated that MIR22HG was mainly localized in the cytoplasm (Fig. 1c). Further analysis via fluorescence in situ hybridization also confirmed that MIR22HG was located in the cytoplasm (Fig. 1d).
Fig. 1. Expression and subcellular distribution of MIR22HG.
aMIR22HG expression in 36 pairs of cancerous and adjacent normal breast tissues detected by real-time PCR, with β-actin as internal control. b MIR22HG expression in normal breast epithelial cell line MCF10A and breast cancer cell lines MCF7, MDA-MB-231, BT549, and SKBR3 detected by real-time PCR, with β-actin as internal control. c MIR22HG expression in subcellular fractions of MDA-MB-231 cells detected by real-time PCR, with U6 and GAPDH as nuclear and cytoplasmic markers, respectively. d Fluorescence in situ hybridization analysis revealed that MIR22HG (red) was mainly located in the cytoplasm. DAPI-stained nuclei are in blue (in 63*oil lens). *P < 0.05, **P < 0.01. All experiments were repeated thrice.
Loss-of-function of MIR22HG promotes breast cancer cell proliferation and migration in vitro
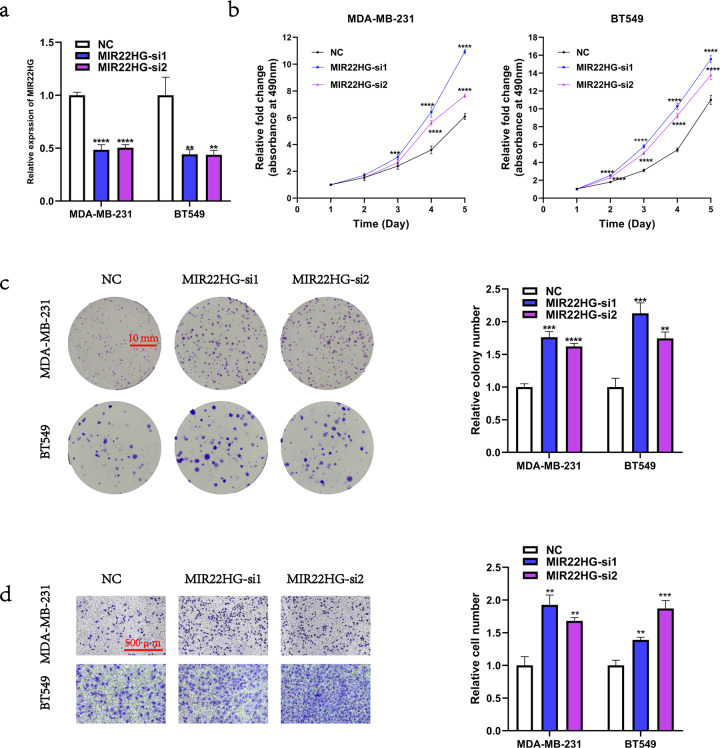
To determine the biological function of MIR22HG in breast cancer cells, we transfected MDA-MB-231 and BT549 cells with MIR22HG-siRNAs. Considering the off-target effect of siRNAs, the MIR22HG-si1 and MIR22HG-si2 were used. The knockdown efficiency of MIR22HG-si1 and MIR22HG-si2 were detected by real-time PCR (Fig. 2a). After MIR22HG knockdown, the changes in the cell proliferation ability were detected by MTT assays and colony formation assays, while the changes in the cell migration ability were analyzed by Transwell assays. Notably, our results showed that loss-of-function of MIR22HG enhanced the proliferation (Fig. 2b, c) and migration (Fig. 2d) abilities of breast cancer cells.
Fig. 2. Loss-of-function of MIR22HG promotes the proliferation and migration of breast cancer cells.
a Knockdown efficiency was detected by real-time PCR after transfection of MDA-MB-231 and BT549 cells with siRNAs targeting MIR22HG (MIR22HG-si1 and MIR22HG-si2) or negative control (NC) for 48 h. b, c MTT assay (5-day period) and colony formation assay (18-day period) were used to determine the viability of MDA-MB-231 and BT549 cells after MIR22HG knockdown. d Transwell assay (20-h period, 3 × 104 cells per well) was performed to evaluate the migration ability of MDA-MB-231 and BT549 cells after MIR22HG knockdown (100 × magnification). **P < 0.01, ***P < 0.001, ****P < 0.0001. All experiments were repeated thrice.
Gain-of-function of MIR22HG inhibits breast cancer cell proliferation and migration in vitro and in vivo
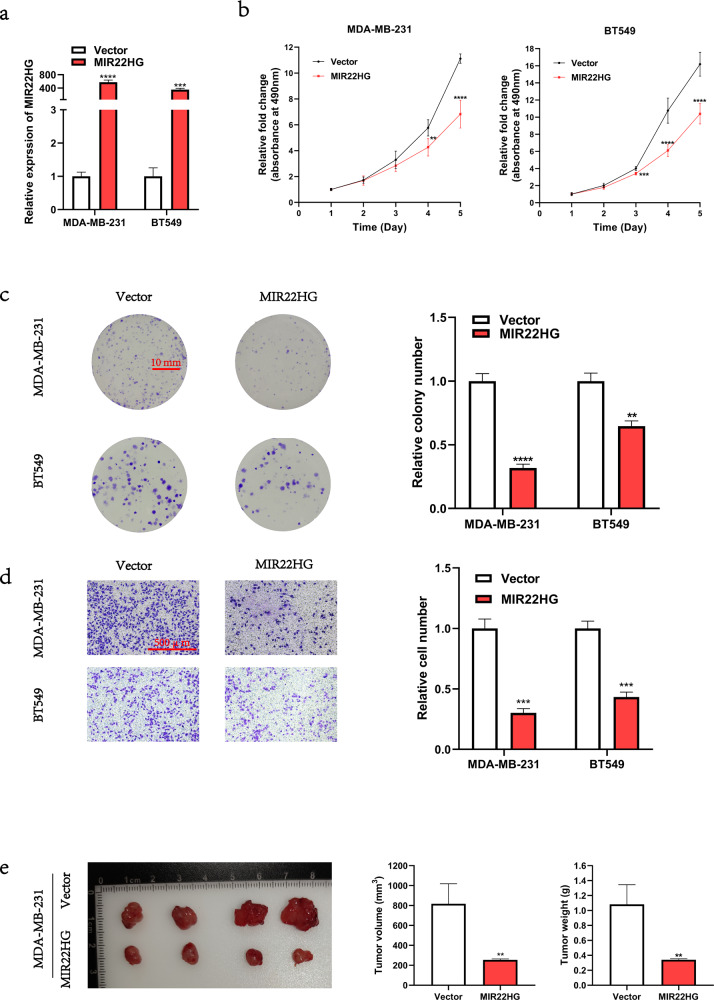
In addition to MIR22HG knockdown, we transfected MDA-MB-231 and BT549 cells with MIR22HG plasmids to achieve overexpression. The overexpression efficiency was detected by real-time PCR (Fig. 3a). Results showed that MIR22HG overexpression suppressed the proliferation (Fig. 3b, c) and migration (Fig. 3d) of breast cancer cells. Furthermore, we constructed MDA-MB-231 cells that stably expressed MIR22HG and a subcutaneous xenograft model to explore the biological function of MIR22HG in vivo. Compared to wild-type MDA-MB-231 cells, the tumorigenicity of MDA-MB-231 cells with stable MIR22HG expression was suppressed (Fig. 3e). Collectively, our results suggest that gain-of-function of MIR22HG inhibited the proliferation of breast cancer cells both in vitro and in vivo.
Fig. 3. Gain-of-function of MIR22HG inhibits the proliferation and migration of breast cancer cells.
a Overexpression efficiency of MIR22HG in MDA-MB-231 and BT549 cells after transfection with MIR22HG pcDNA3.1 plasmid for 48 h. b, c MTT assay (5-day period) and colony formation assay (18-day period) were performed to determine the viability of MDA-MB-231 and BT549 cells after transfection. d Transwell assay (20-h period, 3 × 104 cells per well) was used to evaluate the migration ability of MDA-MB-231 and BT549 cells after MIR22HG overexpression (100× magnification). e A subcutaneous xenograft model was utilized to explore the biological function of MIR22HG in vivo. The MDA-MB-231 cells that stably expressed MIR22HG were subcutaneously injected into 6-week-old BALB/c nu/nu female mice. After 30 days, the tumors were collected, and the tumor volume and weight were measured. **P < 0.01, ***P < 0.001, ****P < 0.0001. All experiments were repeated thrice.
MIR22HG functions as an miR-629-5p sponge in breast cancer cells
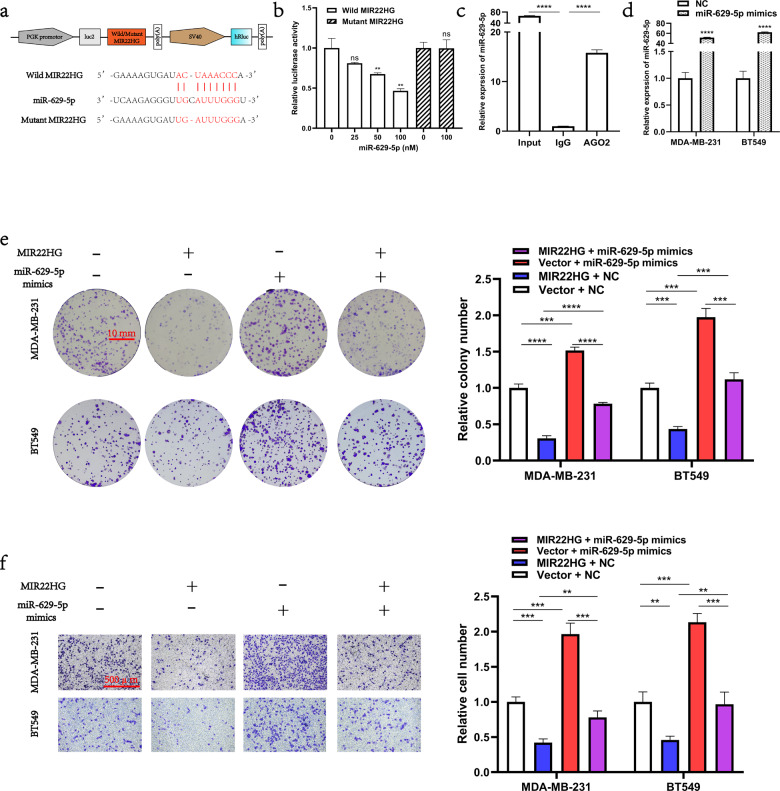
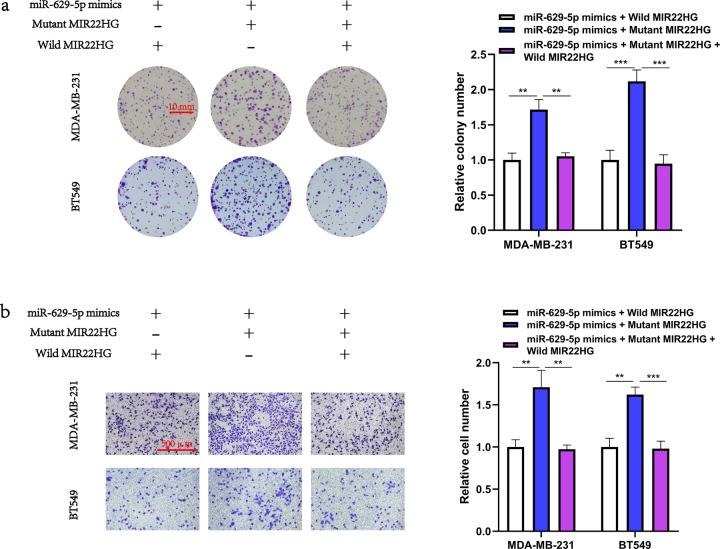
Since MIR22HG was mainly located in the cytoplasm, we hypothesized that MIR22HG may function as a miRNA sponge in breast cancer cells. Hence, we searched the starBase database [11] (http://starbase.sysu.edu.cn/) and discovered that miR-629-5p was a potential target of MIR22HG. To validate the relationship between MIR22HG and miR-629-5p, we constructed the pmirGLO–MIR22HG–wild-type and pmirGLO–MIR22HG–mutant vectors for subsequent luciferase reporter assay (Fig. 4a). Our results indicated that miR-629-5p reduced the luciferase activity of the pmirGLO–MIR22HG–wild-type vector and failed to bind with the mutant MIR22HG (Fig. 4b). In contrast, the immunoprecipitation assay demonstrated that AGO2 was able to pull down both MIR22HG and miR-629-5p (Fig. 4c). Taken together, these results suggest that MIR22HG may act as a competing endogenous RNA molecule to suppress miR-629-5p. We then investigated the function of miR-629-5p in breast cancer cells. The MDA-MB-231 and BT549 cells were transfected with miR-629-5p mimics, and the overexpression efficiency of miR-629-5p mimics was detected by real-time PCR (Fig. 4d). We discovered that miR-629-5p overexpression promoted the proliferation and migration of both MDA-MB-231 and BT549 cells. Notably, results of the rescue experiment suggest that MIR22HG impaired the function of miR-629-5p in breast cancer cells (Fig. 4e, f). Furthermore, the colony formation assay and Transwell assay results of breast cancer cells co-transfected with miR-629-5p and wild-type/mutant MIR22HG revealed that only the wild-type MIR22HG impaired the function of miR-629-5p in breast cancer cells (Fig. 5a, b). Collectively, our results suggest that MIR22HG act as a miRNA sponge that can inhibit function of miR-629-5p in breast cancer cells.
Fig. 4. MIR22HG functions as an miR-629-5p sponge in breast cancer cells.
a Schematic diagram of the interaction between miR-629-5p and wild-type/mutant MIR22HG. b Dual-luciferase assays in HEK293T cells co-transfected with wild-type/mutant MIR22HG and miR-629-5p mimics (25, 50, and 100 nM) or NC. At 48 h after transfection, the 293 T cells were collected for the detection of firefly luciferase activity, with Renilla luciferase activity as internal control. c AGO2 immunoprecipitation assay was performed using HEK293T cells to detect the expression of miR-629-5p associated with AGO2. The AGO2-specific antibody was applied to precipitate the AGO2-RNA complex after MIR22HG overexpression in 293T cells. Total RNA was extracted, and real-time PCR was performed to determine the expression of miR-629-5p associated with AGO2, using IgG-specific antibody as negative control. d Overexpression efficiency of miR-629-5p was analyzed by real-time PCR after MDA-MB-231 and BT549 cells were transfected with miR-629-5p mimics or NC. e The effects of MIR22HG and miR-629-5p co-transfection on the proliferation of MDA-MB-231 and BT549 cells were evaluated by colony formation assay (18-day period). f The effects of MIR22HG and miR-629-5p co-transfection on the migration of MDA-MB-231 and BT549 cells were evaluated by Transwell assay (20-h period, 3 × 104 cells per well, 100× magnification). **P < 0.01, ***P < 0.001, ****P < 0.0001. All experiments were repeated thrice.
Fig. 5. Only wild-type MIR22HG functions as an miR-629-5p sponge in breast cancer cells.
a Breast cancer cells co-transfected with miR-629-5p and wild-type/mutant MIR22HG. Colony formation assay was used to assess the proliferation ability of MDA-MB-231 and BT549 cells. b Transwell assay was performed to assess the migration ability of MDA-MB-231 and BT549 cells. **P < 0.01, ***P < 0.001. All experiments were repeated thrice.
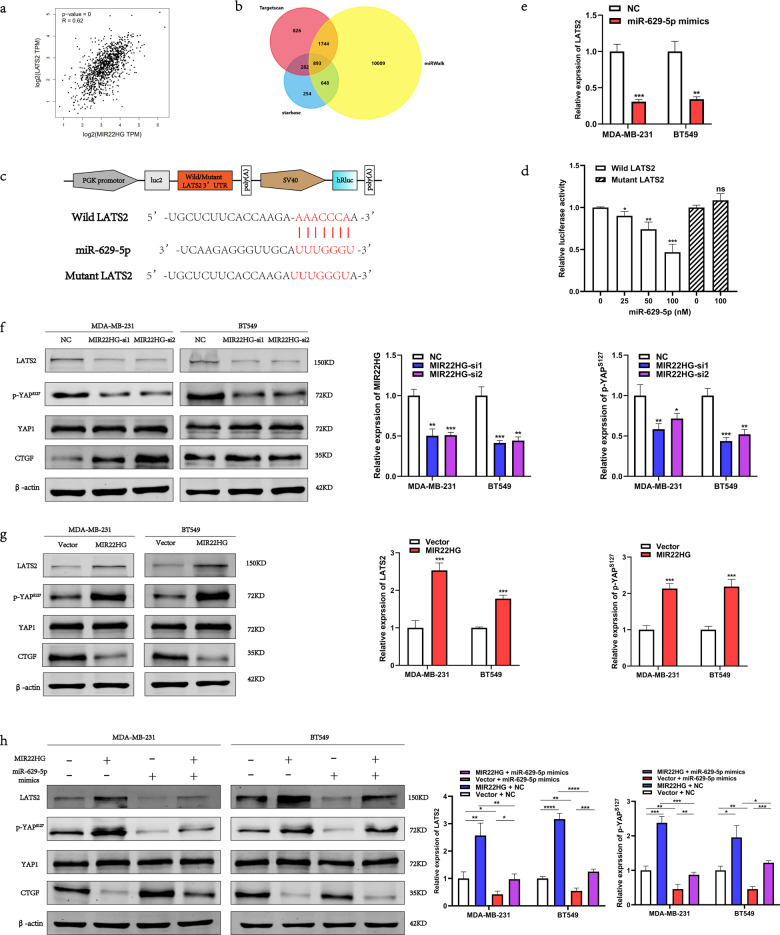
MIR22HG regulates LATS2 expression via inhibition of miR-629-5p
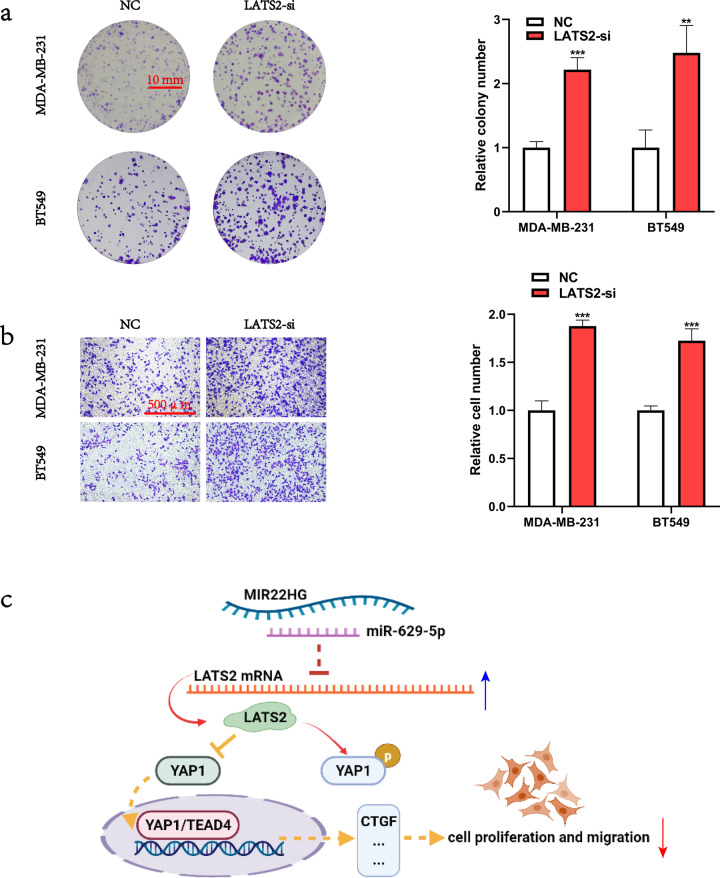
To identify the downstream targets of MIR22HG and miR-629-5p, we analyzed the breast cancer data from The Cancer Genome Atlas (TCGA) database and found that LATS2 expression was positively correlated with MIR22HG expression [12] (Fig. 6a). Additionally, utilization of the miRWalk [13] (http://mirwalk.umm.uni-heidelberg.de/), starBase [11] (http://starbase.sysu.edu.cn/), and Targetscan [14] (http://www.targetscan.org/) databases revealed LATS2 as a potential downstream target of miR-629-5p (Fig. 6b). To confirm the relationship between miR-629-5p and LATS2, we constructed the pmirGLO–LATS2 3′-UTR–wild-type and pmirGLO–LATS2 3′-UTR–mutant vectors (Fig. 6c) and then performed luciferase reporter assay. Our results demonstrated that miR-629-5p can bind with LATS2 3′-UTR (Fig. 6d), which promoted the degradation of LATS2 mRNA (Fig. 6e). We also examined the changes in the expression of LATS2 protein via Western blotting and found that miR-629-5p inhibited LATS2 expression, while MIR22HG slightly counteracted the miR-629-5p-induced decrease in LATS2 expression. As a key member of the Hippo-YAP1 signaling pathway, LATS2 can function as an upstream kinase of the transcriptional cofactor YAP1, which facilitates YAP1 phosphorylation and inactivates the transcriptional activity of the YAP1/TEAD4 complex. Hence, we analyzed the changes in the expression of Hippo pathway signals—CTGF, phosphorylated YAP1, and total YAP1—following MIR22HG knockdown or overexpression. The results showed that MIR22HG promoted YAP1 phosphorylation and slightly counteracted the miR-629-5p-induced decrease in YAP1 phosphorylation, while no changes were observed in total YAP1 expression (Fig. 6f–h). Finally, investigation of the biological function of LATS2 in breast cancer revealed that the siRNA-induced knockdown of LATS2 enhanced the proliferation and migration of breast cancer cells (Fig. 7a, b), indicating that LATS2 was also a tumor suppressor in breast cancer. Taken together, our results suggest that MIR22HG acts as a miR-629-5p sponge that can inhibit the proliferation and migration of breast cancer cells and stabilize LATS2 expression.
Fig. 6. MIR22HG regulates LATS2 expression via inhibition of miR-629-5p activity.
a The expression correlation between MIR22HG and LATS2 based on the Gene Expression Profiling Interactive Analysis 2 (GEPIA2) database. b Venn diagram showing the overlapping target genes of miR-629-5p predicted using the miRWalk, starBase, and TargetScan databases. c Schematic diagram of the interaction between miR-629-5p and wild-type/mutant LATS2 3′-UTR. d Dual-luciferase assays of HEK293T cells co-transfected with wild-type/mutant LATS2 3′-UTR and miR-629-5p mimics (25, 50, and 100 nM) or NC. At 48 h after transfection, the 293 T cells were collected for the detection of firefly luciferase activity, with Renilla luciferase activity as internal control. e Real-time PCR was performed to detect the changes in LATS2 expression in MDA-MB-231 and BT549 after transfection with miR-629-5p mimics for 48 h. f, g Expression of Hippo-YAP1 signals, namely LATS2, YAP1, phosphorylated YAP1 (S127), and CTGF, was assessed by Western blotting after MIR22HG knockdown or overexpression. h Western blot analysis revealed that miR-629-5p inhibited the expression of LATS2, while MIR22HG was found to slightly counteract the miR-629-5p-induced reduction in LATS2 expression. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All experiments were repeated thrice.
Fig. 7. LATS2 knockdown promoted the proliferation and migration of breast cancer cells.
a Colony formation assay was used to determine the viability of MDA-MB-231 and BT549 cells after LATS2 knockdown. b Transwell assay was performed to evaluate the migration ability of MDA-MB-231 and BT549 cells after LATS2 knockdown (100× magnification). c A scheme of the proposed molecular mechanisms. MIR22HG stabilizes LATS2 by counteracting the miR-629-5p-induced degradation of LATS2 mRNA, consequently resulting in the promotion of LATS2-dependent YAP1 phosphorylation and YAP1 inactivation. ***P < 0.001. All experiments were repeated thrice.
Discussion
To determine the function of MIR22HG in breast cancer, we investigated 36 pairs of cancerous tissues and adjacent normal breast tissues via real-time PCR. The results showed that MIR22HG expression was lower in cancerous tissues than in normal tissues. The subcellular distribution of lncRNAs determines their biological function [15]. For example, BCRT1, which is mainly located in the cytoplasm, can competitively bind with miR-1303 [16]. In contrast, BX111, which is primarily located in the nucleus, binds with transcription factor YB1 to regulate the expression of its downstream target genes [17]. In the present study, we discovered that MIR22HG was mainly distributed in the cytoplasm, indicating that MIR22HG may act as a competitive endogenous RNA molecule. Subsequent bioinformatic analysis revealed that MIR22HG can bind with miR-629-5p. Furthermore, the luciferase reporter assay and AGO2 immunoprecipitation assay results confirmed that MIR22HG can act as a miRNA sponge and directly bind with miR-629-5p. Functional analysis via siRNA-induced knockdown of MIR22HG resulted in the enhanced proliferation and metastasis of breast cancer cells, while MIR22HG overexpression inhibited cancer cell proliferation and metastasis. In the Hippo-YAP1 signaling pathway, LATS2 plays a critical role in the phosphorylation and inactivation of the transcription cofactor YAP1/TAZ. The phosphorylated YAP1 is degraded in the cytoplasm by the ubiquitin-proteasome system, consequently inhibiting the expression of YAP1 target genes that are needed for cell proliferation and migration [18]. Taking these into consideration, our results suggest that LATS2 also functions as a tumor suppressor in breast cancer and that miR-629-5p can promote the proliferation and migration of breast cancer cells by targeting LATS2 mRNA. Additionally, MIR22HG can stabilize LATS2 expression by counteracting the miR-629-5p-induced degradation of LATS2 mRNA (Fig. 7c). Therefore, our research reveals the biological function and molecular mechanism of MIR22HG in breast cancer cells and provides novel insights and therapeutic targets for future studies on breast cancer treatment.
Materials and methods
Tissue samples
The 36 pairs of cancerous tissues and adjacent normal breast tissues used in this study were collected from the Department of Breast and Thyroid Surgery, Shanghai Tenth People’s Hospital (Shanghai, China). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the ethics board of Shanghai Tenth People’s Hospital (No. 2020-KN174-01). All study participants were contacted by telephone to obtain verbal informed consent. After collection via surgery, the cancerous and normal tissues were cryopreserved in liquid nitrogen for temporary storage.
Cell culture
The human embryonic kidney cell line HEK293T, human breast cancer cell lines MCF7, MDA-MB-231, BT549, and SKBR3, and human normal breast epithelial cell line MCF10A were purchased from the Chinese Academy of Science, Shanghai (China). The MCF7, MDA-MB-231, and 293 T cells were cultured in DMEM medium with high glucose (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), while the BT549 and MCF10A cells were cultured in RPMI-1640 medium (Gibco, USA) and mammary epithelial cell medium (Procell, Wuhan, China), respectively. The growth media were added with 10% fetal bovine serum (Gibco, USA) and 100 U/mL penicillin and 100 μg/mL streptomycin (Servicebio, Wuhan, China). A 0.25% trypsin-EDTA solution (Servicebio, China) was also applied for cell passage cultivation. All cell lines were cultivated in a cell incubator at 37 °C and 5% CO2.
Cell transfection
The MIR22HG-siRNAs (MIR22HG-si1, Sense: 5′-GAGUAGAAGGCUCAAACAACC-3′ and Antisense: 5′-UUGUUUGAGCCUUCUACUCCU-3′; MIR22HG-si2, Sense: 5′-CAGGAAAUUCAUAAAAGAAAU-3′ and Antisense: 5′-UUCUUUUAUGAAUUUCCUGUG-3′), pcDNA3.1-MIR22HG plasmid, miR-629-5p mimics, large tumor suppressor 2 (LATS2)-siRNA (LATS2-si, Sense: 5′-GAGUGAUAAUCUUCAAAAUGA-3′ and Antisense: 5′-AUUUUGAAGAUUAUCACUCUC-3′), negative control (NC) siRNA (Sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ and Antisense: 5′-ACGUGACACGUUCGGAGAATT-3′), and pcDNA3.1-vector were transfected into MDA-MB-231 and BT549 cells using Lipofectamine® 3000 reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). All sequences were synthesized by Generay Biotechnology (Shanghai, China).
RNA extraction and cDNA synthesis
Total RNA was extracted from MCF10A, MDA-MB-231, BT549, SKBR3, and 293 T cells and from cancerous tissues and paired adjacent normal breast tissues using TRIzol® Reagent (Invitrogen, USA). For first strand cDNA synthesis, 1000 ng total RNA was reverse-transcribed using Hifair® III 1st Strand cDNA Synthesis SuperMix for qPCR (Yeasen, Shanghai, China) and miRNA 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China), following the manufacturers’ instructions.
Subcellular fractionation
The nuclear and cytoplasmic separation in cells were performed using PARIS Kit (Ambion, Life Technologies, Grand Island, NY, USA), following the manufacturer’s protocols.
Fluorescence in situ hybridization
The subcellular localization of MIR22HG in MDA-MB-231 cells was analyzed using Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China), according to the manufacturer’s instructions. The Cy3-labeled MIR22HG probes were designed and synthesized by RiboBio (China). Representative images were obtained using a confocal microscope (Carl Zeiss Jena, Germany).
Real-time PCR
Real-time PCR was performed using Hieff UNICON® qPCR SYBR Green MasterMix (Yeasen, China), with β-actin as internal control for normalizing MIR22HG and LATS2 expression and U6 as internal control for normalizing miR-629-5p expression. All primers were synthesized by Generay Biotechnology (China). The primers used for qPCR include the following: MIR22HG (Forward: 5′-CCATACATTGCGTGTGGGAG-3′, Reverse: 5′-TTCGTAGGTCAAATGACATGGAG-3′); β-actin (Forward: 5′-CATGTACGTTGCTATCCAGGC-3′, Reverse: 5′-CTCCTTAATGTCACGCACGAT-3′); Hsa_miR-629-5p (Forward: 5′-GCGTGGGTTTACGTTGGG-3′, Reverse: 5′-AGTGCAGGGTCCGAGGTATT-3′, RT: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTTCT-3′); and U6 (Forward: 5′-GGAGACACGCAAACGGAAG-3′, Reverse: 5′-AGTGCAGGGTCCGAGGTATT-3′, RT: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTGGCG-3′). The 2−ΔΔCt method was employed to analyze the mRNA expression levels.
MTT assay
The transfected MDA-MB-231 and BT549 cells were placed in 96-well plates at a density of 1 × 103 cells/well. Cell viability was assessed at 24, 48, 72, 96, and 120 h of incubation. A 200-μL complete medium was used for cell incubation, with 20 μL MTT (Yeasen, China) added to each well at set time points. After 6 h incubation at 37 °C and 5% CO2, the supernatants were removed, and 150 μL DMSO (Sangon Biotech, Shanghai, China) was added to each well to dissolve formazan. The absorbance was quantified at a wavelength of 490 nm.
Colony formation assay
First, the transfected MDA-MB-231 and BT549 cells were placed in 6-well plates at a density of 5 × 102 cells/well. Second, the cells were incubated for 7–14 days at 37 °C and 5% CO2, with the medium changed every 3 days. Third, the medium was removed after colonies formed, and cells were washed thrice with phosphate buffered saline (Servicebio, China) and then fixed with 95% ethanol for 10 min. Finally, the clones were stained with 0.1% crystal violet (Yeasen, China). Representative images were obtained using a camera.
Transwell assay
The transfected cells (3 × 104) were placed in the upper chamber of Transwell® (Corning Incorporated, Corning, NY, USA) and incubated in serum-free medium, while medium supplemented with 10% fetal bovine serum was added to the lower chamber. After 18 h, the cells that moved to the lower chamber were fixed with 4% paraformaldehyde and stained with 0.05% crystal violet. Representative images were collected using a light microscope (Nikon, Japan).
Western blot analysis
Proteins were extracted with RIPA lysis buffer (Yeasen, China), and the concentration was measured by BCA Protein Quantification Kit (Yeasen, China). The total protein (30 μg) was resolved by polyacrylamide gel electrophoresis (PAGE), and the separated proteins were transferred to a nitrocellulose membrane (Pall, Port Washington, NY, USA). Following transfer, the membrane was blocked with 5% non-fat milk for 1 h and incubated with the primary antibodies of LATS2 (5888 T, Cell Signalling Technologies [CST], Danvers, MA, USA), Yes-associated protein 1 (YAP1; A1002, ABclonal, Wuhan, China), p-YAP1S127(13008 T, CST, USA), CTGF (A11456, ABclonal, China), and β-actin (AC026, ABclonal, China) for 12–18 h at 4 °C. The membrane was washed thrice with Tris Buffered saline-Tween and then incubated with the secondary antibody IRDye® 800CW Goat anti-Rabbit IgG (925-32211, Li-Cor Biosciences, Lincoln, NE, USA) at room temperature for 1 h. The immunoreactive protein bands on the membrane were scanned using Odyssey scanning system (Li-Cor Biosciences, USA).
Dual-luciferase reporter assay
The dual activity of firefly and Renilla luciferase were detected using Dual-Luciferase Reporter Gene Assay Kit (Promega, Madison, WI, USA), following the manufacturer’s protocol.
AGO2 immunoprecipitation assay
The AGO2 (10686-1-AP, Proteintech, Chicago, IL, USA) and IgG antibodies (AC005, ABclonal, China) were utilized to observe the interaction between MIR22HG and miR-629-5p using RNA Immunoprecipitation (RIP) Kit (BersinBio, Guangzhou, China).
Xenograft mouse model
The MDA-MB-231 cells (1 × 107) and transfected cells with stable MIR22HG expression were suspended in 150 μL DMEM (without fetal bovine serum and penicillin/streptomycin) and subcutaneously injected into 6-week-old BALB/c nu/nu female mice. Mice were divided into 2 groups randomly with 4 mice in each group, which were not blinded to investigators. After 30 days, the mice were euthanized, and the tumors were collected. The tumor volume and weight were subsequently measured. All animal experiments were approved by the Animal Care and Use Committee of the Shanghai Tenth People’s Hospital (No. SHDSYY-2020-0600).
Statistical analysis
The data were presented as means ± standard deviation (SD) and analyzed using GraphPad Prism (version 9.0; GraphPad, San Diego, CA, USA). The variance was similar between the groups that are being statistically compared. Student’s t test was used for two-group comparisons, with P-values < 0.05 considered as significantly different.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No.82073204) and Shanghai Municipal Health Commission (No.202040157).
Author contributions
(I) Conception and design: X.D., D.Y., and L.F.; (II) Administrative support: L.F.; (III) Provision of study materials or patients: X.D., L.F.; (IV) Collection and assembly of data: X.D., D.Y.; (V) Data analysis and interpretation: All authors; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors.
Data availability
All data included in this study are available upon request by contact with the corresponding author.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the ethics board of Shanghai Tenth People’s Hospital (No. 2020-KN174-01). All study participants were contacted by telephone to obtain verbal informed consent. All animal experiments were approved by the Animal Care and Use Committee of the Shanghai Tenth People’s Hospital (No. SHDSYY-2020-0600).
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Dr Francesca Bernassola
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaochong Deng, Danrong Ye.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Crudele F, Bianchi N, Reali E, Galasso M, Agnoletto C, Volinia S. The network of non-coding RNAs and their molecular targets in breast cancer. Mol Cancer. 2020;19:61. doi: 10.1186/s12943-020-01181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–15. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu Y, Bao L, Chen Y, Wang C, Luo M, Zhang B, et al. HIF2-induced long noncoding RNA RAB11B-AS1 promotes hypoxia-mediated angiogenesis and breast cancer metastasis. Cancer Res. 2020;80:964–75. doi: 10.1158/0008-5472.CAN-19-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin X, Xu XE, Jiang YZ, Liu YR, Sun W, Guo YJ, et al. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci Adv. 2019;5:eaat9820. doi: 10.1126/sciadv.aat9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua K, Deng X, Hu J, Ji C, Yu Y, Li J, et al. Long noncoding RNA HOST2, working as a competitive endogenous RNA, promotes STAT3-mediated cell proliferation and migration via decoying of let-7b in triple-negative breast cancer. J Exp Clin cancer Res. 2020;39:58. doi: 10.1186/s13046-020-01561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang DY, Zou XJ, Cao CH, Zhang T, Lei L, Qi XL, et al. Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for hepatocellular carcinoma. Theranostics. 2018;8:3751–65. doi: 10.7150/thno.22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Shao T, Song M, Xie Y, Zhou J, Yin J, et al. MIR22HG acts as a tumor suppressor via TGFβ/SMAD signaling and facilitates immunotherapy in colorectal cancer. Mol Cancer. 2020;19:51. doi: 10.1186/s12943-020-01174-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han M, Wang S, Fritah S, Wang X, Zhou W, Yang N, et al. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain. 2020;143:512–30. doi: 10.1093/brain/awz406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–w60. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PloS One. 2018;13:e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005.. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Goede OM, Nachun DC, Ferraro NM, Gloudemans MJ, Rao AS, Smail C, et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell. 2021;184:2633–2648. doi: 10.1016/j.cell.2021.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer. 2020;19:85. doi: 10.1186/s12943-020-01206-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811–28. doi: 10.1038/s41388-018-0382-1. [DOI] [PubMed] [Google Scholar]
- 18.Dey A, Varelas X, Guan KL. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov. 2020;19:480–94. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data included in this study are available upon request by contact with the corresponding author.