Highlights
-
•
FGF9 induced cell proliferation, EMT, migration, and invasion of mouse Lewis lung cancer (LLC) cells, in vitro.
-
•
FGF9 interacted with FGFR1 and activated FAK, AKT, and ERK/MAPK signal pathways, induced the expression of EMT key proteins (N-cadherin, vimentin, snail, MMP2, MMP3 and MMP13) and reduced the expression of E-cadherin.
-
•
FGF9 promoted liver metastasis of subcutaneous inoculated LLC tumor with tumor growth, angiogenesis, EMT and M2-macrophage infiltration in the tumor microenvironment.
-
•
The FGF9/LLC syngeneic animal model provides a useful tool for the mechanism studies of liver metastasis which is the worst prognostic factor for lung cancer patients with distant organ metastasis.
Keywords: FGF9, FGFR1, Lewis lung carcinoma, Epithelial-to-mesenchymal transition/EMT, M2 macrophage infiltration, Liver metastasis
Abstract
Fibroblast growth factors 9 (FGF9) modulates cell proliferation, differentiation and motility for development and repair in normal cells. Abnormal activation of FGF9 signaling is associated with tumor progression in many cancers. Also, FGF9 may be an unfavorable prognostic indicator for non-small cell lung cancer patients. However, the effects and mechanisms of FGF9 in lung cancer remain elusive. In this study, we investigated the FGF9-induced effects and signal activation profiles in mouse Lewis lung carcinoma (LLC) in vitro and in vivo. Our results demonstrated that FGF9 significantly induced cell proliferation and epithelial-to-mesenchymal transition (EMT) phenomena (migration and invasion) in LLC cells. Mechanism-wise, FGF9 interacted with FGFR1 and activated FAK, AKT, and ERK/MAPK signal pathways, induced the expression of EMT key proteins (N-cadherin, vimentin, snail, MMP2, MMP3 and MMP13), and reduced the expression of E-cadherin. Moreover, in the allograft mouse model, intratumor injection of FGF9 to LLC-tumor bearing C57BL/6 mice enhanced LLC tumor growth which were the results of increased Ki67 expression and decreased cleaved caspase-3 expression compared to control groups. Furthermore, we have a novel finding that FGF9 promoted liver metastasis of subcutaneous inoculated LLC tumor with angiogenesis, EMT and M2-macrophage infiltration in the tumor microenvironment. In conclusion, FGF9 activated FAK, AKT, and ERK signaling through FGFR1 with induction of EMT to stimulate LLC tumorigenesis and hepatic metastasis. This novel FGF9/LLC allograft animal model may therefore be useful to study the mechanism of liver metastasis which is the worst prognostic factor for lung cancer patients with distant organ metastasis.
Introduction
Lung cancer has been the second most commonly diagnosed malignancy next to breast cancer and the leading cause of cancer death in both genders worldwide [1]. The 5-year survival rate for lung cancer of all stages combined is 19%, and for lung cancer patients with metastasis is only 5% [1]. Lung cancer is generally classified as two major histological subtypes: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) which comprise about approximately 90% and 10% of all lung malignancies, respectively. Approximately 30-40% of NSCLC patients and 66% of SCLC patients present with stage IV metastasis disease at the time of diagnosis. [2]. Distant metastasis is very common in lung cancer, accounting for 36.1% and 40.1% of NSCLC and SCLC patients, respectively. The most common site of single organ metastasis is bone (22.3%), followed by the lung (17.2%), brain (15.2%), and liver (10%) [2,3]. However, the patients with lung or bone metastases have the best survival rates, whereas, the patients with liver metastases have the poorest survival rate [2], [3], [4].
To overcome this deadly disease, a lot of efforts have been made in the recent decades to explore the molecular mechanisms in lung cancer occurrence and progression. Several crucial regulatory genes have been identified by comprehensive genome analysis tools, including the TP53 and RB1 in SCLC [5], epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), Kirsten rat sarcoma viral oncogene homolog (KRAS), v-raf murine sarcoma viral oncogene homolog B (BRAF), human epidermal growth factor receptor 2 (HER2), mesenchymal epithelial transition (MET), rearranged during transfection (RET), phosphatase and tensin homolog (PTEN), and fibroblast growth factor receptor (FGFR) in NSCLC [5], [6], [7]. Recently, fibroblast growth factors (FGFs) and their receptors, FGFRs, attract researchers’ great attention due to the repeated reports of FGFR somatic activating mutations and gene amplifications in lung cancer [5]. FGFs are known to interact with and activate FGFRs, which subsequently trigger downstream proteins activation, such as phosphatidylinositol-3-kinase/protein kinase B (PI3K/AKT) and the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathways, promoting lung tumor cell proliferation and inhibiting lung tumor cell apoptosis [8,9]. FGFR1 is the most commonly amplified gene (2.25%) and frequently elevated in lung, breast, and colon cancers [10,11]. Mutation in FGFR1 has been observed in lung, colon and breast cancers, and fused with TACC3 gene (FGFR1-TACC3) in glioblastomas, gliomas and bladder cancers [11], [12], [13]. FGFR2 amplification is less frequent (0.34%) and overexpressed in breast cancer, gastric cancer, and intrahepatic cholangiocarcinomas [10,11]. FGFR2 and FGFR3 mutations are more prevalent (1.36% and 1.83%, respectively). And, FGFR2 fusions are the highest frequency of FGFR fusions. As compared with the other FGFR, several reported partners involved in FGFR2 fusion genes, whose biological activity has not been fully characterized, have been identified mainly in cholangiocarcinoma, colorectal cancer and lung cancer, including FGFR2-AHCYL, FGFR2-BICC1, FGFR2-PPHLN1, and FGFR2-TACC3 fusions [14,15]. FGFR3 is affected by mutations in metastatic urothelial carcinomas; FGFR3-TACC3 fusion is present in glioblastomas, gliomas and bladder cancer [12,13,16]. FGFR4 gene amplification and mutation are rare (0.16%) [17,18]. The specific FGFs and FGFRs, including FGF2, FGF9, FGF10, FGFR1, and FGFR2, have been reported that their expressions in NSCLC cell lines in vitro and in patient-derived lung cancer tissues in vivo are elevated along with increased malignant and a worse prognosis [8,9,[19], [20], [21]]. Furthermore, FGFR1 has been reported to interact with EGFR, especially mutated-EGFR, in lung cancer and subsequence activate oncogenic signaling [22,23]. A combination of EGFR and FGFR inbibition is a potential clinical strategy for preventing and overcoming tumorigenicity and drug resistance in EGFR-mutated NSCLC [22,23].
FGF9 is essential for lung development and lung tissue recovery from hypoxia-induced injury [24,25]. During the early pseudoglandular stage, FGF9 is produced in the mesothelium and pulmonary epithelium. Mesothelial FGF9 promotes mesenchyme proliferation/growth via activating mesenchymal FGF signaling, WNT2A expression and WNT/β‐catenin signaling, whereas epithelial FGF9 regulates lung epithelial specification and branching [26,27]. Fgf9-/- mice have severely impaired lung development, and thus the reduced mesenchyme and decreased branching of airways result in a smaller lung size and early postnatal death [28,29]. Abnormal FGF9 expression and aberrant activation of FGF9/FGFR signaling axis are frequently found associated with different kinds of diseases, developmental disorders and cancers, such as lung cancer [20,24], gastric cancer [30], testicular cancer [31,32], ovarian cancer [33], and colon cancer [34]. In addition, FGF9 could enhance cell proliferation and invasive ability of prostate cancer cells [35], testicular cancer cells [31] and ovarian cancer cells [33]. Overexpression of FGF9 can be attenuated by miRNA-140-5p and miR-26a to reduce the tumor growth and metastasis in hepatocellular carcinoma and gastric cancer, respectively [36]. Clinically, Wang and his colleagues observed that FGF9 was highly expressed in lung cancer patients, and disturbing the function of FGF9 could reduce the development of lung cancer [20,24]. Furthermore, elevated FGF9 expression is associated with poor prognosis in NSCLC patients [20].
Cancer metastasis involves a complicated sequential series of events. The epithelial-to-mesenchymal transition (EMT) and mesenchymal-to-epithelial transition (MET) have been clearly defined as critical events for metastasis of primary and secondary carcinomas [37,38]. EMT increases cell migration, invasion, and extravasation, and further facilitates metastasis in multiple cancer types [37,38]. FGF9 has been reported to promote bone metastasis in the transgenic adenocarcinoma of the mouse prostate (TRAMP) animal model. Overexpression of FGF9 leads to invasive prostatic adenocarcinoma (PCa), which accelerates PCa progression [39]. The tumor microenvironment (TME) in the stromal compartment of FGF9-overexpressed tumor tissues from TRAMP mice would change, such as hyperproliferation and hypercellularity [39]. Clinically, high FGF9 expression in colorectal cancer specimens of patients was correlated with poor overall survival. There is a correlation between FGF9 and EMT markers, including reduction of E-cadherin and upregulation of vimentin [40]. Also, the expression or mutation of FGF9 correlates with β-catenin in colorectal, ovarian, and endometrial carcinomas [41], [42], [43]. In addition, matrix metalloproteinases (MMPs) has been reported to associate with tumor cell proliferation, immune response, angiogenesis, invasion and metastasis [37,44]. It has been reported that rhFGF9 and rhFGF20 could promote cell proliferation and migration in Huh7 hepatoma cells by activating the ERK and nuclear factor NF-κB pathways and increasing the expression of MMP26 [45]. However, the effect of FGF9 expression on initiation and progression of lung cancer and its underlying mechanisms are still unknown.
To study whether overexpression of FGF9 contributes to initiation and progression of lung cancer, mouse Lewis lung carcinoma (LLC) cells, a tumor that spontaneously developed as an epidermoid carcinoma in the lung of a C57BL mouse [46], were exploited. Here, we show that FGF9, through FGFR1, could enhance LLC proliferation, EMT, cell migration and invasion by activating MAPK pathways, and further promote liver metastasis.
Materials and methods
Cell line and treatments
The mouse Lewis lung carcinoma (LLC) cell line was purchased from Bioresource Collection and Research Center (BCRC No. 60475) of the Food Industry Research and Development Institute (Hsinchu, Taiwan), which was originally obtained from American Type Culture Collection (ATCC) (Rockville, CT, USA). LLC cells were cultured in Dulbecco's modified Eagle's medium containing 0.5 mM sodium pyruvate, 2.5 mM L-glutamine, and 1.2 g/L sodium bicarbonate, supplemented with 10% fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. 100 μg/mL streptomycin and 100 I.U./mL penicillin were added, if necessary. All information of chemicals and materials used in this study are listed in Table S1.
Cell proliferation assay
LLC cells were seeded in 96-well plates at a density of 6,000 cells per well with 100 μl culture medium. After 18 h of serum free starvation, the cells were treated with 0 (BSA vehicle control), 1, 5, 25, 50 and 100 ng/ml recombinant human FGF9 in the medium containing 1% FBS for 12, 24, 48 and 72 h, respectively. FGF9 stock solution: 20 μg/ml FGF9 in PBS with 0.1% BSA). MTT (0.5 mg/ml) was added at different time points and incubated at 37°C for 4 h. Then, the MTT medium was discarded and 50 μl DMSO was added per well to dissolve the crystals by shaking the plate for 20 min in the dark. The OD values in each treatment were determined using an ELISA reader at 570 nm (Molecular Devices, San Jose, CA, USA) [31,47].
Migration assay
A. Wound healing migration assay
LLC cells were seeded into ibidi 2-well culture inserts in 12-well dishes and allowed to reach a confluent monolayer. After 24 h, the culture inserts were removed. The wound width was monitored over time by microscopy and photographed.
B. The transwell migration and invasion assay
The 6.5 mm Corning transwell chambers with 8.0 μm pore size polycarbonate membrane filter were used. After cells were serum-starved for 18 h, 5 × 105 cells per well were suspended in 100 μl serum-free RPMI and seeded into the upper chamber of a 24 well plate. Then, 600 μl medium containing 10% FBS was added to the well. After 48 h incubation, the migrated cells were fixed by 4% paraformaldehyde for 20 min and then stained by a 0.5% crystal violet solution in 70% methanol for 20 min. Stained cells were photographed by an inverted light microscope. The quantification of microscopic images was analyzed using ImageJ software.
Western blotting
Treated LLC cells were washed by ice-cold PBS and lysed by ice-cold lysis buffer (Table S1). After centrifugation at 12,000 x g for 12 min at 4°C, supernatants were collected and stored at -20°C. Twenty-five μg total protein was separated by 10% SDS-PAGE and transferred onto polyvinylidence difluoride (PVDF) membranes. The PVDF membranes with transferred protein were blocked with 5% non-fat milk for 1 h, and then incubated with the primary antibody in TBS buffer (Table S1) for 16-18 h at 4°C. After washing with TBS buffer containing 0.1% Tween 20, the signal was detected with horseradish peroxidase-conjugated secondary antibody and visualized with chemiluminescence HRP substrate. The target proteins were quantitated by a computer-assisted image analysis system (UVP bioImage system software, UVP Inc., CA, USA). Protein level was quantitated by using ImageJ software (NIH, Bethesda, MD, USA) and the amount of GAPDH in each lane was detected as a loading control. All information of antibodies used in this study are listed in Table S2.
Lentiviral shRNA silencing of FGFR1-R4
The pCMV-ΔR8.91, pMD.G and all pLKO-based shRNA clones for FGFR1-4 and non-silencing shRNA (scrambled sequence) were obtained from the National RNAi Core Facility at Academia Sinica (Taipei, Taiwan). All shRNA plasmids used in this study are described in Table S3. Lentivirus preparations were performed according to the supplier's protocols. LLC cells were infected with lentivirus in the presence of 8 μg/ml polybrene. For stable cell lines, cells were selected by 5.5 μg/ml puromycin 48 h after infection and maintained in growth medium containing 5.5 μg/ml puromycin.
In vivo animal study
Six-week-old male C57BL/6 mice were purchased from National Cheng Kung University (NCKU) Animal Center (Tainan, Taiwan). Mice received a subcutaneous (s.c.) injection of 5 × 105 LLC cells in the right hip. Five days after tumor inoculation, tumor-bearing mice were randomly assigned to three groups: FGF9, BSA (vehicle control) and control (no treatment). The 100l of 25 ng/ml FGF9 or 0.00125 % BSA were locally injected into the LLC cell-forming tumor mass daily for 16 days. FGF9 injecting solution was freshly prepared by diluting FGF9 stock solution (20 μg/ml FGF9 in PBS with 0.1% BSA). Tumor size and body weight were measured daily. The tumor volume was calculated by the formula: 0.52 × length × width × width [48]. LLC tumor tissues were fixed by a 4% paraformaldehyde solution after the mice were sacrificed. All experiments were performed in accordance with relevant guidelines and regulations, which were approved by the Institutional Animal Care and Use Committee (IACUC) of NCKU (IACUC approval no.: 105102).
Immunohistochemistry assay
Formalin-fixed paraffin-embedded tumor tissue sections were dewaxed in xylene, dehydrated in ethanol, and washed in PBS. Endogenous peroxidase activity was blocked by incubation with 0.3% H2O2 followed by washing with PBS. Sodium citrate buffer was used to retrieve the antigen for 50 min at 120°C autoclave. The sections were blocked with 2% nonfat milk for 1 h and were incubated overnight at 4°C with primary antibodies. Signal was visualized using HRP-conjugated secondary antibody and the chromogenic substrate 3,3′-diaminobenzidine. The sections were then counterstained with hematoxylin, dehydrated with a graded ethanol series, cleared in xylene, and mounted with coverslip using mounting solution. Negative controls were performed in each IHC assay by replacing the primary antibodies with a corresponding non-specific IgG.
Statistical analysis
Statistical analysis was performed by using GraphPad Prism 6 software (La Jolla, CA, USA). The p<0.05 was considered to be statistically significant in this study. The specific statistical tests used are indicated in each figure legend.
Results
FGF9 promoted the proliferation, migration and invasion of LLC cells
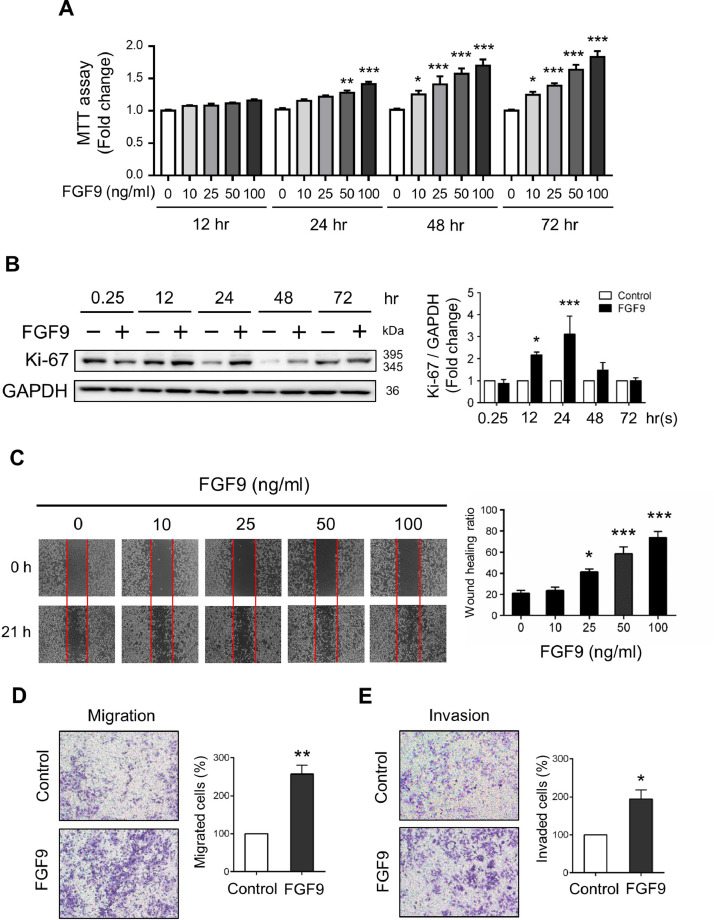
In order to evaluate the effect of FGF9 on the cell proliferation of Lewis lung carcinoma cells (LLC cells), the MTT assay was performed. After FGF9 treatment at concentrations ranging from 0-100 ng/ml for 12, 24, 48 or 72 h, LLC cell proliferation significantly increased in time and dose-dependent manners. FGF9 at 50 and 100 ng/ml significantly induced LLC cell proliferation after 24-, 48- and 72 h exposure, respectively, whereas low doses of FGF9 at 10 and 25 ng/ml could also significantly induce cell proliferation after 48- and 72 h exposure (Fig. 1A). In addition, the expression of Ki-67, a standard marker of cell proliferation that is commonly used to evaluate the growth fraction of a cell population [49], significantly increased at 12 and 24 h after FGF9 treatment (Fig. 1B).
Fig. 1.
FGF9 induced LLC cell proliferation, migration and invasion. (A) MTT assay for the cell proliferation of 0, 10, 25, 50 or 100 ng/ml FGF9-treated LLC cells for 12, 24, 48 and 72 h, respectively. (B) Western blot analysis and quantification of Ki-67 expression in LLC cells treated with 0 (Control) or 50 ng/ml FGF9 for 0.25, 12, 24, 48 and 72 h, respectively. (C) Wound healing cell migration assay for LLC cells treated with 0, 10, 25, 50 and 100 ng/ml FGF9 for 21 h (left panel). The rate of wound closure was quantified as a migration index (right panel) by measuring the area of migrated cells across the baseline (red line in left panel). (D) Migration assay and (E) Matrigel invasion assay with quantified analyses of 0 (Control) or 50 ng/ml FGF9-treated LLC cells using the ibidi and a Transwell system were illustrated, respectively. All values are presented as the mean±SEM; (A) n=6, (B) n=4, (C-E) n=3. The data were analyzed by (A) two-way ANOVA with Sidak's multiple comparisons post-tests, (B, C) one-way ANOVA with Tukey's multiple comparisons post-tests for, and (D) and (E) Student t test; *p<0.05, **p<0.01, and ***p<0.001 vs. the Control (0 ng/ml FGF9) group.
To further assess the effects of FGF9 on LLC cell migration and invasion, examining which is the key determinant in the malignant progression and metastasis of cancer, in vitro wound healing and transwell assays were performed in LLC cells with or without FGF9 treatment. As shown in Fig. 1C, wound healing assays revealed that 25, 50 and 100 ng/ml FGF9-treated LLC cells exhibited rapid wound closure. Furthermore, a significant promotion of cell motility and invasion was observed in LLC cells at 24 h after 50 ng/ml FGF9 treatments compared with the control cells (Fig. 1D and 1E). These results demonstrate that treatment with FGF9 caused significant increase of cell proliferative, migratory and invasive capability in LLC cells.
FGF9 activated FAK, AKT, and MAPK/ERK signaling pathways in LLC cells
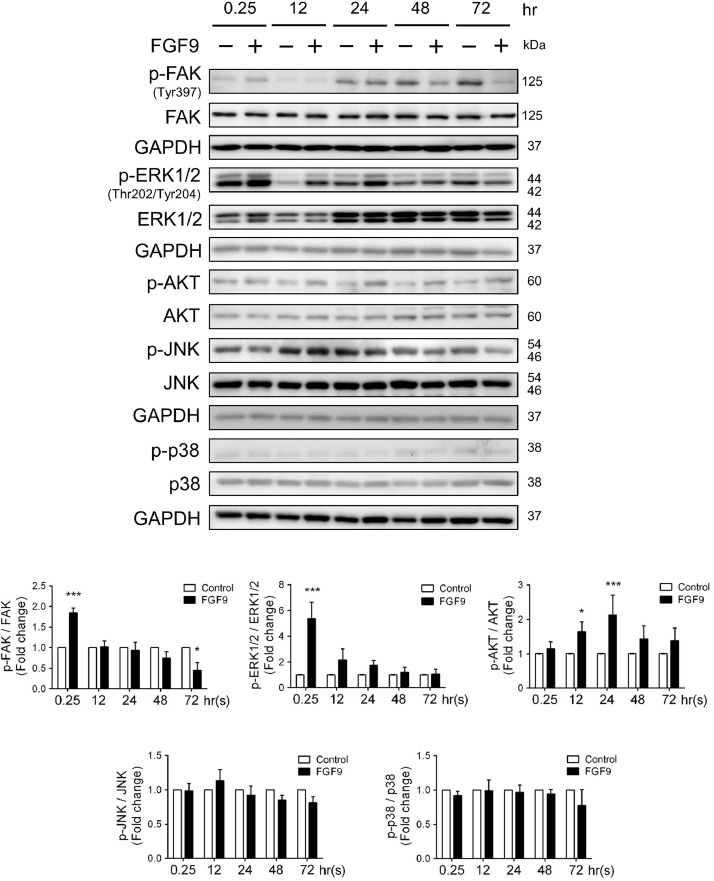
Focal adhesion kinase (FAK), a cytoplasmic protein tyrosine kinase, has long been known as a regulator of cell adhesion, migration, proliferation and survival, and also serves an important role in the EMT process [50]. FAK has been reported to activate the PI3K/AKT signaling cascade, which subsequently lead to the upregulation of EMT marker proteins, including N-cadherin, vimentin and Snail [51,52]. Additionally, several studies have shown that PI3K/Akt and MAPK pathways are the downstream molecules of FGFRs which is activated by FGF9 [53,54]. The PI3K/Akt signaling pathway is one of the well-characterized pathway with regard to the signal transduction and biological processes, such as cell proliferation, anti-apoptosis and cell survival [55]. In addition, the MAPK signaling pathway is known to play a crucial role in the development of lung cancer and its downstream molecules (ERK, JNK, and p38) are associated with tumor growth, migration, and invasion. To further characterize the mechanism underlying the promotion of LLC cell proliferation, migration and invasion by FGF9, we examined the expression of FAK, ERK, AKT, JNK, and p38 using the western blotting analysis. As shown in Fig. 2, while FGF9 had no effect on the phosphorylation of JNK and p38, FGF9 induced the phosphorylation of FAK, AKT, and ERK1/2 compared with the vehicle control, which suggest that FGF9 positively regulated FAK, AKT, and ERK/MAPK signaling pathways related to cell proliferation.
Fig. 2.
FGF9 activated FAK, ERK, and AKT pathways in LLC cells. Western blot analysis and quantification of protein expression of p-FAK, total FAK, p-ERK1/2, total ERK1/2, p-AKT, total AKT, p-JNK, total JNK, p-p38, and total p38 in LLC cells treated with 0 (Control) or 50 ng/ml FGF9 for 0.25, 12, 24, 48 and 72 h, respectively, are illustrated. Quantification values are represented as the mean ± SEM; n=3. The data were analyzed by two-way ANOVA with Sidak's multiple comparisons post-tests. *p<0.05 and ***p<0.001 vs. the Control group at each time point.
FGF9 promoted epithelial-mesenchymal transition in LLC cells
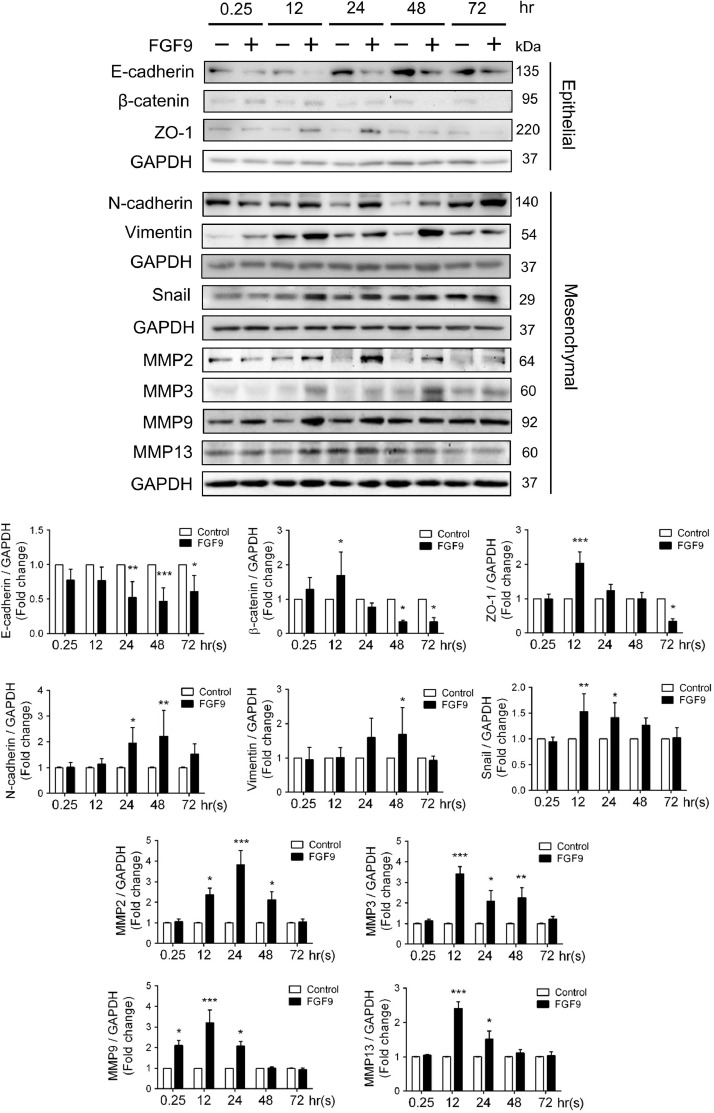
Epithelial-mesenchymal transition (EMT) is a highly dynamic process through which epithelial cancer cells convert into more aggressive phenotypes with increased cell migration and invasion ability [56]. Also, EMT has been reported to be associated with tumor metastasis and is characterized by loss of epithelial markers (E-cadherin, zonula occludens-1 and β-catenin) and gain of mesenchymal markers (N-cadherin, vimentin and MMPs) resulting in the acquisition of the migratory and invasive properties [56]. Thus, we examined the expression levels of EMT markers and MMPs to explore their correlation with FGF9. The results showed that the level of E-cadherin greatly decreased in FGF9-treated LLC cells. β-catenin and ZO-1 were also decreased after FGF9 treatment for 48 and 72 h, respectively. In contrast, the levels of N-cadherin, vimentin, and snail were markedly increased in FGF9-treated group (Fig. 3). In addition, the expression of MMP2, MMP3, MMP9 and MMP13 were significantly induced by FGF9 (Fig. 3), whereas the expression of MMP1 and MMP7 were not affected by FGF9 in LLC cells (Fig. S1). Collectively, these results indicate that FGF9 promoted LLC migration and invasion by inducing EMT and MMPs expressions.
Fig. 3.
FGF9 promoted EMT in LLC cells. Western blot analysis and quantification of protein expression of EMT related proteins: E-cadherin, β-Catenin, ZO-1, N-cadherin, vimentin, Snail, MMP2, MMP3, MMP9, and MMP13 in LLC cells treated with 0 (Control) or 50 ng/ml FGF9 for 0.25, 12, 24, 48 and 72 h, respectively, are illustrated. Quantification values are represented as the mean ± SEM; n=3. The data were analyzed by two-way ANOVA with Sidak's multiple comparisons post-tests. *p<0.05, **p<0.01, and ***p<0.001 vs. the Control group at each time point.
Lentiviral shRNA silencing of FGFR1 inhibited FGF9-induced ERK1/2 phosphorylation, LLC cell proliferation, EMT markers, migration and invasion
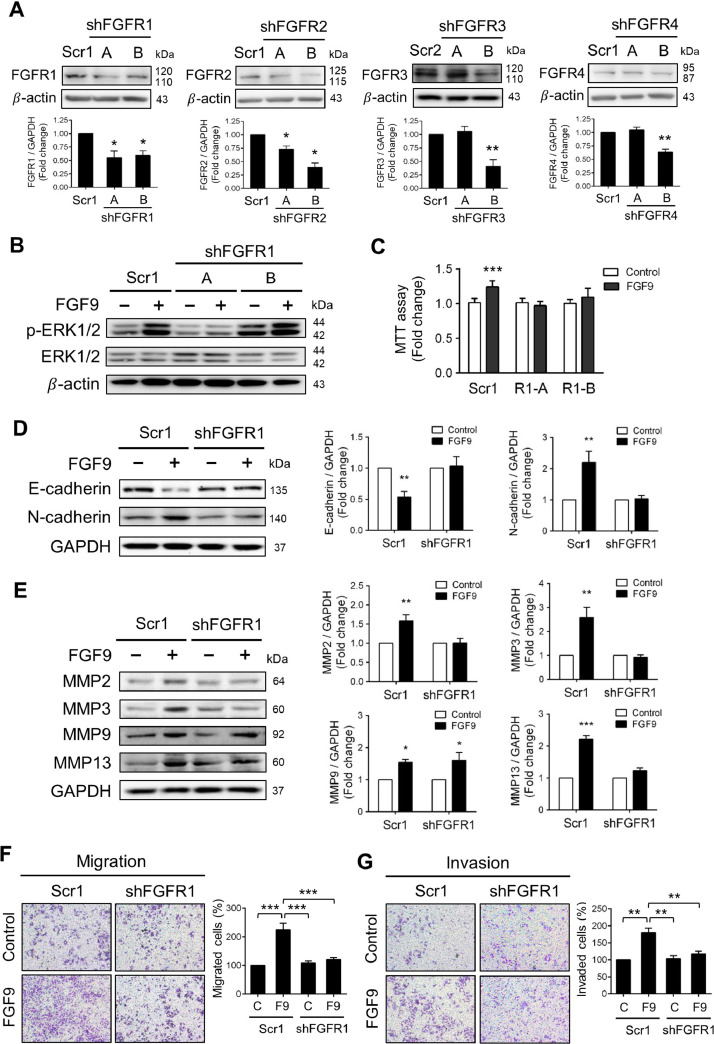
It is well-known that FGFs bind to different FGFRs, FGFR1-FGFR4, to activate multiple intracellular signaling cascades [57]. To identify the responsible receptor(s) for FGF9 signaling in LLC cells, a lentiviral vector-based shRNA system was performed to silence FGFR1-4. The Western blotting analysis and quantification shows a significant knockdown (kd) of FGFR1, FGFR2, FGFR3 and FGFR4 protein levels by the shFGFR1A, shFGFR1B, shFGFR2A, shFGFR2B, shFGFR3B and shFGFR4B sequence in comparison to its corresponding scramble control (Fig. 4A). Each shRNA was effective in suppressing its target protein expression without affecting the expression of other FGFRs (Figs. 4A, S2).
Fig. 4.
Lentiviral shRNA silencing of FGFR1 inhibited FGF9-induced ERK1/2 phosphorylation, cell proliferation, EMT markers, migration and invasion in LLC cells. (A) Western blot analysis and quantification of gene silencing verification at protein level in LLC cells transfected with a specific shRNA against FGFR1-4. Two different shRNA targeted sequences were used for each FGF receptor (Table S3). Cells transfected with a non-silencing shRNA sequence (scrambled sequence) in the PLKO.TRC1-puro (Scr1) or PLKO.TRC2-puro (Scr2) vectors were used as control as indicated. (B) The FGFR1 knockdown (shFGFR1) LLC cells and their scrambled control were treated with 0 (Control) or 50 ng/ml FGF9 for 0.25 h. The protein expression levels of ERK1/2 and p-ERK1/2 were analyzed by Western blot assay. (C) MTT assay for cell proliferation of shFGFR1 LLC cells treated with 0 (Control) or 50 ng/ml FGF9 for 24 h. (D) Western blot analysis for the expression of E-cadherin and N-cadherin in shFGFR1 LLC cells treated with 0 (Control) or 50 ng/ml FGF9 for 24 h. (E) Western blot analysis for the expression of MMP2, MMP3, MMP9 and MMP13 in shFGFR1 LLC cells treated with 0 (Control) or 50 ng/ml FGF9for 24 h. (C) The migration and (D) matrigel invasion assays with 0 (Control) or 50 ng/ml FGF9 treatment in shFGFR1-LLC cells and their scramble control (Scr1) using transwell system. All values are represented as the mean ± SEM; (B) n=6; (C) and (D) n=4. The data were analyzed by two-way ANOVA with Sidak's multiple comparisons post-tests; *p<0.05, **p<0.01 and ***p<0.001 vs. the Control group.
Knockdown of FGFR1 led to the inhibition of FGF9-induced ERK phosphorylation in LLC cells (Fig. 4B). In contrast, the ERK phosphorylation still markedly elevated in FGFR2-, FGFR3- and FGFR4-kd LLC cells after FGF9 treatment (Fig. S3A). Furthermore, the cell proliferation is blocked by FGFR1 knockdown after FGF9 treatment (Fig. 4C), whereas FGFR2, R3 and R4 knockdowns had no significant difference in cell proliferation after FGF9 treatment compared with the Control treatment (Fig. S3B). These results indicate that FGF9 activated ERK1/2 signaling pathway through FGFR1 to induce LLC cell proliferation.
In addition, the EMT related markers, cell migration and invasion abilities were further evaluated in FGFR1-kd LLC cells. As shown in Fig. 4D and 4E, FGFR1 knockdown rescued the E-cadherin inhibition and blocked the expression of N-cadherin, MMP2, MMP3, and MMP13, but not MMP9, after FGF9 treatment. Moreover, FGFR1 knockdown inhibited the migration and invasion of LLC cells after FGF9 exposure (Fig. 4F and 4G). These results indicate that changes in the biological functions, including migration and invasion, of LLC cells induced by FGF9-FGFR1 may be via MMPs (MMP2, MMP3, and MMP13) overexpression, extracellular matrix (ECM) degradation, and EMT.
FGF9 stimulated tumor growth in allograft model with LLC cell induction
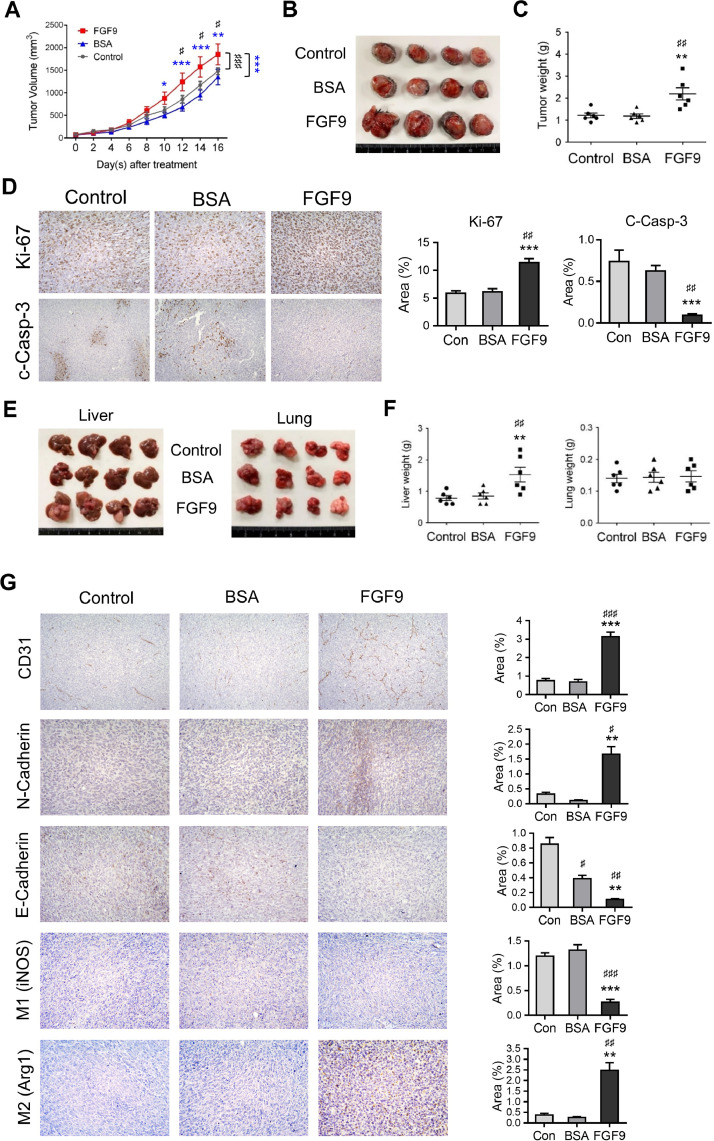
Since FGF9 could promote LLC cell proliferation and EMT in vitro, the effect of FGF9 on lung cancer in vivo was further investigated by using male C57BL/6 mice as the allograft model. LLC cells were subcutaneously injected at the flank of the mice. Five days after LLC cell inoculation, mice were given daily injections of 50 ng/ml FGF9 or BSA (vehicle, 0.001%) for 10 days. Results showed that the tumor volumes of FGF9 group were significantly higher than that of BSA (vehicle) and Control (no treatment) groups (Fig. 5A and 5B), and the tumor weights significantly increased with FGF9 treatment (Control group, 1.22±0.11 g; BSA group, 1.18±0.10 g; and FGF9 group, 2.19±0.28 g) (Fig. 5C).
Fig. 5.
FGF9 stimulated tumor growth, angiogenesis, EMT, M2 macrophage infiltration and tumor metastasis of lung carcinoma in vivo. LLC cells subcutaneously injected C57BL/6 mice were treated with 25 ng/ml FGF9 or 0.00125% BSA (vehicle control) once daily for 16 days. Mice in Control (Con) group were subcutaneously inoculated with LLC cells and received no treatment. (A) Tumor growth curves were plotted against time. (B) Photograph and (C) weights of excised subcutaneous LLC tumors from the Control, BSA and FGF9 group mice at the time mice were sacrificed. (D) Immunohistochemical assays of Ki-67 and cleaved Caspase-3 (c-Casp3) expressions in the LLC subcutaneous injected tumors from various groups (original magnification, Ki67: × 200; c-Casp3: x100). (E) Photograph and (F) weight of excised liver and lung tissues of the FGF9, BSA or Control (Con) mice. (G) Immunohistochemical assays of CD31, E-cadherin, and N-cadherin, and M1 (iNOS) and M2 (Arg1) expressions in the LLC subcutaneous tumors from FGF9, BSA or Control mice (original magnification, CD31: x100, E-Cadherin, N-Cadherin, M1 and M2: x200). Values are represented as the mean ± SEM; n=6. The data were analyzed by two-way ANOVA with Sidak's multiple comparisons post-tests in (A) or one-way ANOVA with Tukey's multiple comparisons post-tests in (C), (D), (F) and (G); ♯p<0.05, ♯♯p<0.01 and ♯♯♯p<0.001 vs. the Control group; *p<0.05, **p<0.01 and ***p<0.001 vs. the BSA group.
To further investigate the tumor-growth promoting effects of FGF9 in vivo, immunohistochemistry (IHC) examinations of Ki-67 and cleaved caspase-3 were performed. Data showed that FGF9 exposure resulted in significantly increased Ki-67 expression and decreased cleaved caspase-3 expression in the LLC tumor tissue (Fig. 5D). These results suggest that FGF9 promoted tumor growth, in vivo, by increasing cell proliferation and decreasing cell death; the apoptosis.
FGF9 promoted EMT marker expressions and liver metastasis of LLC tumor, in vivo
Compared to BSA and Control group mice, liver metastasis significantly increased in FGF9-treated mice (Fig. 5E and 5F). However, there were no significant differences in lung metastasis between FGF9, BSA and Control groups (Fig. 5E and 5F). In addition, the IHC expressions of CD31 and N-cadherin in LLC tumor were significantly increased in FGF9-treaated group as compared to BSA and Control groups (Fig. 5G), whereas the E-cadherin was significantly repressed in LLC tumor with FGF9 exposure. These in vivo observations further support that FGF9 induced the expression of N-cadherin and inhibited the expression of E-cadherin, in vitro (Fig. 3). Thus, FGF9 significantly promoted tumor growth both in in vitro cellular system and in vivo animal model related to tumorigenesis. All these findings strongly suggest that FGF9 might regulate EMT and promote metastasis in lung cancer.
Interplay between cancer epithelial cells and the surrounding immune cells do shape the TME to promote cancer progression [[58], [59], [60]]. Tumor associated macrophages (TAMs) have been reported to involve in the regulation of EMT process [61]. TAMs originate from circulating monocytes which are recruited to the tumor site shaped the TAM phenotype by tumor derived factors, such as vascular endothelial growth factor A (VEGF-A), colony-stimulating factor-1 (CSF-1) and CC chemokine ligand 2 (CCL2), in the tumor microenvironment. These and other factors in the tumor microenvironment promote TAMs toward tumor-supportive M2-polarized macrophages, although M1-polarized macrophages with anti-tumor activity were also reported in some types of cancer [62,63]. M1 macrophages exert tumoricidal activity by secreting pro-inflammatory cytokines, nitro oxide and reactive oxygen species (ROS), presenting antigens and activating cytotoxic T lymphocytes. In contrast, M2 macrophages are responsible for supporting angiogenesis and suppressing immunoresponse in TME, and thus promote tumor progression and distant organ metastasis [59,60]. There are two major arginine metabolic pathways in macrophages. Arginine is a precursor and can be metabolized either by inducible nitric oxide synthase (iNOS; NOS2), which is highly expressed in M1 macrophages, to nitric oxide and citrulline or hydrolyzed by the enzyme arginase 1 (Arg1), which is abundantly expressed in M2 macrophages, to ornithine and urea. Therefore, iNOS and Arg1 are commonly used as one of markers for defining the M1 and M2 polarization of macrophages [[63], [64], [65]].
Here, in the subcutaneous inoculated LLC tumor, the M2 macrophage infiltration was increased in FGF9 treatment group, whereas the M1 macrophages was decreased after FGF9 treatment (Fig. 5G). These results imply that FGF9 may contribute to macrophage recruitment and M2 polarization to further promote liver metastasis.
Discussion
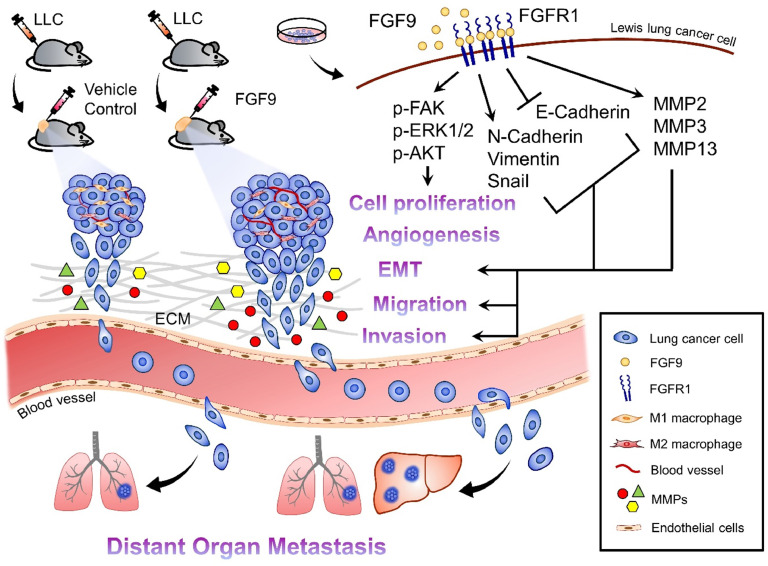
In this study, we explored the role of FGF9 in tumor progression and metastasis in lung cancer. We determined that FGF9 activated the FAK, AKT, ERK/MAPK pathways; promoted lung cancer cell proliferation, migration and invasion; and changed the expressions of EMT markers and MMPs in lung cancer cells. We also found that FGF9 promoted the tumor growth and metastasis of subcutaneous injected tumor in vivo. The major novelty of this study is that FGF9 promoted the liver metastasis of lung cancer cells through FGFR1 via ERK/MAPK signaling pathways and inflammation (Fig. 6).
Fig. 6.
Schematic graph illustrates how FGF9 induces LLC tumorigenesis related to tumor microenvironment changes and liver metastasis. FGF9 induced LLC cell proliferation, migration, invasion, in vitro, and promoted tumor growth, angiogenesis, M2 macrophage infiltration, EMT, and liver metastasis, in vivo, by increasing the expressions of p-FAK, p-AKT, p-ERK1/2, N-cadherin, vimentin and MMPs, and decreasing E-cadherin expression. This novel FGF9/LLC allograft animal model may therefore be useful to study the mechanism of liver metastasis which is the worst prognostic factor for lung cancer patients with distant organ metastasis.
Mechanism wise, we demonstrated that FGF9 could induce cell proliferation, EMT, migration, and invasion in lung cancer cells by regulating the ERK/MAPK pathway, ECM proteins and MMPs. A number of studies have reported that MAPK cascades (ERK1/2, JNK1/2, and p38) are involved in cell growth, migration and MMPs activity in various cancers, including lung, colorectal, ovarian and prostate cancers [66,67]. It has been shown that the MMPs are regulated by MAPK pathways and implicated in regulating ECM degradation and remodeling, which is associated with EMT to promote cell migration and invasion in cancers [68]. Those observations are parallel with ours that FGF9 promoted the proliferation, migration and invasion in LLC cells (Fig. 1). We also demonstrated that FGF9 could interact with FGFR1 and trigger activation of the downstream signaling pathways, including the inhibition of E-cadherin and the upregulations of N-cadherin, MMP2, MMP3 and MMP13, and subsequently increased cell migration and invasion in vitro (Figs. 2, and –4).
In our animal study, IHC staining results showed that the TME of subcutaneous LLC tumor tissue was changed, including hypercellularity and hyperproliferation (Fig. 5). Changes in TME, such as EMT, anti-anoikis, angiogenesis, lymphangiogenesis and immunesupesssion, are complicated processes that take place in the TME during cancer progression [59,69]. TAMs, the major component of the TME, can be polarized into tumor-inhibiting M1 macrophages or tumor-promoting M2 macrophages [59]. Clinically, TAMs are frequently detected in the lung cancer specimens [70]. Accumulating evidences from in vivo animal models and in vitro studies suggest that M2 macrophages could promote tumor growth, suppress antitumor immunity, and induce angiogenesis, lymphangiogenesis and metastasis [62,71]. In this study, we found that FGF9 promoted the recruitment of M2 macrophage rather than M1 macrophage in LLC tumor tissues (Fig. 5G). In addition, CD31 expression was higher in FGF9 group in which might be related to M2 recruitment (Fig. 5G), since it has been reported that the recruitment of M2 could be induced by FGF9 to promote angiogenesis involved in tumorigenesis [71]. Furthermore, upregulation of MMPs by TAMs was linked to the promotion of EMT, angiogenesis and metastasis [69]. Indeed, studies have shown that MMPs are TME proteins facilitating tumor cell migration through the extracellular matrix [69,72].
In the present study, subcutaneous inoculation of LLC cells in C57BL/6 mice was used to investigate the subcutaneous growth and distant organ metastasis of the lung cancer cells. Cancer allograft and xenograft models, classified as subcutaneous (SC) and orthotopic transplanted (OT) tumor models, are widely used to study tumorigenesis, tumor progression and/or to assess response to therapy. SC models are commonly used to investigate tumorigenesis or anticancer activity because of their high reproducibility and ease of monitoring tumor growth and therapeutic response. In recent years, many effort has been made to develop more clinically relevant models by the use of orthotopic transplantation of tumor in rodents. OT models are considered to reproduce the tumor microenvironment, which is thought to be more clinically relevant than SC models, and emulate important biological features of cancer progression, angiogenesis, metastasis, therapeutic sensitivity and drug resistance. The differences between SC and OT models might be associated with different tumor microenvironments [73,74]. However, the detailed mechanisms are still unclear.
Mouse Lewis lung carcinoma is highly metastatic in immunocompetent syngeneic murine background of C57BL/6 mouse, which is a good animal model of tumor metastasis for studying the efficacy of chemotherapeutic agents, the targeted therapies, and the mechanisms of tumor growth and/or metastasis because of the true immune and toxicity responses. In addition, this model is used for investigating the interaction of the tumor and the immune system which cannot be ignored in vivo. LLC is known to metastasize to the lung by subcutaneous and intravenous injection in mice [75,76]. It has been only reported that activation of BMP2 signaling promotes bone metastasize of tail vein-injected LLCs in C57BL/6 mice [76]. Here, our novel finding is that FGF9 could enhance liver metastasis with the subcutaneous LLC tumor (Fig. 5). Clinically, liver metastasis is a well-known worse prognostic indicator in lung cancer patients [4,77]. However, unlike bone or brain metastasis, few studies focused on liver metastasis of lung cancer. It remains unclear why poor clinical outcomes are associated with liver metastasis. Further efforts should be made to reveal the mechanism and find out effective treatment for liver metastasis of lung cancer [2,3]. Here, our novel finding provided a good animal model for studying the mechanisms of liver metastasis of lung cancer in immunocompetent syngeneic background.
Taken together, FGF9 could bind FGFR1 to promote LLC cell proliferation, migration and invasion with the increasing expressions of EMT proteins (N-cadherin, vimentin, MMP2, MMP3 and MMP13) and the decreasing expression of E-cadherin possibly through the activation of MAPK pathway. Moreover, FGF9 could promote the subcutaneous LLC tumor growth with the recruitment of M2 macrophages, angiogenesis and EMT in the TME, and the liver metastasis in vivo. In conclusion, FGF9 triggers ERK signal through FGFR1 with the aberrant TME and induction of EMT to stimulate LLC hepatic tumorigenesis and metastasis.
Table S1. Chemicals and materials used in this study.
CRediT authorship contribution statement
Ming-Min Chang: Conceptualization, Project administration, Methodology, Investigation, Validation, Formal analysis, Writing – original draft. Su-Zhen Wu: Resources, Investigation, Funding acquisition. Shang-Hsun Yang: Resources, Writing – review & editing. Chia-Ching Wu: Resources, Writing – review & editing. Chia-Yih Wang: Supervision, Writing – review & editing. Bu-Miin Huang: Supervision, Conceptualization, Project administration, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors have no conflict of interest.
Acknowledgments
Funding
This research was funded by Ministry of Science and Technology, Taiwan, Republic of China (grant number: MOST 105-2320-B-006-001, and MOST 106-2320-B-006-001, MOST 110-2320-B-006-025 MY3 to BMH; grant number: MOST 106-2811-B-006-014, MOST 107-2811-B-006-0519, MOST 109-2811-B-006-555 to MMC; and grant number: CLFHR11025 to SZW and BMH). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Acknowledgments
We are grateful for the support from the Core Research Laboratory, College of Medicine, National Cheng Kung University.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101208.
Contributor Information
Chia-Yih Wang, Email: b89609046@gmail.com.
Bu-Miin Huang, Email: bumiin@mail.ncku.edu.tw.
Appendix. Supplementary materials
Table S2. Antibodies used in this study.
Table S3. Lentiviral shRNA plasmids used for FGFRs knockdown.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Li J., Zhu H., Sun L., Xu W., Wang X. Prognostic value of site-specific metastases in lung cancer: a population based study. J. Cancer. 2019;10(14):3079–3086. doi: 10.7150/jca.30463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin B.D., Jiao X.D., Liu J., Liu K., He X., Wu Y., Ling Y., Duan X.P., Qin W.X., Wang Z. The effect of liver metastasis on efficacy of immunotherapy plus chemotherapy in advanced lung cancer. Crit. Rev. Oncol. Hematol. 2020;147 doi: 10.1016/j.critrevonc.2020.102893. [DOI] [PubMed] [Google Scholar]
- 4.Shiroyama T., Suzuki H., Tamiya M., Tamiya A., Tanaka A., Okamoto N., Nakahama K., Taniguchi Y., Isa S.I., Inoue T. Clinical characteristics of liver metastasis in nivolumab-treated patients with non-small cell lung cancer. Anticancer Res. 2018;38(8):4723–4729. doi: 10.21873/anticanres.12779. [DOI] [PubMed] [Google Scholar]
- 5.Peifer M., Fernandez-Cuesta L., Sos M.L., George J., Seidel D., Kasper L.H., Plenker D., Leenders F., Sun R., Zander T. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 2012;44(10):1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villalobos P., Wistuba II. Lung cancer biomarkers. Hematol. Oncol. Clin. N. Am. 2017;31(1):13–29. doi: 10.1016/j.hoc.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamberti G., Andrini E., Sisi M., Rizzo A., Parisi C., Di Federico A., Gelsomino F., Ardizzoni A., Beyond EGFR. ALK and ROS1: current evidence and future perspectives on newly targetable oncogenic drivers in lung adenocarcinoma. Crit. Rev. Oncol. Hematol. 2020;156 doi: 10.1016/j.critrevonc.2020.103119. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y., Lu T., Li Z., Lu S. FGFR1 regulates proliferation and metastasis by targeting CCND1 in FGFR1 amplified lung cancer. Cell Adh. Migr. 2020;14(1):82–95. doi: 10.1080/19336918.2020.1766308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K., Ji W., Yu Y., Li Z., Niu X., Xia W., Lu S. FGFR1-ERK1/2-SOX2 axis promotes cell proliferation, epithelial-mesenchymal transition, and metastasis in FGFR1-amplified lung cancer. Oncogene. 2018;37(39):5340–5354. doi: 10.1038/s41388-018-0311-3. [DOI] [PubMed] [Google Scholar]
- 10.Consortium A.P.G. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Facchinetti F., Hollebecque A., Bahleda R., Loriot Y., Olaussen K.A., Massard C., Friboulet L. Facts and new hopes on selective FGFR inhibitors in solid tumors. Clin. Cancer Res. 2020;26(4):764–774. doi: 10.1158/1078-0432.CCR-19-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Stefano A.L., Fucci A., Frattini V., Labussiere M., Mokhtari K., Zoppoli P., Marie Y., Bruno A., Boisselier B., Giry M. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin. Cancer Res. 2015;21(14):3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh D., Chan J.M., Zoppoli P., Niola F., Sullivan R., Castano A., Liu E.M., Reichel J., Porrati P., Pellegatta S. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sia D., Losic B., Moeini A., Cabellos L., Hao K., Revill K., Bonal D., Miltiadous O., Zhang Z., Hoshida Y. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nat. Commun. 2015;6:6087. doi: 10.1038/ncomms7087. [DOI] [PubMed] [Google Scholar]
- 15.Lowery M.A., Ptashkin R., Jordan E., Berger M.F., Zehir A., Capanu M., Kemeny N.E., O'Reilly E.M., El-Dika I., Jarnagin W.R. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin. Cancer Res. 2018;24(17):4154–4161. doi: 10.1158/1078-0432.CCR-18-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mollica V., Rizzo A., Montironi R., Cheng L., Giunchi F., Schiavina R., Santoni M., Fiorentino M., Lopez-Beltran A., Brunocilla E. Current strategies and novel therapeutic approaches for metastatic urothelial carcinoma. Cancers (Basel) 2020;12(6) doi: 10.3390/cancers12061449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Luca A., Esposito Abate R., Rachiglio A.M., Maiello M.R., Esposito C., Schettino C., Izzo F., Nasti G., Normanno N. FGFR fusions in cancer: from diagnostic approaches to therapeutic intervention. Int. J. Mol. Sci. 2020;21(18) doi: 10.3390/ijms21186856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor J.G.t., Cheuk A.T., Tsang P.S., Chung J.Y., Song Y.K., Desai K., Yu Y., Chen Q.R., Shah K., Youngblood V. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J. Clin. Invest. 2009;119(11):3395–3407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Zhang S., Wei L., Wang Z., Ma W., Liu F., Qian Y. FGF2 and FGFR2 in patients with idiopathic pulmonary fibrosis and lung cancer. Oncol. Lett. 2018;16(2):2490–2494. doi: 10.3892/ol.2018.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohgino K., Soejima K., Yasuda H., Hayashi Y., Hamamoto J., Naoki K., Arai D., Ishioka K., Sato T., Terai H. Expression of fibroblast growth factor 9 is associated with poor prognosis in patients with resected non-small cell lung cancer. Lung Cancer. 2014;83(1):90–96. doi: 10.1016/j.lungcan.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Liao R.G., Jung J., Tchaicha J., Wilkerson M.D., Sivachenko A., Beauchamp E.M., Liu Q., Pugh T.J., Pedamallu C.S., Hayes D.N. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73(16):5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quintanal-Villalonga A., Molina-Pinelo S., Cirauqui C., Ojeda-Marquez L., Marrugal A., Suarez R., Conde E., Ponce-Aix S., Enguita A.B., Carnero A. FGFR1 cooperates with EGFR in lung cancer oncogenesis, and their combined inhibition shows improved efficacy. J. Thorac. Oncol. 2019;14(4):641–655. doi: 10.1016/j.jtho.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 23.Raoof S., Mulford I.J., Frisco-Cabanos H., Nangia V., Timonina D., Labrot E., Hafeez N., Bilton S.J., Drier Y., Ji F. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene. 2019;38(37):6399–6413. doi: 10.1038/s41388-019-0887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C.K., Chang H., Chen P.H., Chang J.T., Kuo Y.C., Ko J.L., Lin P. Aryl hydrocarbon receptor activation and overexpression upregulated fibroblast growth factor-9 in human lung adenocarcinomas. Int. J. Cancer. 2009;125(4):807–815. doi: 10.1002/ijc.24348. [DOI] [PubMed] [Google Scholar]
- 25.Pogach M.S., Cao Y., Millien G., Ramirez M.I., Williams M.C. Key developmental regulators change during hyperoxia-induced injury and recovery in adult mouse lung. J. Cell. Biochem. 2007;100(6):1415–1429. doi: 10.1002/jcb.21142. [DOI] [PubMed] [Google Scholar]
- 26.Yin Y.J., Wang F., Ornitz D.M. Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development. Development. 2011;138(15):3169–3177. doi: 10.1242/dev.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Y., Ornitz D.M. FGF9 and FGF10 activate distinct signaling pathways to direct lung epithelial specification and branching. Sci. Signal. 2020;13(621) doi: 10.1126/scisignal.aay4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colvin J.S., White A.C., Pratt S.J., Ornitz D.M. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128(11):2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 29.White A.C., Xu J., Yin Y., Smith C., Schmid G., Ornitz D.M. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133(8):1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- 30.Ren C., Chen H., Han C., Fu D., Wang F., Wang D., Ma L., Zhou L., Han D. The anti-apoptotic and prognostic value of fibroblast growth factor 9 in gastric cancer. Oncotarget. 2016;7(24):36655–36665. doi: 10.18632/oncotarget.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang M.M., Lai M.S., Hong S.Y., Pan B.S., Huang H., Yang S.H., Wu C.C., Sun H.S., Chuang J.I., Wang C.Y. FGF9/FGFR2 increase cell proliferation by activating ERK1/2, Rb/E2F1, and cell cycle pathways in mouse Leydig tumor cells. Cancer Sci. 2018;109(11):3503–3518. doi: 10.1111/cas.13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang M.M., Hong S.Y., Yang S.H., Wu C.C., Wang C.Y., Huang B.M. Anti-cancer effect of cordycepin on FGF9-induced testicular tumorigenesis. Int. J. Mol. Sci. 2020;21(21) doi: 10.3390/ijms21218336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharya R., Chaudhuri S.Ray, Roy S.S. FGF9-induced ovarian cancer cell invasion involves VEGF-A/VEGFR2 augmentation by virtue of ETS1 upregulation and metabolic reprogramming. J. Cell. Biochem. 2018;119(10):8174–8189. doi: 10.1002/jcb.26820. [DOI] [PubMed] [Google Scholar]
- 34.Gan J., Hu C.E. MicroRNA-486-5p targets FGF9 and inhibits colorectal cancer proliferation, migration and invasion. Int. J. Clin. Exp. Pathol. 2016;9(5):5258–5266. [Google Scholar]
- 35.Teishima J., Shoji K., Hayashi T., Miyamoto K., Ohara S., Matsubara A. Relationship between the localization of fibroblast growth factor 9 in prostate cancer cells and postoperative recurrence. Prostate Cancer Prostatic Dis. 2012;15(1):8–14. doi: 10.1038/pcan.2011.48. [DOI] [PubMed] [Google Scholar]
- 36.Deng M., Tang H.L., Lu X.H., Liu M.Y., Lu X.M., Gu Y.X., Liu J.F., He Z.M. miR-26a suppresses tumor growth and metastasis by targeting FGF9 in gastric cancer. PLoS One. 2013;8(8):e72662. doi: 10.1371/journal.pone.0072662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fares J., Fares M.Y., Khachfe H.H., Salhab H.A., Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Target Ther. 2020;5(1):28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuelten C.H., Parent C.A., Montell D.J. Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat. Rev. Cancer. 2018;18(5):296–312. doi: 10.1038/nrc.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y., Jin C., Hamana T., Liu J., Wang C., An L., McKeehan W.L., Wang F. Overexpression of FGF9 in prostate epithelial cells augments reactive stroma formation and promotes prostate cancer progression. Int. J. Biol. Sci. 2015;11(8):948–960. doi: 10.7150/ijbs.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L., Zhang C., Li Y., Zhang Y., Lei Y. DJ-1 promotes epithelial-to-mesenchymal transition via enhancing FGF9 expression in colorectal cancer. Biol. Open. 2020;9(5) doi: 10.1242/bio.051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Zhang Y., Qin X., Wang Y., Fu J. FGF9 promotes cisplatin resistance in colorectal cancer via regulation of Wnt/beta-catenin signaling pathway. Exp. Ther. Med. 2020;19(3):1711–1718. doi: 10.3892/etm.2019.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hendrix N.D., Wu R., Kuick R., Schwartz D.R., Fearon E.R., Cho K.R. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66(3):1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 43.Abdel-Rahman W.M., Kalinina J., Shoman S., Eissa S., Ollikainen M., Elomaa O., Eliseenkova A.V., Butzow R., Mohammadi M., Peltomaki P. Somatic FGF9 mutations in colorectal and endometrial carcinomas associated with membranous beta-catenin. Hum. Mutat. 2008;29(3):390–397. doi: 10.1002/humu.20653. [DOI] [PubMed] [Google Scholar]
- 44.Altorki N.K., Markowitz G.J., Gao D., Port J.L., Saxena A., Stiles B., McGraw T., Mittal V. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat. Rev. Cancer. 2019;19(1):9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S., Lin H., Zhao T., Huang S., Fernig D.G., Xu N., Wu F., Zhou M., Jiang C., Tian H. Expression and purification of an FGF9 fusion protein in E. coli, and the effects of the FGF9 subfamily on human hepatocellular carcinoma cell proliferation and migration. Appl. Microbiol. Biotechnol. 2017;101(21):7823–7835. doi: 10.1007/s00253-017-8468-1. [DOI] [PubMed] [Google Scholar]
- 46.Sugiura K., Stock C.C. Studies in a tumor spectrum. III. The effect of phosphoramides on the growth of a variety of mouse and rat tumors. Cancer Res. 1955;15(1):38–51. [PubMed] [Google Scholar]
- 47.Wu K.H., Ho C.T., Chen Z.F., Chen L.C., Whang-Peng J., Lin T.N., Ho Y.S. The apple polyphenol phloretin inhibits breast cancer cell migration and proliferation via inhibition of signals by type 2 glucose transporter. J. Food Drug Anal. 2018;26(1):221–231. doi: 10.1016/j.jfda.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomayko M.M., Reynolds C.P. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother. Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 49.Jurikova M., Danihel L., Polak S., Varga I., Ki67 PCNA, proteins MCM. Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118(5):544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Parsons J.T., Martin K.H., Slack J.K., Taylor J.M., Weed S.A. Focal adhesion kinase: a regulator of focal adhesion dynamics and cell movement. Oncogene. 2000;19(49):5606–5613. doi: 10.1038/sj.onc.1203877. [DOI] [PubMed] [Google Scholar]
- 51.Wang R., Yu Z., Chen F., Xu H., Shen S., Chen W., Chen L., Su Q., Zhang L., Bi J. miR-300 regulates the epithelial-mesenchymal transition and invasion of hepatocellular carcinoma by targeting the FAK/PI3K/AKT signaling pathway. Biomed. Pharmacother. 2018;103:1632–1642. doi: 10.1016/j.biopha.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Mui K.L., Bae Y.H., Gao L., Liu S.L., Xu T., Radice G.L., Chen C.S., Assoian R.K. N-cadherin induction by ECM stiffness and FAK overrides the spreading requirement for proliferation of vascular smooth muscle cells. Cell Rep. 2015;10(9):1477–1486. doi: 10.1016/j.celrep.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manning B.D., Cantley L.C. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lei Y.Y., Wang W.J., Mei J.H., Wang C.L. Mitogen-activated protein kinase signal transduction in solid tumors. Asian Pac. J. Cancer Prev. 2014;15(20):8539–8548. doi: 10.7314/apjcp.2014.15.20.8539. [DOI] [PubMed] [Google Scholar]
- 55.Liu R., Chen Y., Liu G., Li C., Song Y., Cao Z., Li W., Hu J., Lu C., Liu Y. PI3K/AKT pathway as a key link modulates the multidrug resistance of cancers. Cell Death Dis. 2020;11(9):797. doi: 10.1038/s41419-020-02998-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18(2):128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 57.Eswarakumar V.P., Lax I., Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16(2):139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 58.Katsuta E., Rashid O.M., Takabe K. Clinical relevance of tumor microenvironment: immune cells, vessels, and mouse models. Hum. Cell. 2020;33(4):930–937. doi: 10.1007/s13577-020-00380-4. [DOI] [PubMed] [Google Scholar]
- 59.Lin Y., Xu J., Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019;12(1):76. doi: 10.1186/s13045-019-0760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Q., Guo N., Zhou Y., Chen J., Wei Q., Han M. The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm. Sin. B. 2020;10(11):2156–2170. doi: 10.1016/j.apsb.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su S., Liu Q., Chen J., Chen J., Chen F., He C., Huang D., Wu W., Lin L., Huang W. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25(5):605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 62.Riabov V., Gudima A., Wang N., Mickley A., Orekhov A., Kzhyshkowska J. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front. Physiol. 2014;5:75. doi: 10.3389/fphys.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian B.Z., Pollard J.W. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mantovani A., Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol. 2010;22(2):231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 65.Martinez F.O., Helming L., Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 66.Marchese V., Juarez J., Patel P., Hutter-Lobo D. Density-dependent ERK MAPK expression regulates MMP-9 and influences growth. Mol. Cell. Biochem. 2019;456(1-2):115–122. doi: 10.1007/s11010-019-03496-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y.J., Pan W.W., Liu S.B., Shen Z.F., Xu Y., Hu L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020;19(3):1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das A., Monteiro M., Barai A., Kumar S., Sen S. MMP proteolytic activity regulates cancer invasiveness by modulating integrins. Sci. Rep. 2017;7(1):14219. doi: 10.1038/s41598-017-14340-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gonzalez-Avila G., Sommer B., Garcia-Hernandez A.A., Ramos C. Matrix metalloproteinases' role in tumor microenvironment. Adv. Exp. Med. Biol. 2020;1245:97–131. doi: 10.1007/978-3-030-40146-7_5. [DOI] [PubMed] [Google Scholar]
- 70.Quatromoni J.G., Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am. J. Transl. Res. 2012;4(4):376–389. [PMC free article] [PubMed] [Google Scholar]
- 71.Hegab A.E., Ozaki M., Kagawa S., Hamamoto J., Yasuda H., Naoki K., Soejima K., Yin Y., Kinoshita T., Yaguchi T. Tumor associated macrophages support the growth of FGF9-induced lung adenocarcinoma by multiple mechanisms. Lung Cancer. 2018;119:25–35. doi: 10.1016/j.lungcan.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 72.Brassart-Pasco S., Brezillon S., Brassart B., Ramont L., Oudart J.B., Monboisse J.C. Tumor microenvironment: extracellular matrix alterations influence tumor progression. Front. Oncol. 2020;10:397. doi: 10.3389/fonc.2020.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang W., Fan W., Rachagani S., Zhou Z., Lele S.M., Batra S.K., Garrison J.C. Comparative study of subcutaneous and orthotopic mouse models of prostate cancer: vascular perfusion, vasculature density, hypoxic burden and BB2R-targeting efficacy. Sci Rep. 2019;9(1):11117. doi: 10.1038/s41598-019-47308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fung A.S., Lee C., Yu M., Tannock I.F. The effect of chemotherapeutic agents on tumor vasculature in subcutaneous and orthotopic human tumor xenografts. BMC Cancer. 2015;15:112. doi: 10.1186/s12885-015-1091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gomez-Cuadrado L., Tracey N., Ma R., Qian B., Brunton V.G. Mouse models of metastasis: progress and prospects. Dis. Model Mech. 2017;10(9):1061–1074. doi: 10.1242/dmm.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang F., Cao Y., Wu G., Chen J., CaihongWang W.Lin, Lan R., Wu B., Xie X., Hong J. BMP2 signalling activation enhances bone metastases of non-small cell lung cancer. J. Cell. Mol. Med. 2020;24(18):10768–10784. doi: 10.1111/jcmm.15702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu C., Li F., Jiao S.C. Prognostic factors for survival of patients with extensive stage small cell lung cancer–a retrospective single institution analysis. Asian Pac. J. Cancer Prev. 2012;13(10):4959–4962. doi: 10.7314/apjcp.2012.13.10.4959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Antibodies used in this study.
Table S3. Lentiviral shRNA plasmids used for FGFRs knockdown.