Abstract
The yeast retrotransposon Ty5 preferentially integrates into regions of silent chromatin. Ty5 cDNA also recombines with homologous sequences, generating tandem elements or elements that have exchanged markers between cDNA and substrate. In this study, we demonstrate that Ty5 integration depends upon the conserved DD(35)E domain of integrase and cis-acting sequences at the end of the long terminal repeat (LTR) implicated in integrase binding. cDNA recombination requires Rad52p, which is responsible for homologous recombination. Interestingly, Ty5 cDNA recombines at least three times more frequently with substrates in silent chromatin than with a control substrate at an internal chromosomal locus. This preference depends upon the Ty5 targeting domain that is responsible for integration specificity, suggesting that localization of cDNA to silent chromatin results in the enhanced recombination. Recombination with a telomeric substrate occasionally generates highly reiterated Ty5 arrays, and mechanisms for tandem element formation were explored by using a plasmid-based recombination assay. Point mutations were introduced into plasmid targets, and recombination products were characterized to determine recombination initiation sites. Despite our previous observation of the importance of the LTR in forming tandem elements, recombination cannot simply be explained by crossover events between the LTRs of substrate and cDNA. We propose an alternative model based on single-strand annealing, where single-stranded cDNA initiates tandem element formation and the LTR is required for strand displacement to form a looped intermediate. Retrotransposons are increasingly found associated with chromosome ends, and amplification of Ty5 by both integration and recombination exemplifies how retroelements can contribute to telomere dynamics.
The cDNA generated by reverse transcription during retrotransposon and retrovirus replication can enter the genome by two pathways: it can integrate by using the element-encoded integrase or it can recombine with preexisting elements by using the recombination system of the host (6, 20, 32, 45). Entry into the genome, regardless of the mechanism, alters the host’s genetic material. This can have immediate negative consequences, for example, by generating deleterious mutations. Over evolutionary time, however, some retroelement-induced mutations have likely benefited the host by contributing to the genetic variability that is acted upon by natural selection. In addition, there is increasing evidence that retroelements may contribute to specific cellular processes. The clearest example is the role played by retroelements and reverse transcription in telomere maintenance (24).
The evolution of linear chromosomes has presented a particular difficulty for chromosome replication. Chromosome termini become shorter after each round of DNA replication due to the inability to completely replicate chromosome ends. For most organisms, telomerase extends chromosome ends by using telomeric RNA as a template for reverse transcription (5, 15, 23, 26, 33, 34, 47, 53). A clear link between retrotransposons and telomerases has recently been revealed by the cloning of the telomerase catalytic subunit from Euplotes, yeast, and humans (26, 33, 34). Amino acid sequence analysis of the catalytic subunit indicates that it is related to retrotransposon reverse transcriptases. This suggests that during the evolution of linear chromosomes, a reverse transcriptase may have been borrowed from cellular retrotransposons and used to maintain chromosome ends (11).
In some instances, retrotransposons play a direct role in counterbalancing the telomeric sequence loss that occurs as a consequence of DNA replication. Drosophila melanogaster telomeres, for example, are made up of the non-long terminal repeat retrotransposons HeT-A and TART (24). Telomere extension occurs through preferential integration of these elements onto chromosome ends (3, 46). In addition, an increasing number of retroelements have been identified in the telomeric and subtelomeric regions of other species. These include the SART1 and TRAS1 elements of silkworms, the Zepp elements of Chlorella, and the Ty5 retrotransposons of Saccharomyces (17, 37, 50, 54). The presence of these elements at telomeres suggests that they may contribute to telomere maintenance.
Recombination can also compensate for the telomere shortening that results from DNA replication. Amplification of chromosome end sequences can occur through recombination between telomeric or subtelomeric repeats (27, 29, 39, 41, 51). Recombinational amplification of yeast subtelomeric repeats can overcome telomerase defects and suppress the decreased life span phenotype typically associated with such mutations (30, 31). This amplification requires the host’s homologous recombination system, namely, the RAD52 gene product.
Our laboratory works on the Ty5 retrotransposons of Saccharomyces, which integrate preferentially into silent chromatin (54, 55). Silent chromatin encompasses yeast telomeres and the silent mating loci and is important in mediating Ty5’s target preference (22, 56). Ty5 can recognize domains of silent chromatin, and a single amino acid change at the border of integrase and reverse transcriptase abolishes target specificity (14). We have previously shown that in addition to integration, Ty5 cDNA recombines at high frequencies with homologous substrates (20). In this study, we demonstrate that Ty5 cDNA also recombines preferentially with substrates located in silent chromatin. The preferential amplification of Ty5 at the telomeres through both integration and recombination demonstrates how retrotransposons can contribute to telomere dynamics.
MATERIALS AND METHODS
Strains.
Yeast strains used in this study were YPH499 (MATa GAL trp1Δ63 ura3-52 leu2Δ1 his3Δ200 lys2-801 ade2-101), W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3 trp1-1 ura3-1), and their isogenic derivatives. The Escherichia coli strain XL1-blue (Stratagene) was used for recombinant DNA manipulations.
Plasmids.
Several plasmids (CEN based) carrying either wild-type or mutant Ty5 elements were used to measure recombination: pNK254 (wild-type Ty5), pNK255 [DD(35)E mutation; in-611], pNK530 [DD(35)E and targeting domain double mutant; in-611,1094], pNK535 (polypurine tract [PPT] mutant), pNK536 (U3-tip mutant), and pNK537 (Ty5-HIS3). For coding-sequence mutations, the number refers to the modified amino acid in Ty5’s single open reading frame (e.g., in-611). The strains used to calculate cDNA recombination frequencies included YPH499, rad derivatives of YPH499, and W303-1A strains containing various Ty5 insertions (19, 54, 55).
Plasmid pNK254 contains Ty5 under the transcriptional control of GAL1-10 upstream activating sequences and a his3AI selectable marker (GAL1-Ty5- his3AI) (20). The Ty5 elements with mutations in the DD(35)E domain (pNK255), the PPT (pNK535), or the U3 tip (pNK536) were constructed by PCR-based site-directed mutagenesis of Ty5 subclones (pSZ125, integrase; pNK532, 3′-end sequences) (8). Primer DVO238 (5′-GTCGGATCTGTTCGTGCAGCCAATGGTACAGAATT-3′) was used to change the second aspartate residue of the DD(35)E domain of integrase and the flanking threonine residue to alanine; primer DVO685 (5′-TTCAGTTATCCCCCCTTGTTGAATG-3′) was used to change the putative Ty5 PPT GGGGGGA immediately upstream of the Ty5 3′ LTR to CCCCCCT; primer DVO686 (5′-ATGGGGGGACCTTGAATGTG-3′) was used to change the first two nucleotides in the Ty5 3′ LTR from TG to CC. All mutated fragments were confirmed by DNA sequencing and used to replace the corresponding wild-type regions of pNK254. pNK530, which contains the DD(35)E mutation (in-611) and the Ty5 integration targeting domain mutation (in-1094) (14), was constructed by replacing the XhoI-HpaI fragment of pXW137 with the corresponding fragment from pNK255. pNK538 is a derivative of pNK255 that lacks the artificial intron in HIS3; it was obtained by rescue of a His+ plasmid generated by Ty5 cDNA recombination.
The recombination substrates used to determine the role of the Ty5 LTR in cDNA recombination were constructed by cloning Ty5 fragments into pRS426 (9) from pNK318 (full-length Ty5 in pRS426) (20) or pXW25 (GAL1-Ty5-neoAI [14a]) (see Fig. 6). pNK396 contains the 1.6-kb EcoRI fragment of pNK318, pNK395 contains the 1.0-kb EcoRI-NruI fragment of pNK318, pNK394 contains the 0.6-kb EcoRI-NruI fragment of pNK318, pNK317 contains the Ty5 LTR, pNK322 contains the 0.9-kb NruI-SacII fragment of pNK318, pNK323 contains the 1.9-kb NruI-SacII fragment of pXW25 (which includes neoAI), and pNK397 contains the 1.7-kb NruI-EcoRI fragment of pXW25.
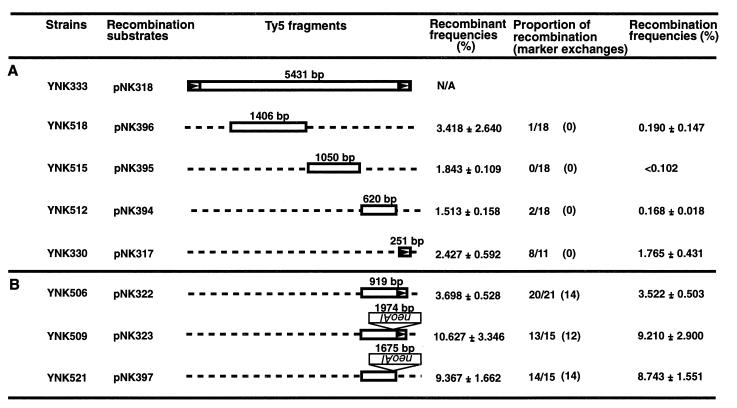
FIG. 6.
Ty5 LTR is critical for tandem element formation but not for marker exchange. The recombination frequencies for different Ty5 substrates were calculated by the assay shown in Fig. 1B. (A) Internal Ty5 sequences or the LTR were used as recombination substrates; only recombination that generated tandem elements was observed. All Ty5 fragments are drawn to scale with the exception of pNK318. (B) Ty5 3′-end sequences with or without an LTR (arrowheads) were used as recombination substrates; marker exchanges were the major class of products.
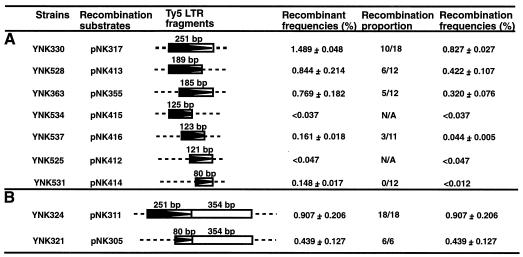
The LTR deletion constructs were made by a PCR-based method with the LTR clone pNK317 as a template (see Fig. 7). Each fragment was PCR amplified by the primers noted in parentheses: pNK413 contains the first 189 bp of the LTR (DVO278 [5′-CCGCTCGAGTGTTGAATGTGATAACCCA-3′] and DVO329 [5′-CGGGATCCTATATGTTATGTAAATG-3′]), pNK355 contains the last 185 bp of the LTR (DVO279 [5′-CCGCTCGAGTAATGTTTTAGACAAG-3′] and DVO190 [5′-TGGATCCTGTTGACGTAGTGAATTA-3′]), pNK415 contains the first half of the LTR (DVO278 and DVO328 [5′-CGGGATCCTTAAGTACTGTCGGATC-3′]), pNK416 contains the internal half of the LTR (DVO279 and DVO329), pNK412 contains the second half of the LTR (primers DVO327 [5′-CGGGATCCATAGTTTCTGTGTACAAG-3′] and DVO190), and pNK414 contains the last one-third of the LTR (DVO214 [5′-CCCTCGAGCATTTACATAACATATAGAAAG-3′] and DVO190). Additional recombination substrates included pNK311, which contains the XhoI-BamHI fragment of pNK318, and pNK305, which contains the XhoI-BamHI fragment of pNK318 that has a deletion of the first 170 bp of the LTR (the Ty5 promoter) (19, 54).
FIG. 7.
Deletion analysis determines the Ty5 LTR sequences (arrowheads) important in mediating tandem element formation. The recombination frequencies for different Ty5 substrates were calculated by the assay shown in Fig. 1B. (A) LTR deletion constructs were used as recombination substrates. (B) LTR sequences coupled with the immediately adjacent internal sequences were used as recombination substrates.
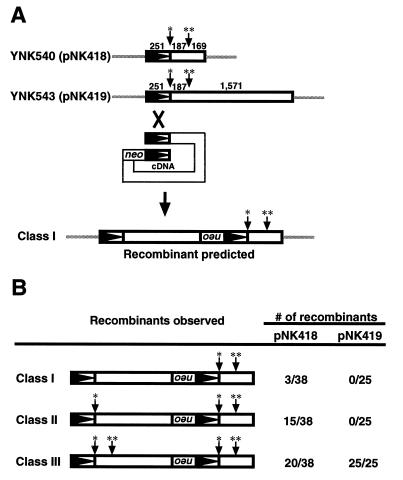
The Ty5 subclones that contain point mutations were constructed as follows (see Fig. 8). The BamHI site was first generated at the junction between the 5′ LTR and the internal region by cloning the BamHI fragment of pNK354 into pNK317 to generate pNK411, and the internal HindIII site of pNK411 was filled in with Klenow to generate pNK418 (2). pNK419 was constructed by replacing the XhoI-BamHI fragment of pNK349 with that from pNK418.
FIG. 8.
Assay to determine the initiation site for tandem element formation. (A) Two point mutations were created in two Ty5 recombination substrates (pNK418 and pNK419). A single asterisk indicates the site of addition of a BamHI site; a double asterisk indicates the location at which a HindIII site was destroyed. Based on our previous model, recombination between the LTRs (arrowheads), which is facilitated by internal sequence homology, would generate class I products that have both markers in the 3′ element (20). (B) The observed recombination products for substrates pNK418 and pNK419 (designated class I, II, and III). The distribution of markers was determined by restriction mapping of the 5′ elements and by DNA sequencing of the 3′ elements.
Strain construction.
One-step gene disruption (42) was used to make the rad derivatives of YPH499. pRR46 contains the RAD1 gene with the region from −212 to +3853 replaced by LEU2 (the kind gift of L. Prakash and S. Prakash) (40). This plasmid was digested with BamHI before YPH499 was transformed by the lithium acetate method (2). Leu+ transformants were confirmed to be rad1 by testing their UV sensitivity and by Southern blot analysis. pSM21 contains the RAD52 gene with a TRP1 insertion (the kind gift of L. Prakash and S. Prakash). After digestion of pSM21 with BamHI and transformation of YPH499, Trp+ transformants were confirmed to be rad52 by Southern blot analysis. The rad1 rad52 strain was constructed by transforming the rad1 strain with BamHI-digested pSM21. Trp+ transformants were confirmed to be rad52 by Southern blot analysis.
Recombination assays.
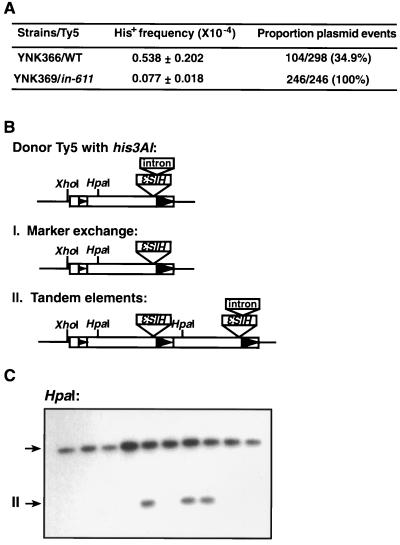
In assays used to study the effect of integrase, the PPT, the LTR end sequences (U3 tip), or RAD genes on Ty5 cDNA recombination, the plasmid-borne Ty5 elements served as both the donor and recipient elements. pNK254 (wild-type Ty5), pNK255 (in-611), pNK535 (PPT mutant), and pNK536 (U3-tip mutant) were transformed into YPH499 and its derivative rad strains. Three independent transformants were used for all analyses. Transposition assays were conducted as previously described (20). His+ colonies (62 to 300) were used to calculate the proportion of plasmid events in all strain combinations. This was calculated by dividing the number of colonies that did not grow on synthetic complete medium without histidine and with 5-FOA (SC-H–5-FOA) by the number of colonies that grew on SC-H medium (see Fig. 1A). Ten individual plasmid events generated by the Ty5 integrase DD(35)E mutant (in-611) in the wild-type strain were then subjected to Southern blot analysis or plasmid rescue followed by restriction mapping to determine whether they were derived by recombination.
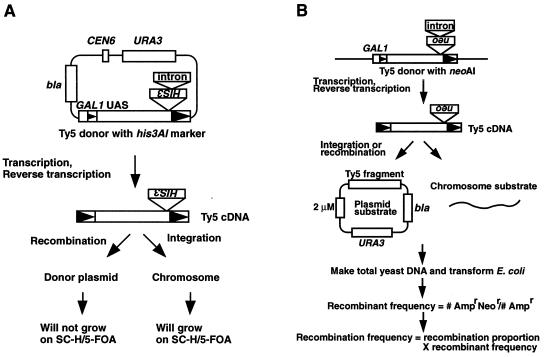
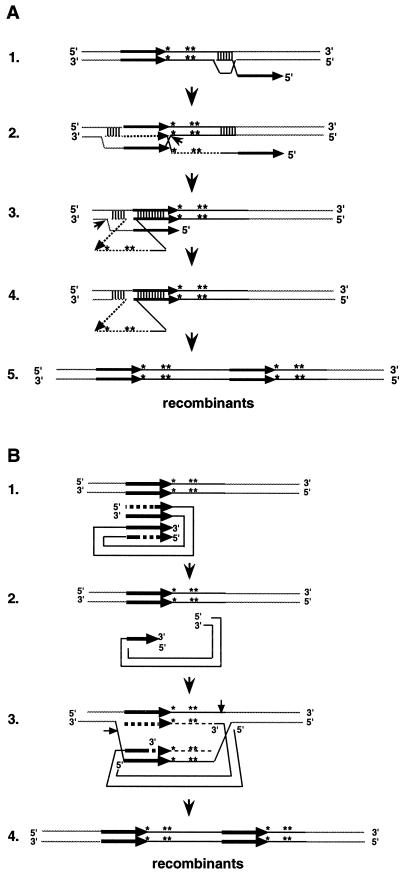
FIG. 1.
Recombination assays used in this study (originally described in reference 20). (A) Transcription of a plasmid-borne GAL1-Ty5-his3AI element is induced by growing cells in galactose medium. After transcription and reverse transcription, the Ty5 cDNA with its functional HIS3 gene either integrates into chromosomes or recombines with plasmid substrates. This generates His+ colonies. Integration and recombination events are distinguished by whether the His+ cells can grow on SC-H–5-FOA plates. (B) Transcription and reverse transcription of a chromosomal GAL1-Ty5-neoAI element gives rise to Ty5 cDNA carrying the neo gene. This cDNA can either integrate or recombine with chromosomal or plasmid targets. Plasmid recombinants containing both the neo and bla genes will confer an Ampr Neor phenotype when introduced into bacteria; all plasmids will confer an Ampr phenotype. Recombination frequencies are calculated as the product of the recombinant frequency and the proportion of recombinants due to recombination. The arrowheads inside the elements represent LTRs.
To determine whether Ty5 cDNA could recombine with Ty5 substrates located within silent chromatin, W303-1A strains with de novo Ty5 insertions at different chromosomal locations were used: W3 (internal region of chromosome XI); W9 (HMR); and W2, W77, and W84 (chromosome III left telomere) (54, 55). To facilitate the identification of new integration or recombination events by genetic selection, the functional HIS3 genes in these Ty5 insertions were replaced with his3AI to generate strains YNK570 (W3-AI), YNK566 (W9-AI), YNK567 (W2-AI), YNK568 (W77-AI), and YNK569 (W84-AI) (19). pNK254 (wild-type Ty5), pNK255 (in-611), and pNK530 (in-611,1094) were then introduced into these strains to serve as cDNA donor elements. Transposition assays were conducted, and recombination and integration events were selected by reconstitution of the functional HIS3 gene. Chromosomal and plasmid events were distinguished by whether they could grow on SC-H–5-FOA plates.
Southern blot or PCR analysis was used to discriminate between chromosomal recombination and integration events. For Southern blots, DNAs were digested with enzymes that cut once within Ty5; strains generating one band were scored as marker exchanges, strains with the original band and a 6.5-kb band were scored as tandem elements, and strains with the original band and a novel band were scored as integration events. Putative recombination events were confirmed by additional enzyme digestions. For PCR analysis, Ty5 primers that flank the his3AI marker were used: DVO445 (5′-CAGAATCATTCAAAGCACATAG-3′) and DVO496 (5′-CTTGTCTAAAACATTACTGAAACAAT-3′). Strains whose DNA gave a PCR product without the intron (1.15 kb) were scored as marker exchanges; strains whose DNA generated a band with the intron (1.25 kb) were scored as gene conversion of the chromosomal his3-11,15 locus; strains whose DNA yielded two bands (1.15 and 1.25 kb) were scored as having either integrated or tandem elements. Tandem elements were distinguished from integration events by the presence of a PCR product (1.5 kb) by using primer DVO445 and the Ty5 GAG primer DVO497 (5′-GGGATTAGATAGATTAATTATGGTCTCT-3′).
The above-mentioned chromosomal substrates were tested for their effectiveness in recombining with linear DNA (transplacement). Plasmid pNK538, containing GAL1-Ty5-HIS3, was digested with XhoI and NotI and the 6.5-kb Ty5-HIS3 fragment was gel purified and transformed into each of the strains. These strains also carried the Ty5-containing plasmid pNK255 (in-611). The linear Ty5-HIS3 DNA could recombine with either the plasmid (pNK255) or chromosomal substrate to generate His+ colonies. These two events were distinguished by whether the His+ cell could grow on SC-H–5-FOA plates. The ratio between chromosomal and plasmid events served as a measure of the chromosomal substrate’s effectiveness in recombination with linear DNA.
The strain with a telomeric insertion (YNK568; W77-AI) was used to determine whether Ty5 cDNA recombination occurs during mating. This telomeric element thus served as both the cDNA donor and recipient. YNK568 was induced to transpose by exposure to the α-factor mating pheromone as described previously (19). Transposition and recombination events were selected on SC-H plates. A total of 32 His+ colonies were analyzed by Southern blotting. Using the enzyme HpaI, which only cuts once in Ty5, recombination events were scored as having only the original Ty5 fragment (marker exchange) or the original Ty5 fragment and a 6.5-kb band (tandem elements). Putative recombination events were further confirmed by Southern analysis with different enzymes.
The cDNA donor element and recombination substrates were separated in the assays used to determine the role of the LTR in cDNA recombination (see Fig. 1B). The donor GAL1-Ty5-neoAI was integrated into the chromosomal LEU2 locus to generate strain YNK364 (20). The plasmid substrates described above were then introduced into this strain. Transposition assays were conducted as described previously (20), and recombination and integration events were selected on YPD-G418 plates. Total yeast DNA was prepared and transformed into E. coli. The recombinant frequency for a particular plasmid substrate (both integration and recombination events) was calculated by dividing the number of Ampr Kanr colonies by the total number of Ampr colonies. Several recombinants were characterized by restriction mapping and DNA sequence analysis to determine whether the recombinants were derived from recombination or integration events. Recombination frequencies were calculated as the product of recombinant frequencies and the recombination proportion.
RESULTS
Effect of Ty5 integrase and Rad52p on Ty5 cDNA recombination.
We previously developed a Ty5 transposition assay in S. cerevisiae which used a functional Ty5 element from Saccharomyces paradoxus (54). This element carries a HIS3 marker gene, which is rendered nonfunctional by the presence of an inactivating intron (his3AI). Transposition events are selected after reconstitution of a functional HIS3 gene by transcription, intron splicing, and reverse transcription of Ty5 mRNA (Fig. 1). In addition to integration, Ty5 cDNA also recombines at high frequency with homologous substrates (20). Two classes of recombination products are recovered: elements that have exchanged markers between the cDNA and the substrate and tandem elements. To determine the relationship of the integration and recombination pathways and to determine the host recombination system(s) involved in Ty5 cDNA recombination, Ty5 integrase and RAD gene mutants were characterized for their effects on Ty5 cDNA entry into the genome.
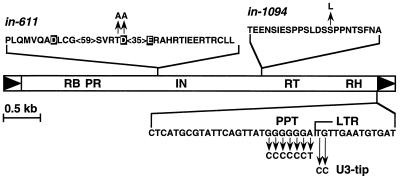
Integrase is responsible for the integration of retroelement cDNA into target DNA, and a portion of its catalytic domain is conserved among all retroviruses, retrotransposons, and even some bacterial transposons (43). The catalytic domain is characterized by two invariant aspartates, the second of which is separated by 35 residues from a glutamate [DD(35)E] (Fig. 2). In studies with retroviruses, such as human immunodeficiency virus type 1, avian Rous sarcoma virus, and the yeast retrotransposon Ty3, mutations in any of these three residues dramatically reduce integration (21, 48). Ty5 integrase was mutated by changing to alanine the second aspartate residue of the DD(35)E domain and an adjacent threonine residue (in-611 [Fig. 2]). Transposition assays were conducted (as described in Fig. 1A), and a sevenfold decrease in the overall frequency of His+ cell formation was observed (Fig. 3A). If HIS3 is carried on the URA3-based donor plasmid, the cells cannot grow on SC-H–5-FOA medium because the 5-FOA is converted by Ura3p to a toxic substance that kills the host cell (7). Recombinant plasmids, therefore, were scored as His+ 5-FOAs papillae; for all of the 246 His+ events, the HIS3 gene was plasmid associated. Characterization of 10 individual plasmid events by Southern blot analysis and restriction mapping indicated that all arose by recombination (Fig. 3C and data not shown). The recombination products fell into two classes (Fig. 3B and C): seven marker exchanges and three tandem elements, which is similar to the ratio generated by wild-type Ty5 elements (20). This indicated that the DD(35)E domain is essential for integration and that integrase mutants generate His+ cells through cDNA recombination with plasmid elements and not with the degenerate native S. cerevisiae elements.
FIG. 2.
Ty5 mutants used in this study. A full-length Ty5 element is shown, with the arrowheads indicating the LTRs. in-611 has a mutation in the DD(35)E domain of integrase; the conserved aspartic acid and glutamic acid residues that define this domain are boxed. in-1094 has a Ser-to-Leu substitution that abolishes targeted integration to silent chromatin (14). U3-tip denotes the first two nucleotide sequences at the 5′ end of the 3′ LTR. The nucleotide changes in the PPT and U3-tip mutants are noted. All mutations were generated by site-directed mutagenesis. RB, RNA binding domain; PR, protease; IN, integrase; RT, reverse transcriptase; RH, RNase H.
FIG. 3.
Structural analysis of recombination events generated by a Ty5 integrase mutant. (A) The overall His+ frequency and the proportion of recombination events are shown for the Ty5 integrase mutant and the wild-type Ty5 element. The data were determined by the assay shown in Fig. 1A. (B) Maps of Ty5 donor element and recombination products. The arrowheads represent LTRs. (C) Southern blot analysis of 10 individual His+ colonies generated by the Ty5 integrase mutant (in-611). The upper arrow indicates the restriction fragment of the donor Ty5 element. For lanes with a single hybridizing restriction fragment, this band represents the his3AI marker that has been converted by marker exchange to a functional HIS3 gene. The second hybridizing restriction fragment in some lanes (lower arrow, labeled “II”) is derived from tandem elements.
Recombination in S. cerevisiae is influenced by some genes involved in DNA repair (38): RAD52, a recombinational repair gene, is responsible for most homologous recombination, and RAD1, an excision repair gene, is involved in direct-repeat recombination (25, 44). To evaluate the roles of RAD1 and RAD52 in Ty5 cDNA recombination, donor plasmids containing either Ty5 or the Ty5 integrase mutant were introduced into wild-type, rad1, rad52, or rad1 rad52 strains. Transposition assays were conducted for each construct and strain combination, and the overall His+ frequencies and the percentage of recombination events were determined (Table 1). The frequency of plasmid events was considered a measure of recombination; the frequency of chromosomal events was considered a measure of integration (20). In the rad52 strain, the overall His+ frequency dropped 2.6-fold for the wild-type Ty5 element, and most His+ events were due to integration. RAD1 mutations, in contrast, did not have much effect on either the overall His+ frequencies or the recombination frequencies. In rad1 rad52 double mutants, the His+ frequency and recombination proportion were similar to those of the rad52 strain. However, in rad52-Ty5 integrase double mutants, the overall His+ frequency showed a synergistic reduction of more than 800-fold. This indicates that integration and RAD52-dependent homologous recombination are the two major pathways by which Ty5 cDNA enters its DNA targets.
TABLE 1.
Recombination of Ty5 cDNA generated by wild-type and integrase mutant (in-611) elements in different rad strainsa
| Strain | Ty5 genotype/strain genotype | His+ frequency (10−5) | His+ frequency relative to wt | Plasmid events/total (%) | Chromosomal events/total (%) |
|---|---|---|---|---|---|
| YNK366 | wt/wt | 8.85 ± 0.30 | 1.00 | 59/105 (56.2) | 46/105 (43.8) |
| YNK372 | wt/rad1 | 7.26 ± 0.12 | 0.82 | 54/145 (37.2) | 91/145 (62.8) |
| YNK378 | wt/rad52 | 3.34 ± 0.05 | 0.38 | 1/119 (0.8) | 118/119 (99.2) |
| YNK384 | wt/rad1 rad52 | 2.54 ± 0.08 | 0.29 | 1/140 (0.7) | 139/140 (99.3) |
| YNK369 | in-611/wt | 1.15 ± 0.02 | 0.13 | 69/70 (98.6) | 1/70 (1.4) |
| YNK375 | in-611/rad1 | 1.24 ± 0.01 | 0.14 | 128/128 (100.0) | 0/128 (0.0) |
| YNK381 | in-611/rad52 | <0.01 | <0.01 | NA | NA |
| YNK387 | in-611/rad1 rad52 | 0.01 ± 0.01 | <0.01 | NA | NA |
His+ frequency was calculated by dividing the His+ cell number by the total Ura+ cell number. The His+ frequency relative to wild type (wt) was calculated by dividing the His+ frequencies of different strains by the His+ frequency of a wild-type strain carrying a wild-type Ty5. The recombination proportion was calculated by dividing the His+ 5-FOAs cell number by the His+ cell number (column 4). The remaining His+ events were chromosomal events. NA, not applicable.
Integration is impaired by mutations in the PPT or the LTR end sequences.
Retroelement mRNA contains a primer binding site adjacent to the 5′ LTR where minus-strand cDNA synthesis initiates from a complementary host tRNA. A PPT adjacent to the 3′ LTR serves as the priming site for plus-strand DNA synthesis. Reverse transcription proceeds through a series of two-strand transfers, ultimately resulting in a linear retroelement cDNA with two flanking LTRs. Integrase recognizes sequences at the ends of the cDNA (10), and the dinucleotide end sequences (TG at the U3 tip and CA at the U5 tip) are nearly invariant among retroviruses and retrotransposons. For Ty1, mutations in the PPT or the U3 and U5 tips largely abolish integration and result in elevated frequencies of recombination (45).
The putative PPT (GGGGGGA) immediately upstream of the Ty5 3′ LTR was changed to the corresponding pyrimidines (CCCCCCT) (Fig. 2). These changes should impair plus-strand priming and block reverse transcription after minus-strand cDNA synthesis. In a separate construct, mutations were introduced in the U3 tip by changing the dinucleotide TG to CC at the 5′ end of the 3′ LTR (Fig. 2). Since the U3 sequence in the 3′ LTR is used as a template during reverse transcription for the synthesis of both LTRs, cDNA synthesized from the mutant would be expected to have CC instead of TG at the 5′ ends of both LTRs. Plasmids containing the mutant Ty5 elements (pNK535 for the PPT mutant and pNK536 for the U3-tip mutant) were transformed into wild-type and rad52 strains. The transposition and recombination frequencies were calculated as described above. As with the integrase mutant, the overall His+ frequencies dropped fourfold in the wild-type strains, and more than 90% of the His+ events were due to plasmid recombination (Table 2). In rad52 strains, there was a synergistic reduction in His+ frequency of at least 50-fold. These data support our initial observation that Ty5 uses both integration and recombination to enter the genome; recombination becomes the primary pathway if integration is crippled either by mutating Ty5 integrase or the cDNA end sequences to which integrase likely binds or by preventing plus-strand DNA synthesis.
TABLE 2.
Effects of Ty5 PPT and U3-tip mutations on integration and recombinationa
| Strain | Ty5 genotype/strain genotypeb | His+ frequency (10−5) | His+ frequency relative to wt | Plasmid events/total (%) | Chromosomal events/total (%) |
|---|---|---|---|---|---|
| YNK366 | wt/wt | 6.23 ± 1.20 | 1.00 | 28/80 (35.0) | 52/80 (65.0) |
| YNK959 | PPT mutant/wt | 1.74 ± 0.29 | 0.28 | 76/79 (96.2) | 3/79 (3.8) |
| YNK960 | U3-tip mutant/wt | 1.76 ± 0.52 | 0.28 | 77/82 (93.9) | 5/82 (6.1) |
| YNK378 | wt/rad52 | 2.99 ± 0.88 | 0.48 | 1/89 (1.1) | 88/89 (98.9) |
| YNK961 | PPT mutant/rad52 | 0.06 ± 0.06 | 0.01 | 1/29 (3.4) | 28/29 (96.6) |
| YNK962 | U3-tip mutant/rad52 | 0.10 ± 0.05 | 0.02 | 1/26 (3.8) | 25/26 (96.2) |
His+ frequency and the proportion of plasmid and chromosomal events were determined as described in footnote a to Table 1.
U3 tip, the first two nucleotides at the 5′ end of the 3′ LTR.
Ty5 cDNA recombines at higher frequencies with substrates associated with silent chromatin.
Although Ty5 cDNA recombines at high frequency with plasmid substrates, we have never observed recombination with native S. cerevisiae Ty5 sequences. This could be due to two reasons: native Ty5 elements are bound in silent chromatin, which may render them inaccessible to the host’s recombination system, or native Ty5 sequences may be too degenerate to serve as effective substrates with the S. paradoxus element used in the assays. To distinguish between these possibilities, Ty5 insertions (100% identical to the donor element) were introduced at different chromosomal locations and used as recombination substrates (W9-AI, HMR; W77-AI, telomere; W3-AI, control [Fig. 4A]) (19). The control substrate, W3-AI, is transcriptionally active, whereas the substrates in silent chromatin (W9-AI and W77-AI) are transcriptionally repressed (19). Plasmids carrying donor elements were then introduced into these strains, and transposition assays were conducted. Transposition or recombination events were selected by the reconstitution of the HIS3 marker gene. Chromosomal and plasmid events were distinguished by whether the His+ cells could grow on SC-H–5-FOA medium. Products of recombination with chromosomal substrates were then identified by Southern blot analysis (data not shown).
FIG. 4.
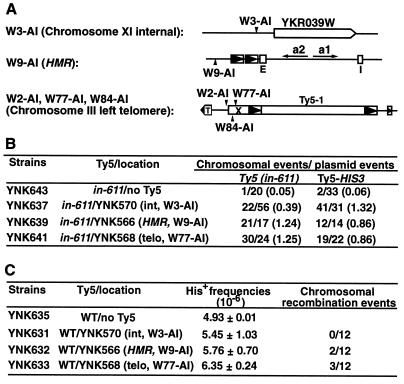
Ty5 cDNA recombines preferentially with substrates located within silent chromatin. (A) Three strains with Ty5 insertions at different chromosomal locations. These Ty5 insertions contain the his3AI marker to facilitate selection of recombination and integration events. YKR039W is an anonymous open reading frame. Ty5-1 is an endogenous Ty5 insertion with flanking LTRs depicted as boxes with arrowheads. Solo endogenous Ty5 LTRs are shown at HMR. E and I designate the HMR transcriptional silencers; T designates the telomeric TG1–3 repeats; X designates the subtelomeric X repeat. (B) Recombination frequency with chromosomal substrates and a donor element with an integrase mutation. A plasmid with a Ty5 integrase mutant (in-611) was introduced into the above-mentioned strains. Transposition assays were conducted, and the chromosomal and plasmid recombination events were distinguished by whether His+ cells could grow on SC-H–5-FOA plates. (C) Recombination frequency with chromosomal substrates and a wild-type (WT) donor element. int, internal; telo, telomeric.
A Ty5 element with an integrase mutation (in-611) was initially used as the cDNA donor, since the majority of His+ events generated by this construct are due to recombination. The effectiveness of a particular chromosomal element as a recombination substrate was evaluated by the ratio of chromosomal recombination events to plasmid recombination events; that is, the number of plasmid recombinants served as an internal reference. Interestingly, we found that Ty5 cDNA recombines approximately 3.2-fold more frequently with substrates associated with silent chromatin than with the internal substrate, W3-AI (1.24 for W9-AI and 1.25 for W77-AI versus 0.39 for W3-AI) (Fig. 4B). This was not because the internal substrate is a recombinational cold spot; when linear Ty5 DNA carrying a functional HIS3 gene was transformed into the above-mentioned strains, it recombined at slightly higher frequencies with the internal substrate than with substrates in silent chromatin. The parental strain, W303-1A, which does not contain any chromosomal Ty5 substrates, was used as a negative control; in this strain the majority of His+ events arose through recombination with plasmid substrates. This demonstrates that Ty5 cDNA can recombine efficiently with homologous substrates in silent chromatin and that degenerate endogenous Ty5 sequences are not effective recombination substrates.
To determine whether the preferential recombination with substrates in silent chromatin was influenced by the in-611 mutation, cDNA recombination for a wild-type Ty5 donor element was also evaluated. The overall frequency of His+ cell formation was comparable to that of the parental strain (Fig. 4C). Twelve independent chromosomal events for each strain were subjected to Southern blot analysis to distinguish between integration and recombination events (data not shown). As with the integrase mutant (in-611), the wild-type Ty5 cDNA recombined preferentially with substrates associated with silent chromatin (2 of 12 for W9, 3 of 12 for W77, and 0 of 12 for W3). This suggests that silent chromatin directs cDNA recombination.
Preferential recombination of Ty5 cDNA with telomeric substrates requires the Ty5-encoded domain that mediates integration specificity.
Mutations in a single amino acid (amino acid 1094) near the boundary of Ty5 integrase and reverse transcriptase abolish preferential integration of Ty5 to regions of silent chromatin (Fig. 2) (14). Although it is not yet known whether this mutation lies within integrase or reverse transcriptase, we have designated this allele as in-1094 because of its integration phenotype. Since this amino acid is essential for integration specificity, it may also contribute to the observed preference of Ty5 cDNA to recombine with substrates in silent chromatin. To test this hypothesis, a Ty5 double mutant (in-611,1094) that contained both the DD(35)E mutation (in-611) and the targeting domain mutation (in-1094) was constructed. Transposition and recombination frequencies were tested for the in-611,1094 mutant, and they did not differ significantly from that of the in-611 single mutant (data not shown). Plasmids carrying the integrase double mutant were transformed into the strains with Ty5 substrates at the telomeres (W2-AI, W77-AI, and W84-AI [Fig. 4A]) or in the internal region on chromosome XI (W3-AI [Fig. 4A]). For comparison, the assays were conducted in parallel with in-611. Note that in contrast to the experiments described in the previous section, two additional telomeric recombination substrates were tested. Transposition assays were conducted, and chromosomal and plasmid His+ events were distinguished by whether the His+ cells could grow on SC-H–5-FOA plates. Chromosomal events were further analyzed by a PCR assay to determine whether they were the result of gene conversion of the endogenous his3-11,15 locus by the HIS3-containing cDNA, recombination with the chromosomal Ty5 substrates, or integration.
As in the previous experiments, the effectiveness of chromosomal substrates for recombination was measured by using the frequency of plasmid recombination events as an internal reference. In this experiment, however, the ratio of His+ chromosomal events to plasmid events was further adjusted by multiplying this ratio by the proportion of chromosomal recombination events as determined by the PCR analysis (Table 3); this effectively excluded events due to gene conversion of his3-11,15 and integration. The data confirmed that for in-611, Ty5 cDNA recombines preferentially with telomeric substrates (at least 3.8-fold higher frequency) compared to the substrate at the internal chromosomal locus. Again, this preference was not because the internal substrate is a recombination cold spot; a transplacement experiment conducted with the linear Ty5-HIS3 DNA demonstrated that the internal substrate recombined at a frequency comparable to that of the telomeric substrates. In contrast to in-611, the in-611,1094 double mutant did not show elevated recombination at the telomeres. This indicates that preferential cDNA recombination with substrates in silent chromatin is mediated by the Ty5 targeting domain.
TABLE 3.
Preferential recombination of Ty5 cDNA with telomeric substrates requires the domain responsible for Ty5 integration specificity
| Strain | Ty5 location | No. of chromosomal recombination events/no. of plasmid events (frequency)a
|
||
|---|---|---|---|---|
| Ty5 in-611
|
Ty5 in-611,1094
|
|||
| TP assay | TF with Ty5-HIS3 | TP assay | ||
| W303-1A | No Ty5 | 1/22 (0.05) | 2/33 (0.06) | 0/24 (0.00) |
| YNK570 | Internal; W3-AI | 11/18 [4/10] (0.24) | 41/31 (1.32) | 13/12 [4/10] (0.43) |
| YNK567 | Telomere; W2-AI | 27/14 [7/10] (1.35) | 14/19 (0.74) | 25/29 [8/9] (0.77) |
| YNK568 | Telomere; W77-AI | 45/24 [9/10] (1.69) | 19/32 (0.59) | 18/39 [9/10] (0.42) |
| YNK569 | Telomere; W84-AI | 26/23 [8/10] (0.90) | 28/14 (2.00) | 21/37 [9/10] (0.51) |
TP, transposition; TF, transformation; in-611, DD(35)E mutant of Ty5 integrase; in-611,1094, DD(35)E and targeting domain double mutant; Ty5-HIS3, Ty5 element with the functional HIS3 gene. The number in brackets is the proportion of chromosomal recombination based on PCR analysis; this proportion was multiplied by the ratio of chromosomal events to plasmid events to give the frequency.
It is interesting to note that for strains with Ty5 substrates at the telomeres, most of the chromosomal events (≥90%) were due to either cDNA recombination with the chromosomal Ty5 substrates or gene conversion of the his3-11,15 locus. However, for the internal Ty5 substrate W3-AI, only 60% of the events were due to recombination and the other 40% were due to integration. This difference in the observed frequency of integration events is consistent with differences in the basal transcriptional activity of the substrate elements. We have previously shown that W3-AI has low levels of basal Ty5 transcription and transposition (19). This contrasts with Ty5 elements in silent chromatin, which are transcriptionally repressed and do not spawn additional transposition events.
Ty5 cDNA recombines with a telomeric substrate when exposed to mating pheromones.
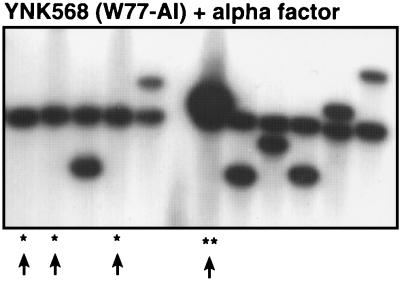
In their native state, the transcription of most Ty5 elements is silenced by telomeric chromatin (19). Since transcription is required for transposition, this would prevent cDNA synthesis and subsequent integration or recombination. Transcriptional activation of elements bound in silent chromatin, however, can be achieved by exposure to mating pheromones, indicating that Ty5 normally transposes during mating (19). Transcription and transposition of a telomeric Ty5 element (W77-AI) was induced by exposure to mating pheromone (α-factor) to test whether Ty5 cDNA can recombine with this homologous telomeric substrate during pheromone activation. Thirty His+ colonies were randomly picked and subjected to Southern blot analysis with an integrase-specific probe. Recombination events were scored as either marker exchange (the presence of only the original Ty5 band) or tandem elements (the original band plus a 6.5-kb band). Integration events were scored as the original band plus a novel band. Of the 30 events characterized, 13 were due to recombination (eight marker exchanges and five tandem elements) (Fig. 5 and data not shown). Two of the five tandem elements were present in multiple copies (Fig. 5, lane **). This suggests that during mating, when Ty5 transcription and transposition naturally occur, Ty5 cDNA also recombines at high frequencies with telomeric elements.
FIG. 5.
Recombination of a telomeric Ty5 induced by mating pheromones. His+ colonies were selected after inducing transcription and transposition of a telomeric element (W77-AI) by exposure to α-factor. Results of a representative Southern hybridization analysis with 11 randomly picked His+ colonies are shown. One asterisk indicates marker exchanges, and two asterisks indicate tandem elements; unlabeled lanes indicate integration events. Note that the tandem elements appear to contain multiple Ty5s, based on hybridization intensity.
Ty5 LTR is important for tandem element formation but not for marker exchange.
Recombination between telomeric and subtelomeric repeats plays a role in telomere maintenance (30, 31, 39, 51). For the S. cerevisiae Y′ elements, recombinational amplification can overcome telomerase defects (30). The enhanced recombination of Ty5 cDNA at the telomeres suggests that it could play a similar role, particularly when sequences are amplified through the formation of tandem or multiple-tandem elements. In a previous study, we found that the Ty5 LTR was critical for generating tandem elements (20). Internal Ty5 sequences, although not good recombination substrates, could facilitate tandem element formation when coupled with a Ty5 LTR. We proposed that tandem elements were formed through recombination between the LTRs of the cDNA and the substrate. In our models, internal homology facilitated base pairing and therefore the formation of tandem products.
Confirmation of the critical role played by the Ty5 LTR in forming tandem elements is shown in Fig. 6A. Plasmids carrying previously untested recombination substrates (Ty5 internal fragments or a Ty5 LTR) were introduced into a strain with an integrated GAL1-Ty5-neoAI element (YNK364). Recombination frequencies for each substrate were calculated as described previously (Fig. 1B). All substrates lacked sequences flanking the marker gene, and the only recombination products observed were tandem elements. The LTR was a much better substrate than internal sequences: the recombination frequency for the LTR substrate was at least 9.3-fold higher than for the internal sequence substrates (1.765 for pNK317 compared to 0.190 for pNK396), even though the internal sequences were more than 2.5 times longer.
To determine whether the Ty5 LTR is also important for marker exchange, we tested recombination substrates carrying 3′-end sequences with or without a LTR. For substrates with sequences on either side of the artificial intron, most recombination products were marker exchanges. Recombination frequencies for Ty5 3′-end sequences with and without the LTR were very similar (Fig. 6B). This indicates that the Ty5 LTR is not involved in marker exchange, which is likely mediated by homologous sequences flanking the neoAI marker.
LTR sequences critical for tandem element formation.
Deletion analysis was used to determine whether specific Ty5 LTR sequences are important in forming tandem elements. LTR deletion constructs were used as recombination substrates in the assay shown in Fig. 1B. When one-fourth of the Ty5 LTR was deleted from either the 5′ or 3′ end, recombination frequencies dropped only 2- to 3-fold (2.6-fold for pNK355 and 2.0-fold for pNK413) (Fig. 7A). This indicated that neither the 5′- nor the 3′-LTR end sequence (65 and 61 bp) is essential for forming tandem elements. However, when one-half or more of the LTR was deleted, the recombination frequencies dropped markedly (at least 18.8-fold for pNK416). This suggests that either these fragments fall below the minimal length required for recombination or they lack essential features that mediate recombination.
To distinguish between these two possibilities, the substrate from the 3′ end of the LTR in pNK414 was further analyzed. This 80-bp LTR fragment was fused to 354 bp of internal flanking sequence (pNK305), and the entire 251-bp LTR was also fused to the same internal sequence as a control (pNK311). Assays were conducted to calculate the recombination frequencies. As shown in Fig. 7B, the substrate in pNK305 mediated tandem element formation at a frequency only twofold lower than that mediated by the substrate in pNK311. This indicates that the 80 bp of LTR in pNK414 likely failed to mediate tandem element formation because it fell below the minimal length for an efficient processing segment for recombination and not because it lacked some essential sequences. It is interesting to note that since all of the Ty5 promoter is deleted in pNK305 (19), basal Ty5 transcription does not greatly affect recombination.
Despite its importance in recombination, tandem element formation cannot be explained by crossover events initiated within the Ty5 LTR.
The importance of the LTR in mediating tandem element formation suggests that recombination is initiated within LTR sequences (20). To determine the recombination initiation sites, point mutations were introduced into two recombination substrates (pNK418 and pNK419) such that a restriction site was gained (BamHI [Fig. 8A]) or destroyed (HindIII [Fig. 8A]). The lengths of the internal Ty5 sequences of these substrates differed. Recombination frequencies with both substrates were first determined and found to be comparable to those with the nonmutagenized substrates (data not shown). Recombination products were then analyzed by restriction mapping and DNA sequencing to determine the pattern of inheritance of the mutations in the tandem elements. If recombination was initiated within the Ty5 LTR, as proposed by our previous models (20), class I products that have both mutations at their 3′ ends (Fig. 8A) would be obtained. Surprisingly, and in contrast to our prediction, most products from pNK418 and all products from pNK419 had both mutations in their 5′ ends. Sequence analysis of seven randomly selected products for pNK418 and four for pNK419 revealed that all carried both mutations at their 3′ ends as well (Fig. 8B). Therefore, despite its critical role in mediating high-frequency tandem element formation, recombination cannot be explained by simple crossover events between the substrate and a full-length Ty5 cDNA.
DISCUSSION
Reverse transcription plays an important role in replenishing sequence loss that occurs at the ends of linear chromosomes after replication: telomerase can template sequences onto chromosome ends by reverse transcription, as can transposition through reverse transcription of the D. melanogaster telomeric HeT-A and TART retrotransposons. Recombination between telomeric and subtelomeric repeats can also amplify chromosome end sequences. The yeast Ty5 retrotransposons use both transposition and cDNA recombination to modify telomeric regions.
Ty5 cDNA recombination requires Rad52p.
The integrase DD(35)E domain, which is conserved among all retroviruses and retrotransposons, is the catalytic domain of Ty5 integrase. Chromosomal integration events were abolished when the second conserved aspartic acid of the DD(35)E domain and its flanking threonine residue were mutated. The Ty5 integration pathway was also blocked by mutating LTR end sequences (U3 tip), the sites where Ty5 integrase likely binds, or by mutating the PPT. The PPT serves as the priming site for plus-strand cDNA synthesis, and mutations block the formation of double-stranded cDNA, the normal substrate for integration. For all mutations that blocked the integration pathway, cDNA recombination predominated. It is interesting to note that for the PPT mutant, the frequency of cDNA recombination was similar to that of wild-type elements. This suggests that single-stranded, minus-strand cDNA is sufficient to mediate cDNA recombination (see the discussion below). Our observations are very similar to those made when the Ty1 integration pathway was disrupted by mutating either Ty1 integrase, the cDNA termini, or the PPT (45). Mutation of the DD(35)E domain has also been shown to affect in vivo and in vitro integration of Ty3, human immunodeficiency virus type 1, and avian sarcoma virus (21, 48).
Ty5 cDNA recombination is dependent on Rad52p, which is involved in most homologous-recombination events (38). In contrast, mutations in RAD1 had no effect on Ty5 cDNA recombination. RAD1 is involved in direct-repeat recombination and excises nonhomologous sequences between substrates (12, 44); this is not a feature of Ty5 cDNA recombination. Recombination of Ty1 cDNA also depends on the homologous recombination proteins RAD51 and RAD52 but not RAD1 (35, 36, 45). RAD1, however, has recently been shown to affect the recombination of Ty1 cDNA with substrates that are actively transcribed (35).
Ty5 cDNA recombination and silent chromatin.
We have previously shown that silent chromatin is required for Ty5’s integration specificity (56). Here we demonstrate that Ty5 also preferentially recombines with homologous substrates in silent chromatin. To more readily recover recombination events, our initial experiments were conducted with a Ty5 integrase mutant (in-611) as a cDNA donor. For substrates located at the telomeres or the silent mating locus HMR, we observed at least a 3.2-fold enhancement in recombination relative to that of a substrate at an internal chromosomal site. Because our internal substrate is transcriptionally active, it can generate integration events, and therefore our values are likely underestimates. The preferential recombination with substrates in silent chromatin was confirmed with a wild-type Ty5 donor element. In addition, the enhancement did not occur simply because our control was a recombinational cold spot. When transformed into yeast cells, linear DNA containing Ty5-HIS3 recombined at slightly higher frequencies with the internal substrate. The preferential recombination was also not due to the transcriptional silencing of substrate elements in silent chromatin, as Ty5 substrates lacking a promoter were approximately as effective as wild-type elements in forming tandem elements or carrying out marker exchanges.
The preference of Ty5 to integrate into regions of silent chromatin requires a Ty5-encoded targeting domain (14). We found that this targeting domain is also required for preferential recombination with telomeric substrates. We hypothesize that the targeting domain interacts with protein components of silent chromatin, which in turn localize Ty5 cDNA to silent regions. An increase in the local concentration of Ty5 cDNA may explain both the integration and recombination target biases. A role for integrase in bringing the cDNA and substrate into proximity is supported by a phenotype observed for elements carrying the U3-tip and PPT mutations. Both of these mutations likely impair the binding of integrase to the cDNA, and neither mutant shows preferential recombination with telomeric substrates (20a). Thus, there appear to be two pathways for Ty5 cDNA recombination: one pathway is integrase dependent, and these events occur preferentially in regions of silent chromatin; the other pathway is integrase independent, shows no site preference, and can occur through either single- or double-stranded cDNA.
To determine whether the recombination substrate bias occurs when Ty5 naturally replicates, we measured cDNA recombination with a telomeric element that serves as both cDNA donor and substrate. When transcription and transposition of this telomeric element is induced by mating pheromones, Ty5 cDNA frequently recombines with this insertion to form either tandem elements (5 of 30) or elements that have exchanged markers (8 of 30). Thus, Ty5 cDNA occurs frequently when native Ty5 elements are replicating. It is interesting to note that some tandem elements are present in multiple copies. This has the net effect of increasing the size of the subtelomeric sequences, which may counter telomeric sequence loss that occurs naturally during DNA replication.
Mechanisms of tandem element formation.
Although the Ty5 LTR is required for the formation of tandem elements, it is not involved in marker exchange. This suggests that the two classes of recombination products arise by different mechanisms. For marker exchange, only homology flanking the marker gene is important, whereas for tandem element formation, LTR sequences are required in addition to internal domain homology between the cDNA and substrate. Deletion analysis indicated that substrates missing one-fourth of the LTR at either the 5′ end or the 3′ end could still form tandem elements efficiently; the sequences missing in these two LTR fragments, therefore, are not essential for recombination. Further deletion (≥126 bp of the 251-bp LTR) reduced recombination to background levels. This is probably because the length of the LTR substrate fell below what has previously been observed for the minimal efficient segment for recombination in yeast (89 to 250 bp) (18, 49) and not because an essential LTR feature was lost. In support of this, when an 80-bp LTR substrate was extended by including adjacent internal Ty5 sequences, the frequency of tandem element formation was restored. Since this 80 bp did not include essential Ty5 promoter sequences, basal transcription was not involved in recombination. In summary, it is the LTR sequences that mediate recombination and not transcription or protein factors that bind the Ty5 LTR.
Although Ty5 LTR sequences are required for tandem element formation, this process cannot be explained by the simple crossover models we proposed earlier (20). Two point mutations were introduced into Ty5 substrates to determine the recombination initiation sites. If recombination initiates within the LTR, most products would contain both mutations in the 3′ element; this product was never observed in the 25 products analyzed for pNK419 and was only observed for three of the 38 products analyzed for pNK418 (Fig. 8, class I). In contrast, most products carried both markers in both the 5′ and 3′ elements (Fig. 8, class III). To explain this observation, we propose that recombination is initiated by single-stranded cDNA based on the single-strand annealing model (13, 16) (Fig. 9A). Single-stranded cDNA, perhaps generated during minus-strand cDNA synthesis, invades the substrate element and is extended into flanking sequences. The length of the cDNA and the site of invasion determine whether it picks up zero, one, or two mutations (the acquisition of two mutations is illustrated in Fig. 9A). The 3′ LTR in the invading single-stranded cDNA then displaces the 5′ LTR and forms a looped intermediate. The resolution of this intermediate structure by the host recombination and repair system would generate tandem elements. This model is consistent with our observation that internal sequences facilitate tandem element formation, in this case by helping to mediate strand invasion of the partial cDNA. The Ty5 LTR is essential for tandem element formation, because it is required for strand displacement to form the looped intermediate. Ty5 elements likely produce abundant single-stranded donor cDNA; Southern hybridizations designed to detect cDNA reveal a smear of variously sized products, suggesting the presence of partial cDNA intermediates (20a). This may be because Ty5 reverse transcriptase is inherently inefficient, which is consistent with low transposition activity of Ty5 relative to other yeast retrotransposons. Furthermore, the efficient cDNA recombination observed with the PPT mutant supports the idea that single-stranded cDNA mediates cDNA recombination.
FIG. 9.
Models for tandem element formation. The models illustrate the formation of class III recombination products (Fig. 8), although the same models can also generate class I and class II products. Class I and II products are formed, depending on the site of strand invasion (A) or the extent of recession (B). A single asterisk indicates the BamHI mutation; two asterisks indicate the HindIII mutation. Thick arrows represent LTRs; thin dotted lines represent newly synthesized DNA; dashed thick arrows represent recessed LTR sequences. (A) Step 1. The 3′ end of partial-minus-strand Ty5 cDNA (single stranded) invades the substrate. Step 2. branch migration and strand displacement continue through the 5′ LTR. The Holliday junction is cleaved (small arrowhead). Step 3. The LTR at the 5′ end of the invading cDNA displaces the newly synthesized LTR at the 3′ end. This forms a looped intermediate. The 3′ OH at the site of the Holliday junction is ligated to the 5′ end of the cDNA. The strand with the free 5′ end, released from Holliday junction cleavage, is trimmed and ligated (small arrowhead). Step 4. The mismatch repair system cleaves the top strand of the mismatched region. DNA repair is carried out, with the bottom strand as a template, followed by ligation. Step 5. Recombination products are formed, with markers in both the 5′ and 3′ elements. (B) Step 1. The cDNA is recessed beyond the region containing the two markers. Step 2. The 3′ end of the cDNA invades the substrate, and DNA synthesis repairs the gap. Step 3. The Holliday structure is resolved; the small arrows indicate the sites of cleavage. Step 4. Recombination products are formed, with markers in both elements.
An alternative model to explain our observations is based on the crossover models we proposed previously (Fig. 9B) (20). It differs, however, in that it requires recession of both strands of the linear cDNA. Recession must remove all of the 5′ LTR sequences and some internal sequences. The extent of recession will determine whether the resultant tandem element has one (class II), both (class III), or no (class I) mutations in the 5′ element. After recession, the cDNA invades the substrate and is resolved to form tandem elements. We do not favor this model, however, because it requires greater recession rates at the 5′ LTR of the cDNA than at the 3′ LTR (3′-LTR sequences must be retained for strand invasion).
Our model for recombinational amplification of Ty5 elements may have implications for the proliferation of other LTR retrotransposons. Tandem arrays of Ty1 and the Schizosaccharomyces pombe Tf1 element have previously been described (1, 52), including a report of large Ty1 arrays at the silent mating locus HML (52). Although most tandem Ty5s contain two back-to-back elements, in this study we observed instances where telomeric Ty5 substrates generated highly reiterated arrays. This is similar to the tandem repeats of non-LTR retrotransposons observed at D. melanogaster telomeres (3, 4, 24, 46) and to the tandem arrays of Y′ subtelomeric repeats observed in S. cerevisiae (28). Telomeres are clearly dynamic sites of DNA synthesis and loss. Amplification of Saccharomyces Ty5 elements by both integration and recombination provides an alternative means of replenishing and modulating chromosome end sequences.
ACKNOWLEDGMENTS
We thank L. Prakash and S. Prakash for plasmid constructs and Giovanni Bosco for helpful comments on the manuscript.
This work was supported by an American Cancer Society Grant (VM145) to D.F.V. and by Hatch Act and State of Iowa funds.
Footnotes
This is journal paper J-17884 of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, project 3383.
REFERENCES
- 1.Atwood A, Lin J-H, Levin H L. The retrotransposon Tf1 assembles virus-like particles that contain excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol. 1996;16:338–346. doi: 10.1128/mcb.16.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1987. [Google Scholar]
- 3.Biessmann H, Champion L E, O’Hair M, Ikenaga K, Kasravi B, Mason J M. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biessmann H, Mason J M, Ferry K, d’Hulst M, Valgeirsdottir K, Traverse K L, Pardue M L. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell. 1990;61:663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn E. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D, Sandmeyer S B. Yeast transposable elements. In: Broach J, Jones E, Pringle J, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1991. pp. 193–261. [Google Scholar]
- 7.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen B, Przybyla A E. An efficient site-directed mutagenesis method based on PCR. BioTechniques. 1994;17:657–659. [PubMed] [Google Scholar]
- 9.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Eichinger D J, Boeke J D. A specific terminal structure is required for Ty1 transposition. Genes Dev. 1989;4:324–330. doi: 10.1101/gad.4.3.324. [DOI] [PubMed] [Google Scholar]
- 11.Eickbush T H. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 12.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 13.Fishman-Lobell J, Rudin N, Haber J E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gai X, Voytas D F. A single amino acid change in the yeast retrotransposon Ty5 abolishes targeting to silent chromatin. Mol Cell. 1998;1:1051–1055. doi: 10.1016/s1097-2765(00)80105-7. [DOI] [PubMed] [Google Scholar]
- 14a.Gai, X., and D. F. Voytas. Unpublished data.
- 15.Greider C W, Blackburn E H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 16.Haber J E. Exploring the pathways of homologous recombination. Curr Opin Cell Biol. 1992;4:401–412. doi: 10.1016/0955-0674(92)90005-w. [DOI] [PubMed] [Google Scholar]
- 17.Higashiyama T, Noutoshi Y, Fujia M, Yamada T. Zepp, a LINE-like retrotransposon accumulated in the Chlorella telomeric region. EMBO J. 1997;16:3715–3723. doi: 10.1093/emboj/16.12.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ke N, Irwin P A, Voytas D F. The pheromone response pathway activates transcription of Ty5 retrotransposons located within silent chromatin of Saccharomyces cerevisiae. EMBO J. 1997;16:6272–6280. doi: 10.1093/emboj/16.20.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ke N, Voytas D F. High frequency cDNA recombination of the Saccharomyces retrotransposon Ty5: the LTR mediates formation of tandem elements. Genetics. 1997;147:545–556. doi: 10.1093/genetics/147.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Ke, N., and D. F. Voytas. Unpublished data.
- 21.Kirchner J, Sandmeyer S B. Ty3 integrase mutants defective in reverse transcription or 3′-end processing of extrachromosomal Ty3 DNA. J Virol. 1996;70:4737–4747. doi: 10.1128/jvi.70.7.4737-4747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levis R W, Ganesan R, Houtchens K, Tolar L A, Sheen F M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 25.Liefshitz B, Parket A, Maya R, Kupiec M. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics. 1995;140:1199–1211. doi: 10.1093/genetics/140.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 27.Louis E J, Haber J E. Mitotic recombination among subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics. 1990;124:547–559. doi: 10.1093/genetics/124.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louis E J, Haber J E. The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis E J, Haber J E. The subtelomeric Y′ repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics. 1990;124:533–545. doi: 10.1093/genetics/124.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundblad V, Blackburn E H. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 31.McEachern M J, Blackburn E H. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 32.Melamed C, Nevo Y, Kupiec M. Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1613–1620. doi: 10.1128/mcb.12.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 35.Nevo-Caspi Y, Kupiec M. Induction of Ty recombination in yeast by cDNA and transcription: role of the RAD1 and RAD52 genes. Genetics. 1996;144:947–955. doi: 10.1093/genetics/144.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevo-Caspi Y, Kupiec M. Transcriptional induction of Ty recombination in yeast. Proc Natl Acad Sci USA. 1994;91:12711–12715. doi: 10.1073/pnas.91.26.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okazaki S, Ishikawa H, Fujiwara H. Structural analysis of TRAS1, a novel family of telomeric repeat-associated retrotransposons in the silkworm, Bombyx mori. Mol Cell Biol. 1995;15:4545–4552. doi: 10.1128/mcb.15.8.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J, Jones E, Pringle J, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1991. pp. 407–521. [Google Scholar]
- 39.Pluta A F, Zakian V A. Recombination occurs during telomere formation in yeast. Nature. 1989;337:429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds P J, Prakash L, Prakash S. Nucleotide sequence and functional analysis of the RAD1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1012–1020. doi: 10.1128/mcb.7.3.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth C W, Kobeski F, Walter M F, Biessmann H. Chromosome end elongation by recombination in the mosquito Anopheles gambiae. Mol Cell Biol. 1997;17:5176–5183. doi: 10.1128/mcb.17.9.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–210. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 43.Rowland S-J, Dyke K G H. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 44.Schiestl R H, Prakash S. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol Cell Biol. 1988;8:3619–3626. doi: 10.1128/mcb.8.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheen F-M, Levis R W. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci USA. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 48.Skalka A M. Retroviral DNA integration: lessons for transposon shuffling. Gene. 1993;135:175–182. doi: 10.1016/0378-1119(93)90063-9. [DOI] [PubMed] [Google Scholar]
- 49.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi H, Okazaki S, Fujiwara H. A new family of site-specific retrotransposons, SART1, is inserted into telomeric repeats of the silkworm, Bombyx mori. Nucleic Acids Res. 1997;25:1578–1584. doi: 10.1093/nar/25.8.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S-S, Zakian V A. Telomere-telomere recombination provides an express pathway for telomere acquisition. Nature. 1990;345:456–458. doi: 10.1038/345456a0. [DOI] [PubMed] [Google Scholar]
- 52.Weinstock K G, Mastrangelo M F, Burkett T J, Garfinkel D J, Strathern J N. Multimeric arrays of the yeast retrotransposon Ty. Mol Cell Biol. 1990;10:2882–2892. doi: 10.1128/mcb.10.6.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu G L, Bradley J D, Attardi L D, Blackburn E H. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 54.Zou S, Ke N, Kim J M, Voytas D F. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 55.Zou S, Kim J M, Voytas D F. The Saccharomyces retrotransposon Ty5 influences the organization of chromosome ends. Nucleic Acids Res. 1996;24:4825–4831. doi: 10.1093/nar/24.23.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou S, Voytas D F. Silent chromatin determines target preference of the retrotransposon Ty5. Proc Natl Acad Sci USA. 1997;94:7412–7416. doi: 10.1073/pnas.94.14.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]