INTRODUCTION
Collecting data at the point-of-care is a critical task for many clinical studies, a process made more feasible by the advent of electronic medical records (EMRs).1 However, creating data entry structures generally requires EMR programming by information technology (IT) specialists,2 resulting in delays and costs that are prohibitive for smaller studies and investigators with limited funding.
We developed an alternative strategy to enter and extract structured data from free-text EMR notes, taking advantage of templates that make data parsing tractable. Most EMRs, including the two largest US vendors (Epic and Cerner), allow users to create and share templates within their notes. Within such templates, specific fields are available for the user to choose from a list of options (an enumeration data type) that populates a specific portion of the text when selected. We describe here our method for programmatically extracting structured data from notes created with dedicated templates.
METHODS
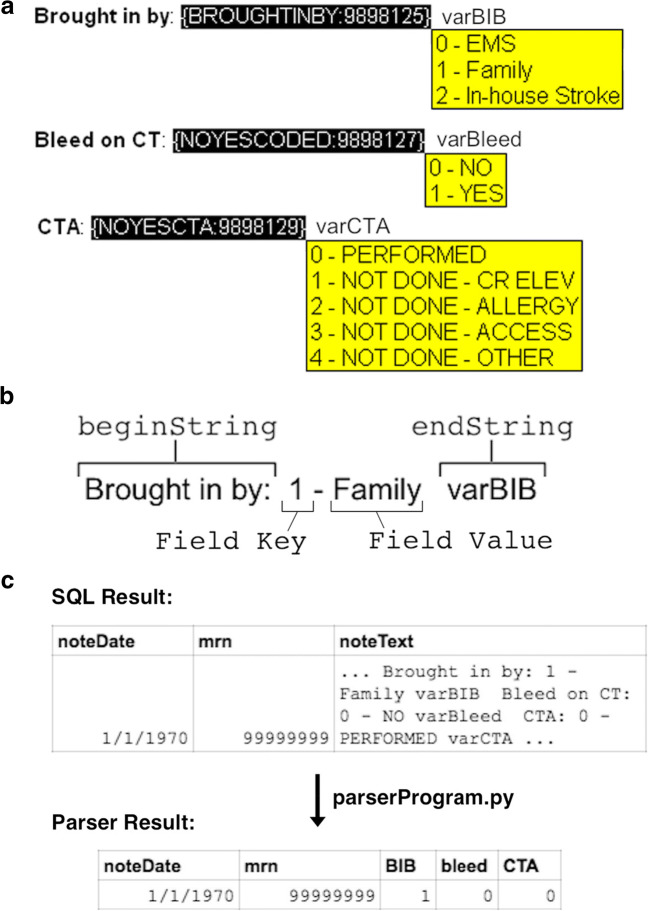
Our technique involves three steps (which we illustrate in the Epic EMR (Epic Systems, Verona, WI)): (1) construct a text template (“SmartPhrase”) containing a unique string identifier tag and embedded list enumerations (“SmartLists”) to allow data entry directly into notes, (2) query a back-end relational database (“Clarity”) to capture notes containing the unique text string tag, and (3) parse the captured notes to extract data into structured form using a Python script employing regular expressions to identify the necessary fields (Fig. 1). Our SQL and Python code are available under an open-source MIT License at https://github.com/alexanderflint/structured-data-from-notes.
Fig. 1.
Method for text-based data storage and extraction of structured data. a) Example showing 3 fields from the front-end text entry template. For each field, a pulldown list enumeration is provided to the user with the ability to select exactly one response. b Each field and flanking text has a similar structure that enables parsing. After user selection from a pulldown menu, the text generated has two unique strings encoding flanking text (beginString and endString), and the intervening text generated by the pulldown selection has a Field Key that consists of one or more integer text characters and a Field Value that consists of non-integer text characters. c The parsing program takes in the result of our SQL query, which consists of multiple rows of data, one for each note, and extracts each integer key from each text field, identified by the flanking text surrounding the data field of interest
We tested this approach in a study of stroke treatment in the 21-hospital Kaiser Permanente Northern California (KPNC) health system. To test performance, 7 text data extraction builds (templates) were created with varying number of data elements (1 to 11), varying number of users (3 to 20), and varying number of hospital centers (1 to 21). Clarity was queried with Teradata SQL Assistant v13.11 to capture notes based on a unique text string present in each template.
Selected users were granted access to the SmartPhrases and given brief feedback in their intended use. After initial roll-out, no additional user feedback was provided so that we could determine user-generated error rates in the absence of reinforcement. This project was judged to not meet the regulatory definition of research by the KPNC Research Determination Official.
RESULTS
Our method used minimal computing resources. Querying 1217 notes from 17,331,944 stored notes took 72 seconds and further data parsing took < 2 seconds. The usable-field rate was high (20,989/21,709 fields = 96.7%), with lower usable-field rates associated with larger numbers of centers, users, and data fields (Table 1).
Table 1.
Template deployments, data captured, and usable-field rates
| Template | Number of centers | Number of users | Number of fields | Total notes | Total fields | Total usable fields | % Usable fields (95% CI) |
|---|---|---|---|---|---|---|---|
| Stroke Hub | 20 | 16 | 11 | 1217 | 13,387 | 12,761 | 95.3% (95.0–95.7%) |
| Cancelled Stroke | 20 | 16 | 2 | 884 | 1768 | 1737 | 98.2% (97.5–98.8%) |
| SAH Data | 1 | 3 | 2 | 89 | 178 | 178 | 100.0% (97.9–100%) |
| ICH Data | 1 | 3 | 5 | 151 | 755 | 755 | 100.0% (99.5–100%) |
| EST Data | 1 | 3 | 10 | 88 | 863 | 852 | 98.7% (97.7–99.4%) |
| Mood Screen | 1 | 20 | 9 | 524 | 4716 | 4664 | 98.9% (98.6–99.2%) |
| mRS Data | 1 | 2 | 1 | 42 | 42 | 42 | 100.0% (91.6–100%) |
| Totals | 21,709 | 20,989 | 96.7% (96.4–96.9%) |
Stroke Hub = template for acute stroke telemedicine data entry; Cancelled Stroke = template for telemedicine data entry when acute stroke codes are cancelled; SAH Data = template for Comprehensive Stroke Center (CSC) subarachnoid hemorrhage (SAH) data entry; ICH Data = template for CSC intracerebral hemorrhage (ICH) data entry; EST Data = template for CSC endovascular stroke treatment (EST) data entry; Mood Screen = template for CSC depression screening data entry; mRS Data = template for CSC modified Rankin Scale (mRS) data entry for patients who underwent EST
Usable fields are defined as any field containing valid information (i.e., no text manipulation by the user that led to unexpected parsing results or blank fields (e.g., line deletion, deletion or editing of flanking text, or deletion or editing of field text))
DISCUSSION
We describe a simple generalizable method for storing user-generated clinical data within the EMR that can be extracted as structured data for quality and research purposes. This system facilitates data collection that would otherwise be unavailable to many investigators and clinicians.
Extracting structured data from the EMR generally requires expensive and time-consuming EMR programming or the use of natural language processing.3, 4 With our method—and some careful planning and EMR familiarity—any user can construct a structured data entry schema with only modest programming support.
Our system has several advantages, including the use of regular EMR notes, minimal development time/costs, and generalizability to any type of note or report. Usable-field rates are high at baseline, but this aspect could likely be further improved by user reinforcement and additional training.
Disadvantages include limited data types (i.e., integer key and string value, although some variations are possible (e.g., date/time fields)), a lack of real-time data validation, and possible data entry problems caused by user manipulation of the text template. Institutional imperatives regarding allowable progress-note content must be followed with this or any other system.
In summary, we provide a flexible solution to a vexing problem for many research and quality-improvement initiatives by facilitating entry and extraction of structured data in EMR notes. Our experience demonstrates the feasibility of this approach.
Compliance with Ethical Standards
Conflict of Interest
All authors declare an absence of any conflicts of interest relevant to this work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cowie MR, Blomster JI, Curtis LH, et al. Electronic health records to facilitate clinical research. Clin Res Cardiol. 2017;106(1):1–9. doi: 10.1007/s00392-016-1025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush RA, Kuelbs CL, Ryu J, Jian W, Chiang GJ. Structured data entry in the electronic medical record: perspectives of pediatric specialty physicians and surgeons. J Med Syst. 2017;41(5):75. doi: 10.1007/s10916-017-0716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim BJ, Merchant M, Zheng C, et al. A natural language processing program effectively extracts key pathologic findings from radical prostatectomy reports. J Endourol. 2014;28(12):1474–1478. doi: 10.1089/end.2014.0221. [DOI] [PubMed] [Google Scholar]
- 4.Sippo DA, Warden GI, Andriole KP, et al. Automated extraction of BI-RADS final assessment categories from radiology reports with natural language processing. J Digit Imaging. 2013;26(5):989–994. doi: 10.1007/s10278-013-9616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]