Abstract
The growth of long and polarized ameloblast-like cells has long been heralded as a major prerequisite for enamel tissue engineering. In this study, we have designed three-dimensional bioreactor/scaffold microenvironments to propagate and assess the ability of cervical loop derivatives to become long and polarized ameloblast-like cells. Our studies demonstrated that cervical loop/periodontal progenitor coculture in a growth-factor-enriched medium resulted in the formation of ameloblast-like cells expressing high levels of amelogenin and ameloblastin. Coculture of cervical loop cells with dental pulp cells on tailored collagen scaffolds enriched with leucine-rich amelogenin peptide (LRAP) and early enamel matrix resulted in singular, elongated, and polarized ameloblast-like cells that expressed and secreted ameloblastin and amelogenin enamel proteins. Bioreactor microenvironments enriched with enamel matrix and LRAP also proved advantageous for the propagation of HAT-7 cells, resulting in a ∼20-fold higher expression of amelogenin and ameloblastin enamel proteins compared with controls growing on plain scaffolds. Together, studies presented here highlight the benefits of microgravity culture systems combined with ameloblast-specific microenvironments and tailored scaffolds for the growth of ameloblast-like cells.
Keywords: amelogenesis, ameloblasts, bioreactor, differentiation, LRAP
Introduction
In their natural environment, cells receive nutrients and structural support from all three dimensions, stretch and contract within the limits of surrounding tissues, and respond to signals and spatial feedback from all sides, top and bottom [1]. As part of a multicellular organism, cells experience gravity in varying angles and orientations with every turn and movement of the animal they live in [2]. In mammals, cells have evolved to thrive in three-dimensional (3D) surroundings for ∼65 million years [3]. For any living cell, the sudden switch from such a rich organismal microenvironment to the confined terrain of a two-dimensional (2D) culture dish must present a radical change in living conditions. At the bottom of a 2D dish, the effects of gravity are strictly unidimensional, signals are depleted within a limited time span, and the supply in nutrients and oxygen and the removal of waste remain static until the next change of medium occurs. Considering this elemental change in living conditions, the predominance of in vitro cell culture as a means of biomedical inquiry has been remarkable, including the prominent role of 2D culture systems in cell biological research. The success of many cell culture protocols is tightly linked to established cell lines, such as NIH 3T3 fibroblasts, keratinocytes, and >2,000 human cell lines from the American Type Culture Collection (ATCC). However, some cell types are less amenable to 2D culture conditions, including many differentiated epithelial cells such as ameloblasts.
Ameloblasts are the principal cell type responsible for the secretion of tooth enamel extracellular matrix and enamel ion mineral transport [4]. Perhaps due to their high level of specialization and interdependency, ameloblast cell culture models have proven to be challenging [5,6]. So far, five different ameloblast-like cell lines have been reported in the literature, in addition to various protocols for the culture of primary ameloblast-like cells. Primary enamel organ cells grown on feeder cell layers express amelogenin, ameloblastin, MMP20, kallikrein 4, and other enamel-related proteins [7]. In addition to enamel organ primary cell culture protocols, three of the five known enamel organ-derived cell lines have been reported to mimic ameloblast-like qualities, the mouse ameloblast-lineage cell line (ALC), the rat dental epithelial cell line (HAT-7), and the mouse LS8 cell line [8]. Two other cell lines have been established but not used frequently: the porcine PABSo-E cell line [9] and the rat SF2-24 cell line [10]. However, the majority of these cells have lost their distinctive polarized cell shape in culture.
In recent years, technologies originally developed by the National Aviation and Space Administration (NASA) to study the effects of microgravity on cultured cells have been applied to solve many of the problems of traditional 3D cell culture as they relate to transport and fluid exchange [11,12]. These bioreactor technologies have resulted in the development of a rotating wall vessel as a single axis clinostat consisting of a fluid-filled, cylindrical, horizontally rotating culture vessel [13,14]. Cells placed in these environments assemble into tissue-like aggregates with high mass transport of nutrients, oxygen, and wastes [14]. As a result, bioreactor technologies have been frequently employed to culture cells that are incompatible with other culture technologies [14].
In this study, we have successfully generated a 3D bioreactor-based ameloblast-like cell culture model based on mouse incisor cervical loop enamel organ cells. We have used this model to examine some of the scaffolds, microenvironments, and coculture conditions necessary to facilitate the differentiation of polarized, elongating, and enamel protein secreting ameloblast-like cells, and we have compared bioreactor-grown ameloblast-like cells with established ameloblast-like cell lines such as HAT-7 cells. The results of our study provide new insights into the microenvironments necessary to support successful growth of ameloblast-like cells.
Materials and Methods
Bioreactor
Cells were cultured in a rotatory cell culture system or Rotary Cell Culture System (RCCS)-4 from Synthecon, Inc. (Houston, TX). This system contains four high-aspect ratio vessels (HARVs) with disk-shaped oxygenator membranes in the vessel wall used in conjunction with a RCCS rotator base (Fig. 1). The RCCS generates a 3D in vitro microgravity environment as a result of low shear force and high mass transfer.
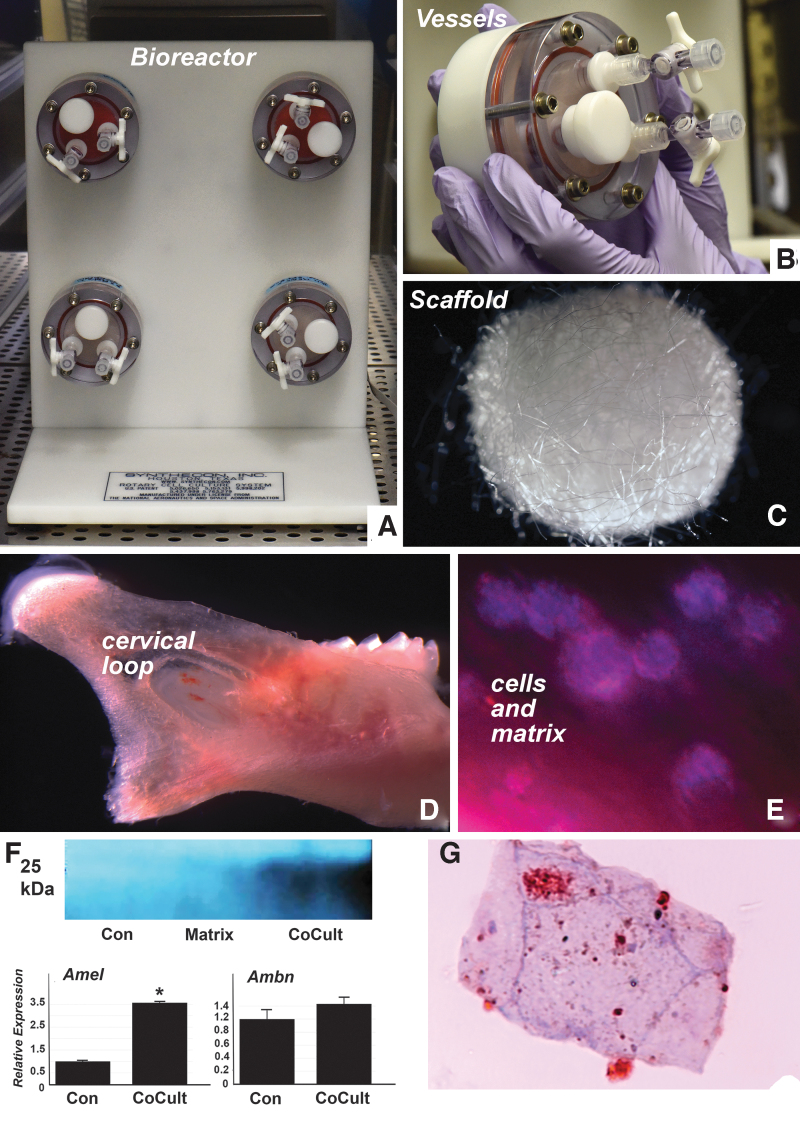
FIG. 1.
3D bioreactor culture model (A) with four vessel capacity. Each vessel (B) holds 10 mL of media. (C) Example of one of the PGA scaffolds that were used in this study. Cells for bioreactor studies were harvested from the cervical loop area of a mouse incisor. The position of the cervical loop relative to the mouse mandible is illustrated in (D). The micrograph in (E) illustrates some of the cervical loop cells cultured with scaffold and matrix but without coculture cell populations. (F) RT-PCR and western blot analysis demonstrated significantly increased amelogenin and ameloblastin expression and elevated amelogenin protein levels in the coculture group as compared with the control and the matrix-enriched groups. (G) Is a cross-section through an ameloblast-like cell body revealing extracellular enamel matrix deposits and intracellular amelogenin-stained vesicles using immunohistochemistry. Note the unique cross-section microanatomy of a paraffin-sectioned cell grown in a 3D culture environment. *P ≤ 0.05. 3D, three dimensional; PGA, polyglycolic acid; RT-PCR, reverse transcriptase polymerase chain reaction.
Cervical loop preparation and 3D bioreactor cell culture
All animal procedures were performed in accordance with TAMU College of Dentistry IACUC 2019-0237 CD regulations. Cervical loop cells were dissected from ten 6 days postnatal (DPN) C57/BL-6J wild-type mouse mandibular incisors. Cells were pooled and digested using collagenase/dispase (Roche, Basel, Switzerland) at 37°C for 30 min to obtain a single cell suspension. This suspension was then passed through a 70 μm cell strainer and centrifuged. After washing with phosphate buffered saline (PBS), cells were counted using a hemocytometer. Cells were then aliquoted for culture in our four 10 mL HARV vessels, representative of the following groups: (1) keratinocyte medium serum-free medium with growth factor supplements (Gibco, Maryland) and additional 10% fetal bovine serum, 1% penicillin/streptomycin and 100 μg/mL ascorbic acid as control group and (2) addition of growth factors and extracellular matrix proteins, including 0.03 μg/mL BMP 2 (R&D systems, MN), 0.03 μg/mL BMP-4 (R&D systems), 10 ng/mL human recombinant FGF (Peprotech, Cranbury, NJ) and 15 ng/mL hEGF (Gibco), 200 μg/mL Matrigel (Corning Life Sciences, New York), 5 μg/mL Laminin (Corning) and 5 μg/mL Fibronectin (Corning) to the control medium for the experimental group.
Coculture studies
The four cervical loop cell suspensions described above provided the basis for our coculture studies using either human periodontal progenitors (passage 4) or human dental pulp progenitors (passage 3). For coculture studies, 1 × 104 cells were added to the bioreactor coculture group. Cervical loop cell monolayers seeded onto scaffolds in the bioreactor without the presence of any mesenchymal support cells served as the control group. The experimental group comprised of cervical loop cells cocultured with either periodontal progenitors or dental pulp progenitors. Immortalized HAT-7 cells originating from a cervical loop epithelium of a rat incisor were used in place of cervical loop cells and cocultured with dental pulp progenitors to determine the feasibility of maintaining an established ameloblast-like cell line in a 3D culture system. All culture experiments were conducted for 10 days, and the samples were harvested for further molecular analysis and histology.
Scaffolds
Multiple experiments were carried out to determine scaffold viability. The majority of data in this study were obtained using either a polyglycolic acid (PGA)-based scaffold or a collagen-based scaffold, both of which yielded valuable data. In a first set of experiments (Fig. 2), six BIOST-Disks biostructure-matrix PGA scaffolds with 5 mm diameter (Synthecon, Inc.) were added to each vessel to provide support for cell attachment and growth. Cells were cultured in the bioreactor for 10 days with a medium change after every 48 h. After extensive testing of various scaffolds, a scaffold termed SpongeCol (Advanced Biomatrix) was chosen as the most compatible scaffold suitable for our experiments (data shown in Figs. 2 and 3). A second strategy entailed a preparation of 500 μL of Fibricol gel enriched with 1 mg of leucine-rich amelogenin peptide (LRAP) (Peptide 2.0, Chantilly, VA), 1 mg of lyophilized early-stage enamel matrix prepared from porcine teeth [15], 5 μg/mL laminin, and 5 μg/mL of fibronectin. These additional components were added separately and together to the SpongeCol scaffold, and placed in the bioreactor together with our cells immediately upon coating. For a third set of studies, we used a combination of LRAP, matrix, and 10 μL special AT-rich sequence-binding protein-1 (SATB1) (Origene, MD) together with the SpongeCol scaffold coated with a collagen I based gel (FibriCol, Advanced Biomatrix). After 10 days of culture, scaffolds were harvested, fixed, and processed for paraffin embedding. Alternatively, samples were frozen for molecular analysis using liquid nitrogen.
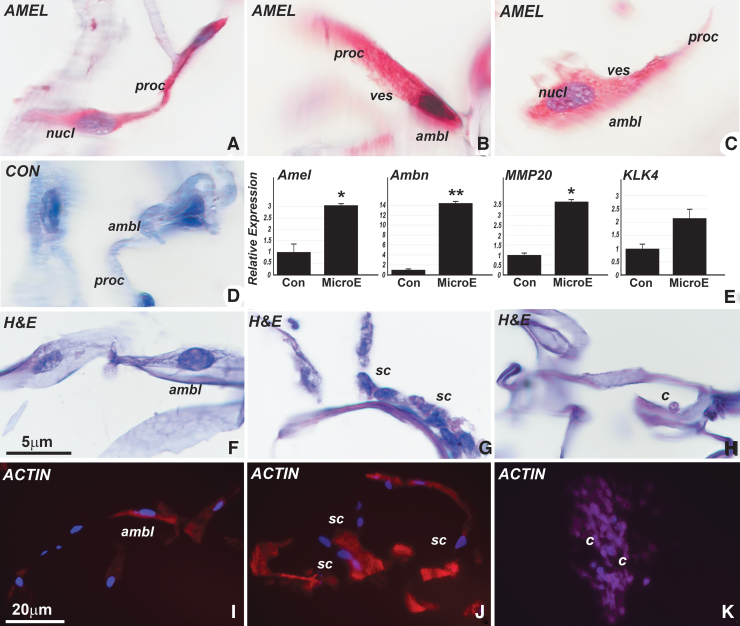
FIG. 2.
Differentiation of ameloblast-like cells from cervical loop stem cells cocultured with dental pulp progenitors after addition of LRAP and early-stage enamel matrix. (A–C) Are differentiated ameloblast-like cells (ambl) as evidenced by their elongated and polarized cell shape and their positive immunoreactivity for amelogenin. Note the conical cell process resembling Tomes' process (proc) and the vesicular intracellular structures resembling secretory vesicles (ves). (D) Represents a control for the immunoreaction. (E) Is an RT-PCR gene expression analysis demonstrating elevation of enamel-related gene products amelogenin, ameloblastin, MMP20, and kallikrein 4, when comparing treatment with the LRAP/matrix/SATB1-coated collagen scaffold (MicroE) to a nontreated control group (Con). This differentiating microenvironment resulted in the formation of two distinct cell populations attached to the scaffold surfaces, with the majority of cells exhibiting ameloblast-like shapes (ambl) (F), while another cell type contained small and rounded cells (sc) that did not stain for amelogenin (G). (I, J) Illustrates the cytoskeleton of the elongated (I) versus the rounded (J) cell populations through phalloidin staining. (H, K) Are control cervical loop cocultured cells that were not subjected to differentiation conditions. These cultures lacked ameloblast-like elongated cells and instead featured small, rounded cells (c) as predominant cell population. *P ≤ 0.05; **P ≤ 0.01. LRAP, leucine-rich amelogenin peptide; SATB1, special AT-rich sequence-binding protein-1.
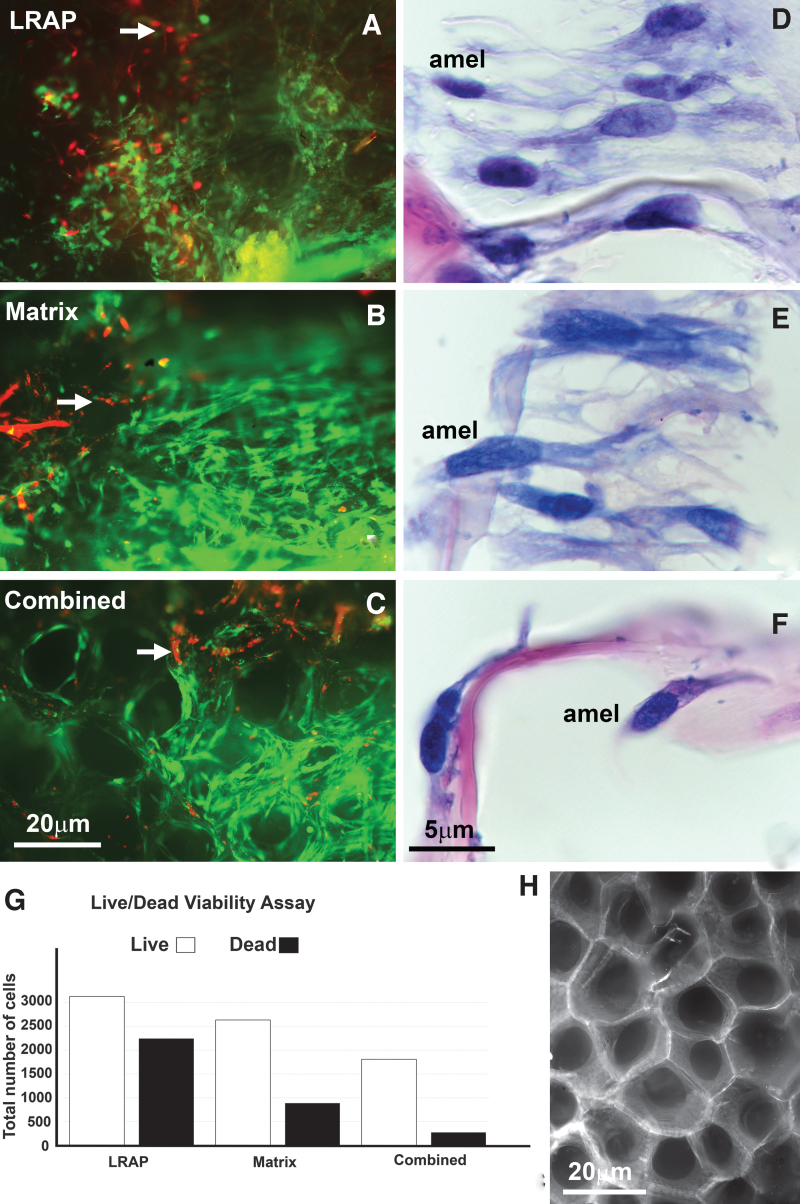
FIG. 3.
Viability assay and paraffin sections of ameloblast-like cells differentiated from cervical loop stem cells cocultured with dental pulp cells using a collagen-based scaffold coated with LRAP, early enamel matrix protein, and with a combination of differentiation factors. (A–C) Are cervical loop cells cocultured with dental pulp cells visualized through live/dead stain where red represents dead cells and green represents live cells after 10 days of culture. (A) Cocultured cells on a SpongeCol scaffold coated with LRAP, (B) Cells cocultured on a SpongeCol scaffold coated with early enamel matrix (matrix), and (C) Cells grown on the SpongeCol scaffold coated with a combination of LRAP, early matrix, and SATB1 (combined). The elongated and polarized structures constitute the architectural framework of the scaffold environment. (D–F) H&E-stained paraffin sections of elongated and polarized cells indicative of polarized ameloblasts in all three scaffold coating groups, LRAP, enamel matrix, and a combination thereof (D–F). Note the parallel organization and tight packing of ameloblast-like cells in (D, E). (G) Is a graph representing the total number of live/dead cells in each of the three coating groups based on the Life/Dead viability assay. (H) Is a micrograph of the highly successful SpongeCol scaffold used for the coculture resulting in elongated and polarized ameloblast-like cells. H&E, hematoxylin and eosin.
Viability assay
Scaffolds cultured with cervical loop and dental pulp cells cultured with LRAP, early enamel matrix, and a combination thereof were harvested after 10 days of culture. These scaffolds were incubated with live/dead stain (Live dead stain kit; Invitrogen, Carlsbad, CA) for 30 min before imaging with a Leica DMR light microscope (Nuhsbaum, IL).
Immunofluorescence and immunohistochemistry
For immunohistochemistry, paraffin slides were deparaffinized and rehydrated. Antigen retrieval was performed by incubating the paraffin-fixed samples in 10 mM sodium citrate buffer (pH 6) for 30 min at 60°C. After antigen retrieval, samples were blocked for 10 min with a blocking buffer supplied with the kit, incubated using mouse monoclonal antiamelogenin antibody (F-11: sc-365284; SantaCruz, Dallas, TX) for 2 h at room temperature (RT) and washed with a phosphate-buffered saline TRIS (PBST) washing buffer. After washing, the samples were incubated with a broad spectrum secondary antibody for 40 min, followed by another wash. Samples were further treated with a horseradish peroxidase substrate for 40 min. Immunohistochemistry was performed using a broad spectrum immunohistochemistry staining kit and an AEC substrate kit (Life Technologies, CA), and stained sections were analyzed using a Leica DMR light microscope (Nuhsbaum). For immunofluorescence, paraffin sections were deparaffinized and rehydrated, permeabilized with 0.1% Triton X for 10 min and incubated with 1% bovine serum albumin (BSA)/PBS for 30 min at 37°C. Samples were incubated in primary antibody against amelogenin (mouse, 1:500; Santa Cruz Biotechnology) for 2 h followed by a wash in PBST buffer. The samples were further incubated in AlexaFluor 488 goat antimouse (A-11029; Invitrogen) for 30 min and mounted with 4′,6-diamidino-2-phenylindole (DAPI) antifade mounting medium (Invitrogen). All stained sections were analyzed using a Leica DMR light microscope (Nuhsbaum).
Protein extraction and western blot analysis
After 10 days culture, cells assembled on scaffolds were subjected to protein extraction in radioimmunoprecipitation assay (RIPA) buffer. The Pierce BCA protein assay kit (ThermoScientific, Waltham, MA) was used to determine the protein concentration for different groups. Protein samples were loaded with 10 μg protein per well on 4% to 20% sodium dodecyl sulfate–polyacrylamide gels and were subjected to gel electrophoresis at 150 V for 58 min. A semidry transfer system was used to transfer proteins from the gel to a polyvinylidene difluoride (PVDF) membrane at 18 V for 40 min. The PVDF membrane was further blocked for 1 h with 5% nonfat dry milk in Tris buffered saline with Tween-20 (TBST), incubated with mouse monoclonal antiamelogenin primary antibody (1:500; Santa Cruz) overnight at 4°C, followed by washing in TBST thrice for 15 min at RT and incubated with antimouse IgG horseraddish peroxidase (HRP)-conjugated secondary antibody (1:2,000; Cell Signaling) for 1 h. After the final wash in TBST thrice for 15 min, the HRP signal was detected using a chemiluminescent substrate (Thermo Scientific).
Paraffin embedding and hematoxylin and eosin staining
Scaffolds with cells harvested after 10 days of culture in the bioreactor were fixed in 10% formalin for 1 day, and processed for regular paraffin sectioning after dehydrating through a graded series of ethanol and xylene. The samples were further embedded in paraffin and sectioned into 5 μm thin paraffin sections using a microtome. For hematoxylin and eosin (H&E) staining, deparaffinized and rehydrated sections were dipped in hematoxylin for 20 s, followed by a gentle wash with running water for 10 min, and dipped in eosin for 2 min. These sections were further placed under gently running water for 10 min, dehydrated again by quick dips in a series of ethanol solutions (50% for 5 min, 70% for 5 min, 80% for 5 min, 90% for 5 min, and 100% 2 × 5 min), and cleared in xylene before mounting with cover slips using a toluene-based mounting solution, Permount (Fisher Scientific, Hampton, NH).
Actin staining
Paraffin sections were deparaffinized, rehydrated, and permeabilized with 0.1% Triton-100. Sections were further stained with rhodamine-conjugated phalloidin (Invitrogen) for 20 min and mounted with antifade DAPI mounting medium. Phalloidin-stabilized microfilaments were then captured under a Leica DMRX fluorescent microscope (Nuhsbaum).
RNA extraction and real-time polymerase chain reaction
Total RNAs were extracted from the cells cultured with the scaffolds using the MicroRNA Easy kit (Qiagene, MD). Relative mRNA levels of amelogenin (Amel), ameloblastin (Ambn), matrix metalloproteinase 20 (MMP20), and kallikrein-related peptidase 4 (KLK4) were examined using a quantitative 2-step reverse transcriptase polymerase chain reaction (RT-PCR) assay and a SYBR green master mix. Sequence-specific primers were designed for this study (Table 1) and applied to a CFX96 Real Time system (Bio-Rad, Hercules, CA). The reaction conditions were as follows: 2 min at 50°C (1 cycle), 10 min at 95°C (1 cycle), 15 s at 95°C, and 1 min at 60°C (40 cycles). Samples were normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The relative expression level was computed using the 2−ΔΔCt analysis method, where GAPDH was used as an internal reference [16]. The relative expression changes were expressed in the form of bar graphs as shown in Figs. 2–4.
Table 1.
Ameloblast-Like Cells Grown in Three-Dimensional Bioreactors: Reverse Transcriptase Polymerase Chain Reaction Primers Used
| RT PCR mouse primer name | Primer sequence |
|---|---|
| Amel F | CTCTGCCTCCACTGTTCTCC |
| Amel R | ACTTCTTCCCGCTTGGTCTT |
| Ambn F | GTCCAGAAGGCTCTCCACTG |
| Ambn R | GTCATTGGGGAAAGCAAGAA |
| MMP20 F | AGGGACGAAGAGAGCTGTGA |
| MMP20 R | AACCTTCAATCACCCTCACG |
| Klk4 F | GCCTGGCATACCAAGTGTCT |
| Klk4 R | TCATGTGGGCCTTGTAGTCA |
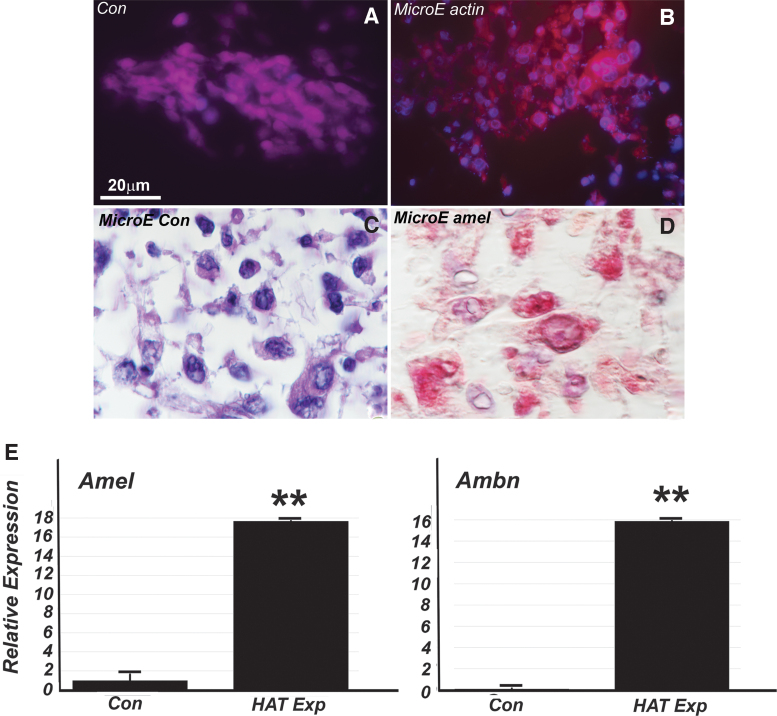
FIG. 4.
Immortalized HAT-7 cells cocultured with dental pulp progenitors in a 3D bioreactor. Addition of differentiation conditions (LRAP, early-stage enamel matrix) yielded cuboidal cells with an augmented cytoplasm (B–D vs. A). The cytoplasm of differentiated cuboidal cells reacted with amelogenin antibodies (D vs. C) and stained for actin/phalloidin (B vs. A). RT-PCR demonstrated a significant 16-fold increased expression of amelogenin and ameloblastin in the coculture group compared with the control (E). **P ≤ 0.01.
Statistical analysis
To determine whether the relative gene expression levels were significantly different, one-way analysis of variance was conducted using the SPSS software, and data were expressed as mean ± standard deviation. The significance levels were set as P < 0.05, and all experiments were repeated thrice to ensure reproducibility. To analyze the data for viability assay, a Live dead quantification macro was applied as part of the ImageJ software package.
Results
Three-dimensional growth of cervical loop cells on BIOST PGA scaffolds cocultured with periodontal ligament progenitors in a growth-factor-enriched medium resulted in the formation of ameloblast-like cells expressing and secreting amelogenin
There are no functioning ameloblasts associated with fully erupted molar teeth of adult mice or rats. However, the continuously erupting rodent incisor generates ameloblast progenitor cells from cervical loop cells throughout the life of the animal [17]. In this study, we have prepared cervical loop cells from 6 days postnatal mouse incisors as a source for ameloblast progenitors in bioreactor culture environments. After 10 days of culture, cervical loop cells differentiated into a conglomerate of cells and secreted matrix on scaffold surfaces (Fig. 1), establishing the feasibility of growing ameloblast progenitors in bioreactors. Coculture of cervical loop cells with periodontal ligament (PDL) progenitors resulted in cell assemblies that reacted positively for amelogenin in immunoreactions and western blot (Fig. 1F, G). The cocultured cells also reacted positively for amelogenin and ameloblastin through RT-PCR, and a western blot demonstrated that amelogenin was significantly elevated only in the coculture experiment (Fig. 1), demonstrating that in addition to cervical loop cells, mesenchymal coculture populations such as periodontal progenitors are necessary for amelogenin expression or secretion.
Cervical loop/dental pulp coculture grown on a scaffold enriched with enamel matrix and LRAP resulted in the formation of singular, elongated, and polarized ameloblast-like cells expressing and secreting ameloblastin and amelogenin enamel proteins
Previous attempts to grow ameloblasts in vitro have not resulted in cells that morphologically resemble the typical ameloblast cell shape consisting of elongated and polarized cell bodies featuring amelogenin-rich secretory vesicles [4]. To differentiate cervical loop cells into ameloblast-like cells, we have pursued a three-pronged strategy, including (1) scaffold coating with SATB1 protein, a known regulator of ameloblast polarization [18]; (2) addition of LRAP peptide based on the role of LRAP in ameloblast differentiation [19]; and (3) addition of lyophilized early-stage enamel matrix as the initial matrix secreted by Tomes' processes [15]. Among these strategies, only the addition of LRAP (1 mg/mL) and/or early-stage enamel matrix (1 mg/mL) resulted in the differentiation of polarized, elongated, and amelogenin secreting ameloblasts (Fig. 2A–C), while SATB1 alone did not result in ameloblast-like morphologies (data not shown). The enamel matrix and LRAP-enriched microenvironment resulted in the differentiation of two different cell populations attached to the scaffold surfaces, with the majority of cells exhibiting ameloblast-like polarized cell shapes (Fig. 2A–D, F) that immunoreacted with amelogenin (Fig. 2A–C), while a second cell population contained small and rounded cells that did not stain for amelogenin (Fig. 2G). The ameloblast-like cell population was distinguished through conical cell processes resembling Tomes' processes and intracellular vesicles similar to secretory vesicles (Fig. 2A–C). RT-PCR analysis confirmed a significant 3-fold increase in the expression of amelogenin, a 14-fold increase in ameloblastin expression, a 3.5-fold increase in the expression of MMP-20, and a 2-fold increase in KLK4 expression in the differentiating microenvironment group versus a control group lacking differentiation conditions (Fig. 2E). The elongated and polarized ameloblast-like cells exhibited distinct actin/phalloidin cytoskeletal staining, especially when compared with the rounded cell populations from the cervical loop cocultured cells that were not subjected to differentiation conditions (Fig. 2I vs. J). Illustrating the essential role of scaffold coating for ameloblast-like cell differentiation, only the LRAP/matrix-coated scaffolds resulted in the differentiation of elongated, polarized, and amelogenin secreting cells, while noncoated scaffolds yielded a population of small and rounded cells (Fig. 2H, K). In addition, the only scaffold successful in supporting ameloblast-like cell shapes was a prefabricated scaffold termed “SpongeCol,” while other scaffolds such as the PGA bioreactor scaffold (Synthecon), gelatin disks, or graphene sheet bioreactor scaffolds (Advanced Biomatrix) did not yield ameloblast-like cells (not shown).
Effect of LRAP, early enamel matrix protein, and a combination thereof confirmed the role of LRAP and early enamel matrix in the elongation and polarization of cocultured cervical loop/dental pulp cells
To understand the stand-alone effect of each component, SpongeCol scaffolds were prepared using LRAP coating, early enamel matrix coating, and a coating with LRAP, enamel matrix, and SATB1 combined. Elongated cells with nuclei in a polarized position were observed when cervical loop and dental pulp cells were cocultured with a collagen-based scaffold coated with either LRAP, early enamel matrix protein, or a combination of differentiating factors LRAP, matrix, and SATB1 (Fig. 3D–F). A viability assay (Fig. 3A–C) revealed the highest percentage of live versus dead cells in the LRAP/enamel matrix combination group (86.85%), compared with 74.82% in the enamel matrix-coated group and 58.82% in the LRAP group alone, demonstrating that scaffold treatment with a combination of LRAP, matrix, and SATB1 was most effective in ensuring ameloblast survival (Fig. 3G). In addition to maintaining cell survival, all three scaffolds resulted in the formation and differentiation of elongated and polarized ameloblast-like cells (Fig. 3D–F), which were parallelly aligned (Figs. 3D, E).
HAT-7 cells cultured in a scaffold enriched with enamel matrix, LRAP, and SATB1 formed aggregates of cuboidal cells that secreted amelogenin and ameloblastin
To verify the applicability of 3D bioreactor culture strategies for established enamel cell lines, HAT-7 cells were treated in an identical manner to cervical loop cells and cultured for 10 days. Cultured HAT-7 cells demonstrated positive immunoreactivity of actin and the enamel protein amelogenin but were neither elongated nor polarized (Fig. 4).
Discussion
The present set of studies was conducted to achieve a seemingly daunting task: the growth of ameloblast-like cells in vitro. In this set of studies, ameloblast-like cells were defined as polarized, elongated, and enamel protein secreting cells. Here, we report the results of four different types of 3D bioreactor-based experiments: (1) to grow cervical loop cells in a bioreactor, (2) to coculture cervical loop cells with PDL progenitors in a growth-factor-enriched matrix, (3) to coculture cervical loop/dental pulp cells on a scaffold enriched with enamel matrix and LRAP, and (4) to determine the feasibility of HAT-7 enamel organ cell culture in a bioreactor environment. In general, all four approaches accomplished cervical loop/dental epithelium-derived cell growth, with only one group (group iii) resulting in the formation of ameloblast-like cells. Together, these studies highlight the suitability of 3D bioreactor environments for the propagation and in vitro culture of cervical loop/enamel organ epithelial cells.
In our studies, a commercially available collagen sponge scaffold termed SpongeCol with an interpenetrating columnar porous architecture (Advanced Biomatrix) proved superior over other scaffold compositions or designs for the growth of polarized and elongated ameloblast-like cells. In recent years, scaffolds have become increasingly important as an integral part of strategies to control the microenvironment inside of bioreactors [20,21]. Specifically, chemical composition and physical factors such as surface topography, spatial configuration, and mechanical behavior are known factors that promote stem cell survival, function, and proliferation, and these factors can be influenced through manufacturing design controls, such as porosity, pore size control, composition, and biodegradability [20–22]. Our present studies have identified Advanced Biomatrix SpongeCol as the most favorable scaffold design for the growth of ameloblast-like cells. However, it still needs to be determined whether these advantageous properties are due to the biophysical properties of the scaffold, or its biochemical composition containing collagens or both.
During our initial studies, some of our combinations of cervical loop cells with mesenchymal cells resulted in the formation of multinucleated cells. When cocultured, these cell populations secreted enamel proteins such as amelogenin or ameloblastin, but their morphology was distinctly tube shaped. The formation of multinucleated syncytia is a frequent effect of microgravity culture systems and ideal for the study of muscle physiology, including studies of bioengineered heart and skeletal muscle [23,24]. It has been demonstrated that microgravity increases polyploidy, perhaps through nuclear localization of Yes-Associated protein 1 [25]. While amelogenesis bioreactor systems featuring multinucleated cells are entirely permissive for enamel protein synthesis or matrix secretion, cellular properties need to be improved for advanced tissue engineering strategies. The hallmark of this study is the fabrication of mononuclear, polarized, and elongated ameloblast-like cells as achieved by adding tailored differentiation inducing agents and a suitable scaffold. This accomplishment proved to be a major step toward the engineering of secretory enamel organ-derived environments with cell shapes and function resembling those of natural ameloblasts.
The formation of polarized, elongated, and amelogenin secreting ameloblasts in our cervical loop/dental pulp coculture models on SpongeCol scaffolds became the highlight of the studies presented here. Neither the cervical loop cells alone nor their combination with mesenchymal cells nor the addition of SATB1 resulted in fully differentiated cells. Only LRAP and other components of the early-stage enamel matrix in combination with dental pulp cell coculture and sponge-gel scaffolds were effective in promoting the growth of polarized and elongated ameloblast-like cells. The importance of the alternatively spliced amelogenin isoform LRAP on ameloblast differentiation, cell polarity, Tomes' process formation, and amelogenin secretion has been established in previous studies [26]. In this study, we are harnessing this knowledge for the engineering of ameloblast-like cells. Another trigger for ameloblast polarization was the early-stage enamel matrix, which is intimately associated with Tomes' process and contains other differentiation signals or protein fragments in addition to LRAP [15]. Other factors contributing to the success of our ameloblast differentiation approach might include dental pulp cells as coculture cells and the collagen scaffold, both of which mimic components of the natural tooth composite environment [4]. However, one of the candidate factors presumed to trigger ameloblast cell polarity and directional amelogenin secretion, SATB1, did not cause ameloblast-like cell polarity and elongation in our bioreactor studies, vouching for the unique role of LRAP and the early enamel matrix as engineering tools for ameloblast-like cell polarization.
The bioreactor environment also proved advantageous for the culture of an already established cell line, the HAT-7 dental epithelial cell line [7,27]. Our studies demonstrated that LRAP and enamel matrix induction on SpongeCol scaffolds signaled HAT-7 cells to form cuboidal cells with occasionally small processes visible. In this environment, HAT-7 cells displayed amelogenin and actin immunoreactivity, with a highly significant 16- to 18-fold higher level of amelogenin and ameloblastin compared with controls grown on plain scaffolds. This was a remarkable outcome establishing HAT-7 as a superior mammalian cell line for the expression of major enamel proteins. Yet, HAT-7 cells shape did not polarize significantly, suggesting that polarized and elongated cell shapes are not necessary for enamel protein secretion per se, even though they may be important for directional tissue growth and protein deposition.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
Generous support for these studies was provided by NIDCR grant no. UG-3-DE028869.
References
- 1.Ruprecht V, Monzo P, Ravasio A, Yue Z, Makhija E, Strale PO, Gauthier N, Shivashankar GV, Studer V, Albiges-Rizo C and Viasnoff V. (2017). How cells respond to environmental cues—insights from bio-functionalized substrates. J Cell Sci 130:51–61 [DOI] [PubMed] [Google Scholar]
- 2.Helmstetter CE. (1997). Gravity and the orientation of cell division. Proc Natl Acad Sci U S A 94:10195–10198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin RD, Soligo C and Tavaré S. (2007). Primate origins: implications of a Cretaceous ancestry. Folia Primatol (Basel) 78:277–296 [DOI] [PubMed] [Google Scholar]
- 4.Pandya M and Diekwisch TGH. (2019). Enamel biomimetics-fiction or future of dentistry. Int J Oral Sci 11:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Yan X, Pandya M, Luan X and Diekwisch TG. (2016). Daughters of the enamel organ: development, fate, and function of the stratum intermedium, stellate reticulum, and outer enamel epithelium. Stem Cells Dev 25:1580–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein OD, Duverger O, Shaw W, Lacruz RS, Joester D, Moradian-Oldak J, Pugach MK, Wright JT, Millar SE, et al. (2017). Meeting report: a hard look at the state of enamel research. Int J Oral Sci 9:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto A, Harada H, Saito M and Taniguchi A. (2011). Induction of enamel matrix protein expression in an ameloblast cell line co-cultured with a mesenchymal cell line in vitro. In Vitro Cell Dev Biol Anim 47:39–44 [DOI] [PubMed] [Google Scholar]
- 8.Chen LS, Couwenhoven RI, Hsu D, Luo W and Snead ML. (1992). Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch Oral Biol 37:771–778 [DOI] [PubMed] [Google Scholar]
- 9.DenBesten PK, Gao C, Li W, Mathews CH and Gruenert DC. (1999). Development and characterization of an SV40 immortalized porcine ameloblast-like cell line. Eur J Oral Sci 107:276–281 [DOI] [PubMed] [Google Scholar]
- 10.Arakaki M, Ishikawa M, Nakamura T, Iwamoto T, Yamada A, Fukumoto E, Saito M, Otsu K, Harada H, Yamada Y and Fukumoto S. (2012). Role of epithelial-stem cell interactions during dental cell differentiation. J Biol Chem 287:10590–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duray P, Hatfill S and Pellis N. (1997). Tissue culture in microgravity. Sci Med 4:46–55 [PubMed] [Google Scholar]
- 12.Unsworth BR and Lelkes PI. (1998). Growing tissues in microgravity. Nat Med 4:901–907 [DOI] [PubMed] [Google Scholar]
- 13.Hammond TG and Hammond JM. (2001). Optimized suspension culture: the rotating-wall vessel. Am J Physiol Renal Physiol 281:F12–F25 [DOI] [PubMed] [Google Scholar]
- 14.Navran S. (2008). The application of low shear modeled microgravity to 3-D cell biology and tissue engineering. Biotechnol Annu Rev 14:275–296 [DOI] [PubMed] [Google Scholar]
- 15.Pandya M, Lin T, Li L, Allen MJ, Jin T, Luan X and Diekwisch TGH. (2017). Posttranslational amelogenin processing and changes in matrix assembly during enamel development. Front Physiol 8:790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ and Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 17.Smith CE and Warshawsky H. (1975). Cellular renewal in the enamel organ and the odontoblast layer of the rat incisor as followed by radioautography using 3H-thymidine. Anat Rec 183:523–561 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Zheng L, Le M, Nakano Y, Chan B, Huang Y, Torbaty PM, Kohwi Y, Marcucio R and Habelitz S. (2019). SATB1 establishes ameloblast cell polarity and regulates directional amelogenin secretion for enamel formation. BMC Biol 17:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stahl J, Nakano Y, Horst J, Zhu L, Le M, Zhang Y, Liu H, Li W and Den Besten P. (2015). Exon4 amelogenin transcripts in enamel biomineralization. J Dent Res 94:836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selden C and Fuller B. (2018). Role of bioreactor technology in tissue engineering for clinical use and therapeutic target design. Bioengineering (Basel) 5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi T, Huang S, Liu G, Li T, Kang Y, Luo Y and Wu J. (2018). Bioreactor synergy with 3D scaffolds: new era for stem cells culture. ACS Appl Bio Mater 1:193–209 [DOI] [PubMed] [Google Scholar]
- 22.Ahmed S, Chauhan VM, Ghaemmaghami AM and Aylott JW. (2019). New generation of bioreactors that advance extracellular matrix modelling and tissue engineering. Biotechnol Lett 41:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoppel WL, Kaplan DL and Black LD. (2016). Electrical and mechanical stimulation of cardiac cells and tissue constructs. Adv Drug Deliv Rev 96:135–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rimington RP, Capel AJ, Chaplin KF, Fleming JW, Bandulasena HCH, Bibb RJ, Christie SDR and Lewis MP. (2019). Differentiation of bioengineered skeletal muscle within a 3D printed perfusion bioreactor reduces atrophic and inflammatory gene expression. ACS Biomater Sci Eng 5:5525–5538 [DOI] [PubMed] [Google Scholar]
- 25.Arun RP, Sivanesan D, Patra B, Varadaraj S and Verma RS. (2019). Simulated microgravity increases polyploid giant cancer cells and nuclear localization of YAP. Sci Rep 9:10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl J, Nakano Y, Kim SO, Gibson CW, Le T and DenBesten P. (2013). Leucine rich amelogenin peptide alters ameloblast differentiation in vivo. Matrix Biol 32:432–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawano S, Morotomi T, Toyono T, Nakamura N, Uchida T, Ohishi M, Toyoshima K and Harada H. (2002). Establishment of dental epithelial cell line (HAT-7) and the cell differentiation dependent on Notch signaling pathway. Connect Tissue Res 43:409–412 [DOI] [PubMed] [Google Scholar]