Abstract
The China National Center for Food Safety Risk Assessment (CFSA) uses the Foodborne Disease Monitoring and Reporting System (FDMRS) to monitor outbreaks of foodborne diseases across the country. However, there are problems of underreporting or erroneous reporting in FDMRS, which significantly increase the cost of related epidemic investigations. To solve this problem, we designed a model to identify suspected outbreaks from the data generated by the FDMRS of CFSA. In this study, machine learning models were used to fit the data. The recall rate and F1-score were used as evaluation metrics to compare the classification performance of each model. Feature importance and pathogenic factors were identified and analyzed using tree-based and gradient boosting models. Three real foodborne disease outbreaks were then used to evaluate the best performing model. Furthermore, the SHapley Additive exPlanation value was used to identify the effect of features. Among all machine learning classification models, the eXtreme Gradient Boosting (XGBoost) model achieved the best performance, with the highest recall rate and F1-score of 0.9699 and 0.9582, respectively. In terms of model validation, the model provides a correct judgment of real outbreaks. In the feature importance analysis with the XGBoost model, the health status of the other people with the same exposure has the highest weight, reaching 0.65. The machine learning model built in this study exhibits high accuracy in recognizing foodborne disease outbreaks, thus reducing the manual burden for medical staff. The model helped us identify the confounding factors of foodborne disease outbreaks. Attention should be paid not only to the health status of those with the same exposure but also to the similarity of the cases in time and space.
Keywords: foodborne disease outbreaks, machine learning, foodborne disease
Introduction
Foodborne diseases are caused by eating food contaminated with pathogenic bacteria, viruses, parasites, natural toxins, or even chemical residues (Horwitz, 1977). Foodborne diseases threaten people's health and cause economic losses globally every year (Todd, 1997; Li et al., 2020). In 2015, the World Health Organization indicated that foodborne diseases have caused a heavy burden on a global scale. Approximately 600 million cases of foodborne diseases occur worldwide annually, causing 420,000 deaths (Oliver, 2019). Therefore, research on foodborne disease monitoring and prediction is necessary.
A foodborne disease outbreak refers to the occurrence of two or more foodborne disease cases with common exposure and similar symptoms or more than one death record (Murphree et al., 2012). The basic conditions for identifying foodborne disease outbreaks are as follows: common exposure to food and multiple people with similar symptoms or one or more deaths. Factors that should be considered to identify foodborne diseases are numerous and complex (Bryan, 1978; Brown et al., 2017), and researchers have analyzed the burden of foodborne diseases in humans, food animals, and food from the perspective of foodborne pathogens (Paudyal et al., 2018), such as Salmonella serovars (Ferrari et al., 2019), Staphylococcus aureus (Jia et al., 2020), and Toxoplasma gondii (Zhang et al., 2019b).
At present, researchers have begun to apply data mining (Thakur et al., 2010), machine learning, deep learning, and other technologies to solve the problems of monitoring and predicting foodborne diseases. Research on foodborne disease monitoring (Wu and Chen, 2018) has helped to track the causes of diseases and prevent further expansion of disease trends. Xiao et al. (2015) proposed detection methods to automatically detect local foodborne disease outbreaks and sporadic foodborne disease outbreaks. Zhang et al. (2019a) integrated big data of different aspects and designed a detection model for foodborne diseases and risk assessment. Sadilek et al. (2012, 2013, 2018) collected Twitter data to monitor the sanitary conditions of eateries and the health of customers. In addition, social media data have also been used to monitor foodborne disease outbreaks (Zhang et al., 2017; Effland et al., 2018).
The prediction of foodborne diseases primarily focuses on predicting the future trends of a certain aspect of the disease. Wang et al. (2018) considered the reporting delay of the Foodborne Surveillance Database of the China National Center for Food Safety Risk Assessment (CFSA) and applied a Bayesian hierarchical model to predict the true daily number of patients using the daily number of patients visited. In terms of disease risk prediction, researchers have used deep learning methods to predict the short-term development trend of influenza-like diseases (Wu et al., 2018b; Adhikari et al., 2019) and foodborne diseases. Chen et al. (2019) developed a regularization-based eXtreme Gradient Boosting (XGBoost) approach, which can be used to predict the trend of foodborne diseases.
It can be seen that the research on foodborne disease outbreaks has drawn the attention of researchers. However, these methods are not fully applicable to the problem that should be solved. In our problem scenario, the Foodborne Disease Monitoring and Reporting System (FDMRS) of CFSA conducted preliminary screening and integration of case data based on rules, such as common food and common eating places, to obtain suspected outbreaks. We counted the number of real and false foodborne disease outbreaks in 2019 generated by FDMRS based on the screening rules. After review by medical staff, there were 6084 (80%) misjudged outbreaks. From this point of view, the accuracy of suspected outbreaks obtained through screening rules is only 20%, and a large number of manual reviews by medical staff are still required. Therefore, the problem that should be solved is to establish and train a classification model, reduce the occurrence of misjudgments and missed judgments of the system, and reduce the burden of manual judgment by medical staff. Simultaneously, we can analyze the importance of each feature extracted from the data and provide medical workers with guidance and suggestions from the data perspective.
Materials and Methods
Dataset
The data used in this study were obtained from the FDMRS of CFSA, which is a national-level technical agency responsible for food safety risk assessment in China. The dataset comprises 2 parts: 3866 foodborne disease outbreaks obtained from the epidemiological survey in 2019, which were confirmed by laboratory test results, and 7619 suspected foodborne disease outbreaks in 2019 selected by FDMRS through screening rules without laboratory testing. These suspected outbreaks have been reviewed by medical staff and divided into confirmed and excluded suspected outbreak groups.
In the dataset, there are one-to-many relationships between the outbreaks and cases. Detailed information on the case data can be obtained from the FDMRS database. The confirmed outbreaks from epidemiological surveys can be directly used as positive samples for the classification model. In all suspected outbreaks, the confirmed suspected outbreaks are regarded as positive samples, whereas the excluded suspected outbreaks are regarded as negative samples.
Features
In this study, the features that we considered included case information, exposure information, symptoms, and diagnosis results, as shown in Supplementary Table S4. The subsequent feature extraction and feature importance analyses were based on these features. For some features that can be subdivided, such as symptoms, diagnostic results, and food processing methods, a corresponding comparison table is also provided, as shown in Supplementary Table S5.
Model design
In this section, we introduce the problem definition, model pipeline, model methods, and performance evaluation metrics. In addition, data preprocessing, feature extraction, and other model training details are presented in Supplementary Document D1 owing to the limitation of the number of words.
Problem definition
In our work, we abstract the problem of distinguishing whether a suspected foodborne disease outbreak is a real outbreak as a classification problem and then build a classification model. The model takes the suspected foodborne disease outbreaks as input and outputs the discrimination result and probability. The discrimination result represents whether the suspected outbreak is a real outbreak (label = 1) or not (label = 0). The output probability indicates the likelihood predicted by the model that the suspected outbreak is a real outbreak.
Model pipeline
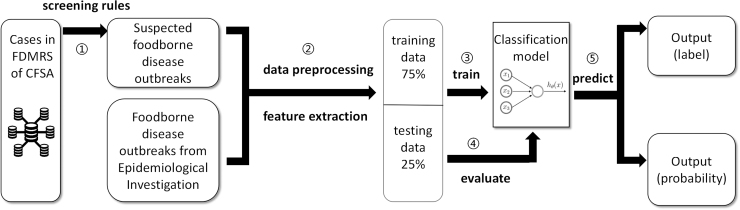
The model pipeline in Figure 1 can be summarized as follows: (1) The FDMRS of CFSA integrates the cases in the database to form suspected outbreaks of foodborne diseases through some screening rules. (2) The suspected outbreaks and the real outbreaks obtained from epidemiological investigations are combined as the entire dataset, and data preprocessing and feature extraction are performed on the dataset. We divided the dataset into training data (75%) and testing data (25%). (3) Multiple classification models are trained using the training data, and parameter tuning is performed on them. (4) The recall rate and F1-score were used to evaluate the model on the testing data. (5) The best performing model obtained from the previous step was selected to make predictions. The model output can be a probability or classification label (0 or 1).
FIG. 1.
Model pipeline of foodborne disease outbreak monitoring and identification in the study. The whole process can be divided into two parts: data preparation, preprocessing, and feature extraction, which are used to obtain the training and test sets of the model, and model training, evaluation, and prediction.
Model methods and evaluation metrics
In this study, support vector machine (Cortes and Vapnik, 1995), logistic regression (Wright, 1995), naive Bayes (Murphy, 2006), decision tree (DT) (Quinlan, 1986), random forest (RF) (Breiman, 2001), gradient boosting DT (Friedman, 2001), adaptive boosting (Adaboost) (Freund and Schapire, 1996), and XGBoost (Chen and Guestrin, 2016) models were used to fit the data.
We used the recall rate and F1-score to measure the classification performance of the models on the test set. The formulas are as follows:
Detailed explanations of these machine learning algorithms and evaluation metrics are provided in Supplementary Document SD1.
Results
Model results
Table 1 shows the recall rate and F1-score of each machine learning model used in our study. As shown in Table 1, the XGBoost model had the highest recall rate (0.9699) and the highest F1-score (0.9582). It can be seen from Table 1 that the performance of the classification model is not significantly different between the test and training sets. The results for the test set were slightly lower than those of the training set, which was normal and acceptable. From this point of view, our model does not have the problem of overfitting. We use the XGBoost model as an example to analyze the confusion matrix* of the model, which is shown in Supplementary Table S1. According to the confusion matrix, the missed outbreaks are only 3% of all positive cases, and 97% of all positive cases were correctly predicted, which is the recall rate.
Table 1.
Recall and F1-Score of the Models in the Training Set and Test Set
| Model | Training set |
Test set |
||
|---|---|---|---|---|
| Recall | F1-score | Recall | F1-score | |
| SVM | 0.9648 | 0.9516 | 0.9641 | 0.9506 |
| LR | 0.9603 | 0.9490 | 0.9599 | 0.9488 |
| NB | 0.9480 | 0.9426 | 0.9524 | 0.9425 |
| DT | 0.9743 | 0.9567 | 0.9616 | 0.9469 |
| RF | 0.9656 | 0.9533 | 0.9641 | 0.9517 |
| GBDT | 0.9767 | 0.9672 | 0.9666 | 0.9574 |
| Adaboost | 0.9782 | 0.9674 | 0.9674 | 0.9554 |
| XGBoost | 0.9715 | 0.9560 | 0.9699 | 0.9582 |
Adaboost, adaptive boosting; DT, decision tree; GBDT, gradient boosting decision tree; LR, logistic regression; NB, naive Bayes; RF, random forest; SVM, support vector machine; XGBoost, eXtreme Gradient Boosting.
In addition to taking categories as the output of the XGBoost model, we also attempt to output the classification results of the XGBoost model in the form of probability. Probability can indicate how likely it is that a suspected foodborne disease outbreak is a real outbreak. By traversing all possible classification thresholds, the threshold value of the maximum F1-score achieved is 0.5341; that is, when the output probability of a suspected outbreak is greater than 0.5341, the outbreak is considered a real foodborne disease outbreak; otherwise, it is excluded.
Feature importance analysis
Interpretable machine learning (Doshi-Velez and Kim, 2017) plays an increasingly important role in the medical field. If we use machine learning models to provide suggestions for risk prediction and diagnostic decisions, but fail to provide reasonable explanations, it is difficult to convince people to accept the results (Ahmad et al., 2018). In most cases, researchers focus more on how the model makes decisions and the influence of each feature in the model on the final decision than the single classification result obtained by the model. Feature importance (Grabczewski and Jankowski, 2005) is a tool for feature selection and for improving the interpretability of the model. In this study, we set the Gini index (Gastwirth, 1972; Lerman and Yitzhaki, 1984) as an indicator to evaluate the importance of features.
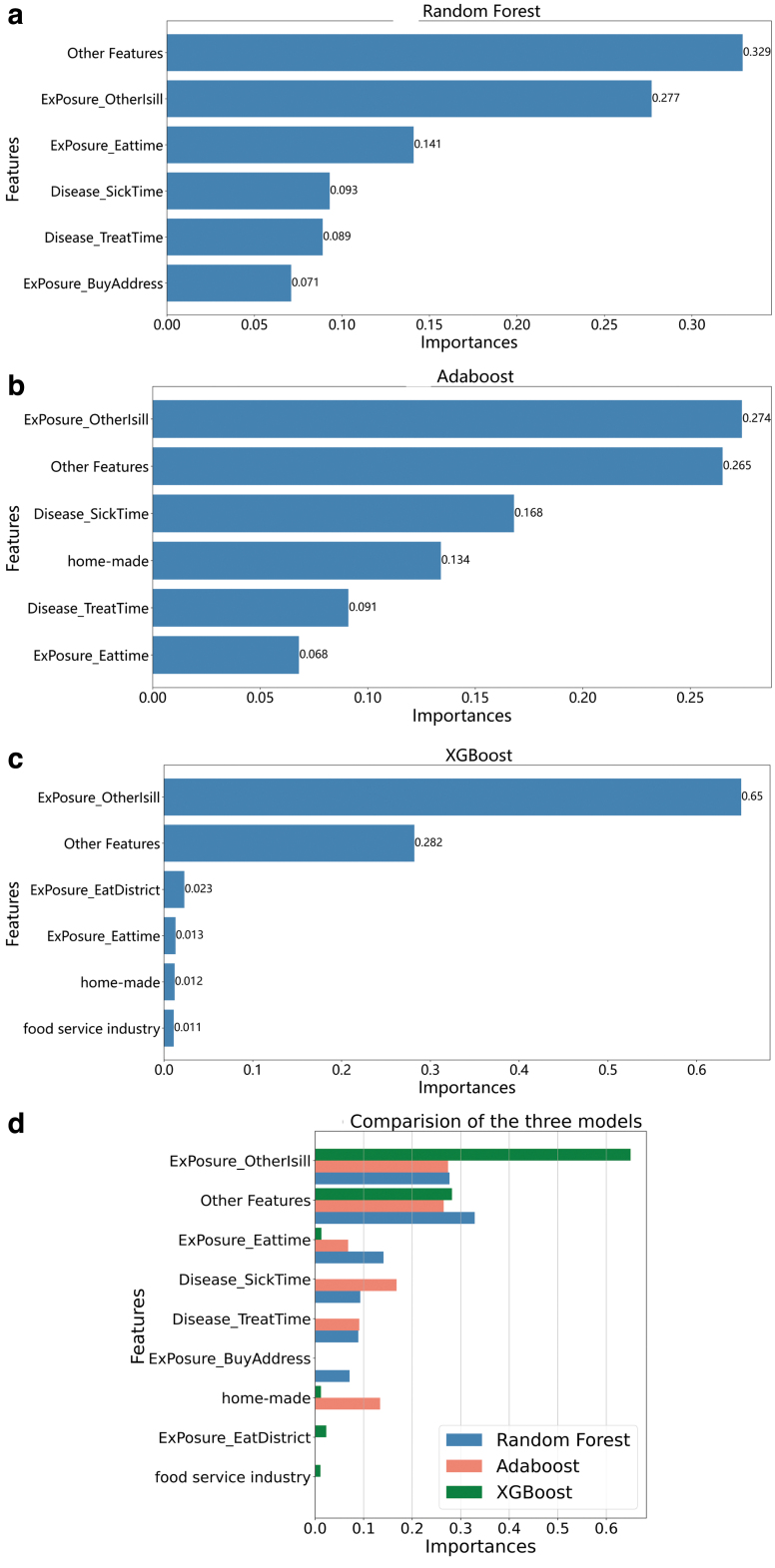
It can be seen from Table 1 that the classification models based on the tree structure and gradient boosting algorithms, such as RF, Adaboost, and XGBoost, attain better performance. Therefore, these three representative models were selected for the feature importance analysis. Figure 2 illustrates the importance of features in the RF, Adaboost, and XGBoost models. Because many features are used as model input, we add up features that account for a very small proportion and denote them as Other Feature. An analysis of the importance of features is presented in the Discussion section.
FIG. 2.
Feature importance of random forest (a), adaptive boosting (b), and eXtreme Gradient Boosting (c) models and comparison of the three models (d). The abscissa represents the feature importance calculated using the Gini index as an indicator. The ordinate is the name of the feature we considered in the model. Because there are many features that are used as model input, we add up features that account for a very small proportion and denote them as Other Feature. Color images are available online.
Identification of pathogenic factors
To understand the impact of various pathogenic factors on foodborne disease outbreaks, we analyzed the proportion of various pathogenic factors in the outbreaks, and the results are shown in Supplementary Figure S3. Among the 158 outbreaks that can be found for the pathogenic factors, most outbreaks were caused by Vibrio parahaemolyticus (41.1%), Salmonella (33.5%), Norovirus (13.9%), diarrheagenic Escherichia coli (7.0%), and others (4.5%).
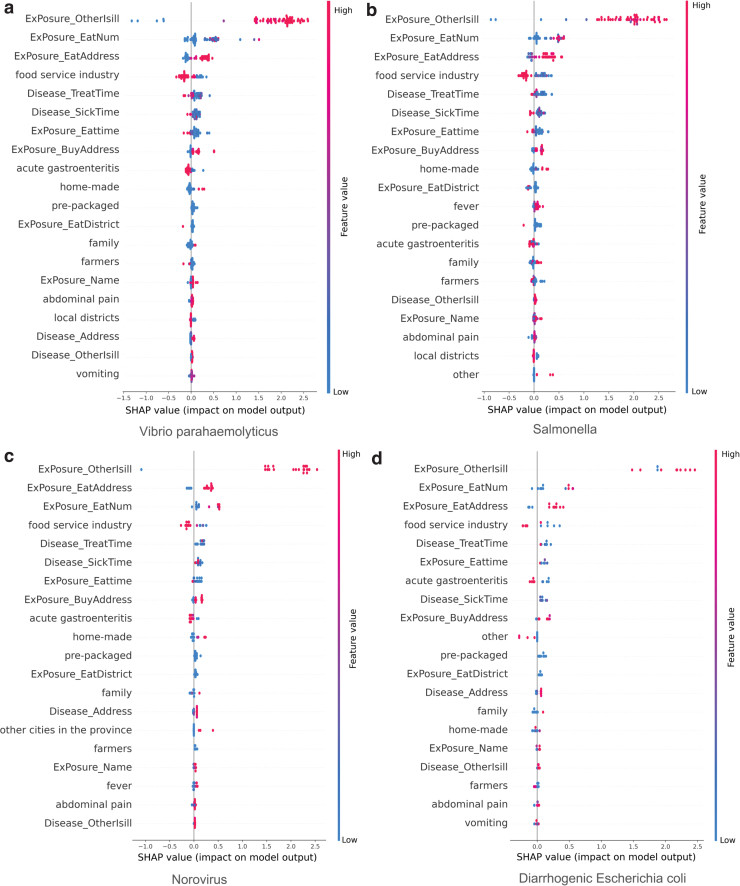
In addition, we integrated outbreaks caused by the same pathogenic factors. The differences in factors influencing foodborne disease outbreaks caused by different pathogenic factors in the XGBoost model were compared, as shown in Figure 3. The SHapley Additive exPlanation (SHAP) value (Lundberg and Lee, 2017; Lundberg et al., 2018) was used as an indicator to measure the importance of features. Figure 3 shows the top 20 features that play a decisive role in model classification. The larger the value, the greater the positive influence on the model output; and the smaller the value, the greater the negative influence on the model. Different colors represent the values of the feature. For example, if the feature ExPosure_Otherisill of a sample point has a large value, this feature has a positive effect on judging a suspected outbreak as a real outbreak. The analysis is presented in the Discussion section.
FIG. 3.
Impact of features on the XGBoost model output in outbreaks caused by Vibrio parahaemolyticus (a), Salmonella (b), Norovirus (c), and diarrheagenic Escherichia coli (d). In the analysis of pathogenic bacteria, the SHAP value is used as an indicator to measure the importance of features. The figure shows the top 20 features that play a decisive role in model classification. The ordinate is the name of the feature, and the abscissa is the size of the SHAP value. The larger the value, the greater the positive influence on the model output; and the smaller the value, the greater the negative influence on the model. All data sample points are displayed in the figure for each feature dimension; therefore, the distribution of data points under a certain feature can be seen in the figure. The different colors represent the value of the feature and size of the feature value. For example, if the feature ExPosure_Otherisill of a sample point has a large value, it means that this feature has a positive effect on judging a suspected outbreak as a real outbreak. SHAP, SHapley Additive exPlanation; XGBoost, eXtreme Gradient Boosting. Color images are available online.
Model validation in real cases
Because the XGBoost model has the highest recall rate and F1-score, we chose the XGBoost model to validate the model performance on real cases. Three confirmed, typical, foodborne disease outbreaks in 2019 were taken as examples to assess the classification effect. Compared with the traditional feature importance (Grabczewski and Jankowski, 2005) and permutation importance (Altmann et al., 2010), the SHAP value (Lundberg and Lee, 2017; Lundberg et al., 2018) can not only reflect the relationship between the features and the predicted results but also reflect the positive and negative effects on a single sample. Therefore, the SHAP value was used to explain the prediction results of these three outbreaks. Specific information on the outbreak cases is presented in Supplementary Table S2. The classification results for the three foodborne disease outbreaks are listed in Table 2.
Table 2.
Prediction Results of Three Foodborne Disease Outbreaks by the eXtreme Gradient Boosting Model
| Outbreak ID | Prediction result (label) | Prediction result (probability) | The meaning of the prediction |
|---|---|---|---|
| Shandong_OUTBREAK_11058 | 1 | 0.9352 | Confirmed as a foodborne disease outbreak |
| Guangdong_OUTBREAK_112 | 1 | 0.9076 | Confirmed as a foodborne disease outbreak |
| Yunnan_OUTBREAK_6014 | 1 | 0.9344 | Confirmed as a foodborne disease outbreak |
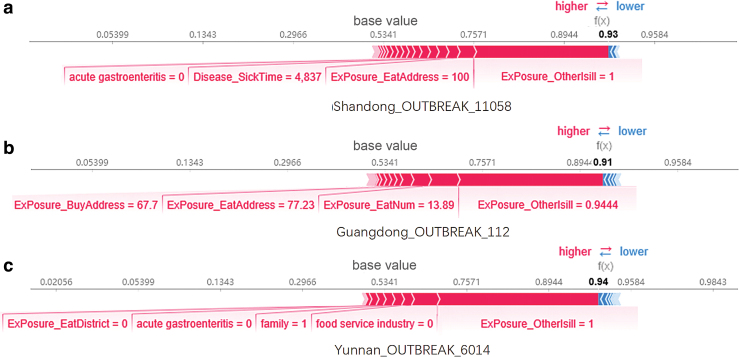
For these three foodborne disease outbreaks, the effects of the features on the final classification results are shown in Figure 4. The prediction results of the three foodborne disease outbreaks were 0.93, 0.91, and 0.94, respectively. The base value is 0.5341, which is the threshold value with the maximum F1-score generated by traversing the thresholds. The red color indicates the feature that increases the predicted value, and blue color indicates the feature that reduces the predicted value. The names and values of the features are also marked in the figure at the bottom of the bar. The figure indicates that the feature of whether other people with the same exposure were sick (ExPosure_OtherIsill) in the three foodborne outbreaks contributes the most to the identification of outbreaks as real ones.
FIG. 4.
Impact of features on predicted results of the three real foodborne disease outbreaks. The prediction results of the three foodborne disease outbreaks are 0.93, 0.91, and 0.94, respectively, indicating how likely it is that the outbreak is real. The base value is 0.5341, which is the threshold of the maximum F1-score generated by traversing the threshold. Red denotes features that increase the predicted value, whereas blue denotes features that decrease the predicted value. The name of the function and value of the function are also marked in the figure at the bottom of the bar. Color images are available online.
Discussion
In this study, we used a variety of machine learning models to fit the data and compared the classification performance of each model, with the recall rate and F1-score as evaluation metrics. In our problem, we focus on whether the model has few missed judgments and how many positive examples are predicted correctly, which is the recall rate. Therefore, in our problem scenario, the recall rate is more important than the F1-score when F1-scores of the models are not significantly different.
In the model validation section, the results are well interpretable: except for the feature of whether other people were sick, other features with high contribution were determined by the characteristics and specific situation of the outbreak. For example, Yunnan_OUTBREAK_6014, the third outbreak in Figure 4, was caused by consumption of homemade liquor; thus, in Figure 4c, we can see that homemade (family = 1) also contributes to the prediction results.
Analysis of pathogenic factors indicated that V. parahaemolyticus and Salmonella are the two main pathogenic bacteria. We find that pathogenic factors of the cases in the same outbreak are similar, which also proves that the real outbreaks are credible and reasonable. Simultaneously, the influencing factors differed slightly in terms of onset symptoms. In outbreaks caused by V. parahaemolyticus, Norovirus, and diarrheagenic E. coli, acute gastroenteritis accounts for a higher proportion, whereas fever is a major feature of outbreaks caused by Salmonella.
As for feature importance, it can be seen that the feature importance values obtained by the three models have similarities and differences. The similarity is that whether others are sick is the most important feature among the three classification models. The difference is that a few features in AdaBoost and XGBoost have extremely high weights, while the distribution of feature importance in RF is more balanced. This difference can be explained by the underlying implementation of the algorithm. Adaboost (Freund and Schapire, 1996) and XGBoost (Chen and Guestrin, 2016) can change the distribution of data during the training process, resulting in proportions of some features becoming increasingly higher after the iteration of the models. Multiple DTs in RF (Breiman, 2001) are constructed by random sampling of samples and features, and the results of multiple DTs are summarized as the final results. The algorithm does not change the data distribution of the training samples; therefore, the weights of some features will not be too high.
We compared the importance of the features of the three models in Figure 2d. In addition to features with the highest weight, other features with higher weights are the features of the time dimension (ExPosure_Eattime, Disease_SickTime, and Disease_TreatTime) and space dimension (ExPosure_BuyAddress and ExPosure_EatAddress). Therefore, when identifying foodborne disease outbreaks, more attention should be paid to the similarity of cases in the time and space dimensions.
Simultaneously, many aspects still need to be improved. First, expanding the size of the dataset may lead to more objective experimental results. Second, foodborne disease outbreaks have regional characteristics. For example, foodborne disease outbreaks in Yunnan province are related to toxic wild mushrooms (Zhao et al., 2018), whereas outbreaks in coastal provinces, such as Shandong, are related to aquatic animals (Wu et al., 2018a). Therefore, for future work, classification models can be trained separately for each province or for a finer-grained division to discover the similarities and differences in foodborne disease outbreaks in various regions.
Conclusions
In this study, we abstract the problem of distinguishing whether a suspected foodborne disease outbreak is a real foodborne disease outbreak as a classification problem and build multiple classification models.† A comparison of the classification performances of different models shows that the XGBoost model has the best performance with a recall of 0.9699 and an F1-score of 0.9582. Considering the interpretability of the model, importance of the features of the models and pathogenic bacteria involved in the outbreak cases were analyzed. In addition, we verified our model on real cases to improve the credibility of our method. In the future, our classification model will be applied to the big data analysis and early warning platform of CFSA, which is still under construction, to improve the accuracy of the existing outbreak identification system to a certain extent, thereby reducing the burden of manual judgment for some medical workers.
Supplementary Material
Acknowledgments
The authors thank the Foodborne Disease Monitoring and Reporting System of the China National Center for Food Safety Risk Assessment (CFSA) for providing data support. The authors thank Yunchang Guo and Jing Zou at CFSA for answering questions related to domain knowledge and the CFSA system. The authors would like to thank Editage (www.editage.cn) for English language editing of the manuscript.
Disclosure Statement
No competing financial interests exist.
Funding Information
This research is supported by the National Key Research and Development Plan under grant number 2017YFC1601504 and the Natural Science Foundation of China under grant number 61836013.
Supplementary Material
Details for the confusion matrix can be found at https://en.wikipedia.org/wiki/Confusion_matrix
Code and data set are available at https://github.com/tent97/FBDOIdentification
References
- Adhikari B, Xu X, Ramakrishnan N, et al. Epideep: Exploiting embeddings for epidemic forecasting. In: Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. New York, NY, USA: Association for Computing Machinery, 2019: pp. 577–586 [Google Scholar]
- Ahmad MA, Eckert C, Teredesai A. Interpretable machine learning in healthcare In: Proceedings of the 2018 ACM international Conference on Bioinformatics, Computational Biology, and Health Informatics. New York, NY, USA: Association for Computing Machinery, 2018, pp. 559–560 [Google Scholar]
- Altmann A, Toloşi L, Sander O, et al. . Permutation importance: A corrected feature importance measure. Bioinformatics 2010;26:1340–1347 [DOI] [PubMed] [Google Scholar]
- Breiman L.Random forests. Mach Learn 2001;45:5–32 [Google Scholar]
- Brown LG, Hoover ER, Selman CA, et al. . Outbreak characteristics associated with identification of contributing factors to foodborne illness outbreaks. Epidemiol Infect 2017;145:2254–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan FL.Factors that contribute to outbreaks of foodborne disease. J Food Prot 1978;41:816–827 [DOI] [PubMed] [Google Scholar]
- Chen S, Xu J, Chen L, et al. . A regularization-based eXtreme Gradient Boosting approach in foodborne disease trend forecasting. Stud Health Technol Inform 2019;264:930–934 [DOI] [PubMed] [Google Scholar]
- Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, NY, USA: Association for Computing Machinery, 2016, pp. 785–794 [Google Scholar]
- Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273–297 [Google Scholar]
- Doshi-Velez F, Kim B. Towards a rigorous science of interpretable machine learning. arXiv Preprint 2017;arXiv:1702..08608. [Google Scholar]
- Effland T, Lawson A, Balter S, et al. . Discovering foodborne illness in online restaurant reviews. J Am Med Inform Assoc 2018;25:1586–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari RG, Rosario DKA, Cunha-Neto A, et al. . Worldwide epidemiology of Salmonella serovars in animal-based foods: A meta-analysis. Appl Environ Microbiol 2019;85:e00591-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund Y, Schapire RE. Experiments with a new boosting algorithm. ICML 1996;96:148–156 [Google Scholar]
- Friedman JH.Greedy function approximation: A gradient boosting machine. Ann Stat 2001;29:1189–1232 [Google Scholar]
- Gastwirth JL.The estimation of the Lorenz curve and Gini index. TRev Econ Stat 1972;54:306–316 [Google Scholar]
- Grabczewski K, Jankowski N. Feature selection with decision tree criterion. In: Fifth International Conference on Hybrid Intelligent Systems (HIS'05). Rio de Janeiro, Brazil: IEEE, 2005, p. 6 [Google Scholar]
- Horwitz MA.Specific diagnosis of foodborne disease. Gastroenterology 1977;73:375–381 [PubMed] [Google Scholar]
- Jia K, Fang T, Wang X, et al. . Antibiotic resistance patterns of Staphylococcus aureus isolates from retail foods in mainland China: A meta-analysis. Foodborne Pathog Dis 2020;17:296–307 [DOI] [PubMed] [Google Scholar]
- Lerman RI, Yitzhaki S. A note on the calculation and interpretation of the Gini index. Econ Lett 1984;15:363–368 [Google Scholar]
- Li W, Pires SM, Liu Z, et al. . Surveillance of foodborne disease outbreaks in China, 2003–2017. Food Control 2020;118:107359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg SM, Erion GG, Lee SI. Consistent individualized feature attribution for tree ensembles. arXiv Preprint 2018;arXiv:1802..03888. [Google Scholar]
- Lundberg S, Lee SI. A unified approach to interpreting model predictions[J]. arXiv preprint arXiv:1705.07874, 2017 [Google Scholar]
- Murphree R, Garman K, Phan Q, et al. . Characteristics of foodborne disease outbreak investigations conducted by Foodborne Diseases Active Surveillance Network (FoodNet) sites, 2003–2008. Clin Infect Dis 2012;54(Suppl 5):S498–S503 [DOI] [PubMed] [Google Scholar]
- Murphy KP.Naive Bayes classifiers. Univ Br Columbia 2006;18:60 [Google Scholar]
- Oliver SP.Foodborne pathogens and disease special issue on the National and International PulseNet Network. Foodborne Pathog Dis 2019;16:439–440 [DOI] [PubMed] [Google Scholar]
- Paudyal N, Pan H, Liao X, et al. . A meta-analysis of major foodborne pathogens in Chinese food commodities between 2006 and 2016. Foodborne Pathog Dis 2018;15:187–197 [DOI] [PubMed] [Google Scholar]
- Quinlan JR.Induction of decision trees. Mach Learn 1986;1:81–106 [Google Scholar]
- Sadilek A, Brennan S, Kautz H, et al. nEmesis: Which restau-rants should you avoid today?. In: Proceedings of the AAAI Conference on Human Computation and Crowdsourcing. Palm Springs, California, USA: AAAI, 2013, p. 1 [Google Scholar]
- Sadilek A, Caty S, DiPrete L, et al. . Machine-learned epidemiology: Real-time detection of foodborne illness at scale. NPJ Digit Med 2018;1:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadilek A, Kautz H, Silenzio V. Predicting disease transmission from geo-tagged micro-blog data. In: Proceedings of the AAAI Conference on Artificial Intelligence. Toronto, Ontario, Canada: AAAI, 2012, p. 26 [Google Scholar]
- Thakur M, Olafsson S, Lee JS, et al. . Data mining for recognizing patterns in foodborne disease outbreaks. J Food Eng 2010;97:213–227 [Google Scholar]
- Todd ECD. Epidemiology of foodborne diseases: A worldwide review. World Health Stat Q 1997;50 30–50 [PubMed] [Google Scholar]
- Wang X, Zhou M, Jia J, et al. . A Bayesian approach to real-time monitoring and forecasting of Chinese foodborne diseases. Int J Environ Res Public Health 2018;15:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RE. Logistic regression. In LG Grimm & PR Yarnold (Eds.), Reading and understanding multivariate statistics. American Psychological Association. 1995, pp. 217–244 [Google Scholar]
- Wu G, Yuan Q, Wang L, et al. Epidemiology of foodborne disease outbreaks from 2011 to 2016 in Shandong Province, China. Medicine (Baltimore) 2018a;97:e13142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Chen J. Food safety monitoring and surveillance in China: Past, present and future. Food Control 2018;90:429–439 [Google Scholar]
- Wu Y, Yang Y, Nishiura H, et al. Deep learning for epidemi-ological predictions. In: The 41st International ACM SIGIR Conference on Research & Development in Information Retrieval. New York, NY, USA: Association for Computing Machinery, 2018b, pp. 1085–1088 [Google Scholar]
- Xiao X, Ge Y, Guo Y, et al. Automated detection for probable homologous foodborne disease outbreaks. In: Pacific-Asia Conference on Knowledge Discovery and Data Mining. Cham: Springer, 2015, pp. 563–575. [Google Scholar]
- Zhang K, Arablouei R, Jurdak R. Predicting prevalence of influenza-like illness from geo-tagged tweets. In: Proceedings of the 26th International Conference on World Wide Web Companion. International World Wide Web Conferences Steering Committee, Republic and Canton of Geneva, CHE. 2017, pp. 1327–1334 [Google Scholar]
- Zhang M, Guo D, Hu J, et al. Risk prediction and assessment of foodborne disease based on big data. In: Proceedings of the 5th ACM SIGSPATIAL International Workshop on the Use of GIS in Emergency Management. 2019. a, pp. 1–6 [Google Scholar]
- Zhang XX, Ren WX, Tan QD, et al. Meta-analysis of Toxoplasma gondii in pigs intended for human consumption in Mainland China. Acta Trop 2019b;198:105081 [DOI] [PubMed] [Google Scholar]
- Zhao J, Min X, Zhang Q, et al. . Analysis on foodborne disease outbreaks in Yunnan Province from 2013 to 2017. J Kunming Med Univ 2018;39:118–123 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.