Abstract
Objective
To develop a radiomics risk score based on dynamic contrast-enhanced (DCE) MRI for prognosis prediction in patients with glioblastoma.
Materials and Methods
One hundred and fifty patients (92 male [61.3%]; mean age ± standard deviation, 60.5 ± 13.5 years) with glioblastoma who underwent preoperative MRI were enrolled in the study. Six hundred and forty-two radiomic features were extracted from volume transfer constant (Ktrans), fractional volume of vascular plasma space (Vp), and fractional volume of extravascular extracellular space (Ve) maps of DCE MRI, wherein the regions of interest were based on both T1-weighted contrast-enhancing areas and non-enhancing T2 hyperintense areas. Using feature selection algorithms, salient radiomic features were selected from the 642 features. Next, a radiomics risk score was developed using a weighted combination of the selected features in the discovery set (n = 105); the risk score was validated in the validation set (n = 45) by investigating the difference in prognosis between the “radiomics risk score” groups. Finally, multivariable Cox regression analysis for progression-free survival was performed using the radiomics risk score and clinical variables as covariates.
Results
16 radiomic features obtained from non-enhancing T2 hyperintense areas were selected among the 642 features identified. The radiomics risk score was used to stratify high- and low-risk groups in both the discovery and validation sets (both p < 0.001 by the log-rank test). The radiomics risk score and presence of isocitrate dehydrogenase (IDH) mutation showed independent associations with progression-free survival in opposite directions (hazard ratio, 3.56; p = 0.004 and hazard ratio, 0.34; p = 0.022, respectively).
Conclusion
We developed and validated the “radiomics risk score” from the features of DCE MRI based on non-enhancing T2 hyperintense areas for risk stratification of patients with glioblastoma. It was associated with progression-free survival independently of IDH mutation status.
Keywords: Glioblastoma, Progression, Dynamic contrast enhanced MRI, Ktrans, Ve, Vp, Radiomics
INTRODUCTION
Glioblastoma is a highly malignant brain tumor that has a high recurrence rate and a median survival after multimodality treatment of approximately 15 months [1]. Glioblastomas are highly vascularized tumors that are associated with enhanced vascular permeability, which can contribute to angiogenesis and edema [2]. The blood-brain barrier starts to be disrupted with gliomagenesis, endothelial cells from normal vessels are roughly separated from the vessel's main structure and form new angiogenic spots [3]. The endothelial cells migrate forming new vessels and destroying normal vascular structures to arrive at the tumor. Besides tumors secrete a lot of different molecules that change the normal microenvironment [3]. In glioblastoma, the morphological changes of blood vessels include disruption of tight junctions and the formation of fenestrations. In addition, the basal lamina thickness is changed, the width of the perivascular space and the number of pericytes related to the vessels is increased [3]. Recently, perfusion-weighted MRI has revealed increased vascularity when the tumor aggresses and progresses, and a radiomics model using dynamic susceptibility contrast MRI has been shown to successfully predict the survival of glioblastoma patients [4]. Moreover, dynamic contrast-enhanced (DCE) MRI is a noninvasive perfusion-weighted MRI technique that can measure the density, integrity, and leakiness of tissue vasculature, or “permeability imaging,” which can provide additional information. The method is based on measurements and mathematical models of how a tracer perfuses through such vessels. Vessels in normal tissues may be characterized by a range of parameters that measure blood flow, vessel permeability, and tissue volume fractions (i.e., fractions of a given sample of tissue that can be attributed to intravascular or extravascular space) [5]. Previous studies have revealed that histological grading, differentiation of pseudo-progression from true progression, and prognosis after surgical resection followed by standard therapy can be predicted using DCE MRI [6,7,8,9]. However, these measures can only demonstrate the group difference (e.g., between progression and non-progression) using a single statistical parameter, such as median and 99th percentile volume transfer constant (Ktrans), which suggests the possibility of developing a good prognostication model using DCE MRI.
Because radiomics extract high-dimensional features from image metrics such as intensity distribution, spatial relationships, and textural heterogeneity, which may reflect the underlying pathophysiology, including intratumoral heterogeneity, better than the single parameter approach, several radiomic prognostication models have been successfully applied to glioblastoma using MRI [10,11,12]. Recently, Park et al. [4] demonstrated that a prognostication model using combined radiomic features obtained from diffusion- and perfusion-weighted MRI showed improved performance over the model using only conventional MRI in glioblastoma.
To date, few studies have analyzed the radiomic features obtained from DCE MRI for prognosis prediction in glioblastoma. To this end, we developed a “radiomics risk score” based on DCE MRI for prognosis prediction in glioblastoma.
MATERIALS AND METHODS
Patients
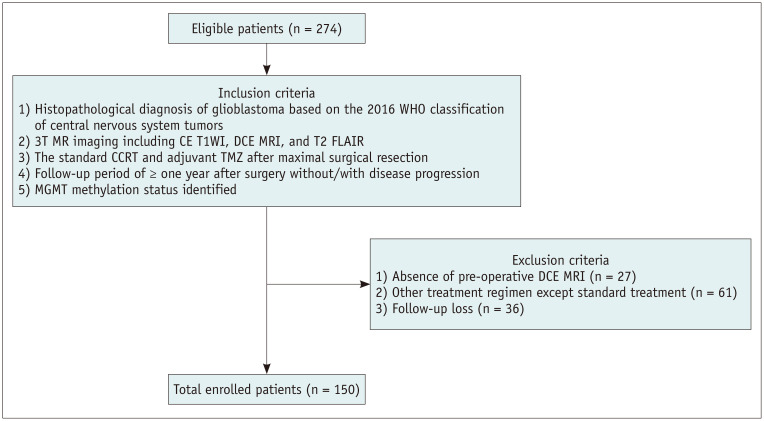
The Institutional Review Board of Seoul National University Hospital approved this retrospective study and waived the requirement for informed consent (IRB No. 2006-144-1134). We identified 274 patients with glioblastoma from January 2011 to July 2019 at Seoul National University Hospital from the radiology report database. The inclusion criteria were as follows: patients 1) with a histopathologic diagnosis of glioblastoma based on the 2016 World Health Organization (WHO) classification of central nervous system tumors; 2) who underwent preoperative 3-Tesla (3T) MRI 24–48 hours before surgery, including CE T1-weighted imaging (T1WI), DCE MRI, and T2-weighted fluid attenuated inversion recovery (FLAIR) imaging; 3) who underwent standard concomitant chemoradiotherapy [1] with temozolomide (TMZ) and six cycles of adjuvant TMZ after maximal surgical resection of the contrast-enhancing region; 4) who had a follow-up period ≥ 1 year after surgery with or without disease progression; and 5) patients in whom O6-methylguanine-DNA methyltransferase (MGMT) methylation status was identified. We specified the extent of resection as maximal (near-total or gross-total) resection because we focused on investigating the effect of non-enhancing T2 hyperintense areas of glioblastoma for prognosis, excluding the recurrence from gross residual tumor [7,13]. The exclusion criteria were as follows: patients 1) with incomplete preoperative DCE MRI (n = 27), 2) who underwent a treatment regimen other than standard treatment (n = 61), or 3) patients with follow-up loss (n = 36).
All patients were classified into disease progression and non-progression groups according to the Response Assessment in Neuro-Oncology criteria 1 year after the completion of adjuvant TMZ, following a previous study [9]. Patients with any of the following were considered to have disease progression: 1) > 25% increase in the sum of the products of the perpendicular diameters of the enhancing lesions with the smallest tumor measurement; 2) a significant increase in non-enhancing T2-weighted FLAIR lesions, not attributable to other non-tumor causes; 3) any new lesions; and 4) clinical deterioration not attributable to non-tumor causes such as a steroid decrease. Thus, disease progression was confirmed by either imaging or histopathologic diagnosis. In cases where there was an evident increase in enhancing lesions that did not meet the criteria of progression, short-term (i.e., 1–2 months) follow-up imaging was performed. Progression-free survival (PFS) was calculated from the date of initial diagnosis to the date of progression. If there was no evidence of disease at the last follow-up, PFS was calculated between the date of the initial diagnostic imaging and the last follow-up. We referred to the electronic medical records at our institution to distinguish progression from non-progression.
For all patients, clinical characteristics, including age, sex, isocitrate dehydrogenase (IDH) mutation status, and methylation status of the MGMT tumor promoter, were recorded. The tissue-based diagnoses and genetic analyses are detailed in the Supplemen.
Imaging Protocol
All MR images were acquired using a 3T scanner (Verio, Trio, or Skyra; Siemens) using a 32-channel head coil. For tumor segmentation, the T1-weighted three-dimensional (3D) magnetization-prepared rapid acquisition gradient echo (MPRAGE) sequence before and after administration of gadobutrol (at a dose of 0.1 mmoL/kg of body weight; Gadovist; Bayer) and FLAIR imaging were used for the enrolled patients. We performed transverse T1WI with the following parameters: repetition time (TR), 558 ms; echo time (TE), 9.8 ms; flip angle (FA), 70°; matrix, 384 × 187; field of view (FOV), 175 × 220; section thickness, 5 mm; and number of excitations (NEX) of 1. The parameters for 3D MPRAGE were as follows: a TR of 1500 ms, a TE of 1.9 ms, an FA of 9°, a matrix of 256 × 232, a FOV of 220 × 250, a section thickness of 1 mm, and a NEX of 1. The parameters for transverse FLAIR were as follows: a TR of 9000 ms; a TE of 97 ms; a TI of 2500 ms; an FA of 130°; a matrix of 384 × 348; a FOV of 199 × 220; a section thickness of 5 mm; and a NEX of 1. Transverse T2WI was performed using the following parameters: a TR of 5160 ms; a TE of 91 ms; an FA of 124–130°; a matrix of 640 × 510–580; a FOV of 199 × 220; a section thickness of 5 mm; and a NEX of 3.
DCE MRI was conducted with 3D gradient-echo T1WI after the intravenous administration of gadobutrol at a dose of 0.1 mmoL/kg of body weight, at a rate of 4 mL/s using a power injector (Spectris MedRad). After contrast, a 30 mL saline bolus was injected at the same injection rate. For each section, 40 images were acquired at intervals equal to the TR. The scan parameters of DCE MRI were as follows: a TR of 2.8 ms, a TE of 1.0 ms, a FA of 10°, a matrix of 192 × 192, a FOV of 240 × 240, a section thickness of 3 mm, a voxel size of 1.25 × 1.25 × 3 mm3, a pixel bandwidth of 789 Hz, and a total acquisition time of 5 minutes 8 seconds.
Image Processing and Analysis
The MR data, including CE T1WI, FLAIR imaging, and DCE MRI, were transferred from the PACS workstation to a personal computer and were processed using the Nordic ICE a software package (v4.1.2; NordicNeuroLab). DCE MRI analysis was based on the two-compartment pharmacokinetic model suggested by Tofts and Kermode, and the perfusion analysis method was used to calculate the pharmacokinetic parameters, including the volume transfer constant between the plasma and extravascular extracellular space (Ktrans), extravascular extracellular space volume per unit volume of tissue (Ve), and the blood plasma volume per unit volume of tissue (Vp) [13]. More specifically, deconvolution with the arterial input function (AIF) was performed using a two-compartment pharmacokinetic model. The AIF was selected from the tumor-supplying arteries near the tumor. Each parameter was calculated using a fixed T1 measurement of 1000 ms.
Before drawing regions of interest (ROIs), we reconstructed the CE T1WI from the sagittal to axial plane, acquired Ktrans, Ve, and Vp maps based on DCE MRI, and resampled the size of the CE T1WI and FLAIR images using one of the maps (Ktrans, Ve, and Vp) as a reference. Two ROIs were drawn: 1) the enhancing tumor without cystic or necrotic regions CE T1WI, and 2) the non-enhancing T2 high signal intensity (SI) areas excluding cystic or necrotic areas. In all ROIs, the vessels were carefully excluded. All ROIs were drawn semi-automatically using threshold segmentation, seed growing, and manually, if needed [14]. Tumor segmentation procedures were performed using the NordicICE software tool (v4.1.2). The ROIs were drawn by a radiologist supervised by an expert radiologist (with 17 years of neuro-oncology imaging experience).
In our research, we used the software 3D Slicer 4.11.0 and the Pyradiomics package for extraction of the radiomic features. For each patient, a total of 642 features were obtained: 107 features, including 18 first-order, 14 shape-based, 24 Gy-level co-occurrence matrix (GLCM), 16 Gylevel size zone matrix (GLSZM), 16 Gy-level run length matrix (GLRLM), 5 neighboring gray tone difference matrix (NGTDM), and 14 Gy level dependence matrix (GLDM), were obtained from each of the Ktrans, Ve, and Vp maps. Both enhancing region and non-enhancing T2 high SI region masks were used, which resulted in six map-region combinations per patient. The details of the radiomic features are described in the Supplement.
Radiomic Feature Selection
The patients were randomly split into the discovery set (n = 105) and validation set (n = 45) at a 7:3 ratio. The radiomic feature selection using the discovery set included three steps: first, features with more than 10% outliers were discarded, and outliers were defined when the largest and smallest 10% value of features were larger than the median ± median absolute deviation (MAD) [15]. Standardization or z-score normalization was performed for each feature. Here, we used the mean and standard deviation of the discovery set to standardize the validation set to prevent data leakage, which may lead to a decrease in the generalized model performance [16,17]. Second, univariable Cox regression analysis was performed to identify significant features. Next, a Cox-least absolute shrinkage selector operator (Cox-LASSO) model was developed to select the final radiomic features in the prediction of PFS using the glmnet R package. LASSO is a popular regularization method for selecting features from multiple variables, which penalizes the coefficients of variables, shrinking some of them to zero in the regression model, and leaving others with non-zero coefficients [18]. To determine the hyperparameter λ in LASSO, 10-fold cross-validation was performed using cv.glmnet.
Radiomics Risk Score and Risk Stratification
A radiomics risk score model was developed using a weighted combination of the final selected features in the discovery set. Next, patients were stratified into either a high- or low-risk group using the cutoff value of the radiomics risk scores obtained by maximally selected rank statistics using the maxstat (R package) [19,20] in the discovery set, which was also applied to the validation set [17]. In other words, the “radiomics risk” was present when the radiomics risk score of the patient was higher than the cutoff value.
Progression Free Survival (PFS) Analysis
A multivariable Cox regression analysis was performed using the following clinical variables as covariates, such as age, sex, IDH mutation status, and MGMT methylation status, in addition to the risk group variable derived from the radiomics risk score. To validate the developed radiomics risk score, the radiomics risk score was calculated using the values of selected features of the validation set, weighted by the coefficients obtained from the discovery set. Using the maximally selected rank statistics, the cutoff value of the radiomics risk score was obtained from the discovery set, and the validation set was stratified into either a high- or low-risk group using the same cutoff value.
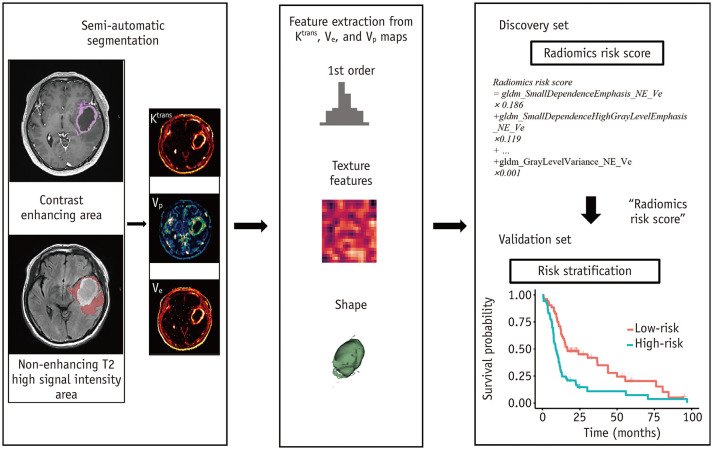
All statistical analyses were performed using R-3.6.3 (R-core Team), and a p value of 0.05 was considered significant. The overall process from feature extraction to risk group stratification is shown in Figure 1.
Fig. 1. Overall process of radiomics risk score model construction and risk stratification.
Ktrans = volume transfer constant, Ve = fractional volume of extravascular extracellular space, Vp = fractional volume of vascular plasma space
RESULTS
Patient Characteristics
150 patients were enrolled in this study (Fig. 2) (92 [61.3%] male; mean age, 60.5 ± 13.5 years old; and 137 [91.3%] IDH-wildtype) (Table 1). The patients were divided into the disease progression group (n = 61, 40.7%) and the non-progression group (n = 89, 59.3%) 1 year after surgery. The median PFS was 11.1 months (range, 0.57–97.0 months). Patients with MGMT promoter methylation status were more frequently observed in the non-progression group than in the progression group (61 of 89 vs. 20 of 61, respectively, p < 0.001). The patient characteristics are detailed in Table 1. There were no significant clinicopathological differences in IDH (p = 1.000), MGMT (p = 0.253), sex (p = 1.000), age (p = 0.149), or number of recurrences (p = 0.093) between the discovery and validation sets (Supplementary Table 1).
Fig. 2. Inclusion/exclusion criteria for the study population.
CCRT = concomitant chemoradiotherapy, CE T1WI = contrast-enhanced T1-weighted imaging, DCE = dynamic contrast-enhanced, MGMT = O6-methylguanine-DNA methyltransferase, TMZ = temozolomide, T2 FLAIR = T2-weighted fluid attenuated inversion recovery, WHO = World Health Organization
Table 1. Clinicopathologic Characteristics of the Study Population.
| Characteristics | Total (n = 150) | Progression at 1 Year (n = 61) | Non-Progression at 1 Year (n = 89) | P | |
|---|---|---|---|---|---|
| Age, years | 60.5 ± 13.5 | 61.5 ± 13.03 | 59.8 ± 13.9 | 0.444* | |
| Radiation dose, Gy | 55.2 ± 9.2 | 53.0 ± 11.7 | 56.7 ± 7.3 | 0.032* | |
| Sex | 0.237† | ||||
| Male | 92 | 41 | 51 | ||
| Female | 58 | 20 | 38 | ||
| Methylated MGMT promoter | < 0.001† | ||||
| Positive | 81 | 20 | 61 | ||
| Negative | 69 | 41 | 28 | ||
| IDH1/2 mutation | 0.075† | ||||
| Positive | 13 | 2 | 11 | ||
| Negative | 137 | 59 | 78 | ||
Data are number of patients except for age and radiation dose which are presented as mean ± standard deviation. *Calculated using an unpaired Student's t test, †Calculated using chi-square or Fisher's exact test. IDH = isocitrate dehydrogenase, MGMT = O6-methylguanine-DNA methyltransferase
Selected Features and Radiomics Risk Score
As a result of discarding outliers using three MADs, 589 of 642 features were left. After univariable Cox regression, 76 significant features were left, and 16 features were selected using a Cox-LASSO model in the prediction of PFS. The intermediate results of the Cox-LASSO procedure are shown in Supplementary Figure 1, and a heatmap of radiomic features using hierarchical clustering with dendrogram is shown in Supplementary Figure 2 for the 76 features selected from univariable Cox regression. The intermediate results in the procedure of the Cox-LASSO model using the training set are illustrated in Supplementary Figure 3. The 16 features selected included 7 features from Ktrans, and the other 9 features from the Ve map of the non-enhancing T2 high SI region: one NGTDM feature, three GLDM features, one first-order feature, five GLRLM features, three shape features, one GLCM feature, and two GLSZM features (Table 2). A radiomics risk score model was developed using a linear combination of the 16 final selected features, with coefficients obtained from the Cox-LASSO model using the discovery set (Eq. 1).
Table 2. Radiomics Features Selected for Radiomics Risk Score.
| No.* | Radiomic Features† | Coefficients‡ |
|---|---|---|
| 1 | gldm_SmallDependenceEmphasis_NE_Ve | 0.186 |
| 2 | gldm_SmallDependenceHighGrayLevelEmpha sis_NE_Ve | 0.119 |
| 3 | firstorder_Energy_NE_Ve | 0.114 |
| 4 | ngtdm_Complexity_NE_Ve | 0.105 |
| 5 | glrlm_ShortRunHighGrayLevelEmphasis_NE_ktrans | 0.100 |
| 6 | glrlm_ShortRunLowGrayLevelEmphasis_NE_ktrans | 0.096 |
| 7 | glszm_SmallAreaEmphasis_NE_ktrans | 0.091 |
| 8 | shape_SurfaceArea_NE_ktrans | 0.086 |
| 9 | glrlm_ShortRunEmphasis_NE_ktrans | 0.082 |
| 10 | glszm_SmallAreaHighGrayLevelEmphasis_NE_ktrans | 0.071 |
| 11 | glrlm_RunLengthNonUniformity_NE_ktrans | 0.053 |
| 12 | glrlm_ShortRunHighGrayLevelEmphasis_NE_Ve | 0.044 |
| 13 | shape_Maximum3DDiameter_NE_Ve | 0.043 |
| 14 | glcm_JointAverage_NE_Ve | 0.029 |
| 15 | shape_MajorAxisLength_NE_Ve | 0.005 |
| 16 | gldm_GrayLevelVariance_NE_Ve | 0.001 |
*Features are given in the descending order of coefficients, †Each part of the feature label indicates the class, the name of features, subregion of tumor, and the pharmacokinetic parametric maps from which the features were derived, in order, ‡Coefficients are given to the third decimal place. firstorder = first order features, glcm = gray level co-occurrence matrix features, gldm = gray level dependence matrix features, glrlm = gray level run length matrix, glszm = gray level size zone matrix features, Ktrans = volume transfer constant, NE = subregion of nonenhancing T2 hyperintense lesion, ngtdm = neighbouring gray tone difference matrix features, shape = shape-based features, Ve = volume of the extravascular extracellular space
| Radiomics risk score = gldm_SmallDependenceEmphasis_NE_Ve × 0.186 + gldm_SmallDependenceHighGrayLevelEmphasis_NE_Ve × 0.119 + firstorder_Energy_NE_Ve × 0.114 +... + glcm_JointAverage_NE_Ve × 0.029 + shape_MajorAxisLength_NE_Ve × 0.005 + gldm_GrayLevelVariance_NE_Ve × 0.001 (Eq. 1) |
Patient Stratification Using the Radiomics Risk Score
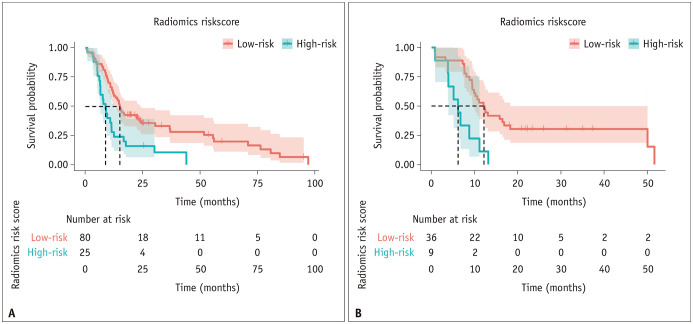
For each discovery and validation set, the radiomics risk score was computed using the 16 selected features obtained from the discovery set. The radiomics risk score stratified high- and low-risk group in both the discovery and validation sets (both p < 0.001 by the log-rank test) (Fig. 3). A cutoff value of 0.374 was obtained using maximally selected rank variables (M = 3.277; p = 0.017). The intermediate results are presented in Supplementary Figure 4.
Fig. 3. Risk group stratification using the radiomics risk score for prediction of progression-free survival: discovery (A) and validation set (B).
Multivariable Cox-Regression Analysis Adding Clinical Variables
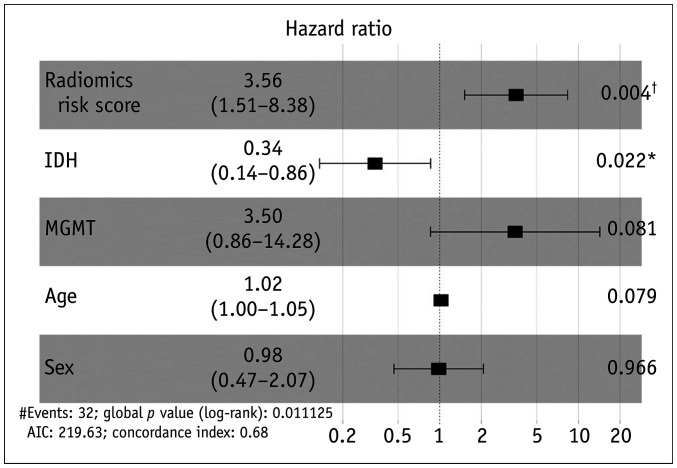
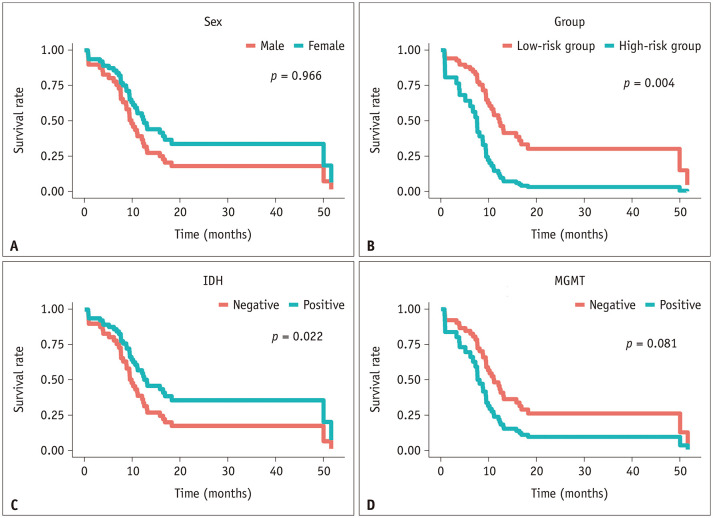
A multivariable Cox regression analysis was performed using the following clinical variables as covariates, such as age, sex, IDH mutation status, and MGMT methylation status, in addition to the “radiomics risk score”. Among the five variables, the presence of a high radiomics risk score and IDH mutation were significant variables (hazard ratio [HR], 3.56; p = 0.004 and HR, 0.34; p = 0.022, respectively). In other words, the high radiomics risk score group showed an approximately 3.5-fold stronger association with progression than the low risk score group, and the IDH mutation group showed an approximately 3-fold stronger association with progression than the wildtype group in a different direction. A forest plot of the multivariable Cox regression analysis is shown in Figure 4. The stratification of the survival curves for each single variable is plotted in Figure 5.
Fig. 4. Forest plot to show the results of the multivariable Cox proportional hazards regression using clinical and radiomics variables (n = 45).
*p < 0.05, †p < 0.01. IDH = isocitrate dehydrogenase, MGMT = O6-methylguanine-DNA methyltransferase
Fig. 5. Survival curves of the multivariable Cox-regression analysis for risk stratification using each variable of the validation set: sex (A), radiomics risk group (B), IDH mutation status (C), and MGMT methylation status (D).
IDH = isocitrate dehydrogenase, MGMT = O6-methylguanine-DNA methyltransferase
DISCUSSION
We developed and validated the “radiomics risk score” from Ktrans and Ve maps of DCE MRI to show the diagnostic capability in the prediction of the high-risk patient group in glioblastoma. In particular, we found that our approaches could be useful for the following: first, the textural features of the Ktrans and Ve maps based on non-enhancing T2 high SI areas were crucial for risk stratification in the recurrence of glioblastoma (p < 0.001 in both the training and validation sets; log-rank test). Second, the developed radiomics risk score was independent of IDH mutation status in Cox regression analysis (HR, 3.56; p = 0.004 and HR = 0.34, p = 0.022, respectively), thereby providing an additional surrogate marker for progression in glioblastoma. The final 16 selected radiomic features out of 642 features were obtained from non-enhancing T2 hyperintense lesions: nine features (gldm Small Dependence Emphasis showing the largest coefficient) from the from Ve map; and the other 7 features (glrlm Short Run High Gray Level Emphasis showing the largest coefficient) from the Ktrans map. In other words, the textural features obtained from the non-enhancing T2 high SI areas of the Ktrans and Ve maps that significantly stratified the low- and high-risk groups (Eq. 1 and Table 2) independent of IDH mutation status, were the most important risk factors. Thus, radiomic features can predict the prognosis of glioblastoma, reflecting the intra-tumoral heterogeneity of “permeability” of non-enhancing T2 hyperintense areas using DCE MRI. This is possible because textural features provide a measure of the spatial arrangement of the intra-lesional voxel intensities, which are obtained by calculating the statistical inter-relationships between neighboring voxel intensities [21].
Our finding that radiomic features from non-enhancing T2 hyperintense lesions were more important than contrast-enhancing lesions in predicting prognosis/progression, especially when total resection was performed, is consistent with previous studies [13,22]. More specifically, non-enhancing T2 high SI regions on the tumor margin have been described as “peritumoral edema.” Non-enhancing T2 high SI lesions are a mixture of infiltrating tumor cells and vasogenic edema, where fluid penetrates into the parenchymal extracellular space [22]. Schoenegger et al. [23] showed that the extent of edema was an independent prognostic factor in patients with glioblastoma. However, it is not possible to differentiate infiltrating neoplasms from vasogenic edema using conventional imaging sequences [22].
Recent studies have shown that perfusion characteristics obtained from perfusion-weighted MRI predict not only survival/progression, but also crucial tumor characteristics such as genetic mutations in glioblastoma [9,14,24]. Among perfusion-weighted MR techniques, DCE MRI images the perfusion characteristics of the tumors that cannot be provided in the conventional MRI sequence, thereby reflecting the exchange between the vasculature in the tissue and interstitium, or “leakage space” [13]. Pharmacokinetic parameters obtained from DCE MRI represent the tissue permeability; Ktrans, the volume transfer constant between the plasma and the extravascular extracellular space, has the capability to histologically grade gliomas, because higher grade gliomas show enhanced angiogenesis, resulting in immature vessels of higher permeability [13,25]; and Ve, the extravascular extracellular space volume per unit volume of tissue, which is another a predictor of progression in patients with high-grade glioma [26]. Kim et al. [13] showed that the analysis of DCE MR parameters of non-enhancing T2 high SI lesions could predict the progression of glioblastoma. The 99th percentile value (PV) Ktrans of non-enhancing T2 high SI lesions in glioblastoma is a candidate imaging biomarker for the prediction of early disease progression after standard treatment. Moreover, another permeability parameter, the 97th PV Ve, could also differentiate non-progression and progression [13]. As a result, infiltrative tumor cells in non-enhancing areas, residing outside the enhancing portion, could serve as surrogate markers of the aggressiveness and prognosis of glioblastoma. Hwang et al. [7] also reported that an increased preoperative median Ktrans from non-enhancing T2 high SI lesions was associated with poor survival in patients with gross total resection followed by standard therapy, using multivariable Cox regression analysis. In our study, the radiomics risk score obtained from Ktrans and Ve maps of non-enhancing T2 high SI lesions showed diagnostic capability in the risk stratification of glioblastoma, which is also consistent with the findings of previous studies.
Radiomics extracts high-dimensional features from image metrics such as intensity distribution, spatial relationships, and textural heterogeneity, which may reflect the underlying pathophysiology, including intratumoral heterogeneity. Several radiomics prognostication models for glioblastoma have been developed and validated using radiomic features obtained from MRI [10,11,12]. However, the high dimensionality (d = 642) of radiomics data, or much higher dimension of data compared to the number of subjects in the discovery set (n = 105), causes the “curse of dimensionality,” leading to “overfitting” of the model to the discovery set. In other words, the model developed using radiomics data may show good performance only in the discovery set, but not in the validation set, or show low “generalizability” [27]. To overcome this issue, we used the Cox-LASSO model to select the salient radiomic features for predicting PFS. LASSO is a popular regularization method for selecting features from multiple variables because it penalizes and imposes the coefficients of some variables to shrink toward zero in the regression model. This leaves the other variables with non-zero coefficients, selects important features [18], which also leads to relieve the “curse of dimensionality” due to the high dimensionality in radiomics data analysis [28].
The IDH1/2 mutation is known to be one of the most important molecular biomarkers of glioblastoma, with diagnostic, prognostic, and predictive roles. Indeed, a previous study demonstrated that the IDH-mutant group showed better survival compared to the IDH-wildtype group [29]. IDH mutation appears to be a significant marker of positive chemosensitivity in secondary glioblastoma [30], and maximal surgical resection of the IDH-mutant group has been shown to provide a survival benefit [31]. Based on these results, glioblastomas are categorized according to the 2016 CNS WHO classification into 1) primary glioblastomas, IDH-wildtype (approximately 90% of cases), and 2) secondary glioblastomas, IDH-mutant (approximately 10% of cases). Secondary glioblastomas show evidence of progression from lower-grade tumors, whereas primary glioblastomas present as advanced cancers at diagnosis [32]. IDH1/2 mutations facilitate easier formation of the tumor microenvironment and increase invasiveness [33]. Previous research has demonstrated that the radiomics model built with multiregional features from multiparametric MRI has the potential to preoperatively detect IDH1 mutation status in patients with glioblastomaa [34,35]. Moreover, in the study by Tan et al. [36], multivariable Cox regression analysis identified the radiomics signature, age, and IDH as independent risk factors in the prediction of high-grade glioma. Our study showed that the developed radiomics risk score increased the risk of progression in patients with glioblastoma, independent of IDH mutation status (HR = 3.56, p = 0.004; HR = 0.34, p = 0.022, respectively).
The present study has several limitations. First, the study was retrospective and the number of patients was limited for sufficient extraction of salient radiomic features, which warrants further improvement of the radiomics risk score with a larger number of patients. Second, the time-consuming, multi-staged workflow, including manual ROI drawing, discourages the application of the developed radiomics risk score in clinical practice. The use of the automated segmentation technique could effectively relieve the problem. Third, radiomic features are sensitive to imaging parameters and systems, which may cause the radiomics models to fail in multi-centered and prospective applications. “Harmonization” of multi-centered radiomics could improve the generalizability, and is our next research topic. Fourth, although there was no significant clinicopathologic difference between the discovery and validation sets, the discovery and validation sets were randomly split, instead of using a stratified split, which might improve the model performance.
In conclusion, we developed and validated the radiomics risk score obtained from DCE MRI and imaging perfusion characteristics for the risk stratification of progression in glioblastoma. The radiomics risk score was mainly extracted from non-enhancing T2 hyperintense areas rather than contrast-enhancing areas. It was associated with PFS independently of IDH mutation, providing an additional surrogate marker of progression in glioblastoma.
Acknowledgments
We thank Yurim Kang, BS, and Seong Yeong Lee, BS, for their invaluable assistance with data collection and analysis.
Footnotes
This study was supported by a grant from the Korea Healthcare technology R&D Projects, Ministry for Health, Welfare & Family Affairs (HI16C1111), by the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2016M3C7A1914002), by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2020R1A2C2008949 and NRF-2020R1A4A1018714), by Creative-Pioneering Researchers Program through Seoul National University (SNU), and by the Institute for Basic Science (IBS-R006-A1).
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
- Conceptualization: Seung Hong Choi.
- Data curation: Elena Pak, Chul-Kee Park, Tae Min Kim, Sung-Hye Park, Joo Ho Lee, Soon-Tae Lee, Inpyeong Hwang.
- Formal analysis: Tae Jin Yun, Ji-Hoon Kim.
- Funding acquisition: Seung Hong Choi.
- Investigation: Elena Pak, Kyu Sung Choi.
- Methodology: Kyu Sung Choi, Elena Pak, Koung Mi Kang, Roh-Eul Yoo.
- Software: Elena Pak, Kyu Sung Choi.
- Supervision: Seung Hong Choi, Chul-Ho Sohn.
- Writing—original draft: Elena Pak, Kyu Sung Choi.
- Writin—review & editing: Seung Hong Choi.
Supplement
The Supplement is available with this article at https://doi.org/10.3348/kjr.2020.1433.
Clinicopathologic Characteristics of Discovery and Validation Set
Illustration of the feature selection process.
Heatmap of radiomic features using hierarchical clustering with a dendrogram for selected features from univariate Cox-regression.
Intermediate results of the Cox-LASSO model using the training set.
The intermediate result of calculating the cutoff value of the radiomics risk score obtained by maximally selected rank statistics using maxstat (R package); the cutoff value was 0.374 (M = 3.277; p = 0.017).
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Treps L, Edmond S, Harford-Wright E, Galan-Moya EM, Schmitt A, Azzi S, et al. Extracellular vesicle-transported Semaphorin3A promotes vascular permeability in glioblastoma. Oncogene. 2016;35:2615–2623. doi: 10.1038/onc.2015.317. [DOI] [PubMed] [Google Scholar]
- 3.Dubois LG, Campanati L, Righy C, D'Andrea-Meira I, Spohr TC, Porto-Carreiro I, et al. Gliomas and the vascular fragility of the blood brain barrier. Front Cell Neurosci. 2014;8:418. doi: 10.3389/fncel.2014.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park JE, Kim HS, Jo Y, Yoo RE, Choi SH, Nam SJ, et al. Radiomics prognostication model in glioblastoma using diffusion- and perfusion-weighted MRI. Sci Rep. 2020;10:4250. doi: 10.1038/s41598-020-61178-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yankeelov TE, Gore JC. Dynamic contrast enhanced magnetic resonance imaging in oncology: theory, data acquisition, analysis, and examples. Curr Med Imaging Rev. 2009;3:91–107. doi: 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas AA, Arevalo-Perez J, Kaley T, Lyo J, Peck KK, Shi W, et al. Dynamic contrast enhanced T1 MRI perfusion differentiates pseudoprogression from recurrent glioblastoma. J Neurooncol. 2015;125:183–190. doi: 10.1007/s11060-015-1893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang I, Choi SH, Park CK, Kim TM, Park SH, Won JK, et al. Dynamic contrast-enhanced MR imaging of nonenhancing T2 high-signal-intensity lesions in baseline and posttreatment glioblastoma: temporal change and prognostic value. AJNR Am J Neuroradiol. 2020;41:49–56. doi: 10.3174/ajnr.A6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dijken BRJ, van Laar PJ, Smits M, Dankbaar JW, Enting RH, van der Hoorn A. Perfusion MRI in treatment evaluation of glioblastomas: clinical relevance of current and future techniques. J Magn Reson Imaging. 2019;49:11–22. doi: 10.1002/jmri.26306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo RE, Choi SH, Kim TM, Park CK, Park SH, Won JK, et al. Dynamic contrast-enhanced MR imaging in predicting progression of enhancing lesions persisting after standard treatment in glioblastoma patients: a prospective study. Eur Radiol. 2017;27:3156–3166. doi: 10.1007/s00330-016-4692-9. [DOI] [PubMed] [Google Scholar]
- 10.Kickingereder P, Burth S, Wick A, Götz M, Eidel O, Schlemmer HP, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280:880–889. doi: 10.1148/radiol.2016160845. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, Vallières M, Bai HX, Su C, Tang H, Oldridge D, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19:862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim R, Choi SH, Yun TJ, Lee ST, Park CK, Kim TM, et al. Prognosis prediction of non-enhancing T2 high signal intensity lesions in glioblastoma patients after standard treatment: application of dynamic contrast-enhanced MR imaging. Eur Radiol. 2017;27:1176–1185. doi: 10.1007/s00330-016-4464-6. [DOI] [PubMed] [Google Scholar]
- 14.Kim JY, Yoon MJ, Park JE, Choi EJ, Lee J, Kim HS. Radiomics in peritumoral non-enhancing regions: fractional anisotropy and cerebral blood volume improve prediction of local progression and overall survival in patients with glioblastoma. Neuroradiology. 2019;61:1261–1272. doi: 10.1007/s00234-019-02255-4. [DOI] [PubMed] [Google Scholar]
- 15.Leys C, Ley C, Klein O, Bernard P, Licata L. Detecting outliers: do not use standard deviation around the mean, use absolute deviation around the median. J Exp Soc Psychol. 2013;49:764–766. [Google Scholar]
- 16.Chollet F. Deep learning with python. New York: Manning; 2018. [Google Scholar]
- 17.Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on gadoxetic acid–enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res. 2019;25:3847–3855. doi: 10.1158/1078-0432.CCR-18-2861. [DOI] [PubMed] [Google Scholar]
- 18.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2011;39:1–13. [PMC free article] [PubMed] [Google Scholar]
- 19.Liang C, Huang Y, He L, Chen X, Ma Z, Dong D, et al. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget. 2016;7:31401–31412. doi: 10.18632/oncotarget.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beig N, Bera K, Prasanna P, Antunes J, Correa R, Singh S, et al. Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in glioblastoma. Clin Cancer Res. 2020;26:1866–1876. doi: 10.1158/1078-0432.CCR-19-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2:36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathore S, Akbari H, Doshi J, Shukla G, Rozycki M, Bilello M, et al. Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning. J Med Imaging (Bellingham) 2018;5:021219. doi: 10.1117/1.JMI.5.2.021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoenegger K, Oberndorfer S, Wuschitz B, Struhal W, Hainfellner J, Prayer D, et al. Peritumoral edema on MRI at initial diagnosis: an independent prognostic factor for glioblastoma? Eur J Neurol. 2009;16:874–878. doi: 10.1111/j.1468-1331.2009.02613.x. [DOI] [PubMed] [Google Scholar]
- 24.Choi KS, Choi SH, Jeong B. Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. Neuro Oncol. 2019;21:1197–1209. doi: 10.1093/neuonc/noz095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung SC, Yeom JA, Kim JH, Ryoo I, Kim SC, Shin H, et al. Glioma: application of histogram analysis of pharmacokinetic parameters from T1-weighted dynamic contrast-enhanced MR imaging to tumor grading. AJNR Am J Neuroradiol. 2014;35:1103–1110. doi: 10.3174/ajnr.A3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulyte A, Katsaros VK, Liouta E, Stranjalis G, Boskos C, Papanikolaou N, et al. Prognostic value of preoperative dynamic contrast-enhanced MRI perfusion parameters for high-grade glioma patients. Neuroradiology. 2016;58:1197–1208. doi: 10.1007/s00234-016-1741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trunk GV. A problem of dimensionality: a simple example. IEEE Trans Pattern Anal Mach Intell. 1979;1:306–307. doi: 10.1109/tpami.1979.4766926. [DOI] [PubMed] [Google Scholar]
- 28.Verleysen M, François D. The curse of dimensionality in data mining and time series prediction. In: Cabestany J, Prieto A, Sandoval F, editors. Computational intelligence and bioinspired systems. Berlin, Heidelberg: Springer; 2005. pp. 758–770. [Google Scholar]
- 29.Stupp R, Brada M, van den Bent MJ, Tonn JC, Pentheroudakis G ESMO Guidelines Working Group. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25:iii93–iii101. doi: 10.1093/annonc/mdu050. [DOI] [PubMed] [Google Scholar]
- 30.SongTao Q, Lei Y, Si G, YanQing D, HuiXia H, XueLin Z, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103:269–273. doi: 10.1111/j.1349-7006.2011.02134.x. [DOI] [PubMed] [Google Scholar]
- 31.Beiko J, Suki D, Hess KR, Fox BD, Cheung V, Cabral M, et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol. 2014;16:81–91. doi: 10.1093/neuonc/not159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13:345. doi: 10.1007/s11910-013-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, Yu J, Tu L, Huang N, Li H, Luo Y. Isocitrate dehydrogenase mutations in glioma: from basic discovery to therapeutics development. Front Oncol. 2019;9:506. doi: 10.3389/fonc.2019.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li ZC, Bai H, Sun Q, Zhao Y, Lv Y, Zhou J, et al. Multiregional radiomics profiling from multiparametric MRI: identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018;7:5999–6009. doi: 10.1002/cam4.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh KL, Chen CY, Lo CM. Radiomic model for predicting mutations in the isocitrate dehydrogenase gene in glioblastomas. Oncotarget. 2017;8:45888–45897. doi: 10.18632/oncotarget.17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan Y, Mu W, Wang XC, Yang GQ, Gillies RJ, Zhang H. Improving survival prediction of high-grade glioma via machine learning techniques based on MRI radiomic, genetic and clinical risk factors. Eur J Radiol. 2019;120:108609. doi: 10.1016/j.ejrad.2019.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinicopathologic Characteristics of Discovery and Validation Set
Illustration of the feature selection process.
Heatmap of radiomic features using hierarchical clustering with a dendrogram for selected features from univariate Cox-regression.
Intermediate results of the Cox-LASSO model using the training set.
The intermediate result of calculating the cutoff value of the radiomics risk score obtained by maximally selected rank statistics using maxstat (R package); the cutoff value was 0.374 (M = 3.277; p = 0.017).