Abstract
Transmigration of leukocytes across blood vessels walls is a critical step of the immune response. Transwell assays examine transmigration properties in vitro by counting cells passages through a membrane; however, the difficulty of in situ imaging hampers a clear disentanglement of the roles of adhesion, chemokinesis, and chemotaxis. We used here microfluidic Transwells to image the cells’ transition from 2D migration on a surface to 3D migration in a confining microchannel and measure cells longitudinal forward-thrusting force in microchannels. Primary human effector T lymphocytes adhering with integrins LFA-1 (αLβ2) had a marked propensity to transmigrate in Transwells without chemotactic cue. Both adhesion and contractility were important to overcome the critical step of nucleus penetration but were remarkably dispensable for 3D migration in smooth microchannels deprived of topographic features. Transmigration in smooth channels was qualitatively consistent with a propulsion by treadmilling of cell envelope and squeezing of cell trailing edge. Stalling conditions of 3D migration were then assessed by imposing pressure drops across microchannels. Without specific adhesion, the cells slid backward with subnanonewton forces, showing that 3D migration under stress is strongly limited by a lack of adhesion and friction with channels. With specific LFA-1 mediated adhesion, stalling occurred at around 3 and 6 nN in 2 × 4 and 4 × 4 μm2 channels, respectively, supporting that stalling of adherent cells was under pressure control rather than force control. The stall pressure of 4 mbar is consistent with the pressure of actin filament polymerization that mediates lamellipod growth. The arrest of adherent cells under stress therefore seems controlled by the compression of the cell leading edge, which perturbs cells front-rear polarization and triggers adhesion failure or polarization reversal. Although stalling assays in microfluidic Transwells do not mimic in vivo transmigration, they provide a powerful tool to scrutinize 2D and 3D migration, barotaxis, and chemotaxis.
Significance
Leukocyte extravasations from blood and lymph vessels are crucial steps of the immune response. We developed here reductionist approaches with microfluidic Transwells to measure transmigration efficiency and stall forces in vitro. With human primary T lymphocytes, transmigration occurred without chemotactic cues and cells arrested under pressure rather than force control. Transmigration in constrictions with smooth walls was consistent with a propulsion by rear contractility and envelope treadmilling clutched by adhesion or friction to walls. Interestingly, pressure did not directly challenge the strength of the propulsion machinery but the growth of lamellipods, which in turn impaired cell polarization and adhesion. Our method should be invaluable to decipher further the transmigration of other immune and cancer cells versus various biomechanical and biochemical cues.
Introduction
During immune surveillance, leukocytes patrol the whole organism via the blood or lymph systems and reach specific lymphoid organs or infected tissues by arresting on and crossing through the inner walls of blood and lymphatic vessels, a phenomenon called “diapedesis” or “transmigration.” Similarly, during cancer spreading, tumor cells circulate in the blood system and eventually form a metastasis in new tissue by arresting on vessel walls and transmigrating through the endothelium (1). Although transmigration is a crucial process in immunology and cancer, its precise biomechanical and biochemical mechanisms remain partially understood (2). Before transmigration, adhesion mediated by integrins allows cells to resist the blood flow and to crawl on the endothelium until they reach specialized exit points, called also “transmigratory cups” or “portals” (3, 4, 5, 6, 7). These sites are arguably composed of endothelial cells with vertical microvilli-like projections enriched in integrin ligand molecules, among which ICAM-1, ligand of integrin LFA-1, was shown to be particularly important for transmigration (4, 5, 6,8). Leukocyte transmigration is therefore thought to be fostered by both guidance cues, including chemokines such as CCL19 and CCL21, for lymphocytes to enter lymph nodes (9) and adhesion cues such as integrin ligands (3,10). Besides chemical properties, mechanical and geometrical properties of the microenvironment play crucial roles in transmigration. As cells undergo profound deformations during their passage through the endothelium, the organization of the migration machinery is highly challenged, and the stiff nucleus was reported to impose a limit of pore size for cells passage without damage (11, 12, 13, 14). In the end, transmigration in vivo is mediated by numerous ill-controlled biochemical and biomechanical parameters, and we developed here quantitative in vitro approaches to control environmental conditions and examine transmigration mechanisms.
Transmigration can be considered as a transition from two-dimensional (2D) migration on a surface to three-dimensional (3D) migration in a confining environment surrounding the entire cells volume. Integrin-mediated crawling of leukocytes (15, 16, 17) and cancer cells (18, 19, 20) in 2D and 3D is well established, and amoeboid migration without adhesion or with integrin-independent adhesion was reported in both 2D (21) and 3D (16,22, 23, 24) conditions. At the molecular level, most migration models rely on actin cytoskeleton polymerization and contractility (24, 25, 26), and ion pumping was proposed as an alternative propulsion source for tumor cells in synthetic microchannels (27). Mechanistically, two main modes of migration groups can be distinguished that are based on either membrane treadmilling or cell deformations. Membrane treadmilling seems functional for cells swimming in a fluid (21,28), as well as for cells crawling on a substrate and cells confined in smooth channels deprived of topological features, both with and without adhesion (21,23,29). The relevance of membrane treadmilling for migration in the extracellular matrix is, in turn, less clear. In contrast, cell deformations by blebbing, contractions, or lamellipod projections are arguably functional for swimming, but clearly relevant in serrated microchannels with topographic features and in the extracellular matrix (22,30, 31, 32), in which they also allow propulsion in the absence of adhesion (22,30). Altogether, the state of the art of 2D and 3D migration mechanisms remains under debate, and working out a precise mechanism of transmigration that combines both 2D and 3D aspects is highly challenging. Our goal here is therefore limited to shed light on some mechanical aspects of the transition from 2D to 3D environments, such as adhesion, confinement, and propulsion force.
In vitro studies of transmigration are often probed with commercial Boyden chambers or Transwell assays (33), which consist of counting the number of cells crossing a microporous membrane separating two chambers. The effects of chemokines or adhesion molecules are tested by changing membrane coatings and soluble signals across the membrane. However, visualization of cells in situ is difficult and rarely performed (11), which hampers a precise analysis of cells phenotypes during the multistep process of transmigration. Here, we developed a method combining a microfluidic version of Transwell assays with optical microscopy and pressure control across the “microfluidic membrane.” The method allowed us to observe the transmigration process at single cell level and to assess forces implied during transmigration. The microfluidic devices were composed of horizontal confining microchannels separating two nonconfining chambers, and flows across the microfluidic membrane were regulated using pressure controllers and visualization of suspended nanotracers. Although these microfluidic devices also allow the application of chemical gradients (34, 35, 36, 37), we focused here on mechanical aspects of transmigration in absence of chemical cue. With primary human effector T lymphocytes, transmigration in microfluidic Transwells occurred spontaneously without chemokines gradients. Adhesion and actomyosin contractility were both crucial to squeeze the cells’ cytoplasm and nucleus into pores, but interestingly, not for fast 3D motility in smooth open channels. The control of pressure across the microfluidic Transwells showed that cell gripping in channels was far less efficient by friction than by integrin-mediated adhesion. Furthermore, the arrest of adherent cells was under pressure control rather than force control, which was consistent with an arrest of lamellipod growth. Altogether, transmigration in smooth microchannels may be explained by membrane treadmilling propulsion and rear squeezing and cell arrest under pressure by a destabilization of cell polarization.
Materials and methods
Cells
Whole blood from healthy adult donors was obtained from the “Établissement Français du Sang.” Peripheral blood mononuclear cells were recovered from the interface of a Ficoll-Paque PLUS (GE Healthcare, Pittsburgh, PA) gradient. T lymphocytes were isolated from peripheral blood mononuclear cells with Pan T cell isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany), then activated for 2 days with T Cell TransAct (Miltenyi Biotec), a polymeric nanomatrix conjugated to humanized CD3 and CD28 agonists. T lymphocytes were subsequently cultivated in Roswell Park Memorial Institute medium 1640 (Gibco by Thermo Fisher Scientific, Waltham, MA) supplemented with 25 mM GlutaMax (Gibco by Thermo Fisher Scientific), 10% fetal bovine serum (Gibco by Thermo Fisher Scientific) at 37°C, 5% CO2 in the presence of IL-2 (50 ng/mL; Miltenyi Biotec) and used 6–10 days after stimulation. For experiments with blebbistatin, cells resuspended in 50 μM blebbistatin solution prepared in culture medium. Before experiments, cells were concentrated to obtain 10 million cell/mL and were allowed to settle for 1 h in the incubator before injecting them into the microchannels, where they settled again for 30 min at 37°C before image acquisition.
Microfluidic channels and surface treatments
PDMS microchannels were fabricated using standard soft lithography. A positive mold was created with a negative photoresist SU-8 3000 (Microchem, Westborough, MA) on silicon wafers (Siltronix, Archamps, France), then replicas were molded in polydimethylsiloxane (PDMS) elastomer (Sylgard 184; Dow Corning, Midland, MI) and sealed on glass cover slides via plasma activation (Harrick Plasma, Ithaca, NY). The device is composed of two large channels with two inlets and two outlets punched with a 1.2 mm puncher (Harris Uni-Core; Sigma-Aldrich, St. Louis, MO). After sealing, the channels were incubated with 3-aminopropyltriethoxysilane (Sigma-Aldrich) for 2 h at 4°C, then rinsed with milliQ water, followed by drying under vacuum for 10 min and heating for 10 min on a 95°C hot plate. For adhesion experiments, the channels were incubated with 10 μg/mL human ICAM-1 (R&D Systems, Minneapolis, MN) for 1 h at room temperature, followed by a blocking solution containing 2.5% bovine serum album (BSA) (w/v; Axday, Dardilly, France) and 2.5% Pluronic acid F-108 (w/v; BASF, Ludwigshafen, Germany) in phosphate-buffered saline (PBS) for 30 min at room temperature. For adhesion-free surfaces, the channel was directly incubated with 5% F-127 Pluronic for 30 min at room temperature. Filling the channels with ICAM or Pluronic solutions was preceded by putting the device under vacuum for 5 min to later allow the liquid to enter the lengthy confinement channels by replacing trapped air. For hybrid experiments with adhesive large channels, nonadhesive, and confined channels, the channels were coated in two steps. First, large channels were treated with ICAM-1, and the confinement channels were kept dry by not putting the device in vacuum before ICAM incubation. Second, the whole device was passivated by rinsing and treating with Pluronic solution. The channels were finally rinsed with PBS and then culture media. For experiments with adhesive or nonadhesive confined channels, patterns were prepared by optical patterning. We used an inverted microscope (TI Eclipse; Nikon, Champigny-sur-Marne, France) coupled to an ultraviolet laser source and a Digital Micromirror Device (Primo; ALVEOLE, Paris, France) (38). Ultraviolet was projected on PEG-coated substrates in presence of a soluble photoactivator (PLPP; ALVEOLE, Paris, France) to activate the PEG layer where we want to make the surface adherent. Samples were then rinsed with PBS solution and incubated with protein A for 1 h at room temperature, then rinsed with PBS and passivated with BSA 2%, followed by incubation with ICAM-1 for 1 h at room temperature, and finally rinsed with PBS and passivated with BSA 2% for 15 min at room temperature before the device is rinsed with PBS and media (39).
Microfluidic setup and pressure control
The device is composed of two large channels of 67 μm height connected by confinement channels of 500 μm length and 2 or 4 μm height depending on the experiment. The large channels are nonconfining for cells and allow 2D migration. The confinement channels allow a 3D migration in sets of microchannels of variable widths (w = 2, 4, 6, and 8 μm).
Cells were loaded in one of the large channels, allowing transmigration in one direction to reduce the probability of more than one cell transmigrating in the same channel. To control drift and apply pressure, the four inlets and outlets were connected to a microfluidic pressure control system (MFCS-EZ; Fluigent, Le Kremlin-Bicêtre, France). Fluorescent beads of 200 nm in diameter (F8809; Thermo Fisher Scientific) were used to abrogate any drift and control equilibrium and pressure variation experiments.
Imaging and data analysis
Experiments were performed on an inverted Zeiss Z1 automated microscope (Carl Zeiss, Oberkochen, Germany) equipped with a CoolSnap HQ CCD camera (Photometrics, Tucson, AZ) and piloted by μManager1.4. Plan-Apochromat 10×/0.3, 20×/0.8, and 63×/1.4 objectives were used for bright-field and fluorescent modes, and a Neofluar 63×/1.25 antiflex was used for reflection interference contrast microscopy (RICM) mode, in combination with a narrow band-pass filter (λ = 546 nm ± 12 nm). Bright-field images for cell tracking were taken every 10 s, and RICM images were taken every 3 s. 3D imaging was performed on cells stained with a lipophilic tracer DiO (Invitrogen, Carlsbad, CA) or with anti-HLA-ABC (Biolegend, San Diego, CA). Live 3D images were recorded using a Zeiss inverted microscope equipped with a Yokogawa CSUX1 spinning-disk and controlled by Volocity software (UltraView VOX; Perkin-Elmer, Waltham, MA). Cells were tracked using the FIJI plugin Trackmate (40). Tracks were exported, and further analysis and plots were performed using GraphPad Prism Software (La Jolla, CA).
Results
Transmigration occurs without chemotaxis
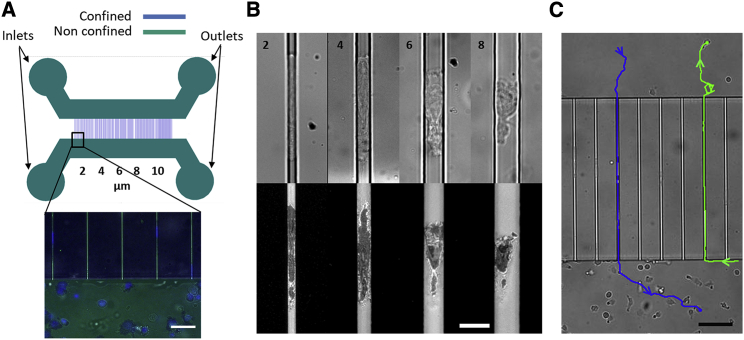
Microfluidic Transwell devices consisted of two large independent channels, in which cells were not confined, separated by a comb of narrow microchannels in lieu of the membrane in usual Transwell assays (Fig. 1 A). Although the thickness of the membrane in Transwell assays is around 20 μm, we used here channels of 500 μm length to allow the measurement of 3D migration properties, such as speed and forward-thrusting force. The transition between the unconfined migration zone (large channels) and confined migration zone (narrow channels) consisted of orthogonally crossing parallelepipedic channels. These channels with straight walls are later called smooth channels, in opposition to serrated channels used in the literature to study cell migration with the help of topographic features (30). The influence of the geometry of the transition zone was not investigated here, except for the size of the cross section of the confining channels. In channels of cross section 8 × 4 μm2, cells were confined between two planes by the lower and upper surfaces of the channel, but not necessarily by the lateral walls, which they may contact one at a time intermittently (Fig. 1 B). For channels of cross sections 6 × 4 μm2, cells were touching all four surrounding walls at all times, but most of them did not occupy the whole cross section of the channel (Fig. 1 B). For channels of cross sections 4 × 4 and 4 × 2 μm2, cells filled the cross section of channels and formed a plug (Fig. 1 B). In what follows, we will call 2D migration the case of cells in contact with a single surface and 3D migration the case of cells surrounded by four walls. The devices were either coated by ICAM-1, which mediated specific adhesion with integrins LFA-1, or Pluronic F127 to deter adhesion (21,23). After seeding, cells either swam or crawled with a random 2D motion on the nonadherent or adherent, respectively, surface of large channels. Upon random encounters with the entry of narrow microchannels or pores, cells had the opportunity to explore the pore with their protruding leading edge and then to choose between fulfilling entry or escaping away from the pore (Videos S1 and S2). Interestingly, cells were able to transmigrate across microchannels of all cross sections (Fig. 1 B), and transmigration occurred both ways (Fig. 1 C; Videos S1 and S2). These observations suggest that the migration machinery of effector T lymphocytes allows spontaneous transition between 2D and 3D migration, so that lymphocytes are capable of exploring efficiently both 2D and 3D without chemotactic guidance. This capacity is typically important for lymphocytes to scan thoroughly lymph nodes and inflamed tissues. The evidence of efficient transmigration without chemotaxis underlines also that Transwell assays are much more than chemotaxis assays. Transmigration depends on adhesion, migration, deformability, and chemokinesis, and the observation of cells in Transwells provides a powerful way to disentangle the roles of these multiple factors on transmigration.
Figure 1.
Transmigration without chemotaxis. (A) Microfluidic Transwell device for transmigration and 3D migration. (Top) Microfluidic device composed of two large nonconfining channels h = 67 μm with one inlet and one outlet each (green) connected by sets of narrow confining channels of variable widths (h = 2 or 4 μm; w = 2, 4, 6, 8, and 10 μm) (blue). (Bottom) Superimposition of bright-field (green), reflection interference contrast microscopy (RICM) (black), and fluorescence images revealing adhesive 2D and 3D migration of cells on ICAM-1-treated surface with cells nuclei stained using DAPI (blue). Scale bars, 20 μm. (B, top) Bright-field and (bottom) RICM images of T cells migrating in channels of variable widths (h = 4 μm; w = 2, 4, 6, and 8 μm) showing migration at different confinement levels. Scale bars, 10 μm. (C) T cells spontaneously transmigrate on ICAM-1-coated channels. Bright-field image showing manual tracks of two cells migrating in two different directions (blue: top to bottom, green: bottom to top). Scale bars, 40 μm. See also Videos S1 and S2. To see this figure in color, go online.
Primary human effector T cells transmigrating for several hours in an ICAM-1 coated Transwell device with 4 × 4 μm2 microchannels in Bright field mode.
RICM image sequence of primary human effector T cells migrating on 2D surface and transmigrating in smooth 4 × 4 μm2 ICAM-1 coated microchannels (Part 1). Bright field (left) and RICM (right) image sequence of a primary human effector T cell entering a smooth 4 × 4 μm2 ICAM-1 coated microchannel (Part2).
Cells have an intrinsic propensity to penetrate 3D confining environments
Whether cells switch between different migration modes in 2D or 3D environments is still debated and not easy to sort out. However, it is possible to determine which environment is more favorable for cell migration by estimating the probability of transition between 2D and 3D, or the transmigration efficiency of cells. For this purpose, we considered all the events of cells encountering the entry of a pore, and determined the probability of transmigration, PTM, as the ratio between the numbers of events leading to a complete entry of cell in the pore versus the total number of encounters between a cell and a pore entry:
| (1) |
To consider events of transmigration by self-propulsion of cells and avoid bias due to residual flow pushing cells in or out of channels, flows were precisely cancelled by controlling the pressure at the entries of the device and by observing the motion of submicrometric fluorescent beads suspended in solution. The uncertainty of pressure across the device to obtain no flow conditions was as low as 0.001 mbar, which corresponds to a maximal net force on cell of 1.6 pN in channels of cross section 4 × 4 μm2. The probability of transmigration PTM decreased with confinement and eventually vanished for the smallest pore entries of cross section 2 × 2 μm2 (Fig. 2 A), and the time of entry of cells achieving full penetration increased with smaller pore size (Fig. 2 B). For channels of cross sections 4 × 4, 6 × 4, and 8 × 4 μm2, the probability was higher than 50%, which supports that the transition of migration from 2D to 3D is not the exception but the preferential choice of lymphocytes. The idea that a confining pore is necessarily an obstacle for migration is therefore misleading. A better view seems to be that polarized effector lymphocytes, which are knowingly equipped to migrate in 2D and 3D, have a marked propensity to dive into anfractuosities of cross sections as small as 16 μm2. This propensity may be an important asset to explore thoroughly microporous environments such as infected tissues.
Figure 2.
Effect of pore size on transmigration probability. (A) Probability of transmigration versus the cross section of pore. (B) Cell entry time for full penetration versus the cross section of pore. Each point corresponds to one cell. (C and D) Probability of cell entry in function of cell protrusion length already penetrated in the pore (C) and cell volume already penetrated in the pore (D). Ncell = 75 (2 × 2 μm), 80 (2 × 4 μm), 91(4 × 2 μm), 97 (4 × 4 μm), 115 (4 × 6 μm), and 94 (4 × 8 μm). Nexp = 3. To see this figure in color, go online.
Nucleus deformation slows down and eventually hampers transmigration
To analyze further the process of transmigration, we then performed acquisitions at higher magnification to measure the length of cell protrusion inside the pore, l, and determine the probability for a cell to fully penetrate a pore as a function of its protrusion length, (l). The probability (l) corresponds to the ratio of the number of cells that eventually achieve penetration in the pore versus all the cells reaching a penetration length l.
| (2) |
The dependence of (l) vs. l is reported in Fig. 2 C for different pore sizes. The initial value for l = 0, (0), corresponds to PTM of Fig. 2 A. (0) decreases with smaller pores, as previously noted. For all pore sizes, increases with l and eventually reaches 100%. This general increase means that the deeper the cell penetration into the pore, the higher the probability to achieve full entry. The competition between 2D and 3D migration during transmigration is continuously tilting to the advantage of 3D migration with the penetration length l. Interestingly, the transition is strongly nonlinear with l. The curves of Fig. 2 C for different pore sizes display a pattern in three phases, called thereafter phases I, II, and III. In phase I, the probability remains stable with l, meaning that the entry of the leading edge in phase I has no critical input into the final outcome. In phase II, (l) increases steadily with l and reaches a value close to 90%. Beyond phase II, the outcome of the cell seems almost settled and penetration irreversible. In phase III, the rest of the cell body had just to enter the pore and fulfill cell entry. The boundaries of the three phases are significantly different when was plotted versus the length of cell that has entered the pore, l (Fig. 2 C). In contrast, by plotting (l) versus the volume of cell that entered the pore,, the boundaries between the three phases collapsed at similar positions for all pores sizes (Fig. 2 D). This suggests that the different entry phases are driven by similar critical fractions of cell volume in the pore. It is known from the literature that a critical step for a cell to enter a constriction consists of squeezing the nucleus into the constriction because the nucleus is the stiffest part of the cell. With an average diameter of 5 μm, the nucleus is deformed in all pores here because the maximal height of microchannels was 4 μm. To precise the role of the nucleus deformation in the different phases of entry, we then stained the nucleus (Video S3) and determined the volume of the cell in the pores before the entry of the nucleus in the pore, and the volume of cell body in the pore after completion of nucleus entry in the pore. On average, 25% of cell volume entered the pore before the initiation of nucleus entry, and 66% of cell volume was in the pore right after completion of nucleus entry. These volume fractions were calculated considering an average cell volume of 370 μm3, as assessed from the average cell diameter of 8.7 μm measured in microscopy with round inactive cells. The nucleus-related boundaries matched well with the boundaries of phases I, II, and III in Fig. 2 D. This supports that phase I corresponds to the entry of the cell leading edge before the nucleus, phase II to the entry of the nucleus, and phase III to the entry of the trailing edge. Altogether, the probability of transmigration depends strongly on the pore size, and the nucleus deformation is controlling transmigration probability and dynamics of cell entry in confining pores. Before the onset of nucleus entry, the choice to transmigrate remains independent of penetration length, whereas after full nucleus entry, transmigration is mostly irreversible.
T cells with DAPI stained nuclei migrating on ICAM-1 coated devices with 4 × 4 μm2 microchannels (Part 1) and 4 × 2 μm2 (Part 2). Images are superimpositions of bright field (grey), RICM (red for 4 × 4 μm2 and green for 4 × 2 μm2) and DAPI fluorescence (blue) images. Dark zones in RICM reveal adhesive fingerprint in 2D, and close contact or adhesive fingerprint in 3D of cells.
Speed and treadmilling machinery are largely similar in 3D and in 2D
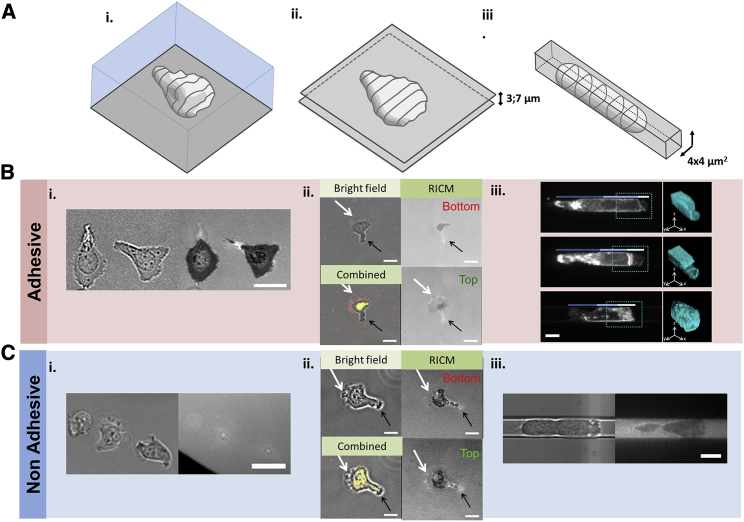
To compare further migration in 2D or 3D, we then monitored cells speed (Fig. 3) and cell morphology in various geometrical conditions (Fig. 4). Cell speed had an optimum with confinement (Fig. 3 A), which shows that apparent strong perturbations of cell morphology are not detrimental to propulsion. More precisely, on an adherent 2D substrate, cells displayed an adherent and widely spread lamellipod (Fig. 4 Bi) and crawled at 20 μm/min, arguably propelled by treadmilling of their envelope linked to the substrate (21). In a channel of cross section 4 × 8 μm2, cells were squeezed between the upper and lower walls distant by 4 μm, but not by the lateral walls, and speed was slightly higher. This case is similar to cells squeezed between two plates, for which we confirmed a significant increase of speed (Fig. S1). Cells in contact with two adherent surfaces had the opportunity to develop two lamellipods, which may explain the difference with cells crawling with a single lamellipod on a single surface. We therefore performed an original experiment of dual RICM microscopy to image cells contact area on both plates. This approach revealed that cells developed only a single lamellipod at a time, either on the upper or lower plate (Fig. 4 Bii), which suggests that the coexistence of two lamellipods is unstable. Cell conformations are, in the end, similar between two plates and on a 2D substrate, and the slight speed difference cannot be linked to an evident morphology difference. In channels of cross sections 4 × 6 and 4 × 4 μm2, cells were confined between four walls, and the average speed in these 3D conditions was significantly higher than for cells crawling in 2D (Fig. 3 A), as previously observed for Jurkat cells and lymphocytes CD4+ (34). Cell morphology here was strongly perturbed as compared to the cases of cells on 2D substrates or between two plates. Instead of a thin widely spread lamellipod, cell front seemingly formed a plug occupying the cross section of the channel, as previously observed with neutrophils (Fig. 4 Biii; (41)). Nevertheless, sheet-like protrusions reminiscent of lamellipods were still detectable, which suggests that treadmilling machinery is still propulsive, potentially together with other confinement specific mechanisms (41). Finally, in the 4 × 2 μm2 channels, the average speed was significantly lower than in 2D and other 3D cases, which suggests that extreme deformations are detrimental for migration. Concerning nonadherent cases (Fig. 4 C), the contact of cells with the environment was almost null on 2D substrate and increased for cells squeezed in plate and then in microchannels. However, cell polarization and envelope treadmilling were conserved in all cases despite strong global morphology changes (21). Altogether, these data support further that envelope treadmilling can participate in propulsion in 3D than in 2D and explain why 3D and 2D environments favor speeds of similar magnitude.
Figure 3.
Effect of confinement, contractility, and adhesion on 3D speed and probability of transmigration. (A) Cell speed in microchannels of different cross sections and on a surface (2D) without and with 50 μM blebbistatin. Data represent means and SEM, ncells > 90 per condition, p value of t-test <0.01 (∗∗), <0.0001 (∗∗∗∗), and >0.1 (ns), Student’s t-test. (B) Probability of transmigration versus the cross section of pore for control cells in fully adherent device (control, solid dots), cells treated with 50 μM blebbistatin in fully adherent device (blebbistatin, star dots), control cells in hybrid adherent device (hybrid, open dots), and controls cells in fully nonadherent device (no adhesion, crossed round dots). Ncells > 80 per data point. Nexp = 3. To see this figure in color, go online.
Figure 4.
Morphologies of migrating cells from 2D to 3D migration. (A) Schematic of cell morphology and (B and C) images of cells in different topological environments and in adhesive (B) and nonadhesive (C) conditions. (i) Cells not confined on a substrate (2D migration) (scale bar, 20 µm), (ii) cells confined between two plates (scale bar, 10 µm), and (iii) cells confined in a narrow microchannel (3D migration) (scale bar, 4 µm); bright-field and RICM images are in grayscale except in combined images of (ii), in which RICM is in red for lower plate and green for upper plate. Confocal microscopy of cells stained with DiO (top and middle) and with anti-HLA (bottom) is shown. (iii) Images are reported with average z projection (left) and 3D (right). Tricolored lines: white, light blue, and dark blue indicate positions of lamellipod, nucleus, and back of the cell, respectively; blue dashed squares indicate the zone represented in 3D mainly for the front leading edge. In adherent conditions, cells display a single wide adherent lamellipod in cell front on a plate and between two plates and short adherent protrusions forming a bulky cell front in microchannels. In nonadherent conditions, cells have similar morphologies with nonadherent and less-spread lamellipods. To see this figure in color, go online.
Adhesion is required outside the pore for transmigration
Lymphocytes can migrate with and without adhesion on 2D substrate (21) and in 3D environments (30,42). We recently proposed that envelope treadmilling and frictional coupling with a fluid or solid environment could explain motility in suspension and in confinement (21). To test the role of adhesion and friction in the transition between 2D and 3D migration, we performed two types of experiments in Transwell of pore sizes 4 × 2 and 4 × 4 μm2. In a first case, the devices were entirely treated with antiadhesive coatings of Pluronic F-127. Cells swam in the collector channels and never penetrated spontaneously into pores (Fig. 3 B). Deformation of cell body and nucleus into pores was therefore not possible without adhesion. In a second case, we made a hybrid treatment for the microfluidic Transwell devices by coating the large collector channels with adhesive ICAM-1 proteins and the confining microchannels with antiadhesive Pluronic F-127 (Fig. S2). This hybrid coating was achieved in two steps: first, by treating the large collector channels with ICAM-1 while keeping the confinement channels dry, and second, by treating the whole device with a passivating solution. In channels of cross section 2 × 2 and 4 × 2 μm2, cells crawled in the collector channels and attempted frequently to enter pores by sending a projection. However, penetration was stopped when the nucleus arrived at the entrance of the pore (Video S4) and the probability of transmigration was null (Fig. 3 B). The difference with fully adherent devices shows that adhesion in pores is participating in the process of transmigration. However, in channels of larger cross section (4 × 4, 4 × 6, and 4 × 8 μm2), the probability of transmigration was not null and rose to 50% (Fig. 3 B). Altogether, adhesion is important for transmigration but is not strictly necessary, as penetration in a nonadherent pore is possible if cells adhere outside the pore and pores are not too narrow.
Primary effector human T cells transmigration in device with large channels treated with adherent ICAM-1 coatings and microchannels of cross section 4 × 2 μm2 treated with non-adherent Pluronic F127 coating. Transmigration is aborted just before complete entry.
Contractility is important for pore penetration, but not for 3D migration, in smooth channels
To test the internal cellular mechanism involved in transmigration, we then performed perturbation experiments with blebbistatin, a potent inhibitor of actomyosin contractility. In terms of propulsion, cell speed showed a marked optimum with confinement. Furthermore, whereas 2D speed decreased from 20 to 12 μm/min with 50 μM blebbistatin, speed remained surprisingly unaffected for 3D confined conditions in channels of cross section 4 × 6, 4 × 4, and 4 × 2 μm2 (Fig. 3 A). This result means that actin contractility does not play a major role in the propulsion of lymphocytes in a smooth channel, at least in the absence of opposing force. In contrast, the addition of blebbistatin significantly decreased the probability of transmigration for all pore sizes (Fig. 3 B). Actomyosin contractility is therefore marginal for 3D migration in open smooth channels but crucial for penetration in confining pores.
Lymphocytes develop a propulsion force of 3 nN in confining channels of cross section 4 × 2 μm2
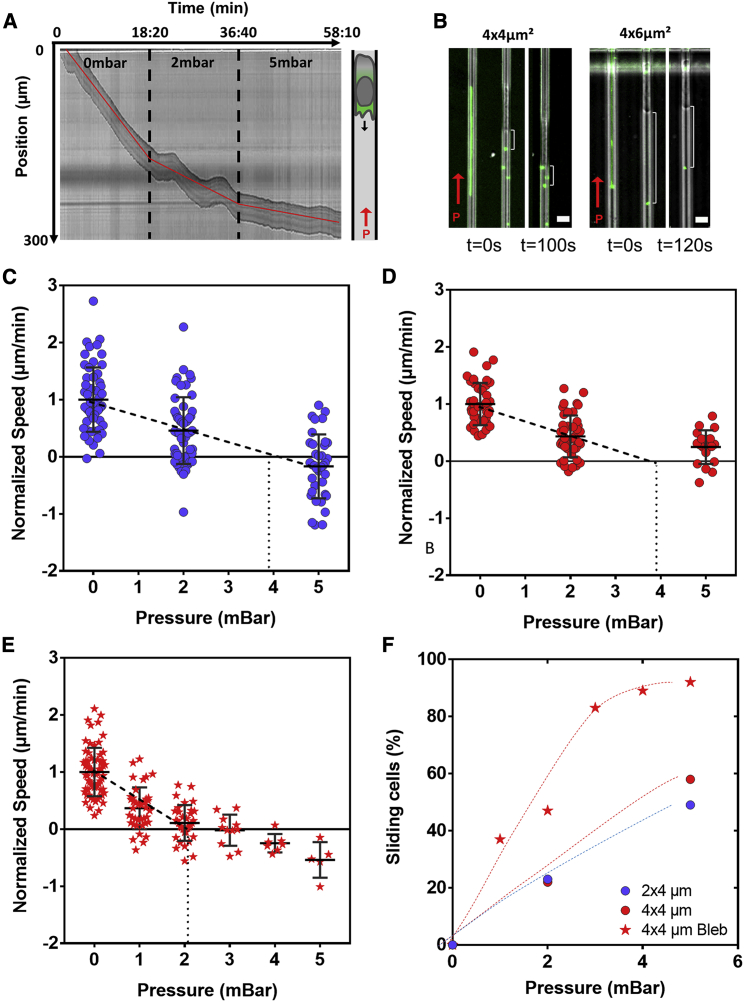
Previous results showed that translocation of cell body in a constriction is a critical step of transmigration. To determine the force that cells can develop to pull themselves into a pore, we then imposed different pressures across the microfluidic Transwell device and monitored the speed of cells under opposing pressure (Fig. 5 A; Video S5). To control the force acting on cells with pressure, it is important that cells act as a watertight plug in the channels, i.e., fluid leakages around the cell in channel corners (43) and through the cells by fluid transport mediated via aquaporin or ion channels (27) must be negligible. We used nanoparticles suspended in the medium to check flow around cells in channels under pressure. Particles were always immobile around cells in channels of cross section 2 × 4 and 4 × 4 μm2, but not in channels of cross section 6 × 4 μm2 or more (Fig. 5 B). Therefore, stall forces were only measured in 2 × 4 and 4 × 4 μm2 channels. In absence of opposing force, cell speed in channel was oscillating with a period around 125 ± 20 s (Fig. S3). To get relevant mean values, we therefore monitored cell speed for at least 400 s for each given pressure step. In channels of cross section 2 × 4 μm2 (Fig. 5 C), the average speed decreased from 12 to 5 μm/min with an opposing pressure of 2 mbar (or force of 1.6 nN) and reversed to negative values at 5 mbar. Furthermore, the fraction of cells detaching from the channels and sliding backward at high speed under pressure increased from 0 to 25 and 50% for pressure of 0, 2, and 5 mbar, respectively (Fig. 5 F). By linear extrapolation of average speed values at 0 and 2 mbar, for which a majority of cells remained adherent, stalling conditions correspond to 4 mbar or 3.2 nN.
Figure 5.
Confined migration against pressure. (A) Representative kymograph of a cell migrating in 4 × 4 μm2 ICAM-1-treated channel under 0, 2, and 5 mbar opposing pressure (left) as schematized on the right; cell migration direction is toward the bottom as indicated by the black arrow and under opposing pressure as indicated by the red arrow. Dashed black lines mark the beginning of pressure exertion, and red lines are visual representation of the changing slope (see also Video S5). (B) Superimposition of phase contrast and fluorescence images of fluorescent nanobeads and cells in 4 × 4 μm2 (left) and 4 × 6 μm2 (right) ICAM-1-coated channels under pressure = 2 mbar exerted in the direction of the red arrow (scale bars, 10 μm). Fluorescent signal shows line tracks of beads movement in channels free of cells (right, in both channel sizes) and beads pushed by cell against pressure as cell advances against pressure in 4 × 4 μm2 channels over a period of 120 s. White accolades show constant cell-bead distance reflecting a plug-like behavior (middle and left in 4 × 4 μm2) and bead movement with pressure revealed by smaller cell-bead distance over a period of 100 s, reflecting fluid leakage. (C and D) Normalized speed for cells migrating in 2 × 4 and 4 × 4 μm2 channels respectively, under 0, 2, and 5 mbar opposing pressure. Linear extrapolation of average speed values (dashed lines) shows that a stall condition is found at 4 mbar (n > 50). (E) Normalized speed of blebbistatin-treated cells migrating in 4 × 4 μm2 channels under 0–5 mbar opposing pressure. Linear extrapolation of average speed values (dashed lines) shows that stall condition is found at 2 mbar (n = 66). (F) Percentage of sliding control cells in 2 × 4 μm2 channels (violet circles) and control (red circles) and blebbistatin-treated cells (stars) in 4 × 4 μm2 channels. To see this figure in color, go online.
Bright field image sequence of a primary human effector T cell transmigrating in a 4 × 4 μm2 ICAM-1 coated microchannel exposed to opposing pressure of 2 and 5 mbar. White arrows indicate the direction of opposing pressure (small arrow: 2 mbar, large arrow: 5 mbar). White horizontal lines indicate cell location when the pressure of 2 and 5 mbar are applied. Time in mins. Scale bars, 20 μm.
Stalling conditions are under pressure control rather than force control
To discriminate the role of pressure or force on stalling conditions, we then performed experiments in channels of cross section 4 × 4 μm2 (Fig. 5 D). By linear extrapolation of average speed values at 0 and 2 mbar, for which a majority of cells remained adherent, the stalling conditions corresponded to 4 mbar or 6.4 nN. It was not possible to vary further the channel size because of pressure leakage in larger channels and altered migration in smaller channels. However, the results with two channel sizes support that stalling conditions are under pressure control rather than under force control. Interestingly, the growth of a lamellipod mediated by polymerizing actin filaments is also under pressure control because the number of filaments growing against the membrane in cell front is increasing linearly with the surface of the cell front. Furthermore, the stalling force of a few nanonewtons measured in our experiments are consistent with the push of a polymerizing actin gel (44,45). All of this suggests that stalling conditions correspond to the arrest of the polymerization-mediated lamellipod growth.
Contractility increases stall force
To test the role of actomyosin contractility on stalling conditions, we then performed experiments with 50 μM blebbistatin. Stall pressure in channels of cross section 4 × 4 μm2 significantly decreased twofold with blebbistatin (Fig. 5 E). Hence, although actomyosin contractility was hardly influencing the speed of lymphocytes in open channels in absence of opposing force, contractility plays a strong role when it comes to resisting an opposing force. Interestingly, contractility was also crucial for the penetration into pores, i.e., to overcome the resistance of cell/nucleus squeezing. Altogether, these results suggest that contractility is dispensable for reaching maximal speed in smooth channels in the absence of force but essential to maintaining efficient propulsion against a resistance to motion.
Adhesion is unstable for stalled cells
Previous stalling experiments were performed in channels with adhesive ICAM-1 coatings. Despite favorable adhesive conditions, some cells under pressure detached and then slid out of channels at high speed (Fig. 5 F). The percentage of detaching cells for a given pressure was similar in channels of cross section 4 × 2 and 4 × 4 μm2, although the adhesion area was twice smaller and the applied force twice stronger in channels of cross section 4 × 4 μm2. The difference of the cell adherent fingerprint does not explain cell detachment, and cell detachments in stalling conditions must imply a decrease of adhesiveness itself. Interestingly, although cell detachments with blebbistatin occurred at lower pressure, they mostly occurred in stalling conditions. Because blebbistatin itself does not significantly alter adhesiveness (Fig. S4), stalling condition again seems linked to adhesiveness failure.
Adhesion is crucial for 3D migration in a smooth channel against a force
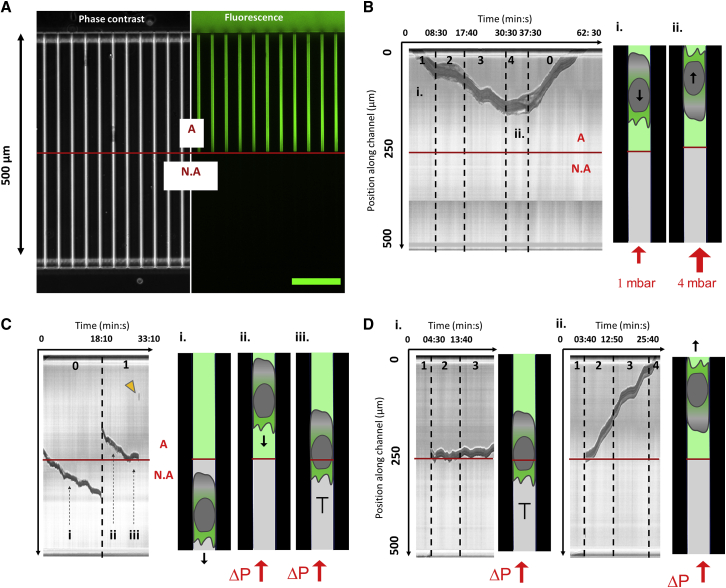
To further test the role of adhesion in propulsion against a force, we then performed stalling experiments with devices patterned with different adhesive conditions. The first half of the microchannels was coated with ICAM-1, and the second half was not (Fig. 6 A). This adhesive pattern was implemented using a “subtractive printing protocol” based on the light-induced multiple adsorption (10). When pressure was applied on cells in the adherent zone, they pursued progression up to the stall pressure of 4 mbar. Beyond the stall pressure, cells arrested, detached, or reversed polarization. This latter case of polarization reversal is illustrated in Fig. 6 B (and Video S6). Without pressure, cells pursued progression beyond the frontier between adhesive and nonadhesive zones (Fig. 6 C; Video S6), which is consistent with the capacity of cells to perform 3D migration with and without adhesion. In contrast, when pressure was applied, cells in nonadherent zones slid backward at high speed even for a pressure as low as 1 mbar (Fig. 6 C; Video S6). Detached cells were either washed out of the microchannel or reattached in the adherent zone, as in Fig. 6 C. In this latter case, cells could resume their motion against flow, which shows that polarization orientation was conserved upon sliding. They then started a new cycle of crawling against pressure, arrest at the adhesive/non adhesive (A/NA) frontier and potential detachment (Fig. 6 C; Video S6). Interestingly, swimming cells in suspension could also be pushed by pressure into microchannels from the nonadherent entry (Fig. 6 D). They were then sliding at high speed in the nonadherent zone and at times arrested in the adherent zone where they initiated migration. Because suspended cells entered with random orientations, the ones with polarization facing pressure were stalled at the A/NA frontier (Fig. 6 Di; Video S6), whereas the ones with opposite polarization simply crawled out of the channels (Fig. 6 Dii; Video S6). Altogether, these data show that cell without specific adhesion with walls can sustain a fast motility but not resist a force.
Figure 6.
Propulsion at the frontier between adhesive and nonadhesive channels. (A) Patterning of adhesion in microfluidic Transwell. (Left) Phase contrast and (right) fluorescence images showing 4 × 4 μm2 channels with upper adhesive and bottom nonadhesive zones. Fluorescent signal of FITC-protein A on which ICAM-1 molecules can bind assesses presence of specific adhesion (marked “A”) in the upper part of the channel and absence of specific adhesion (marked NA) in the lower part of the channel. Scale bar, 100 µm. (B) Polarization reversion by pressure in adherent channels. (Left) Representative kymograph of a cell advancing in adhesive zone under pressure. Phases of the cell advancing against 1 mbar and stalled at 4 mbar are pointed by the marks (i) and (ii). (Right) Schematic of a cell advancing against 1 mbar in phase (i) and advancing in opposite direction after polarization reversal in phase (ii) (see also Video S6). (C) Stalling and detachment at adhesion-nonadhesion frontier. (Left) Representative kymograph of a cell crossing the frontier between adherent and nonadherent zones in absence of pressure (i), sliding backward and reattaching in the adherent zone in phase (ii), and stalled at the frontier A/NA under 1 mbar (iii), then detaching. Yellow arrow shows the cell sliding out of the channel. (Right) Schematic of a cell progressing without pressure in nonadherent channel in phase (i) reattached in advancing against 1 mbar in phase (ii) and stalled at the frontier in phase (iii) (see also Video S6). (D) Transition between passive push and active crawling in confinement. Representative kymographs of cells pushed in nonadherent channels at 2 mbar and arresting at the A/NA frontier are given. Cell polarization pointed either toward increasing pressure and cells being stalled at the A/NA frontier (i) or toward decreasing pressure and cells crawling in adherent conditions out of the channels. The red line marks the limit between the adherent and nonadherent zones (see also Video S6). To see this figure in color, go online.
Part 1- Polarization reversion by pressure in adherent channels (Fig. 6 B). Part 2 - Stalling and detachment at adhesion/non-adhesion frontier. The cell crosses the frontier between adherent and non-adherent zones in absence of pressure. When a 1 mbar pressure is applied, the cell slides backwards, re-attaches in the adherent zone, is stalled at the A/NA frontier and eventually detaches (Fig. 6 C). Part 3- Transition between passive push and active crawling in confinement. Cells pushed in non-adherent channels at 2 mbar, arrest at the A/NA frontier. Depending on their intrinsic polarization, they resume crawling against pressure and are stalled at the A/NA frontier (left) (Fig. 6 Di), or crawl in adherent zones out of the channels (right) (Fig. 6 Dii).
Discussion
Microfluidic Transwells allow one to disentangle the effects of chemotaxis, chemokinesis, deformability, and adhesion. Emigration from blood vessels and transendothelial migration of immune and cancer cells are critical steps of inflammatory responses and metastasis dissemination. In immunology, there is no innate or adaptive response unless leukocytes cross blood vessels. The mechanisms regulating transmigration include complex biochemical and mechanical processes that are difficult to decipher in vivo (46). In vitro reductionist approaches are therefore instrumental for deciphering core mechanisms, although extrapolations to in vivo situations are always arguable (20). Here, we quantified cells deformations and propulsion during cell penetration in smooth microchannels using videomicroscopy and microfluidic Transwell devices (27,34,35,47). As compared to regular Transwell assays, our method allows one to clearly distinguish the contribution of adhesion, autonomous migration, deformation, or chemotaxis. Although chemical gradients can also be applied (34,35,37), we focused here on the mechanical stimuli of transmigration and showed that T lymphocytes have an intrinsic capability to transmigrate and explore confined spaces without chemotactic cues, which may be valuable for scanning lymph nodes and infected tissues in vivo.
Envelope treadmilling is conserved independently of adhesion and confinement conditions
A hallmark of cell transmigration is the drastic change of cell shape, knowing that cell shape itself is tightly linked to migration mechanisms (20,48). The leading edge of amoeboid cells crawling on an adherent surface displays typically a thin sheet-like lamellipod (49,50), whose flatness results both from a templating effect of the substrate and from the tendency of ARP2/3-mediated actin polymerization to form sheet-like protrusions in leukocytes leading edges (21,51). Cells confined between two plates and in tubes have a squeezed cell body; however, their leading edge also displayed a single lamellipod on the upper or lower plate, alternatively (Fig. 4). For cells confined in tubes, the leading edge was bulkier, as reported for neutrophils (41), but flat lamellipod-like extensions were still present. The main structural elements of cell polarization were conserved in confined and unconfined configurations and with or without adhesion. Besides, previous studies attested that the retrograde treadmilling of cortical actin was conserved in cells leading edge with flat (21) or bulky morphologies (23,30,41). Altogether, propulsion by envelope treadmilling is operational during all steps of cells transmigration.
3D migration in smooth channels is consistent with propulsion by envelope treadmilling
Although one study defended propulsion by ion channels in 3D confining channels (27), most propulsion mechanisms are based on actomyosin dynamics. The mechanisms that take advantage of topographic features of the environment (22,30) are not operant in our channels with smooth walls. Propulsion by blebbing (22,25,52) is also not relevant in our assays because effector T lymphocytes did not bleb. In turn, propulsion by a compressive effect of actin polymerization in bulky lamellipods (41) and propulsion by envelope treadmilling coupled to adhesion or friction to walls (21,23,28,53) are potentially functional. Propulsion by envelope treadmilling was particularly relevant here to explain that cell speed hardly changed despite strong perturbations of cells conformation. Within this model, cell speed is indeed largely set by an envelope retrograde flow speed that is conserved in 2D and 3D migration.
3D migration speed increases with moderate confinement and depends hardly on actomyosin contractility in smooth open channels
Treadmilling of cortical actin was reported to be activated by actin polymerization (15,21,54) and/or contractility (23,28,55) with different cellular models. For leukocytes, treadmilling seems to result from a combination of actin polymerization in leading edge and actomyosin contractility in trailing edge (56). Although contractility was repeatedly found important for amoeboid propulsion, as for keratocytes (57) in 2D and for lymphocytes in 2D and 3D environments (34,42,58), our results with human primary T lymphocytes showed a contractility effect on 2D, but not 3D, speed. The studies reporting an effect of contractility on 3D lymphocyte migration (34) concerned devices coated by VCAM-1, a ligand of integrin VLA-4, whereas our devices were coated by ICAM-1, a ligand of integrin LFA-1. The trailing edge of lymphocytes is adherent on VCAM-1 and detached on ICAM-1 (50,59, 60, 61). A specific effect of contractility on speed in VCAM-1 devices may result from a specific need to detach cells trailing edges. Overall, the speed of T lymphocytes in smooth and open microchannels seems mainly controlled by actin polymerization when the trailing edge is not adherent. Interestingly, this predominance of polymerization versus contractility on propulsion was also observed for swimming T lymphocytes (21) and mesenchymal cells (HOS and NIH-3T3) in microchannels (62). The increase of lymphocyte speed with moderate confinement is also consistent with previous studies (34).
Transmigration is conditioned by the critical penetration of nucleus
Measurements of transmigration probability revealed a process in three distinct phases. The first phase of exploration of pore entries by the lamellipod is crucial to initiate transmigration but has no impact on the completion of the transmigration. In contrast, the second phase of the penetration of the nucleus is critical, transmigration being successful in 90% of cases when nucleus was in the pore. This critical role of nucleus is consistent with many previous results on cancer cells and leukocytes (11,13,16,17,25,26,63,64). The lower limit pore size of 4–8 μm2 determined in our microfluidic Transwells is also consistent with data from regular Transwell assays (11). Finally, the third phase of entry of the trailing edge has hardly any impact on transmigration probability, as also reported with dendritic cells (26).
Transmigration is mediated by adhesive envelope treadmilling and contractility
Whereas adhesion was repeatedly found dispensable for lymphocyte 3D migration (10,16,21,24), it played a crucial role for transmigration in our microfluidic Transwells. Cell penetration was less probable when the pores were nonadherent but still possible if the exterior was adherent. This is consistent with propulsion by envelope treadmilling, for which momentum transfer is more efficient with adhesion than with friction. Hence, during the first and second phases, the propulsion of the cell front is lessened in a nonadherent pore, but treadmilling in cell rear outside the pore still pushes the cell (Fig. 7 A). When the whole device is nonadherent, propulsion by envelope treadmilling is altered in the cell front and rear. Concerning actin contractility, it had hardly any effect on speed in smooth channels, whereas its impact on the transmigration probability was significant. At minimum, contractility may intervene in transmigration either by fueling the treadmilling machinery or by squeezing the back of the cell, which may generate an intracellular pressure gradient and internal forward flow (14,16,65). In both cases, blebbistatin would directly favor cell progression and indirectly nucleus penetration in a pore. Contractility may also be involved via a positive signaling forward loop, in which nucleus deformations would trigger myosin activity and facilitate nuclear deformation (66,67). In turn, one study proposed that nucleus deformation was promoted by actin polymerization and ARP2/3-dependent mechanisms (12). Identification of the relevant mechanisms will require further investigations.
Figure 7.
Envelope treadmilling motility in Transwell and stalling experiments. (A) Envelope treadmilling and Transwell. Treadmilling of cell envelope (yellow dash lines) is propulsive in cell front and rear in all adhesive channels (i), mainly in cell rear in hybrid adhesive channels (ii) and hardly at all in nonadhesive channels (iii). + and − signs indicate where envelope is clutched respectively by adhesion and friction. In hybrid channels, cells do not penetrate if adhesive clutching fails before inclusion of nucleus. (B) Envelope treadmilling and pressure stalling. (i) Cell in open channel without opposing pressure. The gray to green gradient sketches that polarized lymphocytes host antonymic controls signal of actomyosin and integrins like Sharpin and RhoA in cell rear and Rac and Rap-1 in cell front. Black arrow shows direction of migration, green arrow the pressure exerted by actin polymerization, and dashed yellow arrow the direction of envelope treadmilling. (ii) With opposing pressure (red arrows), cells slow down. (iii) At stall pressure, lamellipod growth and cell are arrested. The hatched filling shows that polarization establishment is frustrated by front arrest and front-rear antonymic signal mixed. Propulsion and adhesion in this situation are jeopardized. (iv) If unstable stalling conditions are overcome, polarization can be reverted by pressure, which never happens spontaneously with adherent primary effector human T lymphocytes. To see this figure in color, go online.
Forces developed by migrating cells in 2D and 3D range between piconewtons and tens of nanonewtons
Measurements of forces at the cellular level are instrumental to examine cell migration. For 2D migration, traction force microscopy and micropillars deformations yield the traction forces exerted by cells on their substrate. For mesenchymal cells, traction forces were mainly linked to the contraction of actin stress fibers between mature focal adhesions and reached 50–100 nN across cell edges (68,69). For amoeboid cells, which migrate much faster than mesenchymal cells, mature focal adhesion and stress fibers have less time to develop, and cells can migrate without interaction with a substrate (21). Weaker traction forces could therefore be expected, but the sparse available data report forces in the range of 20–50 nN for keratocytes (70), Dictyostelium (71), neutrophils (72), and dendritic cells (73). Unfortunately, there are no data available for T lymphocytes. Very few studies have focused on the forward-thrusting force of migrating cells in 2D. Oliver et al. (74) and Prass et al. (49) applied forces on a whole cell using micropipettes or atomic force microscopy tips and force sensors, which yielded stall forces of 20–40 nN with amoeboid keratocytes. These high stall forces are consistent with observations of lymphocytes crawling under flow, for which speed was unaffected by the global forces around several nanonewtons (75). In contrast, Bohnet et al. (76) showed that a few piconewtons per micrometer applied by a flow with a micropipette stopped the growth of a lamellipod. This discrepancy between the stall forces in the piconewton range for the lamellipod and in the nanonewton range for the whole cell is puzzling. Bohnet et al. (76) suggest that the limiting factor for motion in their assays may be adhesion rather than polymerization-driven growth of the lamellipod.
Concerning transmigration and 3D migration, literature is very scarce, and we distinguish three types of forces: squeezing forces perpendicular to motion, traction forces parallel to cell motion, and forward-thrusting force (Fig. 8 A). Squeezing forces of transmigrating cells were consistently measured around 10–50 nN (77, 78, 79, 80). Interestingly, traction forces parallel to cell motion measured with micropillars for mesenchymal cells (HOS and NIH-3T3) in microchannels (62) were decreasing with confinement, and inhibition of contractility decreased forces in wide channels, but not in confining channels. These results are reminiscent of the fading effect of myosin inhibitor (blebbistatin) on speed in our smooth assays. This suggests that contractility may not be required for high speed in confining smooth channels for both amoeboid and mesenchymal cells. Importantly, we also showed that this conclusion for free 3D migration was not valid for frustrated 3D migration, e.g., when a force (a pressure) or an obstacle (a constriction) was opposing cell motion. Concerning longitudinal forward-thrusting force (Fig. 8 A), a pioneer study by Usami et al. (81) reported measurements with neutrophils. Their method with micropipettes was conceptually similar to our microfluidic assay, but their results differ significantly. They found stall forces of 33 nN (and stall pressures of 17 mbar) in 5 μm diameter tubes, whereas we had a maximal force of 7 nN (and pressure of 4 mbar) in 4 × 4 μm2 channels. More surprisingly, they report no change of forward-thrusting force when the micropipette was coated or not with adhesion molecules, whereas we observed a drastic difference based on whether the channels were adherent or not. It is difficult to determine whether these differences arise from the analysis of different cell types (lymphocytes versus neutrophils) or from different experimental conditions (e.g., different surface coating and topologies of channels with rectangular versus circular cross sections). In any case, it is important to identify the origin of the force determined in stalling experiments.
Figure 8.
Forces during transmigration. (A) Schematic of the different cellular forces (red arrows) measured experimentally during 3D migration and transmigration: squeezing forces were measured around 10–50 nN (77, 78, 79, 80) for leukocytes, longitudinal traction forces around 20–50 nN (62) for mesenchymal cells, and longitudinal pushing forces around 3–6 nN here and 30 nN in other studies (81) for leukocytes. (B) Schematics of cellular deformations and forces during transmigration and migration in a matrix. (Top) Transmigration is a transition/competition between 2D (green arrow) and 3D (orange arrow) propulsion. Cell deformation occurs mainly during pore penetration, and actomyosin contractility plays a role in pore penetration but hardly in 3D propulsion. (Bottom) Migration in a matrix combines 3D propulsion and strong deformations, and contractility plays a role in global motion. To see this figure in color, go online.
Piconewtons are required for free migration and nanonewtons for frustrated migrations
Stall force experiments do not necessarily probe the strength of the motility machinery because different factors can frustrate motion before the limits of the propulsion machinery are reached. For instance, stall force was drastically reduced in nonadherent devices, and the limiting factor was then clearly a lack of gripping between substrates and cells. The understanding of nonadhesion amoeboid migration has not reached a consensus. Some studies reported that cells cannot propel in channels with smooth walls when walls are nonadherent (frictionless) or cells are deficient for integrin adhesion, even without opposing pressure (23,30). The conclusion is then that interactions between a cell and a solid can be weak enough to hamper motion. In contrast, other studies showed that leukocytes can swim in suspension (21,28,82), i.e., by friction between the cell treadmilling envelope and the external fluid. This implies that migration is always possible in solid without adhesion because friction with a solid cannot be lower than with a fluid. The propulsion by envelope treadmilling therefore explains the fast motion of cells in nonadherent tubes, but the poor resistance to an opposing pressure remains puzzling. In absence of adhesion and applied pressure, the force opposing cell motion is due to fluid displacement. For a nonconfined cell (swimming or 2D crawling), fluid resistance can be estimated by the Stokes equation, Funconfined = 6πμRv, where μ is the viscosity of the fluid (∼10−3 Pa ⋅ s), R the radius of the cell (∼4 μm), and v the cell speed (∼10 μm/min). This leads to Funconfined ∼10−5 nN, which is an extremely weak force, typically 30- to 100-fold weaker than the force of a single motor of dynein (83) or myosin molecule (84) (3 pN) and of a single filament of polymerizing actin (1 pN) (85,86). For a cell confined in a tube or channel, fluid resistance corresponds to the pressure drop of the fluid pushed by the cell in the channel, Fconfined = RlLvs, where Rl is the lineic hydrodynamic resistance of the channel, L the length of the channel, v the cell speed, and s the cross section of the channel. For a 4 × 4 μm2 channel with a length of 500 μm, Fconfined ∼2.10−3 nN. The fluidic resistance is larger in a tube than in suspension but remains more than 1000 times smaller than the force developed by cells for motility (1–100 nN) (50). These calculations explain why a minimal friction between a fluid and the treadmilling envelope of cells is sufficient to propel cell swimming (21) and cell migration into a nonadherent tube at high speed. However, adhesive gripping is required to allow cells to resist the 1000 times higher forces exerted by a counterpressure. In the end, stalling experiments probe the friction or adhesion of cell with their environment when cells are nonadherent and not the strength of the propulsion machinery. This conclusion for 3D migration is in line with the proposition of Bohnet et al. (76) for 2D migration, in which stall forces of piconewtons were also attributed to adhesion failure.
Stalling conditions under pressure control at 4 mbar are consistent with arrest of lamellipod growth
In the presence of adhesive ICAM-1 coating, gripping of cells to walls was not the limiting factor of cells progression against a force, and cell arrest was consistent with lamellipod growth arrest. The forward-thrusting force of polymerization in the lamellipod can be estimated by considering a force per growing filament of 2–7 pN (44,87) and a density of filament of 100–250 per μm2 (49,87). This yields a pressure of 2–17.5 mbar (or a force of 1.6–14 nN in a 4 × 2 μm2 channel, and 3.2–28 nN in a 4 × 4 μm2), which is consistent with the stall pressure measured here. Another argument is that stalling conditions were found under pressure control rather than under force control. This is consistent with the stalling conditions set by lamellipod frustration because lamellipod growth is also under pressure control (the pressure of actin filaments pushing on the membrane).
Cell arrest under pressure is consistent with a destabilization of cell polarization
The lamellipod growth can be stopped by the stall pressure of 4 mbar, but why is the whole cell also stopped? We argued that propulsion by protrusion dynamics (88) is not operant in our assays because the smoothness of channels hampers protrusions formation, whereas propulsion by envelope treadmilling (21,23,28) is operational in 2D and 3D. Envelope treadmilling is indeed consistent with gradual transmigration efficiency with adhesion (Fig. 7 A) and with the stability of cell speed within various environmental conditions. However, envelope treadmilling can develop forces of tens of nanonewtons (50,75,81), which is not consistent with a stall force of the few nanonewtons to stop cells. A crucial point is that the arrest of lamellipod growth was found to perturb the font-rear polarization of cells. Polarized lymphocytes are schematically composed of two poles with antonymic biosignaling programs. For instance, Rac-1 and Rap-1 signaling in cell front are known to foster polymerization of actin and activation of integrin LFA-1, respectively, whereas Rho and Sharpin signaling in cell rear foster actin contractility and inhibition of integrin LFA-1, respectively (Fig. 7 Bi). Frustrating the growth of the frontal compartment directly challenges the spatial separation of poles and therefore the regulation of cytoskeleton dynamics and integrin affinity (Fig. 7 Bii and iii). In this view, stalling does not correspond to the limit of the propulsion machinery strength but to the limit of lamellipod growth that then destabilizes the propulsive machinery. In accord with this model, hindrance of lamellipod growth was previously reported to alter adhesion (Fig. 5 F; (20)). We also observed that cells overcoming adhesiveness instability at stall pressure could reverse their polarity under pressure (Figs. 6 B and 7 Biv), which never occurred spontaneously in confined channels. This latter effect strongly supports that the stall pressure directly interferes with cell front-rear polarization.
Transwell assays shed light on 3D migration in microporous environments
Although amoeboid cell speed is generally contractility dependent for 3D migration in polymeric scaffolds (the extracellular matrix in vivo or collagen gels in vitro), 3D migration in smooth synthetic channels was found to be contractility independent. To reconcile these observations, one should consider that 3D migration in polymeric scaffolds involves intertwined events (Fig. 8) of 3D migration without cell deformations (independent of contractility in our assays) and penetration into constriction of sections as low as 10 μm2 (11,89) (dependent of contractility in our assays). In addition, inhibition of adhesion generally decreases cell speed in collagen (11), whereas 3D migration in our smooth channels hardly depended on specific adhesion. In fact, adhesion was required for penetration in pores in our assays, which also supports a crucial role of adhesion for 3D migration in a polymeric scaffold. Finally, the decrease of speed in matrices of finer porosity is in line with our observations of an increase of the penetration time in narrow pores and a decrease of speed in highly confining channels (cross section below 8 μm2). Altogether, microfluidic Transwells shed light on the 3D migration mechanism in polymeric scaffolds and porous materials by distinguishing between pore penetration and 3D migration without deformation (Fig. 8).
Stall force assays shed light on mechanisms of barotaxis and nuclear gauge to find path of least resistance
Amoeboid cells do not degrade the extracellular matrix, and it has been proposed that they use strategies to select paths of least resistance (26,56,90). One strategy consists in using their bulkiest compartment, the nucleus, as a mechanical gauge to discriminate pore size (26,66,67). Nucleus rigidity is clearly the main limiting factor for penetration into pores, and our data can be explained by a passive selection of the path of least resistance, with the nucleus simply hampering the penetration of cells in smallest pores. However, mechanotransduction triggered by nucleus deformation was recently revealed in several cell types (66,67) and shown to increase actomyosin contractility and to facilitate migration in confined spaces. It would be interesting to investigate whether this mechanism is also functional in human effector T lymphocytes during transmigration. Another strategy to select the path of least resistance is barotaxis, i.e., the ability of cells to detect and choose a path opposing lowest pressure. Barotaxis events were attested in vitro in synthetic microchannels (56,90). As discussed above, the pressure opposing cell motion due to the column of fluid pushed in channels is on the order of piconewtons, and at the light of our results, such a weak stimulus may bias the motility of nonadherent cells, but not of adherent cells. Accordingly, Moreau et al. (56) observed barotaxis only in the absence of adhesion. In contrast, Prentice et al. (90) observed barotaxis in both adherent and nonadherent channels. Because their devices were coated with fibronectin that favors adhesion of the cell rear and deadhesion of the cell front (50,61), one may speculate that a detached lamellipod might be sensitive to a piconewton stimulus to select a channel of least resistance.
About the limits of synthetic channels assays to assess barotaxis and stalling in vivo
Synthetic channels fail to mimic the physiological properties of endothelium or extracellular matrix by many aspects. Synthetic channels have notably watertight walls prone to establishing pressure drop across a confined cell, whereas the 3D environments in vivo are generally porous and allow a pressure release in several directions around the cell. Hence, although assays in synthetic channels are powerful for testing cell motility under stress, they teach little about barotaxis or stall pressure in vivo. In turn, stalling experiments by pressure may be relevant to mimic cells stalling by membrane tension in vivo because external pressure and membrane tension frustrate lamellipod growth in a similar way. An increase of membrane tension occurs typically in vivo when cells are slowed down by an obstacle or unable to detach their trailing edge.
Finally, microfluidic Transwell assays and measurement of forward-thrusting force have shed new light on the mechanical aspects of lymphocyte transmigration, and similar investigations complemented with chemokines, different extracellular matrix ligands, different geometries, and rigidities of channel will be instrumental in the future to decipher further the transmigration properties of leukocytes or cancer cells.
Author contributions
L.A. performed experiments and analysis of transmigration in channels without adhesion, 3D imaging of cells in microchannels with the help of S.B., and transmigration against a pressure. C.G. performed and analyzed quantitative experiments for transmigration efficiency in adherent channels. P.N. performed experiments between two plates and analyzed transmigration data in adherent channels. M.B.-P. managed the control of cells culture. M.-P.V. participated in experiments and analysis. O.T. designed the project, supervised experiments, participated in analysis, and wrote the manuscript.
Acknowledgments
Romain Gassiat, Anna Devaux, and Ronan Lavalley performed experiments during their master internships.
This work was supported by Agence Nationale de la Recherche (RECRUTE - ANR-15-CE15-0022; ILIAAD ANR-18-CE09-0029), LABEX INFORM, Région Sud, Turing Centre for Living Systems, and Excellence Initiative of Aix-Marseille University–A∗MIDEX, a French “Investissements d’Avenir” programme. We are also grateful to the Cell Culture Platform facility (Luminy TPR2-INSERM) and the PICSL microscopy platform of the CIML (ImagImm) supported by the French National Research Agency through the “Investissements d’Avenir” program (France-BioImaging, ANR-10-INBS-04).
Editor: Alexander Dunn.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.03.037.
Supporting material
References
- 1.Strilic B., Offermanns S. Intravascular survival and extravasation of tumor cells. Cancer Cell. 2017;32:282–293. doi: 10.1016/j.ccell.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Nourshargh S., Alon R. Leukocyte migration into inflamed tissues. Immunity. 2014;41:694–707. doi: 10.1016/j.immuni.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Carman C.V., Springer T.A. A transmigratory cup in leukocyte diapedesis both through individual vascular endothelial cells and between them. J. Cell Biol. 2004;167:377–388. doi: 10.1083/jcb.200404129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sumagin R., Sarelius I.H. Intercellular adhesion molecule-1 enrichment near tricellular endothelial junctions is preferentially associated with leukocyte transmigration and signals for reorganization of these junctions to accommodate leukocyte passage. J. Immunol. 2010;184:5242–5252. doi: 10.4049/jimmunol.0903319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massena S., Phillipson M. In: Hematology - Science and Practice. Lawrie C., editor. InTech; 2012. Intravascular Leukocyte Chemotaxis: The Rules of Attraction; pp. 229–252. [Google Scholar]
- 6.Sumagin R., Prizant H., Sarelius I.H. LFA-1 and Mac-1 define characteristically different intralumenal crawling and emigration patterns for monocytes and neutrophils in situ. J. Immunol. 2010;185:7057–7066. doi: 10.4049/jimmunol.1001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S., Voisin M.-B., Nourshargh S. Venular basement membranes contain specific matrix protein low expression regions that act as exit points for emigrating neutrophils. J. Exp. Med. 2006;203:1519–1532. doi: 10.1084/jem.20051210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park E.J., Peixoto A., Shimaoka M. Distinct roles for LFA-1 affinity regulation during T-cell adhesion, diapedesis, and interstitial migration in lymph nodes. Blood. 2010;115:1572–1581. doi: 10.1182/blood-2009-08-237917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulz O., Hammerschmidt S.I., Förster R. Chemokines and chemokine receptors in lymphoid tissue dynamics. Annu. Rev. Immunol. 2016;34:203–242. doi: 10.1146/annurev-immunol-041015-055649. [DOI] [PubMed] [Google Scholar]
- 10.Luo X., Seveau de Noray V., Theodoly O. Lymphocytes perform reverse adhesive haptotaxis mediated by LFA-1 integrins. J. Cell Sci. 2020;133:jcs242883. doi: 10.1242/jcs.242883. [DOI] [PubMed] [Google Scholar]
- 11.Wolf K., Te Lindert M., Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiam H.-R., Vargas P., Piel M. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 2016;7:10997. doi: 10.1038/ncomms10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedl P., Wolf K., Lammerding J. Nuclear mechanics during cell migration. Curr. Opin. Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGregor A.L., Hsia C.-R., Lammerding J. Squish and squeeze-the nucleus as a physical barrier during migration in confined environments. Curr. Opin. Cell Biol. 2016;40:32–40. doi: 10.1016/j.ceb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkins R.J., Piel M., Voituriez R. Pushing off the walls: a mechanism of cell motility in confinement. Phys. Rev. Lett. 2009;102:058103. doi: 10.1103/PhysRevLett.102.058103. [DOI] [PubMed] [Google Scholar]
- 16.Lämmermann T., Bader B.L., Sixt M. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 17.Krummel M.F., Friedman R.S., Jacobelli J. Modes and mechanisms of T cell motility: roles for confinement and Myosin-IIA. Curr. Opin. Cell Biol. 2014;30:9–16. doi: 10.1016/j.ceb.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf K., Mazo I., Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahai E., Marshall C.J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 20.Charras G., Sahai E. Physical influences of the extracellular environment on cell migration. Nat. Rev. Mol. Cell Biol. 2014;15:813–824. doi: 10.1038/nrm3897. [DOI] [PubMed] [Google Scholar]
- 21.Aoun L., Farutin A., Theodoly O. Amoeboid swimming is propelled by molecular paddling in lymphocytes. Biophys. J. 2020;119:1157–1177. doi: 10.1016/j.bpj.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tozluoğlu M., Tournier A.L., Sahai E. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat. Cell Biol. 2013;15:751–762. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- 23.Bergert M., Erzberger A., Paluch E.K. Force transmission during adhesion-independent migration. Nat. Cell Biol. 2015;17:524–529. doi: 10.1038/ncb3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paluch E.K., Aspalter I.M., Sixt M. Focal adhesion-independent cell migration. Annu. Rev. Cell Dev. Biol. 2016;32:469–490. doi: 10.1146/annurev-cellbio-111315-125341. [DOI] [PubMed] [Google Scholar]
- 25.Lämmermann T., Sixt M. Mechanical modes of ‘amoeboid’ cell migration. Curr. Opin. Cell Biol. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Renkawitz J., Kopf A., Sixt M. Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature. 2019;568:546–550. doi: 10.1038/s41586-019-1087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stroka K.M., Jiang H., Konstantopoulos K. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014;157:611–623. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Neill P.R., Castillo-Badillo J.A., Gautam N. Membrane flow drives an adhesion-independent amoeboid cell migration mode. Dev. Cell. 2018;46:9–22.e4. doi: 10.1016/j.devcel.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruprecht V., Wieser S., Heisenberg C.-P. Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell. 2015;160:673–685. doi: 10.1016/j.cell.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reversat A., Gaertner F., Sixt M. Cellular locomotion using environmental topography. Nature. 2020;582:582–585. doi: 10.1038/s41586-020-2283-z. [DOI] [PubMed] [Google Scholar]
- 31.Charras G., Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 32.Leithner A., Eichner A., Sixt M. Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat. Cell Biol. 2016;18:1253–1259. doi: 10.1038/ncb3426. [DOI] [PubMed] [Google Scholar]
- 33.Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J. Exp. Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung W.-C., Chen S.-H., Konstantopoulos K. Distinct signaling mechanisms regulate migration in unconfined versus confined spaces. J. Cell Biol. 2013;202:807–824. doi: 10.1083/jcb.201302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong Z., Balzer E.M., Konstantopoulos K. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLoS One. 2012;7:e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeon N.L., Dertinger S.K.W., Whitesides G.M. Generation of solution and surface gradients using microfluidic systems. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 37.Shamloo A., Ma N., Heilshorn S.C. Endothelial cell polarization and chemotaxis in a microfluidic device. Lab Chip. 2008;8:1292–1299. doi: 10.1039/b719788h. [DOI] [PubMed] [Google Scholar]
- 38.Strale P.-O., Azioune A., Studer V. Multiprotein printing by light-induced molecular adsorption. Adv. Mater. 2016;28:2024–2029. doi: 10.1002/adma.201504154. [DOI] [PubMed] [Google Scholar]
- 39.Pasturel A., Strale P.-O., Studer V. A generic widefield topographical and chemical photopatterning method for hydrogels. bioRxiv. 2018 doi: 10.1101/370882. [DOI] [Google Scholar]
- 40.Tinevez J.-Y., Perry N., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Wilson K., Lewalle A., Charras G. Mechanisms of leading edge protrusion in interstitial migration. Nat. Commun. 2013;4:2896. doi: 10.1038/ncomms3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobelli J., Friedman R.S., Krummel M.F. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA-regulated adhesions. Nat. Immunol. 2010;11:953–961. doi: 10.1038/ni.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preira P., Valignat M.-P., Théodoly O. Single cell rheometry with a microfluidic constriction: quantitative control of friction and fluid leaks between cell and channel walls. Biomicrofluidics. 2013;7:24111. doi: 10.1063/1.4802272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abraham V.C., Krishnamurthi V., Lanni F. The actin-based nanomachine at the leading edge of migrating cells. Biophys. J. 1999;77:1721–1732. doi: 10.1016/S0006-3495(99)77018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcy Y., Prost J., Sykes C. Forces generated during actin-based propulsion: a direct measurement by micromanipulation. Proc. Natl. Acad. Sci. USA. 2004;101:5992–5997. doi: 10.1073/pnas.0307704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller W.A. How endothelial cells regulate transmigration of leukocytes in the inflammatory response. Am. J. Pathol. 2014;184:886–896. doi: 10.1016/j.ajpath.2013.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balzer E.M., Tong Z., Konstantopoulos K. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keren K., Pincus Z., Theriot J.A. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prass M., Jacobson K., Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J. Cell Biol. 2006;174:767–772. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valignat M.-P., Nègre P., Theodoly O. Lymphocytes can self-steer passively with wind vane uropods. Nat. Commun. 2014;5:5213. doi: 10.1038/ncomms6213. [DOI] [PubMed] [Google Scholar]
- 51.Fritz-Laylin L.K., Riel-Mehan M., Mullins R.D. Three-dimensional actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. bioRxiv. 2017 doi: 10.1101/120444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergert M., Chandradoss S.D., Paluch E. Cell mechanics control rapid transitions between blebs and lamellipodia during migration. Proc. Natl. Acad. Sci. USA. 2012;109:14434–14439. doi: 10.1073/pnas.1207968109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Seyda N., Seveau V., Theodoly O. Human neutrophils swim and phagocytise bacteria. Biol. Cell. 2021;113:28–38. doi: 10.1111/boc.202000084. Published online November 6, 2020. [DOI] [PubMed] [Google Scholar]
- 54.Renkawitz J., Schumann K., Sixt M. Adaptive force transmission in amoeboid cell migration. Nat. Cell Biol. 2009;11:1438–1443. doi: 10.1038/ncb1992. [DOI] [PubMed] [Google Scholar]
- 55.Hawkins R.J., Poincloux R., Voituriez R. Spontaneous contractility-mediated cortical flow generates cell migration in three-dimensional environments. Biophys. J. 2011;101:1041–1045. doi: 10.1016/j.bpj.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moreau H.D., Blanch-Mercader C., Lennon-Duménil A.-M. Macropinocytosis overcomes directional bias in dendritic cells due to hydraulic resistance and facilitates space exploration. Dev. Cell. 2019;49:171–188.e5. doi: 10.1016/j.devcel.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 57.Fournier M.F., Sauser R., Verkhovsky A.B. Force transmission in migrating cells. J. Cell Biol. 2010;188:287–297. doi: 10.1083/jcb.200906139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morin N.A., Oakes P.W., Kim M. Nonmuscle myosin heavy chain IIA mediates integrin LFA-1 de-adhesion during T lymphocyte migration. J. Exp. Med. 2008;205:195–205. doi: 10.1084/jem.20071543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith A., Stanley P., Hogg N. The role of the integrin LFA-1 in T-lymphocyte migration. Immunol. Rev. 2007;218:135–146. doi: 10.1111/j.1600-065X.2007.00537.x. [DOI] [PubMed] [Google Scholar]
- 60.Pouwels J., De Franceschi N., Ivaska J. SHARPIN regulates uropod detachment in migrating lymphocytes. Cell Rep. 2013;5:619–628. doi: 10.1016/j.celrep.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hornung A., Sbarrato T., Valignat M.-P. A bistable mechanism mediated by integrins controls mechanotaxis of leukocytes. Biophys. J. 2020;118:565–577. doi: 10.1016/j.bpj.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raman P.S., Paul C.D., Konstantopoulos K. Probing cell traction forces in confined microenvironments. Lab Chip. 2013;13:4599–4607. doi: 10.1039/c3lc50802a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davidson P.M., Denais C., Lammerding J. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell. Mol. Bioeng. 2014;7:293–306. doi: 10.1007/s12195-014-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas D.G., Yenepalli A., Egelhoff T.T. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J. Cell Biol. 2015;210:583–594. doi: 10.1083/jcb.201502039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maxian O., Mogilner A., Strychalski W. Computational estimates of mechanical constraints on cell migration through the extracellular matrix. PLoS Comput. Biol. 2020;16:e1008160. doi: 10.1371/journal.pcbi.1008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venturini V., Pezzano F., Ruprecht V. The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science. 2020;370:eaba2644. doi: 10.1126/science.aba2644. [DOI] [PubMed] [Google Scholar]
- 67.Lomakin A.J., Cattin C.J., Piel M. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science. 2020;370:eaba2894. doi: 10.1126/science.aba2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trichet L., Le Digabel J., Ladoux B. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc. Natl. Acad. Sci. USA. 2012;109:6933–6938. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munevar S., Wang Y., Dembo M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys. J. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J., Leonard M., Jacobson K. Traction forces generated by locomoting keratocytes. J. Cell Biol. 1994;127:1957–1964. doi: 10.1083/jcb.127.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lombardi M.L., Knecht D.A., Lee J. Traction force microscopy in Dictyostelium reveals distinct roles for myosin II motor and actin-crosslinking activity in polarized cell movement. J. Cell Sci. 2007;120:1624–1634. doi: 10.1242/jcs.002527. [DOI] [PubMed] [Google Scholar]
- 72.Smith L.A., Aranda-Espinoza H., Hammer D.A. Neutrophil traction stresses are concentrated in the uropod during migration. Biophys. J. 2007;92:L58–L60. doi: 10.1529/biophysj.106.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ricart B.G., Yang M.T., Hammer D.A. Measuring traction forces of motile dendritic cells on micropost arrays. Biophys. J. 2011;101:2620–2628. doi: 10.1016/j.bpj.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oliver T., Dembo M., Jacobson K. Traction forces in locomoting cells. Cell Motil. Cytoskeleton. 1995;31:225–240. doi: 10.1002/cm.970310306. [DOI] [PubMed] [Google Scholar]
- 75.Valignat M.-P., Theodoly O., Lellouch A.C. T lymphocytes orient against the direction of fluid flow during LFA-1-mediated migration. Biophys. J. 2013;104:322–331. doi: 10.1016/j.bpj.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bohnet S., Ananthakrishnan R., Verkhovsky A.B. Weak force stalls protrusion at the leading edge of the lamellipodium. Biophys. J. 2006;90:1810–1820. doi: 10.1529/biophysj.105.064600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rabodzey A., Alcaide P., Ladoux B. Mechanical forces induced by the transendothelial migration of human neutrophils. Biophys. J. 2008;95:1428–1438. doi: 10.1529/biophysj.107.119156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Labernadie A., Trepat X. Sticking, steering, squeezing and shearing: cell movements driven by heterotypic mechanical forces. Curr. Opin. Cell Biol. 2018;54:57–65. doi: 10.1016/j.ceb.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 79.Yeh Y.-T., Serrano R., Lasheras J.C. Three-dimensional forces exerted by leukocytes and vascular endothelial cells dynamically facilitate diapedesis. Proc. Natl. Acad. Sci. USA. 2018;115:133–138. doi: 10.1073/pnas.1717489115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Molino D., Quignard S., Fattaccioli J. On-chip quantitative measurement of mechanical stresses during cell migration with emulsion droplets. Sci. Rep. 2016;6:29113. doi: 10.1038/srep29113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Usami S., Wung S.L., Chien S. Locomotion forces generated by a polymorphonuclear leukocyte. Biophys. J. 1992;63:1663–1666. doi: 10.1016/S0006-3495(92)81745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barry N.P., Bretscher M.S. Dictyostelium amoebae and neutrophils can swim. Proc. Natl. Acad. Sci. USA. 2010;107:11376–11380. doi: 10.1073/pnas.1006327107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schnitzer M.J., Visscher K., Block S.M. Force production by single kinesin motors. Nat. Cell Biol. 2000;2:718–723. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
- 84.Finer J.T., Simmons R.M., Spudich J.A. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 85.Footer M.J., Kerssemakers J.W.J., Dogterom M. Direct measurement of force generation by actin filament polymerization using an optical trap. Proc. Natl. Acad. Sci. USA. 2007;104:2181–2186. doi: 10.1073/pnas.0607052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kovar D.R., Pollard T.D. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mogilner A., Oster G. Polymer motors: pushing out the front and pulling up the back. Curr. Biol. 2003;13:R721–R733. doi: 10.1016/j.cub.2003.08.050. [DOI] [PubMed] [Google Scholar]
- 88.Reversat A., Merrin J., Sixt M. Adhesion-free cell migration by topography-based force transduction. bioRxiv. 2019 doi: 10.1101/793919. [DOI] [Google Scholar]
- 89.Wolf K., Alexander S., Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prentice-Mott H.V., Chang C.-H., Shah J.V. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. Proc. Natl. Acad. Sci. USA. 2013;110:21006–21011. doi: 10.1073/pnas.1317441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primary human effector T cells transmigrating for several hours in an ICAM-1 coated Transwell device with 4 × 4 μm2 microchannels in Bright field mode.
RICM image sequence of primary human effector T cells migrating on 2D surface and transmigrating in smooth 4 × 4 μm2 ICAM-1 coated microchannels (Part 1). Bright field (left) and RICM (right) image sequence of a primary human effector T cell entering a smooth 4 × 4 μm2 ICAM-1 coated microchannel (Part2).
T cells with DAPI stained nuclei migrating on ICAM-1 coated devices with 4 × 4 μm2 microchannels (Part 1) and 4 × 2 μm2 (Part 2). Images are superimpositions of bright field (grey), RICM (red for 4 × 4 μm2 and green for 4 × 2 μm2) and DAPI fluorescence (blue) images. Dark zones in RICM reveal adhesive fingerprint in 2D, and close contact or adhesive fingerprint in 3D of cells.
Primary effector human T cells transmigration in device with large channels treated with adherent ICAM-1 coatings and microchannels of cross section 4 × 2 μm2 treated with non-adherent Pluronic F127 coating. Transmigration is aborted just before complete entry.
Bright field image sequence of a primary human effector T cell transmigrating in a 4 × 4 μm2 ICAM-1 coated microchannel exposed to opposing pressure of 2 and 5 mbar. White arrows indicate the direction of opposing pressure (small arrow: 2 mbar, large arrow: 5 mbar). White horizontal lines indicate cell location when the pressure of 2 and 5 mbar are applied. Time in mins. Scale bars, 20 μm.
Part 1- Polarization reversion by pressure in adherent channels (Fig. 6 B). Part 2 - Stalling and detachment at adhesion/non-adhesion frontier. The cell crosses the frontier between adherent and non-adherent zones in absence of pressure. When a 1 mbar pressure is applied, the cell slides backwards, re-attaches in the adherent zone, is stalled at the A/NA frontier and eventually detaches (Fig. 6 C). Part 3- Transition between passive push and active crawling in confinement. Cells pushed in non-adherent channels at 2 mbar, arrest at the A/NA frontier. Depending on their intrinsic polarization, they resume crawling against pressure and are stalled at the A/NA frontier (left) (Fig. 6 Di), or crawl in adherent zones out of the channels (right) (Fig. 6 Dii).