Abstract
SARS-CoV-2 variants emerged in late 2020, and at least three variants of concern (B.1.1.7, B.1.351, and P1) have been reported by WHO. These variants have several substitutions in the spike protein that affect receptor binding; they exhibit increased transmissibility and may be associated with reduced vaccine effectiveness. In the present work, we report the identification of a potential variant of interest, harboring the mutations T478K, P681H, and T732A in the spike protein, within the newly named lineage B.1.1.519, that rapidly outcompeted the preexisting variants in Mexico and has been the dominant virus in the country during the first trimester of 2021.
Viral mutation is a natural and expected event generated during genomic replication and interaction with the host, resulting in the establishment of genetic groups, also called lineages. The latter differ from each other by specific mutations that accumulate over time, causing the appearance of variants. A variant can be defined as a virus with specific genetic mutations that differ from the original virus, in some cases reflecting the adaptation of SARS-CoV-2 to its new human host. Although the majority of mutations in the SARS-CoV-2 genome are expected to be neutral or deleterious, some mutations can confer a selective advantage and may be associated with enhanced fitness, increased infectivity, and/or immune evasion [1–3]. Importantly, the emergence and spread of variants associated with changes in transmission, virulence, and/or antigenicity can impact the evolution of the COVID-19 pandemic and might require appropriate public health actions and surveillance [4].
New SARS-CoV-2 variants are spreading rapidly around the world, becoming a public health concern. As of February 23, 2021, the Pan-American Health Organization (PAHO)/World Health Organization (WHO) and Global Initiative on Sharing All Influenza Data (GISAID) reported the appearance of at least three variants of concern (VOCs) with characteristics that have implications for public health. Variant B.1.1.7 was identified for the first time in the United Kingdom in September 2020 [4, 5], and by December 2020 it represented 43% of the genomes sequenced, increasing to 82% in January 2021 and to 94% in February 2021 [6]. This variant is of growing concern, since it has been shown to be significantly more transmissible than other variants [7], and it is likely to have increased severity, based on hospitalization and fatality rates. Variant B.1.3 51 was detected for the first time in South Africa, in 64% (261 of 411 genomes) of the sequences reported in December 2020, increasing to 75% (99 of 132 genomes) in the next month [6]. Epidemiological data analysis estimated that this VOC is 50% more transmissible than the previous circulating variants. Finally, the P.1 variant was detected for the first time in Brazil in 47% (61 of 130) of the viral genomes in December 2020, increasing to 74% (111 of 150 genomes) in the next month [6, 8]. Of relevance, it has shown reduced neutralization by convalescent and post-vaccination sera [9]. These SARS-CoV-2 VOCs have acquired some of the same spike protein mutations independently, particularly E484K, N501Y, S477N, and K417T, which have been associated with increased viral transmission and/or decreased sensitivity to antibody neutralization [10].
In Latin America, with the exception of P.1 and P.2 observed in Brazil, no other variants with the potential for rapid expansion have been reported so far [11]. Here, we report the identification of a potential VOI harboring the mutations T478K, P681H, and T732A in the spike protein, within the newly named lineage B.1.1.519, derived from the B.1.1.222 lineage, that rapidly outcompeted the preexisting variants in Mexico and has been the dominant virus in the country during 2021.
Derived from genomic surveillance carried out in Mexico, 2,692 genomic sequences were obtained in this study and are part of the 3,156 sequences deposited in the GISAID from March 1, 2020 to March 21, 2021. As a result of the analysis of this set of sequences, we observed the presence of 91 Phylogenetic Assignment of Named Global Outbreak (PANGO) lineages, with B.1.1.519 (37.8%), B.1 (13.9%), B.1.1.222 (10.3%), B.1.1 (5.7%), B.1.609 (5.6%), and B.1.243 (4.5 %) being the most prevalent. Libraries for whole genome sequencing of SARS-CoV-2 were generated using the protocol developed by the ARTIC Network (https://artic.network/2-protocols.html) or a long-amplicon-based method (https://pubmed.ncbi.nlm.nih.gov/32222995/).
A striking observation was the detection of the B.1.1.519 lineage in the USA, derived from B.1.1.222, which harbors the mutation T478K in the spike protein. This variant had not been detected in Mexico before October 2020, when it was found in Mexico City, and phylogeographic analysis suggested that the B.1.1.519 variant emerged around mid-September 2020 [10]. In November 2020, 13% (16/123) of the characterized cases of COVID-19 were caused by this variant, and in December, this proportion increased to 29.3% (97/331). In January 2021, the percentage of B.1.1.519 rose to 51.5% (229/445), increasing in incidence to 73.6% (808/1098) in February. On the other hand, a decreasing frequency of the B.1 lineage that had predominated in Mexico in 2020 was observed, going from 36.27% (284/783) between March and September to 2.37% (26/1098) in February 2021.
In Mexico, since the identification of B.1.1.519 in November 2020, a total of 6419 genomic sequences have been reported with this lineage. The majority of them are from Mexico City and are spread throughout all country.
A detailed analysis of the samples from Mexico City indicated that, in November, this variant was present in 17.8% (13/73) of the cases, while in December 2020, this proportion increased to 47.5% (47/99). In January 2021, the variant was detected in 77.5% (138/178) of the cases and by February in 90.9% (349/384). This significant increase in the frequency of B.1.1.519 in Mexico City showed that it outcompeted preexisting variants between October 2020 and February 2021, and this increase was also observed in other regions of the country (Fig. 1), representing more than 50% of the characterized viruses in some states during the first trimester of 2021. In particular, the variant was highly prevalent in Baja California Sur (51.3%, 20/39), Guerrero (70%, 21/30), Hidalgo (72.2%, 13/18), Morelos (67.3%, 33/49), State of Mexico (83.5%, 76/91), Oaxaca (51.3%, 19/37), Puebla (78.5%, 77/98), Queretaro (70.8%, 34/48), San Luis Potosi (70%, 35/50), and Veracruz (69.5%, 80/115).
Fig. 1.

Relative frequency of variant B.1.1.519 from October 2020 to February 2021. The monthly numbers of complete genome sequences (n) obtained from samples collected in Mexico City and other states of Mexico are indicated below each bar.
This variant has also been detected in 17 countries on all five continents. In the Americas, it has been reported in Canada and the USA, and recently in Brazil, Chile, Aruba, Martinique, and Curazao [12]. However, this variant currently is not predominant in these countries.
The overall genome analysis of the viruses in the B.1.1.519 lineage showed the presence of 20 mutations in total, compared to the Wuhan-Hu-1 reference genome sequence (NCBI accession number MN908947). Eleven of these mutations are non-synonymous, and four of them are present in the spike protein. Notably, a T478K mutation is present in the receptor binding domain (RBD), where mutations have been shown to reduce the activity of some monoclonal antibodies [9]. All amino acid and nucleotide changes are listed in Table 1.
Table 1.
Amino acid and nucleotide changes
| Mutations | Gene | |
|---|---|---|
| Nucleotide | Amino acid | |
| C203T | – | |
| C222T | – | |
| C241T | – | |
| C3037T | – | ORF1a |
| C3140T | P141S | |
| C10029T | T492I | |
| C10954T | – | |
| A11117G | I49V | |
| C12789T | ||
| C14408T | P323L | ORF1b |
| T19839C | – | |
| C21306T | – | |
| C22995A | T478K | Spike |
| A23403G | D614G | |
| C23604A | P681H | |
| A23756G | T732A | |
| G28881A | – | N |
| G28882A | R203K | |
| G28883C | G204R | |
| C29197T | – | |
The main amino acid substitutions are shown in bold
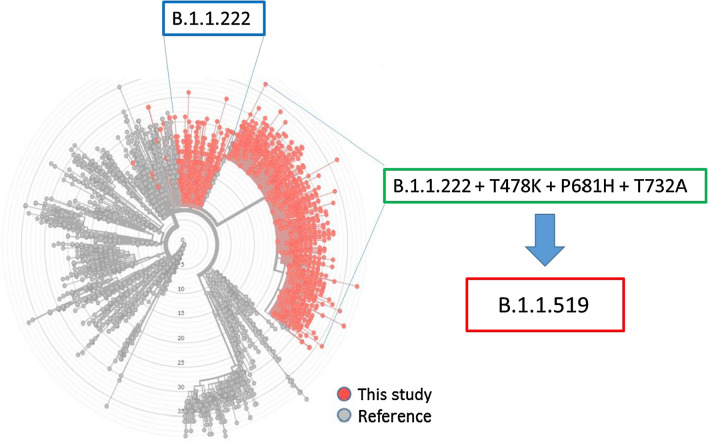
The current B.1.1.519 lineage, represented by the vast majority of the reported Mexican sequences, was first identified as a B.1.1.222 lineage. However, the presence of the mutations T478K, P681H, and T732A clearly differentiated it from this lineage, which does not contain these mutations, giving rise to the B.1.1.519 lineage. A phylogenomic analysis of genomic sequences using the Nextstrain tool showed that the viruses in the lineage B.1.1.519 (B.1.1.1.222+T478K+P681H+T732A) group independently of the lineage B.1.1.222 sequences, strongly suggesting that this variant should be classified as a variant of interest (VOI) (Fig. 2). On the other hand, viruses in the lineage B.1.1.519 are already grouped by the GISAID platform in an independent clade, invariably harboring the three mutations mentioned above.
Fig. 2.
Phylogenomic analysis of SARS CoV-2 sequences obtained in this study (red dots) and of reference sequences (gray dots), showing the clustering of variant B.1.1.519 (red box) independently of the lineage B.1.1.222 (blue box). Viruses in the B.1.1.519 lineage were initially classified within the B.1.1.222 lineage, and they contain the mutations T478K, P681H, and T732A (green box). The phylogenomic tree was powered a CC-BY-4.0 license and attribution of nextstrain.org.
An in silico analysis using different potent structures of related strains suggested that the position of the T478K mutation in the S protein is involved in antibody recognition and the receptor binding site [13]. In a deep mutational scanning of the SARS-CoV-2 receptor binding domain, the T478K mutation did not have a significant effect on folding or binding to human angiotensin-converting enzyme 2 (ACE2) [14]. However, this mutation may be involved in immune evasion, particularly escape from antibody neutralization [15]. The P681H mutation is one of the mutations found in the B.1.1.7 variant detected in the UK. According to the definitions described in the document issued by the WHO "Covid-19 Weekly Epidemiological Update" of February 25, 2021, with the special edition of "Proposed Working Definitions of SARS-CoV-2 Variants of Interest and Variants of Concern", we can consider the lineage B.1.1.519 a potential variant of interest [16].
Finally, two variants with interesting features were identified in this study: first, 13 sequences belonging to the B.1.1.222 lineage without the T478K mutation, but harboring the T732A mutation and the 69-70 deletion in the spike protein, the latter being a characteristic mutation of the B.1.1.7 VOC first detected in the UK; and second, 11 sequences corresponding to four lineages differing from B.1.1.519 (B.1, B.1.1.222, B.1.1.322, and B.1.323) but containing the same T478K, P681H, and T732A mutations in the spike glycoprotein that are present in the variant B.1.1.519. Keeping track of the incidence of these two variants is recommended during genomic surveillance.
So far, we do not have experimental evidence to determine if the mutations described here could be associated with changes in transmission, virulence, and/or antigenicity or if they could have an impact on the severity of disease, reinfection rates, or vaccine effectiveness. For this reason, the importance of a genomic surveillance system, epidemiological studies, and experiments to assess the neutralization of viruses in lineage B.1.1.519 or any new variants are crucial for investigating the possible biological impact of the mutations in the context of public health. Fortunately, all COVID-19 virus variants that have emerged so far respond to the available, approved vaccines to some extent.
Acknowledgements
The authors express their gratitude to Vanessa Rivero, Ariadna Medina, Joaquin Quiroz, Sergio Rangel, Mayra Jiménez, David Fragoso, Nancy Miñoz, Edgar Mendieta, Fabiola Garcés and Adnan Araiza from InDRE for sequencing and bionformatical approach; to Ricardo Grande, Francisco Pulido and Gloria Vazquez from the “Unidad de Secuenciación Masiva y Bioinformática” of the “Laboratorio Nacional de Apoyo Tecnológico a las Ciencias Genómicas” (CONACyT no. 260481) for their support in sequencing services and also to the working team for molecular biology of the Central Laboratory of Epidemiology IMSS, for technical assistance. We thank all of the staff of the Technological Development and Molecular Research Unit of the Virology Department and of the Sample Control and Services Department at InDRE for technical assistance. The findings and conclusions in this report are those of the authors and do not necessarily represent the official opinion of the Ministry of Health of Mexico.
Funding
This work was partially supported by the grants “Epidemiología Genómica de los Virus SARS-CoV-2 Circulantes en México” and "Caracterización de la diversidad viral y bacteriana" from the National Council for Science and Technology (CONACyT) of Mexico to C.F.A. and J.A.V.P., respectively, and also grants 057, 047, and 072 from the Ministry of Education, Science, Technology and Innovation (SECTEI) of Mexico City to C.F.A., L.H.M., and A.H.M., respectively.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abril Paulina Rodríguez-Maldonado, Alberto Cedro-Tanda, Joel Armando Vázquez-Pérez, Blanca Taboada and Celia Boukadida contributed equally to this work. The order of the other authors was randomly assigned.
Contributor Information
Alfredo Hidalgo-Miranda, Email: ahidalgo@inmegen.gob.mx.
Carlos F. Arias, Email: arias@ibt.unam.mx
José Ernesto Ramírez-González, Email: ernesto.ramirez@salud.gob.mx.
References
- 1.Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Nie J, Wu J, et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epidemiological update: occurrence of variants of SARS-CoV-2 in the Americas—20 January 2021. https://iris.paho.org/bitstream/handle/10665.2/53219/EpiUpdate20January2021_eng.pdf?sequence=1&isAllowed=y. Accessed 29 Mar 2021
- 5.Weekly epidemiological update—23 February 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update---23-february-2021. Accessed 29 Mar 2021
- 6.(2021) Tracking of variants. https://www.gisaid.org/hcov19-variants/. Accessed 29 Mar 2021
- 7.Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg305. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faria N, Morales I, Candido D et al (2021). Genomic characterization of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological.org.in. https://bit.ly/3qx9aEU. Accessed 29 Mar 2021
- 9.Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, Liu L, Kwong PD, Huang Y, Shapiro L, Ho DD. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lasek-Nesselquist E, Pata J, Schneider E, George K. A tale of three SARS-CoV-2 variants with independently acquired P681H mutations in New York State. MedRxiv. 2021 doi: 10.1101/2021.03.10.21253285. [DOI] [Google Scholar]
- 11.Resende PC, Gräf T, Paixão ACD, et al. A potential SARS-CoV-2 variant of interest (VOI) harboring mutation E484K in the Spike protein was identified within lineage B.1.1.33 circulating in Brazil. Viruses. 2021;13:724. doi: 10.3390/v13050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GISAID (2021) https://www.gisaid.org/. Accessed 29 Mar 2021
- 13.(2021) CoVsurver: mutation analysis of hCoV-19. https://www.gisaid.org/epiflu-applications/covsurver-mutations-app/. Accessed 29 Mar 2021
- 14.Starr TN, Greaney AJ, Hilton SK, et al. deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, VanBlargan LA, Bloyet LM, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weekly epidemiological update—25 February 2021. https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update. Accessed 29 Mar 2021