Abstract
The regulatory elements that restrict transcription of genes encoding contractile proteins specifically to either slow- or fast-twitch skeletal muscles are unknown. As an initial step towards understanding the mechanisms that generate muscle diversity during development, we have identified a 128-bp troponin I slow upstream element (SURE) and a 144-bp troponin I fast intronic element (FIRE) that confer fiber type specificity in transgenic mice (M. Nakayama et al., Mol. Cell. Biol. 16:2408–2417, 1996). SURE and FIRE have maintained the spatial organization of four conserved motifs (3′ to 5′): an E box, an AT-rich site (A/T2) that binds MEF-2, a CACC site, and a novel CAGG motif. Troponin I slow (TnIs) constructs harboring mutations in these motifs were analyzed in transiently and stably transfected Sol8 myocytes and in transgenic mice to assess their function. Mutations of the E-box, A/T2, and CAGG motifs completely abolish transcription from the TnI SURE. In contrast, mutation of the CACC motif had no significant effect in transfected myocytes or on the slow-specific transcription of the TnI SURE in transgenic mice. To assess the role of E boxes in fiber type specificity, a chimeric enhancer was constructed in which the E box of SURE was replaced with the E box from FIRE. This TnI E box chimera, which lacks the SURE NFAT site, confers essentially the same levels of transcription in transgenic mice as those conferred by wild-type SURE and is specifically expressed in slow-twitch muscles, indicating that the E box on its own cannot determine the fiber-type-specific expression of the TnI promoter. The importance of the 5′ half of SURE, which bears little homology to the TnI FIRE, in muscle-specific expression was analyzed by deletion and linker scanning analyses. Removal of the 5′ half of SURE (−846 to −811) results in the loss of expression in stably transfected but not in transiently expressing myocytes. Linker scanning mutations identified sequences in this region that are necessary for the function of SURE when integrated into chromatin. One of these sites (GTTAATCCG), which is highly homologous to a bicoid consensus site, binds to nuclear proteins from several mesodermal cells. These results show that multiple elements are involved in the muscle-specific activity of the TnIs promoter and that interactions between upstream and downstream regions of SURE are important for transcription in the context of native chromatin.
Skeletal muscle commitment, differentiation, and maturation are largely controlled by the transcriptional regulation of genes encoding channels, receptors, metabolic enzymes, and muscle-specific contractile proteins (8, 52). Whereas our understanding of the transcription factors regulating the commitment and differentiation of myocytes has advanced significantly in the past years, little is known about the factors that regulate fiber type diversity during muscle maturation. During postnatal development, following the innervation of muscles by motoneurons, distinct isoforms of myosin heavy chains and other contractile proteins accumulate in myofibers. These isoforms determine the rates of force generation, the relaxation rates, and the fatigability of the myofibers.
Transcription is the major regulatory mechanism known to restrict the expression of genes encoding contractile proteins to specific types of muscle fibers during development. Thus, one approach to understanding the molecular mechanisms controlling muscle diversification and plasticity is to identify the DNA regulatory sequences that confer fiber-type-specific expression of contractile protein genes. Toward this end, we have used the regulation of troponin I (TnI) genes as a model to elucidate the mechanisms that generate fiber diversification. In the adult, three troponin isoforms, which are presumed to have originated from a common ancestral gene, are specifically expressed in slow (TnIs), fast (TnIf), and cardiac (TnIc) muscles (28). The different isoforms of TnI, in combination with troponins C and T, participate in the formation of a complex that is involved in the regulation of calcium-mediated interactions during muscle contraction (63). As with many other skeletal muscle-specific genes, transcription of TnI genes is initially activated during myoblast differentiation (3, 12, 34, 61). Embryonic and fetal myofibers coexpress the TnIs and TnIf isoforms, but as muscles are innervated by motoneurons and mature, expression of the slow- and fast-muscle genes is confined to type I and II fibers, respectively (27).
We have previously used transfected myocyte cell cultures to identify regions in the TnIs gene that are important for its regulation in the differentiated myotubes (3). Subsequently, somatic gene transfer experiments performed in rats as well as analyses using transgenic mice identified a slow upstream regulatory element (SURE) in human (13) and rat (43) TnIs genes that specifically confers expression to slow-twitch muscles and a fast intronic regulatory element (FIRE) in the quail TnIf gene that directs transcription in fast-twitch muscles (43). Comparison of these elements identified four spatially conserved motifs: an E box, an AT-rich site, a CACC element, and a CAGG motif, of which the first three are also found in numerous other regulatory regions of genes encoding skeletal muscle proteins. The E box interacts with various skeletal muscle-specific regulatory factors of the basic helix-loop-helix (bHLH) family such as MyoD (15), myogenin (19, 59), myf-5 (5), and MRF-4 (6, 41, 48), which have been shown to be important for activating transcription of muscle genes during myogenesis and differentiation (7, 20, 58). The AT-rich site binds MEF2/RSRF factors that are more widely expressed than the myogenic bHLH factors and which are important for the regulation of muscle genes encoding structural proteins and transcription factors (18, 24, 29, 46, 49, 60). The importance of the CACC element in the transcription of muscle genes is not clear, although a cDNA encoding a novel winged-helix protein binding to the CACC/SP1 motif was recently isolated from mouse (4). The CAGG motif present in the TnIs and TnIf enhancers has not been described previously, but a similar site, known as a MEF-3 site, has been found in many other muscle promoters (29, 44).
Since these sites are conserved in both TnI SURE and FIRE, which direct slow- and fast-fiber-type-specific transcription, respectively, it is possible that these sites are important only for their early developmental expression during myogenesis and differentiation. However, these sites may also play a critical role in the slow- and fast-fiber-type-specific expression of troponins in the mature muscle by their interaction with fiber-specific transcription factors. Alternatively, the nonhomologous sequences in SURE and FIRE may harbor elements responsible for the restriction of TnIs and TnIf gene expression to the slow and fast adult muscle, respectively. Thus, to address these possibilities, we have performed transcriptional analyses with constructs containing mutated TnIs promoter fragments as well as a chimeric enhancer containing sequences from both SURE and FIRE in myotube cell cultures and transgenic mice. Since muscle cells in culture serve as excellent models to study gene regulation during development but fail to manifest the specific contractile properties of adult muscles, we have used myotube cell cultures and transgenic mice to study different aspects of TnI regulation. While analyses in cells have allowed us to characterize DNA sequences that regulate the tissue-specific and developmental induction of the TnIs gene, studies with transgenic mice enabled the identification of cis-acting sequences that confer fiber-specific transcription. Based on these studies, we present evidence in this paper that of the four conserved motifs present in SURE, the E-box, MEF-2, and CAGG sites are indispensable for developmental as well as fiber-specific transcription of the rat TnIs gene and that the substitution of the E box in SURE with a similar sequence present in FIRE does not alter the ability of the TnIs promoter to direct slow-fiber-specific expression in the adult muscle.
MATERIALS AND METHODS
Cells and transfections.
Sol8 myoblasts (42) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum and antibiotics (200 U of penicillin per ml and 200 μg of streptomycin per ml). For transient transfections, Sol8 cells were plated in six-well tissue culture dishes at a density of 2 × 105 cells per well. The next day, DNA-calcium phosphate precipitates (25) were added directly to the culture medium. After 16 to 20 h, cells were washed twice with phosphate-buffered saline (PBS) and transferred into differentiation medium (DMEM supplemented with 5% horse serum). Cells in each well were cotransfected with 2.5 μg of luciferase reporter plasmid and 0.1 μg of pRL-SV40 (Promega). For stable transfections, 106 cells per 10-cm-diameter dish were plated and cotransfected with 10 μg of reporter construct and 2 μg of a plasmid conferring hygromycin resistance (pHyg) (56) by using the DNA-calcium phosphate technique. Cells were replated 24 h later, and hygromycin (Sigma) was added the next day to the growth medium at a concentration of 300 μg/ml. Following 3 or 4 weeks of selection, pools of resistant colonies were replated, and 2 days later they were transferred to differentiation medium. Both transient- and stably transfected Sol8 myotubes were harvested 72 h after being switched to the differentiation medium.
Plasmids.
All the reporter constructs were made by using either the promoterless pCAT Basic or pGL3 Basic vectors from Promega. Plasmids TnIs500CAT and TnIs500SURECAT as well as plasmids harboring point mutations in the core motifs (A/T1, CAGG, CACC, A/T2, and E box) of SURE have been described previously (9, 43). The 500-bp upstream sequence of the TnIs gene was also cloned into SmaI-cleaved pGL3 Basic vector to generate TnIs500LUC.
The recombinant PCR method (30) was used to generate E box Chimera, a SURE-FIRE chimeric construct in which the sequence −868 to −759 in SURE is fused 5′ to positions +664 to +633 of the FIRE sequence and cloned into the SstI-NheI-cleaved plasmid Tn500LUC. The plasmid TnIs95LUC was generated by cloning a PCR fragment containing a 95-bp upstream sequence of the rat TnIs gene into the XhoI-HindIII-cleaved pGL3 Basic vector. SURE, FIRE, and two deletion mutants lacking the sequences −868 to −778 from SURE and +776 to +703 from FIRE, respectively, were also amplified by PCR and subcloned into the SstI-NheI-cleaved plasmid TnIs95LUC to generate the constructs TnIs95SURELUC, TnIs95FIRELUC, SUREΔ−868/−778, and FIREΔ+776/+703, respectively. All the constructs generated by PCR were verified by sequencing.
Mutation of SURE by linker scanning.
By using asymmetric PCR and a single mutant primer (45) the 5′ half of SURE was mutated 6 bp at a time with an EcoRI site (GAATTC). The final PCR products were subcloned into the SstI-NheI-cleaved plasmid Tn500LUC and verified by sequencing.
Transgenic mice.
The isolation of fragments for the injection of embryos to generate transgenic mice was performed essentially as described previously (3, 43). The chloramphenicol acetyltransferase (CAT) reporter plasmids Tns500A/T1, Tns500CAGG, Tns500CACC, and Tns500Ebox were digested with HindIII and BamHI to isolate a fragment containing specific TnI sequences linked to the CAT reporter gene plus the simian virus 40 (SV40) large t-antigen intron and polyadenylation site. Two of the four Tns500CACC lines and three of the four Tns500Ebox transgenic mice generated were also described previously (43). The plasmid Ebox Chimera was digested with SstI and BamHI to isolate a fragment containing specific TnI sequence linked to the luciferase reporter gene plus SV40 late poly(A) signal. To prepare DNA for microinjection, constructs were digested to remove plasmid sequences, electrophoretically purified on agarose gels, electroeluted, and purified with ELUTIP-D columns (Schleicher & Schuell). Transgenic mice were prepared by previously described methods (31). Mice were generated, and the subsequent lines were propagated in an FVB/N background. Putative founders and their offspring were screened by Southern or slot blot analysis of tail DNA by using a CAT or luciferase probe. Adult transgenic mice were used to analyze tissue- and muscle-type-specific expression of CAT or luciferase activity. A variety of tissues including brain, liver, kidney, and heart as well as skeletal muscles from the body wall, intercostal area, diaphragm, tongue, and hind limbs were collected for the preparation of extracts. Since the reporter activity was restricted to skeletal muscles, only the extracts prepared from the hind-limb crural muscles of transgenic mice were analyzed.
Analysis of CAT and luciferase activities.
Sol8 cell extracts for CAT analysis were prepared by resuspending the collected cells in 100 μl of 0.25 M Tris-HCl (pH 8) and repeated freeze-thawing. Transgenic mouse tissue extracts were prepared essentially as described previously (17). Briefly, the tissues were sonicated in 0.25 M Tris-HCl (pH 8.0) containing 0.5 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochlorine], 2 mg of leupeptin/ml, 2 mg of aprotinin/ml, and 1 mg of pepstatin A/ml for 10 s with a Branson Sonifier 450 (microtip setting, 5; 50% efficiency). The homogenates were centrifuged at 12,000 × g for 10 min at 4°C, and the supernatants were collected for analysis. For each sample, the protein concentration was determined by the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, Ill.) with bovine serum albumin (BSA) as the standard. The CAT assays (23) were performed with the supernatants at 37°C for 1 or 3 h. The reaction products were separated by thin-layer chromatography and were quantitated with a Molecular Dynamics PhosphorImager.
To assay for luciferase activity, Sol8 myotubes were washed once with PBS and harvested with Promega passive lysis buffer. The Dual-Luciferase Reporter Assay System (Promega) was used to measure luciferase and renilla activity with a Berthold luminometer. For analysis of transgenic mice, tissues were collected in liquid nitrogen, pulverized, and immediately homogenized in Promega lysis buffer plus protease inhibitors (Complete Protease Inhibitor Cocktail; Boehringer Mannheim). Luciferase activity in the extracts was normalized for the renilla activity in the case of transiently transfected cells and for protein concentration in the case of both stably transfected cells and tissues isolated from transgenic mice.
Electrophoretic mobility shift assays (EMSAs).
The double-stranded complementary oligonucleotides of the following sequences were used in the electrophoretic mobility shift assays (the EcoRI [GAATTC] mutant sequences are shown in boldface): SURE −850/−808WT, 5′-ATA ATA GCT ACC GGA TTA ACA TAG CAG GCA TTG TCT TTC TCT G-3′; SURE −850/−808LS#3, 5′-ATA AGA ATT CCC GGA TTA ACA TAG CAG GCA TTG TCT TTC TCT G-3′; SURE −850/−808LS#4, 5′-ATA ATA GCT AGA ATT CTA ACA TAG CAG GCA TTG TCT TTC TCT G-3′; SURE −850/−808LS#5, 5′-ATA ATA GCT ACC GGA TGA ATT CAG CAG GCA TTG TCT TTC TCT G-3′; SURE −850/−808LS#6, 5′-ATA ATA GCT ACC GGA TTA ACA TGA ATT CCA TTG TCT TTC TCT G-3′; SURE −850/−808LS#7, 5′-ATA ATA GCT ACC GGA TTA ACA TAG CAG GGA ATT CCT TTC TCT G-3′; SURE −850/−808LS#8, 5′-ATA ATA GCT ACC GGA TTA ACA TAG CAG GCA TTG TGA ATT CCT G-3′; SURE −842/−815, 5′-TAC CGG ATT AAC ATA GCA GGC ATT GTC T-3′; SURE −844/−827, 5′-GCT ACC GGA TTA ACA TAG-3′; SURE −832/−815, 5′-ACA TAG CAG GCA TTG TCT-3′.
Nuclear extracts from Sol8 myoblasts and myotubes as well as from HepG2, 3T3, and rat cerebellar granule cells were prepared by the method of Dignam et al. (16) and modified according to the procedure described by Ausubel et al. (2). Whole-tissue extracts from rat soleus (SOL) and extensor digitorium longus (EDL) muscles were prepared by the sonication of tissues essentially as described by Kornhauser et al. (35). Protein concentrations were determined by BCA assay (Pierce) with BSA as a standard. For EMSAs, the double-stranded oligonucleotides were labeled with polynucleotide kinase and [γ-32P]dATP (6,000 Ci/mmol; NEN). Five to ten micrograms of nuclear extract obtained from various cell lines or 100 μg of whole-tissue extracts from rat muscle was mixed with binding buffer (20 mM HEPES [pH 7.9], 50 mM KCl, 4 mM MgCl2, 4% Ficoll, 5% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol, 2 μg of poly(dI-dC), and 32P-labeled probe (10,000 cpm) and incubated on ice for 15 min. For the competition assay, 10 pmol of unlabeled competitor oligonucleotide was used along with the labeled probe. The DNA-protein complexes were resolved by electrophoresis at 4°C on a 5% polyacrylamide–2.5% glycerol gel in 0.5× Tris-borate-EDTA buffer (51) and visualized by autoradiography.
RESULTS
TnIs and TnIf reporter constructs.
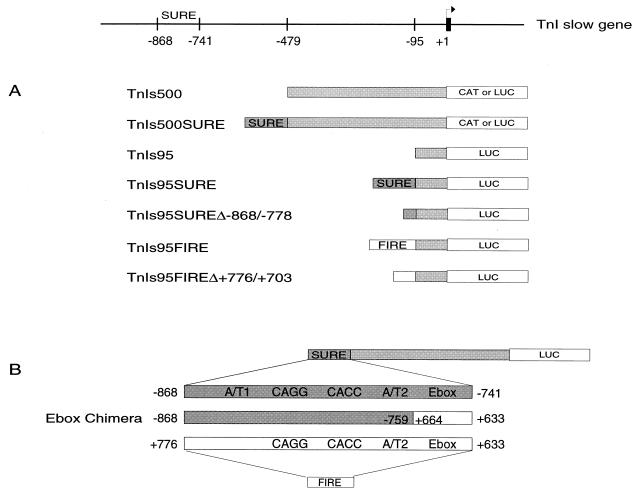
We have previously identified a SURE in the rat TnIs gene that is necessary to convey slow-twitch muscle specificity and a FIRE from the quail TnI gene that confers expression in fast-twitch muscles (9, 43). Sequence comparison of these two elements revealed the presence of four conserved motifs, namely, an E box, an AT-rich site (A/T2), a CACC site, and a novel site we called CAGG. An additional AT-rich motif, which we denoted A/T1, is conserved within the SUREs of the rat and human TnIs genes but is not present in the FIRE sequence of the quail TnIf gene (Fig. 1). In the present study, we have generated mutant TnIs reporter constructs to evaluate the functional role of these conserved motifs in transfected cultured myocytes and transgenic mice. In addition, a chimeric enhancer construct, denoted Ebox Chimera, in which the E-box element of SURE is replaced with the corresponding sequence from FIRE, was generated (Fig. 1) to analyze the role of E-box motifs in the fiber-type-specific expression of the TnIs promoter. The complete sequences of both SURE and FIRE were previously published (9, 43).
FIG. 1.
Organization of the TnIs reporter constructs used in the studies. A schematic diagram of the 5′ flanking region of the TnIs gene is shown at the top. (A) TnIs reporter constructs used for cell transfections and the generation of transgenic mice. TnIs500 contains 479 bp of the TnIs promoter inserted upstream of either the CAT or the luciferase reporter coding sequence. TnIs500SURECAT and TnIs500SURELUC were constructed by the insertion of the 128-bp SURE fragment (−868 to −741) into the TnIs500 reporter. TnIs95LUC contains 95 bp of the basal TnIs promoter upstream of the luciferase reporter coding sequence, TnIs95SURE and TnIs95FIRE were generated by the insertion of SURE and the 144-bp FIRE fragment (+776 to +663 of quail TnIf intronic sequence), respectively, upstream of the TnIs95LUC basal promoter. The deletion constructs TnIs95SURE Δ−868/−778 and TnIs95 FIRE Δ+776/+703 were made by PCR amplification of fragments −778 to −741 of SURE and +703 to +633 of FIRE, respectively, and subsequent insertion into the SstI-NheI-cleaved TnIs95LUC. (B) Schematic representation of the SURE-FIRE TnI E-box Chimera construct. Ebox Chimera was made by using the PCR ligation technique as described in Materials and Methods. The −868 to −759 sequence of SURE was joined to the +664 to +633 sequence of FIRE and inserted 5′ of the luciferase reporter. Open boxes, FIRE sequences; shaded boxes, SURE sequences.
Transcriptional analysis of the conserved SURE motifs in stably transfected myocytes.
The mouse C2C12 and Sol8 cell lines have provided an excellent system for studying muscle-specific gene regulation during myocyte differentiation. In most instances, these cells faithfully recapitulate the pattern of gene regulation observed in primary cell cultures of mammalian muscle and express a battery of skeletal-muscle-specific genes. The genes encoding TnIs and TnIf isoforms are not expressed in the undifferentiated myoblasts. However, their expression is quickly induced within 24 h after the transfer of myoblasts into differentiation medium that contains reduced levels of growth factors (3, 28). We had previously shown that CAT reporter constructs containing a 500-(TnIs500) or 250 (TnIs250)-bp upstream region of the TnIs gene conferred low levels of cell-type-specific transcription in transiently transfected C2C12 myoblasts (3). However, as shown in Fig. 2, the TnIs500 construct is inactive when stably transfected into Sol8 myocytes. This result is consistent with the observations made previously with transgenic mice where both TnIs500 and TnIs250 promoters failed to direct reporter gene expression in embryonic, fetal, and adult muscles (43). The addition of the 128-bp SURE to the TnI500 promoter increased transcription by approximately threefold in transiently transfected cells (9). More importantly, it restored the ability of the TnIs500 promoter to direct transcription in stably transfected cell lines (Fig. 2) and in transgenic mice (43). Since the results obtained from the stable transfections paralleled those observed with transgenic mice, we have chosen the former as our initial assay for examining the functional significance of the conserved SURE motifs in the muscle-specific expression of the TnIs gene.
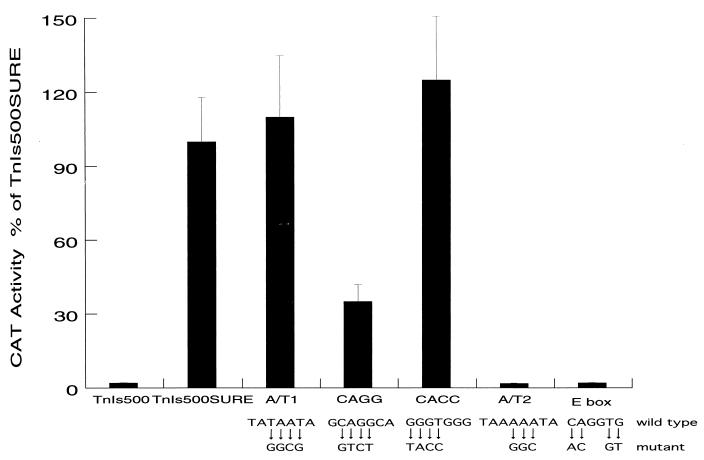
FIG. 2.
Mutational analysis of the conserved sites of SURE in stably transfected Sol8 myocytes. The transcriptional activities conferred by constructs harboring distinct mutations in each of the conserved motifs in TnI SURE were analyzed in stably transfected Sol8 myotubes. The levels of CAT reporter activity were measured in whole-cell extracts prepared from cells transfected with the A/T1, CAGG, CACC, A/T2, and E-box mutant constructs and compared to reporter levels conferred by the wild-type TnIs500SURE construct. The wild-type sequences from each of these motifs, and the mutations made therein, are shown at the bottom. For each construct, two independent DNA preparations were used to make the CaPO4 precipitates, and a total of six pools of stably transfected myotubes were analyzed per construct. The extracts (50 μg) were assayed for CAT activity at 37°C for 3 h under conditions of linear enzymatic activity. The values shown are means, and error bars indicate standard deviations.
Nuclear proteins from differentiated myotubes have been shown to bind the conserved A/T1, CACC, and A/T2 motifs in TnI SURE (9) and the E-box sequence in TnI FIRE (39). Supershift experiments with antibodies have demonstrated that myogenic bHLH factors bind E box and MEF-2 binds to the A/T2 site (9, 39, 43). To test the functional importance of the five conserved motifs, each of these sites was mutated in the context of TnIs500SURE (Fig. 2) and reporter constructs were stably transfected into Sol8 cells. As shown in Fig. 2, mutation of either the A/T2 site or the E box completely abolished the function of the TnI SURE in the Sol8 myotubes, while mutation of the CAGG motif reduced enhancer activity by 70% relative to that of the wild-type element. In contrast, mutations of either the A/T1 or CACC motif had no significant effect on reporter expression in the stably transfected cells. Thus, both the E box and A/T2 motif, which were found to bind the myogenic bHLH factors and MEF-2, respectively, are absolutely required for the function of SURE during myoblast differentiation.
Mutations in the conserved motifs affect SURE function in transgenic mice.
Since mammalian muscle cells in culture fail to mature and manifest fiber-type-specific properties, transgenic mice were used to study the functional requirement of the conserved TnI SURE motifs in specifically directing transcription in slow-twitch muscles. A total of 17 independent transgenic mouse lines were generated to study different mutations, and in all cases at least four independent “tail-positive” lines (i.e., DNA was successfully integrated into the genome) were analyzed for each construct (Fig. 3). The data obtained from hind-limb crural muscles, which include the slow-twitch (SOL) and the fast-twitch tibialis (TAS), gastrocnemius (GAS), and EDL muscles, from independent lines of mice are shown separately in Fig. 3. The muscles were collected from 6- to 8-week-old mice since the maturation of muscles and the manifestation of fiber-type-specific properties develop by 3 to 4 weeks of age.
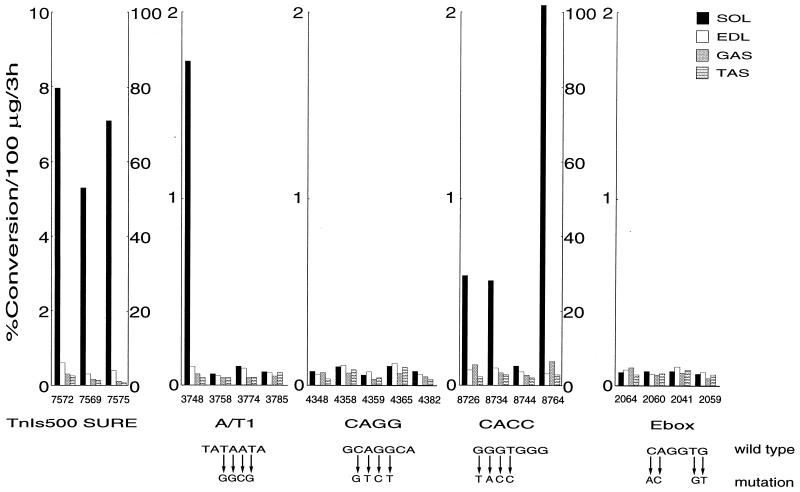
FIG. 3.
Analysis of the effects of mutations in the conserved sites of SURE in muscles of transgenic mice. CAT analyses were performed on extracts made from different muscles on transgenic mice. Independent transgenic mouse lines (indicated by number at the bottom of each bar graph) were generated with the wild-type TnIs500SURE fragment and fragments harboring mutations in the conserved A/T1, CAGG, CACC, and E-box motifs (see Fig. 2). The CAT assays were performed with 100 μg of extract at 37°C for 3 h under conditions of linear enzymatic activity. Since the values for CAT activities in the extracts of transgenic line 7575 of TnIs500SURE and 8764 of CACC were very high in relation to those of the other lines, these results are shown on a different scale.
We have previously demonstrated that all three of the transgene lines harboring the TnI500SURECAT construct exhibited reporter activity specifically in slow-twitch muscles (43), and recently we found that three of four lines made with TnIs95SURELUC also showed the same specificity (data not shown). Thus, six of the seven tail-positive transgenic mouse lines expressed either CAT or luciferase activity when driven by the wild-type SURE. This is consistent with the expected frequency of expression in tail-positive mice, because the transgene fragment integrates 10 to 15% of the time into heterochromatin and fails to express (31). As shown in Fig. 3, none of the four transgenic lines harboring mutations in the SURE E box have detectable CAT reporter activity in any of the muscles tested. These results are consistent with experiments performed with both transiently (9) and stably (Fig. 2) transfected myocytes. Similar results were also obtained for transgenes that carried the mutated SURE CAGG motif. In this case, none of the five lines exhibited CAT reporter activity in any of the muscles tested (Fig. 3). These results are in agreement with those obtained with the stably transfected Sol8 cells, where this mutation was found to diminish CAT reporter activity by 70%. Since mutations in the A/T2 site were completely detrimental to the SURE activity in the stably transfected Sol8 myocytes, we have not analyzed its effects on promoter expression in transgenic mice. However, mutations in the A/T1 motif, unlike in the stably transfected Sol8 myocytes, had a negative effect on the function of SURE in transgenic mice. Only one of four transgenic lines expressed the reporter, and as is the case for the wild-type SURE, CAT activity was restricted to the slow-twitch SOL (Fig. 3). The CACC mutation showed the least effects on TnI SURE function as three of the four transgenic lines expressed CAT preferentially in the slow-twitch SOL (Fig. 3). Thus, our results show that most of the conserved motifs in the TnI SURE are important for directing transcription in adult skeletal muscle. However, from these analyses we could not determine if any of these sites also had the ability to direct slow-muscle-specific expression. Since the role of myogenin bHLH factors in generating muscle diversity has been a matter of controversy (8), we have generated a TnI SURE-FIRE chimera enhancer to examine the involvement of the E-box motif in the determination of fiber-specific expression of the TnI genes (see below).
E-box and NFAT sequences in SURE do not determine fiber type specificity.
We have exploited the fact that the linear arrangement of the four motifs is conserved in both SURE and FIRE to generate chimeric enhancer constructs. Using this approach, we have analyzed the role of E boxes in the determination of the fiber-type-specific transcription of the TnIs gene. As shown in Fig. 1, the construct Ebox Chimera was made by replacing the E box (plus flanking sequences) from SURE with the FIRE E-box sequence, a process which also results in the removal of the NFAT site (10) from the SURE. The experiments presented earlier established that the SURE E box is completely indispensable for enhancer function as tested in either transiently or stably transfected myocytes and in transgenic mice (Fig. 2 and 3) (9). For this reason, the TnI Ebox Chimera was initially tested in transfected Sol8 myotubes where it was found to be as active as the wild-type TnI SURE (data not shown). These results indicate that the FIRE E box can functionally substitute for the SURE E box. To test if the E-box elements were directly involved in the fiber-specific expression of the TnI promoter, six independent transgenic lines were generated with the TnI Ebox Chimera fragment. As shown in Fig. 4, in five of the six transgenic lines the E-box chimera enhancer conferred higher levels of transcription in slow-twitch muscle (SOL) than in the fast-twitch EDL, GAS, and TAS muscles; one line did not express the transgene. The relative levels of reporter activities in different muscles of the Ebox Chimera transgenic mice were indistinguishable from those in the mice harboring the wild-type TnI SURE. These results demonstrate that, even though the FIRE E box functionally substitutes for the E box in SURE, the TnIf E box is not sufficient to redirect transcription to fast-twitch fibers. In addition, the absence of the NFAT site in the chimera construct does not affect slow-twitch-specific activity of the enhancer. To our knowledge, this constitutes the first direct demonstration that the E-box motif, although essential for transcriptional activity of many muscle genes, does not determine the fiber-type-specific expression of skeletal muscle genes.
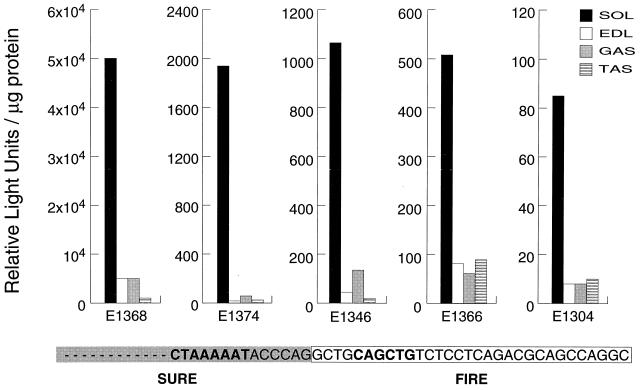
FIG. 4.
The E box from FIRE functionally substitutes for the SURE E box but does not change the fiber-type specificity of TnI SURE. Transgenic mouse lines were generated with a fragment from the TnI Ebox Chimera construct, in which the E box from SURE was swapped for the FIRE E box (Fig. 1). Luciferase activities assayed in extracts made from different hind-limb muscles of five TnI Ebox Chimera transgenic lines were normalized for protein concentration. The sequence of the TnI Ebox Chimera construct, at the boundary between SURE (shaded box) and FIRE (open box), is shown at the bottom; the E-box and A/T-2 motifs are shown in boldface.
The upstream half of SURE is required for transcription in stable transfectants.
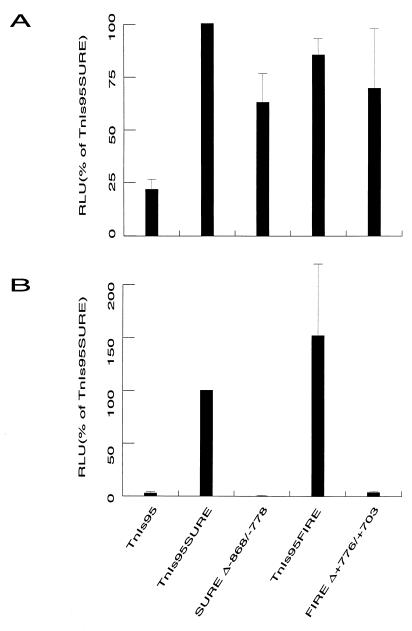
The rat and human TnI SUREs share over 85% homology throughout the entire 128-bp enhancer (9), while homology with the quail TnI FIRE is mostly limited to the four conserved motifs. Although the cis-acting elements present in the downstream halves of SURE (9) and FIRE (39) have been characterized, not much is known about the regulatory sequences in the 5′ end of SURE, which exhibits little homology to FIRE. Hence, we were interested in determining the functional importance of sequences residing in the 5′ region of the TnI enhancers. As shown in Fig. 5, the constructs SUREΔ −868/−778 and FIREΔ +776/+703, which lack all the sequences upstream of the A/T2 and E-box elements in SURE and FIRE, respectively (see Fig. 1), reduce transcription levels by approximately 30% in the transiently transfected Sol8 myotubes (Fig. 5A). In stark contrast, these constructs completely fail to activate transcription in the stably transfected myotubes (Fig. 5B), suggesting that the upstream elements within SURE and FIRE are essential for enhancer function when integrated into chromatin.
FIG. 5.
The 5′ halves of SURE and FIRE are important for transcriptional activity in stably transfected Sol8 myotubes. The transcriptional activities conferred by the constructs TnI95 (negative control), TnI95SURE (positive control), SURE Δ−868/−778 (5′ half deleted), and FIRE Δ+776/+703 (5′ half deleted) (see Fig. 1) were analyzed in transiently (A) and stably (B) transfected Sol8 myotubes. The luciferase reporter activities assayed in the extracts were normalized either to renilla activity expressed from an internal control plasmid (transient transfections) or for protein concentration (stable transfections). Values are means (n ≥ 6); error bars indicate standard deviations.
The upstream region of SURE binds nuclear factors that are not restricted to muscle cells.
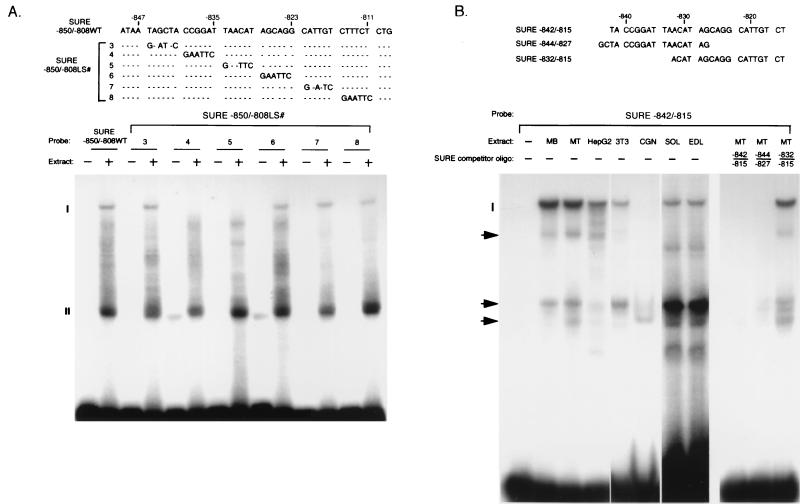
Sequences in the downstream halves of the rat TnI SURE (9) and the quail TnI FIRE (39), extending from the CACC sites to the E boxes, have been shown to bind nuclear factors from Sol8 and 23A2 myofiber cells, respectively. The myogenic bHLH factors bind to the E box, MEF-2 binds to the A/T2 site, and the binding pattern observed with the CACC site is similar to that seen with the myoglobin CACC which binds to Sp1, CB40, and the winged-helix myocyte nuclear factor-1 (4). We had previously shown that the A/T1 site present in the upstream region of SURE bound to factors present in Sol8 myotube nuclear extracts (9). In this study, because of their importance for the expression of the TnIs promoter in stably transfected myocytes (Fig. 5B), we have analyzed the sequences between −850 and −808 of SURE for their ability to interact with the nuclear proteins present in different cell lines and tissues. A 32P-labeled double-stranded oligonucleotide that includes the sequence between −850 to −808 has been used as the wild-type control (SURE −850/−808WT). A series of six oligonucleotides, each with a 6-bp nested EcoRI site mutation, spanning the sequence between −850 and −808 (from SURE −850/−808LS#3 to SURE −850/−808LS#8), were generated to identify the sequences involved in interaction with the nuclear proteins. As seen from the EMSAs performed with Sol8 myotube nuclear extracts (Fig. 6A), the wild-type SURE −850/−808WT sequence bound two prominent complexes, I and II. However, the faster-moving complex II seems to be nonspecific in nature since the appearance of this band is not affected by any of the mutations that were introduced along the length of the probe sequence (Fig. 6A, SURE −850/−808LS#3 to SURE −850/−808LS#8). In contrast, complex I is formed as a result of specific interactions and requires sequences between −840 and −827 of SURE, since only mutations that alter this region in the wild-type oligonucleotide prevented its formation in the presence of Sol8 nuclear extracts (Fig. 6A, SURE −850/−808LS#4 and SURE −850/−808LS#5). The involvement of the sequences between −840 and −829 of SURE in the formation of complex I is also evident from the observation that only the sequence between −844 and −829 (SURE −844/−827), in contrast to the oligonucleotide containing the CAGG motif (SURE −832/−815), is able to compete for the complexes formed by SURE −842/−815 in Sol8 myotube nuclear extracts (Fig. 6B). Interestingly, as described in the next section, the sequence that is responsible for the formation of complex I is also found to be necessary for the expression of SURE in Sol8 myocytes (Fig. 7B, TnIsLS#4 and TnIsLS#5). Comparison of this sequence to known cis elements in the GCG transcription factor database identified a site with a strong consensus (GTTAATCCG) to the Drosophila bicoid binding site (55). To examine the tissue-specific expression of the nuclear factor(s) that bound to this sequence, EMSAs with the SURE sequence from −842 to −815, which contained both the consensus bicoid element and the CAGG motif, were performed with nuclear and whole-cell extracts obtained from various cell lines and tissues. These included mesodermally derived Sol8 myocytes, 3T3 fibroblasts, endodermally derived hepatoma HepG2 cells, the ectodermally derived cerebellar granule cells, and the rat SOL (slow-twitch) and EDL (fast-twitch) muscles. As shown in Fig. 6B, binding to the consensus bicoid sequence was not restricted to the extracts from the differentiated Sol8 myocytes and SOL, and EDL muscle fibers but was also found in the nuclear proteins from undifferentiated myoblasts and HepG2 and 3T3 cells. However, the cerebellar granule cells, except for a minor complex, lacked this binding activity, indicating that the factor(s) involved in the interaction with the bicoid site of SURE, even though present in the nonmuscle cells of mesodermal and endodermal origin, is not ubiquitous. A predominant faster-moving bicoid site binding complex is observed in the muscle extracts compared with the HepG2 and 3T3 extracts (Fig. 6B). However, this complex is equally abundant in both SOL and EDL muscles, suggesting that it may not be involved in the fiber-specific transcription of the TnI genes.
FIG. 6.
EMSAs of nuclear protein binding to SURE upstream sequences. (A) Sequences between −840 and −829 of SURE interact with nuclear extracts from Sol8 myotubes. 32P-labeled double-stranded oligonucleotides with the wild-type sequence (SURE −850/−808WT) and GAATTC linker mutations (SURE −850/−808LS#3 to SURE −850/−808LS#8) are shown at the top (dashes indicate sequence identities between the wild-type and mutant oligonucleotides). The probe oligonucleotides were incubated on ice in the absence (−) or in the presence (+) of 5 μg of nuclear extracts from Sol8 myotubes. The positions of complexes I and II are indicated on the left. (B) The complexes that bind the −840 and −829 sequence are not restricted to muscle cells. Sequences of the SURE −842/−815, SURE −844/−827, and SURE −832/−815 oligonucleotides used in the EMSAs are shown at the top. Nuclear extracts (10 μg) from undifferentiated Sol8 myoblasts (MB), differentiated Sol8 myotubes (MT), HepG2 hepatoma cells, 3T3 fibroblasts, and cerebellar granule cells (CGN), as well as whole-tissue extracts (100 μg) from SOL and EDL hind-limb skeletal muscles were incubated with the 32P-labeled SURE −842/−815 oligonucleotide and analyzed as described above. For competition assays, a 100-fold molar excess of unlabeled SURE −842/−815, SURE −844/−827, or SURE −832/−815 was used. The SOL and EDL lanes were exposed to X-ray film for 72 h, and the other lanes were autoradiographed for 24 h.
FIG. 7.
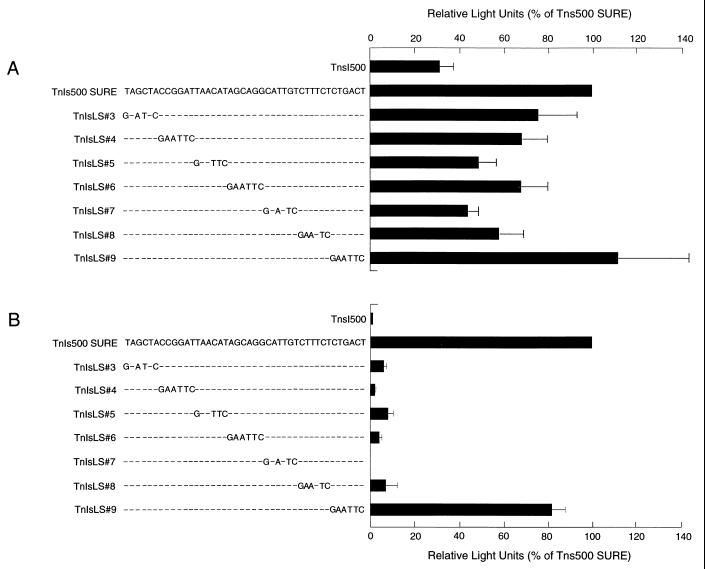
Linker scanning mutations through the 5′ end of TnI SURE. An EcoRI recognition sequence (GAATTC) was used to generate a series of linker scanning mutations in the upstream region of SURE (TnIsLS#3 to TnIsLS#9). Dashes indicate sequence identities between the wild-type and the mutant oligonucleotides. These constructs and wild-type TnIs500SURE were used to transfect Sol8 myocytes. Luciferase reporter activities were assayed in extracts made from Sol8 myotubes and normalized either for the renilla used as an internal control in the transient transfections (A) or for protein concentration in the stable transfections (B). Values are means (n ≥ 6); error bars indicate standard deviations.
Sequences necessary for SURE function are identified by linker scanning mutations.
The effects of the series of progressive 6-bp mutations, which correspond to the sequences used in the EMSAs described above, were tested for SURE function in transfected myocytes. As shown in Fig. 7, mutations in the sequences residing between −846 and −811 only moderately reduced luciferase activity in transiently transfected myotubes. However, the same mutations caused a more dramatic decrease in TnIs promoter expression in the stably transfected cells (Fig. 7B). In the transiently transfected myotubes, the linker scanning mutations LS#3 to LS#8 reduced reporter activity to between 30 to 50% of that of the levels observed with the wild-type TnI SURE. In contrast, mutations LS#3 to LS#8 reduced the levels of reporter gene expression to less than 10% of the levels seen in the cells transfected with the TnI SURE construct. The linker scanning mutation TnsLS#6, which covers the core of the CAGG motif in addition to the upstream flanking bases, was more effective in reducing reporter levels than the point mutations restricted exclusively to the CAGG core (see Fig. 2). Not all the sequences in this region are essential for enhancer function. The construct harboring the LS#9 mutation, mapping to a site located between the CAGG and CACC motifs, conferred approximately 80% of the wild-type activity expressed in stably transfected myotubes. These results corroborate those obtained from the deletion experiments (Fig. 5B), demonstrating the importance of the 5′ sequences for the enhancer activity of SURE in stably transfected myotubes, and suggest that cooperation between the upstream and downstream sequences of SURE is essential for transcription of the TnIs gene in its chromosomal context.
DISCUSSION
Transcriptional analyses presented in this study demonstrate that multiple sequence elements present within SURE determine the early developmental as well as the slow-fiber-type-specific activity of the TnIs promoter. Of these elements, four sites, an E box, an AT-rich element, a CACC sequence, and a CAGG motif, are also conserved in the enhancer element of the quail TnIf gene (39, 61). In addition, SURE contains a second AT-rich sequence and a bicoid consensus site in its 5′ end that are not present in FIRE. Mutations in any of the conserved sites, with the exception of CACC, result in the loss of muscle-specific SURE activity in adult mice, suggesting that interactions among the factors binding to these sites might be essential for its enhancer function. Moreover, since some of these elements are present in both SURE and FIRE, the fiber-specific activity is possibly controlled by the existence of distinct factors that interact with the nonconserved regions within these enhancers.
The importance of the combinatorial involvement of some of these sites in the regulation of many other skeletal muscle genes has been well documented. In the quail TnIf IRE, which we refer to as FIRE, mutations in any single element among the E-box, AT-rich, and CACC sequences abrogate MyoD-induced enhancer activity in myofiber cell cultures (39). The presence of CCAC and AT-rich elements was shown to be essential for expression of the human myoglobin promoter in Sol8 myotubes (26). Similarly, the requirement for a novel site, referred to as MEF-3, along with AT-rich MEF-2 and CACC elements, for transcription of the slow/cardiac TnC gene (44) and of MEF-3 and MEF-2 for the regulation of rat aldolase distal promoter (pM) activity (29) has been demonstrated in muscle cells. However, evidence for the involvement of these elements in the regulation of fiber-type-specific gene expression in the adult muscle has not been established, although MEF-3 in conjunction with NFI was implicated in the activity of rat aldolase pM in a subset of fast-twitch muscles (54).
In order to characterize the discrete elements responsible for both the myocyte-specific and the slow-twitch-fiber-specific activity of the TnIs promoter in its chromatin context, we performed mutational analyses of conserved and non-conserved regions of SURE in stably transfected Sol8 myotubes and transgenic mice. Three elements that are present in the 5′ half of SURE, which we refer to as the A/T1, bicoid-like, and CAGG motifs, were also shown to be necessary for the muscle-specific activity of the enhancer (Fig. 3 and 7). Moreover, since the deletion of these sites, along with CACC, from SURE diminished enhancer activity only in stably transfected myocytes and not in the transiently expressing cells (Fig. 5), this region might play a critical role in establishing the open chromatin structure needed for active transcription. In addition, the deleterious effect of the A/T1 mutation in transgenic mice (Fig. 3) suggests that this site and the sequences flanking it might be important for TnIs promoter activity during early development. Within this region, A/T1 and an adjacent bicoid consensus site (GTTAATCCG) bind nuclear proteins from Sol8 myotubes (see above and reference 9). Although the identity of the factor(s) binding to these sequences is not established, recently a similar AT-rich element in the murine IIB myosin heavy chain gene promoter, named mAT2 site, has been shown to bind Oct-1, a protein that contains a POU binding domain (36). Similarly, sequences related to the GTTAATCCG site, mutation of which results in the loss of SURE activity in the stably transfected Sol8 myotubes (Fig. 4), have been demonstrated to bind the bicoid subfamily of homeobox proteins in Drosophila and mice (37, 40). Oct-1 and bicoid-related proteins are suggested to play an important role in cell lineage specification and pattern formation, respectively, during the embryonic stages of development (11, 14, 21, 38). Experiments involving the analysis of transgenes carrying bicoid site mutations should help in the evaluation of the role of this sequence in the fiber-type-specific expression of the TnIs promoter.
We could not detect any direct interaction between the CAGG sequence and the nuclear extracts from myocytes, although this site is essential for SURE activity in both myocytes and transgenic mice. It is possible that this site interacts with complexes formed on other motifs in the TnIs promoter, and thus the complexes bound to this sequence might not be detected in the absence of those elements. Another possibility is that the phosphorylation status of the protein(s) in the extract might dictate its interaction with the CAGG element. Alternatively, this site and the sequences flanking it might play an important role in the chromatin remodeling mediated by the MyoD family of transcription factors during myogenic lineage determination (22), a hypothesis that is not inconsistent with the observation that E-box mutations result in a complete loss of SURE activity (Fig. 2 and 3). A possible requirement for interaction between the proximal sequences (which include E-box and A/T2 elements) and the distal sequences (which contain A/T1, bicoid consensus, and CAGG motifs) of SURE within the chromosomal context is also indicated by the fact that the SURE construct which includes only the proximal sequences is insufficient to support TnIs promoter activity in the stably transfected Sol8 myotubes (Fig. 5B).
Since the interactions between the factors binding to multiple elements within SURE are necessary for its function, it was important that the order and spacing among these sites were preserved for further characterization of fiber-type-specific regulatory elements in the TnIs enhancer. Using a SURE-FIRE fusion construct, we have examined the role that the E boxes present in SURE and FIRE play in the determination of TnI expression in slow and fast muscles, respectively. Functional differences among various E-box elements, based on variations both within the consensus binding site and the flanking sequences, have been demonstrated in the context of many other muscle-specific gene promoters (1, 62). Differential expression of myogenic factors that interact with E-box sequences has been demonstrated for slow and fast muscles (32, 33, 57). In addition, simultaneous mutation of three E boxes in the muscle creatine kinase promoter reduce transcription in slow-twitch SOL but not in fast-twitch skeletal muscles (53). Based on these findings, it has been proposed that the bHLH factors may regulate the slow- and fast-fiber-type-specific expression of the contractile genes in the muscle (32, 33, 53). However, several lines of evidence that indicated a negative correlation between the transcriptional activation of these genes and the expression of bHLH factors in the skeletal muscle have also been presented (8). Thus, in order to study if the E-box elements of slow and fast TnI enhancers determined the fiber type specificity of their respective promoters by virtue of their ability to bind different bHLH factors, we analyzed the expression of an E-box chimera in transgenic mice (Fig. 4). This construct, in which the E box within SURE was replaced with the equivalent sequence from FIRE, was still transcriptionally active only in the slow-muscle fibers, similar to the wild-type TnI SURE, indicating that the FIRE E box is able to functionally substitute for the SURE E element and that the E box on its own cannot determine the fiber-type-specific activity of the TnI enhancer sequences. These results are in agreement with the observations that mutations of E-box sequences in the MLC1f/3f (47) and aldolase A (50) promoters do not affect fiber-type-specific expression in transgenic mice.
Recently, Chin et al. (10) have shown that expression of the human TnIs and myoglobin promoters in transfected cells is controlled by the calcineurin- and NFAT-mediated signaling pathway. Based on their results, it was proposed that the NFAT transcription factors regulate slow-fiber-type specificity of muscle gene expression. However, as we have shown in this study, a SURE-FIRE E-box chimera construct, in which the NFAT site of SURE (−760/−753) is removed, continues to exhibit slow-fiber-specific expression. In addition, FIRE, which contains at least two consensus NFAT sites (+770/+763 and +756/+749), directs fast-fiber-specific transcription and is not expressed in slow muscles. The various analyses we have presented in this study, performed with both myocytes and transgenic mice, provide strong evidence for the conclusion that the interaction between factors binding to multiple cis elements within SURE are essential for conferring slow-fiber-type specificity to the TnIs promoter and that the fiber-specific expression of SURE does not require the NFAT binding site.
ACKNOWLEDGMENT
We are grateful to Daniel Abebe for his expert technical assistance.
REFERENCES
- 1.Apone S, Hauschka S D. Muscle gene E-box control elements. Evidence for quantitatively different transcriptional activities and the binding of distinct regulatory factors. J Biol Chem. 1995;270:21420–21427. doi: 10.1074/jbc.270.36.21420. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M. Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology. 2nd ed. Brooklyn, N.Y: Greene Publishing Associates; 1992. [Google Scholar]
- 3.Banerjee-Basu S, Buonanno A. cis-acting sequences of the rat troponin I slow gene confer tissue- and development-specific transcription in cultured muscle cells as well as fiber type specificity in transgenic mice. Mol Cell Biol. 1993;13:7019–7028. doi: 10.1128/mcb.13.11.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassel-Duby R, Hernandez M D, Yang Q, Rochelle J M, Seldin M F, Williams R S. Myocyte nuclear factor, a novel winged-helix transcription factor under both developmental and neural regulation in striated myocytes. Mol Cell Biol. 1994;14:4596–4605. doi: 10.1128/mcb.14.7.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold H H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989;8:701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun T, Winter B, Bober E, Arnold H H. Transcriptional activation domain of the muscle-specific gene regulatory protein myf5. Nature. 1990;346:663–665. doi: 10.1038/346663a0. [DOI] [PubMed] [Google Scholar]
- 7.Buckingham M. Making muscle in mammals. Trends Genet. 1992;8:144–148. doi: 10.1016/0168-9525(92)90373-C. [DOI] [PubMed] [Google Scholar]
- 8.Buonanno A, Rosenthal N. Molecular control of muscle diversity and plasticity. Dev Genet. 1996;19:95–107. doi: 10.1002/(SICI)1520-6408(1996)19:2<95::AID-DVG1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 9.Calvo S, Stauffer J, Nakayama M, Buonanno A. Transcriptional control of muscle plasticity: differential regulation of troponin I genes by electrical activity. Dev Genet. 1996;19:169–181. doi: 10.1002/(SICI)1520-6408(1996)19:2<169::AID-DVG9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark M E, Mellon P L. The POU homeodomain transcription factor Oct-1 is essential for activity of the gonadotropin-releasing hormone neuron-specific enhancer. Mol Cell Biol. 1995;15:6169–6177. doi: 10.1128/mcb.15.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corin S J, Juhasz O, Zhu L, Conley P, Kedes L, Wade R. Structure and expression of the human slow twitch skeletal muscle troponin I gene. J Biol Chem. 1994;269:10651–10659. [PubMed] [Google Scholar]
- 13.Corin S J, Levitt L K, O’Mahoney J V, Joya J E, Hardeman E C, Wade R. Delineation of a slow-twitch-myofiber-specific transcriptional element by using in vivo somatic gene transfer. Proc Natl Acad Sci USA. 1995;92:6185–6189. doi: 10.1073/pnas.92.13.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dailey L, Yuan H, Basilico C. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol Cell Biol. 1994;14:7758–7769. doi: 10.1128/mcb.14.12.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis R L, Weintraub H, Lassar A B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 16.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 17.Donoghue M J, Merlie J P, Rosenthal N, Sanes J R. Rostrocaudal gradient of transgene expression in adult skeletal muscle. Proc Natl Acad Sci USA. 1991;88:5847–5851. doi: 10.1073/pnas.88.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edmondson D G, Cheng T C, Cserjesi P, Chakraborty T, Olson E N. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol Cell Biol. 1992;12:3665–3677. doi: 10.1128/mcb.12.9.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edmondson D G, Olson E N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989;3:628–640. doi: 10.1101/gad.3.5.628. . (Erratum 4:1450, 1990.) [DOI] [PubMed] [Google Scholar]
- 20.Emerson C P., Jr Skeletal myogenesis: genetics and embryology to the fore. Curr Opin Genet Dev. 1993;3:265–274. doi: 10.1016/0959-437x(93)90033-l. [DOI] [PubMed] [Google Scholar]
- 21.Finkelstein R, Perrimon N. The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature. 1990;346:485–488. doi: 10.1038/346485a0. . (Comments.) [DOI] [PubMed] [Google Scholar]
- 22.Gerber A N, Klesert T R, Bergstrom D A, Tapscott S J. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 23.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gossett L A, Kelvin D J, Sternberg E A, Olson E N. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham F L, Van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 26.Grayson J, Williams R S, Yu Y T, Bassel-Duby R. Synergistic interactions between heterologous upstream activation elements and specific TATA sequences in a muscle-specific promoter. Mol Cell Biol. 1995;15:1870–1878. doi: 10.1128/mcb.15.4.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hallauer P L, Hastings K E, Peterson A C. Fast skeletal muscle-specific expression of a quail troponin I gene in transgenic mice. Mol Cell Biol. 1988;8:5072–5079. doi: 10.1128/mcb.8.12.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hastings K E, Koppe R I, Marmor E, Bader D, Shimada Y, Toyota N. Structure and developmental expression of troponin I isoforms. cDNA clone analysis of avian cardiac troponin I mRNA. J Biol Chem. 1991;266:19659–19665. [PubMed] [Google Scholar]
- 29.Hidaka K, Yamamoto I, Arai Y, Mukai T. The MEF-3 motif is required for MEF-2-mediated skeletal muscle-specific induction of the rat aldolase A gene. Mol Cell Biol. 1993;13:6469–6478. doi: 10.1128/mcb.13.10.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 31.Hogan B. Manipulating the mouse embryo: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 32.Hughes S M, Koishi K, Rudnicki M, Maggs A M. MyoD protein is differentially accumulated in fast and slow skeletal muscle fibres and required for normal fibre type balance in rodents. Mech Dev. 1997;61:151–163. doi: 10.1016/s0925-4773(96)00631-4. [DOI] [PubMed] [Google Scholar]
- 33.Hughes S M, Taylor J M, Tapscott S J, Gurley C M, Carter W J, Peterson C A. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- 34.Konieczny S F, Emerson C P., Jr Complex regulation of the muscle-specific contractile protein (troponin I) gene. Mol Cell Biol. 1987;7:3065–3075. doi: 10.1128/mcb.7.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornhauser J M, Nelson D E, Mayo K E, Takahashi J S. Regulation of jun-B messenger RNA and AP-1 activity by light and a circadian clock. Science. 1992;255:1581–1584. doi: 10.1126/science.1549784. [DOI] [PubMed] [Google Scholar]
- 36.Lakich M M, Diagana T T, North D L, Whalen R G. MEF-2 and oct-1 bind to two homologous promoter sequence elements and participate in the expression of a skeletal muscle-specific gene. J Biol Chem. 1998;273:15217–15226. doi: 10.1074/jbc.273.24.15217. [DOI] [PubMed] [Google Scholar]
- 37.Lamonerie T, Tremblay J J, Lanctot C, Therrien M, Gauthier Y, Drouin J. Ptx1, a bicoid-related homeobox transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 38.Lanctot C, Lamolet B, Drouin J. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development. 1997;124:2807–2817. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 39.Lin H, Yutzey K E, Konieczny S F. Muscle-specific expression of the troponin I gene requires interactions between helix-loop-helix muscle regulatory factors and ubiquitous transcription factors. Mol Cell Biol. 1991;11:267–280. doi: 10.1128/mcb.11.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, Yuan D, Diepold K, Scarborough T, Ma J. The Drosophila morphogenetic protein Bicoid binds DNA cooperatively. Development. 1996;122:1195–1206. doi: 10.1242/dev.122.4.1195. [DOI] [PubMed] [Google Scholar]
- 41.Miner J H, Wold B. Herculin, a fourth member of the MyoD family of myogenic regulatory genes. Proc Natl Acad Sci USA. 1990;87:1089–1093. doi: 10.1073/pnas.87.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulle C, Benoit P, Pinset C, Roa M, Changeux J P. Calcitonin gene-related peptide enhances the rate of desensitization of the nicotinic acetylcholine receptor in cultured mouse muscle cells. Proc Natl Acad Sci USA. 1988;85:5728–5732. doi: 10.1073/pnas.85.15.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama M, Stauffer J, Cheng J, Banerjee-Basu S, Wawrousek E, Buonanno A. Common core sequences are found in skeletal muscle slow- and fast-fiber-type-specific regulatory elements. Mol Cell Biol. 1996;16:2408–2417. doi: 10.1128/mcb.16.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parmacek M S, Ip H S, Jung F, Shen T, Martin J F, Vora A J, Olson E N, Leiden J M. A novel myogenic regulatory circuit controls slow/cardiac troponin C gene transcription in skeletal muscle. Mol Cell Biol. 1994;14:1870–1885. doi: 10.1128/mcb.14.3.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrin S, Gilliland G. Site-specific mutagenesis using asymmetric polymerase chain reaction and a single mutant primer. Nucleic Acids Res. 1990;18:7433–7438. doi: 10.1093/nar/18.24.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 47.Rao M V, Donoghue M J, Merlie J P, Sanes J R. Distinct regulatory elements control muscle-specific, fiber-type-selective, and axially graded expression of a myosin light-chain gene in transgenic mice. Mol Cell Biol. 1996;16:3909–3922. doi: 10.1128/mcb.16.7.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhodes S J, Konieczny S F. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 1989;3:2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- 49.Salminen M, Spitz F, Fiszman M Y, Demignon J, Kahn A, Daegelen D, Maire P. Myotube-specific activity of the human aldolase A M-promoter requires an overlapping binding site for NF1 and MEF2 factors in addition to a binding site (M1) for unknown proteins. J Mol Biol. 1995;253:17–31. doi: 10.1006/jmbi.1995.0532. [DOI] [PubMed] [Google Scholar]
- 50.Salminen M, Lopez S, Maire P, Kahn A, Daegelen D. Fast-muscle-specific DNA-protein interactions occurring in vivo at the human aldolase A M promoter are necessary for correct promoter activity in transgenic mice. Mol Cell Biol. 1996;16:76–85. doi: 10.1128/mcb.16.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 52.Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 53.Shield M A, Haugen H S, Clegg C H, Hauschka S D. E-box sites and a proximal regulatory region of the muscle creatine kinaes gene differentially regulate expressioni in diverse skeletal muscle and cardiac muscle of transgenic mice. Mol Cell Biol. 1996;16:5058–5068. doi: 10.1128/mcb.16.9.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spitz F, Salminen M, Demignon J, Kahn A, Daegelen D, Maire P. A combination of MEF3 and NFI proteins activates transcription in a subset of fast-twitch muscles. Mol Cell Biol. 1997;17:656–666. doi: 10.1128/mcb.17.2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 56.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voytik S L, Przyborski M, Badylak S F, Konieczny S F. Differential expression of muscle regulatory factor genes in normal and denervated adult rat hindlimb muscles. Dev Dyn. 1993;198:214–224. doi: 10.1002/aja.1001980307. [DOI] [PubMed] [Google Scholar]
- 58.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Truner D, Rupp R, Hollenberg S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 59.Wright W E, Sassoon D A, Lin V K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 60.Yu Y T, Breitbart R E, Smoot L B, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 61.Yutzey K E, Kline R L, Konieczny S F. An internal regulatory element controls troponin I gene expression. Mol Cell Biol. 1989;9:1397–1405. doi: 10.1128/mcb.9.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yutzey K E, Konieczny S F. Different E-box regulatory sequences are functionally distinct when placed within the context of the troponin I enhancer. Nucleic Acids Res. 1992;20:5105–5113. doi: 10.1093/nar/20.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zot A S, Potter J D. Structural aspects of troponin-tropomyosin regulation of skeletal muscle contraction. Annu Rev Biophys Biophys Chem. 1987;16:535–559. doi: 10.1146/annurev.bb.16.060187.002535. [DOI] [PubMed] [Google Scholar]