Abstract
The wood sorrel family, Oxalidaceae, is mainly composed of annual or perennial herbs, a few shrubs, and trees distributed from temperate to tropical zones. Members of Oxalidaceae are of high medicinal, ornamental, and economic value. Despite the rich diversity and value of Oxalidaceae, few molecular markers or plastomes are available for phylogenetic analysis of the family. Here, we reported four new whole plastomes of Oxalidaceae and compared them with plastomes of three species in the family, as well as the plastome of Rourea microphylla in the closely related family Connaraceae. The eight plastomes ranged in length from 150,673 bp (Biophytum sensitivum) to 156,609 bp (R. microphylla). Genome annotations revealed a total of 129–131 genes, including 83–84 protein-coding genes, eight rRNA genes, 37 tRNA genes, and two to three pseudogenes. Comparative analyses showed that the plastomes of these species have minor variations at the gene level. The smaller plastomes of herbs B. sensitivum and three Oxalis species are associated with variations in IR region sizes, intergenic region variation, and gene or intron loss. We identified sequences with high variation that may serve as molecular markers in taxonomic studies of Oxalidaceae. The phylogenetic trees of selected superrosid representatives based on 76 protein-coding genes corroborated the Oxalidaceae position in Oxalidales and supported it as a sister to Connaraceae. Our research also supported the monophyly of the COM (Celastrales, Oxalidales, and Malpighiales) clade.
Keywords: Oxalidaceae, Plastome, Oxalidales, Gene loss, COM clade, Phylogeny
Highlights
-
•
Plastomes from the genus Biophytum (Oxalidaceae) and the family Connaraceae are reported for the first time.
-
•
Little plastome variation was observed among genera of Oxalidaceae.
-
•
Several divergence hotspot regions were identified in the Oxalidaceae species.
-
•
The systematic position of Oxalidaceae was confirmed as sister to Connaraceae within Oxalidales.
1. Introduction
The wood sorrel family, known as Oxalidaceae, is composed of annual or perennial herbs, a few shrubs and trees, and consists of eight genera and about 600 accepted species (The Plant List, 2013) widely distributed in tropical, subtropical, and temperate habitats (Sá et al., 2019). The genera Averrhoa L., Biophytum DC., and Oxalis L. represent the largest groups of Oxalidaceae, with approximately five, 80, and 500 species, respectively (The Plant List, 2013). Most members of Oxalidaceae are important economic crops that possess a multitude of bioactive compounds with high medicinal value, and fancy fruits or flowers utilized for ornamental decoration, such as star fruit (Averrhoa carambola L.), bilimbi (A. bilimbi L.), Biophytum sensitivum (L.) DC., and Oxalis corymbosa DC. (Singh et al., 2017; Sá et al., 2019). Understanding the evolutionary and biological issues of this taxon requires phylogenetic analysis.
Oxalidaceae has often been included in Geraniales and once was suggested to have a close relationship with the Geraniaceae family based on morphology (Mathew, 1958; Stevens et al., 2004). Similarly, the family Connaraceae was once classified into Sapindales based on the characteristics of its leaves and fruits (Paim et al., 2020). However, phylogenetic studies have indicated that the two families both belong to Oxalidales and are sister to each other (APG IV, 2016). Oxalidales belongs to the ‘COM clade’, which also includes Celastrales and Malpighiales and has been broadly recognized as a well-defined monophyletic group (APG IV, 2016; Valencia-D et al., 2020). Nevertheless, several recent studies based on nuclear genes suggested contradictory relationships (Zhao et al., 2016; Zeng et al., 2017; Yang et al., 2020). Despite support for monophyly of each of the orders within COM, the phylogenetic relationships among the orders of this group remain ambiguous (APG IV, 2016; Valencia-D et al., 2020).
Phylogenetic studies of Oxalidaceae are relatively scarce and the number of molecular informative sites used in analyses is limited (Heibl and Renner, 2012; Aoki et al., 2017). Most phylogenetic studies of Oxalidaceae have focused on Oxalis species, which have high morphological variation, phenotypic plasticity, and a wide geographical range (Vaio et al., 2016; Aoki et al., 2017; Moura et al., 2020); in contrast, few studies have been carried out on Averrhoa and Biophytum. Even so, phylogenetic analyses of Oxalis have been mainly based on several plastid non-coding regions (petA-psbJ, trnL-trnF, trnS-trnG, and trnT-trnL), nuclear ribosomal internal transcribed spacer (ITS) sequences or low-copy nuclear genes, and some results showed incongruences or low resolution (Oberlander et al., 2011; Schmickl et al., 2015; Aoki et al., 2017). Therefore, the intrageneric phylogenetic relationships of Oxalis are still ambiguous. For these reasons, more genomic information on Oxalidaceae species is urgently needed to improve the knowledge about their genetic structure and further elucidate their detailed phylogenetic relationships. However, there are few plastomes of Averrhoa and Oxalis species publicly available and no plastome of Biophytum species has been sequenced. Likewise, no study has reported plastome data of Connaraceae species, although these data are necessary for studying relationships between Oxalidaceae and Connaraceae.
Compared with traditional DNA markers, genome-wide data sets have the advantage of providing information to effectively resolve difficult phylogenetic questions at different taxonomic levels (Barrett et al., 2016). The gene content and structure of flowering plant plastomes are highly conserved, having about 120–160 kilobases (kb) and a quadripartite structure that includes two inverted repeats (IRs), a large single-copy (LSC) region, and a small single-copy (SSC) region (Ruhlman and Jansen, 2014; Mower and Vickrey, 2018). However, structural variations in plastomes of angiosperms have been found, including inversions (Mower and Vickrey, 2018), IR boundary shifts, and gene duplications (Zhu et al., 2016). Comparative plastome studies may help estimate sequence divergence and evolutionary pathways related to gene loss, duplication, and transfer events, as well as identify species and elucidate phylogenetic relationships (Wu and Chaw, 2016).
The main objectives of this study are to (1) explore the variation and utility of the plastomes in Oxalidaceae, as well as identify Oxalidaceae-specific genome features; and (2) establish the phylogenetic position of Oxalidaceae and Connaraceae. To achieve these objectives, we sequenced and assembled the plastomes of four Oxalidaceae species (Averrhoa carambola, A. bilimbi, Biophytum sensitivum, and Oxalis corymbosa) and one Connaraceae species, Rourea microphylla (Hook. et Arn.) Planch., which are reported here for the first time. We compared these newly sequenced plastomes with three additional Oxalidaceae plastomes (A. carambola NC_033350.1, O. corniculata L., and O. drummondii A. Gray). Finally, we used 76 protein-coding genes to reconstruct the phylogeny of 55 superrosid species, testing hypotheses regarding relationships among COM orders. This work may provide basic plastid phylogenomic data for Oxalidaceae and Connaraceae, supporting future genomics research.
2. Materials and methods
2.1. Plant materials and DNA extraction
Healthy and fresh leaves of Averrhoa bilimbi and B. sensitivum were sampled from adult plants in Hainan (China), A. carambola and O. corymbosa were collected from Guangdong (China), and R. microphylla was collected from Fujian (China) (Table S1). The samples were put in silica gel immediately after collection for desiccation. All voucher specimens were deposited in the herbarium of the Fujian Agriculture and Forestry University, Fuzhou, China. Total DNA was isolated from silica-dried leaf materials using a modified CTAB method (Doyle and Doyle, 1987).
2.2. Plastid genome sequencing, assembly, and annotation
The purified DNA samples were sheared into fragments with an average length of 350 bp for library preparation following the manufacturer's guidelines (Illumina, San Diego, CA, USA). Paired-end (PE) sequencing of 150 bp was carried out on an Illumina Hiseq-2500 platform (Illumina Inc.) at the Beijing Genomics Institute (Shenzhen, China). The quality of the raw PE reads was verified by the FastQC v.0.11.7 tool (Andrews, 2010) with the parameter set as Q ≥ 25 to obtain high-quality clean reads. De novo assembly of the plastomes was performed by the GetOrganelle pipeline (Jin et al., 2018). The published plastome sequences of A. carambola (NC_033350) and O. drummondii (NC_043802) were served as references. The filtered de Bruijn graphs file “gfa” was visualized and edited by Bandage v.0.8.1 (Wick et al., 2015) and the complete plastome sequence paths were manually selected. All PE reads were mapped to the reference genomes using the Bowite2 v.2.2.5 (Langmead and Salzberg, 2012) plugin in GENEIOUS v.11.1.5 (Kearse et al., 2012) to verify quality and correct assembly errors. The assembled plastomes were annotated with Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al., 2004), and then manually corrected by comparison with the references mentioned above using GENEIOUS v.11.1.5 (Kearse et al., 2012). Protein-coding genes with one or more frame shift mutations or premature stop codons were annotated as pseudogenes. Transfer RNA (tRNA) genes were further verified using the online tRNAscan-SE 1.21 service (Schattner et al., 2005) with default parameters. All fully annotated complete plastid genome sequences were uploaded to the NCBI GenBank database (Table 1). Circular plastome maps were generated using the online software OGDRAW v.1.3.1 (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al., 2019).
Table 1.
Complete plastome features of the seven Oxalidaceae accessions and one Connaraceae accession.
| Species | aAverrhoa bilimbi | aAverrhoa carambola | Averrhoa carambola | aOxalis corymbosa | Oxalis corniculata | Oxalis drummondii | aBiophytum sensitivum | aRourea microphylla |
|---|---|---|---|---|---|---|---|---|
| Genome size (bp) | 156,045 | 155,982 | 155,965 | 152,145 | 152,189 | 152,112 | 150,673 | 156,609 |
| LSC (bp) | 87,111 | 87,222 | 87,217 | 84,145 | 84,426 | 84,340 | 82,783 | 87,267 |
| SSC (bp) | 17,432 | 17,496 | 17,496 | 17,048 | 16,989 | 16,914 | 16,798 | 17,690 |
| IRs (bp) | 25,751 | 25,632 | 25,626 | 25,476 | 25,387 | 25,429 | 25,546 | 25,826 |
| Total number of genes (unique) | 131 (114) | 131 (114) | 131 (114) | 129 (112) | 129 (112) | 129 (112) | 129 (112) | 131 (114) |
| Protein-coding gene number (unique) | 83 (77) | 83 (77) | 83 (77) | 82 (76) | 82 (76) | 82 (76) | 82 (76) | 83 (77) |
| tRNA gene number (unique) | 37 (30) | 37 (30) | 37 (30) | 37 (30) | 37 (30) | 37 (30) | 37 (30) | 37 (30) |
| rRNA gene number (unique) | 8 (4) | 8 (4) | 8 (4) | 8 (4) | 8 (4) | 8 (4) | 8 (4) | 8 (4) |
| Duplicated genes in IR | 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 |
| Pseudogene | infA, rpl32, ycf1 | infA, rpl32, ycf1 | infA, rpl32, ycf1 | infA, ycf1 | infA, ycf1 | infA, ycf1 | infA, ycf1 | infA, rpl32, ycf1 |
| Overall GC content (%) | 36.42 | 36.54 | 36.54 | 36.72 | 36.70 | 36.49 | 36.89 | 36.78 |
| GC content in LSC (%) | 34.13 | 34.28 | 34.28 | 34.47 | 34.40 | 34.11 | 34.76 | 34.60 |
| GC content in SSC (%) | 30.07 | 30.24 | 30.24 | 30.29 | 30.34 | 29.96 | 30.48 | 30.69 |
| GC content in IR (%) | 42.44 | 42.53 | 42.52 | 42.59 | 42.65 | 42.62 | 42.47 | 42.56 |
| GenBank number | MT522015 | MT522016 | NC_033350.1 | MT522018 | NC_051971.1 | NC_043802.1 | MT522017 | MT537171 |
Five newly sequenced plastomes.
2.3. Comparative genome analysis and nucleotide variation analysis
Visual inspection of rearrangements in the eight plastomes (A. bilimbi, A. carambola, A. carambola NC_033350, B. sensitivum, O. corymbosa, O. corniculata, O. drummondii, and R. microphylla) was carried out by progressive Mauve v.2.4.0 (Darling, 2004) with the default “seed families” and default values for all other parameters. The IR/SC boundaries of the plastomes of Oxalidaceae species and R. microphylla were compared using the IRscope software (https://irscope.shinyapps.io/irapp/) (Amiryousefi et al., 2018). Whole-genome alignment of the eight plastomes of Oxalidales was performed and plotted with the mVISTA program (http://genome.lbl.gov/vista/mvista/submit.shtml) (Frazer et al., 2004), which uses the Shuffle-LAGAN model; A. carambola NC_033350 served as the reference. Nucleotide diversity (Pi) was evaluated using DnaSP v.6.12 software (Rozas et al., 2017).
2.4. Codon usage analysis
Codon usage and relative synonymous codon usage (RSCU) values were estimated using Codon W (University of Nottingham, Nottingham, UK) (http://codonw.sourceforge.net/) (Peden, 1999). Repeat sequences and protein-coding regions (CDS) shorter than 300 bp were eliminated from the codon usage calculations to avoid sampling errors, given that short CDS generally result in large estimation errors for codon usage (Rosenberg et al., 2003). Finally, we analyzed 52 CDS from the plastome of O. corymbosa and 53 CDS each from the plastomes of A. bilimbi, A. carambola, A. carambola NC_033350, B. sensitivum, O. corniculata, O. drummondii, and R. microphylla.
2.5. Repeat sequence analysis
The length and location of forward, palindromic, reverse, and complement repeats in the plastomes of Oxalidaceae species and R. microphylla were detected by the REPuter program (https://bibiserv.cebitec.uni-bielefeld.de/reputer) (Bielefeld University, Bielefeld, Germany) (Kurtz et al., 2001). Parameters for repeat identification were set as follows: (1) hamming distance = 3; (2) repeat size ≥ 30 bp; and (3) maximum computed repeats of 90 bp. The positions and types of microsatellites (SSRs) were determined by the microsatellite identification tool MISA (available online: https://webblast.ipk-gatersleben.de/misa/) (Beier et al., 2017) with thresholds of 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides, respectively.
2.6. Phylogenetic analyses
To infer the detailed phylogenetic position of the five newly sequenced species (A. bilimbi, A. carambola, B. sensitivum, O. corymbosa, and R. microphylla) in rosids, we downloaded 55 complete plastomes representing the superrosids lineage of angiosperms from the NCBI GenBank database (Table S2). Paeonia obovata NC_026076.1 (Paeoniaceae) and Heuchera richardsonii NC_042923.1 (Saxifragaceae) were set as the outgroups. Seventy-six protein-coding genes (rpl32 and rps16 were not included because they are missing genes or pseudogenes) were extracted from the 60 plastomes described above using PhyloSuite v.1.1.16 (Zhang et al., 2020). Our analyses were based on protein-coding genes, given their relatively slow rate of evolution. The sequences were aligned using MAFFT v.7.221 (Katoh and Standley, 2013) with manual adjustments when necessary. The phylogenetic relationships were analyzed by maximum likelihood (ML) and Bayesian inference (BI) using the CIPRES Science Gateway web server (available online: http://www.phylo.org/) (Miller et al., 2010). ML analysis was performed by RAxML-HPC2 on XSEDE 8.2.10 with the GTRGAMMA model and 1000 bootstrap replicates (Stamatakis et al., 2008). BI was implemented with MrBayes v.3.2.6 (Ronquist et al., 2012) and the best substitution model (GTR + I + Γ) was determined by the Akaike information criterion (AIC) in jModeltest v.2.1.10 (Darriba et al., 2012). The Markov chain Monte Carlo (MCMC) algorithm was run for 2,000,000 generations, with one tree sampled every 1000 generations until convergence. The first 25% of trees were discarded as burn-in, and the remainder was used to construct majority-rule consensus trees. ML and BI trees were plotted using FigTree v.1.4.2 (Rambaut, 2012).
3. Results
3.1. Plastome features
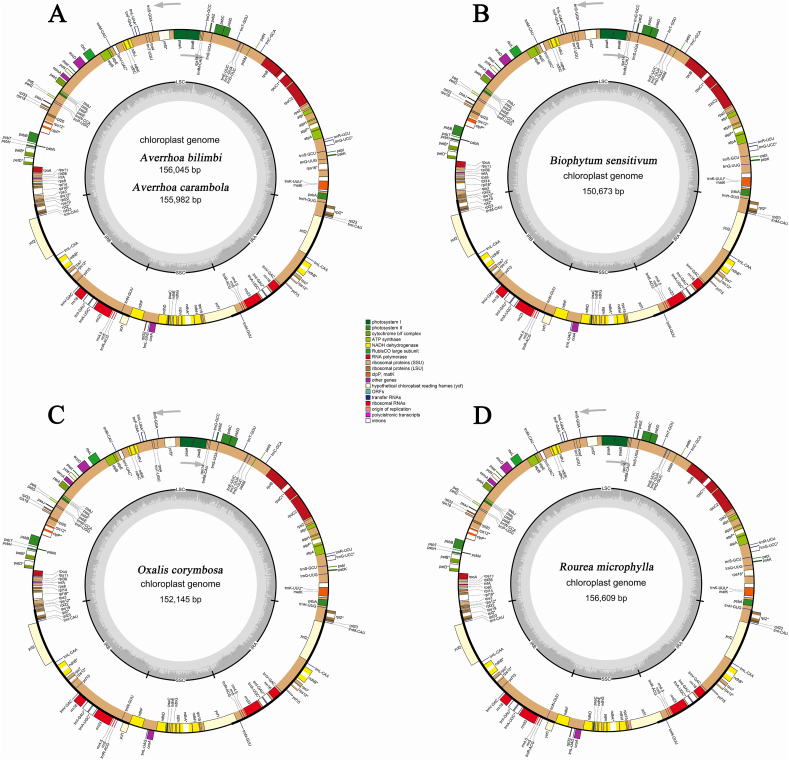
The complete plastome sequences for the four Oxalidaceae species and one Connaraceae species investigated in this study possess the typical quadripartite structure of most angiosperm plastid genomes (Fig. 1). A comparison with the three previously published Oxalidaceae plastomes (A. carambola NC_033350, O. corniculata, and O. drummondii) showed that the Oxalidaceae species differ in sequence length (Table 1). The plastome length of Averrhoa and Oxalis species are approximately 156 kb and 152 kb, respectively. B. sensitivum has the smallest length of 150,673 bp, whereas R. microphylla has the longest (156,609 bp). There was a significant relationship between the LSC region and whole plastome length, although each of the structural regions was not distinctly associated with each other (Fig. S1). The GC content of the IR regions (42.44–42.65%) is visibly greater than that of the LSC (34.11–34.76%) and SSC (29.96–30.69%) regions (Table 1), mainly due to the high GC content of four rRNA genes (rrn23, rrn16, rrn5, and rrn4.5).
Fig. 1.
Plastome maps of four Oxalidaceae species and one Connaraceae species. (A) Two Averrhoa species. (B) Biophytum sensitivum. (C) Oxalis corymbosa. (D) Rourea microphylla. Genes on the inside and outside of the large circle are transcribed clockwise and counterclockwise, respectively. Genes are color-coded on the basis of their functions. The dark gray and lighter gray in the inner circle show the GC content and A/T content, respectively.
In Averrhoa species and R. microphylla, 131 genes (114 unique genes) were detected, including 83 protein-coding genes, eight rRNA genes, 37 tRNA genes, and three pseudogenes (infA, rpl32, and ycf1). Of these genes, 15 have one intron, while clpP, rps12, and ycf3 possess two introns each (Tables S3 and S4). The plastomes of Oxalis species and Biophytum sentivivum harbored fewer genes, including intron-containing genes, due to the deletions of rpl32 and rps16 in the plastomes. Notably, in the B. sentivivum plastome, an intron was lost from the clpP gene. Additionally, pseudogenizations (infA and ycf1) have occurred in the Oxalis species and B. sentivivum. No rearrangements in gene organization were found in the analyzed plastomes (Fig. S2).
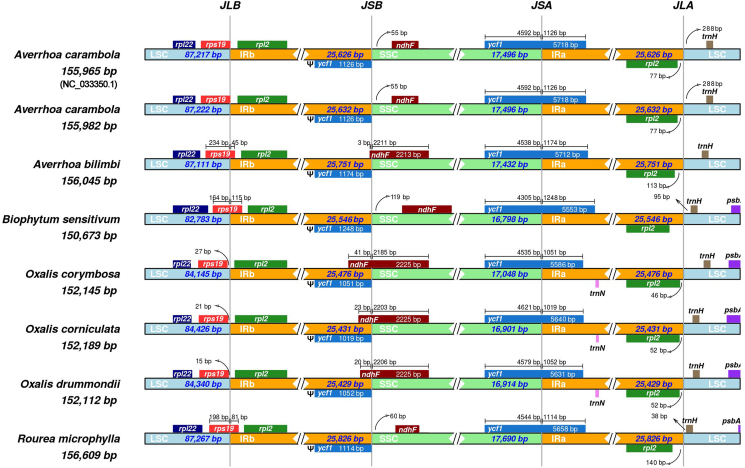
3.2. Expansion and contraction of IRs
Comparative sequence analysis of the Oxalidaceae species and R. microphylla indicated some variations at the IR/SC boundary regions (Fig. 2). In A. bilimbi, B. sensitivum, and R. microphylla, the rps19 gene is located within the LSC/IRb boundary, with 45–115 bp spanning into the IRb region, indicating an expansion of the IR in these three species. However, in the Oxalis species, the rps19 gene is entirely located in the LSC region with 15–27 bp away from the LSC/IRb boundary. In Oxalis species and A. bilimbi, the ndhF gene is located in the SSC/IRb boundary, but in A. carambola, B. sensitivum, and R. microphylla, it is located entirely within the SSC region. In all plastomes, the SSC/IRa boundary is situated in the ycf1 protein-coding gene, and the fragment located in the IRa region ranges from 1019 bp (O. corniculata) to 1248 bp (B. sensitivum).
Fig. 2.
Comparison of the SC/IR boundaries in the plastomes of Oxalidaceae species and Rourea microphylla. JLB, LSC/IRb boundary; JSB, SSC/IRb boundary; JSA, SSC/IRa boundary; JLA, LSC/IRa boundary. Distance in this figure is not to scale. Ψ represents pseudogene.
3.3. Codon usage analysis
The total number of codons for protein-coding genes of the plastomes ranged from 21,210 in O. corymbosa to 21,368 in A. bilimbi (Table S5). Further codon analysis showed that the eight plastomes have similar codon constituents and close RSCU values. Leucine (Leu: 10.39%–10.57%) and isoleucine (Ile: 8.71%–8.90%) are the most encoded amino acids in all plastomes, whereas cysteine (Cys: 1.08%–1.13%) is the least (Fig. S3). The majority of amino acid codons have a bias, although codons AU(T)G and U(T)GG, which encod methionine (Met) and tryptophan (Trp) respectively, both show no codon preferences (RSCU = 1.00). Additionally, most types of preferred synonymous codons (RSCU > 1.00) possessed A- or U-ending codons, except UUG, which encodes trnL-CAA. In protein-coding genes of the plastomes, 70.84%–72.00% of all codons end with A and/or U, which indicates a bias for A/U(T) bases (Table S5).
3.4. Repeat sequence analysis
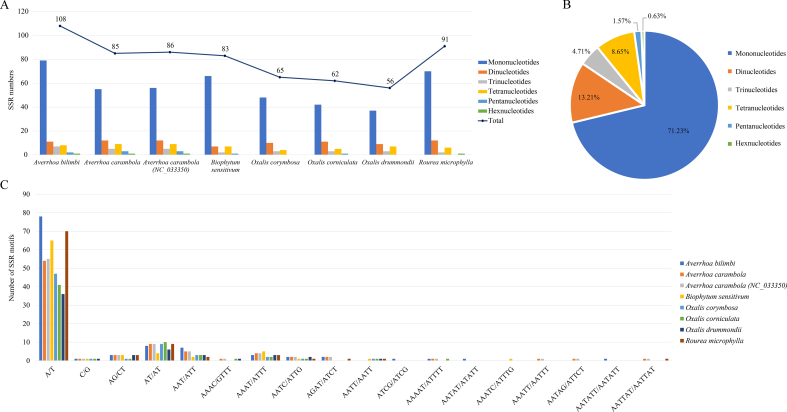
Four categories of repeats (forward, palindromic, reverse, and complement repeats) were identified in the plastomes of Oxalidaceae species and R. microphylla (Fig. S4). There are 364 repeats in the eight plastomes, including 172 (47.25%) forward repeats, 171 (46.98%) palindromic repeats, 16 (4.40%) reverse repeats, and five (1.37%) complement repeats. Of the Oxalidaceae species, the highest number of repeats was found in O. corniculata (56) and the lowest number in O. drummondii (36). We artificially divided all repeats into five categories (30–39 bp, 40–49 bp, 50–59 bp, 60–64 bp, and > 64 bp) based on their length. Of these, 261 (71.70%) have lengths of 30–39 bp, followed by 64 (17.58%) with lengths of 40–49 bp, whereas only nine (2.47%) are longer than 64 bp. A total of 636 SSRs were detected in the eight Oxalidales plastomes, ranging from 56 (O. drummondii) to 108 (A. bilimbi) per plastome (Fig. 3). A. carambola and B. sensitivum have a similar number of SSRs, as do O. corymbosa and O. corniculata. Six SSR types (mono-, di-, tri-, tetra-, penta- and hexa-nucleotide repeats) all appeared in Averrhoa species. Mononucleotide repeats are most abundant (71.23% of the total SSRs), followed by dinucleotide repeats (13.21%), whereas hexanucleotide repeats are very rare among these plastomes. For all plastomes analyzed, SSRs are located mainly in the LSC and IGS (Fig. S5).
Fig. 3.
Analyses of simple sequence repeats (SSR) in the plastomes of Oxalidaceae species and Rourea microphylla. (A) Number of SSRs and their types. (B) Percentage of SSR types. (C) Number of SSR motifs in the eight plastomes.
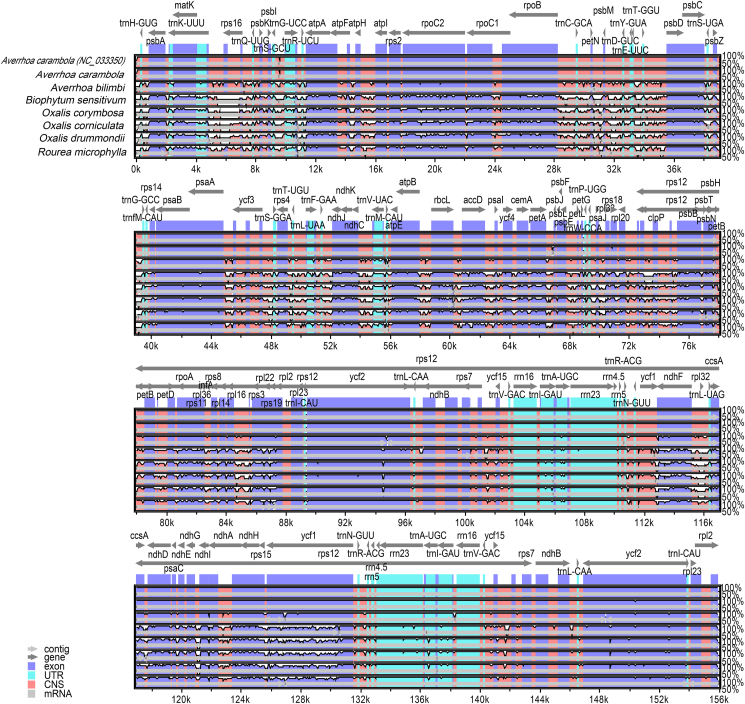
3.5. Sequence divergence analysis
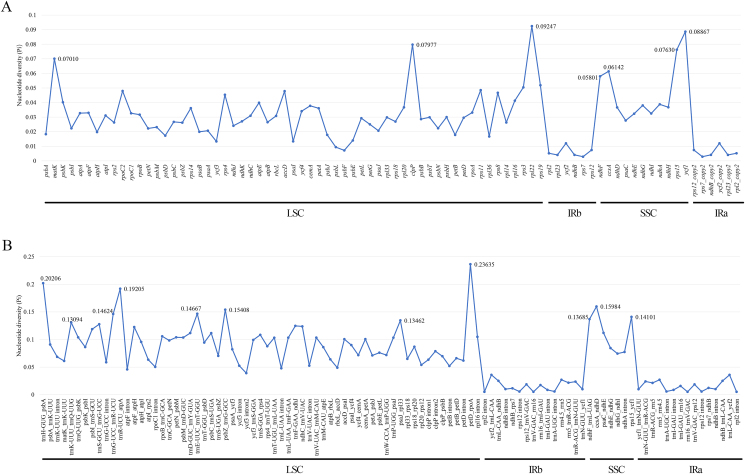
mVISTA was used to align plastome sequences with A. carambola NC_033350 as a reference (Fig. 4). Alignment showed that plastomes from the same genus have low sequence divergence. Protein-coding genes are more conserved than non-coding regions (particularly the IGS); similarly, IR regions are more conserved than SC regions. The average nucleotide variability (Pi) values for non-coding regions were approximately twice as high or higher than those for coding regions (Fig. 5 and Fig. S6). The Pi values for the most fragments of IR regions were relatively low. Pi values were higher in aligned Oxalis plastomes than in aligned Averrhoa species, indicating that variation between Oxalis species was generally higher (Fig. S6). Across coding regions of the Oxalidaceae species, seven hypervariable regions (rpl22, ycf1, clpP, rps15, matk, ccsA and ndhF) were observed (Fig. 5A). The Pi value of eleven non-coding regions was ≥ 0.13094, indicating high variation. Of these, eight (trnH-GUG_psbA, trnK-UUU_trnQ-UUG, trnG-UCC_trnR-UCU, trnR-UCU_atpA, trnE-UUC_trnT-GGU, psbZ_trnG-GCC, psaJ_rpl33, and petD_rpoA) were within the LSC region and three (ndhF_trnL-UAG, ccsA_ndhD, and rps15_ycf1) were in the SSC region (Fig. 5B).
Fig. 4.
Genome alignment of plastomes of Oxalidaceae species and Rourea microphylla using Averrhoa carambola NC_033350 as a reference. The vertical scale indicates the percent identity, ranging from 50% to 100%. Coding regions are marked in purple, and conserved non-coding sequences (CNS) are marked as red. The horizontal axis indicates the coordinates within the plastome.
Fig. 5.
The nucleotide diversity (Pi) values of the aligned Oxalidaceae plastomes. (A) Protein-coding genes. (B) Non-coding regions.
3.6. Phylogenetic analyses
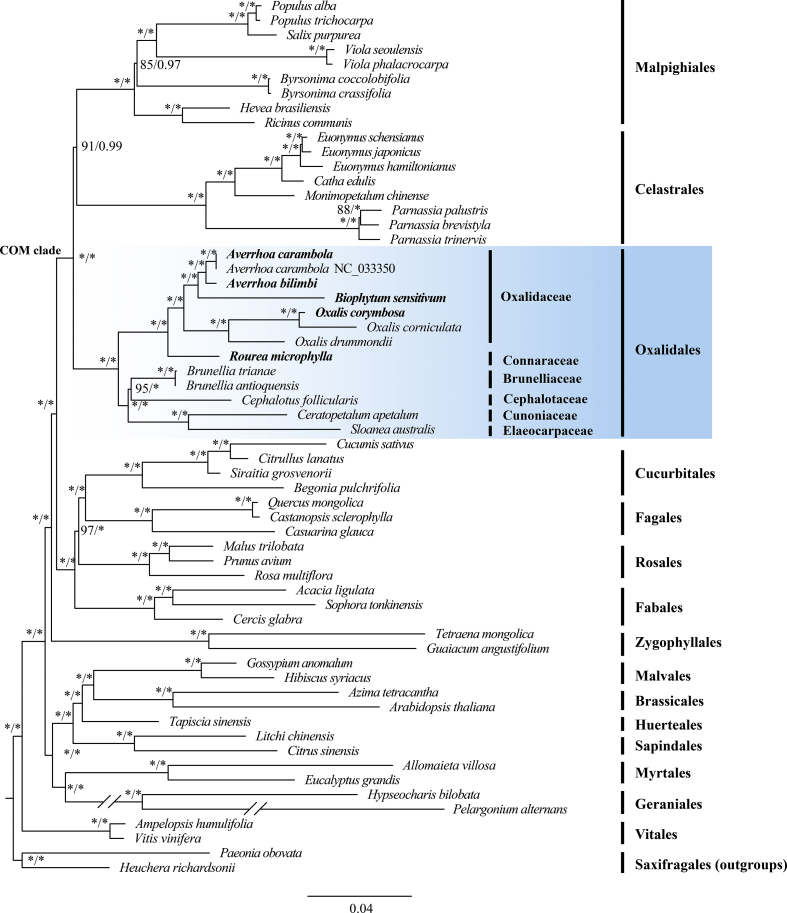
Both the ML and BI phylogenetic trees based on 76 protein-coding genes strongly indicated that the species of the newly obtained plastomes are all included in the COM clade (Fig. 6). The Oxalidaceae species, A. bilimbi, A. carambola, B. sensitivum, O. corymbosa, O. corniculata, and O. drummondii, formed a cluster closed to the Connaraceae species, R. microphylla, with strong support (100/1.00). More specifically, B. sensitivum was placed near the two Averrhoa species (100/1.00). The three Oxalis species gathered into one clade, with O. corymbosa and O. corniculata clustered together (100/1.00). Within the order Oxalidales, Brunelliaceae, Cephalotaceae, Cunoniaceae, and Elaeocarpaceae formed a clade, which was a sister clade to the group of Oxalidaceae-Connaraceae (100/1.00); the sister relationship between Brunelliaceae and Cephalotaceae was well supported (95/1.00). Additionally, the phylogenetic trees indicated the COM clade was monophyletic and showed the (O (C, M)) topology (91/0.99).
Fig. 6.
Phylogenetic trees from maximum likelihood (ML) and Bayesian inference (BI) analyses of 59 species based on 76 protein-coding genes. Numbers near the nodes are ML bootstrap support values (left of the slashes) and Bayesian posterior probabilities (right of the slashes). Asterisks (∗) show the node has 100% bootstrap or 1.00 posterior probability. Paeonia obovata and Heuchera richardsonii were used as outgroups.
4. Discussion
4.1. Variation of plastome sequences
Plastome sizes in the Oxalidaceae species and R. microphylla fall well within the normal ranges of land plant plastomes (120–160 kb) (Jansen and Ruhlman, 2012; Mower and Vickrey, 2018), but exhibit moderate differences among different genera. The plastome lengths of Averrhoa species (~156 kb) are longer than those of the Oxalis species (~152 kb) and B. sensitivum (150,673 bp). One of the reasons for differences in plastome lengths is the expansion and contraction of the SC/IR boundaries (Jansen and Ruhlman, 2012; Ruhlman and Jansen, 2014; Mower and Vickrey, 2018). In this study, the floating of SC/IR boundaries among Oxalidaceae species may be caused by IR contraction/expansion. B. sensitivum has the shortest plastome length, whereas its IR regions are longer than those of the Oxalis species. The variations in length reflect the expansions of IRs and the contractions of SCs.
Differences in plastome sizes are also related to gene spacer region variation, the loss or gain of genes, and introns, which might represent a common pattern throughout plastid genome evolution (Jansen et al., 2007; Jansen and Ruhlman, 2012; Ruhlman and Jansen, 2014; Mower and Vickrey, 2018). In our study, several species (O. corymbosa, O. corniculata, O. drummondii, and B. sensitivum) lacked the genes rpl32 and rps16. The deletions of rpl32 and rps16 in Oxalis have also been reported by Schmickl et al. (2015), and may suggest that the gene loss event is a common feature in this genus. The breadth of the IR boundary shifts in land plants has been explored and demonstrated by Zhu et al. (2016). The rpl32 gene is located near the SSC/IR boundary. Thus, the loss of the rpl32 gene may be correlated with the shifting of the IR boundary. The rpl32 and rps16 genes are also absent in many species of Malpighiales (e.g., Salicaceae, Podostemaceae, and Violaceae) (Jansen et al., 2007; Menezes et al., 2018; Bedoya et al., 2019). Some studies have shown that rpl32 (Park et al., 2015, 2020) and rps16 (Ueda et al., 2008; Park et al., 2020) missing in the plastome of some species have likely been transferred to the nuclear genome. Comparative genome studies have suggested that the transfer of genes from plastids to the nucleus is a continually evolving process (Park et al., 2015). However, further studies are required to determine whether the two genes missing from the plastomes of Oxalis species and B. sensitivum were transferred to the nuclear genome or completely lost.
Our study also indicates that intergenic region variation and intron loss promote variation in plastome size. For instance, comparative sequence analysis showed that intergenic regions vary the most in the plastomes investigated, and that intron numbers for the Oxalis species and B. sensitivum are both smaller than those in the Averrhoa species and R. microphylla. In B. sensitivum, the clpP gene has lost an intron region, a loss which has also been reported in Inga (Fabaceae) (Dugas et al., 2015). Research has previously shown that Acacia clpP CDS has an accelerated rate of synonymous and nonsynonymous mutation, which indicates the presence of a functional nuclear-encoded copy of this gene, at least in some mimosoid taxa (Williams et al., 2015). The clpP gene is thought to be important for the development and function of plastids, especially for plastids with high levels of gene expression (Shikanai et al., 2001). Further investigation should indicate whether clpP plays a vital role in Biophytum plastids or has been transferred to the nuclear genome.
We detected a commonly degraded plastid gene infA in the Oxalidaceae species analyzed and R. microphylla (Connaraceae). We regarded this gene as a pseudogene due to the existence of premature stop codons. The pseudogenization or absence of the infA gene, which is considered one of the most variable plastid genes in angiosperms, has also been reported in a great number of land plants, and has been shown to have frequently been transferred to and stayed in the nucleus (Millen et al., 2001). We also discovered that the protein-coding sequence of the gene ycf1 is interrupted by the SSC/IRa boundary, creating a pseudogene version of ycf1, which has been previously reported (Menezes et al., 2018; Bedoya et al., 2019). In addition, the rpl32 gene that was lost in the Oxalis species and B. sensitivum was pseudogenized in the Averrhoa species, which suggests that rpl32 may be dispensable in Oxalidaceae species.
Trees tend to have lower Pi values compared with herbs (Valencia-D et al., 2020). Similarly, our results showed that Pi values for both the coding and non-coding regions of the Averrhoa species were considerably lower than those of the Oxalis species (Fig. S6), indicating slighter variations between the Averrhoa species. The genus Oxalis presents many identification difficulties (Moura et al., 2020); however, only four plastid DNA sequences (petA-psbJ, trnL-trnF, trnS-trnG, and trnT-trnL) have been used in previous phylogenetic studies (Oberlander et al., 2011; Vaio et al., 2016; Aoki et al., 2017). Among the three Oxalis species studied here, these four regions showed a relatively low Pi of 0.08140 or less (Fig. S6). Thus, for phylogenetic studies of Oxalis, we recommend using regions of the plastome that show higher Pi values of regions (>0.10), such as petD_rpoA, trnH-GUG_psbA, psbI_trnS-GCU, rps 15_ycf1, psbZ_trnG-GCC, ndhC_trnV-UAC, and ccsA_ndhD. In addition, we identified seven coding regions and 11 intergenic regions with the highest variation among the Oxalidaceae species. Several of these regions (e.g., matK, ycf1, clpP, ndhF, rpl22, trnH-GUG_psbA, trnE-UUC_trnT-GGU, psaJ_rpl33 and ccsA_ndhD) have been confirmed in other seed plant plastomes (Ren et al., 2020; Wang et al., 2021; Tang et al., 2021). These hotspot regions may serve as potential molecular markers for species identification, assessment of genetic diversity, and research into the phylogeny of Oxalidaceae.
4.2. Codon usage and repeat sequence analysis
Codon usage bias plays an indispensable role in plastid genome evolution and affects gene function and protein expression (Quax et al., 2015). Codon usage in plastomes is usually biased toward codons ending in A or T (Morton, 1998). This bias was also observed in the plastomes of Oxalidaceae species and R. microphylla. Codon usage bias in plastid genes may be driven by natural selection during the plastome evolutionary process (Jansen and Ruhlman, 2012).
Large repeat sequences might have important roles in plastome sequence divergence and rearrangements (Weng et al., 2014). Of the 364 repeats identified in this study, the most common were short repeats between 30 and 39 bp, which is consistent with many rearranged plastomes (Ren et al., 2020; Zhao et al., 2020). SSRs can be used for plant molecular identification, as well as research on genetic diversity and population genetics (Provan et al., 2001). The most abundant SSRs in the plastomes of Oxalidaceae species and R. microphylla (Connaraceae) are short A/T repeats, whereas G/C mononucleotide repeats are extremely rare across the plastomes. This phenomenon is consistent with many angiosperm plastomes and may be the result of plastome’ bias towards A/T (Ren et al., 2020; Zhao et al., 2020). The majority of SSRs are located in the LSC region, probably because the LSC is longer than the SSC and IR regions. The position of the repeats has been correlated to the occurrence of induced mutation events (Abdullah et al., 2020), which is reflected in our study by the finding that repeats are mostly distributed in hypervariable non-coding regions rather than in coding regions. The SSRs detected in the plastomes could be used as potential resources for further studies on genetic diversity of some important economic plants of Oxalidaceae.
4.3. Phylogenetic analyses
The phylogenetic trees obtained in this study are largely similar to the updated version of the Angiosperm Phylogeny Group (APG) system (APG IV, 2016). Although morphological features have suggested Oxalidaceae is closely related to the Geraniaceae of Geraniales (Mathew, 1958; Stevens et al., 2004), our phylogenetic analyses based on protein-coding genes provide robust support for the close sister relationship with Connaraceae within Oxalidales (Fig. 6), which is in accordance with previous molecular-based phylogenetic studies using limited genes (Heibl and Renner, 2012; Sun et al., 2016). The sister relationship of Oxalidaceae and Connaraceae is also supported by floral structure (e.g., dimorphic and trimorphic heterostyly, hemianatropous to orthotropous ovules) (Matthews and Endress, 2006). Our analysis indicated that the Oxalidaceae species, B. sensitivum, is more closely related to the Averrhoa species than to the Oxalis species, which has been suggested in a previous study (Heibl and Renner, 2012). Considering the limited genomic sources of Oxalidaceae, more phylogenetic information about this family is necessary. The hotspot regions (e.g., petD_rpoA, trnH-GUG_psbA, psbI_trnS-GCU, rps 15_ycf1, psbZ_trnG-GCC, ndhC_trnV-UAC, and ccsA_ndhD) of the Oxalis species detected in this work will be helpful in resolving intrageneric relationships within Oxalis.
Previous studies have indicated that the Oxalidaceae-Connaraceae clade forms a monophyletic group with the four families, Brunelliaceae, Cephalotaceae, Cunoniaceae, and Elaeocarpaceae (Heibl and Renner, 2012; Sun et al., 2016). Our results confirmed the monophyletic group with full support in the phylogenetic trees. In addition, the monophyly of the four families described above was also ascertained, which corroborates previous studies that relied on limited numbers of chloroplast, nuclear or mitochondrial genes (Heibl and Renner, 2012; Sun et al., 2016). However, phylogenetic relationships within the group remain ambiguous (see Table S6). In our study, Brunelliaceae and Cephalotaceae, which share isomerous, apetalous flowers with two whorls of stamens and lack special mucilage cells (Matthews and Endress, 2006), were sister to each other and together sister to Cunoniaceae and Elaeocarpaceae (Fig. 6). The phylogenetic relationships among the four families were more highly resolved than those generated by the small number of nuclear and plastid genes employed in previous studies (Heibl and Renner, 2012; Sun et al., 2016). Further research, with expanded taxon sampling, is required to identify the phylogenetic relationship among these four families.
The position of the COM clade within rosids has long been problematic in angiosperm phylogeny (APG IV, 2016; Sun et al., 2016; Gonçalves et al., 2019). Our study showed that COM is monophyletic with strong support, a result that is consistent with most phylogenetic analyses based on plastid data (e.g., Gonçalves et al., 2019; Valencia-D et al., 2020) but conflicts with several studies using mitochondrial genes (Zhu et al., 2007; Qiu et al., 2010) or nuclear data sets (Zhao et al., 2016; Zeng et al., 2017; Yang et al., 2020). This phylogenetic discordance may be related to an ancient episode of hybridization followed by plastid capture during the rapid radiation of Rosidae (Ruhfel et al., 2014; Sun et al., 2015, 2016). Within the COM group, the relationships among the three orders are controversial (see Table S7). In this study, Celastrales was sister to Malpighiales, which together were sister to Oxalidales. Our plastome phylogenomic analysis based on 76 CDS has provided better resolution of the phylogenetic tree of the COM clade compared with previous studies that used nuclear genome fragments, plastid fragments, or mitochondrial matR gene (Moore et al., 2011; Soltis et al., 2011). Even so, deep phylogeny evolutionary analyses based on more molecular data from different types of datasets (plastome data, nuclear data, or a combination of organelle and nuclear data) are needed to clarify the highly complex evolutionary history of the COM group.
5. Conclusions
In this study, we generated four new plastome sequences for Oxalidaceae and report the first plastome of the Connaraceae species, R. microphylla. Comparative analysis revealed that the smaller plastomes of Oxalis species and B. sensitivum are associated with IR contraction/expansion, gene loss (rps16 and rpl32), intergenic region variation or intron loss (clpP); and that Oxalis species plastome sequences vary more than those of Averrhoa species. We also identified several regions in the Oxalis plastomes with high variation (e.g., petD_rpoA, trnH-GUG_psbA, psbI_trnS-GCU, rps15_ycf1, and psbZ_trnG-GCC) that can be potentially used as markers for phylogenetic analysis. Our phylogenetic analyses revealed that B. sensitivum is more closely related to Averrhoa species than to Oxalis species and confirmed that Oxalidaceae is sister to Connaraceae within Oxalidales.
Author contributions
XPL, SSW and JWZ conceived and designed the experiments. XPL, YMZ and XDT collected samples, and CRL, YTZ and HZ performed the experiments. XPL, YMZ and XDT analyzed the data. XPL wrote the manuscript. ZJL, SSW and JWZ modified the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors have no competing interests to declare.
Acknowledgements
We are grateful to Mingtao Jiang who helped collect plant material and Dingkun Liu of Fujian Agriculture and Forestry University for his constructive suggestions. This work was sponsored by the Disciplinary Professional Construction Project of College of Art & College of Landscape Architecture, Fujian Agriculture and Forestry University (YSYL-bdpy-2, YSYL-bdpy-1).
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.04.004.
Contributor Information
Shasha Wu, Email: shashawu1984@126.com.
Junwen Zhai, Email: zhai-jw@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abdullah M.F., Shahzadi I., Ali Z. Correlations among oligonucleotide repeats, nucleotide substitutions and insertion-deletion mutations in chloroplast genomes of plant family Malvaceae. J. Systemat. Evol. 2020 doi: 10.1111/jse.12585. [DOI] [Google Scholar]
- Amiryousefi A., Hyvönen J., Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- Andrews S. Babraham Institute; Cambridge, United Kingdom: 2010. FastQC: a Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Aoki S., Ohi-Toma T., Li P. Phylogenetic, cytological and morphological comparisons of Oxalis subsect. Oxalis (Oxalidaceae) in East Asia. Phytotaxa. 2017;324:266–278. [Google Scholar]
- APG IV An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181:1–20. [Google Scholar]
- Barrett C.F., Baker W.J., Comer J.R. Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol. 2016;209:855–870. doi: 10.1111/nph.13617. [DOI] [PubMed] [Google Scholar]
- Bedoya A.M., Ruhfel B.R., Philbrick C.T. Plastid genomes of five species of riverweeds (Podostemaceae): structural organization and comparative analysis in Malpighiales. Front. Plant Sci. 2019;10:1035. doi: 10.3389/fpls.2019.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier S., Thiel T., Münch T. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A.C.E. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Dugas D.V., Hernandez D., Koenen E.J. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Sci. Rep. 2015;5:16958. doi: 10.1038/srep16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves D.J., Simpson B.B., Ortiz E.M. Incongruence between gene trees and species trees and phylogenetic signal variation in plastid genes. Mol. Phylogenet. Evol. 2019;138:219–232. doi: 10.1016/j.ympev.2019.05.022. [DOI] [PubMed] [Google Scholar]
- Greiner S., Lehwark P., Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heibl C., Renner S.S. Distribution models and a dated phylogeny for Chilean Oxalis species reveal occupation of new habitats by different lineages, not rapid adaptive radiation. Syst. Biol. 2012;61:823–834. doi: 10.1093/sysbio/sys034. [DOI] [PubMed] [Google Scholar]
- Jansen R.K., Cai Z., Raubeson L.A. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.K., Ruhlman T.A. In: Genomics of Chloroplasts and Mitochondria. Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes) Bock R., Knoop V., editors. Springer; Dordrecht: 2012. Plastid genomes of seed plants; pp. 103–126. [Google Scholar]
- Jin J.J., Yu W.B., Yang J.B. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 2018:256479. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Choudhuri J.V., Ohlebusch E. Reputer: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew P.M. Cytology of Oxalidaceae. Cytologia. 1958;23:200–210. [Google Scholar]
- Matthews M.L., Endress P.K. Floral structure and systematics in four orders of rosids, including a broad survey of floral mucilage cells. Plant Syst. Evol. 2006;260:199–221. [Google Scholar]
- Menezes A.P.A., Resende-Moreira L.C., Buzatti R.S.O. Chloroplast genomes of Byrsonima species (Malpighiaceae): comparative analysis and screening of high divergence sequences. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-20189-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen R.S., Olmstead R.G., Adams K.L. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proc. Gatew. Comput. Environ. 2010;14:1–8. [Google Scholar]
- Moore M.J., Hassan N., Gitzendanner M.A. Phylogenetic analysis of the plastid inverted repeat for 244 species: insights into deeper-level angiosperm relationships from a long, slowly evolving sequence region. Int. J. Plant Sci. 2011;172:541–558. [Google Scholar]
- Morton B.R. Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J. Mol. Evol. 1998;46:449–459. doi: 10.1007/pl00006325. [DOI] [PubMed] [Google Scholar]
- Moura A.I., Oliveira Y.R., da Silva P.H. Karyotype inconsistencies in the taxonomy of the genus Oxalis (Oxalidaceae) Iheringia Ser. Bot. 2020;75 [Google Scholar]
- Mower J.P., Vickrey T.L. In: Chaw S.M., Jansen R.K., editors. vol. 85. Academic Press; Amsterdam (The Netherlands) and New York: 2018. Structural diversity among plastid genomes of land plants; pp. 2–382. (Plastid Genome Evolution). Elsevier. [Google Scholar]
- Oberlander K.C., Dreyer L.L., Bellstedt D.U. Molecular phylogenetics and origins of southern African Oxalis. Taxon. 2011;60:1667–1677. [Google Scholar]
- Paim L.F.N.A., Toledo C.A.P., da Paz J.R.L. Connaraceae: an updated overview of research and the pharmacological potential of 36 species. J. Ethnopharmacol. 2020;261:112980. doi: 10.1016/j.jep.2020.112980. [DOI] [PubMed] [Google Scholar]
- Park S., An B., Park S. Recurrent gene duplication in the angiosperm tribe Delphinieae (Ranunculaceae) inferred from intracellular gene transfer events and heteroplasmic mutations in the plastid matK gene. Sci. Rep. 2020;10:2720. doi: 10.1038/s41598-020-59547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Jansen R.K., Park S. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 2015;15:40. doi: 10.1186/s12870-015-0432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden J.F. University of Nottingham; Nottingham, UK: 1999. Analysis of Codon Usage. Ph.D. Thesis. [Google Scholar]
- Provan J., Powell W., Hollingsworth P.M. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol. Evol. 2001;16:142–147. doi: 10.1016/s0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- Qiu Y.L., Li L.B., Wang B. Angiosperm phylogeny inferred from sequences of four mitochondrial genes. J. Syst. Evol. 2010;48:391–425. [Google Scholar]
- Quax T.E., Claassens N.J., Söll D. Codon bias as a means to fine-tune gene expression. Mol. Cell. 2015;59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. University of Edinburgh; Edinburgh: 2012. FigTree v1.4.2: Molecular Evolution, Phylogenetics and Epidemiology.http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- Ren T., Li Z.X., Xie D.F. Plastomes of eight Ligusticum species: characterization, genome evolution, and phylogenetic relationships. BMC Plant Biol. 2020;20:519. doi: 10.1186/s12870-020-02696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M.S., Subramanian S., Kumar S. Patterns of transitional mutation biases within and among mammalian genomes. Mol. Biol. Evol. 2003;20:988–993. doi: 10.1093/molbev/msg113. [DOI] [PubMed] [Google Scholar]
- Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Ruhfel B.R., Gitzendanner M.A., Soltis P.S. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 2014;14:23. doi: 10.1186/1471-2148-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T.A., Jansen R.K. In: Chloroplast Biotechnology. Maliga P., editor. vol. 1132. Humana Press; Totowa, NJ: 2014. The plastid genomes of flowering plants; pp. 3–38. (Methods in Molecular Biology (Methods and Protocols)). [DOI] [PubMed] [Google Scholar]
- Sá R.D., Vasconcelos A.L., Santos A.V. Anatomy, histochemistry and oxalic acid content of the leaflets of Averrhoa bilimbi and Averrhoa carambola. Rev. Bras. Farmacogn. 2019;29:11–16. [Google Scholar]
- Schattner P., Brooks A.N., Lowe T.M. The tRNAscan–SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl R., Liston A., Zeisek V. Phylogenetic marker development for target enrichment from transcriptome and genome skim data: the pipeline and its application in southern African Oxalis (Oxalidaceae) Mol. Ecol. Res. 2015;16:1124–1135. doi: 10.1111/1755-0998.12487. [DOI] [PubMed] [Google Scholar]
- Shikanai T., Shimizu K., Ueda K. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 2001;42:264–273. doi: 10.1093/pcp/pce031. [DOI] [PubMed] [Google Scholar]
- Singh K., Gambhir L., Berma P.D. Antimicrobial effects of leaf extracts from Oxalis corymbosa against pathogenic bacterial and fungal isolates. World J. Pharmaceut. Res. 2017;6:1267–1278. [Google Scholar]
- Soltis D.E., Smith S.A., Cellinese N. Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 2011;98:704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stevens P.F., Luteyn J., Oliver E. In: Kubitzki K., editor. vol. 6. Berlin/Heidelberg, Springer; 2004. Flowering plants, Dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales. Ericaceae; pp. 145–194. (The Families and Genera of Vascular Plants). [Google Scholar]
- Sun M., Naeem R., Su J.X. Phylogeny of the Rosidae: a dense taxon sampling analysis. J. Syst. Evol. 2016;54:363–391. [Google Scholar]
- Sun M., Soltis D.E., Soltis P.S. Deep phylogenetic incongruence in the angiosperm clade Rosidae. Mol. Phylogenet. Evol. 2015;83:156–166. doi: 10.1016/j.ympev.2014.11.003. [DOI] [PubMed] [Google Scholar]
- The Plant List, 2013. Version 1.1. Published on the internet. Available at: http://www.theplantlist.org/(accessed: 10 April, 2020).
- Tang H., Tang L., Shao S. Chloroplast genomic diversity in Bulbophyllum section Macrocaulia (Bl.) Aver. (Orchidaceae, Epidendroideae, Malaxideae): insights into species divergence and adaptive evolution. Plant Divers. 2021 doi: 10.1016/j.pld.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M., Nishikawa T., Fujimoto M. Substitution of the gene for chloroplast rps16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008;25:1566–1575. doi: 10.1093/molbev/msn102. [DOI] [PubMed] [Google Scholar]
- Vaio M., Gardner A., Speranza P. Phylogenetic and cytogenetic relationships among species of Oxalis section Articulatae (Oxalidaceae) Plant Systemat. Evol. 2016;302:1253–1265. [Google Scholar]
- Valencia-D J., Murillo-A J., Orozco C.I. Complete plastid genome sequences of two species of the Neotropical genus Brunellia (Brunelliaceae) Peer J. 2020;8 doi: 10.7717/peerj.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Moore M.J., Wang H. Plastome evolution and phylogenetic relationships among Malvaceae subfamilies. Gene. 2021;765 doi: 10.1016/j.gene.2020.145103. [DOI] [PubMed] [Google Scholar]
- Weng M.L., Blazier J.C., Govindu M. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 2014;31:645–659. doi: 10.1093/molbev/mst257. [DOI] [PubMed] [Google Scholar]
- Wick R.R., Schultz M.B., Zobel J. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.V., Boykin L.M., Howell K.A. The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.S., Chaw S.M. Large-scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in conifers II (Cupressophytes) Genome Biol. Evol. 2016;8:3740–3750. doi: 10.1093/gbe/evw278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Yang L., Su D., Chang X. Phylogenomic insights into deep phylogeny of angiosperms based on broad nuclear gene sampling. Plant Commun. 2020;1:100027. doi: 10.1016/j.xplc.2020.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Zhang N., Zhang Q. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol. 2017;214:1338–1354. doi: 10.1111/nph.14503. [DOI] [PubMed] [Google Scholar]
- Zhang D., Gao F., Jakovlić I. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Zhao F., Li B., Drew B.T. Leveraging plastomes for comparative analysis and phylogenomic inference within Scutellarioideae (Lamiaceae) PLoS One. 2020;15 doi: 10.1371/journal.pone.0232602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Li X., Zhang N. Phylogenomic analyses of large-scale nuclear genes provide new insights into the evolutionary relationships within the rosids. Mol. Phylogenet. Evol. 2016;105:166–176. doi: 10.1016/j.ympev.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Zhu A., Guo W., Gupta S. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016;209:1747–1756. doi: 10.1111/nph.13743. [DOI] [PubMed] [Google Scholar]
- Zhu X.Y., Chase M.W., Qiu Y.L. Mitochondrial matR sequences help to resolve deep phylogenetic relationships in rosids. BMC Evol. Biol. 2007;7:217. doi: 10.1186/1471-2148-7-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.