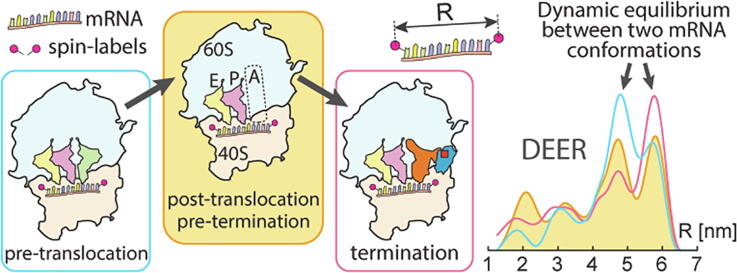
Graphical abstract
Highlights
-
•
DEER reveals the conformational variability of mRNA at the certain translation steps.
-
•
Elongation and termination complexes exist in 2 conformations in dynamic equilibrium.
-
•
The conformations of mRNA in 40S channel undergo no major change during termination.
Abstract
The conformation of mRNA in the region of the human 80S ribosome decoding site was monitored using 11-mer mRNA analogues that bore nitroxide spin labels attached to the terminal nucleotide bases. Intramolecular spin–spin distances were measured by DEER/PELDOR spectroscopy in model complexes mimicking different states of the 80S ribosome during elongation and termination of translation. The measurements revealed that in all studied complexes, mRNA exists in two alternative conformations, whose ratios are different in post-translocation, pre-translocation and termination complexes. We found that the presence of a tRNA molecule at the ribosomal A site decreases the relative share of the more extended mRNA conformation, whereas the binding of eRF1 (alone or in a complex with eRF3) results in the opposite effect. In the termination complexes, the ratios of mRNA conformations are practically the same, indicating that a part of mRNA bound in the ribosome channel does not undergo significant structural alterations in the course of completion of the translation. Our results contribute to the understanding of mRNA molecular dynamics in the mammalian ribosome channel during translation.
1. Introduction
Cell viability basically depends on the synthesis of proteins on ribosomes, specialized organelles that translate the genetic program from the sequences of codons in mRNAs to the sequences of amino acid residues in the polypeptide chains of proteins. Ribosomes are very complex ribonucleoprotein machineries, whose structure and functions have many conserved features in all kingdoms of life. They are composed of small and large subunits responsible for the decoding of mRNA codons and the synthesis of peptide bonds, respectively. During polypeptide chain elongation, the decoding is performed with the participation of tRNA molecules, which carry specific amino acid residues for the polypeptide synthesis and contain anticodons to recognize the corresponding mRNA codons. Ribosomes have three tRNA binding sites: aminoacyl (A) site for incoming aminoacyl-tRNA, peptidyl (P) site for tRNA with nascent peptide chain, and exit (E) site for used discharged tRNA. The crucially important stage of translation, ensuring the formation of normal-sized proteins, is its termination, which occurs when one of three mRNA stop codons (UAA, UAG or UGA) arrives to the ribosomal A site. In eukaryotes, these codons are recognized by the polypeptide release factor eRF1. The latter, acting together with GTPase eRF3, triggers the hydrolysis of the complex-ester bond between the P site tRNA and the synthesized polypeptide at the ribosomal peptidyl transferase center (PTC) and thereby provides the release of the polypeptide from the ribosome (for review, see [1]).
Near-atomic models of eukaryotic ribosomes, including those from higher organisms, and their complexes with ligands, such as mRNAs, tRNAs and translation factors, are currently available from the high resolution cryo-electron microscopy (cryo-EM) and X-ray crystallography studies [2], [3], [4], [5], [6], [7], [8]. These models, together with the data on chemical probing of the rRNA structures in various types of complexes of human ribosomes with release factors [9] have provided general idea on the molecular mechanisms of translation termination. Briefly, stop codon is recognized by specific conserved peptides of the N-terminal domain of eRF1, but universally conserved GGQ tripeptide in the middle domain of eRF1, which is responsible for triggering the P site peptidyl-tRNA hydrolysis, is positioned far away from PTC. The hydrolysis of GTP by eRF3, occurring after the stop codon recognition, induces large conformational rearrangements in the ribosomal complex, leading to the placement of the GGQ tripeptide in the PTC, which results in the release of the synthesized polypeptide. At that, eRF3•GDP does not dissociate from the complex and leaves it only at the next stage of termination, at which eRF1 still remains bound to the ribosome. Near-atomic models of ribosomal translation complexes built from cryo-EM data give detailed information on the structural organization of the complexes, but they are static and dependent on the three-dimensional classification procedure used for the selection of structurally homogeneous translational complexes. Therefore, these models leave aside the variations that may arise due to the conformational dynamics of the complexes at certain translation steps. Such variations could be explored using approaches capable of providing dynamic information complementary to that ensured by the near-atomic models of ribosomal complexes.
One of the fruitful approaches that have recently been applied for studying the interplay between the human ribosome and its RNA ligands is electron paramagnetic resonance (EPR) using synthetic RNAs that carry one or two spin labels at the desired locations [10], [11], [12], [13]. In particular, the utilization of RNAs bearing two spin labels simultaneously enables measurements of the intramolecular spin–spin distances depending on the RNA conformation, which is determined by the type of ribosomal complex. The application of such kind of RNAs has allowed us to compare conformations of free RNA and RNA fixed as mRNA analogue in the ribosomal mRNA-binding channel by the interaction with a cognate tRNA, and to show that these conformations are similar to each other [10]. Recently, we have suggested an approach based on the use of doubly-spin-labeled RNAs having sequences prone to form folded structures, which become unfolded when bound to ribosomes as mRNA analogues, to monitor the portion of ribosome-bound RNA in complexes mimicking various elongation steps [11]. It has proven useful to evaluate the stabilizing effect of interactions of mRNA with tRNA at various ribosomal sites on the mRNA binding. Here, we applied this approach to examine possible differences between the conformations of mRNA in the decoding region of the human ribosome at the elongation and termination stages of translation. We found that mRNA in the ribosome decoding site can adopt two alternative conformations, which differ in their straightening, and that the distribution of mRNA between these conformations depends on the type of complex.
2. Materials and methods
2.1. Doubly spin labeled derivatives of RNAs
The undecaribonucleotides 5′-UGUAGACUUCA-3′ (mRNA-sense) and 5′-UCUUGAUUAGA-3′ (mRNA-stop) carrying ethylene diamine spacers simultaneously at the C5 and C8 atoms of the 5′-terminal uridine and 3′-terminal adenosine residues, respectively, were synthesized and purified as described [14]. The 3D structures of these oligoribonucleotides were predicted using the RNAComposer [15] on the online RNA structure modeling server (http://rnacomposer.cs.put.poznan.pl) in an interactive mode.
The 3-carboxy-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-oxyl succinimidyl ester (NHS-M2) spin label was prepared as described in [16] without deuteration [10], [11]. The doubly spin-labeled RNA derivatives (MR-sense and MR-stop) labeled at the 5′- and 3′-terminal bases were obtained from the corresponding undecaribonucleotides, mRNA-sense and mRNA-stop, by treatment with NHS-M2, purified and characterized as in [11], [12].
3. Ribosomes, tRNAs and termination factors
40S and 60S ribosomal subunits from human placenta were isolated and characterized as described [17], [11] and stored in liquid nitrogen in solution in D2O at a concentration of 25 pmol/μl as 5 μl aliquots for a single thawing operation. 80S ribosomes were obtained from the subunits re-activated in binding buffer A (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 13 mM MgCl2 and 0.5 mM EDTA in D2O) at 37 °C for 10 min by mixing their solutions so that the ratio 40S:60S was 1:1.3. The activity of 80S ribosomes in the poly(U)-dependent binding of [14C]Phe-tRNAPhe was about 70% (approximately 1.4 mol of Phe-tRNAPhe was bound per mol of 80S ribosomes). tRNAPhe and tRNAVal (both about 1300 pmol/A260 unit) from E. coli were the kindly provided by Dr. V.I. Katunin (National Research Center “Kurchatov Institute” B.P. Konstantinov St. Petersburg Nuclear Physics Institute). A yeast tRNAAspGUC transcript obtained by in vitro T7 transcription [18] was used as tRNAAsp. Prior to use for binding to the ribosomes, tRNAs were re-activated by incubation in buffer A at 37 °C for 5 min. Recombinant human polypeptide chain release factors eRF1 (full-length) and eRF3 (lacking the nonessential N-terminal 138 amino acids) were obtained according to [19], [20], [21]. The GTPase activity of eRF3 was determined by the formation of 32P-inorganic phosphate stimulated by 80S ribosomes and eRF1, as described in [22], [23]. The turnover rate was 18 mmol of GTP hydrolyzed • mol−1 of eRF3 • s−1 at 25 °C.
The eRF1 and eRF3 preparations were stored in a buffer A at concentrations of 130 and 400 pmol/μl, respectively, in liquid nitrogen.
4. Ribosomal complexes
Complexes of 80S ribosomes with MR-sense or MR-stop containing tRNAAsp and tRNAVal bound at the P and E sites, respectively, as well as the complex with MR-sense, where in addition to the above tRNA molecules, tRNAPhe was bound at the A site, were obtained and characterized according to [11]. Typically, the mixture in a final volume of 13–15 μl contained 112 pmol of 80S ribosomes, 100 pmol of MR-sense or MR-stop, 270 pmol of tRNAAsp and 450 pmol of tRNAVal. The mixture for preparation of complex with MR-sense and tRNAPhe at the A site additionally contained 450 pmol of tRNAPhe. To obtain 80S ribosomal complexes, where the A site was occupied with eRF1 alone or as eRF1•eRF3•GMPPNP or eRF1•eRF3•GTP, the complex with MR-stop and tRNAs at the P and E sites was incubated at 23 °C for 1 h with either 200 pmol of eRF1 in 1.5 μl or with a mixture containing 200 pmol of eRF1, 200 pmol of eRF3 and 20 mM GTP or GMPPNP in 2.5 μl, pre-incubated for 5 min before use. The eRF1 and eRF3 contents in these 80S ribosomal complexes were examined by dot-blot hybridization with the use of the respective polyclonal rabbit antibodies (#NBP1-57565 and #NBP1-33024 from Novus Biologicals) applied to the fractions of the complexes purified by sucrose density gradient centrifugation as described [9], [24]. The binding levels of tRNAs targeted to the E site or the A site were examined by nitrocellulose filtration technique using the respective 5′-32P-labeled tRNAs, similar to how it has been done in our previous studies [10], [11], [12].
5. EPR experiments
DEER experiments were performed at 50 K temperature and Q-band (34 GHz) using a Bruker Elexsys E580 EPR spectrometer equipped with a commercial Q-band solid-state 150 W amplifier at 10 dB attenuation (15 W), an EN5107D2 resonator and an Oxford Instruments temperature control system. Samples of ca. 10 μl volume in quartz tubes (OD 1.65 mm, ID 1.15 mm) were shock frozen in liquid nitrogen prior to being placed into a pre-cooled resonator. To measure dipolar oscillations, we applied a four-pulse DEER sequence with eight-step pulse position cycle (Δd1 = 16 ns) to average the deuteron modulation and a two–step phase cycle of the first pulse to remove the baseline offset [25]. The employed microwave power of 15 W allowed the observer pulse lengths to be 12–14 ns for π/2 and 20–26 ns for π pulses, pump pulse length was kept 22 ns. Time delays d1 = 340 ns, d2 = 4500 ns, tgate = 66–68 ns, pump pulse time increment was 8 ns, shot repetition time was 5 μs. The pump pulse was applied at the maximum of nitroxide spectrum; the echo observation was done 60 MHz away at the nitroxide central line. Time-domain dipolar evolution traces were processed using Tikhonov regularization [26] and deep neural network [27] embedded in DeerAnalysis2019 and multi-gaussian Monte Carlo approach [13].
6. Results and discussion
6.1. Model mRNAs and ribosomal complexes
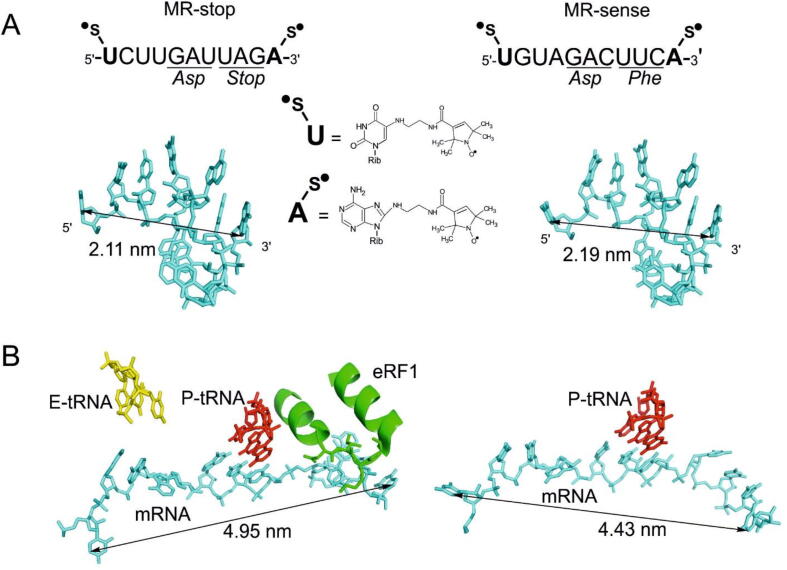
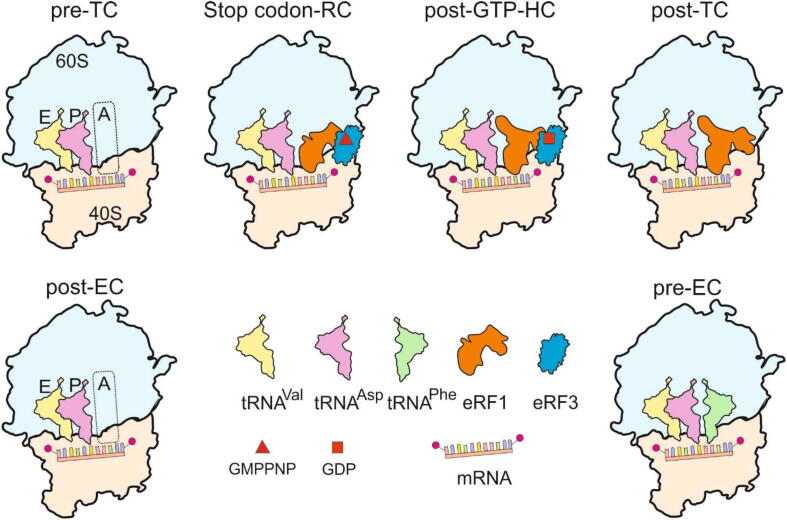
Here, we utilized two doubly nitroxide spin-labeled undecaribonucleotide mRNA analogues, MR-sense and MR-stop, containing in the middle GAC or GAU triplets coding for Asp, followed by either UUC triplet coding for Phe or stop codon UAG (Fig. 1A). This allowed us to obtain simplified complexes imitating either the elongating 80S ribosome (complexes with MR-sense mimicking post-translocation and pre-translocation ones, post-EC and pre-EC, respectively) or its states corresponding to various stages of translation termination, including the pre-termination state. It should be especially noted that short (6–12nucleotideslong) mRNA analogues have proven to be adequate tools for studying various ribosomal translation complexes using different approaches (reviewed in ref. 28), including X-ray crystallograpy [29], [30], and the data obtained with the application of longer mRNA analogues have in general confirmed those gained with short ones (see ref. 28). Nitroxide spin labels inserted into such mRNA analogues do not interfere with their ability to specifically bind to human ribosomes, which is completely dependent on the codon-anticodon interaction between mRNA and the P site tRNA cognate to the desired mRNA codon (see ref. 28), and do not affect significantly Kd values of the respective mRNA analogues [10]. Model termination complexes, very similar to those used here, have been fruitfully applied for studying various aspects of translation termination by means of site-directed cross-linking [24], [31], [32] and chemical footprinting [9], which justifies the utilization of such complexes assembled on the MR-stop in the current study. The adenosine residue following the UAG stop codon allows the formation of a quadruplet/U-turn structure in the A-site peculiar for termination complexes [7]. When preparing the complexes, we used the well-known feature of ribosomes to stably bind tRNA at the P site at elevated Mg2+ concentrations without translation factors, which ensured the fixation of mRNA at the ribosome channel by codon-anticodon interaction between the P site tRNA and an mRNA triplet targeted to this site (for review, see [28]). Accordingly, in our study, the GAC or GAU triplet was directed to the P site by tRNAAsp, while the A site was programmed with either the UUC triplet (in MR-sense) or the stop codon UAG (in MR-stop), which enabled an occupation of the A site with either tRNAPhe or with release factors, respectively. In the latter case, the complex with the empty A site represents the pre-terminating ribosome (pre-termination complex, pre-TC), and the complex with eRF1•eRF3•GMPPNP at the A site corresponds to the state, in which the stop codon is recognized by eRF1 (stop codon recognition complex, stop codon-RC). The complex formed using eRF1•eRF3•GTP targeted to the A site, in which GTPase eRF3 is activated by the ribosome and GTP is converted to GDP, imitates the state after GTP hydrolysis before eRF3•GDP leaves the ribosome (post-GTP hydrolysis complex, post-GTP-HC). Finally, the state with eRF1 alone at the A site (post-termination complex, post-TC) represents a post-termination step when eRF3•GDP is already dissociated from the ribosome. All of the above complexes are shown schematically in Fig. 2.
Fig. 1.
Structures of doubly spin-labeled 11-mers designed as the mRNA analogues. (A) The sequences of mRNA analogues MR-stop and MR-sense with marked positions of spin labels (s•) and the triplets targeted to the P site (left) and the A site (right). Below the sequences, the predicted spatial structures of the MR-stop and MR-sense in free form and the calculated distances between nucleotide atoms bearing spin-labeled groups. (B) The structures of 11-mer fragments of mRNAs bound at the 40S subunit mRNA channel extracted from the cryo-EM-derived structures of the mammalian ribosomal termination complex containing eRF1 and eRF3 [8]; PDB ID:5LZT) (left) and post-translocation complex [36], PDB ID:6Y2L) (right). The extraction was performed so that the nucleotides in positions 5–7 of the mRNA fragments corresponded to the P site codon. The mRNA, anticodons of the E and P site-bound tRNAs, and the eRF1 fragment containing the NIKS motif are shown in cyan, yellow, red and green, respectively. Arrows indicate the distances between atoms in the bases of the terminal nucleotides of the 11-mer fragments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Types of the complexes studied. Upper line: pre-TC, pre-termination complex; stop codon-RC, stop codon recognition complex; post-GTP-HC, a complex formed after GTP hydrolysis before eRF3 dissociation (note that in this complex, GTP is converted to GDP by the GTPase activity of eRF3, and the state after GTP hydrolysis is shown when eRF3•GDP remains bound to the ribosome); post-TC, post-termination complex. Lower line: post-EC, post-translocational elongation complex; pre-EC, pre-translocational elongation complex. The Figure does not display the difference between pre-TC and post-EC, since it only concerns the mRNA trinucleotide sequences at the A site.
The conditions for the stable binding of mRNA analogues and tRNAs at the desired 80S ribosomal sites when obtaining the pre-TC with MR-stop or the post-EC and the pre-EC with MR-sense were almost the same as those used in our earlier studies with the application of similar mRNA analogues [10], [11]. The principles of the formation of model ribosomal complexes, in which the desired codons of mRNA analogues are positioned at the ribosomal P and A sites, were the same as in our previous studies. They are based on the well-known phenomenon that without translation factors, tRNA has maximum affinity for the P site of 80S ribosomes, considerably greater than those for other tRNA binding sites (for review, see [28], [33]). Thus, the P site is occupied with the desired mRNA codon duplexed with the cognate tRNA, which places the next downstream mRNA codon at the A site, allowing its specific recognition by the cognate tRNA, whose affinity for the A site is much higher than that of non-cognate tRNA [34]. Cognate and non-cognate tRNAs bind at the E-site of 80S ribosomes with the occupied P-site to a similar extent (e.g., see ref. 11). In the current study, we confirmed the expected pre-EC, post-EC and pre-TC compositions under the conditions used by estimating the binding levels of mRNA analogues and tRNAs targeted to the P, A or E sites utilizing the corresponding 32P-labeled ligands (for detail, see Fig. S11 and the respective comments in SI). The similar binding levels of the P site tRNAAsp and MR-stop in termination complexes were inferred from the eRF1 binding data (see below), given that the factor does not bind to ribosomes when the A site is not occupied with a stop codon [35], and that short mRNA analogues do not enter the mRNA channel without codon-anticodon interaction with the P site tRNA (see [28]).
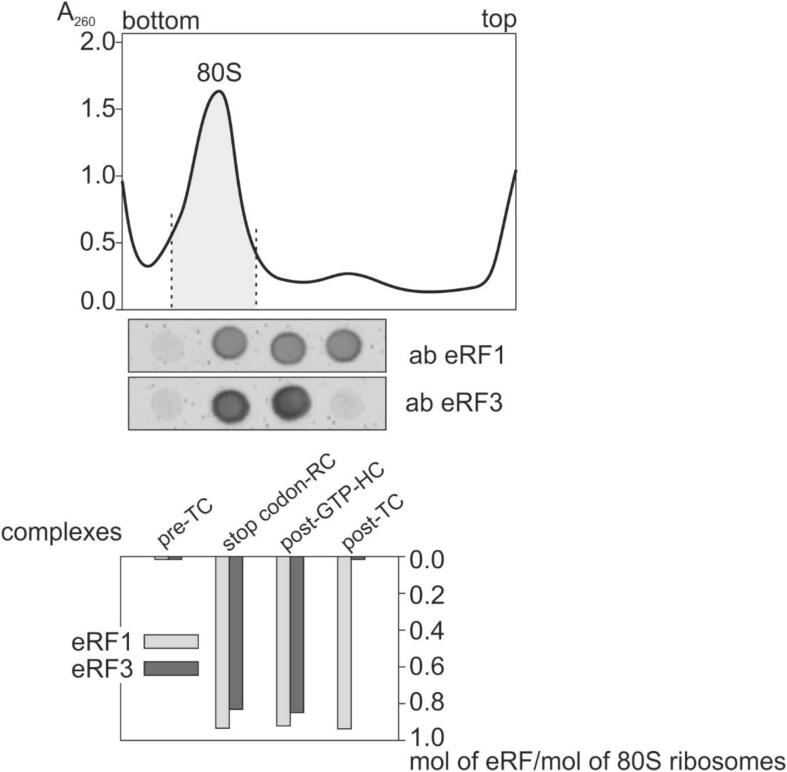
The approach to the assembly of simplified model termination complexes from the pre-TC and eRF1 or its complexes with eRF3 was the same as in our previous studies [9], [24]. The use of deacylated tRNA instead of peptidyl-tRNA when assembling the pre-TC has been justified by the finding that the binding of eRF1 alone or together with eRF3 to the such kind of the pre-TC, caused protections of the 28S rRNA nucleotides at the PTC, indicating that the middle domain of eRF1 with its GGQ motif did indeed arrive to this center, i.e., adopted a functionally relevant position [9]. We believe that GTP is actually hydrolyzed into GDP in the post-GTP-HC, since earlier studies on the chemical footprinting of 28S rRNA in model termination complexes have revealed specific footprints in the ribosomal GTPase associated center with a complex similar to the post-GTP-HC, which have been not observed with a one analogous to the stop codon-RC, where no GTP hydrolysis occurred [9]. Here, we examined the binding levels of eRFs in each of the termination complexes shown in Fig. 2 using dot blot hybridization applied to fractions of the respective 80S ribosomal complexes purified by sucrose density gradient centrifugation. The quantitative analysis of the eRF1 and eRF3 contents in the above fractions (Fig. 3) confirmed their expected compositions with practically stoichiometric extents of binding of the factors to 80S ribosomes. Besides, it reaffirmed the data obtained earlier [9] that eRF3 did not leave the ribosome after hydrolysis of GTP in the complex post-GTP-HC.
Fig. 3.
Analysis of eRF1 and eRF3 binding levels in the model termination ribosomal complexes (see Fig. 2). Determination of the release factors’ contents in the purified ribosomal complexes using the dot blot assay with the subsequent quantification by scanning the respective signals displayed on the membrane with the help of calibration curves obtained with definite amounts of eRF1 or eRF3 developed by the same assay (Fig. S11). The eRFs binding levels were normalized to a fraction of active ribosomes of about 70%, which was evaluated by measuring the activity of 80S ribosomes in codon-dependent aminoacyl-tRNA binding; the relative error in estimating the binding levels was within 15%. A typical sedimentation profile of the 80S ribosomal complexes in the sucrose density gradient is presented in the upper part of the Figure, and the fraction taken for the dot blot assay is shown. The relevance of the main ribosome peak in the sedimentation profile to the 80S couples was confirmed by a parallel sedimentation analysis of the mixture of 40S and 60S ribosomal subunits pre-incubated under association conditions.
It should be noted that all complexes contain a tRNA molecule at the E site, since it is usually present there in the real translating ribosome, and this increases the stability of the termination and post-termination complexes [37]. In our study, tRNAVal was targeted to the E site in the above complexes, which was not cognate to the E site mRNA triplet in the complexes with MR-stop (Fig. 2), because the occurrence of codon-anticodon interaction at this site was questionable. Moreover, in our previous EPR study, we have not found substantial differences in the arrangements of mRNA at the ribosomal channel for complexes assembled with cognate and non-cognate tRNAs at the E site [11]. It is also worth noting that the pre-EC with three tRNA molecules simultaneously bound at the P, A and E sites of the same 80S ribosome is functionally relevant. This stems from the data of the cryo-EM study, in which the pre-translocation state with all these sites occupied with tRNA molecules is visualized among many other intermediates in ex vivo-derived human polysomes [38]. Finally, it should be noted that in some portion of the complexes obtained in this study, the E site was occupied with tRNAPhe instead of tRNAVal, since the former was taken in the same excess over ribosomes as the latter, and, therefore, a portion of tRNAPhe, unbound at the A site, could compete with tRNAVal for the E site, like any other non-cognate tRNA (for detail, see Fig. S11 and the respective comments in SI). We believe that the nature of tRNA at the E site does not contribute substantially to the structure of the ribosomal complex under investigation.
7. Rearrangements of the mRNA structure in the ribosomal complexes
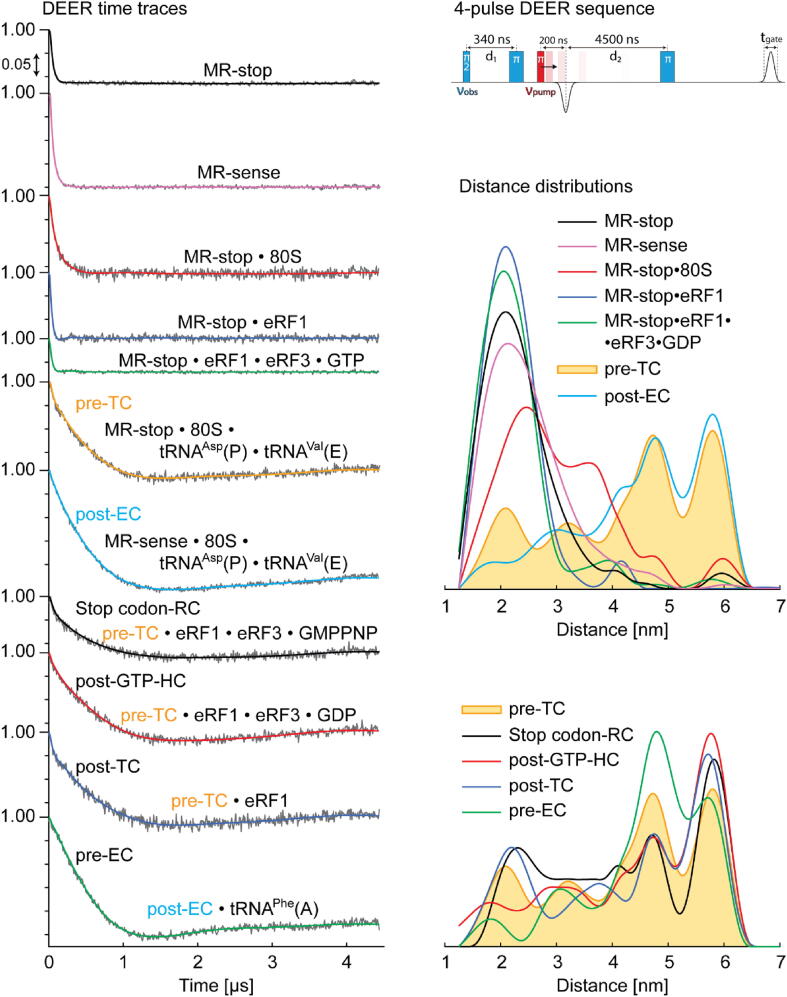
Both mRNA analogues, MR-sense and MR-stop, used in this study are expected to be folded, which should result in short interspin distances when they are free (Fig. 1A) [11], [12]. Two MR-sense molecules can also form imperfect duplex with the expected separation of 2–2.5 nm between the terminal nucleotides (Fig. 1D) that were indicated by additional line broadening observed in the continuous wave (CW) EPR spectrum at 300 K (Fig. S3 in SI). Within ribosomal complexes, mRNAs unfold and the duplexes dissociate, so the interspin distances are supposed to be considerably larger (Fig. 1B), similar to what has been observed with the mRNA analogues utilized in our previous studies [11], [12]. As before, we applied here DEER (PELDOR) to examine distances between two nitroxide spin labels on the 5′- and 3′-termini of the mRNA analogues free and bound to ribosomes in various complexes. Fig. 4 shows the DEER time traces and distance distributions obtained using the Tikhonov regularization method with MR-stop and MR-sense. As expected, with free mRNA analogues (in the absence of ribosomes), one main state with the interspin distance RDEER < 2.6 nm was observed (Fig. 4). With binary mixtures of mRNA analogues with ribosomes, additional peaks with RDEER in the region 2.6 – 4.0 nm were detected. We assigned these peaks, by analogy with the previous studies [11], [12], to partially unfolded RNA in its labile ribosomal complexes involving the 40S subunit protein uS3. Similar RDEER values have been also observed for unstable binary complexes of tRNAs with short mRNA analogues [11].
Fig. 4.
DEER time traces (left) and corresponding distance distributions (right). The inset on top right shows DEER pulse sequence applied and the time delays used. Background corrected experimental time traces are shown in grey, DeerAnalysis2019 Tikhonov fits are colored according to the distance distributions colors. Note that GTP in the post-GTP-HC formed using eRF1•eRF3•GTP was actually transformed to GDP at the time of the DEER recording.
Finally, the states of mRNA corresponding to its unfolding, with large interspin distances in the regions 4.0–5.2 nm and > 5.2 nm, were seen only with the ribosomal complexes in which mRNA analogues were fixed in the 40S subunit binding channel by the interaction of the GAC (MR-sense) or GAU (MR-stop) triplets with the P site tRNAAsp (Fig. 4). Mixing of MR-stop with eRF(s) in the absence of ribosome did not lead to any unfolding of mRNA, which preserved RDEER < 2.6 nm. The shares of labile binary complexes of the mRNA analogues with tRNAs or ribosomes (which could not be discriminated by the RDEER values) depended on the type of mixture. However, in all cases, the total share of these complexes together with that of the free mRNA analogue was less than the total share of the ribosomal complexes under investigation (Table 1), which, in turn, was close to the fraction of active ribosomes (Fig. S11).
Table 1.
Integral of the peaks in the distance distributions obtained using Tikhonov regularization for the studied ribosomal complexes. See SI for information on uncertainty.
| Sample | integral in the range [%] |
||
|---|---|---|---|
| <4.0 nm | 4.0–5.2 nm | >5.2 nm | |
| pre-TC | 36 | 34 | 30 |
| Stop codon-RC | 42 | 25 | 33 |
| post-GTP-HC | 31 | 27 | 42 |
| post-TC | 38 | 24 | 38 |
| post-EC | 29 | 36 | 35 |
| pre-EC | 21 | 47 | 32 |
Comprehensive analysis using different approaches to DEER data processing (Tikhonov regularization, multi-gaussian Monte Carlo and deep neural network, see details in SI) results in similar distance distribution functions with two different peaks in the regions RDEER > 4.0, clearly showing that unfolded mRNAs in the ribosome channel exist in two alternative conformations. A less extended RNA conformation, with the mean RDEER value about 4.7 nm, has been reported earlier [11], [12] with similar 11-mer mRNA analogues, and in general it fits to the structure of mRNA in available atomic models of 80S ribosomal complexes corresponding to various stages of translation (e.g., see [39], [40]). The more extended mRNA conformation with interspin distance about 5.8 nm was revealed for the first time in this work, and it was observed for all types of the ribosomal complexes under investigation (Fig. 4). Such long distance became detectable due to the power upgrade of the Q-band microwave bridge from 1 W up to 15 W, which drastically increased the sensitivity and the range of accessible distances, thus making it possible to obtain more detailed information on the systems under study. Note that for the measured DEER traces, background correction brings significant uncertainty in determining long distances, so only the integral of the peak assigned to the more extended mRNA is analyzed. The effects of the DEER trace length and the baseline correction on the distance distribution functions are discussed in more detail in SI.
Differences in the distance distributions and in the integrated peaks areas reflect the transformations in conformations of the mRNA analogues bound to the ribosome in the studied complexes. Observable resolution of peaks at 4.7 and 5.8 nm was verified via background correction analysis and processing DEER data using Tikhonov regularization, neural network and Monte Carlo multi-gaussian search (Fig. S9 in SI). Integral values were validated as well. In order to characterize conformational transformations, we analyzed the weights of conformations, i.e. the corresponding peak integrals. Although different approaches give variable peak integrals for the same complex, with each processing method the ratios between peak integrals for different complexes together with all the trends are preserved (Fig. S10 in SI).
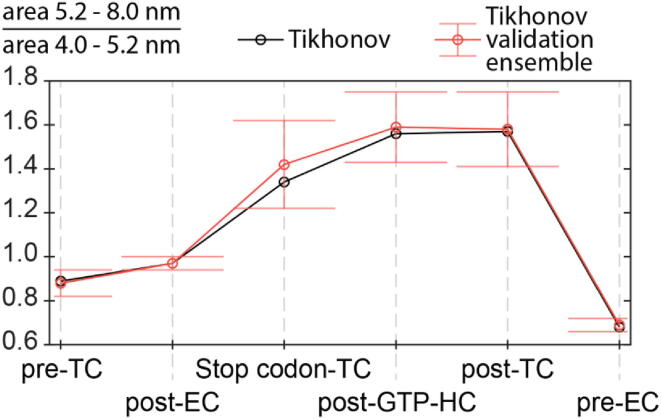
Analysis of the peak integrals (Table 1) shows that the ratio (Fig. 5) of conformations adopted by the mRNA analogue depends on the type of complex. We observed reliable differences in the ratios between the pre-TC with the empty A site and the termination complexes, where this site was occupied with release factor(s). In particular, in the presence of the ligand(s) at the A site, the ratio shifts to an increased contribution of the more extended mRNA conformation.
Fig. 5.
Ratios of area 5.2–8.0 nm to area 4.0–5.2 nm obtained using Tikhonov regularization for the studied ribosomal complexes. See SI for information on Tikhonov validation ensemble.
To clarify whether the effect of the A site bound eRF(s) on mRNA conformation is specific to termination complexes or is caused by the binding of any kind of ligand interacting with the A site mRNA codon, we compared the results obtained with these complexes and with the pre-EC, where the A site was occupied with tRNAPhe (Fig. 2). Although the MR-stop and MR-sense sequences used for the assembly of model termination and elongation complexes, respectively, are not identical (they differ from each other by the codons targeted to the A and E sites), the comparison is correct, because the distance distributions of the complexes with vacant A site for MR-stop (pre-TC) and MR-sense (post-EC) are very similar (Fig. 4). This comparison shows that the occupation of the A site with tRNAPhe results in a significant decrease in the relative proportion of the more extended mRNA form (see Fig. 4, Fig. 5 and Table 1), the effect opposite to that caused by the binding of eRF(s) at the A site. Moreover, the relative share of the latter mRNA form in the pre-EC is the fewest among all the studied complexes.
The eRF1, which directly interacts with mRNA stop codon at the A site, generally is considered as an aminoacyl-tRNA mimic [5]. Its evolutionary conserved NIKS and YxCxxF motifs in the N-terminal domain bind at the ribosome decoding site, playing the role similar to that of tRNA anticodon, and the universal GGQ tripeptide in the middle domain appears at the PTC after GTP hydrolysis as 3′-terminus of tRNA, where it triggers the hydrolysis of the peptidyl-tRNA ester bond [5], [21], [31], [32], [40], [41], [42] (Fig. 2). The findings from our study demonstrate that the binding of eRF1 (alone or in a complex with eRF3) at the A site affects mRNA conformation in a manner opposite to that of tRNA molecule, increasing the relative share of the more extended mRNA form in the complex, which indicates an essential difference between the A site interaction of eRF1 and a tRNA molecule. Since both mRNA conformations occur in the pre-TC, one can assume that eRF1 prefers to bind to the pre-TC with the more extended mRNA conformation, thereby shifting the equilibrium towards the latter. Alternatively, it is possible that eRF1 binds to the pre-TC with the less extended mRNA conformation, and this binding induces rearrangements resulting in the mRNA adopting the more extended conformation, which may be due to the conformational dynamics of the ribosome itself, e.g. translocation/back-translocation, head tilt and swivel movements, etc (see ref. [43], [44]). We do not currently have enough data to choose between these possibilities, and clarifying of the above issues is the next frontier of investigations.
The results obtained in general are in accordance with cryo-EM data showing that the geometries of the A site mRNA codons interacting with either eRF1 or the aminoacyl-tRNA anticodon differ significantly from each other [4], [7]. In particular, in the elongation complexes, the A site codon belongs to one chain of the double-helix structure, while in the termination complexes its conformation resembles the geometry of the U-turn motif [7]. As mentioned in the Introduction, cryo-EM-derived models do not provide information on the conformational dynamics of translation complexes at any particular translation step, since these models display only one kind of the structure, which eventually depends on the procedure of 3D classification applied for the selection of structurally homogeneous translational complexes. Besides, information on several mRNA conformations in ribosomal complexes is lost in cryo-EM models, because some mRNA nucleotides in the 40S subunit binding channel remain unresolved due to their conformational flexibility. Our findings gained by DEER/PELDOR for the first time show that both elongation and termination complexes exist in two alternative conformations that are likely to be in dynamic equilibrium. It should be noted that the interspin distances in the mRNA analogues measured in this study (Fig. 4) do not coincide with the distances between mRNA bases in positions corresponding to those of the spin-labeled nucleotides in the MR-stop and MR-sense calculated from cryo-EM-derived models of ribosomal complexes of the respective types (Fig. 1B). This is not a discrepancy, because the spin labels are separated from nucleotide bases in the mRNA analogues by the linkers, whose length and geometry contribute to the interspin distances observed in this study.
Because of the conformational flexibility of mRNA in the decoding region of the ribosome, the ratio between its conformations depends on the type of complex (pre-translocation, post-translocation or termination one). However, the differences in the ratios of mRNA conformations between the complexes are not dramatic; moreover, they are barely pronounced between particular termination complexes (see Table 1). The latter indicates that the distribution of mRNA between two alternative conformations does not significantly change during translation termination. Therefore, we can conclude that the conformational status of mRNA in the 40S subunit binding channel does not undergo major alterations during termination, at least, until eRF1 leaves the ribosome.
Noteworthy, if we consider intra-mRNA distances, corresponding to those studied in our work, in cryo-EM-derived models of mammalian 80S ribosomal elongation and termination complexes, one can find regularities in general similar to those obtained with the use of MR-stop and MR-sense. For instance, one can see from Fig. 1B that in the termination complex containing eRF1 (the left model), the distance between the mRNA sites under investigation is significantly larger than that in the post-translocation complex with vacant A site (the right model). This correlates with our data on a higher ratio of the more extended mRNA form to the less extended one in the post-TC as compared to that in the post-EC (Fig. 4, Table 1). A similar comparison of the distances in the pre-translocation (pdb 5LZS, [36]) and post-translocation (pdb 6Y2L, [8]) complexes can reveal the smaller distance in the first, which also correlates with our results showing a lower ratio of the more extended mRNA form to the less extended one in the pre-EC versus the post-EC (Fig. 4, Table 1).
Thus, the application of DEER/PELDOR allowed us to obtain new important details concerning the conformational variability of mRNA in 80S ribosomal complexes. Our findings contribute to the further development of an understanding of the functional and dynamic aspects of translation in mammals, which has so far been mainly based on static near-atomic models of various ribosomal complexes, obtained using cryo-EM and X-ray crystallography. It is possible that the nature of the stop codon and/or the adjacent nucleotide residue following it, as well as the specific features of the mRNA sequence bound at the ribosomal channel, affect the dynamics of mRNA in the termination complexes and, accordingly, the ratio between its conformations, and therefore the study this issue could be the topic of the next frontier of research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors are grateful to Igor Kirilyuk for providing the spin label MTSL The work was supported by the Ministry of Science and Education of the Russian Federation (grant 14.W03.31.0034).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.08.024.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Petry S., Weixlbaumer A., Ramakrishnan V. The termination of translation. Curr. Opin. Struct. Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Taylor D., Unbehaun A., Li W., Das S., Lei J., Liao H.Y. Cryo-EM structure of the mammalian eukaryotic release factor eRF1–eRF3-associated termination complex. Proc. Natl. Acad. Sci. USA. 2012;109:18413–18418. doi: 10.1073/pnas.1216730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Preis A., Heuer A., Barrio-Garcia C., Hauser A., Eyler D.E., Berninghausen O. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014;10:59–65. doi: 10.1016/j.celrep.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown A., Shao S., Murray J., Hegde R.S., Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524(7566):493–496. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F. The crystal structure of human eukaryotic release factor eRF1 – Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100(3):311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 6.Muhs M., Hilal T., Mielke T., Skabkin M.A., Sanbonmatsu K.Y., Pestova T.V. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol. Cell. 2015;57:422–432. doi: 10.1016/j.molcel.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matheisl S., Berninghausen O., Becker T., Beckmann R. Structure of a human translation termination complex. Nucleic Acids Res. 2015;43(18):8615–8626. doi: 10.1093/nar/gkv909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao S., Murray J., Brown A., Taunton J., Ramakrishnan V., Hegde R.S. Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell. 2016;167(5):1229–1240.e15. doi: 10.1016/j.cell.2016.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulygin K.N., Bartuli Y.S., Malygin A.A., Graifer D.M., Frolova L.Y., Karpova G.G. Chemical footprinting reveals conformational changes of 18S and 28S rRNAs at different steps of translation termination on the human ribosome. RNA. 2016;22(2):278–289. doi: 10.1261/rna.053801.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malygin A.A., Graifer D.M., Meschaninova M.I., Venyaminova A.G., Krumkacheva O.A., Fedin M.V. Doubly spin-labeled RNA as an EPR reporter for studying multicomponent supramolecular assemblies. Biophys. J. 2015;109:2637–2643. doi: 10.1016/j.bpj.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malygin A.A., Graifer D.M., Meschaninova M.I., Venyaminova A.G., Timofeev I.O., Kuzhelev A.A. Structural rearrangements in mRNA upon its binding to human 80S ribosomes revealed by EPR spectroscopy. Nucleic Acids Res. 2018;46:897–904. doi: 10.1093/nar/gkx1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malygin A.A., Krumkacheva O.A., Graifer D.M., Timofeev I.O., Ochkasova A.S., Meschaninova M.I., Venyaminova A.G., Fedin M.V., Bowman M.K., Karpova G.G., Bagryanskaya E.G. Exploring the interactions of short RNAs with the human 40S ribosomal subunit near the mRNA entry site by EPR spectroscopy. Nucleic Acids Res. 2019;47:11850–11860. doi: 10.1093/nar/gkz1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timofeev I.O., Krumkacheva O.A., Fedin M.V., Karpova G.G., Bagryanskaya E.G. Refining spin-spin distance distributions in complex biological systems using multi-gaussian monte carlo analysis. Appl. Magn. Reson. 2018;49:265–276. [Google Scholar]
- 14.Repkova M.N., Ivanova T.M., Komarova N.I., Meshchaninova M.I., Kuznetsova M.A., Venyaminova A.G. H-phosphonate synthesis of oligoribonucleotides containing modified bases. I. Photoactivatable derivatives of oligoribonucleotides with perfluoroarylazide groups in heterocyclic bases. Russ. J. Bioorgan. Chem. 1999;25:690–701. [Google Scholar]
- 15.Popenda M., Szachniuk M., Antczak M., Purzycka K.J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hankovszky H.O., Hideg K., Tigyi J. Nitroxides. II. 1-Oxyl-2,2,5,5-tetramethylpyrroline-3-carboxylic acid derivatives. Acta Chim. Acad. Sci. Hungaricae. 1978;98:339–348. [Google Scholar]
- 17.Matasova N.B., Myltseva S.V., Zenkova M.A., Graifer D.M., Vladimirov S.N., Karpova G.G. Isolation of ribosomal subunits containing intact rRNA from human placenta: Estimation of functional activity of 80S ribosomes. Anal. Biochem. 1991;198(2):219–223. doi: 10.1016/0003-2697(91)90416-q. [DOI] [PubMed] [Google Scholar]
- 18.Chavatte L., Frolova L., Kisselev L., Favre A. The polypeptide chain release factor eRF1 specifically contacts the s4UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem. 2001;268:2896–2904. doi: 10.1046/j.1432-1327.2001.02177.x. [DOI] [PubMed] [Google Scholar]
- 19.Frolova L.Y., Simonsen J.L., Merkulova T.I., Litvinov D.Y., Martensen P.M., Rechinsky V.O. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem. 1998;256(1):36–44. doi: 10.1046/j.1432-1327.1998.2560036.x. [DOI] [PubMed] [Google Scholar]
- 20.Frolova L.Y., Merkulova T.I., Kisselev L.L. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA. 2000;6(3):381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frolova L., Seit-Nebi A., Kisselev LEV. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8(2):129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frolova L.Y., Le Goff X., Zhouravleva G., Davydova E., Philippe M., Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 23.Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125(6):1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Bulygin K.N., Graifer D.M., Hountondji C., Frolova L.Y., Karpova G.G. Exploring contacts of eRF1 with the 3'-terminus of the P site tRNA and mRNA stop signal in the human ribosome at various translation termination steps. Biochim. Biophys. Acta - Gene Regul. Mechan. 2017;1860(7):782–793. doi: 10.1016/j.bbagrm.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Pannier M., Veit S., Godt A., Jeschke G., Spiess H.W. Dead-time free measurement of dipole–dipole interactions between electron spins. J. Mag. Res. 2011;213:316–325. doi: 10.1016/j.jmr.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 26.Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J. DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- 27.Worswick S.G., Spencer J.A., Jeschke G., Kuprov I. Deep neural network processing of DEER data. Sci. Adv. 2018;4(8):eaat5218. doi: 10.1126/sciadv.aat5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graifer D.M., Karpova G.G. Photoactivatable RNA derivatives as tools for studying the structural and functional organization of complex cellular ribonucleoprotein machineries. RSC Adv. 2013;3:2858–2872. [Google Scholar]
- 29.Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 30.Korostelev A., Trakhanov S., Laurberg M., Noller H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126(6):1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 31.Bulygin K.N., Khairulina Y.S., Kolosov P.M., Ven'yaminova A.G., Graifer D.M., Vorobjev Y.N. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA. 2010;16(10):1902–1914. doi: 10.1261/rna.2066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulygin K.N., Khairulina Y.S., Kolosov P.M., Ven'yaminova A.G., Graifer D.M., Vorobjev Y.N. Adenine and guanine recognition of stop codon is mediated by different N domain conformations of translation termination factor eRF1. Nucleic Acids Res. 2011;39:7134–7146. doi: 10.1093/nar/gkr376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graifer D.M., Karpova G.G. Interaction of tRNA with eukaryotic ribosome. Int. J. Mol. Sci. 2015;16:7173–7194. doi: 10.3390/ijms16047173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111(5):721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 35.Bulygin K.N., Repkova M.N., Ven’yaminova A.G., Graifer D.M., Karpova G.G., Frolova L.Y., Kisselev L.L. Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes. FEBS Lett. 2002;514:96–101. doi: 10.1016/s0014-5793(02)02304-9. [DOI] [PubMed] [Google Scholar]
- 36.Bhaskar V., Graff-Meyer A., Schenk A.D., Cavadini S., von Loeffelholz O., Natchiar S.K. Dynamics of uS19 C-terminal tail during the translation elongation cycle in human ribosomes. Cell Rep. 2020;31(1):107473. doi: 10.1016/j.celrep.2020.03.037. [DOI] [PubMed] [Google Scholar]
- 37.Susorov D., Mikhailova T., Ivanov A., Sokolova E., Alkalaeva E. Stabilization of eukaryotic ribosomal termination complexes by deacylated tRNA. Nucleic Acids Res. 2015;43:3332–3343. doi: 10.1093/nar/gkv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behrmann E., Loerke J., Budkevich T.V., Yamamoto K., Schmidt A., Penczek P.A. Structural snapshots of actively translating human ribosomes. Cell. 2015;161(4):845–857. doi: 10.1016/j.cell.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budkevich T.V., Giesebrecht J., Behrmann E., Loerke J., Ramrath D.J.F., Mielke T. Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell. 2014;158(1):121–131. doi: 10.1016/j.cell.2014.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frolova L.Yu., Tsivkovskii R.Yu., Sivolobova G.F., Oparina N.Yu., Serpinsky O.I., Blinov V.M. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5(8):1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertram G., Bell H.A., Ritchie D.W., Fullerton G., Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6(9):1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chavatte L., Seit-Nebi A., Dubovaya V., Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korostelev A., Ermolenko D.N., Noller H.F. Structural dynamics of the ribosome. Curr. Opin. Chem. Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noller H.F., Lancaster L., Mohan S., Zhou J. Ribosome structural dynamics in translocation: yet another functional role for ribosomal RNA. Quart. Rev. Biophys. 2017;50 doi: 10.1017/S0033583517000117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.