Abstract
Fluorescence micrographs of the plasma membrane of cells expressing fluorescently labeled G protein-coupled receptors (GPCRs) often exhibit small clusters of pixels (or puncta) with intensities that are higher than those of the surrounding pixels. Although studies of GPCR interactions in uniform membrane areas abound, understanding the details of the GPCR interactions within such puncta as well as the nature of the membrane formations underlying the puncta is hampered by the lack of adequate experimental techniques. Here, we introduce an enhancement of a recently developed method termed fluorescence intensity fluctuation spectrometry, which permits analysis of protein-protein interactions within the puncta in live cell membranes. We applied the novel fluorescence intensity fluctuation data analysis protocol to previously published data from cells expressing human secretin receptors and determined that the oligomer size increases with receptor concentration and duration of treatment with cognate ligand, not only within uniform regions of the membrane (in agreement with previous publications) but also within the puncta. In addition, we found that the number density and fractional area of the puncta increased after treatment with ligand. This method could be applied for probing the evolution in the time of the chain of events that begins with ligand binding and continues with coated pits formation and receptor internalization for other GPCRs and, indeed, other membrane receptors in living cells.
Significance
Recent introduction of the fluorescence intensity fluctuation spectrometry has helped reveal protein-proteins interactions in live cells using fluorescence images. Recent studies have targeted mostly flat plasma membrane portions or homogeneous protein solutions. In this article, we significantly expand fluorescence intensity fluctuation spectrometry to study inhomogeneous regions on the cell membrane appearing as brighter spots or puncta within the larger homogeneous regions of the membrane. We believe that this method provides new insights into the receptor dynamics inside membrane microdomains (puncta) and will eventually allow one to determine the microdomains identity when used in conjunction with specific biochemical assays.
Introduction
A considerable number of G protein-coupled receptors (GPCR) are known to bind either to receptors of their own kind to form homo-oligomers or to different receptors to form hetero-oligomers. Although the relationship between the size of the oligomer and its biological function remains elusive (1, 2, 3, 4), detailed computer simulations (5,6) as well as recent advancements in imaging technology (7, 8, 9, 10) have provided evidence that the oligomeric size of GPCRs is modulated by interactions with ligands; this may eventually allow the long-awaited promise of using protein-protein interactions as drug targets to be realized in practice (11). Nevertheless, many open questions remain regarding the nature of such interactions as well as their physiological relevance, mostly because of experimental challenges caused, among other things, by the heterogeneous structure of cell membranes, including the existence and the dynamic character of membrane folds, invaginations, docked vesicles, and other inhomogeneities.
Several methods have been developed over the past three decades for quantifying membrane-receptor interactions. From this arsenal of techniques, methods based on the collection and analysis of fluorescent signals from fluorescently labeled proteins of interest remain at the forefront for quantifying protein-protein interactions in living cells. The family of fluorescence-based methods can itself be broken down into a number of different approaches—e.g., fluorescence lifetime imaging (12), Förster resonance energy transfer (FRET) (13, 14, 15, 16, 17), fluorescence polarization anisotropy (18), and fluorescence fluctuation spectroscopy (FFS) (7,8,19, 20, 21, 22, 23, 24)—each providing its own set of benefits and drawbacks and differing in the type, extent, and precision of the information provided. Among these methods, those employing FFS to measure molecular brightness have proven to be of significant value for the specific task of probing the existence of an interaction between proteins as they are relatively straightforward to implement.
In the FFS family of methods, the molecular (or molecular complex) brightness is proportional to the product of the molecule’s absorption cross section and quantum yield, which are both intrinsic properties of the fluorescent tag, as well as the number of molecules comprising the oligomer, i.e., the oligomer size. Earlier FFS-based methods provided the average oligomer brightness and an average number of oligomeric structures for an ensemble of molecules measured from either temporal fluctuations in fluorescence intensity, as in the case of the number and brightness analysis (21,25), or fluctuations in fluorescence intensities among pixels within an image, as in the case of spatial intensity distribution analysis (22, 23, 24). These analysis methods work best if the region of interest (ROI) is predominantly populated by an oligomer of only a single size. However, when the ROI contains a mixture of oligomers with different sizes or the relative proportions of the various oligomers vary as a function of, e.g., receptor concentration, only an average oligomer size is obtained. Such an outcome does not provide a complete picture of the dependence of the protein-protein interactions on the receptor concentration. Use of the traditional FFS-based approaches becomes even more challenging when comparing receptors treated with a ligand to untreated receptors, as ligand treatment might result in changes that may go undetected when extracting an average oligomer size over a broad receptor concentration range. In addition, one needs to carefully choose ROIs with homogeneous distributions of intensities or else the combination of fluorescence intensities from homogeneous regions with the comparatively high intensities from membrane inhomogeneities included in the same ROI would result in broad intensity distributions that would be mistaken for drastically increased average oligomer size.
The difficulties mentioned in the previous paragraph have been addressed by the recent introduction of an improved FFS method named fluorescence intensity fluctuation (FIF) spectrometry. This method provides quantitative information on the size and stability of oligomers as a function of protomer concentration (7) and presents an inherent inhomogeneity-filtering property (8). Moreover, if the analyzed set of ROIs contains a mixture of oligomers with different sizes, FIF spectrometry can predict the abundances of the various oligomers comprising the mixture as a function of protomer concentration.
FIF spectrometry is implemented in four steps, as follows: 1) large ROIs within fluorescence images of cells expressing fluorescently labeled molecules of interest are divided into smaller segments with a preset area using a computer algorithm. 2) Intensity distributions, generated from pixel-level intensity values for each ROI segment, are fit with a Gaussian function, and the mean and variance of the fitted Gaussian are used to calculate an effective molecular brightness, εeff, and a concentration, Cm, value for each segment. 3) Histograms of εeff, also termed “brightness spectra,” are compiled from the εeff values of individual segments and are sorted according to user-selected concentration ranges. 4) The εeff spectra sorted according to their concentration ranges may be either visually assessed or mathematically deconvoluted using a monomeric brightness spectrum determined a priori to quantify the abundance of differently sized oligomeric species. Using FIF spectrometry, Stoneman et al. (7) determined that the human wild-type secretin receptor (hSecR) exists as a mixture of different oligomer sizes whose equilibrium may be shifted by receptor concentration or ligand binding, and Vu et al. (26) demonstrated that E-cadherin forms constitutive lateral (cis) dimers at the plasma membrane, whereas Ahmed et al. (27) showed that biased receptor tyrosine kinase ligands induce differential stabilization of receptor tyrosine kinase dimers.
Fluorescence micrographs of cellular membranes populated with fluorescently labeled receptors often contain small groups of pixels, which we call “puncta” herein, presenting intensity values that are significantly higher than the typical intensity level of pixels in the surrounding region. The large intensity fluctuations introduced by puncta (relative to uniform, low-intensity regions) generate artificially broad distributions of intensities within an ROI segment and hence artificially high brightness values. Earlier intensity fluctuation-based methods removed this artifact by painstakingly avoiding such high-intensity puncta or other inhomogeneities during the ROI-generation process. This method is rather tedious and imprecise, especially because for many receptors treated with ligands, the plasma membrane acquires an obvious “punctate” look. Apart from being imprecise, defining what is meant by uniform region is a very subjective act, which may result in inadvertently avoiding fluctuations resulting from the diffusion of molecules, which are the very fluctuations that are required to calculate the brightness.
The challenge of objectively analyzing fluorescence images containing puncta is easily circumvented in FIF spectrometry because of this method’s inherent low-pass filtering capability, which stems from the following (8): 1) fitting the intensity histograms with a Gaussian, as opposed to taking the mean of the distribution, reduces the effect of the high-intensity tail contribution of the puncta to the calculated mean and variance for the ROI segment analyzed. 2) Dividing large ROIs into smaller segments (of which a punctum now represents a significant portion) pushes the calculated brightness of segments that contain puncta to the far right of the brightness distribution, which is cropped at reasonable values of a few times the brightness of a monomer. In a recent publication from our lab (8), the fluorescence images were analyzed using FIF spectrometry before and after removing the fluorescent puncta from the images, and the results in both cases were comparable. This finding demonstrated the robustness of FIF spectrometry, even when applied to analysis of fluorescence images containing clear high-intensity puncta littered throughout the plasma membrane of imaged cells.
Nevertheless, in many situations, the high-intensity puncta appearing in the cell membrane micrographs are the actual objects of interest. High-intensity puncta that are visible within fluorescence images of cell membranes may represent a number of organized structures in the plasma membrane. Such membrane regions (also known as lipid microdomains) include endocytic vesicles, clathrin- or caveolin-coated pits, corrals (which are bordered by actin filaments, tubulin rod junctions and boundaries, or both), and rafts (28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42). Each of these microdomains have a molecular composition and supramolecular architecture that are different from the surrounding membrane areas. For example, rafts and caveolae (caveolin-coated pits/vesicles) contain more cholesterol than a typical membrane patch, whereas most pits and endocytic vesicles contain clathrin networks. Furthermore, each of the membrane domains have specific functionalities. For example, rafts were found to transport proteins along the apical membrane (30, 31, 32,37,41) or take part in endocytosis as caveolae (38, 39, 40), and pits await departure from the membrane via transformation into endocytic vesicles (28,31,33,42,43). Each of these domains will change the motion of proteins along the membrane and within the domain. For example, corals (29,36) permit Brownian motion within its boundaries with a finite probability of proteins to hop in and out of the region, whereas clathrin networks and caveolae trap the proteins in the pit with a reduced ability to move within the pit and a small chance of leaving it (34,38,40, 41, 42).
The vast majority of studies investigating these different membrane domains have focused on the molecular makeup and specific role played by the domain as a whole in the overall function of the membrane. However, the interaction properties of the proteins confined within these domains are less often characterized (29,32). The focus of the work described herein is to modify the FIF spectrometry method to allow identification of the nature of the puncta and provide information on the size of the oligomers entering these puncta relative to those outside the puncta, both in the absence and presence of cognate ligand. We have introduced two new key elements into the analysis, as follows:
-
1)
Before uniform segmentation of the membrane ROIs, the ROIs are subjected to the simple linear iterative clustering (SLIC) algorithm (44,45), which we have previously implemented to identify and remove puncta from the analysis of fluorescence micrographs (8). In this work, the fluorescence intensity recorded in the pixels located within the boundaries of the puncta are collected and analyzed separately.
-
2)
As a single punctum is typically too small for reliable punctum-by-punctum analysis using FIF spectrometry, we combine the pixel content of a number of puncta with the same average intensity and calculate a single molecular brightness value from this cluster of puncta. The remaining steps follow the original FIF spectrometry procedure as described previously (7) and summarized above.
We tested this method using a set of previously obtained (7) fluorescence images of cells expressing hSecR labeled with mEGFP and observed that the size of the oligomer entering the puncta increases with receptor concentration and ligand treatment. This dependence is similar to that observed for uniform (i.e., depunctate) membrane patches. To determine whether the puncta were endocytic vesicles or pre-endocytic structures (i.e., pits) versus exocytic vesicles or entirely different structures, we have also computed the density of puncta per image as it changed with ligand treatment duration. We found that the density of the puncta increased with increasing ligand treatment duration. This observation suggests that the high-intensity puncta may represent pits that trap functional receptor oligomers and bud off into the cell, becoming endocytic vesicles (28,31,33,43), although additional assays are necessary to determine whether other membrane microdomains better explain this behavior.
Materials and methods
Source of data and outline of the methods of analysis
Fluorescence micrographs of the basolateral membrane of Chinese hamster ovary (CHO) cells expressing hSecR fused to monomeric enhanced green fluorescent protein (hSecR-mEGFP) and Flp-In T-REx 293 cells expressing either a monomeric (PM-1-mEGFP) or a tandem-dimer (PM-2-mEGFP) form of mEGFP anchored to the membrane were acquired previously (7) using a two-photon microscope and made available at figshare: https://figshare.com/s/77b90d060901fa8b4cb3. The analysis of the fluorescence images collected both in the absence and the presence of agonist ligand (secretin) was applied at three different levels: whole membrane, depunctate image, and clusters of puncta.
In the whole-membrane analysis, homogeneous and inhomogeneous sections of the basolateral portion of the membrane are not separated from one another; the only precaution has been to draw ROIs inside the cell contour to avoid analyzing portions of the membrane at the edge of the cell outline that are oriented parallel rather than perpendicular to the optical axis. Although whole-membrane analysis has already been applied to a subset of the fluorescence images analyzed in this study, (see Fig. 3 of Stoneman et al. (7)), we still included the results of applying it to the images analyzed in this work as a reference and because a wider range of receptor concentrations were included in the analysis herein.
In the depunctate image analysis, the fluorescence images are first subjected to a puncta identification and removal procedure (see Fig. 1), which is described in the next subsection. After applying the puncta-removal procedure, the images are then analyzed with FIF using the same steps as done in the whole-membrane analysis. The depunctate image analysis has also been applied previously to the same data as used in this study (8). However, two key changes have been made to the analysis procedure when compared with the previous work: 1) more puncta have been removed in this work because of the fact that comparatively lower intensity puncta have been removed from the images (by lowering the threshold used in the SLIC procedure) when compared with the intensity of puncta that were removed in the previous work, and 2) a more efficient and rigorous puncta-removal procedure has been developed in this work.
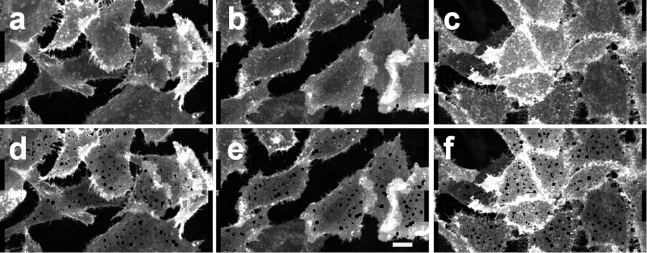
Figure 1.
Typical fluorescence images of CHO cells expressing SecR-mEGFP untreated and treated with secretin, before and after puncta extraction. (a–c) Fluorescence images of the whole basolateral membranes of CHO cells before removal of puncta from the image. (d–f) Images of basolateral membranes of the cells after puncta were removed from the image. The cells were either untreated (a and d) or treated with secretin for 10 min (b and e) and 30 min (c and f). A 10-μm scale bar is indicated by a white rectangle in (e).
In the clustered-puncta analysis, the high-intensity puncta that were removed in the depunctate image analysis approach are themselves analyzed to assess the most prevalent hSecR oligomer sizes occurring within the puncta. To generate large enough statistical ensembles of pixels, the puncta were sorted according to their individual average intensity values and assembled into clusters containing five puncta with similar average intensity, using a procedure that will be expanded upon below. Pixel-level intensity distributions were obtained from each cluster of puncta, and a Cm and a εeff value were extracted for each cluster, which were further analyzed using the standard method of FIF (7). Specifically, the collection of Cm and εeff values obtained from individual clusters were used to generate brightness spectrograms for various concentrations ranges; these spectrograms were then deconvoluted to determine the most prevalent oligomer sizes occurring within the puncta.
Extraction of puncta using SLIC
An automated procedure for identification and removal of high-intensity puncta, i.e., contiguous groups of pixels with higher intensities than their surroundings, from fluorescence images using the SLIC algorithm was described previously (8). In this report, we refined that procedure to collect and further analyze the pixels located within those puncta to quantify the extent of the receptors’ interactions located in the puncta. The main steps of the SLIC algorithm (44) for both identification and retention of the puncta from fluorescence images are listed in Supporting materials and methods, note 2. The SLIC algorithm was modified in this article to improve the accuracy in identifying the puncta. The major modifications were as follows: 1) before applying the SLIC procedure, the fluorescence image was first smoothened using a Wiener filter. 2) The calculation of the distance between pixels and nearby segment center, which incorporates pixel intensity as part of the calculation, was changed to reflect the magnitude of the fluctuations in the segment/punctum and made sensitive to whether the pixel in test has a higher or lower intensity than the intensity of the center of the nearby segment. 3) The thresholding process in which segments are selected or rejected based on the value of their average intensity relative to the average intensity of a subset of the other pixels within the same ROI was adjusted to more accurately identify puncta in the images. In former publications, segments that had an average intensity that was further than three SDs away from the average intensity of the entire ROI were defined as punctum. However, in this work, we selected only the immediate surrounding pixels around the tested segment and calculated the average intensity of only the pixels within this surrounding area as a reference for the threshold process (as opposed to the entire ROI). Segments with a lower average intensity compared with the average intensity of the surrounding pixels plus one SD of the intensity of the surrounding pixels, i.e., , were classified as a punctum for the purpose of subsequent analysis steps.
Concatenating pixel-level intensities of individual puncta into a single cluster of puncta
We determined that the fitting of intensity distributions assembled from the intensity values from pixels of individual puncta was unreliable because of the limited number of pixels available (see Figs. S3 and S4). Therefore, we consolidated the lists of intensity values from pixels of five individual puncta into a single unified list of intensity values (see Supporting materials and methods, note 3 for details on the algorithm), which we call a cluster of puncta. The puncta included in each cluster were selected based on their receptor concentration level and not their spatial proximity. This was done by sorting the puncta according to their receptor concentrations and then grouping together the pixels within the puncta that are close to each other in the sorted list. This concatenation protocol is implemented in steps four and five of the algorithm described in Supporting materials and methods, note 3. Once the clusters of puncta are generated, intensity histograms are constructed from each cluster of puncta. In this article, we make the approximation that the individual puncta are small enough, and thus, any calculation of brightness within a punctum results from intensity fluctuations and not plasma membrane deformations/invaginations, which would change the parameter γ that depends on the local orientation of the membrane relative to the direction of the laser beam (8).
Molecular brightness of monomeric and dimeric standards
To apply FIF spectrometry (7) to the analysis of fluorescence images of hSecR-mEGFP, we first needed to obtain effective brightness distributions from fluorescence images of cells expressing either monomeric (PM-1-mEGFP) or tandem-dimer (PM-2-mEGFP) forms of the fluorescent marker attached to the cell membrane (see Source of data and outline of the methods of analysis); these fluorescence images (reported in Fig. S1 of (7)) were acquired using the same instrument and acquisition settings as used to acquire fluorescence images of cells expressing mEGFP-labeled hSecR. PM-1-mEGFP and PM-2-mEGFP were targeted to the membrane by addition of a palmitoylation-myristoylation sequence, (Met)-Gly-Cys-Ile-Asn-Ser-Lys-Arg-Lys-Asp, to the amino terminus of the A206K mEGFP and the A206K mEGFP tandem dimer. Effective brightness distributions of the PM-1-mEGFP and PM-2-mEGFP were obtained from fluorescence images in which the puncta were first removed using SLIC (as described above and in Supporting materials and methods, note 2 and demonstrated in Fig. S1). By fitting the effective brightness distributions of cells expressing either 1) PM-1-mEGFP, 2) PM-2-mEGFP, or 3) simultaneous fitting of both PM-1-mEGFP and PM-2-mEGFP with a sum of Gaussians in which the position of the maximum of the nth Gaussian is constrained to be n times that of the Gaussian with the lowest mean value (which we refer to as the monomeric Gaussian), we extracted the monomeric brightness distribution, i.e., the mean (or peak position) of the monomeric Gaussian, , and the SD of the monomeric Gaussian, σ. The best-fit value of is used for the calculation of the receptor concentration, Cm, within the segments or cluster of puncta that were collected from images of cells expressing the hSecR-mEGFP constructs. The monomeric distribution as well as the oligomeric distributions resulting from the process described above are also used for unmixing the εeff distributions, or brightness spectrograms, which were assembled from the Cm and εeff values extracted from the segments or clusters of puncta in hSecR-mEGFP-expressing cell images. The calculated values were 60.7, 63.4, and 62.3 with σ of 27.9, 51.1, and 46.5 corresponding to cases (1), (2), and (3), respectively. A more detailed description of the procedure for obtaining is provided in the Supporting materials and methods, note 1. The various values were utilized to determine the oligomer size abundance values along with estimating the error of those values by employing a bootstrapping procedure that is described in the next subsection.
Calculating oligomeric abundances
Each brightness spectrogram may be decomposed to find the fraction of protomers within each of the different sized oligomer species (or the oligomer size fraction) relative to the total number of protomers in the sample. The process for the calculation of the oligomeric species fractions for different receptor concentration ranges using the bootstrapping procedure is performed in several steps, as follows:
-
1)
Assemble all fluorescence images obtained from a particular sample into a single image stack (use the depunctate images for the depunctate image analysis).
-
2)
Identify ROIs for all the images and then divide the ROIs into segments.
-
3)
Calculate the brightness and receptor concentration values for each segment using step 2 from the FIF spectrometry procedure as described in the Introduction or in previous publications (7,8). For the receptor concentration calculation, use the obtained from the simultaneous fitting of both PM-1-mEGFP and PM-2-mEGFP brightness distributions (see above subsection).
-
4)
Assign each brightness and receptor concentration pair to the respective image frame it was extracted from.
-
5)
Randomly choose X frames from the image stack and collect all the brightness and receptor concentration values from these frames into a list. As part of this random process, frames can be chosen by chance more than once or not chosen at all.
-
6)
Construct brightness distributions for different receptor concentration ranges using the list from step 5.
-
7)
Fit the brightness distributions using the oligomeric brightness distributions calculated in the above subsection with the from step 3. The brightness distributions of the oligomers were of a Gaussian shape with a mean of for an oligomer of size n and the same SD, σ, as found when fitting the brightness standard.
-
8)
Use the amplitudes from the fittings in step 7 to calculate the species fractions using the description of the data analysis program section in (7).
-
9)
Repeat steps 5–8 300 times.
-
10)
Repeat steps 3–9 for the other two values obtained from cells expressing either PM-1-mEGFP or PM-2-mEGFP. When repeating step 3, only the receptor concentration is recalculated as the brightness values are not dependent on .
-
11)
Collect the 900 species fraction values for each oligomeric species and each receptor concentration range and calculate the average and SD values for each collection.
-
12)
Plot the average species fraction as a function of receptor concentration using the values calculated in step 11. The error bars for each species fraction value in the plot represent the SD calculated in step 11. The species fraction versus receptor concentration plots are presented in Figs. 2, 3, and S2.
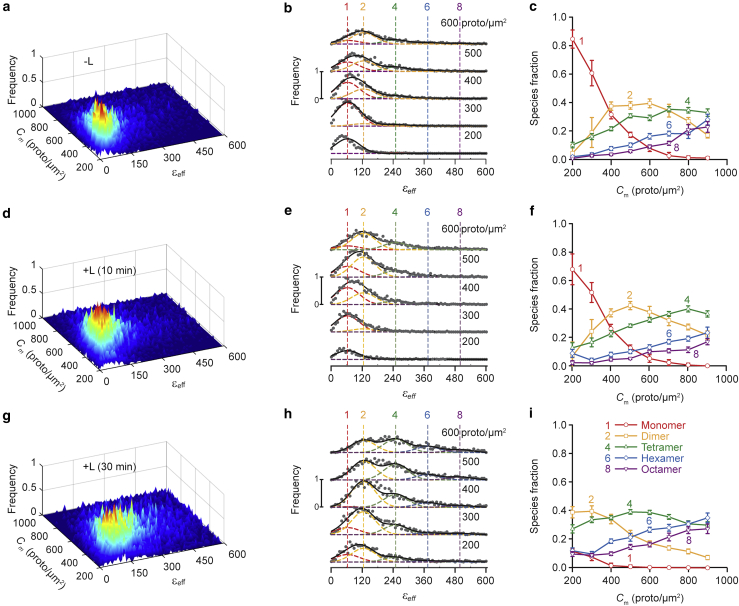
Figure 2.
FIF spectrometry analysis of basolateral membrane patches after excluding puncta (i.e., depunctate membrane analysis). Fluorescence images were obtained using two-photon excitation of cells expressing wild-type hSecR-mEGFP in the absence of agonist ligand (−L) (first row of graphs) or after 10- (second row of graphs) or 30-min (third row of graphs) treatment with 100 nM ligand (+L). (a, d, and g) Surface plots of the εeff occurrence frequency versus receptor concentration of protomers using (a) 13,393, (d) 15,288, and (g) 12,964 total image segments. The maximal segment area was 22 × 22 pixels2. Segment brightness and receptor concentration values were extracted from 82, 80, and 82 images, respectively, each of which contained several cells. (b, e, and h), Stacks of cross sections of surface plots in (a), (d), and (g), respectively, representing brightness distributions for different receptor concentration ranges. Middle range receptor concentration for each cross section (in protomers × μm−2) is indicated above each curve. The vertical dashed lines indicate the peak positions for the brightness spectra of monomers , dimers, and so on, obtained from the simultaneous fitting of the PM-1-mEGFP and PM-2-mEGFP spectrograms used as standards of brightness (Fig. S1). The images of the cells expressing the brightness standards were analyzed in the same manner, i.e., after removing puncta from the images. (c, f, and i) Relative concentration of protomers in each oligomeric species versus total concentration of protomers, as derived from the fitting of the curves in (b), (e), and (h), respectively, with the different Gaussian components representing different oligomeric species. Each data point and its error bar represent the mean ± SD, respectively, of 900 different relative fraction values, obtained via a bootstrapping procedure described in (7) and in the Materials and methods. To see this figure in color, go online.
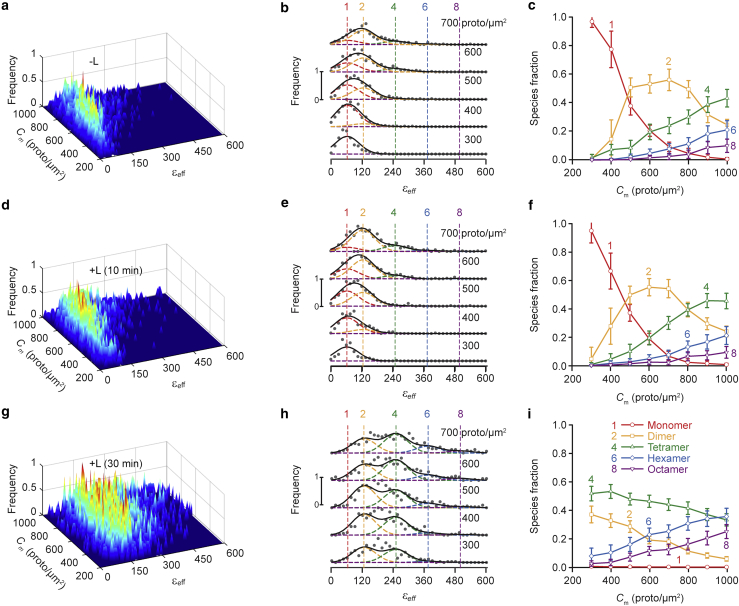
Figure 3.
FIF spectrometry as applied to clusters of puncta extracted from fluorescence images of the basolateral membrane of cells expressing wild-type hSecR fused to mEGFP. The analysis was performed on the same images as in Fig. 2. (a, d, and g). Surface plots of the εeff occurrence frequency versus receptor concentration of protomers using 3103 (a), 3677 (d), and 3704 (g) total puncta clusters. (b, e, and h) Stacks of cross sections taken from the surface plots in (a), (d), and (g), respectively. Middle range receptor concentration for each range (in protomers × μm−2) is indicated above each curve (see explanation in Fig. 2). The vertical dashed lines indicate the peak positions for the brightness spectra of monomers , dimers, and so on (see Materials and methods). The monomeric brightness was extracted from the depunctate areas of the monomer and tandem-dimer standard samples (c, f, and i), Relative receptor concentration of protomers in each oligomeric species versus total receptor concentration of protomers, as derived from fitting of the curves in (b), (e), and (h), respectively, with a sum of different Gaussian components representing different oligomeric species. Each data point and its error bar represent the mean ± SD, respectively, of 900 different relative fraction values, obtained from the statistical “bootstrapping” procedure mentioned in Fig. 2. To see this figure in color, go online.
The bootstrapping procedure as used in this article was reported earlier in Stoneman et al. (7). A modified version of the bootstrapping procedure was also applied to the clustered-puncta analysis method. The modification included an additional step between steps 5 and 6 in which we use the procedure described in the third subsection of Materials and methods. Another difference between the depunctate image analysis and the clustered-puncta analysis is emphasized in step 3. Although for the depunctate image analysis, the segmentation procedure requires a simple grid-like division of each ROI into a square segment of a set size, for the clustered-puncta analysis, we performed the segmentation procedure described in the second subsection of Materials and methods.
Evaluating the goodness-of-fit between an intensity distribution and a single Gaussian function
To test whether a cluster of puncta is a more suitable entity for analysis than an individual punctum, we compared the quality of the fit of a Gaussian function to the intensity distributions obtained from punctum-by-punctum analysis (i.e., analyzing all the puncta in which each punctum is an individual entity) and clustered-puncta analysis (described above). To quantify the “goodness-of-fit” of a Gaussian function to an intensity distribution, we used a reduced χ-square, which is calculated using the following expression:
| (1) |
where j is the index of the intensity bin along the histogram, O is the normalized measured intensity distribution, E is the representation of the Gaussian fit, and DOF stands for the degrees of freedom, defined as:
| (2) |
where Bj is 1 for Ej > 10−3 and 0 otherwise. We utilized the parameter Bj in the calculation of to exclude intensity bins that were further than three times the SD from the mean of the histogram. This thresholding becomes necessary because of the presence of intensity bins that were far from the mean of the distribution but still registered a count of 1 or more (most likely because of the presence of noise or asymmetric distribution of intensities). At the same time, values of Ej far from the mean of the Gaussian are very small, and therefore, the becomes artificially high because of the division by an extremely small value. The fitting of each intensity distribution was accomplished by adjusting the amplitude, mean, and SD of the single Gaussian fitting function to minimize . The values corresponding to the best fit of each individual punctum and cluster of puncta are displayed in the scatterplots of Fig. S4.
Calculation of average puncta density
To monitor possible changes in the population of puncta after ligand treatment, we computed for each sample the average puncta density per image area via the expression
| (3) |
where is the number of puncta within the ith image, is the total image area in pixels (only pixels located within a defined ROI were counted toward the image area), and signifies the average of the quantity it encloses. In addition, we computed the average fractional area of the puncta per image area using the expression
| (4) |
where is the total puncta area (in pixels) within an image.
The SD corresponding to Dpuncta and Fpuncta was also calculated to evaluate the significance of the changes in the two average quantities as a function of ligand treatment duration using a two-tailed Student’s t-test analysis (see Table 1).
Table 1.
Puncta density per image area
| Ligand Treatment | Number of Puncta per Image Area (Average ± SD) | Puncta Area per Image Area (Average ± SD) |
|---|---|---|
| S1 | (2.4 ± 0.2) × 10−3 | (0.120 ± 0.009) × pixel−1 |
| S2 | (2.4 ± 0.3) × 10−3 | (0.120 ± 0.010) × pixel−1 |
| S3 | (3.0 ± 0.2) × 10−3 | (0.150 ± 0.015) × pixel−1 |
| S1 versus S2 t-statistic (confidence level) | 1.94 (5.45%) | 0.18 (85.8%) |
| S1 versus S3 t-statistic (confidence level) | 17.12 (<0.01%) | 14.27 (<0.01%) |
S1, no treatment; S2, 10-min treatment; S3, 30-min treatment.
Results and discussion
As described in detail in the Introduction, FIF spectrometry relies on dividing ROIs containing flat portions of the membrane into small segments to determine the brightness and concentration values corresponding to each segment from intensity fluctuations and assembling multiple such pairs of values (from thousands of ROI segments) into two-dimensional brightness and concentration histograms (or spectrograms). From these spectrograms and using reference brightness values (such as monomeric and dimeric or only monomeric brightness), information on oligomer size distribution within the sample is gleaned directly from the spectrogram or extracted more precisely using quantitative analysis. The primary aim of this work is to extend the FIF spectrometry approach to include the analysis of receptor oligomerization within high-intensity puncta (or spots) that are routinely observed in fluorescence images of cell membranes harboring fluorescently labeled receptors and to compare its results to those obtained from uniform membrane regions, both in the presence and absence of receptor agonist. In addition, we assessed the density of such puncta before and after treatment with ligand to help identify their biological nature. An algorithm has been developed, as described in detail in the Materials and methods and Supporting materials and methods, note 2, which identifies, extracts, and analyzes high-intensity fluorescence puncta using the SLIC method implemented previously (8). We applied this analysis method to a set of fluorescence images obtained from CHO cells expressing hSecR in the presence and absence of agonist ligand (secretin), as described below.
Comparison between depunctate membrane and whole membrane
As a reference for subsequent analysis, we first quantified the oligomerization properties of the receptors in the relatively uniform regions of the plasma membrane obtained after identification and extraction of the puncta from images. Typical fluorescence images of untreated and ligand-treated CHO cell membranes expressing hSecR-mEGFP before and after puncta extraction are presented in Fig. 1 for the visualization of the effectiveness of the algorithm for puncta identification and removal. Oligomerization of hSecR in such uniform regions has been investigated previously for lower receptor concentrations (between 180 and 600 protomers/μm2) relative to those of the receptors found within puncta (8); in this study, we extended our present analysis to higher receptor concentrations: up to 900 or even 1000 protomers/μm2, depending on the availability of data. The results of the new analysis are shown in Fig. 2. As seen in Fig. 2 c, increasing the receptor concentration caused a reduction in the relative proportion of the monomeric to oligomeric species in the cell membrane. A similar trend was observed for cells treated with ligand for various lengths of time (see Fig. 2, f and i). In addition, cell treatment with ligand for 10 min (Fig. 2 f) reduced the relative proportion of monomers, whereas 30 min of treatment (Fig. 2 i) almost completely abolished the monomers, with tetramers becoming the most abundant oligomers. The results obtained for the narrower receptor concentration range of 200–600 protomers/μm2 agree well with the previously published results obtained using the same set of measurements (8). Including membrane regions with higher receptor concentrations allowed us to observe the formation of hexamers and octamers in higher abundances.
The oligomeric species content in depunctate images (Fig. 2) differed only slightly from those of the whole membrane (see Fig. S2), which confirms, once again, that FIF spectrometry has a built-in ability to filter out the puncta and any other strong inhomogeneity in the membrane, as shown in (8). This is a very significant advantage of basic FIF spectrometry (as introduced in previous publications (7,8)) when one is interested in oligomerization within homogeneous regions of membranes only.
Extending the FIF method to investigate hSecR oligomerization in membrane puncta
It is often the case that the addition of ligand to cells harboring various receptors leads to the generation of additional punctate-looking features in the membrane (46, 47, 48). Herein, therefore, the puncta are the focus of our analysis. Because the number of pixels corresponding to typical individual puncta (∼50 pixels per punctum vs. ∼500 pixels for segments of uniform membrane ROIs) are not large enough to generate statistical ensembles for accurately calculating molecular brightness and receptor concentration values (see Figs. S3 and S4), we grouped together puncta with similar average intensities to form clusters of five puncta (or ∼250 pixels), as described in the Materials and methods and Supporting materials and methods, note 3. In addition, because receptors are anchored to the membrane pit, making them immobile, intensity fluctuations corresponding to each punctum would be low, and this reduces one’s ability to extract brightness and concentration information from a single punctum. Therefore, by clustering the puncta together, the intensity fluctuations among the puncta within the cluster can provide a more accurate brightness and concentration calculation and, thus, a more accurate molecular size estimation of the receptor oligomers. Note that the receptor concentration within the cluster of puncta needed to be approximately uniform; otherwise, the molecular brightness calculated for that cluster would reflect more the difference in concentration of the receptors from punctum to punctum within the cluster than the fluctuations in the intensity from pixel to pixel within a punctum. As shown in Figs. S3 and S4, a cluster of five puncta was large enough to provide a smooth intensity histogram and not too large to reduce the resulting number of brightness values (determined from each cluster) significantly. Further details regarding the comparison between the punctum-by-punctum and clustered-puncta analyses is provided in Figs. S3 and S4 as well as in Supporting materials and methods, note 4.
With a reliable procedure at hand for extracting the brightness and receptor concentration values from clusters of puncta, we set out to characterize the nature of the interactions within these structures. To calculate the receptor concentration, we have used the same for the analysis of depunctate (i.e., uniform) membranes and the clusters of puncta. The results obtained from the analysis of the clusters of membrane puncta harboring hSecR are presented in Fig. 3. As seen, there is a common trend in the kinetic curves representing the species fraction versus receptor concentration plots obtained from the depunctate image analysis (Fig. 2), clustered-puncta analysis (Fig. 3), and whole-membrane analysis (Fig. S2); namely, the oligomer size increased both with receptor concentration and duration of treatment with ligand. However, the exact shape of the wire stack plots (Fig. 3, b and e) and the kinetic curves (Fig. 3, c and f) obtained from clustered-puncta analysis differed noticeably from those of the depunctate image or whole-membrane analysis, in three main regards, as follows:
-
1)
The wire stack plots shown in Fig. 3, b and e for the clustered puncta exhibit similar patterns as seen in the depunctate image and whole-membrane analyses; however, they appear to be shifted along the brightness axis toward the lower brightness values for each concentration range (compare to Fig. 2, b and e and Fig. S2, b and e). This shift may be artifactual and could originate from assuming that the puncta are as flat as the adjacent membrane area, which was implicit in our use of the same value for the geometrical factor, γ, for both types of analyses. The value of γ is calculated according to the following:
| (5) |
and is used in the calculation of effective brightness (see Eq. 6) and depends on the point spread function (PSF) of the instrument and its orientation relative to the membrane (20). If, however, a punctum in an image actually corresponds to an invagination (such as a coated pit (38,40)) in the membrane, which at least on its edges has a different orientation relative to the optical axis, one would need to use a different geometrical factor for computing the brightness of receptors within the punctum (pit).
To assess the effect of using an incorrect γ-factor value, we run Monte-Carlo simulations in which molecules were placed at random positions on a two-dimensional lattice and were illuminated with a Lorentzian-Gaussian-shaped laser beam. The intensity was collected for the entire lattice into a single detector. The simulations were repeated 500 times for each segment out of the 1000 segments simulated. The analysis of the brightness was performed using the following equation:
| (6) |
where is the variance of the intensity-dependent noise arising from the detector and can be calculated from separate measurements (see (7)), and Is is the intensity collected for each simulation within a segment (detailed description of the simulation can be found in Supporting materials and methods, note 5). The results of the simulation (see Fig. S5) indicate that the shift of the brightness frequency curves for puncta (Fig. 3) relative to the depunctate membrane curves (Fig. 2) originate from the change in membrane orientation in the puncta, which would happen if the puncta are in fact small membrane invaginations (or coated pits). Note that a fully quantitative test of this hypothesis would require an ability to model the gradual change in orientation of the membrane in a pit from mostly perpendicular to the beam propagation axis (i.e., z axis) in the center of the pit parallel to the z axis at the edge of the pit, as is the case in a semispherical pit.
-
2)
The plots of the species fraction versus receptor concentration for clustered puncta (Fig. 3, c and f) exhibit the same pattern as seen in the depunctate image (Fig. 2, c and f) and whole-membrane analyses (Fig. S2, c and f). However, they appear to be shifted toward higher receptor concentrations. That small shift may have the same underlying cause as mentioned under point (1). Although the concentration does not depend on γ, it does depend on the PSF intersection with the sample as it can be seen from the following equation for the concentration (7):
| (7) |
When the membrane containing the fluorescent molecules is oriented either perpendicular or parallel to the beam propagation axis, the PSF is integrated along x and y (or z for the parallel orientation), and the remaining coordinate (z for perpendicular and y for parallel) is assigned the value of 0. To demonstrate the effect on the brightness spectra as well as on the kinetic behavior of protein interactions when assuming different membrane orientations, we have recalculated the results shown in Fig. 3 using a modified integral in Eq. 7 along with a different γ-factor for recalculating the brightness (see Eq. 6). For that exercise, we assumed that, within the PSF, the membrane oriented perpendicular to the beam propagation axis occupies an area that is 10 times larger than the membrane oriented parallel to the beam axis. Even though the orientation of the membrane was assumed to be predominantly flat and perpendicular to the z axis, the addition of a small fraction of membrane oriented parallel to the beam propagation axis was sufficient to mimic the results shown in Fig. 2 for the depunctate images more closely (compare Fig. S6, b, c, e, and f to Fig. 2, b, c, e, and f). Thus, it is reasonable to assume that the shift in the brightness histograms presented in Fig. 3, b and e may result from the curvature of the puncta, which suggests that they may represent coated pits. The same is true for the shift in concentration as emphasized in Fig. 3, c and f.
-
3)
The species fraction versus concentration plots for each of the different oligomeric sizes seems to reach higher values for clusters of puncta as compared to depunctate membrane areas (compare Figure 2, Figure 3, c, f, and i and 3, c, f, and i). For example, for receptor concentrations between 300 and 330 protomers/μm2, the monomeric fraction was greater than 85% for the case of clusters of puncta as opposed to below 85% for the depunctate membrane case within the range of 200–230 protomers/μm2 (The choice for the different receptor concentration ranges is explained in point (2) above). In addition, the dimeric fraction reached 55% at its peak for the case of clusters of puncta, although it barely reached 40% for depunctate membranes (compare Fig. 2, c and f to Fig. 3, c and f). Furthermore, the tetramers and other high-order oligomers displayed the same behavior as the monomers and dimers. Even after correcting the data for membrane orientation, a reduction in the amplitudes of the species fraction versus concentration peaks (Fig. S6, c and f) was still not seen. The remaining differences may stem from the fact that mostly associated receptors would be driven toward the pits, and thus, one detects mostly higher-order oligomers from the intensity distributions of the puncta (versus simply monomers). This is because the apparent size of the receptor oligomers within the puncta reflects mostly fluctuations in fluorescence intensities from punctum to punctum, which are determined by the size of the oligomers that enter the puncta. As for the oligomers within the puncta themselves, fluctuations in their fluorescence intensities must be comparatively low, if the oligomers are immobilized within the pit once incorporated into it. A more definite conclusion in this regard may not be reached based on intensity fluctuation analysis alone, and it might require an ability to probe interprotomeric distances within the coated pits, using, e.g., FRET.
Determination of the average puncta area and density within the membrane
Having established the similarities between the receptor oligomerization kinetics within high-intensity puncta compared with the oligomerization kinetics within uniform portions of the membranes, we next wanted to assess whether these puncta were indeed coated pits (or endocytic vesicles) as opposed to exocytic vesicles. For that purpose, we quantified the average puncta density per image excluding the pixels outside of the ROIs (in pixels) as well as the average fractional area of the puncta within an image with the same exclusion. Both of these calculations are described in the Calculation of average puncta density section. Table 1 summarizes the results of the average puncta density, expressed as the number of puncta per image area as well as the average area of the puncta per image area. As can be seen, both of these densities were the same between untreated cells and cells treated with secretin for 10 min. However, when the cells were treated with secretin for 30 min, both the number of puncta per image area and the average area of the puncta per image area increased. To test the statistical significance of the differences seen in the average puncta density and average fractional area of the puncta between untreated and treated cells, we performed a two-tailed Student’s t-test with the null hypothesis that there is no difference between the two compared populations. The critical confidence level (i.e., critical p-value) for rejecting the null hypothesis that we have chosen is 1% (below which the null hypothesis is rejected). As it can be seen from Table 1, the differences in both the average number of puncta per image and the fractional area density between untreated and 30-min treated cells were statistically significant.
In the case of ligand-induced internalization, we expect the number of endocytic vesicles to increase, as demonstrated in the literature (46, 47, 48). For example, Liu et al. (46) investigated internalization of EDG-1 receptor fused to GFP treated with Sphingosin-1-Phosphate, Xia et al. (47) have examined the internalization of galanin R2 receptors, and Ward et al. (48) researched the internalization of orexin OX1 and cannabinoid CB1 receptors. Those studies demonstrated ligand-induced internalization by showing a decrease in the signal of the unbound receptors in the flat membrane regions and an increasing density of cytoplasmic vesicles as the ligand treatment duration increased. We have instead quantified the density of the puncta on the membrane itself, which is the location at which endocytic vesicles form, starting as coated pits.
The finding that the increase in the puncta densities suggests that they may correspond to a subpopulation of coated pits and endocytic vesicles that are still docked at the membrane before their internalization. This hypothesis implies that the puncta should present local curvature compared with the uniform membrane regions, which causes their corresponding γ-value to decrease (see above). This in turn leads to an apparent increase in both the concentration of receptors and the maximum of the oligomeric species fraction described in the previous section, which is consistent with the observations made under point (1) above. Nevertheless, as these are all indirect results, biochemical assays, including the use of endocytosis inhibitors (49), should be used to distinguish coated pits and endocytic vesicles from other membrane microdomains.
Conclusions
Fluorescence images of the basolateral membrane of cells expressing fluorescently labeled GPCRs often present groups of pixels with intensities that are higher than those of surrounding pixels. Such puncta may be attributed to endocytic vesicles, pits, rafts, corals, or any other type of bounded membrane regions. Although the study of GPCR interactions within areas of the membrane that do not include, or simply ignore, these puncta has been extensive, the interaction properties of GPCRs within the puncta has remained relatively unexplored. In this report, we have introduced a method designed to enable the study of the interactions of GPCRs within the puncta. This method, which we believe to be novel, is based on the original FIF spectrometry method (7) but incorporates a few essential modifications. The first modification is to utilize a SLIC procedure (8) to identify puncta within the images, but instead of simply discarding the information contained in the pixels within the puncta (as it was done previously), this information is retained and used for further analysis. The second modification to the FIF approach is grouping the puncta into clusters of pixels with similar average concentrations. The clustering of the puncta is a required step as individual puncta are typically too small, i.e., enclose too few pixels, and do not provide the needed statistical information. Once the clusters of puncta are generated, the analysis proceeds with the original FIF spectrometry (7) as applied on the segments used in the depunctate image and whole-membrane analysis.
We tested the modified FIF method using fluorescence microphotographs of cells expressing hSecR fused to mEGFP subjected to two-photon excitation. The same analysis may be done, of course, using single-photon microscopy, such as confocal microscopy. We have first confirmed that within the nonpunctate membrane regions, the hSecR oligomer size increases with an increase in receptor concentration as well as an increase in ligand treatment duration, in agreement with previously published reports covering a subset of the receptor concentration ranges studied here (7,8). Next, we have also confirmed that excluding the puncta from the fluorescence images (depunctate image analysis) did not change the relative proportions of the various oligomer sizes extracted from applying FIF spectrometry when compared with the whole-membrane analysis of the same images. Furthermore, we gauged the level of GPCR interactions within the extracted puncta and compared them to the level of interactions found in the depunctate patches of the membrane. We noted that the calculation of the brightness and receptor concentration is affected by the local curvature of the membrane at the level of the puncta, which when considered, provided mathematically more consistent results.
Because FIF, like any other intensity fluctuation technique, relies on diffusion and because the receptors within various membrane microdomains are likely to be immobile (34,38,40, 41, 42), the oligomers inside these microdomains could have any size (most likely larger than those in the outside, uniform membrane regions) but be unable to convey information on their size through FIFs. Our obtained values for the oligomer size inside the puncta must therefore characterize the size of the oligomers as they are being incorporated within the puncta and thus are added to those already present in there. Results of any technique relying on fluctuations of fluorescence intensities must be regarded with caution when applied to vesicles and pits. Unambiguous evaluation of the oligomer size within membrane pits and vesicles would require such an analysis to be complemented with, e.g., FRET spectrometry analysis (50, 51, 52), as we have also suggested in a recent publication (53).
Author contributions
G.B. designed algorithms, wrote the computer program, and performed data analysis. M.R.S. participated in refining the concepts and analysis methods presented. V.R. conceived and designed the study and supervised the project. G.B., M.R.S., and V.R. wrote the manuscript.
Acknowledgments
This work was partly supported by grants from the National Science Foundation (grants PHY 1126386 and DBI 1919670) as well as the University of Wisconsin–Milwaukee Research Growth Initiative (101X396).
Editor: Baron Chanda.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.06.015.
Supporting material
References
- 1.Milligan G. G-protein-coupled receptor heterodimers: pharmacology, function and relevance to drug discovery. Drug Discov. Today. 2006;11:541–549. doi: 10.1016/j.drudis.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Meyer B.H., Segura J.M., Vogel H. FRET imaging reveals that functional neurokinin-1 receptors are monomeric and reside in membrane microdomains of live cells. Proc. Natl. Acad. Sci. USA. 2006;103:2138–2143. doi: 10.1073/pnas.0507686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurel D., Comps-Agrar L., Pin J.P. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat. Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra A.K., Mavlyutov T., Raicu V. The sigma-1 receptors are present in monomeric and oligomeric forms in living cells in the presence and absence of ligands. Biochem. J. 2015;466:263–271. doi: 10.1042/BJ20141321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y., Basith S., Choi S. Recent advances in structure-based drug design targeting class A G protein-coupled receptors utilizing crystal structures and computational simulations. J. Med. Chem. 2018;61:1–46. doi: 10.1021/acs.jmedchem.6b01453. [DOI] [PubMed] [Google Scholar]
- 6.Miao Y., McCammon J.A. Graded activation and free energy landscapes of a muscarinic G-protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2016;113:12162–12167. doi: 10.1073/pnas.1614538113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoneman M.R., Biener G., Raicu V. A general method to quantify ligand-driven oligomerization from fluorescence-based images. Nat. Methods. 2019;16:493–496. doi: 10.1038/s41592-019-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoneman M.R., Biener G., Raicu V. Reply to: spatial heterogeneity in molecular brightness. Nat. Methods. 2020;17:276–278. doi: 10.1038/s41592-020-0735-x. [DOI] [PubMed] [Google Scholar]
- 9.Ward R.J., Pediani J.D., Milligan G. Chemokine receptor CXCR4 oligomerization is disrupted selectively by the antagonist ligand IT1t. J. Biol. Chem. 2020;296:100139. doi: 10.1074/jbc.RA120.016612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paprocki J., Biener G., Raicu V. In-cell detection of conformational substates of a G protein-coupled receptor quaternary structure: modulation of substate probability by cognate ligand binding. J. Phys. Chem. B. 2020;124:10062–10076. doi: 10.1021/acs.jpcb.0c06081. [DOI] [PubMed] [Google Scholar]
- 11.George S.R., O’Dowd B.F., Lee S.P. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat. Rev. Drug Discov. 2002;1:808–820. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- 12.Wallrabe H., Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Mishra A.K., Gragg M., Park P.S.-H. Quaternary structures of opsin in live cells revealed by FRET spectrometry. Biochem. J. 2016;473:3819–3836. doi: 10.1042/BCJ20160422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margineanu A., Chan J.J., French P.M.W. Corrigendum: screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) Sci. Rep. 2016;6:33621. doi: 10.1038/srep33621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoneman M.R., Paprocki J.D., Raicu V. Quaternary structure of the yeast pheromone receptor Ste2 in living cells. Biochim. Biophys. Acta Biomembr. 2017;1859:1456–1464. doi: 10.1016/j.bbamem.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Stoneman M.R., Raicu N., Raicu V. Fluorescence-based methods for the study of protein-protein interactions modulated by ligand binding. Curr. Pharm. Des. 2020;26:5668–5683. doi: 10.2174/1381612826666201116120934. [DOI] [PubMed] [Google Scholar]
- 17.King C., Stoneman M., Hristova K. Fully quantified spectral imaging reveals in vivo membrane protein interactions. Integr. Biol. 2016;8:216–229. doi: 10.1039/c5ib00202h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jameson D.M., Ross J.A. Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 2010;110:2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian H., Elson E.L. Distribution of molecular aggregation by analysis of fluctuation moments. Proc. Natl. Acad. Sci. USA. 1990;87:5479–5483. doi: 10.1073/pnas.87.14.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Müller J.D., Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys. J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Digman M.A., Dalal R., Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godin A.G., Costantino S., Wiseman P.W. Revealing protein oligomerization and densities in situ using spatial intensity distribution analysis. Proc. Natl. Acad. Sci. USA. 2011;108:7010–7015. doi: 10.1073/pnas.1018658108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ojosnegros S., Cutrale F., Fraser S.E. Eph-ephrin signaling modulated by polymerization and condensation of receptors. Proc. Natl. Acad. Sci. USA. 2017;114:13188–13193. doi: 10.1073/pnas.1713564114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pediani J.D., Ward R.J., Milligan G. Spatial intensity distribution analysis: studies of G protein-coupled receptor oligomerisation. Trends Pharmacol. Sci. 2018;39:175–186. doi: 10.1016/j.tips.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul M.D., Rainwater R., Hristova K. Probing membrane protein association using concentration-dependent number and brightness. Angew. Chem. Int.Engl. 2021;60:6503–6508. doi: 10.1002/anie.202010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vu V., Light T., Leckband D. P120 catenin potentiates constitutive E-cadherin dimerization at the plasma membrane and regulates trans binding. Curr. Biol. 2021;S0960-9822 doi: 10.1016/j.cub.2021.04.061. 00608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed F., Zapata-Mercado E., Hristova K. The biased ligands NGF and NT-3 differentially stabilize trk-A dimers. Biophys. J. 2021;120:55–63. doi: 10.1016/j.bpj.2020.11.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein J.L., Anderson R.G., Brown M.S. Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature. 1979;279:679–685. doi: 10.1038/279679a0. [DOI] [PubMed] [Google Scholar]
- 29.Saxton M.J. Single-particle tracking: effects of corrals. Biophys. J. 1995;69:389–398. doi: 10.1016/S0006-3495(95)79911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 31.Ehrlich M., Boll W., Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 32.Wawrezinieck L., Rigneault H., Lenne P.F. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanyaloglu A.C., von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida A., Sakai N., Yoshimura S.H. Morphological changes of plasma membrane and protein assembly during clathrin-mediated endocytosis. PLoS Biol. 2018;16:e2004786. doi: 10.1371/journal.pbio.2004786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanagawa M., Hiroshima M., Sako Y. Single-molecule diffusion-based estimation of ligand effects on G protein-coupled receptors. Sci. Signal. 2018;11:eaao1917. doi: 10.1126/scisignal.aao1917. [DOI] [PubMed] [Google Scholar]
- 36.Lee S., Tan H.Y., Calvert P.D. Actin filaments partition primary cilia membranes into distinct fluid corrals. J. Cell Biol. 2018;217:2831–2849. doi: 10.1083/jcb.201711104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusumi A., Ike H., Fujiwara T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin. Immunol. 2005;17:3–21. doi: 10.1016/j.smim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Pelkmans L., Helenius A. Endocytosis via caveolae. Traffic. 2002;3:311–320. doi: 10.1034/j.1600-0854.2002.30501.x. [DOI] [PubMed] [Google Scholar]
- 39.Walker J.K., Premont R.T., Shetzline M.A. Properties of secretin receptor internalization differ from those of the beta(2)-adrenergic receptor. J. Biol. Chem. 1999;274:31515–31523. doi: 10.1074/jbc.274.44.31515. [DOI] [PubMed] [Google Scholar]
- 40.Bian J., Zhang S., Liu H. The mechanisms behind decreased internalization of angiotensin II type 1 receptor. Vascul. Pharmacol. 2018;103–105:1–7. doi: 10.1016/j.vph.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Ianoul A., Grant D.D., Pezacki J.P. Imaging nanometer domains of beta-adrenergic receptor complexes on the surface of cardiac myocytes. Nat. Chem. Biol. 2005;1:196–202. doi: 10.1038/nchembio726. [DOI] [PubMed] [Google Scholar]
- 42.Cézanne L., Lecat S., Lopez A. Dynamic confinement of NK2 receptors in the plasma membrane. Improved FRAP analysis and biological relevance. J. Biol. Chem. 2004;279:45057–45067. doi: 10.1074/jbc.M404811200. [DOI] [PubMed] [Google Scholar]
- 43.Pearse B.M. Receptors compete for adaptors found in plasma membrane coated pits. EMBO J. 1988;7:3331–3336. doi: 10.1002/j.1460-2075.1988.tb03204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Achanta R., Shaji A., Süsstrunk S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012;34:2274–2282. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- 45.Achanta R., Shaji A., Süsstrunk S. SLIC superpixels. EPFL Technical Report. 2010;149300:1–15. [Google Scholar]
- 46.Liu C.H., Thangada S., Hla T. Ligand-induced trafficking of the sphingosine-1-phosphate receptor EDG-1. Mol. Biol. Cell. 1999;10:1179–1190. doi: 10.1091/mbc.10.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia S., Kjaer S., Xu Z.-Q.D. Visualization of a functionally enhanced GFP-tagged galanin R2 receptor in PC12 cells: constitutive and ligand-induced internalization. Proc. Natl. Acad. Sci. USA. 2004;101:15207–15212. doi: 10.1073/pnas.0406571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward R.J., Pediani J.D., Milligan G. Ligand-induced internalization of the orexin OX(1) and cannabinoid CB(1) receptors assessed via N-terminal SNAP and CLIP-tagging. Br. J. Pharmacol. 2011;162:1439–1452. doi: 10.1111/j.1476-5381.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dejonghe W., Sharma I., Russinova E. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. 2019;15:641–649. doi: 10.1038/s41589-019-0262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raicu V., Stoneman M.R., Saldin D.K. Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nat. Photonics. 2009;3:107–113. [Google Scholar]
- 51.Raicu V. Extraction of information on macromolecular interactions from fluorescence micro-spectroscopy measurements in the presence and absence of FRET. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018;199:340–348. doi: 10.1016/j.saa.2018.03.075. [DOI] [PubMed] [Google Scholar]
- 52.Raicu V. Ab initio derivation of the FRET equations resolves old puzzles and suggests measurement strategies. Biophys. J. 2019;116:1313–1327. doi: 10.1016/j.bpj.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoneman M.R., Biener G., Raicu V. Proposal for simultaneous analysis of fluorescence intensity fluctuations and resonance energy transfer (IFRET) measurements. Methods Appl. Fluoresc. 2020;8:035011. doi: 10.1088/2050-6120/ab9b68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.