Abstract
Csx/Nkx2.5, a member of the homeodomain-containing transcription factors, serves critical developmental functions in heart formation in vertebrates and nonvertebrates. In this study the putative nuclear localization signal (NLS) of Csx/Nkx2.5 was identified by site-directed mutagenesis to the amino terminus of the homeodomain, which is conserved in almost all homeodomain proteins. When the putative NLS of Csx/Nkx2.5 was mutated a significant amount of the cytoplasmically localized Csx/Nkx2.5 was unphosphorylated, in contrast to the nuclearly localized Csx/Nkx2.5, which is serine- and threonine-phosphorylated, suggesting that Csx/Nkx2.5 phosphorylation is regulated, at least in part, by intracellular localization. Tryptic phosphopeptide mapping indicated that Csx/Nkx2.5 has at least five phosphorylation sites. Using in-gel kinase assays, we detected a Csx/Nkx2.5 kinase whose molecular mass is approximately 40 kDa in both cytoplasmic and nuclear extracts. Mutational analysis and in vitro kinase assays suggested that this 40-kDa Csx/Nkx2.5 kinase is a catalytic subunit of casein kinase II (CKII) that phosphorylates the serine residue between the first and second helix of the homeodomain. This CKII site is phosphorylated in vivo. CKII-dependent phosphorylation of the homeodomain increased Csx/Nkx2.5 DNA binding. Serine-to-alanine mutation at the CKII phosphorylation site reduced transcriptional activity when the carboxyl-terminal repressor domain was deleted. Although the precise biological function of Csx/Nkx2.5 phosphorylation by CKII remains to be determined, it may play an important role, as this CKII phosphorylation site within the homeodomain is fully conserved in all known members of the NK2 family of the homeobox genes.
The homeobox gene products, which contain a highly conserved 60-amino-acid homeodomain, are transcription factors that regulate cell growth, cell fate determination, tissue differentiation, and body patterning (23). Contrary to highly specific biological functions of individual homeobox genes in vivo, in vitro DNA binding studies suggest that most homeoproteins bind to similar short consensus DNA sequences containing the TAAT motif (23, 29). This apparent discrepancy suggests that target gene specificity of each homeoprotein in vivo may be achieved by a combinational interaction with other cofactors (5), small differences in DNA binding affinities to individual target sites among different homeoproteins (19), or posttranscriptional modification of homeobox gene expression, such as translational regulation (43, 48), subcellular localization (41), or protein phosphorylation (22).
A number of transcription factors appear to be regulated by phosphorylation or dephosphorylation (30, 34). Several homeodomain proteins were shown to be phosphorylated at different sites by different protein kinases. The human Cut is phosphorylated by protein kinase C (PKC) and by casein kinase II (CKII) (14, 16), the rat TTF-1 is phosphorylated by MST2 kinase and by PKC (3, 60), the murine Pit-1 (mPit-1) and murine Oct-1 (mOct-1) are phosphorylated by PKA (33, 51), and Drosophila Engrailed and Antennapedia (7, 32) are phosphorylated by CKII. Either DNA binding activity (7, 14, 16, 33, 51) or protein-protein interactions (32) were reported to be regulated by phosphorylation. In addition, phosphorylation of homeoproteins may be regulated by cell cycle (15) or temporally and spatially during development (21, 39).
CKII is a ubiquitously expressed serine/threonine kinase (2, 31). CKII forms 130-kDa heterotetramers, which are composed of two larger catalytic subunits (α and/or α′, with a molecular mass between 36 and 44 kDa) and two smaller β-subunits (molecular mass, 25 kDa), forming α2β2/αα′β2. The consensus phosphorylation site of CKII is (S/T)XX(D/E), where X is any amino acid. Several transcription factors have been reported to be phosphorylated by CKII (2, 31). In a recent study of the Drosophila antennapedia homeobox gene, a mutant Antennapedia whose putative CKII phosphorylation sites were replaced with alanine showed severe defects in thoracic and abdominal development due to its reduced ability to form a complex with another homeoprotein, Extradenticle (32). In addition, CKII phosphorylation is known to regulate the nuclear translocation of some transcription factors. The CKII phosphorylation site of simian virus 40 T antigen is located near the nuclear localization signal (NLS). When this CKII phosphorylation site was mutated, the nuclear translocation was markedly reduced during the early phase (<40 min), although it was not altered during steady state (>12 h) (47). Thus, CKII phosphorylation may control the rate of nuclear import of transcription factors (59).
Murine Csx/Nkx2.5 (36, 38) is a member of the NK2 class of homeobox genes, which are characterized by the presence of a tyrosine residue (Y54) in the third helix of the homeodomain (28). Csx/Nkx2.5 is a murine homologue of the Drosophila tinman gene, which is required for a subdivision of the dorsal mesoderm into the cardiac and visceral mesoderm (6). Homologous Csx/Nkx2.5 genes have been isolated from zebra fish (13), frog (20, 57), chick (50), and human (53, 58) cells. In the mouse, expression of Csx/Nkx2.5 begins as early as 7.5 days post coiticum (dpc) in the precardiac mesoderm and the adjacent endoderm (35, 36, 38). Homozygous Csx/Nkx2.5-targeted mice showed normal heart tube formation and a normal expression pattern of most myofilament genes, but these mice died before 11 dpc due to an arrest of cardiac development prior to the completion of looping (40, 56). Expression of cardiac transcription factors, such as the bHLH gene product eHAND, the cardiac ankyrin repeat protein (CARP), and the MADS domain protein MEF2C, was significantly reduced in Csx/Nkx2.5−/− mutant embryos (4, 56, 61), suggesting that Csx/Nkx2.5 regulates several key transcription factors required for the heart formation beyond the looping stage.
Csx/Nkx2.5 has also been shown to bind to and synergistically transactivate the promoter-enhancer of atrial natriuretic factor (ANF) in association with zinc finger protein GATA4 (18, 37) and the α-cardiac actin promoter in association with serum response factor (11). Csx/Nkx2.5-targeted mice showed the lack of expression of ANF in the cardiac ventricle (56). Thus, ANF is one of the Csx/Nkx2.5 direct target genes in vivo. Csx/Nkx2.5 is also expressed at high levels in other organs, such as spleen, stomach, liver, and anterior larynx, and a subset of cranial skeletal muscles during the fetal stage (35, 38). The function of Csx/Nkx2.5 in the development of noncardiac tissues has not been determined due to early embryonic cardiac lethality of Csx/Nkx2.5−/−.
Relatively little is known about the NLS of homeodomain proteins. In fact, although several hundreds of this gene family have been cloned in many species, NLSs have been reported in only three studies (rat TTF-1, mOct-6, and Saccharomyces cerevisiae MAT-α2) (24, 27, 54). To gain insights into Csx/Nkx2.5 homeodomain protein regulation, we examined the NLS and phosphorylation of Csx/Nkx2.5. We found that Csx/Nkx2.5 is a nuclear phosphoprotein whose NLS requires the amino-terminal basic residues of the homeodomain. Interestingly, when the NLS of Csx/Nkx2.5 is mutated, the cytoplasmically localized Csx/Nkx2.5 is partly unphosphorylated. We further demonstrated that CKII is one of the kinases that phosphorylates the Csx/Nkx2.5 homeodomain in vivo at the highly conserved serine residue between the first and second helix of the homeobox.
MATERIALS AND METHODS
Plasmid construct.
pcDNA3-Csx/Nkx2.5 was mutated by PCR as follows. Initial PCRs were performed with the combination of each forward (F) primer and C2 reverse primer (5′-CGCGGGGTCCCCCAGGCAGGGCT-3′) and the corresponding reverse (R) primer and P4 forward primer (5′-CCAAGTGCTCTCCTGCTTC-3′). Equal amounts of these two PCR products were applied to a second PCR using C2 and P4 primers. BstEII-PflMI PCR fragments were replaced with that of pcDNA3-Csx/Nkx2.5. The following primers were used: pcDNA3-Csx/Nkx2.5163S-A (F, 5′-GCGGTACCTGGCTGCGCCAGAGC-3′; R, 5′-GCTCTGGCGCAGCCAGGTACCGC-3′), pcDNA3-Csx/Nkx2.5 PM1 (F, 5′-GAGAGACCACGCGCAGCGGCTGCAGCGGCCGCAGCTGTGCTCTTCTCGCAG-3′; R, 5′-CTGCGAGAAGAGCACAGCTGCGGCCGCTGCAGCCGCTGCGCGTGGTCTCTC-3′), pcDNA3-Csx/Nkx2.5 PM2 (F, 5′-GCACGGCGGCGACGGGCCGCAGCTGTGCTCTTCTCGCAGGCGCAG-3′; R, 5′-CTGCGAGAAGAGCACAGCTGCGGCCCGTCGCCGCCGTGC-3′), and pcDNA3-Csx/Nkx2.5 PM3 (F, 5′-GAGAGACCACGCGCAGCGGCTGCAGCGAAGCCACGCGTGCTC-3′; R, 5′-GAGCACGCGTGGCTTCGCTGCAGCCGCTGCGCGTGGTCTCTC-3′).
Maltose binding protein (MBP)-Nkx2.5 and MBP-homeodomain (HD) plasmids were kindly provided by R. Schwartz (12). MBP-Csx/Nkx2.5 fusion protein, which includes the full length of Csx/Nkx2.5, was constructed as follows. The PCR fragment amplified with the primers 5′-GGAGCTCGATGTTCCCCAGCCCTGCGCT-3′ (F) and 5′-CTTGCACTTGTAGCGACGGT-3′ (R) and digested with SacI-SphI was replaced with that of the MBP-Nkx2.5 construct. MBP-Csx/Nkx2.5 was digested with PstI and religated to construct MBP-Csx/Nkx2.5ΔC. The XhoI-SacII fragment of MBP-Csx/Nkx2.5 was replaced with that of pcDNA3-Csx/Nkx2.5163S-A to construct MBP-Csx/Nkx2.5163S-A. All the PCR-amplified fragments were sequenced to rule out DNA misincorporations.
Western blotting and immunostaining.
Western blotting and immunostaining were performed as described previously (35). Anti-Csx/Nkx2.5 monoclonal antibody (MAb) (2D10) was used for immunostaining, immunoprecipitation, and Western blotting, and polyclonal antibody (PAb) was used for detection of immunoprecipitated mouse cardiac Csx/Nkx2.5. Anti-MBP PAb (New England Biolabs) was used at a 1:5,000 dilution.
Immunoprecipitation and alkaline phosphatase treatment of Csx/Nkx2.5.
COS cells in 6-cm-diameter plates transfected with pcDNA3-Csx/Nkx2.5 expression vector by using Lipofectamine (GIBCO) were briefly sonicated in 1 ml of phosphatase buffer (50 mM Tris [pH 8.0], 1 mM MgCl2, 0.1 mM ZnCl2, 0.1% sodium dodecyl sulfate [SDS], 10% glycerol, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF]), and centrifuged at 18,900 × g for 10 min. Supernatant (100 μl) was incubated with 20 U of calf intestinal alkaline phosphatase (CIAP) (Promega) for 20 min at 30°C. As a control, the same reaction was performed in the presence of 20 mM Na2HPO4, which competed for the phosphatase reaction.
For gel retardation assays, transfected COS cells on 100-mm-diameter plates were lysed in 100 μl of phosphatase buffer, centrifuged, divided into two tubes, and incubated with or without 10 U of CIAP. The lysates were incubated with 0.1 volume of 1 M of KH2PO4 (pH 7.5) at 4°C for 30 min and centrifuged at 18,900 × g for 20 min to remove SDS (55). Total protein concentration of the supernatants was determined by Bradford assay, Csx/Nkx2.5 protein expression was confirmed by Western blotting, and then equal amounts of protein were used for the gel retardation assay.
To detect Csx/Nkx2.5 protein in mouse cardiac myocytes, 70 mg of neonatal heart was homogenized in 1 ml of RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% SDS, 1% Nonidet P-40 [NP-40], 2 mM EDTA, and 1% deoxycholate). After sonication, the supernatant was immunoprecipitated with 2D10 MAb, or control mouse immunoglobulin G1 (IgG1) bound to 20 μl of protein G beads (Sigma). Beads suspended in 100 μl of buffer (50 mM Tris [pH 7.5], 1 mM MgCl2, 0.1% SDS) were incubated with or without 20 U of CIAP. The samples were then analyzed by Western blotting with Csx/Nkx2.5 PAb.
Expression of Csx/Nkx2.5 protein in bacteria.
MBP fusion proteins were prepared as described by Chen and Schwartz (12). Briefly, 300 ml of cultured Escherichia coli BL21(DE3) (Novagen) was induced with 0.3 mM of IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h at 37°C and collected and lysed by sonication in 10 ml of lysis buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 10 mM MgCl2, 1% Triton X-100, aprotinin [2 μg/ml], pepstatin [0.7 μg/ml], 0.1 mM PMSF, 1 mM DTT, 10% glycerol). After centrifugation, the lysate was incubated with 500 μl of amylose resin (New England Biolabs) for 2 h at 4°C and washed five times with lysis buffer. MBP fusion protein bound to the beads was used directly for the in vitro kinase assay after washing with kinase buffer (25 mM Tris [pH 7.4], 200 mM NaCl, 10 mM MgCl2). Fusion proteins were eluted from the beads by adding lysis buffer containing 10 mM maltose for electrophoretic mobility shift assay (EMSA) and in-gel kinase assays.
Phosphorylation assays.
NIH 3T3 fibroblasts were washed with HBS buffer (25 mM HEPES [pH 7.6], 130 mM NaCl) and washed once with low-salt buffer (25 mM Tris [pH 7.6], 1 mM DTT, 0.1% Triton X-100, 1 mM sodium orthovanadate). Scraped cells were homogenized in a Dounce homogenizer by approximately 10 strokes, with monitoring of the lysis condition by light microscopy. After centrifugation at 250 × g for 5 min, the supernatant was used as the cytoplasmic lysate and pelleted nuclei were extracted in a high-salt buffer (20 mM HEPES [pH 7.6], 450 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 25% glycerol, 0.5 mM PMSF, 1 mM sodium orthovanadate) for 30 min at 4°C. After extraction, the buffer was changed to kinase buffer with a Microcon 10 device (Amicon).
In vitro phosphorylation reactions were performed as follows: approximately 0.2 μg of each purified bacterial fusion protein was incubated with 2 μg of either cytosolic or nuclear extract or 1 U of CKII (Promega) in 20 μl of kinase buffer at 37°C for 15 min. Ten microliters of each sample was boiled in SDS sample buffer and then subjected to SDS-polyacrylamide gel electrophoresis (PAGE).
For detection of 32P-labeled Csx/Nkx2.5, COS cells were transfected with pcDNA3-Csx/Nkx2.5 and then labeled with [32P]orthophosphate at 0.5 mCi/ml for phosphopeptide and phosphoamino acid analysis, in phosphate-free Dulbecco’s modified Eagle’s medium (GIBCO) containing 10% dialyzed fetal calf serum (GIBCO) for 24 h. Cells were lysed with RIPA buffer, precleaned with control mouse IgG1 and protein G, and then immunoprecipitated with 2D10 MAb coupled with protein G.
Phosphoamino acid analysis and phosphopeptide mapping.
Immunoprecipitants of in vivo-labeled Csx/Nkx2.5 or in vitro-phosphorylated MBP-HD fusion protein were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) after SDS-PAGE and then autoradiographed. The membrane containing the radioactive Csx/Nkx2.5 band was hydrolyzed with 6 M HCl at 110°C for 60 min for phosphoamino acid analysis. The hydrolysate was dried (Speedvac; Savant) and dissolved in 5 μl of water and then was electrophoresed in pH 1.9 buffer (7.8% acetic acid, 2.5% formic acid) in the first dimension and pH 3.5 buffer (5% acetic acid, 0.5% pyridine, 0.5 mM EDTA) in the second dimension as described previously (8).
For phosphopeptide mapping, the membrane containing the radioactive Csx/Nkx2.5 band was digested with trypsin (Sigma) overnight. The eluted phosphopeptides (approximately 2,000 cpm) were separated by thin-layer electrophoresis with pH 1.9 buffer for 48 min at 1 kV in the first dimension followed by thin-layer chromatography (TLC) with n-butanol–pyridine–acetic acid–H2O (15:10:3:12) for the second dimension according to the methods described by Boyle et al. (8).
In-gel kinase assay.
MBP (0.2 mg/ml), and MBP-Csx/Nkx2.5 or MBP-HD fusion protein (0.1 mg/ml) were incorporated into SDS–15% PAGE separation gels. NIH 3T3 nuclear extracts (50 μg) were boiled in SDS sample buffer and then loaded on the gels. After SDS-PAGE, gels were washed twice in 20% isopropyl alcohol for 30 min, denatured with 6 M guanidine-HCl in the buffer (40 mM HEPES [pH 7.4], 20 mM NaCl) for 60 min, and renatured gradually by decreasing guanidine-HCl from 3 to 0 M in 40 mM HEPES (pH 7.4)–20 mM NaCl–0.05% NP-40. Renatured gels were incubated in the presence of [γ-32P]ATP (0.25 mCi/ml)–25 μM ATP in 40 mM of HEPES (pH 8.0)–5 mM MgCl2–2 mM DTT–0.1 mM EGTA at room temperature for 1 h and washed with 5% trichloroacetic acid and 1% sodium pyrophosphate.
EMSA.
A double-stranded 21-bp oligonucleotide corresponding to the Csx/Nkx2.5 DNA binding consensus sequence A20 (AGTTAATTG) (12) was produced by annealing 5′-GGGATCCGAGTTAATTG-3′ and 5′-GGCGCAATTAACTCGGA-3′. The ANF Csx/Nkx2.5 binding site, 5′-TCACACCTTTGAAGTGGGGGCCTCTTGAGGCAAATCA-3′ (from position −262 to −226) (37), was produced by annealing 5′-TCACACCTTTGAAGTGGGGGCCTCTTGAGGCAAATC-3′ and 5′-TGATTTGCCTCAAGAGGCCCCCACTTCAAAGGT-3′. Annealed oligonucleotide (2 pM) was incubated with 1 μl of 1 mM deoxynucleoside triphosphate (dATP, dTTP, dCTP), 5 μl of [α-32P]dGTP, and 2 μl of Klenow enzyme in 40 μl for 10 min at 37°C. After addition of 1 μg of 1 mM unlabeled dGTP, the sample was incubated for an additional 5 min. End-labeled ANF Csx/Nkx2.5 binding site 5′-CACCTTTGAAGTGGGGGCCTCTTGAGGCAAAT-3′ (from position −261 to −228) was annealed with 5′-ATTTGCCTCAAGAGGCCCCCACTTCAAAGGTG-3′ and used for EMSA in the absence of CIAP.
Bacterially expressed fusion protein (50 ng) was mixed with 100 μM ATP with 5 U of CKII (Promega) or heat-inactivated CKII (80°C for 10 min) in kinase buffer (total volume, 10 μl) at 37°C for 30 min. One nanogram of fusion protein or 2 μg of COS cell extract and twofold serial dilutions of each protein were mixed with 50,000 cpm of probe–50 μg of bovine serum albumin (BSA)–0.5 μg of poly(dg-dc) in 10 mM HEPES (pH 8.0)–50 mM KCl–1 mM EGTA–10% glycerol–2.5 mM DTT–7 mM MgCl2 in a 15-μl reaction volume for 20 min at room temperature, and separated in 5% polyacrylamide gel with 0.25× Tris-glycine buffer at 15 mA for approximately 20 min. Fourfold serially diluted 2D10 MAb (1 μl) was added for DNA supershift analysis.
Reporter gene assays.
10T1/2 fibroblasts cultured in six-well plates were transfected with 1.5 μg of ANF-luciferase (ANF-luc) reporter construct (−638) (provided by K. R. Chien) or 3xA20-luciferase (3xA20-luc) construct (provided by R. J. Schwartz), 0.5 or 0.25 μg of pcDNA3-Csx/Nkx2.5 expression vector, and 1 μg of murine sarcoma virus β-galactosidase by using the calcium phosphate method. The total plasmid amount was adjusted to 3 μg with pcDNA3 vector plasmid. After glycerol shock with 1× HEPES buffer containing 15% glycerol, cells were cultured for another 48 h, lysed with 300 μl of reporter lysis buffer (Promega), and assayed for luciferase activity (by a Promega assay) and β-galactosidase activity.
Protein-protein interaction.
Bacterially produced glutathione-S-transferase (GST)-GATA4, GST (Pharmacia), and in vitro-transcribed and -translated [35S]methionine-labeled Csx/Nkx2.5 were made as described previously (37). Briefly, in vitro-translated Csx/Nkx2.5 (75,000 cpm) was incubated with 1 μg of GST fusion protein in 400 μl of binding buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 10 mM MgCl2, 1% Triton X-100, aprotinin [2 μg/ml], pepstatin [0.7 μg/ml], 0.1 mM PMSF, 1 mM DTT, and 1% BSA) at 4°C for 2 h. Beads were washed with binding buffer (without BSA) and subjected to SDS-PAGE.
RESULTS
The putative NLS of Csx/Nkx2.5 is in the homeodomain.
We previously found that in most tissues and cells Csx/Nkx2.5 protein is localized in the nucleus (35). In the present study, we searched for the NLS motif (1) of Csx/Nkx2.5 protein and found one consensus NLS-like lysine-arginine-rich cluster with proline at the amino terminus of the homeodomain (see Fig. 9). We mutated this putative NLS, transfected into COS cells, and examined the cellular localization of mutated Csx/Nkx2.5 by immunostaining with anti-Csx/Nkx2.5 MAb (2D10) (Fig. 1Ba, c, e, g, and i) and Hoechst dye (Fig. 1Bb, d, f, h, and j). The wild-type Csx/Nkx2.5 localized exclusively to the nucleus (Fig. 1Ba, arrows) which colocalized with Hoechst dye (Fig. 1Bb, arrows). When we mutated all seven amino acids into alanine (PM1 in Fig. 1A), the mutant Csx/Nkx2.5 localized in the cytoplasm (PM1 in Fig. 1Bc). When the last three amino acids were mutated into alanine (PM2), the majority of mutant protein was cytoplasmically localized (Fig. 1Be). When the first four amino acids were substituted into alanine (PM3), the majority of PM3 protein translocated into the nucleus (Fig. 1Bg), indicating that the four arginine residues around the amino terminus of the homeodomain are not essential for the function of the NLS. Thus, the NLS of Csx/Nkx2.5 requires the amino terminus of the homeodomain and the three-amino-acid cluster KPR at positions 3, 4, and 5 of the homeodomain is necessary to translocate Csx/Nkx2.5 protein into the nucleus. Whether the basic residues at the amino terminus are sufficient to translocate a heterologous protein into the nucleus remains to be determined.
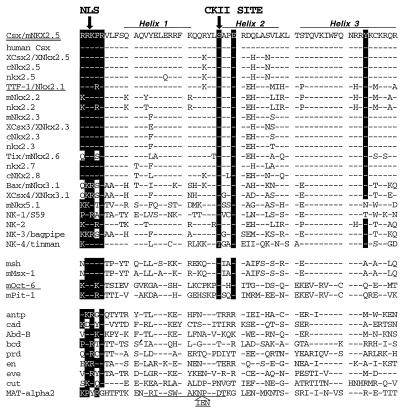
FIG. 9.
Sites of CKII phosphorylation and NLS. The CKII phosphorylation site of Csx/Nkx2.5 is located between the first and second helix of the homeodomain. The consensus CKII site ([S/T]XX[E/D]) is fully conserved in all NK class homeobox proteins (top set) but not in other classes of homeobox proteins with the exception of msh class (msh and mMsx-1) and POU class (mOct-6 and mPit-1) homeoproteins (middle and bottom sets). The NLS of Csx/Nkx2.5 requires the amino-terminal KPR residues of the homeodomain. The consensus NLS is the stretch of arginine and lysine residues and usually includes proline (1). Three other reported NLSs, those of the homeoproteins rat TTF-1/Nkx2.1 (24), mOct-6 (54), and yeast MAT-α2 (27) (underlined), are shown. Tyrosine residues (Y54) characteristic of the NK2 family are also marked. Note that NK-3 and NK-4 belong to the NK2 family whereas NK-1 and Nkx5.1 do not. References for homeodomain sequences are as follows: top set, 28, 46, and 58; middle and bottom sets, 17, 23, and 42.
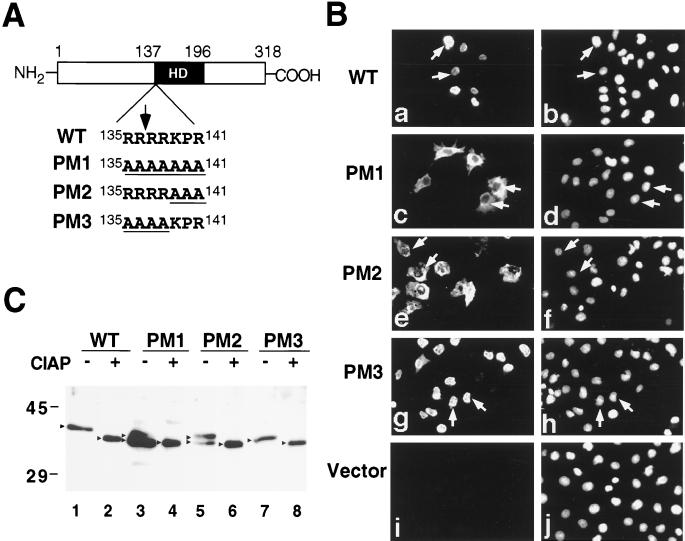
FIG. 1.
NLS of Csx/Nkx2.5. (A) Schematic of NLS of wild-type (WT) Csx/Nkx2.5 and mutant Csx/Nkx2.5 at the amino terminus of the homeodomain (HD). WT Csx/Nkx2.5 contains a seven-amino-acid stretch of arginine-lysine-rich sequence (RRRRKPR). An arrow indicates the homeodomain starting site (R137). Seven amino acids were mutated into polyalanines in PM1. Either the last three amino acids (KPR) (PM2) or the first four amino acids (RRRR) (PM3) are also changed to alanine (underlined). (B) These plasmids were transiently transfected into COS cells and stained with anti-Csx/Nkx2.5 MAb 2D10 (a, c, e, g, and i) and Hoechst dye (b, d, f, h, and j). WT Csx/Nkx2.5 (a) colocalized with Hoechst nuclear staining (b). PM1 and PM2 mutant proteins were predominantly localized in the cytoplasm (c and e). PM3 protein localized in the nucleus (g). (C) Western blotting of the transfected COS cells with 2D10 MAb. Compared with WT Csx/Nkx2.5, which migrated as a single band at approximately 37 kDa (lane 1), cytoplasmically localized PM1 (lane 3) and PM2 (lane 5) migrated as two separate bands. However, after CIAP treatment, these two bands migrated as a single band (lanes 4 and 6), like WT Csx/Nkx2.5 (lane 2) and PM3 (lane 8). Molecular masses (in kilodaltons) are given to the left of the gel.
Nuclearly and cytoplasmically localized Csx/Nkx2.5 is differentially phosphorylated.
To confirm the molecular size of the mutant Csx/Nkx2.5 proteins, each wild-type and mutant protein was analyzed by Western blotting with 2D10 MAb (Fig. 1C). A single 37-kDa band was noted in the nuclearly localized Csx/Nkx2.5 (wild type and PM3) (Fig. 1C, lanes 1 and 7). In contrast, two distinct bands were noted in the cytoplasmically localized Csx/Nkx2.5 (PM1 and PM2) (Fig. 1C, lanes 3 and 5). We examined whether these two bands represent phosphorylated and nonphosphorylated forms of Csx/Nkx2.5. We treated each cell lysate with CIAP and subjected the lysate to Western blotting. After CIAP treatment, all bands shifted into faster-migrating bands (Fig. 1C, lanes 2, 4, 6, 8; Fig. 2A, lane 2) which were close to the size expected for Csx/Nkx2.5 protein (34.1 kDa) based on the deduced amino acid sequence. The band shift was completely inhibited by the addition of excess phosphate, which competed for the phosphatase reaction (Fig. 2A, lane 3). Thus, the nuclearly localized Csx/Nkx2.5 seems to be fully phosphorylated, and the cytoplasmically localized Csx/Nkx2.5 is a mixture of phosphorylated and nonphosphorylated forms.
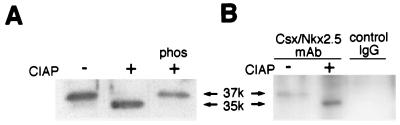
FIG. 2.
Csx/Nkx2.5 phosphorylation in vivo. (A) Western blot of COS cells transiently transfected with the expression vector encoding the wild-type Csx/Nkx2.5. Cell lysates were incubated with (lanes 2 and 3) or without (lane 1) CIAP. Dephosphorylated Csx/Nkx2.5 protein migrated to 35 kDa (native Csx/Nkx2.5, 37 kDa). When the CIAP reaction was inhibited by Na2HPO4 (lane 3), the band shift was completely abolished. (B) Western blot of mouse neonatal heart cells. Cardiac cell lysates were immunoprecipitated with Csx/Nkx2.5 MAb (lanes 1 and 2) or control mouse IgG1 (lane 3) and incubated with (lane 2) or without (lanes 1 and 3) CIAP. After CIAP treatment, the Csx/Nkx2.5 band migrated to 35 kDa (native Csx/Nkx2.5, 37 kDa).
Csx/Nkx2.5 is phosphorylated in vivo.
In order to determine whether the endogenous Csx/Nkx2.5 protein is phosphorylated, Csx/Nkx2.5 was immunoprecipitated from mouse neonatal hearts and analyzed by Western blotting. In the absence of CIAP, endogenous Csx/Nkx2.5 migrated at 37 kDa (Fig. 2B, lane 1). After CIAP treatment, a similar band shift was observed (Fig. 2B, lane 2). These results demonstrate that Csx/Nkx2.5 protein is phosphorylated in transiently transfected COS cells (Fig. 2A) as well as in neonatal heart tissue (Fig. 2B).
In metabolically labeled COS cells transfected with Csx/Nkx2.5 expression plasmid, the phosphorylated 37-kDa Csx/Nkx2.5 band was immunoprecipitated with 2D10 MAb (Fig. 3A, lane 1). No phosphorylated 37-kDa band was observed in COS cells transfected with pcDNA3 vector alone (Fig. 3A, lane 2) or Csx/Nkx2.5-transfected COS cells immunoprecipitated with control IgG1 (Fig. 3A, lane 3).
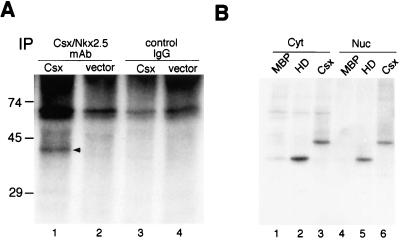
FIG. 3.
Csx/Nkx2.5 is phosphorylated in vivo and in vitro. (A) COS cells transfected with Csx/Nkx2.5 expression vector (lanes 1 and 3) or pcDNA3 vector alone (lanes 2 and 4) were 32P labeled, lysed, and immunoprecipitated (IP) with Csx/Nkx2.5 MAb (lanes 1 and 2) or control IgG1 (lanes 3 and 4). The 37-kDa Csx/Nkx2.5 band was specifically immunoprecipitated with Csx/Nkx2.5 MAb (lane 1) but not with control IgG1 (lane 3). (B) MBP alone (MBP), MBP-homeodomain Csx/Nkx2.5 (HD), and MBP-full length Csx/Nkx2.5 (Csx) were phosphorylated with [γ-32P]ATP and 2 μg of cytoplasmic (Cyt) or nuclear (Nuc) extract of NIH 3T3 cells. Samples were subjected to SDS-PAGE, and phosphorylated proteins were visualized by autoradiography. Both cytoplasmic (lanes 1 to 3) and nuclear lysates (lanes 4 to 6) approximately equally phosphorylated Csx/Nkx2.5 proteins (lane 2 versus lane 5; lane 3 versus lane 6). MBP was not phosphorylated by either cytoplasmic (lane 1) or nuclear (lane 4) extracts.
Csx/Nkx2.5 phosphorylation in vitro.
To determine whether the kinase that phosphorylates Csx/Nkx2.5 resides in the nucleus or the cytoplasm, we prepared nuclear and cytoplasmic extracts from NIH 3T3 cells and tested their ability to phosphorylate bacterially expressed Csx/Nkx2.5 in the presence of [γ-32P]ATP (Fig. 3B). Csx/Nkx2.5 homeodomain was similarly phosphorylated in both cytoplasmic (Fig. 3B, lane 2) and nuclear (Fig. 3B, lane 5) extracts. The full-length Csx/Nkx2.5 protein was also phosphorylated similarly by cytoplasmic (lane 3) or nuclear (lane 6) extracts. MBP, which migrated below the 45-kDa marker, was not significantly phosphorylated by either cytoplasmic or nuclear fractions (lanes 1 and 4). These results suggest that the kinase(s) that phosphorylates Csx/Nkx2.5 is present in both the cytosol and the nucleus.
Phosphorylation of Csx/Nkx2.5 by CKII in vitro.
To determine the molecular mass of the kinase(s) that phosphorylates Csx/Nkx2.5, we performed in-gel kinase assays in the presence of [γ-32P]ATP. NIH 3T3 nuclear extracts were separated by SDS-PAGE which was incorporated with either MBP-HD fusion protein, full-length Csx/Nkx2.5-MBP fusion protein, or MBP alone (Fig. 4A). Both homeodomain alone and full-length Csx/Nkx2.5 fusion proteins were phosphorylated by a kinase with an approximate molecular mass of 40 kDa (Fig. 4A, lanes 1 and 2). The full-length Csx/Nkx2.5 yielded less 32P-labeling than the homeodomain alone due to its lower molar amount for the same weight of protein and overestimation of protein concentration of the intact Csx/Nkx2.5 protein (note the full-length Csx/Nkx2.5 preparation contained more proteolized Csx/Nkx2.5 and nonfused MBP protein, as shown in Fig. 4C, lanes 1 and 2 of the MBP Western blot). No 40-kDa band was detected in the control experiment using MBP as a substrate (Fig. 4A, lane 3). Similar results were obtained when the cytoplasmic lysate was used as the source of kinases (data not shown).
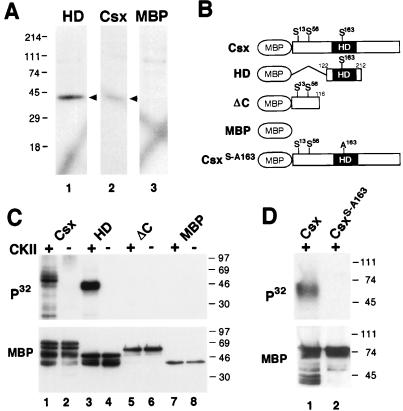
FIG. 4.
Phosphorylation of Csx/Nkx2.5 by CKII within the homeodomain. (A) In-gel kinase assays. NIH 3T3 cell nuclear extracts (25 μg) were separated in a gel containing either the homeodomain-MBP fusion protein (HD), the full-length Csx/Nkx2.5-MBP fusion protein (Csx), or MBP. After denaturation and renaturation, gels were incubated in kinase buffer containing [γ-32P]ATP. One kinase with a molecular mass of ∼40 kDa (arrowheads) phosphorylated the Csx/Nkx2.5 homeodomain, Csx/Nkx2.5 (lane 1), and the full-length Csx/Nkx2.5 (lane 2), but not MBP (lane 3). (B) Schematic representation of Csx/Nkx2.5 mutants carrying the consensus CKII sites and serine-to-alanine substitutions. (C) In vitro CKII phosphorylation of Csx/Nkx2.5. Three different Csx/Nkx2.5 fusion proteins described in panel B were incubated with [γ-32P]ATP in the presence (+) or absence (−) of purified CKII. Each sample was subjected to SDS-PAGE and transferred to a PVDF membrane. Autoradiography (upper panel) revealed that CKII phosphorylated the full-length Csx/Nkx2.5 (lane 1) and the homeodomain (lane 3) but neither the C-terminally deleted mutant (lane 5) nor MBP (lane 7). Loaded fusion proteins are shown by Western blotting of the same PVDF membrane using anti-MBP antibody (lower panel). (D) Serine 163 was mutated into alanine (Csx/Nkx2.5163S-A) and the kinase assay was performed. Lane 1 contains the wild-type Csx/Nkx2.5 and lane 2 contains the mutant Csx/Nkx2.5163S-A. Equal amounts of proteins were subjected to the kinase reaction as shown in the MBP Western blot (lower panel).
We hypothesized that the 40-kDa kinase is CKII, because the catalytic subunit of CKII has a molecular mass of 38 to 42 kDa and Csx/Nkx2.5 contains three consensus CKII sites ([S/T]XX[E/D], where X is any amino acid), including one site (S163) in the homeodomain (Fig. 4B). To determine the CKII phosphorylation site(s), we incubated purified CKII and [γ-32P]ATP with three different Csx/Nkx2.5 fusion proteins: the full-length protein (Csx/Nkx2.5, containing three consensus CKII sites, S13, S56, and S163), Csx/Nkx2.5 homeodomain (HD, containing S163), and a carboxyl-terminal deletion mutant (ΔC, containing S13 and S56) (all shown in Fig. 4B). As shown in Fig. 4C, CKII phosphorylated the full-length Csx/Nkx2.5 (lane 1) and the homeodomain of Csx/Nkx2.5 (lane 3) but neither the C-terminal deletion mutant (lane 5) nor MBP (lane 7). This suggests that the serine residue at amino acid position 163 is the CKII phosphorylation site. To confirm this, we point mutated the serine 163 residue to alanine (Csx/Nkx2.5163S-A) and repeated the CKII kinase assay. As shown in Fig. 4D, CKII phosphorylation was undetectable in the mutant Csx/Nkx2.5163S-A. Interestingly, this CKII phosphorylation sequence ([S/T]XX[D/E]) is completely conserved in all NK family homeobox genes as well as a subset of divergent homeoproteins but not in the Hox class of homeobox genes (see Fig. 9).
Phosphorylation of Csx/Nkx2.5 by CKII in vivo.
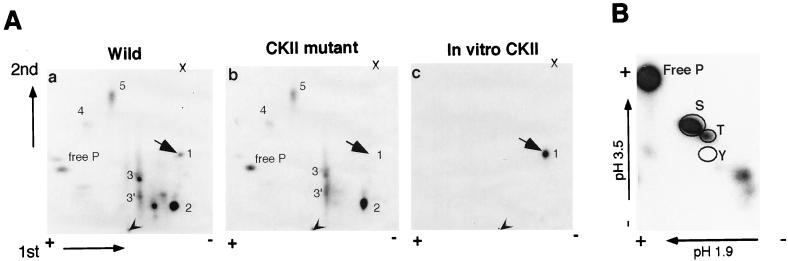
To examine whether serine 163 is phosphorylated in vivo, we metabolically 32P labeled Csx/Nkx2.5 and the CKII mutant protein, Csx/Nkx2.5163S-A, in COS cells. The immunoprecipitant with anti-Csx/Nkx2.5 MAb (2D10) was resolved by SDS-PAGE, transferred to PVDF membrane, and autoradiographed. In vitro CKII-phosphorylated MBP-HD protein was also electrophoresed and autoradiographed as a positive control. These protein bands were cut out, digested with trypsin, and resolved on TLC plates by electrophoresis in the first dimension and by chromatography in the second dimension. Of note, the tryptic digestion sites (after R and K; possible partial digestion at R[D/E], K[D/E], and at the tandem repeat of R and K; nondigestion at [K/R]P) (8) which span serine 163 (sequence from position 160 to 176, (RYLSAPERDQLASVLKL [serine 163 is underlined]) were within the MBP-HD (positions 122 to 212). Thus, the tryptic peptide of the MBP-HD fusion protein phosphorylated in vitro would be resolved at the same position as the wild type Csx/Nkx2.5. The tryptic phosphopeptide mapping of wild-type Csx/Nkx2.5 (Fig. 5Aa), the CKII mutant (Fig. 5Ab), and in vitro CKII-phosphorylated MBP-HD protein (Fig. 5Ac) showed that the in vitro CKII-phosphorylated peptide (Fig. 5Ac, large arrow) corresponded to peptide 1 in the wild-type Csx/Nkx2.5 (Fig. 5Aa, large arrow). This spot in the CKII mutant was barely detectable (Fig. 5Ab, large arrow). These results indicate that serine 163 is phosphorylated in vivo.
Phosphopeptide mapping also indicates that the wild-type Csx/Nkx2.5 may have at least five phosphorylated tryptic peptides (Fig. 5Aa), including one highly phosphorylated site (peptide 2). The two phosphopeptides near peptide 2 which were missing in the CKII mutant may be the partial digestion products of either peptide 2 or peptide 1. Peptide 1 contains the R167-D168 sequence which may cause partial tryptic digestion (see above).
A simultaneous phosphoamino acid analysis of wild-type Csx/Nkx2.5 (Fig. 5B) showed that Csx/Nkx2.5 was phosphorylated predominantly at serine, weakly at threonine, and not at tyrosine. Identification of other phosphorylation sites and responsible kinases require further studies.
FIG. 5.
Phosphopeptide and phosphoamino acid analysis of Csx/Nkx2.5. (A) Tryptic phosphopeptide mapping of wild-type Csx/Nkx2.5 (a), CKII mutant (b), and in vitro CKII-phosphorylated MBP-homeodomain protein (c). Metabolically 32P-labeled Csx/Nkx2.5 and the CKII mutant (Csx/Nkx2.5163S-A) were immunoprecipitated with anti-Csx/Nkx2.5 MAb (2D10), resolved by SDS-PAGE, transferred to a PVDF membrane, and autoradiographed. In vitro CKII-phosphorylated MBP-homeodomain protein was also electrophoresed and autoradiographed. These protein bands were cut out, digested with trypsin, and resolved on TLC plates by electrophoresis in the first dimension and chromatography in the second dimension. In vitro CKII-phosphorylated peptide (c [arrow]) which corresponded to peptide 1 in the wild type (a [arrow]), was markedly reduced in the CKII mutant (b). Arrowheads, sample loaded points; X, lysine marker. (B) Phosphoamino acid analysis of wild-type Csx/Nkx2.5 showed Csx/Nkx2.5 is phosphorylated predominantly in S and weakly in T.
Csx/Nkx2.5 phosphorylation increases DNA binding affinity.
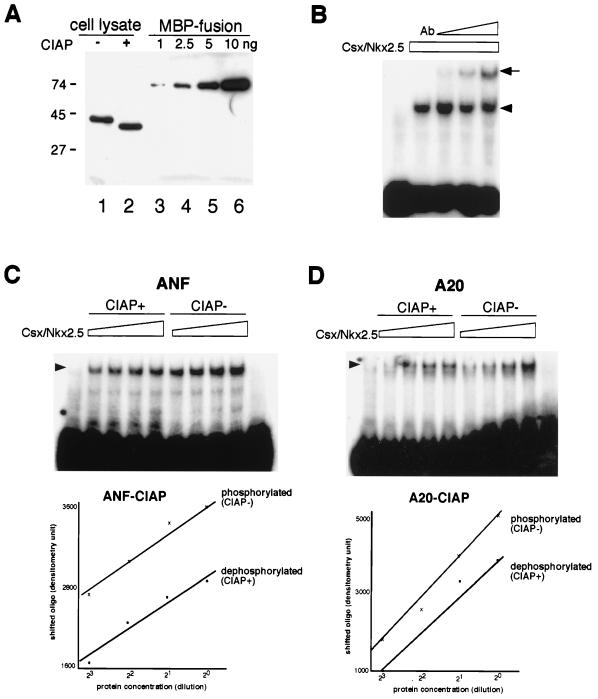
Since the identified CKII phosphorylation site lies within the homeodomain, we examined a potential effect of CKII phosphorylation on DNA binding to a native Csx/Nkx2.5 binding site (TGAAGTG) in the ANF promoter (37) and the A20 binding site (AGTTAATTG) (12) by EMSA. First, the DNA binding affinities of the phosphorylated and nonphosphorylated forms of Csx/Nkx2.5 were examined. We prepared phosphorylated Csx/Nkx2.5 protein lysates from COS cell transfectants in the presence and absence of CIAP (Fig. 6A, lanes 1 and 2). The amount of protein in 5 μl of cell lysates used in lanes 1 and 2 was approximately equivalent to 5 ng of MBP-Csx/Nkx2.5 fusion protein (lane 5). Shown in Fig. 6B, the Csx/Nkx2.5 MAb (2D10) supershifted the protein-DNA complex in a dose-dependent manner, indicating that the shifted band contains Csx/Nkx2.5 protein. Fig. 6C showed the DNA binding of CIAP-treated Csx/Nkx2.5 (lanes CIAP+ [dephosphorylated]) or CIAP-untreated Csx/Nkx2.5 (lanes CIAP− [phosphorylated]), and their twofold serially diluted protein lysate to the ANF binding site. To avoid dephosphorylation of 32P-labeled DNA, we used the filling-in labeling method using Klenow enzyme with [32P]dGTP and deoxynucleoside triphosphate. The shifted bands were scanned and plotted against the protein concentration (lower panel). Dephosphorylation of Csx/Nkx2.5 shifted the plotted line downward, indicating that dephosphorylated Csx/Nkx2.5 had lower DNA binding affinity than phosphorylated protein. Figure 6D shows the results of similar experiment using the A20 binding site.
FIG. 6.
Csx/Nkx2.5 phosphorylation increases DNA binding affinity. Csx/Nkx2.5 protein expressed in COS cells was dephosphorylated with CIAP, and DNA binding activity was compared to that of phosphorylated Csx/Nkx2.5. (A) Transfected-cell lysates were separated by SDS-PAGE and blotted with Csx/Nkx2.5 MAb. The majority of Csx/Nkx2.5 protein was dephosphorylated by CIAP treatment (lane 2). A 5-μl aliquot of each cell lysate (lanes 1 and 2) and 5 ng of MBP-Csx/Nkx2.5 fusion protein contained equivalent amounts of Csx/Nkx2.5 protein. (B) EMSAs with ANF Csx/Nkx2.5 binding site (TGAAGTG). The DNA-Csx/Nkx2.5 protein complex (arrowhead) was supershifted by anti-Csx/Nkx2.5 MAb (arrow) in a dose-dependent manner. ANF probe was end-labeled in this experiment. (C) EMSA with ANF Csx/Nkx2.5 binding site. Lanes CIAP+ contain twofold serially diluted CIAP-treated Csx/Nkx2.5 protein; lanes CIAP− contain untreated Csx/Nkx2.5 protein. The shifted bands were scanned and plotted against protein concentration (lower panel). The plotted line was shifted downward by CIAP treatment, indicating phosphorylated Csx/Nkx2.5 has higher DNA binding affinity than dephosphorylated protein. (D) DNA mobility shift assay (similar to that shown in panel C) using A20 Csx/Nkx2.5 binding site (AGTTAATTG). Phosphorylated protein (lanes CIAP−) showed higher DNA binding affinity than dephosphorylated protein.
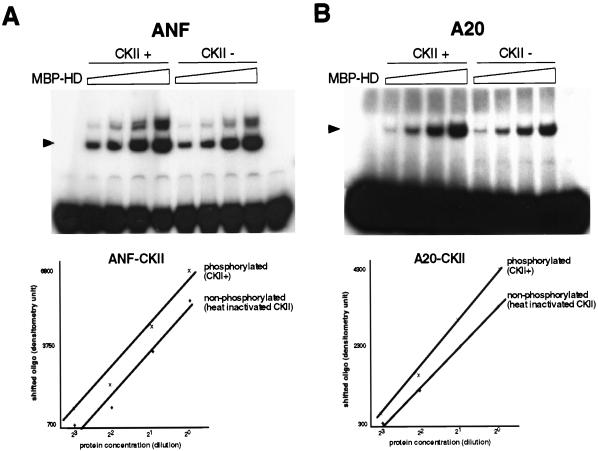
In the above experiments, however, we cannot rule out the effect of dephosphorylation of sites other than the CKII site. In order to determine the effect of CKII phosphorylation on DNA binding affinity more directly, we treated MBP-HD with either CKII or heat-inactivated CKII (Fig. 7A, lanes CKII+ and CKII−, respectively) and then examined their DNA binding affinity. DNA binding affinity for the ANF site appeared to be higher in the CKII-phosphorylated Csx/Nkx2.5 than in the heat-inactivated CKII-treated Csx/Nkx2.5, since CKII phosphorylation shifted the plotted line upward (Fig. 7A, lower panel). Similar results were obtained with the A20 binding site (Fig. 7B). Thus, CKII-phosphorylated Csx/Nkx2.5 had higher DNA binding affinity than the nonphosphorylated form.
FIG. 7.
CKII phosphorylation increases DNA binding affinity. (A) EMSA with ANF Csx/Nkx2.5 binding site. Csx/Nkx2.5 homeodomain fusion protein (MBP-HD) (1 ng) incubated with either CKII (lanes CKII+) or heat-inactivated CKII (lanes CKII−) and their twofold serially diluted proteins were assayed for DNA binding. The shifted bands were scanned and plotted against the protein concentration (lower panel). CKII treatment shifted the plotted line upward, indicating that CKII-phosphorylated protein has higher DNA binding affinity. Slower-migrating bands might represent the dimerized protein. The first lane contains the gel shift with non-fused MBP protein; the last lane contains the free probe without added fusion protein. (B) A result similar to that shown in panel A is observed with A20 Csx/Nkx2.5 binding site.
Mutation of the CKII phosphorylation site reduces transcriptional activity.
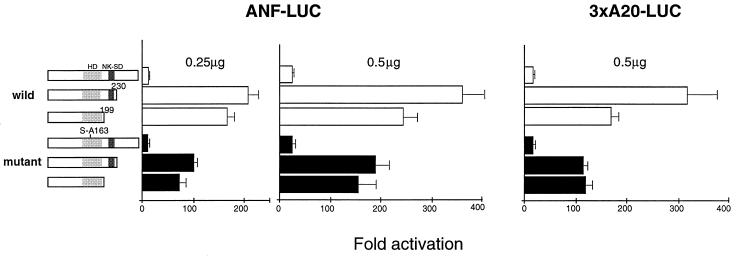
To determine whether CKII phosphorylation of serine 163 regulates transcriptional activity of Csx/Nkx2.5, we tested the ability of the mutant Csx/Nkx2.5163S-A and the wild-type Csx/Nkx2.5 to transactivate the Csx/Nkx2.5-dependent reporter plasmids, ANF-luciferase reporter plasmid (−638) (37) and three copies of A20 Csx/Nkx2.5 binding site fused to the cardiac actin minimal promoter and luciferase reporter gene (12). Prior to these experiments, we confirmed the nuclear localization of the mutant Csx/Nkx2.5163S-A by immunostaining (data not shown). Previous studies reported that the Csx/Nkx2.5 is a modest transactivator by itself due to the presence of a strong repressor domain in the carboxyl terminus (12, 52). Consistent with these previous reports, carboxyl-terminal deletions of Csx/Nkx2.5 [mutant with residues 231 to 318 and 200 to 318 deleted, respectively, Csx/Nkx2.5(1-230) and Csx/Nkx2.5(1-199)] markedly increased the luciferase activity of the ANF reporter gene (Fig. 8, left two panels, open bars) and the A20 reporter gene (Fig. 8, right panel, open bars). When we transfected 0.5 μg of Csx/Nkx2.5(1-230) with ANF-luc reporter plasmid, we noted approximately 350-fold transactivation, which was 15 times higher than that of the full-length protein (Fig. 8, middle panel). The solid bars in Fig. 8 shows transactivation effects of the Csx/Nkx2.5163S-A mutants. Although the full-length Csx/Nkx2.5163S-A did not show significant effect compared to the wild type (see Discussion), Csx/Nkx2.5163S-A(1-230) and Csx/Nkx2.5163S-A(1-199) had only 8.2- and 6.4-fold increases, respectively, on the ANF promoter. Thus, the transactivation function of Csx/Nkx2.5 (C-terminal deletion mutants) was reduced by the mutation at the CKII phosphorylation site by approximately 50%, suggesting that CKII phosphorylation is involved in transcriptional function. A similar result was obtained with the 3xA20-luc reporter gene (right panel).
FIG. 8.
Effect of mutation of CKII phosphorylation site in Csx/Nkx2.5 on transcriptional activation. Transcriptional activation by wild-type Csx/Nkx2.5, CKII mutant, and their carboxyl-terminal deletion mutants. 10T1/2 cells were transfected with expression vectors encoding the full-length or carboxyl-terminal deletion mutants [Csx/Nkx2.5(1-230) and Csx/Nkx2.5(1-199)] with the reporter gene ANF-luc (expression plasmid dose, 0.25 and 0.5 μg) and A20x3-luc (plasmid dose, 0.5 μg). In the wild-type Csx/Nkx2.5 (open bars), carboxyl-terminal deletion mutants (1-230) and (1-199) markedly increased the transcriptional activity on both ANF-luc and 3xA20-luc. When 0.5 μg of Csx/Nkx2.5(1-230) was transfected with the ANF reporter gene (middle panel), luciferase activity increased up to 350-fold, which was 15 times higher than that for the full-length Csx/Nkx2.5. In the CKII mutants (solid bars), similar transactivation by the carboxyl-terminal deletion was observed; however, the fold activation was almost half that of wild-type Csx/Nkx2.5. In full-length Csx/Nkx2.5, wild type and CKII mutants transactivated the reporter gene approximately equally. The fold activation value of luciferase activity is a relative value compared to the cells transfected with the empty expression vector (pcDNA3) and the reporter gene ANF-luc or A20x3-luc. Bars represent means + standard errors.
GATA4 binding activity of Csx/Nkx2.5 is independent of CKII phosphorylation.
Csx/Nkx2.5 and GATA4 have been shown to associate with each other (18, 37, 52). The homeodomain of Csx/Nkx2.5 and the carboxyl-terminal zinc finger of GATA4 are required for Csx/Nkx2.5-GATA4 interaction (37, 52). Accordingly, we tested whether the mutant Csx/Nkx2.5163S-A associates with GATA4. GST-GATA4 fusion protein coupled to Sepharose beads was incubated with equal amounts of in vitro-translated wild-type Csx/Nkx2.5 or Csx/Nkx2.5163S-A protein. Since reticulocyte lysates contain a significant amount of CKII (44), wild-type Csx/Nkx2.5 protein would most likely be phosphorylated at serine 163. Mutant Csx/Nkx2.5163S-A and wild-type Csx/Nkx2.5 associated equally with GST-GATA4 (data not shown).
DISCUSSION
Homeobox genes have diverse functions during development of metazoas, and several hundred genes containing the homeodomain have been isolated from various organisms. Surprisingly however, there have been scant reports on the NLS of homeodomain proteins (24, 27, 54). The putative NLS in the amino terminus of the Csx/Nkx2.5 homeodomain was similar, but its sequence was not identical, to that of two other documented NLSs of the homeodomain proteins, mOct-6 (54) and rat TTF-1 (24). The amino terminus of the homeodomain has highly conserved arginine-lysine-rich NLS-like sequences, particularly at amino acid 2, 3, and 5 of the homeodomain (Fig. 9). Out of 344 homeoproteins examined (9), 303 (88%) have either lysine or arginine at position 2, 91% have lysine or arginine at the position 3, and 98% have arginine at the position 5. The yeast homeodomain protein, MAT-α2, which does not have basic residues at positions 2, 3, and 5, has the NLS within the first helix extending into the first loop (27). Thus, it is possible that the NLS of most, if not all, homeodomain proteins are located at the amino terminus of the homeodomain itself. If this is the case, the homeodomain plays at least three roles: nuclear translocation, DNA binding, and protein-protein interaction.
The NLS is one of the motifs necessary for transporting the protein into the nucleus (25, 26, 45). The NLS interacts with importin-α and then forms complexes with other NLS-binding proteins, such as importin-β, Ran/TC4, and pp15, to target the protein to the nuclear pore (25, 26, 45). Masking the NLS by either phosphorylation, intra- or intermolecular interactions, or cytoplasmic anchoring also regulates nuclear translocation (59). The rate of nuclear translocation may be important in regulating the transcriptional activity of homeodomain proteins, as shown with Drosophila Extradenticle protein, which requires Decapentaplegic and Wingless signals for its graded nuclear translocation in the midgut endoderm (41).
By mutating the putative NLS, we created cytoplasmically localized Csx/Nkx2.5 mutants. Interestingly, a significant portion of cytoplasmically localized Csx/Nkx2.5 was unphosphorylated, compared to nuclear Csx/Nkx2.5, which is serine- and threonine-phosphorylated. Since the ability to phosphorylate Csx/Nkx2.5 is present both in the cytoplasm and nucleus (Fig. 3A), it is possible that nuclearly localized Csx/Nkx2.5 is protected from phosphatase activities. We determined that CKII phosphorylates the serine residue between the first and second helix both in vitro and in vivo. This CKII phosphorylation site is totally conserved among all known NK class homeoproteins and some other classes of homeoproteins (Fig. 9), suggesting that it plays important functional roles. The tyrosine residue (Y54) present in the homeodomain of all NK2 family was not phosphorylated in vivo.
Transcription factors are modulated by phosphorylation at three levels (30, 34). First, phosphorylation of the factor itself or a cytoplasmic anchor protein allows translocation of the transcription factor into the nucleus, where it acts. Second, DNA binding activity of transcription factors can be positively or negatively modulated by phosphorylation. Third, phosphorylation can affect the interaction of a transcription factor with other transcription factors or with the basal transcription machinery. Thus, we tested several possible roles for CKII phosphorylation of Csx/Nkx2.5. First, the mutation in the CKII phosphorylation site (serine 163 to alanine) did not alter the nuclear localization of Csx/Nkx2.5 in transiently transfected cells. Second, we tested whether or not DNA binding was modified by CKII phosphorylation. CKII phosphorylation increased the DNA binding affinity to the ANF promoter sequence, which contained the canonical Csx/Nkx2.5 binding site (TNAAGTG), and to a synthetic target site (A20 [AGTTAATTG]). Third, phosphorylated Csx/Nkx2.5 had higher DNA binding affinity than the dephosphorylated form, for both the ANF site and the A20 site.
Based on the structure of other homeodomain proteins (23), this CKII phosphorylation site does not come in contact with DNA directly; thus, the effect of phosphorylation of this site is probably regulatory. In Engrailed homeoprotein, CKII phosphorylation increased the DNA binding activity, even though the CKII phosphorylation site was mapped to the outside of the homeodomain (7). In contrast, in POU homeoproteins (mOct-1 and mPit-1/GHF-1), protein kinase A phosphorylation at the serine or threonine residue at position 7 of the homeodomain decreased the DNA binding affinity (10, 33, 51).
Full-length Csx/Nkx2.5 has been reported to be a modest transactivator (12, 52). When the carboxyl-terminal repressor domain was deleted, its transcriptional activity was markedly increased, consistent with the previous studies (12, 53). In the CKII mutant, however, the transactivational effect of the carboxyl-terminal deletion was approximately half that of wild-type Csx/Nkx2.5. In contrast, we did not detect significant differences in the transcriptional activity between the full-length wild-type and its CKII mutant Csx/Nkx2.5163S-A. One possible explanation is that the carboxyl terminus of Csx/Nkx2.5 dominantly interferes with the transcriptional function of CKII-phosphorylated homeodomain, perhaps due to a conformational effect of the carboxyl-terminal repressor domain as recently proposed (53).
It should be noted that DNA binding assays or reporter cotransfection assays may reveal but a small part of homeoprotein functions. For example, it has recently been shown that Drosophila Antennapedia homeoprotein is CKII phosphorylated (32). The mutations in CKII sites to alanine did not significantly change DNA binding activity as a monomer but caused severe thoracic and abdominal defects in transgenic flies (32). Interestingly, CKII site mutations in Antennapedia affected its ability to associate with another homeoprotein, Extradentricle, and its ability to bind DNA as a heterodimer. Because Csx/Nkx2.5 associates with GATA4 (18, 37, 53), we examined whether CKII phosphorylation affects this interaction. Although the mutation in the CKII site did not affect GATA4-Csx/Nkx2.5 interaction in vitro, it is possible that interactions with other transcription cofactors may be affected by CKII phosphorylation. Thus, the elucidation of the full biological function of CKII phosphorylation of Csx/Nkx2.5 requires additional studies such as “knock-in” studies of the Csx/Nkx2.5163S-A allele.
CKII activity is regulated temporally and spatially during development (31). The activity of CKII extracted from whole mouse embryos increases three- to fourfold at 12 dpc and decreases to baseline at birth (49). Although the nuclearly localized Csx/Nkx2.5 is constitutively phosphorylated in transfected COS cells and in the neonatal heart, it is possible that this phosphorylation is temporally and spatially regulated during embryonic stages, like the Drosophila homeoprotein Ultrabithorax (21, 39). Indeed, we recently found that Csx/Nkx2.5 is localized in the cytoplasm of a subset of cranial skeletal muscle (35). Whether this cytoplasmic localization results in any differences in Csx/Nkx2.5 phosphorylation remains to be determined. It should be emphasized that CKII is one of the kinases that phosphorylate Csx/Nkx2.5 in vivo. We identified five different tryptic phosphopeptides in Csx/Nkx2.5, and have not identified other serine and threonine phosphorylation sites and their responsible kinases. Further work is necessary to address these issues.
ACKNOWLEDGMENTS
We thank J. Sadoshima, A. Usheva, N. Horikoshi, A. Horwitz, C. Crovello, H. Aoki, and T. Shioi for valuable suggestions and technical help; E. O. Weinberg for critical reading of the manuscript; R. J. Schwartz and C. Y. Chen for pMAL-Csx/Nkx2.5 and A20-luc plasmids; K. R. Chien for the ANF-luc plasmid; and D. Wilson for GST-GATA plasmid.
This work was supported by NIH grant HL51253 (S.I). A part of this work was done during the tenure of American Heart Association-Michigan Affiliate and American Heart Association-Massachusetts Affiliate Fellowships (H.K.)
REFERENCES
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular biology of the cell. 3rd ed. New York, N.Y: Garland Publishing, Inc.; 1994. [Google Scholar]
- 2.Allende J E, Allende C C. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 3.Aurisicchio L, Di Lauro R, Zannini M. Identification of the thyroid transcription factor-1 as a target for rat MST2 kinase. J Biol Chem. 1998;273:1477–1482. doi: 10.1074/jbc.273.3.1477. [DOI] [PubMed] [Google Scholar]
- 4.Biben C, Harvey R P. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
- 5.Biggin M D, McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: the role of DNA binding in functional activity and specificity. Development. 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- 6.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 7.Bourbon H M, Martin-Blanco E, Rosen D, Kornberg T B. Phosphorylation of the Drosophila engrailed protein at a site outside its homeodomain enhances DNA binding. J Biol Chem. 1995;270:11130–11139. doi: 10.1074/jbc.270.19.11130. [DOI] [PubMed] [Google Scholar]
- 8.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 9.Büglin T R. A comprehensive classification of homeobox genes. Oxford, United Kingdom: Oxford University Press; 1994. [Google Scholar]
- 10.Caelles C, Hennemann H, Karin M. M-phase-specific phosphorylation of the POU transcription factor GHF-1 by a cell cycle-regulated protein kinase inhibits DNA binding. Mol Cell Biol. 1995;15:6694–6701. doi: 10.1128/mcb.15.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C Y, Croissant J, Majesky M, Topouzis S, McQuinn T, Frankovsky M J, Schwartz R J. Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev Genet. 1996;19:119–130. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 12.Chen C Y, Schwartz R J. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- 13.Chen J N, Fishman M C. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- 14.Coqueret O, Berube G, Nepveu A. DNA binding by cut homeodomain proteins is down-modulated by protein kinase C. J Biol Chem. 1996;271:24862–24868. doi: 10.1074/jbc.271.40.24862. [DOI] [PubMed] [Google Scholar]
- 15.Coqueret O, Berube G, Nepveu A. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates P21WAF1/CIP1/D1 in S phase. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coqueret O, Martin N, Berube G, Rabbat M, Litchfield D W, Nepveu A. DNA binding by cut homeodomain proteins is down-modulated by casein kinase II. J Biol Chem. 1998;273:2561–2566. doi: 10.1074/jbc.273.5.2561. [DOI] [PubMed] [Google Scholar]
- 17.Double D. Guidebook to the homeobox genes. New York, N.Y: A Sambrook & Tooze Publication at Oxford University Press; 1994. [Google Scholar]
- 18.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekker S C, Jackson D G, von Kessler D P, Sun B I, Young K E, Beachy P A. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 1994;13:3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Y, Izumo S. Cardiac myogenesis: overexpression of XCsx2 or XMEF2 in whole Xenopus embryos induces the precocious expression of XMHC gene. Roux’s Arch Dev Biol. 1995;205:198–202. doi: 10.1007/BF00357766. [DOI] [PubMed] [Google Scholar]
- 21.Gavis E R, Hogness D S. Phosphorylation, expression and function of the Ultrabithorax protein family in Drosophila melanogaster. Development. 1991;112:1077–1093. doi: 10.1242/dev.112.4.1077. [DOI] [PubMed] [Google Scholar]
- 22.Gay N J, Poole S J, Kornberg T B. The Drosophila engrailed protein is phosphorylated by a serine-specific protein kinase. Nucleic Acids Res. 1988;16:6637–6647. doi: 10.1093/nar/16.14.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring W J, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 24.Ghaffari M, Zeng X, Whitsett J A, Yan C. Nuclear localization domain of thyroid transcription factor-1 in respiratory epithelial cells. Biochem J. 1997;328:757–761. doi: 10.1042/bj3280757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 26.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 27.Hall M N, Craik C, Hiraoka Y. Homeodomain of yeast repressor alpha 2 contains a nuclear localization signal. Proc Natl Acad Sci USA. 1990;87:6954–6958. doi: 10.1073/pnas.87.18.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey R P. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi S, Scott M P. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990;63:883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- 30.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 31.Issinger O G. Casein kinases: pleiotropic mediators of cellular regulation. Pharmacol Ther. 1993;59:1–30. doi: 10.1016/0163-7258(93)90039-g. [DOI] [PubMed] [Google Scholar]
- 32.Jaffe L, Ryoo H D, Mann R S. A role for phosphorylation by casein kinase II in modulating Antennapedia activity in Drosophila. Genes Dev. 1997;11:1327–1340. doi: 10.1101/gad.11.10.1327. [DOI] [PubMed] [Google Scholar]
- 33.Kapiloff M S, Farkash Y, Wegner M, Rosenfeld M G. Variable effects of phosphorylation of Pit-1 dictated by the DNA response elements. Science. 1991;253:786–789. doi: 10.1126/science.1652153. [DOI] [PubMed] [Google Scholar]
- 34.Karin M, Hunter T. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- 35.Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extra-cardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- 36.Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y, Shioi T, Kasahara H, Jobe J S, Markham B E, Izumo S. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lints T J, Parsons L M, Hartley L, Lyons I, Harvey R P. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 39.Lopez A J, Hogness D S. Immunochemical dissection of the Ultrabithorax homeoprotein family in Drosophila melanogaster. Proc Natl Acad Sci USA. 1991;88:9924–9928. doi: 10.1073/pnas.88.22.9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyons I, Parsons L M, Hartley L, Li R, Andrews J E, Robb L, Harvey R P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 41.Mann R S, Abu-Shaar M. Nuclear import of the homeodomain protein extradenticle in response to Wg and Dpp signalling. Nature. 1996;383:630–633. doi: 10.1038/383630a0. [DOI] [PubMed] [Google Scholar]
- 42.Meijer D, Graus A, Kraay R, Langeveld A, Mulder M P, Grosveld G. The octamer binding factor Oct6: cDNA cloning and expression in early embryonic cells. Nucleic Acids Res. 1990;18:7357–7365. doi: 10.1093/nar/18.24.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mlodzik M, Gehring W J. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell. 1987;48:465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- 44.Molkentin J D, Li L, Olson E N. Phosphorylation of the MADS-Box transcription factor MEF2C enhances its DNA binding activity. J Biol Chem. 1996;271:17199–17204. doi: 10.1074/jbc.271.29.17199. [DOI] [PubMed] [Google Scholar]
- 45.Pante N, Aebi U. Toward the molecular dissection of protein import into nuclei. Curr Opin Cell Biol. 1996;8:397–406. doi: 10.1016/s0955-0674(96)80016-0. [DOI] [PubMed] [Google Scholar]
- 46.Reecy J M, Yamada M, Cummings K, Sosic D, Chen C-Y, Eichele G, Olson E N, Schwartz R J. Chicken Nkx-2.8: a novel homeobox gene expressed in early heart progenitor cells and pharyngeal pouch-2 and -3 endoderm. Dev Biol. 1997;188:295–311. doi: 10.1006/dbio.1997.8641. [DOI] [PubMed] [Google Scholar]
- 47.Rihs H P, Jans D A, Fan H, Peters R. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 1991;10:633–639. doi: 10.1002/j.1460-2075.1991.tb07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring W J, Jackle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 49.Schneider H R, Reichert G H, Issinger O G. Enhanced casein kinase II activity during mouse embryogenesis. Identification of a 110-kDa phosphoprotein as the major phosphorylation product in mouse embryos and Krebs II mouse ascites tumor cells. Eur J Biochem. 1986;161:733–738. doi: 10.1111/j.1432-1033.1986.tb10501.x. [DOI] [PubMed] [Google Scholar]
- 50.Schultheiss T M, Xydas S, Lassar A B. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121:4203–4214. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- 51.Segil N, Roberts S B, Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- 52.Sepulveda J L, Belaguli N, Nigam V, Chen C Y, Nemer M, Schwartz R J. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiojima I, Komuro I, Mizuno T, Aikawa R, Akazawa H, Oka T, Yamazaki T, Yazaki Y. Molecular cloning and characterization of human cardiac homeobox gene CSX1. Circ Res. 1996;79:920–929. doi: 10.1161/01.res.79.5.920. [DOI] [PubMed] [Google Scholar]
- 54.Sock E, Enderich J, Rosenfeld M G, Wegner M. Identification of the nuclear localization signal of the POU domain protein Tst-1/Oct6. J Biol Chem. 1996;271:17512–17518. doi: 10.1074/jbc.271.29.17512. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki H, Terada T. Removal of dodecyl sulfate from protein solution. Anal Biochem. 1988;172:259–263. doi: 10.1016/0003-2697(88)90440-x. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka, M., and S. Izumo. Unpublished data.
- 57.Tonissen K F, Drysdale T A, Lints T J, Harvey R P, Krieg P A. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 1994;162:325–328. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- 58.Turbay D, Wechsler S B, Blanchard K M, Izumo S. Molecular cloning, chromosomal mapping, and characterization of the human cardiac-specific homeobox gene hCsx. Mol Med. 1996;2:86–96. [PMC free article] [PubMed] [Google Scholar]
- 59.Vandromme M, Gauthier-Rouviere C, Lamb N, Fernandez A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 60.Zannini M, Acebron A, De Felice M, Arnone M I, Martin-Perez J, Santisteban P, Di Lauro R. Mapping and functional role of phosphorylation sites in the thyroid transcription factor-1 (TTF-1) J Biol Chem. 1996;271:2249–2254. doi: 10.1074/jbc.271.4.2249. [DOI] [PubMed] [Google Scholar]
- 61.Zou Y, Evans S, Chen J, Kuo H C, Harvey R P, Chien K R. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]