Abstract
Given a data-set of Ribonucleic acid (RNA) sequences we can infer the phylogenetics of the samples and tackle the information for scientific purposes. Based on current data and knowledge, the SARS-CoV-2 seemingly mutates much more slowly than the influenza virus that causes seasonal flu. However, very recent evolution poses some doubts about such conjecture and shadows the out-coming light of people vaccination. This paper adopts mathematical and computational tools for handling the challenge of analyzing the data-set of different clades of the severe acute respiratory syndrome virus-2 (SARS-CoV-2). On one hand, based on the mathematical paraphernalia of tools, the concept of distance associated with the Kolmogorov complexity and Shannon information theories, as well as with the Hamming scheme, are considered. On the other, advanced data processing computational techniques, such as, data compression, clustering and visualization, are borrowed for tackling the problem. The results of the synergistic approach reveal the complex time dynamics of the evolutionary process and may help to clarify future directions of the SARS-CoV-2 evolution.
Keywords: COVID-19, Kolmogorov complexity theory, Shannon information theory, Multidimensional scaling, Evolutionary dynamics
Introduction
The severe acute respiratory syndrome virus (SARS-CoV-2) is a single-stranded Ribonucleic Acid (RNA) beta-coronavirus presenting a 29,903 nucleotides-long genome. It caused an outbreak in the Chinese city of Wuhan, in December 2019. Subsequently, the new coronavirus has spread worldwide in less than 4 months, and a pandemic situation was declared by the World Health Organization (WHO). The disease was named on February 2020, Coronavirus Disease 2019 (COVID-19), and the disease is now responsible for more than 145,000,000 confirmed cases, and 4,230,000 deaths reported worldwide as of August 3, 2021 (source: World Health Organization https://covid19.who.int). There are significant differences in case fatality rates (proportion of deaths from a specific disease compared to the total number of individuals diagnosed with the disease for a particular period) between countries, possibly related to the efficacy of the measures adopted to limit viral spreading, the demographic pyramid, i.e., the distribution of various age groups in a particular country, and the screening, test and tracing strategy [1].
Using the data from the database of the Global Initiative on Sharing Avian Influenza Data (GISAID) Consortium, major clades of SARS-CoV-2 were identified [2]. The initial RNA genome (accession number: NC 045512.2), identified in Wuhan is named as clade ‘L’. This root clade suffered mutations due to numerous factors, and some new clades emerged with stable mutations: clade ‘G’ (presenting an alteration of the spike protein S-D614G was the first dominant variant) and its two derivatives ‘GH’ (with ORF3a-Q57H mutation) and ‘GR’ (affected by RG203KR mutation); clade ‘V’ (variant of the ORF3a coding protein NS3-G251), and clade ‘S’ (variant ORF8-L84S). Other alleles or combinations different from the previously described clades are classified as clade ‘O’ [2]. These data confirmed, until now, the relatively low variability presented by SARS-CoV-2 compared to other respiratory viruses [3]. The different fatality rates, speed of transmission, and infectiousness profiles observed in different countries were probably not related to differences in virulence of the clades and their characteristic mutations. However, very recently, some new mutations were found with potential epidemiological consequences. For example, 12 human cases were identified in September 2020 in North Jutland with a unique variant called ‘cluster 5’, a combination of mutations that have been not previously described. All 12 cases were linked to the mink farming industry or the local community [4]. However, the clinical presentation, severity, and duration of COVID-19, and transmission among those infected were similar to that of other circulating SARS-CoV-2 viruses. The variant cluster 5 has not been detected since September despite extensive sequencing and data sharing and is thought to be a dead-end, owing to the very restricted spreading and infectiousness to humans [4].
More disturbing is the new variant strain of SARS-CoV-2 that contains 23 mutations, eight of which are in the spike protein the virus uses to bind to and enter human cells. The spike protein is also the focus of most COVID-19 vaccines that are now being administered in numerous countries. Moreover, the diagnostics tests of COVID-19 are also based on the protein sequence found on the Wuhan reference strain spike. Therefore, their efficacy can be changed by these genomic variations. The appearance of these mutations can also lead to immunological resistance and vaccine escape [5]. The new British variant was reported for the first time in December 2020 and has become highly prevalent globally and responsible for another COVID-19 wave in numerous countries [6]. Based on these mutations, this variant has been predicted to potentially be more quickly transmissible than other circulating strains of SARS-CoV-2. The variant is referred to as SARS-CoV-2 VUI 202012/01 (i.e., Variant Under Investigation, year 2020, month 12, variant 01), or B.1.1.7, as the lineage of the clade GR was classified on GISAID. Variant B.1.1.7 presents increased transmissibility and increased virus load. However, apparently, there is no association with more severe disease [7] although the increased infectiousness can lead to more deaths due to the strain of the health systems of the affected countries. In Mid-November in South Africa, a new lineage of the clade GH has emerged but shared one of the mutations described in the British variant. The South African virus variant (B.1.351), known as ‘triple variant’ is distinct from the UK variant, but both contain an unusually high number of mutations, with potential functional significance, compared to other SARS-CoV-2 lineages, and it can, apparently, partially escape to some available vaccines [8]. Several other variants were described more recently in several countries. Examples are the variants P1 and P2 in Brazil, variants B.1.429 and B.1.526 found in the USA, and yet more recently, a variant first sequenced in India (variant B.1.167) [9]. In April 2021, the South Africa, Brazil, and India variants caused or are causing large disease outbreaks in their respective countries with increased excess deaths due to rupture of the hospital capacities.
The SARS-CoV-2 B.1.1.7 variant was already detected at the end of December 2020 in France, Denmark, Holland, and Italy [10]. In 31 of May, the WHO has assigned simple, easy to say and remember labels for the most important variants of SARS-CoV-2, using letters of the Greek alphabet (see Tracking SARS-CoV-2 variants at ).
The most important variants of concern according to this new classification are: Alpha, corresponding to lineage B.1.1.7, found in the United Kingdom in September 2020; Beta, corresponding to lineages B.1.351, B.1.351.2, B.1.351.3 present in South Africa in May 2020; Gamma, corresponding to lineages P.1, P.1.1, P.1.2, sequenced in Brazil, in November 2020; and, Delta, corresponding to lineages B.1.617.2, AY.1, AY.2 and AY.3 found in India in October 2020 (see Table 1).
Table 1.
Currently designated SARS-CoV-2 variants of concern
| WHO label | Lineages | Clade | Date of designation |
|---|---|---|---|
| Alpha | B.1.1.7 | GRY | 18-Dec-2020 |
| Beta | B.1.351, B.1.351.2, B.1.351.3 | GH/501Y.V2 | 18-Dec-2020 |
| Gamma | P.1, P.1.1, P.1.2 | GR/501Y.V3 | 11-Jan-2021 |
| Delta | B.1.617.2, AY.1, AY.2, AY.3 | G/478K.V1 | 11-May-2021 |
In the Summer of 2021, the SARS-CoV-2 Delta variant is becoming dominant in Europe, North America and other parts of the world and is responsible for a new wave, mainly in the unvaccinated population [11]. It is highly transmissible and it also appears to affect vaccine effectiveness and breakthrough infections in vaccinated individuals appear to be more frequent with this variant [11]. It was reported that the viral load is 1000 times higher for Delta compared with previous variants of the initial wave of infections and may have a faster replication rate, a reduced incubation period, and greater viral shedding [12]. The Delta variant was found to be approximately 64% more transmissible than the Alpha variant that was dominant in the waves of the end of December 2020 and first months of 2021. On the other hand, Alpha was already estimated to be 50% more transmissible than the D614G strain, responsible for the first wave in the beginning of 2020 [13].
The so-called coronavirus ‘waves’ can be more contagious and spread faster than the initial ones due to the new variants that can also present other epidemiological problems. There is no definitive evidences that the various variants are associated with an higher disease severity. However, there is a clear risk that future epidemic ‘waves’ may be larger and, therefore, associated with greater burden for the health systems and society due to the lockdowns. Therefore, with this work we try to understand SARS-CoV-2 new variants and the relations with the already known virus clades using new strategies for finding the relationships among them [14–16]. It is important to understand the recent evolution of the virus partially responsible for the so-called ‘waves’ [17]. With this regard computational techniques associated with mathematical tools are a promising strategy to tackle genomic data-sets. This approach was tested in [18, 19] using the Kolmogorov complexity and Shannon information theories, associated with clustering techniques [20, 21] such as the Multidimensional Scaling (MDS). Given its successful application in a primary set of 133 items, encompassing a variety of virus, this paper extends the study to a more challenging problem, namely of analyzing and comparing the SARS-CoV-2 mutations. For that purpose a new data-set of 307 virus including RNA information from the beginning of the spread up to the day of writing this paper is selected. Furthermore, based on the aforementioned mathematical tools, we include a larger set of indices to allow a more complete comparison. In the case of the Kolmogorov complexity we consider four indices [22, 23], normalized information distance, Compression-based Dissimilarity Measure, Chen-Li Metric and Compression-based Cosine (that are abbreviated by the acronyms NCD, CDM, CLM and CosS). In the scope of the Shannon information theory we consider also four metrics, namely the Jaccard, Jensen-Shannon, Jeffreys and Topsøe distances (denoted as , , and ) [24, 25]. A third type and distinct type of assessment is also included and consists of the Hamming distance (), widely used in information theory [26].
Following these ideas the rest of the paper is organized as follows. Section 2 introduces the fundamental tools. The main mathematical concepts involved with distance, Kolmogorov complexity, Shannon information and Hamming metric are summarized. Additionally, the computational tools such as data compression and MDS are also included. Section 3 describes and analyses the data-set of very close, but distinct, information for a large number of RNA sequences. Section 4 develops a synergistic performance of a variety of measures associated with the MDS clustering and visualization. The results shed light on the dynamics of the evolutionary process of the SARS-CoV-2 lineages. Finally, Sect. 5 presents the conclusions.
Fundamental tools
Distance
A function stands for the distance between two objects x and y if satisfies three axioms [27], namely identity if , symmetry and triangle inequality . These axioms imply the non-negativity (or separation condition) . On the other hand, they allow the definition of a plethora of different functions, with distinct pros and cons [24, 25]. Based on these notions, several algorithms [28–30] were adopted for comparing data sequences [26, 31–33]. However, users must have in mind that the selection of a set of distances for a given application requires some experience and that a number of numerical trials are usually necessary before finding the ‘best’ ones [34–37].
In the final part of the paper, the Appendix presents the mathematical and algorithmic fundamentals of the distances used in this paper for assessing the genetic information.
Data compression
Compression data algorithms can be classified as ‘lossless’ and ‘lossy’. Lossless compression algorithms are typically used for archival or other high fidelity purposes and reduce the size of files without losing any information in the file, which means that we can reconstruct the original data from the compressed file. Lossy compression algorithms reduce the size of files by discarding the less important information in a file, which can significantly reduce file size but also affect file quality.
In this paper we used the BZip2 compression algorithm which is based on Burrows-Wheeler transform [38]. This compressor has the extension BZ2 designating a pure data compression format not providing file archival feature. In this algorithm the speed is somewhat slower than for the compressor LZW (extension .Z) and Deflate (extension .zip and .gz) compression algorithms [39]. These employ the classic Deflate algorithm (even if correctly implemented Bzip2 algorithm can be easily made parallel, and benefit of recent multi-core CPU), but faster than more powerful compression schemes as in RAR format, 7Z format, and new ZIPX format. The compression ratio, also, is usually intermediate between the older Deflate-based ZIP/GZ files and modern RAR, 7Z and ZIPX formats [40].
Multidimensional scaling
Let us consider a group of N objects , , in a q-dim space. The MDS is a computational method for [41] that re-organizes them in a structure where the objects are represented by points trying to highlight the similarities between them in the sense of a predefined distance [42]. The process starts by calculating a dimensional matrix, , with for and , , giving object to object distances [43]. In a second phase, the MDS calculates the point coordinates in a -dim space, trying to mimic the original distances. The MDS technique includes a numerical iterations for optimizing a cost function, often called Stress, that compares the distances and , so that the index is minimized.
The MDS points have coordinates that yield a symmetric matrix of distances that approximate D. The MDS results are interpreted based on the clusters, and eventually of patterns, of points [18, 44]. Therefore, similar objects are represented by nearby points, and the opposite for dissimilar objects. Different distances produce distinct MDS maps and it is up to the user to choose the metrics that reflect better the characteristics of the objects under analysis. By other words, the different distances are correct from the mathematical point of view, but the association of each metrics with the MDS algorithm may produce disparate patterns in the plots. In some cases the emerging patterns, although different, lead to similar conclusions. In other cases, some distances reflect better (or worst) the information embedded in the dataset and the selection of the ‘best’ metric depends on a trial and error set of tests based on the user experience.
Dataset description
The RNA is commonly sequenced indirectly by copying it into the complementary DNA (cDNA). Then the cDNA is amplified and analyzed using a number of DNA sequencing methods. The sequences of the RNA are published in the databases presenting the bases, adenine (A), cytosine (C), guanine (G), and thymine (T). Some symbols, such as N (unspecified or unknown nucleoside), R, (unspecified purine nucleoside), Y (unspecified pyrimidine nucleoside) and others, permeate only a small percentage of the information and are not considered [45]. The information about the GS was collected in the Global Initiative on Sharing Avian Influenza Data (GISAID) available at https://www.gisaid.org/. The information regarding the sequences, serial, clade/variant and country are listed in the Tables 5, 6, 7, 8 and 9. The genetic information is organized in 8 clades {GH, GR, O, GV, G, L, S, V} with 10 elements each, making a total of 80 cases. The recent advent of new variants correspond to the remaining 227 additional items as follows:
South Africa, with 10 cases for the variant ‘South Africa Triple Variant’, denoted as TV-ZA
Denmark, with 10 cases for the variant ‘Mink Cluster V’, denoted as CL5-DK
England, with 10 cases for the variant VUI2020/01, denoted as VUI-GB
Italy, with 10 cases for the variant VUI2020/01, denoted as VUI-IT
Denmark, with 10 cases for the variant VUI2020/01, denoted as VUI-DK
Portugal, with 10 cases for the variant VUI2020/01, denoted as VUI-PT
USA, with 10 cases for the variant VUI2020/01, denoted as VUI-US
- a mixture of several cases scattered along the world to give an higher spatial diversity
- Ireland, with 7 cases for the variant VUI2020/ 01, denoted as VUI-IE
- Japan, with 6 cases for the variant VUI2020/01, denoted as VUI-JP
- Australia, with 5 cases for the variant VUI2020/ 01, denoted as VUI-AU
- Singapore, with 5 cases for the variant VUI 2020/01, denoted as VUI-SG
- Israel, with 4 cases for the variant VUI2020/01, denoted as VUI-IL
- South Korea, with 3 cases for the variant VUI2020/01, denoted as VUI-KR
- Norway, with 3 cases for the variant VUI2020/ 01, denoted as VUI-NO
- France, with 2 cases for the variant VUI2020/01, denoted as VUI-FR
- Germany, with 2 cases for the variant VUI2020/ 01, denoted as VUI-DE
- Spain, Gibraltar, Hong Kong, India, Luxembourg, Switzerland and Sweden, with 1 case each, for the variant VUI2020/01, denoted simply as VUI
Japan with 10 cases for the variant B.1.1.28, denoted as B.1.1.28-JP
Brazil with 10 cases for the variant B.1.1.28, denoted as B.1.1.28-BR
Brazil with 10 cases for the variant P.1.1.28, denoted as P.1-BR
Brazil with 10 cases for the variant P.2, denoted as P.2-BR
South Africa with 10 cases for the variant B.1.351, denoted as B.1.351-ZA
USA with 10 cases for the variant B.1.427, denoted as B.1.427-US
California, USA, with 10 cases for the variant B.1.429, denoted as B.1.429 -US
New York, USA, with 10 cases for the variant B.1.526, denoted as B.1.526-US
India with 10 cases for the first variant VUI B.1.617, denoted as VUI B.1.617.1-IN
India with 10 cases for the second variant VUI B.1.617, denoted as VUI B.1.617.2-IN
India with 10 cases for the third variant VUI B.1.617, denoted as VUI B.1.617.3-IN.
In synthesis, we have collected a first set of 80 GS of the SARS-CoV-2 virus obtained in several countries during a first period of the outbreak. The second set includes 227 recent GS. The smaller number of cases the recent genomic data for some countries is limited to the data set available at the time of writing this paper. All ASCII files have approximately 30 kBytes.
Table 5.
Information about the GS
| i | Serial | Clade | Variant | Country | Acronym |
|---|---|---|---|---|---|
| 1 | hCoV-19/England/MILK-ACC1C7/2020EPI_ISL_6293162020-10-21 | GH | England | GH | |
| 2 | hCoV-19/England/MILK-AC843B/2020EPI_ISL_6293342020-10-21 | GH | England | GH | |
| 3 | hCoV-19/England/MILK-AC7D31/2020EPI_ISL_6293582020-10-21 | GH | England | GH | |
| 4 | hCoV-19/England/MILK-ACE3A1/2020EPI_ISL_6293612020-10-21 | GH | England | GH | |
| 5 | hCoV-19/England/MILK-ACE4F9/2020EPI_ISL_6293662020-10-21 | GH | England | GH | |
| 6 | hCoV-19/England/MILK-ACC3D0/2020EPI_ISL_6293882020-10-21 | GH | England | GH | |
| 7 | hCoV-19/England/MILK-ACC2B5/2020EPI_ISL_6293992020-10-21 | GH | England | GH | |
| 8 | hCoV-19/England/MILK-ACCA9F/2020EPI_ISL_6294192020-10-21 | GH | England | GH | |
| 9 | hCoV-19/England/MILK-AC850B/2020EPI_ISL_6294502020-10-21 | GH | England | GH | |
| 10 | hCoV-19/USA/TX-DSHS-1223/2020EPI_ISL_6316832020-03-25 | GH | USA | GH | |
| 11 | hCoV-19/Germany/NW-HHU-269/2020EPI_ISL_6313572020-09-23 | GR | Germany | GR | |
| 12 | hCoV-19/Germany/NW-HHU-271/2020EPI_ISL_6313592020-09-23 | GR | Germany | GR | |
| 13 | hCoV-19/Germany/NW-HHU-274/2020EPI_ISL_6313622020-09-25 | GR | Germany | GR | |
| 14 | hCoV-19/Germany/NW-HHU-275/2020EPI_ISL_6313632020-09-25 | GR | Germany | GR | |
| 15 | hCoV-19/Germany/NW-HHU-276/2020EPI_ISL_6313642020-09-26 | GR | Germany | GR | |
| 16 | hCoV-19/Germany/NW-HHU-242/2020EPI_ISL_6313752020-09-15 | GR | Germany | GR | |
| 17 | hCoV-19/Germany/NW-HHU-248/2020EPI_ISL_6313812020-09-19 | GR | Germany | GR | |
| 18 | hCoV-19/Germany/NW-HHU-253/2020EPI_ISL_6313862020-09-27 | GR | Germany | GR | |
| 19 | hCoV-19/USA/WI-WSLH-200603/2020EPI_ISL_6313982020-07-13 | GR | USA | GR | |
| 20 | hCoV-19/Italy/APU-IZSPB-187PT/2020EPI_ISL_7228952020-08-28 | GR | Italy | GR | |
| 21 | hCoV-19/Denmark/DCGC-1430/2020EPI_ISL_6224742020-04-27 | O | Denmark | O | |
| 22 | hCoV-19/Denmark/DCGC-1934/2020EPI_ISL_6225282020-04-27 | O | Denmark | O | |
| 23 | hCoV-19/Denmark/DCGC-1948/2020EPI_ISL_6225312020-04-20 | O | Denmark | O | |
| 24 | hCoV-19/Denmark/DCGC-1896/2020EPI_ISL_6225632020-07-06 | O | Denmark | O | |
| 25 | hCoV-19/New Zealand/20CV0297/2020EPI_ISL_6227942020-03-27 | O | New Zealand | O | |
| 26 | hCoV-19/New Zealand/20CV0302/2020EPI_ISL_6227972020-03-29 | O | New Zealand | O | |
| 27 | hCoV-19/New Zealand/20CV0303/2020EPI_ISL_6227982020-03-30 | O | New Zealand | O | |
| 28 | hCoV-19/England/204290933/2020EPI_ISL_6228622020-10-17 | O | USA | O | |
| 29 | hCoV-19/USA/NY-NYCPHL-000063/2020EPI_ISL_6315572020-03-11 | O | USA | O | |
| 30 | hCoV-19/USA/NY-NYCPHL-000136/2020EPI_ISL_6316272020-04-02 | O | USA | O | |
| 31 | hCoV-19/Germany/NW-HHU-234/2020EPI_ISL_6313672020-09-14 | GV | Germany | GV | |
| 32 | hCoV-19/Germany/NW-HHU-235/2020EPI_ISL_6313682020-09-14 | GV | Germany | GV | |
| 33 | hCoV-19/Germany/NW-HHU-236/2020EPI_ISL_6313692020-09-15 | GV | Germany | GV | |
| 34 | hCoV-19/Germany/NW-HHU-237/2020EPI_ISL_6313702020-09-15 | GV | Germany | GV | |
| 35 | hCoV-19/Germany/NW-HHU-238/2020EPI_ISL_6313712020-09-15 | GV | Germany | GV | |
| 36 | hCoV-19/Germany/NW-HHU-239/2020EPI_ISL_6313722020-09-15 | GV | Germany | GV | |
| 37 | hCoV-19/Germany/NW-HHU-240/2020EPI_ISL_6313732020-09-15 | GV | Germany | GV | |
| 38 | hCoV-19/Germany/NW-HHU-241/2020EPI_ISL_6313742020-09-15 | GV | Germany | GV | |
| 39 | hCoV-19/Germany/NW-HHU-243/2020EPI_ISL_6313762020-09-15 | GV | Germany | GV | |
| 40 | hCoV-19/Germany/NW-HHU-255/2020EPI_ISL_6313882020-09-30 | GV | Germany | GV | |
| 41 | hCoV-19/England/QEUH-AD061C/2020EPI_ISL_6299792020-10-23 | G | England | G | |
| 42 | hCoV-19/England/MILK-ACD278/2020EPI_ISL_6300072020-10-21 | G | England | G | |
| 43 | hCoV-19/England/CAMC-AAFF66/2020EPI_ISL_6311492020-10-23 | G | England | G | |
| 44 | hCoV-19/England/MILK-ABBDC7/2020EPI_ISL_6311832020-10-21 | G | England | G | |
| 45 | hCoV-19/Germany/NW-HHU-244/2020EPI_ISL_6313772020-09-29 | G | Germany | G | |
| 46 | hCoV-19/USA/WI-WSLH-200700/2020EPI_ISL_6314572020-09-22 | G | USA | G | |
| 47 | hCoV-19/USA/WI-WSLH-200726/2020EPI_ISL_6314822020-07-02 | G | USA | G | |
| 48 | hCoV-19/USA/WI-WSLH-200727/2020EPI_ISL_6314832020-07-02 | G | USA | G | |
| 49 | hCoV-19/USA/WI-WSLH-200728/2020EPI_ISL_6314842020-07-02 | G | USA | G | |
| 50 | hCoV-19/USA/TX-DSHS-1219/2020EPI_ISL_6316792020-03-28 | G | USA | G | |
| 51 | hCoV-19/Lithuania/MR-LUHS-Eilnr30/2020EPI_ISL_6030902020-04-06 | L | Lithuania | L | |
| 52 | hCoV-19/USA/CA-QDX-2362/2020EPI_ISL_6046562020-03-16 | L | USA | L | |
| 53 | hCoV-19/USA/CA-QDX-2383/2020EPI_ISL_6046622020-03-15 | L | USA | L | |
| 54 | hCoV-19/USA/UT-QDX-2315/2020EPI_ISL_6047222020-03-16 | L | USA | L | |
| 55 | hCoV-19/Ireland/NVRL-20G31886/2020EPI_ISL_6050732020-09-22 | L | Ireland | L | |
| 56 | hCoV-19/Henan/HN04/2020EPI_ISL_6059302020-02-24 | L | Henan | L | |
| 57 | hCoV-19/Hong Kong/HKPU-0442/2020EPI_ISL_6101752020-03-26 | L | Hong Kong | L | |
| 58 | hCoV-19/USA/NM-UNM-00797/2020EPI_ISL_6102612020-05-06 | L | USA | L | |
| 59 | hCoV-19/USA/WI-WSLH-200589/2020EPI_ISL_6313912020-04-03 | L | USA | L | |
| 60 | hCoV-19/USA/TX-DSHS-1208/2020EPI_ISL_6316692020-03-18 | L | USA | L | |
| 61 | hCoV-19/Denmark/DCGC-1430/2020EPI_ISL_6224742020-04-27 | S | Denmark | S | |
| 62 | hCoV-19/Denmark/DCGC-1934/2020EPI_ISL_6225282020-04-27 | S | Denmark | S | |
| 63 | hCoV-19/Denmark/DCGC-1948/2020EPI_ISL_6225312020-04-20 | S | Denmark | S | |
| 64 | hCoV-19/Denmark/DCGC-1896/2020EPI_ISL_6225632020-07-06 | S | Denmark | S | |
| 65 | hCoV-19/New Zealand/20CV0297/2020EPI_ISL_6227942020-03-27 | S | New Zealand | S | |
| 66 | hCoV-19/New Zealand/20CV0302/2020EPI_ISL_6227972020-03-29 | S | New Zealand | S | |
| 67 | hCoV-19/New Zealand/20CV0303/2020EPI_ISL_6227982020-03-30 | S | New Zealand | S | |
| 68 | hCoV-19/England/204290933/2020EPI_ISL_6228622020-10-17 | S | England | S | |
| 69 | hCoV-19/USA/NY-NYCPHL-000063/2020EPI_ISL_6315572020-03-11 | S | USA | S | |
| 70 | hCoV-19/USA/NY-NYCPHL-000136/2020EPI_ISL_6316272020-04-02 | S | USA | S |
Table 6.
Information about the GS
| i | Serial | Clade | Variant | Country | Acronym |
|---|---|---|---|---|---|
| 71 | hCoV-19/England/LIVE-1D2EC0/2020EPI_ISL_6120172020-03-31 | V | England | V | |
| 72 | hCoV-19/England/LIVE-1D2EDF/2020EPI_ISL_6120752020-03-30 | V | England | V | |
| 73 | hCoV-19/England/PORT-2D21A8/2020EPI_ISL_6132832020-03-22 | V | England | V | |
| 74 | hCoV-19/England/PORT-2D220F/2020EPI_ISL_6132852020-03-22 | V | England | V | |
| 75 | hCoV-19/USA/NY-NYCPHL-000032/2020EPI_ISL_6315282020-03-12 | V | England | V | |
| 76 | hCoV-19/USA/NY-NYCPHL-000133/2020EPI_ISL_6316242020-04-02 | V | England | V | |
| 77 | hCoV-19/England/LIVE-1D2BC9/2020EPI_ISL_6124142020-03-29 | V | England | V | |
| 78 | hCoV-19/England/LIVE-1D2C02/2020EPI_ISL_6124152020-03-29 | V | England | V | |
| 79 | hCoV-19/England/LIVE-1D2D2D/2020EPI_ISL_6124182020-03-30 | V | England | V | |
| 80 | hCoV-19/England/LIVE-1D2D4B/2020EPI_ISL_6124192020-03-31 | V | England | V | |
| 81 | hCoV-19/South Africa/N00352/2020EPI_ISL_7120842020-10-30 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 82 | hCoV-19/South Africa/N00336/2020EPI_ISL_7120852020-10-30 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 83 | hCoV-19/South Africa/N00334/2020EPI_ISL_7120862020-10-28 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 84 | hCoV-19/South Africa/N00349/2020EPI_ISL_7120872020-11-03 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 85 | hCoV-19/South Africa/N00337/2020EPI_ISL_7120882020-11-03 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 86 | hCoV-19/South Africa/N00350/2020EPI_ISL_7120892020-10-28 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 87 | hCoV-19/South Africa/N00348/2020EPI_ISL_7120902020-10-30 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 88 | hCoV-19/South Africa/N00343/2020EPI_ISL_7120912020-10-28 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 89 | hCoV-19/South Africa/N00338/2020EPI_ISL_7120952020-11-03 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 90 | hCoV-19/South Africa/N00344/2020EPI_ISL_7120962020-11-04 | GH | South Africa Triple Variant | South Africa | TV-ZA |
| 91 | hCoV-19/Denmark/DCGC-4467/2020EPI_ISL_6156522020-09-14 | GR | Mink Cluster V | Denmark | CL5-DK |
| 92 | hCoV-19/Denmark/DCGC-4468/2020EPI_ISL_6156532020-09-14 | GR | Mink Cluster V | Denmark | CL5-DK |
| 93 | hCoV-19/Denmark/DCGC-4472/2020EPI_ISL_6156572020-09-14 | GR | Mink Cluster V | Denmark | CL5-DK |
| 94 | hCoV-19/Denmark/DCGC-4476/2020EPI_ISL_6156612020-09-14 | GR | Mink Cluster V | Denmark | CL5-DK |
| 95 | hCoV-19/Denmark/DCGC-4306/2020EPI_ISL_6157432020-09-07 | GR | Mink Cluster V | Denmark | CL5-DK |
| 96 | hCoV-19/Denmark/DCGC-3152/2020EPI_ISL_6159722020-08-31 | GR | Mink Cluster V | Denmark | CL5-DK |
| 97 | hCoV-19/Denmark/DCGC-3542/2020EPI_ISL_6163352020-08-31 | GR | Mink Cluster V | Denmark | CL5-DK |
| 98 | hCoV-19/Denmark/DCGC-3631/2020EPI_ISL_6164022020-08-31 | GR | Mink Cluster V | Denmark | CL5-DK |
| 99 | hCoV-19/Denmark/DCGC-2934/2020EPI_ISL_6166952020-08-24 | GR | Mink Cluster V | Denmark | CL5-DK |
| 100 | hCoV-19/Denmark/DCGC-2965/2020EPI_ISL_6167272020-08-24 | GR | Mink Cluster V | Denmark | CL5-DK |
| 101 | hCoV-19/England/ALDP-C3B5E3/2020EPI_ISL_7318502020-12-08 | GR | VUI2020/01 | England | VUI-GB |
| 102 | hCoV-19/England/ALDP-C3BAF3/2020EPI_ISL_7318522020-12-10 | GR | VUI2020/01 | England | VUI-GB |
| 103 | hCoV-19/England/ALDP-C3AA4F/2020EPI_ISL_7318542020-12-09 | GR | VUI2020/01 | England | VUI-GB |
| 104 | hCoV-19/England/ALDP-C3C9F5/2020EPI_ISL_7318572020-12-10 | GR | VUI2020/01 | England | VUI-GB |
| 105 | hCoV-19/England/ALDP-C3CEAB/2020EPI_ISL_7318652020-12-10 | GR | VUI2020/01 | England | VUI-GB |
| 106 | hCoV-19/England/ALDP-C3D0D1/2020EPI_ISL_7318662020-12-10 | GR | VUI2020/01 | England | VUI-GB |
| 107 | hCoV-19/England/ALDP-C3C05A/2020EPI_ISL_7318722020-12-10 | GR | VUI2020/01 | England | VUI-GB |
| 108 | hCoV-19/England/ALDP-C3B10D/2020EPI_ISL_7318772020-12-08 | GR | VUI2020/01 | England | VUI-GB |
| 109 | hCoV-19/England/ALDP-C3BEAC/2020EPI_ISL_7318782020-12-10 | GR | VUI2020/01 | England | VUI-GB |
| 110 | hCoV-19/England/MILK-BFF537/2020EPI_ISL_7318932020-12-02 | GR | VUI2020/01 | England | VUI-GB |
| 111 | hCoV-19/Italy/LAZ-AMC3-5015/2020EPI_ISL_7179782020-12-14 | GR | VUI2020/01 | Italy | VUI-IT |
| 112 | hCoV-19/Italy/CAM-TIGEM-174/2020EPI_ISL_7367862020-12-21 | GR | VUI2020/01 | Italy | VUI-IT |
| 113 | hCoV-19/Italy/CAM-TIGEM-181/2020EPI_ISL_7367872020-12-21 | GR | VUI2020/01 | Italy | VUI-IT |
| 114 | hCoV-19/Italy/CAM-INMI-117/2020EPI_ISL_7369962020-12-20 | GR | VUI2020/01 | Italy | VUI-IT |
| 115 | hCoV-19/Italy/CAM-INMI-118/2020EPI_ISL_7369972020-12-20 | GR | VUI2020/01 | Italy | VUI-IT |
| 116 | hCoV-19/Italy/ABR-TE351971/2020EPI_ISL_7380452020-12-18 | GR | VUI2020/01 | Italy | VUI-IT |
| 117 | hCoV-19/Italy/ABR-TE353967/2020EPI_ISL_7380462020-12-18 | GR | VUI2020/01 | Italy | VUI-IT |
| 118 | hCoV-19/Italy/ABR-TE353968/2020EPI_ISL_7380472020-12-18 | GR | VUI2020/01 | Italy | VUI-IT |
| 119 | hCoV-19/Italy/ABR-TE353969/2020EPI_ISL_7380482020-12-18 | GR | VUI2020/01 | Italy | VUI-IT |
| 120 | hCoV-19/Italy/APU-IZSPB-400PT/2020EPI_ISL_7451922020-12-21 | GR | VUI2020/01 | Italy | VUI-IT |
| 121 | hCoV-19/Denmark/DCGC-9482/2020EPI_ISL_6685982020-11-09 | GR | VUI2020/01 | Denmark | VUI-DK |
| 122 | hCoV-19/Denmark/DCGC-9581/2020EPI_ISL_6685992020-11-09 | GR | VUI2020/01 | Denmark | VUI-DK |
| 123 | hCoV-19/Denmark/DCGC-9642/2020EPI_ISL_6686002020-11-09 | GR | VUI2020/01 | Denmark | VUI-DK |
| 124 | hCoV-19/Denmark/DCGC-13966/2020EPI_ISL_7112292020-11-30 | GR | VUI2020/01 | Denmark | VUI-DK |
| 125 | hCoV-19/Denmark/DCGC-14038/2020EPI_ISL_7112302020-11-30 | GR | VUI2020/01 | Denmark | VUI-DK |
| 126 | hCoV-19/Denmark/DCGC-14208/2020EPI_ISL_7112312020-11-30 | GR | VUI2020/01 | Denmark | VUI-DK |
| 127 | hCoV-19/Denmark/DCGC-14596/2020EPI_ISL_7112322020-11-23 | GR | VUI2020/01 | Denmark | VUI-DK |
| 128 | hCoV-19/Denmark/DCGC-15593/2020EPI_ISL_7132352020-11-23 | GR | VUI2020/01 | Denmark | VUI-DK |
| 129 | hCoV-19/Denmark/DCGC-15226/2020EPI_ISL_7132692020-11-23 | GR | VUI2020/01 | Denmark | VUI-DK |
| 130 | hCoV-19/Denmark/DCGC-18854/2020EPI_ISL_7482802020-12-07 | GR | VUI2020/01 | Denmark | VUI-DK |
| 131 | hCoV-19/Portugal/PT2101/2020EPI_ISL_7381012020-12-18 | GR | VUI2020/01 | Portugal | VUI-PT |
| 132 | hCoV-19/Portugal/PT2102/2020EPI_ISL_7381022020-12-18 | GR | VUI2020/01 | Portugal | VUI-PT |
| 133 | hCoV-19/Portugal/PT2104/2020EPI_ISL_7381032020-12-17 | GR | VUI2020/01 | Portugal | VUI-PT |
| 134 | hCoV-19/Portugal/PT2105/2020EPI_ISL_7381042020-12-17 | GR | VUI2020/01 | Portugal | VUI-PT |
| 135 | hCoV-19/Portugal/PT2106/2020EPI_ISL_7381052020-12-17 | GR | VUI2020/01 | Portugal | VUI-PT |
| 136 | hCoV-19/Portugal/PT2107/2020EPI_ISL_7381062020-12-15 | GR | VUI2020/01 | Portugal | VUI-PT |
| 137 | hCoV-19/Portugal/PT2108/2020EPI_ISL_7381072020-11-09 | GR | VUI2020/01 | Portugal | VUI-PT |
| 138 | hCoV-19/Portugal/PT2110/2020EPI_ISL_7381082020-12-07 | GR | VUI2020/01 | Portugal | VUI-PT |
| 139 | hCoV-19/Portugal/PT2112/2020EPI_ISL_7381092020-12-19 | GR | VUI2020/01 | Portugal | VUI-PT |
| 140 | hCoV-19/Portugal/PT2093/2020EPI_ISL_7381102020-12-20 | GR | VUI2020/01 | Portugal | VUI-PT |
Table 7.
Information about the GS
| i | Serial | Clade | Variant | Country | Acronym |
|---|---|---|---|---|---|
| 141 | hCoV-19/USA/CO-CDPHE-2100156850/2020EPI_ISL_7518002020-12-24 | GR | VUI2020/01 | USA | VUI-US |
| 142 | hCoV-19/USA/CA-SEARCH-5574/2020EPI_ISL_7518012020-12-29 | GR | VUI2020/01 | USA | VUI-US |
| 143 | hCoV-19/USA/FL-CDC-STM-P012/2020EPI_ISL_7555932020-12-19 | GR | VUI2020/01 | USA | VUI-US |
| 144 | hCoV-19/USA/CA-CDC-STM-P017/2020EPI_ISL_7555942020-12-20 | GR | VUI2020/01 | USA | VUI-US |
| 145 | hCoV-19/USA/CA-CDC-STM-P019/2020EPI_ISL_7555952020-12-21 | GR | VUI2020/01 | USA | VUI-US |
| 146 | hCoV-19/USA/CA-CDC-STM-P025/2020EPI_ISL_7555962020-12-20 | GR | VUI2020/01 | USA | VUI-US |
| 147 | hCoV-19/USA/CA-CDPH-UC301/2020EPI_ISL_7559402020-12-20 | GR | VUI2020/01 | USA | VUI-US |
| 148 | hCoV-19/USA/CA-CDPH-UC302/2020EPI_ISL_7559412020-12-20 | GR | VUI2020/01 | USA | VUI-US |
| 149 | hCoV-19/Ireland/D-NVRL-20IRL10513/2020EPI_ISL_7353842020-12-20 | GR | VUI2020/01 | Ireland | VUI-IE |
| 150 | hCoV-19/Ireland/KK-NVRL-20IRL10079/2020EPI_ISL_7353852020-12-19 | GR | VUI2020/01 | Ireland | VIU-IE |
| 151 | hCoV-19/Ireland/D-NVRL-20IRL09344/2020EPI_ISL_7353862020-12-18 | GR | VUI2020/01 | Ireland | VIU-IE |
| 152 | hCoV-19/Ireland/D-NVRL-20IRL10553/2020EPI_ISL_7353872020-12-20 | GR | VUI2020/01 | Ireland | VIU-IE |
| 153 | hCoV-19/Ireland/D-NVRL-20IRL10860/2020EPI_ISL_7353882020-12-21 | GR | VUI2020/01 | Ireland | VIU-IE |
| 154 | hCoV-19/Ireland/D-NVRL-20IRL10527/2020EPI_ISL_7353892020-12-20 | GR | VUI2020/01 | Ireland | VIU-IE |
| 155 | hCoV-19/Ireland/D-NVRL-20IRL10603/2020EPI_ISL_7353902020-12-20 | GR | VUI2020/01 | Ireland | VIU-IE |
| 156 | hCoV-19/Netherlands/NH-RIVM-20227/2020EPI_ISL_7285662020-12-05 | GR | VUI2020/01 | Netherlands | VUI-NL |
| 157 | hCoV-19/Netherlands/NH-RIVM-20432/2020EPI_ISL_7285682020-12-13 | GR | VUI2020/01 | Netherlands | VUI-NL |
| 158 | hCoV-19/Netherlands/ZH-EMC-1148/2020EPI_ISL_7341662020-12-19 | GR | VUI2020/01 | Netherlands | VUI-NL |
| 159 | hCoV-19/Netherlands/NH-RIVM-20631/2020EPI_ISL_7475212020-12-16 | GR | VUI2020/01 | Netherlands | VUI-NL |
| 160 | hCoV-19/Netherlands/NH-RIVM-20635/2020EPI_ISL_7475222020-11-29 | GR | VUI2020/01 | Netherlands | VUI-NL |
| 161 | hCoV-19/Netherlands/NH-RIVM-20652/2020EPI_ISL_7475242020-12-21 | GR | VUI2020/01 | Netherlands | VUI-NL |
| 162 | hCoV-19/Japan/IC-0413/2020EPI_ISL_7354392020-12 | GR | VUI2020/01 | Japan | VUI-JP |
| 163 | hCoV-19/Japan/IC-0419/2020EPI_ISL_7354402020-12 | GR | VUI2020/01 | Japan | VUI-JP |
| 164 | hCoV-19/Japan/IC-0421/2020EPI_ISL_7354412020-12 | GR | VUI2020/01 | Japan | VUI-JP |
| 165 | hCoV-19/Japan/IC-0422/2020EPI_ISL_7354422020-12 | GR | VUI2020/01 | Japan | VUI-JP |
| 166 | hCoV-19/Japan/IC-0423/2020EPI_ISL_7354432020-12 | GR | VUI2020/01 | Japan | VUI-JP |
| 167 | hCoV-19/Japan/IC-0424/2020EPI_ISL_7368912020-12 | GR | VUI2020/01 | Japan | VUI-JP |
| 168 | hCoV-19/Australia/NSW3456/2020EPI_ISL_6783862020-11-30 | GR | VUI2020/01 | Australia | VUI-AU |
| 169 | hCoV-19/Australia/NSW3647/2020EPI_ISL_7177112020-12-09 | GR | VUI2020/01 | Australia | VUI-AU |
| 170 | hCoV-19/Australia/SAP593/2020EPI_ISL_7329612020-12-19 | GR | VUI2020/01 | Australia | VUI-AU |
| 171 | hCoV-19/Australia/NSW1242/2020EPI_ISL_7408732020-12-09 | GR | VUI2020/01 | Australia | VUI-AU |
| 172 | hCoV-19/Australia/SAP595/2020EPI_ISL_7525982020-12-26 | GR | VUI2020/01 | Australia | VUI-AU |
| 173 | hCoV-19/Singapore/1453/2020EPI_ISL_7281892020-12-08 | GR | VUI2020/01 | Singapore | VUI-SG |
| 174 | hCoV-19/Singapore/1468/2020EPI_ISL_7540752020-12-21 | GR | VUI2020/02 | Singapore | VUI-SG |
| 175 | hCoV-19/Singapore/1489/2020EPI_ISL_7540762020-12-22 | GR | VUI2020/03 | Singapore | VUI-SG |
| 176 | hCoV-19/Singapore/1467/2020EPI_ISL_7540822020-12-19 | GR | VUI2020/04 | Singapore | VUI-SG |
| 177 | hCoV-19/Singapore/1466/2020EPI_ISL_7540832020-12-17 | GR | VUI2020/05 | Singapore | VUI-SG |
| 178 | hCoV-19/Israel/CVL-7075/2020EPI_ISL_7334982020-12-16 | GR | VUI2020/01 | Israel | VUI-IL |
| 179 | hCoV-19/Israel/CVL-46880/2020EPI_ISL_7372022020-12-20 | GR | VUI2020/01 | Israel | VUI-IL |
| 180 | hCoV-19/Israel/CVL-46754/2020EPI_ISL_7372032020-12-20 | GR | VUI2020/01 | Israel | VUI-IL |
| 181 | hCoV-19/Israel/CVL-46879/2020EPI_ISL_7372042020-12-20 | GR | VUI2020/01 | Israel | VUI-IL |
| 182 | hCoV-19/South Korea/KDCA0001/2020EPI_ISL_7381392020-12-22 | GR | VUI2020/01 | South Korea | VUI-KR |
| 183 | hCoV-19/South Korea/KDCA0002/2020EPI_ISL_7381412020-12-22 | GR | VUI2020/01 | South Korea | VUI-KR |
| 184 | hCoV-19/South Korea/KDCA0003/2020EPI_ISL_7381422020-12-22 | GR | VUI2020/01 | South Korea | VUI-KR |
| 185 | hCoV-19/Norway/7115/2020EPI_ISL_7383132020-12-21 | GR | VUI2020/01 | Norway | VUI-NO |
| 186 | hCoV-19/Norway/7129/2020EPI_ISL_7383142020-12-13 | GR | VUI2020/01 | Norway | VUI-NO |
| 187 | hCoV-19/France/CVL-SC719/2020EPI_ISL_7353912020-12-18 | GR | VUI2020/01 | France | GR-FR |
| 188 | hCoV-19/France/HDF-IPP11311/2020EPI_ISL_7548422020-12-21 | GR | VUI2020/01 | France | GR-FR |
| 189 | hCoV-19/Germany/NW-RKI-I-0026/2020EPI_ISL_7517992020-12-07 | GR | VUI2020/01 | Germany | GR-DE |
| 190 | hCoV-19/Germany/DE-BW-ChVir21528/2020EPI_ISL_7541742020-12-21 | GR | VUI2020/01 | Germany | GR-DE |
| 191 | hCoV-19/Spain/MD-H12-02-5372/2020EPI_ISL_7379302020-12-23 | GR | VUI2020/01 | Spain | VUI |
| 192 | hCoV-19/Gibraltar/205000662/2020EPI_ISL_7099572020-12 | GR | VUI2020/01 | Gibraltar | VUI |
| 193 | hCoV-19/Hong Kong/CM20000424/2020EPI_ISL_7335732020-12-07 | GR | VUI2020/01 | Hong Kong | VUI |
| 194 | hCoV-19/India/KA-NIMH-SEQ-8/2020EPI_ISL_7472442020-12-22 | GR | VUI2020/01 | India | VUI |
| 195 | hCoV-19/Luxembourg/LNS2043044/2020EPI_ISL_7555892020-12-24 | GR | VUI2020/01 | Luxembourg | VUI |
| 196 | hCoV-19/Switzerland/ZH-UZH-IMV130/2020EPI_ISL_7511932020-12-18 | GR | VUI2020/01 | Switzerland | VUI |
| 197 | hCoV-19/Sweden/20-53787/2020EPI_ISL_7381332020-12-20 | GR | VUI2020/01 | Sweden | VUI |
| 198 | hCoV-19/Japan/IC-0500/2020EPI_ISL_7687092020-12 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 199 | hCoV-19/Japan/IC-0524/2020EPI_ISL_7792072020-12 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 200 | hCoV-19/Japan/IC-0526/2020EPI_ISL_7792092020-12 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 201 | hCoV-19/Japan/IC-0533/2020EPI_ISL_7792162020-12 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 202 | hCoV-19/Japan/IC-0566/2020EPI_ISL_7792452020-09 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 203 | hCoV-19/Japan/IC-0567/2020EPI_ISL_7792462020-09 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 204 | hCoV-19/Japan/IC-0561/2021EPI_ISL_7926802021-01-02 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 205 | hCoV-19/Japan/IC-0562/2021EPI_ISL_7926812021-01-02 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 206 | hCoV-19/Japan/IC-0563/2021EPI_ISL_7926822021-01-02 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
| 207 | hCoV-19/Japan/IC-0564/2021EPI_ISL_7926832021-01-02 | GR | B.1.1.28 | Japan | B.1.1.28-JP |
Table 8.
Information about the GS
| i | Serial | Clade | Variant | Country | Acronym |
|---|---|---|---|---|---|
| 208 | hCoV-19/Brazil/AM-20890117JP/2020EPI_ISL_8014012020-09-15 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 209 | hCoV-19/Brazil/AM-20890261MV/2020EPI_ISL_8014022020-05-03 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 210 | hCoV-19/Brazil/AM-20892948LS/2020EPI_ISL_8014032020-05-12 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 211 | hCoV-19/Brazil/AM-L70-CD1731/2020EPI_ISL_8048172020-12-17 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 212 | hCoV-19/Brazil/AM-L70-CD1729/2020EPI_ISL_8048182020-12-17 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 213 | hCoV-19/Brazil/AM-L70-CD1715/2020EPI_ISL_8048252020-12-15 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 214 | hCoV-19/Brazil/AM-L70-CD1717/2020EPI_ISL_8048262020-12-16 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 215 | hCoV-19/Brazil/AM-L70-CD1735/2020EPI_ISL_8048302020-12-21 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 216 | hCoV-19/Brazil/AM-L71-CD1748/2020EPI_ISL_8048342020-12-23 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 217 | hCoV-19/Brazil/AM-L71-CD1741/2020EPI_ISL_8048412020-12-22 | GR | B.1.1.28 | Brazil | B.1.1.28-BR |
| 218 | hCoV-19/Brazil/AM-L70-CD1716/2020EPI_ISL_8048152020-12-16 | GR | P.1 | Brazil | P.1-BR |
| 219 | hCoV-19/Brazil/AM-L70-CD1718/2020EPI_ISL_8048192020-12-16 | GR | P.1 | Brazil | P.1-BR |
| 220 | hCoV-19/Brazil/AM-L70-CD1739/2020EPI_ISL_8048202020-12-21 | GR | P.1 | Brazil | P.1-BR |
| 221 | hCoV-19/Brazil/AM-L70-CD1733/2020EPI_ISL_8048212020-12-21 | GR | P.1 | Brazil | P.1-BR |
| 222 | hCoV-19/Brazil/AM-L70-CD1722/2020EPI_ISL_8048232020-12-17 | GR | P.1 | Brazil | P.1-BR |
| 223 | hCoV-19/Brazil/AM-L70-CD1721/2020EPI_ISL_8048242020-12-17 | GR | P.1 | Brazil | P.1-BR |
| 224 | hCoV-19/Brazil/AM-L70-CD1723/2020EPI_ISL_8048322020-12-17 | GR | P.1 | Brazil | P.1-BR |
| 225 | hCoV-19/Brazil/AM-L71-CD1743/2020EPI_ISL_8048332020-12-22 | GR | P.1 | Brazil | P.1-BR |
| 226 | hCoV-19/Brazil/AM-L70-CD1727/2020EPI_ISL_8048352020-12-18 | GR | P.1 | Brazil | P.1-BR |
| 227 | hCoV-19/Brazil/AM-L71-CD1740/2020EPI_ISL_8048432020-12-21 | GR | P.1 | Brazil | P.1-BR |
| 228 | hCoV-19/Brazil/AP-IEC-177409/2021EPI_ISL_9185612021-01-10 | GR | P.2 | Brazil | P.2-BR |
| 229 | hCoV-19/Brazil/SP-1076/2021EPI_ISL_9406282021-01-22 | GR | P.2 | Brazil | P.2-BR |
| 230 | hCoV-19/Brazil/RS-CEVS-83356/2020EPI_ISL_9436042020-11-20 | GR | P.2 | Brazil | P.2-BR |
| 231 | hCoV-19/Brazil/RS-CEVS-82920/2020EPI_ISL_9436072020-11-20 | GR | P.2 | Brazil | P.2-BR |
| 232 | hCoV-19/Brazil/RS-CEVS-82973/2020EPI_ISL_9436082020-11-18 | GR | P.2 | Brazil | P.2-BR |
| 233 | hCoV-19/Brazil/RS-CEVS-83139/2020EPI_ISL_9436102020-11-18 | GR | P.2 | Brazil | P.2-BR |
| 234 | hCoV-19/Brazil/RS-CEVS-83150/2020EPI_ISL_9436122020-11-19 | GR | P.2 | Brazil | P.2-BR |
| 235 | hCoV-19/Brazil/TO-1185/2020EPI_ISL_9439842020-10-26 | GR | P.2 | Brazil | P.2-BR |
| 236 | hCoV-19/Brazil/TO-1187R2/2020EPI_ISL_9439862020-12-22 | GR | P.2 | Brazil | P.2-BR |
| 237 | hCoV-19/Brazil/GO-1190R2/2020EPI_ISL_9439892020-12-18 | GR | P.2 | Brazil | P.2-BR |
| 238 | hCoV-19/South Africa/KRISP-K008261/2021EPI_ISL_9441692021-01-04 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 239 | hCoV-19/South Africa/KRISP-K008262/2021EPI_ISL_9441702021-01-04 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 240 | hCoV-19/South Africa/KRISP-K008263/2021EPI_ISL_9441712021-01-04 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 241 | hCoV-19/South Africa/KRISP-K008265/2021EPI_ISL_9441722021-01-05 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 242 | hCoV-19/South Africa/KRISP-K008267/2021EPI_ISL_9441732021-01-04 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 243 | hCoV-19/South Africa/KRISP-K008269/2021EPI_ISL_9441742021-01-01 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 244 | hCoV-19/South Africa/KRISP-K008271/2021EPI_ISL_9441752021-01-04 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 245 | hCoV-19/South Africa/KRISP-K008274/2021EPI_ISL_9441762021-01-04 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 246 | hCoV-19/South Africa/KRISP-K008276/2021EPI_ISL_9441772021-01-05 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 247 | hCoV-19/South Africa/UFS-VIRO-NGS-163/2020EPI_ISL_9496322020-12-23 | GH | B.1.351 | South Africa | B.1.351-ZA |
| 248 | hCoV-19/USA/CA-CZB-17547/2020EPI_ISL_9556002020-12-16 | GH | B.1.427 | USA | B.1.427-US |
| 249 | hCoV-19/USA/CA-CZB-17562/2020EPI_ISL_9556092020-12-16 | GH | B.1.427 | USA | B.1.427-US |
| 250 | hCoV-19/USA/CA-CZB-17583/2020EPI_ISL_9556242020-12-18 | GH | B.1.427 | USA | B.1.427-US |
| 251 | hCoV-19/USA/CA-CZB-17584/2020EPI_ISL_9556252020-12-18 | GH | B.1.427 | USA | B.1.427-US |
| 252 | hCoV-19/USA/CA-CZB-17586/2020EPI_ISL_9556262020-12-18 | GH | B.1.427 | USA | B.1.427-US |
| 253 | hCoV-19/USA/CA-CZB-17587/2020EPI_ISL_9556272020-12-18 | GH | B.1.427 | USA | B.1.427-US |
| 254 | hCoV-19/USA/CA-CZB-19323/2020EPI_ISL_9557132020-12-28 | GH | B.1.427 | USA | B.1.427-US |
| 255 | hCoV-19/USA/CA-CZB-19357/2020EPI_ISL_9557422020-12-30 | GH | B.1.427 | USA | B.1.427-US |
| 256 | hCoV-19/USA/CA-CZB-19363/2020EPI_ISL_9557482020-12-29 | GH | B.1.427 | USA | B.1.427-US |
| 257 | hCoV-19/USA/CA-CZB-19394/2021EPI_ISL_9557752021-01-04 | GH | B.1.427 | USA | B.1.427-US |
| 258 | hCoV-19/USA/CA-RD581-CV0013/2021EPI_ISL_10688622021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 259 | hCoV-19/USA/CA-RD581-CV0016/2021EPI_ISL_10688652021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 260 | hCoV-19/USA/CA-RD581-CV0017/2021EPI_ISL_10688662021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 261 | hCoV-19/USA/CA-RD581-CV0018/2021EPI_ISL_10688672021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 262 | hCoV-19/USA/CA-RD581-CV0019/2021EPI_ISL_10688682021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 263 | hCoV-19/USA/CA-RD581-CV0020/2021EPI_ISL_10688692021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 264 | hCoV-19/USA/CA-RD581-CV0021/2021EPI_ISL_10688702021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 265 | hCoV-19/USA/CA-RD581-CV0024/2021EPI_ISL_10688712021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 266 | hCoV-19/USA/CA-RD581-CV0026/2021EPI_ISL_10688732021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 267 | hCoV-19/USA/CA-RD581-CV0027/2021EPI_ISL_10688742021-02-05 | GH | B.1.429 | California | B.1.429 -US |
| 268 | hCoV-19/USA/NY-NP-3987/2020EPI_ISL_10807992020-12 | GH | B.1.526 | New York | B.1.526-US |
| 269 | hCoV-19/USA/NY-NP-4053/2020EPI_ISL_10808002020-12 | GH | B.1.526 | New York | B.1.526-US |
| 270 | hCoV-19/USA/NY-NP-4698/2021EPI_ISL_10808012021-01 | GH | B.1.526 | New York | B.1.526-US |
| 271 | hCoV-19/USA/NY-NP-4916/2021EPI_ISL_10808022021-02 | GH | B.1.526 | New York | B.1.526-US |
| 272 | hCoV-19/USA/NY-NP-4974/2021EPI_ISL_10808032021-01 | GH | B.1.526 | New York | B.1.526-US |
| 273 | hCoV-19/USA/NY-NP-5102/2021EPI_ISL_10808042021-02 | GH | B.1.526 | New York | B.1.526-US |
| 274 | hCoV-19/USA/NY-NP-5107/2021EPI_ISL_10808052021-02 | GH | B.1.526 | New York | B.1.526-US |
| 275 | hCoV-19/USA/NY-NP-5113/2021EPI_ISL_10808062021-02 | GH | B.1.526 | New York | B.1.526-US |
| 276 | hCoV-19/USA/NY-NP-5140/2021EPI_ISL_10808072021-02 | GH | B.1.526 | New York | B.1.526-US |
| 277 | hCoV-19/USA/NY-NP-5197/2021EPI_ISL_10808082021-02 | GH | B.1.526 | New York | B.1.526-US |
Table 9.
Information about the GS
| 278 | hCoV-19/India/MH-NEERI-NGP-32956/2021EPI_ISL_13603522021-01-30 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 279 | hCoV-19/India/MH-NEERI-NGP-34282/2021EPI_ISL_13603592021-02-08 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 280 | hCoV-19/India/MH-NEERI-NGP-34605/2021EPI_ISL_13603612021-02-10 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 281 | hCoV-19/India/MH-NEERI-NGP-34972/2021EPI_ISL_13603642021-02-13 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 282 | hCoV-19/India/MH-NEERI-NGP-36241/2021EPI_ISL_13603702021-02-19 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 283 | hCoV-19/India/MH-NEERI-NGP-36444/2021EPI_ISL_13603712021-02-20 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 284 | hCoV-19/India/MH-NEERI-NGP-37270/2021EPI_ISL_13603752021-02-24 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 285 | hCoV-19/India/MH-NEERI-NGP-39017/2021EPI_ISL_13603822021-03-03 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 286 | hCoV-19/India/ILSGS00182/2021EPI_ISL_13720112021-02-26 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| 287 | hCoV-19/India/ILSGS00308/2020EPI_ISL_13720932020-12-01 | G | VUI B.1.617 | India | VUI B.1.617.1-IN |
| i | Serial | Clade | Variant | Country | Acronym |
| 288 | hCoV-19/India/MH-ILSGS01660/2021EPI_ISL_23429642021-04-04 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 289 | hCoV-19/India/JH-ILSGS01687/2021EPI_ISL_23429722021-04-07 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 290 | hCoV-19/India/JH-ILSGS01694/2021EPI_ISL_23429732021-04-08 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 291 | hCoV-19/India/JH-ILSGS01695/2021EPI_ISL_23429742021-04-10 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 292 | hCoV-19/India/JH-ILSGS01701/2021EPI_ISL_23429752021-04-14 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 293 | hCoV-19/India/JH-ILSGS01706/2021EPI_ISL_23429762021-04-14 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 294 | hCoV-19/India/JH-ILSGS01715/2021EPI_ISL_23429772021-04-13 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 295 | hCoV-19/India/JH-ILSGS01718/2021EPI_ISL_23429782021-04-12 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 296 | hCoV-19/India/JH-ILSGS01736/2021EPI_ISL_23429792021-04-08 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 297 | hCoV-19/India/OR-ILSGS01781/2021EPI_ISL_23429802021-04-20 | G | VUI B.1.617.2 | India | VUI B.1.617.2-IN |
| 298 | hCoV-19/India/MH-SEQ-221_S66_R1_001/2021EPI_ISL_19398912021-03-22 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| _S66_R1_001/2021EPI_ISL_19398912021-03-22 | |||||
| 299 | hCoV-19/India/GJ-ICMR-NIV-INSAGOG-GSEQ-3117/2021EPI | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| _ISL_19704982021-04-23 | |||||
| 300 | hCoV-19/India/MH-NCCS-119281/2021EPI_ISL_21070582021-03-22 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| 301 | hCoV-19/India/WB-1931300249962/2021EPI_ISL_21327152021-03-23 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| 302 | hCoV-19/India/AP-CCMB-CIA1346/2021EPI_ISL_22316612021-04-21 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| 303 | hCoV-19/India/MH-ILSGS01543/2021EPI_ISL_23429162021-03-24 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| 304 | hCoV-19/India/MH-ILSGS01566/2021EPI_ISL_23429252021-03-24 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| 305 | hCoV-19/India/MH-ILSGS01585/2021EPI_ISL_23429332021-03-29 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| 306 | hCoV-19/India/MH-ILSGS01643/2021EPI_ISL_23429492021-04-03 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
| 307 | hCoV-19/India/MH-ILSGS01658/2021EPI_ISL_23429622021-04-04 | G | VUI B.1.617.3 | India | VUI B.1.617.3-IN |
The GS exhibit very small differences and, therefore, are difficult to distinguish. We can first characterize them by their length L that varies between minimum and maximum values of and symbols. Moreover, we have an average and standard deviation of and symbols, respectively. This small variability in the size is relevant for the reliability when comparing strings with the Kolmogorov- and Hamming-based metrics.
As mentioned before, the viral RNA information is represented by ASCII files with the four nitrogenous bases. Therefore, we can consider the grouping of , consecutive symbols. For simplicity, we denote the corresponding sub-strings by and obtain the statistics listed in Tables 2, 3 and 4. The term ‘others’ stands for a small number of other symbols distinct of the four nucleotide bases. Therefore, occasionally we find the symbols N, M, S, R, Y, K, H, V, and W, that abbreviate aNy (A, C, G or T), aMino (A or C), Strong interaction (3 H-bonds, G or C), puRine (A or G), pYrimidine (C or T), Keto (G or T), not-G (A, C or T), not-T (A, C or G), and Weak interaction (2 H-bonds, A or T), respectively.
Table 2.
Statistics of 1-tuples of symbols in the GS
| Symbol | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| A | 7352 | 8952 | 8805.5 | 204.5 | T | 7854 | 9609 | 9476.7 | 224.9 |
| C | 4497 | 5491 | 5409.8 | 126.9 | G | 4870 | 5863 | 5787.5 | 132.4 |
| Other | 0 | 5037 | 293.0 | 666.6 |
Table 3.
Statistics of 2-tuples of symbols in the GS
| l | l | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | 1207 | 1469 | 1405.3 | 43.8 | CA | 848 | 1053 | 1028.4 | 27 | TA | 945 | 1205 | 1174.3 | 32.4 | GA | 641 | 836 | 798.5 | 28.5 |
| AC | 834 | 1040 | 998.8 | 25 | CC | 353 | 448 | 434.1 | 12 | TC | 588 | 763 | 693.8 | 44.5 | GC | 489 | 606 | 579 | 17.9 |
| AT | 936 | 1200 | 1139.2 | 39.6 | CT | 829 | 1067 | 1020.3 | 29.6 | TT | 1312 | 1620 | 1589.3 | 40 | GT | 829 | 1030 | 989.4 | 30.3 |
| AG | 727 | 887 | 853.6 | 24.4 | CG | 180 | 235 | 219.2 | 11.2 | TG | 1073 | 1322 | 1278.6 | 32 | GG | 457 | 574 | 534 | 18.3 |
| other | 0 | 2525 | 148.9 | 336.3 |
Table 4.
Statistics of 3-tuples of symbols in the GS
| AAA | 242 | 329 | 290.8 | 19.2 | CAA | 193 | 246 | 230.3 | 10.2 | TAA | 77 | 345 | 271.8 | 85.7 | GAA | 85 | 255 | 166.2 | 54.6 |
| AAC | 169 | 216 | 203.7 | 9.6 | CAC | 98 | 194 | 156.9 | 28.4 | TAC | 154 | 233 | 208.5 | 19.6 | GAC | 66 | 143 | 110.9 | 27.3 |
| AAT | 164 | 317 | 242.4 | 44.8 | CAT | 116 | 178 | 164.3 | 15.2 | TAT | 112 | 282 | 217 | 65.5 | GAT | 50 | 214 | 140.2 | 58.9 |
| AAG | 97 | 248 | 174.9 | 62.5 | CAG | 76 | 188 | 130.4 | 43.8 | TAG | 76 | 211 | 145 | 45.3 | GAG | 70 | 121 | 92.1 | 12.5 |
| ACA | 214 | 291 | 258.1 | 18.4 | CCA | 90 | 139 | 112.5 | 12.6 | TCA | 143 | 190 | 178.9 | 8.1 | GCA | 82 | 169 | 117.7 | 21.8 |
| ACC | 89 | 151 | 130.4 | 18.8 | CCC | 28 | 45 | 39.7 | 5 | TCC | 48 | 88 | 68.7 | 9.3 | GCC | 39 | 73 | 60.5 | 10.3 |
| ACT | 142 | 292 | 213.3 | 42 | CCT | 80 | 149 | 108.3 | 19.4 | TCT | 117 | 219 | 173.8 | 30.8 | GCT | 75 | 266 | 158.3 | 58.6 |
| ACG | 36 | 66 | 55.9 | 7.2 | CCG | 13 | 30 | 25.8 | 4.2 | TCG | 25 | 43 | 35 | 4.4 | GCG | 16 | 39 | 26.6 | 5.7 |
| ATA | 93 | 188 | 144.5 | 29.6 | CTA | 90 | 269 | 170.3 | 71 | TTA | 191 | 363 | 280.7 | 56.6 | GTA | 102 | 175 | 146.8 | 24.6 |
| ATC | 88 | 123 | 111.7 | 6.5 | CTC | 60 | 116 | 92.2 | 16.5 | TTC | 120 | 211 | 180.5 | 29.2 | GTC | 56 | 105 | 88.7 | 15.9 |
| ATT | 166 | 289 | 243.8 | 38.4 | CTT | 170 | 275 | 234.5 | 30 | TTT | 247 | 387 | 338.3 | 39.5 | GTT | 139 | 311 | 219.1 | 52.4 |
| ATG | 95 | 315 | 207.9 | 89.8 | CTG | 64 | 266 | 152.7 | 78.6 | TTG | 149 | 364 | 255.3 | 79.7 | GTG | 78 | 279 | 167.9 | 81.1 |
| AGA | 112 | 277 | 221 | 54.4 | CGA | 19 | 42 | 32.9 | 3.8 | TGA | 61 | 306 | 241.7 | 81.6 | GGA | 59 | 119 | 95.4 | 19.2 |
| AGC | 47 | 141 | 111.9 | 31 | CGC | 24 | 41 | 34 | 6.2 | TGC | 81 | 270 | 204.6 | 61.2 | GGC | 44 | 97 | 74.8 | 18.9 |
| AGT | 127 | 202 | 174.2 | 22.8 | CGT | 44 | 68 | 54.8 | 5.7 | TGT | 193 | 370 | 306.7 | 58.8 | GGT | 52 | 246 | 136.8 | 62.4 |
| AGG | 71 | 136 | 111.3 | 17.7 | CGG | 16 | 33 | 25.9 | 4.5 | TGG | 90 | 264 | 205.1 | 57.2 | GGG | 30 | 52 | 46.6 | 6.4 |
| other | 0 | 1689 | 100.4 | 225.6 |
Figure 1 shows the cumulative numbers of cases for sub-strings including symbols in the GS.
Fig. 1.
Cumulative numbers of cases for sub-strings including symbols in the GS
As a complementary analysis of the relationship between GS we use the VOSviewer software tool [46–50] for constructing and visualizing bibliometric networks (https://www.vosviewer.com/). The program was built having in mind scientometric applications, but can be used in the present case if we consider the associations of symbols as keywords in a standard technical text. For that purpose we construct -tuples of consecutive symbols in the GS in order to have ‘words’ (i.e., sub-strings) with consecutive symbols. We considered the VOSviewer options ‘Full counting’, ‘Minimum number of occurrences of a term = 5’ and ‘Number of terms selected = 28’. For example, Fig. 2 shows the network for the sub-strings with consecutive symbols in the genomic sequence. For the other virus the results are of the same type. We observe the very small relevance of ‘phrases’ with several triplets and, on the other hand, the complex network relationship between triplets.
Fig. 2.
Network of the sub-strings with symbols extracted from the GS of case
From the Tables 2, 3 and 4 and Figs. 1 and 2 we verify that the case of symbols represent a good compromise between complexity and accurate description of the information content. This conclusion follows previous observations with genetic data [19, 51–53].
The existence of the symbols classified as ‘other’ and the variation of size in the GS can be considered as a kind of noise. However, as verified with the previous tests they reflect in very small numbers. Also, in most cases we considered about 10 GS for each type of virus. Therefore, we can proceed with the analysis of the genetic information knowing its robustness against possible volatility in the data.
The mathematical description of the viral information is based on the Kolmogorov, Shannon and Hamming perspectives implemented by the distances , and presented in the Appendix. The MDS clustering and visualization is used to unravel relationships between the data and to identify possible patterns. In the case of the Kolmogorov complexity we consider the compressor BZip2 (https://www.zlib.net). In the case of the Shannon information, we start by calculating the 64-bin histograms (i.e., the triplets {AAA, AAC, , GGT, GGG}) for the triplets () of the nitrogenous bases and then we calculate the distances between them. The resulting matrix D, dimensional, that is processed by MDS using the Matlab command cmdscale.
Data-set analysis: clustering results
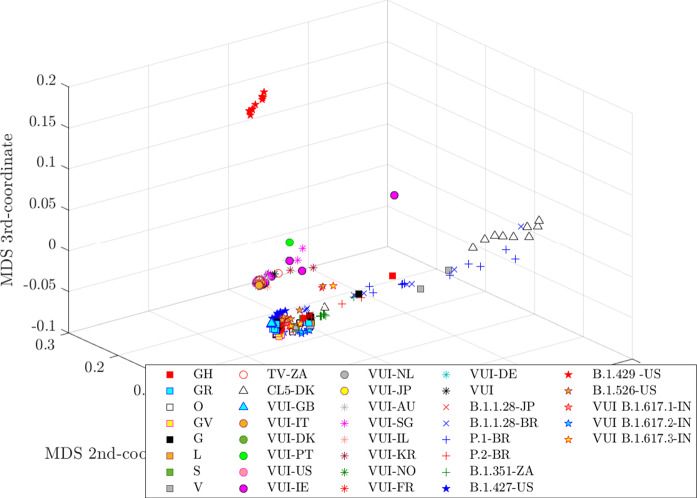
The distances following the Kolmogorov theory yield almost similar MDS plots. This behavior was also observed in previous studies with distinct data-sets [54]. In what concerns the distances based on the Shannon theory we note that produces a slightly different plot from the group , which return charts having just small differences. On the other hand, the distance leads to a very different chart. Therefore, for parsimony, for these sets of distances we depict just the plots for the NCD, , and .
The MDS charts produced by the four distances are represented in Fig. 3, 4, 5 and 6, respectively. In all cases the MDS plots require a careful rotation to get the correct 3-dimensional perspective and assessment, since the planar projections in the figures are not totally capable of depicting their structure.
Fig. 3.
One perspective of the MDS 3-dim locus for the set of virus using the normalized information distance, NCD, and BZip2
Fig. 4.
One perspective of the MDS 3-dim locus for the set of virus using the Jaccard distance, , based on the sub-strings with consecutive symbols
Fig. 5.
One perspective of the MDS 3-dim locus for the set of virus using the Jensen-Shannon divergence, , based on the sub-strings with consecutive symbols
Fig. 6.
One perspective of the MDS 3-dim locus for the set of virus using the Hamming distance, , based on the sub-strings with consecutive symbols
Several MDS loci reveal different clusters, but, in general we do not have a clear group for each variant of the virus. The Kolmogorov- and the Shannon-based metrics show some clusters, but with a mixture of many variants, while the Hamming scheme gives the worst map in the perspective of clustering. Nonetheless, we can ask a different and more relevant question which is how to assess the ‘dynamics’ of the evolutionary process. We must note that the available information just reflects ‘time samples’ of the variants, that is, the date where the procedure for collecting, identifying and recording the GS took place. Consequently, we do not have a precise control of the time elapsed between the real mutation and the laboratory measurement. Nonetheless, we can have a good idea of the dynamical behavior if we include time information in the MDS plots, even if some ‘noise’ is present in the time information.
Figures 7, 8 , 9 and 10 depict the MDS plots with the time information represented by the colors of the marks. The colorbar with the interval corresponds to the period between the dates 2020/Feb/24 and 2021/Apr/23. We observe some improvement over the initial set of experiments, but we have still some ‘noisy’ behavior of the time flow.
Fig. 7.
MDS 3-dim locus exhibiting the dynamics in time of the virus evolution during the period 2020/Feb/24-2021/Apr/23 for the set of virus using the normalized information distance, NCD, and BZip2
Fig. 8.
MDS 3-dim locus exhibiting the dynamics in time of the virus evolution during the period 2020/Feb/24-2021/Apr/23 for the set of virus using the Jaccard distance, , based on the sub-strings with consecutive symbols
Fig. 9.
MDS 3-dim locus exhibiting the dynamics in time of the virus evolution during the period 2020/Feb/24-2021/Apr/23 for the set of virus using the Jensen-Shannon divergence, , based on the sub-strings with consecutive symbols
Fig. 10.
MDS 3-dim locus exhibiting the dynamics in time of the virus evolution during the period 2020/Feb/24-2021/Apr/23 for the set of virus using the Hamming distance, , based on the sub-strings with consecutive symbols
It is well known that each distance captures a given characteristic of the phenomenon under analysis. Therefore, we wander if some measure associating several distances could reveal better the patterns embedded in the datataset. In this line of thought we can design a ‘generalized’ distance by weighting several of the previous distances. If we consider the distances NCD, , and , then we can define the new metric:
| 1 |
where , , and are weight factors for the distances NCD, , and , respectively, so that .
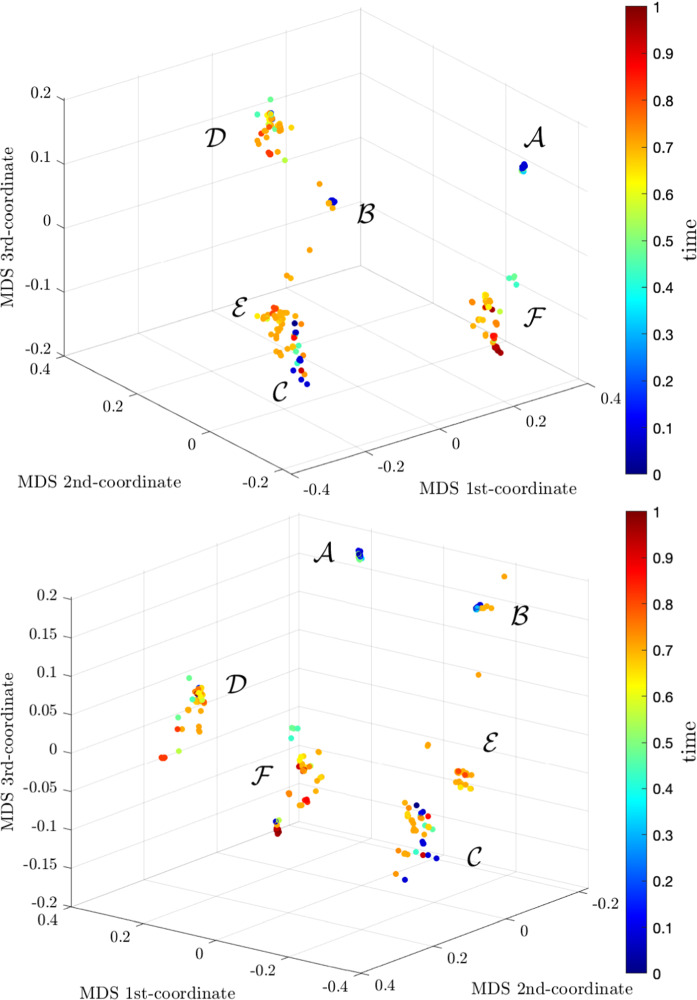
We can adjust the numerical values of the wight factor to reflect the importance of each distance for improving the MDS representation in the sense of providing a more clear visualization of the dynamical effect. In our case, after some experiments, and having in mind the dynamics in time, we consider the weights , , and . Figure 11 shows the MDS plot where we observe the emergence of five clusters denoted by the symbols to . The time ‘arrow’ follows somehow the pattern .
Fig. 11.
Two perspectives of the MDS 3-dim locus exhibiting the dynamics in time of the virus evolution during the period 2020/Feb/24-2021/Apr/23 for the set of virus using the Generalized distance, . The time ‘arrow’ follows the pattern
We verify (i) the first and second clusters, and , exhibit a very low scattering in contrast with the others, (ii) the existence of some noise in the evolutionary process, (iii) that time flows in discrete steps in the MDS representation, that is, the time evolution is not continuous, (iv) that we can have some evolutionary bifurcations in time as it is visible for , and (v) clusters for different time instants may be close in the MDS locus. A deep reflection upon these results seems consistent with our present knowledge of the evolutionary process. In fact, we can interpret and justify the previous assertions as follows:
the initial strains of virus had very similar characteristics, contrary to the more recent variants that reveal an increasing variability, which agrees with the first observation
evolution and, in particular, mutations, occur randomly which justifies the second conclusion
mutations do not emerge continuously in time and, in fact, they are the result of a multitude of issues such as environmental, social, geographical and economical factors. Therefore, new variants that lead to a relevant number of infections are expected to emerge without a clear time pattern which is in accordance with the third consideration
the infection spreads in very different space locations and it is reasonable to expect to have several important variants at the same time, particularly when the number of infected people grows considerably. These considerations support the forth remark
the close location of some clusters for non- consecutive time samples (e.g., and ) can be interpreted as the MDS representation the infection ‘waves’, which explain the fifth observation.
It is important to analyze the effect of the clustering algorithm, the dimension of the visualization space and the type of representation. In this perspective, the Generalized distance (1) and, consequently, the same matrix D used for the MDS scheme, is now used with the set of programs Phylip [55, 56] available at http://evolution.genetics.washington.edu/phylip.html. This program is used in in phylogenetics for displaying the evolutionary relationships among various biological objects. The program allows 2-dim representations where the final objects are the ‘leafs’ of some type of tree. The algorithm neighbor processes the matrix D and produces the data clustering. The programs drawgram and drawtree are used for obtaining the graphical representations in the form of dendrograms and trees, respectively.
Figures 12 and 13 shows the dendrogram and tree generated by Phylip for the generalized distance, , respectively. The time evolution is represented by colors as before.
Fig. 12.
Dendrogram representation of the SARS-COV2 time dynamics during the period 2020/Feb/24-2021/Apr/23 for GS using the generalized distance,
Fig. 13.
Tree representation of the SARS-COV2 time dynamics during the period 2020/Feb/24-2021/Apr/23 for GS using the generalized distance,
The 2-dim graphical representations are easier to visualize in the sense that the user does not needs to rotate and shift the plot. However, the lack of the third dimension leads to a clustering somewhat inferior to the one performed by the 3-dim MDS. Nonetheless, we can still see some clusters. In the case of the dendrogram we observe a simple evolution from top (initial period) to bottom (final period) with 3 main clusters. The two clusters at the top do not show a precise separation of the periods of time since with have overlapping object for the initial 60% of the total period. In the case of a graphical output my means of a tree we have a more intricate pattern, with the objects placed in the form of two ‘arcs’. The outer arc covers the period of time from beginning up to approximately 75%, while the inner arc corresponds to the rest, that is, to the more recent 25% GS. Moreover, the initial period of time of about 30% is place it the top of the outer arc.
In summary, the strategy followed in this paper is consistent with present-day understanding about the SARS-CoV-2 genome and the adoption of several distinct distances allows users to have a complementary interpretation of the information embedded in the data-set.
Conclusions
The information of 307 RNA viruses available in a public database was explored by means of an association of mathematical and computational tools. The notions Kolmogorov complexity, Shannon information and Hamming distance, on the perspective of analytic tools, and the ideas of compression and MDS algorithms, on the point of view of computational tools, were considered. Three sets of indices of dissimilarity were adopted, allowing a broad comparison of the results, with four distances based on the BZip2 compression, four distances using 2-dimensional histograms of consecutive triplets, and one metric using the Hamming distance between triplets of bases. From these, the MDS algorithm allowed an efficient clustering and 3-dim visualization. The MDS plots revealed pros and cons of the alternative distances adopted for assessing the set of viruses. The problem at stake proved to pose a considerable challenge and no clear clusters emerged for the virus variants included in the dataset. This motivated a new question, namely the relevance of clustering the variants when thinking about its evolutionary dynamics, since many variants have minor differences between themselves. The idea of assessing the dynamics in time lead to the design of a generalized distance taking advantage of the characteristics of distinct metrics and allowing a adjustment to the phenomenon by means of weight factors. The results showed interesting dynamical effects in terms of forming clear clusters in time. Besides the analytic tools, several algorithmic mechanisms were explored, such as the Matlab, VOSviwer and Phylip programs, which illustrate the diversity and richness of present day computational strategies. The synergistic perspectives provided by distinct processing tools and graphical representations allow comparing the genomic data and provide a computational strategy for exploring future viral outbreaks. The association of analytic and computational techniques may help interpreting the phylogeny of these new strain outbreaks, associate its dynamics and selective pressures, and give additional insight for the quick development and testing of tailored countermeasures. The computational analysis of the SARS-CoV-2 genome may have a role in the early detection of potential variants of concern and help in the characterization of the risk posed to global public health. This is important as it may contribute to the global monitoring of SARS-CoV-2 variants and to improve the search for a more effective response to the COVID-19 pandemic.
Acknowledgements
The authors thank all those who have contributed and shared sequences to the GISAID database available at https://www.gisaid.org/.
Appendix
This appendix describes the metrics adopted in the paper. The Kolmogorov complexity and Shannon information theories are revisited and several distances are recalled. Additionally, the Hamming distance is also included.
Kolmogorov complexity
The Kolmogorov complexity tackles the assessment of information without considering probabilistic notions [57]. Given a string x, the Kolmogorov complexity can be defined as ‘the length of the shortest program that, given an empty string , can compute x and then halts’. Consequently, the Kolmogorov complexity can be somehow viewed as the length of the compressed form of the string x.
The information distance between two strings [22, 23] leads to concepts such as, normalized information distance (NCD) [58, 59], Chen-Li Metric (CLM) [60–62], Compression-based Dissimilarity Measure (CDM) [63] and Compression-based Cosine (CosS) [54], given by:
| 2a |
| 2b |
| 2c |
| 2d |
The operator C(x) represents the length (measured as a number of bits) of string x after being compressed by a given algorithm . The concatenation of two strings, x and y, is denoted as xy and, therefore, C(xy) gives the number of bits when compressing together x and y. Moreover, the notation C(x|y) stands for the length of x when conditionally compressed with a model built on string y.
Sculley and Brodley [54] noted that the compression has an associated feature space, . Therefore, they map a string x into a vector and . After some simplifications, we verify that the four compression distances (2) use the expression since we can write
| 3a |
| 3b |
| 3c |
| 3d |
The four indices reduce to the form 1, where the normalizing term varies for each case.
Shannon information
In the scope of the Information theory [64], the so-called information content I of an event with probability is defined as:
| 4 |
where X denotes a discrete random variable.
The expected value of the information, E(I), named Shannon entropy [65, 66], becomes:
| 5 |
that follows the four Khinchin axioms [67, 68].
In the case of a two-dimensional distribution we have the joint entropy and mutual information given by:
| 6 |
| 7 |
Several distances can be defined from these mathematical tools. In the follow-up we adopt the Jaccard and Jensen-Shannon distances, and respectively.
The Jaccard distance represents a set-theoretic interpretation of information based on the normalized distance and is formulated as:
| 8 |
The Jensen-Shannon divergence is the symmetric version of the Kullback-Leibler divergence and is given by:
| 9 |
where .
The expression of can be rewritten as:
| 10 |
In the Shannon’s entropy family we include also the Jeffreys () and Topsøe () expressions given by:
| 11 |
| 12 |
Hamming distance
We adopt also a distance based on standard concepts for comparing the symbols forming the genetic sequences (GS). In information processing, the Hamming metric between two strings of equal length is given by the number of positions at which the corresponding symbols are different [26, 69]. Therefore, we start by considering triplets of consecutive symbols in each string. This means that we form sub-strings with successive symbols in the DNA strand, so that for a genetic sequence x with length we obtain of -tuples. The assessment of similarity between the k-th character, , in the i-th pair of sub-strings (i.e., the and symbols in the two sub-strings i extracted from the x and y sequences), is then based on the concept of Hamming metric:
| 13 |
where the indices k and i stand for the k-th symbol in the i-th sub-string (, and ), respectively. In general the strings x and y have slightly different lengths and we simply adopt the length . For calculating the total distance between two strings we test the ‘normalized’ version of the Hamming distance:
| 14 |
where and the term ‘normalization’ means that the final values are given in relation to the total number of compared sub-strings, .
Data availability
The reformatted data that support the findings of this study are available in http://ave.dee.isep.ipp.pt/jtm/FILES/datasets/Archive.zip.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
J. A. Tenreiro Machado, Email: jtm@isep.ipp.pt.
J. M. Rocha-Neves, Email: joaorochaneves@hotmail.com
Filipe Azevedo, Email: fta@isep.ipp.pt.
J. P. Andrade, Email: jandrade@med.up.pt
References
- 1.Dowd JB, Andriano L, Brazel DM, Rotondi V, Block P, Ding X, Liu Y, Mills MC. Demographic science aids in understanding the spread and fatality rates of COVID-19. Proc. Natl. Acad. Sci. 2020;117(18):9696–9698. doi: 10.1073/pnas.2004911117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020 doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92(5):522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mallapaty S. COVID mink analysis shows mutations are not dangerous - yet. Nature. 2020;587(7834):340–341. doi: 10.1038/d41586-020-03218-z. [DOI] [PubMed] [Google Scholar]
- 5.Hamed SM, Elkhatib WF, Khairalla AS, Noreddin AM. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci. Rep. 2021 doi: 10.1038/s41598-021-87713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rambaut, A., Loman, N., Pybus, O., Barclay, W., Barrett, J., Carabelli, A., Connor, T., Peacock, T., Robertson, D.L., (on behalf of COVID-19 Genomics Consortium UK (CoG-UK)9, E.V.: Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (2020). [Online; posted 18-December-2020]
- 7.Frampton, D., Rampling, T., Cross, A., Bailey, H., Heaney, J., Byott, M., Scott, R., Sconza, R., Price, J., Margaritis, M., Bergstrom, M., Spyer, M.J., Miralhes, P.B., Grant, P., Kirk, S., Valerio, C., Mangera, Z., Prabhahar, T., Moreno-Cuesta, J., Arulkumaran, N., Singer, M., Shin, G.Y., Sanchez, E., Paraskevopoulou, S.M., Pillay, D., McKendry, R.A., Mirfenderesky, M., Houlihan, C.F., Nastouli, E.: Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. The Lancet Infectious Diseases (2021). 10.1016/s1473-3099(21)00170-5 [DOI] [PMC free article] [PubMed]
- 8.Tegally, H., Wilkinson, E., Giovanetti, M., Iranzadeh, A., Fonseca, V., Giandhari, J., Doolabh, D., Pillay, S., San, E.J., Msomi, N., Mlisana, K., von Gottberg, A., Walaza, S., Allam, M., Ismail, A., Mohale, T., Glass, A.J., Engelbrecht, S., Van Zyl, G., Preiser, W., Petruccione, F., Sigal, A., Hardie, D., Marais, G., Hsiao, M., Korsman, S., Davies, M.A., Tyers, L., Mudau, I., York, D., Maslo, C., Goedhals, D., Abrahams, S., Laguda-Akingba, O., Alisoltani-Dehkordi, A., Godzik, A., Wibmer, C.K., Sewell, B.T., Lourenço, J., Alcantara, L.C.J., Pond, S.L.K., Weaver, S., Martin, D., Lessells, R.J., Bhiman, J.N., Williamson, C., de Oliveira, T.: Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv (2020). 10.1101/2020.12.21.20248640. https://www.medrxiv.org/content/early/2020/12/22/2020.12.21.20248640
- 9.Long SW, Olsen RJ, Christensen PA, Subedi S, Olson R, Davis JJ, Saavedra MO, Yerramilli P, Pruitt L, Reppond K, Shyer MN, Cambric J, Finkelstein IJ, Gollihar J, Musser JM. Sequence analysis of 20, 453 severe acute respiratory syndrome coronavirus 2 genomes from the Houston metropolitan area identifies the emergence and widespread distribution of multiple isolates of all major variants of concern. Am. J. Pathol. 2021 doi: 10.1016/j.ajpath.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, N., Barnard, R.C., Jarvis, C.I., Kucharski, A.J., Munday, J.D., Pearson, C.A., Russell, T.W., Tully, D.C., Abbott, S., Gimma, A., Waites, W., Wong, K.L., van Zandvoort, K., working group, C., Eggo, R.M., Funk, S., Jit, M., Atkins, K.E., Edmunds, W.J.: Estimated transmissibility and severity of novel SARS-CoV-2 variant of concern 2020/12/01 in England. https://cmmid.github.io/topics/covid19/uk-novel-variant.html (2020). [First online: 23-12-2020, Last update: 03-03-2021]
- 11.Brown, C.M., Vostok, J., Johnson, H., Burns, M., Gharpure, R., Sami, S., Sabo, R.T., Hall, N., Foreman, A., Schubert, P.L., Gallagher, G.R., Fink, T., Madoff, L.C., Gabriel, S.B., MacInnis, B., Park, D.J., Siddle, K.J., Harik, V., Arvidson, D., Brock-Fisher, T., Dunn, M., Kearns, A., Laney, A.S.: Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings – Barnstable county, Massachusetts, July 2021. MMWR. Morbidity and Mortality Weekly Report 70(31) (2021). 10.15585/mmwr.mm7031e2 [DOI] [PMC free article] [PubMed]
- 12.Li, B., Deng, A., Li, K., Hu, Y., Li, Z., Xiong, Q., Liu, Z., Guo, Q., Zou, L., Zhang, H., Zhang, M., Ouyang, F., Su, J., Su, W., Xu, J., Lin, H., Sun, J., Peng, J., Jiang, H., Zhou, P., Hu, T., Luo, M., Zhang, Y., Zheng, H., Xiao, J., Liu, T., Che, R., Zeng, H., Zheng, Z., Huang, Y., Yu, J., Yi, L., Wu, J., Chen, J., Zhong, H., Deng, X., Kang, M., Pybus, O.G., Hall, M., Lythgoe, K.A., Li, Y., Yuan, J., He, J., Lu, J.: Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 delta variant (2021). 10.1101/2021.07.07.21260122
- 13.Allen, H., Vusirikala, A., Flannagan, J., Twohig, K.A., Zaidi, A., Groves, N., Lopez-Bernal, J., Harris, R., Charlett, A., Dabrera, G., Kall, M.: Increased household transmission of COVID-19 cases associated with SARS-CoV-2 variant of concern B.1.617.2: a national case-control study (2021). https://www.gov.uk/government/collections/new-sars-cov-2-variant
- 14.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Marc GP, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Özlem, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. New Eng. J. Med. 2020 doi: 10.1056/nejmoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voysey, M., Clemens, S.A.C., Madhi, S.A., Weckx, L.Y., Folegatti, P.M., Aley, P.K., Angus, B., Baillie, V.L., Barnabas, S.L., Bhorat, Q.E., Bibi, S., Briner, C., Cicconi, P., Collins, A.M., Colin-Jones, R., Cutland, C.L., Darton, T.C., Dheda, K., Duncan, C.J.A., Emary, K.R.W., Ewer, K.J., Fairlie, L., Faust, S.N., Feng, S., Ferreira, D.M., Finn, A., Goodman, A.L., Green, C.M., Green, C.A., Heath, P.T., Hill, C., Hill, H., Hirsch, I., Hodgson, S.H.C., Izu, A., Jackson, S., Jenkin, D., Joe, C.C.D., Kerridge, S., Koen, A., Kwatra, G., Lazarus, R., Lawrie, A.M., Lelliott, A., Libri, V., Lillie, P.J., Mallory, R., Mendes, A.V.A., Milan, E.P., Minassian, A.M., McGregor, A., Morrison, H., Mujadidi, Y.F., Nana, A., O’Reilly, P.J., Padayachee, S.D., Pittella, A., Plested, E., Pollock, K.M., Ramasamy, M.N., Rhead, S., Schwarzbold, A.V., Singh, N., Smith, A., Song, R., Snape, M.D., Sprinz, E., Sutherland, R.K., Tarrant, R., Thomson, E.C., Török, M.E., Toshner, M., Turner, D.P.J., Vekemans, J., Villafana, T.L., Watson, M.E.E., Williams, C.J., Douglas, A.D., Hill, A.V.S., Lambe, T., Gilbert, S.C., Pollard, A.J., Aban, M., Abayomi, F., Abeyskera, K., Aboagye, J., Adam, M., Adams, K., Adamson, J., Adelaja, Y.A., Adlou, S., Ahmed, K., Akhalwaya, Y., Akhalwaya, S., Alcock, A., Ali, A., Allen, E.R., Allen, L., Almeida, T.C.D.S.C., Alves, M.P., Amorim, F., Andritsou, F., Anslow, R., Appleby, M., Arbe-Barnes, E.H., Ariaans, M.P., Arns, B., Arruda, L., Awedetan, G., Azi, P., Azi, L., Babbage, G., Bailey, C., Baker, K.F., Baker, M., Baker, N., Baker, P., Baldwin, L., Baleanu, I., Bandeira, D., Bara, A., Barbosa, M.A., Barker, D., Barlow, G.D., Barnes, E., Barr, A.S., Barrett, J.R., Barrett, J., Bates, L., Batten, A., Beadon, K., Beales, E., Beckley, R., Belij-Rammerstorfer, S., Bell, J., Bellamy, D., Bellei, N., Belton, S., Berg, A., Bermejo, L., Berrie, E., Berry, L., Berzenyi, D., Beveridge, A., Bewley, K.R., Bexhell, H., Bhikha, S., Bhorat, A.E., Bhorat, Z.E., Bijker, E., Birch, G., Birch, S., Bird, A., Bird, O., Bisnauthsing, K., Bittaye, M., Blackstone, K., Blackwell, L., Bletchly, H., Blundell, C.L., Blundell, S.R., Bodalia, P., Boettger, B.C., Bolam, E., Boland, E., Bormans, D., Borthwick, N., Bowring, F., Boyd, A., Bradley, P., Brenner, T., Brown, P., Brown, C., Brown-O-Sullivan, C., Bruce, S., Brunt, E., Buchan, R., Budd, W., Bulbulia, Y.A., Bull, M., Burbage, J., Burhan, H., Burn, A., Buttigieg, K.R., Byard, N., Puig, I.C., Calderon, G., Calvert, A., Camara, S., Cao, M., Cappuccini, F., Cardoso, J.R., Carr, M., Carroll, M.W., Carson-Stevens, A., de M. Carvalho, Y., Carvalho, J.A., Casey, H.R., Cashen, P., Castro, T., Castro, L.C., Cathie, K., Cavey, A., Cerbino-Neto, J., Chadwick, J., Chapman, D., Charlton, S., Chelysheva, I., Chester, O., Chita, S., Cho, J.S., Cifuentes, L., Clark, E., Clark, M., Clarke, A., Clutterbuck, E.A., Collins, S.L., Conlon, C.P., Connarty, S., Coombes, N., Cooper, C., Cooper, R., Cornelissen, L., Corrah, T., Cosgrove, C., Cox, T., Crocker, W.E., Crosbie, S., Cullen, L., Cullen, D., Cunha, D.R., Cunningham, C., Cuthbertson, F.C., Guarda, S.N.F.D., da Silva, L.P., Damratoski, B.E., Danos, Z., Dantas, M.T., Darroch, P., Datoo, M.S., Datta, C., Davids, M., Davies, S.L., Davies, H., Davis, E., Davis, J., Davis, J., Nobrega, M.M.D., Kalid, L.M.D.O., Dearlove, D., Demissie, T., Desai, A., Marco, S.D., Maso, C.D., Dinelli, M.I., Dinesh, T., Docksey, C., Dold, C., Dong, T., Donnellan, F.R., Santos, T.D., dos Santos, T.G., Santos, E.P.D., Douglas, N., Downing, C., Drake, J., Drake-Brockman, R., Driver, K., Drury, R., Dunachie, S.J., Durham, B.S., Dutra, L., Easom, N.J., van Eck, S., Edwards, M., Edwards, N.J., Muhanna, O.M.E., Elias, S.C., Elmore, M., English, M., Esmail, A., Essack, Y.M., Farmer, E., Farooq, M., Farrar, M., Farrugia, L., Faulkner, B., Fedosyuk, S., Felle, S., Feng, S., Silva, C.F.D., Field, S., Fisher, R., Flaxman, A., Fletcher, J., Fofie, H., Fok, H., Ford, K.J., Fowler, J., Fraiman, P.H., Francis, E., Franco, M.M., Frater, J., Freire, M.S., Fry, S.H., Fudge, S., Furze, J., Fuskova, M., Galian-Rubio, P., Galiza, E., Garlant, H., Gavrila, M., Geddes, A., Gibbons, K.A., Gilbride, C., Gill, H., Glynn, S., Godwin, K., Gokani, K., Goldoni, U.C., Goncalves, M., Gonzalez, I.G., Goodwin, J., Goondiwala, A., Gordon-Quayle, K., Gorini, G., Grab, J., Gracie, L., Greenland, M., Greenwood, N., Greffrath, J., Groenewald, M.M., Grossi, L., Gupta, G., Hackett, M., Hallis, B., Hamaluba, M., Hamilton, E., Hammersley, D., Hanrath, A.T., Hanumunthadu, B., Harris, S.A., Harris, C., Harris, T., Harrison, T.D., Harrison, D., Hart, T.C., Hartnell, B., Hassan, S., Haughney, J., Hawkins, S., Hay, J., Head, I., Henry, J., Herrera, M.H., Hettle, D.B., Hill, J., Hodges, G., Horne, E., Hou, M.M., Houlihan, C., Howe, E., Howell, N., Humphreys, J., Humphries, H.E., Hurley, K., Huson, C., Hyder-Wright, A., Hyamns, C., Ikram, S., Ishwarbhai, A., Ivan, M., Iveson, P., Iyer, V., Jackson, F., Jager, J.D., Jaumdally, S., Jeffers, H., Jesudason, N., Jones, B., Jones, K., Jones, E., Jones, C., Jorge, M.R., Jose, A., Joshi, A., Júnior, E.A., Kadziola, J., Kailath, R., Kana, F., Karampatsas, K., Kasanyinga, M., Keen, J., Kelly, E.J., Kelly, D.M., Kelly, D., Kelly, S., Kerr, D., de Ávila Kfouri, R., Khan, L., Khozoee, B., Kidd, S., Killen, A., Kinch, J., Kinch, P., King, L.D., King, T.B., Kingham, L., Klenerman, P., Knapper, F., Knight, J.C., Knott, D., Koleva, S., Lang, M., Lang, G., Larkworthy, C.W., Larwood, J.P., Law, R., Lazarus, E.M., Leach, A., Lees, E.A., Lemm, N.M., Lessa, A., Leung, S., Li, Y., Lias, A.M., Liatsikos, K., Linder, A., Lipworth, S., Liu, S., Liu, X., Lloyd, A., Lloyd, S., Loew, L., Ramon, R.L., Lora, L., Lowthorpe, V., Luz, K., MacDonald, J.C., MacGregor, G., Madhavan, M., Mainwaring, D.O., Makambwa, E., Makinson, R., Malahleha, M., Malamatsho, R., Mallett, G., Mansatta, K., Maoko, T., Mapetla, K., Marchevsky, N.G., Marinou, S., Marlow, E., Marques, G.N., Marriott, P., Marshall, R.P., Marshall, J.L., Martins, F.J., Masenya, M., Masilela, M., Masters, S.K., Mathew, M., Matlebjane, H., Matshidiso, K., Mazur, O., Mazzella, A., McCaughan, H., McEwan, J., McGlashan, J., McInroy, L., McIntyre, Z., McLenaghan, D., McRobert, N., McSwiggan, S., Megson, C., Mehdipour, S., Meijs, W., Mendonça, R.N., Mentzer, A.J., Mirtorabi, N., Mitton, C., Mnyakeni, S., Moghaddas, F., Molapo, K., Moloi, M., Moore, M., Moraes-Pinto, M.I., Moran, M., Morey, E., Morgans, R., Morris, S., Morris, S., Morris, H.C., Morselli, F., Morshead, G., Morter, R., Mottal, L., Moultrie, A., Moya, N., Mpelembue, M., Msomi, S., Mugodi, Y., Mukhopadhyay, E., Muller, J., Munro, A., Munro, C., Murphy, S., Mweu, P., Myasaki, C.H., Naik, G., Naker, K., Nastouli, E., Nazir, A., Ndlovu, B., Neffa, F., Njenga, C., Noal, H., Noé, A., Novaes, G., Nugent, F.L., Nunes, G., O-Brien, K., O-Connor, D., Odam, M., Oelofse, S., Oguti, B., Olchawski, V., Oldfield, N.J., Oliveira, M.G., Oliveira, C., Oosthuizen, A., O-Reilly, P., Osborne, P., Owen, D.R., Owen, L., Owens, D., Owino, N., Pacurar, M., Paiva, B.V., Palhares, E.M., Palmer, S., Parkinson, S., Parracho, H.M., Parsons, K., Patel, D., Patel, B., Patel, F., Patel, K., Patrick-Smith, M., Payne, R.O., Peng, Y., Penn, E.J., Pennington, A., Alvarez, M.P.P., Perring, J., Perry, N., Perumal, R., Petkar, S., Philip, T., Phillips, D.J., Phillips, J., Phohu, M.K., Pickup, L., Pieterse, S., Piper, J., Pipini, D., Plank, M., Plessis, J.D., Pollard, S., Pooley, J., Pooran, A., Poulton, I., Powers, C., Presa, F.B., Price, D.A., Price, V., Primeira, M., Proud, P.C., Provstgaard-Morys, S., Pueschel, S., Pulido, D., Quaid, S., Rabara, R., Radford, A., Radia, K., Rajapaska, D., Rajeswaran, T., Ramos, A.S.F., Lopez, F.R., Rampling, T., Rand, J., Ratcliffe, H., Rawlinson, T., Rea, D., Rees, B., Reiné, J., Resuello-Dauti, M., Pabon, E.R., Ribiero, C.M., Ricamara, M., Richter, A., Ritchie, N., Ritchie, A.J., Robbins, A.J., Roberts, H., Robinson, R.E., Robinson, H., Rocchetti, T.T., Rocha, B.P., Roche, S., Rollier, C., Rose, L., Russell, A.L.R., Rossouw, L., Royal, S., Rudiansyah, I., Ruiz, S., Saich, S., Sala, C., Sale, J., Salman, A.M., Salvador, N., Salvador, S., Sampaio, M., Samson, A.D., Sanchez-Gonzalez, A., Sanders, H., Sanders, K., Santos, E., Guerra, M.F.S., Satti, I., Saunders, J.E., Saunders, C., Sayed, A., van der Loeff, I.S., Schmid, A.B., Schofield, E., Screaton, G., Seddiqi, S., Segireddy, R.R., Senger, R., Serrano, S., Shah, R., Shaik, I., Sharpe, H.E., Sharrocks, K., Shaw, R., Shea, A., Shepherd, A., Shepherd, J.G., Shiham, F., Sidhom, E., Silk, S.E., da Silva Moraes, A.C., Silva-Junior, G., Silva-Reyes, L., Silveira, A.D., Silveira, M.B., Sinha, J., Skelly, D.T., Smith, D.C., Smith, N., Smith, H.E., Smith, D.J., Smith, C.C., Soares, A., Soares, T., Solórzano, C., Sorio, G.L., Sorley, K., Sosa-Rodriguez, T., Souza, C.M., Souza, B.S., Souza, A.R., Spencer, A.J., Spina, F., Spoors, L., Stafford, L., Stamford, I., Starinskij, I., Stein, R., Steven, J., Stockdale, L., Stockwell, L.V., Strickland, L.H., Stuart, A.C., Sturdy, A., Sutton, N., Szigeti, A., Tahiri-Alaoui, A., Tanner, R., Taoushanis, C., Tarr, A.W., Taylor, K., Taylor, U., Taylor, I.J., Taylor, J., te Water Naude, R., Themistocleous, Y., Themistocleous, A., Thomas, M., Thomas, K., Thomas, T.M., Thombrayil, A., Thompson, F., Thompson, A., Thompson, K., Thompson, A., Thomson, J., Thornton-Jones, V., Tighe, P.J., Tinoco, L.A., Tiongson, G., Tladinyane, B., Tomasicchio, M., Tomic, A., Tonks, S., Tran, N., Tree, J., Trillana, G., Trinham, C., Trivett, R., Truby, A., Tsheko, B.L., Turabi, A., Turner, R., Turner, C., Ulaszewska, M., Underwood, B.R., Varughese, R., Verbart, D., Verheul, M., Vichos, I., Vieira, T., Waddington, C.S., Walker, L., Wallis, E., Wand, M., Warbick, D., Wardell, T., Warimwe, G., Warren, S.C., Watkins, B., Watson, E., Webb, S., Webb-Bridges, A., Webster, A., Welch, J., Wells, J., West, A., White, C., White, R., Williams, P., Williams, R.L., Winslow, R., Woodyer, M., Worth, A.T., Wright, D., Wroblewska, M., Yao, A., Zimmer, R., Zizi, D., Zuidewind, P.: Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet (2020). 10.1016/s0140-6736(20)32661-1
- 16.Knoll MD, Wonodi C. Oxford-AstraZeneca COVID-19 vaccine efficacy. Lancet. 2020 doi: 10.1016/s0140-6736(20)32623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machado, J.A.T., Lopes, A.M.: Rare and extreme events: the case of COVID-19 pandemic. Nonlinear Dyn. 100(3), 2953–2972 (2020). 10.1007/s11071-020-05680-w [DOI] [PMC free article] [PubMed]
- 18.Lopes AM, Andrade JP, Machado JT. Multidimensional scaling analysis of virus diseases. Comput. Methods Progr. Biomed. 2016;131:97–110. doi: 10.1016/j.cmpb.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machado JAT, Rocha-Neves JM, Andrade JP. Computational analysis of the SARS-CoV-2 and other viruses based on the Kolmogorov’s complexity and Shannon’s information theories. Nonlinear Dyn. 2020;101(3):1731–1750. doi: 10.1007/s11071-020-05771-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machado JT, Lopes AM. A computational perspective of the periodic table of elements. Commun. Nonlinear Sci. Num. Simul. 2019;78:104883. doi: 10.1016/j.cnsns.2019.104883. [DOI] [Google Scholar]
- 21.Machado JT, Lopes AM. Multidimensional scaling and visualization of patterns in prime numbers. Commun. Nonlinear Sci. Num. Simul. 2020;83:105128. doi: 10.1016/j.cnsns.2019.105128. [DOI] [Google Scholar]
- 22.Bennett CH, Gács P, Li M, Vitányi P, Zurek WH. Information distance. IEEE Trans. Inf. Theory. 1998;44(4):1407–1423. doi: 10.1109/18.681318. [DOI] [Google Scholar]
- 23.Fortnow, L., Lee, T., Vereshchagin, N.: Kolmogorov complexity with error. In: Durand, B., Thomas, W. (eds.) STACS 2006–23rd Annual Symposium on Theoretical Aspects of Computer Science, Marseille, France, February 23–25, 2006. Lecture Notes in Computer Science, pp. 137–148. Springer, Berlin, Heidelberg (2006)
- 24.Cha, S.: Taxonomy of nominal type histogram distance measures. In: Proceedings of the American Conference on Applied Mathematics, pp. 325–330. Harvard, Massachusetts, USA (2008)
- 25.Deza MM, Deza E. Encyclopedia of distances. Berlin, Heidelberg: Springer-Verlag; 2009. [Google Scholar]
- 26.Hamming RW. Error detecting and error correcting codes. Bell Syst. Tech. J. 1950;29(2):147–160. doi: 10.1002/j.1538-7305.1950.tb00463.x. [DOI] [Google Scholar]
- 27.Cilibrasi R, Vitany PMB. Clustering by compression. IEEE Trans. Inf. Theory. 2005;51(4):1523–1545. doi: 10.1109/TIT.2005.844059. [DOI] [Google Scholar]
- 28.Yin C, Chen Y, Yau SST. A measure of DNA sequence similarity by Fourier transform with applications on hierarchical clustering complexity for DNA sequences. J. Theor. Biol. 2014;359:18–28. doi: 10.1016/j.jtbi.2014.05.043. [DOI] [PubMed] [Google Scholar]
- 29.Kubicova V, Provaznik I. Relationship of bacteria using comparison of whole genome sequences in frequency domain. Inf. Technol. Biomed. 2014;3:397–408. doi: 10.1007/978-3-319-06593-9_35. [DOI] [Google Scholar]
- 30.Glunčić M, Paar V. Direct mapping of symbolic DNA sequence into frequency domain in global repeat map algorithm. Nucleic Acids Res. 2013 doi: 10.1093/nar/gks721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hautamaki V, Pollanen A, Kinnunen T, Aik K, Haizhou L, Franti L. A comparison of categorical attribute data clustering methods. Springer: Berlin; 2014. pp. 53–62. [Google Scholar]
- 32.Hu, L.Y., Huang, M.W., Ke, S.W., Tsai, C.F.: The distance function effect on k-nearest neighbor classification for medical datasets. Springer Plus 5,(2016). 10.1186/s40064-016-2941-7 [DOI] [PMC free article] [PubMed]
- 33.Aziz M, Alhadidi D, Mohammed N. Secure approximation of edit distance on genomic data. BMC Med Genom. 2017 doi: 10.1186/s12920-017-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yianilos, P.N.: Normalized forms of two common metrics. Tech. Rep. Report 91–082-9027-1, NEC Research Institute (1991)
- 35.Yu, J., Amores, J., Sebe, N., Tian, Q.: A new study on distance metrics as similarity measurement. In: IEEE International Conference on Multimedia and Expo, pp. 533–536 (2006). 10.1109/ICME.2006.262443
- 36.Guyon I, Gunn S, Nikravesh M, Zadeh LA, editors. Feature extraction: foundations and applications. Berlin: Springer; 2008. [Google Scholar]
- 37.Russel R, Sinha P. Perceptually based comparison of image similarity metrics. Perception. 2011;40:1269–1281. doi: 10.1068/p7063. [DOI] [PubMed] [Google Scholar]
- 38.Burrows, M., Wheeler, D.J.: A block sorting lossless data compression algorithm. Technical Report 124, Digital Equipment Corporation (1994)
- 39.Welch T. A technique for high-performance data compression. Computer. 1984;17(6):8–19. doi: 10.1109/mc.1984.1659158. [DOI] [Google Scholar]
- 40.Kodituwakku S. Comparison of lossless data compression algorithms for text data. Indian J. Comput. Sci. Eng. 2010;1(4):416–425. [Google Scholar]
- 41.Saeed N, Haewoon Imtiaz, Saqib M. A survey on multidimensional scaling. ACM Comput. Surv. (CSUR) 2018;51(3):47. doi: 10.1145/3178155. [DOI] [Google Scholar]
- 42.Hartigan JA. Clustering algorithms. London: Wiley; 1975. [Google Scholar]
- 43.Tenreiro Machado JA, Galhano AM. Multidimensional scaling visualization using parametric similarity indices. Entropy. 2015;17(4):1775–1794. doi: 10.3390/e17041775. [DOI] [Google Scholar]
- 44.Machado JAT. Relativistic time effects in financial dynamics. Nonlinear Dyn. 2014;75(4):735–744. doi: 10.1007/s11071-013-1100-8. [DOI] [Google Scholar]
- 45.Liébecq, C. (ed.): IUPAC-IUBMB Joint Commission on Biochemical Nomenclature and Nomenclature Commission of IUBMB. In: Biochemical Nomenclature and Related Documents. Portland Press (1992)
- 46.van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2009;84(2):523–538. doi: 10.1007/s11192-009-0146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waltman L, van Eck NJ, Noyons EC. A unified approach to mapping and clustering of bibliometric networks. J. Inf. 2010;4(4):629–635. doi: 10.1016/j.joi.2010.07.002. [DOI] [Google Scholar]
- 48.van Eck, N.J., Waltman, L.: Visualizing bibliometric networks. In: Measuring Scholarly Impact, pp. 285–320. Springer International Publishing (2014). 10.1007/978-3-319-10377-8_13
- 49.Perianes-Rodriguez A, Waltman L, van Eck NJ. Constructing bibliometric networks: a comparison between full and fractional counting. J. Inf. 2016;10(4):1178–1195. doi: 10.1016/j.joi.2016.10.006. [DOI] [Google Scholar]
- 50.van Eck NJ, Waltman L. Citation-based clustering of publications using CitNetExplorer and VOSviewer. Scientometrics. 2017;111(2):1053–1070. doi: 10.1007/s11192-017-2300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machado JAT. Shannon information and power law analysis of the chromosome code. Abstr. Appl. Anal. 2012;2012:1–13. doi: 10.1155/2012/439089. [DOI] [Google Scholar]
- 52.Machado JAT, Costa AC, Quelhas MD. Can power laws help us understand gene and proteome information? Adv. Math. Phys. 2013;2013:1–10. doi: 10.1155/2013/917153. [DOI] [Google Scholar]
- 53.Machado JT. Fractional order description of DNA. Appl. Math. Model. 2015;39(14):4095–4102. doi: 10.1016/j.apm.2014.12.037. [DOI] [Google Scholar]
- 54.Sculley, D., Brodley, C.: Compression and machine learning: a new perspective on feature space vectors, p. 332. IEEE (2006). 10.1109/dcc.2006.13
- 55.Felsenstein, J.: PHYLIP (phylogeny inference package), version 3.5 c. Joseph Felsenstein (1993)
- 56.Tuimala J. A primer to phylogenetic analysis using the PHYLIP package. Finland: CSC - Scientific Computing Ltd.; 2006. [Google Scholar]
- 57.Kolmogorov A. Three approaches to the quantitative definition of information. Int. J. Comput. Math. 1968;2(1–4):157–168. doi: 10.1080/00207166808803030. [DOI] [Google Scholar]
- 58.Li M, Chen X, Li X, Ma B, Vitanyi P. The similarity metric. IEEE Trans. Inf. Theory. 2004;50(12):3250–3264. doi: 10.1109/tit.2004.838101. [DOI] [Google Scholar]
- 59.Li, M., Chen, X., Li, X., Ma, B., Vitanyi, P.: The similarity metric. In: Proceedings of the 14th Annual ACM-SIAM Symposium on Discrete Algorithms, pp. 863–872 (2004)
- 60.Chen, X., Kwong, S., Li, M.: A compression algorithm for DNA sequences and its applications in genome comparison. In: Genome Informatics: Proceedings of the 10th Workshop on Genome Informatics, pp. 51–61 (1999) [PubMed]
- 61.Li M, Badger JH, Chen X, Kwong S, Kearney P, Zhang H. An information-based sequence distance and its application to whole mitochondrial genome phylogeny. Bioinformatics. 2001;17(2):149–154. doi: 10.1093/bioinformatics/17.2.149. [DOI] [PubMed] [Google Scholar]
- 62.Chen X, Francia B, Li M, McKinnon B, Seker A. Shared information and program plagiarism detection. IEEE Trans. Inf. Theory. 2004;50(7):1545–1551. doi: 10.1109/tit.2004.830793. [DOI] [Google Scholar]
- 63.Keogh, E., Lonardi, S., Ratanamahatana, C.A.: Towards parameter-free data mining. In: Proceedings of the 2004 ACM SIGKDD international conference on Knowledge discovery and data mining, pp. 206–215. ACM Press (2004). 10.1145/1014052.1014077
- 64.Shannon CE. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27(3):623–656. doi: 10.1002/j.1538-7305.1948.tb00917.x. [DOI] [Google Scholar]
- 65.Gray RM. Entropy and information theory. New York: Springer-Verlag; 2011. [Google Scholar]
- 66.Beck C. Generalised information and entropy measures in physics. Contemp. Phys. 2009;50(4):495–510. doi: 10.1080/00107510902823517. [DOI] [Google Scholar]
- 67.Khinchin AI. Mathematical foundations of information theory. New York: Dover; 1957. [Google Scholar]
- 68.Jaynes ET. Information theory and statistical mechanics. Phys. Rev. 1957;106(6):620–630. doi: 10.1103/PhysRev.106.620. [DOI] [Google Scholar]
- 69.Pilcher CD, Wong JK, Pillai SK. Inferring HIV transmission dynamics from phylogenetic sequence relationships. PLoS Med. 2008;5(3):e69. doi: 10.1371/journal.pmed.0050069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The reformatted data that support the findings of this study are available in http://ave.dee.isep.ipp.pt/jtm/FILES/datasets/Archive.zip.