Abstract
Background
Antibiotic treatment decisions in severely ill patients must often be made in the absence of microbiologic results. The recently Food and Drug Administration–cleared BioFire FilmArray Pneumonia Panel (PN) detects 15 bacteria semiquantitatively, 3 atypical pneumonia bacteria, 8 viruses, and 7 antimicrobial resistance markers by multiplex PCR in ~1 hour in the laboratory. Previous reports have shown that the PN Panel bacterial detections are highly accurate, even when routine culture had no growth.
Methods
Consecutive bronchoalveolar lavage and endotracheal specimens submitted for culture between June and September 2018 from 270 patients with sufficient clinical and laboratory data were tested with the PN Panel. Patients were divided into 3 groups: (1) both culture and PN Panel positive, (2) PN Panel positive but culture uninformative (no growth or normal flora), and (3) patients with no PN Panel detections.
Results
Groups 1 and 2 had significantly higher maximum temperatures on the day of culture (P = .00036, analysis of variance [ANOVA] with Bonferroni correction), higher levels of an inflammatory response as measured by percent polymorphonuclear leukocytes in bronchoalveolar lavage (P = .00025, ANOVA with Bonferroni correction), and gram stain report of white blood cells, as previously reported [1].
Conclusions
Both group 1 (culture and PN Panel positive), and group 2 (PN Panel positive but culture uninformative) had higher levels of host response inflammatory responses compared with group 3, which had no targets detected, suggesting that PN Panel detections need to be interpreted in the clinical context, even if cultures are discordant. Depending on laboratory turnaround time, there could be opportunities for improved diagnosis and antibiotic stewardship.
Keywords: BioFire Pneumonia, Panel, bronchoalveolar lavage % PMNs, Clinical Pulmonary Infection Score (CPIS), gram stain white blood cells, multiplex bacterial PCR
The diagnosis of hospital- and/or ventilator-associated pneumonia in the intensive care unit setting has a great deal of clinical uncertainty, and as a result, antibiotic treatment decisions may need to be made before culture results are known. The recently Food and Drug Administration–cleared BioFire FilmArray Pneumonia Panel (PN) detects 15 bacteria semiquantitatively, 3 atypical pneumonia bacteria, 8 viruses, and 7 antimicrobial resistance markers by multiplex PCR in about 1 hour in the laboratory. We compared this multiplex PCR assay with routine microbiology results and corresponding clinical data for 270 unique hospitalized patients. Hospital-acquired pneumonia and ventilator-associated pneumonia have high mortality and account for much of the antibiotic usage in intensive care units. Because of the rapid (≈1 h) lab turnaround time for the PN Panel, it has the potential to influence antibiotic choice and even empower possible de-escalation in real time when culture specimens are submitted.
Previous studies have shown that the PN Panel detects >98% of bacteria that are subsequently identified in the microbiology laboratory, with extremely high negative predictive value [1–4]. However, at least 20%–25% additional patients have positive molecular test results that are not confirmed by culture [1–4], leaving the clinician uncertain about how to assess their importance. Furthermore, the panel is semiquantitative and reports the common bacteria in bins of 104, 105, 106, and ≥107 copies/mL. We and others [1–4] have found that there is a strong general relationship between PN Panel copy number and semiquantitative culture results, but because molecular copy number could include nonviable organisms, it may not reflect the same results as culture that relies on viable organisms.
Here, we show that both bacterial targets detected by the PN Panel but not grown in culture and intermediate or low copy numbers show similar relationships to clinical variables (eg, peak temperature on the day of culture, gram stain white blood cells [WBC], bronchoalveolar lavage [BAL] polymorphonuclear leukocytes [PMNs]) as those of the highest copy numbers themselves and those positive by both culture and PN Panel. Our intent was to understand the performance of the PN Panel in relation to clinical measures and outcome variables as detailed in the “Methods,” rather than exploring or endorsing its use to diagnose hospital-acquired or ventilator-associated pneumonia.
METHODS
This retrospective study was carried out in the clinical microbiology laboratory at the University of Florida Health Shands Hospital, Gainesville, Florida, a large tertiary care hospital (>1000 beds) serving North Central Florida. Patients underwent a bronchoscopy or were intubated and had endotracheal suction specimens submitted for culture and susceptibility as part of their routine standard of care between June 11 and September 24, 2018. The study was approved by our Institutional Review Board (IRB# 2018–01834). Consecutive unique BAL fluids and endotracheal aspirates were frozen at –70°C within 18 hours of receipt in the laboratory, until tested with the PN Panel. Data submitted by the manufacturer to the FDA showed no significant alteration in PN Panel performance from freezing and thawing (https://www.accessdata.fda.gov/cdrh_docs/reviews/K180966.pdf). A total of 396 unique patient specimens, having both culture results from the microbiology laboratory and PCR results from the PN Panel, were included and have been previously reported [1]. Clinical data were obtained from the patients’ electronic medical records (Epic Systems, Madison, WI, USA) and were available for a subset of 270 patients, as the rest were either outpatients or in a long-term acute care hospital that has a separate medical record. The patients were 158 (58.7%) male and had a mean age (interquartile range, range) of 53.3 ± 20.2 (40–68, 0–88) years in males and 52.0 ± 23.8 (0–97, 40–68) years in females (n = 111).
Microbiologic Methods
Cultures were performed by standard methods, and all isolates were identified and antimicrobial susceptibility testing performed by VITEK MS mass spectrometry and by Vitek II (AST GP 72, AST GP 78, AST GN 73, and AST XN06; bioMerieux, Durham, NC, USA). All specimens were run on the PN Panel per the manufacturer’s instructions using the FilmArray 2.0 instrument (BioFire Diagnostics, Salt Lake City, UT, USA) in our research laboratory. As all specimens were collected from the lower respiratory tract, contamination by epithelial cells was reported for clinical consideration/information but not used to reject specimens that were obtained from a deep source in the lung. All cultures were plated using the standard quadrant technique onto blood, chocolate, and MacConkey agar (Becton Dickinson, Sparks, MD, USA), incubated in 5% CO2 at 36°C, and examined for growth over the next 24–48 hours. Growth from endotracheal aspirates and nonquantitative BAL specimens was reported semiqualitatively based on the following criteria: <10 colonies “few,” 1+ for growth in the first quadrant, 2+ for growth in the second quadrant, and so forth. Potential pathogens with growth on plates were only reported if they were a sole pathogen or if their growth was at least 2+ or exceeded the growth of normal respiratory flora. Normal respiratory flora was reported as “normal flora,” “normal oral flora,” or “normal respiratory flora,” along with the growth of potential pathogens. Direct gram stains were performed on all specimens, and the presence of WBCs was reported semiquantitatively as no WBC, few, moderate, many, or 1+, 2+, 3+, 4+, depending on the technologist. Under laboratory policy, 1+ or “rare” corresponds to <1 WBC per low power field (lpf), 2+ or “few” is equivalent to 1–9 WBC/lpf, 3+ or “moderate” corresponds to 10–25 WBC/lpf, and 4+ or “many” corresponds to >25 WBC/lpf. The presence of bacteria in the gram stain was reported in a similar manner, along with appropriate morphologic characteristics.
Gram stains for bacteria along with WBC from unconcentrated BALs were reported as described above for endotracheal aspirates. Specimens ordered for quantitative counts were inoculated onto standard media (see above) using a 0.001-mL calibrated loop and were incubated at 36°C in 5% CO2. Quantitative growth was reported as CFU/mL and ranged from no growth to <1000 CFU/mL, 1000–10 000 CFU/mL, and the actual colony count/mL for those with growth of 104 and above. All potential pathogens with growth of 104 or greater were identified as described above. Those that were not ordered quantitatively were plated using standard quadrant plating and reported as no growth, few, 1+, 2+, 3+, and 4+, as described above.
Bronchoalveolar Lavage Polymorphonuclear Leukocytes
Bronchoalveolar lavage percentage PMNs were counted in the clinical hematology laboratory using standard Wright’s stain and counting procedures, when ordered by the patient’s physician.
Radiographic Analysis
Chest and thoracic computed tomography scan reports between 5 days before and 5 days after the day of collection of the culture were reviewed by 2 authors (K.R. and K.C.) independently of the culture and PN Panel results. Based on the radiologist’s findings, interpretations, and progression of infiltrates over time, each independently estimated a bacterial pneumonia likelihood score from 1 to 5, where 1 = definitely no pneumonia, 2 = pneumonia unlikely, 3 = pneumonia indeterminate, 4 = pneumonia likely, and 5 = pneumonia definitely found.
Clinical Pulmonary Infection Score
The Clinical Pulmonary Infection Score (CPIS) was calculated from https://www.mdapp.co/clinical-pulmonary-infection-score-cpis-calculator-236/ [5] and modified slightly as follows because of the retrospective analysis of the data: Purulent tracheal secretions were considered present (score = 2) if the gram stain was reported as having ≥3+, 4+, many, or moderate white blood cells on the gram stain report (as defined above); only the radiographic pneumonia likelihood score of 4 or 5 by either investigator was included as a score of 2, and a score of 1 was given for positive cultures (we did not attempt to match the gram stains with culture growth, as no organisms were seen in 152 and mixed morphology was reported in 74). In addition to CPIS score, discharge diagnosis codes were collected for PN Panel correlation analysis.
Data Analysis
Clinical variables were analyzed by multiple independent 1-way analysis of variance (ANOVA) for an association with the PN bacterial copy number and with growth in culture, specifically colony counts for BALs and semiquantitative growth on plate cultures (eg, no growth, few, 1+, 2+, etc.): maximum FiO2 on the day of culture, maximum temperature on the day of culture (day 0), BAL % Polys, intensive care unit (ICU) length of stay (LOS; days), average temperature, ventilator days, hospital LOS, procalcitonin, blood WBC on date of culture, minimum pO2 all on the day of culture. P values from ANOVA were adjusted with Bonferroni correction. The categorical variables (discharge billing coded for pneumonia on ventilator on the day of culture, mortality, receiving antibiotics on the day of culture) were analyzed with the chi-square linear-by-linear test (chi-square), with the Bonferroni correction.
RESULTS
Multiple independent 1-way ANOVAs showed a statistically significant association with the PN Panel bacterial copy number for 5/14 variables tested, as shown in Table 1, and for 4 variables based on microbiology laboratory culture results. However, for the PN Panel after Bonferroni correction for multiple comparisons, only the peak temperature on the day of culture collection, discharge coding for pneumonia, and BAL % Polys remained highly statistically significant, while none of the variables remained statistically significant for culture growth results.
Table 1.
Pneumonia Panel Copy Number and Semi-quantitative Conventional Microbiology Results: Correlation with Clinical and Outcome Variables
| Clinical Correlate | Pneumonia Panel Copy # | Bonferroni Correction | Conventional Microbiology | Bonferroni Correction |
|---|---|---|---|---|
| ICU length of stay, d | P = .006 | NS | P = .017 | NS |
| Pneumonia coded on discharge | P = .000288 | P = .004 | P = .013 | NS |
| BAL % Polys | P = .00009 | P = .00025 | ||
| Peak temperature on day of culture | P = .00003 | P = .00084 | ||
| On ventilator day of culture | P = .012 | NS | ||
| Procalcitonin on day of culture | P = .008 | NS | ||
| On antibiotics on day of culture | P = .011 | NS |
Abbreviations: BAL, bronchoalveolar lavage; ICU, intensive care unit; NS, not statistically significant.
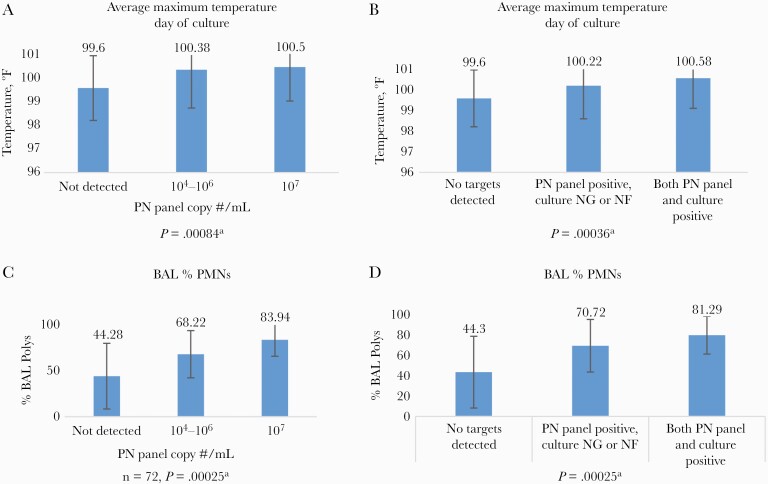
To explore these relationships in more detail, we grouped the patients in 2 ways as shown in Figure 1A–D, by copy number/mL (no targets detected, 104–106, and ≥107) and by culture/PN Panel results (ie, no targets detected, culture with no growth or normal respiratory flora/PN Panel positive, and culture positive/PN Panel positive). These groups were compared for their average maximum temperature on the day of culture and for the % PMNs in their BAL, with statistically significant differences across groups (temperature P = .00084, 1-way ANOVA with Bonferroni correction). Intermediate PN Panel copy numbers 104–106 copies/mL and those with uninformative culture results (ie, no growth or normal flora) that were positive by the PN Panel had temperatures and BAL % PMNs that were intermediate between those with no targets detected and those with either maximum PN Panel copy number (≥107) or matching culture results and PN Panel detections.
Figure 1.
A and B show the relationship of the group average maximum temperature in ºF on the day of the culture, when the patients were divided by copy number (A) and by PN Panel targets detected (B) when cultures were either uninformative (no growth or normal flora) or were consistent between the PN Panel detection and the culture result. C and D show % BAL PMNs reported from the Hematology lab when the patients were divided into the same respective groups as in (A) and (B). aMultiple 1-way ANOVA, with Bonferroni correction for multiple variables. Abbreviations: ANOVA, analysis of variance; BAL, bronchoalveolar lavage; PMNs, polymorphonuclear leukocytes.
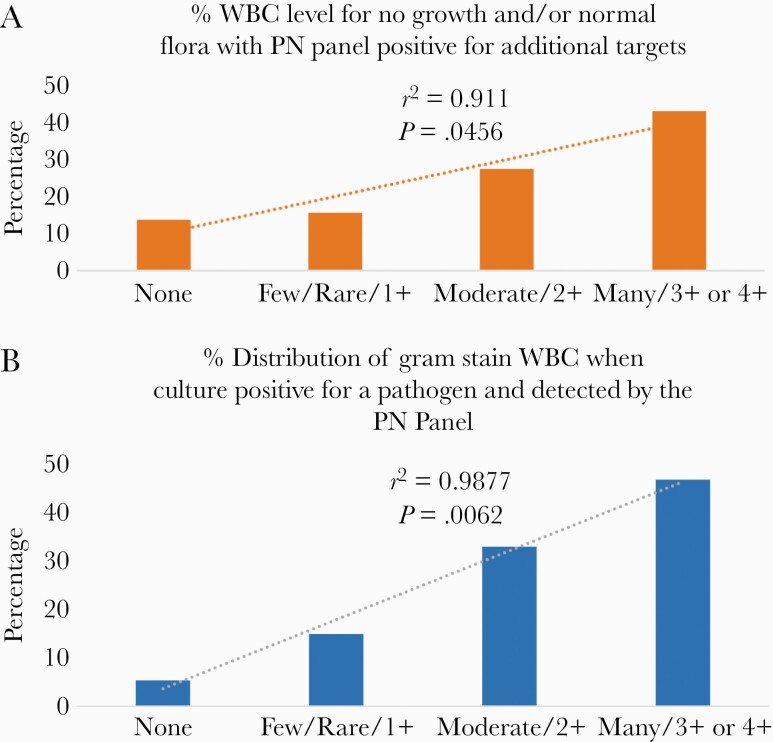
Previously, we showed a strong relationship between WBC reported on the gram stain and PN Panel copy number [1]. These results were extended in Figure 2A and B showing a statistically significant linear correlation for the distribution of gram stain WBC when the PN Panel and culture were both positive (r2 = 0.9877; P = .0062) and a similar linear relationship but with lower slope for gram stain WBC when the culture was uninformative but a target was detected by the PN Panel (r2 = 0.911; P = .0456). PN Panel–negative results showed no statistical relationship with the level of gram stain WBC (data not shown).
Figure 2.
A, The distribution of semiquantitative reported gram stain WBCs when the culture had no growth or normal flora only and the PN was positive for at least 1 target. B, The distribution of semiquantitative reported gram stain WBC when the culture grew the same organism as detected by the PN Panel. In both cases, there is a statistically significant linear relationship that was not seen when the PN Panel had no detections or when the culture either had no growth or normal flora only. Abbreviation: WBC, white blood cells.
In view of the strong relationship of percentage BAL PMNs, gram stain WBC, and PN Panel results, it was important to determine whether the percent BAL PMNs correlated with the gram stain report. Gram stains reported as none, rare, or few had an average BAL % PMNs of 45%, while those with 2+ or moderate WBC averaged 72% and those with 3+, 4+, or many WBC averaged 80% BAL % PMNs, respectively (P = .002578, ANOVA).
There were no differences in average maximum temperature on the day of culture or WBC described on gram stain for those patients who grew a gram-positive vs a gram-negative organism (100.4°F ± 1.33°F vs 100.7°F ± 1.47°F, respectively; P = NS), but those patients who grew any bacteria (that were included in the PN Panel) did have a significantly higher maximum average temperature than those who had a virus (with or without a concomitant bacterial detection) detected by the panel (100.6°F ± 1.45°F vs 99.79°F ± 1.22°F, respectively; P = .0036, t test). Likewise, high levels of WBC on gram stain (moderate, many, 2+, 3+, or 4+) were found in 8/24 (33%) with virus detections compared with 74/95 (78%) of those with no viruses detected whose cultures grew bacteria (P = .000073, chi-square with Yates correction).
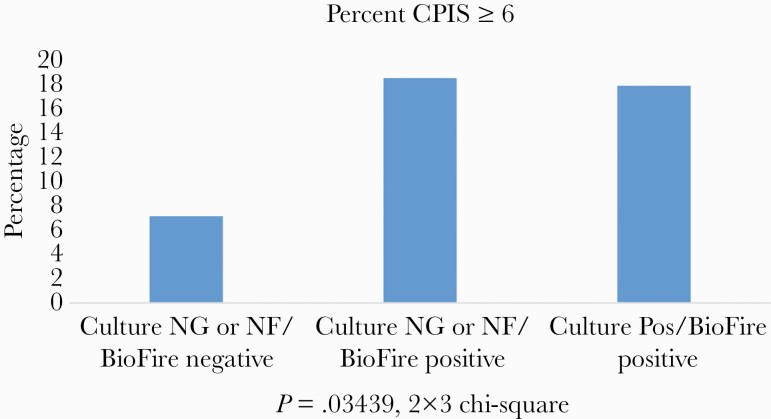
In Figure 3, the CPIS score was calculated for the groups with (1) no targets detected, (2) culture uninformative (no growth or normal flora) but PN Panel positive, and (3) culture positive/PN Panel positive. A positive score was as defined as ≥6 based on reference 9 and as described in the “Methods.” Both the uninformative culture/PN Panel positive and the culture positive/PN Panel positive groups had a statistically higher proportion with a positive CPIS score than the PN Panel negative patients (P = .03439, chi-square).
Figure 3.
CPIS score was calculated as described in the “Methods,” and the percentage of those scores considered positive (≥6; https://www.mdapp.co/clinical-pulmonary-infection-score-cpis-calculator-236/) for the patient groups are shown: (1) PN Panel negative, (2) PN Panel positive/culture no growth or normal flora in culture, and (3) PN Panel positive/culture positive (for the same organism). Abbreviations: CPIS, Clinical Pulmonary Infection Score; NG, no growth; NF, normal flora.
Thirty-seven patients whose molecular detection and culture were both positive for the same organism also had 1 or more additional molecular bacterial targets detected, but this group was not different in terms of increased ICU LOS or higher levels of other clinical variables (data not shown).
DISCUSSION
The PN Panel copy number was strongly statistically associated with percent PMNs in the BAL fluid and average maximum temperature on the day of culture. An equally strong relationship was also shown between the PN Panel categorized into group with (1) no detections, (2) positive PN Panel bacterial detection but no growth in culture, and (3) positive detection with growth of the corresponding organism in culture. PN Panel copy number was also significantly related to the discharge coding diagnosis of pneumonia. Other outcome variables (see list in “Methods”) were either nonsignificant or became nonsignificant after Bonferroni correction for multiple comparisons (shown in Table 1). When the same analysis was applied to the microbiologic results as reported in the clinical record, the individually significant relationships were no longer statistically significant after correction for multiple comparisons. These findings suggest that the PN Panel copy number and PN Panel–positive results independent of growth in culture correlate better with the basic host response to infection, temperature, and polymorphonuclear leukocytes at the presumptive site of infection than traditional microbiologic culture results.
In an earlier study, we found the gram stain WBC report to be strongly related to PN Panel copy number [1]. Here we have extended those findings, showing that the gram stain WBC reported was linearly related to positive PN Panel detections both when culture confirmed and even when the PN Panel had a positive bacterial detection but growth was not found in culture. As the percentage of BAL PMNs was one of the strongest variables found in the multiple independent 1-way ANOVA, it was important to confirm that the gram stain readings did in fact correlate with the percent BAL PMNs, as was found to be the case. In this context, Choi et al. [6] reported that total WBC in BALs showed a sensitivity of 83.3% and receiver operating characteristics curve (ROC) of 0.855 in differentiating between bacterial and viral pneumonia, while the ROC of BAL % PMNs was 77 in a previous report by the same group [7].
Several recent studies have also documented the findings of a relatively high number of targets detected by the PN Panel that were not recovered in culture [1–4]. In our previous report, we were able to confirm a number of these with an in-house PCR, but in a very large study comparing the BioFire PN Panel with conventional culture results. Murphy et al. [2] found additional targets in 875 sputum and lower respiratory tract specimens from 1764 patients with valid PN Panel tests. Of these, about 25.1% were considered to have been true positives but with colony counts below their quantitative plating method cutoff of 3.5 × 103 CFU/mL, and the vast majority of the rest (74.5%) were confirmed as true positives by alternative molecular test methods. The current study extends this work—not only are the additional molecular detections “real,” they also show relationships similar to culture-confirmed molecular detections to such clinical measures as day of culture average maximum temperature, WBC reported on gram stain, BAL % PMNs, and CPIS score.
Since its original description, the CPIS has been reported to show modest correlation with ventilator-associated pneumonia [5, 8–11]. Most recently, a modified version was used to differentiate between ventilator-associated tracheitis and ventilator-associated pneumonia with 60% sensitivity and 70% specificity, using the ≥6 cutoff score we used [12]. However, the modifications they introduced probably do not allow exact correspondence of their scores and ours. In the current study, some of the CPIS measures such as volume and purulence of pulmonary secretions were modified to use the gram stain WBC reported, which had the advantage of being semiquantitative. Although gram stain interpretations are inherently subjective and may vary from technician to technician, estimating tracheal secretion purulence in real time is also inherently subjective and has shown interobserver variability [9, 11]. The radiographic data were interpreted by 2 of us (K.R. and K.C.) using the radiographic reports without reference to other study data, that is, blinded to the PN Panel and culture results. Anyone calculating a CPIS in real time would also have to use the radiologists’ readings and interpretations, so the authors’ estimation of the likelihood of pneumonia is analogous but had the advantage of knowing whether infiltrates described initially persisted or cleared. Nonetheless, the key point in this study is that the CPIS was compared across patient groups defined by their PN Panel and culture results alone, with no reason to favor 1 group over another. Thus, finding statistically higher numbers of patients with CPIS scores ≥6 https://www.mdapp.co/clinical-pulmonary-infection-score-cpis-calculator-236/ [5] for both the PN Panel positive/culture no growth or normal flora and PN Panel positive/culture positive (for the same organism) compared with the PN Panel negative group is suggestive that PN Panel detections may be clinically relevant, even if not confirmed by culture.
It is important to point out that the CPIS is in no way a gold standard for the diagnosis of hospital-acquired or ventilator-associated pneumonia. Two recent reports compared the CPIS with National Healthcare Safety Network (NHSN) criteria and found comparable but low levels of sensitivity for both the CPIS and NHSN measures. Gunalan et al. [13] reported 34.1% of 273 patients using CPIS, but only 12.1% were scored as having VAP by NHSN criteria. Agreement between the 2 measures was “fairly good” with a kappa score of 0.42, and all patients classified as having pneumonia by NHSN were also found by CPIS. Sensitivity and specificity were not reported in this study. In a smaller study of 85 ICU patients, Rahimibashar et al. [14] found a sensitivity of 54.2% for the NHSN criteria and 68.75% for the CPIS, using clinical diagnosis as the “gold standard.” These levels are comparable to the 77% sensitivity reported by Fabregas et al. [15] comparing the CPIS score with immediate postmortem histologic diagnosis of pneumonia as the gold standard.
If laboratories adopt the PN Panel, infectious diseases specialists, intensive care physicians, and other providers could rightly ask how—and whether—to treat bacterial targets detected that are not also grown in the microbiology laboratory. Although our study was not designed to look at this question, we note that patients in this group had on average intermediate levels of maximum temperature, BAL % PMNs, and semiquantitative gram stain WBC (Figures 1 and 2, respectively) between the PN Panel “no targets detected” and both culture and the PN Panel positive for the same organism. As fever and BAL or endotracheal WBC levels are both measures of host response to infection, it seems reasonable to conclude that culture-negative but PN Panel–positive detections should not be dismissed as insignificant or as colonization, but considered with clinical correlation and judgment of the potential significance. In principle, this is not different from interpreting and correlating conventional microbiologic results, for example, 1 + Escherichia coli, which clinicians do routinely. Although issues like this are complex and have a degree of uncertainty, there are advantages to both positive and negative results from the PN Panel being reported in “real” time. Examples include discontinuation of vancomycin, if methicillin-resistant Staphylococcus aureus is not detected, receiving an unexpected high copy number for S. aureus or Pseudomonas aeruginosa, or having a molecular detection of Acinetobacter spp. when the gram stain shows only gram-positive cocci.
One of the major advantages of the PN Panel is the rapid turnaround time to a result, especially for larger institutions that can perform testing 24/7. It would be reasonable to ask if the more rapid turnaround time (which in our laboratory is about 3 hours after receipt of the specimen for similar multiplex panels) would actually improve patient outcomes and quality of care. In support of this, for example, 1 patient in this study was a 75-year-old woman resuscitated after a massive heart attack and cardiac arrest. Because of a history of a penicillin allergy, she was treated with aztreonam for 3 days before a culture of Staphylococcus aureus was reported. Then the penicillin allergy was re-evaluated, and the patient treated with cephalosporin. The PN Panel result was ≥107 copies of S. aureus and would have been available within hours. While studies have not been reported yet showing patient benefit from the PN Panel itself, a recent study with the multiplex gastroenteritis panel was estimated to have led to a shorter length of stay by 0.5 days and overall lower health care costs to the hospital of $293.61/patient [16]. In a meta-analysis, Kuti et al. showed that delayed appropriate treatment for pneumonia was associated with a worse outcome than appropriate therapy [17].
In conclusion, there were very strong relationships between the PN Panel results and the important host inflammatory responses of temperature and WBC level from the likely site of infection. This was true for both PN Panel copy number/mL per se and for those cases with noninformative cultures (no growth or normal flora) but positive PN Panel detections, suggesting that PN Panel detections need to be interpreted in the clinical context, even if cultures are discordant. Depending on laboratory turnaround time capacity, rapidly available results both positive and negative could potentially reduce diagnostic uncertainty and lead to improved antibiotic stewardship.
Acknowledgments
We gratefully acknowledge the support of the staff in the UF Health Shands Hospital clinical microbiology laboratory and from the UF Health Department of Pathology, Immunology and Laboratory Medicine at the University of Florida, Gainesville, FL, USA.
Financial support. This work was supported by a grant from BioFire Diagnostics, Inc. (Salt Lake City, UT, USA).
Potential conflicts of interest. Dr. Rand has received speaker fees from BioFire Diagnostics. Drs. Rand and Beal have received unrelated grant funding from BioFire Diagnostics. Brianne Couturier, Beth Lingenfelter, Cory Rindlisbacher, and Jay Jones are or were employed by BioFire Diagnostics. To the best of our knowledge, there are otherwise no authors with a potential conflict of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. The design of the work has been approved by the University of Florida Health Science Institutional Review Board IRB# 2018-01834.
References
- 1.Rand KH, Beal SG, Cherabuddi K, et al. . Performance of a semiquantitative multiplex bacterial and viral PCR panel compared with standard microbiological laboratory results: 396 patients studied with the Biofire Pneumonia Panel. Open Forum Infect Dis 2021; 8:ofaa560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy CN, Fowler R, Balada-Llasat JM, et al. . Multicenter evaluation of the Biofire® Filmarray® Pneumonia/Pneumonia Plus Panel for the detection and quantification of agents of lower respiratory tract infection. J Clin Microbiol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchan BW, Windham S, Balada-Llasat JM, et al. . Practical comparison of the BioFire® FilmArray® Pneumonia Panel to routine diagnostic methods and potential impact on antimicrobial stewardship in adult hospitalized patients with lower respiratory tract infections. J Clin Microbiol. 2020; 58:e00135–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webber DM, Wallace MA, Burnham CA, Anderson NW. Evaluation of the BioFire® FilmArray® Pneumonia Panel for detection of viral and bacterial pathogens in lower respiratory tract specimens in the setting of a tertiary care academic medical center. J Clin Microbiol. 2020; 58:e00343–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical Pulmonary Infection Score (CPIS) MDApp. Available at: https://www.mdapp.co/clinical-pulmonary-infection-score-cpis-calculator-236/. Accessed 12 December 2020.
- 6.Choi SH, Hong SB, Hong HL, et al. . Usefulness of cellular analysis of bronchoalveolar lavage fluid for predicting the etiology of pneumonia in critically ill patients. PLoS One 2014; 9:e97346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huh JW, Lim CM, Koh Y, et al. . Diagnostic utility of the soluble triggering receptor expressed on myeloid cells-1 in bronchoalveolar lavage fluid from patients with bilateral lung infiltrates. Crit Care 2008; 12:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pugin J, Auckenthaler R, Mili N, et al. . Diagnosis of ventilator-associated pneumonia by bacteriologic analysis of bronchoscopic and nonbronchoscopic “blind” bronchoalveolar lavage fluid. Am Rev Respir Dis 1991; 143:1121–9. [DOI] [PubMed] [Google Scholar]
- 9.Schurink CAM, Nieuwenhoven CAV, Jacobs JA, et al. . Clinical pulmonary infection score for ventilator-associated pneumonia: accuracy and inter-observer variability. Intensive Care Med 2004; 30:217–24. [DOI] [PubMed] [Google Scholar]
- 10.Shan J, Chen HL, Zhu JH. Diagnostic accuracy of clinical pulmonary infection score for ventilator-associated pneumonia: a meta-analysis. Respir Care 2011; 56:1087–94. [DOI] [PubMed] [Google Scholar]
- 11.Zilberberg MD, Shorr AF. Ventilator-associated pneumonia: the clinical pulmonary infection score as a surrogate for diagnostics and outcome. Clin Infect Dis 1988; 51:S131–5. [DOI] [PubMed] [Google Scholar]
- 12.Gaudet A, Martin-Loeches I, Povoa P, et al. ; TAVeM study group. Accuracy of the clinical pulmonary infection score to differentiate ventilator-associated tracheobronchitis from ventilator-associated pneumonia. Ann Intensive Care 2020; 10:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunalan A, Sistla S, Sastry AS, Venkateswaran R. Concordance between the National Healthcare Safety Network (NHSN) surveillance criteria and clinical pulmonary infection score (CPIS) criteria for diagnosis of ventilator-associated pneumonia (VAP). Indian J Crit Care Med 2021; 25: 296–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahimibashar F, Miller AC, Yaghoobi MH, Vahedian-Azimi A. A comparison of diagnostic algorithms and clinical parameters to diagnose ventilator-associated pneumonia: a prospective observational study. BMC Pulm Med 2021; 21:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fàbregas N, Ewig S, Torres A, et al. . Clinical diagnosis of ventilator associated pneumonia revisited: comparative validation using immediate post-mortem lung biopsies. Thorax 1999; 54:867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beal SG, Tremblay EE, Toffel S, et al. . A gastrointestinal PCR panel improves clinical management and lowers health care costs. J Clin Microbiol 2017; 56:e01457–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuti EL, Patel AA, Coleman CI. Impact of inappropriate antibiotic therapy on mortality in patients with ventilator-associated pneumonia and blood stream infection: a meta-analysis. J Crit Care 2008; 23:91–100. [DOI] [PubMed] [Google Scholar]