Abstract
The yeast ENA1/PMR2A gene encodes a cation extrusion ATPase in Saccharomyces cerevisiae which is essential for survival under salt stress conditions. One important mechanism of ENA1 transcriptional regulation is based on repression under normal growth conditions, which is relieved by either osmotic induction or glucose starvation. Analysis of the ENA1 promoter revealed a Mig1p-binding motif (−533 to −544) which was characterized as an upstream repressing sequence (URSMIG-ENA1) regulated by carbon source. Its function was abolished in a mig1 mig2 double-deletion strain as well as in either ssn6 or tup1 single mutants. A second URS at −502 to −513 is responsible for transcriptional repression regulated by osmotic stress and is similar to mammalian cyclic AMP response elements (CREs) that are recognized by CREB proteins. This URSCRE-ENA1 element requires for its repression function the yeast CREB homolog Sko1p (Acr1p) as well as the integrity of the Ssn6p-Tup1p corepressor complex. When targeted to the GAL1 promoter by fusing with the Gal4p DNA-binding domain, Sko1p acts as an Ssn6/Tup1p-dependent repressor regulated by osmotic stress. A glutathione S-transferase–Sko1 fusion protein binds specifically to the URSCRE-ENA1 element. Furthermore, a hog1 mitogen-activated protein kinase deletion strain could not counteract repression on URSCRE-ENA1 during osmotic shock. The loss of SKO1 completely restored ENA1 expression in a hog1 mutant and partially suppressed the osmotic stress sensitivity, qualifying Sko1p as a downstream effector of the HOG pathway. Our results indicate that different signalling pathways (HOG osmotic pathway and glucose repression pathway) use distinct promoter elements of ENA1 (URSCRE-ENA1 and URSMIG-ENA1) via specific transcriptional repressors (Sko1p and Mig1/2p) and via the general Ssn6p-Tup1p complex. The physiological importance of the relief from repression during salt stress was also demonstrated by the increased tolerance of sko1 or ssn6 mutants to Na+ or Li+ stress.
The study of adaptation mechanisms during salinity stress in the yeast Saccharomyces cerevisiae has revealed several components of sensing and signal transduction pathways, as well as target genes whose expression is activated upon salt stress (for review see references 18, 45, and 46). Increased expression of the ENA1 gene has been found to represent a crucial cellular response after salt challenge. The ENA/PMR2 gene cluster of S. cerevisiae contains a tandem array of nearly identical genes encoding P-type ATPases involved in the extrusion of Na+ and Li+ ions from the cytoplasm (17, 59). Active export of these toxic ions is a crucial cellular process to avoid deleterious intracellular Na+ and Li+ concentrations. Mutants lacking the first ENA gene in the gene cluster (ena1) are hypersensitive to salt stress (17). The ENA1 gene is highly regulated at the transcriptional level, and its expression is increased strongly in response to salt stress (12) and glucose starvation (1, 41).
Salt induction of ENA1 expression depends on both the calcineurin pathway and the high-osmolarity glycerol (HOG) mitogen-activated protein (MAP) kinase pathway (28). The first pathway is activated by high concentrations of either Na+ or Ca2+ and is dependent on the phosphoprotein phosphatase calcineurin (28, 33). A calcium signalling pathway composed of calmodulin (7), the calcineurin heterodimeric enzyme (6, 23), and the zinc finger transcriptional activator Crz1/Tcn1/Hal8p (31, 32, 50) has been reported to contribute to the resistance of yeast to elevated concentrations of several cations (Na+, Li+, and Mn2+). Therefore, Ca2+/calmodulin signalling may act, at least in part, through the transcriptional activation of ion transporter genes such as ENA1.
The HOG pathway responds to moderate concentrations of osmotic agents and rapidly activates via a multistep phosphorelay mechanism the Hog1p MAP kinase by Tyr phosphorylation (2, 26, 39). Although a great number of genes have been found to need HOG signalling for their osmotic up-regulation, the mechanism of gene activation through phosphorylated Hog1p kinase is still unknown. In Schizosaccharomyces pombe, the basic leucine zipper (bZIP) transcriptional activator Atf1p has been identified as a direct phosphorylation target of the Hog1p homolog MAP kinase Sty1p (49, 51, 60). Activated Atf1p, in turn, can bind directly to UASs (upstream activating sequences) located in various stress-regulated promoters and then trigger gene expression (60). In S. cerevisiae, stress response promoter elements (STREs) represent UASs that respond to a great variety of stresses (22, 27, 43) and are bound by the zinc finger activators Msn2p and Msn4p (30, 42). Recent work, however, indicates that osmotic induction of several genes including ENA1 occurs by the release from transcriptional repression (29) and involves the general repressor complex Ssn6p-Tup1p. In the case of the HAL1 gene, an upstream repressing sequence (URS) regulated by osmotic stress has been identified (29). This mechanism based on regulated repressors bound to URSs is similar to the one operating in carbon source regulation.
A great number of yeast genes, including ENA1, are derepressed under glucose starvation conditions, and for many of them the inactivation of the general repressor complex Mig1p-Ssn6p-Tup1p (21, 36, 54) through the protein kinase Snf1p (3, 53) has been reported as an important mechanism of glucose-regulated transcriptional control.
In this work, we analyzed the promoter of the ENA1 gene and found that transcriptional regulation during osmotic stress as well as during glucose starvation occurs through a repression mechanism dependent on the Ssn6p-Tup1p general corepressor. Signalling through general glucose repression occurs through a Mig1/2p binding site (URSMIG-ENA1), whereas osmotic stress signalling through the HOG pathway is mediated through a cyclic AMP (cAMP) response element (CRE)-like sequence (URSCRE-ENA1) that is bound by the bZIP transcriptional factor Sko1p.
MATERIALS AND METHODS
Strains and growth conditions.
The S. cerevisiae strains used in this work are listed in Table 1. Gene disruptions using the loxp-KAN MX-loxp cassette were carried out as described previously (16). All null mutations were verified by genomic PCR. YPD (or YPGal) contained 2% glucose (or 2% galactose), 2% peptone, and 1% yeast extract. Synthetic medium (SD) contained 2% glucose, 0.67% yeast nitrogen base without amino acids (Difco), and the amino acids purine and pyrimidine bases required by the strain of interest. Yeast cells were transformed as described elsewhere (15). The growth of yeast strains under different osmotic and salt stress conditions was assayed by spotting dilutions of saturated cultures onto YPD plates containing the indicated concentrations of osmotic agents or salts.
TABLE 1.
Strains of S. cerevisiae used in this work
| Strain | Genotype | Reference |
|---|---|---|
| W303-1A | MATa can1-100 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 ade2-1 | 57 |
| MAP5 | W303-1A with tup1::loxp-KAN-loxp | This work |
| MAP6 | W303-1A with ssn6::loxp-KAN-loxp | This work |
| MAP12 | W303-1A with mig1::loxp-KAN-loxp | This work |
| MAP19 | W303-1A with sko1::loxp-KAN-loxp | This work |
| MAP20 | W303-1A with bcy1::LEU2 | This work |
| MAP21 | W303-1A with mig2::loxp-KAN-loxp | This work |
| MAP24 | W303-1A with mig1::loxp mig2::loxp-KAN-loxp | This work |
| MAP28 | W303-1A with mig1::loxp mig2::loxp sko1::loxp-KAN-loxp | This work |
| MAP32 | W303-1A with hog1-Δ1::TRP1 | This work |
| MAP33 | W303-1A with hog1-Δ1::TRP1 sko1::loxp-KAN-loxp | This work |
| YPH499 | MATa ura3 leu2 his3 trp1 lys2 ade2 | 2 |
| JBY10 | YPH499 with hog1-Δ1::TRP1 | 2 |
| SKY683 | W303-1A with cnb1::LEU2 | 11 |
| SFY526 | MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 gal4-542 gal80-538 URA3::GAL1-lacZ | 1a |
| MAP34 | SFY526 with ssn6::loxp-KAN-loxp | This work |
| MAP35 | SFY526 with tup1::loxp-KAN-loxp | This work |
Construction of plasmids.
To analyze the ENA1 promoter, different stretches from the 5′ upstream region were amplified by PCR, generating PstI and XhoI restriction sites at the ends. The fragments were then inserted into the CYC1-lacZ reporter construct pJS205 (44) that was digested with PstI and XhoI. An empty vector control (pMP206) was generated by XhoI/SalI digestion of pJS205 to remove the same CYC1 promoter region as in all the insertion constructs and subsequent self-ligation. The URSMIG-ENA1-CYC1-lacZ plasmid pMP222 was constructed by inserting the double-stranded oligonucleotide AGCTATTTTGCGGGGCATCGAT (giving HindIII-compatible ends after hybridization; the original ENA1 sequence is underlined) into pMP206. Similarly, the URSCRE-ENA1-CYC1-lacZ plasmid pMP224 was constructed by inserting the double-stranded oligonucleotide AGCTATCGATTATTTCCTACTTCTATGACGTTT (the original ENA1 sequence is underlined) into pMP206 digested with HindIII. Constructs pMP226 (CRE-CYC1-lacZ) and pMP227 (mutant CRE [CRE*]-CYC1-lacZ) were obtained by insertion of double-stranded oligonucleotides ENACRE1/2 AGCTATCGATCTATGACGTTT (for pMP226; the original CRE sequence from ENA1 is underlined) or ENACRE3/4 AGCTATCGATCTATGAT*GTTT (for pMP227; the point mutation within CRE is indicated by the asterisk) into pMP206 digested with HindIII. In all cases, the number and orientation of inserted oligonucleotides were determined by sequencing. A GAL4DBD-SKO1 fusion plasmid (pMP235) was obtained by inserting a PCR fragment (SmaI/SalI) containing nearly the entire SKO1 gene (coding region for amino acids 4 to 647) in the two-hybrid vector pGBT9 (Clontech, Palo Alto, Calif.).
β-Galactosidase assay.
Transformed yeast strains were grown selectively until saturation in the appropriate SD liquid media and were diluted into YPD. Logarithmically growing cells (optical density at 660 nm of 0.5 to 0.8) were then transferred to fresh YPD, YPGal, or YPD with NaCl, KCl, or sorbitol, and β-galactosidase activity was determined after 1 h (0.3 M NaCl and KCl, 0.5 M sorbitol) or 4 h (YPGal, YPD with 0.8 M NaCl or KCl). The enzyme assay was performed as described elsewhere (14). Results presented are mean values obtained from at least three independent transformants measured in duplicate.
Purification of GST-Sko1p and gel retardation.
The entire reading frame of SKO1 was obtained by PCR using genomic DNA as template generating a NcoI restriction site around the ATG start site and a SalI restriction site after the stop site of SKO1. The fragment was inserted into the bacterial glutathione S-transferase (GST) expression vector pGEX-KG (Pharmacia Biotech) digested with NcoI and XhoI. Expression and purification by affinity chromatography of the full-length GST-Sko1 protein were performed as recommended by the manufacturer. For the protein-DNA binding studies, the double-stranded oligonucleotides ENACRE1/2 and ENACRE3/4 were 32P-labeled by the Klenow fill-in reaction and purified by polyacrylamide gel electrophoresis. In the binding assays, approximately 1 μg of GST-Sko1p was incubated with 0.5 ng of labeled probe in the presence of 0.5 μg of poly(dI-dC)–10 mM HEPES (pH 7.4)–15% glycerol–0.1 mM EDTA–20 mM NaCl–4 mM MgCl2–2 mM dithiothreitol at room temperature for 20 min. Binding reaction mixtures were directly loaded onto a 4% polyacrylamide gel in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA).
RESULTS
Deletion analysis of the ENA1 promoter.
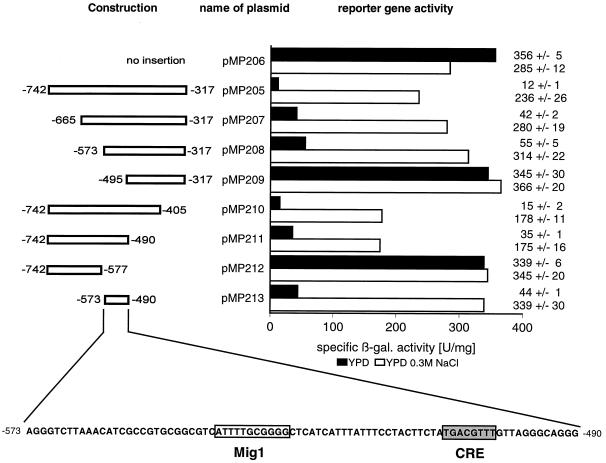
A preliminary analysis of the ENA1 promoter has shown that the region up to −752 (translational start point is +1) is responsible for the osmotically induced, calcineurin-independent expression of ENA1 (1). To identify sequences in this upstream control region of ENA1 that are important for its regulated expression, we investigated a set of PCR-generated segments of the ENA1 promoter in a CYC1-lacZ test vector under basal (YPD) and induced (0.3 M NaCl) conditions (Fig. 1). The control construct pMP206 gave under both conditions high levels of β-galactosidase activity due to the CYC1 TATA box-mediated expression. Insertion of a large upstream region of ENA1 (−317 to −742) resulted in a strong decrease of expression under normal growth conditions, while β-galactosidase levels again reached control values after osmotic shock. This result indicated that regulation of the ENA1 gene consists mainly of a derepression (rather than an activation) process. We also compared the induction of the artificial fusion of ENA1 (−317 to −742) with the CYC1 TATA box in construct pMP205 with that of an entire ENA1-lacZ fusion (up to −1380), but neither the induction capacity nor the kinetics of induction by 0.3 M NaCl was found to be significantly different (data not shown). Therefore, the promoter region from −317 to −742 was subjected to further deletion analysis (Fig. 1, plasmids pMP207 to pMP213). Subsequent removal of 5′ or 3′ sequences resulted in a total loss of regulation when the region from −490 to −573 (referred to below as URSENA1) was affected (plasmids pMP209 and pMP212). Moreover, this region alone was able to change the constitutive control into a salt-regulated promoter by its function as a repressor under basal conditions (plasmid pMP213). Interestingly, the only STRE sequence found in the ENA1 upstream region (AGGGG; −651 to −647) did not contribute to the regulation because its removal caused no loss of the responsiveness to stress (compare pMP207 and pMP208 in Fig. 1), and a promoter version containing STRE but not URSENA1 was no longer inducible by NaCl (pMP212). Assuming that URSENA1 contained the relevant protein-binding sites for the stress-regulated expression, we examined this 84-bp region by computer analysis using the MatInspector program (40). Within the URS element, two putative recognition sequences for transcription factors were found. Nucleotides −544 to −534 (ATTTTGCGGGG) perfectly match the consensus binding sequence of the yeast transcriptional repressor Mig1p (24), and nucleotides −509 to −502 (TGACGTTT) showed a similarity to mammalian CREs (4, 35).
FIG. 1.
Deletion analysis of the ENA1 promoter. Segments from the ENA1 upstream region indicated at the left were inserted into a CYC1-lacZ reporter. β-Galactosidase (β-gal.) specific activity (nanomoles per minute per milligram) was determined in transformed wild-type cells (W303-1A) after growth without (YPD) or with (YPD–0.3 M NaCl) salt. Absolute values including the standard deviation are given at the right. The sequence of URSENA1 (−490 to −573) is depicted at the bottom.
The ENA1 promoter contains functional Mig1p-binding and CRE sites.
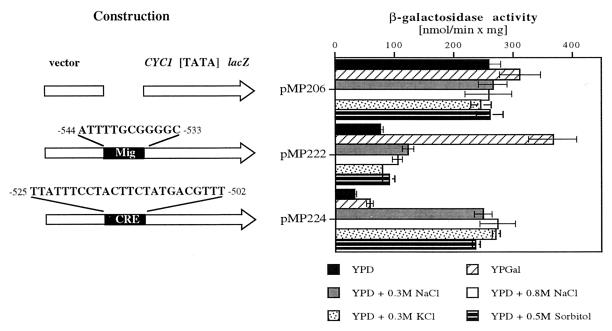
Preliminary characterization of the ENA1 control region qualified the region from −490 to −573 as a URS element. Since we found that the repression effect through URSENA1 was counteracted by osmotic stress (0.3 and 0.8 M of either NaCl or KCl) as well as by glucose starvation (galactose or ethanol as the carbon source [data not shown]), we now addressed the question of whether the response to these different environmental changes is triggered by distinct promoter motifs. By testing the two promoter elements separately, we found that both are efficiently repressing transcription under basal conditions (Fig. 2). However, they respond to completely different stimuli. A URSMIG-ENA1-regulated reporter gene was exclusively derepressed by glucose starvation (pMP222 with YPGal) but not by osmotic induction, while a URSCRE-ENA1-regulated reporter gene responded exclusively to osmotic stress (pMP224 with NaCl, KCl, and sorbitol) but not to glucose starvation. From these results, we conclude that glucose derepression and hyperosmotic shock induce ENA1 expression independently via (at least) two distinct URS elements, URSMIG-ENA1 and URSCRE-ENA1, whose separation allowed us now to investigate the function of transcription factors and signalling components that would specifically affect these repression elements.
FIG. 2.
URSMIG-ENA1 and URSCRE-ENA1 are functional repressor elements of the ENA1 promoter. Oligonucleotides containing the indicated sequences of the ENA1 promoter were inserted into a CYC1-[TATA]-driven lacZ reporter. β-Galactosidase activity was determined after growth of transformed cells (W303-1A) under basal (YPD), glucose-derepressed (YPGal), salt stress (YPD–0.3 M NaCl, YPD–0.8 M NaCl), or osmotic stress (YPD–0.3 M KCl or YPD–0.5 M sorbitol) conditions.
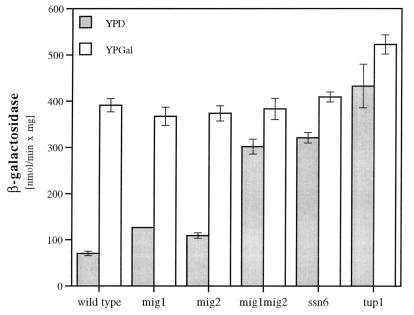
Repression through URSMIG-ENA1 depends on the function of Mig1p and Mig2p.
To characterize the roles of the two zinc finger repressors, Mig1p and Mig2p, that have been already reported to interact with the GC box motif (25, 36), we tested the repression effect of URSMIG-ENA1 in Δmig1, Δmig2, and Δmig1 Δmig2 mutant strains. In the absence of either MIG1 or MIG2, repression of a URSMIG-ENA1-regulated reporter (pMP222) was only partially lost, while the absence of both genes caused nearly the complete loss of regulation occurring on this promoter element (Fig. 3). This result indicated that both homologous repressors, Mig1p and Mig2p, contribute nearly equally to glucose repression on the ENA1 promoter. A similar effect of complete deregulation of URSMIG-ENA1 as for the Δmig1 Δmig2 mutant was observed in the absence of components of the general repressor complex SSN6-TUP1 (Fig. 3).
FIG. 3.
URSMIG-ENA1 function depends on MIG1, MIG2, SSN6, and TUP1. Repressed (growth in YPD) and derepressed (growth in YPGal) expression of a URSMIG-ENA1-CYC1-lacZ fusion gene (pMP222) was measured in transformed wild-type (W303-1A) and various mutant (MAP12, Δmig1; MAP21, Δmig2; MAP24, Δmig1 Δmig2; MAP6, Δssn6; MAP5, Δtup1) strains.
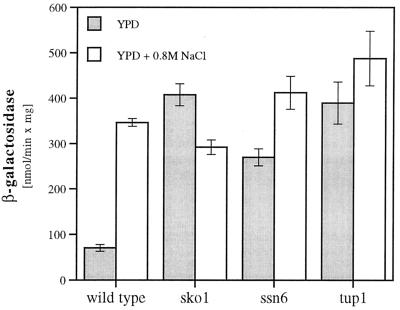
Regulation through URSCRE-ENA1 requires the functions of the bZIP repressor Sko1p (Acr1p) and the corepressors Ssn6p and Tup1p.
The yeast SKO1 (ACR1) gene has been found to encode a bZIP transcriptional repressor that binds to CRE sequences, although the physiological role of the protein remained undetermined (37, 56). We therefore examined whether the URSCRE-ENA1 promoter element would need Sko1p for its repression function. Indeed, as depicted in Fig. 4, a Δsko1 mutant showed a complete loss of regulation through the CRE-like ENA1 sequence, indicating that Sko1p is the CRE-interacting protein responsible for the osmotically regulated repression of ENA1. The URSCRE-ENA1 motif, like the URSMIG-ENA1 element, was dependent on a functional Ssn6p-Tup1p general repressor complex since Δssn6 or Δtup1 mutants were defective in repression of a URSCRE-ENA1-CYC1-lacZ reporter plasmid (Fig. 4). Although both negative cis elements of the ENA1 gene were dependent on Ssn6p-Tup1p, glucose and osmotic signalling were strictly separated on the level of the DNA-binding transcription factors, since the Δsko1 mutation did not affect URSMIG-ENA1-regulation, nor were Δmig1 mutants defective in URSCRE-ENA1 regulation (data not shown). Furthermore, we tested whether URSCRE-ENA1 was able to repress activated transcription. We found that when placed upstream or downstream of UASRap1 (binding site for the transcriptional activator Rap1p), the CRE of ENA1 was a functional repressor regulated by osmotic stress and dependent on Sko1p and Ssn6p (data not shown).
FIG. 4.
URSCRE-ENA1 function depends on SKO1, SSN6, and TUP1. Repressed (growth in YPD) and derepressed (growth in YPD–0.8 M NaCl) expression of a URSCRE-ENA1-CYC1-lacZ fusion gene (pMP224) was measured in transformed wild-type (W303-1A) and various mutant (MAP19, Δsko1; MAP6, Δssn6; MAP5, Δtup1) strains.
Sko1p binds to the CRE-like sequence of ENA1.
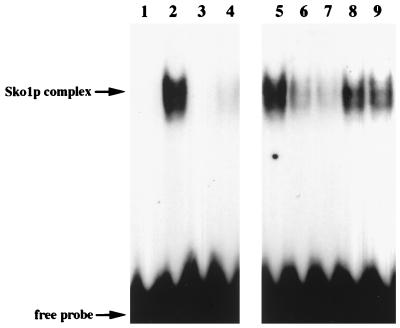
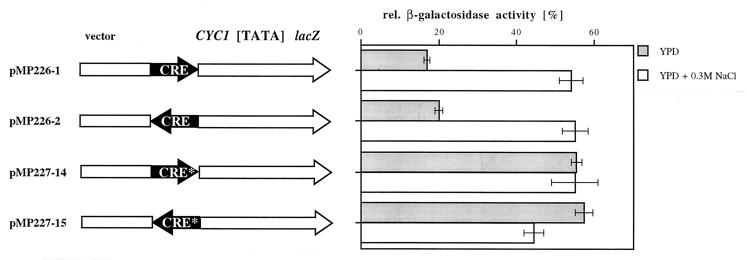
To test whether Sko1p can directly and specifically interact with the CRE motif of the ENA1 promoter, we performed gel retardation assays using oligonucleotides containing the entire CRE sequence or a point-mutated version of the binding motif changing the core sequence ACGT to ATGT (designated CRE*). As shown in Fig. 5, Sko1p bound specifically to the CRE sequence (lane 2) but not to CRE* (lane 4). Moreover, the Sko1p complex was efficiently competed by the use of nonlabeled CRE but not the CRE* sequence (Fig. 5, lanes 6 to 9). The same oligonucleotides were also tested for transcriptional repression by insertion into the CYC1-lacZ reporter system. As shown in Fig. 6, the 12 nucleotides representing the original CREENA1 motif repressed transcription under nonstress conditions independently of their orientation to the transcription start, while the CRE* sequence was not functional. The repression effect was counteracted by elevated concentrations of either NaCl (Fig. 6), KCl, or sorbitol (data not shown). Taken together, the results indicated that the binding of Sko1p to its CRE target sequence is responsible for ENA1 repression that is relieved by osmotic shock.
FIG. 5.
Sko1p binds to CREENA1 in vitro. Purified GST-Sko1p was incubated with labeled CRE (TGACGTTT) or CRE* (TGATGTTT). Lanes: 1, labeled CRE without added protein; 2, labeled CRE with added GST-Sko1p; 3, labeled CRE* without added protein; 4, labeled CRE* with added GST-Sko1p; 5, as lane 2; 6 (and 7), competition with 20× (and 50×) excess of CRE; 8 (and 9), competition with 20× (and 50×) excess of CRE*.
FIG. 6.
Binding of Sko1p correlates with the repression of CREs. Oligonucleotides that were used for Sko1p-binding assays (Fig. 5) were tested for repression ability in a CYC1-lacZ reporter. Constructions indicating the orientation of oligonucleotide insertion are given at the left. The repression effect of each oligonucleotide is depicted at the right as a percentage of the activity of the control plasmid without any insertion.
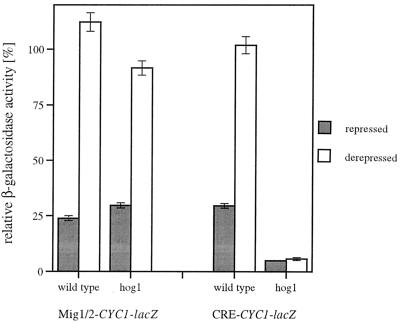
Derepression of URSCRE-ENA1 requires signalling through the Hog1p MAP kinase.
Induction of ENA1 expression during salt stress has been reported to be dependent on various signalling pathways (28). We therefore attempted to relate the CRE-mediated osmotic regulation to one (or more) of the known signal transduction pathways. We tested a variety of regulatory mutants for their effect on a URSCRE-ENA1-CYC1-lacZ gene. No significant change in the repression/derepression behavior was found for mutants bearing Δcnb1, defective in calcineurin phosphatase activity, or Δbcy1, defective in signalling through protein kinase A (PKA) by constitutively activating PKAs (data not shown). However, a dramatic effect was observed with a Δhog1 mutant with impaired HOG MAP kinase signalling. As can be seen in Fig. 7, the Δhog1 strain repressed a CRE-regulated reporter gene even more strongly under basal conditions and was unable to remove repression during osmotic shock, while a Mig1p-binding site-regulated control was not affected. These results strongly suggested that the Sko1p-mediated repression on the CRE sequence is a target of osmotic sensing through the HOG pathway.
FIG. 7.
Derepression of URSCRE-ENA1 is dependent on the Hog1p MAP kinase. A URSMIG-ENA1-CYC1-lacZ (pMP222) and a URSCRE-ENA1-CYC1-lacZ (pMP224) reporter were assayed in wild-type (YPH499) and Δhog1 mutant (JBY10) cells under repressed (YPD) and derepressed (YPGal for pMP222; YPD–0.3 M NaCl for pMP224) conditions. The degree of repression is given as relative β-galactosidase activity compared to the nonregulated empty CYC1-lacZ vector.
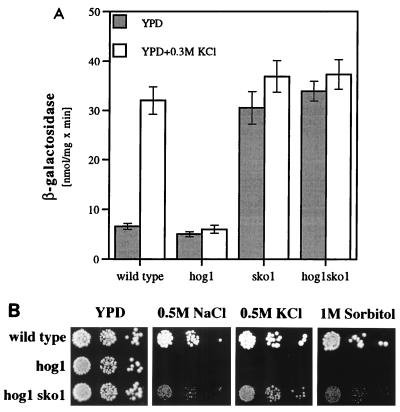
Disruption of SKO1 suppresses Δhog1 mutant phenotypes.
To test whether the Hog1p kinase acts through the Sko1p repressor, we investigated epistatic effects of the loss of SKO1 function over the hog1 disruption. We examined the expression of an integrated ENA1-lacZ fusion gene in Δhog1 and Δsko1 single mutants and in Δhog1 Δsko1 double mutants. As shown in Fig. 8A, osmotic induction of ENA1 by KCl is absent in hog1 mutants. On the other hand, a strong increase in basal ENA1 expression (YPD without salt) is observed in a Δsko1 strain. This elevated expression cannot be further induced by osmotic shock (0.3 M KCl). With respect to the expression of ENA1, the loss of SKO1 completely compensates for the hog1 deletion since the Δhog1 Δsko1 double mutant showed the same elevated expression levels as the Δsko1 strain. Therefore, we conclude that in the case of the osmotic up-regulation of ENA1, signalling through the HOG MAP kinase pathway acts on the Sko1p transcriptional repressor. This was also confirmed by the finding that additional deletion of SKO1 in a Δhog1 background partially suppressed the osmotic sensitivity of Δhog1 mutants (Fig. 8B). Again, this result clearly qualified Sko1p as a downstream effector of the HOG pathway.
FIG. 8.
Disruption of SKO1 acts in an epistatic manner over the loss of Hog1p function. (A) Expression of an integrative ENA1-lacZ gene (pFR70i) was measured under repressed (YPD) and derepressed (YPD–0.3 M KCl) growth conditions in strains W303-1A (wild type), MAP32 (Δhog1), MAP19 (Δsko1), and MAP33 (Δhog1 Δsko1). (B) Growth of wild-type strain W303-1A, MAP32 (Δhog1), and MAP33 (Δhog1 Δsko1) on high-osmolarity media.
A Gal4DBD-Sko1 fusion protein confers osmotic regulation to the GAL1 promoter that depends on Ssn6p and Tup1p function.
To prove that Sko1p contains transcriptional repression activity that is regulated by external osmolarity, we targeted the protein to the heterologous promoter of the GAL1 gene by expressing a GAL4DBD-SKO1 fusion. As depicted in Table 2 the fusion protein repressed the GAL1-lacZ reporter under normal growth conditions (YPD), while this down-regulation was counteracted by osmotic shock (0.3 M NaCl). This led to a strong osmotic regulation of the GAL1 promoter (10-fold) that is normally not regulated by salt. Moreover, the differential transcriptional control through Gal4-Sko1p is largely abolished in either ssn6 or tup1 mutants (Table 2), indicating that Sko1p function is dependent on the Ssn6-Tup1p corepressor complex.
TABLE 2.
A Gal4DBD-Sko1 fusion protein confers osmotic regulation to the heterologous GAL1 promotera
| Strain | GAL4DBD-SKO1 | β-Galactosidase sp act (nmol/min/mg)
|
Induction factor | |
|---|---|---|---|---|
| YPD | 0.3 M NaCl | |||
| Wild type | − | 1.08 ± 0.19 | 1.40 ± 0.10 | 1.3 |
| + | 0.24 ± 0.06 | 2.39 ± 0.63 | 10 | |
| ssn6 | − | 1.51 ± 0.20 | 1.26 ± 0.11 | 0.8 |
| + | 1.36 ± 0.16 | 2.74 ± 0.32 | 2.0 | |
| tup1 | − | 3.51 ± 0.49 | 2.93 ± 0.39 | 0.8 |
| + | 4.86 ± 0.58 | 9.25 ± 0.86 | 1.9 | |
The GAL4DBD-SKO1 fusion plasmid pMP235 was expressed in wild-type (SFY526) and in Δssn6 (MAP34) and Δtup1 (MAP35) mutant strains. Expression of the integrated GAL1-lacZ reporter was assayed after growth in YPD and YPD–0.3 M NaCl. Strains without the GAL4-SKO1 fusion contained the empty vector pGBT9.
ENA1 expression is modulated by multiple repressors.
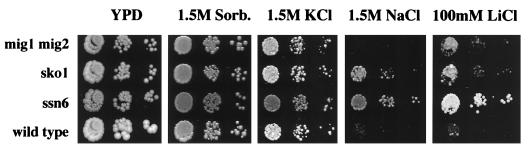
To test the extent to which the repressors Mig1/2p and Sko1p contribute to ENA1 regulation, we measured the transcriptional regulation in the whole promoter context, using an integrative ENA1-lacZ fusion. In general, the loss of one of the three repressors caused a partial derepression of ENA1 under normal growth conditions and concomitantly a drop in the level of induction observed upon glucose starvation or salt shock (Table 3). This was in agreement with the speculative role of all three repressors influencing the basal expression of ENA1. As was found for the specific regulation of URSMIG-ENA1, the whole ENA1 promoter was under the additive control of both repressors Mig1p and Mig2p since the double mutant showed an even higher degree of derepression than either of the single mutants. Although induction by both galactose and NaCl was reduced in a Δmig1 Δmig2 strain, specific involvement of Mig1/2p in salt induction is unlikely because a separate URSMIG-ENA1 clearly was not derepressed by salinity. The loss of SKO1 caused an increase in basal expression, and the inducibility of ENA1 upon salt stress was severely diminished whereas induction upon glucose starvation was only slightly affected. This was in agreement with the previous finding that Sko1p is responsible for the part of repression that is susceptible to osmotic stress. However, Δsko1 mutants still showed a fourfold derepression upon severe Na+ stress, and a Δsko1 Δmig1 Δmig2 strain, although dramatically impaired for ENA1 transcriptional regulation, still responded to a high sodium shock, indicating that part(s) of the salt-regulated repression is not regulated by Sko1p. Therefore, we also derepressed ENA1 transcription by osmotic stress by using 0.8 M KCl, which does not activate calcineurin signalling (28). Under these conditions, the wild type increases ENA1 expression fivefold, while no derepression occurs in the sko1 null mutant, which has derepressed ENA1 expression levels under normal conditions (Table 3). Similar results were obtained with moderate concentrations of NaCl (0.3 M [data not shown]), indicating that Sko1p mediates osmotic induction whereas upon high-sodium challenge, the ENA1 gene is additionally up-regulated by calcineurin-mediated activation. The most severe phenotype with respect to ENA1 expression was exhibited by a Δssn6 mutant strain, which lost all responsiveness due to the total derepression of the gene under nonstress conditions (Table 3). By applying severe salt stress under glucose derepression conditions (YPGal plus 0.8 M NaCl), the ENA1-lacZ gene was completely derepressed in the strains tested up to levels comparable to the expression observed in the Δssn6 strain (data not shown). We also tested whether the elevated ENA1 expression in the various mutants had consequences for the survival under osmotic and salt stress conditions. While a Δmig1 Δmig2 mutant strain grew only slightly better under conditions of elevated Li+ concentrations, the Δsko1 and Δssn6 mutant strains clearly showed a greater resistance to high concentrations of Na+ and Li+ (Fig. 9). Under osmotic stress conditions (1.5 M sorbitol and 1.5 M KCl), all strains grew similarly. According to their different degrees of derepression of the ENA1 gene (Table 3), Δssn6 mutants were more resistant to severe salt stress than Δsko1 mutants, as was particularly evident for Li+ resistance. However, loss of the glucose-regulated repression in the Δmig1 Δmig2 double mutant did not give rise to a clear salt resistance despite activating ENA1 expression even more strongly than the Δsko1 mutation. This result suggested that the Sko1p-Ssn6p/Tup1p-mediated regulation may also play an important role in the repression of other salt stress defense genes that are not affected by glucose repression.
TABLE 3.
ENA1 expression is modulated by multiple repressorsa
| Strain | β-Galactosidase sp act (nmol/min/mg of protein)
|
Derepression factor
|
|||||
|---|---|---|---|---|---|---|---|
| YPD | YPGal | YPD–0.8 M NaCl | YPD–0.8 M KCl | Galactose | 0.8 M NaCl | 0.8 M KCl | |
| Wild type | 6.3 ± 0.6 | 40.9 ± 1.7 | 81.6 ± 4.1 | 32.0 ± 2.8 | 6.5 | 13.0 | 5.1 |
| Δmig1 | 15.4 ± 0.9 | 64.1 ± 7.6 | 130.7 ± 17.1 | ND | 4.2 | 8.5 | ND |
| Δmig2 | 19.8 ± 1.8 | 93.2 ± 10.5 | 192.4 ± 21.5 | ND | 4.7 | 9.7 | ND |
| Δmig1 Δmig2 | 64.8 ± 0.9 | 138.6 ± 13.9 | 287.5 ± 28.9 | ND | 2.1 | 4.4 | ND |
| Δsko1 | 30.5 ± 3.3 | 155.4 ± 17.6 | 126.3 ± 19.3 | 36.9 ± 3.2 | 5.1 | 4.1 | 1.2 |
| Δsko1 Δmig1 Δmig2 | 101.3 ± 11.3 | 185.9 ± 19.8 | 364.5 ± 42.8 | ND | 1.8 | 3.6 | ND |
| Δssn6 | 316.7 ± 27.8 | 399.0 ± 46.1 | 423.7 ± 45.9 | ND | 1.2 | 1.3 | ND |
Transcriptional regulation of ENA1 was monitored by using an integrative ENA1-lacZ fusion (pFR70i) under repressed (YPD) and derepressed (YPGal, YPD–0.8 M NaCl, or YPD–0.8 M KCl) growth conditions. The strains used were W303-1A, MAP12, MAP21, MAP24, MAP19, MAP28, and MAP6 (see Table 1 for genotypes). ND, not determined.
FIG. 9.
Mutants Δsko1 and Δssn6 are resistant to Na+ and Li+ stress. Growth of mutant strains MAP24 (Δmig1 Δmig2), MAP19 (Δsko1), and MAP6 (Δssn6) on high-osmolarity media is compared with that of the wild type (W303-1A).
DISCUSSION
Increased expression of the ENA1 sodium and lithium extrusion ATPase in S. cerevisiae is a crucial adaptation to salt stress. We have shown that the underlying regulatory mechanism to adjust ENA1 expression is based mainly on a negative control through multiple DNA-binding repressors that act together with the general corepressor complex Tup1p-Ssn6p. Deletion of SSN6 has dramatic consequences with respect to the transcriptional regulation of ENA1, since the gene is nearly expressed to the maximal level under nonstress conditions and only a marginal up-regulation upon salt stress or glucose starvation is left. Similar results were obtained for a tup1 null mutant (data not shown). This is in complete agreement with the recent finding that the Ssn6p-Tup1p complex is involved in the osmotic regulation of stress-regulated genes (29). To different extents, transcriptional repression modulates the expression of other salt-inducible genes such as GPD1, CTT1, and ALD2, whose basal transcription is clearly increased in a Δssn6 background (29). In the case of these genes, a further activation during osmotic shock can be observed. Therefore, the overall regulation on these promoters is composed of a negative component keeping transcript levels low during normal growth and a positive component that activates transcription only when cells are confronted with high salinity or other stresses. A positively acting STRE with the core AGGGG has been found to be responsible for transcriptional activation as a response to a great variety of stresses (22, 27, 43). STREs can be regarded as the promoter elements that trigger the cellular multistress response, as they can be activated by osmotic, oxidative, and heat stress, as well as by nutrient starvation. Osmotic induction of STRE is dependent on signalling through the HOG MAP kinase pathway (43), and generally STRE regulation is negatively affected by the RAS-cAMP pathway (27). The two zinc finger proteins Msn2p and Msn4p directly bind and activate STRE sequences and are important determinants of the multistress resistance of yeast cells (30, 42). Clearly the ENA1 gene is not regulated by STREs. Only one sequence that matches the core sequence AGGGG (positions −651 to −647) can be found in the ENA1 upstream region, but this promoter region is dispensable for stress induction (Fig. 1 and reference 1). Moreover, in a Δmsn2Δmsn4 mutant background, ENA1 transcriptional regulation upon salt shock and glucose starvation is not affected compared to the wild type (1, 39a). Together with the finding that in repression-deficient (Δssn6 or Δtup1) mutants ENA1 transcription is nearly fully activated without any exposure to osmotic stress, it has to be concluded that stimulation of ENA1 expression by osmotic stress acts through the inactivation of repression. According to this concept, we present the identification of two separated promoter elements that mediate negative regulation via two different signalling systems, the general glucose repression pathway and the HOG MAP kinase pathway. The ENA1 promoter is activated by carbon source starvation and osmotic stress independently, and both stimuli additively increase ENA1 expression (1, 39a). The following results presented in this work demonstrate that repression by glucose is carried out by the binding of the zinc finger repressors Mig1p and Mig2p to the URSMIG-ENA1 element (−533 to −544): (i) a separate URSMIG-ENA1 element in single copy confers carbon source-regulated repression to a reporter gene; (ii) Δmig1 and Δmig2 single mutants (partially) and Δmig1Δmig2 double mutants (totally) lose repression of a URSMIG-ENA1-regulated reporter gene; and (iii) Δmig1, Δmig2, and Δmig1Δmig2 mutants show increased basal levels of expression of an integrated ENA1-lacZ gene. Obviously the two homologous repressor proteins Mig1p and Mig2p can recognize the same promoter region within ENA1. The same observation, although with a minor contribution of Mig2p, has been made for the glucose-regulated SUC2 gene (25). The mechanism by which Mig1p-mediated repression is removed upon carbon source starvation occurs through its phosphorylation by the Snf1p protein kinase (38, 38a, 53) and subsequent nuclear export (9). Accordingly, the induction of ENA1 by glucose derepression is absent in an snf1 null mutant (1).
Osmotic induction of many genes requires the HOG MAP kinase signalling cascade, and for CTT1 (encoding cytosolic catalase) and HSP12 (encoding a small heat shock protein), the importance of activated STREs has been demonstrated to be crucial for this process (43, 55). Nevertheless, it remains to be determined how the activated HOG pathway finally stimulates STRE-driven transcription via its binding factors Msn2p and Msn4p. Although ENA1 is a HOG-dependent gene, its differential expression during osmotic shock is completely independent of Msn2p/4p and STREs. A very different mechanism must be proposed to explain how the active Hog1p MAP kinase can modulate ENA1 expression. We provide several lines of evidence that this mechanism consists of the repressing activity of the bZIP transcription factor Sko1p, which recruits the Ssn6p-Tup1p corepressor by binding to the URSCRE-ENA1 motif: (i) a single copy of the URSCRE-ENA1 promoter element (−513 to −502) confers repression to a CYC1-lacZ reporter that is abolished under osmotic stress conditions; (ii) in the sko1, ssn6, and tup1 null mutants the URSCRE-ENA1-mediated repression is completely absent; (iii) a GST-Sko1 fusion protein binds specifically to the CREENA1 element in vitro; (iv) a Gal4DBD-Sko1 fusion protein acts as a Ssn6p/Tup1p-dependent repressor of the GAL1 promoter that is counteracted by osmotic stress; (v) in a hog1 null mutant, repression occurring on URSCRE-ENA1 is constitutive and cannot be overcome during salt treatment; (vi) osmotic stress-sensitive phenotypes of Δhog1 mutants can be rescued by additional deletion of SKO1; and (vii) Δsko1 mutants have increased basal ENA1 expression and are hyperresistant to Na+ and Li+ stress. Therefore, we propose a model of osmotic gene induction that implies the inactivation of a Sko1p-Ssn6p/Tup1p repressor complex by the activity of the Hog1p kinase. This scenario has an interesting parallelism to stress signalling in fission yeast and higher eukaryotes, where transcription factors of the bZIP family are phosphorylation targets of MAP kinase cascades (19, 49, 52, 60). In the specific case of S. pombe, a signal transduction pathway that is activated by a broader spectrum of adverse environmental conditions (osmotic, oxidative, heat, and UV stress) than in the case of the S. cerevisiae HOG pathway has been identified. It contains the Hog1p-homologous MAP kinase Sty1p (Spc1p) (20, 34, 47) that is activated by the MAP kinase kinase Wis1p (48, 58). As a final signalling target in S. pombe, the pleiotropic bZIP activator Atf1p has been identified (49, 51, 60). Activated Atf1p promotes the transcription from various target promoters, including those for stress response genes (gpd1+, fbp1+, and ctt1+) and genes required for sexual differentiation (ste11+), through binding to CRE motifs. Although the output of stress signalling in fission yeast was considered the primary Atf1p-mediated gene activation event, recent evidence suggests that transcriptional repression plays an important role in stress regulation of S. pombe (8).
Our genetic data strongly implicate the bZIP repressor Sko1p as a possible target of signalling through the HOG pathway of S. cerevisiae. Originally the SKO1 (ACR1) gene was identified as a multicopy suppressor of lethal PKA overexpression (37) and by the loss of repression mediated through ATF/CREB sites in a sko1 mutant (56). A contribution of Sko1p to the repression of SUC2 (encoding invertase) has been reported (37), although this effect was much weaker than that exhibited by Mig1p. Although a putative PKA target motif (KRRMS) within Sko1p has been described, the relationship of this transcriptional repressor to cAMP signalling has not been described. The in vitro binding of Sko1p homodimers to the canonical CRE (TGACGTCA) and related sequences has been shown (37, 56), but the physiological function of Sko1p remained undetermined. We have demonstrated that Sko1p mediates repression to the ENA1 gene that is counteracted upon osmotic shock by a mechanism dependent on Hog1p. Whether the Sko1p-CRE interaction also plays an important role in the regulation of other HOG-dependent genes should be investigated by identifying relevant CRE-like sequences in the various promoters. Although we demonstrate that Sko1p confers repression that is released by osmotic stress, a contribution of CRE-binding activators (56) to osmotic control cannot be excluded. We found no influence of signalling through PKA on the responsiveness of URSCRE-ENA1 to osmotic shock. In a bcy1 null mutant with constitutive PKA activity, derepression upon salt shock remained unaffected although in general the expression levels were markedly decreased both in the CYC1-lacZ control and in the URSCRE-ENA1-regulated reporter (data not shown). This points to a more general modulation through PKA on both basal and stress-induced expression, as has been described for regulation of the whole ENA1 promoter (28). However, the effect of a hog1 null mutation on the Sko1p- and Ssn6p-Tup1p-regulated CRE was dramatic, and further experimental approaches will be focused on the likely interaction of Sko1p with the corepressor complex and the mechanism of signal transduction from activated Hog1p MAP kinase to Sko1p. In addition to the data presented in this work, a connection between HOG signalling and transcriptional repression has been established by the finding that also ssn6 or tup1 null mutations can partially suppress the osmotic stress-sensitive phenotype of Δhog1 mutant cells (29), a result very similar to those presented in this work for the sko1 null mutation. Interestingly, the induction of the HOG-independent HAL1 gene (14) by severe osmotic stress also occurs through the release from Ssn6p-Tup1p repression (29). In this case, a protein complex formed on a negative HAL1 promoter element is abolished under stress conditions. Neither the DNA-binding protein nor the signal-transducing pathway operating in this case has been identified.
In addition to the negative mechanisms of regulation described in this work, ENA1 is also subjected to positive control. A third signal transduction pathway involving Ca2+/calmodulin-calcineurin contributes to the salt inducibility of the ENA1 gene (28, 33). Signalling through calcineurin is activated by high Na+ and Ca2+ concentrations and leads to the transcriptional activation of genes in addition to ENA1 such as PMC1 and PMR1, encoding Ca2+-ATPases (5), and FKS2, encoding a subunit of glucan synthase (10, 13). Very recently a calcineurin-dependent zinc-binding transcription factor, Crz1p (Tcn1p, Hal8p), that acts as an activator of transcription has been identified (31, 32, 50). The binding of Crz1p to a calcineurin-dependent UAS element in the FKS2 promoter has been described (50). Δcrz1 mutants show decreased ENA1 expression and are hypersensitive to salt stress (32). Most likely, calcineurin signalling results in ENA1 transcriptional activation through the binding of Crz1p. The UAS element responsible for this calcineurin-dependent activation in the ENA1 upstream control region has not been identified. However, a promoter region (positions −752 to −853 in ENA1) that responds to high Ca2+ therefore could include a binding site for Crz1p has been described (1).
We have demonstrated that Sko1p-mediated repression explains the osmotic regulation of the ENA1 gene since under conditions that do not activate the calcineurin pathway (0.8 M KCl or 0.3 M NaCl), the sko1 null mutant cannot further increase ENA1 expression. By additionally stimulating calcineurin signalling (0.8 M NaCl), the ENA1 promoter is further activated independently of HOG- and Sko1p-mediated repression (Table 3). Taken together, these findings allow us to propose a model to explain how different environmental signals are integrated on the ENA1 gene: (i) glucose starvation activates the Snf1p protein kinase that subsequently inhibits Mig1/2p-Ssn6p-Tup1p-mediated repression on the URSMIG-ENA1 element; (ii) the high Na+ or Ca2+ signal is triggered by the calcineurin phosphatase and subsequent activation of Crz1p (Tcn1p, Hal8p), which enhances transcription from a so far unidentified UASENA1; (iii) osmotic induction through the HOG pathway operates by counteraction of Sko1p-Ssn6p-Tup1p-mediated repression on the URSCRE-ENA1 element. The variety of regulatory events that influence transcription from the ENA1 promoter region render this gene a very productive and complex model of stress signalling.
ACKNOWLEDGMENTS
We thank A. Pascual-Ahuir and J. A. Márquez for very helpful collaboration, P. Kötter (Frankfurt am Main, Germany) for providing primers to create tup1, ssn6, and mig1 disruption cassettes, M. C. Gustin (Houston, Tex.) for strains YPH499 and JBY10, and A. Rodriguez-Navarro (Madrid, Spain) for plasmid pFR70i.
M.P. is supported by the European TMR program (ERB-FMRX-CT96-0007).
REFERENCES
- 1.Alepuz P M, Cunningham K W, Estruch F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol Microbiol. 1997;26:91–98. doi: 10.1046/j.1365-2958.1997.5531917.x. [DOI] [PubMed] [Google Scholar]
- 1a.Bartel P L, Chien C-T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 2.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 3.Celenza J L, Carlson M. A yeast gene that is essential for the release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 4.Comb M, Birnberg N C, Seasholtz A, Herbert E, Goodman H M. A cyclic AMP- and phorbol ester-inducible DNA element. Nature. 1986;323:353–356. doi: 10.1038/323353a0. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham K W, Fink G R. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J Cell Biol. 1994;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cyert M S, Kunisawa R, Kaim D, Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci USA. 1991;88:7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis T N, Urdea M S, Masiarz F R, Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986;47:423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- 8.Degols G, Russell P. Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:3356–3363. doi: 10.1128/mcb.17.6.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeVit M J, Waddle J A, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng W K, Faucette L, McLaughlin M M, Cafferkey R, Koltin Y, Morris R A, Young P R, Johnson R K, Livi G P. The yeast FKS1 gene encodes a novel membrane protein, mutations in which confer FK506 and cyclosporin A hypersensitivity and calcineurin-dependent growth. Gene. 1994;151:61–71. doi: 10.1016/0378-1119(94)90633-5. [DOI] [PubMed] [Google Scholar]
- 11.Ferrando A, Kron S J, Rios G, Fink G R, Serrano R. Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol Cell Biol. 1995;15:5470–5481. doi: 10.1128/mcb.15.10.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garciadeblás B, Rubio F, Quintero F J, Bañuelos M A, Haro R, Rodríguez-Navarro A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol Gen Genet. 1993;236:363–368. doi: 10.1007/BF00277134. [DOI] [PubMed] [Google Scholar]
- 13.Garrett-Engele P, Moilanen B, Cyert M S. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H+ ATPase. Mol Cell Biol. 1995;15:4103–4114. doi: 10.1128/mcb.15.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaxiola R, de Larrinoa I F, Villalba J M, Serrano R. A novel and conserved salt induced protein is an important determinant of salt-tolerance in yeast. EMBO J. 1992;11:3157–3164. doi: 10.1002/j.1460-2075.1992.tb05392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/ssDNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 16.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haro R, Garciadeblas B, Rodriguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 18.Hohmann S. Shaping up: the response of yeast to osmotic stress. In: Hohmann S, Mager W H, editors. Yeast stress responses. R. G. Austin, Tex: Landes Co.; 1997. pp. 101–134. [Google Scholar]
- 19.Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf H J, Herrlich P. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato T, Okazaki K, Murakami H, Stettler S, Fantes P, Okavama H. Stress signal, mediated by a HOG1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett. 1996;378:207–212. doi: 10.1016/0014-5793(95)01442-x. [DOI] [PubMed] [Google Scholar]
- 21.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi N, McEntee K. Identification of cis and trans components of a novel heat shock regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:248–256. doi: 10.1128/mcb.13.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Ishii S, Tokai M, Tsutsumi H, Ohke O, Akada R, Tanaka K, Tsuchiya E, Fukui S, Miyakawa T. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol Gen Genet. 1991;227:52–59. doi: 10.1007/BF00260706. [DOI] [PubMed] [Google Scholar]
- 24.Lundin M, Nehlin J O, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutfiyya L L, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 27.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Márquez J A, Serrano R. Multiple transduction pathways regulate the sodium-extrusion gene PMR2/ENA1 during salt stress in yeast. FEBS Lett. 1996;382:89–92. doi: 10.1016/0014-5793(96)00157-3. [DOI] [PubMed] [Google Scholar]
- 29.Márquez J A, Pascual-Ahuir A, Proft M, Serrano R. The Ssn6-Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG-dependent and -independent genes. EMBO J. 1998;17:2543–2553. doi: 10.1093/emboj/17.9.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-Pastor M T, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 31.Matheos D P, Kingsbury T J, Ahsan U S, Cunningham K W. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendizabal I, Rios G, Mulet J M, Serrano R, de Larrinoa I F. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 1998;425:323–328. doi: 10.1016/s0014-5793(98)00249-x. [DOI] [PubMed] [Google Scholar]
- 33.Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo J M. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- 34.Millar J B A, Buck V, Wilkinson M G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- 35.Montminy M R, Sevarino K A, Wagner J A, Mandel G, Goodman R H. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilm’s tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nehlin J O, Carlberg M, Ronne H. Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res. 1992;20:5271–5278. doi: 10.1093/nar/20.20.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Östling J, Carlberg M, Ronne H. Functional domains in the Mig1 repressor. Mol Cell Biol. 1996;16:753–761. doi: 10.1128/mcb.16.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Östling J, Ronne H. Negative control of the Mig1p repressor by the Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- 39.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 39a.Proft, M. Unpublished results.
- 40.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rios G, Ferrando A, Serrano R. Mechanisms of salt tolerance conferred by overexpression of the HAL1 gene in Saccharomyces cerevisiae. Yeast. 1997;13:515–528. doi: 10.1002/(sici)1097-0061(199705)13:6<515::aid-yea102>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schüller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schüller H-J, Hahn A, Tröster F, Schütz A, Schweitzer E. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 1992;11:107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano R. Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int Rev Cytol. 1996;165:1–52. doi: 10.1016/s0074-7696(08)62219-6. [DOI] [PubMed] [Google Scholar]
- 46.Serrano R, Márquez J A, Rios G. Crucial factors in salt stress tolerance. In: Hohmann S, Mager W H, editors. Yeast stress responses. R. G. Austin, Tex: Landes Co.; 1997. pp. 147–169. [Google Scholar]
- 47.Shiozaki K, Russell P. Cell cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- 48.Shiozaki K, Russell P. Counteractible roles of protein phosphatase 2C (PP2C) and a MAP kinase kinase homolog in the osmoregulation of fission yeast. EMBO J. 1995;14:492–502. doi: 10.1002/j.1460-2075.1995.tb07025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 50.Stathopoulos A M, Cyert M. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, Comb M J. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1997;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 53.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 55.Varela J C S, Praekelt U M, Meacock P A, Planta R J, Mager W H. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol. 1995;15:6232–6245. doi: 10.1128/mcb.15.11.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vincent A C, Struhl K. ACR1, a yeast ATF/CREB repressor. Mol Cell Biol. 1992;12:5394–5405. doi: 10.1128/mcb.12.12.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eucaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 58.Warbrick E, Fantes P. The Wis1 protein kinase is a dose-dependent regulator of mitosis in Schizosaccharomyces pombe. EMBO J. 1991;10:4291–4299. doi: 10.1002/j.1460-2075.1991.tb05007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wieland J, Nitsche A M, Strayle J, Steiner H, Rudolph H K. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkinson M G, Samuels M, Takeda T, Toone W M, Shieh J-C, Toda T, Millar J B A, Jones N. The Atf transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]