Abstract
Iron is an essential micronutrient for all eukaryotic organisms because it participates as a redox cofactor in multiple metabolic processes. Iron bioavailability is highly restricted due to the low solubility of its oxidized form, frequently leading to iron deficiency anemia. The baker’s yeast Saccharomyces cerevisiae is used as a model organism for iron homeostasis studies, but also as a food supplement and fermentative microorganism in the food industry. Yeast cells use the vacuolar Ccc1 transporter to detoxify and store excess iron in the vacuoles. Here, we modulate CCC1 expression and properties to increase iron extraction from the environment. We show that constitutive expression of full-length CCC1 is toxic, whereas deletion of its cytosolic amino-terminal (Nt) domain (NtΔCCC1) rescues this phenotype. Toxicity is exacerbated in cells lacking AFT1 transcription factor. Further characterization of NtΔCcc1 protein suggests that it is a partially functional protein. Western blot analyses indicate that deletion of Ccc1 Nt domain does not significantly alter GFP-Ccc1 protein stability. A functional full-length GFP-Ccc1 protein localized to particular regions of the vacuolar membrane, whereas GFP-NtΔCcc1 protein was evenly distributed throughout this endogenous membrane. Interestingly, expression of NtΔCCC1 increased the accumulation of endogenous iron in cells cultivated under iron-sufficient conditions, a strategy that could be used to extract iron from media that are not rich in iron.
Keywords: yeast, Saccharomyces cerevisiae, iron, Ccc1
1. Introduction
Iron is an essential micronutrient for all eukaryotic organisms. Its redox and coordination properties allow iron to participate as a cofactor in a wide range of metabolic processes including the synthesis of DNA, proteins, and lipids, mitochondrial respiration and photosynthesis, and oxygen transport. Despite being one of the most abundant elements in the Earth’s crust, iron bioavailability is extremely restricted in aerobic environments as it is mostly present in its oxidized from (Fe3+), which is highly insoluble at physiological pH. According to the World Health Organization, iron deficiency anemia (IDA) is one of the most prevalent human nutritional disorders (even present in developed countries) with high impact and potentially fatal health consequences in children, women of fertile age, pregnant women, and the elderly (reviewed in [1,2]). Consequences of IDA include diminished learning ability and poor physical growth of infants, fatigue and reduced work productivity in adults, greater susceptibility to infections, and increased risk of death associated with pregnancy and childbirth. Multiple strategies are used to prevent and treat IDA including diversification of the diet and supplementation or food fortification with particular iron forms (reviewed in [3]).
The baker’s yeast Saccharomyces cerevisiae contributed significantly to the identification and characterization of many components of the human, plant, and fungal iron homeostatic networks. Moreover, this yeast is used in multiple biotechnological applications including fermentative processes to obtain different food products (e.g., bread, wine, and beer), as a nutritional supplement since it is rich in proteins, vitamins, and fiber, and as a factory for the production of compounds of interest. Iron-enriched yeasts are currently being evaluated as promising sources of iron to prevent and palliate iron deficiency in humans and animals [4]. In fact, iron-enriched baker’s yeasts are efficient in helping animals to recover from iron deficiency [5]. However, when present in excess, iron can engage in Fenton redox reactions that promote the formation of reactive oxygen species (ROS), which damage cells at the level of proteins, lipids, and DNA [6]. Therefore, yeast cells must coordinate iron acquisition, distribution, utilization, detoxification, and storage.
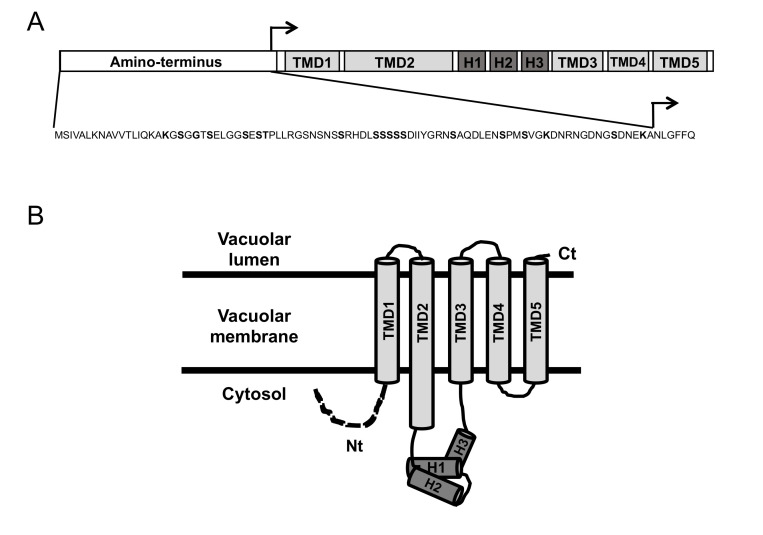
Opposite to animals, yeast cells do not express ferritin; instead they accumulate iron in the vacuole for detoxification and storage purposes [7]. Iron can enter yeast vacuoles by endocytosis and via Ccc1, a protein that localizes to the vacuolar membrane and transports iron into its lumen [8,9]. Since yeast cells cannot excrete iron, Ccc1 constitutes the main yeast iron detoxification and resistance factor. Thus, yeast cells lacking CCC1 (ccc1Δ mutants) are highly sensitive to elevated levels of environmental iron [8,10]. With the exception of animals, Ccc1 homologs are present in many organisms, including bacteria, protists, fungi, and plants, where they are denoted VIT1 proteins (reviewed in [11]). The resolution of the crystal structure of Eucalyptus grandis VIT1 ortholog (EgVIT1) made possible the identification of essential and conserved motifs required for metal transport, as well as other species-specific domains [11,12]. According to EgVIT1 structure and protein alignments, Ccc1/VIT1 proteins, including yeast Ccc1, possess five transmembrane domains (TMDs), a metal-binding loop containing three alpha helixes (H1–H3) between TMD2 and TMD3 that faces the cytosol, a cytosolic amino-terminal (Nt) region, and a vacuolar carboxyl terminus (Figure 1) [12]. Structure-function data in EgVIT1 also suggest that Ccc1/VIT1 proteins dimerize and function as H+ antiporters for Fe2+ [12]. The Ccc1/VIT1 cytosolic Nt domain is not conserved and displays different properties depending on the species [11]. Yeast possesses a cytosolic Ccc1 Nt with serine and lysine residues that have been identified in proteomic studies as phosphorylated and ubiquitylated, respectively [13,14]. Moreover, Malaysian wild S. cerevisiae strains harboring a mutation in their Nt region (G22R) are sensitive to high iron concentrations [15]. Thus, although according to conservation features the Nt domain of yeast Ccc1 seems dispensable, it may be important for regulatory functions.
Figure 1.
Schematic representation of Ccc1/VIT1 domain organization (A) and membrane topology (B), based on EgVIT1 structure and protein alignments [11,12]. A and B: Grey rectangles/cylinders represent transmembrane domains (TMDs); dark grey rectangles/cylinders represent three helices. A: White rectangles represent the Nt domain, whose amino acid sequence is indicated below. Bold letters indicate phosphorylated and ubiquitylated residues, as well as mutation G22R in Malaysian yeast strains [13,14,15]. Black arrow indicates where NtΔCcc1 protein starts.
Multiple transcription factors contribute to upregulation of CCC1 mRNA levels in response to iron excess (reviewed in [11,16,17]). A member of the yeast activator protein (Yap) subfamily of transcriptional factors denoted Yap5 associates to specific cis elements within CCC1 promoter and activates its transcription in response to iron excess [10,18,19,20]. Moreover, the low-glucose sensor Snf1 also contributes to CCC1 mRNA upregulation in high iron conditions by activating the general stress transcription factors Mns2 and Msn4 [21]. In response to iron deficiency, yeast Aft1 and Aft2 transcription factors activate iron acquisition via the Fet3-Ftr1 high-affinity iron transport system, iron mobilization from the vacuole, and a metabolic remodeling mediated by the mRNA-binding protein Cth2 that post-transcriptionally limits CCC1 expression [17,18,22,23,24].
Previous studies demonstrated that CCC1 overexpression promotes iron uptake and accumulation into the vacuole [8,9,25]. Although this strategy seems appropriate to extract iron from the environment and enrich yeast cells with iron, we show here that constitutive expression of CCC1 is toxic to yeast cells cultivated in iron replete conditions. Interestingly, deletion of the Ccc1 Nt domain rescues toxicity, allowing a more efficient extraction of iron from the surrounding milieu. A potential function of Ccc1 Nt is discussed.
2. Materials and Methods
2.1. Yeast Strains
Yeast strains used in this study (Invitrogen) include BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0), ccc1Δ (BY4741 ccc1::KanMX6), ccc1Δ (BY4743 ccc1::KanMX6/ccc1::KanMX6), fet3Δ (BY4741 fet3::KanMX6), ftr1Δ (BY4741 ftr1::KanMX6), aft1Δ (BY4741 aft1::KanMX6), and cth2Δ (BY4741 cth2::KanMX6).
2.2. Plasmids
Plasmids used in this work include empty vectors p416TEF [26], p416GAL, and pUG36 (kindly provided by U. Güldener and J. H. Hegemann, Heinrich Heine University Düsseldorf, Germany), and the corresponding vectors with either CCC1 or NtΔCCC1 (lacking the first 87 Nt amino acids) coding sequences cloned into BamHI and XhoI. The pUG36 vector expresses either CCC1 or NtΔCCC1 coding sequences under the control of the methionine-regulated MET17 promoter and adds a GFP tag at the Nt end.
2.3. Growth Conditions
Yeasts were cultivated at 30 °C in synthetic complete (SC) medium lacking uracil (SC-Ura) or lacking both uracil and methionine (SC-Ura-Met). Glucose was substituted with galactose in SC-Ura/Galactose medium. Iron was added to SC media at the indicated concentrations from a 100 mM Fe(NH4)2(SO4)2 (FAS, Merck-Sigma) stock solution. Stock solutions were prepared in 0.1M HCl to facilitate iron solubility and were used fresh. A control with the equivalent HCl concentration was always prepared. The Fe2+-specific chelator ferrozine (Fz, Merck-Sigma) was added to SC plates from a 100 mM stock solution freshly prepared in water. To grow yeast cells in liquid media, SC-Ura or SC-Ura-Met precultures were reinoculated at an optical density at 600 nm (OD600nm) of 0.2 and then incubated at 30 °C and 190 rpm in 50 mL flasks overnight. Alternatively, for duplication time measurements, yeast cells were inoculated in 96-well plates at an OD600nm of 0.2 in 270 μL of liquid SC-Ura medium with 500 μM FAS, and the OD600nm was determined in a Spectrostar Nano absorbance microplate reader (BMG Labtech, Ortenberg, Germany) every 30 min for 36 h at 28 °C. Duplication times were calculated using the online tool GCAT (Growth Curve Analysis Tool) as described in [27]. To perform growth assays in solid media, cells from overnight precultures were diluted to an OD600nm of 0.1 and then spotted directly and after 1:10 and 1:100 dilutions in sterile water. Plates were incubated at 28 °C for 3 to 6 days and were then photographed.
2.4. Iron Measurements
Yeast cells were collected and treated as previously described, and a colorimetric assay was used to determine the endogenous levels of iron [28,29]. To determine yeast dry weight (DW), cells corresponding to a total OD600nm x mL of 30 were collected, washed, transferred to a previously weighed Eppendorf tube, and dried at 50 °C for 3 days. Finally, yeast DW was obtained by subtracting the total weight from the Eppendorf tube weight. To determine the iron that yeast cells extracted from the surrounding medium, endogenous iron levels were divided by the volume of medium in which these cells were growing.
2.5. RNA Analyses
To determine the expression levels of particular mRNAs, total RNA was extracted and reverse transcription-quantitative PCR (RT-qPCR) was performed as previously described [30]. ACT1 mRNA values were used to normalize data. Data and error bars represent the averages and standard deviations from three independent biological experiments. Specific primer pairs for FTR1, CTH2, and ACT1 were used.
2.6. Protein Analyses
To determine Ccc1 and NtΔCcc1 protein stability, exponentially growing cells were cultivated in SC-Ura-Met without (iron-sufficient conditions) or with 3 mM FAS (high-iron conditions) for 1 h (modified from [31]). CCC1 synthesis was halted by adding methionine to a final concentration of 3 mM and cells were harvested at the indicated time points. Yeast proteins were extracted by the alkali method and processed and analyzed by western blot, as previously detailed [32,33]. Anti-GFP (Roche) and anti-Pgk1 (Thermo Fisher Scientific, Waltham, MA, USA) were used to detect GFP-Ccc1/GFP-NtΔCcc1 and Pgk1 proteins, respectively.
2.7. Fluorescence Microscopy
Images were captured on an Eclipse 90i Nikon microscope (Nikon Corporation, Tokio, Japan) using a digital camera (Nikon DS-5Mc, Tokio, Japan). To stain vacuolar membranes, cells were incubated for 15 min in 8 μM of FM4-64, washed, and incubated for an additional hour.
2.8. Statistical Analyses
Tailed T-student tests were used to determine statistical significance. Significant differences (p-value < 0.05) are indicated by different letters, whereas at least one letter being the same indicates that there are no significant differences.
3. Results
3.1. The Toxicity of a Constitutively Expressed CCC1 Is Rescued by Iron Addition
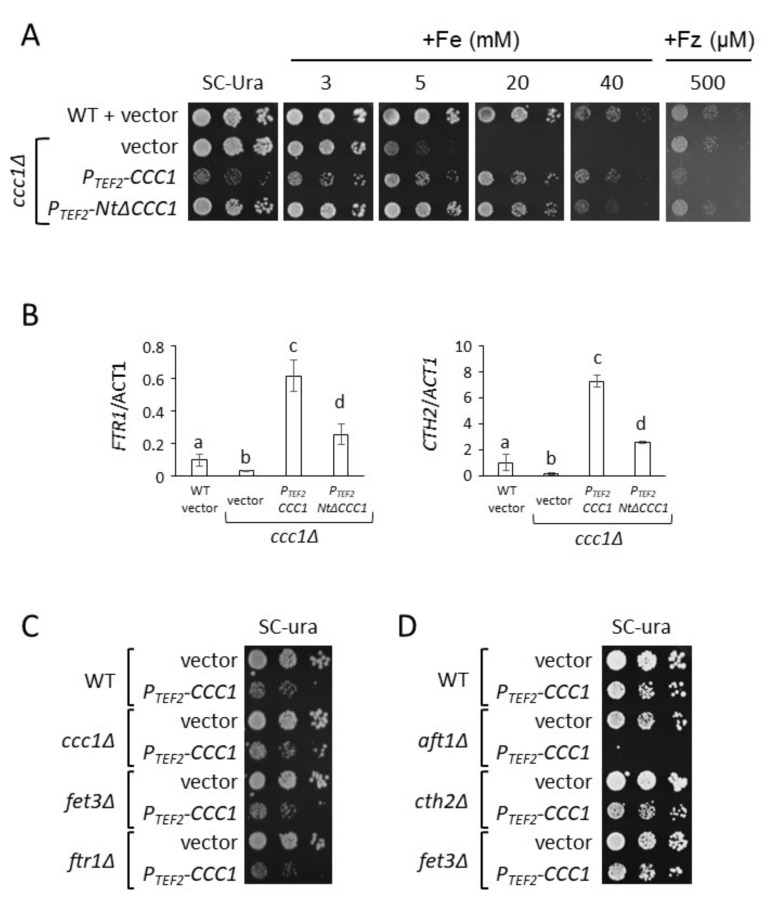
Previous studies demonstrated that overexpression of the vacuolar iron transporter CCC1 increases iron uptake and total iron content, leading to its accumulation in the vacuole [8,9,25]. Given that iron is naturally detoxified and stored in the vacuole, we considered that overexpression of CCC1 could be an appropriate strategy to increase yeast endogenous iron content. Therefore, we expressed the CCC1 coding sequence under the control of the constitutive and high-expression promoter of TEF2 gene (PTEF2-CCC1) in a ccc1Δ strain. Since CCC1 deletion causes high cellular sensitivity to extracellular iron, we expected that CCC1 overexpression would provide iron resistance [8,10]. To test yeast growth, we performed spot growth assays in solid media with increasing concentrations of ferrous ammonium sulfate (FAS). We used wild-type (WT) cells and ccc1Δ mutants as controls. As previously described, ccc1Δ cells (ccc1Δ + vector) grow similarly to WT cells (WT + vector) under iron-sufficient conditions (SC-Ura); however, they are more sensitive to an increase in extracellular iron, which limits ccc1Δ growth at 5 mM FAS and fully abrogates growth at 20 mM FAS (Figure 2A) [8]. Surprisingly, we observed that overexpression of CCC1 with TEF2 promoter was extremely toxic under normal iron-sufficient conditions, whereas growth was recovered upon addition of iron to the growth medium, resulting in higher resistance than ccc1Δ cells and only slightly less tolerance than WT cells (Figure 2A). These results indicate that altering the expression of CCC1 under iron replete conditions could be detrimental to cells.
Figure 2.
Toxicity of CCC1 constitutive expression under iron-sufficient conditions is rescued by iron addition or by deleting the Ccc1 cytosolic amino-terminal region. BY4741 (WT) cells were transformed with p416TEF (vector), and ccc1Δ cells were transformed with p416TEF (vector), p416TEF-CCC1 (PTEF2-CCC1), and p416TEF-NtΔCCC1 (PTEF2-NtΔCCC1) plasmids. (A) Expression of CCC1 altered growth in media with different iron availability. Cells were cultivated overnight in SC-Ura and then spotted on SC-Ura media containing different concentrations of FAS (Fe) or ferrozine (Fz). (B) FTR1 and CTH2 mRNA levels. Yeast cells were cultivated in SC-Ura + 500 μM FAS for 6 h. Total RNA was extracted and the levels of particular mRNAs were determined by RT-qPCR. ACT1 was used to normalize data. Three independent biological replicates were performed and the average and standard deviation were determined relative to the WT. Different letters above the bars represent statistically significant differences (p-value < 0.05). (C,D) Deletion of the high-affinity iron uptake system did not rescue CCC1 overexpression toxicity. BY4741 (WT), ccc1Δ, fet3Δ, ftr1Δ, aft1Δ, and cth2Δ cells transformed with p416TEF (vector) or p416TEF-CCC1 (PTEF2-CCC1) were spotted on SC-Ura. Plates were incubated at 30 °C and photographed.
A potential explanation for the increase in iron acquisition and accumulation in the vacuole previously described for CCC1-overexpressing cells is an activation of the iron regulon due to decreased cytosolic and mitochondrial iron levels. Indeed, we observed that PTEF2-CCC1 cells increased the expression of the members of the iron regulon, including the Aft1 targets FTR1 and CTH2, as compared with WT cells (Figure 2B). Given that deletion of FET3, which encodes for an essential component of the cell surface Fet3-Ftr1 high-affinity iron transport complex, was shown to fully remove iron uptake of CCC1-overexpressing cells, we ascertained whether deletion of either FET3 or FTR1 would rescue its toxicity under iron-sufficient conditions [25]. As seen in Figure 2C, none of the deletions improved growth of PTEF2-CCC1 cells, suggesting that other reasons beyond increased iron uptake and accumulation are responsible for CCC1 toxicity under normal conditions (see Discussion). Consistent with this finding, decreasing extracellular bioavailability with the addition of an Fe2+-specific chelator (ferrozine, Fz) did not rescue growth of PTEF2-CCC1 cells (Figure 2A, Fz). Previous data indicated that increased expression of CTH2 under normal growth conditions also leads to cellular growth defects [33,34]. Since we observed that CCC1 overexpression increases CTH2 mRNA levels 7-fold (Figure 2B), we assessed whether deletion of CTH2 rescued CCC1 overexpression toxicity. As shown in Figure 2D, deletion of CTH2 did not promote growth when CCC1 is overexpressed, which indicates that the increased levels of CTH2 are not the reason for toxicity. Next, we explored whether deletion of AFT1 transcription factor, which would simultaneously limit iron uptake, iron mobilization from the vacuole, and CTH2 expression, rescued the CCC1 overexpression phenotype. Remarkably, we observed that instead of recovering growth, deletion of AFT1 dramatically increased the toxicity of CCC1 overexpression (Figure 2D). This result and the lack of rescue by FTR1, FET3, and CTH2 deletions contradict the hypothesis that the underlying reason for CCC1 toxicity is an increase in iron uptake, and strongly suggest that a decrease in cytosolic iron bioavailability, due to higher transport of iron into the vacuole mediated by Ccc1, is the most likely reason for this phenotype.
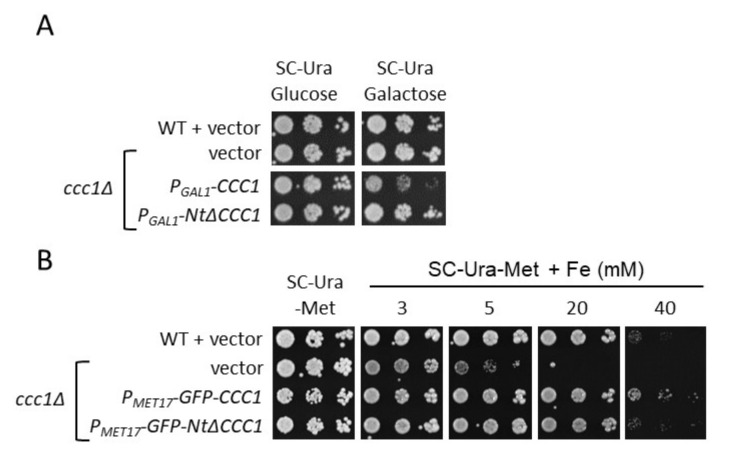
To solve the toxicity caused by CCC1 overexpression, we decided to test the regulatable promoters of GAL1, which is repressed by glucose and activated in media with galactose as the single carbon source (PGAL1-CCC1 cells), and MET17, which is repressed by methionine (PMET17-GFP-CCC1 cells). PGAL1-CCC1 cells exhibited a strong toxicity when spotted on a galactose medium (SC-Ura/Galactose) (Figure 3A). This phenotype was probably due to excessive CCC1 expression, as no growth defect was observed in SC-Ura (Figure 3A). Furthermore, PMET17-GFP-CCC1 cells were cultivated in methionine-containing conditions to inhibit CCC1 expression and then spotted on SC-Ura plates lacking methionine (SC-Ura-Met), which allows CCC1 expression. Under these conditions, we did not observe any growth defect as compared with WT cells (Figure 3B), probably due to lower expression levels of MET17-driven expression as compared with GAL1 and TEF2 promoters. We noticed that PMET17-GFP-CCC1 cells were as resistant to high iron concentrations as WT cells, which supports the notion that the GFP-Ccc1 tagged protein is functional (Figure 3B). Interestingly, iron resistance of PMET17-GFP- NtΔCCC1 cells was lower than that of PMET17-GFP-CCC1 cells, suggesting that the Ccc1 Nt region is important for function (Figure 3B). Moreover, these results indicate that the promoter that drives CCC1 expression is critical for appropriate growth.
Figure 3.
Growth of yeast cells expressing CCC1 with regulatable promoters. (A) Expression of CCC1, but not NtΔCCC1, with GAL1 promoter is toxic. BY4741 (WT) cells transformed with p416TEF (vector), and ccc1Δ cells transformed with p416TEF (vector), p416GAL-CCC1 (PGAL1-CCC1), and p416GAL-NtΔCCC1 were cultivated overnight in SC-Ura and then spotted on normal SC-Ura (SC-Ura Glucose) and SC-Ura without glucose but with 2% galactose (SC-Ura Galactose). (B) Expression of CCC1 with MET17 promoter rescues toxicity. BY4743 (WT) cells were transformed with p416TEF (vector), and ccc1Δ cells were transformed with p416TEF (vector), pUG36-CCC1 (PMET17-GFP-CCC1), and pUG36-NtΔCCC1 (PMET17-GFP-NtΔCCC1). Cells were cultivated overnight in SC-Ura (which contains methionine) to inhibit CCC1 expression, and then spotted on SC-Ura-Met media with different concentrations of FAS (Fe). Plates were incubated at 30 °C and photographed.
3.2. Deletion of CCC1 Amino-Terminal Region Rescues Toxicity by Limiting Iron-Resistance Capacity
Yeast CCC1 protein contains an Nt region that faces the cytosol (Figure 1), and whose function has not been described. According to genome-wide data, this Nt domain contains multiple serine residues that are phosphorylated and lysine residues that are ubiquitylated [13,14]. To test whether this region was important for Ccc1 function under high-iron conditions or contributed to its toxicity under iron-sufficient conditions, we constructed a Ccc1 protein lacking its 87 Nt amino acids (NtΔCcc1). Remarkably, expression of NtΔCCC1 with either TEF2, GAL1, or MET17 promoter did not alter cell growth in iron-sufficient conditions (Figure 2A and Figure 3). A potential explanation for this result could be that deletion of the Nt region fully removes Ccc1 function, but this was not the case as both PTEF2-NtΔCCC1 and PMET17-NtΔCCC1 constructs conferred an iron resistant phenotype only slightly lower than that of the cells expressing CCC1 under the control of the same promoter (Figure 2A and Figure 3B). Therefore, in contrast to full-length Ccc1, overexpression of NtΔCCC1 does not cause cellular toxicity.
3.3. Cultures of NtΔCCC1 Cells Are More Efficient in Extracting Iron from Media
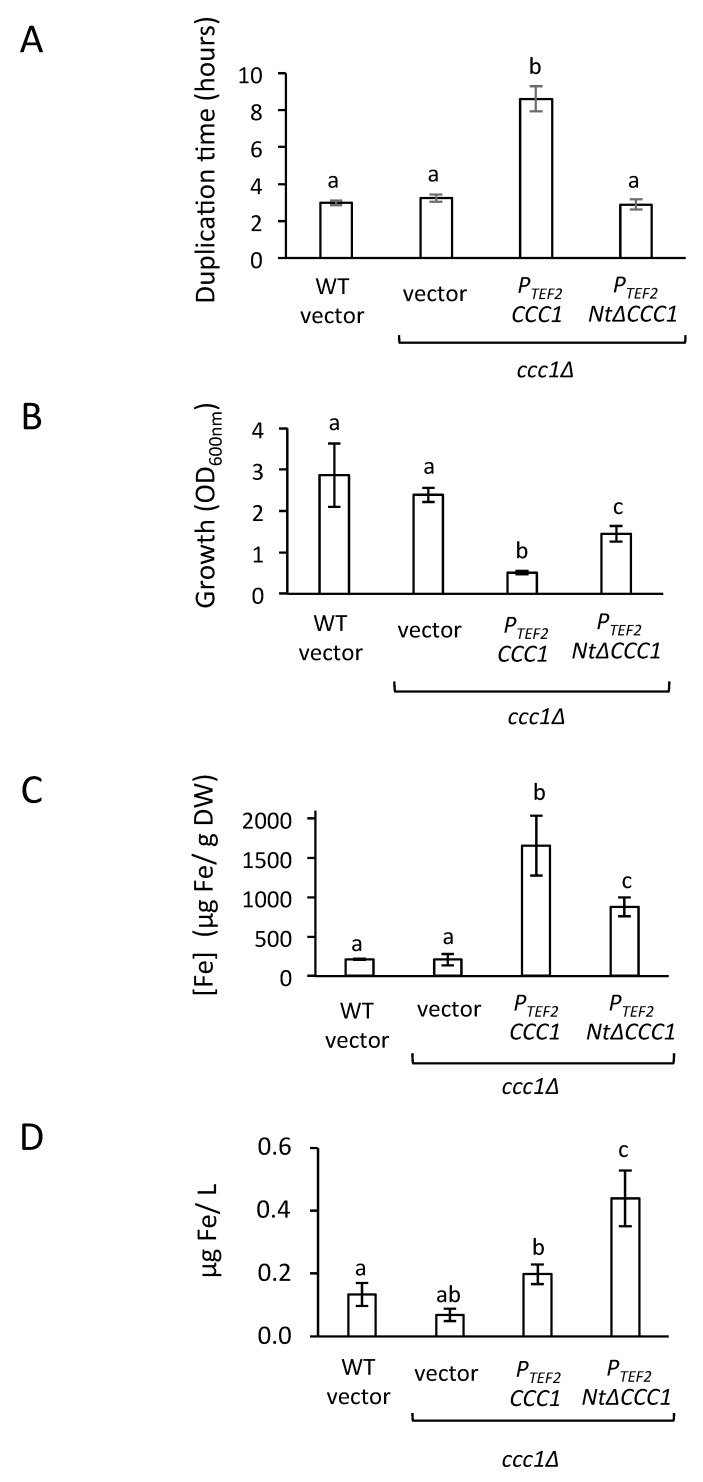
To further study the contribution of the Ccc1 Nt region to iron acquisition and growth, we first cultivated WT, ccc1Δ, and cells expressing either PTEF2-CCC1 or PTEF2-NtΔCCC1 for 36 h in a microplate reader and calculated the duplication times from the resulting growth curves (Figure 4A and Figure S1). While we observed similar duplication times for WT, ccc1Δ, and PTEF2-NtΔCCC1 cells, PTEF2-CCC1 cells showed a 3-fold increase in duplication time, underscoring an important growth defect, in accordance with the results observed in solid media. Although PTEF2-NtΔCCC1 cells showed a growth curve similar to those of WT and ccc1Δ, a small growth defect was indicated by the lower maximum OD600nm achieved. To further characterize this differential growth, we then cultivated WT, ccc1Δ, and cells expressing either PTEF2-CCC1 or PTEF2-NtΔCCC1 for six hours in a liquid medium containing 500 μM FAS. We observed that WT and ccc1Δ cells grew properly, whereas PTEF2-CCC1 cells again displayed a dramatic growth defect (Figure 4B). Remarkably, although PTEF2-NtΔCCC1 cells grew better than PTEF2-CCC1 cells did, they also exhibited a significant growth decrease as compared with WT and ccc1Δ cells (Figure 4B). Next, we measured cellular iron accumulation and observed an 8-fold and 4-fold increase in iron accumulation in PTEF2-CCC1 and PTEF2-NtΔCCC1 cells, respectively, as compared with WT cells (Figure 4C). These results suggest that CCC1-overexpressing cells could be used to extract and accumulate iron from the environment. However, when we determined total iron extracted per volume of medium, we observed that PTEF2-CCC1 cells did not improve the capacity of WT or ccc1Δ cells to remove iron from the environmental milieu, mostly due to their low growth (Figure 4D). Interestingly, PTEF2-NtΔCCC1 cells extracted twice as much iron from the extracellular medium as the rest of the cells assayed did (Figure 4D). These results suggest that overexpressing an Nt-truncated Ccc1 protein could be an interesting strategy to more efficiently obtain iron from media.
Figure 4.
Overexpression of NtΔCCC1 improves iron extraction from media. (A) Yeast cells described in Figure 2 were inoculated at an OD600nm of 0.2 in SC-Ura with 500 μM FAS in a 96-well microplate reader and duplication times were calculated from the resulting growth curves as described in Materials and Methods. (B–D) Yeast cells described in Figure 2 were cultivated in SC-Ura + 500 μM FAS for 6 h, and OD600nm (B), μg Fe/g dry weight (DW) (C), and μg Fe/L of medium (D) were determined. Three independent biological replicates were performed. The average and standard deviation were determined. Different letters above the bars represent statistically significant differences (p-value < 0.05).
3.4. Ccc1 Amino-Terminus Modulates Subcellular Localization
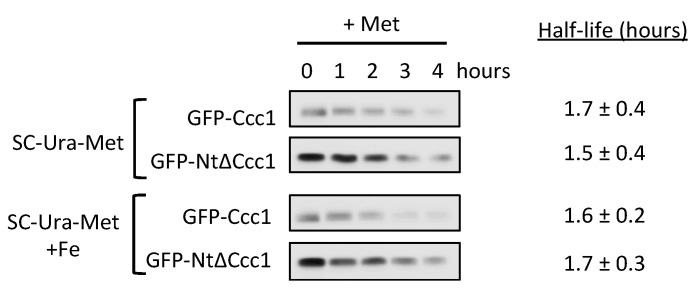
The decreased resistance to high-iron conditions of yeast cells expressing NtΔCCC1 could be due to decreased stability or mislocalization of the truncated protein. Moreover, Ccc1 Nt contains serine and lysine residues that, according to genome-wide studies, are phosphorylated and ubiquitylated, respectively (Figure 1) [13,14]. To ascertain if this was the case, we used yeast cells expressing GFP-tagged Ccc1 or NtΔCcc1 proteins under the control of MET17 promoter, previously assayed for growth in iron-containing media (Figure 3B). It is important to highlight that incorporation of an Nt tag, such as GFP, precludes any change in protein stability due to differences in the initial amino acids of the protein, allowing direct conclusions to be made from the Nt region. Thus, we investigated protein stability by stopping CCC1 and NtΔCCC1 expression with the addition of methionine and determining protein levels at different time points. As shown, Ccc1 protein stability was not altered by the deletion of its Nt domain, neither under normal growth conditions nor after addition of iron to the medium (Figure 5 and Figure S2). These results indicate that the deleted Nt domain does not significantly alter Ccc1 protein stability under these growth conditions.
Figure 5.
GFP-Ccc1 and GFP-NtΔCcc1 protein stability in normal and high-iron conditions. Yeast ccc1Δ cells transformed with either pUG36-CCC1 (PMET17-GFP-CCC1) or pUG36-NtΔCCC1 (PMET17-GFP-NtΔCCC1) plasmids were cultivated in SC-Ura-Met to exponential phase and 3 mM FAS was added (or not) for 1 h. Expression was stopped by addition of 3 mM methionine, and aliquots were isolated at the indicated time points. Total proteins were extracted, and Ccc1 protein levels were determined by immunoblotting with anti-GFP antibody. Equal amounts of total proteins were loaded in each lane. Three independent biological experiments were performed, and GFP-Ccc1 and GFP-NtΔCcc1 protein half-lives was determined. The average half-life and its standard deviation are presented.
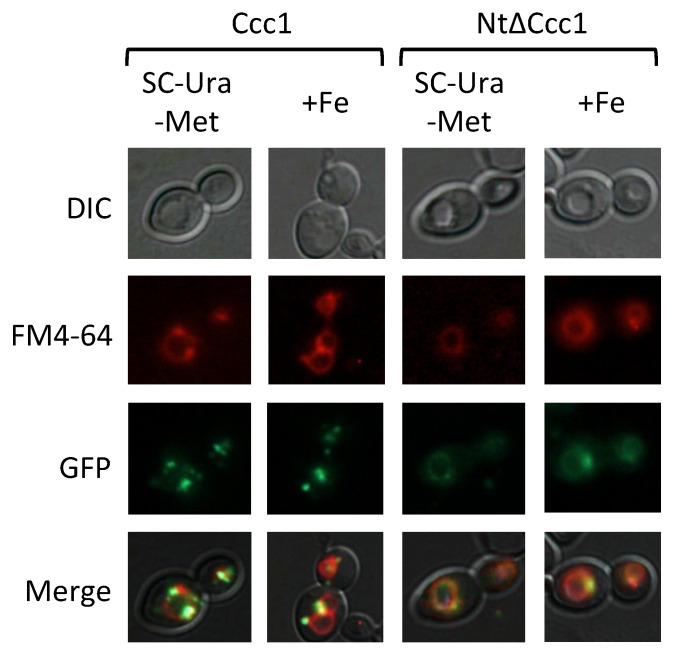
To ascertain whether deletion of Ccc1 Nt region alters cellular distribution, we determined the subcellular localization of GFP-Ccc1 and GFP-NtΔCcc1 proteins by fluorescence microscopy under iron-sufficient and high-iron conditions (1 h in 3 mM FAS). Differential interference contrast (DIC) was used to visualize vacuoles, since they appear as clear indentations, whereas the lipophilic dye FM4-64 selectively localized to vacuolar membranes after appropriate incubation. We observed that full length Ccc1 protein colocalized with FM4-64 under both iron-sufficient and high-iron conditions, particularly at specific regions of the vacuolar membrane (Figure 6). NtΔCcc1 protein displayed a more uniform distribution throughout the vacuolar membrane, as compared with wild-type Ccc1 under both iron-sufficient and high-iron conditions (Figure 6). These results suggest that the Ccc1 Nt domain is important in modulation of Ccc1 protein localization.
Figure 6.
Subcellular localization of Ccc1 and NtΔCcc1 proteins in normal (SC-Ura-Met) and high-iron (+Fe) conditions. Yeast cells were cultivated as described in Figure 5, and visualized under Nomarski (DIC) and GFP fluorescence optics. Vacuolar membranes were stained with the styryl dye FM4-64 as indicated in Materials and Methods. Merge shows the overlap of the different signals.
4. Discussion
An important drawback of increasing intracellular iron concentrations is the toxicity caused by ROS generation. Here, we decided to explore the potential of the main yeast iron resistance factor, Ccc1, to increase endogenous iron by storing it in an unharmful form within the vacuole. For this purpose, we overexpressed CCC1 and observed that it led to low growth and cellular viability when cells were cultivated in normal iron replete media (Figure 2 and Figure 3). Since increased expression of CCC1 stimulates iron uptake and accumulation due to iron regulon activation (Figure 2), we postulated that the reason for Ccc1 toxicity could be iron acquisition [8,9,25]. In fact, previous genetic and biochemical analyses demonstrated that iron toxicity was due to the accumulation of cytosolic iron [35]. Moreover, the toxicity caused by the expression of a constitutively active Aft1 transcription factor was shown to be, at least partially, rescued by deletion of FET3 [36]. However, deletion of the components of the high-affinity iron uptake system Fet3-Ftr1 at the cell surface did not rescue the growth of CCC1-overexpressing cells (Figure 2). We investigated whether the upregulation of CTH2, which was previously shown to limit growth, was responsible for CCC1 overexpression toxicity, but no rescue was observed in a cth2Δ mutant (Figure 2) [33,34]. Unexpectedly, we observed that deletion of the AFT1 transcriptional factor increased CCC1 overexpression toxicity (Figure 2). Aft1 is mainly implicated in increasing cytosolic iron levels by promoting iron uptake and mobilization from the vacuole. Therefore, these results are consistent with CCC1 causing a decrease in the bioavailability of iron to the cytosol and other organelles, including mitochondria, whose pools are highly interconnected [37,38]. Indeed, the activation of FTR1 and CTH2 that we observed (Figure 2A) is probably a consequence of decreased iron-sensing by Aft1 transcription factor. In any case, we cannot discard other non-exclusive possibilities for such toxicity. For instance, CCC1 overexpression could cause a decrease in vacuolar lumen acidification, since Ccc1 seems to be a H+/Fe2+ antiporter [12]. In this sense, recent data demonstrated that the loss of vacuolar acidification leads to oxidative damage of mitochondrial Fe/S cluster biosynthesis components and activities and the development of an age-related mitochondrial dysfunction [39,40]. Importantly, Ccc1 overexpression toxicity was not observed in high iron media (Figure 2 and Figure 3). This result is consistent with the natural expression pattern of CCC1, which is induced in response to elevated extracellular iron, and reinforces the hypothesis that low cytosolic and mitochondrial iron levels could be the reason for its toxicity [18,20]. However, more detailed studies are necessary to fully elucidate the molecular reason for the toxicity of Ccc1 protein when overexpressed under iron replete conditions.
The comparison of Ccc1/VIT1 protein sequences unveiled that their cytosolic Nt domain was not conserved during evolution and displays particular characteristics depending on the species [11]. In the case of fungi, including S. cerevisiae, Ccc1 Nt is rich in proline and serine residues [11]. The fact that genome-wide studies show that these serine residues could be phosphorylated prompted us to explore Ccc1 Nt function by constructing and expressing an NtΔCcc1 protein lacking its first 87 amino acids [13,14]. Overexpression of NtΔCCC1 did not confer toxicity under any of the promoters assayed (Figure 2 and Figure 3). This was probably due a decrease in NtΔCcc1 functionality since it did not produce the same resistance to high iron media as full length Ccc1 did (Figure 2 and Figure 3). We observed that both GFP-tagged Ccc1 and NtΔCcc1 proteins localized to the vacuolar membrane, although full-length Ccc1 preferentially accumulated in small regions of the vacuolar membrane (Figure 6). It is tempting to speculate that phosphorylation of serine residues within the Ccc1 Nt domain could modulate subcellular distribution of the protein. It was proposed that Ccc1/VIT1 proteins dimerize, but no aggregation of such proteins has been described [12]. Further studies are necessary to decipher the contribution of the Nt domain to Ccc1 function in metal transport. Importantly, NtΔCcc1-expressing cells were still able to upregulate iron incorporation while maintaining considerable growth (Figure 4). Consequently, NtΔCcc1 cells were able to incorporate at least 2-fold higher levels of iron from iron replete media than WT, ccc1Δ, and CCC1-overexpressing cells were.
5. Conclusions
Iron deficiency is a prevalent nutritional disorder whose prevention requires the development of novel strategies that increase iron intake. Baker’s yeast S. cerevisiae is widely used in food technology to produce bakery derived products but also as a food supplement. Therefore, greater knowledge of yeast iron homeostasis and its ability to increase its capacity to acquire and store iron could prove meaningful. Here, we have observed that overexpression of the vacuolar iron importer Ccc1 is toxic to yeast cells. Remarkably, overexpression of an Nt-truncated version of Ccc1 (NtΔCcc1) did not alter growth and led to an increase in the extraction of iron from the environmental medium. Therefore, this truncated version of Ccc1 protein could be used to design strategies aimed to increase the capacity of yeast cells to incorporate iron from non-iron rich media. Additional studies aiming to enrich yeast cells with iron could contribute to its utilization as a tool to palliate iron deficiency.
Acknowledgments
We are grateful to the members of the Iron Homeostasis laboratory for technical and scientific assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12081120/s1, Figure S1: Overexpression of CCC1 induced a decrease in the growth rate. Yeast cells described in Figure 2 were inoculated at an OD600nm of 0.2 in liquid SC-Ura medium supplemented with 500 µM FAS. The OD600nm was recorded every 30 minutes with a Spectrostar Nano absorbance 96-plate reader for 36 h at 28 °C. The average curve and standard deviation of at least three independent biological replicates are shown. Figure S2. GFP-Ccc1 and GFP-Nt Ccc1 protein stability quantification and loading controls. (A) Quantification of GFP-Ccc1 and GFP-Nt Ccc1 protein levels. The average of three different samples and their standard deviation is presented. (B) Western blot for Pgk1 protein levels. (C) Ponceau staining.
Author Contributions
Conceptualization, S.P.; methodology, R.S.-D., M.T.M.-P. and S.P.; data curation, R.S.-D.; writing—original draft preparation, S.P.; writing—review and editing, M.T.M.-P. and R.S.-D.; visualization, S.P. and R.S.-D.; supervision, M.T.M.-P. and S.P.; funding acquisition, M.T.M.-P. and S.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a predoctoral contract ACIF/2018/077 (to R.S.-D.) and grant PROMETEO/2020/014 from the Regional Government of Valencia (Generalitat Valenciana), grant BIO2017-87828-C2-1-P and PID2020-116940RB-I00 from the Spanish Ministry of Science, Innovation and Universities, and FEDER funds (ESF, European Social Fund).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in Digital.CSIC (https://digital.csic.es, accessed on 13 July 2021) at DOI: http://dx.doi.org/10.20350/digitalCSIC/13962 (accessed on 13 July 2021).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zimmermann M.B., Hurrell R.F. Nutritional iron deficiency. Lancet. 2007;370:511–520. doi: 10.1016/S0140-6736(07)61235-5. [DOI] [PubMed] [Google Scholar]
- 2.Zimmermann M.B. Global look at nutritional and functional iron deficiency in infancy. Hematology. 2020;2020:471–477. doi: 10.1182/hematology.2020000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanvisens N., Puig S. Biology of Starvation in Humans and Other Organisms. Nova Science Publishers; New York, NY, USA: 2011. Causes and consequences of nutritional iron deficiency in living organisms; pp. 245–276. [Google Scholar]
- 4.Pas M., Piskur B., Sustaric M., Raspor P. Iron enriched yeast biomass--a promising mineral feed supplement. Bioresour. Technol. 2007;98:1622–1628. doi: 10.1016/j.biortech.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Kyyaly M.A., Powell C., Ramadan E. Preparation of iron-enriched baker’s yeast and its efficiency in recovery of rats from dietary iron deficiency. Nutrition. 2015;31:1155–1164. doi: 10.1016/j.nut.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 6.Eid R., Arab N.T., Greenwood M.T. Iron mediated toxicity and programmed cell death: A review and a re-examination of existing paradigms. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:399–430. doi: 10.1016/j.bbamcr.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Raguzzi F., Lesuisse E., Crichton R.R. Iron storage in Saccharomyces cerevisiae. FEBS Lett. 1988;231:253–258. doi: 10.1016/0014-5793(88)80742-7. [DOI] [PubMed] [Google Scholar]
- 8.Li L., Chen O.S., McVey Ward D., Kaplan J. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 2001;276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- 9.Cockrell A., McCormick S.P., Moore M.J., Chakrabarti M., Lindahl P.A. Mossbauer, EPR, and modeling study of iron trafficking and regulation in Δccc1 and CCC1-up Saccharomyces cerevisiae. Biochemistry. 2014;53:2926–2940. doi: 10.1021/bi500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rietzschel N., Pierik A.J., Bill E., Lill R., Muhlenhoff U. The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters. Mol. Cell. Biol. 2015;35:370–378. doi: 10.1128/MCB.01033-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorribes-Dauden R., Peris D., Martinez-Pastor M.T., Puig S. Structure and function of the vacuolar Ccc1/VIT1 family of iron transporters and its regulation in fungi. Comput. Struct. Biotechnol. J. 2020;18:3712–3722. doi: 10.1016/j.csbj.2020.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato T., Kumazaki K., Wada M., Taniguchi R., Nakane T., Yamashita K., Hirata K., Ishitani R., Ito K., Nishizawa T., et al. Crystal structure of plant vacuolar iron transporter VIT1. Nat. Plants. 2019;5:308–315. doi: 10.1038/s41477-019-0367-2. [DOI] [PubMed] [Google Scholar]
- 13.Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteom. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swaney D.L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N.J., Villen J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H.N., Mostovoy Y., Hsu T.Y., Chang A.H., Brem R.B. Divergence of iron metabolism in wild Malaysian yeast. G3. 2013;3:2187–2194. doi: 10.1534/g3.113.008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., Ward D.M. Iron toxicity in yeast: Transcriptional regulation of the vacuolar iron importer Ccc1. Curr. Genet. 2018;64:413–416. doi: 10.1007/s00294-017-0767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos-Alonso L., Romero A.M., Martinez-Pastor M.T., Puig S. Iron Regulatory Mechanisms in Saccharomyces cerevisiae. Front. Microbiol. 2020;11:582830. doi: 10.3389/fmicb.2020.582830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L., Bagley D., Ward D.M., Kaplan J. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol. Cell. Biol. 2008;28:1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Miao R., Bertram S., Jia X., Ward D.M., Kaplan J. A role for iron-sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J. Biol. Chem. 2012;287:35709–35721. doi: 10.1074/jbc.M112.395533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pimentel C., Vicente C., Menezes R.A., Caetano S., Carreto L., Rodrigues-Pousada C. The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS ONE. 2012;7:e37434. doi: 10.1371/journal.pone.0037434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Kaplan J., Ward D.M. The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast. J. Biol. Chem. 2017;292:15577–15586. doi: 10.1074/jbc.M117.802504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philpott C.C., Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urbanowski J.L., Piper R.C. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 1999;274:38061–38070. doi: 10.1074/jbc.274.53.38061. [DOI] [PubMed] [Google Scholar]
- 24.Puig S., Askeland E., Thiele D.J. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 25.Chen O.S., Kaplan J. CCC1 suppresses mitochondrial damage in the yeast model of Friedreich’s ataxia by limiting mitochondrial iron accumulation. J. Biol. Chem. 2000;275:7626–7632. doi: 10.1074/jbc.275.11.7626. [DOI] [PubMed] [Google Scholar]
- 26.Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 27.Bukhman Y.V., DiPiazza N.W., Piotrowski J., Shao J., Halstead A.G.W., Bui M.D., Xie E., Sato T.K. Modeling Microbial Growth Curves with GCAT. BioEnergy Res. 2015;8:1022–1030. doi: 10.1007/s12155-015-9584-3. [DOI] [Google Scholar]
- 28.Martinez-Garay C.A., de Llanos R., Romero A.M., Martinez-Pastor M.T., Puig S. Responses of Saccharomyces cerevisiae Strains from Different Origins to Elevated Iron Concentrations. Appl. Environ. Microbiol. 2016;82:1906–1916. doi: 10.1128/AEM.03464-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamarit J., Irazusta V., Moreno-Cermeno A., Ros J. Colorimetric assay for the quantitation of iron in yeast. Anal. Biochem. 2006;351:149–151. doi: 10.1016/j.ab.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Sanvisens N., Romero A.M., An X., Zhang C., de Llanos R., Martinez-Pastor M.T., Bano M.C., Huang M., Puig S. Yeast Dun1 kinase regulates ribonucleotide reductase inhibitor Sml1 in response to iron deficiency. Mol. Cell. Biol. 2014;34:3259–3271. doi: 10.1128/MCB.00472-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou P. Determining protein half-lives. Methods Mol. Biol. 2004;284:67–77. doi: 10.1385/1-59259-816-1:067. [DOI] [PubMed] [Google Scholar]
- 32.Kushnirov V.V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 33.Romero A.M., Martinez-Pastor M., Du G., Sole C., Carlos M., Vergara S.V., Sanvisens N., Wohlschlegel J.A., Toczyski D.P., Posas F., et al. Phosphorylation and Proteasome Recognition of the mRNA-Binding Protein Cth2 Facilitates Yeast Adaptation to Iron Deficiency. mBio. 2018;9:e01694-18. doi: 10.1128/mBio.01694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson M.J., Lai W.S., Taylor G.A., Blackshear P.J. Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: Zinc finger-mediated impairment of cell growth. Gene. 1996;174:225–233. doi: 10.1016/0378-1119(96)00084-4. [DOI] [PubMed] [Google Scholar]
- 35.Lin H., Li L., Jia X., Ward D.M., Kaplan J. Genetic and biochemical analysis of high iron toxicity in yeast: Iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J. Biol. Chem. 2011;286:3851–3862. doi: 10.1074/jbc.M110.190959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos-Alonso L., Wittmaack N., Mulet I., Martinez-Garay C.A., Fita-Torro J., Lozano M.J., Romero A.M., Garcia-Ferris C., Martinez-Pastor M.T., Puig S. Molecular strategies to increase yeast iron accumulation and resistance. Metallomics. 2018;10:1245–1256. doi: 10.1039/C8MT00124C. [DOI] [PubMed] [Google Scholar]
- 37.Li L., Kaplan J. A mitochondrial-vacuolar signaling pathway in yeast that affects iron and copper metabolism. J. Biol. Chem. 2004;279:33653–33661. doi: 10.1074/jbc.M403146200. [DOI] [PubMed] [Google Scholar]
- 38.Li L., Murdock G., Bagley D., Jia X., Ward D.M., Kaplan J. Genetic dissection of a mitochondria-vacuole signaling pathway in yeast reveals a link between chronic oxidative stress and vacuolar iron transport. J. Biol. Chem. 2010;285:10232–10242. doi: 10.1074/jbc.M109.096859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H., Miller P.W., Johnson D.L., Laribee R.N. The Ccr4-Not complex regulates TORC1 signaling and mitochondrial metabolism by promoting vacuole V-ATPase activity. PLoS Genet. 2020;16:e1009046. doi: 10.1371/journal.pgen.1009046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes C.E., Coody T.K., Jeong M.Y., Berg J.A., Winge D.R., Hughes A.L. Cysteine Toxicity Drives Age-Related Mitochondrial Decline by Altering Iron Homeostasis. Cell. 2020;180:296–310.e18. doi: 10.1016/j.cell.2019.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are openly available in Digital.CSIC (https://digital.csic.es, accessed on 13 July 2021) at DOI: http://dx.doi.org/10.20350/digitalCSIC/13962 (accessed on 13 July 2021).