Abstract
We conducted this study to investigate the isolation frequency and phenotypic antibiotic resistance pattern of Staphylococcus aureus isolated from rodents, chickens, humans, and household soils. Specimens were plated onto mannitol salt agar (Oxoid, Basingstoke, UK) and incubated aerobically at 37 °C for 24 h. Presumptive colonies of S. aureus were subjected to Gram staining, as well as catalase, deoxyribonuclease (DNAse), and coagulase tests for identification. Antibiotic susceptibility testing was performed by using the Kirby–Bauer disc diffusion method on Mueller–Hinton agar (Oxoid, Basingstoke, UK). The antibiotics tested were tetracycline (30 μg), erythromycin (15 μg), gentamicin (10 μg), ciprofloxacin (5 μg), clindamycin (2 μg), and amoxicillin-clavulanate (20 μg/10 μg). The S. aureus strain American Type Culture Collection (ATCC) 25,923 was used as the standard organism. We found that 483 out of 956 (50.2%) samples were positive for S. aureus. The isolation frequencies varied significantly between samples sources, being 52.1%, 66.5%, 74.3%, and 24.5%, respectively, in chickens, humans, rodents, and soil samples (p < 0.001). S. aureus isolates had high resistance against clindamycin (51.0%), erythromycin (50.9%), and tetracycline (62.5%). The overall prevalence of multidrug-resistant (MDR) S. aureus isolates was 30.2%, with 8.7% resistant to at least four different classes of antibiotics.
Keywords: Staphylococcus aureus, antibiotic resistance, humans, chickens, rodents, soil
1. Introduction
Staphylococcus aureus is both an opportunistic pathogen and a commensal microbe that colonizes a wide range of hosts, including humans, livestock, wild ungulates, and the environment [1,2,3]. S. aureus is also a leading cause of different infections in humans that range from minor skin infections to life-threatening diseases, such as pneumonia, osteomyelitis, endocarditis, and sepsis [2,4,5]. In farm animals, S. aureus causes mastitis in dairy animals [6,7] and septic arthritis in chickens [8], resulting in economic losses due to mortality and reduced production [9]. The pathogenicity of S. aureus is influenced by two important features: its ability to resist more than three classes of antibiotics [10] and the capacity to produce several toxins [2,11]. Multidrug-resistant bacteria have increased worldwide, resulting in the sharing of their genes with commensal microorganisms in humans, animals, and the environment and endangering public health [12]. Rodents have been extensively documented to carry and transmit different zoonotic pathogens, including S. aureus, to humans and livestock [3,13,14]. Commensal rodents colonized with pathogens have been widely reported to invade chicken [15,16] and human houses [17,18,19], exposing them to bacterial infections. Different studies in Tanzania have documented the interaction of rodents with humans in households, predisposing them to rodent-borne zoonotic diseases [20,21,22]. Rodent infestation in human settlements has been frequently reported in Karatu, where interactions of rodents with humans and livestock are very common, making it a plague focus area [20,21,23,24,25]. However, studies on the occurrence and pattern of multidrug-resistant (MDR) S. aureus among humans, rodents, and the environment in the area are missing. Therefore, this study aimed to determine the occurrence of MDR S. aureus isolates in humans, rodents, chickens, and soils in the households of Karatu in northern Tanzania.
2. Materials and Methods
2.1. Study Area
The study was conducted in the Karatu district in the northern zone of Tanzania between June 2020 and March 2021. Karatu is located between latitudes 3°10′ and 4°00′ S and longitude 34°47′ to 59.99′ E. The district has a population of 230,166 people comprised of 117,769 men and 112,397 women, with an average of five people per household. Karatu has an altitude range of 1000 to 1900 m above sea level with two wet seasons annually (short rains between October and December and long rains from March to June).
2.2. Sampling Strategy
The study population comprised of households keeping local chickens, while the sampling frame was the list of these households. Five wards, Karatu, Endabash, Endamarariek, Mbulumbulu, and Rhotia, were purposively selected based on the population density (at least 16,000 people), number of households with chickens, and household size of at least five people. Households were randomly selected from a list provided by a livestock field officer at the ward level by using a table of random numbers. At the household level, permission from the head of the household was granted first before trapping the rodents where areas for trapping in the surrounding environments relied on signs of rodents’ activities. For each household, one adult human (18 years and above) and one mature (seven months) scavenging chicken were involved in microbiological sampling to get one nasal swab and one cloaca swab, respectively. Furthermore, at least one rodent (in-house rat, peri-domestic rat, or both) could be captured, and one soil sample was collected per household. The selection of adult humans and mature chickens was based on the assumption that old individuals have been exposed to the interaction with rodents for a longer time than young ones, and hence are more likely to facilitate the sharing of infections.
2.3. Trapping of Rodents for Sample Collection
Live trapping of rodents was carried out using modified Sherman traps baited with peanut butter. An average of 100 traps (50 in houses and 50 in outside environments) were deployed per trap night for five consecutive nights in each ward. Each captured rodent was subjected to humane killing by using di-ethyl-ether and deep pharyngeal swabs, and the intestines were aseptically collected from the carcasses.
2.4. Collection of Samples from Humans, Chickens, and Soil
A total of 956 samples were collected from 286 households in the Karatu district wards. Of these, 286 were from chickens, 284 from humans, and 285 from soil (Table 1). Sterile cotton swabs were used to collect cloaca swabs from randomly picked scavenging chickens and human nasal swabs in households. Soil samples were randomly collected from five points in the household yards and mixed to compose one pooled soil sample [26]. Thereafter, cloaca and human nasal swabs were stored in sterile containers at −4°C and transported using Cary Blair transport medium and trypticase soy broth medium (Oxoid, Basingstoke, UK), respectively, to the Tanzania Veterinary Laboratory Agency (TVLA)–Arusha laboratory for processing within four hours after collection.
Table 1.
Isolation frequencies of Staphylococcus aureus from different sample sources.
| Types of Sample Sources | Number of Samples n (%) |
Positive Samples n (%) |
Chi-Squared | p-Value |
|---|---|---|---|---|
| Chickens | 286 (29.9) | 149 (52.1) | X2 = 83.849, df = 3 | <0.001 |
| Humans | 284 (29.7) | 189 (66.5) | ||
| Rodents | 101 (10.6) | 75 (74.3) | ||
| Soil | 285 (29.8) | 70 (24.5) | ||
| Total | 956 (100.0) | 483 (50.5) |
2.5. Culture, Isolation, and Identification of S. aureus Isolates
Specimens were plated onto mannitol salt agar (Oxoid, Basingstoke, UK) and incubated aerobically at 37 °C for 24 h. Presumptive colonies of S. aureus were subjected to Gram staining, as well as catalase, deoxyribonuclease (DNAse), and coagulase tests for identification.
2.6. Antibiotic Susceptibility Testing of S. aureus Isolates
An antibiotic susceptibility test was performed by using the Kirby–Bauer disc diffusion method on Mueller–Hinton agar (Oxoid, Basingstoke, UK) with commercially available discs, as described by [27]. The antibiotics tested were tetracycline (30 μg), erythromycin (15 μg), gentamicin (10 μg), ciprofloxacin (5 μg), clindamycin (2 μg), and amoxicillin-clavulanate (20 μg/10 μg). Pure colonies of the identified lactose fermenters were emulsified into 5 mL of sterile saline. The suspensions were adjusted to achieve a turbidity equivalent to 0.5 McFarland standard solutions, emulsified using sterile cotton swabs onto a Mueller–Hinton agar plate, and incubated at 37 °C for 16 to 18 h. After incubation, the inhibition zone of each antimicrobial agent was measured, and the results were interpreted according to the standards of [27]. S. aureus strain American Type Culture Collection (ATCC) 25,923 was used as the standard organism. An isolate was considered to be multidrug-resistant (MDR) if it was non-susceptible to three or more drugs from different classes of antibiotics [28].
2.7. Statistical Analyses
Isolation frequencies of S. aureus and the antibiotic resistance pattern of isolates were entered into Microsoft Excel version 2010 (Microsoft Corporation, Redmond, WA, USA) and their percentages were calculated by descriptive statistics. The association between categorical variables was analysed by using a chi-squared (Fisher’s exact and Pearson’s) test. Statistical significance was accepted at p < 0.05.
3. Results
3.1. Isolation of Staphylococcus aureus from the Samples
Overall, 483 samples out of 956 (50.5%) had S. aureus. Significant variation in isolation frequencies was observed between the types of samples, being higher in rodents (74.3%) compared to soil (24.5%) samples (p < 0.001) (Table 1).
3.2. Antibiotic Susceptibility Testing (AST) Results of the S. aureus Isolates
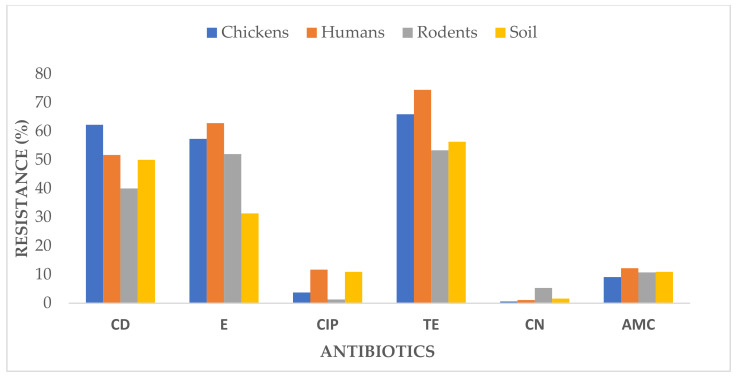
The overall resistance rates were 51.0% to clindamycin, 50.9% to erythromycin, 6.9% to ciprofloxacin, 62.5% to tetracycline, 2.2% to gentamycin, and 10.7% to amoxicillin-clavulanate. The specific resistance rates are shown in Table 2 and Figure 1.
Table 2.
Antibiotic resistance pattern of Staphylococcus aureus isolates from chicken, human, rodent, and soil samples.
| Sample Type | Antibiotics, n (%) | Overall R | Chi-Squared Test | |||||
|---|---|---|---|---|---|---|---|---|
| Clindamycin | Erythromycin | Ciprofloxacin | Tetracycline | Gentamycin | Amoxicillin-Clavulanate | |||
| Overall R | 51.0 % | 50.9 % | 6.9 % | 62.5 % | 2.2 % | 10.7 % | ||
| Chickens | ||||||||
| R | 102 (62.2) | 94 (57.3) | 6 (3.7) | 108 (65.9) | 1 (0.6) | 15 (9.1) | 33.1 % | 247.61, df = 5, p < 0.001 |
| I | 16 (9.8) | 21 (12.8) | 17 (10.4) | 13 (7.9) | 0 (0.0) | 2 (1.2) | ||
| S | 46 (28.0) | 49 (29.9) | 141 (86.0) | 43 (26.2) | 163 (99.4) | 147 (89.6) | ||
| Subtotal | 164 (100.0) | 164 (100.0) | 164 (100.0) | 164 (100.0) | 164 (100.0) | 164 (100.0) | ||
| Humans | ||||||||
| R | 93 (51.7) | 113 (62.8) | 21 (11.7) | 134 (74.4) | 2 (1.1) | 22 (12.2) | 35.7 % | 243.1, df = 5, p < 0.001 |
| I | 12 (6.7) | 16 (8.9) | 18 (10.0) | 13 (7.2) | 9 (5.0) | 6 (3.3) | ||
| S | 75 (41.7) | 51 (28.3) | 141 (78.3) | 33 (18.3) | 169 (93.9) | 152 (84.4) | ||
| Subtotal | 180 (100.0) | 180 (100.0) | 180 (100.0) | 180 (100.0) | 180 (100.0) | 180 (100.0) | ||
| Rodents | ||||||||
| R | 30 (40.0) | 39 (52.0) | 1 (1.3) | 40 (53.3) | 4 (5.3) | 8 (10.7) | 27.1 % | 79.74, df = 5, p < 0.001 |
| I | 10 (13.3) | 11 (14.7) | 8 (10.7) | 9 (12.0) | 1 (1.3) | 5 (6.7) | ||
| S | 35 (46.7) | 25 (33.3) | 66 (88.0) | 26 (34.7) | 70 (93.3) | 62 (82.7) | ||
| Subtotal | 75 (100.0) | 75 (100.0) | 75 (100.0) | 75 (100.0) | 75 (100.0) | 75 (100.0) | ||
| Soil | ||||||||
| R | 32 (50.0) | 20 (31.3) | 7 (10.9) | 36 (56.3) | 1 (1.6) | 7 (10.9) | 26.8 % | 61.21, df = 5, p < 0.001 |
| I | 6 (9.4) | 7 (10.9) | 4 (6.3) | 6 (9.4) | 0 (0.0) | 1 (1.6) | ||
| S | 26 (40.6) | 37 (57.8) | 53 (82.8) | 22 (34.4) | 63 (98.4) | 56 (87.5) | ||
| Subtotal | 64 (100.0) | 64 (100.0) | 64 (100.0) | 64 (100.0) | 64 (100.0) | 64 (100.0) | ||
R = resistant, I = intermediate, and S = susceptible.
Figure 1.
Resistance of S. aureus isolates against the antibiotics; CD = clindamycin, E = erythromycin, CIP = ciprofloxacin, TE = tetracycline, CN = gentamycin, and AMC = amoxicillin-clavulanate.
3.3. Prevalence of Multidrug-Resistant Isolates of S. aureus in Different Types of Samples
About 146 out of 483 isolates (30.2%) were resistant to at least three different classes of antibiotics. The population of MDR S. aureus was composed of 70 (14.5%), 51 (10.6%), 15 (3.1%), and 10 (2.1%) isolates from chicken, human, rodent, and soil samples, respectively (Table 3). The MDR rates varied significantly for isolates from chickens, humans, rodents, and soil (p < 0.001). In all types of samples, none of the MDR S. aureus isolates were resistant to all six classes of antibiotics.
Table 3.
MDR rates of S. aureus isolates from different types of samples.
| Type of Sample Source | Number of Antibiotic Classes to Which the Isolates Were Resistant, n (%) | Chi-Squared | p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | Total Isolates |
MDR Isolates (3–6 Classes) |
|||
| Overall | 81 (16.8) | 74 (15.3) | 182 (37.7) | 104 (21.5) | 32 (6.6) | 10 (2.1) | 0 (0.0) | 483 (100.0) | 146 (30.2) | ||
| Chickens | 34 (42.0) | 15 (20.3) | 45 (24.7) | 61 (58.7) | 9 (58.7) | 0 (0.0) | 0 (0.0) | 164 (34.0) | 70 (14.5) | 143.66 df = 3 | p < 0.001 |
| Humans | 12 (14.8) | 31 (41.9) | 86 (47.3) | 30 (28.8) | 14 (43.8) | 7 (70.0) | 0 (0.0) | 180 (37.3) | 51 (10.6) | 195.12 df = 3 | p < 0.001 |
| Rodents | 19 (23.5) | 16 (21.6) | 25 (13.7) | 8 (7.7) | 7 (21.9) | 0 (0.0) | 0 (0.0) | 75 (15.5) | 15 (3.1) | 51.47 df = 6 | p < 0.001 |
| Soil | 16 (19.8) | 12 (16.2) | 26 (14.3) | 5 (4.8) | 2 (6.3) | 3 (30.0) | 0 (0.0) | 64 (13.3) | 10 (2.1) | 57.84 df = 6 | p < 0.001 |
3.4. Prevalence of MDR S. aureus in Samples from Different Wards in the Study Area
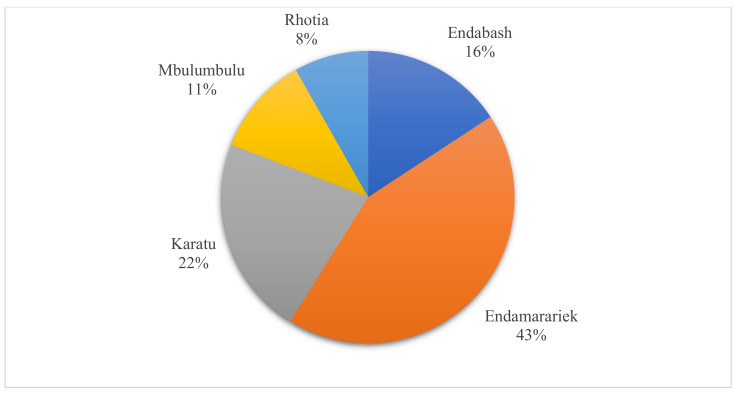
Most of the MDR S. aureus isolates (43.2%) were found in samples from Endamarariek, followed by Karatu (21.9%) and Endabash (15.8%), while a few MDR isolates were observed in samples from Mbulumbulu (11.0%) and Rhotia (8.2%) (Figure 2). The occurrence of MDR isolates varied significantly in samples from the Endabash, Karatu, Endamarariek (p < 0.001), Mbulumbulu (p < 0.006), and Rhotia (p < 0.005) wards (Table 4).
Figure 2.
Distribution of MDR S. aureus isolates in different wards of Karatu.
Table 4.
Prevalence of MDR S. aureus isolates in different samples by wards.
| Wards | MDR Isolates from Different Sample Sources n (%) | Chi-Squared | p-Value | ||||
|---|---|---|---|---|---|---|---|
| Chickens | Humans | Rodents | Soil | Total | |||
| Overall MDR | 70 (14.5) | 51 (10.6) | 15 (3.1) | 10 (2.1) | 146 (30.2) | ||
| Endabash | 16 (3.3) | 3 (0.6) | 3 (0.6) | 1 (0.2) | 23 (4.8) | 24.826 df = 3 | <0.001 |
| Endamarariek | 18 (3.7) | 30 (6.2) | 10 (2.1) | 3 (0.6) | 61 (12.6) | 29.508 df = 3 | <0.001 |
| Karatu | 20 (4.1) | 8 (1.7) | 0 (0.0) | 4 (0.8) | 32 (6.6) | 28 df = 3 | <0.001 |
| Mbulumbulu | 8 (1.7) | 7 (1.4) | 1 (0.2) | 1 (0.2) | 17 (3.5) | 12.5 df = 3 | 0.0059 |
| Rhotia | 8 (1.7) | 3 (0.6) | 1 (0.2) | 1 (0.2) | 13 (2.7) | 12.667 df = 3 | 0.0054 |
| Chi-squared | 9.1429 | 55.962 | 22 | 8.25 | |||
| p-Value | 0.0576 | <0.001 | 0.0002 | 0.0828 | |||
3.5. Phenotypic Patterns of MDR S. aureus Isolates
As shown in Table 5, MDR S. aureus isolates displayed variable resistance patterns, where CD-E-TE was the most common that appeared in chicken (34.2%), human (14.4%), rodent (3.4%), and soil (1.4%) isolates. CD-E-TE-AMC was also common in chicken (6.2%), human (6.2%), and rodent (2.7%) isolates, but not in the soil isolates. Patterns showing resistance to five different classes of antibiotics were CD-E-TE-CN-AMC found in soil and rodent (0.7%) samples and CD-E-CIP-TE-AMC in soil (1.4%) and human (4.8%) samples. However, none of the MDR S. aureus isolates were resistant to all antibiotic classes.
Table 5.
Phenotypic resistance patterns of MDR S. aureus isolates from chickens, humans, rodents, and soil samples.
| Source of Samples (N = 146) | Number of Isolates (n) | Occurrence (%) | Antibiotic Resistance Patterns | Number of Antibiotic Classes |
|---|---|---|---|---|
| Chickens | 50 | 34.2 | CD, E, TE | 3 |
| (n = 70) | 3 | 2.1 | CD, CIP, TE | |
| 1 | 0.7 | E, CIP, TE | ||
| 5 | 3.4 | E, TE, AMC | ||
| 9 | 6.2 | CD, E, TE, AMC | 4 | |
| 1 | 0.7 | CD, E, CIP, TE | ||
| 1 | 0.7 | CD, E, TE, CN | ||
| Humans | 21 | 14.4 | CD, E, TE | 3 |
| (n = 51) | 3 | 2.1 | CD, CIP, TE | |
| 2 | 1.4 | CD, E, CIP | ||
| 1 | 0.7 | CD, E, AMC | ||
| 1 | 0.7 | CD, TE, AMS | ||
| 1 | 0.7 | E, TE, AMC | ||
| 9 | 6.2 | CD, E, TE, AMC | 4 | |
| 1 | 0.7 | E, CIP, TE, CN | ||
| 3 | 2.1 | CD, E, CIP, TE | ||
| 2 | 1.4 | CD, CIP, TE, AMC | ||
| 7 | 4.8 | CD, E, CIP, TE, AMC | 5 | |
| Rodents | 5 | 3.4 | CD, E, TE | 3 |
| (n = 15) | 1 | 0.7 | CD, E, AMC | |
| 1 | 0.7 | CIP, CN, AMC | ||
| 1 | 0.7 | CD, TE, AMC | ||
| 4 | 2.7 | CD, E, TE, AMC | 4 | |
| 2 | 1.4 | CD, E, TE, CN | ||
| 1 | 0.7 | CD, E, TE, CN, AMC | 5 | |
| Soil | 2 | 1.4 | CD, E, TE | 3 |
| (n = 10) | 1 | 0.7 | E, TE, AMC | |
| 1 | 0.7 | CD, TE, AMC | ||
| 1 | 0.7 | CD, CIP, TE | ||
| 2 | 1.4 | CD, E, CIP, TE | 4 | |
| 2 | 1.4 | CD, E, CIP, TE, AMC | 5 | |
| 1 | 0.7 | CD, E, TE, CN, AMC | ||
| Total | 146 | 100.0 |
AMC = amoxicillin-clavulanate, TE = tetracycline, E = erythromycin, CD = clindamycin, CIP = ciprofloxacin, and CN = gentamycin.
4. Discussion
This is the first study to investigate the carriage of S. aureus in chickens, humans, rodents, and soils in a household environment in Tanzania. Overall, the isolation frequency of S. aureus was 50.5%. We observed significant variations in isolation frequencies among sample sources, where rodents had more S. aureus (74.3%), and soil had the lowest (24.5%). The presence of drug-resistant bacteria in soil serves as a potential reservoir of antibiotic resistomes, which encompasses all types of antibiotic resistance genes (ARGs) that can spread to humans and animals and to a wider environment [29,30]. Rodents carrying different zoonotic pathogens have been frequently reported to invade human residences in Karatu [20,21,22]. Overall, the isolates exhibited high resistance to clindamycin (51.0%), tetracycline (62.5%), and erythromycin (50.9%). These antibiotics are commonly used in humans and poultry production in the study area, and their frequent use and misuse can significantly contribute to increased resistance [6,18,31,32,33]. In this community, there is frequent use and misuse of the drugs in food animals, including poultry, mainly tetracycline and erythromycin [34,35]. Farmers in rural areas of Tanzania have been treating their chickens with antibiotics without diagnosis or prescriptions from veterinarians [35].
Our study observed that 146 out of 483 (30.2 %) isolates were MDR, including 14.5% chicken, 10.6% human, 3.1% rodent, and 2.1% soil isolates. The higher prevalence of MDR S. aureus in humans and poultry can be associated with the extensive use of drugs in human medicine and poultry in the community [36]. Lower multidrug resistance rates in rodents could be because these animals are not direct consumers of antibiotics, as is the case for humans and chickens. Their exposure to drugs is indirect, depending on contact with human and chicken wastes when dropped in the household environment, as explained in other studies [37,38]. Our findings are in keeping with those of Vitale et al. [32], showing that S. aureus derived from humans were more resistant to antibiotics compared with those of animal origin.
In our study, most MDR isolates were found in the Endamarariek ward (12.6%), which is basically a rural area compared to Karatu (6.6%), an urban and district headquarter. These variations could be due to differences in the levels of awareness and use of antibiotics between the wards. Endamarariek is a rural area with a scarcity of veterinarians, where farmers mostly treat their chickens based on experiences using home-stored antibiotics and those purchased from village shops with a low level of control. Such variations can also explain why we found more MDR S. aureus isolates (2.1%) in rodent samples from Endamarariek compared to Karatu samples (0%). Among MDR patterns, CD-E-TE, standing for clindamycin, erythromycin, and tetracycline, was displayed in most of the isolates (34.2 %), and the pattern is identical for humans, chickens, and rodents. Different studies on the resistance profiles of S. aureus have reported similar patterns as well [7,18,32,33,39,40]. Erythromycin and tetracycline are the most commonly used antibiotics in this area, since they are cheap and can be purchased over the counter without a prescription [41].
Limitation of the Study
Despite our findings being useful in the control of antimicrobial resistance in Tanzania, a genetic characterization of antibiotic resistance and virulence factors of S. aureus could provide additional information to compliment the phenotypic approach.
5. Conclusions
These results suggest a potential role of the interaction of humans, chickens, and rodents in cross-transmission of MDR S. aureus among them, with the possibility of causing human and animal infections that are difficult to treat. Unfortunately, treatment alternatives are very limited due to the few types of antibiotics in the studied area and the economic reality. Therefore, necessary interventions, such as continuous educative campaigns on effective cleanliness in households, safe disposal of animal wastes, and rodent control strategies, are urgently needed.
Author Contributions
Collection of samples, laboratory processing, and data analysis V.S.S., supervision and verification of the analytical methods M.I.M. and G.M. Manuscript developed by V.S.S. with input from M.I.M. and G.M. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by the World Bank to the African Centre of Excellence for Innovative Rodent Pest Management & Biosensor Technology Development (IRPM&BTD) at Sokoine University of Agriculture (SUA).
Institutional Review Board Statement
The ethical clearance for the study was issued by the National Institute for Medical Research (NIMR) of Tanzania (NIMR/HQ/R.8a/Vol.IX/3386). The permission to work in the study area was sought from the regional administrative office (RAS-Arusha).
Informed Consent Statement
Informed verbal consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gao J., Ferreri M., Yu F., Liu X., Chen L., Su J., Han B. Molecular types and antibiotic resistance of Staphylococcus aureus isolates from bovine mastitis in a single herd in China. Ir. Vet. J. 2012;192:550–552. doi: 10.1016/j.tvjl.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 2.Wang W., Baloch Z., Jiang T., Zhang C., Peng Z., Li F., Fanning S., Ma A., Xu J. Enterotoxigenicity and antimicrobial resistance of Staphylococcus aureus isolated from retail food in China. Front. Microbiol. 2017;8:2256. doi: 10.3389/fmicb.2017.02256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalaf S.K., Nagham M., Abdulkarim J., Jenan M. Isolation of methicillin resistant Staphylococcus aureus (MRSA) from Rattus rattus from Adhamiyah district in Baghdad governorate. MRVSA. 2015;4:9–23. [Google Scholar]
- 4.Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., Fowler Jr V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodríguez-Lázaro D., Ariza-Miguel J., Diez-Valcarce M., Fernández-Natal I., Hernández M., Rovira J. Foods confiscated from non-EU flights as a neglected route of potential methicillin-resistant Staphylococcus aureus transmission. Int. J. Food Microbiol. 2015;209:29–33. doi: 10.1016/j.ijfoodmicro.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Cvetnić L., Samardžija M., Duvnjak S., Habrun B., Cvetnić M., Jaki Tkalec V., Đuričić D., Benić M. Multi Locus Sequence Typing and spa typing of Staphylococcus aureus isolated from the milk of cows with subclinical mastitis in Croatia. Microorganisms. 2021;9:725. doi: 10.3390/microorganisms9040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massawe H., Mdegela R., Kurwijila L. Antibiotic resistance of Staphylococcus aureus isolates from milk produced by smallholder dairy farmers in Mbeya Region, Tanzania. Int. J. One Health. 2019;5:31–37. doi: 10.14202/IJOH.2019.31-37. [DOI] [Google Scholar]
- 8.Marcon A.V., De Oliveira G., Caldara F.R., Garcia R.G., Matins R., Marcon A., Crone C., Assunción A.S.d.A. Bacteriological and Histopathological Evaluation of Articulations of Chickens Diagnosed with Arthritis. Rev. Bras. 2019;21 doi: 10.1590/1806-9061-2018-0805. [DOI] [Google Scholar]
- 9.Nazia M.K., Durrani N., Kamboh A., Lakho S., Rind R., Abro S., Soomro N. Prevalence of septic arthritis caused by Staphylococcus aureus in poultry birds at Tandojam. Trop Anim. Health Prod. 2015;3:73–77. [Google Scholar]
- 10.Jackson C.R., Davis J.A., Barrett J.B. Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from retail meat and humans in Georgia. J. Clin. Microbiol. 2013;51:1199–1207. doi: 10.1128/JCM.03166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kmieciak W., Szewczyk E.M. Coagulase-positive species of the genus Staphylococcus–taxonomy, pathogenicity. Postępami Mikrobiol. 2019;56:233–244. doi: 10.21307/PM-2017.56.2.233. [DOI] [Google Scholar]
- 12.Subramanya S.H., Bairy I., Metok Y., Baral B.P., Gautam D., Nayak N. Detection and characterization of ESBL-producing Enterobacteriaceae from the gut of subsistence farmers, their livestock, and the surrounding environment in rural Nepal. Sci. Rep. 2021;11:2091. doi: 10.1038/s41598-021-81315-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himsworth C.G., Miller R.R., Montoya V., Hoang L., Romney M.G., Al-Rawahi G.N., Kerr T., Jardine C.M., Patrick D.M., Tang P. Carriage of methicillin-resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus) PLoS ONE. 2014;9:e87983. doi: 10.1371/journal.pone.0087983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge J., Zhong X.-s., Xiong Y.-q., Qiu M., Huo S.-t., Chen X.-j., Mo Y., Cheng M.-j., Chen Q. Methicillin-resistant Staphylococcus aureus among urban rodents, house shrews, and patients in Guangzhou, southern China. BMC Vet. Res. 2019;15:1–8. doi: 10.1186/s12917-019-2012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henzler D., Opitz H. The role of mice in the epizootiology of Salmonella enteritidis infection on chicken layer farms. Avian Dis. 1992;36:625–631. doi: 10.2307/1591757. [DOI] [PubMed] [Google Scholar]
- 16.Lapuz R., Tani H., Sasai K., Shirota K., Katoh H., Baba E. The role of roof rats (Rattus rattus) in the spread of Salmonella Enteritidis and S. Infantis contamination in layer farms in eastern Japan. Epidemiol. Infect. 2008;136:1235–1243. doi: 10.1017/S095026880700948X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morzunov S.P., Rowe J.E., Ksiazek T.G., Peters C.J., St. Jeor S.C., Nichol S.T. Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J. Virol. 1998;72:57–64. doi: 10.1128/JVI.72.1.57-64.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabiee M.H., Mahmoudi A., Siahsarvie R., Kryštufek B., Mostafavi E. Rodent-borne diseases and their public health importance in Iran. PLOS Negl. Trop. Dis. 2018;12:e0006256. doi: 10.1371/journal.pntd.0006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witmer G.W., Shiels A.B. Ecology and Management of Terrestrial Vertebrate Invasive Species in the United States. CRC Press; Boca Raton, FL, USA: 2017. Ecology, impacts, and management of invasive rodents in the United States; pp. 193–220. [Google Scholar]
- 20.Kilonzo B., Mbise T., Mwalimu D., Kindamba L. Observations on the endemicity of plague in Karatu and Ngorongoro, northern Tanzania. Tanzan. J. Health Res. 2006;8:1–6. doi: 10.4314/thrb.v8i1.14262. [DOI] [PubMed] [Google Scholar]
- 21.Makundi R.H., Massawe A.W., Mulungu L.S., Katakweba A., Mbise T.J., Mgode G. Potential mammalian reservoirs in a bubonic plague outbreak focus in Mbulu District, northern Tanzania, in 2007. Mammalia. 2008;72:253–257. doi: 10.1515/MAMM.2008.038. [DOI] [Google Scholar]
- 22.Ziwa M., Matee M., Hang’ombe B., Lyamuya E., Kilonzo B.S. Plague in Tanzania: An overview. Tanzan. J. Health Res. 2013;15:252–258. doi: 10.4314/thrb.v15i4.7. [DOI] [PubMed] [Google Scholar]
- 23.Makundi R.H., Massawe A.W., Borremans B., Laudisoit A., Katakweba A. We are connected: Flea–host association networks in the plague outbreak focus in the Rift Valley, northern Tanzania. Wildl. Res. 2015;42:196–206. doi: 10.1071/WR14254. [DOI] [Google Scholar]
- 24.Theonest N.O., Carter R.W., Amani N., Doherty S.L., Hugho E., Keyyu J.D., Mable B.K., Shirima G.M., Tarimo R., Thomas K.M., et al. Molecular detection and genetic characterization of Bartonella species from rodents and their associated ectoparasites from northern Tanzania. PLoS ONE. 2019;14:e0223667. doi: 10.1371/journal.pone.0223667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haule M., Lyamuya E.E., Hang’ombe B.M., Matee M.I. Investigation of Fleas as Vectors in the Transmission of Plague during a Quiescent Period in North-Eastern, Tanzania. [(accessed on 20 June 2021)];2013 Available online: http://www.suaire.sua.ac.tz/handle/123456789/1037.
- 26.Lupindu A.M., Dalsgaard A., Msoffe P.L., Ngowi H.A., Mtambo M.M., Olsen J.E. Transmission of antibiotic-resistant Escherichia coli between cattle, humans and the environment in peri-urban livestock keeping communities in Morogoro, Tanzania. Prev. Vet. Med. 2015;118:477–482. doi: 10.1016/j.prevetmed.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute . CLSI-Performance Standards for Antimicrobial Susceptibility Testing. Volume 40. CLSI; Wayne, PA, USA: 2020. p. 293. [Google Scholar]
- 28.Magiorakos A.-P., Srinivasan A., Carey R.t., Carmeli Y., Falagas M.t., Giske C.t., Harbarth S., Hindler J.t., Kahlmeter G., Olsson-Liljequist B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 29.D’Costa V.M., Griffiths E., Wright G.D. Expanding the soil antibiotic resistome: Exploring environmental diversity. Curr. Opin. Microbiol. 2007;10:481–489. doi: 10.1016/j.mib.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Woolhouse M., Ward M., Van Bunnik B., Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. B Biol. Sci. 2015;370:20140083. doi: 10.1098/rstb.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akanbi O.E., Njom H.A., Fri J., Otigbu A.C., Clarke A.M. Antimicrobial susceptibility of Staphylococcus aureus isolated from recreational waters and beach sand in Eastern Cape Province of South Africa. Int. J. Environ. Res. Public Health. 2017;14:1001. doi: 10.3390/ijerph14091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitale M., Galluzzo P., Buffa P.G., Carlino E., Spezia O., Alduina R. Comparison of antibiotic resistance profile and biofilm production of Staphylococcus aureus isolates derived from human specimens and animal-derived samples. Antibiotics. 2019;8:97. doi: 10.3390/antibiotics8030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amoako D.G., Somboro A.M., Abia A.L., Molechan C., Perrett K., Bester L.A., Essack S.Y. Antibiotic resistance in Staphylococcus aureus from poultry and poultry products in uMgungundlovu District, South Africa, using the “Farm to Fork” approach. Microb. Drug Resist. 2020;26:402–411. doi: 10.1089/mdr.2019.0201. [DOI] [PubMed] [Google Scholar]
- 34.Caudell M.A., Quinlan M.B., Subbiah M., Call D.R., Roulette C.J., Roulette J.W., Roth A., Matthews L., Quinlan R.J. Antimicrobial use and veterinary care among agro-pastoralists in Northern Tanzania. PLoS ONE. 2017;12:e0170328. doi: 10.1371/journal.pone.0170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rugumisa B., Call D.R., Mwanyika G.O., Subbiah M., Buza J. Comparison of the Prevalence of Antibiotic-Resistant Escherichia Coli Isolates from Commercial-Layer and Free-Range Chickens in Arusha District, Tanzania. [(accessed on 20 June 2021)];2016 Available online: http://dspace.nm-aist.ac.tz/handle/123456789/457.
- 36.Odsbu I., Khedkar S., Lind F., Khedkar U., Nerkar S.S., Orsini N., Tamhankar A.J., Stålsby Lundborg C. Trends in resistance to extended-spectrum cephalosporins and carbapenems among Escherichia coli and Klebsiella spp. isolates in a district in Western India during 2004–2014. Int. J. Environ. Res. Public Health. 2018;15:155. doi: 10.3390/ijerph15010155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gómez P., González-Barrio D., Benito D., García J.T., Viñuela J., Zarazaga M., Ruiz-Fons F., Torres C. Detection of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in wild small mammals in Spain. J. Antimicrob. Chemother. 2014;69:2061–2064. doi: 10.1093/jac/dku100. [DOI] [PubMed] [Google Scholar]
- 38.Silva V., Gabriel S.I., Borrego S.B., Tejedor-Junco M.T., Manageiro V., Ferreira E., Reis L., Caniça M., Capelo J.L., Igrejas G. Antimicrobial Resistance and Genetic Lineages of Staphylococcus aureus from Wild Rodents: First Report of mecC-Positive Methicillin-Resistant S. aureus (MRSA) in Portugal. Animals. 2021;11:1537. doi: 10.3390/ani11061537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdeen E.E., Mousa W.S., Abdelsalam S.Y., Heikal H.S., Shawish R.R., Nooruzzaman M., Soliman M.M., Batiha G.E., Hamad A., Abdeen A. Prevalence and Characterization of Coagulase Positive Staphylococci from Food Products and Human Specimens in Egypt. Antibiotics. 2021;10:75. doi: 10.3390/antibiotics10010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oladipo A.O., Oladipo O.G., Bezuidenhout C.C. Multi-drug resistance traits of methicillin-resistant Staphylococcus aureus and other Staphylococcal species from clinical and environmental sources. J. Water Health. 2019;17:930–943. doi: 10.2166/wh.2019.177. [DOI] [PubMed] [Google Scholar]
- 41.Mdegela R.H., Mwakapeje E.R., Rubegwa B., Gebeyehu D.T., Niyigena S., Msambichaka V., Nonga H.E., Antoine-Moussiaux N., Fasina F.O. Antimicrobial Use, Residues, Resistance and Governance in the food and agriculture sectors, Tanzania. Antibiotics. 2021;10:454. doi: 10.3390/antibiotics10040454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.