Abstract
Immune-checkpoint inhibitors (ICIs) play a key role in the treatment of advanced stage colorectal cancer (CRC) patients featuring a deficient DNA mismatch repair (dMMR) system or a high microsatellite instability (MSI-H) profile. However, beyond the established role in CRC patients, ICIs have highly proven efficacy in other solid tumors featuring MSI-H/dMMR status represented by endometrial, gastric, ovarian, prostatic, and pancreatic carcinomas (EC, GC, OC, PrC, and PaC). Our aim was to compare the concordance rates among the Idylla™ MSI test, TapeStation 4200, and immunohistochemical (IHC) analysis in assessing MSI-H/dMMR status in EC, GC, OC, PrC, and PaC patients. The Sanger sequencing-based Titano MSI test was used in discordant cases. One hundred and eighty-five cases (n = 40 PrC, n = 39 GC, n = 38 OC, n = 35 PaC, and n = 33 EC) were retrospectively selected. MMR protein expression was evaluated by IHC. After DNA quality and quantity evaluations, the IdyllaTM and TapeStation 4200 platforms were adopted for the evaluation of MSI status. Remarkably, compared to IHC, the Idylla™ platform achieved a global concordance rate of 94.5% (154/163) for the microsatellite stable (MSS)/proficient MMR (pMMR) cases and 77.3% (17/22) for the MSI-H/dMMR cases. Similarly, a global concordance rate of 91.4% (149/163) and 68.2% (15/22) for MSS/pMMR and MSI-H/dMMR cases was also identified between IHC and the TapeStation 4200 microfluidic system. In addition, a global concordance of 93.1% (148/159) and 69.2% (18/26) for MSS/pMMR and MSI-H/dMMR cases was observed between the Idylla™ and TapeStation 4200 platforms. Discordant cases were analyzed using the Titano MSI kit. Overall, our data pinpointed a central role for molecular techniques in the diagnostic evaluation of dMMR/MSI-H status not only in CRC patients but also in other types of solid tumors.
Keywords: predictive molecular pathology, IHC, fully automated RT-PCR, microfluidic, MMR, MSI, immunotherapy, immune checkpoint inhibitors (ICIs)
1. Introduction
DNA mismatch repair (MMR) is a highly conserved system responsible for restoring mismatching errors, such as single base mismatches, or small insertions and deletions. When these errors are not corrected, owing to a flawed MMR system, genomic steadiness is disrupted during DNA replication and recombination—a phenomenon that eventually gives rise to multiple cancer-associated mutations [1,2,3,4,5]. Generally, a deficient MMR (dMMR) system, is triggered by germline (e.g., Lynch syndrome), somatic, and epigenetic changes, which, in turn, result in the inactivation of MMR genes [1,3,6,7,8,9]. Thus, identifying MMR defective genes is crucial for cancer patient management.
The status of the MMR system is generally assessed by immunohistochemical analysis of the four proteins involved in MMR: namely, MLH1, MSH2, MSH6 and PMS2 [1,10]. However, evaluation and interpretation of immunohistochemical results may at times be challenging because of intra- and inter-observer variability and pre-analytical and analytical issues [1]. In this scenario, evaluation of microsatellite instability (MSI) through molecular approaches, such as polymerase chain reaction (PCR), genomic sequencing, and capillary electrophoresis, has been proposed as a valuable alternative to overcome some of the issues inherent to immunohistochemistry (IHC) [1,11].
In brief, during DNA replication, microsatellites, or short tandem mono or dinucleotide repeated sequences, which are widely disseminated in both coding and non-coding regions of the genome, may be susceptible to errors that are usually corrected by the MMR complex [12]. Generally, the most widely adopted procedure for MSI evaluation relies on the Sanger sequencing-based Bethesda panel, which covers two mononucleotide (BAT-25 and BAT-26) and three dinucleotide (D5S346, D2S123, and D17S250) repetitions [13] or, alternatively, a panel covering five poly-A mononucleotide repeats (BAT-25, BAT-26, NR-21, NR-24, NR-27) [14]. However, recent studies have demonstrated that the fully automated PCR high-resolution melt curve analysis (IdyllaTM, Biocartis, Mechelen, Belgium) [11,15,16,17,18,19,20,21,22,23,24,25,26] and the automated microfluidic electrophoretic run chip-based assay (TapeStation 4200, Agilent Technologies, Santa Clara, CA, USA) are easier to use, faster, and less expensive than Sanger sequencing [11,27,28].
Clinically, MMR/MSI testing has attracted increasing interest following Food and Drug Administration (FDA) approval in 2017 of the anti-programmed cell death protein 1 (PD-1) immune checkpoint monoclonal antibody pembrolizumab for MSI high (MSI-H)/dMMR patients with unresectable or metastatic solid tumors, regardless of age and histotype [29,30].
Beyond the established role of immune checkpoint inhibitors (ICIs) in the management of colorectal cancer (CRC) patients, these anticancer drugs have proven highly effective in other solid tumors featuring MSI-H/dMMR status. Among these, endometrial cancer (EC) displays a higher incidence of MSI-H status in endometrioid histotypes (40%) than in serous ones (2%) [31]. Overall, about 30% of primary ECs, and 13% to 30% of recurrent ECs, are MSI-H/dMMR [32]. Regarding the response rates to ICIs, MSI-H/dMMR advanced EC patients show higher response rates (from 27% to 57%) compared to microsatellite stable (MSS) patients (from 3% to 23%) [32].
A positive response to ICIs has also been reported in gastric carcinoma (GC) patients, who in 20% of cases feature MSI-H [33]. Indeed, MSI-H GC patients show a higher objective response rate (ORR) and disease control rate (DCR) to ICIs than MSS GC patients [34]. On the other hand, a lower frequency (12%) of MSI-H/dMMR status has been seen in unselected ovarian cancers (OCs) [35,36,37]. Regarding treatment response, more studies are needed to validate the efficacy of ICIs in MSI-H/dMMR OCs, suggesting that for this type of cancer, a better treatment option would be a combination therapy of ICIs and other anticancer drugs [38,39]. In intraductal papillary mucinous neoplasm (IPMN) of the pancreas, the incidence of MSI-H/dMMR is lower, dropping to 6.9% [40]. The lowest incidence rates of MSI-H/dMMR status have, instead, been observed in prostatic (PrC, about 3%) and pancreatic (PaC, about 1%) cancers [41,42]. However, despite such low incidence, ICIs have proven highly effective in both PrC and PaC patients featuring an MSI-H/dMMR status [41,42].
Thus, this evidence strongly underlines the importance of expanding the evaluation of MSI-H/dMMR status to different types of solid tumors. Although our research group has demonstrated the feasibility of adopting the fully automated PCR high-resolution melt curve analysis (IdyllaTM) and the automated microfluidic electrophoretic run chip-based assay (TapeStation 4200) to assess MSI-H/dMMR status in CRC patients [11], little is yet known about their performance in other types of tumors. In an attempt to fill this knowledge gap, we compared the concordance rates between these two molecular approaches and IHC analysis in assessing MSI-H/dMMR status in EC, GC, OC, PrC, and PaC patients. The Sanger sequencing-based Titano MSI test (Diatech Pharmacogenetics, Jesi, Italy) was used to confirm discordant cases.
2. Materials and Methods
2.1. Study Design
For our comparative analysis, we retrospectively selected a total of n = 199 formalin fixed paraffin embedded (FFPE) (n = 40 PrC, n = 40 GC, n = 39 OC, n = 40 PaC, and n = 40 EC) tumor samples, analyzed by IHC for MMR protein expression at the Fondazione IRCCS Casa Sollievo della Sofferenza (San Giovanni Rotondo, Italy) from 2015 to 2020. The percentage of neoplastic cells and the pathological status of each patient were evaluated by two expert pathologists. Briefly, for the molecular evaluation of MSI, tumor and a corresponding normal tissue specimen were collected after revision of original hematoxylin-and-eosin (H&E)-stained sections. All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Declaration of Helsinki (http://www.wma.net/en/30publications/10policies/b3/ last accessed 25 May 2021).
2.2. Immunohistochemical Analysis
Expression of MMR proteins (MLH1, PMS2, MSH2, and MSH6) was evaluated by IHC, as previously reported [11].
Briefly, 3-µm thick FFPE tissue sections were deparaffinized in xylene, rehydrated in graded alcohols, washed in double-distilled water, and pretreated with DAKO solution (EnVision FLEX Target Retrieval Solution, High pH 50×) at 97 °C. The slides were then incubated with primary monoclonal antibodies against MLH1 (clone ES05 diluted 1:50, DAKO), PMS2 (clone EP51 diluted 1:40, DAKO), MSH2 (clone FE11 diluted 1:50, DAKO), and MSH6 (clone EP49 diluted 1:50, DAKO) for 30 min. The analysis was performed on the automated platform Autostainer Link 48 (Dako, Carpinteria, CA, USA) according to the manufacturer’s instructions. The antigen-antibody reaction was inspected with the EnVision FLEX kit with diaminobenzidine as chromogen.
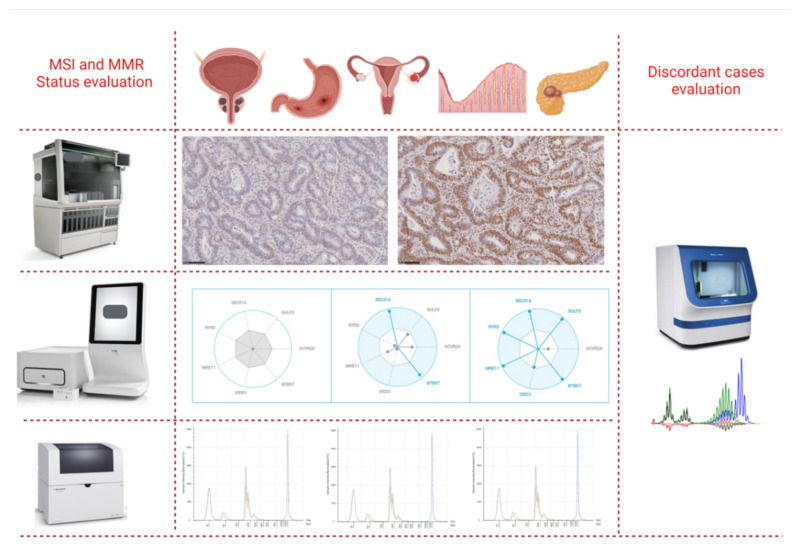
MMR protein expression was categorized as (i) retained (i.e., proficient MMR; pMMR), when a moderate to strong nuclear protein expression was detected in tumor cells as well as in internal control; and (ii) lost (i.e., dMMR), when a complete loss of nuclear expression in tumor cells was observed, but retained in normal cells [43]. Tumor samples showing absence of immunoreactions in internal controls were classified as “inadequate” for IHC evaluation and analysis and excluded from the study. A total of 185 cases were submitted to molecular approach (Figure 1).
Figure 1.
We retrospectively selected a total of n = 185 formalin fixed paraffin embedded (n = 40 prostate, n = 39 gastric, n = 38 ovarian, n = 35 pancreatic, and n = 33 endometrial) tumor samples, previously analyzed by immunohistochemistry for mismatch repair protein expression. In all samples, microsatellite instability (MSI) status was evaluated by Idylla™ MSI test and TapeStation 4200. Discordant cases were further analyzed by Titano MSI test analysis.
2.3. DNA Extraction and Qualification
DNA extraction and qualification were performed as previously reported [11].
Overall, four 5-µm thick sections obtained from tumor tissues and corresponding normal mucosae were adopted. The neoplastic cell area was manually microdissected. Then, DNA was extracted with the Mini Amp kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Finally, it was eluted in 30 µL of DNAse- and RNAse-free water (Thermo Fisher Scientifics, Waltham, MA, USA) and stored at −20 °C until DNA was qualified by the TapeStation 4200 microfluidic platform. In particular, the genomic ladder and sample buffer (Agilent Genomic ScreenTape, Agilent Technologies) were performed on the Genomic Screen Tape device (Agilent Technologies) according to the manufacturer’s instructions (Figure 1).
2.4. Microfluidic Analysis for MSI Status Evaluation
TapeStation 4200 analysis was performed as previously reported [11].
Five amplification reaction mixtures were prepared to analyze patients’ MSI status by using the Bethesda panel, starting from 20 ng of extracted DNA on the TapeStation 4200 system, as previously validated [11]. Briefly, for each patient, 1 µL of each amplified product from both tumor and normal tissues of each reaction mixture and 3 µL of D1000 Buffer (Agilent Technologies) were automatically charged on a solid device comprising 16 nanocapillaries (D1000 ScreenTape) and analyzed on the TapeStation 4200 platform. Results were inspected with proprietary software (TapeStation Analysis Software, Agilent Technologies) (Figure 1).
2.5. Idylla™ MSI Assay
The Idylla™ MSI test was performed as previously reported [11].
This test consists of a fully automated RT-qPCR system able to detect microsatellite instability directly from FFPE human cancer tissue sections. Overall, a total of four 5-micron thick slides obtained from tumor specimens were used for MSI analysis, according to the manufacturer’s instructions. Briefly, tumor areas were scraped from each sample with a sterile blade and inserted into an MSI cartridge, as previously described [11]. Overall, DNA was automatically extracted by a combination of HIFU, enzymatic/chemical digestion and heat, and then amplified into 5 PCR chambers. The Idylla MSI assay then analyzed homopolymers in 7 biomarkers (ACVR2A, BTBD7, DIDO1, MRE11, RYR3, SEC31A, and SULF2). Fluorescent signals, generated by fluorescently labeled molecular beacon probes, were automatically inspected by proprietary software able to analyze a minimum allele frequency of 10% and to calculate a probability score (MSI score) for all the tested biomarkers. Overall, samples with ≥2 of the 7 mutated biomarkers and <2 of the 7 mutated biomarkers were classified as MSI-H and MSS, respectively (Figure 1).
2.6. Discordant Cases Evaluted with the Titano MSI Test
The Titano MSI test analysis was performed as previously reported [11]. Briefly, the Titano MSI test (Diatech Pharmacogenetics) was used to investigate discordant cases between IHC and the 2 molecular approaches adopted in the study. DNA obtained from tumor and corresponding normal mucosa samples for each case were analyzed with MSI Titano kit. In brief, this test detects MSI status in cancer tissues through multiplex amplifications with fluorescent primers and subsequent DNA fragment analysis on an automated sequencer. An input of optimal 20 ng of extracted DNA is generally required to analyze variations in the number of microsatellite loci of 10 different molecular targets (BAT25, BAT26, D2S123, D17S250, D5S346, BAT40, D18S58, NR21, NR24, TGFβRII). In essence, variations are identified by comparing the peak profiles generated by capillary electrophoresis runs of tumor and corresponding normal tissue samples from each patient (Figure 1).
3. Results
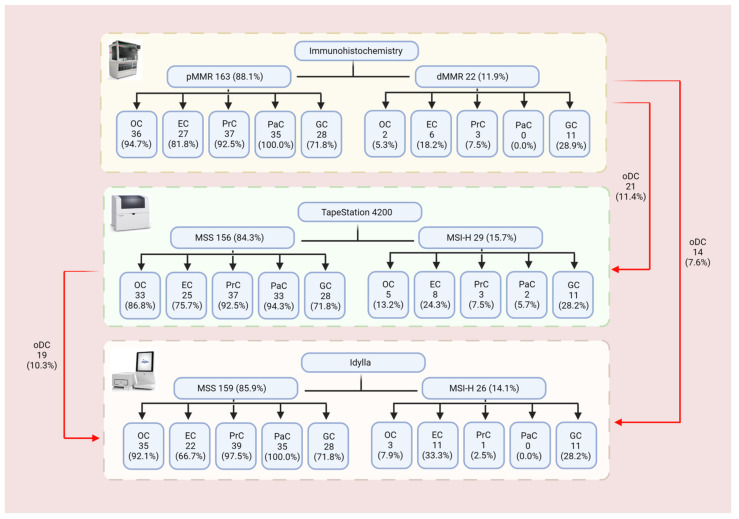
Overall, IHC analysis was successfully carried out in n = 185/199 (93.0%) formalin fixed paraffin embedded (FFPE) tumor samples (n = 40/40 PrC, n = 39/40 GC, n = 38/39 OC, n = 35/40 PaC, and n = 33/40 EC). DNA qualification revealed a median DNA concentration of 70.9 (ranging from 2.0 to 424.0 ng/µL) for the tumor samples and a median DNA concentration of 40.4 ng/µL (ranging from 0.1 to 60.0 ng/µL) for the normal samples. In detail, a median of 137.0 ng/µL (ranging from 13.2 to 414.0 ng/µL) and 38.2 ng/µL (ranging from 2.3 to 60.0 ng/µL), 90.8 ng/µL (ranging from 6.6 to 343.0 ng/µL) and 49.2 ng/µL (ranging from 9.5 to 60.0 ng/µL), 32.7 ng/µL (ranging from 2.1 to 179.0 ng/µL) and 31.8 ng/µL (ranging from 3.1 to 60.0 ng/µL), 31.0 ng/µL (ranging from 2.4 to 182.0 ng/µL) and 49.3 ng/µL (ranging from 6.3 to 60.0 ng/µL), 64.5 ng/µL (ranging from 4.4 to 187.0 ng/µL) and 35.7 ng/µL (ranging from 0.1 to 60.0 ng/µL) was evaluated for OC, EC, PrC, PaC, and GC tumor and normal mucosa specimens, respectively. In addition, a DNA integrity number (DIN) median value of 3.6 (ranging from 1.2 to 5.9) was globally observed in 183/185 (98.9%) cases. In particular, our analyses revealed a DIN median value of 4.3 (ranging from 1.9 to 5.9) for OC, of 3.4 (ranging from 1.9 to 5.6) for EC, of 3.8 (ranging from 2.3 to 5.3) for PrC of 2.8 (ranging from 1.2 to 4.8) for PaC, and of 3.7 (ranging from 2.4 to 5.3) for GC patients. IHC analysis highlighted an overall proficient MMR (pMMR) and dMMR status in 163/185 (88.1%) and 22/185 (11.9%) cases, respectively. In detail, 36/38 (94.7%) and 2/38 (5.3%), 27/33 (81.8%) and 6/33 (18.2%), 37/40 (92.5%) and 3/40 (7.5%), and 28/39 (71.8%) and 11/39 (28.9%) pMMR and dMMR cases were identified in OC, EC, PrC, and GC patients, respectively. All cases showed a pMMR profile in PaC samples.
Similarly, the Idylla™ platform and TapeStation 4200 system globally detected an overall MSS profile in 159/185 (85.9%) and 156/185 (84.3%) cases, respectively, whereas a global MSI-H profile was identified in 26/185 (14.1%) and in 29/185 (15.7%) cases, respectively. Regarding, the MSS status detected by the TapeStation 4200 system, 35/156 (22.4%) cases displayed a low MSI (MSI-L) status. Moreover, an MSI-H profile was respectively detected in 3/38 (7.9%) and in 5/38 (13.2%) of OC patients; in 11/33 (33.3%) and in 8/33 (24.3%) of EC patients; in 1/40 (2.5%) and 3/40 (7.5%) of PrC patients; in 11/39 (28.2%) and in 11/39 (28.2%) of GC patients by using the Idylla™ and TapeStation 4200 platforms, respectively. The remaining cases showed an MSS profile. Among PaC cases, Idylla™ detected an MSS profile in all cases, whereas TapeStation 4200 detected 33/35 (94.3%) MSS cases and 2/35 (5.7%) MSI-H cases. Regarding the MSS cases, TapeStation 4200 detected 9/38 (23.7%), 2/33 (6.1%), 13/40 (32.5%), 6/39 (15.4%), and 5/35 (14.3%) MSI-L cases in OC, EC, PrC, GC, and PaC, respectively. Remarkably, compared to IHC, the Idylla™ platform achieved a global concordance rate of 94.5% (154/163) for the MSS/pMMR cases and 77.3% (17/22) for the MSI-H/dMMR cases. Similarly, a global concordance rate of 91.4% (149/163) and 68.2% (15/22) for MSS/pMMR and MSI-H/dMMR cases was also identified between IHC and the TapeStation 4200 microfluidic system. In addition, a global concordance of 93.1% (148/159) and 69.2% (18/26) for MSS/pMMR and MSI-H/dMMR cases was also observed between the Idylla™ and TapeStation 4200 platforms. Concordant results between the five histological groups are reported in Table 1, Table 2, Table 3, Table 4 and Table 5 and Figure 2 and Figure 3.
Table 1.
Summary of molecular analysis of MMR/MSS status for endometrial cancer patients.
| ID | % Neoplastic Cells (I Evaluation) | % Neoplastic Cells (II Evaluation) | DNA Amount (ng/µL) | DIN | IHC | Idylla™ | TapeStation 4200 |
|---|---|---|---|---|---|---|---|
| 1 | 90.00 | 80.00 | 205.00 | 3.50 | pMMR | MSI-H | MSI-H |
| 2 | 40.00 | 60.00 | 15.30 | 3.30 | pMMR | MSS | MSS |
| 3 | 70.00 | 70.00 | 7.99 | 2.20 | pMMR | MSS | MSS |
| 4 | 90.00 | 70.00 | 77.70 | 4.30 | pMMR | MSS | MSS |
| 5 | 25.00 | 30.00 | 172.00 | 5.60 | pMMR | MSS | MSS |
| 6 | 80.00 | 60.00 | 9.91 | 2.60 | dMMR | MSS | MSI-H |
| 7 | 70.00 | 60.00 | 133.00 | 2.90 | pMMR | MSS | MSS |
| 8 | 90.00 | 70.00 | 101.00 | 2.80 | pMMR | MSS | MSS |
| 9 | 80.00 | 70.00 | 6.58 | 1.90 | pMMR | MSI-H | MSS |
| 10 | 80.00 | 60.00 | 284.00 | 3.90 | pMMR | MSI-H | MSI-H |
| 11 | 80.00 | 70.00 | 14.70 | 2.10 | pMMR | MSS | MSS |
| 12 | 90.00 | 30.00 | 63.80 | 4.00 | pMMR | MSI-H | MSI-H |
| 13 | 70.00 | 70.00 | 67.80 | 4.30 | pMMR | MSS | MSS |
| 14 | 70.00 | 60.00 | 343.00 | 4.50 | pMMR | MSS | MSS |
| 15 | 80.00 | 60.00 | 19.50 | 3.30 | pMMR | MSS | MSS |
| 16 | 60.00 | 80.00 | 163.00 | 4.00 | pMMR | MSS | MSS |
| 17 | 80.00 | 60.00 | 31.60 | 4.20 | pMMR | MSS | MSS |
| 18 | 80.00 | 80.00 | 78.50 | 3.30 | pMMR | MSS | MSS |
| 19 | 70.00 | 60.00 | 253.00 | 4.90 | pMMR | MSS | MSS |
| 20 | 90.00 | 70.00 | 131.00 | 3.50 | dMMR | MSI-H | MSI-H |
| 21 | 80.00 | 70.00 | 119.00 | 2.10 | pMMR | MSS | MSS |
| 22 | 90.00 | 70.00 | 106.00 | 4.30 | dMMR | MSI-H | MSS |
| 23 | 50.00 | 60.00 | 103.00 | 3.80 | pMMR | MSS | MSS |
| 24 | 90.00 | 70.00 | 14.50 | 4.30 | pMMR | MSI-H | MSI-H |
| 25 | 90.00 | 60.00 | 29.40 | 3.30 | pMMR | MSS | MSS |
| 26 | 90.00 | 70.00 | 15.80 | 2.70 | dMMR | MSI-H | MSS * |
| 27 | 90.00 | 60.00 | 156.00 | 4.50 | pMMR | MSI-H | MSI-H |
| 28 | 70.00 | 60.00 | 78.90 | 2.80 | dMMR | MSS | MSS |
| 29 | 80.00 | 70.00 | 121.00 | 3.40 | pMMR | MSS | MSS |
| 30 | 90.00 | 50.00 | 45.10 | 3.20 | pMMR | MSS | MSS |
| 31 | 70.00 | 50.00 | 7.58 | 2.70 | pMMR | MSS | MSS |
| 32 | 80.00 | 80.00 | 16.80 | 3.20 | pMMR | MSI-H | MSS * |
| 33 | 60.00 | 60.00 | 6.84 | 2.40 | dMMR | MSI-H | MSI-H |
Note: * MSI-L. Abbreviations: DIN: DNA integrity number; dMMR: deficient mismatch repair; ID: identification number; IHC: immunohistochemistry; MSI-H: high microsatellite instability; MSI-L: low microsatellite instability MSS: microsatellite stable; pMMR: proficient mismatch repair.
Table 2.
Summary of molecular analysis of MMR/MSS status for ovarian cancer patients.
| ID | % Neoplastic Cells (I Evaluation) | % Neoplastic Cells (II Evaluation) | DNA Amount (ng/µL) | DIN | IHC | Idylla™ | TapeStation 4200 |
|---|---|---|---|---|---|---|---|
| 1 | 35.00 | 20.00 | 117.00 | 5.90 | pMMR | MSS | MSS |
| 2 | 70.00 | 70.00 | 414.00 | 5.10 | pMMR | MSS | MSS |
| 3 | 80.00 | 40.00 | 89.40 | 5.80 | pMMR | MSS | MSS * |
| 4 | 80.00 | 60.00 | 118.90 | 5.30 | pMMR | MSS | MSS * |
| 5 | 80.00 | 70.00 | 374.00 | 5.10 | pMMR | MSS | MSS |
| 6 | 80.00 | 70.00 | 163.00 | 5.80 | pMMR | MSS | MSS |
| 7 | 40.00 | 30.00 | 48.00 | 4.60 | pMMR | MSS | MSS * |
| 8 | 90.00 | 60.00 | 254.00 | 5.30 | pMMR | MSS | MSS |
| 9 | 80.00 | 80.00 | 199.00 | 5.80 | pMMR | MSS | MSI-H |
| 10 | 90.00 | 70.00 | 83.50 | 4.70 | pMMR | MSS | MSI-H |
| 11 | 90.00 | 40.00 | 46.10 | 3.90 | dMMR | MSI-H | MSI-H |
| 12 | 90.00 | 60.00 | 25.40 | 3.80 | pMMR | MSS | MSS |
| 13 | 90.00 | 60.00 | 221.00 | 4.20 | pMMR | MSS | MSS |
| 14 | 90.00 | 80.00 | 191.00 | 3.90 | pMMR | MSS | MSS * |
| 15 | 70.00 | 50.00 | 48.30 | 2.40 | pMMR | MSI-H | MSS * |
| 16 | 90.00 | 60.00 | 147.00 | 4.60 | pMMR | MSS | MSS |
| 17 | 80.00 | 70.00 | 230.00 | 4.40 | pMMR | MSS | MSS |
| 18 | 90.00 | 60.00 | 112.00 | 4.50 | pMMR | MSS | MSS |
| 19 | 50.00 | 40.00 | 18.70 | 4.20 | pMMR | MSS | MSS |
| 20 | 90.00 | 80.00 | 93.80 | 3.70 | pMMR | MSS | MSS |
| 21 | 80.00 | 40.00 | 25.10 | 2.60 | pMMR | MSS | MSS * |
| 22 | 90.00 | 60.00 | 48.30 | 3.90 | pMMR | MSS | MSS * |
| 23 | 70.00 | 50.00 | 324.00 | 5.10 | pMMR | MSS | MSS |
| 24 | 90.00 | 70.00 | 424.00 | 4.30 | pMMR | MSS | MSI-H |
| 25 | 70.00 | 60.00 | 130.00 | 5.20 | pMMR | MSS | MSS |
| 26 | 80.00 | 60.00 | 79.40 | 5.20 | pMMR | MSS | MSS |
| 27 | 60.00 | 20.00 | 24.50 | 5.30 | pMMR | MSS | MSS |
| 28 | 80.00 | 60.00 | 41.80 | 5.70 | pMMR | MSS | MSS |
| 29 | 50.00 | 20.00 | 13.20 | 2.90 | pMMR | MSS | MSS |
| 30 | 90.00 | 70.00 | 30.80 | 3.60 | dMMR | MSI-H | MSS |
| 31 | 90.00 | 70.00 | 322.00 | 5.70 | pMMR | MSS | MSS * |
| 32 | 80.00 | 50.00 | 126.00 | 3.80 | pMMR | MSS | MSS |
| 33 | 60.00 | 60.00 | 218.00 | 5.10 | pMMR | MSS | MSS |
| 34 | 80.00 | 70.00 | 235.00 | 3.60 | pMMR | MSS | MSS |
| 35 | 90.00 | 70.00 | 45.90 | 2.20 | pMMR | MSS | MSS |
| 36 | 80.00 | 70.00 | 94.40 | 3.70 | pMMR | MSS | MSS * |
| 37 | 80.00 | 50.00 | 21.80 | 1.90 | pMMR | MSS | MSS |
| 38 | 90.00 | 80.00 | 13.40 | 2.00 | pMMR | MSS | MSI-H |
Note: * MSI-L. Abbreviations: DIN: DNA integrity number; dMMR: deficient mismatch repair; ID: identification number; IHC: immunohistochemistry; MSI-H: high microsatellite instability; MSI-L: low microsatellite instability MSS: microsatellite stable; pMMR: proficient mismatch repair.
Table 3.
Summary of molecular analysis of MMR/MSS status for pancreas cancer patients.
| ID | % Neoplastic Cells (I Evaluation) | % Neoplastic Cells (II Evaluation) | DNA Amount (ng/µL) | DIN | IHC | Idylla™ | TapeStation 4200 |
|---|---|---|---|---|---|---|---|
| 1 | 80.00 | 30.00 | 11.60 | 2.50 | pMMR | MSS | MSS |
| 2 | 70.00 | 20.00 | 6.10 | 2.10 | pMMR | MSS | MSS |
| 3 | 85.00 | 50.00 | 3.00 | 2.40 | pMMR | MSS | MSS |
| 4 | 95.00 | 70.00 | 10.00 | 2.70 | pMMR | MSS | MSS |
| 5 | 90.00 | 30.00 | 3.30 | 2.30 | pMMR | MSS | MSS |
| 6 | 80.00 | 50.00 | 4.80 | 2.20 | pMMR | MSS | MSS |
| 7 | 90.00 | 70.00 | 5.80 | 2.00 | pMMR | MSS | MSS * |
| 8 | 95.00 | 40.00 | 14.60 | 2.00 | pMMR | MSS | MSS |
| 9 | 80.00 | 50.00 | 3.10 | 2.00 | pMMR | MSS | MSS |
| 10 | 95.00 | 60.00 | 8.10 | 2.00 | pMMR | MSS | MSS |
| 11 | 80.00 | 30.00 | 19.00 | 2.10 | pMMR | MSS | MSS |
| 12 | 90.00 | 70.00 | 114.00 | 2.40 | pMMR | MSS | MSS |
| 13 | 95.00 | 60.00 | 12.60 | 3.70 | pMMR | MSS | MSS |
| 14 | 95.00 | 50.00 | 56.70 | 3.00 | pMMR | MSS | MSS |
| 15 | 60.00 | 30.00 | 4.90 | 4.00 | pMMR | MSS | MSS * |
| 16 | 20.00 | 20.00 | 60.30 | 2.70 | pMMR | MSS | MSS |
| 17 | 90.00 | 80.00 | 146.00 | 3.30 | pMMR | MSS | MSI-H |
| 18 | 60.00 | 30.00 | 15.20 | 1.70 | pMMR | MSS | MSS |
| 19 | 70.00 | 60.00 | 4.50 | 1.80 | pMMR | MSS | MSS |
| 20 | 80.00 | 60.00 | 22.80 | 2.10 | pMMR | MSS | MSS |
| 21 | 90.00 | 70.00 | 2.40 | 0.00 | pMMR | MSS | MSS * |
| 22 | 20.00 | 40.00 | 3.20 | 1.20 | pMMR | MSS | MSS |
| 23 | 40.00 | 20.00 | 4.00 | 2.70 | pMMR | MSS | MSS |
| 24 | 90.00 | 70.00 | 182.00 | 4.80 | pMMR | MSS | MSI-H |
| 25 | 25.00 | 30.00 | 12.70 | 3.10 | pMMR | MSS | MSS * |
| 26 | 80.00 | 30.00 | 8.40 | 3.30 | pMMR | MSS | MSS |
| 27 | 80.00 | 50.00 | 28.60 | 3.50 | pMMR | MSS | MSS |
| 28 | 90.00 | 70.00 | 42.40 | 3.20 | pMMR | MSS | MSS |
| 29 | 90.00 | 80.00 | 18.40 | 3.20 | pMMR | MSS | MSS |
| 30 | 20.00 | 10.00 | 43.50 | 4.70 | pMMR | MSS | MSS |
| 31 | 80.00 | 50.00 | 4.50 | 3.00 | pMMR | MSS | MSS |
| 32 | 80.00 | 20.00 | 14.80 | 1.90 | pMMR | MSS | MSS * |
| 33 | 70.00 | 40.00 | 131.00 | 3.60 | pMMR | MSS | MSS |
| 34 | 90.00 | 70.00 | 27.70 | 4.80 | pMMR | MSS | MSS |
| 35 | 80.00 | 40.00 | 36.10 | 2.80 | pMMR | MSS | MSS |
Note: * MSI-L. Abbreviations: DIN: DNA integrity number; dMMR: deficient mismatch repair; ID: identification number; IHC: immunohistochemistry; MSI-H: high microsatellite instability; MSI-L: low microsatellite instability MSS: microsatellite stable; pMMR: proficient mismatch repair.
Table 4.
Summary of molecular analysis of MMR/MSS status for prostate cancer patients.
| ID | % Neoplastic Cells (I Evaluation) | % Neoplastic Cells (II Evaluation) | DNA Amount (ng/µL) | DIN | IHC | Idylla™ | TapeStation 4200 |
|---|---|---|---|---|---|---|---|
| 1 | 80.00 | 50.00 | 12.20 | 4.00 | pMMR | MSS | MSS |
| 2 | 85.00 | 70.00 | 18.90 | 4.10 | pMMR | MSS | MSS * |
| 3 | 95.00 | 60.00 | 34.30 | 4.00 | pMMR | MSS | MSS * |
| 4 | 95.00 | 60.00 | 21.40 | 3.10 | pMMR | MSS | MSS * |
| 5 | 70.00 | 50.00 | 15.10 | 4.00 | dMMR | MSS | MSI-H |
| 6 | 85.00 | 40.00 | 4.09 | 3.00 | pMMR | MSS | MSS * |
| 7 | 85.00 | 40.00 | 16.30 | 3.90 | pMMR | MSS | MSS |
| 8 | 95.00 | 70.00 | 27.50 | 4.00 | pMMR | MSS | MSS |
| 9 | 95.00 | 70.00 | 56.10 | 4.20 | pMMR | MSS | MSS |
| 10 | 85.00 | 30.00 | 25.20 | 4.30 | pMMR | MSS | MSS |
| 11 | 85.00 | 70.00 | 14.10 | 3.50 | pMMR | MSS | MSI-H |
| 12 | 90.00 | 80.00 | 52.70 | 4.40 | pMMR | MSI-H | MSS * |
| 13 | 90.00 | 60.00 | 23.60 | 3.80 | pMMR | MSS | MSS |
| 14 | 80.00 | 60.00 | 13.80 | 4.10 | pMMR | MSS | MSS * |
| 15 | 90.00 | 70.00 | 42.10 | 3.10 | pMMR | MSS | MSS |
| 16 | 85.00 | 60.00 | 13.30 | 3.20 | pMMR | MSS | MSS |
| 17 | 90.00 | 80.00 | 23.10 | 3.70 | pMMR | MSS | MSS |
| 18 | 85.00 | 50.00 | 7.07 | 3.80 | pMMR | MSS | MSS |
| 19 | 90.00 | 70.00 | 58.30 | 3.90 | pMMR | MSS | MSS* |
| 20 | 85.00 | 70.00 | 25.90 | 3.40 | pMMR | MSS | MSS |
| 21 | 85.00 | 50.00 | 51.00 | 5.00 | dMMR | MSS | MSS |
| 22 | 80.00 | 60.00 | 38.20 | 4.70 | pMMR | MSS | MSS |
| 23 | 90.00 | 80.00 | 31.80 | 3.80 | pMMR | MSS | MSS |
| 24 | 90.00 | 80.00 | 52.40 | 3.70 | pMMR | MSS | MSS |
| 25 | 85.00 | 60.00 | 92.20 | 4.70 | dMMR | MSS | MSS |
| 26 | 85.00 | 80.00 | 30.80 | 3.70 | pMMR | MSS | MSS |
| 27 | 95.00 | 80.00 | 104.00 | 3.80 | pMMR | MSS | MSS * |
| 28 | 90.00 | 60.00 | 22.40 | 4.00 | pMMR | MSS | MSS |
| 29 | 90.00 | 70.00 | 13.40 | 2.80 | pMMR | MSS | MSS * |
| 30 | 90.00 | 60.00 | 10.90 | 2.70 | pMMR | MSS | MSS |
| 31 | 95.00 | 70.00 | 37.70 | 4.00 | pMMR | MSS | MSS |
| 32 | 90.00 | 60.00 | 38.50 | 3.90 | pMMR | MSS | MSS |
| 33 | 90.00 | 50.00 | 14.20 | 3.00 | pMMR | MSS | MSS |
| 34 | 90.00 | 50.00 | 18.90 | 2.60 | pMMR | MSS | MSS * |
| 35 | 95.00 | 70.00 | 23.30 | 4.60 | pMMR | MSS | MSS * |
| 36 | 80.00 | 50.00 | 9.67 | 3.40 | pMMR | MSS | MSS * |
| 37 | 90.00 | 50.00 | 179.00 | 5.30 | pMMR | MSS | MSS |
| 38 | 85.00 | 50.00 | 2.04 | 2.30 | pMMR | MSS | MSS |
| 39 | 90.00 | 70.00 | 14.50 | 3.80 | pMMR | MSS | MSI-H |
| 40 | 85.00 | 60.00 | 14.50 | 3.40 | pMMR | MSS | MSS * |
Note: * MSI-L. Abbreviations: DIN: DNA integrity number; dMMR: deficient mismatch repair; ID: identification number; IHC: immunohistochemistry; MSI-H: high microsatellite instability; MSI-L: low microsatellite instability MSS: microsatellite stable; pMMR: proficient mismatch repair.
Table 5.
Summary of molecular analysis of MMR/MSS status for gastric cancer patients.
| ID | % Neoplastic Cells (I Evaluation) | % Neoplastic Cells (II Evaluation) | DNA Amount (ng/µL) | DIN | IHC | Idylla™ | TapeStation 4200 |
|---|---|---|---|---|---|---|---|
| 1 | 80.00 | 20.00 | 17.20 | 2.90 | pMMR | MSS | MSS * |
| 2 | 90.00 | 70.00 | 60.00 | 3.70 | pMMR | MSS | MSS |
| 3 | 80.00 | 60.00 | 65.80 | 4.80 | pMMR | MSS | MSS |
| 4 | 80.00 | 40.00 | 4.40 | 0.00 | pMMR | MSS | MSS |
| 5 | 90.00 | 60.00 | 145.00 | 5.30 | pMMR | MSS | MSS * |
| 6 | 90.00 | 50.00 | 12.30 | 3.20 | pMMR | MSS | MSS |
| 7 | 90.00 | 70.00 | 66.30 | 2.80 | pMMR | MSS | MSS |
| 8 | 90.00 | 70.00 | 92.60 | 5.50 | dMMR | MSI-H | MSI-H |
| 9 | 70.00 | 20.00 | 34.40 | 2.70 | pMMR | MSS | MSS |
| 10 | 90.00 | 60.00 | 84.60 | 3.30 | pMMR | MSS | MSS |
| 11 | 85.00 | 70.00 | 33.30 | 3.20 | dMMR | MSI-H | MSI-H |
| 12 | 70.00 | 30.00 | 88.30 | 3.80 | pMMR | MSS | MSS |
| 13 | 80.00 | 70.00 | 92.20 | 2.70 | dMMR | MSI-H | MSI-H |
| 14 | 60.00 | 40.00 | 50.40 | 4.60 | pMMR | MSS | MSS |
| 15 | 90.00 | 50.00 | 27.90 | 2.50 | dMMR | MSI-H | MSI-H |
| 16 | 95.00 | 60.00 | 10.50 | 3.70 | dMMR | MSI-H | MSS |
| 17 | 90.00 | 20.00 | 4.80 | 2.80 | pMMR | MSS | MSS * |
| 18 | 60.00 | 20.00 | 25.20 | 3.60 | pMMR | MSS | MSS |
| 19 | 90.00 | 70.00 | 68.00 | 2.40 | pMMR | MSS | MSS |
| 20 | 80.00 | 20.00 | 51.80 | 3.60 | pMMR | MSS | MSS |
| 21 | 90.00 | 60.00 | 187.00 | 4.20 | dMMR | MSI-H | MSI-H |
| 22 | 80.00 | 40.00 | 16.30 | 3.90 | pMMR | MSS | MSS |
| 23 | 90.00 | 70.00 | 109.00 | 3.00 | pMMR | MSS | MSS |
| 24 | 90.00 | 60.00 | 22.10 | 3.20 | pMMR | MSS | MSS |
| 25 | 80.00 | 30.00 | 45.70 | 3.60 | pMMR | MSS | MSS * |
| 26 | 85.00 | 60.00 | 10.30 | 3.80 | dMMR | MSI-H | MSI-H |
| 27 | 95.00 | 60.00 | 120.60 | 3.80 | dMMR | MSI-H | MSI-H |
| 28 | 90.00 | 50.00 | 111.00 | 4.20 | pMMR | MSS | MSS |
| 29 | 80.00 | 60.00 | 25.40 | 4.30 | pMMR | MSS | MSS |
| 30 | 85.00 | 70.00 | 41.30 | 3.10 | pMMR | MSS | MSS * |
| 31 | 85.00 | 70.00 | 57.30 | 3.50 | dMMR | MSI-H | MSI-H |
| 32 | 80.00 | 40.00 | 120.00 | 5.30 | pMMR | MSS | MSS |
| 33 | 95.00 | 70.00 | 64.40 | 4.40 | dMMR | MSI-H | MSI-H |
| 34 | 90.00 | 60.00 | 56.80 | 4.70 | dMMR | MSI-H | MSI-H |
| 35 | 95.00 | 80.00 | 85.90 | 2.50 | pMMR | MSS | MSI-H |
| 36 | 70.00 | 30.00 | 159.00 | 3.50 | pMMR | MSS | MSS |
| 37 | 90.00 | 50.00 | 93.20 | 4.40 | pMMR | MSS | MSS |
| 38 | 80.00 | 50.00 | 94.80 | 5.20 | pMMR | MSS | MSS * |
| 39 | 65.00 | 30.00 | 60.40 | 3.90 | pMMR | MSS | MSS |
Note: * MSI-L. Abbreviations: DIN: DNA integrity number; dMMR: deficient mismatch repair; ID: identification number; IHC: immunohistochemistry; MSI-H: high microsatellite instability; MSI-L: low microsatellite instability MSS: microsatellite stable; pMMR: proficient mismatch repair.
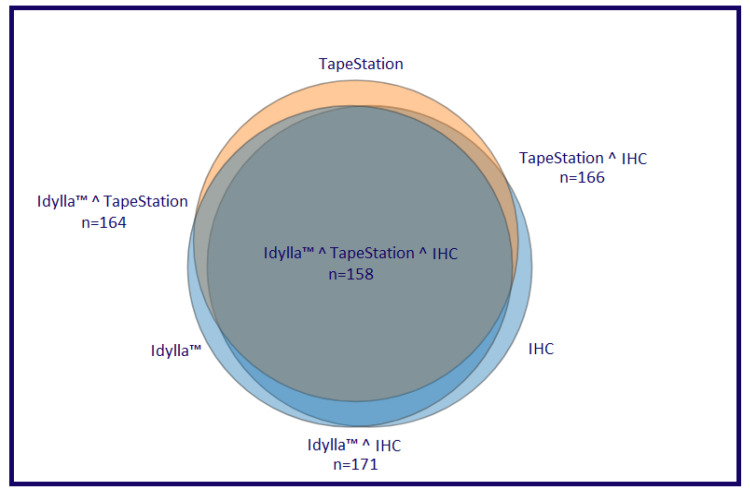
Figure 2.
Venn diagram representing the concordance rate among the technologies adopted in the present study. Briefly, dark gray circle represents fully concordant cases among the three different assays; dark gray plus dark orange circles represent the concordant cases between TapeStation 4200 and IHC; dark gray and dark blue circles represent the concordant cases between IdyllaTM and IHC.
Figure 3.
Flow chart summarizing results obtained with the three different assays. Abbreviations: dMMR: deficient mismatch repair; EC: endometrial cancer; GC: gastric cancer; MSI-H: high microsatellite instability; MSS: microsatellite stable; OC: ovarian cancer; oDC: overall discordant cases; PaC: pancreatic cancer; pMMR: proficient mismatch repair; PrC: prostatic cancer.
Discordant cases among the TapeStation 4200 system, IdyllaTM platform, and IHC approach were successfully analyzed by applying the Titano MSI kit. The results showed that in 14 out of 27 (51.8%) cases, the Titano kit and IHC showed concordant results (n = 10 MSS/pMMR and n = 4 MSI-H/dMMR); whereas in 13 out 27 cases (48.2%), a discordant result was reported (n = 7 MSS/dMMR and n = 6 MSI-H/pMMR). As far as the comparison between the Titano MSI kit and IdyllaTM is concerned, overall, 22 concordant cases (81.5%; n = 13 MSS and n = 9 MSI-H) were detected. Only five discordant cases (n = 4 MSS for Titano MSI kit and MSI-H for IdyllaTM and n = 1 MSI-H for Titano MSI kit and MSS for IdyllaTM) were reported. The results showed that in 11 out of 27 (40.7%) cases, the Titano MSI kit and TapeStation 4200 showed concordant results (n = 6 MSS and n = 5 MSI-H); whereas in 16 out 27 cases (59.3%) a discordant result was reported (n = 11 MSS for Titano MSI kit and MSI-H for TapeStation 4200 and n = 5 MSI-H for Titano MSI kit and MSS for TapeStation 4200). The results are summarized in Table 6.
Table 6.
Comparative analysis of the discordant cases.
| Case ID | Titano | IHC | IdyllaTM | TapeStation 4200 |
|---|---|---|---|---|
| EC 1 | MSI-H | pMMR | MSI-H | MSI-H |
| EC 6 | MSI-H | dMMR | MSS | MSI-H |
| EC 11 | MSS * | pMMR | MSI-H | MSS |
| EC 12 | MSS * | pMMR | MSI-H | MSI-H |
| EC 14 | MSI-H | pMMR | MSI-H | MSI-H |
| EC 28 | MSI-H | dMMR | MSI-H | MSS |
| EC 30 | MSI-H | pMMR | MSI-H | MSI-H |
| EC 32 | MSI-H | dMMR | MSI-H | MSS * |
| EC 33 | MSI-H | pMMR | MSI-H | MSI-H |
| EC 34 | MSS* | dMMR | MSS | MSS |
| EC 38 | MSI-H | pMMR | MSI-H | MSS * |
| OC 9 | MSS | pMMR | MSS | MSI-H |
| OC 10 | MSS | pMMR | MSS | MSI-H |
| OC 15 | MSS * | pMMR | MSI-H | MSS * |
| OC 24 | MSS | pMMR | MSS | MSI-H |
| OC 31 | MSS | dMMR | MSI-H | MSS |
| OC 39 | MSS | pMMR | MSS | MSI-H |
| PaC 19 | MSS | pMMR | MSS | MSI-H |
| PaC 27 | MSS | pMMR | MSS | MSI-H |
| PrC 5 | MSS | dMMR | MSS | MSI-H |
| PrC 11 | MSS | pMMR | MSS | MSI-H |
| PrC 12 | MSI-H | pMMR | MSI-H | MSS * |
| PrC 21 | MSS | dMMR | MSS | MSS |
| PrC 25 | MSS | dMMR | MSS | MSS |
| PrC 39 | MSS | pMMR | MSS | MSI-H |
| GC 16 | MSI-H | dMMR | MSI-H | MSS |
| GC 36 | MSS | pMMR | MSS | MSI-H |
Note: * MSI-L. Abbreviations: dMMR: deficient mismatch repair; EC: endometrial carcinoma; GC: gastric carcinoma; ID: identification number; IHC: immunohistochemistry; MSI-H: high microsatellite instability; MSI-L: low microsatellite instability; MSS: microsatellite stable; OC: ovarian carcinoma; PaC: pancreatic carcinoma; PrC: prostatic carcinoma; pMMR: proficient mismatch repair.
4. Discussion
The crucial role of MMR/MSI analysis for ICI administration followed the FDA approval in 2017 of the anti-PD-1 immune checkpoint monoclonal antibody pembrolizumab for MSI-H/dMMR patients with unresectable or metastatic solid tumors, regardless of age and histotype [29,30]. However, beyond the clear role in the management of CRC patients, the efficacy of ICIs in other solid tumors featuring MSI-H/dMMR status, including ECs, OCs, PaCs, PrCs, and GCs, has been demonstrated [31,32,33,34,35,36,37,38,39,40,41,42]. Thus, it is pivotal that MSI-H/dMMR status in different types of solid tumors be evaluated.
Although IHC remains the gold standard approach for MSI biomarker analysis in solid tumors, our data indicate that both Idylla and TapeStation molecular platforms are able to analyze MSI status just as efficiently as IHC. Indeed, a high concordance rate was observed between the Idylla™ platform and IHC (94.5% and 77.3% for the for MSS/pMMR and MSI-H/dMMR cases, respectively), and between the TapeStation 4200 microfluidic system and IHC (91.4% and 68.2% for the MSS/pMMR and MSI-H/dMMR cases, respectively). Of note, overall high concordance rates were also seen between the TapeStation 4200 and Idylla™ platforms (93.1% and 69.2% for MSS and MSI-H cases, respectively). In addition, in line with previous data, GCs (28.9%) and ECs (27.0%) showed the highest rates of dMMR cases, followed by PrCs (7.5%) and OCs (5.3%). In our experience, all PaC cases showed a pMMR profile. Similarly, MSI-H status was more frequent in GCs (28.2% and 28.2%, by IdyllaTM and TapeStation 4200, respectively) and ECs (33.3% and 24.3%, by IdyllaTM and TapeStation 4200, respectively) than in OCs (7.3% and 13.2%, by IdyllaTM and TapeStation 4200, respectively), PrCs (2.5% and 7.5%, by IdyllaTM and TapeStation 4200, respectively), and PaCs (0.0% and 5.7%, by IdyllaTM and TapeStation 4200, respectively).
Hence, our data clearly highlight that molecular analysis represents a valid upfront approach to evaluate MSI status in various types of cancer patients who would definitely benefit from ICI treatments. Indeed, we have demonstrated that MMR/MSI testing has acquired a central role not only in CRC management but also in the clinical stratification of different solid tumors [29,30]. In fact, patients displaying MSI-H/dMMR status respond well to immunotherapy regimens [29,30]. It is against this background that molecular analysis could represent a valuable strategy to overcome the well-known limitations of IHC, including intra- and inter-observer variability and pre-analytical and analytical issues [1]. In particular, a recent study has highlighted that a pMMR IHC profile may be retained in cases featuring antigenically intact non-functioning proteins despite the presence of an MSI-H status [44]. In addition, the high global concordance (91.4% and 68.2% for MSS/pMMR and MSI-H/dMMR cases, respectively) observed between the fully automated PCR high-resolution melt curve analysis (IdyllaTM) and the automated microfluidic electrophoretic run chip-based assay highlights that these user-friendly, rapid, and sensitive molecular techniques are interchangeable and, therefore, all equally efficient. Moreover, in all cases they seemed to outperform the gold standard IHC approach in selecting MSI-H/dMMR advanced patients for ICI treatments. Remarkably, for each solid tumor analyzed, they were both able to identify a higher percentage of patients eligible for ICI administration than did IHC. Although these results clearly indicate the practical advantages of integrating these approaches into clinical practice, further studies are definitely warranted to confirm whether their implementation in clinical practice might actually help oncologists to choose the best treatment option for their patients.
Among the 27 discordant cases, 13 tumors showed different IHC and Titano profiling. Twelve of these tumors were endometrial and prostate adenocarcinomas. Of note, different molecular mechanisms characterize MSI in the different organs and may impact microsatellite instability testing and MMR IHC [45]. Focusing on EC, high agreement between IHC and MSI analysis has been described. However, several factors can affect this view: (i) MSH6 negative tumors may show MSS or MSI-L at MSI analysis [46]; (ii) POLE mutated tumors may present an MSI profile due to the accumulation of mutations in microsatellite loci despite preserved MMR protein expression; (iii) subclonal loss of MMR protein expression is present in around 3% of EC cases; a minimal microsatellite shift characterizes MSI EC and may interfere with the diagnostic interpretation of the results [47]; (iv) the use of obsolete IHC evaluation criteria may impact MMR evaluation [48]; and (v) the different PCR-based approaches are affected by intrinsic limitations that adequate training of the user and a high quality sample can overcome [15]. Overall, our data pinpointed a central role for molecular techniques in the diagnostic evaluation of MSI-H/dMMR status not only in CRC patients but also in other types of solid tumors. Future efforts should be focused on designing and developing clinical trials to validate the actual ability of molecular techniques to stratify patients with different types of solid tumors, including breast cancer [49,50], who may benefit from ICIs.
Acknowledgments
We thank Paola Merolla for editing the manuscript.
Author Contributions
Conceptualization, U.M., G.T. and P.G.; methodology, U.M., P.P. (Paola Parente), F.P., C.D.L., P.P. (Pasquale Pisapia), R.S., M.N., G.G., G.R., F.C., C.B., E.V., A.I., C.C. (Claudia Covelli), M.B., C.C. (Celeste Clemente), G.P., A.D., F.S., M.F., G.T., P.G.; software, U.M., P.P. (Paola Parente), F.P., C.D.L., P.P. (Pasquale Pisapia), R.S., M.N., G.G., G.R., F.C., C.B., E.V., A.I., C.C. (Claudia Covelli), M.B., C.C. (Celeste Clemente), G.P., A.D., F.S., M.F., G.T, P.G.; validation, U.M., P.P. (Paola Parente), F.P., C.D.L., P.P. (Pasquale Pisapia), R.S., M.N., G.G., G.R., F.C., C.B., E.V., A.I., C.C. (Claudia Covelli), M.B., C.C. (Celeste Clemente), G.P., A.D., F.S., M.F., G.T., P.G.; formal analysis, U.M., P.P. (Paola Parente), F.P., C.D.L., P.P. (Pasquale Pisapia), R.S., M.N., G.G., G.R., F.C., C.B., E.V., A.I., C.C. (Claudia Covelli), M.B., C.C. (Celeste Clemente), G.P., A.D., F.S., M.F., G.T., P.G.; investigation, U.M., P.P. (Paola Parente), F.P., C.D.L., P.P. (Pasquale Pisapia), R.S., M.N., G.G., G.R., F.C., C.B., E.V., A.I., C.C. (Claudia Covelli), M.B., C.C. (Celeste Clemente), G.P., A.D., F.S., M.F., G.T, P.G.; resources, G.T., P.G.; data curation, U.M., P.P. (Paola Parente), F.P., C.D.L., P.P. (Pasquale Pisapia), R.S., M.N., G.G., G.R., F.C., C.B., E.V., A.I., C.C. (Claudia Covelli), M.B., C.C. (Celeste Clemente), G.P., A.D., F.S., M.F., G.T, P.G.; writing—original draft preparation, U.M., F.P., P.P. (Pasquale Pisapia), and G.T.; writing—review and editing, U.M., P.P. (Paola Parente), F.P., C.D.L., P.P. (Pasquale Pisapia), R.S., M.N., G.G., G.R., F.C., C.B., E.V., A.I., C.C. (Claudia Covelli), M.B., C.C. (Celeste Clemente), G.P., A.D., F.S., M.F., G.T, P.G.; visualization, U.M., P.P. (Paola Parente), F.P., C.D.L., P.P. (Pasquale Pisapia), R.S., M.N., G.G., G.R., F.C., C.B., E.V., A.I., C.C. (Claudia Covelli), M.B., C.C. (Celeste Clemente), G.P., A.D., F.S., M.F., G.T., P.G.; supervision, G.T., P.G.; project administration, G.T., P.G.; funding acquisition, M.F., G.T. All authors have read and agreed to the published version of the manuscript.
Funding
1. Monitoraggio ambientale, studio ed approfondimento della salute della popolazione residente in aree a rischio—In attuazione della D.G.R. Campania n.180/2019 to G.T. 2. POR Campania FESR 2014–2020 Progetto “Sviluppo di Approcci Terapeutici Innovativi per patologie Neoplastiche resistenti ai trattamenti—SATIN” to G.T. 3. This work was partly supported by a grant from the Italian Health Ministry and Veneto Region research program NET-2016–02363853 to M.F. The funding agencies had no role in the design and performance of the study.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
Umberto Malapelle has received personal fees (as consultant and/or speaker bureau) from Boehringer Ingelheim, Roche, MSD, Amgen, Thermo Fisher Scientifics, Eli Lilly, Diaceutics, GSK, Merck, and AstraZeneca, unrelated to the current work. Elena Vigliar has received personal fees (as consultant and/or speaker bureau) from Diaceutics, unrelated to the current work. Matteo Fassan has received personal fees (as consultant and/or speaker bureau) from Roche, Diaceutics, GSK, and Astellas Pharma and research grants from Astellas Pharma, QED Therapeutics, and Macrophage Pharma, unrelated to the current work. Giancarlo Troncone reports personal fees (as speaker bureau or advisor) from Roche, MSD, Pfizer, and Bayer, unrelated to the current work. Paolo Graziano received personal fees (as consultant and/or speaker bureau) from Eli Lilly, AstraZeneca, Pfizer, Novartis, Boehringer Ingelheim, Roche, and MSD, unrelated to the current work. The other Authors have nothing to disclose.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Luchini C., Bibeau F., Ligtenberg M.J.L., Singh N., Nottegar A., Bosse T., Miller R., Riaz N., Douillard J.Y., Andre F., et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: A systematic review-based approach. Ann. Oncol. 2019;30:1232–1243. doi: 10.1093/annonc/mdz116. [DOI] [PubMed] [Google Scholar]
- 2.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fassan M., Scarpa A., Remo A., De Maglio G., Troncone G., Marchetti A., Doglioni C., Ingravallo G., Perrone G., Parente P., et al. Current prognostic and predictive biomarkers for gastrointestinal tumors in clinical practice. Pathologica. 2020;112:248–259. doi: 10.32074/1591-951X-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li K., Luo H., Huang L., Luo H., Zhu X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int. 2020;20:16. doi: 10.1186/s12935-019-1091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evrard C., Tachon G., Randrian V., Karayan-Tapon L., Tougeron D. Microsatellite Instability: Diagnosis, Heterogeneity, Discordance, and Clinical Impact in Colorectal Cancer. Cancers. 2019;11:1567. doi: 10.3390/cancers11101567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leclerc J., Vermaut C., Buisine M.P. Diagnosis of Lynch Syndrome and Strategies to Distinguish Lynch-Related Tumors from Sporadic MSI/dMMR Tumors. Cancers. 2021;13:467. doi: 10.3390/cancers13030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morak M., Schackert H.K., Rahner N., Betz B., Ebert M., Walldorf C., Royer-Pokora B., Schulmann K., von Knebel-Doeberitz M., Dietmaier W., et al. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur. J. Hum. Genet. 2008;16:804–811. doi: 10.1038/ejhg.2008.25. [DOI] [PubMed] [Google Scholar]
- 8.Bao F., Panarelli N.C., Rennert H., Sherr D.L., Yantiss R.K. Neoadjuvant therapy induces loss of MSH6 expression in colorectal carcinoma. Am. J. Surg. Pathol. 2010;34:1798–1804. doi: 10.1097/PAS.0b013e3181f906cc. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein J.B., Wu W., Borras E., Masand G., Cuddy A., Mork M.E., Bannon S.A., Lynch P.M., Rodriguez-Bigas M., Taggart M.W., et al. Can Microsatellite Status of Colorectal Cancer Be Reliably Assessed after Neoadjuvant Therapy? Clin. Cancer Res. 2017;23:5246–5254. doi: 10.1158/1078-0432.CCR-16-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piciotti R., Venetis K., Sajjadi E., Fusco N. Mismatch Repair Status Characterization in Oncologic Pathology: Taking Stock of the Real-World Possibilities. J. Mol. Pathol. 2021;2:93–100. doi: 10.3390/jmp2020009. [DOI] [Google Scholar]
- 11.Malapelle U., Parente P., Pepe F., De Luca C., Cerino P., Covelli C., Balestrieri M., Russo G., Bonfitto A., Pisapia P., et al. Impact of Pre-Analytical Factors on MSI Test Accuracy in Mucinous Colorectal Adenocarcinoma: A Multi-Assay Concordance Study. Cells. 2020;9:2019. doi: 10.3390/cells9092019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baretti M., Le D.T. DNA mismatch repair in cancer. Pharmacol. Ther. 2018;189:45–62. doi: 10.1016/j.pharmthera.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Umar A., Boland C.R., Terdiman J.P., Syngal S., de la Chapelle A., Rüschoff J., Fishel R., Lindor N.M., Burgart L.J., Hamelin R., et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goel A., Nagasaka T., Hamelin R., Boland C.R. An optimized pentaplex PCR for detecting DNA mismatch repair-deficient colorectal cancers. PLoS ONE. 2010;5:e9393. doi: 10.1371/annotation/572bb6d3-0315-40b1-a6d7-ce818809b5ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siemanowski J., Schömig-Markiefka B., Buhl T., Haak A., Siebolts U., Dietmaier W., Arens N., Pauly N., Ataseven B., Büttner R., et al. Managing Difficulties of Microsatellite Instability Testing in Endometrial Cancer-Limitations and Advantages of Four Different PCR-Based Approaches. Cancers. 2021;13:1268. doi: 10.3390/cancers13061268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ukkola I., Nummela P., Pasanen A., Kero M., Lepistö A., Kytölä S., Bützow R., Ristimäki A. Detection of microsatellite instability with Idylla MSI assay in colorectal and endometrial cancer. Virchows Arch. 2021 doi: 10.1007/s00428-021-03082-w. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velasco A., Tokat F., Bonde J., Trim N., Bauer E., Meeney A., de Leng W., Chong G., Dalstein V., Kis L.L., et al. Multi-center real-world comparison of the fully automated Idylla™ microsatellite instability assay with routine molecular methods and immunohistochemistry on formalin-fixed paraffin-embedded tissue of colorectal cancer. Virchows Arch. 2021;478:851–863. doi: 10.1007/s00428-020-02962-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farmkiss L., Hopkins I., Jones M. Idylla microsatellite instability assay versus mismatch repair immunohistochemistry: A retrospective comparison in gastric adenocarcinoma. J. Clin. Pathol. 2020 doi: 10.1136/jclinpath-2020-207033. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 19.Gilson P., Levy J., Rouyer M., Demange J., Husson M., Bonnet C., Salleron J., Leroux A., Merlin J.L., Harlé A. Evaluation of 3 molecular-based assays for microsatellite instability detection in formalin-fixed tissues of patients with endometrial and colorectal cancers. Sci. Rep. 2020;10:16386. doi: 10.1038/s41598-020-73421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mindiola-Romero A.E., Green D.C., Al-Turkmani M.R., Godwin K.N., Mackay A.C., Tafe L.J., Ren B., Tsongalis G.J. Novel Biocartis Idylla™ cartridge-based assay for detection of microsatellite instability in colorectal cancer tissues. Exp. Mol. Pathol. 2020;116:104519. doi: 10.1016/j.yexmp.2020.104519. [DOI] [PubMed] [Google Scholar]
- 21.Pécriaux A., Favre L., Calderaro J., Charpy C., Derman J., Pujals A. Detection of microsatellite instability in a panel of solid tumours with the Idylla MSI Test using extracted DNA. J. Clin. Pathol. 2021;74:36–42. doi: 10.1136/jclinpath-2020-206581. [DOI] [PubMed] [Google Scholar]
- 22.Bourhis A., De Luca C., Cariou M., Vigliar E., Barel F., Conticelli F., Marcorelles P., Nousbaum J.B., Robaszkiewicz M., Samaison L., et al. Evaluation of KRAS, NRAS and BRAF mutational status and microsatellite instability in early colorectal carcinomas invading the submucosa (pT1): Towards an in-house molecular prognostication for pathologists? J. Clin. Pathol. 2020;73:741–747. doi: 10.1136/jclinpath-2020-206496. [DOI] [PubMed] [Google Scholar]
- 23.Zwaenepoel K., Holmgaard Duelund J., De Winne K., Maes V., Weyn C., Lambin S., Dendooven R., Broeckx G., Steiniche T., Pauwels P. Clinical Performance of the Idylla MSI Test for a Rapid Assessment of the DNA Microsatellite Status in Human Colorectal Cancer. J. Mol. Diagn. 2020;22:386–395. doi: 10.1016/j.jmoldx.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Lee M., Chun S.M., Sung C.O., Kim S.Y., Kim T.W., Jang S.J., Kim J. Clinical Utility of a Fully Automated Microsatellite Instability Test with Minimal Hands-on Time. J. Pathol. Transl. Med. 2019;53:386–392. doi: 10.4132/jptm.2019.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X., Xu J., Li L., Mu X., Wang Y., Li X. Evaluation of a Fully Automated Idylla Test System for Microsatellite Instability in Colorectal Cancer. Clin. Colorectal. Cancer. 2019;18:e316–e323. doi: 10.1016/j.clcc.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Samaison L., Grall M., Staroz F., Uguen A. Microsatellite instability diagnosis using the fully automated Idylla platform: Feasibility study of an in-house rapid molecular testing ancillary to immunohistochemistry in pathology laboratories. J. Clin. Pathol. 2019;72:830–835. doi: 10.1136/jclinpath-2019-205935. [DOI] [PubMed] [Google Scholar]
- 27.Pepe F., Smeraglio R., Vacirca D., Malapelle U., Barberis M., Troncone G. Microsatellite instability evaluation by automated microfluidic electrophoresis: An update. J. Clin. Pathol. 2017;70:90–91. doi: 10.1136/jclinpath-2016-204200. [DOI] [PubMed] [Google Scholar]
- 28.Odenthal M., Barta N., Lohfink D., Drebber U., Schulze F., Dienes H.P., Baldus S.E. Analysis of microsatellite instability in colorectal carcinoma by microfluidic-based chip electrophoresis. J. Clin. Pathol. 2009;62:850–852. doi: 10.1136/jcp.2008.056994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcus L., Lemery S.J., Keegan P., Pazdur R. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clin. Cancer Res. 2019;25:3753–3758. doi: 10.1158/1078-0432.CCR-18-4070. [DOI] [PubMed] [Google Scholar]
- 30.FDA Grants Accelerated Approval to Pembrolizumab for First Tissue/Site Agnostic Indication. [(accessed on 14 April 2021)]; Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-pembrolizumab-first-tissuesite-agnostic-indication.
- 31.Cancer Genome Atlas Research Network. Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R., et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green A.K., Feinberg J., Makker V. A Review of Immune Checkpoint Blockade Therapy in Endometrial Cancer. Am. Soc. Clin. Oncol. Educ. Book. 2020;40:1–7. doi: 10.1200/EDBK_280503. [DOI] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C., Zhang F., Zhou N., Gu Y.M., Zhang Y.T., He Y.D., Wang L., Yang L.X., Zhao Y., Li Y.M. Efficacy and safety of immune checkpoint inhibitors in advanced gastric or gastroesophageal junction cancer: A systematic review and meta-analysis. Oncoimmunology. 2019;8:e1581547. doi: 10.1080/2162402X.2019.1581547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal T., Permuth-Wey J., Kumar A., Sellers T.A. Systematic review and meta-analysis of ovarian cancers: Estimation of microsatellite-high frequency and characterization of mismatch repair deficient tumor histology. Clin. Cancer Res. 2008;14:6847–6854. doi: 10.1158/1078-0432.CCR-08-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreira L., Balaguer F., Lindor N., de la Chapelle A., Hampel H., Aaltonen L.A., Hopper J.L., Le Marchand L., Gallinger S., Newcomb P.A., et al. EPICOLON Consortium. Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308:1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deshpande M., Romanski P.A., Rosenwaks Z., Gerhardt J. Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability. Cancers. 2020;12:3319. doi: 10.3390/cancers12113319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita H., Nakayama K., Ishikawa M., Ishibashi T., Nakamura K., Sawada K., Yoshimura Y., Tatsumi N., Kurose S., Minamoto T., et al. Relationship between Microsatellite Instability, Immune Cells Infiltration, and Expression of Immune Checkpoint Molecules in Ovarian Carcinoma: Immunotherapeutic Strategies for the Future. Int. J. Mol. Sci. 2019;20:5129. doi: 10.3390/ijms20205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marabelle A., Le D.T., Ascierto P.A., Di Giacomo A.M., De Jesus-Acosta A., Delord J.P., Geva R., Gottfried M., Penel N., Hansen A.R., et al. Efficacy of Pembrolizumab in Patients with Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results from the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020;38:1–10. doi: 10.1200/JCO.19.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupinacci R.M., Goloudina A., Buhard O., Bachet J.B., Maréchal R., Demetter P., Cros J., Bardier-Dupas A., Collura A., Cervera P., et al. Prevalence of Microsatellite Instability in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology. 2018;154:1061–1065. doi: 10.1053/j.gastro.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Ghidini M., Lampis A., Mirchev M.B., Okuducu A.F., Ratti M., Valeri N., Hahne J.C. Immune-Based Therapies and the Role of Microsatellite Instability in Pancreatic Cancer. Genes. 2020;12:33. doi: 10.3390/genes12010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abida W., Cheng M.L., Armenia J., Middha S., Autio K.A., Vargas H.A., Rathkopf D., Morris M.J., Danila D.C., Slovin S.F., et al. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol. 2019;5:471–478. doi: 10.1001/jamaoncol.2018.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarode V.R., Robinson L. Screening for Lynch Syndrome by Immunohistochemistry of Mismatch Repair Proteins: Significance of Indeterminate Result and Correlation with Mutational Studies. Arch. Pathol. Lab. Med. 2019;143:1225–1233. doi: 10.5858/arpa.2018-0201-OA. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy A.J., Capo-Chichi J.M., Spence T., Grenier S., Stockley T., Kamel-Reid S., Serra S., Sabatini P., Chetty R. Heterogenous loss of mismatch repair (MMR) protein expression: A challenge for immunohistochemical interpretation and microsatellite instability (MSI) evaluation. J. Pathol. Clin. Res. 2019;5:115–129. doi: 10.1002/cjp2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shia J. The diversity of tumours with microsatellite instability: Molecular mechanisms and impact upon microsatellite instability testing and mismatch repair protein immunohistochemistry. Histopathology. 2021;78:485–497. doi: 10.1111/his.14271. [DOI] [PubMed] [Google Scholar]
- 46.Stelloo E., Jansen A.M.L., Osse E.M., Nout R.A., Creutzberg C.L., Ruano D., Church D.N., Morreau H., Smit V.T.H.B.M., van Wezel T., et al. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann. Oncol. 2017;28:96–102. doi: 10.1093/annonc/mdw542. [DOI] [PubMed] [Google Scholar]
- 47.Wu X., Snir O., Rottmann D., Wong S., Buza N., Hui P. Minimal microsatellite shift in microsatellite instability high endometrial cancer: A significant pitfall in diagnostic interpretation. Mod. Pathol. 2019;32:650–658. doi: 10.1038/s41379-018-0179-3. [DOI] [PubMed] [Google Scholar]
- 48.Singh N., Wong R., Tchrakian N., Allen S.G., Clarke B., Gilks C.B. Interpretation of mismatch repair protein expression using obsolete criteria results in discrepancies with microsatellite instability and mutational testing results. Mod. Pathol 2020, 33, 871–879. Mod. Pathol. 2021;34:1031–1032. doi: 10.1038/s41379-020-00680-y. [DOI] [PubMed] [Google Scholar]
- 49.Signorelli D., Giannatempo P., Grazia G., Aiello M.M., Bertolini F., Mirabile A., Buti S., Vasile E., Scotti V., Pisapia P., et al. Patients Selection for Immunotherapy in Solid Tumors: Overcome the Naïve Vision of a Single Biomarker. Biomed. Res. Int. 2019;2019:9056417. doi: 10.1155/2019/9056417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sivapiragasam A., Ashok Kumar P., Sokol E.S., Albacker L.A., Killian J.K., Ramkissoon S.H., Huang R.S.P., Severson E.A., Brown C.A., Danziger N., et al. Predictive Biomarkers for Immune Checkpoint Inhibitors in Metastatic Breast Cancer. Cancer Med. 2021;10:53–61. doi: 10.1002/cam4.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.