Abstract
The Saccharomyces cerevisiae MRE11 gene is required for the repair of ionizing radiation-induced DNA damage and for the initiation of meiotic recombination. Sequence analysis has revealed homology between Mre11 and SbcD, the catalytic subunit of an Escherichia coli enzyme with endo- and exonuclease activity, SbcCD. In this study, the purified Mre11 protein was found to have single-stranded endonuclease activity. This activity was absent from mutant proteins containing single amino acid substitutions in either one of two sequence motifs that are shared by Mre11 and SbcD. Mutants with allele mre11-D56N or mre11-H125N were partially sensitive to ionizing radiation but lacked the other mitotic phenotypes of poor vegetative growth, hyperrecombination, defective nonhomologous end joining, and shortened telomeres that are characteristic of the mre11 null mutant. Diploids homozygous for the mre11-H125N mutation failed to sporulate and accumulated unresected double-strand breaks (DSB) during meiosis. We propose that in mitotic cells DSBs can be processed by other nucleases that are partially redundant with Mre11, but these activities are unable to process Spo11-bound DSBs in meiotic cells.
DNA double-strand breaks (DSBs) are potentially lethal lesions that occur spontaneously during normal cellular processes, such as replication, or by treatment of cells with DNA-damaging agents. DSBs are potent stimulators of recombination and serve to initiate a variety of recombination events including meiotic recombination (9, 57), Saccharomyces cerevisiae mating type switching (55), and rearrangement of the T-cell receptor and immunoglobulin loci (48). Eukaryotic cells use two pathways for the repair of DSBs, homologous recombination and nonhomologous end joining (NHEJ). Repair of DSBs by homologous recombination requires the presence of homologous duplex DNA elsewhere in the genome and generally occurs with high fidelity. In contrast, the NHEJ pathway requires little, if any, sequence homology and is potentially mutagenic. Although yeast cells are capable of repairing breaks by either pathway, homologous recombination is favored. In birds and mammals, both pathways appear to contribute to ionizing radiation resistance of somatic cells (5, 12, 17).
Analysis of the severe combined immunodeficiency (scid) mouse has provided important clues about the mechanism of end joining. The scid mouse lacks mature T and B cells due to a defect in joining coding ends created by RAG1 and RAG2 during V(D)J recombination. The fact that cell lines derived from the scid mouse show sensitivity to ionizing radiation implies that the end-joining step of V(D)J recombination occurs by a general cellular pathway for DSB repair (6). Analysis of other X-ray-sensitive rodent cell lines identified three complementation groups that were defective in V(D)J recombination. XRCC5 mutant cells lack the 86-kDa subunit of the heterodimeric DNA-binding protein Ku, which is the regulatory subunit for the DNA-dependent protein kinase DNA-PK (46, 60). Subsequent biochemical studies revealed a defect in the DNA-PK catalytic subunit in scid and V3 cells (28, 30). The product of the XRCC4 gene, which is also required for V(D)J recombination, has recently been shown to interact with and stimulate the activity of DNA ligase IV, suggesting a direct role in end joining (20, 31). Studies of illegitimate recombination and NHEJ in yeast have shown the requirement for a large number of genes, including RAD50, XRS2, MRE11, HDF1 (which encodes a protein with homology to Ku70), HDF2, SIR2, SIR3, SIR4, and DNL4 (34, 36, 50, 64, 65, 68). Of these genes, only RAD50, MRE11, and XRS2 are required for radiation resistance in yeast (1, 18, 23). Mutation of the other genes involved in these pathways does not lead to radiation sensitivity except in a rad52 background (54, 65, 68). Homologs of MRE11 and RAD50 have been identified in mammals (15, 43), and protein localization studies suggest that they play a direct role in the repair of DSBs (32, 38). Furthermore, the human analog of XRS2, NBS1, is mutated in Nijmegen breakage syndrome, a disease associated with radiation sensitivity, immunodeficiency, and chromosomal instability (10, 66).
In yeast, the repair of DSBs by homologous recombination requires genes of the RAD52 epistasis group (RAD50 to 57, RAD59, MRE11, and XRS2), most of which were identified by their role in repair of ionizing radiation-induced DNA damage (1, 3, 23, 42). Initial steps in the repair process involve processing of DNA ends to produce 3′ single-stranded tails, the substrates for homologous pairing and strand invasion (9, 58, 67). RAD51, RAD52, RAD54, RAD55, and RAD57 all appear to act during the homologous pairing stage of the reaction after exonucleolytic processing (40, 53, 56). The identity of the nuclease (or nucleases) involved in this processing step has remained elusive, but recent studies suggest Mre11 as a candidate for this activity. The MRE11 gene was identified by its essential function in meiotic recombination and subsequently shown to be required for ionizing radiation resistance (1). mre11 null mutants have phenotypes identical to those of rad50 and xrs2 mutants, including poor mitotic growth, a delay in mating type switching, elevated rates of spontaneous mitotic recombination between heteroalleles in diploids, short telomeres, and a defect in the formation of meiosis-specific DSBs (2, 7, 23, 24, 26, 29, 63). Johzuka and Ogawa (26) have shown that Mre11 interacts with both Rad50 and Xrs2, consistent with the similarity of the mutant phenotypes. Although a role in the formation of meiosis-specific DSBs seems incongruent with a role in processing DSBs, nonnull alleles of both RAD50 and MRE11 have been identified which separate these two meiotic functions (2, 37, 63). In rad50S cells, meiosis-specific DSBs are formed, but Spo11 remains covalently attached to 5′ ends, preventing resection (9, 27). Further evidence in support of the model that the Mre11-Rad50-Xrs2 complex functions in DSB processing has come from physical analysis of mating type switching. The DSB produced by the HO endonuclease is processed to generate 3′ single-stranded tails. This processing reaction occurs more slowly in rad50, xrs2, and mre11 null mutants (24, 63).
Sequence alignments have revealed similarity between Mre11 and several prokaryotic nucleases, including E. coli SbcD, T4 gp47, and T5 gpD12 (51). The regions of highest homology include four sequence motifs, termed a phosphoesterase signature, that are shared by a variety of phosphoesterases, including serine/threonine protein phosphatases (51, 69). Although the SbcD protein alone exhibits single-stranded endonuclease activity, in association with SbcC, the complex shows ATP-dependent double-stranded exonuclease activity (13, 14). The SbcC protein is defined as a member of the structural-maintenance-of-chromosomes (SMC) family, which includes Rad50 (51). The human Mre11 (hMre11) protein alone and in association with hRad50 has 3′-to-5′ exonuclease and endonuclease activities, confirming the significance of the sequence homology with SbcD (41).
A point mutation within one of the Mre11 phosphoesterase motifs was reported to confer defective Mre11 nuclease activity. This mutation conferred similar phenotypes in mitotic cells to the mre11 null mutation, and in meiosis, DSBs were formed but were not processed (11, 63). In this study, we have generated mutations within two of the other phosphoesterase motifs and characterized the resulting mutants for nuclease activity by both in vivo and in vitro assays. Our results demonstrate that the conserved phosphoesterase motifs are essential for single-stranded DNA endonuclease activity. However, the mitotic phenotypes conferred by these mutations are strikingly different from those conferred by null mutations of MRE11. We conclude that the endonuclease activity is essential for DSB processing in meiosis, but not for end joining or telomere maintenance.
MATERIALS AND METHODS
Media, growth conditions, and genetic methods.
Rich medium, synthetic complete (SC) medium lacking the appropriate amino acid or nucleic acid base, and sporulation medium were prepared as described previously (52). Raffinose (2%) was substituted for glucose as a nonrepressing carbon source in SC medium that was used for induction of HO or MRE11. Transcription from the GAL1 promoter was induced by the addition of 1/10 volume of 20% galactose to the growth medium. Yeast cells were grown at 30°C unless otherwise stated. Transformations were performed by the lithium acetate method (22). Sporulation was carried out as described previously (52). The percent sporulation was determined by microscopic analysis of cells scraped from the sporulation plates and suspended in water. Percent sporulation values are the averages of at least two independent cultures of each strain, and no fewer than 200 cells were counted per sample. For SPO13 strains, cells with three or four visible spores were considered sporulated; for spo13 strains, cells with two visible spores were considered sporulated. Tetrad and dyad dissection was carried out as described previously (52).
Yeast strains.
The strains used for this study were derived from W303 or SK1 and are listed in Table 1. Strains LSY568, LSY569, LSY649, and LSY650 were constructed by one-step replacement (49) of W303-1A, W303-1B, AMP464, and AMP268, respectively, with a PCR fragment to generate a deletion-disruption allele of MRE11. PCR was performed on a LEU2-containing template (pRS405) with the following primers: 5′-TTCGTAGTACTGACCATAATTATGTCATTCTCATCAATAcgactacgtcgtaaggcccg and 5′-GACTATGGACTATCCTGATCCAGACACAATAAGGATTTTActcgaggagaacttctagta. Uppercase letters indicate the bases in MRE11 sequences, and lowercase letters indicate the bases in LEU2 sequences. Leu+ transformants that showed sensitivity to ionizing radiation were analyzed by PCR using primers complementary to sequences within the LEU2 gene (5′-GGTGATGCTGTCGCCGAAG) and the MRE11 3′ noncoding region (5′-TACTATGACGTTTATTCAGG) to verify disruption of MRE11. The chromosomal mre11-H125N allele was generated by a PCR-based allele replacement method (16). The adaptamer primers for MRE11 were obtained from Research Genetics and the plasmid pRS414:mre11-D56N,H125N used as a template; the primers for URA3 were as described previously (16). The only yeast transformant that was obtained by this method contained a nontandem duplication of mre11-H125N and mre11-D56N separated by the URA3 gene. The strain containing the duplication marked by URA3 was used for crosses. Subsequently, the URA3 gene and one copy of mre11 were removed by intrachromosomal recombination selected on medium containing 5-fluoroorotic acid. Eighty-eight percent of colonies that grew on medium containing 5-fluoroorotic acid had the weak ionizing radiation sensitivity characteristic of the nuclease defect. For two of these strains, LSY716A and LSY726A, the chromosomal mre11 gene was PCR amplified, sequenced, and shown to contain the mre11-H125N mutation. All other strains were generated by crossing the appropriate haploid strains and dissecting tetrads from the resulting diploids to obtain haploid segregants of the required genotype.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotypea or description | Source |
|---|---|---|
| SK-1 derivatives | ||
| AMP268 | MATα his4-G | A. Mitchell |
| AMP464 | MATa his4-N | A. Mitchell |
| LSY675 | Diploid of AMP268 × AMP464 | This study |
| LSY674 | MATa his4-N mre11::LEU2 | This study |
| LSY649 | MATα his4-G mre11::LEU2 | This study |
| LSY660 | Diploid of LSY674 × LSY649 | This study |
| W303 derivatives | ||
| W303-1A | MATa | R. Rothstein |
| W303-1B | MATα | R. Rothstein |
| W303 | Diploid of W303-1A × W303-1B | This study |
| YBL2 | MATα rad27::TRP1 | 59 |
| YAR70 | MATα spo13::hisG-URA3-hisG | A. Rattray |
| HKY677-11C | MATamei4::ADE2 | H. Klein |
| MAY1-21A | MATα leu2-K::ADE2-URA3::leu2-R rad50S::URA3 | H. Klein |
| MAY1-21D | MATa leu2-K rad50S::URA3 | H. Klein |
| R877 | MATα hdf1::LEU2 | R. Rothstein |
| LSY525-1C | MATa mei4::ADE2 spo13::hisG-URA3-hisG | This study |
| LSY525-3A | MATα mei4::ADE2 spo13::hisG-URA3-hisG | This study |
| LSY568 | MATamre11::LEU2 | This study |
| LSY569 | MATα mre11::LEU2 | This study |
| LSY730 | Diploid of LSY568 × LSY569 | This study |
| LSY716 | MATamre11-H125N::URA3::mre11-D56N | This study |
| LSY716A | MATa mre11-H125N | This study |
| LSY726 | MATα mre11-H125N::URA3::mre11-D56N | This study |
| LSY726A | MATα mre11-H125N | This study |
| LSY729 | Diploid of LSY716 × LSY726 | This study |
| LSY731 | MATamei4::ADE2 | This study |
| MATα mei4::ADE2 | ||
| LSY732 | MATaspo13::hisG-URA3-hisG | This study |
| MATα spo13::hisG-URA3-hisG | ||
| LSY733 | MATamre11::LEU2spo13::hisG-URA3-hisG | This study |
| MATα mre11::LEU2 spo13::hisG-URA3-hisG | ||
| LSY734 | MATamre11-H125Nspo13::hisG-URA3-hisG | This study |
| MATα mre11-H125N spo13::hisG-URA3-hisG | ||
| LSY735 | MATamre11-H125Nspo13::hisG-URA3-hisGmei4::ADE2 | This study |
| MATα mre11-H125N spo13::hisG-URA3-hisG mei4::ADE2 | ||
| LSY738 | MATaspo13::hisG-URA3-hisGmei4::ADE2 | This study |
| MATα spo13::hisG-URA3-hisG mei4::ADE2 | ||
| LSY741 | MATarad50S::URA3leu2-K | This study |
| MATα rad50S::URA3 leu2-K::ADE2-URA3::leu2-R | ||
| LSY736 | MATa hdf1::LEU2 mre11::LEU2 | This study |
| LSY737 | MATa hdf1::LEU2 mre11-H125N | This study |
| Other | ||
| YDS371 | MATa leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407 GAL+ | D. Shore |
SK-1 derivatives have the genotype ura3 leu2::hisG trp1::hisG lys2 ho::LYS2 and W303 derivatives have the genotype leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15; differences from these genotypes are listed in the table.
Plasmids.
pRS414:MRE11 was generated by cloning a 3.1-kb BamHI/NruI fragment from plasmid pKJ1101 into the polylinker of pRS414. This plasmid was used for the construction of MRE11 mutations with the Quick Change Site-Directed Mutagenesis kit from Stratagene. The oligonucleotides 5′-TTGTACAGTCCGGTAATCTTTTTCACGTGAA and 5′-TTCACGTGAAAAAGATTACCGGACTGTACAA were used to generate the D56N mutation, and 5′ GCATATCAGGTAATAATGATGATGCGTCGGG and 5′CCCGACGCATCATCATT ATTACCTGATATGC were used to generate the H125N mutation. Two rounds of site-directed mutagenesis were performed to create plasmid pRS414:mre11-D56N,H125N.
The MRE11 open reading frame (ORF) was amplified by PCR from pRS414:MRE11 using two primers, one containing a BamHI site, a FLAG peptide-encoding sequence, and the beginning of the MRE11 ORF (5′-AGCTAGGATCCGACTACAAGGATGACGATGACAAGCATGGACTATCCTGATCCAGACA) and the second containing the 3′ end of the MRE11 ORF and a HindIII site (5′-GTAGCTAAGCTTTCGCGAAGGCAAGCCCTTGG). The resulting PCR fragment was digested with BamHI and HindIII and cloned into the expression vector pEGKT, a yeast glutathione S-transferase (GST) fusion vector (35). GST-mre11-D56N was constructed in a similar fashion except that pRS414:mre11-D56N was used as a template for the PCR. GST-mre11H-125N was made by site-directed mutagenesis of GST-MRE11 using the primers described above for generation of the mre11-H125N allele.
pBM272-HO was constructed by insertion of a 2.5-kb HindIII fragment containing the HO gene into the HindIII site of pBM272 (25), adjacent to the GAL1 promoter.
γ-Irradiation survival assays.
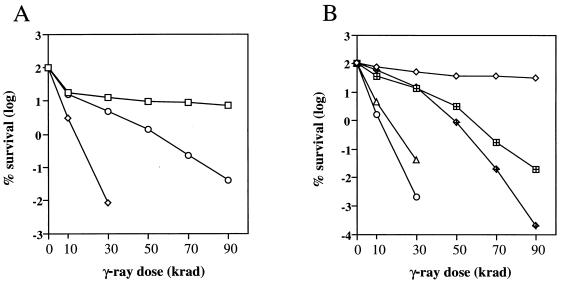
Cells were grown in liquid medium to mid-log phase. The cultures were serially diluted (five 10-fold serial dilutions), and aliquots of each dilution were plated on solid medium. The plates were irradiated in a Gammacell-220 containing 60Co (Atomic Energy of Canada) for the designated dose. The dose rate of the Gammacell-220 was 79 rads/s. The plates were incubated for 3 days before survivors were counted. Each strain was assayed three separate times, and the mean survival for each dose is presented in Fig. 3.
FIG. 3.
Radiation sensitivity of mre11 strains. (A) Complementation of ionizing radiation sensitivity of mre11 strains (LSY568) by plasmids expressing wild-type or mutant alleles of MRE11. Symbols: □, wild-type or mre11Δ strain with pRS414:MRE11; ◊, mre11Δ; ○, mre11Δ strain with pRS414:mre11-D56N or pRS414:mre11-H125N or pRS414:mre11-D56N,H125N. (B) Ionizing radiation sensitivity of haploid and diploid strains. Strains containing chromosomal mre11 alleles were used for this experiment. MRE11 (W303-1A) and MRE11/MRE11 (W303) (⋄), mre11Δ (LSY568) (○), mre11Δ/mre11Δ (LSY730) (▵), mre11-H125N (LSY716A) (⊞), and mre11-H125N/mre11-H125N (LSY729) ( ) strains were used.
Physical analysis of mating type switching.
Strains to be tested were transformed to Ura+ with plasmid pBM272-HO. Ura+ plasmid-containing transformants were grown in 5 ml of SC medium lacking uracil (SC-Ura) for 24 h. Cells were harvested, washed with water and used to inoculate 250 ml of SC medium with raffinose and without uracil [SC(raffinose)-Ura]. Cultures were grown to an optical density at 600 nm of 0.4 to 0.5 and 27.5 ml of 20% galactose were added. One hour after addition of galactose, the cultures were harvested and resuspended in 250 ml of SC(glucose)-Ura. Samples (50 ml) were removed prior to HO induction and at 1-h intervals after induction for DNA analysis. Cells were harvested by centrifugation and washed with water, and the cell pellets were frozen in liquid nitrogen. DNA was extracted, digested with BamHI and StyI, and DNA fragments were separated by electrophoresis through 1% alkaline agarose gels. DNA fragments were transferred to nylon membranes and hybridized with a 300-bp PCR fragment generated by amplification of MAT sequences distal to the HO-cut site. The cultures were maintained in SC medium even after HO induction, because the growth rate difference between MRE11 and mre11 strains was less than in rich medium.
Mitotic recombination assay.
Diploid LSY660 was transformed to Trp+ using plasmid pRS414, pRS414:MRE11, or pRS414:mre11-H125N. At least 13 independent Trp+ transformants were grown to mid-log phase in SC-Trp medium and then serially diluted with water. Aliquots from the appropriate dilutions were plated on SC-Trp medium to determine the number of cells and on SC-His medium to determine the number of His+ prototrophs. The recombination frequency was derived from the number of His+ prototrophs/total number of cells, and the median value is given.
Plasmid end joining.
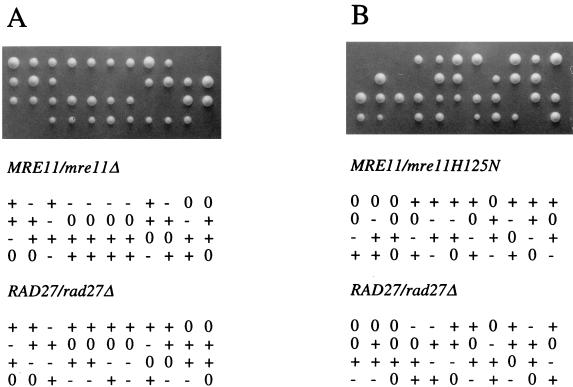
Precise end joining was assayed as described previously (8, 34). In brief, plasmid pRS413 was digested with EcoRI and 200 ng of linear or uncut DNA was used to transform each strain. The ratio of His+ transformants obtained with cut or uncut DNA was determined for each strain. The assay was repeated two more times, and the average value for each strain is presented in Fig. 5.
FIG. 5.
Synthetic lethality of rad27 mre11 double mutants. Viability and genotype of spores derived from a RAD27/rad27Δ mre11Δ/MRE11 diploid (A) or a RAD27/rad27Δ mre11-H125N/MRE11 diploid (B). In each case, + indicates the wild-type locus, − indicates a mutant locus, and 0 indicates a dead spore.
Analysis of telomere lengths.
Genomic DNA was isolated from 5-ml saturated cultures and digested with XhoI. The products were examined by Southern analysis with pYT14 as a hybridization probe (44). Wild-type strains yield a terminal restriction fragment of ∼1.3 kb which includes ∼400 bp of the G1–3T telomeric repeat (44).
DSB detection during meiosis.
Diploids were induced to undergo meiosis as previously described (9). Samples (20 ml) were removed at 2-h intervals from 0 to 8 h after induction of meiosis, mixed with 20 ml of 100% ethanol, and stored at −20°C. DNA was extracted as previously described (9) except that only one phenol-chloroform extraction was performed, followed by extraction with chloroform, and the Sephadex G-50 column purification step was omitted. Twenty-four percent of the total DNA was digested with EcoRI and run on a 0.7% agarose gel. DNA fragments were transferred to Hybond-N membranes for hybridization. The THR4 DNA fragment used for the hybridization probe was generated by PCR using primers 5′-CTAACGCTTCCCAAGTTTAC, corresponding to nucleotides 5 to 24 of the THR4 ORF, and 5′-TTGTCACCAGTGACGTTTTG, corresponding to nucleotides 320 to 301 of the THR4 ORF.
Purification of GST-Mre11.
The GST fusion plasmids were transformed into strain YDS371. One-liter cultures were grown in SC(raffinose)-Ura medium to an optical density at 600 nm of 0.4. Galactose was added to a final concentration of 2%, and growth continued for 12 h. Cells were harvested, washed with water, and resuspended in a volume equal to the wet weight of cells in lysis buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride). An equal volume of glass beads was added, and cells were lysed by vortexing (five 1-min pulses). The lysate was clarified by centrifugation for 1 h at 100,000 × g. The soluble fraction was mixed with 1 ml of glutathione Sepharose 4B (Pharmacia) that had been equilibrated with lysis buffer and gently mixed for 10 min. The beads were collected by centrifugation and washed three times with lysis buffer. The GST fusion protein was recovered from the beads by elution with 10 mM glutathione in 50 mM Tris-HCl, pH 8.0.
Nuclease assays.
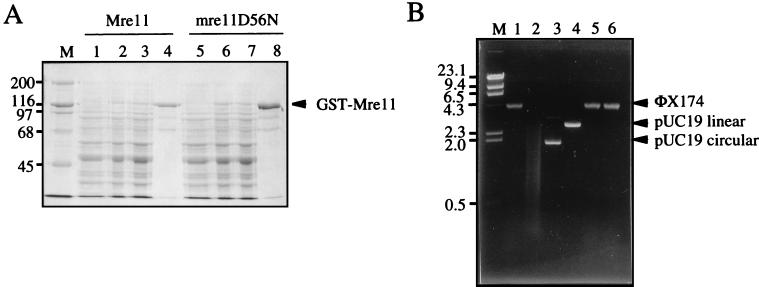
Three hundred nanograms of φX174 viral DNA or 200 ng of pUC19 DNA was incubated for 1 h at 30°C in a 30-μl reaction mixture volume containing 25 mM Tris-HCl [pH 8.0], 0.1 mM dithiothreitol, 2.5 mM MnCl2 or MgCl2, and 100 μg of bovine serum albumin per ml with 1 μg of GST-Mre11 or mutant protein GST-mre11-D56N or GST-mre11-H125N. The reactions were terminated by the addition of EDTA to 5 mM and proteinase K to a final concentration of 50 mg/ml and incubated for 10 min at 37°C. The reaction products were analyzed by agarose gel electrophoresis. Although 1 μg of GST-Mre11 (or mutant proteins) was used for the experiment shown in Fig. 2, nuclease activity was still detected using 100 ng of GST-Mre11. Excess protein was used to ensure that the mutants were devoid of a weak nuclease activity.
FIG. 2.
Mre11 has a single-stranded DNA endonuclease activity. (A) GST-Mre11 was purified from yeast by affinity chromatography of crude extracts on glutathione Sepharose. Lane M, molecular mass markers (in kilodaltons); lanes 1 and 5, uninduced soluble fraction; lanes 2 and 6, induced soluble fraction; lanes 3 and 7, unbound fraction from glutathione Sepharose; lanes 4 and 8, bound fraction from glutathione Sepharose eluted with 10 mM glutathione. (B) Endonuclease activity of Mre11. Lane M, λ DNA digested with HindIII, lane 1, φX174 viral DNA; lane 2, φX174 viral DNA with Mre11 and Mn2+; lane 3, pUC19 circular DNA with Mre11 and Mn2+; lane 4, pUC19 linear DNA with Mre11 and Mn2+; lane 5, φX174 viral DNA with Mre11 and Mg2+, φX174 viral DNA with mre11-D56N and Mn2+. The migration positions of molecular size markers (in kilobases) are given to the left.
RESULTS
The phosphoesterase motifs of Mre11 are required for nuclease activity.
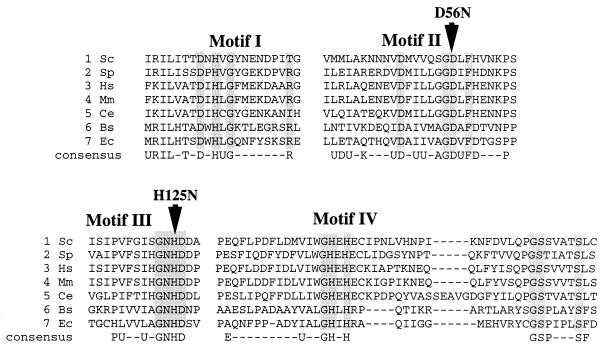
The sequence homology between Mre11 and SbcD suggests that Mre11 is a nuclease (Fig. 1). To test this hypothesis, the Mre11 protein was purified as a GST fusion and then tested for activity on a variety of DNA substrates. The fusion protein included a FLAG epitope tag inserted immediately before the first codon of Mre11 and was expressed from the galactose-inducible GAL1 promoter in yeast. The fusion protein was shown to complement the ionizing radiation sensitivity of an mre11 null mutant, even under noninducing conditions, indicating that the GST moiety does not interfere with Mre11 function. The fusion protein was purified from crude cell extracts by affinity chromatography using a glutathione Sepharose matrix (Fig. 2A). The glutathione eluate contained one major species of approximately 120 kDa that cross-reacted with anti-FLAG antibodies (data not shown). Most of the lower-molecular-mass species observed in the glutathione eluate also cross-reacted with the antibody, suggesting that these were likely to correspond to proteolytic breakdown products. The glutathione eluate was used directly for nuclease assays. Nuclease activity was observed only for φX174 single-stranded DNA (Fig. 2B). No activity was apparent with duplex linear or circular substrates in the presence or absence of ATP. However, using a 5′-end-labeled 39-bp linear duplex substrate, a weak 3′-to-5′ exonuclease activity was detected (data not shown). The exonuclease activity observed was comparable to the weak activity identified for hMre11 in the absence of hRad50 (41). The nuclease activity was optimal with Mn2+ as a cofactor, and no activity was observed with Mg2+. To ensure that the activity observed was due to Mre11 rather than a contaminating protein, two mutant proteins were also purified and shown to lack the single-stranded DNA endonuclease activity. The mutants used contained point mutations at D56 in phosphoesterase motif II and H125 in phosphoesterase motif III (Fig. 1) and were purified by the same procedure used for the wild-type protein (Fig. 2; only mre11-D56N is shown). The failure to detect nuclease activity by either of these proteins confirmed that the activity was due to Mre11 and that motifs II and III are important for activity.
FIG. 1.
Alignment of eukaryotic Mre11 sequences with prokaryotic SbcD sequences. The aligned sequences are divided into blocks corresponding to the mostly highly conserved motifs (motifs I to IV). Sequences of S. cerevisiae (Sc) Mre11, Schizosaccharomyces pombe (Sp) Rad32 (Mre11), Homo sapiens (Hs) Mre11, Mus musculus (Mm) Mre11, Caenorhabditis elegans (Ce) Mre11, Bacillus subtilis (Bs) SbcD, and E. coli (Ec) SbcD are shown. The arrows indicate the locations of the point mutations D56N and H125N. Gaps introduced to optimize alignment are indicated by dashes.
DSB processing is impaired in the mre11-D56N and mre11-H125N mutants.
To determine the role of the Mre11 nuclease activity in vivo, the two point mutations described above were generated within the MRE11 gene carried on a single-copy-number yeast plasmid. These plasmids were then used to transform an mre11 deletion strain to test for complementation of the mre11 phenotypes. Yeast transformants containing either mutant allele showed mitotic growth rates equivalent to those observed by transformants containing the wild-type MRE11 gene. In contrast, the doubling time of the mre11 null mutant was increased by 50%. Although the mre11 null mutant showed extreme sensitivity to ionizing radiation, mre11-D56N and mre11-H125N mutations conferred only partial sensitivity (Fig. 3A). At 50 kilorads, a dose sufficient for greater than 105-fold killing of the null mutant, the point mutant strains showed only a 10-fold reduction in survival compared to the wild type. If each mutation had resulted in partial retention of nuclease activity, which was not detected by the biochemical assay, then we predicted that overexpression of the mutant proteins might completely restore resistance to γ-rays. However, high-copy-number plasmids containing the mre11 point mutations complemented the null mutant to the same level as observed with the single-copy-number plasmids (data not shown). We also constructed a double mutant containing the D56N and H125N alleles, and this mutant showed the same survival as each of the single mutants (Fig. 3A). These results are consistent with the hypothesis that residues Asp 56 and His 125 form part of the catalytic site for nuclease activity and mutation of either residue destroys the same function of the protein. The mre11-H125N allele was substituted for the wild-type chromosomal allele of MRE11, and the sensitivity of the resulting strain was identical to that of the strains containing plasmid-borne alleles (Fig. 3B). Strains containing the chromosomal mre11-H125N allele were used for all of the experiments described below, except for determination of spontaneous mitotic recombination frequencies (see Materials and Methods).
Diploids homozygous for the mre11, rad50, or xrs2 mutation show increased radiation resistance compared with haploids, a phenomenon known as diploid-specific repair. This is thought to reflect a role for these genes in sister chromatid repair, whereas in diploid cells lesions can be repaired, albeit inefficiently, from a homolog. Diploids containing the mre11-H125N allele were found to be more γ-ray sensitive than haploids (Fig. 3B). One interpretation of this result is that the Mre11-Rad50-Xrs2 complex is still present and lesions are not diverted to the homolog for repair. If lesions are not diverted to the homolog for recombinational repair in the mre11-H125N strain, then we would expect to eliminate the hyperrecombination phenotype that is observed for mre11Δ diploids. The frequency of His+ prototroph formation was determined for diploid strains heteroallelic at the his4 locus. The mre11-H125N strain showed the same frequency of prototrophs as the wild-type strain did (5.0 × 10−6 and 4.5 × 10−6, respectively). Consistent with previous observations, the mre11Δ strain exhibited a hyperrecombination phenotype for heteroallelic recombination (1.0 × 10−4 His+ prototroph).
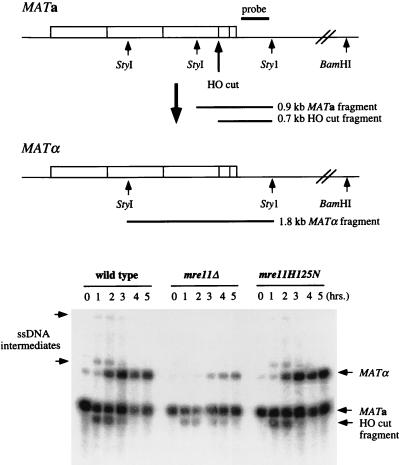
To determine whether the mutants were defective in the repair of a single well-defined DSB, a mating type switching assay was performed. The repair of an HO endonuclease-induced DSB was monitored at the DNA level after induction of HO endonuclease for 1 h. To measure resection of the 5′ end at the MAT locus, as well as the formation of switched products, the samples were digested with StyI and BamHI and separated on alkaline agarose gels. Because single-stranded DNA is resistant to digestion by restriction endonucleases and the 3′ end remains intact, the amount of DNA resected can be monitored by the appearance of high-molecular-weight StyI/BamHI fragments (67). The appearance of a 1.8-kb StyI fragment is indicative of repair of the DSB from the HMLα locus. The wild-type and mre11-H125N strains showed identical kinetics of repair and equivalent amounts of single-stranded DNA intermediates, indicating no obvious defect in resection of the 5′ end at the MAT locus (Fig. 4). In contrast, in the mre11 null mutant, the cut product was still visible 5 h after induction of HO. Repaired products were formed, but their appearance was delayed compared to that for the wild type. This result is similar to that observed previously for mre11 null mutants (63). Unlike the mre11-H125N strain, single-stranded DNA intermediates were barely detectable in the mre11Δ strain.
FIG. 4.
Kinetics of mating type switching. A schematic representation of the MATa and MATα loci indicating the locations of BamHI and StyI sites and the hybridization probe is shown at the top of the figure. HO endonuclease produces a 0.7-kb fragment from the 0.9-kb MATa StyI fragment. A 1.8-kb StyI fragment is produced when the mating type switches from MATa to MATα. Extensive 5′-to-3′ degradation from the HO-cut site through the distal BamHI and StyI sites leads to the appearance of high-molecular-weight BamHI/StyI fragments. Double digestions with StyI and BamHI were performed because the StyI site at position 5450 (67) is absent in W303 strains. DNA was isolated from cultures prior to galactose induction (0-h time point) and at 1-h intervals after HO induction. The arrows to the left of the gel indicate the high-molecular-weight fragments generated by nuclease digestion from the HO-cut site. ssDNA, single-stranded DNA.
Synthetic lethality of mre11 rad27 double mutants.
The RAD27 gene encodes a nuclease that functions to process Okazaki fragments during lagging strand DNA synthesis (61). Strains containing a deletion of RAD27 are viable, but viability is dependent on homologous recombination (59). To determine whether the Mre11 nuclease activity is essential for processing lesions generated in a rad27 mutant, diploids heterozygous for RAD27 and MRE11 were generated, and following sporulation, tetrads were dissected to obtain haploid progeny. The high spore inviability observed for crosses involving either mre11Δ or mre11-H125N suggested that the double mutant combination with rad27 was inviable (Fig. 5). This was confirmed by replica plating dissection plates to selective media to score for the presence of markers diagnostic for each mutation. None of the viable spores contained both the rad27 and mre11 mutations.
End joining and telomere maintenance are normal in the mre11-H125N strain.
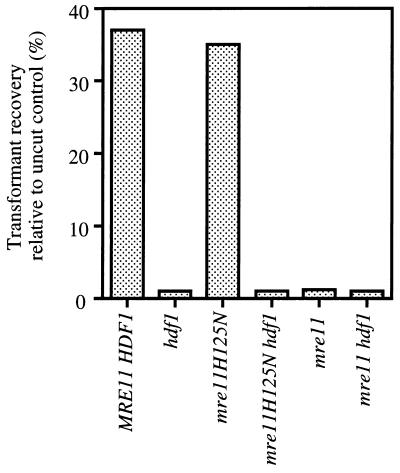
Since MRE11 is required for both homologous and NHEJ pathways, it seemed possible that some of the residual repair of ionizing radiation-induced DNA damage observed in the mre11-H125N strain was due to the NHEJ pathway. To determine the effect of loss of nuclease function on NHEJ, a plasmid rejoining assay was used. In this assay, the efficiency of NHEJ is determined by the percent transformants obtained with a cut plasmid relative to an uncut control plasmid. The plasmid utilized contained CEN and ARS elements for stable maintenance in yeast and the HIS3 selectable marker. The plasmid was digested with EcoRI, which cuts within a region of the plasmid with no homology to the yeast genome to avoid repair events by homologous recombination. The proportion of transformants recovered with cut plasmid relative to uncut plasmid in wild-type and mre11-H125N strains was equivalent, indicating proficient end joining (Fig. 6). In contrast, the hdf1 (Ku70-deficient) and mre11Δ mutants showed a 30-fold reduction in the recovery of transformants using the cut plasmid relative to the uncut control plasmid. The hdf1 mre11Δ and hdf1 mre11-H125N double mutants showed the same low level of end joining, indicating that these genes are in the same pathway for NHEJ.
FIG. 6.
The Mre11 nuclease activity is not required for end joining. End-joining assays were as described in Materials and Methods.
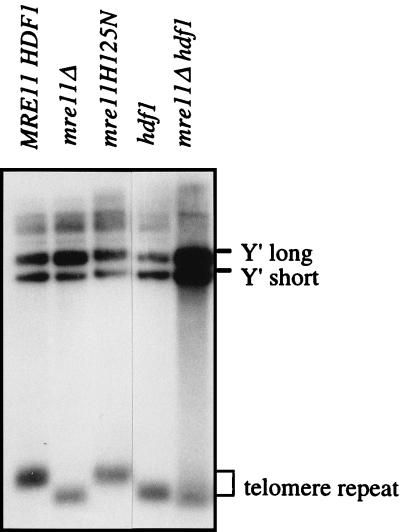
rad50, mre11, and xrs2 mutants have shortened telomeres and a senescence phenotype, indicating a defect in telomere maintenance (7, 29). The length of telomeric DNA fragments from mre11 mutants was examined to determine whether the nuclease activity of Mre11 is required for telomere maintenance. Wild-type and mre11-H125N strains had telomeres of similar length, whereas the telomeres of mre11Δ and hdf1 strains were shorter (Fig. 7). The length of the telomeres from a hdf1 mre11Δ double mutant was similar to those of the two single mutants, but the intensity of the telomere signal was less than the Y′ signal, suggesting a more severe telomere defect in the double mutant.
FIG. 7.
The Mre11 nuclease activity is not required for telomere maintenance. Genomic DNA isolated from each of the indicated strains was digested with XhoI to liberate a terminal fragment of ∼1.3 kb. The positions of the Y′ and telomere fragments are indicated to the right of the autoradiogram.
The Mre11 nuclease is required for meiosis.
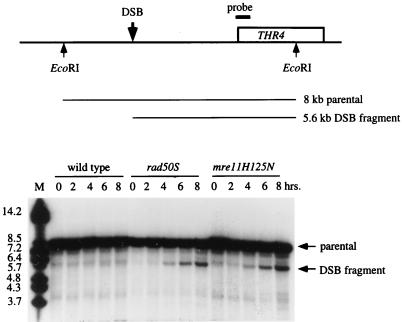
Diploids homozygous for the chromosomal mre11-H125N or mre11Δ alleles failed to sporulate in the W303 background (Table 2). To determine the step in meiosis at which the block occurred, an epistasis analysis was performed using spo13 and mei4 mutations in combination with the chromosomal mre11-H125N allele. The spo13 mutation allows cells to bypass meiosis I and rescues the spore inviability of mutants that are blocked prior to DSB formation. However, mutants that are defective in the repair of DSBs are not rescued by spo13. The sporulation defect of the mre11Δ diploid was rescued by spo13, but the spo13 mre11-H125N homozygous diploid failed to sporulate. The latter phenotype is similar to that observed for the mre11S and rad50S mutations, suggesting that meiosis-specific DSBs are formed but are not repaired in the mre11-H125N strain. Because a mei4 mutation prevents the formation of meiosis-specific DSBs, it was expected to rescue the sporulation defect of the spo13 mre11-H125N strain. The triple mutant showed normal levels of dyads, of which 98% were viable. This analysis suggested that the nuclease activity of Mre11 is essential for processing meiosis-specific DSBs. To test this, DSB formation was monitored at the THR4 locus in W303 derivatives (Fig. 8). In both rad50S and mre11-H125N strains, a discrete band of 5.6 kb, corresponding to the location of the THR4 hot spot, was initially detected at 4 h and persisted through 8 h. In wild-type cells, DSBs are processed to produce 3′ single-stranded tails that are characterized by a smear rather than a discrete band. DSB fragments were not detected in the wild type because the strain background used, W303, does not have the highly synchronous meiosis required to detect DSB intermediates during meiosis. The detection of DSB fragments in the mre11-H125N strain is further evidence supporting the hypothesis that DSBs are not processed in the absence of the Mre11 nuclease.
TABLE 2.
The nuclease activity of Mre11 is required for meiosis
| Strain | % Sporulation | % Spore viability |
|---|---|---|
| MRE11 | 50 | 99 |
| mre11Δ | <1 | |
| mre11-H125N | <1 | |
| spo13 | 45 | 75 |
| mei4 | <1 | |
| spo13 mei4 | 30 | 81 |
| mre11Δ spo13 | 43 | 80 |
| mre11-H125N spo13 | <1 | |
| mre11-H125N spo13 mei4 | 46 | 98 |
FIG. 8.
Meiosis-specific DSBs at the THR4 locus. The physical map of the THR4 region of chromosome III indicating the EcoRI sites, location of the probe, and approximate site of the DSB is shown at the top of the figure. A time course (in hours) of meiosis of wild-type, rad50S, and mre11-H125N strains is shown at the bottom. DNA samples from each time point were digested with EcoRI. The positions of the parental and DSB fragments are indicated to the right of the autoradiogram, and the migration positions of molecular size markers (in kilobases) are given to the left.
DISCUSSION
The phosphoesterase motifs present in Mre11 are important for nuclease activity.
Mre11 contains sequence motifs that are shared by a variety of phosphoesterases, including serine/threonine protein phosphatases and nucleases (51, 69). The importance of these sequence motifs in protein phosphatases has been established by mutational and structural studies (19, 21, 69). To evaluate the role of these motifs in Mre11 function, conservative amino acid substitutions were generated within motifs II and III. The sites mutated, D56 and H125, correspond to residues of lambda phosphatase that are essential for catalysis (69). Furthermore, the crystal structure of mammalian protein phosphatase 1 identified the equivalent residues as part of the catalytic site (19). The aspartic acid residue of protein phosphatase 1 equivalent to D56 is directly involved in coordination of the two manganese ions at the catalytic site and is therefore predicted to play an essential role in catalysis. The mre11-D56N and mre11-H125N mutant proteins lack the single-stranded endonuclease associated with the wild-type Mre11 protein, consistent with the predicted role of these residues in catalysis (Fig. 2).
Role of the Mre11 nuclease in processing DSBs.
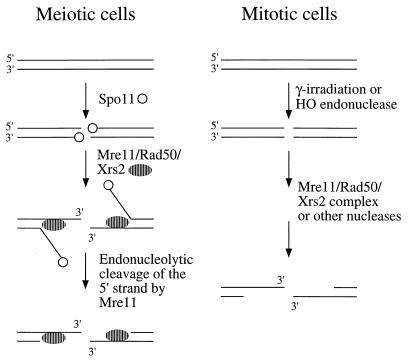
The Mre11-Rad50-Xrs2 complex has been suggested to function in processing DSBs based on two lines of evidence. First, yeast strains containing a null allele of rad50 or xrs2 show a delay in mating type switching and the extent of exonucleolytic processing of 5′ ends is reduced. Second, diploid strains containing rad50S, mre11S, or mre11-58 alleles accumulate unprocessed DSBs during meiosis. In this study, we show that nuclease-defective alleles of MRE11 confer weak sensitivity to ionizing radiation and no delay in mating type switching. Therefore, the delay in mating type switching observed in mre11Δ mutants does not appear to be due to loss of the Mre11 endonuclease activity. These data suggest that either the Mre11 nuclease does not process DSBs in mitotic cells or that other nucleases can efficiently process breaks in the absence of the Mre11 nuclease. The mre11-58 mutation is a His-to-Tyr substitution of a conserved histidine residue (H213) within one of the phosphodiesterase motifs of MRE11 (63). The mre11-58 protein has been shown to lack the nuclease activities associated with the wild-type Mre11 protein and also fails to interact with Rad50 (40a). Thus, the more severe mitotic phenotype of the mre11-58 strain compared with that of the mre11-H125N strain may be due to disruption of the Mre11-Rad50-Xrs2 complex in addition to the defect in the nuclease function of Mre11. The only mitotic phenotype shared by mre11Δ and mre11-H125N strains is lethality in combination with rad27. This result suggests that the Mre11 nuclease is required to process lesions generated in a rad27 strain. The failure of redundant nucleases to efficiently process these lesions may be because the level of damage is too high or because the nature of the lesions prevents the activity of other nucleases. For example, some of the lesions may be flap structures that can be processed only by an endonuclease. During meiosis, unprocessed DSBs accumulated in both the mre11-H125N and rad50S diploid strains. This result, in combination with the spo13 epistasis analysis, indicates that the Mre11 nuclease activity is required for DSB processing in meiosis. Meiosis-specific DSBs differ from breaks produced by HO endonuclease by the covalent attachment of a protein, Spo11, to the 5′ ends. Our results suggest that the Mre11 nuclease is essential when 5′ ends are blocked, either by the attachment of a protein, an unusual structure, or possibly base damage induced by ionizing radiation (Fig. 9).
FIG. 9.
Models for Mre11 function. In meiotic cells, the 5′ ends at DSB sites remain bound by the Spo11 protein, preventing access of exonucleases. The Mre11-Rad50-Xrs2 complex binds to break sites and unwinds the ends to reveal single-stranded DNA for cleavage by Mre11. This cleavage removes Spo11 and the 5′ strand, resulting in the formation of a 3′ single-stranded tail. When DSBs are produced by ionizing radiation or by the HO endonuclease, the 5′ ends are generally free and accessible to other nucleases in addition to the Mre11-Rad50-Xrs2 complex.
The E. coli SbcD protein has a single-stranded endonuclease activity but in the presence of SbcC functions as an ATP-dependent exonuclease (13, 14). The SbcC protein contains conserved nucleotide binding motifs and two coiled-coil domains that are characteristic of the SMC family of proteins. Rad50, which is known to associate with Mre11, is also a member of the SMC family. The Walker A nucleotide binding site is essential for Rad50 function, and the purified protein shows ATP-dependent DNA binding (2, 47). These findings suggest that the Rad50-Mre11 complex may also be an ATP-dependent exonuclease. Alternatively, Rad50 or some other factor might unwind duplex ends to provide a single-stranded region for the Mre11 endonucleolytic activity (Fig. 9).
Biochemical analysis of the SbcCD complex has shown that the polarity of exonuclease degradation is 3′ to 5′ (29a). Similarly, the hMre11 protein alone, or in association with hRad50 and p95, has 3′-to-5′ exonuclease and single-stranded endonuclease activities (41, 62). However, in yeast, DSBs are processed to generate 3′ single-stranded tails, a reaction thought to require the activity of a 5′-to-3′ nuclease. The function of the 3′-to-5′ activity of Mre11 is currently unclear. It has been suggested that the hMre11 nuclease functions with DNA ligase I in the imprecise end-joining pathway (41). This pathway for the repair of extrachromosomal DSBs has been well characterized in mammalian cells, and the types of junctions generated are similar to those produced by hMre11 and ligase I in vitro.
Role of Mre11 in vegetative cells.
Strains containing the mre11-D56N or mre11-H125N mutation have quite different phenotypes in vegetative cells compared to strains containing an mre11 null mutation. The mre11 null mutation confers poor vegetative growth, hypersensitivity to ionizing radiation, increased rates of heteroallelic recombination, a defect in illegitimate recombination, and short telomeres. In contrast, the mre11-H125N strain has only weak sensitivity to ionizing radiation and is not defective in the other processes listed above. Based on the phenotypic differences between mre11 null and mre11-H125N strains, we suggest that the Mre11-Rad50-Xrs2 complex has important functions in mitotic cells other than nucleolytic processing of DSBs. The requirement for this complex in maintenance of telomeres and NHEJ suggests that it interacts with DNA ends directly or in association with the Ku heterodimer. Recent studies have shown that although the efficiency of precise end joining is decreased to a similar level by mutation of HDF1, HDF2, RAD50, MRE11, or XRS2, there are qualitative differences (7). Repaired products recovered from hdf1 or hdf2 strains contain large deletions, whereas products obtained from rad50, mre11, and xrs2 strains are usually precise. These observations imply that Ku protects ends from nuclease degradation while end joining takes place. Since end-joined products obtained from the mre11 hdf1 double mutant are indistinguishable from those obtained from hdf1 strains, it seems unlikely that the Mre11 nuclease is responsible for the degradation that occurs in hdf1 strains. Moore and Haber (36) examined the repair of HO-induced DSBs generated at the MATα locus in the absence of homologous recombination. In this system, precise end-joining events create substrates for further cleavage by HO and aberrant end-joining events are selected as survivors. Most of the survivors from wild-type strains expressing HO through the cell cycle had small (2-bp) insertions at the HO-cut site. In rad50, xrs2, and mre11 strains, the efficiency of repair was reduced almost 100-fold and most of the events were deletions of 3 bp, but some larger deletions (>700 bp) were also detected. These results are consistent with the view that the nuclease activity of Mre11 does not play a significant role in end joining. Instead, the function of the Mre11-Rad50-Xrs2 complex may be to bring together broken ends and/or to recruit other factors involved in joining.
The Mre11-Rad50-Xrs2 complex is proposed to function in telomere maintenance by facilitating telomere replication. It has been suggested that the nuclease activity of the Mre11 complex may be important to generate a single-stranded DNA substrate for telomerase (39). However, our demonstration of normal length telomeres in the mre11-H125N strain suggests additional functions for the complex at telomeres.
Meiotic functions of the Mre11-Rad50-Xrs2 complex.
The Mre11-Rad50-Xrs2 complex has at least two functions in meiotic cells. First, it is required for the formation of meiosis-specific DSBs. There are at least seven other genes required for DSB formation in meiotic cells, but of these genes, only SPO11 has been shown to be directly involved (27). Spo11 catalyzes DSB formation by a transesterification mechanism that results in covalent attachment of the protein to 5′ ends through a tyrosine residue (4, 27). How Spo11 is targeted to sites at which DSBs occur is currently unknown. In rad50S mutants, Spo11 remains covalently bound to the 5′ ends at DSB sites, preventing resection (2, 27). Because unprocessed DSBs, indistinguishable from those generated in rad50S strains, accumulate in mre11S, mre11-58, mre11-H125N, and com1/sae2Δ mutants, it is assumed that Spo11 remains bound to break sites in these mutants as well (33, 37, 45, 63). This suggests that the second function of Mre11 and Rad50 is in a step subsequent to DSB formation. The removal of Spo11 from 5′ ends could occur by a direct reversal of the esterification reaction or by endonucleolytic removal of Spo11 attached to single-stranded DNA (27). The observations that Mre11 has a single-stranded DNA endonuclease activity and that nuclease-defective alleles of mre11 result in retention of Spo11 on 5′ ends are consistent with the hypothesis that Mre11 is directly involved in removal of Spo11 from DSB sites. By this endonucleolytic mode, the 5′ ends could be resected a variable distance from the DSB site. Continued resection of 5′ ends could occur by the action of the Mre11-Rad50-Xrs2 complex or by a different nuclease (Fig. 9).
In summary, Mre11 is an endonuclease and the sequence motifs characteristic of this class of proteins are important for nuclease activity. The nuclease activity of Mre11 plays an essential role in meiosis, probably in the removal of Spo11 protein from 5′ ends of DSB sites. However, in mitotic cells, the nuclease activity is dispensable for normal vegetative growth, mating type switching, NHEJ, and telomere maintenance. We propose that in mitotic cells, DSBs can be processed by other nucleases that are partially redundant with Mre11, but these activities are unable to process Spo11-bound DSBs in meiotic cells.
ACKNOWLEDGMENTS
We thank members of the Symington laboratory and W. Holloman for critical reading of the manuscript. We thank H. Tsubouchi and H. Ogawa for providing plasmid pKJ1101, H. Smith for construction of pBM272-HO, and H. Klein, A. Mitchell, A. Rattray, R. Rothstein, and D. Shore for providing yeast strains.
This work was supported by Public Health Service grants GM41784 and 2 T32 CA09503 from the National Institutes for Health and the National Cancer Institute, respectively.
REFERENCES
- 1.Ajimura M, Leem S-H, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 3.Bai Y, Symington L S. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 4.Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. An atypical topoisomerase II from archae with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 5.Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J-M. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/−mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 6.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 7.Boulton S J, Jackson S P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulton S J, Jackson S P. Saccharomyces cerevisiaeKu70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 9.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 10.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M, Yates J R, Hays L, Morgan W F, Petrini J H J. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 11.Chepurnaya O V, Kozhin S A, Peshekhonov V T, Korolev V G. RAD58 (XRS4)—a new gene in the RAD52epistasis group. Curr Genet. 1995;28:274–279. doi: 10.1007/BF00309787. [DOI] [PubMed] [Google Scholar]
- 12.Chu G. Double strand break repair. J Biol Chem. 1997;272:24097–24100. doi: 10.1074/jbc.272.39.24097. [DOI] [PubMed] [Google Scholar]
- 13.Connelly J C, de Leau E S, Okely E A, Leach D R F. Overexpression, purification and characterization of the SbcCD protein from Escherichia coli. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- 14.Connelly J C, Leach D R F. The sbcC and sbcD genes of Escherichia coliencode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 15.Dolganov G M, Maser R S, Novikov A, Tosto L, Chong S, Bressan D A, Petrini J H J. Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdeniz N, Mortensen U H, Rothstein R. Cloning-free PCR-based allele replacement methods. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essers J, Hendriks R W, Swagemakers S M A, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H J, Kanaar R. Disruption of mouse RAD54reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 18.Game J, Mortimer R K. A genetic study of X-ray sensitive mutants in yeast. Mutat Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg J, Huang H, Kwon Y, Greengard P, Nairn A, Kuriyan J. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature. 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 20.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson T E, Mann M, Lieber M R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 21.Huang H-B, Horiuchi A, Goldberg J, Greengard P, Nairn A C. Site-directed mutagenesis of amino acid residues of protein phosphatase 1 involved in catalysis and inhibitor binding. Proc Natl Acad Sci USA. 1997;94:3530–3535. doi: 10.1073/pnas.94.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov E, Korolev V, Fabre F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics. 1992;132:651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov E L, Sugawara N, White C I, Fabre F, Haber J. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeney S, Giroux C N, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 28.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 29.Kironmai K M, Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 29a.Leach, D. Personal communication.
- 30.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day R S, Barron G M, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Otevrel T, Gao Y, Cheng H-L, Seed B, Stamato T D, Taccioli G E, Alt F W. The XRCC4gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 32.Maser R S, Monsen K J, Nelms B E, Petrini J H J. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKee A H Z, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–815. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne G T, Jin S, Shannon K, Weaver D T. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 36.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nairz K, Klein F. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelms B E, Maser R S, MacKay J F, Lagally M G, Petrini J H J. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 39.Nugent C I, Bosco G, Ross L O, Evans S K, Salinger A P, Moore J K, Haber J E, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 40.Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman E H, Ogawa H. RecA-like recombination proteins in eukaryotes: functions and structures of RAD51genes. Cold Spring Harbor Symp Quant Biol. 1993;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- 40a.Ogawa, T., and H. Ogawa. Personal communication.
- 41.Paull T T, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 42.Petes T D, Malone R E, Symington L S. Recombination in yeast. I. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. [Google Scholar]
- 43.Petrini J H J, Walsh M E, DiMare C, Chen X-N, Korenberg J R, Weaver D T. Isolation and characterization of the human MRE11homologue. Genomics. 1995;29:80–86. doi: 10.1006/geno.1995.1217. [DOI] [PubMed] [Google Scholar]
- 44.Porter S E, Greenwell P W, Ritchie K B, Petes T D. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rathmell W K, Chu G. A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol Cell Biol. 1994;14:4741–4748. doi: 10.1128/mcb.14.7.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raymond W E, Kleckner N. RAD50 protein of S. cerevisiaeexhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roth D B, Nakajima P B, Menetski J P, Bosma M J, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 49.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 50.Schiestl R H, Zhu J, Petes T D. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharples G J, Leach D R F. Structural and functional similarities between the SbcCD proteins of Escherichia coliand the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1220. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 52.Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 53.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiaeis a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 54.Siede W, Friedl A A, Dianova I, Eckardt-Schupp F, Friedberg E C. The Saccharomyces cerevisiaeKu autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics. 1996;142:91–102. doi: 10.1093/genetics/142.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strathern J N, Klar A J S, Hicks J B, Abraham J A, Ivy J M, Nasmyth K A, McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MATlocus. Cell. 1982;31:183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 56.Sugarawa N, Ivanov E L, Fishman-Lobell J, Ray B L, Haber J E. DNA structure-dependent requirements for yeast RADgenes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 57.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 58.Sun H, Treco D, Szostak J. Extensive 3′-overhang, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 59.Symington, L. S. Homologous recombination is essential for the viability of rad27 mutants. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 60.Taccioli G, Gottlieb T M, Blunt T, Priestly A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Ku80: product of the XRCC5gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 61.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27is distinct from mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 62.Trujillo K M, Yuan S-S F, Lee E Y-H, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 63.Tsubouchi H, Ogawa H. A novel mre11mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsukamoto Y, Kato J, Ikeda H. Budding yeast Rad50, Mre11, Xrs2, and Hdf1, but not Rad52, are involved in the formation of deletions on a dicentric plasmid. Mol Gen Genet. 1997;255:543–547. doi: 10.1007/s004380050527. [DOI] [PubMed] [Google Scholar]
- 65.Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 66.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, Nowak N J, Stumm M, Weemaes C M R, Gatti R A, Wilson R K, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 67.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson T E, Grawunder U, Lieber M R. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 69.Zhuo S, Clemens J C, Stone R L, Dixon J E. Mutational analysis of a Ser/Thr phosphatase. J Biol Chem. 1994;269:26234–26238. [PubMed] [Google Scholar]