Abstract
Simple sequence repeat telomeric DNA is maintained by a specialized reverse transcriptase, telomerase. The integral RNA subunit of telomerase contains a template region that determines the sequence added to chromosome ends. Aside from providing the template, little is known about the role of the telomerase RNA. In addition, no hypotheses have been suggested to account for the striking evolutionary divergence in size and sequence between telomerase RNAs of ciliates, yeasts, and mammals. We show that the two- to threefold increase in size of the mammalian telomerase RNAs relative to ciliate telomerase RNAs is due to the presence of an extra domain resembling a box H/ACA small nucleolar RNA (snoRNA). The human telomerase RNA (hTR) H/ACA domain is essential in vivo for hTR accumulation, hTR 3′ end processing, and telomerase activity. By substituting the U64 box H/ACA snoRNA for the hTR H/ACA domain, we demonstrate that a heterologous snoRNA can function to promote chimeric RNA accumulation and 3′ end processing but not telomerase activity. In addition, we show that maturation of full-length hTR and its assembly into active telomerase occur from an mRNA promoter-driven RNA polymerase II transcript but not from a U6 snRNA promoter-driven RNA polymerase III transcript. Finally, we show that a small percentage of hTR is associated with nucleoli. These results have implications for the biogenesis and structure of hTR and the human telomerase ribonucleoprotein complex. They also expand the structural and functional diversity of the box H/ACA snoRNA motif.
Telomerase is a ribonucleoprotein (RNP) reverse transcriptase responsible for adding one strand of simple sequence DNA repeats to the ends of linear chromosomes. De novo repeat addition by telomerase is required to balance the loss of telomeric repeats that results from incomplete replication of the lagging strand and possibly nucleolytic cleavage (19). In most human somatic cells, a lack of telomerase activity is correlated with proliferation-dependent telomere shortening (21). The attrition of telomeric DNA to an as yet undefined “critical state” has been proposed to trigger cellular senescence and thus limit proliferative lifespan (20). As predicted by this model, the activation of human telomerase and the coincident reinstatement of telomere maintenance can forestall cellular senescence (for an example, see reference 8). Extracts of cancer cells, germline cells, and immortalized cultured cells are predominantly telomerase positive, and telomeres in these cells are stably maintained (21). However, the presence of telomerase activity in cell extracts does not absolutely predict the maintenance of telomere length in the corresponding cells (2), and there are telomerase-negative immortalized cell lines with stable telomeres (10, 33).
The RNA component of telomerase has been characterized in a variety of species, including ciliates, yeasts, and mammals (18). Ciliate telomerase RNAs are RNA polymerase III (Pol III) transcripts of ∼160 to ∼190 nucleotides (nt). Although these RNAs have little primary sequence identity, they possess a conserved secondary structure as initially predicted by phylogenetic analysis (38). Telomerase RNAs from the budding yeasts Kluyveromyces lactis and Saccharomyces cerevisiae are much larger than their ciliate counterparts (∼1,300 nt [31, 40]). They are transcribed by Pol II and processed at their 3′ ends from polyadenylated precursor forms (11). The mature telomerase RNAs of human and mouse cells are transcripts of 451 and 397 nt, respectively, and have been predicted to be products of Pol II mRNA-type promoters (4, 7, 13, 25, 45). The lack of structural and phylogenetic information for nonciliate telomerase RNAs, combined with their low sequence identities and disparities in length, has hindered progress in understanding the structure and function of these RNAs outside the template region.
The telomerase RNAs are intimately associated with an incompletely defined collection of protein components. Telomerase proteins have been characterized predominantly in the ciliates, by biochemical methods, and in yeasts, by genetic methods (37). Mammalian homologs of some telomerase components have also been identified: TEP1, a homolog of the Tetrahymena thermophila p80 protein (22, 35), and hTERT, the reverse transcriptase-like catalytic subunit (34). Reciprocal coimmunoprecipitation of TEP1 and hTERT indicates that both are associated with active telomerase (23). Although telomerase activity produced by coupled in vitro transcription-translation in rabbit reticulocyte lysate requires the addition of only human telomerase RNA (hTR) and hTERT (6, 44), the number of proteins in the endogenous mammalian telomerase RNP has not been determined. The mass of the active telomerase RNP from HeLa cell nuclear extracts (∼1,000 kDa [39]) or partially purified from rat S100 extracts (>1,000 kDa [35]) suggests that additional telomerase and telomerase-associated proteins remain to be identified.
Numerous families of small RNAs have been discovered, including small nucleoplasmic RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). Functions of these classes of RNAs include mRNA splicing (U1, U2, and U4 to U6 snRNAs), mRNA and rRNA processing (U7 snRNA; U3 and U8 snoRNAs), and site selection for RNA modification by methylation of the 2′ hydroxyl group (box C/D snoRNAs) or by pseudouridine formation (box H/ACA snoRNAs). The box H/ACA snoRNAs were most recently recognized as a small RNA family by virtue of an ACA trinucleotide located 3 nt upstream of the mature snoRNA 3′ end (41). In addition to this ACA box, they have the consensus H box sequence (5′-ANANNA-3′) but have no other primary sequence identity. Despite this lack of primary sequence conservation, the H and ACA boxes are embedded in an evolutionarily conserved hairpin-hinge-hairpin-tail core secondary structure with the H box in the single-stranded hinge region and the ACA box in the single-stranded tail (5, 16). Most box H/ACA snoRNAs specify sites of pseudouridine formation in rRNA (15, 36). Although box H/ACA snoRNAs are associated with higher-order nucleolar structures (16), little is known about the composition of the presumed box H/ACA snoRNPs. Two S. cerevisiae proteins, Gar1p and Cbf5p, have been shown to associate specifically with box H/ACA snoRNAs (9, 16, 17). Of these two, only Cbf5p, a putative pseudouridine synthase, is required for H/ACA snoRNA stability (29). Mammalian homologs of the putative pseudouridine synthase (dyskerin or NAP57 [24, 32]) but not Gar1p have been identified.
Because the mechanisms governing hTR expression, processing, and assembly into an active telomerase RNP were unknown, we began our investigation of telomerase RNP biogenesis by studying recombinant hTR transcripts in transiently transfected human 293 cells, an adenovirus-transformed embryonic kidney cell line with high levels of endogenous hTR and telomerase activity. Our results reveal novel requirements for the production of hTR and active telomerase RNP including an RNA polymerase selectivity for precursor RNA synthesis and an integral hTR domain resembling a box H/ACA snoRNA. Our findings also expand the known structural and functional diversity of the box H/ACA motif-containing RNA family.
MATERIALS AND METHODS
Constructs.
Throughout this work the following restriction enzymes are abbreviated as indicated: ApaLI, A; EcoRI, E; NlaIII, N; SacI, S; SmaI, Sm; and XbaI, X. The hTR SE restriction fragment was generated by PCR amplification of human genomic DNA with 5′-CCGGGAGCTCAGCGCACCGGGTTGCGGAGGG-3′ and 5′-GAATTCAGCACACTGGC-3′ and cloned into pBS/KS+ after digestion with E and S (phTR4). The sequence tag was generated by blunt-end ligation of a double-stranded linker (coding strand, 5′-GCTGATATAACCTTCAGGGG-3′) into the BalI or StuI restriction site of phTR4. Restriction fragments from phTR4 for Pol II expression constructs were cloned into the pRc/CMV vector (Invitrogen) at HindIII or NotI following filling in of recessed ends with the Klenow fragment of Escherichia coli DNA polymerase I (Klenow). Mutagenesis was performed by using the DpnI protocol as previously described (14). The ACA box restoration of the SN construct was performed by subcloning the StuI-tagged SN restriction fragment from phTR4 into the filled KpnI site of the pRSETA vector (Invitrogen), followed by subcloning of the E-PstI restriction fragment into the pRc/CMV vector at the HindIII site as described above. The U64 coding region, engineered by annealing complementary primers and extending with Klenow, was cloned into pBS/KS+ at the Sm restriction site. Chimeric hTR-U64 constructs were generated by replacing the SmE restriction fragment of wild-type-template (WT) and altered-template (AT) versions of the Pol II SE construct with a U64-containing restriction fragment. For hTR Pol III expression constructs, the SE restriction fragment (sequence tagged at BalI) or XE restriction fragment (sequence tagged at StuI) was cloned into a filled BglII site at Tetrahymena telomerase RNA (tTR) nt 20 in pU6tTR, a construct containing the 159-nt tTR sequence (including 16 nt of the Tag sequence) with its intrinsic Pol III transcriptional termination signal behind the human U6 snRNA promoter (28) in the pUC119 vector. Untagged SE and SSm restriction fragments (T41A, T101A) were engineered in phTR4 downstream of the U6 promoter (from pU6tTR including tTR nt 1 to 24) and with a downstream (dT)8 transcriptional termination signal at either the E or Sm site.
Transient transfection, total RNA preparation, and Northern blot analyses.
Calcium phosphate-mediated transfections and total RNA preparation by acid guanidine thiocyanate-phenol chloroform extraction were performed as described previously (1). Each transfection was performed a minimum of two times. Where indicated, an in vitro-transcribed recovery control RNA (RC) was added after the initial guanidine solublization (tTR nt 1 to 110 for transient transfections; hTR nt 1 to 80 plus 330 nt of pBS/KS+ polylinker sequence for subcellular fractionation). RNA was separated by electrophoresis on denaturing polyacrylamide gels (7 M urea, 0.6× Tris-borate EDTA [TBE], 4% acrylamide:bisacrylamide [19:1]). αhTR hybridization probes used in this study included 5′-end-radiolabeled h4 (5′-ACCACCCCUCCCAGG-3′) and h5 (5′-GCCUACGCCCUUCUCAGUUAGG-3′) 2′-O-methyl (2′-OMe) RNA oligonucleotides with 5′ biotinylated (dT)6 and (dA)4 5′ leader sequences, respectively; αTag (5′-CCCCUGAAGGUUAUAUCAGC-3′) 2′-OMe RNA oligonucleotide; DNA oligonucleotides complementary to hTR nt 487 to 463 (h6), nt 443 to 419 (h3), nt 235 to 211 (h2), and nt 205 to 182 (h1); and random hexamer-radiolabeled SE restriction fragment. Additional 5′ end-labeled DNA oligonucleotides used in this study were αhU3 snoRNA (5′-ACCACTCAGACCGCGTTCTCTCCCTCTCAC-3′), αhU2 snRNA (5′-AAGCTCCTATTCCAACTCCTAGTTC-3′), and αhSRP RNA (5′-CGGTTCACCCCTCCTTAGGCAACC-3′).
Whole-cell extract preparation.
Trypsinized cells were washed in ice-cold buffer Z (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 15% glycerol [vol/vol], 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml), pelleted, resuspended in buffer Z plus 0.3% Nonidet P-40 (vol/vol) and sonicated for 20 s with a Heat Systems microtip on setting 2. Following addition of NaCl to a final concentration of 200 mM, sonicates were cleared either in a microcentrifuge for 5 min and then in a Beckman TLA100.3 rotor for 45 min at 45,000 rpm or in a microcentrifuge for 30 min.
Telomerase activity assay.
Telomerase activity was detected by the PCR-based telomere repeat amplification protocol (TRAP [27]) with minor alterations: extension was performed at 30°C for 1 h with 5 μl of extract diluted in buffer Z in a final volume of 47.5 μl; hot-start PCR was performed beneath a layer of mineral oil at 94°C with the addition of 2.5 μl containing Taq polymerase, [α-32P]dGTP, and WT-specific primer C3TA2 (5′-CCGCGCCCTAACCCTAACCCTA-3′) or AT-specific primer A2C4 (5′-AACCCCAACCCCAACCCC-3′), C4A2 (5′-CAACCCCAACCCCAACCCCAA-3′), or C2A2C2 (5′-CCAACCCCAACCCCAACC-3′). RNase A-treated samples were incubated with 0.5 μg of RNase A for 10 min at 30°C prior to addition of the extension mix. Products were analyzed by electrophoresis in 1× TBE in a native 10% polyacrylamide gel.
Subcellular fractionation.
Subcellular fractionation was performed as described previously (42, 43) with minor alterations: HeLa cells trypsinized off plates at approximately 90% confluency were lysed by the addition of 0.3% Nonidet P-40 (vol/vol); sonication was performed with a Heat Systems microtip on setting 2 in 0.35 M sucrose–0.25 mM MgCl2. The nucleolar pellet was obtained by centrifugation at 4,000 rpm for 12 min in a Beckman SW50.1 rotor. For sucrose gradient fractionation, nucleoli were prepared as described above from 2 × 108 HeLa cells grown in suspension, resuspended in 100 μl of 0.88 M sucrose–0.05 mM MgCl2, layered over a 10-ml 1.0 to 2.5 M sucrose gradient containing 0.5 mM MgCl2, and sedimented for 20 min at 3,000 rpm in a Beckman SW40Ti rotor.
RESULTS
The 3′ end of hTR has putative box H/ACA snoRNA-like motifs.
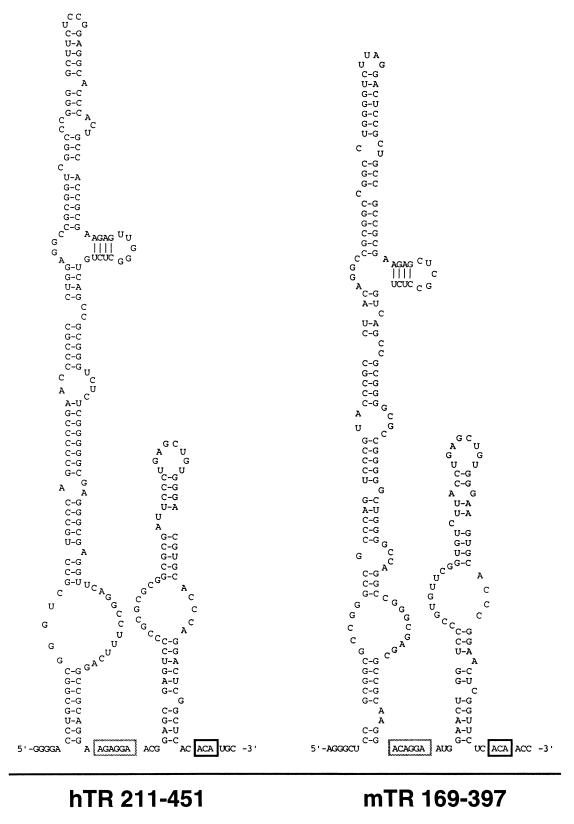
We noticed that the 3′ end of mature hTR (45) has an ACA trinucleotide 3 nt upstream of its 3′ end. In addition, the 3′ region of hTR contains a single H box consensus sequence (5′-AGAGGA-3′). This region also has the potential to fold into the characteristic hairpin-hinge-hairpin-tail core box H/ACA snoRNA secondary structure, with the H and ACA boxes located in single-stranded hinge and tail regions, respectively (Fig. 1). Our hTR gene sequence differs from that initially reported (13) but is identical to a second GenBank sequence submitted subsequent to our sequencing (accession no. U85256). Numbering in this report is based on the latter sequence.
FIG. 1.
Potential secondary structures of the 3′ ends of hTR and mTR. The structures (hTR nt 211 to 451 and mTR nt 169 to 397 as indicated) are based on the conserved hairpin-hinge-hairpin-tail secondary structure of box H/ACA snoRNAs and a sequence comparison between hTR and mTR. H and ACA box primary sequence elements are contained within the light and dark boxes, respectively.
Comparison with the murine telomerase RNA (mTR) (7) suggests that the snoRNA-like features of hTR are evolutionarily conserved. The mTR 3′ end (nt 169 to 397 as numbered in reference 25) has ∼76% sequence identity with the corresponding region of hTR (nt 211 to 451) and includes consensus H (5′-ACAGGA-3′) and ACA box sequences. Although the 3′ end of mTR has not been mapped precisely, the primary sequence identity between mTR and hTR genes drops sharply downstream of the conserved ACA trinucleotide. The “ACA plus 3 nt” rule governing box H/ACA snoRNA 3′ end processing (5) predicts mature mTR to end at position 397. The H and ACA box motifs of mTR, like those of hTR, can be modeled into a consensus box H/ACA core secondary structure (Fig. 1).
The secondary structure models presented in Fig. 1 are intended to illustrate the potential of these regions to fold into a domain with conserved box H/ACA snoRNA-like features. Physical and genetic methods combined with phylogenetic data will be required to determine the details of the hTR and mTR structures. However, as predicted by our functional data (see below), a conserved box H/ACA snoRNA-like domain is also found in the phylogenetic comparative analysis of telomerase RNA from many other mammalian species (12).
Another common structural feature of box H/ACA snoRNAs is the presence of an internal loop structure in one or both hairpins that can direct pseudouridine formation by hybridization with substrate RNAs (15). Confirmation of the presence of a genuine pseudouridylation pocket requires the identification of a modification target sequence. Without more extensive phylogenetic comparison of mammalian telomerase RNAs and some indication of a modification target sequence, we cannot yet predict whether hTR directs pseudouridine modification.
Recombinant hTR production requires H and ACA box sequence elements.
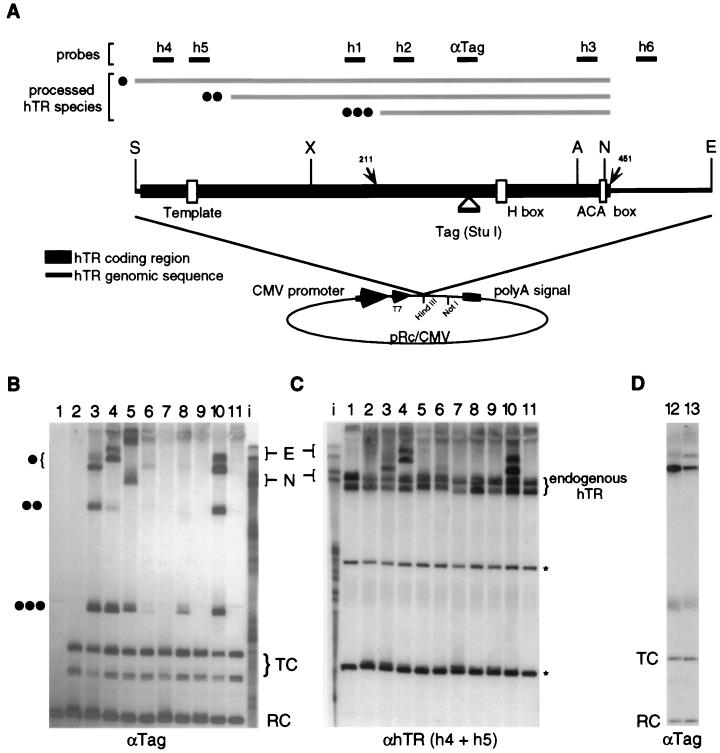
To investigate the role of the putative box H/ACA snoRNA-like motifs in hTR, we expressed sequence-tagged versions of recombinant hTR by transient transfection in 293 cells and assayed their stability and processing. In previous studies, recombinant hTR was expressed either from its endogenous promoter (13) or from the Pol II U2 snRNA promoter (30). A comparative analysis of hTR and mTR promoters concluded that they represent mRNA-type promoters rather than snRNA-type Pol II promoters (25). We expressed recombinant hTR with the cytomegalovirus Pol II mRNA promoter and a downstream bovine growth hormone polyadenylation signal (Fig. 2A). A region of the hTR gene including RNA nt 1 to 451 and an additional 109 nt of 3′ genomic sequence (the SE restriction fragment) (Fig. 2A) was engineered with a 20-nt sequence tag to facilitate the discrimination of recombinant and endogenous species. Expression of recombinant hTR from the SE restriction fragment construct resulted in the accumulation of a sequence-tagged hTR species with an electrophoretic mobility and oligonucleotide hybridization profile indicative of wild-type 3′ end maturation (Fig. 2B, lane 3, single circle). Note that full-length hTR (endogenous, recombinant, or in vitro transcribed) migrates as a doublet in a denaturing polyacrylamide gel; the significance of the doublet and the cause of the observed variation in the relative intensities of the bands are unknown. The correctly processed, recombinant hTR accumulates at the level characteristic of endogenous telomerase RNA (∼1,000 copies/cell [4]; Fig. 2C lanes 3 to 4) in a population of transfected cells. This could reflect higher recombinant hTR levels in fewer cells (low-efficiency transfection) or a restriction on recombinant hTR accumulation similar to that on the endogenous form. Although transfection efficiency was not monitored in each experiment, high-efficiency transfection of an alkaline phosphatase reporter construct under similar transfection conditions in pilot assays supports the latter conclusion.
FIG. 2.
hTR expression by Pol II. (A) Schematic of hTR Pol II expression system indicates relative positions of restriction endonuclease sites, template residues, H and ACA box residues, sequence tag insertion site (StuI), and probes used in Northern blot analyses (h1 to h6, αTag). Processed hTR species are indicated by gray bars and correspond according to the number of filled circles with those in panel B. (B and C) Northern blot analysis of total RNA prepared from transiently transfected 293 cells probed with αTag oligonucleotide (B) or stripped and reprobed with h4 and h5 oligonucleotides (C). Lane 1, mock transfection; lane 2, empty pRc/CMV vector; lanes 3 to 11, hTR restriction fragments cloned into pRc/CMV HindIII site (except lane 4 [into NotI site]). Restriction fragments: lanes 3 and 4, SE; lane 5, XE (note for panel C that construct does not contain the sequences complementary to h4 or h5); lane 6, SN; lane 7, SA; lanes 8 to 11, SE with the following mutations in the ACA and H boxes: ACA→TCA, ACA→TGT, AGAGGA→ATATTA, and AGAGGA→TGTGGT, respectively (mutated residues are underlined). Filled circles indicate processed hTR species in lane 3. Lane i contains in vitro-transcribed hTR standards: E and N are transcripts from the sequence-tagged SE fragment in pRc/CMV driven by the T7 promoter and terminated approximately at the E and N restriction sites, respectively. A 110-nt RNA RC was included during sample preparation to verify loading equivalents. In lanes 2 to 11, the U6tTR construct was included as a control for relative transfection efficiency (TC). Cross-reacting endogenous non-hTR RNAs are indicated by an asterisk. (D) Northern blot analysis performed as described above and probed with αTag oligonucleotide. Lane 12, SE; lane 13, SN with restored ACA box. Note that both endogenous and recombinant hTR, as well as hTR transcribed in vitro by T7 RNA polymerase, migrate as doublets in a denaturing polyacrylamide gel.
Two faster-migrating recombinant RNAs also accumulated from the SE restriction fragment hTR expression construct. Mapping by oligonucleotide hybridization (Fig. 2 and data not shown) indicated that both were processed near the wild-type 3′ end (hybridization with h3 but not h6); additional processing occurred between the template residues and the 5′ end of the putative H/ACA domain (hybridization with h1 but not h4 or h5) or near the 5′ end of the putative H/ACA domain (hybridization with h2 but not h1). The migration of the latter RNA marked by three circles was similar to that of an in vitro-transcribed RNA corresponding to the putative hTR H/ACA domain (data not shown). Thus, although an hTR H/ACA domain molecule is not detected as a stable endogenous form of hTR in wild-type 293 cells, it accumulates as an independently stable domain in transfected 293 cells that express various forms of recombinant hTR (Fig. 2B lanes 3 to 6, 8, and 10 and data not shown).
All known human box H/ACA snoRNAs are processed from introns at their 5′ and 3′ ends. We examined the potential for 5′ end processing of mature recombinant hTR by adding extra sequence between the transcriptional start site and the 5′ end of hTR. This was accomplished by cloning the SE restriction fragment into a polylinker restriction site further downstream of the cytomegalovirus promoter (NotI, Fig. 2A). Expression of hTR from this construct resulted in the accumulation of a slower-migrating mature hTR species, suggesting that the 5′ end of the transcript including additional vector polylinker sequence is intact (Fig. 2B, lane 4). We also tested the processing of an hTR transcript lacking the telomerase template region but containing the H/ACA domain. Expression of hTR nt 160 to 560 from the XE construct resulted in the accumulation of a species processed near its wild-type 3′ end (lane 5), suggesting that nt 1 to 160 are not essential for hTR 3′ end processing.
To examine the dependence of hTR accumulation and 3′ end processing on the putative H and ACA box elements, we constructed two hTR 3′ end deletions and a series of H and ACA box mutations. In human and yeast box H/ACA snoRNAs, mutation of conserved H or ACA box residues prevents accumulation of mature RNAs (5, 16). Engineering of the SN construct removed the predicted ACA box but left an ACA sequence 2 nt upstream of the wild-type box (Fig. 1). This resulted in the accumulation of mature hTR, although much less was accumulated than was observed with the SE construct (Fig. 2B, compare lanes 6 and 3). When the altered spacing of the ACA box relative to the putative upstream stem structure was restored in the SN construct, the accumulation of mature hTR was equal to that observed from the SE construct (Fig. 2D). Thus, hTR 3′ end processing site specificity is independent of the genomic sequence 3′ of the ACA box, consistent with the processing of yeast box H/ACA snoRNAs (5).
In the SA construct, deletion of the 3′-terminal 23 nt including the ACA box prevented accumulation of any detectable tagged hTR species (Fig. 2B, lane 7). A point mutation in the first ACA box position (A446T) in the SE construct dramatically reduced recombinant hTR accumulation (lane 8), while mutation of the ACA box to TGT rendered it undetectable (lane 9). Similarly, mutation of the conserved H box residues (AGAGGA→TGTGGT) prevented hTR accumulation (lane 11), while mutation of the nonconserved residues (AGAGGA→ATATTA) had no effect (lane 10) (mutated residues are underlined). Thus, as for the snoRNA family, the H and ACA box motifs of hTR are important for RNA stability and 3′ end processing.
We noticed the accumulation of high-molecular-weight hTR-containing species from a variety of our Pol II expression constructs, especially those lacking hTR nt 1 to 160 (lane 5) or with certain H or ACA box defects (lanes 6, 8, and 11). Although the relative amounts of these hTR species varied between transfections, production of these RNAs from a Pol II promoter but not from a Pol III promoter (see below) suggests that they may be polyadenylated precursor forms of hTR. Although no such intermediates of endogenous hTR biogenesis have been reported, their presence may be transient and more readily detectable when RNAs are overexpressed or compromised at their 5′ or 3′ ends.
Recombinant hTR is incorporated into an active telomerase RNP.
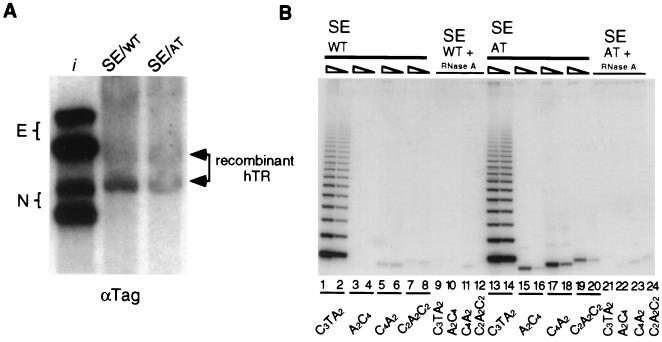
The RNA components of various RNPs are stabilized in vivo by assembly with associated proteins. Thus, it seemed likely that the recombinant hTR molecules observed were assembled into telomerase RNPs. To directly address whether sequence-tagged, recombinant hTR was assembled into an active telomerase RNP, we mutated template residues in the SE construct to create an altered template version of the RNA. By changing the template at two positions (T47C, T53C), telomeric repeat synthesis can be altered from the wild-type human repeat sequence TTAGGG to the Tetrahymena repeat sequence TTGGGG (mutated residues are underlined). To assay telomerase activity, a telomerase-dependent, single-stranded primer elongation step is followed by PCR amplification with primers specific for either the WT or AT repeat sequence. PCR amplification of TTGGGG repeats generates a single PCR product rather than a ladder of products (13). This pattern of product DNA can result from differences in the PCR amplification stage of the reaction and is not necessarily indicative of the relative processivity of the AT versus WT telomerase activities.
Extracts were prepared from 293 cells transfected with WT and AT versions of the SE hTR construct in parallel. Accumulation of AT and WT recombinant sequence-tagged forms of hTR in transfected cells is shown in Fig. 3A. Telomerase activity was detected by PCR amplification of telomerase products specific for WT or AT sequences. Wild-type activity was present in both extracts due to endogenous telomerase, which served as a positive control for extraction (Fig. 3B, lanes 1, 2, 13 and 14). In contrast, AT telomerase activity was detected only in AT extracts. Three primers specific for AT repeats amplified single products of varying lengths according to their template permutation with respect to the elongation primer (lanes 15 to 20). Both WT and AT telomerase activities were sensitive to pretreatment of extract with RNase A, as expected if activity is dependent on hTR (lanes 9 to 12 and 21 to 24). Low levels of PCR products that were detected in both RNase A-treated and untreated samples (lanes 5 to 8, 11, 12, 23, and 24) were primer specific and telomerase independent and thus considered to be background. We conclude that the sequence-tagged, recombinant hTR expressed from a Pol II mRNA promoter and processed at its 3′ end like a box H/ACA snoRNA can be incorporated into an active telomerase RNP.
FIG. 3.
Recombinant hTR incorporation into an active telomerase RNP. (A) Northern blot analysis of total RNA prepared from cells transiently transfected with WT and AT versions of the sequence-tagged SE construct (arrowheads). The blot was probed with αTag oligonucleotide. Lane i contains in vitro-transcribed hTR standards E and N. (B) TRAP activity assay of extracts prepared from WT and AT transiently transfected 293 cells. Oligonucleotides amplifying WT (C3TA2) or AT (A2C4, C4A2, C2A2C2) telomerase extension products were used as indicated. For lanes 9 to 12 and 21 to 24, extracts were incubated with RNase A prior to the telomerase extension reaction. Each pair of lanes corresponds to 20- and 100-fold dilutions of whole-cell extracts.
A heterologous box H/ACA snoRNA is sufficient for hTR stability and 3′ end processing but not for telomerase activity.
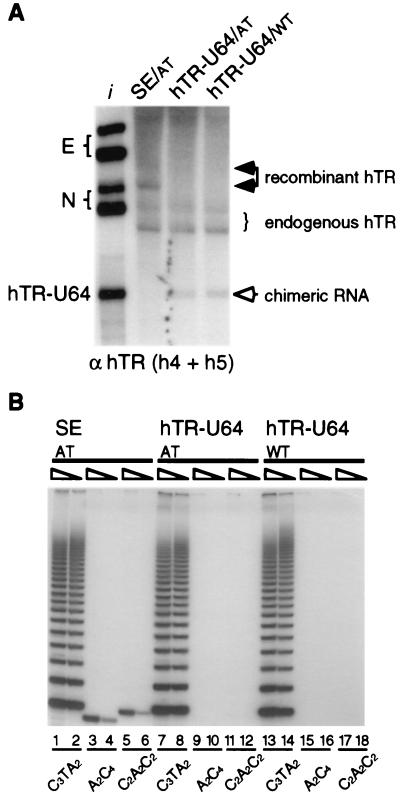
Because the hTR H/ACA domain is not required for a minimal level of telomerase activity by in vitro reconstitution of nuclease-treated native telomerase (3) or by in vitro assay with reticulocyte lysate-expressed hTERT (6), we reasoned that another box H/ACA snoRNA might substitute for the hTR H/ACA domain in vivo. To test this, we replaced the hTR H/ACA domain with the well-characterized human U64 box H/ACA snoRNA (16) in our Pol II expression system. An RNA with an electrophoretic mobility and oligonucleotide hybridization profile indicative of a 3′ end-processed hTR-U64 chimera accumulated in transfected cells (Fig. 4A and data not shown). This demonstrates that a heterologous box H/ACA snoRNA can confer stability on the telomerase template region and direct 3′ end processing of a chimeric RNA.
FIG. 4.
Processing, accumulation, and activity of a chimeric hTR-U64 RNA. (A) Northern blot analysis of total RNA prepared from cells transiently transfected with the following constructs: sequence-tagged AT SE (solid arrowheads), AT hTR-U64, and WT hTR-U64 (open arrowhead). The blot was probed with h4 and h5 oligonucleotides. Lane i contains in vitro-transcribed hTR E and N and chimeric hTR-U64 RNA standards. (B) TRAP activity assay of extracts prepared from transiently transfected 293 cells. Oligonucleotides amplifying WT (C3TA2) or AT (A2C4, C2A2C2) telomerase extension products were used as indicated. Each pair of lanes corresponds to 5- and 25-fold dilutions of whole-cell extracts.
Because 5′ end processing of U64 out of the hTR-U64 chimera would produce a U64 species similar in size to the endogenous RNA, we were unable to address whether some chimera was lost to U64 5′ end processing. However, it is clear that at least some percentage of U64 remained covalently attached to an hTR template region, a novel result considering that endogenous U64 is processed from an intron of the ribosomal protein S4 transcript at both its 5′ and 3′ ends (16).
Because we could detect a stable, 3′ end-processed hTR-U64 chimera, we assayed for telomerase activity of a chimeric RNA-containing RNP using an AT version of the transfection construct. Unlike other AT hTRs previously tested, the AT hTR-U64 chimera did not generate a functional telomerase RNP. Telomerase activity directed by the altered template was undetectable in extracts of cells transfected with the AT version of the hTR-U64 chimera (Fig. 4B, lanes 9 to 12). As a control for the extraction of AT chimeric RNA relative to an AT hTR control with demonstrable AT activity, we verified that the abundance of these RNAs in extracts reflected their abundance in whole cells (data not shown). Thus, although U64 snoRNA is sufficient to direct 3′ end processing and stability of a chimeric RNA containing the hTR template region, it is insufficient for activity in the chimeric RNA-containing RNP. There are several possible explanations for the inactivity of the chimera. For example, U64 RNA or an associated protein could prevent telomerase activity in a dominant fashion. Alternately, sequence elements of the hTR 3′ end that are not part of the conserved snoRNA-like core may be required for telomerase activity.
Expression of hTR by Pol III generates only the H/ACA domain.
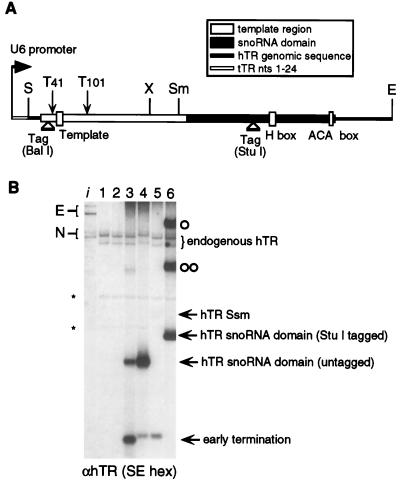
Ciliate telomerase RNAs are transcripts of Pol III, whereas all known human box H/ACA snoRNAs are encoded within introns of Pol II transcripts (41). To investigate whether hTR processing and accumulation require expression by Pol II, we placed the hTR SE fragment under the control of the human Pol III U6 snRNA promoter (Fig. 5A). Although the majority of transcripts from this construct terminated near runs of six or five T’s in the hTR gene (nt 38 to 43, 99 to 103) constituting Pol III transcriptional termination signals, two additional hTR species, including the hTR H/ACA domain, accumulated (Fig. 5B, lane 3). Thus, even though full-length hTR transcripts were made, processing events prevented accumulation of mature hTR (Fig. 5B and data not shown). Expression from the Pol III SE construct with mutated internal Pol III termination signals (T41A, T101A) resulted in the increased accumulation of the hTR H/ACA domain relative to the early termination products, but again no full-length hTR accumulated (Fig. 5B, lane 4). As a control, the T41A and T101A mutations in the Pol II SE construct did not prevent accumulation of mature hTR (data not shown).
FIG. 5.
hTR expression by Pol III. (A) Schematic of hTR Pol III expression system indicates relative positions of restriction endonuclease sites template residues, H and ACA box residues, sequence tag insertion sites (BalI and StuI), and T→A mutations. (B) Northern blot analysis of total RNA prepared from transiently transfected 293 cells probed with the random hexamer-labeled SE restriction fragment (SE hex). Lane 1, mock transfection; lane 2, empty pRc/CMV vector; lane 3, SE sequence tagged at BalI; lane 4, untagged SE (T41A, T101A); lane 5, untagged SSm (T41A, T101A); lane 6, XE sequence tagged at StuI. Unprocessed (○) and 3′ end-processed (○○) hTR species present in lane 6 are indicated. Lane i contains in vitro-transcribed hTR standards E and N. Non-hTR cross-hybridizing RNAs are indicated by an asterisk. Relative transfection efficiency and RNA recovery were monitored as described in the legend to Fig. 2 (data not shown).
We also tested expression of the template region alone, without the H/ACA domain, in the Pol III expression system. Expression from a T41A, T101A version of the SSm restriction fragment construct (nt 1 to 205) produced extremely low levels of stable RNA (Fig. 5B, lane 5). In contrast, deletion of the template region SX restriction fragment (nt 1 to 160) from a Pol III expression construct allowed high levels of three hTR species to accumulate (Fig. 5B, lane 6). Based on their relative mobilities and oligonucleotide hybridization profiles, these species are predicted to be the following: an unprocessed, poly(U)-terminated precursor, a species processed only at its 3′ end near the ACA box, and the hTR H/ACA domain. We conclude that hTR is not a transcript of Pol III and, more interestingly, cannot be stably expressed as a transcript of Pol III under the control of the U6 snRNA promoter. The presence of the template region may actually destabilize Pol III transcripts, because partially 5′ processed molecules were observed for transfections with the XE construct but not the SE construct.
Subcellular fractionation of hTR reveals a partial nucleolar association.
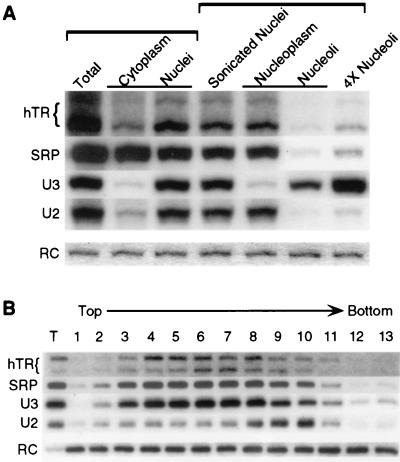
Human box H/ACA snoRNAs assayed by subcellular fractionation are associated almost entirely with nucleoli, as their name would suggest. To determine if hTR is enriched in nucleoli, we examined the relative amounts of hTR in subcellular fractions of telomerase-positive HeLa cells (Fig. 6A). The majority of total hTR was recovered in the nuclear fraction, although a significant proportion was recovered in the cytoplasmic fraction as well (leakage of components from nuclei during their isolation is not uncommon). Purified nuclei were further fractionated into nucleoplasm and nucleoli by a sonication procedure that favors complete nuclear disruption at the expense of nucleolar integrity. The majority of nuclear hTR was recovered in the nucleoplasmic fraction, whereas a small percentage (7 to 8% of total sonicated nuclear hTR) was reproducibly recovered in the nucleolar fraction. Total RNA from these subcellular fractions was probed for U3 snoRNA to control for recovery of nucleoli, and for U2 snRNA and signal recognition particle (SRP) RNA to control for contamination of purified nucleoli with nonnucleolar RNAs (Fig. 6A). The nucleolar fraction contained 50 to 65% of total sonicated nuclear U3 snoRNA and 2 to 5% each of total sonicated nuclear U2 and SRP RNAs, suggesting that the amount of the hTR detected in the nucleolar fraction is significant. We have observed similar results using another transformed cell line (293) and two primary fibroblast lines (IMR90 and F65) which lack detectable telomerase activity but express low levels of hTR (data not shown). We conclude that despite its H/ACA domain, hTR is either not predominantly nucleolar or is associated with the nucleolus in a weak or peripheral manner that does not readily survive nucleolar purification.
FIG. 6.
Localization of hTR by subcellular fractionation. (A) Northern blot analysis of RNA prepared from subcellular fractions of HeLa cells. The blot was probed with h4 and h5 oligonucleotides to detect endogenous hTR and the 410-nt RC RNA added during sample preparation to verify loading equivalents and then with oligonucleotides complementary to U3 snoRNA, U2 snRNA, and SRP RNA. Brackets above lanes indicate cell equivalent loadings among the Total, Cytoplasm, and Nuclei lanes and among the Sonicated Nuclei, Nucleoplasm, and Nucleoli lanes. 4× Nucleoli represents 4 cell equivalents of nucleolar RNA relative to the Sonicated Nuclear group. (B) Northern blot analysis of RNA prepared from sucrose gradient fractions following sedimentation of crude HeLa nucleoli, probed as described for panel A. A fraction of total nucleolar RNA is present in lane T.
If the hTR present in our crude nucleolar fraction is indeed associated with nucleoli, it should cofractionate with nucleoli upon further purification. To test this, crude nucleoli were resolved on a 1.0 to 2.5 M sucrose gradient (Fig. 6B). We observed the cosedimentation of hTR with U3 snoRNA. The majority of contaminating U2 snRNA did not cosediment with hTR or U3 snoRNA, indicating as expected that it is not associated with nucleoli. Although SRP RNA, the RNA component of the SRP, is predominantly cytoplasmic (Fig. 6A), recent studies have demonstrated it to be transiently associated with nucleoli (26). This is consistent with our observation that the small percentage of SRP RNA recovered in the nucleolar fraction cosedimented with hTR and U3 snoRNA (Fig. 6B). Taken together, these data suggest that the association of a small percentage of hTR with nucleoli is specific.
DISCUSSION
Identification of a box H/ACA snoRNA-like domain in hTR.
Within all telomerase RNAs, the template for DNA synthesis is contained in less than 10% of the total RNA sequence. Little is known about the function of the remainder of the molecule, even for the relatively compact ciliate RNAs. This deficiency in the state of our knowledge is more evident in light of the apparent expansion of telomerase RNA length in yeast and mammal cells. Surprisingly, the evolutionary “expansion” for mammalian telomerase RNAs appears to be the result of fusion with a preexisting RNA motif: a box H/ACA snoRNA.
We have identified a domain at the 3′ end of hTR that shares some, but not all, of the demonstrated characteristics of the box H/ACA family of snoRNAs. The hTR H/ACA domain contains both primary sequence motifs (H and ACA boxes) and the potential to adopt the conserved hairpin-hinge-hairpin-tail core secondary structure common to all box H/ACA snoRNAs. In addition, the dependence on intact H and ACA boxes for stability and 3′ end processing is shared by hTR and box H/ACA snoRNAs. In contrast, the 5′ end processing typical of box H/ACA snoRNAs must be abrogated by the hTR H/ACA domain in order to produce a functional telomerase RNA that includes the template region. Stable nucleolar localization, a property of all known box H/ACA snoRNAs, may also be lost in order to allow telomerase to interact with telomeres in the nucleoplasm, although a nucleolar phase of RNP biogenesis could be required. In addition, hybridization to target RNA for site selection of pseudouridine modification may or may not be conserved in hTR. Although this is a demonstrated property of most box H/ACA snoRNAs, there remain examples in both yeast and human cells of box H/ACA snoRNAs without known modification targets. Thus, it is not known whether the direction of pseudouridine synthesis is common to all box H/ACA snoRNAs.
The functions of conserved box H/ACA snoRNA sequence motifs in hTR processing, telomerase RNP assembly, and directly or indirectly in telomerase activity reveal new cellular roles for this RNA motif. The fusion of the hTR H/ACA domain with the template region indicates that the motif can direct the 3′ end processing and stabilization of a 5′ extended RNA species. Although hTR is not predominantly nucleolar by subcellular fractionation, the hTR H/ACA domain may accomplish a transient nucleolar localization of hTR that promotes RNA folding, RNA modification, or RNP assembly. Thus, fusion to a snoRNA-like domain may provide the hTR template region a better opportunity to assemble into a functional RNP in the context of the mammalian nucleus.
Although mammals, yeasts, and ciliated protozoa all have telomerase RNAs and box H/ACA snoRNAs, to date only mammals are known to have combined the two. It is possible that the yeast K. lactis will provide another example of this coupling: the mature telomerase RNA is processed at its 3′ end from a polyadenylated precursor 138 nt upstream of the poly(A) tail and 3 nt after an ACA trinucleotide (based on the 1,267-nt RNA described in reference 11, with nt 1 at position 675 of the GenBank accession no. U31465 sequence [31]). We anticipate that phylogenetic analyses of a broad range of species will be most useful in revealing the evolutionary relationship between telomerase RNAs and snoRNAs.
hTR accumulation and processing.
We show that hTR can be expressed from a Pol II mRNA promoter, processed at its 3′ end by an activity that requires box H/ACA snoRNA signature motifs, and assembled into an active telomerase RNP. Although expressed from a strong promoter, recombinant full-length hTR accumulated to levels similar to the low-copy-number endogenous hTR (∼1,000 copies per cell [4]) in a population of transfected cells. This suggests that one limitation of the maximum attainable telomerase activation in vivo is set by the level of hTR accumulation. The presence of the H/ACA domain alone as a product of both recombinant Pol II and Pol III expression may be a consequence of overexpression of the recombinant hTR precursor. With atypically high levels of precursor, the recombinant hTR transcript may be recognized by the box H/ACA snoRNA 5′ and 3′ end processing machineries. Interestingly, the hTR H/ACA domain excised from full-length hTR could accumulate to a level well above that of endogenous hTR when expressed in the Pol III system (Fig. 5B, lanes 4 and 6) but not in the Pol II system (Fig. 2).
The enzymatic machinery involved in box H/ACA snoRNA 3′ end processing has not been identified. However, it seems likely that hTR, the hTR H/ACA domain, and box H/ACA snoRNAs are all processed by a common mechanism. Each RNA requires the canonical H and ACA box sequence elements for accumulation and 3′ end processing, and both hTR and the box H/ACA snoRNAs are trimmed to precisely 3 nt downstream of the ACA motif. The 3′ nucleolytic processing activities recruited to hTR and box H/ACA snoRNA precursors could depend on RNA structure or RNA binding proteins either unique to hTR or the snoRNAs or shared between them.
The potential for shared RNP protein components raises the possibility that some mammalian telomerase proteins are not unique to the telomerase RNP. Because ciliate telomerase RNAs do not utilize a snoRNA 3′ end processing mechanism, human homologs of the ciliate telomerase proteins (TEP1 and hTERT) are likely to be telomerase specific and associated with the template region rather than the H/ACA domain of hTR. However, it is possible that proteins common to all known box H/ACA snoRNAs are associated with telomerase RNA. In S. cerevisiae, these proteins include the box H/ACA snoRNP-specific Gar1p and the putative pseudouridine synthase Cbf5p. Although there is no known mammalian homolog of Gar1p, human and rat homologs of the putative pseudouridine synthase Cbf5p have been identified (24, 32). In humans, mutations in this gene cause dyskeratosis congenita, an X-linked disorder characterized by skin abnormalities, gastrointestinal lesions, bone marrow failure, and a predisposition to cancer (24). Given that telomerase may be required for normal cellular function in highly proliferative tissues, it is tempting to speculate that a telomerase deficiency can contribute to the symptoms observed in these patients.
The inability of Pol III to produce stable, full-length hTR may indicate a strict polymerase selectivity for reasons beyond the obvious presence of Pol III termination signals early in the transcript. We considered the possibility that only Pol II transcripts were able to assemble into telomerase RNP because only these transcripts were protected from degradation at their 5′ ends by a particular cap structure. However, some 5′ end processing must have occurred to generate the observed H/ACA domain alone. In addition, stable products of Pol III, albeit not full length, did accumulate (Fig. 5B, lane 6). Thus, it may be that the Pol II precursor or the act of transcription itself provides information relevant to the telomerase RNP biogenesis pathway in vivo.
Telomerase RNP biogenesis.
We favor the hypothesis that telomerase RNP biogenesis, like the biogenesis of ribosomes and snRNPs, occurs in an obligatory multistep assembly pathway. The in vivo requirement for the H/ACA domain and the subcellular fractionation profile of hTR are most easily reconciled with a model in which some stage of RNP biogenesis occurs in the nucleolus, followed by transit of the RNP to the nucleoplasm. Recent localization studies with fluorescent SRP RNA in mammalian cell nuclei demonstrated that microinjected SRP RNA transiently localizes to nucleoli before transport to the cytoplasm (26). The coincident sucrose gradient fractionation profiles of nucleolear U3 RNA, SRP RNA, and hTR raise the possibility that telomerase RNP biogenesis has a nucleolar phase as well. If the nucleolar population of hTR reflects an intermediate in telomerase RNP biogenesis, it may have properties different from the nucleoplasmic RNP pool. Unfortunately, because human telomerase activity assays from purified nucleolar fractions are not reliably quantitative (data not shown), we were unable to establish whether the nucleolar population of mature hTR was predominantly associated with an active or inactive telomerase RNP. The incomplete nucleolar association of hTR relative to box H/ACA snoRNAs may result from the absence of a snoRNP component in the telomerase RNP, or the presence of a dominant relocalization factor.
It will be of interest to determine what specific roles transcription by Pol II, the H/ACA domain, and the nucleolus play in the assembly of human telomerase and to define which components and pathways are shared by telomerase and the box H/ACA snoRNPs.
ACKNOWLEDGMENTS
We thank T. Cech, R. Freiman, P. Kaufman, T. Pederson, D. Rio, and members of our laboratory for critical reading of the manuscript and R. Bell and A. Fisher for tissue culture assistance.
This work was supported by University of California and National Institutes of Health grants to K.C. and a National Science Foundation predoctoral fellowship to J.R.M.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moore D, Seidman F, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 2.Autexier C, Greider C W. Telomerase and cancer: revisiting the telomere hypothesis. Trends Biol Sci. 1996;21:387–391. [PubMed] [Google Scholar]
- 3.Autexier C, Pruzan R, Funk W, Greider C. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 4.Avilion A A. Ph.D. thesis. Stony Brook: State University of New York; 1995. [Google Scholar]
- 5.Balakin A, Smith L, Fournier M. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 6.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 7.Blasco M A, Funk W, Villeponteau B, Greider C W. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- 8.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C-P, Morin G, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 9.Bousquet-Antonelli C, Henry Y, Gelugne J-P, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan T, Marusic L, Bacchetti S, Namba M, Reddel R. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum Mol Gen. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- 11.Chapon C, Cech T, Zaug A. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J.-L., M. A. Blasco, and C. W. Greider. Personal communication.
- 13.Feng J, Funk W D, Wang S, Weinrich S L, Avilion A A, Chiu C, Adams R R, Chang E, Allsopp R C, Yu J, Le S, West M, Harley C B, Andrews W H, Greider C W, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 14.Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 16.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 17.Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greider C W. Telomerase biochemistry and regulation. In: Blackburn E H, Greider C W, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 35–68. [Google Scholar]
- 19.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;66:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 20.Harley C B. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 21.Harley C B, Villeponteau B. Telomeres and telomerase in aging and cancer. Curr Op Genet Dev. 1995;5:249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 22.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Program E, Bass M B, Arruda I, Robinson M O. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 23.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S K, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heiss N S, Knight S W, Vulliamy T J, Klauck S M, Wiemann S, Mason P J, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 25.Hinkley C, Blasco M, Funk W, Feng J, Villeponteau B, Greider C, Herr W. The mouse telomerase RNA 5′-end lies just upstream of the telomerase template sequence. Nucleic Acids Res. 1998;26:532–536. doi: 10.1093/nar/26.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson M R, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L C, Coviello G M, Wright W E, Weinrich S L, Shay J W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2014. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 28.Kunkel G R, Pederson T. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev. 1988;2:196–204. doi: 10.1101/gad.2.2.196. [DOI] [PubMed] [Google Scholar]
- 29.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol Cell Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McEachern M J, Blackburn E H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 32.Meier U T, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murnane J P, Sabatier L, Marder B A, Morgan W F. Telomere dynamics in an immortal cell line. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 36.Ni J, Tien A, Fournier M. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 37.Nugent C I, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 38.Romero D P, Blackburn E H. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 39.Schnapp G, Rodi H-P, Rettig W J, Schnapp A, Damm K. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 1998;26:3311–3313. doi: 10.1093/nar/26.13.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 41.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 42.Tyc K, Steitz J. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warner J. Distribution of newly formed ribosomal proteins in HeLa cell fractions. J Cell Biol. 1979;80:767–772. doi: 10.1083/jcb.80.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichsteiner S, Kim N W, Trager J B, Taylor R D, Carlos R, Andrews W H, Wright W E, Shay J W, Harley C B, Morin G B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 45.Zaug A, Lingner J, Cech T. Method for determining RNA 3′ ends and application to human telomerase RNA. Nucleic Acids Res. 1996;24:532–533. doi: 10.1093/nar/24.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]