Abstract
U4 snRNA release from the spliceosome occurs through an essential but ill-defined Prp38p-dependent step. Here we report the results of a dosage suppressor screen to identify genes that contribute to PRP38 function. Elevated expression of a previously uncharacterized gene, SPP381, efficiently suppresses the growth and splicing defects of a temperature-sensitive (Ts) mutant prp38-1. This suppression is specific in that enhanced SPP381 expression does not alter the abundance of intronless RNA transcripts or suppress the Ts phenotypes of other prp mutants. Since SPP381 does not suppress a prp38::LEU2 null allele, it is clear that Spp381p assists Prp38p in splicing but does not substitute for it. Yeast SPP381 disruptants are severely growth impaired and accumulate unspliced pre-mRNA. Immune precipitation studies show that, like Prp38p, Spp381p is present in the U4/U6.U5 tri-snRNP particle. Two-hybrid analyses support the view that the carboxyl half of Spp381p directly interacts with the Prp38p protein. A putative PEST proteolysis domain within Spp381p is dispensable for the Spp381p–Prp38p interaction and for prp38-1 suppression but contributes to Spp381p function in splicing. Curiously, in vitro, Spp381p may not be needed for the chemistry of pre-mRNA splicing. Based on the in vivo and in vitro results presented here, we propose that two small acidic proteins without obvious RNA binding domains, Spp381p and Prp38p, act in concert to promote U4/U5.U6 tri-snRNP function in the spliceosome cycle.
Pre-mRNA splicing occurs through a pair of transesterification reactions catalyzed by a complex enzyme, the spliceosome (reviewed in references 19, 23, and 26). Each round of intron removal progresses through an evolutionarily conserved cycle of subunit addition, catalysis, and subunit release from the surface of the splicing substrate. The great specificity of splicing is derived largely from the precise interaction of requisite intron consensus sequences with the dynamic spliceosomal structure.
In vitro, ATP-independent interactions between the U1 small nuclear ribonucleoprotein (snRNP) particle and the pre-mRNA 5′ splice site and branchpoint regions occur to form a complex resistant to competing substrate challenge (reviewed in reference 34). This structure, the commitment complex, stimulates the subsequent ATP-dependent addition of the U2 snRNP to the pre-mRNA branchpoint. The resulting U1–U2–pre-mRNA prespliceosome is then bound by the U4/U6.U5 tri-snRNP particle to form the spliceosome. Before pre-mRNA 5′ splice site cleavage (chemical step 1 in splicing), the extensive U4/U6 intermolecular snRNA helices are unwound through a poorly understood ATP-dependent maturation step. U4 snRNA may then be released from the spliceosome (9, 18, 29), and pre-mRNA splicing progresses (47).
The number of ATP hydrolysis events that occur in spliceosome assembly is unknown. At least seven proteins with sequence similarity to a class of RNA-dependent helicases interact with the splicing complex (the DExD/H-box proteins, reviewed in references 8 and 38). It is generally believed that ATP hydrolysis by the DExD/H-box proteins drive conformational changes within the splicing complex, such as the resolution of certain temporally restricted snRNA-snRNA and pre-mRNA–snRNA base pairing interactions (1, 16, 17, 27, 37, 43, 46). Even though Prp38p, a U4/U6.U5-specific protein, does not have a DExD/H-box motif, it is required for the resolution of the U4/U6 helices within the spliceosome (45). Prp38p likely functions indirectly in this process, perhaps by facilitating substrate presentation or through the recruitment or activation of a helicase activity.
In this study, we used the conditional lethal mutant prp38-1 to screen for spliceosomal factors that interact with Prp38p. Elevated expression of the single gene SPP381 (for the suppressor of prp38-1) was found to suppress the temperature-sensitive growth and splicing defects of the prp38-1 mutant. The genetic and biochemical data presented suggest that the SPP381-mediated prp38-1 suppression occurs through direct contact between the two gene products. Like Prp38p, Spp381p is a small, acidic protein component of the U4/U6.U5 tri-snRNP particle. Curiously, Spp381p contains a PEST proteolysis motif that appears important for pre-mRNA splicing. This report identifies an unusual new component of the splicing apparatus that functions in concert with the Prp38p spliceosome maturation factor to support cellular pre-mRNA processing.
MATERIALS AND METHODS
Yeast strains.
Yeast strains used were MGD353-46D (MATα leu2-3,112 trp1-289 ura3-52 his cyhr), MGD353-13D (MATa leu2-3,112 trp1-289 ura3-52 arg4 ade2), MGD407 (MATα/a leu2-3,112 trp1-289 ura3-52; diploid of MGD353-46D × MGD353-13D), ts192 (MATα prp38-1 leu2-3,112 trp1-289 ura3-52 his cyhr), YPB2 (MATa leu2-3,112 ade2-101 his3-200 lys2-801 trp1-901 ura3-52 gal4-542 gal80-538 LYS2::GAL1-HIS3 URA3::GAL-lacZ canr), SUL1 [MATa spp381::LEU2 leu2-3,112 trp1-289 ura3-52 pBM150 (GAL1::SPP381HA)], SUL2 [MATα prp38-1 leu2-3,112 trp1-289 ura3-52 his YEplac112 (TRP1 SPP381)], SLU3 (MATa ura3-52 leu2-3,112 spp381::LEU2 his), KBY2 [MATα prp38::LEU2 trp1-289 ura3-52 leu2-3,112 pYCplac22 (TRP1 PRP38HA)], SLY27 [MATα leu2-3,112 trp1-289 ura3-52 arg4 prp39::LEU2 pYCplac22 (TRP1 PRP39HA)], and JXY6 [MATa prp38::LEU2 trp1-289 ura3-52 leu2-3,112 ade2 YCplac22 (TRP1 prp38-2)].
Identification of suppressor plasmids.
Standard protocols were used for yeast culture, transformation, sporulation, and tetrad dissection (15). The YEp13-based yeast genomic library was obtained from the American Type Culture Collection (ATCC stock no. 37323). Approximately 1 μg of the YEp13 library was used to transform strains ts192. Suppressors were selected on synthetic medium lacking leucine at 37°C for 48 h. Putative suppressor plasmids were recovered from lysates (28) prepared from colony-purified yeast. This DNA was amplified in Escherichia coli TG1 and reintroduced into the ts192 yeast host to test for plasmid-linked suppression. DNA sequence analysis on the recovered plasmids was performed with primers that flank the YEp13 BamHI cloning site (5′GCG CCG GTG ATG CCG GCC ACG AT3′ and 5′CTA CTT GGA GCC ACT ATC GAC TAC3′). A Southern blot of XbaI/PstI-digested plasmid DNA was hybridized with probes consisting of the PRP38 and SPP381 open reading frames (ORFs) in order to identify plasmids with related DNA inserts. The isolation of mutant alleles prp38-1 and prp38-2 has been described previously (5, 45).
Subcloning and gene disruption.
Plasmid pBM150 (SPP381HA) was created by placing the SPP381HA ORF downstream of the GAL1 promoter of plasmid pBM150 (14). This was done by inserting an SPP381HA PCR fragment flanked by BamHI (upstream primer, 5′TCC CAC GGA TCC ATG AGT TTT AGA CAT TTC AAG AGG3′ [the BamHI site is underlined, and the translational initiation codon is boldfaced]) and SalI (downstream primer, 5′TCC CAC GTC GAC TTA AGC GTA GTC TGG AAC GTC GTA TGG GTA TAT AAC CGA ATA TTC AGT TTC TTC3′ [the SalI site is underlined and the hemagglutinin {HA} epitope is boldfaced]) restriction sites into BamHI/SalI-digested pBM150 plasmid DNA. Protein sequence analysis for PEST elements was performed with the public domain software available at www.at.embnet.org/embnet/tools/bio/PESTfind.
Disruption of the SPP381 ORF was performed in two steps by standard recombinant DNA techniques (36). First a 2.6-kbp EcoRI fragment of library plasmid YEp13-7 was subcloned into vector YEp112 to create the SPP381-bearing plasmid YEplac112-7A. This plasmid was digested with BamHI and BstEII to release SPP381 sequences from −261 to +820 near the 3′ end of the SPP381 coding sequence. This sequence was replaced with a 315-bp BamHI-BstEII-digested fragment created by PCR that extends from the BamHI site to a novel BstEII site introduced at position +42 by PCR (upstream primer, 5′ATA TTT ATA ACG CTA AGA TGA3′; downstream primer, 5′CCA CAC GGT GAC CAG ATC TTG AGC TTG TGT CAA GTC TCC T3′ [the BstEII site is underlined]). The resulting construct, spp381Δ42-820, removes all coding sequences between positions 42 and 820 of the 876-nucleotide SPP381 ORF. The 1.6-kbp LEU2 gene was inserted by placing a 1.6-kbp BamHI fragment from plasmid YDpL (4) into the BglII site at the spp381Δ42-820 deletion boundary. This plasmid was then cleaved with BamHI and BstUI to release the spp381::LEU2 fragment for transformation into the diploid yeast strain MGD407. The hemizygous disruptants were selected on medium lacking leucine. After sporulation, yeast cells were dissected on 1% yeast extract–2% BactoPeptone (YP) agar with 2% glucose (15) and incubated at 30°C. The presence of the correct genomic disruption in the haploid offspring was confirmed by PCR with primers upstream (5′ATA TTT ATA ACG AGA TGA3′) and downstream (5′GTA CTG TAT TTC TGC TAG ATT G3′) of the SPP381 gene. Parallel experiments were performed with the hemizygous spp381::LEU2 disruptant transformed with pBM150 (GAL1::SPP381HA). In this case, however, 2% galactose was used in the YP agar for yeast tetrad dissection. Growth assays were performed on YP-glucose and YP-galactose media, and MGD353-46D was used as the wild-type parent. The PEST sequence deletion was introduced by PCR into the GAL1::SPP381HA and YEplac112-7A backgrounds with primers 5′CCT TTA CCG AGG CCA TTA TTT ATG3′ and 5′TCC TGT ACC ATT TAA AAT TTC GCC3′. Northern assays for pre-mRNA splicing were performed as previously described (5). Ethidium bromide included in the sample loading buffer allowed the stained rRNA bands to be used as controls for equivalent sample loading, transfer efficiency, and RNA integrity.
Two-hybrid analysis.
The two-hybrid plasmids pACT and pAS2 have been described previously (24) (note that pAS2 was previously referred to as pAS1). Two-hybrid constructs of SPP381 were prepared as BamHI-BamHI (or BamHI-BglII) fragments inserted into the BglII site of pACT and the BamHI site of pAS2. BamHI (GGATCC) and BglII (AGATCT) restriction sites were introduced by PCR and are underlined in the sequences described. Upstream and downstream primers for full-length SPP381 fusion were 5′GGA TCC TAA TGA GTT TTA GAC ATT TCA AGG3′ and 5′GGA TCC TAA CTG AAA GGC ATG TGG GTT TG3′, respectively. The SPP381(1–145) construct was prepared by using the upstream primer paired with the internal primer 5′AGG ATC CAA CTA TTG ATT AGC TTT GTC GAT3′. The SPP381(146–292) construct was prepared by pairing the downstream primer with the internal primer 5′AGG ATC CAA GTG GCA AAG AAC TAG GAA. The full-length PRP38 fusion, PRP38(1–242), was prepared similarly by using the upstream primer 5′CTT AGA TCT GAC TAC AAT GGC TGT CAA TG3′ and the downstream primer 5′TTG GGA TCC TCG GGT GAA ATT GCA AAT GAC3′. The internal primers 5′AGG ATC CAA TCA AGC AAT AAT ATA TTT CGA3′ and 5′AGG ATC CAA TTG CAA CTG GTT TAT GCG3′ were used for PRP38(1–121) and PRP38(122–242) amplification, respectively. β-Galactosidase assays were carried out as described previously (15) except that the yeast cells were permeabilized by immersion in liquid nitrogen for 1 min prior to the assay.
Yeast extracts, immune precipitation, and Western blotting.
Yeast extracts were prepared by the ground cell pellet method of Umen and Guthrie (41). To prepare yeast extracts depleted of Spp381HAp, a 10-ml saturated culture of strain SUL1 grown in YP-galactose was added to 2 liters of YP-glucose and grown at 30°C for 17.5 h prior to harvest. Immune precipitations were carried out essentially as described previously (21). Twenty microliters of yeast extract (at approximately 20 μg of protein per μl of extract) was mixed with 40 μl of a 50% slurry of protein A+G agarose (Oncogene, Inc.) bound with the HA.11 antibody (Babco) or with the mAb63 control antibody as described previously (25). To this mixture was added 3 μl of 100 mM dithiothreitol, 50 U of RNasin (Promega), and HNT (20 mM HEPES [pH 7.9], 100 mM NaCl, 12.5 mM MgCl2, 0.05% Triton X-100) to 150 μl. The tubes were incubated with constant rotation at room temperature for 30 min. The unbound extract was removed by centrifugation at 4,000 × g for 1 min. The beads were then washed six times with 300 μl of HNT. Precipitations at 50 mM NaCl were carried out at this salt level in both the binding and the wash steps. All other samples were bound at 100 mM NaCl, and the salt was adjusted between 100 and 400 mM in the wash buffers. snRNAs bound to the antibody matrix were released by the addition of 100 μl of 1× PK buffer (100 mM Tris-HCl [pH 7.5], 12.5 mM EDTA, 150 mM NaCl, 1% sodium dodecyl sulfate, 2 mg of proteinase K/ml) for 10 min at 37°C. The samples were phenol extracted and precipitated with ethanol prior to Northern blot analysis with the previously described snRNA probe set (5). Glycerol gradients (10 to 35%) were run and assayed for snRNA content by immune precipitation as previously described (25). Approximately 150 μl (20 mg of protein content/ml) of total extract protein was resolved and separated into 22 fractions of 0.5 ml each. One-third of each fraction was either assayed directly for snRNA content or precipitated with the HA.11 antibody and then scored for snRNA.
Western blotting was performed on 60 μg of total extract protein resolved on a 10 or 12% discontinuous polyacrylamide gel prepared with a 4% stacking phase. Proteins were electroblotted to an Immobilon P membrane (Millipore) in a mini-V 8-10 apparatus (Gibco/BRL) with transfer buffer (24.8 mM Tris-HCl–192 mM glycine) adjusted to 10% (vol/vol) with methanol. Immune detection was carried out with the anti-HA antibody HA.11 (Babco) diluted 1:1,500 in phosphate-buffered saline containing 5% nonfat dry milk. The secondary antibody was a goat anti-mouse immunoglobulin G (heavy and light chains)-alkaline phosphatase conjugate (Gibco/BRL) developed with the 5-bromo-4-chloro-3-indolylphosphate (BCIP)–nitroblue tetrazolium chloride (NBT) chromogenic substrate mixture as recommended by the supplier (Gibco/BRL).
RESULTS
Selection of prp38-1 dosage suppressors.
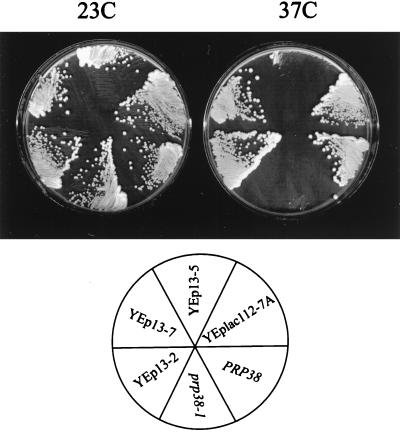
A yeast genomic library based on the high-copy-number plasmid YEp13 was used to select plasmids that reduced the temperature-sensitive growth defect caused by the prp38-1 mutation in yeast strain ts192 (5). From approximately 20,000 yeast transformants, 11 that showed plasmid-dependent colony formation at 37°C were identified. DNA restriction site analysis and hybridization studies on the recovered plasmids revealed that three distinct types of genomic inserts were recovered. Plasmids with type-A inserts (e.g., YEp13-2) or type-B inserts (e.g., YEp13-7) relieved the prp38-1 growth defect with high efficiency, while plasmids with type-C inserts (e.g., YEp13-5) suppressed it much more weakly (Fig. 1). The wild-type allele of PRP38 was present in both of the identified type-A plasmids. DNA sequence analysis showed that the type-C insert DNA was not present in the published yeast genome database or in other publicly held databases. Plasmids of categories A and C were not studied further.
FIG. 1.
Identification of dosage suppressors of the Ts prp38-1 mutation. Yeast cultures were streaked on nonselective medium and incubated at 23 and 37°C. The colony sizes of the untransformed prp38-1 mutant and wild-type (PRP38) yeast strains are compared with those of the prp38-1 mutant transformed with the indicated multi-copy plasmids. The plasmids encoded a functional PRP38 gene (YEp13-2), a weak suppressor of prp38-1 (YEp13-5), or an efficient suppressor of prp38-1 (YEp13-7 and YEplac112-7A).
Each of the seven type-B plasmids contained identical or overlapping regions of the right arm of yeast chromosome II. Two genes of defined function, RPB5 and RIB7, were present in these library segments. RPB5 encodes the 27-kDa subunit common to the three nuclear RNA polymerases (44). RIB7 encodes an activity required for riboflavin biosynthesis (7). Besides these genes, two uncharacterized ORFs, YBR151w and YBR152w, were present. Of these, YBR152w had the capacity to code for a small acidic protein (Fig. 2). Embedded within its amino terminus are two serine-rich elements similar to a sequence found in the carboxyl terminus of the Prp38p protein (5). The second of the YBR152w-encoded serine-rich elements contains an exceptionally strong match to the PEST protein degradation signal (amino acids 56 through 95). Prp38p does not contain the PEST motif. YBR152w was subcloned free of the adjacent genes and assayed for suppression in the high-copy-number shuttle vector YEplac112 (13). The YEplac112-7A subclone efficiently suppressed the ts192 temperature-sensitive growth defect, confirming that YBR152w contained the suppressor activity (Fig. 1). This dosage suppressor of prp38-1 was renamed SPP381, for suppressor of prp38-1.
FIG. 2.
Predicted amino acid sequence encoded by ORF YBR152w (SPP381). The acidic serine-rich elements common to Prp38p and Spp381p are underlined. The putative PEST sequence is represented by bold italics. Spp381p has a predicted molecular mass of 33.8 kDa and a predicted pI of 5.4. In comparison, Prp38p is a 28-kDa protein with a predicted pI of 5.0 (5).
Suppression by SPP381 is gene restricted, as evidenced by the fact that library plasmid YEp13-7 did not relieve the Ts growth defects of randomly selected prp2, prp5, prp11, prp9, prp17, prp19, prp20, or prp39 mutants. Conceivably, elevated Spp381p levels might influence the abundance, localization, or activity of the Prp38-1p protein. Alternatively, the increased abundance of Spp381p may bypass the cellular requirement for Prp38p. The latter possibility was ruled out with the demonstration that SPP381 on plasmid YEplac112-7A did not suppress a prp38::LEU2 null allele (reference 35) (data not shown). Thus, enhanced expression of SPP381 supports, but does not supplant, PRP38 activity.
Efficient pre-mRNA splicing is restored by enhanced SPP381 expression.
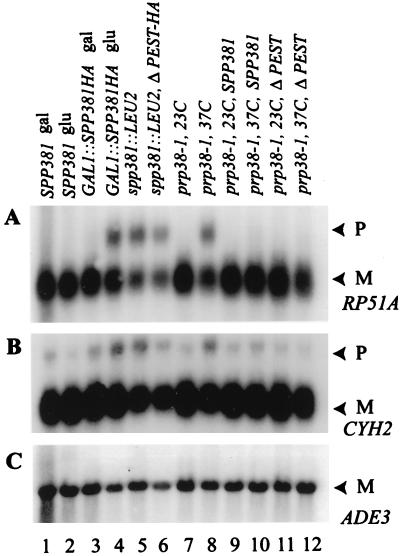
A Northern blot of cellular RNA was used to score for the impact of plasmid-borne SPP381 expression on pre-mRNA splicing efficiency (Fig. 3). The RP51A pre-mRNA–to–mRNA ratio of the wild-type parental strain was compared with that of the untransformed prp38-1 mutant and the prp38-1 mutant transformed with various suppressor plasmid constructs. RNA was extracted from cultures grown continuously at the permissive temperature of 23°C and from cultures shifted to the restrictive temperature of 37°C for 2.5 h. When assayed at the permissive temperature, all cultures showed abundant amounts of spliced RP51A mRNA and little pre-mRNA (Fig. 3, lanes 1 to 7). In contrast, the pre-mRNA/mRNA ratio increased greatly with the temperature shift in the untransformed mutant (Fig. 3, lane 13) and in mutants transformed with the weak suppressor (lane 9) or a randomly chosen library plasmid (lane 12). Plasmid-based expression of the wild-type PRP38 gene (Fig. 3, lane 8) or enhanced expression of SPP381 (lanes 10 and 11) decreased the pre-mRNA/mRNA ratio in the mutant strain to near-wild-type levels (lane 14). By this measure, enhanced SPP381 expression reverses the pre-mRNA processing defect caused by the prp38-1 mutation.
FIG. 3.
Influence of suppressor gene expression on cellular pre-mRNA splicing. RNA was isolated from the indicated yeast cultures grown continuously at 23°C and after a 2-h shift to 37°C. The hybridization probe consisted of exon and intron sequences of the yeast RP51A gene, and the positions of pre-mRNA (P) and mRNA (M) are noted. The cultures were from untransformed wild-type yeast (PRP38), the untransformed mutant strain ts192 (prp38-1), and the prp38-1 mutant after transformation with the indicated plasmids containing the PRP38 gene (YEp13-2), a weak suppressor of prp38-1 (YEp13-5), or an efficient suppressor of ts192 (YEp13-7 and YEplac112-7A). Plasmid YEp13-R was a negative-control plasmid from the YEp13 library that did not suppress the prp38-1 mutation.
SPP381 is an important but not an essential gene for normal yeast growth.
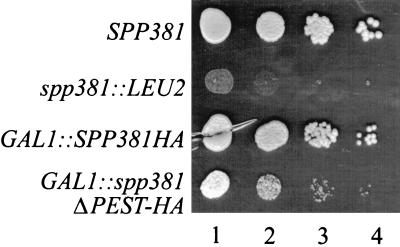
The SPP381 gene was disrupted by replacement of approximately 90% of its coding sequence with the LEU2 selectable marker. When diploid yeast cells heterozygous for this mutation were sporulated, equal numbers of colonies of two distinct sizes were observed in the meiotic offspring. PCR and Southern blot analyses showed that colonies visible after 2 days of incubation on the tetrad dissection plate all possessed the uninterrupted SPP381 allele. Yeast cells that formed visible colonies only after 6 to 7 days of incubation all possessed the spp381::LEU2 disruption. In liquid medium, the generation time of the spp381::LEU2 disruptant cultures was approximately seven times that of the wild-type siblings (10 to 11 versus 1.75 h). This mutant was not obviously heat or cold sensitive, as colony formation appeared equivalently poor at 17, 23, and 37°C compared with that of the SPP381 strain (Fig. 4 and data not shown). The limited SPP381 coding sequence still present in this strain did not contribute to the viability of this mutant, since a disruptant in which HIS3 replaced all sequences between the SPP381 translational initiation and termination codons showed equivalent results (10a). In both cases, the spp381::LEU2 slow-growth defect was specifically reversed by expression of the SPP381 ORF from the GAL1 promoter as an HA fusion construct (GAL1::SPP381HA) (Fig. 4 and data not shown). Given the approximate sevenfold decrease in the growth rate observed for the spp381::LEU2 mutant, SPP381 is clearly an important, albeit not an essential, gene in yeast. Deletion of the putative PEST motif from GAL1::SPP381HA (or from SPP381) greatly reduced its ability to complement spp381::LEU2, although such strains did grow slightly better than the mutant bearing the null allele alone (Fig. 4). Thus, the proposed PEST sequence contributes to Spp381p biological activity in vivo.
FIG. 4.
Comparison of growth in SPP381 and spp381::LEU2 mutant yeast cells. Yeast cultures were grown to saturation in nonselective broth with 2% galactose. Each strain was adjusted to a culture density at 600 nm of 0.150. The presence of equivalent cell numbers in each culture was confirmed microscopically. Serial 10-fold dilutions (positions 1 to 4) were spotted in 5-μl volumes on galactose-containing agar medium and incubated for 4 days at 30°C. The strains used were the wild-type parent (SPP381), the untransformed spp381::LEU2 mutant, and the spp381::LEU2 mutant transformed with the GAL1::SPP381HA fusion gene or its ΔPEST-HA derivative.
Spp381p contributes to cellular pre-mRNA splicing.
A direct contribution of Spp381p to pre-mRNA splicing might underlie the genetic interaction between prp38-1 and SPP381. To address this, RNA samples isolated from a wild-type strain and from the spp381::LEU2 disruptant strain were analyzed by Northern blotting (Fig. 5). Indicative of decreased splicing efficiency, yeast with the spp381::LEU2 disruption showed a greatly elevated ratio of RP51A pre-mRNA to mRNA (Fig. 5A, lane 5) compared with the wild-type strain (Fig. 5A, lanes 1 and 2). Primer extension carried out with an RP51A exon II primer showed that most of this intron-bearing RNA was pre-mRNA rather than lariat intermediate, indicating that splicing was inhibited before 5′ splice site cleavage. As a control for the specificity, a spp381::LEU2 strain transformed with the GAL1::SPP381HA fusion gene was assayed in parallel. The spp381::LEU2 splicing defect was specifically reversed by GAL1::SPP381HA under conditions that induced transcription of this fusion gene (i.e., growth on galactose). Similar to what has been reported for other splicing factors (see references 21 and 25 and references therein), splicing was inhibited 10 to 15 h after transcriptional repression of GAL1::SPP381HA (Fig. 5A, lanes 3 and 4, and data not shown). While results with the CYH2 gene were somewhat less pronounced, transcripts of the CYH2 gene also showed decreased levels of mRNA and increased levels of pre-mRNA in the spp381::LEU2 mutant background (Fig. 5B). The more modest CYH2 defect suggests that not all introns are equally dependent upon Spp381p protein for excision. We note also that, compared with RP51A, the splicing of CYH2 precursors was somewhat less sensitive to Prp38-1p temperature inactivation (Fig. 5A and B, lanes 7 and 8). No reproducible changes with the intronless ADE3 mRNA, rRNA, or trimethylguanosine-capped snRNAs were associated with the spp381::LEU2 mutation (Fig. 5C) (unpublished data). Thus, the growth impediment of the spp381::LEU2 mutant can be accounted for by decreased pre-mRNA splicing efficiency in the absence of Spp381p. Unlike the prp38-1 mutation, the spp381::LEU2 splicing defect was not suppressed by overexpression of PRP38 on a high-copy-number plasmid or as a GAL1::PRP38 fusion gene. Together, these data provide strong evidence that PRP38 and SPP381 make important and independent contributions to cellular pre-mRNA splicing.
FIG. 5.
Contribution of SPP381 to the efficiency of pre-mRNA splicing in vivo. RNA was isolated from wild-type yeast (lanes 1 and 2) and the spp381::LEU2 disruptant before (lane 5) and after transformation with GAL1::SPP381HA (lanes 3 and 4) or with GAL1::spp381ΔPEST-HA (lane 6). Galactose (gal) or glucose (glu) was used to activate or repress the GAL1 fusion constructs as indicated. Lanes 7 to 12, RNA from the untransformed prp38-1 mutant (lanes 7 and 8) and the same strain after transformation with the high-copy-number (i.e., YEp112) plasmid bearing SPP381 (lanes 9 and 10) or its ΔPEST (YEplac112-based) derivative (lanes 11 and 12). The RNA was recovered from cultures grown continuously at the permissive temperature for prp38-1 (23°C) or after 2.5 h at the restrictive temperature (37°C). The positions of pre-mRNA (P) and spliced mRNA (M) are indicated by arrowheads. (A) Hybridization with an RP51A-specific intron-plus-exon probe. (B) Hybridization with a CYH2-specific intron-plus-exon probe. (C) Hybridization with an ADE3 gene body probe.
Yeast that expressed GAL1::ΔPEST-HA spliced RP51A pre-mRNA poorly (Fig. 5A and B, lanes 6) consistent with its weak complementation of the spp381::LEU2 mutation. Curiously, however, the high-copy-number ΔPEST construct continued to suppress the temperature-dependent splicing defect of the prp38-1 mutant (Fig. 5A and B, lanes 7 to 12). Thus, Spp381p retained an aspect of its biological activity even in the absence of the PEST sequence. In addition, the splicing deficiency of ΔPEST suggests that Spp381p may contribute to splicing through a step independent of its proposed interaction with Prp38p.
Spp381p is found in the U4/U5.U6 tri-snRNP particle.
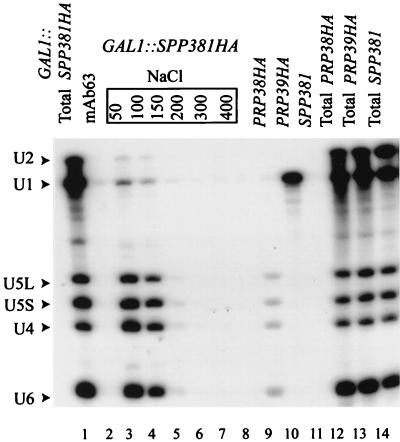
Prp38p is one of a small group of proteins uniquely associated with the U4/U6.U5 tri-snRNP particle (45). Immune precipitation was used to determine whether the genetically interacting protein, Spp381p, was likewise associated with snRNP. In all extracts tested, the total (i.e., unfractionated) snRNA levels were equivalent (Fig. 6, lanes 1 and 12 to 14). Extracts prepared from the GAL1::SPP381HA and control strains were incubated with the anti-HA antibody HA.11 or with the irrelevant control antibody mAb63. A Northern blot of the immune pellets washed at 100 mM NaCl revealed that, as with Prp38HAp (Fig. 6, lane 9), the U4, U5, and U6 snRNAs specifically coprecipitated with Spp381HAp (lane 4). In contrast, U1 snRNA was recovered with the HA-tagged U1 snRNP protein, Prp39p (Fig. 6, lane 10). The snRNA precipitation with Spp381HAp was salt sensitive; the levels of U4, U5, and U6 snRNAs were greatly reduced at 150 mM NaCl (Fig. 6, lanes 3 to 8). Nevertheless, the U4, U5, and U6 snRNA recovery was specific, as evidenced by the fact that almost no U-snRNAs were recovered at 100 mM NaCl from an untagged extract (Fig. 6, lane 11) or when the irrelevant monoclonal antibody, mAb63, was used with the GAL1::SPP381HA extract (lane 2). The lower level of snRNA recovery with the Prp38HAp extract (compared with that observed for Spp381HAp) may reflect a lower Prp38HAp abundance or reduced antibody accessibility. This was not due to a lower affinity of Prp38HAp for the U4/U6.U5 particle, however, as this interaction is stable to at least 200 mM NaCl (45).
FIG. 6.
Coprecipitation of snRNA with HA-tagged proteins. Extracts of untagged yeast (lanes 11 and 14) and HA-tagged Spp381HAp (lanes 1 to 8), Prp38HAp (lanes 9 and 12), and Prp39HAp (lanes 10 and 13) were immune precipitated with the HA-specific antibody HA.11 (lanes 3 to 10) or the irrelevant antibody mAb63 (lane 2). The immune pellets were fractionated on a denaturing 5% polyacrylamide gel and transferred to a membrane, and the blot was hybridized with probes specific for the spliceosomal snRNAs (indicated by arrowheads). Immune pellets in lanes 3 to 8 were washed with buffer containing the indicated levels of NaCl; all other pellets were washed with buffer adjusted to 100 mM NaCl. For comparison of relative snRNAs, nonprecipitated extract RNAs (Total) were resolved in parallel (lanes 1 and 12 to 14).
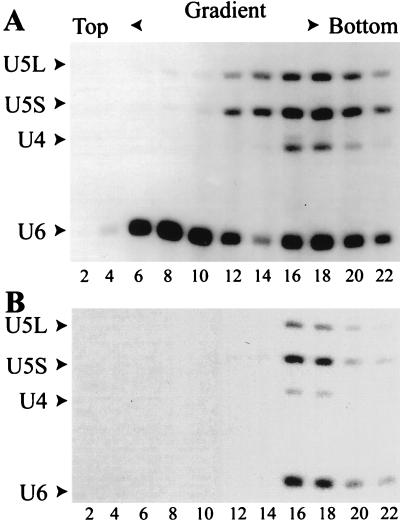
Based on the immune precipitation results, it appeared that Spp381p was present in the U4/U6.U5 tri-snRNP particle. This prediction was tested by glycerol gradient fractionation of the Spp381HAp splicing extract (Fig. 7). As described previously (6, 45), this fractionation technique resolves the free U6 snRNP particles (Fig. 7A, fractions 6 to 10) and free U5 snRNP particles (Fig. 7A, fractions 12 and 14) from the U4/U6.U5 tri-snRNP particles (fractions 16 and 18). The U4, U5, and U6 snRNAs coprecipitate efficiently with the HA.11 antibody only from the tri-snRNP fractions (Fig. 7B, fractions 16 and 18), even though both U6 and U5 are abundantly present elsewhere in the gradient. The cofractionation and coprecipitation results provide clear evidence for Spp381HAp association with the U4/U6.U5 tri-snRNP particle.
FIG. 7.
Cofractionation of Spp381HAp with the U4/U6.U5 tri-snRNP particle. The yeast splicing extract was fractionated on a linear 10 to 35% glycerol gradient. (A) Even-numbered fractions were assayed by Northern blotting for the presence of U5 (the long and short forms, U5L and U5S, respectively), U4, and U6 snRNA. (B) Spp381HAp was immune precipitated from the fractions presented in panel A with the anti-HA antibody HA.11 and was assayed for coassociated snRNAs by Western blotting with the same antibody.
Prp38p and Spp381p interact in vivo.
The dosage suppression and snRNA precipitation studies showed that Spp381p and Prp38p interact genetically and associate with the same biochemical complex. A two-hybrid analysis was next performed to investigate the possibility that these two proteins associate directly. Simultaneous expression of Spp381p and Prp38p as Gal4 fusion products led to strong transactivation of the lacZ and HIS3 reporter genes in the host strain (Table 1 and data not shown). A Spp381–Gal4 binding domain plasmid did not transactivate when paired with an empty activation domain vector or when the PRP38 DNA was inserted in the reverse orientation. As a first step in assessing the domain structure of the Spp381p and Prp38p molecules, fusion constructs consisting of the amino- and carboxyl-terminal halves were used for two-hybrid analysis. For unknown reasons, activation domain-independent stimulation with Spp381p(146–291) was about twofold greater than that observed with full-length Spp381p. Nevertheless, sequences necessary and sufficient for the Prp38p interaction clearly reside in the Spp381p carboxyl terminus, since Spp381p(146–291) and Spp381p(1–291) stimulated Prp38p-dependent transactivation to similar levels. In contrast, Spp381p amino acids 1 to 145 failed to interact with Prp38p. Thus, the putative PEST sequence, although important for the Spp381p function in splicing, is not required for interaction with Prp38p. Spp381p(146–291) did not interact with Prp38p(1–121) or Prp38p(122–242), suggesting that the interacting domain may comprise (or span) both halves of Prp38p. Supporting evidence for a critical N-terminal interaction was provided by the observation that either of two Ts mutations in this region, G66D (prp38-1) and C87Y (prp38-2), greatly reduced the level of transactivation with Spp381p(1–291). Overall, the two-hybrid results support an interaction between the carboxyl terminus of Spp381p and one or more regions of Prp38p.
TABLE 1.
Analysis of interaction between Spp381p and Prp38p using the two-hybrid system
| Protein fusion
|
β-Galactosidase activity (U) (avg ± SD)a | |
|---|---|---|
| Binding domain | Activation domain | |
| Spp381p(1–291) | pACT | 0.77 ± 0.19 |
| Spp381p(1–291) | Prp38p(reverse)b | 1.06 ± 0.10 |
| Spp381p(1–291) | Prp38p(1–242) | 16.99 ± 0.57 |
| Spp381p(1–291) | Prp38-1p(1–242); G66Dc | 1.56 ± 0.12 |
| Spp381p(1–291) | Prp38-2p(1–242); C87Yc | 1.51 ± 0.28 |
| Spp381p(1–291) | Prp38p(1–121) | 0.69 ± 0.10 |
| Spp381p(1–291) | Prp38p(122–242) | 0.68 ± 0.10 |
| Spp381p(1–145) | Prp38p(1–242) | 0.71 ± 0.23 |
| Spp381p(146–291) | Prp38p(1–242) | 15.81 ± 1.57 |
| Spp381p(1–145) | Prp38p(1–121) | 0.65 ± 0.15 |
| Spp381p(1–145) | Prp38p(122–242) | 0.59 ± 0.23 |
| Spp381p(146–291) | Prp38p(1–121) | 3.91 ± 0.29 |
| Spp381p(146–291) | Prp38p(122–242) | 3.60 ± 0.69 |
| Spp381p(146–291) | Prp38p(reverse 1–121)d | 3.66 ± 0.60 |
Averages from three independent assays on yeast cultures grown at 30°C.
The PRP38 fragment was inserted in the reverse orientation in the activation domain vector.
Temperature-sensitive mutant of Prp38p.
The PRP38 fragment encoding the amino terminus was inserted in the reverse orientation in the activation domain vector.
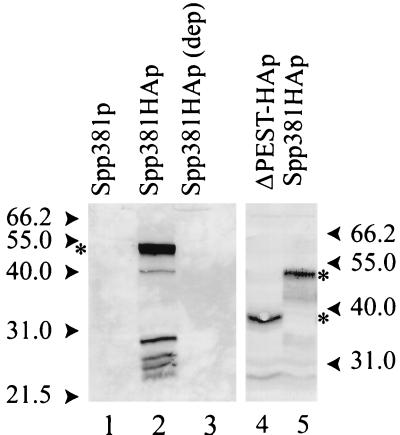
The Spp381HAp fusion protein has a predicted molecular size of approximately 35 kDa. On polyacrylamide gels, however, Spp381HAp migrates as a 51-kDa protein (Fig. 8, lanes 1 and 2). This anomalous migration might be caused by the highly acidic amino terminus of this protein, which also contains numerous possible sites for phosphorylation. Consistent with this, removal of the 4.6-kDa PEST sequence deletion caused an apparent 13-kDa shift to produce a protein with an apparent mass of 38 kDa (Fig. 8, lanes 4 and 5; predicted mass, 30.3 kDa). Equivalent amounts of the Spp381HAp and ΔPEST-HAp derivatives accumulated in cells when expressed from the GAL1 promoter, indicating that under these conditions the PEST sequence contributes little to stability. Curiously, given the importance of Spp381p to in vivo splicing, extracts prepared from the glucose-depleted GAL1::SPP381HA culture (Fig. 8, lane 3) were found to splice RP51A pre-mRNA through both chemical steps in vitro (data not shown). Thus, it appears that either Spp381p is not required in vitro, or it contributes to an activity not assayed under typical in vitro splicing conditions (see Discussion).
FIG. 8.
Identification of Spp381HAp. Sixty micrograms of yeast extract protein was resolved on 12% (lanes 1 to 3) and 10% (lanes 4 and 5) polyacrylamide gels. A Western blot from each gel was hybridized with the anti-HA antibody HA.11 to reveal the presence of Spp381HAp and its ΔPEST-HAp derivative (indicated by asterisks). Numbers on the left and right show the positions of protein molecular weight markers (mid-range; Promega) run in adjacent lanes. The extracts assayed included an untagged yeast strain (lane 1), a GAL1::SPP381HA strain grown continuously in galactose (lanes 2 and 5) or shifted to a glucose-based medium for 18 h (lane 3), and extract prepared from the GAL1::ΔPEST-HA derivative (lane 4).
DISCUSSION
Prp38p was recently shown to be necessary for dissociation of the U4/U6 intermolecular helices (45), an essential maturation step that occurs prior to pre-mRNA 5′ splice site cleavage. In this study, we used the genetic approach of dosage suppression to identify a novel protein, Spp381p, that contributes to Prp38p function in splicing. The genetic and biochemical evidence indicates that Spp381 and Prp38p define a novel class of interacting acidic proteins which promote U4/U6.U5 tri-snRNP activity in the spliceosome cycle.
Multiple models for dosage suppression have been described based on kinetic or thermodynamic contributions of the overexpressed gene product to the process under study (for instance, see references 20 and 32). The fact that extra copies of SPP381 do not suppress a prp38::LEU2 null allele shows convincingly that increased Spp381p levels do not bypass the need for Prp38p in splicing. In principle, the elevated abundance of Spp381p might increase the stability or residual activity of the temperature-sensitive prp38-1 gene product. The latter suggestion appears more likely, since under a variety of conditions, we find no evidence for increased Prp38-1p abundance with enhanced Spp381p expression (unpublished data). While this result and the positive two-hybrid data are consistent with direct contact between Prp38p and Spp381p, we cannot rule out the possibility that a third component mediates this interaction. However, in the absence of data supporting a such a third factor, we favor the view that the genetic suppression occurs due to an increased frequency of Spp381p interaction with a functionally impaired Prp38-1p protein. An Spp381p–Prp38-1p interaction may promote a favorable structural change within Prp38-1p to facilitate the binding of Prp38-1p to the U4/U6.U5 tri-snRNP, promote Prp38-1 interaction with other splicing components, or in some other manner enhance the activity of the essential Prp38p protein.
Since Prp38p appears to be exclusively a U4/U6.U5 tri-snRNP protein (45), it is likely within this particle that Spp381p normally contacts Prp38p. Previously, two uncharacterized proteins similar to Spp381p in size were reported in the yeast tri-snRNP (11). Glycerol gradient fractionation of yeast snRNP complexes presented here demonstrates the presence of Spp381HAp in the U4/U6.U5 tri-snRNP particle and the absence of antibody-accessible Spp381HAp in the free U5 or free U6 snRNP complexes. It remains possible, however, that Spp381HAp binds to the low-abundance U4/U6 di-snRNP precursor (19, 23, 26) or is present in an antibody-inaccessible form in other snRNP complexes. Both the two-hybrid results presented here and the presence of Spp381HAp in an immune pellet prepared with an anti-Prp38p antibody (32) support the view that Spp381p and Prp38p are ubiquitous (as opposed to alternative) components of the U4/U6.U5 tri-snRNP particle. Prp38p and Spp381p are clearly “weakly associated” snRNP proteins, similar in salt sensitivity to the phylogenetically conserved SF3a proteins of the 17S U2 snRNP (reference 3; see also references in reference 19), U1C (39), U1-Prp42p (25), and Prp38p tri-snRNP protein (45). No clear counterpart to Spp381p is known in mammals, although at least one small, highly charged phosphoprotein is a component of the mammalian U4/U6.U5 tri-snRNP particle (12).
What is the role of Spp381p in splicing? In vivo, enhanced SPP381 expression relieves the block to pre-mRNA 5′ splice site cleavage imposed by the loss of Prp38p function. This observation suggests that Spp381p contributes to snRNP rearrangement events that lead to the catalytic activation of the spliceosome. Consistent with this, deletion of the SPP381 gene severely impairs yeast growth and inhibits step 1 in splicing. Surprisingly, we have not detected in vitro splicing defects in extracts prepared from glucose-repressed GAL1::SPP381HA cultures. U4 snRNA is released from the spliceosome, and both chemical steps in splicing occur unimpeded. Since SPP381 affects the efficiency (but not the absolute occurrence) of splicing in vivo, the lack of an obvious in vitro defect may indicate that Spp381p function is not rate limiting for the comparatively slow in vitro reaction. Alternatively, Spp381p function may be important in vitro but its function may not be obvious under standard assay conditions. Ample precedent exists for such behavior by splicing factors. For instance, extracts deficient in Prp22p, Prp24p, or Prp43p can correctly process pre-mRNA through both chemical steps in splicing but are defective in postsplicing steps of mRNA release (Prp22p [10]), U4/U6 snRNA reanealing (Prp24p [30, 42]), and intron release (Prp43p [1]). The spliceosomal precursor pool in the Spp381HAp-depleted culture is an unknown and may increase by the release of endogenous spliceosomes during the 8-h extract preparation protocol. If Spp381p acts as a recycling or dissociation factor, its importance might not be obvious until the spliceosomal precursors become saturated with, or depleted of, the exogenously added pre-mRNA, as shown for Prp24p (30).
It is well established that the removal of a PEST sequence can greatly increase the half-life of an unstable protein (e.g., c-Fos [40]) and that the transfer of a PEST sequence to a naturally stable protein can greatly enhance its turnover (e.g., dihydrofolate reductase [22]; see references 31 and 33 for additional examples of PEST-mediated destabilization). Proteins with functionally defined PEST elements often have PEST values in the range of 4 to 16 (e.g., yeast Gcn4, mammalian Fos, ornithine decarboxylase, Aspergillus NIMA). The 40-amino-acid Spp381p element has a PEST sequence value of +29.8 and ranks higher than all of the 99 PEST elements presented in two recent reviews (2, 31). The presence of a PEST sequence raises the interesting possibility that proteolysis of Spp381p triggers a particular event of the spliceosome cycle. Many PEST-mediated proteolysis events are regulated (31). Perhaps the most obvious role for proteolysis would be to promote spliceosome disassembly following pre-mRNA splicing. A spliceosome-dependent turnover would explain the similar intracellular levels of Spp381HAp and its ΔPEST-HA derivative when these proteins are overexpressed (beyond the needs of splicing) by the GAL1 promoter. Intriguingly, while uncommon in spliceosomal proteins, the DExD/H-box proteins Prp22p, Prp28p, Brr2p, Prp43p, and Prp16p all possess good fits to the PEST consensus (PEST scores of 14.5, 13.4, 11.3, 7.3, and 5.7, respectively). Although highly speculative, it is conceivable that PEST sequence-mediated modification (i.e., phosphorylation or proteolysis subsequent to function rather than dissociation accounts for the “transient” association of members of this group with the spliceosome (see reference 38). Experiments are under way to define the function of Spp381p in splicing and to test the possibility that proteolysis contributes to the spliceosome cycle.
ACKNOWLEDGMENTS
We thank Frances McFarland, Martha Peterson, John Woolford, and our lab colleagues Seyung Chung, Mitch McLean, and Liz Otte for their helpful comments on the manuscript.
This work was supported by an HHMI summer support fellowship to V.B. and by National Institutes of Health grant GM42476 (to B.C.R.).
REFERENCES
- 1.Arenas J E, Abelson J N. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes J A, Gomes A V. PEST sequences in calmodulin-binding proteins. Mol Cell Biochem. 1995;149/150:17–27. doi: 10.1007/BF01076559. [DOI] [PubMed] [Google Scholar]
- 3.Behrens S-E, Tyc K, Kaster B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Beickman, K., and B. C. Rymond. Unpublished data.
- 4.Berben G, Dumont J, Gilliquet V, Bolle P-A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 5.Blanton S, Srinivasan A, Rymond B C. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol Cell Biol. 1992;12:3939–3947. doi: 10.1128/mcb.12.9.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordonne R, Banroques J, Abelson J, Guthrie C. Domains of yeast U4 spliceosomal RNA required for PRP4 protein binding, snRNP-snRNP interactions, and pre-mRNA splicing in vivo. Genes Dev. 1990;4:1185–1196. doi: 10.1101/gad.4.7.1185. [DOI] [PubMed] [Google Scholar]
- 7.Buitrago M J, Gonzalez G A, Saiz J E, Revuelta J L. Mapping of the RIB1 and RIB7 genes involved in the biosynthesis of riboflavin in Saccharomyces cerevisiae. Yeast. 1993;9:1099–1102. doi: 10.1002/yea.320091009. [DOI] [PubMed] [Google Scholar]
- 8.Burgess S M, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci. 1993;18:381–834. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- 9.Cheng S-C, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- 10.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 10a.Dembla-Rajpal, N., and B. C. Rymond. Unpublished data.
- 11.Fabrizio P, Esser S, Kastner B, Lührmann R. Isolation of S. cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science. 1994;264:261–265. doi: 10.1126/science.8146658. [DOI] [PubMed] [Google Scholar]
- 12.Fetzer S, Lauber J, Will C L, Lührmann R. The U4/U6.U5 tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA. 1997;3:344–355. [PMC free article] [PubMed] [Google Scholar]
- 13.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 14.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 16.Kim S-H, Lin R-J. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc Natl Acad Sci USA. 1993;90:888–892. doi: 10.1073/pnas.90.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S-H, Lin R-J. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konarska M M, Sharp P A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 19.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 20.Last R L, Maddock J R, Woolford J L., Jr Evidence for related functions of the RNA genes of Saccharomyces cerevisiae. Genetics. 1987;117:619–631. doi: 10.1093/genetics/117.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lockhart S, Rymond B C. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol Cell Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loetscher P, Pratt G, Rechsteiner M. The C terminus of mouse ornithine decarboxylase confers rapid degradation on dihydrofolate reductase. Support for the PEST hypothesis. J Biol Chem. 1991;266:11213–11220. [PubMed] [Google Scholar]
- 23.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka S, Edwards M C, Bai C, Parker S, Zhang P, Baldini A, Harper J W, Elledge S J. P57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 25.McLean M R, Rymond B C. Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol Cell Biol. 1998;18:353–360. doi: 10.1128/mcb.18.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNA by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–358. [Google Scholar]
- 27.O’Day C L, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 28.Pikielny C W, Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell. 1986;45:869–877. doi: 10.1016/0092-8674(86)90561-1. [DOI] [PubMed] [Google Scholar]
- 29.Pikielny C W, Rymond B C, Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly of yeast splicing complexes. Nature. 1986;324:341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- 30.Raghunathan P L, Guthrie C. A spliceosome recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 31.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 32.Rine J. Gene overexpression in studies of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:239–251. doi: 10.1016/0076-6879(91)94019-9. [DOI] [PubMed] [Google Scholar]
- 33.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–386. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 34.Rosbash M, Seraphin B. Who’s on first? The U1 snRNP-5′ splice site interaction and splicing. Trends Biochem Sci. 1991;16:187–190. doi: 10.1016/0968-0004(91)90073-5. [DOI] [PubMed] [Google Scholar]
- 35.Rymond B C. Convergent transcripts of the yeast PRP38-SMD1 locus encode two essential splicing factors, including the D1 core polypeptide of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1993;90:848–852. doi: 10.1073/pnas.90.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 39.Tang J, Abovich N, Fleming M L, Seraphin B, Rosbash M. Identification and characterization of the yeast homolog of U1 snRNP-specific protein C. EMBO J. 1997;16:4082–4091. doi: 10.1093/emboj/16.13.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsurumi C, Ishida N, Tamura T, Kakizuka A, Nishida E, Okumura E, Kishimoto T, Inagaki M, Okazaki K, Sagata N, Ichihara A, Tanaka K. Degradation of c-Fos by the 26S proteasome is accelerated by c-Jun and multiple protein kinases. Mol Cell Biol. 1995;15:5682–5687. doi: 10.1128/mcb.15.10.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umen J G, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- 42.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Wagner J D O, Guthrie C. Prp16, a DEAH-box splicing factor, unwinds RNA duplexes in vitro. Curr Biol. 1998;8:441–451. doi: 10.1016/s0960-9822(98)70178-2. [DOI] [PubMed] [Google Scholar]
- 44.Woychik N A, Liao S M, Kolodziej P A, Young R A. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 1990;4:313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- 45.Xie J, Beickman K, Otte E, Rymond B C. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 1998;17:2938–2946. doi: 10.1093/emboj/17.10.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu D, Nouraini S, Field D, Tang S J, Friesen J D. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature. 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- 47.Yean S-L, Lin R-J. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol Cell Biol. 1991;11:5571–5577. doi: 10.1128/mcb.11.11.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]