Abstract
A novel cyclin gene was discovered by searching an expressed sequence tag database with a cyclin box profile. The human cyclin E2 gene encodes a 404-amino-acid protein that is most closely related to cyclin E. Cyclin E2 associates with Cdk2 in a functional kinase complex that is inhibited by both p27Kip1 and p21Cip1. The catalytic activity associated with cyclin E2 complexes is cell cycle regulated and peaks at the G1/S transition. Overexpression of cyclin E2 in mammalian cells accelerates G1, demonstrating that cyclin E2 may be rate limiting for G1 progression. Unlike cyclin E1, which is expressed in most proliferating normal and tumor cells, cyclin E2 levels were low to undetectable in nontransformed cells and increased significantly in tumor-derived cells. The discovery of a novel second cyclin E family member suggests that multiple unique cyclin E-CDK complexes regulate cell cycle progression.
The eukaryotic cell cycle is regulated by a family of cyclin-dependent kinases (CDKs) that are cyclically activated to trigger the different phases of the cell cycle in the correct order and at the right time. CDKs are regulated by a number of different proteins, including the cyclins that bind and activate the CDKs to form a serine/threonine kinase holoenzyme complex. In mammals, D-type cyclins in conjunction with cyclins E and A are required for cells to traverse G1 and enter S phase (19, 25, 29). There are three members of the D-type cyclins, two members of the cyclin A family, and, to date, one mammalian cyclin E gene (33). The formation of distinct G1 cyclin-CDK complexes regulates the temporal phosphorylation of specific substrates that drive cells through G1 and into S phase. Cyclin-CDK complexes are negatively regulated by two families of CDK inhibitors (34). The Ink4 family of CDK inhibitors exclusively regulate D-type cyclin-CDK complexes, while the Kip/Cip family regulate D-type cyclins as well as CDK complexes comprised of cyclin E or A. p27Kip1 and p21Cip1 bind and inhibit cyclin E-Cdk2 complexes by acting as competitive inhibitors for substrates and by preventing cyclin-activating kinase (CAK)-mediated phosphorylation and activation of Cdk2 (23, 34).
Cyclin E was originally discovered by its ability to complement the G1 cyclins in budding yeast (16, 18). This observation suggested that cyclin E regulated the progression of cells through the cell cycle. It was later shown that cyclin E-Cdk2 complexes stimulate mammalian cells to traverse G1 and enter S phase by the temporal phosphorylation of specific substrates such as the retinoblastoma tumor suppressor protein (Rb) (17, 20, 33). In fission yeast, a critical size must be reached prior to entry into mitosis, and this is normally regulated by coupling Cdk1 activation to cell growth. Premature activation of Cdk1 in fission yeast causes cells to enter mitosis at a reduced size. A similar effect occurs when cyclin E is overexpressed three- to fourfold in mammalian cells; the cells have a shorter G1 and enter mitosis at a reduced size (25, 29). These results, combined with experiments demonstrating that cyclin E is required for the G1/S transition, suggest that cyclin E catalyzes rate-limiting steps that are essential for entry into S phase. While cyclin E complexes phosphorylate Rb, it is likely that additional substrates exist during late G1. In support of this idea, cyclin E mutants that retain H1 kinase activity but are unable to phosphorylate Rb are still able to accelerate G1 in cells lacking Rb (15). Also, the recent observation that nuclear protein mapped to the AT locus is a substrate for cyclin E-Cdk2, but not cyclin A-Cdk2 complexes, demonstrates that unique substrates exist for cyclin E (42).
Aberrant regulation of cell cycle control is one of the hallmarks of tumorigenesis. The expression and activity of a number of cell cycle regulators is perturbed during tumor progression. Cyclin E is often overexpressed in human breast cancers, where its activity is predictive of tumor aggression and mortality (3, 14, 27). The level of cyclin E in tumors is independent of the proliferation index, suggesting that overexpression of cyclin E may be a cause rather than an effect of tumorigenesis. Researchers have also demonstrated that in tumor-derived cell lines lacking active cyclin D-Cdk4 complexes, cyclin E levels are elevated and cyclin E-Cdk2 complexes are able to function redundantly to phosphorylate and inactivate Rb (7). Thus, cyclin E plays an integral role in regulating transit through the normal cell cycle, and the loss of cyclin E regulation is associated with tumor progression.
We have discovered novel mouse and rat cDNAs that are similar, but not homologous, to cyclin E. We have used these partial DNA sequences to isolate full-length murine and human cDNAs which encode a gene with a characteristic cyclin box motif and 47% similarity to cyclin E. Based on its similarity to cyclin E, we have named this novel gene cyclin E2. The expression of cyclin E2 is regulated in a cell cycle-specific manner in human breast epithelial cells, and cyclin E2 binds Cdk2 in a catalytically active complex. Discovery of a novel cyclin E family member demonstrates that, in addition to the previously characterized cyclin E1, alternative cyclin E2 complexes that modulate cell cycle progression may exist.
MATERIALS AND METHODS
Cells and cell culture.
Saos-2 cells, IMR-90 human fibroblasts, and 293T cells were obtained from the American Type Culture Collection and cultured in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) supplemented with 5 mM glutamine and penicillin-streptomycin. Normal human bronchial and cervical epithelial cells were purchased from Clonetics Corp. (San Diego, Calif.). The immortalized breast epithelial cell line MCF10 was generously provided by Steve Ethier, University of Michigan (5). The lung cancer cell lines H1299, H23, H358, H441, H460, H520, H522, H727, H146, H209, H446, H510A, H526, and H889 and the cervical cancer cells Caski, C-4-I, MS751, SiHa, and C-33-A were all obtained from the American Type Culture Collection. Normal cervical epithelial cells were culture in KBM2 (Clonetics) supplemented with bovine pituitary extract (5.2 mg/ml), hydrocortisone (1 μg/ml), epidermal growth factor (EGF; 2 ng/ml), epinephrine (1 μg/ml), retinoic acid (0.1 ng/ml), transferrin (5 μg/ml), triiodothyronine (6.5 ng/ml), and insulin (5 μg/ml). Normal bronchial epithelial cells were cultured in BEBM (Clonetics) supplemented with bovine pituitary extract (5.2 μg/ml), hydrocortisone (1 μg/ml), EGF (0.5 ng/ml), epinephrine (0.5 mg/ml), transferrin (10 μg/ml), insulin (5 μg/ml), retinoic acid (0.1 ng/ml), and triiodothyronine (5.5 ng/ml). MCF10 cells were cultured in a modified DMEM-F12 (Gibco) supplemented with 10 mM HEPES, 2 mM glutamine, 0.1 mM nonessential amino acids, 0.5 mM ethanolamine, transferrin (5 μg/ml), bovine serum albumin (1 mg/ml), sodium selenite (5.0 ng/ml), triiodothyronine (20 ng/ml), EGF (10 ng/ml), insulin (5 μg/ml), and hydrocortisone (0.5 μg/ml) (DMEM-F12C) (4). For synchronization, the cells were plated and allowed to grow for 2 days. At this time, DMEM-F12C was removed and incubation continued in the supplemented medium described above without the growth factors EGF and insulin. The lung cancer cells were cultured in RPMI (minimal essential medium; Gibco) supplemented with 10 mM HEPES, 2 mM glutamine, 0.1 mM nonessential amino acids, and 10% FBS. The cervical cancer cells were cultured in Earle’s minimal essential medium supplemented with 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 10% FBS. All cells were routinely screened for mycoplasma contamination and maintained at 37°C in an atmosphere of 6.5% CO2.
Transfections, Western blotting, immunoprecipitations, and antibodies.
Cells were lysed in a modified radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 10% glycerol, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate [DOC], 20 mM HEPES [pH 7.4], 100 mM NaCl) containing protease and phosphatase inhibitors (10 μg each of aprotinin, leupeptin, and pepstatin per ml, 50 mM NaF, 1 mM sodium vanadate). Protein concentrations were determined by the Bradford assay, and 50-μg aliquots of cell extracts were electrophoresed on 12% polyacrylamide gels. After being transferred to polyvinylidene difluoride membranes, proteins were detected by incubation in appropriate primary antibodies followed by horseradish peroxidase-conjugated protein A or horseradish peroxidase-conjugated protein A/G (Sigma), and enhanced chemiluminescence detection was performed as described by the manufacturer (Amersham). Immunoprecipitation of lysates from cells was performed as follows. Approximately 200 μl of cell lysates normalized for protein concentrations was incubated at 4°C for 1 h with appropriate dilutions of antibodies, followed by the addition of 50 μl of a 50% slurry of protein A/G-Sepharose (Pharmacia) suspended in modified RIPA buffer minus the SDS and DOC. After rotating for 30 min at 4°C, the beads were pelleted and washed four times with modified RIPA buffer (minus SDS and DOC), then quenched in SDS sample loading buffer, and separated by SDS-polyacrylamide gel electrophoresis followed by Western transfer where indicated. Kinase assays were performed as previously described (17). Histone H1 was obtained from Sigma (St. Louis, Mo.), glutathione S-transferase (GST)–Rb (C-terminal fragment), and GST-p53 were obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Monoclonal antibodies to the hemagglutinin (HA) epitope tag were obtained from Boehringer Mannheim (Indianapolis, Ind.). Monoclonal antibodies to the Myc epitope tag were obtained from Santa Cruz. Antibodies to CD20 were obtained from Becton Dickinson (San Jose, Calif.). Antibodies to green fluorescent protein (GFP) were obtained from Clontech (Palo Alto, Calif.). p27, Cdk2, cyclin A, and cyclin E antibodies were obtained from Santa Cruz. Cyclin E2 rabbit polyclonal antibodies were raised against a 21-mer peptide (H2N-AKQQPQPSQTESPQEAQIIQA-COOH) derived from the N terminus of human cyclin E2. The cyclin E2 antiserum was affinity purified by using a protein A/G-agarose column as specified by the manufacturer (Pierce, Rockford, Ill.).
Cells were transfected with the indicated plasmids, using either the calcium phosphate precipitation method or liposome-mediated delivery according to the manufacturer’s directions. For flow cytometry experiments, 3 μg of CD20 plasmid and 1 μg of HA epitope-tagged Cdk2 plasmid were transfected with 15 μg of CS2+MT (six Myc epitope tags) vector alone or 15 μg of either N-terminal Myc-tagged human cyclin E1 or N-terminal Myc-tagged human cyclin E2. Cells were harvested 40 h later and immunostained with CD20 antibodies and propidium iodide for cell cycle DNA analysis as described previously (15). A minimum of 10,000 CD20-positive or -negative cells were analyzed by flow cytometry, and the percentage of cells in the G1, S, and G2/M phases was determined by using MacCycle AV software (Phoenix Flow Systems, San Diego, Calif.).
Cloning and plasmid construction.
An Amgen internal EST (expressed sequence tag) database containing cDNAs from more than 200 different libraries was searched with a cyclin box profile. The profile consisted of several compiled sequences containing a cyclin box. A murine EST, Bmme-7-133, obtained from this search was identified and used to search a public database containing human DNA sequences. Several human ESTs matching the 5′ end of the murine Bmme-7-133 clone were identified. One of these ESTs, R84331, was used to screen cDNA libraries and obtain additional 3′ sequence which included a stop codon. 5′ (GAA GAG AAT GTC AAG ACG AAG AAG CC) and 3′ (CAG TTC TAC CCA ATC TTG GTG AAT) PCR primers were then designed to clone the full-length human cyclin E2 gene. Human fetal lung and thymus Marathon libraries (Clontech) were used as templates. The conditions for the PCR were as follows: 2.5 μl of cDNA, 5 pmol of each primer, 0.5 μl of 50× deoxynucleoside triphosphates (dNTPs), 0.5 μl of Pfu polymerase (Stratagene, La Jolla, Calif.), 2.5 μl of 10× PCR buffer to a final volume of 25 μl. The cycle parameters were 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 3 min. The resulting approximately 1.3-kb DNA band was resolved on an agarose gel, purified, and subcloned into pCR2.1 (InVitrogen, Carlsbad, Calif.). Digestion of two independent clones with the restriction enzyme EcoRI revealed one fragment of approximately 1.3 kb and one fragment of approximately 1.2 kb. The sequence analysis of the two fragments revealed a full-length human cyclin E2 clone (the 1.3-kb fragment) and a human cyclin E2 splice variant clone (cyclin E2SV; the 1.2-kb fragment) with an in-frame deletion of 135 bp (lacking bases 496 to 631 of the full-length clone) in the cyclin box. RNase protection analysis was performed with human thymus and testes RNA as the template to confirm the presence of the human cyclin E2 splice variant and full-length cyclin E2 in normal tissues.
To clone murine cyclin E2, the murine EST Bmme-7-133 was used to design 5′ (ATT TAA GCT GGG CAT GTT CAC AGG A) and 3′ (GTC TTC AGC TTC ACT GGA CTC ACA CTT) PCR primers. A mouse brain Marathon library cDNA (Clontech) was used as the PCR template. The conditions for the PCR were as follows: 5 μl of cDNA, 20 pmol of each primer, 0.5 μl of 50× dNTPs, 0.5 μl of AmpliTaq (Roche Molecular Systems, Inc., Branchburg, N.J.), and 5 μl of 10× PCR buffer in a final volume of 50 μl. Reaction parameters were 94°C for 2 min, 35 cycles of 94°C for 20 s, 60°C for 30 s, and 72°C for 50 s, and a hold at 72°C for 7 min after the last cycle. The PCR product was a fragment of about 723 bp containing residues 308 to 1031. This fragment was cloned into pCR2.1, and an approximately 723-bp insert was digested with EcoRI, which generated fragments of 153 bp (residues 879 to 1031) and 570 bp (residues 308 to 878). The 570-bp fragment was gel purified and used as a DNA probe to screen a Uni-Zap mouse testes XR library (Stratagene catalog no. 937308). The library was plated out to approximately 106 PFU, and duplicate plaque lifts were prepared by using charged nylon membranes (Bio-Rad, Hercules, Calif.). The mouse cyclin E2 570-bp probe was radiolabeled by using a Rediprimer kit (Amersham Life Science, Arlington Heights, Ill.) according to the manufacturer’s protocol. The specific activity of the probe was 109 cpm/μg. Hybridization of the filters was conducted overnight at 42°C in standard hybridization solution containing 2.5× Denhardt’s solution, 50% formamide, 0.1% SDS, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.1 mg of salmon sperm DNA per ml. After hybridization, the filters were washed twice in 2× SSC at room temperature for 15 min, three times in 2× SSC at 50°C for 30 min, and then once in 0.2× SSC plus 0.5% SDS at 42°C for 30 min. The filters were exposed to film at −70°C for 3 days, using intensifying screens. Six positive clones were identified from this hybridization screen. The insert of each clone was subcloned into the vector pBluescript (Stratagene) by using standard techniques, and each insert was sequenced. GFP fusion constructs were generated by PCR subcloning of full-length and splice variant cyclin E2 into eGFPN1 plasmid (Clontech) at 5′ SalI and 3′ BamHI sites. Human cyclin E1 and human cyclin E2 were subcloned into a modified pcDNA3.1 vector containing six N-terminal Myc epitope tags. HA-Cdk2, p27, and CD20 plasmids were kindly provided by J. Roberts (Fred Hutchinson Cancer Research Center, Seattle, Wash.).
Yeast strains and plasmids.
The Saccharomyces cerevisiae CLN1 CLN2 CLN3 triple-deletion mutant was kindly provided by M. Tyers (38). Yeast were grown in yeast extract-peptone-galactose or appropriate selective medium with galactose or glucose. Human cyclin E1, cyclin E2, and cyclin E2SV were PCR amplified and cloned into SalI-EcoRI restriction sites in plasmid pY10. Plasmid pY10 contains 2-mm sequences for replication in yeast, the TRP1 gene for selection in yeast, the BLA gene for selection in Escherichia coli, and the ADH1 promoter for constitutive expression in yeast (37). DNA sequences were confirmed by double-stranded DNA sequencing.
RNA extraction and Northern blotting.
Total RNA was prepared by lysing cell monolayers in guanidinium isothiocyanate and centrifuging them over a 5.7 M CsCl cushion as described previously (8). RNA (20 μg) was electrophoresed on denaturing formaldehyde gels, transferred to MagnaNT membranes and cross-linked with UV. The probes used for Northern analyses included a 2.0-kb EcoRI fragment containing the entire cyclin E1 cDNA, a 330-bp fragment of cyclin E2, and an 800-bp PstI fragment from p36B4 (21). All probes were labeled with [α-32P]dCTP to a specific activity of approximately 109 cpm/μg of DNA, using a random-primed labeling kit (Boehringer Mannheim). Human tissue mRNA blots used in cyclin E2 Northern blot analysis were obtained from Clonetics.
Nucleotide sequence accession number. The GenBank accession numbers for murine and human cyclin E2 are AF106691 and AF106690, respectively.
RESULTS
Cloning of human and murine cyclin E2.
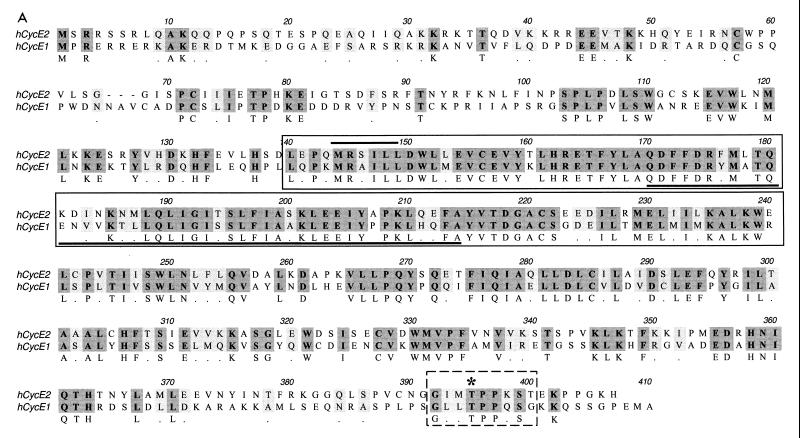
To identify cDNAs that encode potentially novel cyclins, we searched an EST database with a cyclin fold profile and discovered rat and murine ESTs that were similar but not homologous to cyclin E. The rat and murine sequences were compared and discovered to be orthologs from the two species. The DNA sequences were then used to search the public EST database, where several more homologous human and mouse ESTs were identified. From this sequence information, PCR primers were designed and 3′ RACE (rapid amplication of cDNA ends) reactions performed with a human fetal lung cDNA library as the template. A 1,300-bp cDNA sequence containing an open reading frame of 1,212 bp was obtained. This cDNA encoded a 404-amino-acid protein with a predicted molecular weight of 46,758 (Fig. 1A). The encoded protein shares 47% overall similarity to human cyclin E and contains a cyclin box motif that is characteristic of all cyclins. The next most closely related mammalian cyclin is cyclin B1, with 23% similarity at the protein level. There is 70% identity between the cyclin box present in cyclin E and the corresponding domain in cyclin E2. We propose that the mammalian cyclin E described previously be termed cyclin E1 and that the novel cyclin E be termed cyclin E2. During our initial screen of the human fetal lung cDNA library, we isolated cyclin E2SV, an in-frame splice variant of human cyclin E2 lacking bp 496 to 631 of the full-length cyclin E2. Cyclin E2SV encodes a protein missing 45 amino acids (residues 167 to 211) within the cyclin box domain (Fig. 1A). RNase protection analysis confirmed the presence of cyclin E2 splice variant mRNA in normal human thymus (data not shown). The full-length murine cyclin E2 cDNA was also cloned and found to have 94% identity with human cyclin E2. Murine cyclin E2 shares an overall similarity of 45% with murine cyclin E1 and is approximately 100 amino acids shorter at its C terminus than murine cyclin E1 (Fig. 1B).
FIG. 1.
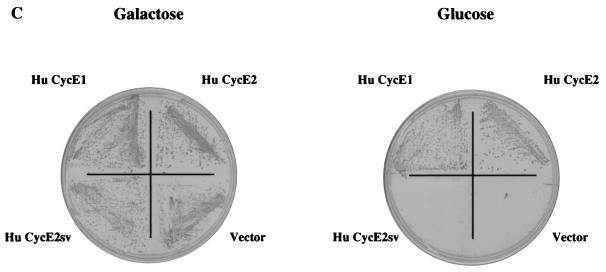
Sequence alignment (using the Clustal alignment program) and functional activity of cyclin E2 and cyclin E1. (A) Alignment of full-length human cyclin E2 protein with human cyclin E1 protein. The cyclin box is boxed with an unbroken line, the MRSILL sequence is overlined, and the peptide sequence missing from cyclin E2SV is underlined. The C-terminal PEST sequence motif is outlined with a dashed line, and the CDK consensus threonine phosphorylation site is starred. (B) Clustal alignment of murine cyclin E2 and murine cyclin E1. The cyclin box motif is boxed, and the MRSILL sequence is overlined. (C) Functional complementation of yeast G1 cyclins by human cyclin E2. S. cerevisiae MTY618 has a deletion of the CLN1, CLN2, and CLN3 genes and is kept alive by an integrated GAL-CLN3 gene. On galactose medium, CLN3 is transcribed and the cells live; on glucose, the GAL promoter is shut off and the cells fail to grow. Yeast expressing either human cyclin E1 (Hu CycE1) or human cyclin E2 (Hu CycE2) are able to grow on glucose, while yeast harboring vector alone or the human cyclin E2SV (Hu CycE2sv) fail to grow on glucose.
Cyclin E1 was originally isolated by its ability to complement loss of G1 cyclins in S. cerevisiae (16, 18). To test whether cyclin E2 also complements loss of G1 cyclins, we examined whether it rescued the lethality associated with a CLN1 CLN2 CLN3 triple mutant. The viability of the triple-mutant cells is maintained when Cln3 is conditionally expressed under the control of the GAL promoter. This strain grows in medium containing galactose but fails to grow in the presence of glucose where Cln3 is not expressed. Human cyclin E2 rescued triple CLN yeast mutants grown in medium containing glucose, whereas cyclin E2SV was unable to complement the conditional lethality defect (Fig. 1C). Cyclin E1 was also able to rescue yeast grown on glucose (Fig. 1C). These results indicate that cyclin E2 can complement a G1 cyclin defect in yeast and that amino acids 167 to 211 are required for its activity. The ability of cyclin E2 to rescue yeast lacking G1 cyclins also demonstrates that it can presumably bind and activate yeast Cdc28 kinase.
Cyclin E2 is expressed in human cancer cells.
The expression of cyclin E2 was analyzed in normal human tissues by Northern blot analysis. The cyclin E2 probe specifically recognized a 2.8-kb mRNA transcript, which is 600- to 700-bp larger than the transcript size for human cyclin E1 (data not shown). The highest levels of cyclin E2 were found in adult testes, thymus, and brain. Lower but significant expression was also observed in the placenta, spleen, and colon (data not shown). The tissue distribution of murine cyclin E2 mRNA in the adult mouse was similar to that observed for human cyclin E2 (data not shown). With the exception of brain, the overall expression of cyclin E2 mRNA correlates with tissues that contain proliferating cells. This finding suggests that cyclin E2 may play a tissue-specific role in regulating cell division.
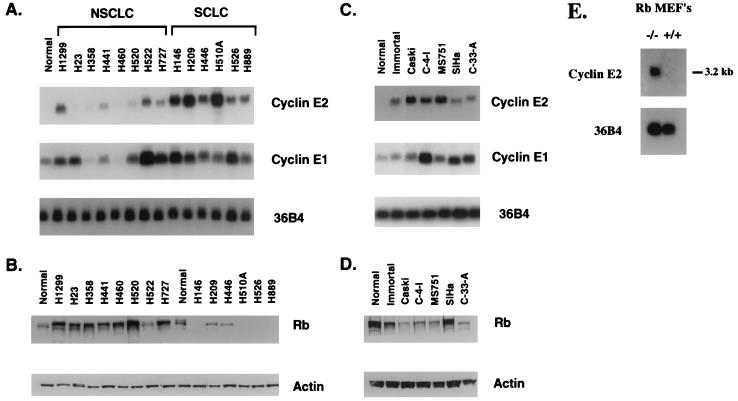
The differential expression of cyclin E1 and cyclin E2 mRNAs was examined in a panel of cells derived from human lung cancers (Fig. 2A). A low level of cyclin E1 mRNA, but no detectable cyclin E2 mRNA, was present in proliferating normal bronchial epithelial cells. Cell lines derived from different types of non-small-cell lung cancers (NSCLCs) demonstrated variable levels of both cyclin E1 and E2 mRNA. H1299 (large-cell carcinoma), H522 (adenocarcinoma), and H727 (carcinoid) had relatively high levels of cyclin E2 mRNA. In contrast, the six cell lines derived from small-cell lung carcinomas (SCLCs) all expressed relatively high levels of cyclin E2 mRNA. We have also demonstrated by reverse transcription-PCR analysis that full-length cyclin E2 mRNA and its splice variant are present in primary lung tumors (data not shown). Because mutations in the Rb gene are common in SCLCs (11), we sought to determine if there was a correlation between Rb protein levels and cyclin E2 mRNA expression in the same panel of lung cancers. Western blot analysis indicated that Rb was present at normal to increased levels in all of the NSCLCs but low or completely absent in the SCLCs (Fig. 2B). Together, these data suggest that, analogous to cyclin E1, cyclin E2 mRNA may also be repressed in cells having a functional Rb protein.
FIG. 2.
Northern blot analysis of cyclin E2 and cyclin E1 in human lung and cervical epithelial cells. (A) RNAs isolated from normal bronchial epithelial cells (Normal), NSCLC cell lines H1299, H23, H358, H441, H460, H520, H522, and H727, and SCLC cell lines H146, H209, H446, H510A, H526, and H889 were electrophoresed, transferred, and hybridized with probes for cyclin E1 and cyclin E2. 36B4 was included as an internal control for loading efficiency. (B) Western blot analysis of Rb in the same panel of lung cancer cells as used for panel A. Total protein extracts (50 μg) from exponentially growing cells was electrophoresed, transferred, and subsequently probed with antibodies against Rb. The blots were also probed with actin antibody as an internal control for loading efficiency. (C) RNAs isolated from normal cervical epithelial cells (Normal), immortalized cervical epithelial cells expressing the HPV E6 and E7 genes (Immortal), and the cervical cancer cells Caski, C-4-I, MS751, SiHa, and C-33-A were prepared and treated as described for panel A. (D) Total protein extracts (50 μg) from the cervical cells used for panel C were examined for Rb and actin expression as described for panel B. (E) RNA isolated from proliferating primary Rb+/+ MEFs and Rb−/− MEFs was electrophoresed, transferred, and hybridized with a probe specific for murine cyclin E2. 36B4 was included as an internal control for loading efficiency.
Approximately 93% of human cervical cancers and its precursor lesions contain integrated copies of the human papillomavirus (HPV) genome (1, 39). Because one of the HPV genes, E7, binds to and inactivates the Rb protein (13), one would expect cyclin E2 mRNA levels to also be increased in cervical cancer cells. Cyclin E2 mRNA was undetectable in normal human cervical epithelial cells (Fig. 2C). Cervical epithelial cells that were immortalized with a retrovirus expressing the HPV E6 and E7 genes and all cervical cancer cells examined showed moderate to high levels of cyclin E2 mRNA. The increased expression of cyclin E2 mRNA correlated well with loss of Rb protein in the same cells (Fig. 2D).
To further corroborate an association between loss of Rb function and increased cyclin E2 mRNA expression, we examined cyclin E2 levels in cells that were genetically null for Rb. Northern blot analysis of RNA harvested from primary mouse embryo fibroblasts (MEFs) showed that cyclin E2 mRNA was not present in Rb+/+ MEFs but was expressed in the Rb−/− MEFs (Fig. 2E). While Rb knockout MEFs do have subtle cell cycle defects they are not transformed by most criteria. Rb−/− MEFs do not grow in soft agar, and they arrest following serum starvation. Moreover, Rb−/− p21−/− double-knockout MEFs do not form tumors in nude mice (2, 10). The presence of cyclin E2 in Rb−/− MEFs suggests that in some cases E2F complexes may transcriptionally regulate the expression of cyclin E2.
Cyclin E2 was present at very low levels in normal breast epithelial cells, and its expression was elevated in all breast cancer cells examined (data not shown). Because the Rb protein is intact in most human breast cancers, these results suggest that other unknown factors likely play a role in determining cyclin E2 mRNA levels in cancer cells. Overall, our data demonstrate that cyclin E2 is consistently elevated in tumor-derived cells compared to nontransformed proliferating cells. The consistency of cyclin E2 overexpression in human cancer cells and its lack of expression in normal proliferating cells suggests that it may be a surrogate marker for cell transformation.
Cyclin E2 forms a catalytically active complex with Cdk2.
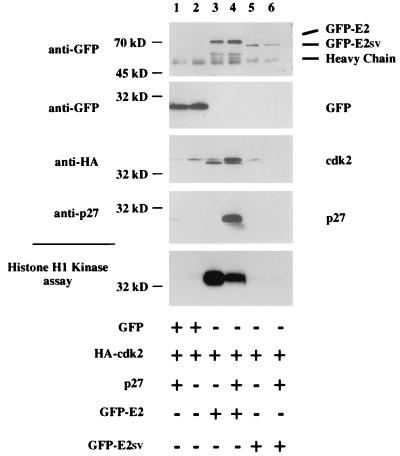
Cyclin E1 binds Cdk2 to form a serine/threonine kinase holoenzyme complex (17). The cyclin subunit imparts substrate specificity to the complex since cyclin A-Cdk2 complexes recognize substrates, such as lamin B, that are not phosphorylated by cyclin E1-Cdk2 complexes (12). To determine if cyclin E2 bound Cdk2, we examined the ability of cyclin E2 to form an active cyclin E2-Cdk2 complex in transiently transfected cells. 293T cells were transfected with HA epitope-tagged Cdk2, and plasmids expressing GFP vector alone or GFP fused to full-length human cyclin E2 (GFP-E2) or cyclin E2SV (GFP-E2SV). GFP protein alone and GFP-E2SV did not bind Cdk2, while cyclin E2 bound Cdk2 in a catalytically active complex (Fig. 3, lane 3). The ability of cyclin E2 to bind Cdk2 in an active complex suggests that, besides cyclin E1-Cdk2 and cyclin A-Cdk2 complexes, functional cyclin E2-Cdk2 complexes that may regulate G1 progression exist. These results also demonstrate that amino acids 167 to 211, which reside within the cyclin box, are required for cyclin E2 to bind Cdk2. It is possible that the inability of the splice variant to rescue yeast lacking G1 cyclins (Fig. 1C) is due to its deficiency in CDK binding. These same GFP fusion constructs were used to demonstrate that full-length human cyclin E2 were exclusively localized to the nucleus in transiently transfected proliferating COS-7 cells (data not shown). Interestingly, GFP-E2SV was present in both the nucleus and cytoplasm, suggesting that an intact cyclin box, and perhaps CDK binding, may be required for exclusive nuclear localization.
FIG. 3.
Cyclin E2 forms a catalytically active complex with Cdk2. 293T cells were transiently transfected with empty GFP vector (lanes 1 and 2) or a vector expressing GFP-E2 (lanes 3 and 4) or GFP-E2SV (lanes 5 and 6). Where indicated, HA-Cdk2 (lanes 1 to 6) and p27Kip1 (lanes 1, 4 and 6) plasmids were cotransfected. Immunoprecipitations were performed with GFP antibodies followed by either Western blotting or kinase assays. The top four panels represent Western blot analyses, and the bottom panel represents histone H1 kinase assays performed on the GFP immunoprecipitates.
To determine if a CDK inhibitor recognized cyclin E2-Cdk2 complexes, we coexpressed p27Kip1 with cyclin E2-Cdk2 complexes. Figure 3 demonstrates that p27Kip1 (and p21Cip1 [data not shown]) bound and inactivated cyclin E2-Cdk2 complexes. The binding of p27Kip1 to cyclin E2-Cdk2 complexes shifted the mobility of Cdk2 to its slower-migrating inactive form (Fig. 3, lane 4). Previous reports have demonstrated that p27Kip1 prevents CAK from phosphorylating and activating cyclin E1-Cdk2 complexes (23). Our data suggest that p27Kip1 may play a similar role in regulating cyclin E2-Cdk2 complexes. It is also possible that p27Kip1 stabilizes cyclin E2-Cdk2 complexes containing the slower-migrating inactive form of Cdk2.
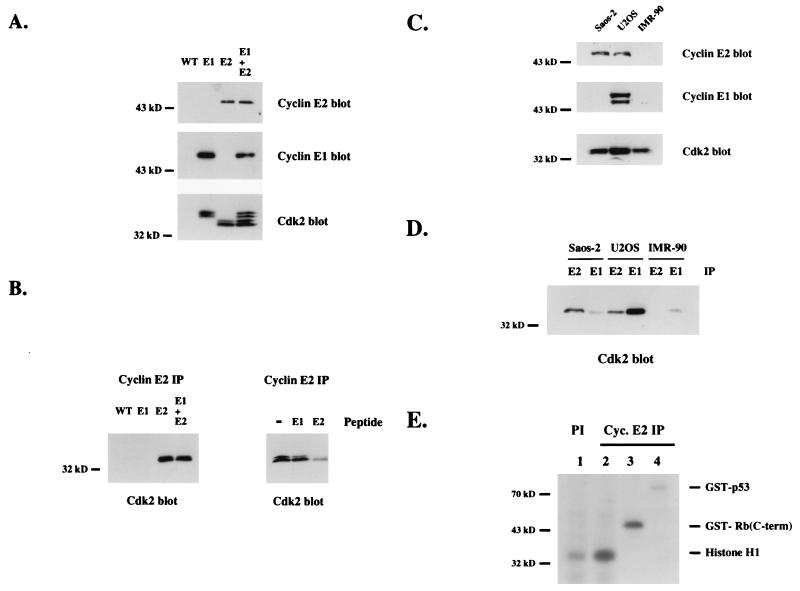
To examine endogenous cyclin E2 protein expression, polyclonal antibodies were generated against an N-terminal human cyclin E2 peptide antigen. The specificity of this antibody was tested on insect cell-expressed recombinant cyclin E2 protein. Cyclin E2 antibodies specifically recognized recombinant cyclin E2, and not cyclin E1, in both straight Western blots and immunoprecipitations (Fig. 4A and B). For the immunoprecipitation analysis, we distinguished between Cdk2 bound to cyclin E2 versus cyclin E1 by coexpressing a slower-migrating, epitope-tagged Cdk2 with cyclin E1 (Fig. 4A). Both the cyclin E1-Cdk2 and cyclin E2-Cdk2 complexes were catalytically active in histone H1 kinase assays (data not shown).
FIG. 4.
Endogenous cyclin E2 binds Cdk2. (A) His-tagged human cyclin E2-Cdk2 complexes and His-tagged human cyclin E1-HA-Cdk2 complexes were expressed in insect cells, purified by nickel chromatography, and analyzed by cyclin E2, cyclin E1, and Cdk2 Western blotting. Extracts made from wild-type virus (WT) were compared to cyclin E1 (E1) and cyclin E2 (E2) complexes alone or mixed at a 1:1 ratio (E1 + E2). (B) The same recombinant cyclin-Cdk2 complexes as used for panel A were immunoprecipitated with cyclin E2 polyclonal antiserum and analyzed by Cdk2 western blots (left), or recombinant cyclin E2-Cdk2 complexes were immunoprecipitated with cyclin E2 antiserum alone (−) or antiserum plus competing N-terminal cyclin E1 peptides (E1) or N-terminal cyclin E2 peptides (E2) and analyzed by Cdk2 Western blot analysis (right). (C) Western blot analysis of cyclin E2, cyclin E1, or Cdk2 in proliferating Saos-2, U2OS, and IMR-90 cells. (D) Extracts made from proliferating Saos-2, U2OS, and IMR-90 cells were immunoprecipitated (IP) with either cyclin E2 (E2) or cyclin E1 (E1) antiserum and analyzed by Cdk2 Western blot analysis. (E) Kinase assays were performed on anti-cyclin (Cyc.) E2 (lanes 2 to 4) and preimmune (lane 1) immunoprecipitates from proliferating Saos-2 cell extracts. Purified recombinant proteins comprised of histone H1 (lanes 1 and 2), GST fused to a C-terminal (C-term) fragment of Rb (lane 3), and GST fused to full-length p53 (lane 4) were used as substrates. GST protein alone was not phosphorylated by cyclin E2 immunoprecipitates (data not shown).
To determine if endogenous cyclin E1 and E2 were differentially expressed, we examined their expression in human osteosarcoma cells versus the human diploid fibroblast cell line IMR-90. IMR-90 and U2OS cells contain a functional Rb protein, and Saos-2 cells lack Rb. Cyclin E2 was expressed in both osteosarcoma cell lines but not in the fibroblasts. Cyclin E1 was detected only in U2OS cells. Presumably, cyclin E1 was below the limits of detection in Saos-2 cells and diploid fibroblasts by straight Western blot analysis (Fig. 4C). Immunoprecipitation analysis showed that cyclin E2-Cdk2 complexes were present in both osteosarcoma cells lines. As expected from the Western blot profile, cyclin E1-Cdk2 complexes were detected only in U2OS cells (Fig. 4D). In U2OS cells where both cyclin E1 and cyclin E2 were expressed, more Cdk2 was bound to cyclin E1 than to cyclin E2. These results suggest that in U2OS cells, cyclin E1 is expressed at a higher level than cyclin E2 or that cyclin E2 has additional CDK partners that compete for Cdk2 binding.
The endogenous cyclin E2 complexes present in Saos-2 cells are catalytically active and capable of phosphorylating histone H1 and GST-Rb but not GST-p53 (Fig. 4E). Transfected cyclin E2-Cdk2 complexes also recognized histone H1 (Fig. 3) and Rb (data not shown) as substrates but were unable to phosphorylate p53. While cyclin E1-Cdk2 complexes have been shown to phosphorylate p53 (9, 28), in our hands neither cyclin E1 (data not shown) nor cyclin E2 (Fig. 4E) complexes phosphorylated p53 protein. Our results are similar to those of a previous report demonstrating that cyclin A-Cdk2 and cyclin B-Cdk1, but not cyclin E1-Cdk2, phosphorylate p53 (40).
Cyclin E2-associated kinase activity is cell cycle regulated.
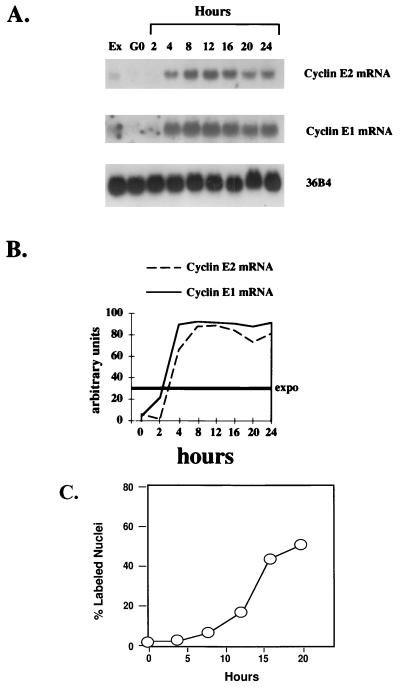
CDKs are positively regulated through their interactions with cyclins, and the formation of cyclin-CDK complexes is regulated by serum mitogens, extracellular nutrients, and cell anchorage. In some cases, the abundance of cyclins is modulated through transcriptional and translational pathways in a cell cycle-regulated manner (6, 18, 22). The expression of CDKs remains relatively constant throughout the cell cycle. However, the periodic expression of cyclins and their association with CDKs activates the kinase activity of the complexes at specific times during the cell cycle. To determine if cyclin E2 levels were cell cycle regulated, cyclin E2 mRNA was analyzed in immortalized MCF10 breast epithelial cells traversing the cell cycle. Cyclin E2 mRNA levels were low in quiescent cells (Fig. 5A). The expression of cyclin E2 increased 4 to 8 h after stimulation with growth factors, peaked at the G1/S phase boundary (12 h), and showed a slight decline as cells entered into G2 and mitosis (Fig. 5). In contrast, cyclin E1 mRNA expression peaked at mid to late G1 (4 and 8 h) and remained high as cells traversed S phase and entered mitosis (Fig. 5).
FIG. 5.
Cyclin E2 mRNA expression is cell cycle regulated. (A) Northern blot analysis of cyclin E2 and cyclin E1 in immortalized MCF10 breast epithelial cells. Quiescent normal mammary epithelial cells (G0) were restimulated to enter the cell cycle by the addition of growth factors, and RNA was harvested at the time points indicated; 20 μg of total RNA was loaded in each lane, and Northern blotting was performed with probes specific for human cyclin E2, human cyclin E1, and 36B4 (internal control for loading efficiency). Lane 1 represents cyclin expression in exponentially proliferating MCF10 cells (Ex). (B) Relative intensities of bands determined by densitometric scanning. The intensity of the bands is depicted on the y axis as arbitrary units, 0 indicates quiescent cells, and Expo. indicates the cyclin expression levels in exponentially growing cells, which averaged between 25 to 30% of the peak expression level. (C) Cumulative [3H]thymidine uptake into DNA determined in 24-well cultures that were plated and synchronized as described in Materials and Methods. At least 300 individual cells were examined to obtain data for each time point.
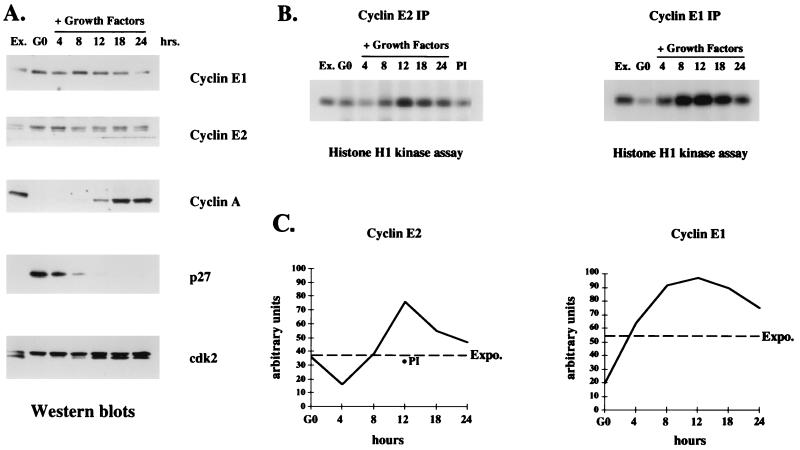
Cyclin mRNA expression is often not predictive of cyclin protein levels. Therefore, we determined the levels of cyclin E2 protein and its associated kinase activity in MCF10 cells at different phases of the cell cycle. The expression profile of cyclin E2 was compared with those of other cell cycle-regulated proteins known to function during G1 and S phase. Interestingly, cyclin E2 protein levels did not parallel its mRNA expression. Both cyclin E1 and cyclin E2 proteins were present in G0-arrested, G1- and S-phase cells, with a slight decrease observed as cells exited S phase and entered into G2 and mitosis (Fig. 6A). These results concur with previous reports showing that cyclin E1 mRNA expression does not correlate with its protein levels (35). In contrast to cyclins E1 and E2, cyclin A expression was tightly cell cycle regulated, with low levels in quiescent cells and a gradual increase as cells traversed S phase and entered G2/M (Fig. 6A). p27Kip1 levels were elevated in quiescent epithelial cells, suggesting that it may inhibit the activity of constitutively expressed cyclin E protein in nonproliferating epithelial cells. However, as cells progressed through G1 and entered S phase, p27Kip1 levels decreased sharply, thereby enabling Cdk2 complexes to be recognized by CAK (Fig. 6A). In support of this idea, the mobility of Cdk2 shifted to the faster-migrating CAK-phosphorylated active form as cells entered late G1 and S phase (Fig. 6A). Alternatively, the increased expression of cyclin A during late G1 may promote the formation of cyclin A-Cdk2 complexes that are in turn recognized by CAK.
FIG. 6.
Cyclin E2-associated kinase activity is cell cycle regulated. (A) Western blot analyses with cell extracts harvested at the times indicated following growth factor stimulation of quiescent human MCF10 cells. Ex., exponentially proliferating cells; G0, quiescent cells at time zero. The membranes were probed with antibodies for cyclin E1, cyclin E2, cyclin A, p27Kip1, and Cdk2 expression. (B) Histone H1 kinase assays performed with cyclin E1 and cyclin E2 immunoprecipitates (IP) from the same extracts as used in the Western blots depicted in panel A. PI indicates kinase activity present in preimmune serum immunoprecipitates from MCF10 extracts harvested 12 h after growth factor addition. (C) Relative intensities of phosphorylated histone H1 bands determined by densitometric scanning. The intensity of each band is depicted on the y axis in arbitrary units, 0 indicates quiescent cells, and Expo. indicates the cyclin-associated kinase activity in exponentially growing cells. For the cyclin E2 analysis, the level of kinase activity in preimmune immunoprecipitates at 12 h is depicted with a filled circle.
The cyclin E2-associated kinase activity was twofold higher than in G0 cells 12 h after growth factor stimulation when cells traversed late G1 and entered S phase (Fig. 6B and C; Fig. 5C). The cyclin E2-associated kinase activity detected in exponentially proliferating MCF10 cells was 2.5 times lower than the activity found in Saos-2 cells (Fig. 6B and C). This difference likely reflects the lower level of cyclin E2 protein present in proliferating MCF10 epithelial cells than in Saos-2 cells. The cell cycle-regulated kinase activity for cyclin E2 complexes suggests that cyclin E2 functions during late G1 and S phase. Cyclin E1-associated kinase activity was also examined in the same synchronized cell population. The results indicate a gradual increase in cyclin E1-associated kinase activity during mid-G1, with a fivefold peak increase occurring 12 h after growth factor addition followed by a decline in activity during late S phase and G2/M (Fig. 6B and C). These results are similar to those reported previously for cyclin E1-associated kinase activity in human breast epithelial cells (35) and further suggest that cyclin E1 and cyclin E2 kinase activities function at similar points during the cell cycle.
Cyclin E2 overexpression shortens G1.
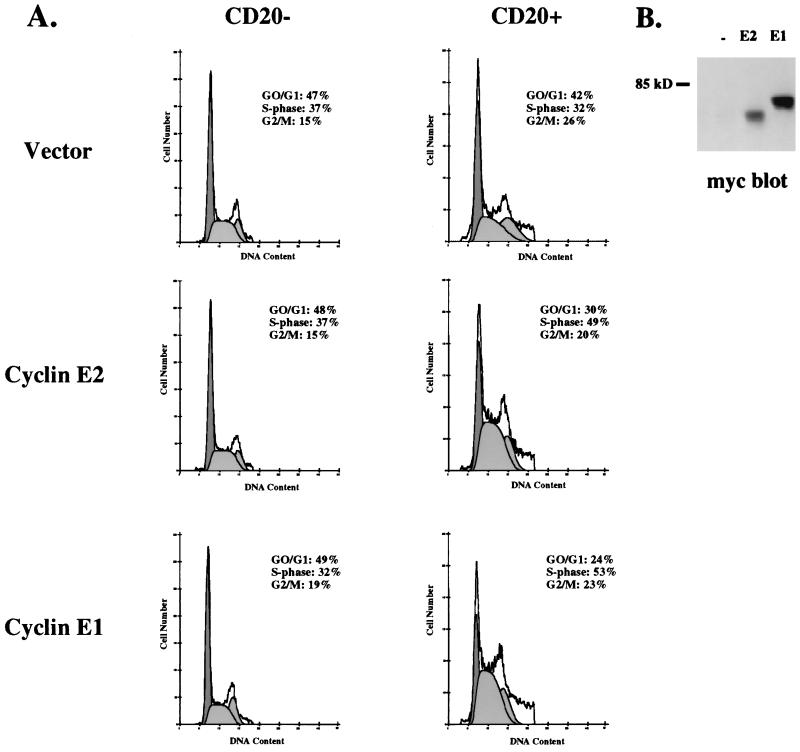
Previous reports have shown that overexpression of cyclin E1 accelerates G1, increases the percentage of cells in S phase, and decreases cell size (26, 29). The inappropriate activation of cyclin E1 complexes shortens G1 by presumably phosphorylating substrates at an earlier point during G1, thereby forcing cells to enter S phase prematurely. Since the kinase activity associated with cyclin E2 complexes peaks during mid- to late G1, it is possible that cyclin E2 also regulates G1 progression. To test this idea, Myc epitope-tagged cyclin E2 was transiently overexpressed in proliferating Saos-2 cells and the distribution of cells within the cell cycle was determined by flow cytometry. As expected, cyclin E1 decreased the percentage of cells in G1 and increased the number of cells in S phase, a profile that is typically associated with acceleration of G1 (Fig. 7A). While slightly less cyclin E2 was expressed (Fig. 7B), it also decreased the percentage of cells in G1 and increased the number of cells in S phase. The results from three independent experiments showed that cyclin E2 decreased the percentage of cells in G1 by an average of 11% ± 4% and increased the percentage of cells in S phase by an average of 13% ± 3%. These results demonstrate that cyclin E2 can regulate the cell cycle and suggests that it may be rate limiting for G1 progression.
FIG. 7.
Cyclin E2 overexpression decreases G1 length. (A) Proliferating Saos-2 cells were transiently transfected with CD20 plasmid plus vector alone, Myc-tagged cyclin E1, or Myc-tagged cyclin E2. Cells were harvested 40 h later, and the gated CD20− and CD20+ populations were analyzed by flow cytometry to determine their cell cycle profile. (B) Extracts made from the total population of cells transfected with vector alone (−), cyclin E2 (E2), or cyclin E1 (E1) plasmids were analyzed by Myc western blotting. All samples for flow cytometry and Western blot analysis were derived from the same experiment.
DISCUSSION
Identification of genes that regulate progression of cells through the cell cycle is crucial in understanding the pathways that regulate cell division. At least five different human cyclin families are expressed during the G1 or S phase of the cell cycle. Of these, the D-type cyclins, cyclin E, and cyclin A have all been implicated in regulating transit through G1 and entry into S phase. Multiple cyclins are likely required to convey and integrate the diverse array of extracellular and intracellular signals that regulate G1 progression. In this study, we have identified a novel cyclin, cyclin E2, that associates with Cdk2 in a catalytically active complex. Cyclins E1 and E2 have a high degree of amino acid identity within their cyclin boxes and decreased similarity in regions outside this domain. Notably, cyclin E2 possesses an MRSILL motif within the amino-terminal region of the cyclin box instead of the characteristic MRAILL motif present in mammalian cyclin E1 (16, 18). The crystal structure of cyclin A-Cdk2-p27 complexes demonstrates that the N-terminal RNLFG sequence of p27 contacts a shallow groove within cyclin A that contains the MRAIL motif (30). A number of other proteins, including p21 and Cdc25A, have also been shown to contact this same MRAIL motif on cyclin E1 (31, 43). The ability of cyclin E2 complexes to phosphorylate substrates and to bind p27Kip1 demonstrates that replacement of alanine with serine within the MRAIL motif does not appear to alter the activity of cyclin E2 complexes. Like cyclin E1, cyclin E2 has a carboxy-terminal PEST sequence motif that targets proteins for degradation. In addition, threonine 392 lies within a conserved CDK consensus phosphorylation site. Mutation of the analogous threonine 380 in cyclin E1 prevents its phosphorylation by Cdk2 and increases its stability (4, 41).
Cyclin E2-associated Rb and histone H1 kinase activity is present in cyclin E2 immunoprecipitates from proliferating cells. Moreover, cyclin E2-associated kinase activity is regulated in a cell cycle-dependent manner. These data, considered together with experiments demonstrating that cyclin E2 overexpression shortens G1, argue in favor of a role for cyclin E2 in promoting cell cycle progression. The combined evidence suggests that cyclin E2 has properties very similar to those of cyclin E1, a cyclin subunit that regulates progression of cells through G1.
Cyclin E1 is a molecule involved in relaying extracellular growth signals to the nucleus, where cyclin E1 complexes phosphorylate substrates that regulate cell division. The discovery of a second cyclin E family member suggests that additional G1 substrates that are not recognized by cyclin E1 complexes may exist. The differences in substrate specificity between cyclin A-Cdk2 and cyclin E1-Cdk2 complexes lends support to this idea (12, 32). While we have demonstrated that cyclin E2 forms an active complex with Cdk2, it is not clear if this is the only CDK that binds cyclin E2. We have not determined if Cdk3, a cyclin E1 binding partner, binds cyclin E2; however, neither Cdk4 nor Cdk1 was present in cyclin E2 immunoprecipitates (data not shown). Alternatively, the fact that the cyclin box domains within cyclin E2 and cyclin E1 are 70% identical suggests that Cdk2 and perhaps Cdk3 are the only CDKs that bind cyclin E2. If this is the case, then either tissue-restricted expression or substrate specificity may distinguish the functional roles of cyclin E1 and cyclin E2 in regulating cell growth and differentiation.
One of the more striking differences observed between cyclin E1 and cyclin E2 mRNA expression was in normal versus transformed cells. Normal proliferating bronchial epithelial cells expressed cyclin E1 but not cyclin E2 mRNA. In contrast, when a panel of lung cancer cell lines was examined, most of the NSLCs and all of the SCLCs demonstrated high levels of cyclin E2 mRNA. Because most SCLCs have defects in the Rb gene (11), we postulate that cyclin E2 mRNA may be repressed in normal cells by an Rb-dependent mechanism. Further support for this idea derives from the examination of cells genetically null for Rb. In contrast to wild-type MEFs, Rb−/− MEFs expressed cyclin E2 mRNA, suggesting that E2F complexes may play a role in regulating the expression of cyclin E2. Cyclin E1 expression is also upregulated in Rb−/− MEFs compared to wild-type MEFs (10). However, our observation that cyclin E2 mRNA is highly expressed in breast cancer-derived epithelial cells (data not shown) that typically lack Rb mutations suggests that multiple factors in addition to E2F regulate cyclin E2.
The discovery of a novel cyclin E family member shifts the paradigm of a single mammalian cyclin E regulating mid to late G1 progression. While cyclins D, E, and A are involved in regulating progression of cells through G1 and S phase, the cell cycle-regulated activity of cyclin E2-CDK complexes suggests that it also functions during late G1 and early S phase. The three D-type cyclins all appear to regulate cellular events in the early to mid-G1 phase of the cell cycle. Mice lacking cyclin D1 or D2, while viable, have obvious reproductive and developmental defects demonstrating that a complete set of D-type cyclins is required for normal development. Whether both members of the cyclin E family are required for normal development remains to be determined; however, it is possible that overlapping substrate specificity may reduce any potential developmental defects observed in mice lacking either gene alone.
The finding that microinjected active cyclin E1-Cdk2 complexes drives quiescent normal human fibroblasts into S phase supports the idea that cyclin-CDK activation is one of the primary rate-limiting steps in mitogen-stimulated progression of cells through G1. In summary, we have demonstrated that cyclin E2 functions in vivo to regulate the cell cycle. The discovery of a second cyclin E family member raises the possibility that cyclin E1 and cyclin E2 complexes phosphorylate distinct sets of substrates to regulate the progression of cells through G1.
ACKNOWLEDGMENTS
We thank our colleagues at Amgen for comments, helpful discussions, and reagents. We thank the members of the Amgen EST program for constructing the EST database. We also thank M. Tyers (University of Toronto, Toronto, Ontario, Canada) for generously providing the triple-CLN-deletion yeast strain and J. Roberts (Fred Hutchinson Cancer Research Center, Seattle, Wash.), K. Tsai (MIT, Boston, Mass.), and T. Jacks (MIT) for providing materials. We also thank Bethany Sutton and Steven Foster for expert sequence analysis and John Delaney for assistance in recombinant protein expression.
REFERENCES
- 1.Arends M J, Buckley C H, Wells M. Aetiology, pathogenesis and pathology of cervical neoplasias. J Clin Pathol. 1998;51:96–103. doi: 10.1136/jcp.51.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brugarolos J, Bronson R T, Jacks T. p21 is a critical CDK2 regulator essential for proliferation control in Rb-deficient cells. J Cell Biol. 1998;141:503–504. doi: 10.1083/jcb.141.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catzavelos C, Bhattacharya N, Ung Y, Wilson J, Roncari L, Sandhu C, Shaw R, Yeger H, Morava-Protzner I, Kapusta L, Franssen E, Pritchard K, Slingerland J. Decreased levels of the cell-cycle inhibitor p27Kip1 protein: prognostic implications in primary breast cancer. Nat Med. 1997;3:227–230. doi: 10.1038/nm0297-227. [DOI] [PubMed] [Google Scholar]
- 4.Clurman B E, Sheaff R, Thress K, Groudine M, Roberts J M. Cdk2-dependent cyclin E ubiquitination. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 5.Ethier S P, Mahacek M L, Gullick W J, Frank T S, Weber B L. Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites. Cancer Res. 1993;53:627–635. [PubMed] [Google Scholar]
- 6.Evans T, Rosenthal E, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 7.Gray-Bablin J, Zalvide J, Fox M, Knickerbocker C, DeCaprio J, KeyoMarsi K. Cyclin E, a redundant cyclin in breast cancer. Proc Natl Acad Sci USA. 1996;93:15215–15220. doi: 10.1073/pnas.93.26.15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudas J M, Knight G B, Pardee A B. Nuclear posttranscriptional processing of thymidine kinase mRNA at the onset of DNA synthesis. Proc Natl Acad Sci USA. 1988;85:4705–4709. doi: 10.1073/pnas.85.13.4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao M, Lowy A, Kapoor M, Deffie A, Liu G, Lozano G. Mutation of phosphoserine 389 affects p53 function in vivo. J Biol Chem. 1996;217:29380–29385. doi: 10.1074/jbc.271.46.29380. [DOI] [PubMed] [Google Scholar]
- 10.Herrera R, Sah V, Williams B, Makela T, Weinberg R, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point control in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horowitz J M, Park S H, Bogenmann E, Cheng J C, Yandell D W, Kaye F J, Minna J D, Dryja T P, Weinberg R A. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci USA. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton L, Templeton D. The cyclin box and C-terminus of cyclins A and E specify CDK activation and substrate specificity. Oncogene. 1997;14:491–498. doi: 10.1038/sj.onc.1200851. [DOI] [PubMed] [Google Scholar]
- 13.Jones D L. Interactions of the human papillomavirus E7 protein. Semin Cancer Biol. 1996;7:327–337. doi: 10.1006/scbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- 14.Keyomarsi K, O’Leary N, Molnar G, Lees E, Fingert H J, Pardee A B. Cyclin E, a potential prognostic marker for breast cancer. Cancer Res. 1994;54:380–385. [PubMed] [Google Scholar]
- 15.Kelly B, Wolfe K, Roberts J. Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein. Proc Natl Acad Sci USA. 1998;95:2535–2540. doi: 10.1073/pnas.95.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koff A, Cross F, Fisher A, Schumacher J, Leguellec K, Philippe M, Roberts J. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell. 1991;66:1217–1228. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- 17.Koff A, Giordano A, Desai D, Yamashita K, Harper J, Elledge S, Nishimoto T, Morgan D, Franza B, Roberts J. Formation and activation of a cyclin E-Cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 18.Lew D J, Dulic V, Reed S. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 19.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundberg A S, Weinberg R. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masiakowski P, Breathnach R, Bloch J, Gannon F, Krust A, Chambon P. Cloning of cDNA sequences of hormone-regulated genes from the MCF-7 human breast cancer cell line. Nucleic Acids Res. 1982;10:7895–7903. doi: 10.1093/nar/10.24.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minshull J, Golsteyn R, Hill C, Hunt T. The A- and B-type cyclin associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle. EMBO J. 1990;9:2865–2875. doi: 10.1002/j.1460-2075.1990.tb07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan D O. The dynamics of cyclin dependent kinase structure. Curr Opin Cell Biol. 1996;8:767–772. doi: 10.1016/s0955-0674(96)80076-7. [DOI] [PubMed] [Google Scholar]
- 24.Mumberg D, Wick M, Burger C, Haas K, Funk M, Muller R. Cycline Et, a new splice variant of human cyclin E with a unique expression pattern during cell cycle progression and differentiation. Nucleic Acids Res. 1997;25:2098–2105. doi: 10.1093/nar/25.11.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtsubo M, Roberts J. Cyclin-dependent regulation of G1, in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- 26.Ohtsubo M, Theodoras A, Schumacher J, Roberts J, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter P, Malone K, Heagerty P, Alexander G, Gatti L, Firpo E, Daling J, Roberts J. Expression of cell-cycle regulators p27Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med. 1997;3:222–225. doi: 10.1038/nm0297-222. [DOI] [PubMed] [Google Scholar]
- 28.Price B D, Hughes-Davies L, Park S. Cdk2 kinase phosphorylates serine 315 of human p53 in vitro. Oncogene. 1995;11:73–80. [PubMed] [Google Scholar]
- 29.Resnitzky D, Reed S. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo A, Jeffrey P, Patten A, Massague J, Pavletich N. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 31.Saha P, Eichbaum Q, Silberman E, Mayer B, Dutta A. p21Cip1 and Cdc25A: competition between an inhibitor and an activator of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4338–4345. doi: 10.1128/mcb.17.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarcevic B, Lilischkis R, Sutherland R. Different phosphorylation of T-47D breast cancer cell substrates by D1-, D3-, E-, and A-type cyclin-CDK complexes. J Biol Chem. 1997;272:33327–33337. doi: 10.1074/jbc.272.52.33327. [DOI] [PubMed] [Google Scholar]
- 33.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 34.Sherr C, Roberts J. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 35.Slingerland J M, Hengst L, Pan C-H, Alexander D, Stampfer M R, Reed S I. A novel inhibitor of cyclin-cdk activity detected in transforming growth factor beta-arrested epithelial cells. Mol Cell Biol. 1994;14:3683–3694. doi: 10.1128/mcb.14.6.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soule H D, Maloney T M, Wolman S R, Peterson W D, Jr, Brenz R, McGrath C M, Russo J, Pauley R J, Jones R F, Brooks S C. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 37.Thukral S, Blain G, Chang K, Fields S. Distinct residues of human p53 implicated in binding to DNA, simian virus 40 large T antigen, 53BP1, and 53BP2. Mol Cell Biol. 1994;14:8315–8321. doi: 10.1128/mcb.14.12.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: cln3 may be an upstream activator of cln1, cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasudevan D M, Vijayakumar T. Viruses in human oral cancers. J Exp Clin Cancer Res. 1998;17:27–31. [PubMed] [Google Scholar]
- 40.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 41.Won K, Reed S. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu L, Harlow E, Dynlacht D. p107 uses a p21Cip1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]