Abstract
Signaling by phosphorylated species of phosphatidylinositol (PI) appears to regulate diverse responses in eukaryotic cells. A differential display screen for fat- and muscle-specific transcripts led to identification and cloning of the full-length cDNA of a novel mammalian 2,052-amino-acid protein (p235) from a mouse adipocyte cDNA library. Analysis of the deduced amino acid sequence revealed that p235 contains an N-terminal zinc-binding FYVE finger, a chaperonin-like region in the middle of the molecule, and a consensus for phosphoinositide 5-kinases at the C terminus. p235 mRNA appears as a 9-kb transcript, enriched in insulin-sensitive cells and tissues, likely transcribed from a single-copy gene in at least two close-in-size splice variants. Specific antibodies against mouse p235 were raised, and both the endogenously and heterologously expressed proteins were biochemically detected in 3T3-L1 adipocytes and transfected COS cells, respectively. Immunofluorescence microscopy analysis of endogenous p235 localization in 3T3-L1 adipocytes with affinity-purified anti-p235 antibodies documented a punctate peripheral pattern. In COS cells, the expressed p235 N-terminal but not the C-terminal region displayed a vesicular pattern similar to that in 3T3-L1 adipocytes that became diffuse upon Zn2+ chelation or FYVE finger truncation. A recombinant protein comprising the N-terminal but not the C-terminal region of the molecule was found to bind 2.2 mole equivalents of Zn2+. Determination of the lipid kinase activity in the p235 immunoprecipitates derived from 3T3-L1 adipocytes or from COS cells transiently expressing p235 revealed that p235 displayed unique preferences for PI substrate over already phosphorylated PI. In conclusion, the mouse p235 protein determines an important novel class of phosphoinositide kinases that seems to be targeted to specific intracellular loci by a Zn-dependent mechanism.
Research over the past several years strongly implicates polyphosphoinositides as key regulators of diverse responses in eukaryotic cells such as membrane ruffling, secretion, vesicular trafficking, insulin-mediated membrane translocation of the GLUT4 glucose transporter, cell adhesion, chemotaxis, DNA synthesis, and cell cycle (for recent reviews, see references 1, 8, 12, 25, 30, 31, and 50 to 52). Species of phosphatidylinositol (PI) phosphorylated at the D-5 position of the inositol ring have attracted central attention because of several aspects. First, PI 4,5-bisphosphate (P2) is a key precursor of at least three second-messenger molecules, including inositol 1,4,5-trisphosphate (P3), diacylglycerol, and PI 3,4,5-P3. Second, two novel 5′ phosphoinositide species, PI 5-P and PI 3,5-P2, misidentified as PI 4-P and PI 3,4-P2 in previous studies, have been documented in yeast and mammalian cells (14, 40, 53, 57). Until recently, it was thought that the biosynthesis of PI 4,5-P2 involves two consecutive phosphorylation reactions of PI in canonical order: first, PI 4-kinase specifically phosphorylates position 4 of the inositol ring to generate PI 4-P, which is then phosphorylated by PI-4-phosphate 5-kinase [PI(4) P5K] type I or type II at position D-5 to generate PI 4,5-P2 (8, 31). It has now been recognized that this pathway is catalyzed only by the type I enzymes (or PI 5-Ks [51]), which display specificity towards position D-5 of the inositol ring (40) and which, in addition to PI 4-P, can utilize PI 3-P, PI 3,4-P2 (53, 62), and PI (53) as substrates. Type II enzymes (or PIP 4-Ks [51]) possess preferences towards position D-4 (40) and seem to utilize only already phosphorylated PI substrates (53). cDNAs of both types have been isolated and found to define differently sized molecules which, outside the kinase domain, have no homology with each other or with other lipid and protein kinases (31).
While the phosphoinositides’ essential function in intracellular regulation has been extensively documented in a variety of experimental paradigms, the molecular mechanism(s) of their action is still elusive. Interactions of polyphosphoinositides with protein modules such as the pleckstrin homology domain of several proteins appear to contribute to specific protein targeting or protein activation (for a recent review, see reference 51). Very recently a new evolutionarily conserved Zn2+-binding domain, known as FYVE (49) or RING finger (38), has been recognized as a specific protein module for PI phosphorylated exclusively at position D-3 of the inositol ring (7, 17, 38). Thus, specific interaction with protein modules offers a promising concept in deciphering the molecular mechanisms of the phosphoinositides’ role in coordinated intracellular regulation.
In this study we describe the identification, cloning, and characterization of a novel mammalian protein, p235, which harbors two key domains: an N-terminal FYVE finger and a C-terminal PI 5-K homology domain. p235 was detected both biochemically and morphologically in 3T3-L1 adipocytes with specific-antibody preparations. Its distinctive peripheral vesicular pattern of appearance in 3T3-L1 adipocytes as detected by immunofluorescence analysis seems to be conferred by its FYVE finger and a Zn2+-binding mechanism. p235 preferentially utilizes PI and, less effectively, PI 4-P substrates but not PI 3-P or PI 5-P to generate PIP and PI 4,5-P2, respectively. Thus, p235 defines a distinct class of the phosphoinositide kinase family that likely operates at distinct intracellular sites.
MATERIALS AND METHODS
Cell cultures.
Conditions for differentiation of L6 rat myoblasts (a gift from John Lawrence, Jr.) and 3T3-L1 mouse fibroblasts into insulin-sensitive myocytes and adipocytes, respectively, on plates or glass coverslips (for immunofluorescence microscopy analysis) were as previously described (46, 47). MCF-7, HeLa, and COS-7 cells were grown to the densities indicated in the figure legends on plates or glass coverslips in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum (FBS), 50 U of penicillin per ml, and 50 μg of streptomycin sulfate per ml. Chinese hamster ovary (CHO)-K1 cells were grown to confluence on 150-mm-diameter plates in Ham’s F-12 medium containing 10% FBS and the antibiotics mentioned above.
Tissue differential display and cDNA library screening.
A modified reverse transcription-PCR-based differential display protocol with selected primers that preferentially targets mRNAs of moderate to low abundance was used as described in detail elsewhere (21). Particularly, total RNA, derived from four rat tissues, namely, adipose, muscle, brain, and liver, was subjected to first-strand cDNA synthesis with Moloney murine leukemia virus reverse transcriptase (Gibco, BRL) and an antisense (5′-GTGAGATCTGTGCTGGTGTC-3′) primer named above. Fragment amplification by PCR was performed with the antisense primer paired with a sense primer (5′-GATCACAATAAACAGCTGGAG-3′) under 50°C annealing and 72°C extension temperatures for 30 cycles. The radiolabeled oligonucleotide products were separated on 6% polyacrylamide sequencing gel, and a band of 363 bp (fragment 9) was selected on the basis of its unique appearance in fat and muscle but not in brain and liver. After elution, reamplification by PCR, and subcloning in pGEM-T Easy vector (Promega), fragment 9 was subjected to sequence analysis by Dye Deoxy Terminator cycle sequencing (automated sequencer, model 373A; Applied Biosystems). A mouse 3T3-422A adipocyte cDNA library constructed in lambdaZapII vector (a gift of Bruce Spiegelman) was first screened with fragment 9 (probe 1; nucleotides 3710 to 4040 of the mouse p235 sequence) and then with the probes indicated below by following previously published procedures (47). Briefly, 800,000 PFU was plated with the XL1-Blue MRF′ strain of Escherichia coli as a host (Stratagene). The plaques were transferred onto nitrocellulose filters (Schleicher & Schuell), alkali denatured, and fixed for 2 h at 80°C in a vacuum oven. The filters were hybridized with 32P-labeled cDNA probes and washed according to standard protocols as we described previously (47). The selected positive clones were plaque purified and then subjected to in vivo excision of the pBluescript SK+ phagemid from the lambdaZapII vector using ExAssist helper phage (Stratagene) and protocols provided by the manufacturer. To extend the sequences of the isolated positive clones in both the 5′ and 3′ directions, the initial filters were screened for three more rounds under the same conditions with new probes that had been isolated from the preceding screening and positioned closer to the 3′ and 5′ ends of the p235 cDNA sequence. The following probes were used: probe 2, nucleotides 2200 to 3481, derived from clone IRG1; probe 3, nucleotides 5527 to 6000, derived from clone IRG7; and probe 5, nucleotides 347 to 2085, derived from clone N12 (see Fig. 1A). In every new round of screening the positive clones identified in the preceding screen were disregarded.
FIG. 1.
Cloning of p235. (A) Schematic representation of the seven overlapping clones, encompassing the full-length p235l cDNA sequence. The numbers under the lines represent the nucleotide numbers of the full-length p235l nucleotide sequence included in the clones. The initiation and termination codons are noted. The position of the initial PCR fragment, fragment 9, is indicated. (B) Deduced amino acid sequence of p235l. The termination codon is marked by an asterisk.
cDNA sequences were determined at least four times on both strands by Dye Deoxy Terminator cycle sequencing with T3, T7, or cDNA-specific primers. For database sequence similarity searches, Basic Logic Alignment Search Tool (BLAST) was used (2). DNA and protein sequence comparisons were carried out with the software GeneWorks 2.5 and MacVector 6.0.1 (Oxford Molecular Group). Domain searches were performed with “coils” (33) and “motifs” (18).
RNA isolation and Northern blot analysis.
The guanidinium thiocyanate method was used to isolate total RNA from either L6 and 3T3-L1 cells (on the indicated days of the differentiation programs), other cultured cells (after reaching confluence), or mouse and rat tissues as previously described (47). RNAs were dissolved in water, quantified by measuring the absorbancy at 260 nm, and subjected to electrophoresis on agarose gels followed by ethidium bromide staining for analysis of their integrity. For Northern blot analysis total RNA was fractionated on formaldehyde–1% agarose gels, blotted with alkali (50 mM NaOH), and fixed onto Zeta-Probe blotting nylon membranes (Bio-Rad). The blots were hybridized with a p235 cDNA fragment (probe 2, nucleotides 2200 to 3481, derived from clone IRG1 by EcoRI digestion, 32P labeled by random priming to 109 cpm/μg [16]) for 16 h at 60°C in 0.5 M NaH2PO4 (pH 7.2) containing 1 mM Na2EDTA and 7% sodium dodecyl sulfate (SDS), according to the manufacturer’s protocol. Blots were washed twice in 40 mM NaH2PO4 (pH 7.2) containing 1 mM Na2EDTA and 5% SDS at 25°C and twice in the same buffer, but with 1% SDS, at 60°C over a period of 2 to 3 h. RNA levels were detected by autoradiography and quantified by scanning the autoradiograms with a laser densitometer (Molecular Dynamics). Several exposures of each blot were quantified to ensure that the exposures were within the linear range of the film. The amounts of RNA on the blots were controlled by hybridization with radiolabeled 340-bp probe, derived from chicken 18S ribosomal cDNA.
Southern blot analysis.
Genomic DNA was isolated from rat (Sprague-Dawley) and mouse (Swiss Webster) livers by common procedures (42) and digested with restriction endonucleases (EcoRI, EcoRI plus EcoRV, EcoRI plus NcoI, or EcoRI plus HindIII). Digests were resolved on 0.8% agarose gel and alkali transferred to a Zeta-Probe blotting nylon membrane. The genomic Southern blot was hybridized with a radiolabeled 1.2-kbp EcoRI fragment of the clone IRG1 (probe 2, nucleotides 2200 to 3481, 106 cpm/ml, random-priming labeling). Hybridization and subsequent washing conditions were the same as described above for the Northern blot analysis.
Fusion proteins.
A C-terminal region of p235 (amino acids 1684 to 2052) was expressed as a glutathione S-transferase (GST) fusion protein in pGEX5X-3 vector (Pharmacia). The pGEX-C1684-2052 construct was generated by subcloning the BamHI-XhoI fragment of clone K12 into a BamHI-XhoI digest of pGEX5X-3 vector and was confirmed by restriction mapping. An N-terminal region of p235 (amino acids 99 to 473) was expressed as a GST fusion protein in the pGEX-1 vector (Amrad). To construct pGEX-1Nl99-473 the SacI fragment (1.2 kbp) of clone 2N3 was first subcloned in pGEM4Z (Promega) and the EcoRI fragment (1.2 kbp) of the resulting construct was ligated into an EcoRI digest of pGEX-1. E. coli XA-90 was used for transformation. The production, purification, and elution of the GST fusion proteins from glutathione-agarose beads (Sigma) were performed essentially as we previously described (47) except for the bacterial cell lysis, which was performed at 4°C with lysozyme (1 mg/ml; Boehringer Mannheim) and followed by DNase I (0.1 mg/ml; Boehringer Mannheim) digestion. The concentrations and quality of the eluted purified proteins were determined electrophoretically by comparing the intensities of the Coomassie blue-stained protein bands with that of a bovine serum albumin standard (Pierce).
N-terminal or C-terminal portions of p235 were fused with the enhanced green fluorescent protein (EGFP) for fluorescence microscopy studies. To prepare the pEGFP-Ns1-286 construct, the 850-nucleotide HindIII fragment of clone N16 was subcloned into a HindIII digest of pEGFP-C2 (Clontech). A short fragment of 14 amino acids (SLISNSARARVHVE) left from the pBluescript polylinker after the HindIII site preceded the initial Met. The pEGFP-Ns1-180 construct was engineered by digestion of pEGFP-Ns1-286 with PstI and subsequent ligation of the resulting construct. To generate pEGFP-C1684-2052, the BamHI-SalI fragment (2.3 kbp) of cDNA clone K12 was first subcloned into a BamHI-SalI digest of pGEM4Z (Promega). The BamHI-XbaI fragment (2.3 kbp) of the pGEM4Z construct was then subcloned into a BamHI-XbaI digest of pEGFP-C2. All constructs were confirmed by restriction endonuclease mapping.
To engineer a full-length clone from the p235 partial sequence clones, we used convenient restriction sites. Briefly, p235s (6.4 kbp) was constructed in the XbaI- SalI sites of pBluescript II SK(+) by ligating the pregenerated N- and C-terminal fragments in the unique KpnI restriction site at position 2929. The C-terminal fragment (3.6 kbp) was generated in two steps. First, the HindIII-PstI fragment of clone K12 (Fig. 1A) and the HindIII-NcoI fragment of clone IRG7 were ligated into the NcoI- and PstI-digested pGEM-T vector (Promega). Next, the resulting insert, released by NcoI and SalI, was ligated with the KpnI-NcoI fragment of IRG2 into KpnI and SalI-digested pBluescript II SK(+). The N-terminal fragment (2.8 kbp) was also generated in two steps. First, we linked the HindIII-EcoRV fragment of clone N12 to the EcoRV-KpnI fragment of IRG4 in a HindIII-KpnI digest of pBluescript II SK(+). The resulting HindIII-KpnI fragment together with the EcoRI-HindIII segment of clone N16 was ligated into an EcoRI-KpnI digest of pCRII (Invitrogen). Finally, the XbaI-KpnI N terminus was ligated to the KpnI-SalI C terminus in pBluescript II SK(+). The full-length p235s cDNA was tagged at the NH2 terminus with a 9-amino-acid epitope, YPYDVPDYA, derived from influenza virus hemagglutinin (HA) as we previously described (47). The double-stranded oligonucleotide encoding the sequence detailed above after the initial Met and tailed with convenient restriction sites (5′, EcoRI; 3′, XbaI) together with the full-length p235s XbaI-SalI fragment of pBluescript was subcloned into EcoRI and SalI sites of the mammalian expression vector pCMV5. The proper organization of the construct pCMV5-HA-p235s was confirmed by restriction endonuclease mapping.
Transient-expression assay.
COS-7 cells were seeded at 750,000 cells per 100-mm-diameter plate or at 125,000 cells per coverslip (six-well dish). Transfections with pEGFP-Ns1-286, pEGFP-Ns1-180, or pEGFP-C1684-2052 were performed with Lipofectamine (Gibco, BRL) as a transfection reagent by the manufacturer protocol. Fifteen to 24 h after transfections, the cells were either homogenized and fractionated for immunoblotting analysis or processed further for fluorescence microscopy studies. Transfections with pCMV5-HA-p235s were performed by the calcium phosphate precipitation method as we described previously (47). Forty-eight hours posttransfection, the cell lysates were collected and used for immunoprecipitation and Western blotting.
Cell treatment.
Fifteen hours posttransfection with the pEGFP-Ns1-286 construct, COS-7 cells grown on coverslips were serum starved for 3 h. To chelate Zn2+, the cells were treated with 200 μM N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN; Sigma) dissolved in a culture medium as we described previously (43). Following incubation at 37°C for 30 min, the cells were immediately fixed in methanol for fluorescence microscopy studies.
Fluorescence and immunofluorescence microscopy.
COS-7 cells grown on glass coverslips and transiently transfected with the GFP fusion constructs indicated in the figure legends were serum starved for 3 h prior to the experiment. The cells were then washed three times in phosphate-buffered saline (PBS), fixed in methanol for 6 min at −20°C, and washed three more times in PBS. The coverslips were then mounted on slides as indicated below. Immunofluorescence experiments with 3T3-L1 adipocytes were performed as described previously (44, 46). Briefly, 3T3-L1 adipocytes differentiated on glass coverslips and serum deprived for 3 h were washed in PBS, fixed in methanol (6 min at −20°C), rewashed three times in PBS, and permeabilized with 0.5% Triton X-100–1% FBS in PBS for 15 min. Cells were then double stained with affinity-purified anti-p235 peptide antibodies in PBS containing 1% FBS–0.5% Triton X-100 and affinity-purified anti-GLUT4 monoclonal antibody (1F8, kind gift from K. Kandror) by incubating for 90 min at 25°C. The cells were washed and exposed (30 min at 25°C) to fluorescein isothiocyanate-coupled goat anti-rabbit immunoglobulin G (IgG) (Bio Science International) for immunodetection of anti-p235 antibodies and to Texas red-coupled goat anti-mouse IgG (Molecular Probes) for immunodetection of anti-GLUT4 antibody. The cells were thoroughly washed, first in PBS–1% FBS–0.5% Triton X-100 and then only in PBS, and fixed in 4% formaldehyde. The coverslips were mounted on slides with a Slow fade Antifade kit (Molecular Probes). Fluorescence analyses were performed with a confocal microscope (Zeiss model LSM 310) with a 60/1.4 immersion lens.
p235 antibodies, immunoblotting, and immunoprecipitation.
Rabbit polyclonal anti-p235 antibodies (East Acres, Southbridge, Mass.) were directed against a synthetic mouse p235 C-terminal peptide (amino acids 2035 to 2052; purified peptide conjugated to keyhole limpet hemocyanin via either tyrosine or glutaraldehyde [Peptide Synthesis Facility, University of Massachusetts Medical Center, Worcester]; R6951, R6952, and R6953) or against a recombinant GST-p235N1-100 polypeptide (R7066 and R7069). The antisera (dilution, 1:3,000) or protein A-purified IgG was used for immunoblotting or immunoprecipitation, respectively. For Western blot analysis, the samples (20 to 100 μg of protein) solubilized in Laemmli sample buffer (29) were separated by SDS-polyacrylamide gel electrophoresis (PAGE). After transfer onto nitrocellulose filters, the blots were saturated with blocking buffer under previously specified conditions (45) and probed (16 h at 4°C) with the antibodies indicated in the figure legends. After washes, bound antibodies were detected with horseradish peroxidase-bound anti-rabbit IgG (Boehringer) and a chemiluminescence detection kit (DuPont, NEN). Quantitative p235 immunoprecipitation from 3T3-L1 adipocyte lysates prepared in RIPA buffer (50 mM Tris-HCl [pH 8.0] containing 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 1× protease inhibitor cocktail [1 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, 1 μg of pepstatin per ml, 1 mM benzamidine]) was performed with anti-p235 (R6953 and R7069) antisera preadsorbed to protein A-Sepharose CL-4B (Pharmacia) for 16 h at 4°C. Quantitative HA-p235 immunoprecipitation from transiently transfected COS-7 cell lysates, prepared in the buffer described above, was performed with both of the anti-p235 antibodies and with polyclonal anti-HA antibodies. Control immunoadsorption to a corresponding preimmune IgG was run in every experiment. Immunoprecipitates were washed extensively with RIPA buffer and analyzed by Western blotting or subjected to lipid kinase assay.
Denaturation and renaturation of blotted HA-p235.
Anti-HA immunoprecipitates (duplicate samples) derived from lysates of pCMV5-HA-p235- or pCMV5 (control)-transfected COS-7 cells were resolved by SDS–6% PAGE and transferred onto nitrocellulose membranes. Half of the membrane was probed with anti-HA antibodies to visualize the expressed HA-p235 protein and determine its position on the blot. The second half was subjected to a denaturation-renaturation procedure in order to renature p235 kinase activity blotted to the membranes (10). Briefly, the nitrocellulose membrane was soaked in 25 mM HEPES buffer (pH 7.9) containing 6 M guanidine · HCl, 25 mM NaCl, 5 mM MgCl2, and 1 mM dithiothreitol for 30 min at 4°C. The buffer was changed seven consecutive times, each time with a half concentration of the guanidine · HCl and incubation for 15 min at 4°C. After renaturation, strips corresponding to the HA-p235 protein band or the same area from the adjacent control lane were excised from the membrane and subjected to a lipid kinase assay.
Determination of Zn2+ content.
Zinc content was estimated essentially as described elsewhere (19). Briefly, proteins were dialyzed extensively against 10 mM Tris-HCl buffer (pH 8.0) containing 0.2 M NaCl and 5% glycerol and diluted further in the buffer described above to reach concentrations of 1 to 5 μM. p-Hydroxymercuriphenyl sulfonate and 4-(2-pyridylazo)resorcinol (Sigma) solutions were added to the diluted proteins (1 ml) at final concentrations of 200 and 100 μM, respectively. The A500 was read (Beckman model DU-50 spectrophotometer), and the concentration of Zn2+ was calculated by using the extinction coefficient for the 4-(2-pyridylazo)resorcinol-Zn2+ complex, 6.6 × 104 M−1 cm−1 (61).
Lipid kinase assay.
After immunoprecipitation and washings with RIPA buffer, the beads were washed twice with 50 mM HEPES (pH 7.4)–1 mM EDTA–150 mM NaCl, three times with 100 mM Tris-HCl (pH 7.5)–500 mM LiCl, twice with 10 mM Tris-HCl (pH 7.5)–100 mM NaCl–1 mM EDTA, and twice with assay buffer (50 mM Tris-HCl [pH 7.5], 1 mM EGTA, 10 mM MgCl2). The excised nitrocellulose strips corresponding to renatured HA-p235 or the control band were washed in assay buffer. The kinase reaction was carried out in the assay buffer (50-μl final volume) for 15 min at 37°C, and the reaction mixture contained 50 μM ATP, 12.5 μCi of [γ-32P]ATP (6,000 Ci/mmol; Du-Pont, NEN), and 100 μM sonicated lipid substrate (PI [Avanti Polar Lipids Inc.], PI 4-P [Sigma], PI 3-P [Matreya], or PI 5-P [Echelon Research Labs]) as indicated in the legend to Fig. 11. The reaction was stopped with 200 μl of 1 N HCl, and the lipids were extracted with 160 μl of 1:1 (vol/vol) chloroform-methanol. The lower layer was collected and washed twice with 100 μl of 1:1 (vol/vol) methanol–1 N HCl. Aliquots of the resulting organic layer were applied to a preeluted (23) and activated (110°C, 16 h) thin-layer chromatography (TLC) plate (PE SIL G, 250 μm; Whatman). The lipids were separated by a chromatographic solvent system as described previously (23). Lipid standards were detected under UV light following spraying with 0.001% primulin (Sigma). Generated radioactive products were detected by autoradiography and quantified by laser densitometry or liquid scintillation counting of silica gel scrapings corresponding to the spot of interest.
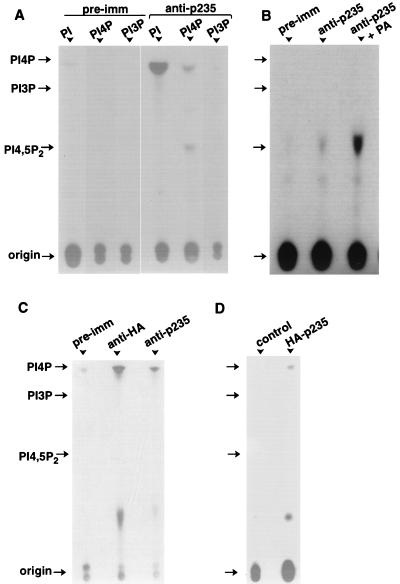
FIG. 11.
Lipid kinase activity of native and heterologously expressed p235. (A and B) Proteins from 3T3-L1 lysates were immunoprecipitated on preimmune (pre-imm) or anti-p235 peptide antibodies as described in Materials and Methods. After various washes, the immunoprecipitates were subjected to lipid kinase assay as described in Materials and Methods, in the presence of the indicated substrates (A) or PI 4-P (B). Where indicated, phosphatidic acid (PA; 140 μM; Avanti Polar Lipids Inc.) was present during the assay. (C) Lysates from COS-7 cells transfected with pCMV5-HA-p235 were immunoprecipitated with preimmune, anti-HA, or anti-p235 IgG as indicated. The immunoprecipitates were assayed for kinase activity as before with PI as a substrate. (D) COS-7 cells, transfected with pCMV5-HA-p235 or the empty vector, were immunoprecipitated with anti-HA IgG, and after SDS-PAGE the proteins were electrotransferred onto a nitrocellulose membrane. Transferred proteins were subjected to renaturation as described in Materials and Methods. Strips from both transfected and control lanes, corresponding to the electrophoretic mobility of HA-p235 (determined by Western blotting run in parallel) were excised and assayed for kinase activity in the presence of PI. Shown are TLC separations of extracted lipids and the positions of the indicated lipid standards and the origin (long arrow).
Nucleotide sequence accession number.
The GenBank accession number for the p235L sequence is AF102777.
RESULTS AND DISCUSSION
Isolation and sequence analysis of p235.
While exploring reverse transcription-PCR-based differential display screens for identifying transcripts that, like the insulin-regulated glucose transporter GLUT4, are expressed in a fat- and muscle-unique fashion, we isolated a candidate cDNA fragment. It predicted a peptide closely related to residues 1211 to 1322 of S. cerevisiae Fab1p, a probable PI(4)P5K implicated in yeast membrane trafficking (59) with no known mammalian counterpart. Screening of a mouse 3T3-F442A adipocyte library isolated a full-length cDNA (composite sequence from seven overlapping partial cDNAs [Fig. 1A]) of a novel mammalian gene encoding a protein of 2,052 amino acids with a calculated molecular weight of 233,040 (Fig. 1B). The predicted ATG initiation codon conforms well to the Kozak consensus sequence (28) for the translation initiation start and is preceded by an in-frame terminator (nucleotide 78), thus supporting the notion that this ATG represents the translation initiator of the p235 gene product (Fig. 1B). We have designated this protein p235l (p235 long form) to reflect the molecular mass of its gene product. A short splice variant with an 11-amino-acid peptide deletion (residues 108 to 118) of the p235 N-terminal region was also isolated and designated p235s (p235 short form) as discussed below.
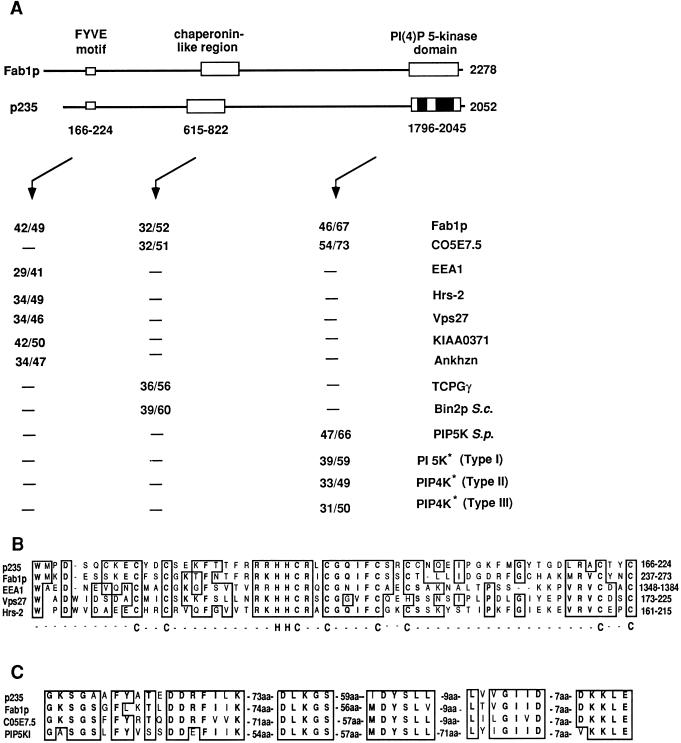
Database analysis of the deduced amino acid sequence revealed that p235 contains domains with potential functional significance, including, in order from its N terminus, a Zn2+-binding FYVE finger (49), a large chaperonin-like region (15), and, spread over the C-terminal portion, a PI(4)P5K homology region (31) (Fig. 2). As shown in Fig. 2A, the overall architecture and size of p235 are thus very similar to those of the 2,279-amino-acid yeast Fab1p (59). The individual domains of p235 demonstrate high degrees of homology to other proteins in the database. Thus, the FYVE motif, located at the N terminus of the molecule and composed of 59 amino acids (residues 166 to 224), displays 41 to 50% overall similarity to the corresponding domains in Fab1p (59), EEA1 (35), Vps27 (39), Vac1 (56), Hrs-2 (4, 27), human KIAA0371 (36), mouse Ankhzn (GenBank accession no. 2914017), and several open reading frames (ORFs) of Caenorhabditis elegans (Fig. 2A and B and data not shown). Although zinc fingers are thought to be an attribute of nuclear DNA-binding proteins, an increasing body of evidence indicates that they have a role in protein-protein (for a recent review, see reference 34) or protein-lipid (7, 17, 38) interactions. The FYVE finger has been previously documented in 11 nonnuclear proteins (49) and is characterized by eight conserved cysteines and two histidines potentially engaged in Zn2+ coordination (Fig. 2B). The present study expands this growing family with several new members, including p235, mouse Ankhzn, human KIAA0371 (Fig. 2A), and several ORFs of C. elegans (not shown), and suggests that additional members can be expected. Intriguingly, among those proteins, several are implicated in membrane trafficking and, at least with respect to EEA1, the best-studied FYVE finger-containing protein thus far, this domain is thought to dictate endosomal localization (35, 37, 49). Thus, FYVE finger presence in p235 suggests a direct or indirect role for the protein in endosome-related membrane trafficking.
FIG. 2.
Sequence analysis of p235. Similarity of mouse p235 to other FYVE finger-containing proteins, molecular chaperones, PI 5-Ks, and PIP 4-Ks. (A) Map of the domain structure of p235 and of related domains in Fab1p. Values are percentages of identity over percentages of similarity between the indicated proteins, the boxed areas, or the black portions of the PI(4)P5K domain (asterisks). The amino acid sequences used for the analysis were obtained from the following GenBank database accession numbers: S. cerevisiae Fab1p, 398498; C. elegans C05e7.5, 1065686; human EEA1, 2135066; rat Hrs-2, 1885385; S. cerevisiae Vps27, 785067; human KIAA0371, 2224683; mouse Ankhzn, 2914017; human TCP1-γ, 1729873; S. cerevisiae (S.c.) Bin2p, 493574; Schizosaccharomyces pombe (S.p.) PIP 5-K, 2894286; human PI 5-Kα (type I), 1743875; human PIP 4-K (type II), 1346720; and human PIP 4-K (type III, now considered to be type II), 1730569. (B) FYVE finger in a conserved Zn2+-binding region. Potential Zn2+-coordinating His-Cys clusters are indicated below the alignment. (C) Alignment showing similarities between subsets of highly conserved motifs with a predicted role in PI 5-K function. Similarities and identities are denoted by boxed areas. aa, amino acids.
A homology to molecular chaperones (chaperonin-like region) is present within the N-terminal half of p235 and spans approximately 200 amino acids (residues 615 to 822) (Fig. 2A). This region of p235 displays more than 50% similarity to the corresponding domains in S. cerevisiae Fab1p (59) and Bin2p (11) and a C. elegans ORF (Fig. 2A). Its apparent high degree of similarity (>55%) to members of the TCP1 protein family, i.e., TCP1-γ protein (55) (Fig. 2A), predicts a function of p235 in assisting in noncovalent protein assembly (for a recent review, see reference 15).
The p235 C terminus (amino acids 1796 to 2045) displays a high sequence similarity to the predicted catalytic region of PI(4)P5Ks in mammals and yeast, including human or murine type I (11, 32) and type II (5, 13) PI(4)P5Ks, S. cerevisiae Fab1p (59) and Mss4p (60), Schizosaccharomyces pombe PI(4)P5K (accession no. 2894286), and a C. elegans ORF (accession no. 1065686) (Fig. 2A). It includes a potential nucleotide-binding motif (GKS), magnesium coordination residues (DLKG), and downstream sequences (6) (Fig. 2C). Intriguingly, outside the predicted kinase domain, p235 shows no homology with the known mammalian PI(4)P5Ks and is distinguished from them in having additional unique sequences on the N-terminal side of the catalytic domain. Likewise, p235 displays no homology to the kinase motifs in other lipid, phosphoinositide, or protein kinases available in the database. Together, these unique features are consistent with the idea that mouse p235 defines a new class of mammalian phosphoinositide 5-K family.
The p235 protein sequence possesses other structural motifs and recognition sites, including three 21-amino-acid regions with the potential (P = 0.89, P = 0.74 and P = 0.92) to form a coiled-coil structure, 4 potential tyrosine kinase phosphorylation sites, 33 potential protein kinase C phosphorylation sites, 8 potential phosphorylation sites for cyclic AMP- and cyclic GMP-dependent protein kinases, 43 potential casein kinase II phosphorylation sites, and 9 potential N-glycosylation sites. These results indicate that p235 is likely influenced by multiple cellular regulators.
p235 message is enriched in insulin-sensitive cells and tissues.
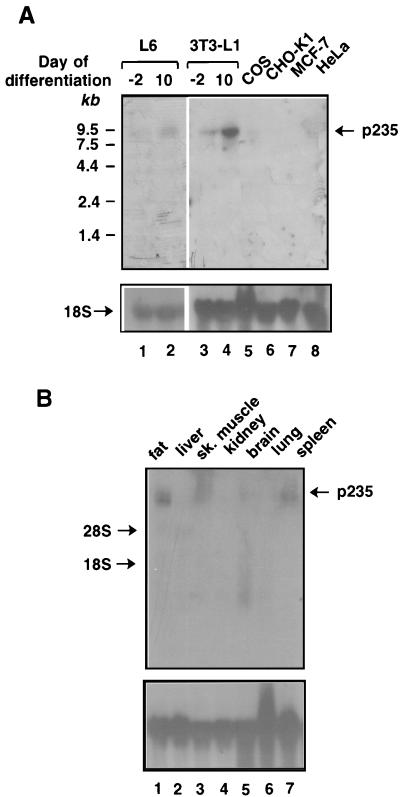
The tissue differential display methodology used to identify p235 predicts its exclusive expression in insulin-sensitive cells and tissues. We determined the levels of p235 mRNA present in insulin-sensitive versus insulin-unresponsive cultured cells (Fig. 3A). Under certain cultured conditions, 3T3-L1 mouse and L6 rat cell lines differentiate from relatively insulin-unresponsive fibroblasts to highly insulin-sensitive adipocytes and myocytes, respectively, expressing the insulin-regulated GLUT4 glucose transporter. Northern blot analysis detected p235 mRNA as a single clear-cut 9-kb transcript, highly abundant in these insulin-sensitive cultured cell models (Fig. 3A). In the respective fibroblastic lines, the p235 transcript was also detected but to a significantly lesser extent (less than threefold) (Fig. 3A). In other cultured cells lacking GLUT4, including COS, CHO, MCF-7, and HeLa cells, the p235 message was undetectable (Fig. 3A). These data indicate that the transcript level of p235 substantially increases upon differentiation into insulin-responsive cells.
FIG. 3.
Northern blot analysis of p235 transcript levels in cultured cells (A) and mouse tissues (B). Each lane was loaded with 20 μg of total RNA isolated from the indicated cultured cells or mouse tissues as described in Materials and Methods. Blots were sequentially hybridized with a 32P-labeled cDNA fragment of p235 corresponding to nucleotides 2200 to 3481 (A and B, upper gels) or a chicken 18S ribosomal cDNA probe (A and B, lower gels) under high-stringency conditions. L6 myocytes and 3T3-L1 adipocytes were used at the indicated day of the differentiation program. The increase of p235 transcript levels in 3T3-L1 adipocytes was threefold (two independent differentiations). sk, skeletal.
The results of Northern blot analysis with total RNA derived from various mouse tissues are depicted in Fig. 3B. This analysis confirmed the presence of p235 mRNA in fat and muscle tissues (Fig. 3B). However, while in the tissues analyzed here, specifically, brain, liver, kidney, and lung, the p235 message was undetectable, a clear-cut 9-kb message was documented for spleen (Fig. 3B). These results indicate that p235 is expressed in a tissue-specific manner but is not absolutely restricted to adipose and muscle.
Genomic Southern blot analysis predicts one p235 gene.
Existence of multiple isoforms of both human and murine type I and type II PI(4)P5Ks (9, 22, 31) prompted us to determine the number of p235-related genes in the mouse genome. To address this issue, genomic DNAs derived from mouse and rat were analyzed by Southern blotting with a probe derived from a region of p235 with a unique sequence (EcoRI fragment of clone IRG1, nucleotides 2200 to 3481 [Fig. 1A]). As demonstrated in Fig. 4, the probe hybridized with single fragments from Sprague-Dawley rat and Swiss Webster mouse DNAs, following genomic DNA fragmentation with EcoRI, either alone or combined with EcoRV, NcoI, and HindIII. The slight variations in the sizes of the DNA fragments between mice and rats detected on the blots are apparently related to characteristic species variations (Fig. 4). These data are consistent with the notion that p235 is a single-copy gene in the mouse and rat genomes.
FIG. 4.
Southern blot analysis of restriction endonuclease-digested genomic DNAs from rat and mouse. Genomic DNA isolated from Sprague-Dawley rat or Swiss Webster mouse liver was digested with EcoRI (lanes 2 and 7), EcoRI plus EcoRV (lanes 3 and 8), EcoRI plus NcoI (lanes 4 and 9), and EcoRI plus HindIII (lanes 5 and 10) or not digested (lanes 1 and 6). Electrophoresis and transfer were performed as described in Materials and Methods. The blot was hybridized with probe 2 (nucleotides 2200 to 3481). A BstE II digest of λ DNA (New England Biolabs) was used as a molecular size standard.
Alternative splicing.
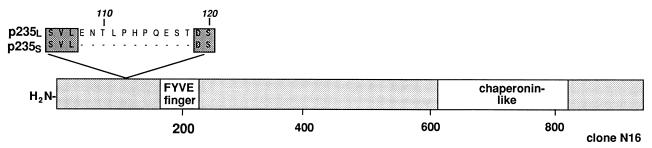
While Southern and Northern blot analyses predict a single-copy gene encoding p235, a variant of the p235 N-terminal region was isolated from the adipocyte cDNA library. Sequence analysis of two additional clones, N16 (2.9 kbp) and N17 (2.7 kbp), demonstrated identity to p235 over the entire region covered by them (amino acids 1 to 955) except for a 33-nucleotide deletion that led to elimination of an 11-amino-acid peptide (ENTLPHPQEST) in the region from residues 108 to 118 of the deduced p235l amino acid sequence (Fig. 1B and 5). This observation suggests that the two clones are products of alternative splicing of the p235 gene thus encoding a short and a long p235 splice variant, called herein p235s and p235l, respectively (Fig. 5). The two forms are likely equally abundant, as judged by the equal numbers of isolated cDNA clones encoding either form, but are indistinguishable in Northern blot analysis due to the small differences between their mRNA sizes (∼0.35%). These results indicate that mouse adipocytes express at least two forms of p235 distinguished by the presence or absence of an 11-amino-acid stretch; the functional significance of this difference is yet to be elucidated.
FIG. 5.
Schematic representation of the 11-amino-acid insert in p235l. The short splice variant, p235s, is predicted by the clone N16 (nucleotides 130 to 2876), which is identical to p235l over its whole sequence from amino acids 1 to 955, with the exception of the indicated deletion of an 11-amino-acid stretch (amino acids 108 to 118). Clone N16 was isolated from the mouse adipocyte cDNA library by hybridization to probe 2 (nucleotides 2200 to 3481) as described in Materials and Methods.
p235 is localized to vesicle structures.
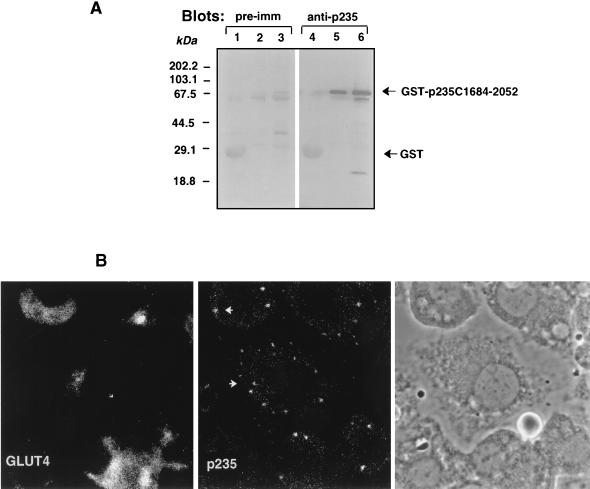
To examine the intracellular localization of the endogenous p235 protein, we first developed specific antibodies against the C-terminal peptide (residues 2035 to 2052). Anti-peptide p235 antibodies specifically and quantitatively recognized a protein fragment of p235 that carries the C terminus, GST–C1684-2052, expressed as a GST fusion protein in a bacterial host (Fig. 6A). In order to achieve specific visualization of the endogenous p235, anti-p235 antiserum (R6953) was affinity purified on this C-terminal protein fragment. Results of immunofluorescence analysis in serum-deprived 3T3-L1 adipocytes with p235 affinity-purified antibodies are illustrated in Fig. 6B. A distinctive peripheral vesicular pattern of the fluorescence staining associated with immunoreactive p235 was observed. A diffuse staining only at a background level was usually documented under these conditions, consistent with the idea of a lack of substantial cytosolic amounts of p235. Some vesicles could be observed in the perinuclear area, where GLUT4 transporter protein was visualized, as demonstrated by double staining with the monoclonal 1F8 anti-GLUT4 antibody (Fig. 6B). However, both proteins displayed different staining patterns; GLUT4 shows typical perinuclear staining as we have documented previously (44), while p235 shows punctate peripheral staining. These observations suggest a lack of significant colocalization between those two proteins in resting 3T3-L1 adipocytes. Preimmune sera or equal amounts of nonimmune IgG showed only low background immunofluorescence under these conditions. Together, these results support the notion that p235 is principally a membrane-bound protein associated with vesicle structures in 3T3-L1 adipocytes. However, the exact definition of p235-containing vesicles remains to be established.
FIG. 6.
Intracellular localization of endogenous p235 in 3T3-L1 adipocytes. (A) Specificities of anti-p235 antibodies. GST or GST–p235-C1684-2052 fusion proteins were expressed in E. coli XA-90 and purified on glutathione-agarose beads as described in Materials and Methods. Aliquots of the beads equivalent to 2 μg of GST (lanes 1 and 4) and 200 and 800 ng of GST–C1684-2052 (lanes 2 and 5 and lanes 3 and 6, respectively) were boiled in Laemmli sample buffer, resolved by SDS-PAGE (10.5% acrylamide; duplicate gels) and analyzed by immunoblotting with preimmune (pre-imm; dilution, 1:500) or anti-p235 (R6953; dilution, 1:3,000) antiserum as indicated. Anti-p235 peptide antiserum, generated against the C-terminal 18 residues (amino acids 2035 to 2052), detects specifically and in a linear relationship the C-terminal p235 protein (lanes 5 and 6). (B) Immunofluorescence microscopy of immunoreactive p235 in 3T3-L1 adipocytes. Cells, grown and differentiated on coverslips, were washed, fixed in methanol, permeabilized, and stained with the anti-p235 antibodies affinity purified on a GST–C1684-2052 protein band cut from the nitrocellulose membrane shown in panel A. For the double labeling, mouse monoclonal anti-GLUT4 antibody (1F8) was used. Immunodetection of anti-p235 was achieved with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG, and that of anti-GLUT4 IgG was achieved with Texas red-conjugated goat anti-mouse IgG. The phase-contrast image depicts cell shape and nuclei of the same field.
p235 N terminus confers the peripheral vesicular pattern.
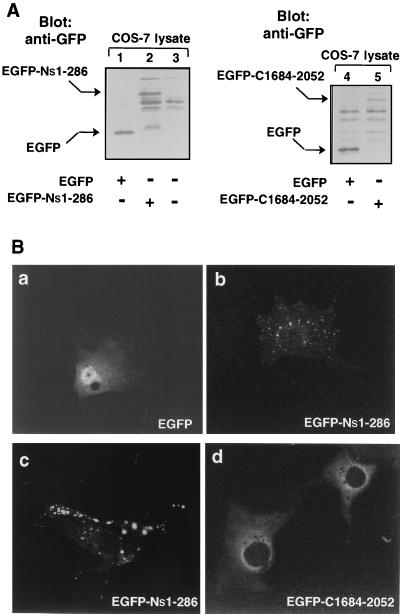
The distinctive peripheral vesicular pattern of immunoreactive p235 on the images illustrated in Fig. 6B suggests the possibility that a membrane-targeting signal is present within p235. A hallmark of the p235 N-terminal amino acid sequence is determined by the presence of a FYVE finger (amino acids 166 to 224). Recent studies indicate that the FYVE finger in EEAI confers localization to early endosomes (49), a characteristic typical of this protein (35, 37). These results imply that the p235 FYVE finger may also direct the molecule to membranes of the endocytic pathway and may account for the peripheral punctate appearance of immunoreactive p235. To test this hypothesis, the p235 N-terminal region (amino acids 1 to 286) was fused with GFP and the resultant new vector, pEGFP-Ns1-286, was used to transiently express the N-terminal portion in COS-7 cells. We first confirmed the expression of the predicted proteins by Western blotting of SDS-PAGE-resolved COS cell lysates. As demonstrated in Fig. 7A, anti-GFP antibodies visualized the p235-Ns1-286 protein fragment as a 70-kDa protein band, consistent with the expected size of the fusion protein. Next, fluorescence microscopy analysis of the transfected cells revealed that while the green fluorescent signals of EGFP-expressing cells were detected in both the cytoplasm and the nucleus, in agreement with other studies (24, 48), those of EGFP–Ns1-286 were found associated exclusively with vesicular structures reminiscent of endosomes (Fig. 7B). No diffuse staining was documented even at substantially high levels of expression of EGFP–Ns1-286 under the conditions of our experiments (Fig. 7B), in line with the observed absence of detectable p235 or p235-Ns1-286 soluble forms (Fig. 6B and data not shown). Conversely, no vesicular appearance of the fluorescence associated with the C-terminal portion of the molecule, residues 1684 to 2052, was observed (Fig. 7B). When expressed as a similar EGFP construct in COS cells, verified by immunoblotting with anti-GFP antibodies (Fig. 7A), the green fluorescent signals of EGFP–C1684-2052 were found perinuclearly along with diffuse staining, a characteristic of its presence in the cytosol (Fig. 7B). Taken together, these data are consistent with the idea that the N-terminal part of the molecule, probably through the FYVE motif, confers the observed peripheral vesicular pattern of p235 intracellular localization.
FIG. 7.
Expression and intracellular localizations of the p235 N-terminal and the C-terminal regions in transfected COS-7 cells. COS-7 cells were transiently transfected with the EGFP–Ns1-286 and EGFP–C1684-2052 cDNAs either on a 35-mm-diameter dish (A) or on coverslips (B) as indicated in Materials and Methods. (A) Cell lysates were collected in RIPA buffer 24 h after transfection and subjected to SDS-PAGE, electrotransfer to nitrocellulose membranes, and immunoblotting with anti-GFP monoclonal antibody (2 μg/ml). (B) Cells were washed and fixed in methanol 15 h after transfection with the indicated constructs. Localization of the expressed proteins was determined by the fluorescence signals of GFP with a standard fluorescein filter of a Zeiss confocal microscope. Characteristic targeting of EGFP–Ns1-286 protein to vesicle structures reminiscent of endosomes is observed at both low (b) and high (c) levels of expression. No punctate peripheral staining is associated with EGFP–C1684-2052 (d), but rather perinuclear and diffuse green fluorescent signals, excluded from the nucleus, are observed.
p235 FYVE motif binds zinc.
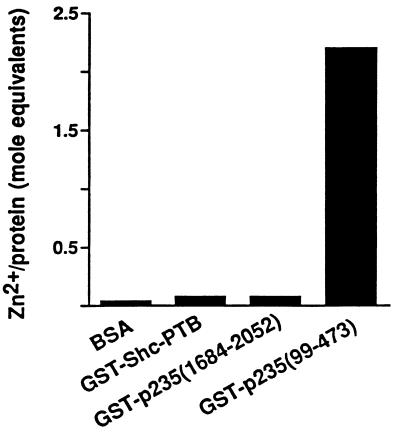
As demonstrated in Fig. 2B, the FYVE fingers in p235 and in other proteins are characterized by eight conserved cysteines and two histidines, all potentially engaged in Zn2+ coordination (49). Both the EEA1 FYVE motif and Hrs-2 have been shown to bind Zn2+, and mutagenesis in the conserved Cys or His within the EEA1 FYVE motif has been shown to decrease Zn2+ content in the mutant proteins (4, 49). To test whether p235 possesses the ability to coordinate zinc and whether its FYVE domain contributes to this property, we expressed the N-terminal (amino acids 99 to 473) and the C-terminal (amino acids 1684 to 2052) regions of p235 as GST fusion proteins. The proteins were then purified on glutathione agarose beads and subjected to a colorimetric assay to determine the zinc contents of the fusion proteins. As illustrated in Fig. 8, while virtually no zinc was found associated with GST–C1684-2052 and the negative controls (bovine serum albumin and the expressed phosphotyrosine-binding (PTB) domain of Shc as a GST fusion protein, GST–Shc1-233), the GST–Nl99-473 fusion protein that carries the FYVE motif was found to contain 2.2 mole equivalents of Zn2+. These results indicate that the p235 protein binds Zn2+, an ability associated with the N-terminal region of the molecule, presumably through the cysteine-rich FYVE finger.
FIG. 8.
Determination of Zn2+ contents in the N-terminal and C-terminal regions of p235. The amounts of Zn2+ associated with the indicated proteins were determined as described in Materials and Methods. After the readout EDTA was added to a final concentration of 2 mM. The difference in the values for A500 measured in the absence and presence of EDTA was used to calculate the mole equivalents of liberated Zn2+. Shown are the results of a representative experiment of two with identical results. BSA, bovine serum albumin.
The peripheral vesicular pattern of EGFP–Ns1-286 is dependent upon Zn2+ presence.
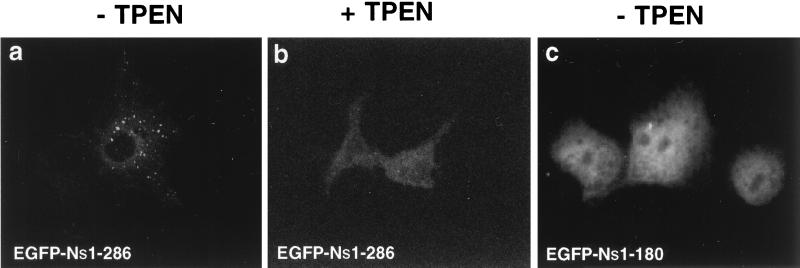
The properties of the N-terminal portion of the p235 molecule of binding Zn2+, demonstrated in Fig. 8, and localizing to vesicular structures reminiscent of endosomes, demonstrated in Fig. 7, may be related. We tested this hypothesis by examining the intracellular localization of EGFP–Ns1-286 in transfected cells subjected to Zn2+ depletion by TPEN treatment. This lipid-soluble cell-permeable chelator exhibits an extraordinarily high affinity for heavy metals, with the highest K association value being for Zn2+ (K association values equal to 1010.27 M−1, 1014.61 M−1, and 1015.58 M−1 for Mn2+, Fe2+, and Zn2+, respectively [3, 20]). As illustrated in Fig. 9, short application of the chelator (30 min) at concentrations that have no noticeable adverse side effects (200 μM [43]) to COS-7 cells, transiently expressing EGFP–Ns1-286, resulted in a profound diffuse staining pattern. By contrast, no change in the fluorescence associated with EGFP was documented (not shown). Thus, Zn2+ depletion causes a pronounced dissociation of EGFP–Ns1-286 from the vesicle compartments to the cytosol. Similar cytosolic distribution was documented in COS-7 cells heterologously transfected with EGFP–Ns1-180, a construct missing the most conserved portion of the FYVE finger (Fig. 9). Together, these results are consistent with the idea that the peripheral vesicular pattern of the green fluorescent signals associated with EGFP–Ns1-286 protein is dependent upon a Zn2+-sensitive binding mechanism and an intact FYVE finger.
FIG. 9.
Effect of zinc depletion or FYVE finger deletion on the peripheral intracellular localization of the p235 N-terminal region in transiently transfected COS-7 cells. Fifteen hours after transfection with EGFP–Ns1-286 (a and b) or EGFP–Ns1-180 constructs (c), the cells were serum starved (3 h) and then treated (b) or not treated at 37°C with TPEN (30 min, 200 μM). The cells were washed and fixed in methanol. Green fluorescent signals were detected with a confocal microscope. Upon TPEN treatment, a dramatic change in the EGFP–Ns1-286 staining pattern from endosome-like to diffuse cytoplasmic was readily observed, similar to the staining pattern of the EGFP–Ns1-180 construct lacking the FYVE finger.
Recent studies, published while this manuscript was under revision, have demonstrated that the FYVE fingers of several proteins in yeast and mammalian cells function as PI 3-P-binding domains and that this interaction confers an endosomal localization (7, 17, 38). Importantly, PI 3-P–FYVE finger interactions appear critically dependent upon an intact Zn2+-binding FYVE finger, as both Zn2+ chelation by TPEN and a point mutation within the FYVE finger completely abolished the binding of EEA1 FYVE finger to PI 3-P-containing liposomes (7). Our data are in agreement with these results, and together they underscore the Zn2+ sensitivity of the FYVE finger’s ability to direct molecules to specific intracellular loci.
Biochemical detection of the endogenous and epitope-tagged p235.
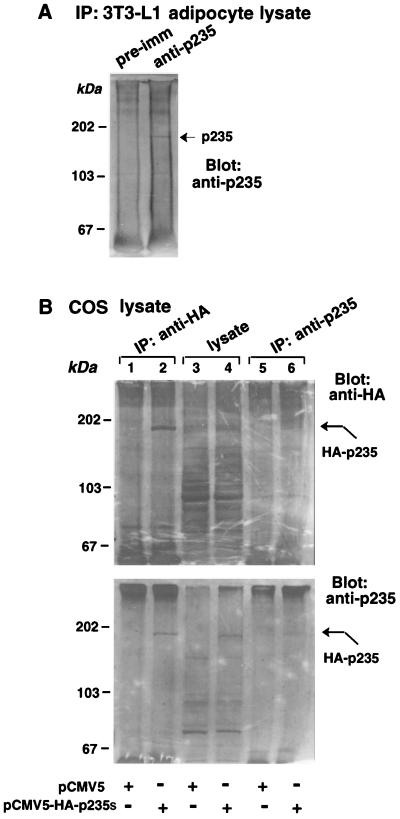
In order to detect p235 protein biochemically, 3T3-L1 adipocyte lysates were immunoprecipitated with either the immune IgG fraction, the crude antiserum of the anti-p235 antibody preparation (R6953), or the corresponding preimmune serum. Immunoprecipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting with the anti-p235 antiserum, whose specificity in such analysis is illustrated in Fig. 6A. A single 180-kDa protein band, not seen with the corresponding preimmune IgG, was detected in the p235 immunoprecipitates (Fig. 10A). An identical protein band has been visualized with the antiserum directed against the N-terminal 100-amino-acid segment (R7069; data not shown) suggesting that the detected band is the authentic adipocytic p235.
FIG. 10.
Detection of endogenous p235 in 3T3-L1 adipocytes (A) and expressed epitope-tagged p235 in COS-7 cells (B) by immunoprecipitation and Western blotting. (A) Proteins from 3T3-L1 lysates were immunoprecipitated (IP) on preimmune (pre-imm) or anti-p235 antibodies (R6953) as described in Materials and Methods. After five washes in RIPA buffer, the immunoprecipitated proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-p235 peptide antiserum (R6953). (B) COS-7 cells were transfected with pCMV5-HA-p235s cDNA or with the empty vector as indicated. Cell lysates were immunoprecipitated with the anti-p235 or anti-HA antiserum. The lysates of transfected cells, together with HA and p235 immunoprecipitates, were sequentially Western blotted with anti-HA and anti-p235 antisera as indicated.
p235 electrophoretic mobility was substantially higher than that predicted by the deduced p235 sequence. To verify that the immunoprecipitated protein band represents p235, we exploited an epitope-tagging approach of full-length p235 combined with heterologous expression. Lysates of COS-7 cells transiently transfected with the HA-epitope-tagged p235s-pCMV5 construct or the empty vector were immunoprecipitated or not with anti-HA and anti-p235 antibodies and subjected to SDS-PAGE. The blots were probed with anti-HA or anti-p235 antiserum (Fig. 10B). Immunoblotting with both probes illustrated that anti-HA IgG immunoprecipitated a single band migrating at ∼180 to 190 kDa only from the HA-p235-transfected cells (Fig. 10B, lane 1 versus lane 2). The same band was clearly visible in the lane with total lysates (Fig. 10B, compare lanes 3 and 4). Similarly, a protein band with the same electrophoretic mobility was selectively detected in the p235 immunoprecipitates derived from HA-p235-transfected cells, as judged by probing with both antibodies (Fig. 10B, lane 5 versus lane 6). Based on the apparent intensity of this band, the immunoprecipitating potency of the anti-p235 antibody was less pronounced than that of anti-HA polyclonal antibody (Fig. 10B, lane 2 versus lane 6). Thus, the mobility of immunoprecipitated heterologously expressed p235 was in the vicinity of the 180-kDa protein band detected in 3T3-L1 adipocytes in similar analyses, supporting the notion that the band documented in Fig. 10A represents the authentic adipocytic protein, which, for reasons that remain to be identified, displays an anomalous mobility by SDS-PAGE.
Lipid kinase activity of endogenous and epitope-tagged p235.
The overall sequence and structural similarity of p235 to yeast Fab1p, a probable PI(4)P5K (59), prompted us to examine whether p235 acts to generate PI 4,5-P2. For this purpose, 3T3-L1 adipocyte lysates were immunoprecipitated with polyclonal anti-p235 C-terminal peptide antibodies or preimmune IgG and the pellets were subjected to lipid kinase assay in the presence of PI 4-P as a substrate. TLC analysis of the lipid products revealed generation, although to a modest level, of PI 4,5-P2 (Fig. 11A). Intriguingly, PI 4,5-P2 generation by p235 was largely activated by phosphatidic acid (6- to 15-fold) (Fig. 11B), similarly to type I PI 5-Ks (22, 23). Surprisingly, when PI was provided as a substrate, the 32P incorporation was about 2 orders of magnitude more efficient than that with PI 4-P (Fig. 11A). The product generated migrated in the vicinity of monophosphorylated PI, i.e., slightly lower (between 1.5 and 3.5 mm) than the PI 4-P standard and higher than the PI 3-P standard, which difference may result from the different natures of the fatty acids in the lipid substrates. No other coproducts were detected with PI as a substrate, suggesting that, unlike PI 5-Ks, which display a concerted activity and convert PI to PI 5-P and PI 4,5-P2 (53), p235 has a striking specificity toward a single OH position of the inositol ring (Fig. 11A). Virtually no activity was detected in the presence of PI 3-P (Fig. 11A) or PI 5-P (data not shown) as substrates.
We next analyzed whether heterologously expressed p235 in COS-7 cells displays a similar ability to convert PI to PIP. The expressed HA-p235 was immunoprecipitated with either anti-HA or anti-p235 antibodies from detergent-solubilized cell lysates as illustrated in Fig. 10B. Aliquots of these same immunoprecipitates were also analyzed for lipid kinase activity. Significant amounts of PIP were routinely produced with both types of the immune IgG but not with the preimmune serum (Fig. 11C). More importantly, the amount of generated PIP closely corresponded to the amount of the immunoprecipitated HA-p235 protein (Fig. 10B, lane 2 versus lane 6), further supporting the notion that p235 is the source of the measured lipid kinase activity. Finally, to definitely determine that the p235 kinase activity is intrinsic, rather than coming from associated protein(s), we took advantage of the reported ability of PI 5-Ks to renature from SDS-polyacrylamide gels (23). Lysates of COS-7 cells transfected with pCMV5-HA-p235s or with the empty vector were subjected to immunoprecipitation with anti-HA antiserum. Immunoprecipitates were resolved by SDS-PAGE, and after transfer, the proteins on nitrocellulose membrane were renatured as described in Materials and Methods. Nitrocellulose strips corresponding to the HA-p235 electrophoretic mobility (detected by Western blotting analysis of duplicate samples run in parallel [Fig. 10B]) from the transfected or nontransfected cells were excised and subjected to lipid kinase assay. As illustrated in Fig. 11D, the kinase activity measured with PI substrate was renatured only in the HA-p235 strip. Noteworthy, p235 protein fragment harboring the predicted catalytic region and expressed as a fusion protein (GST–C1684-2052) in a bacterial host did not display detectable activity under the conditions of our kinase assay, suggesting that additional regions of the molecule are also essential. Together, these results demonstrate that p235 displays the ability to phosphorylate PI species yet its substrate specificity is distinct from both PI 5-Ks (type I) and PIP 4-Ks (type II). The finding that p235 in a unique fashion preferentially generates PIP from PI and, to a lesser extent, PI 4,5-P2 from PI 4-P but not from PI 5-P, indicates that p235 defines a novel class of the phosphoinositide kinase family.
Several lines of evidence suggest that p235 phosphorylates the D-5 rather than the D-4 position. Thus, it displays higher sequence homology to PI 5-Ks than to PIP 4-Ks, and as with PI 5-Ks, the generation of PI 4,5-P2 from PI 4-P is activated by phosphatidic acid (22, 23). Next, only if the D-4 position, and not the D-5 position, is already phosphorylated, p235 generates PI 4,5-P2. Finally, p235 has no homology with any PI 4-Ks presently known, and unlike many of them, p235 PIP-generating activity is only slightly inhibited (<15%) by adenosine at 300 μM (data not shown), a concentration shown to completely inhibit PI 4-K (58). However, more reliable separation techniques than TLC (40) will be required to definitely determine p235’s specificity toward the D-5 position of the inositol ring.
Although the specific function of p235 in the complex regulation of living cells is unknown, p235 likely operates to support the intracellular PIP pool and, to a lesser extent, the PI 4,5-P2 pool. It remains to be elucidated whether the PI 5-P lipid product has a specific function in eukaryotic cell regulation on its own and/or serves as a substrate for further actions of PIP 4-Ks (40) or PI 3-Ks (52) (separately or in concert) to generate PI 4,5-P2, PI 3,4,5-P3, or PI 3,5-P2. Several recent studies demonstrate the importance of a phosphorylated D-5 position in polyphosphoinositides for membrane trafficking events. Thus, studies with yeast indicate a rapid PI 3,5-P2 burst associated with an acute osmotic adaptation thought to involve an acceleration of vesicle trafficking (14). Insulin-regulated action on GLUT4 translocation to the cell surface, associated with a PI 3,4-P2 and PI 3,4,5-P3 increase (26, 41), appears extremely sensitive to a selective dephosphorylation of position 5 of PI 3,4,5-P3, achieved by overexpressing the 5′-inositolphosphatase SHIP in 3T3-L1 adipocytes (54). The role of p235 and its lipid products in membrane trafficking is further reinforced by the findings demonstrating that the loss of function of S. cerevisiae Fab1p, whose phospholipid product remains to be identified, causes multiple defects in vacuole function and morphology (59). Finally, the presence of a FYVE finger in p235, a motif found in several yeast and mammalian cell proteins, such as Hrs-2, Vac1, Vps27, and Fab1p (4, 39, 56, 59), relevant in membrane trafficking, adds further support to the predicted role of p235 and/or its direct lipid product in the mechanisms regulating intracellular membrane trafficking. Intriguingly, mouse Hrs protein undergoes tyrosine phosphorylation in response to growth factors (27), implying a possible hormonal regulation of membrane trafficking events by FYVE motif-containing proteins. It remains to be identified whether p235 PI 5-K is specifically activated by receptor signaling systems, particularly that initiated by the insulin receptor. The cloning of p235 cDNA and the extensive characterization of its gene product, presented here, should facilitate efforts to answer this question.
ACKNOWLEDGMENTS
We thank members of the core facilities involved in this study, including M. Hagen (CMMG Macromolecular Core Facility, Wayne State University [WSU]), K. Moin and L. Mayernik (Morphology Core Facility, WSU), and R. Caraway (Peptide Synthesis Core Facility, University of Massachusetts Medical Center), for their outstanding work. We thank B. Spiegelman for the gift of the adipocyte cDNA library; K. Kandror for anti-GLUT4 antibodies; and M. Czech, L. Kozma, and J. Buxton for the GST–Shc-PTB construct and anti-HA antiserum. The technical help of the graduate students D. Post and D. Draganov and the assistance of S. Doxsey and M. Blomberg in analysis of coiled-coil domains are gratefully appreciated.
This project was supported by an ADA Career Development Award (A.S.).
REFERENCES
- 1.Alb J G, Jr, Kearns M A, Bankaitis V A. Phospholipid metabolism and membrane dynamics. Curr Opin Cell Biol. 1996;8:534–541. doi: 10.1016/s0955-0674(96)80032-9. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Arslan P, Di Virgilio F, Beltrame M, Tsien R Y, Pazzan T. Cytosolic Ca2+ homeostasis in Ehrlich and Yoshida carcinomas. J Biol Chem. 1985;260:2719–2727. [PubMed] [Google Scholar]
- 4.Bean A J, Seifert R, Chen Y A, Sacks R, Scheller R H. Hrs-2 is an ATPase implicated in calcium-regulated secretion. Nature. 1997;385:826–829. doi: 10.1038/385826a0. [DOI] [PubMed] [Google Scholar]
- 5.Boronenkov I V, Anderson R A. The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J Biol Chem. 1995;270:2881–2884. doi: 10.1074/jbc.270.7.2881. [DOI] [PubMed] [Google Scholar]
- 6.Bourne H R, Sanders D A, McCormick F. The GTP-ase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–126. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 7.Burd G C, Emr S D. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter C L, Cantley L C. Phosphoinositide kinases. Curr Opin Cell Biol. 1996;8:153–158. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 9.Castellino A M, Parkers G J, Boronenkov I V, Anderson R A, Chao M V. A novel interaction between the juxtamembrane region of the p55 tumor necrosis factor receptor and phosphatidylinositol-4-phosphate 5-kinase. J Biol Chem. 1997;272:5861–5870. doi: 10.1074/jbc.272.9.5861. [DOI] [PubMed] [Google Scholar]
- 10.Celenza J L, Carlson M. Renaturation of protein kinase activity of protein blots. Methods Enzymol. 1991;200:423–430. doi: 10.1016/0076-6879(91)00158-s. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Sullivan D, Huffaker T. Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc Natl Acad Sci USA. 1994;91:9111–9115. doi: 10.1073/pnas.91.19.9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czech M P. Molecular action of insulin on glucose transport. Annu Rev Nutr. 1995;15:441–471. doi: 10.1146/annurev.nu.15.070195.002301. [DOI] [PubMed] [Google Scholar]
- 13.Divecha N, Truong O, Hsuan J J, Hinchliffe K A, Irvine R F. The cloning and sequence of the C isoform of PtdIns4P 5-kinase. Biochem J. 1995;309:715–719. doi: 10.1042/bj3090715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dove K K, Cooke F T, Douglas M R, Sayers L G, Parker P J, Mitchell R H. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 15.Ellis R J. Steric chaperones. Trends Biochem Sci. 1998;23:43–45. doi: 10.1016/s0968-0004(98)01175-x. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 17.Gaullier J-M, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 18.GenomeNet. Search patterns in protein sequences. 1997. [Google Scholar]
- 19.Giedroc D P, Keating K M, Williams K R, Konigsberg W H, Coleman J E. Gene 32 protein, the single-stranded DNA binding protein from bacteriophage T4, is a zinc metalloprotein. Proc Natl Acad Sci USA. 1986;83:8452–8456. doi: 10.1073/pnas.83.22.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hesketh T R, Smith G A, Moore J P, Taylor M V, Metcalf J C. Free cytosolic calcium concentration and the mitogenic stimulation of lymphocytes. J Biol Chem. 1983;258:4876–4882. [PubMed] [Google Scholar]
- 21.Ikonomov O C, Jacob M H. Differential display protocol with selected primers that preferentially isolates mRNAs of moderate- to low-abundance in a microscopic system. BioTechniques. 1996;20:1030–1042. doi: 10.2144/96206rr01. [DOI] [PubMed] [Google Scholar]
- 22.Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T, Oka Y. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J Biol Chem. 1998;273:8741–8748. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins G H, Fisette P L, Anderson R A. Type I phosphatidylinositol 4-phosphate 5-kinases are specifically stimulated by phosphatidic acid. J Biol Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- 24.Jiang W, Hunter T. Analysis of cell-cycle profiles in transfected cells using a membrane-targeted GFP. BioTechniques. 1997;24:348–354. [PubMed] [Google Scholar]
- 25.Kapeller R, Cantley L C. Phosphatidylinositol 3-kinase. Bioessays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 26.Kelly K L, Ruderman N B. Insulin-stimulated phosphatidylinositol 3-kinase. J Biol Chem. 1993;268:4391–4398. [PubMed] [Google Scholar]
- 27.Komada M, Kitamura N. Growth factor induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol Cell Biol. 1995;15:6213–6221. doi: 10.1128/mcb.15.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Liscovitch M, Cantley L C. Signal transduction and membrane traffic: the PITP/phosphoinositide connection. Cell. 1995;81:659–662. doi: 10.1016/0092-8674(95)90525-1. [DOI] [PubMed] [Google Scholar]
- 31.Loijens J C, Boronenkov I V, Parker G J, Anderson R A. The phosphatidylinositol 4-phosphate 5-kinase family. Adv Enzyme Regul. 1996;36:115–140. doi: 10.1016/0065-2571(95)00005-4. [DOI] [PubMed] [Google Scholar]
- 32.Loijens J C, Anderson R A. Type I phosphatidylinositol 4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J Biol Chem. 1996;271:32937–32943. doi: 10.1074/jbc.271.51.32937. [DOI] [PubMed] [Google Scholar]
- 33.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 34.Mackay J P, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 35.Mu F T, Callaghan J M, Steele-Mortimer O, Stenmark H, Parton R G, Campbell P L, McCluskey J, Yeo J P, Tock E P, Toh B-H. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “finger” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 36.Nagase T, Ishikawa K, Nakajima D, Ohir M, Seki N, Miyajim N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1997;4:141–150. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- 37.Patki V, Virbasius J, Lane W S, Toh B-H, Shpetner H S, Corvera S. Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 1997;94:7326–7330. doi: 10.1073/pnas.94.14.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patki V, Lawe D C, Corvera S, Virbasius J, Chavla A. A functional PtdIns(3)P-binding motif. Nature. 1998;394:433–434. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- 39.Piper R C, Cooper A A, Yang H, Stevens T H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rameh L E, Tolias K F, Duckworth B C, Cantley L C. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;309:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 41.Ruderman N, Kapeller R, White M F, Cantley L C. Activation of phosphatidylinositol-3-kinase by insulin. Proc Natl Acad Sci USA. 1990;87:1411–1415. doi: 10.1073/pnas.87.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T, editors. Analysis and cloning of eukaryotic genomic DNA in molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Shisheva A, Gefel D, Shechter Y. Insulin-like effects of zinc ion in vitro and in vivo. Preferential effects on desensitized adipocytes and induction of normoglycemia in streptozocin-induced rats. Diabetes. 1992;41:982–988. doi: 10.2337/diab.41.8.982. [DOI] [PubMed] [Google Scholar]
- 44.Shisheva A, Buxton J, Czech M. Differential intracellular localizations of GDP-dissociation inhibitor isoforms. Insulin-dependent redistribution of GDP-dissociation inhibitor-2 in 3T3-L1 adipocytes. J Biol Chem. 1994;269:23865–23868. [PubMed] [Google Scholar]
- 45.Shisheva A, Czech M P. Association of cytosolic Rab4 with GDI isoforms in insulin-sensitive 3T3-L1 adipocytes. Biochemistry. 1997;22:6564–6570. doi: 10.1021/bi970202g. [DOI] [PubMed] [Google Scholar]
- 46.Shisheva A, Doxsey S J, Buxton J M, Czech M P. Pericentriolar targeting of GDP-dissociation inhibitor isoform 2. Eur J Cell Biol. 1995;68:143–158. [PubMed] [Google Scholar]
- 47.Shisheva A, Sudhof T C, Czech M P. Cloning, characterization, and expression of a novel GDP-dissociation inhibitor isoform from skeletal muscle. Mol Cell Biol. 1994;14:3459–3468. doi: 10.1128/mcb.14.5.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stearns T. Green fluorescent protein. The green revolution. Curr Biol. 1995;5:262–264. doi: 10.1016/s0960-9822(95)00056-x. [DOI] [PubMed] [Google Scholar]
- 49.Stenmark H, Aasland R, Toh B-H, D’Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J Biol Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- 50.Summers S A, Birnbaum M J. A role for the serine/threonine kinase, Akt, in insulin-stimulated glucose uptake. Biochem Soc Trans. 1997;25:981–988. doi: 10.1042/bst0250981. [DOI] [PubMed] [Google Scholar]
- 51.Toker A. The synthesis and cellular roles of phosphatidylinositol 4,5-bisphosphate. Curr Opin Cell Biol. 1998;10:254–261. doi: 10.1016/s0955-0674(98)80148-8. [DOI] [PubMed] [Google Scholar]
- 52.Toker A, Cantley L C. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 53.Tolias K F, Rameh L E, Ishihara H, Shibassaki Y, Chen J, Prestwich G D, Cantley L C, Carpenter C L. Type I phophatidylinositol-4-phosphate 5-kinases synthesize the novel lipid phosphatidylinositol 3,5-bisphosphate and phosphatidylinositol 5-phosphate. J Biol Chem. 1998;273:18040–18046. doi: 10.1074/jbc.273.29.18040. [DOI] [PubMed] [Google Scholar]
- 54.Vollenweider, P., M. Clodi, T. Imamura, S. Martin, and J. M. Olefsky. 1998. Overexpression of an SH2 containing 5′inositolphosphatase (SHIP) inhibits insulin-induced GLUT4 translocation and growth factor induced actin filament rearrangement. Diabetes 47(Suppl. 1):A332. [DOI] [PMC free article] [PubMed]
- 55.Walkley N A, Demaine A G, Malik A N. Cloning, structure and mRNA expression of human Cctg, which encodes the chaperonin subunit CCT gamma. Biochem J. 1996;313:381–389. doi: 10.1042/bj3130381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weisman L S, Wickner W. Molecular characterization of VAC1, a gene organized for vacuole inheritance and vacuole protein sorting. J Biol Chem. 1992;267:618–623. [PubMed] [Google Scholar]
- 57.Whiteford C C, Brearley C A, Ulug E T. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323:597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong K, Cantley L C. Cloning and characterization of a human phosphatidylinositol 4-kinase. J Biol Chem. 1994;269:28878–28884. [PubMed] [Google Scholar]
- 59.Yamamoto A, DeWald D B, Boronenkov I V, Anderson R A, Emr S D, Koshland D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida S, Ohya Y, Nakano A, Anraku Y. Genetic interactions among genes involved in the STT4-PKC1 pathway of Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:631–640. doi: 10.1007/BF00283416. [DOI] [PubMed] [Google Scholar]
- 61.Yu H, Schreiber S L. Cloning, Zn2+ binding and structural characterization of the guanine nucleotide exchange factor human Mss4. Biochemistry. 1995;34:9103–9110. doi: 10.1021/bi00028a020. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X, Loijens J C, Boronenkov I V, Parker G J, Norris F A, Chen J, Thum O, Prestwich G D, Majerus P W, Anderson R A. Phosphatidylinositol-4-phosphate 5-kinase isozymes catalyze the synthesis of 3-phosphate-containing phosphatidylinositol signaling molecules. J Biol Chem. 1997;272:17756–17761. doi: 10.1074/jbc.272.28.17756. [DOI] [PubMed] [Google Scholar]