Abstract
Cyclin A-Cdk2 complexes bind to Skp1 and Skp2 during S phase, but the function of Skp1 and Skp2 is unclear. Skp1, together with F-box proteins like Skp2, are part of ubiquitin-ligase E3 complexes that target many cell cycle regulators for ubiquitination-mediated proteolysis. In this study, we investigated the potential regulation of cyclin A-Cdk2 activity by Skp1 and Skp2. We found that Skp2 can inhibit the kinase activity of cyclin A-Cdk2 in vitro, both by direct inhibition of cyclin A-Cdk2 and by inhibition of the activation of Cdk2 by cyclin-dependent kinase (CDK)-activating kinase phosphorylation. Only the kinase activity of Cdk2, not of that of Cdc2 or Cdk5, is reduced by Skp2. Skp2 is phosphorylated by cyclin A-Cdk2 on residue Ser76, but nonphosphorylatable mutants of Skp2 can still inhibit the kinase activity of cyclin A-Cdk2 toward histone H1. The F box of Skp2 is required for binding to Skp1, and both the N-terminal and C-terminal regions of Skp2 are involved in binding to cyclin A-Cdk2. Furthermore, Skp2 and the CDK inhibitor p21Cip1/WAF1 bind to cyclin A-Cdk2 in a mutually exclusive manner. Overexpression of Skp2, but not Skp1, in mammalian cells causes a G1/S cell cycle arrest.
The cell cycle is driven by a family of protein kinases called cyclin-dependent kinases (CDKs) (21). The activity of CDKs is tightly regulated by an intricate system of protein-protein interaction and phosphorylation (24). Activation of CDKs requires binding to a cyclin subunit and phosphorylation on the Thr161 residue. Phosphorylation of the Thr14 and Tyr15 residues and binding to protein inhibitors of the p21Cip1/WAF1 and p16INK4A family inhibit the activity of CDKs.
Comparison of the compositions of the proteins that associate with cyclin-CDK complexes in normal and cancer cells reveals proteins that may be important for the deregulation of cyclin-CDK during tumorigenesis (38). Examples of these CDK regulators are the CDK inhibitors p21Cip1/WAF1 and p16INK4A, which are found in cyclin-CDK complexes in normal cells but not in many transformed cells. p21Cip1/WAF1 is induced by the tumor suppressor p53 and is responsible for the inhibition of Cdk2 after DNA damage (27). Lack of these CDK inhibitors in transformed cells may contribute to the loss of normal inhibition of cell cycle progression following DNA damage.
In normal human fibroblasts, cyclin A-Cdk2 exists in a quaternary complex that contains p21Cip1/WAF1 and PCNA (proliferating cell nuclear antigen) (41). But in many transformed cells, p21Cip1/Waf1 and PCNA disappear from the cyclin A-Cdk2 complexes, and instead a substantial fraction of cyclin A-Cdk2 complexes are associated with a 19-kDa protein and a 45-kDa protein (38). The 19- and 45-kDa proteins were subsequently identified and renamed Skp1 and Skp2 (for S-phase kinase-associated protein), respectively (40). Identification of Skp1 homologs from Caenorhabditis elegans, Arabidopsis thaliana, and Saccharomyces cerevisiae indicates that Skp1 is evolutionarily highly conserved (6). Cyclin A-Cdk2 complexes appear to bind to Skp1 indirectly through Skp2 (4, 40), and Skp1 binds to Skp2 via a novel structural motif in Skp2 called the F box (4), which is found in a large number of diverse proteins including cyclin F, Grr1, and Cdc4.
The mRNA level of Skp2 is most abundant during the S phase (40). Microinjection of anti-Skp2 antibodies or Skp2 antisense oligonucleotides into normal human fibroblasts or HeLa cells inhibits entry into S phase (but not S-phase progression) (40), suggesting that Skp2 is an important regulator of S phase. The SKP2 gene has been mapped to chromosome position 5p13, and the SKP1 gene has been mapped to 7q11.2 (7). A close homolog or a pseudogene of SKP1 (Skp1B) was mapped to 12p12 (7). All three of these loci are associated with karyotypic alterations, known amplifications, or suspected tumor suppressor genes.
From a different perspective, Skp1 was identified as a suppressor of cdc4 mutants and as a cyclin F-binding protein (4). In S. cerevisiae, Cdc4, acting in concert with Cdc34 and Cdc53, is involved in the ubiquitin-dependent degradation of multiple cell cycle regulators, including Cln2, Cln3, Far1, and Sic1 (20, 31, 36). Cyclin F was isolated as a suppressor of the G1/S deficiency of a cdc4 mutant (3), suggesting cyclin F may also be involved in the destruction of other cell cycle regulators.
The fact that Skp1 is associated with cyclin F and Cdc4 suggests that Skp1 may also be involved in the ubiquitin-dependent destruction of cell cycle regulators. Indeed, Skp1 is required for ubiquitin-mediated proteolysis of Cln2, Clb5, and the CDK inhibitor Sic1 (4). Skp1, Cdc4, and Cdc53 assemble into a ubiquitin-ligase complex, named SCFCdc4, that can reconstitute ubiquitination of phosphorylated Sic1 in the presence of E1 enzyme, the E2 enzyme Cdc34, and ubiquitin (8, 17, 19, 33). Different skp1 temperature-sensitive mutants arrest cells in either G1 or G2, suggesting a connection between regulation of proteolysis in different stages of the cycle (4, 6). Skp1 was also identified as a subunit of CBF3, a multiprotein complex that binds centromere DNA in vitro (6). Skp1 therefore represents an intrinsic kinetochore protein conserved throughout eukaryotic evolution and may be directly involved in linking kinetochore function with the cell cycle-regulatory machinery.
In this study, we investigated the involvement of Skp1 and Skp2 in regulating cyclin A-Cdk2 activity. We showed that Skp2 can inhibit the kinase activity of cyclin A-Cdk2 and found that Skp2 can block the phosphorylation of Cdk2 by CDK-activating kinase (CAK) and Wee1. Skp2 can also inhibit the kinase activity associated with cyclin A-Cdk2, cyclin E-Cdk2, and Skp2 isolated from mammalian cell extracts. Skp2 was phosphorylated by cyclin A-Cdk2 on residue Ser76, but mutation of nonphosphorylatable mutants of Skp2 can still inhibit the kinase activity of cyclin A-Cdk2 toward histone H1. Consistent with the biochemical data, overexpression of Skp2, but not Skp1, in mammalian cells resulted in cell cycle arrest. The F-box region of Skp2 is important in binding to Skp1, and both the N- and C-terminal regions of Skp2 are involved in binding to cyclin A-Cdk2. These data suggest that apart from the potential action as mediators of ubiquitin-dependent proteolysis of cyclin A, Skp1 and Skp2 may also directly regulate the kinase activity of cyclin A-Cdk2.
MATERIALS AND METHODS
Plasmids.
The human Skp2 clone (Skp2 in pBluescriptSK−) was a gift from David Beach (Cold Spring Habor Laboratory) (40). Glutathione S-transferase (GST)–Skp2 in pGEX-KG and hexahistidine (H6)-tagged Skp2 (Skp2-H6) in pET21d clones were constructed by PCR amplification of Skp2 in pBluescriptSK− with primers 5′-TTCCATGGACGTATTTAAAACTCCCGGGC-3′ (Skp2 forward) and 5′-ATCTCGAGTAGACAACTGGGCTTTTGCAGTGT-3′ (Skp2 reverse), cut with NcoI and XhoI, and ligated into pGEX-KG (Pharmacia, Uppsala, Sweden) and pET21d (Novagen, Madison, Wis.), respectively. Skp2 mutant CΔ230-H6 was made by putting the NcoI-BamHI fragment of GST-Skp2 in pGEX-KG into pET21d. CΔ131-H6 and CΔ161-H6 were made by PCR of GST-Skp2 in pGEX-KG with Skp2 forward primer plus 5′-ACCAGAGACCTCGAGCAGCTCAGG-3′ and 5′ ACCAGTCACCTCGAGGTGCAGATT-3′, respectively; the PCR products were cut with NcoI-XhoI and ligated into pET21d. CΔ96 (without H6 tag) was made by cutting Skp2-H6 in pET21d with StuI-XhoI, Klenow treated, and then religated. NΔ230-H6 was created by putting the BamHI-XhoI fragment of GST-Skp2 in pGEX-KG into pET21b. NΔ161-H6 and NΔ131-H6 were made by PCR of GST-Skp2 in pGEX-KG with Skp2 reverse primer plus 5′-AATCTGCACCCCATGGTGACTGGT-3′ and 5′-CCTGAGCTGCCCATGGTCTCTGGT-3′, respectively; the PCR products were cut with NcoI-XhoI and ligated into pET21d. CΔ65-NΔ160-H6 in pET21d and CΔ65-NΔ231-H6 in pET21d were created by PCR amplification of CΔ161-H6 in pET21d and CΔ231-H6, respectively, with primers 5′-AACATCCCCATGGAACTGCTC-3′ and 5′-CTAGTTATTGCTCAGCGGTGG-3′; the PCR products were cut with NcoI-XhoI and ligated into pET21d.
Site-directed mutants of Skp2 were constructed by a PCR method as described elsewhere (12), using Skp2 forward and Skp2 reverse primers and the oligonucleotides 5′-CACCCGGAGGCCCCCCCGCGGAAA-3′ (S76A [Ser76 changed to alanine]), 5′-GAACATTTCGCCCCTTTTCGT-3′ (S191A [Ser191 changed to alanine]), 5′-CACCCGGAGACCCCCCCGCGGAAACGG-3′ (S76T [Ser76 changed to threonine]), and 5′-GACTCTCTTGCGGATGAGCT-3′ (P113A [conserved proline residue in the F box changed to alanine]) and their antisense forms to introduce the mutations; the PCR products were cut with NcoI-XhoI and ligated into pET21d. The S76A S191A double mutant was created by the same method but using S76A as a starting clone. Skp2 in pcDNA3.1(−) was made by putting the XhoI-EcoRI fragment of Skp2 in pBluescriptSK− into pcDNA3.1(−). FLAG-Skp2 in pUHD-P1 (pUHD-P1 is a variant of pUHD10-3 [9] with the FLAG tag sequence inserted at the N-terminal end of the expressed protein; a gift from K. P. Lu [Beth Israel Deaconess Medical Center, Harvard Medical School]) was made by PCR amplification of GST-Skp2 in pGEX-KG with 5′-GACCCAATGTGCCTGGATGCG-3′ and 5′-TTTCCATGGTCATCACCGAAACGCGCGAG-3′; the PCR products were cut with NcoI and ligated into pUHD-P1. FLAG-tagged Skp2 S76A, S191A, and S76A S191A in pUHD-P1 were made by ligation of the NcoI-BamHI fragments of the respective constructs in pET21d and the BamHI-EcoRI fragment of FLAG-Skp2 in pUHD-P1 into NcoI-EcoRI sites of pUHD-P1.
Skp1 was amplified from human cDNA library by PCR with primers 5′-CACCATGGCTTCAATTAAGTTGCA-3′ and 5′-ACCTCGAGCTTCTCTTCACACCACTGGTT-3′; the PCR product was cut with NcoI-XhoI and put into pGEX-KG (GST-Skp1 in pGEX-KG) and pET21d (Skp1-H6 in pET21d). FLAG-Skp1 in pUHD-P1 was made as was FLAG-Skp2 in pUHD-P1 except that the starting clone was GST-Skp1 in pGEX-KG.
GST-Cdk2 in pGEX-2T and GST–kinase-inactive Cdk2 [Cdk2(K33R)] in pGEX-2T (29), staphylococcal protein A (PA)-cyclin A (14), and GST-Wee1 (26) were as described elsewhere. Human B-cell antigen CD20 in pCMX was a gift from H. Toyoshima (The Salk Institute). FLAG-tagged p21 in pUHD-P1 (18) and p21-H6 in pET21d (27) were as described elsewhere. GST-Cdk5 and GST-p25 were as described previously (23).
Expression and purification of recombinant proteins.
Expression of GST-tagged and histidine-tagged proteins in bacteria and purification with glutathione (GSH)-agarose and nickel-nitrilotriacetic acid agarose chromatography, respectively, were as described elsewhere (28). Protein concentrations were estimated by comparison to bovine serum albumin after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. Digestion of GST fusion proteins with thrombin and inactivation of thrombin were as described elsewhere (26). Coupled transcription-translation reactions in the presence of [35S]methionine in rabbit reticulocyte lysate were performed as instructed by the manufacturer (Promega, Madison, Wis.). PA-cyclin A-GST-Cdk2 complexes were activated by a CAK immunoprecipitate as described elsewhere (29).
Binding assays.
For binding of rabbit reticulocyte lysate-translated proteins, bacterially expressed GST fusion proteins (∼100 ng) were incubated with rabbit reticulocyte lysate programmed to contain 35S-labeled proteins (5 μl) at 30°C for 30 min. GST fusion proteins were then recovered with 15 μl of GSH-agarose in 250 μl of bead buffer (50 mM Tris-HCl [pH 7.4], 5 mM NaF, 250 mM NaCl, 5 mM EDTA, 5 mM EGTA, 0.1% Nonidet P-40, aprotinin [2 μg/ml], benzamidine [15 μg/ml], leupeptin [1 μg/ml], soybean trypsin inhibitor [10 μg/ml]). After incubation at 4°C with end-to-end rotation for 45 min, the beads were washed five times with 250 μl of bead buffer. The samples were then dissolved in 30 μl of SDS sample buffer, and the bound 35S-labeled proteins were detected by SDS-PAGE followed by phosphorimagery analysis (Molecular Dynamics). For binding assays involving only bacterially expressed proteins, purified proteins (∼100 ng of each) were mixed and incubated at 30°C for 30 min in the presence of 10 μg of bovine serum albumin as carrier. The proteins were recovered with GSH-agarose and washed as described above.
Phosphorylation assays.
Histone H1 kinase, CAK, and Wee1 kinase assays were performed as described elsewhere (26). Two-dimensional phosphoamino acid analysis after partial acid hydrolysis was performed on 32PO4-labeled polypeptides after transfer to Immobilon (Millipore) as described elsewhere (13). Phosphoamino acids were separated on cellulose thin-layer chromatography plates (Schleicher & Schuell) by electrophoresis at 1,600 V for 60 min with pH 1.9 buffer, followed by thin-layer chromatography using the phospho-chromatography buffer (35).
Cell culture.
HtTA1 cells, gifts from Hermann Bujard, are HeLa (human cervical carcinoma) cells stably transfected with pUHD15-1 expressing the tTA tetracycline repressor chimera (9) and can express genes cloned into the pUHD-P1 vector in the absence of tetracycline. AG1523 (normal human foreskin diploid) fibroblasts were obtained from the NIA Aging Cell Repository, Institute for Medical Research, Camden, N.J. (used between passage 8 and 25). The chemical carcinogen-transformed human fibroblast cell line HUT12 was a gift from Gertrud Orend (Burnham Institute, La Jolla, Calif.). Cell lines HeLa, HaCaT (immortalized human keratinocytes), 293 (transformed human embryonic kidney cells), HepG2 (human hepatocellular carcinoma cells), H4 (human neuroglioma cells), HLB100 (human breast epithelial cells), MG63 (human osteosarcoma cells), and K562 (human chronic myelogenous leukemia cells) were obtained from the American Type Culture Collection (Rockville, Md.). Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% (vol/vol) calf serum or 10% (vol/vol) fetal bovine serum in a humidified incubator at 37°C in 5% CO2.
Semiconfluent HtTA1 cells (10-cm-diameter plates) were transiently transfected with constructs (20 μg) by the calcium phosphate method (2). Cell extracts were prepared as described elsewhere (28). The protein concentration of cell lysates was measured with a bicinchoninic acid protein assay system (Pierce, Rockford, Ill.), using bovine serum albumin as a standard.
The effect on the cell cycle distribution of transfected DNA was studied with a method similar to one described elsewhere (11). Semiconfluent HtTA1 cells (10-cm-diameter plates) were transiently transfected with constructs (20 μg) together with a vector expressing CD20 surface marker (2 μg) by the calcium phosphate method (2). After transfection (16 h), the cells were washed with phosphate-buffered saline and then grown in fresh medium for another 24 h. For fluorescence-activated cell sorter (FACS) analysis, cells were trypsinized, washed with phosphate-buffered saline, and incubated with fluorescein isothiocyanate-conjugated anti-CD20 monoclonal antibody (DAKO) according to the manufacturer’s instructions. Cells were then fixed in cold 70% ethanol and stained with a solution containing propidium iodide (40 μg/ml) and RNase A (40 μg/ml) at 37°C for 30 min. Cell cycle distribution was analyzed with a FACSVantage machine (Becton Dickinson).
Antibodies and immunological methods.
Rabbit antibodies against Cdk2 and Cdk7 (30), anti-cyclin A monoclonal antibody E72 (28), rabbit anti-cyclin A polyclonal antibodies (22), and rabbit anti-Cdc2 polyclonal and anti-PSTAIRE monoclonal antibodies (25) were as previously described. Anti-cyclin E monoclonal antibodies HE12 (for immunoblotting) and HE172 (for immunoprecipitation) were gifts from Emma Lees. Goat anti-Skp1 antibodies were raised against a peptide corresponding to the C-terminal 20 amino acids of human Skp1 (sc-1568; Santa Cruz Biotechnology). Goat anti-Skp2 antibodies were raised against a peptide corresponding to the N-terminal 19 amino acids of human Skp2 (sc-1567; Santa Cruz Biotechnology). Rat monoclonal antibody YL1/2 against mammalian tubulin was from Julian Gannon and Tim Hunt (Imperial Cancer Research Fund South Mimms, United Kingdom). Monoclonal antibody M2 against FLAG tag was from Eastman Kodak, and purified rabbit polyclonal antibody sc-807 against FLAG tag was from Santa Cruz Biotechnology. Immunoprecipitation and immunoblotting were performed as described elsewhere (27) except for the anti-FLAG tag monoclonal antibody M2, which was used according to the manufacturer’s instructions. For some assays, normal rabbit serum (NRS) was used.
RESULTS
Expression of Skp2 in normal and cancer cells.
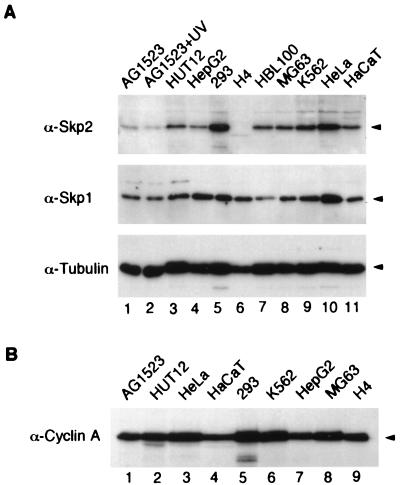
We first investigated which cell lines expressed Skp1 and Skp2. It was demonstrated that the mRNA level of Skp2 is overexpressed in many transformed cells (40), but the total protein levels of Skp2 in cell lines have not been fully investigated. Figure 1A shows that the Skp2 protein level was typically higher in a variety of human transformed and cancer cell lines (lanes 3 to 11) than in normal human diploid fibroblasts (lanes 1 and 2). In contrast, no significant correlation in the level of Skp1 between the normal cells and cancer cells was found (Fig. 1A, middle panel). Figure 1A also shows the relatively constant level of tubulin in these cell extracts as a control (bottom panel). Moreover, contrary to common belief, levels of cyclin A were similar in normal cells and the various cancer cell lines (Fig. 1B). Interestingly, the human neuroglioma cell line H4 contained no detectable Skp2 but did contain Skp1 (lane 6), indicating that overexpression of Skp2 is not a universal feature of transformed cells. The level of Skp2 was not altered after the fibroblasts were treated with UV irradiation (lanes 1 and 2), indicating that the level of Skp2 did not vary after DNA damage.
FIG. 1.
Expression of Skp1 and Skp2 in normal and transformed cells. (A) Cell extracts were prepared from growing normal human fibroblasts (cell line AG1523) (lane 1), AG1523 cells at 24 h after treatment with UV irradiation (lane 2), and HUT12 (lane 3), HepG2 (lane 4), 293 (lane 5), H4 (lane 6), HBL100 (lane 7), MG63 (lane 8), K562 (lane 9), HeLa (lane 10), and HaCaT (lane 11) cells (see Materials and Methods for descriptions of cell lines). The extracts were dissolved in SDS sample buffer, and 10 μg each was subjected to SDS-PAGE on a 17.5% gel. The proteins were transferred onto a membrane and immunoblotted with antibodies against Skp2 (top), Skp1 (middle), and tubulin (bottom). (B) Cell extracts prepared from the indicated cell lines were dissolved in SDS sample buffer, and 10 μg each was subjected to SDS-PAGE on a 17.5% gel. The proteins were transferred onto a membrane and immunoblotted with a monoclonal antibody against cyclin A.
Binding of cyclin A-Cdk2 to Skp2 and p21Cip1/WAF1.
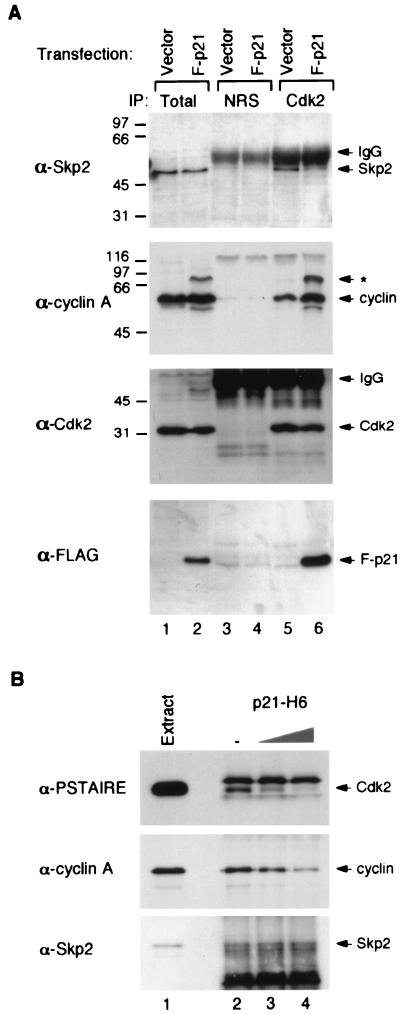
Given that the level of Skp2 is high in most cancer cells, and one feature of many cell lines is the low level of p21Cip1/WAF1 in the cyclin A-Cdk2 complex, we next investigated whether Skp2 and p21Cip1/WAF1 interact with cyclin A-Cdk2 in a mutually exclusive manner. Plasmids that expressed FLAG-tagged p21 or control vector were transiently transfected into HeLa cells, and the relative amount of Skp2 that coimmunoprecipitated with Cdk2 was measured. HeLa cells were used because of the relative abundant level of Skp2 (Fig. 1) and low level of p21Cip1/WAF1 that they contained. Figure 2A shows that FLAG-p21 was expressed in cells transfected with expression plasmid but not with control vector (lanes 1 and 2). Similar levels of endogenous Skp2, cyclin A, and Cdk2 were found in these cells. However, the level of Skp2 that associated with Cdk2 was lower in cells expressing FLAG-p21 (lane 6) than in cells transfected with control vector (lane 5). Coimmunoprecipitation of similar amounts of cyclin A-Cdk2 was confirmed by immunoblotting with antibodies against cyclin A and Cdk2. This finding also shows that overexpression of FLAG-p21 did not disrupt the cyclin A-Cdk2 complex. The expression of FLAG-p21 and its binding to Cdk2 was seen by immunoblotting with the anti-FLAG tag monoclonal antibody M2 (Fig. 2A, bottom panel).
FIG. 2.
Interactions between cyclin A-Cdk2, Skp2, and p21Cip1/WAF1. (A) HtTA1 cells were transfected with either pUHD-P1 vector alone (lanes 1, 3, and 5) or FLAG-p21 in pUHD-P1 (lanes 2, 4, and 6). Cell extracts were prepared, and 60 μg was immunoprecipitated (IP) with either NRS (lanes 3 and 4) or anti-Cdk2 serum (lanes 5 and 6). The immunoprecipitates and total cell extracts (10 μg) (lanes 1 and 2) were subjected to SDS-PAGE on a 17.5% gel, transferred onto a membrane, and immunoblotted with antibodies against Skp2, cyclin A, Cdk2, or FLAG tag to detect FLAG-p21, as indicated at the left. Positions of Skp2, cyclin A, Cdk2, FLAG-p21 (F-p21), and IgG chains are indicated on the right. The asterisk indicates the position of an extra cyclin A band (which can also be immunoprecipitated with Cdk2) seen after transfection with the p21-expressing construct. The positions of molecular size standards (in kilodaltons) are shown on the left. (B) HeLa cell extracts (120 μg) were immunoprecipitated with antiserum against Skp2 (lanes 2 to 4). Buffer (lane 2) or purified recombinant p21-H6 protein expressed in bacteria (1 μg [lane 3] or 5 μg [lane 4]) was incubated with the Skp2 immunoprecipitates at 30°C for 30 min. After washing, the immunoprecipitates were dissolved in SDS sample buffer and subjected to SDS-PAGE on a 17.5% gel. Total cell lysate (10 μg) was loaded in lane 1. The proteins were transferred onto a membrane and immunoblotted with a monoclonal antibody against PSTAIRE to detect Cdk2 (anti-PSTAIRE monoclonal antibody was used instead of anti-Cdk2 polyclonal antibody because it gave a cleaner background of the IgG bands) (top), anti-cyclin A monoclonal antibody E72 (middle), and anti-Skp2 antiserum (bottom).
It is possible that the expression of FLAG-p21 in the above experiment may result in cell cycle arrest and may alter the binding between Skp2 and cyclin A-Cdk2. Therefore, we performed a second set of experiments in which Skp2 was immunoprecipitated from cell extracts derived from asynchronized cells and incubated with purified bacterially expressed p21Cip1/WAF1. The amount of Cdk2 and cyclin A that associated with the Skp2 immunoprecipitates was diminished after the immunoprecipitates were incubated with recombinant p21Cip1/WAF1 (Fig. 2B). These data suggest the possibility that Skp2 and p21Cip1/WAF1 may bind to cyclin A-Cdk2 in a mutually exclusive manner.
Interactions between recombinant Skp2, cyclin A-Cdk2, and Skp1.
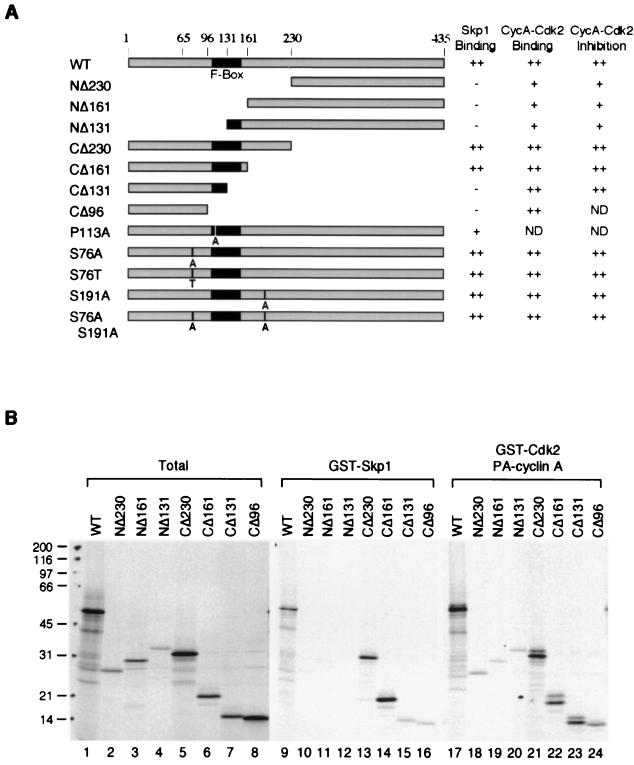
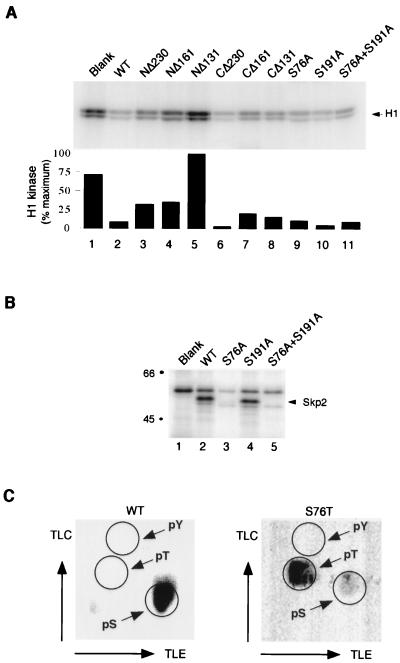
We next investigated which regions of Skp2 are important for binding to cyclin A-Cdk2 and to Skp1. We constructed mutants of Skp2 that were truncated from either the N terminus or the C terminus (Fig. 3A). Clones truncated from the N terminus and the C terminus are indicated by NΔ and CΔ, respectively, followed by the amino acid position to which the clones are truncated. Skp2 and mutants were translated in a coupled transcription-translation rabbit reticulocyte lysate system in the presence of [35S]methionine, and their ability to bind to bacterially expressed Skp1 and cyclin A-Cdk2 complexes was assayed (Fig. 3B; summarized in Fig. 3A). As expected, the F-box region of Skp2 was required for binding to Skp1. The main binding region of Skp2 for cyclin A-Cdk2 was at the N-terminal of the F box, but the C terminus also has affinity for cyclin A-Cdk2. Note that the mobility of Skp2 proteins on SDS-PAGE shifted after binding to cyclin A-Cdk2, due to phosphorylation of Skp2 by cyclin A-Cdk2 (see below). These data also indicate that it is possible to obtain mutants of Skp2 that bind cyclin A-Cdk2 without binding to Skp1 (like CΔ131), which will be useful for future studies of the function of Skp2.
FIG. 3.
Binding of Skp2 to cyclin A-Cdk2 and Skp1. (A) Skp2 wild type (WT) and mutants. The Skp2 truncation and site-directed point mutants were constructed as described in Materials and Methods. The position of the F box (∼112 to 152) is indicated. Abilities to bind GST-Skp1 and GST-Cdk2-cyclin A, and inhibition of cyclin A-Cdk2 kinase activity, are indicated (see text). ND, not determined. CΔ96 does not contain an H6 tag and hence cannot be purified from bacteria for cyclin A-Cdk2 inhibition assay. (B) Binding of Skp1 and cyclin A-Cdk2 to Skp2 truncation mutants. Wild-type Skp2 (WT) or truncated mutants of Skp2 were translated in a coupled transcription-translation rabbit reticulocyte lysate system in the presence of [35S]methionine. The amount of 35S label in the translated proteins was quantitated by SDS-PAGE (17.5% gel) and phosphorimagery (lanes 1 to 8). The proteins were adjusted to the same amount of labeling (the strongest band was about 10% of the input for the binding experiments) and incubated with bacterially expressed GST-Skp1 (lanes 9 to 16) or GST-Cdk2 and PA-cyclin A (lanes 17 to 24). The GST fusion proteins and associated proteins were then precipitated with GSH-agarose and analyzed by SDS-PAGE (17.5% gel) and phosphorimagery. No significant binding to these proteins was detected when GST was used (data not shown). The positions of molecular size standards (in kilodaltons) are shown on the left.
Regulation of cyclin A-Cdk2 kinase activity by Skp2.
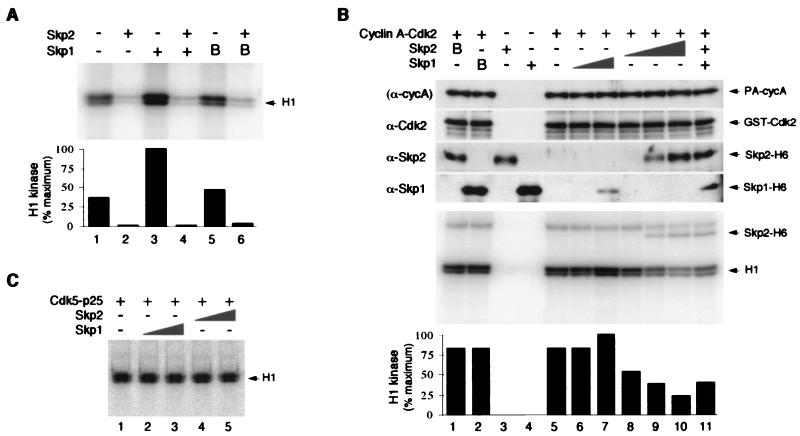
We next studied whether upon binding, the kinase activity of Cdk2 is affected by Skp2 and Skp1. CAK-activated cyclin A-Cdk2 complexes were incubated with bacterially expressed Skp1-H6 and Skp2-H6, and the kinase activity against histone H1 was assayed. The kinase activity of cyclin A-Cdk2 was inhibited by Skp2-H6 under these conditions (Fig. 4A, lane 2). Titration of increasing amounts of Skp2 gradually inhibited the kinase activity of cyclin A-Cdk2 (Fig. 4B, lanes 8 to 10). As a control, denaturation of Skp2-H6 by boiling abolished its ability to inhibit cyclin A-Cdk2 kinase activity (Fig. 4B, lane 1).
FIG. 4.
Regulation of cyclin A-Cdk2 kinase activity by Skp1 and Skp2. (A) Bacterially expressed and CAK-activated PA-cyclin A-GST-Cdk2 complexes (250 nM) were incubated with 250 nM bacterially expressed Skp2-H6 (lanes 2, 4, and 6), Skp1-H6 (lanes 3 and 4), or boiled Skp1-H6 (B) (lanes 5 and 6) at 23°C for 30 min. The kinase activity against histone H1 was then assayed, and phosphorylation was detected by SDS-PAGE followed by phosphorimagery. Quantitation from the phosphorimagery is shown in the lower panel. (B) PA-cyclin A-GST-Cdk2 complexes (250 nM) (lanes 1, 2, and 5 to 11) or buffer (lanes 3 and 4) were incubated with bacterially expressed Skp2-H6 (250 nM [lanes 1, 3, 10, and 11], 25 nM [lane 9], or 2.5 nM [lane 8]) and Skp1-H6 (6.5 μM [lanes 2, 4, 7, and 11] or 650 nM [lane 6]) at 23°C for 30 min. In lanes 1 and 2, Skp2-H6 and Skp1-H6, respectively, were boiled (B) prior to incubation. The kinase activity against histone H1 was then assayed, and phosphorylation was detected by SDS-PAGE (17.5 gel) followed by phosphorimagery. The positions of histone H1 and Skp2-H6 are indicated. Quantitation of histone H1 phosphorylation from the phosphorimagery is shown in the lower panel. The levels of the proteins added were confirmed by immunoblotting with antibodies against Cdk2, Skp2, or Skp2; PA-cyclin A was detected by virtue of the PA tag binding to IgG in the immunoblotting reaction. (C) Skp2 has no effect on p25-Cdk5 kinase activity. GST-p25 (250 nM) and GST-Cdk5 (250 nM) were incubated with Skp1-H6 (650 nM [lane 2] or 6.5 μM [lane 3]) or Skp2-H6 (25 nM [lane 4] or 250 nM [lane 5]) at 23°C for 30 min. The kinase activity against histone H1 was then assayed, and phosphorylation was detected by SDS-PAGE followed by phosphorimagery.
Interestingly, we consistently found that the kinase activity of cyclin A-Cdk2 was stimulated in the presence of Skp1-H6 (Fig. 4A, lane 3). Skp1-H6 alone in the absence of cyclin A-Cdk2 contained no kinase activity (Fig. 4B, lane 4), indicating that the increase in kinase activity was not due to some contaminated kinase in the Skp1 preparation. As a control, denaturation of Skp1-H6 by boiling abolished this stimulation of cyclin A-Cdk2 kinase activity (Fig. 4A, lane 5; Fig. 4B, lane 2). Addition of Skp1-H6 and Skp2-H6 together inhibited the kinase activity of cyclin A-Cdk2 (Fig. 4A, lane 4; Fig. 4B, lane 11), suggesting that the effect of Skp2 was dominant over that of Skp1. The reduction of cyclin A-Cdk2 kinase activity was not due to proteolysis of cyclin A or Cdk2, since the levels of all input proteins were confirmed by immunoblotting at the end of the experiments (representations are shown in Fig. 4B). Interestingly, although Skp2 reduced the kinase activity of cyclin A-Cdk2 toward the substrate histone H1, Skp2 itself was a substrate for cyclin A-Cdk2 in vitro (Fig. 4B, lanes 9 to 11). We found that Skp2 was also phosphorylated in vivo and that the phosphorylation was mapped to one major site (see below).
The regions of Skp2 involved in the inhibition of cyclin A-Cdk2 were similar to that involved in binding to cyclin A-Cdk2 (see Fig. 6A; summarized in Fig. 3). Hence, physical association between Skp2 and cyclin A-Cdk2 was likely to be important for the inhibition of the kinase activity. As a further control, we investigated whether the kinase activity of the neuron-specific Cdk5 can be inhibited by Skp1 and Skp2. Bacterially expressed Cdk5 was activated by binding to the neuronal protein p25 (23). Figure 4C shows that when Cdk5-p25 complexes were incubated with Skp1 or Skp2, the histone H1 kinase activity of Cdk5-p25 was not affected. This result shows that the effects on the kinase activity by Skp1 and Skp2 are specific for cyclin A-Cdk2 and not for Cdk5-p25.
FIG. 6.
Phosphorylation of Skp2 by cyclin A-Cdk2. (A) Inhibition of Cdk2 kinase activity by Skp2 wild type (WT) and mutants. Assay of the inhibition of cyclin A-Cdk2 kinase activity against histone H1 by Skp2 truncation and site-directed point mutants was as described for Fig. 4. Quantitation from the phosphorimagery is shown in the lower panel. (B) Phosphorylation of site-directed Skp2 mutants by cyclin A-Cdk2. Buffer (lane 1), Skp2-H6 (lane 2), and mutants S76A (lane 3), S191A (lane 4), and S76A S191A (lane 5) were incubated with reconstituted PA-cyclin A-GST-Cdk2 in the presence of [γ-32P]ATP. The reactions were terminated by addition of SDS sample buffer, and phosphorylations were analyzed by SDS-PAGE (17.5% gel) followed by phosphorimagery. (C) Phosphoamino acid analysis of Skp2. Skp2-H6 (left) and the S76T mutant (right) were phosphorylated by reconstituted PA-cyclin A-GST-Cdk2 in vitro. The proteins were separated by SDS-PAGE (17.5% gel) and transferred to an Immobilon membrane. The phosphorylated Skp2 bands were cut out and subjected to phosphoamino acid analysis using thin-layer electrophoresis (TLE) in the first dimension and thin-layer chromatography (TLC) in the second dimension, followed by analysis with phosphorimagery. The positions of phosphoamino acids standards are indicated.
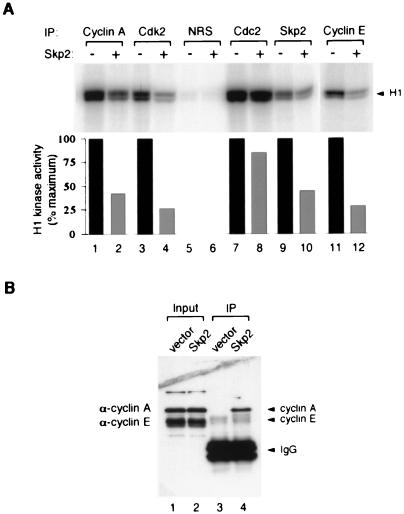
Given that Skp2 was able to inhibit the kinase activity of recombinant cyclin A-Cdk2, we next investigated whether endogenous cyclin-CDK complexes in mammalian cell extracts could also be inhibited by Skp2. Figure 5A shows that when cyclin A, cyclin E, and Cdk2 were immunoprecipitated from the cell lysates and incubated with Skp2-H6, the kinase activities associated with these proteins were reduced. In comparison, the kinase activity associated with Cdc2 changed relatively less after incubation with Skp2. Intriguingly, kinase activity was found to associate with the Skp2 immunoprecipitates, and this was reduced by incubation with more Skp2-H6 (lanes 9 to 10). These findings suggest the possibility that higher stoichiometry of Skp2 is required to inhibit Cdk2, there are two separate binding conformations between Skp2 and Cdk2, or Cdk2 dissociated from the Skp2 upon dilution in the kinase buffer (see Discussion).
FIG. 5.
Inhibition of endogenous cyclin-CDK kinase activity by Skp2. (A) HeLa cell extracts (250 μg) were immunoprecipitated with antiserum against cyclin A or Cdk2, NRS, or serum against Cdc2, Skp2, or cyclin E, as indicated above the lanes. The immunoprecipitates were incubated with buffer (odd-numbered lanes) or 1 μg of bacterially expressed Skp2-H6 (even-numbered lanes) at 30°C for 30 min. The kinase activity against histone H1 was then assayed, and phosphorylation was analyzed by SDS-PAGE (17.5% gel) followed by phosphorimagery. Quantitation of histone H1 phosphorylation from the phosphorimagery is shown in the lower panel; the kinase activity associated with NRS immunoprecipitates in lanes 5 and 6 were very low, and the quantitation is not shown. (B) Binding of cyclin A and cyclin E to Skp2. HeLa cells were transiently transfected with a control vector (lanes 1 and 3) or plasmid expressing FLAG-Skp2 (lanes 2 and 4). Extracts were prepared; 200 μg was immunoprecipitated (IP) with anti-FLAG antiserum and dissolved in 30 μl of SDS sample buffer; 10 μl was loaded onto an SDS–17.5% polyacrylamide gel, transferred onto a membrane, and immunoblotted with the anti-cyclin A monoclonal antibody E72 and anti-cyclin E monoclonal antibody HE12.
Interestingly, cyclin A but not cyclin E was reported to bind to Skp2 in the cell. What is the relative affinity of Skp2 to cyclin A and cyclin E? We transfected either a blank plasmid or a plasmid expressing FLAG-tagged Skp2 into HeLa cells. Cell extracts were prepared, and FLAG-Skp2 was immunoprecipitated; the immunoprecipitates were then immunoblotted with monoclonal antibodies against cyclin A and cyclin E together for comparison (Fig. 5B). We found that cyclin A can easily be detected in the Skp2 immunoprecipitates, but far less cyclin E was detectable in the Skp2 immunoprecipitates, although the two antibodies have similar sensitivities. Similar results were obtained with the endogenous Skp2 in growing HeLa cells (data not shown). Although both cyclins can be inhibited by incubation with excess bacterially expressed Skp2, these data suggest that cyclin A-CDK has a higher affinity for Skp2 than cyclin E-Cdk2 in the cell.
As shown in Fig. 4B, Skp2 was phosphorylated by cyclin A-Cdk2 in vitro. In contrast, recombinant Skp1 was not phosphorylated by cyclin A-Cdk2 under the same conditions (data not shown). Moreover, no difference in the phosphorylation of Skp2 by cyclin A-Cdk2 was observed in the presence or absence of Skp1 (data not shown). Incubation of bacterially expressed Skp2-H6 with cyclin A-Cdk2 in the presence of [γ-32P]ATP resulted in the phospholabeling of Skp2-H6 (Fig. 6B, lane 2). The substrate specificity of Cdk2 is known to be either serine or threonine followed by a proline. Inspection of the sequence of Skp2 reveals three possible Cdk2 phosphorylation sites: Thr6, Ser76, and Ser191. Phosphoamino acid analysis of the phosphorylated Skp2 indicated that the phosphorylation was exclusively in serine residues (Fig. 6C). This ruled out Thr6 as the possible phosphorylation site on Skp2. We next tested whether the Skp2 S76A, S191A, and S76A S191A) mutants could be phosphorylated by cyclin A-Cdk2. Figure 6B shows that nearly all phosphorylation was abolished in the S76A mutant and the S76A S191A double mutant. In contrast, very little change in phosphorylation was seen with the S191A mutant. We next mutated the Ser76 residue in Skp2 to threonine (S76T) and found that this mutant was phosphorylated by cyclin A-Cdk2, and all phosphorylation was now on threonine (Fig. 6C). We found that all phosphorylation mutants of Skp2 can bind to cyclin A-Cdk2 and Skp1 just as wild-type Skp2 can (Fig. 3). These data indicate that the major phosphorylation site in Skp2 in vitro is Ser76.
We tested the S76A, S191A, and S76A S191A mutants described above for their inhibition of cyclin A-Cdk2 and found that all inhibited the kinase activity of cyclin A-Cdk2 toward histone H1 as did wild-type Skp2 (Fig. 6A). This datum indicates that phosphorylation of Skp2 is not required for the inhibition of cyclin A-Cdk2 kinase activity toward histone H1.
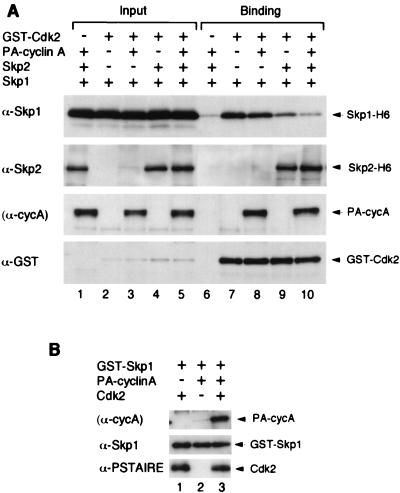
Given that Skp1 can stimulate the activity of cyclin A-Cdk2 as described above, we tested whether there was any direct interaction between Skp1 and cyclin A-Cdk2. Purified proteins expressed in bacteria were used to exclude the possibility that there are adaptor proteins between the complexes. Figure 7A shows that purified Skp1-H6 alone was sufficient to bind to GST-Cdk2 (lane 7), whereas no association between Skp1-H6 and GST control was detected (lane 6). We performed the converse experiments by looking at the binding of Cdk2 and cyclin A to GST-Skp1. Figure 7B indicates that GST-Skp1 can bind directly to Cdk2 (lane 1) but not to cyclin A (lane 2).
FIG. 7.
Direct interaction between Skp1 and cyclin A-Cdk2. (A) Purified bacterially expressed GST (lanes 1 and 6) or GST-Cdk2 (other lanes) was incubated with Skp1-H6 in the presence of Skp2-H6 (lanes 1, 4, 5, 6, 9, and 10) and PA-cyclin A (lanes 1, 3, 5, 6, 8, and 10). Input samples (10% of that used for binding) were loaded in lanes 1 to 5. GST-Cdk2 and associated proteins were precipitated with GSH-agarose (lanes 6 to 10), and the bound proteins were detected by immunoblotting with antibodies against Skp1, Skp2, and GST; PA-cyclin A was detected by virtue of the PA tag binding to IgG in the immunoblotting reaction. (B) Purified bacterially expressed GST-Skp1 was incubated with PA-cyclin A (lanes 2 and 3) and Cdk2 (lanes 1 and 3). GST-Skp1 and associated proteins were precipitated with GSH-agarose, and the bound proteins were detected by immunoblotting with antibodies against Skp1 and PSTAIRE (detected Cdk2), as indicated on the left; in the top panel, PA-cyclin A was detected by virtue of the PA tag binding to IgG in the immunoblotting reaction.
Skp2 can inhibit the phosphorylation of Cdk2 by CAK and Wee1.
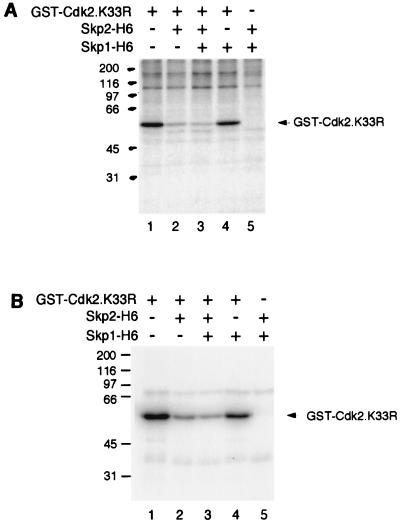
Cdk2 is phosphorylated on Thr160 on the T loop by CAK and on Tyr15 by Wee1. Both of these phosphorylations are important for the regulation of Cdk2 kinase activity. Using the kinase-inactive fusion protein GST-Cdk2(K33R) as a substrate for CAK, we found that Skp2 was able to block the phosphorylation of Thr160 (Fig. 8A, lane 2). Skp2-H6 inhibited the phosphorylation of Cdk2 by CAK either in the presence or in the absence of Skp1-H6 (lanes 2 and 3). In contrast, Skp1-H6 alone did not affect the phosphorylation of GST-Cdk2(K33R) by CAK (lane 4). Similarly, phosphorylation of GST-Cdk2(K33R) by Wee1 was blocked by Skp2, either in the presence or in the absence of Skp1, but not by Skp1 alone (Fig. 8B). In conclusion, Skp2, but not Skp1, was able to block the phosphorylation of cyclin A-Cdk2 Thr160 and Tyr15 by their respective kinases. It is likely that binding of Skp2 to cyclin A-Cdk2 blocks the access of CAK and Wee1 to Cdk2 or changes the conformation of cyclin A-Cdk2 so that they cannot be recognized by CAK and Wee1 (although we cannot rule out the possibility that Skp2 inhibited the kinase activity of CAK and Wee1 directly). This may represent a further mechanism that Skp2 can affect the activity of cyclin A-Cdk2.
FIG. 8.
Inhibition of Cdk2 Thr160 and Tyr15 phosphorylation by Skp2. GST-Cdk2(K33R)-PA-cyclin A (lanes 1 to 4) was mixed with bacterially expressed Skp2-H6 (lanes 2, 3, and 5) and Skp1-H6 (lanes 3 to 5). The reactions were incubated with an immunoprecipitate of CAK (A) or bacterially expressed GST-Wee1 (B) in the presence of [γ-32P]ATP. Phosphorylation was detected by SDS-PAGE (17.5% gel) followed by phosphorimagery analysis. The positions of GST-Cdk2(K33R) are indicated on the right; positions of molecular size standards (in kilodaltons) are shown on the left.
Skp2 can inhibit cell cycle progression.
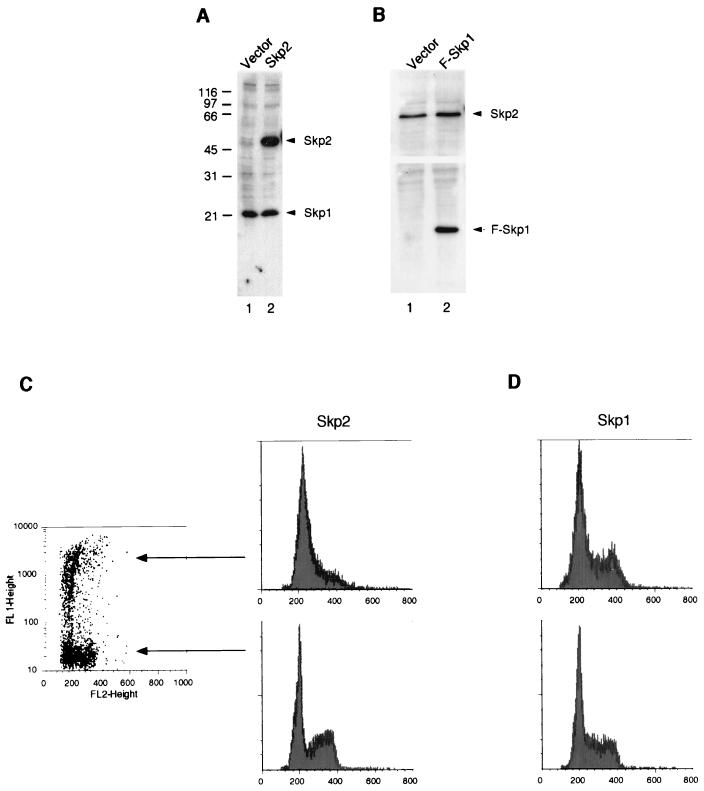
Given that Skp2 can block the kinase activity associated with cyclin A-Cdk2, as well as inhibit the activation of cyclin A-Cdk2 by CAK, we next investigated whether overexpression of Skp1 and Skp2 has any effect on the cell cycle. HeLa cells were transiently transfected with a vector control or Skp2-expressing construct. Figure 9A shows that cell extracts prepared from cells transfected with the Skp2-expressing construct (lane 2) contained higher levels of Skp2 than cells transfected with vector alone (lane 1). As a comparison, the level of the endogenous Skp1 was the same in the two cell extracts. Similarly, Fig. 9B shows the level of overexpressed FLAG-tagged Skp1, and the endogenous Skp2 proteins as a control, in cells transfected with vector alone (lane 1) or FLAG-Skp1 construct (lane 2). To examine the cell cycle distribution of the cells expressing exogenous Skp1 or Skp2, cells were cotransfected with a Skp1/2-expressing construct and a construct expressing the surface marker CD20. After transfection, cells were harvested and the cell cycle distribution of the CD20-positive (transfected) cells was compared to that of the nontransfected cells by FACS analysis (see Materials and Methods). Figure 9C shows that overexpression of Skp2 caused the accumulation of cells with a G1 DNA content. In contrast, the Skp1-expressing construct (Fig. 9D) or vector alone (data not shown) did not affect the cell cycle profile of the transfected cells.
FIG. 9.
Cell cycle arrest after overexpression of Skp2 in mammalian cells. (A) HtTA1 cells were transiently transfected with the same amount of vector pcDNA3.1(−) (lane 1) or Skp2 in pcDNA3.1(−) (lane 2). Cell extracts were prepared and subjected to SDS-PAGE (17.5% gel). Proteins were transferred to a membrane and immunoblotted with antibodies against Skp2 and Skp1. Positions of the endogenous Skp1 and transfected Skp2 are shown on the right; positions of molecular size standards (in kilodaltons) are shown on the left. (B) HtTA1 cells were transiently transfected with the same amount of vector pUHD-P1 (lane 1) or FLAG-Skp1 in pUHD-P1 (F-Skp1; lane 2). Cell extracts were prepared and subjected to SDS-PAGE (17.5% gel). Proteins were transferred to a membrane and immunoblotted with antibodies against Skp2 (top) and FLAG-tag (bottom). Positions of the endogenous Skp2 and transfected FLAG-tagged Skp1 are shown on the right. (C) HtTA1 cells were cotransfected with Skp2 in pcDNA3.1(−) construct and a plasmid expressing CD20. Immediately after harvest, the cells were incubated with a fluorescein isothiocyanate-conjugated anti-CD20 monoclonal antibody, fixed, and stained with propidium iodide. DNA content of the transfected cells (upper portion of the cell population diagram) and nontransfected cells (lower portion of the cell population diagram) was analyzed by FACS. (D) HtTA1 cells were cotransfected with FLAG-tagged Skp1 in pUHD-P1 and a CD20-expressing plasmid as for panel C. FACS analyses of the transfected and nontransfected cells are shown.
DISCUSSION
Skp1 and Skp2 are believed to be important in several different cellular processes. First, the level of Skp2 is high in S phase, and its association with cyclin A at this time may be important for cell cycle regulation. Second, the level of Skp2 is typically elevated in transformed cells; hence, overexpression of Skp2 can either play a role in or be a consequence of transformation. Third, Skp1 is a component of the kinetochore, suggesting that it may link kinetochore function with the cell cycle. Finally, degradation of Sic1 in S. cerevisiae is mediated by the E3 complex containing Skp1 and the F-box protein Cdc4 (SCFCdc4), and it is possible that proteolysis of many key cell cycle regulators is mediated by SCF complexes containing other F-box proteins such as Skp2.
We found that the kinase activity of recombinant cyclin A-Cdk2 can be inhibited by Skp2 in vitro. The kinase activity of endogenous cyclin A-Cdk2 in cell lysates can also be inhibited by Skp2. Other explanations for the inhibition of kinase activity by Skp2 are conceivable; for instance, there could be a phosphatase copurified with Skp2 that removes phosphates from histone H1, or Skp2 could bind to histone H1 and sequester them from Cdk2. The fact that Skp2 binds directly to cyclin A-Cdk2 is consistent with the idea that the inhibition could be due to Skp2 directly. Apart from directly inhibiting cyclin A-Cdk2 kinase activity, Skp2 also inhibited the activation of cyclin A-Cdk2 by Thr160 phosphorylation by CAK. Similarly, the p21Cip1/WAF1 and p16INK4A families of CDK inhibitors also block the phosphorylation by CAK (1). These data are in contrast to a previous report that the kinase activity of cyclin A-Cdk2 was not affected by Skp2 in a study using baculovirus-expressed proteins (40). In contrast to cyclin A-Cdk2, the kinase activity of the neuronal p25-Cdk5 complexes was not affected by Skp2 (Fig. 4). Similarly, it was also found that Cdk5 was not inhibited by p21Cip1/WAF1 and p27Kip1 (16). The parallel between Skp2 and p21Cip1/WAF1 is further seen by the fact that Skp2 and p21Cip1/WAF1 appears to bind to cyclin A-Cdk2 in a mutually exclusive manner (Fig. 2). This may partly explain why Skp2 and p21Cip1/WAF1 are not found together binding to cyclin A-Cdk2 in different cell lines (40). However, in the experiments described here, we do not exclude the possibility that Skp2 interacts with p21Cip1/WAF1 directly.
As for p21Cip1/WAF1 (10, 39), there was kinase activity associated with Skp2 which could be reduced by addition of more Skp2 (Fig. 5). This finding suggests that higher stoichiometry of Skp2 may be required for the inhibition of one molecule of Cdk2 or that Cdk2 dissociates from Skp2 immunoprecipitates at a high rate. Alternatively, there may be two separate binding conformations between cyclin A-Cdk2 and Skp2, one that inhibits the kinase activity and another that does not. The last possibility is supported by the fact that both the N-terminal and C-terminal regions of Skp2 are found to be involved in binding to cyclin A-Cdk2 (Fig. 3). In this connection, p21Cip1/WAF1 also contains two cyclin binding sites, one at the N terminus and the other at the C terminus of the protein.
While Skp2 can inhibit the kinase activity of cyclin A-Cdk2 toward histone H1, Skp2 itself served as a substrate for cyclin A-Cdk2 at the same time (Fig. 4 and 6). We found that Ser76 was the major site in Skp2 phosphorylated by cyclin A-Cdk2, and phosphorylation caused a mobility shift of Skp2 on SDS-PAGE (Fig. 3). Furthermore, S191A, but neither S76A nor S76A S191A, exhibited a mobility shift on SDS-PAGE as did wild-type Skp2 (data not shown). We have evidence that normally in the cell, all endogenous Skp2 is present in the Ser76-phosphorylated, shifted form (unpublished data). This scenario is similar to that for the CDK inhibitors p21Cip1/WAF1 and p27Kip1, which inhibit Cdk2 but are phosphorylated by Cdk2 at the same time (32, 39). This may in part be due to the requirement of multiple molecules or multiple binding sites models of Cdk2 inhibition by p21Cip1/WAF1 described above. Given that Skp2 was phosphorylated by cyclin A-Cdk2, it is possible that when Skp2 bound to cyclin A-Cdk2, Skp2 became the preferred substrate of cyclin A-Cdk2 and thus less phosphate was incorporated into substrates like histone H1. This may be similar to the possibility suggested for the inhibition of cyclin A-Cdk2 by p107 and p130 (37). However, mutants of Skp2 that were not phosphorylated by cyclin A-Cdk2 (S76A and S76A S191A) can still inhibit the kinase activity of cyclin A-Cdk2 toward histone H1 (Fig. 6), suggesting that Skp2 is not a mere substrate that competed with histone H1 in these assays.
We analyzed that regions of Skp2 that are important for binding to Skp1 and cyclin A-Cdk2 (Fig. 3). The regions of Skp2 that are important for binding to cyclin A-Cdk2 were also important for inhibition of the kinase activity, mainly at the region N terminal to the F box, although the C-terminal sequences also have some affinity for cyclin A-Cdk2. As expected, the F-box motif of Skp2 is required for the binding to Skp1. When we mutated the conserved proline residue in the F-box to alanine to create the P113A mutants we observed that binding between Skp1 and Skp2 decreased about 20% (data not shown); in contrast, in the case of another F-box protein, Cdc4, binding to Skp1 of the proline-to-alanine mutant was completely abolished (4). It is interesting that in Skp2, at least one of the binding site for cyclin A-Cdk2 is close to the F box. This finding suggests that Cdk2 may be in close proximity to Skp1 in the complex and that the direct interaction between Skp1 and Cdk2 that we described could also be present within the cyclin A-Cdk2-Skp2-Skp1 complex.
The stimulation effect of Skp1 on Cdk2 kinase activity is intriguing (Fig. 4). Skp1 alone did not contain kinase activity, which suggests that it is unlikely that the increase in kinase activity was due to a contaminated kinase in the Skp1 preparation. Boiling of Skp1 abolished the kinase stimulation, suggesting that the effect was likely to be due to a protein factor and not the buffer. In contrast to Skp2, which associated with both the Cdk2 and cyclin A subunits, Skp1 interacted with Cdk2 but not the cyclin A subunit (Fig. 7). We suspect that the binding between cyclin A-Cdk2 and Skp1 was weaker than that between cyclin A-Cdk2 and Skp2, or between Skp2 and Skp1, because the stimulation of cyclin A-Cdk2 by Skp1 was abolished in the presence of Skp2. There are precedents that proteins can promote the activity of cyclin-CDK complexes. Cdc37 stabilizes and promotes the folding of Cdk4 and Cdk6 (34), leading to the formation of more active cyclin D-CDK complexes. It has also been suggested that at low concentrations, p21Cip1/WAF1, p27Kip1, and p57Kip2 promote the assembly of cyclin D-Cdk4 (15). However, we do not think that Skp1 (or Skp2) promotes the assembly of cyclin A-Cdk2 complex in vitro. Rapid complex formation between cyclin A and Cdk2 was observed when these proteins purified from bacteria were mixed together (reaching a maximum in ∼5 min). In contrast, the binding of Skp1 or Skp2 to cyclin A-Cdk2 was much slower (reached a maximum in ∼30 min). We did not detect any change in the rate of cyclin A-Cdk2 assembly in the presence of Skp1 or Skp2 (unpublished data). One possibility is that some chaperones from bacteria were carried over with the Skp1 preparation, which in turn may assist the folding of Cdk2.
The arrest of the cell cycle after overexpression of Skp2 in mammalian cells was in good agreement with the findings that Skp2 can inhibit the kinase activity of cyclin A-Cdk2 and can block the activation of Cdk2 by CAK. We found that after gel filtration fractionation of HeLa cell extracts, Skp1 consisted of two populations, an apparent monomeric form and a complexed form (unpublished data). This finding suggests that Skp1 is likely to be in excess over its partners like Skp2 in the cell. This may explain why further expression of extra Skp1 in the cell has little effect on the cell cycle distribution. In contrast, gel filtration fractionation suggests that all of the Skp2 molecules are complexed to other proteins in the cell, suggesting that overexpression of Skp2 may disrupt the equilibrium between cyclin-CDK and cyclin-CDK-Skp2. It should be noted that there are ample potential problems underlying these kinds of ectopic expression experiments, where the expression levels of Skp2 and Skp1 must be exceedingly high. One alternative explanation of the action of Skp2 is that the overexpressed Skp2 may bind to Skp1 and cyclin A-Cdk2 separately, and the usual cyclin A-Cdk2-Skp2-Skp1 complexes (which may be required for G1-S transition) are disrupted. Furthermore, if Skp2 is involved in mediating the proteolysis of cyclin A, the destruction of cyclin A upon expression of Skp2 may also arrest the cell cycle. One useful experiment is to express a mutant of Skp2 that is defective in Skp1 or cyclin A-Cdk2 binding in cells and observe the effect on the cell cycle. We found that expression of the Skp2 CΔ96 and CΔ131 mutants (which can bind to cyclin A-Cdk2 but not to Skp1) in mammalian cells has no effect on the cell cycle (data not shown). However, one problem is that both N-terminal and C-terminal regions of Skp2 are involved in binding to cyclin A-Cdk2 (Fig. 3).
Perhaps a more important question is whether the proteolysis of cyclin A is regulated through binding to Skp2 and Skp1 (SCFSkp2). It is at present unclear whether Skp1 and Skp2 are involved in cyclin A destruction. The actions of Skp2 serving as a mediator of cyclin A proteolysis or an inhibitor of cyclin A-Cdk2 kinase activity would presumably serve the same end—turning off cyclin A-Cdk2 kinase activity. It is not inconceivable that the cyclin A that is targeted for destruction would be inhibited by Skp2 before proteolysis actually occurs.
One intriguing question is why Skp2 is generally expressed at higher levels in transformed cells than in normal fibroblasts (Fig. 1). In contrast, the levels of cyclin A and Skp1 do not vary significantly between normal and cancer cells. It should be noted that not all cancer cell lines have elevated levels of Skp2; one example is the human neuroglioma cell line H4 shown in Fig. 1. It is possible that overexpression of Skp2, acting either as an inhibitor of cyclin A-Cdk2 or as a mediator of cyclin A proteolysis, would lead to cyclin A-Cdk2 being turned on later or turned off earlier during S phase and hence may contribute to transformation. Another possibility is that the increase in Skp2 in transformed cells is a mechanism used by the cells to compensate for the loss of negative regulators of cyclin-CDK such as p21Cip1/WAF1. Consistent with this, there is an inverse relationship between the level of p21Cip1/WAF1 and Skp2 in cultured cells (38), and there is a correlation between the higher expression level of cyclin A and that of Skp2 in hepatocellular carcinoma (5).
ACKNOWLEDGMENTS
We are very grateful for David Beach, Hermann Bujard, Emma Lees, Julian Gannon, Tim Hunt, Tony Hunter, Kun Ping Lu, Gertrud Orend, Hideo Toyoshima, and Masakane Yamashita for reagents. We thank Tim Hunt, Tony Hunter, and members of the Poon lab for help and discussions. We also thank Frances Chan for help with the FACS analysis.
This work was supported in part by Research Grants Council grant DAG96/97-SC26 and British Council/Research Grants Council grant JRS96/31 to R.Y.C.P. C.H.Y. is a Sir Edward Youde Memorial Fellow. R.Y.C.P. is a member of the Biotechnology Research Institute.
REFERENCES
- 1.Aprelikova O, Xiong Y, Liu E T. Both p16 and p21 families of cyclin-dependent kinase (CDK) inhibitors block the phosphorylation of cyclin-dependent kinases by the CDK-activating kinase. J Biol Chem. 1995;270:18195–18197. doi: 10.1074/jbc.270.31.18195. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991. [Google Scholar]
- 3.Bai C, Richman R, Elledge S J. Human cyclin F. EMBO J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 5.Chao Y, Shih Y-L, Chiu J-H, Chau G-Y, Lui W Y, Yang W K, Lee S-D, Huang T-S. Overexpression of cyclin A but not Skp2 correlates with the tumor relapse of human hepatocellular carcinoma. Cancer Res. 1998;58:985–990. [PubMed] [Google Scholar]
- 6.Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demetrick D J, Zhang H, Beach D H. Chromosomal mapping of the genes for the human CDK2/cyclin A-associated proteins p19 (SKP1A and SKP1B) and p45 (SKP2) Cytogenet Cell Genet. 1996;73:104–107. doi: 10.1159/000134318. [DOI] [PubMed] [Google Scholar]
- 8.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 9.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L-H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, Fox M P, Wei N. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heuvel S V D, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 12.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis. Oxford, England: IRL Press; 1991. pp. 217–247. [Google Scholar]
- 13.Kamps M P. Determination of phosphoamino acid composition by acid hydrolysis of protein blotted to Immobilon. Methods Enzymol. 1991;201:21–27. doi: 10.1016/0076-6879(91)01005-m. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi H, Steward E, Poon R, Adamczewski J P, Gannon J, Hunt T. Identification of the domains in Xenopus cyclin A required for binding to and activation of p34cdc2 and p32cdk2 protein kinase subunits, and for cell cycle regulated proteolysis. Mol Biol Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 16.Lee M H, Nikolic M, Baptista C A, Lai E, Tsai L H, Massague J. The brain-specific activator p35 allows Cdk5 to escape inhibition by p27Kip1 in neurons. Proc Natl Acad Sci USA. 1996;93:3259–3263. doi: 10.1073/pnas.93.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisztwan J, Marti A, Sutterlüty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45SKP2: evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu K P, Hunter T. Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell. 1995;81:413–424. doi: 10.1016/0092-8674(95)90394-1. [DOI] [PubMed] [Google Scholar]
- 19.Lyapina S A, Correll C C, Kipreos E T, Deshaies R J. Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc Natl Acad Sci USA. 1998;95:7451–7456. doi: 10.1073/pnas.95.13.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathias N, Johnson S L, Winey M, Adams A E, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 22.Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60, and behaves differently from cyclin B. Nature. 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- 23.Poon R Y, Lew J, Hunter T. Identification of functional domains in the neuronal Cdk5 activator protein. J Biol Chem. 1997;272:5703–5708. doi: 10.1074/jbc.272.9.5703. [DOI] [PubMed] [Google Scholar]
- 24.Poon R Y C. Cell cycle control. In: Bertino J R, editor. Encyclopedia of cancer. San Diego, Calif: Academic Press; 1996. pp. 246–255. [Google Scholar]
- 25.Poon R Y C, Chau M S, Yamashita K, Hunter T. The role of Cdc2 feedback loop control in the DNA damage checkpoint in mammalian cells. Cancer Res. 1997;57:5168–5178. [PubMed] [Google Scholar]
- 26.Poon R Y C, Hunter T. Dephosphorylation of Cdk2 Thr160 by the cyclin-dependent kinase-interacting phosphatase KAP in the absence of cyclin. Science. 1995;270:90–93. doi: 10.1126/science.270.5233.90. [DOI] [PubMed] [Google Scholar]
- 27.Poon R Y C, Jiang W, Toyoshima H, Hunter T. Cyclin-dependent kinases are inactivated by a combination of p21 and Thr14/Tyr15 phosphorylation after UV-induced DNA damage. J Biol Chem. 1996;271:13283–13291. doi: 10.1074/jbc.271.22.13283. [DOI] [PubMed] [Google Scholar]
- 28.Poon R Y C, Toyoshima H, Hunter T. Redistribution of the CDK inhibitor p27 between different cyclin · CDK complexes in the mouse fibroblast cell cycle and in cells arrested with lovastatin or UV irradiation. Mol Biol Cell. 1995;6:1197–1213. doi: 10.1091/mbc.6.9.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poon R Y C, Yamashita K, Adamczewski J P, Hunt T, Shuttleworth J. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 1993;12:3123–3132. doi: 10.1002/j.1460-2075.1993.tb05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon R Y C, Yamashita K, Howell M, Ershler M A, Belyavsky A, Hunt T. Cell cycle regulation of the p34cdc2/p33cdk2-activating kinase p40MO15. J Cell Sci. 1994;107:2789–2799. doi: 10.1242/jcs.107.10.2789. [DOI] [PubMed] [Google Scholar]
- 31.Schwob E, Böhm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1/S transition in Saccharomyces cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 32.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 33.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 34.Stepanova L, Leng X, Parker S B, Harper J W. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 35.van der Geer P, Luo K, Sefton B M, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis on cellulose thin-layer plates. In: Celis J E, editor. Cell biology: a laboratory handbook. San Diego, Calif: Academic Press; 1994. pp. 422–450. [Google Scholar]
- 36.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 37.Woo M S, Sanchez I, Dynlacht B D. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol Cell Biol. 1997;17:3566–3579. doi: 10.1128/mcb.17.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y, Zhang H, Beach D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993;7:1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Hannon G J, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A/CDK2 S-phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Xiong Y, Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol Biol Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]