Abstract
We have analyzed the influence of in vivo treatment and in vitro addition of thyroid hormone on in organello mitochondrial DNA (mtDNA) transcription and, in parallel, on the in organello footprinting patterns at the mtDNA regions involved in the regulation of transcription. We found that thyroid hormone modulates mitochondrial RNA levels and the mRNA/rRNA ratio by influencing the transcriptional rate. In addition, we found conspicuous differences between the mtDNA dimethyl sulfate footprinting patterns of mitochondria derived from euthyroid and hypothyroid rats at the transcription initiation sites but not at the mitochondrial transcription termination factor (mTERF) binding region. Furthermore, direct addition of thyroid hormone to the incubation medium of mitochondria isolated from hypothyroid rats restored the mRNA/rRNA ratio found in euthyroid rats as well as the mtDNA footprinting patterns at the transcription initiation area. Therefore, we conclude that the regulatory effect of thyroid hormone on mitochondrial transcription is partially exerted by a direct influence of the hormone on the mitochondrial transcription machinery. Particularly, the influence on the mRNA/rRNA ratio is achieved by selective modulation of the alternative H-strand transcription initiation sites and does not require the previous activation of nuclear genes. These results provide the first functional demonstration that regulatory signals, such as thyroid hormone, that modify the expression of nuclear genes can also act as primary signals for the transcriptional apparatus of mitochondria.
Thyroid hormone regulates the expression of key nucleus encoded mitochondrial genes (23, 37, 38, 47) and the steady-state concentration of all mitochondrial DNA (mtDNA)-encoded mRNAs (mt-mRNAs) (15–17, 32, 44). In rat liver, changes in mt-mRNA levels in response to in vivo treatment with thyroid hormones were well correlated with amounts of the corresponding polypeptides, while mtDNA copy number remained unmodified (33, 47). Based on these observations, transcriptional control has been tentatively proposed as the main mechanism to explain the thyroid hormone effect on mt-mRNA steady-state levels and protein synthesis (33–36).
Two alternative potential mechanisms for the regulation of mtDNA transcription by thyroid hormone have been suggested: through activation of mitochondrial transcription factor A (mtTFA) expression (18, 45), or by a direct action of the hormone through mitochondrial receptors (48). The second alternative is supported by earlier (42, 43) and recent (1, 48) reports that have proposed the existence of T3 (3,3,5-triiodo-l-thyronine) binding proteins in mitochondria, which could interact with cis elements of mtDNA (7, 48). However, the lack of experimental evidence showing thyroid hormone-mediated transcription regulation in the absence of nuclear gene expression prevents any definitive conclusion about whether direct regulation of mtDNA transcription by this hormone occurs (38). Presently it is more generally believed that this transcriptional regulation, if it exists, could be exclusively exerted through the activation of nucleus encoded transcription factors (22, 33, 36, 41, 47).
The influence of thyroid hormone on the steady-state level of mtRNAs, however, represents a more complex situation. Reports from several groups showed a decrease in the in vivo steady-state mRNA/rRNA ratio in hypothyroid versus euthyroid mitochondria (referred to hereafter in this work as hypothyroid and euthyroid mitochondria, respectively). This effect was mainly due to a decrease in the mRNA levels, since rRNA levels were not (17, 32) or were only slightly (44) affected. Therefore, the proposed transcriptional control should be able to discriminate between mRNA and rRNA genes, although different effects of T3 on the stability of mRNAs and rRNAs could also account for this phenomenon.
Several mechanisms could potentially be responsible for the discrimination between mRNA and rRNA synthesis. The mammalian mtDNA H strand encodes the two rRNAs and all but one of the mt-mRNAs, and it is transcribed as two polycistronic molecules (31). The smaller polycistron, responsible for synthesis of the rRNAs, completely overlaps the larger one, responsible for synthesis of the mRNAs. Two models have been proposed to explain the independent regulation of mRNA and rRNA syntheses in mammalian mitochondria. One model is based on the identification of the 5′ end of the transcripts synthesized on a partially reconstructed in vitro transcription system (6, 13), and it proposes that the synthesis of both polycistrons starts at a single initiation site, upstream of the tRNAPhe gene. The polymerase transcribes the rRNA genes more frequently stopping at the 3′ end of 16S rRNA due to the binding to mtDNA of a mitochondrial transcription termination factor (mTERF) (6, 12, 21, 25). Occasionally, the polymerase is able to pass through this termination point and then also synthesize the mRNAs. A second model, based on the identification of the 5′ end of the in vivo synthesized primary transcripts, proposes the existence of two sites for H-strand transcription initiation. Thus, synthesis of the small polycistron starts at the IH1 initiation site, upstream of the tRNAPhe gene (31), which corresponds to the unique initiation site in the previous model. Synthesis of the larger polycistron starts at the IH2 initiation site, at the border between the 12S rRNA and tRNAPhe genes (3, 30, 31). According to this model, when transcription initiates at the IH1 site, it normally stops at the 3′ end of 16S rRNA due to the action of mTERF. In this model, when the polymerase initiates at the IH2 site, it is able to read through the mTERF-dependent termination, transcribing almost the whole mtDNA H strand, including both rRNAs and most of the mRNAs. Thus, in the first model, regulation of the mRNA/rRNA ratio could be achieved only by modulation of the transcription termination at the mTERF binding site. The second model suggests that this ratio could be preferentially modulated by changing transcription initiation between IH1 and IH2. Interestingly, the regulation of the mRNA/rRNA ratio by thyroid hormone represents an occasion to address both models.
To discriminate between the alternative mechanisms proposed for the in vivo-induced changes in the steady-state level of mtRNAs by thyroid hormone, as well as for its selective influence on mRNA versus rRNA synthesis, we took advantage of a highly efficient in organello transcription system (9, 10), using liver mitochondria isolated from rats differing in thyroid status. Thus, we have analyzed the influence of the in vivo treatment and the in vitro addition of thyroid hormone on in organello mtRNA synthesis and stability. In parallel, using transcriptionally active organelles, we have investigated the influence of the hormone on the in organello footprinting patterns at the mtDNA regions involved in the regulation of transcription. The results presented in this report confirm that transcriptional control is the main mechanism underlying the thyroid hormone effect on mtRNA steady-state concentration. In addition, they strongly support a model in which this regulation is achieved by acting at multiple levels and involving both genomes. Here, we show that the action of T3 on control of the mRNA/rRNA ratio is exclusively due to changes in transcriptional rate and is exerted directly on mitochondria, without prior activation of nuclear genes. Our results also suggest that this effect is achieved by selective modulation of the activity of the alternative H-strand transcription initiation sites.
MATERIALS AND METHODS
Animals.
Hypothyroidism was induced in male Wistar rats weighing 200 to 250 g by administration of 0.05% (wt/vol) propylthiouracyl (PTU) in drinking water for 4 to 8 weeks. A subgroup of hypothyroid rats (treated group) were injected intraperitoneally once daily with 20 μg and 3 μg of T4 (3,3′,5,5′-tetraiodo-l-thyronine) and T3, respectively, per 100 g of body weight for 2 days, and the animals were killed 15 h after the second treatment. Hyperthyroidism was induced in euthyroid rats by daily intraperitoneal injections of the same amount of T4 and T3 as before for 5 days, and the rats were used for experimentation on day 6 after initiation of treatment. Control hypothyroid and euthyroid rats were injected for the same time period with the same volume of vehicle (0.9% NaCl-propylene glycol).
In organello RNA synthesis and pulse-chase experiments.
Mitochondria were isolated from rat liver as previously described (9, 10) and incubated at a final mitochondrial protein concentration of 2 mg/ml in 0.5 ml of incubation buffer containing 25 mM sucrose, 75 mM sorbitol, 100 mM KCl, 10 mM K2HPO4, 0.05 mM EDTA, 5 mM MgCl2, 1 mM ADP, 10 mM glutamate, 2.5 mM malate, 10 mM Tris-HCl (pH 7.4), and 1 mg of bovine serum albumin per ml in 1.5-ml Eppendorf tubes (9, 10). For in organello transcription analysis, 20 μCi of [α-32P]UTP (400 to 600 Ci/mmol) was added to the medium, and incubation was maintained at 37°C for 60 min in a rotary shaker (12 rpm). For pulse-chase experiments, isolated mitochondria were prelabeled with [α-32P]UTP for 2 h; then the mitochondrial samples were pelleted at 13,000 × g for 1 min, and the supernatant with the nonincorporated [α-32P]UTP was removed. Then, the mitochondria were resuspended in incubation medium in the presence of a 200-fold excess of unlabeled UTP and incubated for various periods of time before harvesting (9). Mitochondrial nucleic acids were extracted and analyzed by methylmercury hydroxide-agarose gels as previously described (11). Subsequently, the gels were first stained with ethidium bromide and photographed under UV light and then dried and exposed for autoradiography either at −70°C with a DuPont screen intensifier or at room temperature. Amounts of RNA on the autoradiograms were quantified, after selection of the appropriate exposures, with an LKB Ultroscan XL laser densitometer and Gel Scan XL software.
In organello and in vitro footprinting.
For methylation interference assays, mitochondria were incubated for 30 min as described above, then dimethyl sulfate (DMS) was added to a final concentration of 0.1% and allowed to react for 2 min at 37°C, and the mixture was quickly cooled on ice. Then 1 ml of ice cooled incubation buffer was added, and the organelles were spun down for 1 min at 13,000 rpm. This wash was repeated twice, and mtDNA was isolated as described below. For each set of in organello footprinting experiments, a sample of the mitochondrial fraction was treated identically except for the omission of DMS, and mtDNA was extracted. For in vitro DMS treatment of naked mtDNA, 15 to 25 μg of mitochondrial nucleic acids derived from non-DMS-treated organelles was resuspended in 200 μl of Tris-EDTA, DMS was added to a final concentration of 0.1% and allowed to react for 2 min at 37°C, and the mixture was quickly cooled on ice. Then the reaction was stopped by adding 50 μl of DMS stop buffer (1.5 M sodium acetate [pH 7.0], 1 M 2-mercaptoethanol). Finally, mtDNA was recovered by ethanol precipitation (28).
Piperidine cleavage of DNA was performed on pellets containing mtDNA from in organello DMS-treated mitochondrial fraction and on equivalent samples of in vitro DMS-treated total mitochondrial nucleic acids as described previously (28).
Primer extension of the DMS-treated mtDNA.
The following oligodeoxynucleotides, designated according to the numbering system of Gadaleta et al. (14), were used for primer extension: H30-5′-GAATCCATCTAAGCATTTTCAG-3′ (designated P-REV1 in reference 5), H175-5′-TATATTATGTGCTTGATGCCCT-3′, L16108-5′CCCCAAAAACATTAAAGCAAGA-3′ (D-Rat-viv-2 in reference 5), and L16228-5′-GACACAAAATCTTTCCTCCTAA-3′ for footprinting of the transcription initiation areas; H16107-5′TTTGGCATTGAAGTTTCAGGTG-3′ (D-REV2 in reference 5) and L15747-5′-ACTGAAACTTTACAGGCATCTG-3′ for footprinting of the region containing the putative thyroid-responsive element (TRE); and H2761-5′-GATTAGGAGTGTTAGGATATTA-3′ (ND1 in reference 5) and L2542-5′-CCCAGTTACGAAAGGACAAGAG-3′ (16S in reference 5) for footprinting of the rDNA transcription termination region. These oligodeoxynucleotides were 5′-end labeled with [γ-32P]ATP as detailed previously (28).
DMS-treated and piperidine-cleaved mtDNA (0.5 μg) was used as a template for primer extension analysis with the appropriate labeled oligodeoxynucleotide. PCR amplification and electrophoresis of the products through 6% polyacrylamide (19:1 acrylamide/bisacrylamide)–7 M urea sequencing gels in Tris-borate-EDTA buffer were carried out as previously described (28).
DMS is known to methylate mainly guanine and, with lower frequency, adenine residues (27). Cytosine and thymine can also be methylated when in single-stranded DNA, with the alkylation of the latter being favored in alkaline solution (27). In the present work, the majority of methylation reactivity changes were observed at purine sites, although some pyrimidines also showed an altered methylation pattern as reported by others (5, 20, 29). The methylation of cytosine residues in organello was explained by the occurrence of single-stranded DNA segments at those sites as a result of helical distortions caused by protein binding or by RNA polymerase pauses (29). In the case of thymine, it was suggested that the alkaline environment of the mitochondrial matrix would favor its methylation (29).
RESULTS
The transcriptional ability of isolated mitochondria is predetermined in vivo by thyroid hormone.
To investigate the mechanism responsible for the in vivo-induced changes in the steady-state level of mitochondrial mRNAs by thyroid hormones, we performed in organello transcription experiments with isolated rat liver mitochondria purified from rats of four different thyroid states: a control group (euthyroid animals), a group treated for a minimum of 1 month with drinking water containing PTU (17) (hypothyroid animals), a group injected for 5 consecutive days with constant doses of hormone (hyperthyroid animals), and a group consisting of hypothyroid animals injected for 2 days with the same hormone doses (treated animals). The body weight of the animals, monitored to follow the influence of the PTU treatment, reached an average difference of 1.4-fold in favor of the euthyroid animals compared with the hypothyroid ones. Blood samples of all the animals were collected at the moment of sacrifice, and the levels of hormones were analyzed by radioimmunoassay to check their thyroid status (Table 1).
TABLE 1.
Levels of thyroid hormones in blood samples of the experimental animals
| Rat | Concn (mean ± SEM [n = 8])
|
||
|---|---|---|---|
| Total T3 (ng/100 ml) | Total T4 (μg/100 ml) | Free T4 (pg/100 ml) | |
| Euthyroid | 86.7 ± 4.18 | 4.99 ± 0.60 | 15.59 ± 1.84 |
| Hypothyroid | 34.0 ± 3.61 | 0.57 ± 0.02 | 0.98 ± 0.09 |
| Treated | 306 ± 24.4 | 41.37 ± 1.95 | 139 ± 20.27 |
| Hyperthyroid | 418 ± 37.4 | 22.7 ± 2.75 | 76.8 ± 15.85 |
As shown in Fig. 1A, transcription was more active in the organelles isolated from animals with higher plasma concentrations of thyroid hormones. Thus, hypothyroid mitochondria showed a 50% reduction of the radioactivity incorporated into RNA compared with euthyroid organelles. Furthermore, this decrease was partially restored by in vivo treatment of the hypothyroid animals with thyroid hormone. In agreement with this general trend, isolated organelles from hyperthyroid rats were able to accumulate labeled RNA at a higher rate than controls.
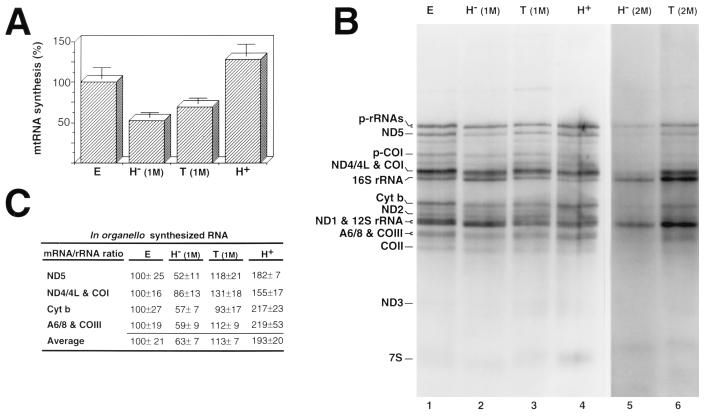
FIG. 1.
In organello RNA synthesis in rat liver mitochondria isolated from animals of different thyroid status. (A) Diagram showing the overall transcriptional rate of mitochondria isolated from euthyroid (E), hypothyroid [1-month PTU treatment; H− (1M)], treated [hypothyroid treated with hormone; T (1M)], and hyperthyroid (H+) animals. Data were normalized to the content of mtDNA in each lane, taking the euthyroid value as 100%, and are given as mean ± standard error of the mean (n = 5). (B) Electrophoretic patterns of the in organello-synthesized RNAs by mitochondria from the indicated thyroid status (lanes 1 to 4) and from hypothyroid rats obtained by 2-month PTU treatment [H− (2M)] and 2-month hypothyroid treatment with hormone [T (2M)] (lanes 5 and 6). Similar amounts of total radioactivity incorporated into RNA were loaded on lanes 1 to 4, and amounts of mtRNA synthesized by equivalent fractions of mitochondria were loaded on lanes 5 and 6. (C) Relative mRNA/rRNA ratio for different representative mRNAs synthesized by mitochondria isolated from rats of the indicated thyroid status. The values (mean ± standard error of the mean n = 5) correspond to the ratio mRNA to all transcribed 16S rRNA genes (see text) and are given as relative to the euthyroid ratio (100%). The differences in mRNA/rRNA ratio between the euthyroid group and the hypothyroid and hyperthyroid groups were significant (P < 0.005) by Student’s t test. COI, COII, and COIII, mRNAs for subunits I, II, and III of cytochrome c oxidase; ND1, ND2, ND3, ND4/4L, and ND5, mRNAs for subunits 1, 2, 3, 4, 4L, and 5 of the NADH dehydrogenase; A6/8, mRNA for subunits 6 and 8 of H+-ATPase; Cyt b, mRNA for apocytochrome b; p-rRNAs, precursors of rRNAs; p-CoI, precursor of COI mRNA.
Besides this general effect of thyroid hormone on mtRNA transcription rate, a fine analysis of the synthesis of discrete RNA species revealed that T3 also promoted a remarkable change in the relative synthesis of mRNAs with respect to rRNAs (Fig. 1B and C). Thus, when the relative labeling of several mRNAs was compared with the labeling of the RNAs (calculated as the radioactivity incorporated into mature 16S rRNA plus the radioactivity incorporated into the rRNA precursor that includes 16S rRNA), an average ∼1.6-fold reduction in the mRNA/rRNA ratio in hypothyroid mitochondria was observed (Fig. 1B, lane 2; Fig. 1C). This reduction was reversed almost completely by in vivo treatment of the hypothyroid rats with the hormone (Fig. 1B, lane 3; Fig. 1C). Moreover, hyperthyroid organelles showed a ∼1.9-fold increase in the mRNA/rRNA ratio relative to the control. Interestingly, the decrease in the mRNA/rRNA ratio was even higher when mitochondria from rats treated for 2 months with PTU were assayed (Fig. 1B, lane 5), and the modification of the mRNA/rRNA ratio was again partially recovered by 2 days of in vivo treatment of the animals with thyroid hormones (Fig. 1B, lane 6; Fig. 1C). It should be mentioned that fractionation of in vivo- and in organello-synthesized mtRNAs by oligo(dT)-cellulose columns allowed us to determine that the ratio of 12S to 16S rRNA was not affected by the thyroid status of the animals (not shown).
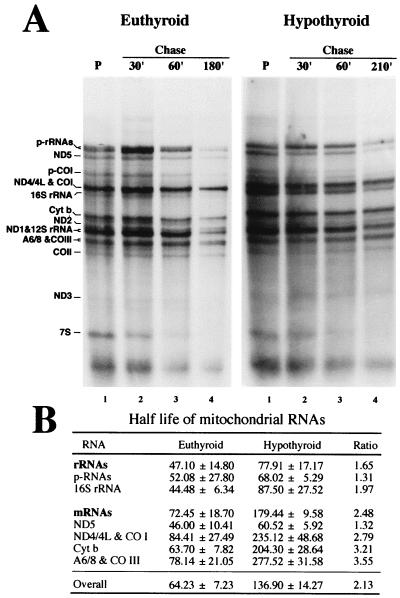
The differences in accumulation of RNA by isolated organelles could be due either to modifications of the transcriptional activity or to alterations of the RNA stability. To distinguish between these two alternatives, we performed pulse-chase experiments and estimated the overall decay and half-lives of RNA synthesized by isolated organelles from euthyroid and hypothyroid rats. Thus, overall half-lives of 64.23 ± 7.23 and 136.9 ± 14.27 min (mean ± standard deviation) found for the RNA synthesized by euthyroid and hypothyroid organelles, respectively (Fig. 2). Therefore, since the stability of the RNA is increased in hypothyroid organelles, the reduced accumulation of labeled RNA in hypothyroid mitochondria must be a consequence of differences in the rate of RNA synthesis. Pulse-chase experiments also allowed estimation of stability for the different species of RNA. Figure 2 shows that after short periods of chase, the radioactivity incorporated in several of the discrete RNA bands increases due to the transformation of previously labeled nascent chains into mature RNAs (9). This is more evident with euthyroid (Fig. 2A, left) than with hypothyroid (Fig. 2A, right) organelles, possibly because of the lower transcription activity in hypothyroid mitochondria. In contrast, after longer periods of chase, the labeling of all RNA species synthesized during the pulse by both types of mitochondria decreases progressively, allowing determination of their stability (Fig. 2B). Interestingly, we found that the half-lives of the mRNAs and rRNAs were increased (1.65- and 2.48-fold, respectively) in hypothyroid mitochondria. Therefore, the alteration in the mRNA/rRNA ratio promoted by thyroid hormone is due to differences in their relative transcriptional rates rather than a consequence of posttranscriptional regulation phenomena. In summary, these results confirm that thyroid hormone promotes differences both in the overall transcriptional rate of mtDNA and in the relative transcriptional rate of the rRNA and mRNA genes.
FIG. 2.
Influence of thyroid status on the stability of the RNAs synthesized by isolated mitochondria. (A) Pulse-chase experiment of in organello RNA synthesis with mitochondria isolated from euthyroid and hypothyroid animals. Pulse (P) was 2 h of incubation in the presence of [α-32P]UTP, after which the mitochondria were pelleted, washed and resuspended in fresh medium with excess of unlabeled UTP, and incubated for the indicated chase periods. Bands are labeled as in Fig. 1. (B) Half-lives (mean ± standard deviation), in minutes, for total RNA (overall) or different mtRNA species calculated from the pulse-chase experiments. Ratio is hypothyroid/euthyroid value.
In vitro addition of thyroid hormone to isolated hypothyroid mitochondria restores the normal mRNA/rRNA proportion.
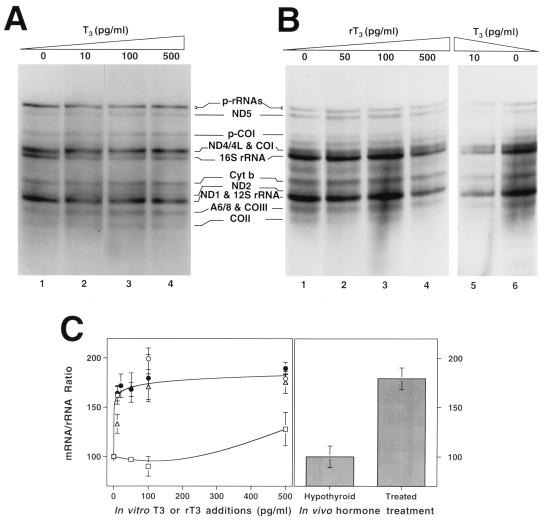
To investigate if the influence of thyroid hormones on mtDNA transcription could be exerted directly on mitochondria, in organello RNA synthesis was carried out with rat liver mitochondria isolated from 1-month hypothyroid rats incubated in the presence of different concentrations of added hormone. As shown in Fig. 3A, addition of increasing concentrations of T3 to the incubation medium did not significantly influence the overall transcriptional rate of mitochondria. However, the hormone strikingly modified the ratio of mRNA/rRNA synthesized by isolated organelles. In agreement with the in vivo hormone effect described above, T3 addition preferentially favored mRNA labeling, increasing the mRNA/rRNA ratio (Fig. 3A and C). This effect was revealed at a very low concentration of T3 (10 pg/ml), reaching a plateau at 50 pg/ml, where the increase of the mRNA/rRNA ratio was 1.8- to 2.0-fold (Fig. 3A and C). Similar results were obtained in three independent experiments (Fig. 3C). Furthermore, when 2-month hypothyroid mitochondria were assayed, a similar increase in the mRNA/rRNA ratio induced by a very low concentration of T3 (10 pg/ml) was again observed (Fig. 3B, lanes 5 and 6). Very interestingly, reverse-T3 (rT3), a much less active analog of T3, was unable to induce any effect on the mRNA/rRNA ratio in the same organelles, even at a 10-fold higher concentration (100 pg/ml). Only when rT3 was added at 500 pg/ml was it able to mimic the effect on the mRNA/rRNA ratio produced by T3 (Fig. 3B, lane 4). It should be noted that the in vitro addition of T3 to hypothyroid mitochondria produced an increase in the mRNA/rRNA ratio quantitatively identical to that observed in hypothyroid animals treated with thyroid hormones (Fig. 3C).
FIG. 3.
Effect of thyroid hormone addition to the incubation medium on in organello mtRNA synthesis. (A and B) Electrophoretic patterns of the in organello-synthesized RNA by isolated mitochondria from hypothyroid rat liver (1-month treatment) incubated in the presence of the indicated T3 concentrations (A) and from hypothyroid rat liver (2-month treatment) incubated in the presence of the indicated concentration of rT3 (lanes 1 to 4) or T3 (lanes 5 and 6) (B). Amounts of RNA synthesized by an equal fraction of mitochondria were loaded per lane in each gel. (C) Comparison of the in vivo effect of T3 and in vitro addition effect of T3 or rT3 on the in organello mRNA/rRNA synthesis ratio. Left, average change in the mRNA/rRNA ratio after addition of T3 in three independent experiments (▵, ○, •) or rT3 (□) to the incubation medium. The differences in the mRNA/rRNA ratio at any concentration of T3 added to the incubation medium compared with no T3 were significant (P < 0.001) by Student’s t test. The difference in mRNA/rRNA ratio at 500 pg of rT3 per ml compared with no addition was significant (P < 0.01) by Student’s t test. Right, effect of the in vivo treatment of hypothyroid animals with T3 on the mRNA/rRNA ratio. The difference was significant (P < 0.01) by Student’s t test. Data are expressed as the mean ± standard error of the mean of the proportion of radioactivity incorporated in the bands for the mRNAs ND5, COI plus ND4/L, Cyt b, and COIII plus A6/8 (see the legend to Fig. 1 for definitions) with respect to the radioactivity incorporated into the band corresponding to the mature 16S rRNA plus the corresponding fraction of 16S rRNA contained in the p-rRNAs band.
The footprinting pattern of the transcription regulatory region of mtDNA is predetermined in vivo by thyroid hormone.
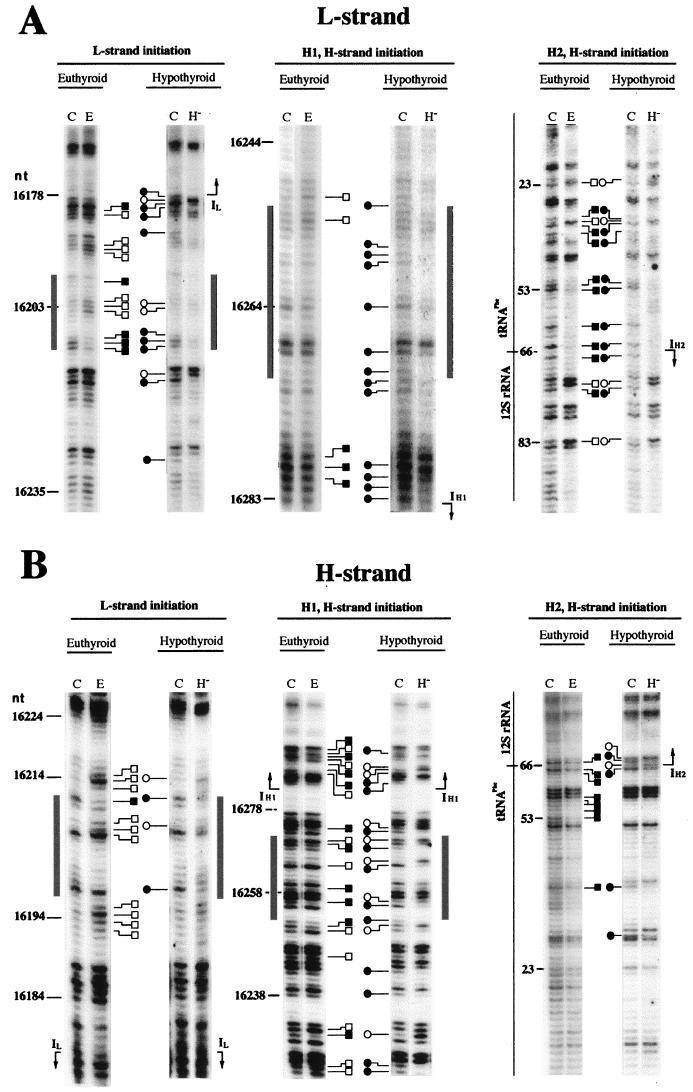
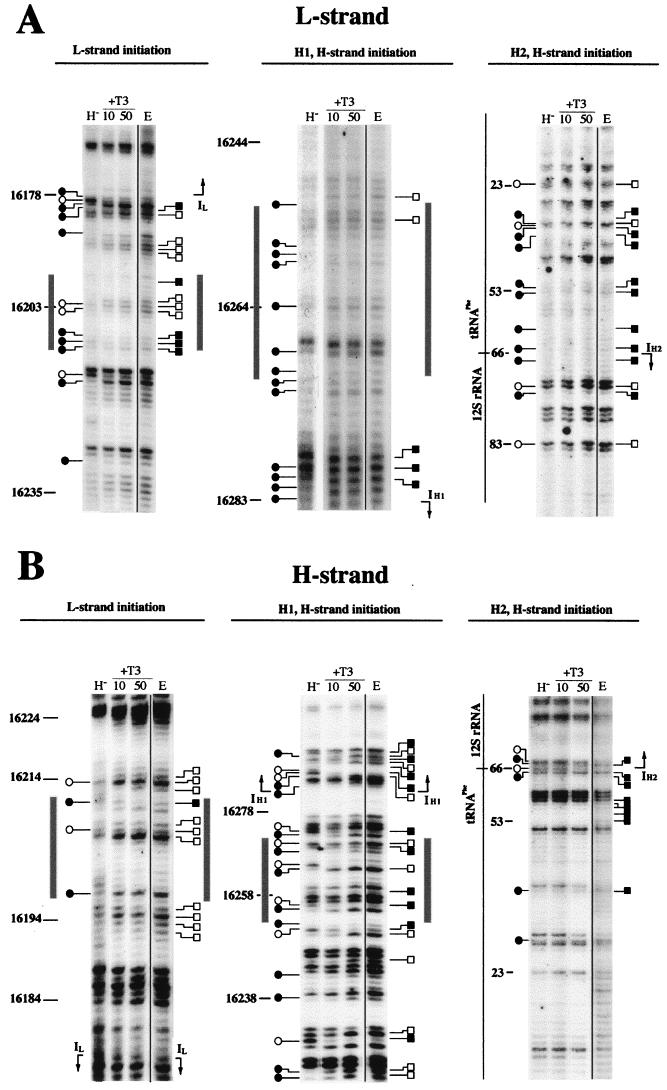
Regulation of the mRNA/rRNA ratio by thyroid hormone could be exerted by selective modulation of the H-strand transcription initiation at IH1 or IH2 (3, 30, 31). Therefore, we first investigated if thyroid hormone modifies the protein-DNA interactions in the transcription initiation area in a way that could be detected by methylation interference analysis. Figures 4 and 5 show the methylation patterns of the L and H strands in the area where the transcription promoters for both strands are located. By comparing the in organello mtDNA methylation patterns of euthyroid (Fig. 4, lanes E) and hypothyroid (lanes H−) animals with those generated by DMS treatment of deproteinized (naked) mtDNA (lanes C), one can identify several regions of altered methylation reactivity. In particular, the nucleotides located around the transcription start sites for the L strand (for IL, nucleotide [nt] 16178) and the H strand (for IH1, nt 16283; for IH2, nt 66) show clear protections and hypermethylations (Fig. 4 and 5A). Additional alterations are concentrated in the regions spanning nt 16197 to 16215 and nt 16252 to 16271 (Fig. 4 and 5A), which correspond to the binding sites of the mitochondrial transcription activator mtTFA (5, 13). Finally, it was possible to observe some methylation alterations in the regions spanning nt 16211 to 16252, between the IL and IH1 promoters, and in the region spanning nt 22 to 41, between the IH1 and IH2 transcription initiation sites (Fig. 4 and 5A).
FIG. 4.
Effect of the in vivo thyroid status on the footprinting pattern at the mtDNA transcription initiation region. Comparison between euthyroid and hypothyroid mitochondria of the in organello footprinting patterns of mtDNA H strand (A) and L strand (B) in the region containing the transcription initiation sites. C, naked mtDNA treated in vitro with DMS; E, in organello DMS-treated mtDNA from euthyroid animals; H−, in organello DMS-treated mtDNA from hypothyroid animals. Hypermethylated bands are represented by open squares for euthyroid and open circles for hypothyroid samples; protected bands are represented by closed squares for euthyroid and closed circles for hypothyroid samples. Gray boxes indicate the proposed mtTFA binding sites. IL, IH1, IH2, transcription initiation sites at the L and the H strands (IL and IH1 according to reference 5; IH2 by analogy with human DNA [29]).
FIG. 5.
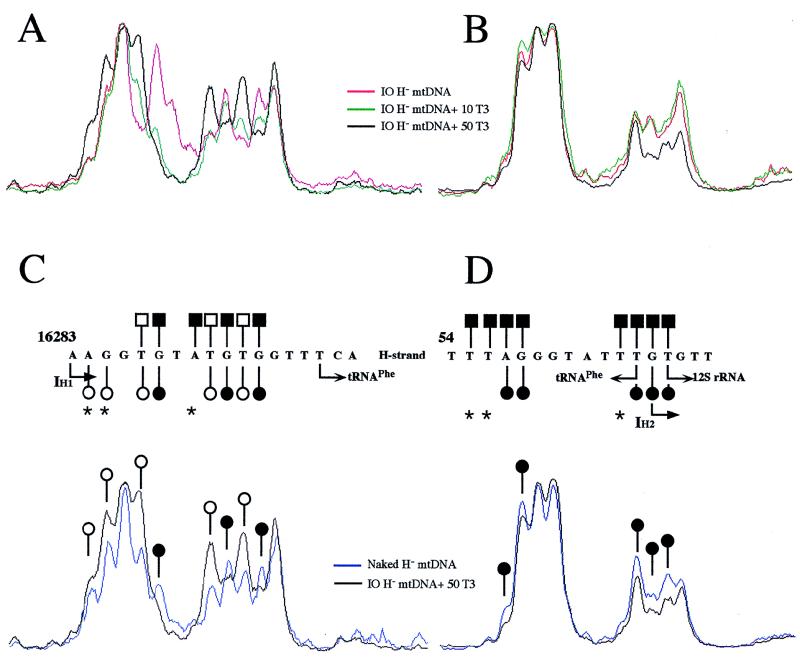
Summary and details of the methylation interference sites from hypothyroid (circles) and euthyroid (squares) mitochondria in the mtDNA transcription initiation region. (A) Sequence of the DMS-sensitive positions within the D-loop region containing the transcription initiation sites. The mtTFA binding areas and the transcription initiation sites (IL, IH1, and IH2) are shown. (B and C) Comparison of densitometric tracings from footprinting analysis, between hypothyroid (bottom) and euthyroid (middle) mitochondria, at the template strand of the H-strand initiation sites IH1 (B) and IH2 (C). A summary of the changes is shown above each panel on the sequence containing each initiation site. IO, in organello DMS-treated mtDNA; other symbols are as described in the legend to Fig. 4.
In summary, most of the protection or hypermethylation phenomena described above appear to occur at positions of known protein-DNA interactions (mtTFA) or at potential sites of similar interactions (RNA polymerase or transcription factor[s]) at or near the positions of RNA synthesis initiation. There is a reasonable correspondence between the mtDNA regions exhibiting methylation alterations shown in Fig. 4 and 5 with those found previously in rat liver (5), bovine brain (19), and human mtDNA (20, 29).
Comparison of the footprinting patterns of euthyroid and hypothyroid mitochondria revealed clear differences (Fig. 4 and 5A). In particular, the areas surrounding the IL, IH1, and IH2 transcription start sites showed the most conspicuous differences between hypothyroid and euthyroid mtDNA. Remarkably, they mostly affected the template strand for each start site, the L strand for IL (Fig. 4A, left panel) and the H strand for IH1 (Fig. 4B, central panel; Fig. 5B) and IH2 (Fig. 4B, right panel; Fig. 5C). In addition, some differences were also evident on both strands in the binding regions of the transcription factor mtTFA. Those mainly consisted in the weakening or loss of the footprinting signal for the mtTFA binding site close to IL in the hypothyroid sample (Fig. 4, left panels). The changes in the mtTFA binding site near the H-strand rRNA initiation site (IH1) were more complex, showing new nucleotides with methylation interference (i.e., nt 16255, 16259, and 16265 [Fig. 4B, central panel]) and reversion of the methylation status in other positions (i.e., nt 16256 and 16264 [Fig. 4B, central panel]) in the hypothyroid sample. These differences suggest either an alteration of the DNA-protein interactions or a structural change in mtDNA at the transcription promoter area determined by the in vivo thyroid state that could be responsible for the changes in the transcription activity described above.
In vitro addition of thyroid hormone to isolated hypothyroid mitochondria restores the euthyroid footprinting patterns of the transcription regulatory region of mtDNA.
Since the in vitro addition of the hormone was able to modify the transcription activity of the organelles, we next analyzed whether this addition would also affect the footprinting patterns of hypothyroid mitochondria in a way that could be correlated with the transcription changes. For this, we performed methylation interference experiments with hypothyroid mitochondria in the presence of different concentrations of thyroid hormone. We observed that the addition of increasing amounts of T3 modified the footprinting pattern of the transcription initiation areas such that it becomes progressively more similar to that obtained from euthyroid organelles (Fig. 6, lanes 10 and 50). This is clearly indicated by the intensification of the footprinting signal on both strands at the mtTFA binding site close to IL (Fig. 6, left panels). Furthermore, the euthyroid-like methylation status of the mtTFA binding site near IH1 is also recovered with the addition of increasing concentrations of the hormone (e.g., nt 16255, 16256, 16263, and 16264; Fig. 6B, central panel). Remarkably, the DMS sensitivity at the L-strand and the two H-strand transcription initiation sites was strongly modified by T3 addition to the incubation medium, and again the euthyroid-like methylation pattern was reached at 50 pg of T3 per ml (Fig. 6, lanes 50).
FIG. 6.
Effect of in vitro thyroid hormone addition to organelles isolated from hypothyroid animals on the footprinting patterns of mtDNA at the transcription initiation region. Shown is comparison of the mtDNA DMS reactivity, at the L strand (A) and H strand (B), between euthyroid (E) and hypothyroid mitochondria incubated without (H−) or with the indicated T3 concentration (10 or 50 pg/ml). For the naked DNA methylation pattern, see Fig. 4. Symbols are as described in the legend to Fig. 4.
The more interesting changes induced by T3 were observed at the two putative H-strand transcription initiation sites (IH1 and IH2), because of their proposed role in the regulation of mRNA and rRNA synthesis (31). Those changes affected only nucleotides on the template strand (Fig. 6B, central and right panels; Fig. 7). A similar phenomenon can be observed at the L-strand promoter (Fig. 6A, left panel). In particular, at the IH1 site, nt T16287, G16288, T16291, G16292, T16293, and G16294 recovered the euthyroid methylation status when hypothyroid organelles were incubated in the presence of 50 pg of T3 per ml (Fig. 7A and C). Similarly, at the IH2 site, nt A57, G58, T65, G66, and T67 recovered the euthyroid methylation status after incubation with thyroid hormone (Fig. 7B and D). However, recovery of the euthyroid footprinting pattern at the IH1 and IH2 sites was not complete. Thus, at the IH1 site, nt A16284 and G16285 were hypermethylated in the presence of 50 pg of T3 per ml and unmodified in the euthyroid patterns, and nt A16290 was unmodified in the presence of 50 pg of T3 per ml and protected in the euthyroid patterns (Fig. 7A and C). On the other hand, at the IH2 site, nt T55, T56, and T64 were protected in the euthyroid patterns and unmodified after incubation of hypothyroid mitochondria with the hormone (Fig. 7B and D). It should be mentioned that only at the IH2 site were methylation patterns of the H-strand naked euthyroid and hypothyroid mtDNAs not identical. Thus, nt G68 showed a clear methylation in the hypothyroid naked mtDNA patterns but only a faint methylation in its euthyroid naked mtDNA counterpart (Fig. 4B, right panel). This observation was consistent between different independent experiments, but its significance is unclear. Nevertheless, the changes in the methylation sensitivity of each nucleotide with respect to the naked control DNA can still be clearly detected. In summary, the addition of T3 to the incubation medium of hypothyroid mitochondria modifies the footprinting patterns of the transcription regulatory regions of mtDNA, substantially restoring the patterns observed in euthyroid mitochondria.
FIG. 7.
Summary of the influence of in vitro T3 addition on the methylation reactivity at the two H-strand transcription initiation sites. Shown are densitometric tracings of the mtDNA footprinting patterns from Fig. 6 at the template strand of the IH1 (A and C) and IH2 (B and D) transcription initiation sites. (A and B) Changes in the methylation reactivity of mtDNA from hypothyroid organelles incubated without (IO [in organello DMS-treated] H− mtDNA) or with the indicated concentrations of T3 (10 and 50 pg/ml). (C and D) Densitometric tracings showing the methylation reactivity of hypothyroid mtDNA incubated in the presence of T3 (50 pg/ml) at the IH1 (C) and IH2 (D) areas. Comparison with the methylation reactivity of euthyroid mtDNA (open and closed squares) is shown above each panel on the sequence containing each initiation site.
The footprinting pattern of the transcription termination region of mtDNA is not influenced by the in vivo thyroid status or by the in vitro addition of thyroid hormone to hypothyroid mitochondria.
Another potential mechanism to regulate the mRNA/rRNA ratio could be by influencing mTERF-dependent transcription termination. The function of mTERF requires its specific binding to an mtDNA motif at the boundary between the rRNA and mRNA genes (6, 12, 21, 25). Therefore, we also analyzed the footprinting pattern of the mtDNA area containing the mTERF binding site in organelles isolated from euthyroid and hypothyroid animals, in the latter case with and without the addition of T3 to the incubation medium (Fig. 8). We observed a strong modification in methylation reactivity with respect to naked DNA (Fig. 8, lanes C) in the in organello DMS-treated samples, regardless of their in vivo thyroid status (lanes E and H−). Nucleotides on both strands were clearly affected in a way very similar to that described previously (5). However, no significant differences between hypothyroid and euthyroid samples were observed (Fig. 8). Moreover, when thyroid hormone was added to the incubation medium of hypothyroid mitochondria, no changes in the methylation reactivity at the mTERF binding site could be detected (lanes 10 and 50). Therefore, we conclude that thyroid hormone does not affect the binding of mTERF to mtDNA.
FIG. 8.
Effect of in vivo thyroid status and in vitro T3 addition on the footprinting pattern of mtDNA at the transcription termination region. Shown are in organello footprinting patterns of rat mtDNA transcription termination areas at the L and H strands. In the sequence summary at the bottom, the gray box indicates the tridecamer sequence where mTERF binds. Symbols are as described in the legend to Fig. 6.
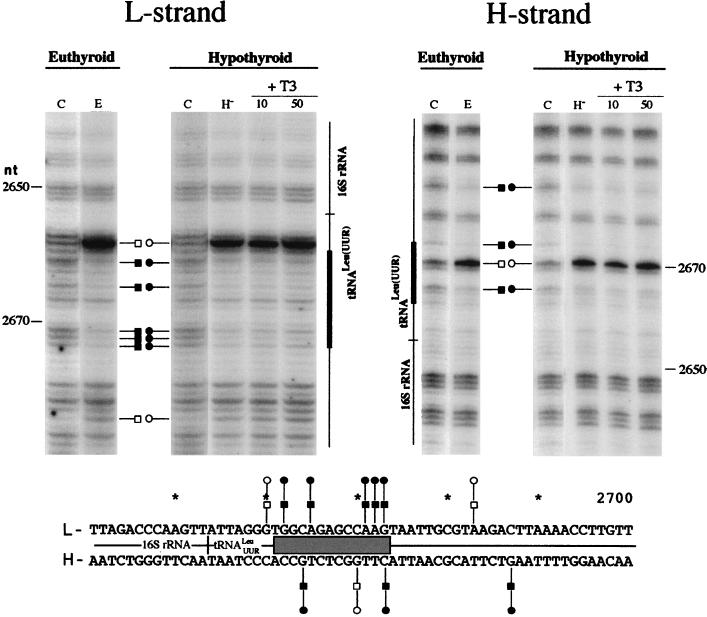
The footprinting pattern of a putative TRE in the mtDNA D-loop region reveals no detectable protein-DNA interactions.
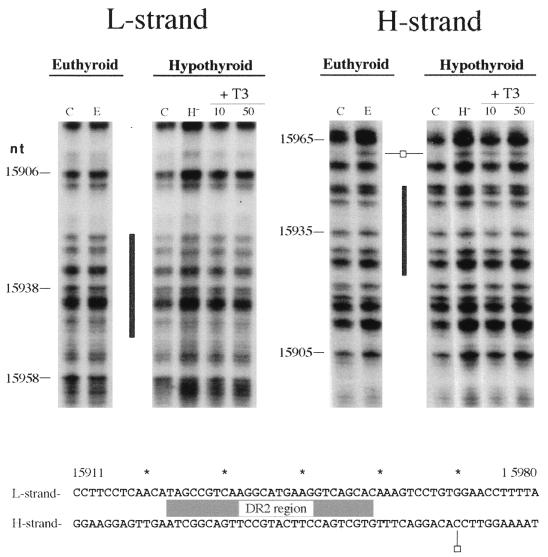
Wrutniack and coworkers (48) have described a mitochondrial matrix 43-kDa protein able to bind T3 and canonical TREs as well as a specific sequence in the rat mtDNA D-loop area near the transcription promoters; this observation was obtained by gel mobility retardation assays. To verify its potential role as a cis element that might mediate the action of thyroid hormone on mitochondrial transcription, we investigated the existence of protein-DNA interactions at this location. We performed footprinting analysis on a 300-bp area (from nt 15768 to 16086), which includes the full oligonucleotide used by Wrutniack et al. (48) in their band shift assays and also includes the conserved sequence block I, a region known to interact with proteins (5). In agreement with previous descriptions (5), we found conspicuous alteration of DMS reactivity at and downstream of the conserved sequence block I region (nt 16001 to 16057) that was not affected by the in vivo thyroid status or by the in vitro addition of the hormone to the incubation medium of hypothyroid mitochondria (data not shown). In contrast, no differences in DMS reactivity between naked mtDNA (Fig. 9, lanes C) and in organello mtDNA (lanes E and H−) were observed within the area containing the proposed TRE (nt 15923 to 15949). We could find only one hypermethylated band, at position 15960 on the H strand, downstream of the proposed TRE-containing region. Moreover, the in vivo thyroid status as well as the in vitro addition of the hormone to the isolated organelles did not affect the mtDNA DMS reactivity properties in this area (Fig. 9).
FIG. 9.
In organello footprinting pattern (top) and sequence summary (bottom) of rat mtDNA in the D-loop area proposed to contain the TRE. The gray box indicates the location of the putative TRE (DR2). Symbols are as described in the legend to Fig. 6.
DISCUSSION
Our results have allowed a deeper understanding of the influence of thyroid hormone on the expression of mtDNA. (i) We have confirmed that T3 exerts in vivo a double effect on mitochondrial transcription: an increase in the overall transcriptional rate, and a differential modulation of the relative mRNA/rRNA transcriptional rate. (ii) Part of this influence, i.e., the change in ratio of mRNA to rRNA synthesis, is exerted directly on mitochondria, without previous modulation of nuclear gene expression, and it is probably achieved by selection of transcription initiation at alternative sites on the H strand.
The observation of a double effect of T3 on mtDNA transcription is fully in agreement with previous reports from different groups that showed a 1.4-fold (2) or 3- to 7-fold (32) increase in transcription activity between rat liver mitochondria isolated from hypothyroid rats and hypothyroid animals treated in vivo with T3. Likewise, a ∼2-fold decrease in the in vitro activity of RNA polymerase partially purified from hypothyroid rat liver mitochondria compared with control mitochondria was also reported (17). Furthermore, our results are also in agreement with previous reports showing a decrease in the mRNA/rRNA ratio of the in vivo steady-state levels of rat liver mtRNAs in hypothyroid versus euthyroid mitochondria. This effect was mainly due to a decrease in the mRNA levels since rRNA levels were not (17, 32) or were only slightly (44) affected. Thus, variations of ∼1.75-fold (17), 2- to 7-fold (32), or 1.5- to 2-fold (44) in the mRNA/rRNA ratio were observed. Although these observations strongly suggested a regulatory action of thyroid hormone on mtDNA transcription, the lack of experimental data on direct measurement of the transcriptional rate and on stability of the different mitochondrial transcripts prevented a definitive conclusion. The reproduction in our in organello transcription system of the effects observed in vivo confirms the suitability of this experimental approach to investigate the action of thyroid hormone on mtDNA transcription. Moreover, the maintenance of in vivo-like conditions for mtDNA transcription, the analysis of the RNA synthesized de novo under controlled conditions, and, in parallel, the possibility of footprinting specific mtDNA areas while transcription is in progress allowed us to obtain an integrated picture of the action of thyroid hormone on mitochondrial transcription.
Thus, we have demonstrated that the thyroid status affects the stability of all mtRNAs, being on average twofold more stable in hypothyroid than in control organelles. The increase in mtRNA stability probably reflects a general mechanism operating in mitochondria when transcription is depressed, rather than a specific regulatory action of thyroid hormone. In fact, an increase in the stability of mtRNAs induced after inhibiting mitochondrial transcription has been described previously for cultured cells (8). If the stability of the RNAs is taken into consideration, the decrease in the rate of RNA synthesis observed in hypothyroid mitochondria is greater than that indicated by the incorporation of [α-32P]UTP into RNA (2.34-fold versus 1.85-fold). In addition, the reduction in the mRNA/rRNA ratio observed in hypothyroid mitochondria must be a consequence of the alteration in the relative rate of synthesis of both types of RNA, since the increase in stability of the mRNAs is slightly (1.5-fold) higher than that of the rRNAs.
Besides the effects on transcription, we have also found that the footprinting patterns at the regions of mtDNA that contain the transcription start sites of both strands are predetermined in vivo by thyroid hormone. This observation strongly suggests that the interaction of the proteins involved in the initiation of transcription with mtDNA is influenced by the hormone levels. In contrast, the in vivo thyroid status does not affect the interaction of mTERF with mtDNA. It should be remarked that thyroid hormone particularly affects the footprinting patterns at the template strand on the L-strand and the two H-strand transcription initiation sites. Therefore, in light of these results, the observed changes in the relative synthesis of mRNA and rRNA are more likely due to the influence of T3 on the selection of the H-strand initiation sites than to transcription termination at the mTERF binding site.
In summary, our results confirm that transcriptional control is the main mechanism underlying the thyroid hormone effect on mtRNA steady-state levels. However, the in vivo experiments discussed above do not allow discrimination between nucleus-dependent or direct regulation of mtDNA transcription by the hormone.
In the nucleus, thyroid hormone stimulates transcription through its interaction with receptors that recognize TREs located in the regulatory region of target genes. In mitochondria, two alternative models of action of thyroid hormone have been proposed. On one side, T3 could activate mitochondrial transcription by an indirect mechanism, the previous activation of mtTFA synthesis (46). In support of this, it has been reported that in hyperthyroid rats the level of the mtTFA mRNA in liver increases in parallel with mtDNA-encoded transcripts (18), and a putative TRE has been found in the promoter region of the mtTFA gene (18). However, the recent development of mtTFA knockout mice shows that there is not a direct correlation between the amount of mtTFA and the steady-state levels of mtRNA (26). On the other hand, a direct action of the hormone through mitochondrial receptors that would be able to interact with TREs present at the regulatory region of mtDNA has been proposed (1, 7, 48). This model is supported by recent reports describing c-Erb-related proteins in rat liver mitochondria (1, 48). Particularly, a 43-kDa protein, immunologically related to c-ErbAα1, which is able to bind specifically and with high affinity to T3 and canonical TREs, has been found in the mitochondrial matrix. Very interestingly, this protein is able to show specific binding, in gel mobility shift assays, to sequences contained in the regulatory region of rat mtDNA (48).
Two lines of evidence indicate that T3 directly influences mtDNA transcription. First, our analysis shows that the modification of the mRNA/rRNA ratio induced in vivo by T3 can be fully reproduced by the direct addition of the hormone to isolated hypothyroid mitochondria. In agreement with this, we also found that the normal DMS interference methylation patterns in the regulatory region of initiation of mtDNA transcription, predetermined in vivo by thyroid hormone, can be substantially reestablished by in vitro addition of the hormone to isolated hypothyroid mitochondria.
The very low amount of hormone that is needed to promote the change in the mRNA/rRNA ratio (10 pg/ml), plus the fact that it is a saturable effect, strongly suggests that this phenomenon can be mediated by a high-affinity T3 receptor present in limited amount in the mitochondria. The specificity of the in vitro action of T3 was tested by monitoring the ability of rT3 to promote the same effect in the relative synthesis of mRNA versus rRNA. It is known that rT3 is able to bind nuclear c-ErbA-type T3 receptors but with a ∼100-fold-lower affinity than T3 (39). However, while the affinity of the receptor for different T3 analogues may vary, the occupancy of the ligand-binding domain by iodothyronine or similar ligands will mimic the T3 effect (4). If a c-ErbA-like receptor is somehow involved in the observed effect of T3 on in organello transcription, then this effect would be mimicked by rT3 but only at a clearly higher concentration. In agreement with this, a change of the mRNA/rRNA ratio qualitatively similar to that produced by 10 pg of T3 per ml was observed only when rT3 was added at 500 pg/ml. This observation could support the existence of c-ErbA-related proteins in rat liver mitochondria as proposed elsewhere (1, 48).
Wrutniak et al. (48) have also found a direct repeat sequence (DR2) within the regulatory region of mtDNA (nt 15923 to 15949) that was proposed as a potential TRE. They have shown that an oligonucleotide containing this sequence is specifically bound by the mitochondrial c-ErbA-related protein (48). We could not verify by DMS methylation interference experiments if the proposed mtDNA TRE was in fact interacting with proteins in functional isolated mitochondria. However, while T3 was able to induce detectable alterations at the binding site of other transcription factors, the absence of DMS reactivity changes at well-characterized TREs on the regulatory region of nuclear genes seems to be a common phenomenon (24). Therefore, on the grounds of the absence of DMS methylation reactivity, we cannot rule out the role of the proposed region of the mtDNA as a true TRE.
Concerning the mechanism by which the relative synthesis of mRNA and rRNA could be affected in mitochondria, differential regulation of the two H-strand initiation sites by T3 seems to us the more plausible explanation. Footprinting analysis reveals that the protein-DNA interactions at the mtTFA binding sites as well as at the transcription start sites are remarkably different in hypothyroid and euthyroid mtDNAs. In parallel, the mRNA/rRNA ratio as well the overall transcriptional rate are modified. The in vitro presence of the hormone is sufficient to recover the way in which transcription factor(s) interact with the mtDNA in euthyroid organelles. The changes in the DMS reactivity induced in vitro by T3 were not accompanied by alterations of the transcriptional activity as a whole but correlated with a reduction in the synthesis of rRNAs in favor of the mRNAs. Thus, the differential accessibility of transcription initiation factor(s) and likely the RNA polymerase to mtDNA, induced by T3, would enhance the election of IH2 as the transcription initiation site for the H strand. Interestingly, this behavior substantially resembles what has been described for the influence of T3 on the regulatory region of the nucleus encoded growth hormone gene. There, the presence of T3 does not seem to be required for, but facilitates, the binding of other transcription factors, such as Sp-1 or Pit-1, to the DNA (24). The alternative explanation for the modulation of the mRNA/rRNA ratio, that is, regulation of the transcription termination of the smaller H-strand-derived polycistron, seems unlikely, because the binding activity of the factor responsible for the termination of transcription (mTERF) remains unaltered between different thyroid statuses and after the in vitro addition of the hormone.
Finally, the fact that the in vitro addition of T3 to the incubation medium was unable to increase the overall transcriptional rate in isolated mitochondria suggests that this effect, promoted in vivo by the hormone, is probably mediated by an indirect mechanism that can involve modulation of nuclear gene expression. As a consequence, the mt-mRNA/rRNA ratio is increased in organello, to the detriment of the synthesis of rRNAs (Fig. 3A and B). Interestingly, when the effect on the mRNA/rRNA ratio is combined with the stimulation in the overall transcription observed after in vivo treatment with the hormones, a net accumulation of mt-mRNAs should be expected (1.6- to 2.3-fold), while the steady-state level of the rRNAs should remain constant or increase only slightly. This is, in fact, what has been described by several groups (17, 32, 44).
In conclusion, the results presented in this report are the first functional demonstration of a direct influence of hormones, particularly thyroid hormone, on the expression of mtDNA. This evidence is fundamental not only for a better understanding of the genetic regulation of the energetic metabolism by thyroid hormone but also to prove the existence of non-nucleus-mediated regulators of mammalian mtDNA transcription (9) and understand the signaling pathways that coordinate gene expression between the mitochondrial and nuclear compartments (40).
ACKNOWLEDGMENTS
We are very grateful to F. Martínez-Azorín for valuable comments and discussion and to Santiago Morales for excellent technical assistance.
This work was supported by grants from the Spanish Dirección General de Investigación Científica y Técnica (PB94-0567 and PB97-1019) and from the Zaragoza City Council.
REFERENCES
- 1.Ardail D, Lerme F, Puymirat J, Morel G. Evidence for the presence of α and β-related T3 receptors in rat liver mitochondria. Eur J Cell Biol. 1993;62:105–113. [PubMed] [Google Scholar]
- 2.Barsano C P, Degroot L J, Getz G S. The effect of thyroid hormone on in vitro rat liver mitochondrial RNA synthesis. Endocrinology. 1977;100:52–60. doi: 10.1210/endo-100-1-52. [DOI] [PubMed] [Google Scholar]
- 3.Bogenhagen D F, Applegate E F, Yoza B K. Identification of a promoter for transcription of the heavy strand of human mtDNA: in vitro transcription and deletion mutagenesis. Cell. 1984;36:1105–1113. doi: 10.1016/0092-8674(84)90061-8. [DOI] [PubMed] [Google Scholar]
- 4.Brent G A, Moore D D, Larsen P R. Thyroid hormone regulation of gene expression. Annu Rev Physiol. 1991;53:17–35. doi: 10.1146/annurev.ph.53.030191.000313. [DOI] [PubMed] [Google Scholar]
- 5.Cantatore P, Daddabbo L, Fracasso F, Gadaleta M N. Identification by in organello footprinting of protein contact sites and of single-stranded DNA sequences in the regulatory region of rat mitochondrial DNA-protein binding sites and single-stranded DNA regions in isolated rat liver mitochondria. J Biol Chem. 1995;270:25020–25027. doi: 10.1074/jbc.270.42.25020. [DOI] [PubMed] [Google Scholar]
- 6.Christianson T W, Clayton D A. A tridecamer DNA sequence supports human mitochondrial RNA 3′-end formation in vivo. Mol Cell Biol. 1988;8:4502–4509. doi: 10.1128/mcb.8.10.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demonacos C V, Karayanni N, Hatzoglou E, Tsiriyiotis C, Spandidos D A, Sekeris C E. Mitochondrial genes as sites of primary action of steroid hormones. Steroids. 1996;61:226–232. doi: 10.1016/0039-128x(96)00019-0. [DOI] [PubMed] [Google Scholar]
- 8.England J M, Costantino P, Attardi G. Mitochondrial RNA and protein synthesis in enucleated African green monkey cells. J Mol Biol. 1978;119:455–462. doi: 10.1016/0022-2836(78)90226-7. [DOI] [PubMed] [Google Scholar]
- 9.Enríquez J A, Fernández-Silva P, Pérez-Martos A, López-Pérez M J, Montoya J. The synthesis of mRNA in isolated mitochondria can be maintained for several hours and is inhibited by high levels of ATP. Eur J Biochem. 1996;237:601–610. doi: 10.1111/j.1432-1033.1996.0601p.x. [DOI] [PubMed] [Google Scholar]
- 10.Enríquez J A, Pérez-Martos A, López-Pérez M J, Montoya J. In organello RNA synthesis system from mammalian liver and brain. Methods Enzymol. 1996;264:50–57. doi: 10.1016/s0076-6879(96)64008-7. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Silva P, Enríquez J A, Montoya J. A simple procedure for recovering the denaturing effect of methylmercury in agarose gel electrophoresis. BioTechniques. 1992;12:480–482. [PubMed] [Google Scholar]
- 12.Fernández-Silva P, Martínez-Azorín F, Micol V, Attardi G. The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J. 1997;16:1066–1079. doi: 10.1093/emboj/16.5.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher R P, Topper J N, Clayton D A. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 14.Gadaleta G, Pepe G, De Candia G, Quagliarelllo C, Sbisà E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 15.Gadaleta M N, Di Reda N, Bove G, Saccone C. Effects of triiodothyronine on rat-liver mitochondrial transcription process. Eur J Biochem. 1975;51:495–501. doi: 10.1111/j.1432-1033.1975.tb03949.x. [DOI] [PubMed] [Google Scholar]
- 16.Gadaleta M N, Minervini G R, Renis M, Giorgi C D, Giovine A. Mitochondrial DNA, RNA and protein synthesis in normal and hypothyroid developing rat liver. Cell Differ. 1986;19:43–49. doi: 10.1016/0045-6039(86)90024-2. [DOI] [PubMed] [Google Scholar]
- 17.Gadaleta M N, Petruzzella V, Fracasso F, Fernández-Sílva P, Cantatore P. Acetyl-l-carnitine increases cytochrome oxidase subunit I mRNA content in hypothyroid rat liver. FEBS Lett. 1990;277:191–193. doi: 10.1016/0014-5793(90)80841-6. [DOI] [PubMed] [Google Scholar]
- 18.Garstka H L, Facke M, Escribano J R, Wiesner R J. Stoichiometry of mitochondrial transcripts and regulation of gene expression by mitochondrial transcription factor A. Biochem Biophys Res Commun. 1994;200:619–626. doi: 10.1006/bbrc.1994.1493. [DOI] [PubMed] [Google Scholar]
- 19.Ghivizzani S C, Madsen C S, Hauswirth W W. In-organello footprinting—analysis of protein binding at regulatory regions in bovine mitochondrial DNA. J Biol Chem. 1993;268:8675–8682. [PubMed] [Google Scholar]
- 20.Ghivizzani S C, Madsen C S, Nelen M R, Ammini C V, Hauswirth W W. In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Mol Cell Biol. 1994;14:7717–7730. doi: 10.1128/mcb.14.12.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess J F, Parisi M A, Bennett J L, Clayton D A. Impairment of mitochondrial transcription termination by a point mutation associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1991;351:236–239. doi: 10.1038/351236a0. [DOI] [PubMed] [Google Scholar]
- 22.Ichikawa K, Hashizume K. Cellular binding proteins of thyroid hormones. Life Sci. 1991;49:1513–1522. doi: 10.1016/0024-3205(91)90323-4. [DOI] [PubMed] [Google Scholar]
- 23.Izquierdo J M, Cuezva J M. Thyroid hormones promote transcriptional activation of the nuclear gene coding for mitochondrial beta-F(1)-ATPase in rat liver. FEBS Lett. 1993;323:109–112. doi: 10.1016/0014-5793(93)81459-d. [DOI] [PubMed] [Google Scholar]
- 24.Kim S W, Ahm I M, Larsen P R. In vivo genomic footprinting of thyroid hormone-responsive genes in pituitary tumor cell lines. Mol Cell Biol. 1996;16:4465–4477. doi: 10.1128/mcb.16.8.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 26.Larsson N-G, Wang J, Wilhelmosson H, Oldfors A, Rustin P, Lewandoski M, Barsh G S, Clayton D A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 27.Lawley P D. Effects of some chemical mutagens and carcinogens on nucleic acids. Prog Nucleic Acid Res Mol Biol. 1966;5:89–131. doi: 10.1016/s0079-6603(08)60232-9. [DOI] [PubMed] [Google Scholar]
- 28.Micol V, Fernández-Silva P, Attardi G. In vivo footprinting of human mitochondrial DNA in cultured cell systems. Methods Enzymol. 1996;264:50–57. doi: 10.1016/s0076-6879(96)64003-8. [DOI] [PubMed] [Google Scholar]
- 29.Micol V, Fernández-Silva P, Attardi G. Functional analysis of in vivo and in organello footprinting of HeLa cell mitochondrial DNA in relationship to ATP and ethidium bromide effects on transcription. J Biol Chem. 1997;272:18896–18909. doi: 10.1074/jbc.272.30.18896. [DOI] [PubMed] [Google Scholar]
- 30.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy strand and light strand transcription in human mitochondrial DNA. Proc Natl Acad Sci USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montoya J, Gaines G L, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 32.Mutvei A, Kuzela S, Nelson B D. Control of mitochondrial transcription by thyroid hormone. Eur J Biochem. 1989;180:235–240. doi: 10.1111/j.1432-1033.1989.tb14638.x. [DOI] [PubMed] [Google Scholar]
- 33.Nagley P. Coordination of gene expression in the formation of mammalian mitochondria. Trends Genet. 1991;7:1–4. doi: 10.1016/0168-9525(91)90002-8. [DOI] [PubMed] [Google Scholar]
- 34.Nelson B D, Joste V, Wielburski A, Rosenqvist U. The effect of triiodothyronine on the synthesis of mitochondrial proteins in isolated rat hepatocytes. Biochim Biophys Acta. 1980;608:422–426. doi: 10.1016/0005-2787(80)90187-2. [DOI] [PubMed] [Google Scholar]
- 35.Nelson B D, Joste A, Mutvei V. Regulation of biosynthesis of the rat liver inner mitochondrial membrane by thyroid hormone. Arch Biochem Biophys. 1984;228:41–48. doi: 10.1016/0003-9861(84)90044-4. [DOI] [PubMed] [Google Scholar]
- 36.Nelson B D. Thyroid hormone regulation of mitochondrial function. Comments on the mechanism of signal transduction. Biochim Biophys Acta. 1990;1018:275–277. doi: 10.1016/0005-2728(90)90266-7. [DOI] [PubMed] [Google Scholar]
- 37.Nelson B D, Luciakova K, Li R G, Betina S. The role of thyroid hormone and promoter diversity in the regulation of nuclear encoded mitochondrial proteins. Biochim Biophys Acta. 1995;1271:85–91. doi: 10.1016/0925-4439(95)00014-u. [DOI] [PubMed] [Google Scholar]
- 38.Pillar T M, Seitz H J. Thyroid hormone and gene expression in the regulation of mitochondrial respiratory function. Eur J Endocrinol. 1997;136:231–239. doi: 10.1530/eje.0.1360231. [DOI] [PubMed] [Google Scholar]
- 39.Samuels H H, Forman B M, Horwitz Z D, Ye Z-S. Regulation of gene expression by thyroid hormone. Annu Rev Physiol. 1989;51:623–639. doi: 10.1146/annurev.ph.51.030189.003203. [DOI] [PubMed] [Google Scholar]
- 40.Shadel G S, Clayton D A. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 41.Söboll S. Thyroid hormone action on mitochondrial energy transfer. Biochim Biophys Acta. 1993;1144:1–16. doi: 10.1016/0005-2728(93)90024-a. [DOI] [PubMed] [Google Scholar]
- 42.Sterling K, Lazarus J H, Milch P O, Sakurada T, Brenner M A. Mitochondrial thyroid hormone receptor: localization and physiological significance. Science. 1978;201:1126–1129. doi: 10.1126/science.210507. [DOI] [PubMed] [Google Scholar]
- 43.Sterling K, Brenner M A, Sakurada T. Rapid effect of triiodothyronine of the mitochondrial pathway in rat liver in vivo. Science. 1980;210:340–342. doi: 10.1126/science.7423197. [DOI] [PubMed] [Google Scholar]
- 44.van Itallie C M. Thyroid hormone and dexamethasone increase the levels of a messenger ribonucleic acid for a mitochondrially encoded subunit but not for a nuclear-encoded subunit cytochrome c oxidase. Endocrinology. 1990;127:55–62. doi: 10.1210/endo-127-1-55. [DOI] [PubMed] [Google Scholar]
- 45.Virbasius J V, Scarpulla R C. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiesner R J. Adaptation of mitochondrial gene expression to changing cellular energy demands. News Physiol Sci. 1997;12:178–184. [Google Scholar]
- 47.Wiesner R J, Kurowski T T, Zak R. Regulation by thyroid hormones of nuclear and mitochondrial genes encoding subunits of cytochrome-c oxidase in rat liver and skeletal muscle. Mol Endocrinol. 1992;6:1458–1467. doi: 10.1210/mend.6.9.1331777. [DOI] [PubMed] [Google Scholar]
- 48.Wrutniak C, Cassar-Malek I, Marchal S, Rascle A, Heusser S, Keller J M, Flechon J, Dauca M, Samarut J, Ghysdael J, Cabello G. A 43-kDa protein related to c-Erb A a1 is located in the mitochondrial matrix of rat liver. J Biol Chem. 1995;270:16347–16354. doi: 10.1074/jbc.270.27.16347. [DOI] [PubMed] [Google Scholar]