Abstract
Systemic lupus erythematosus (SLE) is a heterogeneous multifactorial disease. Upregulated TLR7 signaling is a known risk factor for SLE. Recently, it was shown that specific genetic variants in UNC93B1 affect the physiological regulation of TLR7 signaling and cause characteristic autoimmune phenotypes with monogenic autosomal recessive inheritance in mutant mice and dogs. We therefore hypothesized that homologous variants in the human UNC93B1 gene might be responsible for a fraction of human SLE patients. We analyzed 536 patients of the Swiss SLE Cohort Study for the presence of genetic variants affecting the C-terminal tail of UNC93B1. None of the investigated patients carried bi-allelic UNC93B1 variants that were likely to explain their SLE phenotypes. We conclude that genetic variants affecting the C-terminal tail of UNC93B1 are not a common risk factor for SLE. It cannot be excluded that such variants might contribute to other heritable autoimmune diseases.
Keywords: Homo sapiens, immunology, autoimmunity, candidate gene, TLR7 signaling
1. Introduction
SLE (Systemic Lupus Erythematosus) is a highly complex and heterogenous autoimmune disease with incompletely understood etiopathology [1,2]. Age of onset, specific organs affected, and the severity of the disease are highly variable between patients. SLE is characterized by a breakdown in immune tolerance, which promotes the formation of autoreactive B and T cells, abnormal cytokine production, and the subsequent generation of autoantibodies against DNA- and RNA-based self-antigens [2]. Women are nine times more frequently affected than men, and the incidence of the disease is highest in women of childbearing age [3].
SLE is thought to be caused by interactions between susceptibility genes and environmental factors resulting in an irreversible loss of immunologic self-tolerance. Several GWAS identified more than 100 risk loci for SLE, including associations to the HLA locus and many non-coding and presumably regulatory genome regions [4,5]. The X-chromosomal TLR7 gene encoding toll-like receptor 7 is one of the confirmed risk loci for SLE [5]. Increased TLR7 activity promotes autoimmunity [6,7,8], and there are indications that partial escape of TLR7 from X-chromosome inactivation may contribute to the extreme sex-bias in SLE incidence [9]. A single nucleotide variant in the 3′-UTR, rs3853839, modulates TLR7 expression and has been repeatedly associated with SLE [10,11].
While SLE is a genetically highly complex disease, rare patients exist in which SLE or related autoimmune disorders are caused by single-gene defects. Genes affected in such patients include DNASE1 [12] and TREX1 [13].
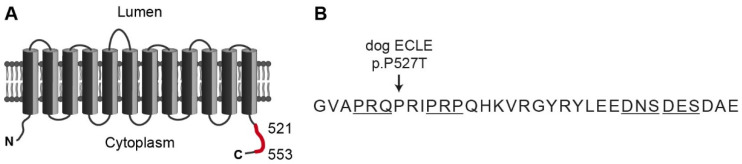
TLR7 is activated by single-stranded RNA and represents a component of the innate immune defense against RNA viruses. The trafficking chaperone UNC93B1 is required for the correct localization of TLR7 to endosomal membranes, and its loss-of-function leads to an immune deficiency [14,15,16]. TLR7 and UNC93B1 form a heterotetrameric complex in a 2:2 stoichiometry in endosomal membranes [17]. Subsequent to TLR7 activation, syndecan binding protein (SDCBP) binds to the C-terminal tail of UNC93B1, which induces the termination of TLR7 signaling [18]. Genetic variants in UNC93B1 that prevent SDCBP binding without affecting other functions of UNC93B1 result in the disruption of this negative feedback loop and consequently overactive TLR7 signaling that will eventually be triggered by endogenous RNA molecules [18,19,20,21]. Unc93b1PKP/PKP mice with a targeted disruption of the SDCBP binding motif develop a fatal systemic inflammation and autoimmune disease [18]. In dogs, a spontaneous missense variant affecting the C-terminal tail of UNC93B1 causes exfoliative cutaneous lupus erythematosus (ECLE) [22]. ECLE starts with skin lesions but typically develops into a systemic form of lupus in the affected dogs [22,23,24,25,26] (Figure 1).
Figure 1.
Topology of the human UNC93B1 protein. (A) UNC93B1 comprises 597 amino acids and contains 12 transmembrane domains. A segment of the C-terminal tail indicated in red is required for the interaction with SDCBP and subsequent dampening of TLR7 signaling [18]. (B) Amino acid sequence from position 521 to 553. Substitution of a highly conserved proline with threonine causes exfoliative cutaneous lupus erythematosus in dogs [22]. Targeted mutagenesis of the four underlined motifs disrupted SDCBP binding in mouse macrophages [18]. A targeted mouse mutant, Unc93b1PKP/PKP, in which the residues corresponding to the human positions 530–532 were replaced by alanines, developed systemic inflammation and autoimmunity [18].
Based on the recent insights about UNC93B1 function and the phenotypes in UNC93B1 mutant mice and dogs, we hypothesized that genetic variants affecting the C-terminal tail of UNC93B1 might also be responsible for SLE or related autoimmune disease in human patients. We therefore investigated the sequence of the last exon of the UNC93B1 gene in patients of the Swiss SLE Cohort Study.
2. Materials and Methods
2.1. Patient Selection and DNA Extraction
This study included 536 patients of the Swiss SLE Cohort Study (SSCS). Of the patients, 457 (85%) were female and 414 (78%) of European descent. In 497 (93%) of patients, SLE was diagnosed after the age of 18 years. Details about this cohort have been reported previously [27,28,29]. Genomic DNA was isolated from ETDA blood samples with the Maxwell RSC Whole Blood Kit using a Maxwell RSC instrument (Promega, Dübendorf, Switzerland).
2.2. UNC93B1 Targeted Sanger Sequencing
A 1000 bp PCR amplicon was amplified with primers AAGGGACAGTGCTGGATGTG (Primer F) and CAGGGCATCCGTGCATCC (Primer R). This amplicon contained 198 bp of the last intron of UNC93B1, 312 bp protein-coding region of the last exon and 490 bp of the 3′-UTR. The protein-coding part corresponded to codons 495 to 597 of the open reading frame. PCRs were performed in 10 μL total volume containing 10 ng template DNA, 5 pmol of each primer, 5 μL AmpliTaqGold360Mastermix, and 1 μL of GC enhancer (Thermo Fisher Scientific, Waltham, MA, USA). A touchdown PCR was performed with an initial denaturation for 10 min at 95 °C, followed by 5 cycles of 30 s denaturation at 95 °C, 30 s annealing at 65 °C with a decrease of 1 °C at each cycle, and 60 s polymerization at 72 °C. Subsequently, 30 cycles of 30 s denaturation at 95 °C, 30 s annealing at 60 °C and 60 s polymerization at 72 °C followed. At the end, a final extension step of 7 min at 72 °C was performed. After treatment with shrimp alkaline phosphatase and exonuclease I, PCR amplicons were sequenced with the forward primer F and reverse primer R1 (AGCTGTGGGGATCTGGAGC) on an ABI 3730 DNA Analyzer (Thermo Fisher Scientific). Sequencher 5.1 software was used to analyze the Sanger sequences (GeneCodes, Ann Arbor, MI, USA).
2.3. Whole Genome Sequencing
An Illumina TruSeq PCR-free DNA library with ~400 bp insert size of patient no. 8 was prepared. We collected 373 million 2 × 150 bp paired-end reads or 33x coverage on a NovaSeq 6000 instrument. Variant calling was performed using a community-developed pipeline from bcbio nextgen v1.1.6a0 (https://github.com/bcbio/bcbio-nextgen, accessed 9 August 2021). In short, the reads were mapped to the human reference genome assembly GRCh38 using BWA-MEM v. 0.7.17 [30]. Variant calling was conducted using GATK HaplotypeCaller v. 3.8 [31] and FreeBayes v. 1.1.0.46 (https://github.com/freebayes/freebayes, accessed 9 August 2021) [32] and the union of quality-filtered calls from both tools was used for downstream analysis. The variants were annotated using Ensembl Variant Effect Predictor (VEP) v. 98.3 [33] using in-house scripts. The short-read alignments of UNC39B1 were visually inspected for structural variants using the integrative genomics viewer (IGV) [34].
2.4. Gene Analysis
Numbering within the human UNC93B1 gene corresponds to the NCBI RefSeq accessions NM_030930.4 (mRNA) and NP_112192.2 (protein).
3. Results
We successfully amplified and sequenced the last exon of the UNC93B1 gene in all 536 investigated patients and identified six coding variants with respect to the reference sequence (Table 1, Table S1).
Table 1.
Details of the six detected UNC93B1 variants.
| dbSNP | HGVS-c | HGVS-p | Alternative Allele Count (Frequency) | gnomAD Allele Frequency |
|---|---|---|---|---|
| rs7149 | c.1557C>G | p.Arg519= | 253 (23.6%) | 16.0% |
| rs576491436 | c.1629G>A | p.Glu543= | 1 (0.1%) | 5 × 10−4 |
| rs1308430306 | c.1651G>A | p.Asp551Asn | 1 (0.1%) | 8 × 10−6 |
| n.a. | c.1724_1725delinsAG | p.Pro575Gln | 8 (0.7%) | n.a. |
| rs2375182 | c.1768G>T | p.Gly590Trp | 4 (0.4%) | 0.4% |
| rs964738111 | c.1777G>A | p.Gly593Arg | 1 (0.1%) | 2 × 10−4 |
Five variants did not affect the known SDCBP binding motif spanning amino acids 521 to 553 and were not investigated further. Only the p.Asp551Asn variant was located in the SDCBP binding domain of UNC93B1. In our cohort, one patient carried one copy of the mutant Asn-allele, which was also present once in the current gnomAD dataset of 128,930 alleles.
This female patient developed the first SLE manifestations including polyarthritis, acute cutaneous lupus, antinuclear, and anti-dsDNA antibodies at 16 years of age. In the following years, she developed alopecia, thrombocytopenia, and in particular, recurrent, difficult to treat acute and chronic skin lupus lesions. Additionally, at 30 years of age, she developed neuropsychiatric manifestations and chronic diffuse pain requiring intensive and chronic treatment complicated by drug allergies and recurrent viral infections. The patient was refractory to multiple SLE-targeted treatments. She died at the age of 44 years for reasons seemingly unrelated to SLE. We performed a whole-genome sequencing experiment on this heterozygous patient and evaluated the entire UNC93B1 gene for the presence of potential additional loss-of-function variants. However, we did not detect anything unusual in the other parts of the gene and concluded that the patient had a fully functional second copy of the UNC93B1 gene.
4. Discussion
In this study, we tested the hypothesis that genetic variants affecting the SDCBP binding domain of UNC93B1 might cause SLE. This hypothesis was developed from the observation that mouse and dog mutants with such variants exhibit severe autoimmune phenotypes [18,22]. The autoimmune phenotype in ECLE affected dogs with UNC93B1 variants shows autosomal recessive inheritance [22]. Extrapolating from dogs, a hypothetical human patient with UNC93B1-related SLE would, therefore, be required to carry either bi-allelic UNC93B1 variants abrogating SDCBP binding or a combination of one mutant allele encoding a UNC93B1 protein with impaired SDCBP binding together with a complete loss-of-function allele on the second chromosome. As we did not find such a genotype in our cohort of 536 analyzed SLE patients, we conclude that variants affecting the SDCBP binding domain of UNC93B1 do not represent a common genetic risk factor for SLE. The majority of the patients in the SSCS cohort are of European descent (78%). Hypothetical UNC93B1 risk alleles might be more common in other populations.
Different from the current knowledge in dogs, the mode of inheritance in Unc93b1 mutant mice is semi-dominant. Homozygous mutant Unc93b1PKP/PKP mice have a very severe autoimmune phenotype and die early as a consequence of their systemic autoimmune disease. Heterozygous Unc93b1WT/PKP mice exhibit only mild signs of autoimmune disease and enhanced TLR7 signaling [18]. Therefore, we cannot exclude the possibility that the heterozygous 551Asn allele observed in one of our patients contributed to her autoimmune disease, probably in combination with other risk factors.
It has to be cautioned that we focused our analysis on the published SDCBP binding domain of UNC93B1 (amino acids 521-553). To the best of our knowledge, the exact three-dimensional structure of the UNC93B1-SDCBP complex is currently unknown. There might be additional important amino acids in the C-terminal tail or other cytosolic parts of UNC93B1 that are also required for SDCBP binding. The gnomAD database and our study provide evidence that several very rare UNC93B1 alleles with amino acid substitutions in the C-terminal tail exist in the human population. Variants affecting other components of this regulatory pathway, for example, the enzymes mediating phosphorylation or ubiquitination of UNC93B1, might also contribute to autoimmune diseases.
Our results do not exclude the possibility that variants affecting the SDCBP binding domain of UNC93B1 might cause or contribute to other autoimmune phenotypes than SLE in human patients. The investigated patients mostly had adult-onset SLE [29]. Unc93b1PKP/PKP mice develop signs of systemic inflammation and antinuclear antibodies very early in life [18]. Homozygous UNC93B1 mutant dogs with ECLE develop a cutaneous form of lupus as juveniles at a few months of age. In most affected dogs, this progresses to a systemic autoimmune disease with frequent involvement of the joints. However, the formation of antinuclear antibodies, a hallmark of SLE, is normally not seen in dogs with ECLE. These phenotypic differences between UNC93B1 mutant mice and dogs impede an accurate prediction of the resulting phenotype in human patients with homologous UNC93B1 variants. We therefore think that further studies investigating the presence of genetic variants affecting the C-terminal tail of UNC93B1 in patient cohorts with other autoimmune phenotypes, including familial cases of extremely rare autoimmune disorders, are warranted.
Acknowledgments
The authors are grateful to all patients who participated in this research. We thank the Next Generation Sequencing Platform of the University of Bern for performing the high-throughput sequencing experiments, and the Interfaculty Bioinformatics Unit of the University of Bern for providing high performance computing infrastructure.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12081268/s1, Table S1: UNC93B1 genotyping results of 536 patients from the SSCS cohort.
Author Contributions
Conceptualization, T.L.; investigation, S.K.; resources, C.R., C.C., M.T., U.H.-D., and J.v.K.; data curation, I.K.; writing—original draft, S.K., C.R., and T.L.; writing—review and editing, S.K., C.R., I.K., C.C., M.T., U.H.-D., J.v.K., and T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Swiss National Science Foundation, grant number 310030_200354.
Institutional Review Board Statement
The Swiss SLE Cohort Study (SSCS) was approved by the SwissEthics review board (PB_2017-01434).
Informed Consent Statement
All patients gave written informed consent according to the guidelines of the Declaration of Helsinki for conducting the research here described.
Data Availability Statement
The data presented in this study is contained within the article and supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rahman A., Isenberg D.A. Systemic lupus erythematosus. N. Engl. J. Med. 2008;358:929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 2.Domeier P.P., Schell S.L., Rahman Z.S. Spontaneous germinal centers and autoimmunity. Autoimmunity. 2017;50:4–18. doi: 10.1080/08916934.2017.1280671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitacre C.C. Sex differences in autoimmune disease. Nat. Immunol. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 4.Goulielmos G.N., Zervou M.I., Vazgiourakis V.M., Ghodke-Puranik Y., Garyfallos A., Niewold T.B. The genetics and molecular pathogenesis of systemic lupus erythematosus (SLE) in populations of different ancestry. Gene. 2018;668:59–72. doi: 10.1016/j.gene.2018.05.041. [DOI] [PubMed] [Google Scholar]
- 5.Fike A.J., Elcheva I., Rahman Z.S.M. The Post-GWAS Era: How to Validate the Contribution of Gene Variants in Lupus. Curr. Rheumatol. Rep. 2019;21:3. doi: 10.1007/s11926-019-0801-5. [DOI] [PubMed] [Google Scholar]
- 6.Deane J.A., Pisitkun P., Barrett R.S., Feigenbaum L., Town T., Ward J.M., Flavell R.A., Bolland S. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27:801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisitkun P., Deane J.A., Difilippantonio M.J., Tarasenko T., Satterthwaite A.B., Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 8.Subramanian S., Tus K., Li Q.Z., Wang A., Tian X.H., Zhou J., Liang C., Bartov G., McDaniel L.D., Zhou X.J., et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc. Natl. Acad. Sci. USA. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Souyris M., Cenac C., Azar P., Daviaud D., Canivet A., Grunenwald S., Pienkowski C., Chaumeil J., Mejía J.E., Guéry J.C. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 2018;3:eaap8855. doi: 10.1126/sciimmunol.aap8855. [DOI] [PubMed] [Google Scholar]
- 10.Shen N., Fu Q., Deng Y., Qian X., Zhao J., Kaufman K.M., Wu Y.L., Yu C.Y., Tang Y., Chen J.Y., et al. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA. 2010;107:15838–15843. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raafat I.I., El Guindy N., Shahin R.M.H., Samy L.A., El Refai R.M. Toll-like receptor 7 gene single nucleotide polymorphisms and the risk for systemic lupus erythematosus: A case-control study. Z. Rheumatol. 2018;77:416–420. doi: 10.1007/s00393-017-0283-7. [DOI] [PubMed] [Google Scholar]
- 12.Yasutomo K., Horiuchi T., Kagami S., Tsukamoto H., Hashimura C., Urushihara M., Kuroda Y. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat. Genet. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 13.Rice G., Newman W.G., Dean J., Patrick T., Parmar R., Flintoff K., Robins P., Harvey S., Hollis T., O’Hara A., et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y.-M., Brinkmann M.M., Paquet M.-E., Ploegh H.L. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 15.Casrouge A., Zhang S.Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N., et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 16.Tabeta K., Hoebe K., Janssen E.M., Du X., Georgel P., Crozat K., Mudd S., Mann N., Sovath S., Goode J., et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat. Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 17.Ishida H., Asami J., Zhang Z., Nishizawa T., Shigematsu H., Ohto U., Shimizu T. Cryo-EM structures of Toll-like receptors in complex with UNC93B1. Nat. Struct. Mol. Biol. 2021;28:173–180. doi: 10.1038/s41594-020-00542-w. [DOI] [PubMed] [Google Scholar]
- 18.Majer O., Liu B., Kreuk L.S.M., Krogan N., Barton G.M. UNC93B1 recruits syntenin-1 to dampen TLR7 signaling and prevent autoimmunity. Nature. 2019;575:366–370. doi: 10.1038/s41586-019-1612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitoh S., Miyake K. Regulatory molecules required for nucleotide-sensing Toll-like receptors. Immunol. Rev. 2009;227:32–43. doi: 10.1111/j.1600-065X.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 20.Majer O., Liu B., Woo B.J., Kreuk L.S.M., Van Dis E., Barton G.M. Release from UNC93B1 reinforces the compartmentalized activation of select TLRs. Nature. 2019;575:371–374. doi: 10.1038/s41586-019-1611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukui R., Saitoh S., Kanno A., Onji M., Shibata T., Ito A., Onji M., Matsumoto M., Akira S., Yoshida N., et al. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity. 2011;35:69–81. doi: 10.1016/j.immuni.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Leeb T., Leuthard F., Jagannathan V., Kiener S., Letko A., Roosje P., Welle M.M., Gailbreath K.L., Cannon A., Linek M., et al. A missense variant affecting the C-terminal tail of UNC93B1 in dogs with exfoliative cutaneous lupus erythematosus (ECLE) Genes. 2020;11:159. doi: 10.3390/genes11020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivry T., Linder K.E., Banovic F. Cutaneous lupus erythematosus in dogs: A comprehensive review. BMC Vet. Res. 2018;14:132. doi: 10.1186/s12917-018-1446-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vroom M.W., Theaker A.J., Rest J.R., White S.D. Case report: Lupoid dermatosis in 5 German short-hair pointer. Vet. Dermatol. 1995;6:93–98. doi: 10.1111/j.1365-3164.1995.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 25.Bryden S.L., White S.D., Dunston S.M., Burrows A.K., Olivry T. Clinical, histopathological and immunological characteristics of exfoliative cutaneous lupus erythematosus in 25 German short-haired pointers. Vet. Dermatol. 2005;16:239–252. doi: 10.1111/j.1365-3164.2005.00468.x. [DOI] [PubMed] [Google Scholar]
- 26.Mauldin E.A., Morris D.O., Brown D.C., Casal M.L. Exfoliative cutaneous lupus erythematosus in German shorthaired pointer dogs: Disease development, progression and evaluation of three immunomodulatory drugs (ciclosporin, hydroxychloroquine, and adalimumab) in a controlled environment. Vet. Dermatol. 2010;21:373–382. doi: 10.1111/j.1365-3164.2010.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chizzolini C., Cohen C.D., Eisenberger U., Hauser T., Hunziker T., Leimgruber A., Pechula M., Ribi C., Stoll T., Trendelenburg M., et al. Towards the Swiss systemic lupus erythematosus cohort study (SSCS) Rev. Med. Suisse. 2009;5:808–811. [PubMed] [Google Scholar]
- 28.Ribi C., Trendelenburg M., Gayet-Ageron A., Cohen C.D., Dayer E., Eisenberger U., Hauser T., Hunziker T., Leimgruber A., Lindner G., et al. The Swiss Systemic lupus erythematosus Cohort Study (SSCS)-I-Cross-sectional analysis of clinical characteristics and treatments across different medical disciplines in Switzerland. Swiss Med. Wkly. 2014;144:w13990. doi: 10.4414/smw.2014.13990. [DOI] [PubMed] [Google Scholar]
- 29.Meier A.L., Bodmer N.S., Wirth C., Bachmann L.M., Ribi C., Pröbstel A.K., Waeber D., Jelcic I., Steiner U.C., Swiss SLE Cohort Study (SSCS) Neuro-psychiatric manifestations in patients with systemic lupus erythematosus: A systematic review and results from the Swiss lupus cohort study. Lupus. 2021;21:9612033211025636. doi: 10.1177/09612033211025636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 20131303.3997v2 [Google Scholar]
- 31.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 20121207.3907 [Google Scholar]
- 33.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study is contained within the article and supplementary material.