Abstract
Metallothioneins are small, highly conserved, cysteine-rich proteins that bind a variety of metal ions. They are found in virtually all eukaryotic organisms and are regulated primarily at the transcriptional level. In humans, the predominant metallothionein gene is hMTIIA, which accounts for 50% of all metallothioneins expressed in cultured human cells. The hMTIIA promoter is quite complex. In addition to cis-acting DNA sequences that serve as binding sites for trans-acting factors such as Sp1, AP1, AP2, AP4, and the glucocorticoid receptor, the hMTIIA promoter contains eight consensus metal response element sequences. We report here the cloning of a novel zinc finger protein with a molecular mass of 120 kDa (PZ120) that interacts specifically with the hMTIIA transcription initiation site. The PZ120 protein is ubiquitously expressed in most tissues and possesses a conserved poxvirus and zinc finger (POZ) motif previously found in several zinc finger transcription factors. Intriguingly, we found that a region of PZ120 outside of the zinc finger domain can bind specifically to the hMTIIA DNA. Using transient-transfection analysis, we found that PZ120 repressed transcription of the hMTIIA promoter. These results suggest that the hMTIIA gene is regulated by an additional negative regulator that has not been previously described.
Metallothioneins (MTs) were initially discovered by biochemists searching for tissue constituents responsible for the natural accumulation of cadmium. They have been reported to occur throughout the animal kingdom, as well as in plants, eukaryotic microorganisms, and prokaryotes (reviewed in references 25 and 35). In humans, MTs have been isolated from the liver (57), cultured cells (41, 59), and the brain (growth inhibitory factor) (75). They are present in four distinguishable forms known as hMTI, hMTII, hMTIII, and hMTIV (reviewed in references 2, 25, 37, and 55).
Different tissues and cell types synthesize different MTs in various levels. However, the hMTIIA gene is responsible for the majority of MTs expressed in most tissues in humans (41). This high level of hMTIIA basal constitutive expression coupled with the gene’s remarkable inducibility makes hMTIIA easily assayable and, therefore, provides an ideal model for studying transcriptional regulation in eukaryotic cells. Although there are reports that hMTIIA may be regulated by gene amplification (47), DNA methylation (33), or posttranscriptional events (60), there is no question that the principal mechanism of regulation lies at the level of transcriptional initiation (44).
The interest in transcriptional regulation of the hMTIIA gene centers on three key issues. First, shortly after the cloning of the hMTIIA gene, there was an explosive increase of interest in the identification of the cis-acting DNA sequences in the hMTIIA promoter that are responsible for basal and induced transcription. Second, many laboratories have intensely pursued the identification of trans-acting proteins that interact with these cis-acting DNA sequences. Finally, there is a strong desire to understand the mechanisms by which these trans-acting proteins activate hMTIIA expression.
The cis-acting DNA sequences that permit expression of the hMTIIA gene have been elucidated in different studies (38, 39). In general, in these studies, a region that corresponds to the 5′ flanking sequence of the hMTIIA gene is linked to a reporter gene and introduced into cultured cells by transfections followed by assays for reporter gene expression. Promoters with 5′ and 3′ deletions, as well as internal deletions, and linker-scanner mutations were similarly tested for their ability to transcribe a reporter gene. By this approach, it was discovered that the hMTIIA gene provides an excellent model for unraveling the complexity of protein-DNA interactions that can occur at a single promoter and that influence the transcription of a gene. Extensive analysis of the 5′ flanking regions of this promoter revealed a variety of trans-acting factors that bind to different upstream cis-acting sites (3, 31, 32, 48, 51, 53, 65). In addition to the several metal response elements, a GC box which is recognized by transcription factor Sp1 is located between nucleotides −57 and −68 relative to the start of transcription. An AP1 binding site is present at nucleotides −96 to −105 (48), and overlapping this site are sequences that can be recognized by AP4. Three binding sites for the transcription factor AP2 are present between −103 and −227 (31). An element that confers glucocorticoid and progesterone responsiveness, a glucocorticoid receptor element, is located between −240 and −270 (39, 40, 66). Further upstream is an interferon response element that may be involved in alpha interferon-induced transcription of the hMTIIA gene (20).
Interestingly, in addition to the many factors that bind the hMTIIA upstream sequence, early studies using the DNase I footprinting procedure detected proteins that cover the transcription initiation site (3, 31), suggesting that a distinct class of transcription factors may play an important role in the regulation of hMTIIA basal or induced transcription. It has been suggested that in Rat 2 fibroblasts, the cadmium- and dexamethasone-responsive elements in the hMTIIA promoter are present in the upstream region and do not require its initiation site (38). However, the role of the initiation region alone in response to other heavy metals, in different cells, or in basal transcriptional regulation is not known. One early study, which pointed to the importance of the hMTIIA initiation region in basal transcription, showed that deletion of this region, together with the TATA box, renders the promoter less active than the wild-type promoter in vitro (48). However, a detailed study of the importance of the hMTIIA initiation site alone and identification of possible factors that may interact with this region were not done.
Recently, using a cell-free in vitro transcription system and in vitro mutagenesis, we have found evidence that the hMTIIA transcriptional initiation site binds both transcriptional activators and transcriptional repressors. Using electrophoretic mobility shift assays (EMSA) and the hMTIIA initiation sequence as a probe, we have identified five sequence-specific DNA-protein complexes from HeLa cells (70). Subsequently, one of the DNA-binding activities was purified, subjected to microsequencing analysis, and determined to be the large component (70 kDa subunit) of replication protein A. This finding is consistent with the findings of previous studies where replication protein A (RPA) was found to interact with different transcriptional activators; interestingly, the 70-kDa subunit of RPA is part of the human RNA polymerase II complex (49). Intriguingly though, our RPA preparation recognized double-stranded hMTIIA initiation sequences at a specificity higher than that at which it recognized single-stranded DNA. Because purified recombinant RPA binds the single-stranded hMTIIA initiation site effectively, a modified form of RPA may bind the double-stranded hMTIIA initiation site. Alternatively, other hMTIIA initiation sequence-binding proteins may influence RPA’s unique DNA-binding property at the hMTIIA promoter. Identification and cloning of these additional factors, therefore, would be a major prerequisite for making definitive progress in the understanding of hMTIIA regulation.
To further examine cellular DNA-binding proteins that interact with the hMTIIA initiation sequence, we have screened an expression library with multimerized hMTIIA initiation sequences as a probe. We report here the isolation of a novel zinc finger protein, PZ120, that binds specifically to the hMTIIA transcription initiation site and represses hMTIIA gene transcription. The PZ120 protein belongs to a family of zinc finger transcription factors that contain a conserved poxvirus and zinc finger (POZ) motif. Surprisingly, we found that PZ120 binds specifically to the hMTIIA initiation sequence in the absence of its zinc fingers. Taken together, our results suggest the existence of an additional, yet novel, mechanism of regulation of the hMTIIA gene.
MATERIALS AND METHODS
Plasmids.
Plasmid p4′5CAT contains the hMTIIA promoter sequence from −286 to +73 linked to the chloramphenicol acetyltransferase (CAT) reporter gene (42, 43). Plasmid pRSV-βgal carries the β-galactosidase gene downstream from the long terminal repeat of the Rous sarcoma virus. pGEM-1021 was constructed by digesting λgt11 clone 1021 with EcoRI and then subcloning the 1.5-kb fragment into a pGEM7Z vector (Promega). pH25-2 contains a 4.5-kb insert excised from the λ clone H25-2 and subcloned into a pBluescript vector. pGEM-PZ120, containing the full-length 5-kb PZ120 cDNA, was generated by ligating the 3′ portion of clone H25-2 to the 5′ portion of clone 1021. The insert from pH25-2, digested with NheI and SacI, was subcloned into pGEM-1021, which was also cut with NheI and SacI. pCMV is identical to the eukaryotic expression vector pcDNAIamp (Invitrogen). PZ120 cDNA from pGEM-PZ120 was digested with EcoRI and subcloned in the 5′-to-3′ direction downstream of the cytomegalovirus (CMV) promoter in pCMV to generate pCMV-PZ120. pCMV-PZ120(−) was produced by subcloning PZ120 cDNA in the reverse orientation into the pCMV vector. To generate the POZ domain deletion expression construct pCMV-PZ120 (Δ3–344), pCMV-PZ120 was digested with SacII and NheI and an 8.9-kb DNA fragment was recovered and then religated in the presence of an adapter (5′-GGATGTCAG-3′ or 5′-CTAGCTGACATCCGC-3′). pCMV-PZ120 (1–344) was produced by digesting pCMV-PZ120 with SacI and NheI, recovering a 6.8-kb DNA fragment, blunting the fragment at both ends with T4 DNA polymerase, and religating with T4 DNA ligase. pGST-PZ120 was constructed by taking the 5-kb EcoRI fragment from pGEM-PZ120 and then ligating this fragment to the vector pGEX4T-3 (Pharmacia) digested with EcoRI. pHis-PZ120 (1–482) was generated by subcloning the 1.5-kb insert of clone 1021 into the pQE9 expression vector (Qiagen). Clone 1021 was first digested with EcoRI, and then the 1.5-kb fragment was ligated to the pQE9 vector with the 6-histidine tag at the N-terminal region. pCEP4F has previously been described (79). pCEP4F-PZ120, pCEP4F-PZ120 (Δ3–344), and pCEP4F-PZ120 (1–344), were constructed by taking inserts respectively from pCMV-PZ120, pCMV-PZ120 (Δ3–344), and pCMV-PZ120 (1–344) and ligating them in frame to the FLAG epitope in pCEP4F. All constructs were confirmed by dideoxy sequencing (62).
Southwestern blot analysis.
Purified proteins or a nuclear extract from HeLa cells was separated on a sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred onto a nitrocellulose membrane. Proteins on the membrane were denatured in a binding buffer (250 mM HEPES [pH 7.9], 30 mM MgCl2, 400 mM KCl) containing 6 M guanidine hydrochloride and gradually renatured in the same buffer with 3, 1.5, 0.75, 0.375, and 0.1875 M guanidine hydrochloride. Subsequently, the membrane was rinsed with the binding buffer and probed with a 32P-labeled concatemerized double-stranded oligodeoxynucleotide containing the hMTIIA initiation site sequence (−7 to +11 relative to the start of transcription [42]), 5′-GCACTCCACCACGCCTCCT-3′, and its complementary strand. Hybridization was done at 4°C for 12 to 16 h, and the membrane was washed three times at 4°C with the binding buffer before exposure in a PhosphorImage screen.
Isolation of PZ120 cDNA.
A HeLa cell λgt11 cDNA library (64) was screened by a standard protocol (61) with 32P-labeled concatemerized double-stranded synthetic oligodeoxynucleotides that contain the hMTIIA initiation sequence, 5′-GCACTCCACCACGCCTCCT-3′, and its complement. Positive plaques on duplicated membranes were amplified and rescreened. Probes used for secondary and tertiary screens contained hMTIIA sequences identical to those used in the primary screen but differed from the primary probe in flanking sequences. A 1.5-kb clone, clone 1021, was isolated, and its cDNA insert was used to rescreen a λgt11 HeLa cDNA library (Clontech) and a λZAPII H1262 fibroblast cDNA library (56). Hybridizations were conducted at 60°C for 16 to 24 h with a solution containing 6× SSC, (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt’s solution, 1% SDS, and 100 μg of denatured salmon sperm DNA per ml. Stringent washes were done in 0.1× SSC–0.1% SDS at 60°C for 1 h. Phage DNA prepared from positive plaques of the HeLa cDNA library were digested with EcoRI and subcloned into pGEM3Z vectors (Promega) for dideoxy sequencing (62). Positive clones from the λZAPII fibroblast library were plaque purified, excised, ligated into pBluescript vectors (Stratagene), and then characterized by dideoxy sequencing. The complete final sequence of the PZ120 cDNA was determined from both DNA strands.
Preparation of lysogenic phage extract.
Host Escherichia coli Y1089 was infected with bacteriophage clone 1021 and plated onto Luria broth (LB) plates containing ampicillin. Lysogenic recombinant bacteriophage λgt11 colonies were picked by selecting those that grew at 30°C but not at 42°C. Individual lysogens were shifted from 30 to 42°C, induced by isopropyl-1-thio-β-d-galactopyranoside (IPTG), and cultured at 37°C. One induced lysogenic culture was lysed by repeated freeze-thaw cycles, and lysates were used in EMSA.
EMSA.
Single-stranded oligodeoxynucleotides corresponding to the transcription initiation sequence of the hMTIIA gene, 5′-GCACTCCACCACGCCTCCT-3′, were labeled individually with [γ32P]ATP and T4 polynucleotide kinase, heated together at 65°C, and allowed to anneal by slow cooling to room temperature. Each 12-μl reaction mixture contained 12 mM HEPES (pH 7.9), 10% glycerol, 5 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol, 0.5 mM EDTA, 50 μg of bovine serum albumin per ml, 0.05% Nonidet P-40, 1 μg of poly(dI-dC), approximately 7 μg of protein extract or 10 ng of purified protein, and 13 ng of radiolabeled DNA. Reaction mixtures were incubated for 10 min at room temperature, separated on 4% nondenaturing acrylamide gels (0.0225 M Tris-borate, 0.0005 M EDTA), dried, and subjected to autoradiography. For competition experiments, excess unlabeled DNAs (competitors) were included in the reaction mixture.
Northern blot analysis.
A human multiple tissue Northern blot was purchased from Clontech Laboratories. The blot contains poly(A)+ RNA that was purified by several passages through oligo(dT) cellulose columns, separated on denatured 1.2% formaldehyde agarose gel, and blotted onto nylon membranes. Each lane of the blot contains approximately 2 μg of a pure poly(A)+ RNA from a specific tissue. A 1.5-kb PZ120 cDNA derived from clone 1021 was labeled with [α-32P]dCTP by random priming. Prehybridization and hybridization were carried out in a solution containing 5× SSPE (0.75 M NaCl, 0.05 M NaH2PO4 · H2O, 0.005 M Na2EDTA [pH 7.4]), 10× Denhardt’s solution, 100 μg of denatured salmon sperm DNA per ml, 50% formamide, and 2% SDS at 42°C. Blots were washed at high stringency before exposure to X-ray film. Hybridization was carried out simultaneously with a human β-actin cDNA to control for differences in loading.
In vitro transcription and translation.
Using a TNT kit (Promega), pGEM-PZ120 was transcribed with SP6 or T7 RNA polymerase and translated with reticulocyte lysates in the presence of [35S]methionine. The product was analyzed on an SDS–10% polyacrylamide gel and then autoradiographed.
Expression and purification of histidine-tagged PZ120 (residues 1 to 482) protein.
pHis-PZ120 (1–482) was transformed into DH5α cells and grown in LB with 100 μg of ampicillin per ml at 37°C until the A600 reached 0.6. The cells were then induced with 0.1 mM IPTG for 5 h and harvested. Cell pellets were resuspended in buffer A (6 M guanidine hydrochloride, 0.1 M sodium phosphate, 0.01 M Tris-HCl [pH 8.0]). Cell debris was removed, and the lysates were passaged through 1 ml of nickel-nitrilotriacetic acid agarose (Qiagen) columns preequilibrated with buffer A. The columns were then washed with buffer A, buffer B (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris-HCl [pH 8.0]), and buffer C (same as buffer B but with pH 6.3), and finally eluted with buffer D (same as buffer B but with pH 5.9) followed by buffer E (same as buffer B but with pH 4.5). Fractions of flowthrough, wash, and elution were collected and assessed by boiling in SDS sample loading buffer, followed by separation through an SDS–10% polyacrylamide gel, and staining with Coomassie blue. Eluted fractions containing the histidine-tagged proteins were subjected to renaturation procedures. Fractions were pooled and sequentially dialyzed in renaturation buffers (0.1 M NaCl, 0.01 M Tris-HCl [pH 8.0]) containing 4, 2, 1, and 0.1 M urea in that order. A final dialysis was done with phosphate-buffered saline (PBS).
Preparation of extracts containing GST or GST-PZ120 fusion proteins.
pGST-PZ120 or the glutathione S-transferase (GST) vector (pGEX4T-3) were transformed into DH5α cells grown in LB with 100 μg of ampicillin per ml at 37°C for 2 h and induced with IPTG. Cells were collected 4 h after induction, resuspended in PBS, and lysed by sonication. After centrifugation, the supernatants were saved as soluble cell extract for EMSA.
Cell culture and transfections.
CV1 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 IU of penicillin per ml, and 100 μg of streptomycin (Life Technologies, Inc.) per ml. Each transfection contained different amounts of an effector plasmid, 5 μg of a CAT reporter plasmid, and 5 μg of a plasmid expressing β-galactosidase by calcium phosphate precipitation (23). Cells were harvested 48 h after transfection, and CAT activity was assayed as described previously (22). β-Galactosidase activity was measured with a Galacto light kit (Tropix) to normalize for transfection efficiency.
Immunofluorescence.
CV1 cells were grown on charged slides inside 100-mm-diameter tissue culture plates for about 24 h and transfected with 20 μg of either pCEP4F, pCEP4F-PZ120, pCEP4F-PZ120 (Δ3–344), or pCEP4F-PZ120 (1–344). Two days later, cells were washed with ice-cold PBS, fixed with 4% paraformaldehyde for 15 min, rinsed again with PBS, permeabilized with 1% glycine–0.5% Triton X-100 in PBS overnight, and then treated with anti-FLAG antibody (Sigma) in 200 μl of PBS containing 1% bovine serum albumin and 0.1% Nonidet P-40. Cells were then incubated for 1 h at room temperature, followed by washing with PBS, and further incubated for 30 min with a 1:200 dilution of sheep anti-mouse immunoglobulin G coupled with fluorescein isothiocyanate (Sigma). Subsequently, cells were subjected to extensive washings with PBS and one drop of DAPI (4′, 6-diamidino-2-phenylindole) anti-fade dye (Vector) was applied to each coverslip before the cells were analyzed under a Leitz Orthoplan microscope equipped with a charge-coupled device camera.
Nucleotide sequence accession number.
The nucleotide sequence of the PZ120 gene appears in the EMBL, GenBank, and DDBJ databases with accession no. U03378.
RESULTS
Identification of a protein that binds the hMTIIA initiation sequence.
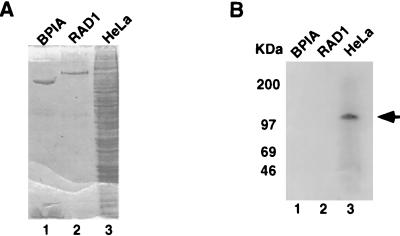
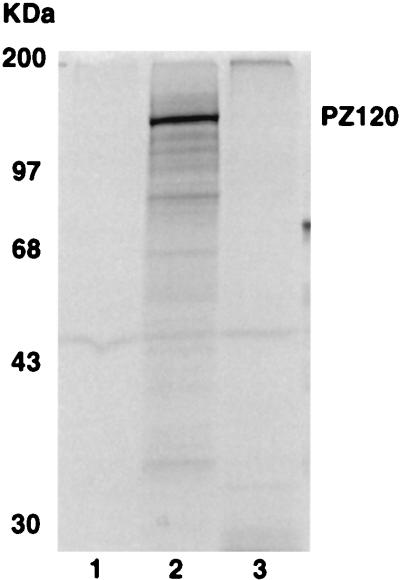
Previously, we and others have identified several specific DNA-binding activities associated with the transcription initiation region of the hMTIIA gene (3, 31, 70). Using standard protein purification procedures, we found that RPA is one component of a complex that binds the hMTIIA transcription initiation sequence (70). To further examine cellular DNA-binding proteins that interact with the hMTIIA initiation sequence, and to study other proteins that bind the hMTIIA transcription start site, we performed a Southwestern blot analysis using the hMTIIA transcription initiation sequence as a probe. As shown in Fig. 1B, a protein of approximately 120 kDa from HeLa cells specifically interacted with the hMTIIA DNA (lane 3). This interaction is highly specific because two purified proteins (lanes 1 and 2), BPIA and RAD1, as well as other proteins present in HeLa cell preparation did not react with the hMTIIA initiation sequence. This result confirms that in addition to RPA, there are other proteins that bind specifically to the hMTIIA initiation sequence. It also points to the feasibility of the identification of additional hMTIIA sequence-binding proteins by direct expression cloning.
FIG. 1.
Southwestern blot analysis of hMTIIA transcription initiation sequence-binding protein. Purified proteins (lanes 1 and 2) or HeLa cell nuclear extract (lane 3) was separated on an SDS-polyacrylamide gel and visualized by Coomassie blue staining (A) or transferred onto a nitrocellulose membrane and probed with the hMTIIA initiation site sequence (B). The arrow indicates a protein that binds specifically to the 32P-labeled hMTIIA transcription initiation DNA sequence. The sizes of molecular mass markers are indicated on the left of the blot.
Cloning of a gene encoding an hMTIIA initiation sequence-binding protein.
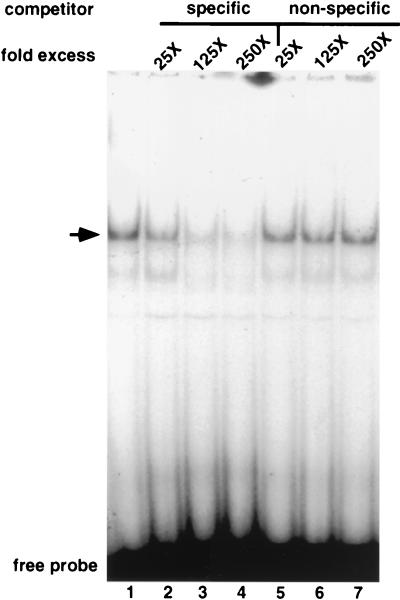
To clone the 120-kDa protein from HeLa cells that binds the hMTIIA sequence, we screened a HeLa λgt11 cDNA expression library with a DNA fragment containing multiple copies of the hMTIIA initiation sequence. As a result, we have identified one phage clone that encodes proteins that associate with the hMTIIA DNA. The binding activity of this clone, referred to as clone 1021, was analyzed by EMSA with lysate prepared from the lysogenic form of the phage. Figure 2 shows that a distinct mobility complex is formed with the hMTIIA transcription initiation sequence (lane 1). This complex is not seen in equivalent lysates from the uninfected E. coli host strain (data not shown). The addition of specific (lanes 2 to 4) and nonspecific (lane 5 to 7) competitor DNA shows that complex is the result of sequence-specific protein binding.
FIG. 2.
EMSA of extracts prepared from induced strains lysogenized with phage clone 1021. The arrow indicates a protein-DNA complex specifically inhibited by the addition of excess hMTIIA initiation sequence (specific competitor [5′-GCACTCCACCACGCCTCCT-3′ and its complement]) but not by the addition of an AP1 oligodeoxynucleotide (nonspecific competitor [5′-GGATGTTATAAAGCATGAGTCAGACACCTCTGGCT-3′ and its complement]).
Analysis of hMTIIA initiation sequence-binding protein (PZ120) cDNA.
The entire 1021 clone was sequenced by the dideoxy method (62), and the predicted amino acid sequence was determined by theoretical translation of the phage clone open reading frame. Since a consensus sequence for initiation of translation (46) was not found in the 5′ end of the cDNA and the reading frame remains open at both the 5′ and 3′ ends, this cDNA may represent only a partial coding sequence.
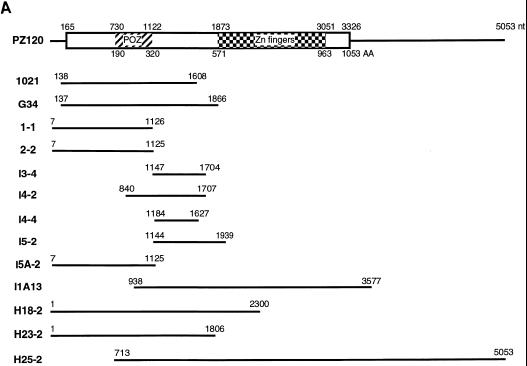
To obtain a full-length cDNA, a different λgt11 cDNA library derived from HeLa cells and a λZAP II cDNA human fibroblast library were screened with a radiolabeled probe corresponding to the 5′ and 3′ ends of clone 1021. Twelve overlapping cDNA clones were isolated from 109 phage plaques (Fig. 3A), and the cDNA sequence and its predicted amino acids are shown in Fig. 3B. Analysis of the amino acid sequence revealed an open reading frame of 1053 amino acids with an in-frame stop codon upstream of the first methionine at nucleotide 100 and in the 3′ end at nucleotide 3323. This indicates that we have obtained a cDNA encoding a full-length hMTIIA transcription initiation sequence-binding protein. Sequence motif searches indicated that this hMTIIA sequence-binding protein contains 12 C2H2 zinc fingers (amino acids 571 to 963 [Fig. 3C]) commonly seen among proteins from the Krüppel family.
FIG. 3.
Nucleotide and protein sequences of PZ120. (A) Schematic representation of the PZ120 cDNAs. The rectangular box denotes the long open reading frame. The locations of the zinc fingers and the POZ domain are indicated by a checkerboard pattern and hatching, respectively. nt, nucleotide; AA, amino acid. (B) The cDNA sequence of PZ120 clones and the predicted protein sequence are shown. Amino acid sequence numbering starts at the putative translation initiation codon. The stop codon is denoted by an asterisk. The POZ domain (amino acids 190 to 320) is underlined by a dotted line, and the zinc finger sequence (amino acids 571 to 963) is underlined by a solid line. An in-frame stop codon upstream of the first methionine is indicated by bold letters, as are the two potential polyadenylation signals at the 3′ untranslated region. (C) Comparison of the different zinc fingers in PZ120.
Sequence comparison of our newly cloned protein in the GenBank and EMBL databases revealed it to be a novel protein with a stretch of amino acids that contains a conserved POZ (also known as Bric à brac-Tramtrack-Broad Complex [BTB] or zinc finger N terminal [ZiN]) motif present in a family of proteins that include the Drosophila transcription factors Tramtrack and Broad Complex, GAGA, ZF5, and ZID, as well as a group of poxvirus proteins. Since the predicted molecular mass of this protein is 120 kDa, we named it PZ120 for POZ-zinc finger protein of 120 kDa.
mRNA and protein analysis of PZ120.
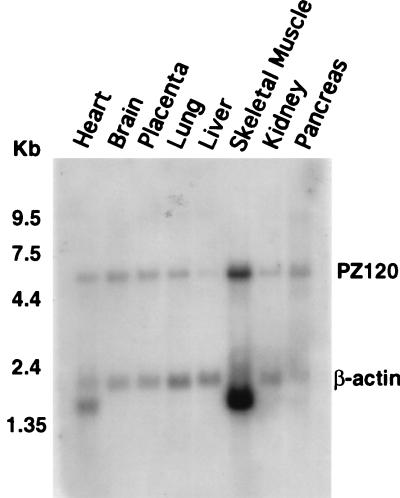
Using Northern blot analysis, we explored the tissue expression pattern of PZ120. As shown in Fig. 4, a message of approximately 6.2 kb was ubiquitously expressed in every tissue (a longer exposure revealed that PZ120 is expressed in the human liver but at a lower level).
FIG. 4.
Northern blot analysis of PZ120 mRNA. The positions of PZ120 and β-actin in the blot are indicated. The sizes of molecular markers are indicated on the left of the blot.
To ensure that the PZ120 cDNA that we have isolated represents a true open reading frame and to determine the actual molecular mass of PZ120, we in vitro transcribed the PZ120 cDNA and translated the resulting cRNA in a rabbit reticulocyte lysate. As shown in Fig. 5, a protein that migrates in SDS-polyacrylamide gel as an approximately 120-kDa polypeptide was produced (lane 2). Thus, the predicted molecular size of PZ120 is consistent with its actual molecular mass. No specific protein was produced by the antisense PZ120 cRNA (lane 3).
FIG. 5.
In vitro transcription and translation of PZ120 cDNA. The vector alone (lane 1) and a construct containing PZ120 full-length cDNA (pGEM-PZ120 [lanes 2 and 3]) were used as templates for coupled in vitro transcription-translation with SP6 (lane 2) or T7 (lane 3) RNA polymerase. The [35S]methionine-labeled protein products were separated by electrophoresis and autoradiographed. The position of PZ120 in the gel in indicated. The sizes of molecular mass markers are indicated on the left of the gel.
Analysis of hMTIIA initiation sequence-binding activity by PZ120.
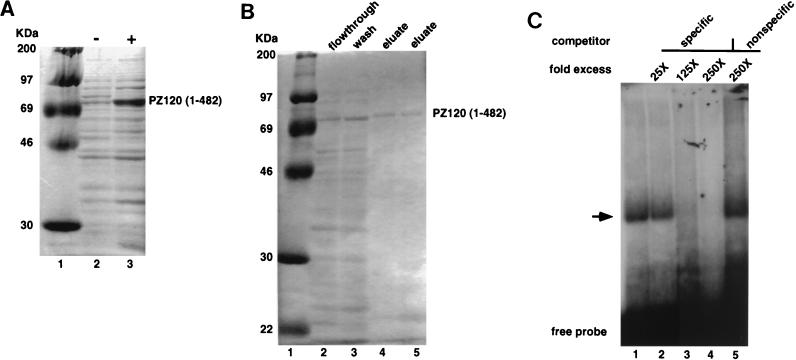
One surprising finding during the cloning of a full-length PZ120 cDNA was the fact that clone 1021, which expresses only the N-terminal portion of PZ120 lacking the zinc finger domain, was sufficient to bind the hMTIIA sequence with high specificity. To confirm this interesting finding, partial PZ120 (amino acids 1 to 482) from clone 1021 was expressed as a histidine fusion protein in E. coli (Fig. 6A), purified by affinity chromatography (Fig. 6B), and tested for its ability to bind to the hMTIIA initiation sequence. Figure 6C displays the results of an EMSA in which a 32P-labeled oligodeoxynucleotide containing the hMTIIA transcription initiation sequence was used as a probe. Currently, we do not know why the predicted 507-amino-acid protein produced by initiation at the first AUG migrates in SDS-polyacrylamide gels with such a high apparent molecular weight. Nevertheless, the fusion protein bound to the labeled probe (lane 1) and the binding could be competed by the addition of excess unlabeled hMTIIA oligodeoxynucleotide (lanes 3 and 4) but not by the addition of an irrelevant AP1 oligodeoxynucleotide (lane 5). Thus, the PZ120 cDNA that we have isolated encodes a protein that binds to the hMTIIA transcription start site and the binding occurs outside of the PZ120 zinc fingers.
FIG. 6.
Expression, purification, and EMSA of histidine-partial PZ120 fusion protein. (A and B) Histidine-PZ120 (amino acids 1 to 482) fusion protein overexpressed in bacteria and analyzed on an SDS-polyacrylamide gel that was culture induced (+) and noninduced (−) with IPTG. The sizes of molecular mass markers are indicated on the left of the gels. (C) Recombinant PZ120 (amino acids 1 to 482) protein binds specifically to its cognate sites in EMSA. An arrow indicates the complex specifically inhibited by the addition of excess oligodeoxynucleotide containing an hMTIIA initiation site (specific competitor) but not by the addition of an AP1 oligodeoxynucleotide (nonspecific competitor). Probes and competitor oligodeoxynucleotide sequences are identical to those used in the experiment shown in Fig. 2.
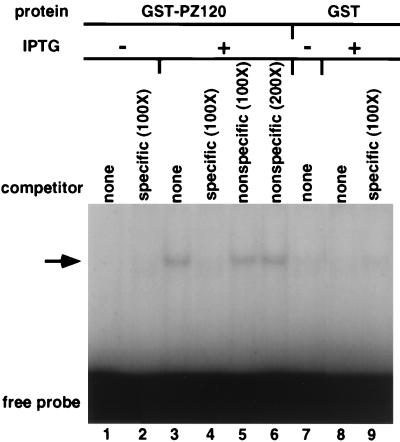
To determine whether a full-length PZ120 can similarly bind the hMTIIA initiation sequence with specificity, we constructed a bacterial expression plasmid that contains the entire coding region of PZ120 fused to the GST protein, induced expression, and prepared an extract containing the full-length protein. As shown in Fig. 7, extract from IPTG-induced, but not noninduced bacteria, contained hMTIIA sequence-binding activity (compare lanes 1 and 3). The binding activity was specific because it could be prevented by addition of excess unlabeled hMTIIA initiation sequence (lane 4) but not by an unrelated oligodeoxynucleotide (lane 5 and 6). Furthermore, no complex was seen from extracts prepared from bacteria transformed with GST expression plasmid alone (lanes 7 to 9).
FIG. 7.
EMSA of a full-length GST-PZ120 fusion protein. Whole-cell lysates from bacteria overexpressing GST or GST-PZ120 protein were used for EMSA. An arrow indicates specific PZ120-hMTIIA DNA complex. Expression and EMSA were carried out as described in Materials and Methods and in the legend to Fig. 2.
Analysis of transcriptional activity mediated by PZ120.
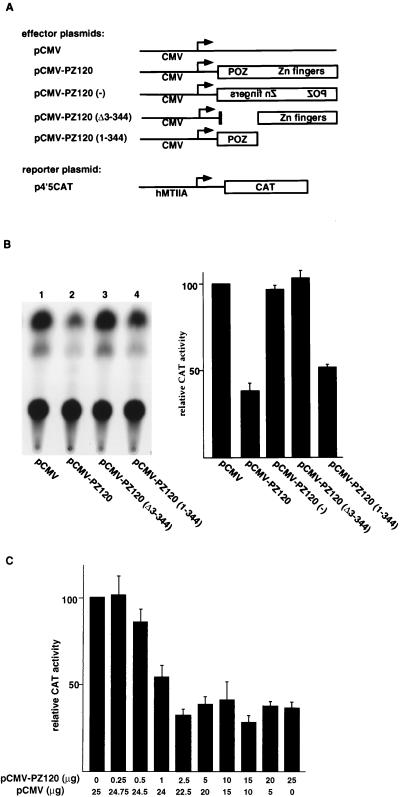
To examine transcriptional effects of hMTIIA by PZ120, we constructed a plasmid that expresses PZ120 under the CMV promoter and cotransfected it into CV1 cells together with a plasmid that contains the hMTIIA promoter upstream of the CAT reporter gene. We found that overexpression of PZ120 inhibited the transcriptional activity from the hMTIIA promoter (Fig. 8B, compare lanes 1 and 2). Repression by PZ120 was dose dependent, with an optimum concentration around 2.5 μg of transfected PZ120 expression plasmid (Fig. 8C). A plasmid containing β-galactosidase expressed under the simian virus 40 promoter was not affected by overexpression of PZ120. Furthermore, cotransfection of a plasmid expressing the PZ120 antisense RNA had no effect on the hMTIIA promoter, arguing that repression by PZ120 is a specific phenomenon. This finding is both exciting and intriguing, for it strongly suggests that PZ120’s binding to the hMTIIA sequence is physiologically relevant.
FIG. 8.
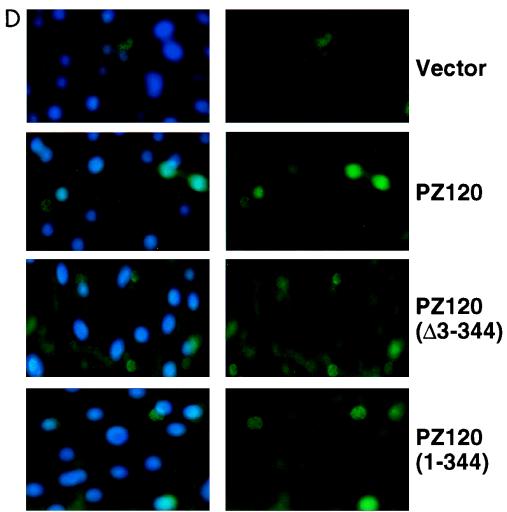
Transcriptional repression by the cloned Z120. (A) Schematic drawings of plasmids used in transient-transfection assays. Bent arrows indicate the direction of transcription. (B and C) Results of transfection assays showing that PZ120 represses transcription when it is targeted to the hMTIIA promoter. All transfections were normalized to equal amounts of DNA with parental expression vectors. (B) The gel at left is a representative autoradiogram of a CAT assay. (B and C) The graphs show relative CAT activity results as the means ± standard deviations of results from three to six separate transfections after normalization with β-galactosidase activity. (D) Detection of PZ120 expression by indirect immunofluorescence in cultured CV1 cells. Each picture shown represents a typical field from several that were imaged. In all images, the fluorescein signal is shown in the right panel and the merged image with DAPI-stained DNA is shown in the left.
Since earlier studies suggested that POZ domains may be important in transcriptional repression, we were encouraged to question the role of the POZ domain in PZ120. Our results indicate that deletion of the POZ domain (amino acids 3 to 344) in PZ120 abolished repression of the hMTIIA promoter by PZ120 (Fig. 8B, lane 3), consistent with the idea that the POZ domain mediates transcriptional repression. Interestingly, a plasmid that expresses the N terminus of PZ120 (amino acids 1 to 344) repressed the hMTIIA promoter efficiently (lane 4). To rule out the possibility that the loss of repression activity by PZ120 lacking amino acids 3 to 344 (Δ3–344) is a reflection of inefficient expression of this mutant protein, we performed immunofluorescence analyses on transfected cells. As shown in Fig. 8D, a FLAG epitope-tagged PZ120 (Δ303–344) protein was clearly expressed. These results, therefore, argue strongly that the portion of PZ120 outside of the zinc finger region is necessary and sufficient for its biological function on the hMTIIA promoter.
DISCUSSION
MTs are important regulatory proteins in many organisms. Because of the high level of hMTIIA expression, coupled with the many protein-DNA interactions that occur at the hMTIIA promoter, the hMTIIA gene has traditionally been an excellent model for the study of transcriptional regulation. Extensive analysis of the 5′ flanking region of this promoter has provided much important information concerning trans-acting factors that bind to different upstream cis-acting sites to influence transcription of this gene. However, since DNA elements surrounding the transcription start sites of many genes play equally crucial roles (for examples, see references 4, 11, 12, 17, 24, 28, 30, 34, 63, 67–69, 71, and 73), a complete picture of how transcriptional regulation is achieved in the hMTIIA gene requires that proteins that bind to the hMTIIA transcription initiation sequence be identified and their mechanisms clearly understood.
The hMTIIA transcription initiation sequence is identical to sequences found in a promoter of the fish Xiphophorus maculatus (19) and in the promoter of the human α-2 macroglobulin gene (50). The hMTIIA initiation sequence also has over 80% identity with a sequence located in the human androgen receptor promoter (72). However, it has not yet been established whether sequences in these other promoters are part of a transcription start site or whether they play any essential role in transcription. Nevertheless, the high conservation of these DNA sequences suggest that the hMTIIA initiation sequence may be important for accurate regulation of transcription of the hMTIIA and other genes and, thus, may represent an individual recognition site for a component of the transcriptional machinery. In fact, we and others have found specific proteins that bind to this site in the hMTIIA promoter (3, 31, 70).
In this paper, we describe the cloning of a novel protein, PZ120, that binds the hMTIIA transcription start site and plays a role in the control of transcription of the hMTIIA gene. At the C terminus of PZ120 are 12 zinc fingers of the C2H2 type that display imperfect tandem repeats of CysX2 CysX3ψX5ψX2HisX3–5His, where ψ indicates hydrophobic residues. Most of the fingers are joined via His-Cys links (TGEKPY/F) similar to those in the developmental control gene Krüppel in Drosophila melanogaster. Sequence comparison of PZ120 with proteins in the GenBank and EMBL databases revealed a protein with a stretch of amino acids that contains a conserved POZ motif present in a family of proteins that includes the Drosophila transcription factors Tramtrack and Broad Complex, GAGA, ZF5, and ZID, as well as a group of poxvirus proteins. In previous studies, the POZ domain acted as a specific protein-protein interaction domain, inhibited DNA binding, and appeared to localize proteins to discrete regions of the nucleus (5, 36). In addition, POZ domain proteins have been associated with a variety of processes, including nucleosome and chromatin disruption, pattern formation, metamorphogenesis, oogenesis, and eye and limb development in Drosophila (16, 18, 21, 26, 74, 76, 77, 80). In humans, two POZ domain zinc finger genes, PLZF and BCL6 (also called LAZ3), are associated with chromosomal translocation breakpoints in acute promyelocytic leukemia and non-Hodgkins’s lymphoma, respectively (6, 9, 10, 45, 52, 78). Finally, several POZ-zinc finger proteins have been proposed to be transcriptional repressors (7, 8, 14, 26, 54, 58, 76), an observation that is clearly consistent with the results of our study. The biological and functional significance of the POZ domain in relation to PZ120 and to hMTIIA regulation is not completely known at this time. However, the conservation of this region among PZ120 and many factors important in the control of development suggests that PZ120 may be a key regulatory protein in humans.
Many studies suggest that proteins that have previously been described as transcription factors may represent only single members of protein complexes that form at DNA sites and function together to regulate transcription. We believe that PZ120 may also require an additional factor(s) for its function. First, in addition to PZ120, we have detected at least four other specific protein-DNA complexes bound to the hMTIIA transcription start site (70). Second, and perhaps the best indication that PZ120 is involved in protein-protein interactions with other cellular factors, is the fact that PZ120 possesses a POZ domain that has previously been shown to mediate protein-protein interactions (1, 5, 15). Recent studies indicate that PZLF and BCL6 associate with SMRT-mSin3-HDAC corepressor complexes and repress transcription by recruiting histone deacetylases (13, 27, 29). Thus, it is conceivable that while PZ120 possesses a unique ability to bind hMTIIA DNA, like other POZ domain transcription factors it merely serves as a platform for assembly of additional components of the transcriptional regulatory system. Work is under way to determine whether PZ120 associates with corepressor complexes and to identify novel cellular proteins that interact with PZ120. Knowledge of these interactions will provide a basic understanding of the mechanisms of PZ120 function and add to our understanding of hMTIIA regulation.
Like the mRNAs of many of the zinc transcription factors previously studied, PZ120 mRNA is ubiquitously expressed. However, we have found that the level of PZ120, a repressor of hMTIIA, is particularly low in the liver. Remarkably, this finding is in good agreement with early reports that among all human organs, hMTIIA is expressed most highly in the liver (41). Therefore, it is conceivable that PZ120 is a tissue-specific regulator of hMTIIA.
One of the most intriguing findings of our study is that PZ120 binds the hMTIIA transcription start site with high specificity in the absence of the zinc finger domain. The initial expression cloning of PZ120 indicated that amino acids 1 to 482, a region devoid of any known DNA-binding motif, is sufficient for hMTIIA DNA binding. Subsequent experiments using recombinant PZ120 protein expressed in bacteria and EMSA confirmed that amino acids 1 to 482 can indeed bind specifically to the hMTIIA transcription initiation sequence. Besides the presence of the POZ domain, a casual inspection of amino acids to 1 to 482 of PZ120 did not reveal any other obvious motif. So far, there is no report that the POZ domain can function in sequence-specific DNA binding. We are currently working to further localize the exact region of PZ120 that can bind hMTIIA DNA. It is conceivable that the POZ domain may actually bind DNA, a possible phenomenon that has not yet been fully investigated. Alternatively, it is possible that binding of the PZ120 POZ domain to DNA is an isolated case. It is also conceivable that a novel DNA-binding domain is present in PZ120 and lies outside of (surrounding) the POZ domain. Finally, it is reasonable to speculate that the PZ120 protein may contain standby DNA-binding domains that are used only when the natural zinc finger DNA-binding domain is not present. So far, we have not been able to detect specific binding of the hMTIIA initiation sequence using recombinant proteins containing the zinc finger portion of PZ120 alone (data not shown). As a result, we currently favor a model where PZ120 is a multifunctional protein that, in addition to using its N-terminal region (with or without the POZ domain) to bind DNA sequences that closely resemble the hMTIIA initiation sequence, uses its C-terminal zinc fingers to bind a completely different set of DNA.
A number of proteins that bind to transcription start sites of many different genes have previously been reported, but very few have been shown to function as repressors. Intriguingly, two hMTIIA initiation sequence-binding proteins that we have identified, RPA (70), and PZ120 in this report, both possess transcriptional repression activity. Despite the many exhaustive studies of the hMTIIA promoter, the exact number of different factors capable of regulating hMTIIA is unclear at this time. Also undetermined is whether all transcription factors that bind the hMTIIA transcription initiation region work by the same mechanism. The identification of negative regulatory factors that bind the hMTIIA transcription start site has provided an excellent starting point from which to address these questions.
ACKNOWLEDGMENTS
We thank Alan Tomkinson for purified RAD1 protein; Mon-Li Chu for the cDNA library; Nikola Valkov for assistance with microscopy; Tom Shenk, Rosalind Jackson, Eden Kahle, and Julia Lee for discussion and critical reading of the manuscript; and the Moffitt Cancer Center Imaging Facility for technical support.
This work was supported by a grant from the National Institutes of Health (ES09262) to E.S.
REFERENCES
- 1.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA-and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 2.Andrews G K. Regulation of metallothionein gene expression. Prog Food Nutr Sci. 1990;14:193–258. . (Review.) [PubMed] [Google Scholar]
- 3.Angel P, Imagawa M, Chiu R, Stein B, Imbra R J, Rahmsdorf H J, Jonat C, Herrlich P, Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 4.Ayer D E, Dynan W S. Simian virus 40 major late promoter: a novel tripartite structure that includes intragenic sequence. Mol Cell Biol. 1988;8:2021–2033. doi: 10.1128/mcb.8.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell V J, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 6.Baron B W, Nucifora G, McCabe N, Espinosa R, Le B M, McKeithan T W. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci USA. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown J L, Wu C. Repression of Drosophila pair-rule segmentation genes by ectopic expression of tramtrack. Development. 1993;117:45–58. doi: 10.1242/dev.117.1.45. [DOI] [PubMed] [Google Scholar]
- 8.Chang C C, Ye B H, Chaganti R S, Dalla F R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S J, Zelent A, Tong J H, Yu H Q, Wang Z Y, Derre J, Berger R, Waxman S, Chen Z. Rearrangements of the retinoic acid receptor alpha and promyelocytic leukemia zinc finger genes resulting from t(11;17)(q23;q21) in a patient with acute promyelocytic leukemia. J Clin Investig. 1993;91:2260–2267. doi: 10.1172/JCI116453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Brand N J, Chen A, Chen S J, Tong J H, Wang Z Y, Waxman S, Zelent A. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Concino M F, Lee R F, Merryweather J P, Weinmann R. The adenovirus major late promoter TATA box and initiation site are both necessary for transcription in vitro. Nucleic Acids Res. 1984;12:7423–7433. doi: 10.1093/nar/12.19.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corden J, Wasylyk A, Buchwalder A, Sassone-Corsi P, Kedinger C, Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980;209:1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- 13.David G, Alland L, Hong S H, Wong C W, DePinho R A, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 14.Deweindt C, Albagli O, Bernardin F, Dhordain P, Quief S, Lantoine D, Kerckaert J P, Leprince D. The LAZ3/BCL6 oncogene encodes a sequence-specific transcriptional inhibitor: a novel function for the BTB/POZ domain as an autonomous repressing domain. Cell Growth Differ. 1995;6:1495–1503. [PubMed] [Google Scholar]
- 15.Dhordain P, Albagli O, Lin R J, Ansieau S, Quief S, Leutz A, Kerckaert J P, Evans R M, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiBello P R, Withers D A, Bayer C A, Fristrom J W, Guild G M. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dierks P, Ooyen A V, Cochran M D, Dobkin C, Reiser J, Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit β-globin gene in mouse 3T6 cells. Cell. 1983;32:695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- 18.Dorn R, Krauss V, Reuter G, Saumweber H. The enhancer of position-effect variegation of Drosophila, E(var)3-93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proc Natl Acad Sci USA. 1993;90:11376–11380. doi: 10.1073/pnas.90.23.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedenreich H, Schartl M. Transient expression directed by homologous and heterologous promoter and enhancer sequences in fish cells. Nucleic Acids Res. 1990;18:3299–3305. doi: 10.1093/nar/18.11.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman R L, Stark G R. Alpha-interferon-induced transcription of HLA and metallothionein genes containing homologous upstream sequences. Nature. 1985;314:637–639. doi: 10.1038/314637a0. [DOI] [PubMed] [Google Scholar]
- 21.Godt D, Couderc J-L, Cramton S E, Laski F A. Pattern formation in the limbs of Drosophila: bric a brac is expressed in both a gradient and wave-length pattern and is required for specification and proper segmentation of the tarsus. Development. 1993;119:799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- 22.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham F, van der Eb A. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–457. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 24.Grosschedl R, Birnstiel M L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci USA. 1980;77:1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamer D H. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- 26.Harrison S D, Travers A A. The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J. 1990;9:207–216. doi: 10.1002/j.1460-2075.1990.tb08097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He L Z, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi P P. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 28.Hen R, Sassone-Corsi P, Corden J, Gaub M P, Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci USA. 1982;79:7132–7136. doi: 10.1073/pnas.79.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong S H, David G, Wong C W, Dejean A, Privalsky M L. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu S L, Manley J L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci USA. 1981;78:820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 32.Imbert J, Culotta V, Furst P, Gedamu L, Hamer D. Regulation of metallothionein gene transcription by metals. Adv Inorg Biochem. 1990;8:139–164. . (Review.) [PubMed] [Google Scholar]
- 33.Jahroudi N, Foster R, Price H J, Beitel G, Gedamu L. Cell-type specific and differential regulation of the human metallothionein genes. Correlation with DNA methylation and chromatin structure. J Biol Chem. 1990;265:6506–6511. [PubMed] [Google Scholar]
- 34.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagi J H. Overview of metallothionein. Methods Enzymol. 1991;205:613–626. doi: 10.1016/0076-6879(91)05145-l. [DOI] [PubMed] [Google Scholar]
- 36.Kaplan J, Calame K. The ZiN/POZ domain of ZF5 is required for both transcriptional activation and repression. Nucleic Acids Res. 1997;25:1108–1116. doi: 10.1093/nar/25.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karin M. Metallothioneins: proteins in search of function. Cell. 1985;41:9–10. doi: 10.1016/0092-8674(85)90051-0. [DOI] [PubMed] [Google Scholar]
- 38.Karin M, Hasligner A, Holtgreve H, Cathala G, Slater E, Baxter J D. Activation of a heterologous promoter in response to dexamethasone and cadmium by metallothionein gene 5′-flanking DNA. Cell. 1984;36:371–379. doi: 10.1016/0092-8674(84)90230-7. [DOI] [PubMed] [Google Scholar]
- 39.Karin M, Haslinger A, Holtgreve H, Richards R I, Krauter P, Westphal H M, Beato M. Characterization of DNA sequences through which cadmium and glucocorticoid hormones induce human metallothionein-IIA gene. Nature. 1984;308:513–519. doi: 10.1038/308513a0. [DOI] [PubMed] [Google Scholar]
- 40.Karin M, Herschman H R. Dexamethasone stimulation of metallothionein synthesis in HeLa cell cultures. Science. 1979;204:176–177. doi: 10.1126/science.432639. [DOI] [PubMed] [Google Scholar]
- 41.Karin M, Herschman H R. Characterization of the metallothioneins induced in HeLa cells by dexamethasone and zinc. Eur J Biochem. 1980;107:395–401. doi: 10.1111/j.1432-1033.1980.tb06042.x. [DOI] [PubMed] [Google Scholar]
- 42.Karin M, Richards R I. Human metallothionein genes—primary structure of the metallothionein-II gene and a related processed gene. Nature. 1982;299:797–802. doi: 10.1038/299797a0. [DOI] [PubMed] [Google Scholar]
- 43.Karin M, Richards R I. Human metallothionein genes: molecular cloning and sequence analysis of the mRNA. Nucleic Acids Res. 1982;10:3165–3173. doi: 10.1093/nar/10.10.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karin M, Andersen R D, Slater E, Smith K, Herschman H R. Metallothionein mRNA induction in HeLa cells in response to zinc or dexamethasone is a primary induction response. Nature. 1980;286:295–297. doi: 10.1038/286295a0. [DOI] [PubMed] [Google Scholar]
- 45.Kerckaert J P, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- 46.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 47.Leadon S A, Snowden M M. Differential repair of DNA damage in the human metallothionein gene family. Mol Cell Biol. 1988;8:5331–5338. doi: 10.1128/mcb.8.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee W, Haslinger A, Karin M, Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SF40. Nature. 1987;325:368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- 49.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 50.Matthijs G, Cassiman J J, Van den Berghe H, Marynen P. Characterization of the human alpha 2-macroglobulin gene promoter: identification of a novel, triple TRE/RARE/ERE response element. Biochem Biophys Res Commun. 1994;202:65–72. doi: 10.1006/bbrc.1994.1894. [DOI] [PubMed] [Google Scholar]
- 51.Mermod N, Williams T J, Tjian R. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vivo. Nature. 1988;332:557–561. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- 52.Miki T, Kawamata N, Hirosawa S, Aoki N. Gene involved in the 3q27 translocation associated with B-cell lymphoma, BCL5, encodes a Krüppel-like zinc-finger protein. Blood. 1994;83:26–32. [PubMed] [Google Scholar]
- 53.Mitchell P J, Wang C, Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 54.Numoto M, Niwa O, Kaplan J, Wong K K, Merrell K, Kamiya K, Yanagihara K, Calame K. Transcriptional repressor ZF5 identifies a new conserved domain in zinc finger proteins. Nucleic Acids Res. 1993;21:3767–3775. doi: 10.1093/nar/21.16.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmiter R D. Molecular biology of metallothionein gene expression. EXS. 1987;52:63–80. doi: 10.1007/978-3-0348-6784-9_4. [DOI] [PubMed] [Google Scholar]
- 56.Pan T C, Zhang R Z, Mattei M G, Timpl R, Chu M L. Cloning and chromosomal location of human alpha 1(XVI) collagen. Proc Natl Acad Sci USA. 1992;89:6565–6569. doi: 10.1073/pnas.89.14.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pulido P, Kagi J H R, Vallee B L. Isolation and some properties of human metallothionein. Biochemistry. 1966;5:1768–1777. doi: 10.1021/bi00869a046. [DOI] [PubMed] [Google Scholar]
- 58.Read D, Levine M, Manley J L. Ectopic expression of the Drosophila tramtrack gene results in multiple embryonic defects, including repression of even-skipped and fushi tarazu. Mech Dev. 1992;38:183–195. doi: 10.1016/0925-4773(92)90052-l. [DOI] [PubMed] [Google Scholar]
- 59.Rudd C J, Herschman H R. Metallothionein in a human cell line: the response of HeLa cells to cadmium and zinc. Toxicol Appl Pharmacol. 1979;47:273–278. doi: 10.1016/0041-008x(79)90321-1. [DOI] [PubMed] [Google Scholar]
- 60.Sadhu C, Gedamu L. Metal-specific posttranscriptional control of human metallothionein genes. Mol Cell Biol. 1989;9:5738–5741. doi: 10.1128/mcb.9.12.5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 62.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 64.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 65.Skroch P, Buchman C, Karin M. Regulation of human and yeast metallothionein gene transcription by heavy metal ions. Prog Clin Biol Res. 1993;380:113–128. . (Review.) [PubMed] [Google Scholar]
- 66.Slater E P, Cato A C, Karin M, Baxter J D, Beato M. Progesterone induction of metallothionein-IIA gene expression. Mol Endocrinol. 1988;2:485–491. doi: 10.1210/mend-2-6-485. [DOI] [PubMed] [Google Scholar]
- 67.Smale S T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 68.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 69.Smale S T, Schmidt M C, Berk A J, Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci USA. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang C M, Tomkinson A E, Lane W S, Wold M S, Seto E. Replication protein A is a component of a complex that binds the human metallothionein IIA gene transcription start site. J Biol Chem. 1996;271:21637–21644. doi: 10.1074/jbc.271.35.21637. [DOI] [PubMed] [Google Scholar]
- 71.Tebb G, Mattaj I W. Positionally exact initiation is required for the formation of a stable RNA polymerase II transcription complex in vivo. EMBO J. 1988;7:3785–3792. doi: 10.1002/j.1460-2075.1988.tb03263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tilley W D, Marcelli M, McPhaul M J. Expression of the human androgen receptor gene utilizes a common promoter in diverse human tissues and cell lines. J Biol Chem. 1990;265:13776–13781. [PubMed] [Google Scholar]
- 73.Tokunaga K, Hirose S, Suzuki Y. In monkey COS cells only the TATA box and the cap site region are required for faithful and efficient initiation of the fibroin gene transcription. Nucleic Acids Res. 1984;12:1543–1558. doi: 10.1093/nar/12.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of gaga transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 75.Uchida Y, Takio K, Titani K, Ihara Y, Tomonaga M. The growth inhibitory factor that is deficient in the Alzheimer’s disease brain is a 68 amino acid metallothionein-like protein. Neuron. 1991;7:337–347. doi: 10.1016/0896-6273(91)90272-2. [DOI] [PubMed] [Google Scholar]
- 76.Xiong W C, Montell C. tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev. 1993;7:1085–1096. doi: 10.1101/gad.7.6.1085. [DOI] [PubMed] [Google Scholar]
- 77.Xue F, Cooley L. Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 78.Ye B H, Lista F, Lo C F, Knowles D M, Offit K, Chaganti R S, Dalla F R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 79.Zhu X, Mancini M A, Chang K H, Liu C Y, Chen C F, Shan B, Jones D, Yang-Feng T L, Lee W H. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol Cell Biol. 1995;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zollman S, Godt D, Prive G G, Couderc J L, Laski F A. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]